ABSTRACT
The porcine reproductive and respiratory syndrome virus (PRRSV) can lead to severe reproductive problems in sows, pneumonia in weaned piglets, and increased mortality, significantly negatively impacting the economy. Post-translational changes are essential for the host-dependent replication and long-term infection of PRRSV. Uncertainty surrounds the function of the ubiquitin network in PRRSV infection. Here, we screened 10 deubiquitinating enzyme inhibitors and found that the ubiquitin-specific proteinase 1 (USP1) inhibitor ML323 significantly inhibited PRRSV replication in vitro. Importantly, we found that USP1 interacts with nonstructural protein 1β (Nsp1β) and deubiquitinates its K48 to increase protein stability, thereby improving PRRSV replication and viral titer. Among them, lysine at position 45 is essential for Nsp1β protein stability. In addition, deficiency of USP1 significantly reduced viral replication. Moreover, ML323 loses antagonism to PRRSV rSD16-K45R. This study reveals the mechanism by which PRRSV recruits the host factor USP1 to promote viral replication, providing a new target for PRRSV defense.
IMPORTANCE
Deubiquitinating enzymes are critical factors in regulating host innate immunity. The porcine reproductive and respiratory syndrome virus (PRRSV) nonstructural protein 1β (Nsp1β) is essential for producing viral subgenomic mRNA and controlling the host immune system. The host inhibits PRRSV proliferation by ubiquitinating Nsp1β, and conversely, PRRSV recruits the host protein ubiquitin-specific proteinase 1 (USP1) to remove this restriction. Our results demonstrate the binding of USP1 to Nsp1β, revealing a balance of antagonism between PRRSV and the host. Our research identifies a brand-new PRRSV escape mechanism from the immune response.
KEYWORDS: PRRSV, USP1, deubiquitination, Nsp1β, immune escape
INTRODUCTION
The porcine reproductive and respiratory syndrome virus (PRRSV) is the etiologic agent of porcine reproductive and respiratory syndrome (PRRS), causing a tremendous economic loss in the swine industry globally (1, 2). PRRSV is a small, enveloped, single positive-stranded RNA virus (3–5). The length of the PRRSV genome ranges from 14.9 to 15.5 kb, and it uses two different transcription methods to express a variety of nonstructural and structural proteins (6). The pathogenicity of PPRSV is associated with its high genetic diversity. The outbreak of high pathogenicity PRRSV (HP-PRRSV) strains in China in 2006 resulted in atypical PRRS pandemics and 20% mortality in pigs (7). Since 2013, novel PRRSV variations, NADC30-like strains, and proven imports from North America and adoption in China have caused PRRS to be common once again (8, 9). Multiple PRRSV variants coexist in Chinese swine herds currently, which has caused the swine industry to suffer enormous financial losses and increased worry about virus recombination with rising pathogenic risk (10). Since neither commercial vaccinations nor conventional tactics can effectively manage PRRS (11–13), therefore, it is critical to understand the pathogenic mechanisms of PRRSV for PRRS prevention and treatment (14–17).
PRRSV acts as an immunosuppressive virus. Type I interferons (IFNs-α/β) are the most effective part of innate immunity against invading viruses. Innate immunity is the first line of defense of the host against infections. At least six PRRSV proteins [nonstructural protein (Nsp)1α, Nsp1β, Nsp2, Nsp4, Nsp11, and N] are now IFN antagonists; several IFNs are inhibited by PRRSV (18, 19). In addition, PRRSV hijacks apoptosis to promote self-replication (20, 21). miRNAs can also positively or negatively regulate PRRSV replication by degrading mRNAs or inhibiting translation (22). PRRSV replication activates the expression of proinflammatory cytokines, which activate the inflammatory storm in the body and aggravate the symptoms of infection (23). Overall, PRRSV is host dependent for replication. Consequently, the host immune system restricts viral replication and eliminates the viruses. To evade recognition by PRRs, PRRSV self-occlusion inhibits innate immunity, alters cytokine responses, and regulates apoptosis to survive and spread within the cell.
The most prevalent family of deubiquitinating enzymes (DUBs) is the ubiquitin-specific proteinases (USPs), which carry out protein turnover and destruction by ubiquitination and deubiquitination (24). USPs primarily participate in various biological activities, such as plasma membrane receptor modulation, cell cycle progression, cell proliferation, and cell differentiation (25). Numerous studies have shown that USPs are associated with the host infection of diverse RNA viruses. USP29 prevents proteasome degradation of ORF9b to promote severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) replication (26). Deubiquitinating enzyme inhibitors prevent Chikungunya virus (CHIKV) replication. Inhibition of the deubiquitinating enzyme ubiquitin C-terminal hydrolase 1 (UCHL1) reduces SARS-CoV-2 replication (27). USP38 suppressed Zika virus (ZIKV) infection because it binds to the ZIKV envelope (E) protein through its C-terminal domain and reduces K48- and K63-linked polyubiquitination (28). According to this research, USPs are crucial in viral infection and host immunological reactions. A crucial component that regulates Fanconi anemia is USP1 (25). However, the mechanism of the USP1 complex in innate antiviral immunity, especially against PRRSV infection, remains unknown.
This study found that the deubiquitinating enzyme inhibitor ML323 inhibited PRRSV infection by downregulating USP1 expression in cells. Overexpression of USP1 also promoted PRRSV infection in cells. Meanwhile, USP1 interacts with Nsp1β, and USP1 promotes PRRSV infection by removing the polyubiquitination of the Nsp1β protein K48 linkage and subsequently stabilizing the expression of Nsp1β protein. The findings showed that PRRSV uses a unique approach to circumvent the host’s innate immune response by using the ubiquitination pathway. USP1 might be a key protein in the PRRSV–host interaction and a potential target for antiviral studies.
RESULTS
ML323 inhibited PRRSV replication
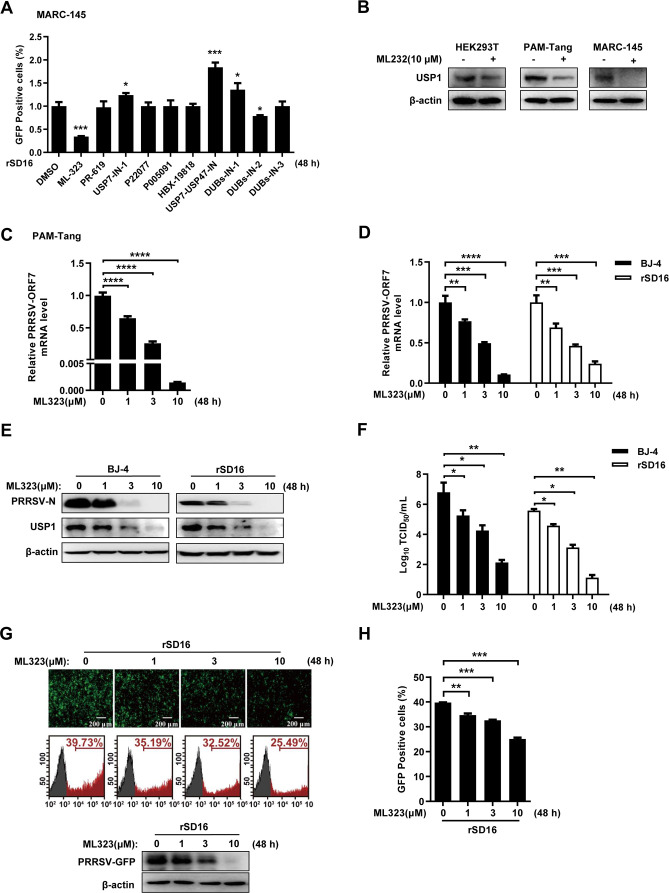
Deubiquitination processes significantly control the number of cellular proteins that behave, act, localize, and remain stable. It has been demonstrated that several viruses exploit cellular ubiquitin-binding systems to efficiently infect cells, promote genome replication, or assemble and release viral offspring (29–33). First, we evaluated 10 deubiquitinating enzyme inhibitors [USP1 (ML-323), USP2 (PR-619), USP7 (USP7-in-1, P22077, P005091, and HBX-19818), USP7-USP47 (USP7-USP47 IN), and USP8 (DUBs-IN-1, DUBs-IN-2, and DUBs-IN-3)]. Fluorescence infusion results showed that the USP1 inhibitor ML323 and the USP8 inhibitor DUBs-IN-1 inhibited rSD16 replication, with ML323 having the most significant inhibitory effect, whereas several other small-molecule compounds either promoted rSD16 replication or had no effect (Fig. 1A). Therefore, we selected ML323 for a detailed study. However, to verify the interspecies variability of ML323, we first carried out the validation in different cell lines and found that human-derived HEK293T, porcine-derived PAM-Tang, and monkey-derived MARC-145 cells exhibited inhibition of USP1 protein expression after ML323 treatment (Fig. 1B). First, we examined the transcription levels of PRRSV ORF7 in the PRRSV host cell line PAM-Tang following treatment with various concentrations (0, 1, 3, and 10 µM) of ML323, and the results showed that ML323 decreased the mRNA levels of PRRSV ORF7 in a concentration-dependent manner (Fig. 1C). Then, we used BJ-4 and rSD16 PRRSV strains to infect cells, and we looked at how different ML323 dosages affected PRRSV replication. The outcomes demonstrated that ML323 significantly and dose-dependently decreased the mRNA expression of PRRSV BJ-4 and rSD16 ORF7 (Fig. 1D). Western blotting analysis showed that ML323 could significantly reduce the expression of the PRRSV N protein and suppress the expression of USP1 (Fig. 1E). PRRSV BJ-4 and rSD16 virus titers were simultaneously dose-dependently decreased by ML323 (Fig. 1F). Consistently, fluorescence microscopy, Western blot, and flow cytometry (Fig. 1G and H) assays further confirmed that ML323 inhibited rSD16 replication. These results indicated that ML323 inhibited PRRSV replication.
Fig 1.
ML323 inhibited PRRSV replication. (A) The cells treated with 10 different small-molecule compounds were collected for 48 h and detected by flow cytometry. (B) Western blot assay for ML323 (10 µM) was performed after treatment with HEK293T, PAM-Tang, and MARC-145 and detected with antibodies against USP1 and β-actin. (C) After pretreatment with ML323, PRRSV BJ-4-infected PAM-Tang cells were identified using 0, 1, 3, and 10 µM ML323, and reverse transcription-quantitative PCR (RT-PCR) was used to evaluate the level of ORF7 mRNA in the cells. (D) MARC-145 cells were pretreated with ML323 at doses of 0, 1, 3, and 10 µM for 4 h and infected with BJ-4 and rSD16 PRRSV (MOI = 10). qPCR was used to detect the PRRSV ORF7 mRNA levels in MARC-145 cells. (E) The treated cells were collected after 48 h, and Western blot analysis was performed with USP1 antibody and PRRSV-N antibody. (F) PRRSV BJ-4 and rSD16 viruses were infected into ML323-pretreated MARC-145 cells, and subviral titers were assayed. (G) The 48-h treated cells were collected and examined by fluorescence microscopy and Western blot. (H) The treated cells collected at 48 h were examined by flow cytometry. The data were presented with significant differences (note: P > 0.05 ns, *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.000 1).
USP1 can enhance PRRSV infection
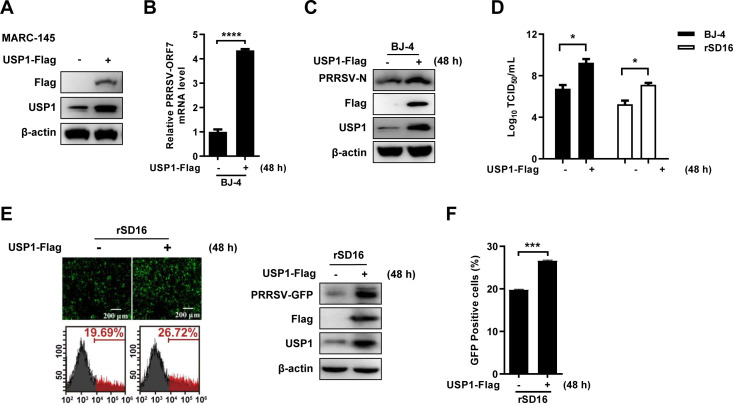
To validate the role of USP1 in PRRSV infection, we constructed a monkey-derived USP1-Flag expression vector and validated it in MARC-145 cells (Fig. 2A). Results from reverse transcription-quantitative PCR (RT-PCR revealed that USP1 ectopic expression markedly increased the quantity of PRRSV BJ-4 ORF7 mRNA (Fig. 2B). Western blot analysis revealed that USP1 promoted PRRSV BJ-4 N protein–protein expression (Fig. 2C). The 50% tissue culture infective dose (TCID50) assay indicated that ectopic expression of USP1 significantly increased the yield of PRRSV BJ-4 and rSD16 progeny virions (Fig. 2D). By utilizing rSD16, the impact of USP1 expression was further validated and shown in fluorescence microscopy, Western blot, and flow cytometry (Fig. 2E and F). According to these findings, USP1 might encourage PRRSV replication.
Fig 2.
USP1 can enhance PRRSV infection. (A) MARC-145 cells were transfected using the monkey USP1-Flag overexpression plasmid, and Western blot was used to detect USP1 and Flag expression. (B) USP1-Flag-transfected MARC-145 cells were infected with PRRSV BJ-4 (MOI = 10), and qPCR was used to detect the mRNA load of ORF7. (C) PRRSV N protein expression test by Western blot. (D) PRRSV BJ-4 and rSD16 viruses were each infected into USP1-Flag-transfected MARC-145 cells, and the titer of the daughter viruses was found. (E and F) Fluorescence microscopy, Western blot, and flow cytometry were performed to detect the rSD16 virus during overexpression of USP1, and the fluorescence content of viral green fluorescent protein (GFP) was compared with that of controls. Data were displayed with noticeable differences (note: P > 0.05 ns, *P < 0.05, ***P < 0.001, and ****P < 0.000 1).
Knockdown of USP1 inhibits PRRSV replication
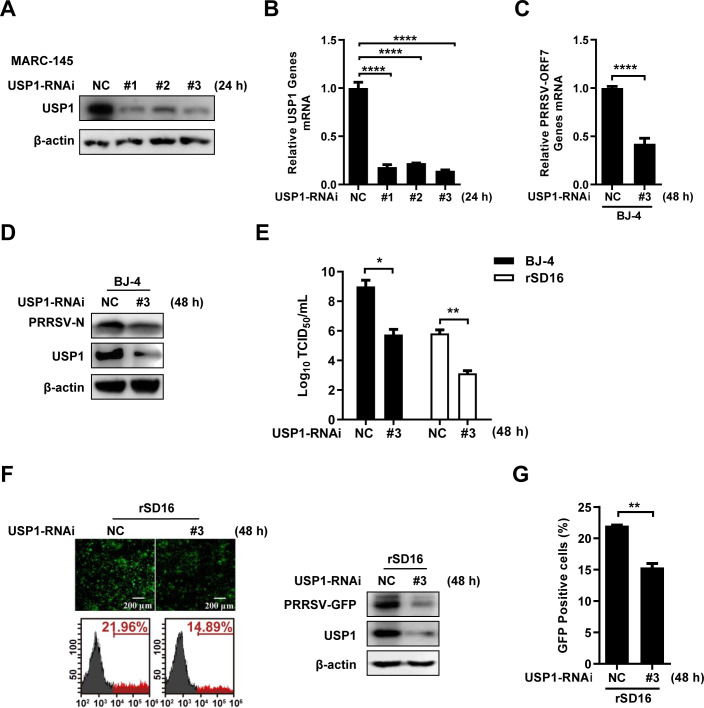
We created three short-interfering RNAs to block USP1 expression in MARC-145 cells to confirm the function of USP1 in PRRSV infection. The results showed that all small-interfering RNA could reduce the USP1 protein level (Fig. 3A) and USP1 mRNA expression (Fig. 3B), with USP1-RNAi#3 demonstrating the highest interference efficiency. RT-qPCR findings after USP1 interference revealed that the mRNA level of ORF7 of the PRRSV BJ-4 strain was dramatically downregulated (Fig. 3C). Western blot showed that siUSP1-transfected cells significantly reduced the N protein level of PRRSV BJ-4 (Fig. 3D). The TCID50 assay showed that the formation of PRRSV BJ-4 and rSD16 progeny virions was significantly reduced upon knockdown of USP1 (Fig. 3E). Similarly, fluorescence microscopy, Western blot, and flow cytometry showed that interference of USP1 significantly inhibited rSD16 replication (Fig. 3F and G). In summary, these data indicate that USP1 positively regulates PRRSV proliferation.
Fig 3.
Knockdown of USP1 inhibits PRRSV infection. (A) MARC-145 cells were transfected with siNC and USP1siRNA#1/2/3, and USP1 protein expression was detected after 36 h. (B) Simultaneous qPCR was performed to detect USP1 mRNA expression. (C and D) PRRSV BJ-4 (MOI=10) was infected with siNC and USP1siRNA#3 after transfection for 24 h, qPCR was used to detect the mRNA load of ORF7 (C). Western blot assay to detect PRRSV N and USP1 (D). (E) Twenty-four hours after transfection, PRRSV BJ-4 and PRRSV rSD16 (MOI=10) were infected, and infectious zygotic viral load was determined by the TCID50 assay. (F and G) The detection of PRRSV-GFP virus with fluorescence microscopy and Western blot and flow cytometry during the USP1 iRNA#3 (F). The fluorescence content of viral-GFP was compared with the control (G). The data were presented with significant differences (P > 0.05 ns, *P < 0.05, **P < 0.01, and ****P < 0.0001).
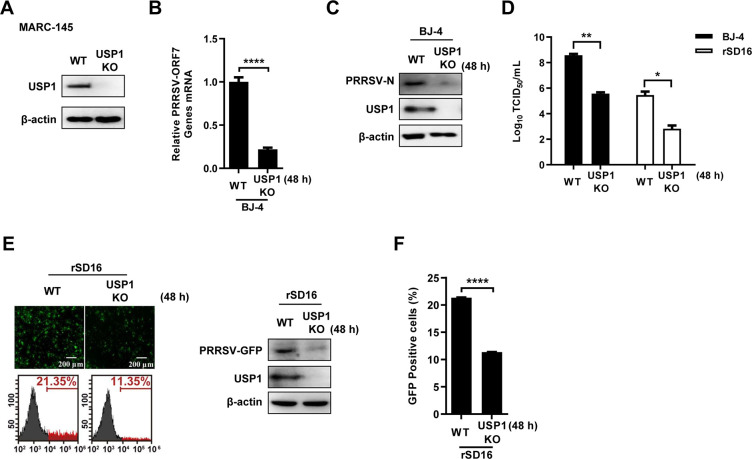
To further confirm the regulatory role of USP1 in PRRSV replication, we generated USP1-deficient MARC-145 cells by CRISPR/Cas9 technology and verified the protein expression of USP1 in cells by Western blot (Fig. 4A). USP1 deficiency decreased PRRSV BJ-4 ORF7 mRNA levels and PRRSV N protein expression after USP1 knockdown cells and control cells were infected with PRRSV (Fig. 4B and C). Similarly, viral titers were significantly reduced in USP1 knockdown cells (Fig. 4D). Fluorescence microscopy, Western blot, and flow cytometry analyses showed that PRRSV-infected cells were significantly reduced in the absence of USP1 (Fig. 4E and F). These outcomes matched the results of the USP1 knockdown model, indicating that USP1 aids PRRSV propagation.
Fig 4.
Knockdown of USP1 inhibits PRRSV infection. (A) USP1 protein expression in wild-type (WT) and USP1-deficient (USP1 KO) MARC-145s was analyzed by immunoprecipitation. (B) When USP1 KO MARC-145 cells were infected with PRRSV BJ-4 (MOI = 10) WT, PRRSV ORF7 transcription was determined by qPCR. (C) The protein levels of PRRSV N in WT and USP1 KO MARC-145 cells. (D) Loads of infectious PRRSV BJ-4 and rSD16 were determined by TCID50. (E and F) Fluorescence microscopy, Western blot, and flow cytometry were used to determine the PRRSV GFP virus in WT and USP1 KO MARC-145 cells. The fluorescence content of viral GFP was compared with that of the WT. The data were presented with significant differences (note: P > 0.05 ns, *P < 0.05, **P < 0.01, and ****P < 0.0001).
USP1 interacts with and stabilizes Nsp1β
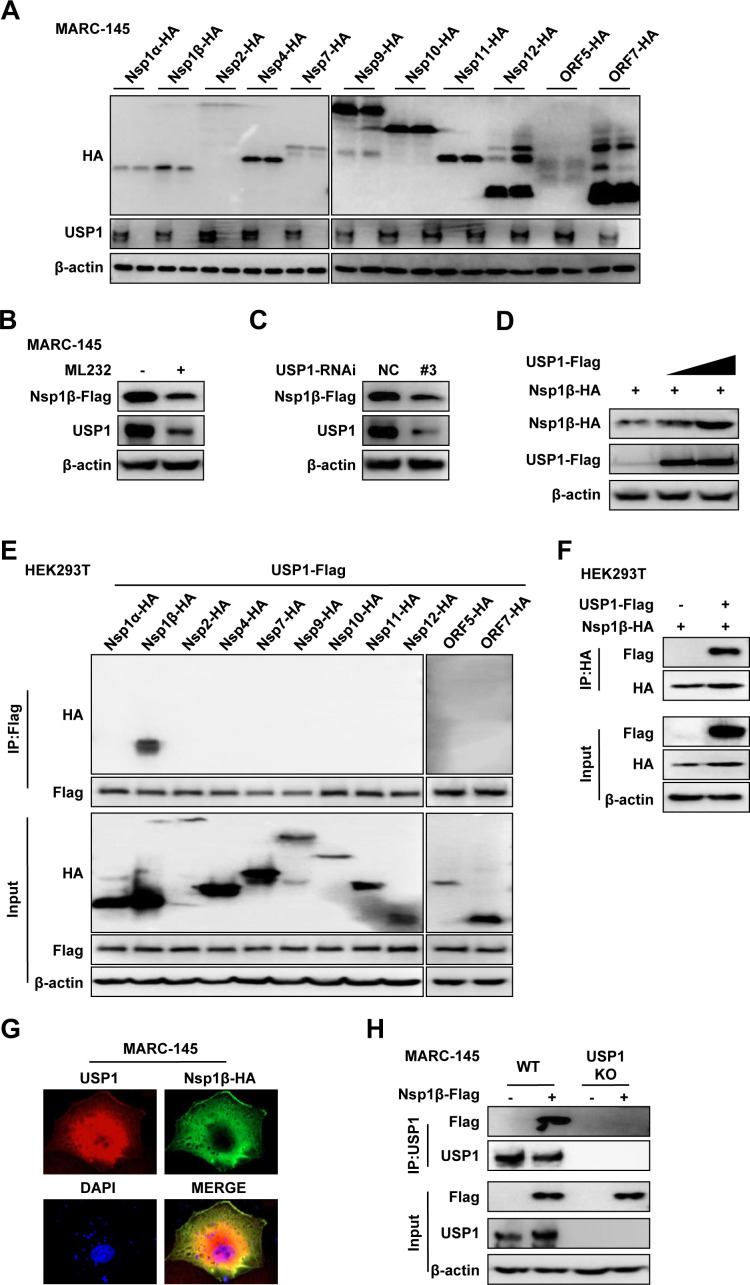
To clarify the relationship between USP1 and Nsp1β and to further explore the mechanism of USP1 in PRRSV infection, we constructed the eukaryotic expression plasmid of PRRSV. PRRSV expression plasmids were transfected in USP1 KO and control MARC-145 cells. The PRRSV protein level was considerably downregulated in USP1 KO MARC-145 cells, according to Western blot data (Fig. 5A). Meanwhile, the expression of Nsp1β was significantly suppressed upon ML323 treatment and knockdown of USP1 (Fig. 5B and C). The results of Nsp1β cotransfection with the USP1 plasmid showed that different doses of USP1 promoted Nsp1β protein expression (Fig. 5D). We confirmed the connection between USP1 and PRRSV proteins in further detail. Then, HEK293T cells were transfected with USP1-Flag and pCAGGS-HA-Nsp1α, 1β, 2, 4, 7, 9, 10, 11, 12, ORF5, and ORF7. According to the Co-IP data (Fig. 5E and F), USP1 interacted with PRRSV Nsp1β, but not with other Nsps. Additionally, confocal microscopy research revealed that in MARC-145 cells, USP1 and Nsp1β co-localized (Fig. 5G), suggesting that USP1 is related to Nsp1β. In addition, co-immunoprecipitation showed that Nsp1β could bind to endogenous USP1 in MARC-145 cells (Fig. 5H). The aforementioned results indicate that USP1 binds to Nsp1β and that USP1 stabilizes the PRRSV nonstructural protein Nsp1β.
Fig 5.
USP1 interacts with Nsp1β and stabilizes its protein. (A) Viral protein expression plasmids (Nsp1a, Nsp1β, Nsp2, Nsp4, Nsp7, Nsp9, Nsp10, Nsp11, Nsp12, ORF5, and ORF7-HA) were transfected in WT and USP1 KO MARC-145 cells. Western blot detects virus protein expression. (B) After MARC-145 cells were pretreated with DMSO and ML323, Nsp1β-Flag transfection was performed for 24 h, and the related protein expression was then discovered. (C) Western blot detection of protein expression in MARC-145 cells after transfection with Nsp1β-Flag for 24 h after transfection with siNC and siUSP1#3. (D) Western blot detection of different concentrations of USP1-Flag (0, 1, and 2 µg) after transfection of MARC-145 cells with Nsp1β-Flag (1 µg). (E) HEK293T cells were co-transfected with PRRSV (Nsp1a, Nsp1β, Nsp2, Nsp4, Nsp7, Nsp9, Nsp10, Nsp11, Nsp12, ORF5 and ORF7) expression plasmids and USP1-Flag for 24 hours. Cell samples were lysed to carry out immunoprecipitation with an anti-Flag pAb. (F) HEK293T cells were cotransfected with USP1-Flag and Nsp1β-HA. Cells for Co-IP assay were taken 24 h after transfection. (G) Nsp1β-HA-transfected cells were observed by confocal microscopy 24 h after transfection. Anti-USP1 and anti-HA antibodies were used to identify endogenous USP1 and Nsp1β, respectively. (H) Immunoprecipitation analysis of USP1-deficient MARC-145 cells transfected with Nsp1β-Flag.
USP1 stabilizes Nsp1β proteins by cleaving the K48 ubiquitination on Nsp1β lysine at position 45
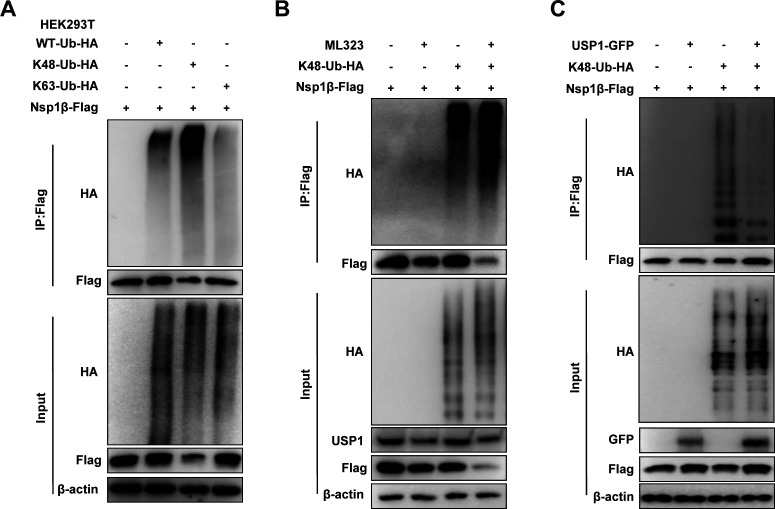
Since USP1 functions as a deubiquitinating enzyme, we next verified the effect of USP1 on Nsp1β protein stability. K48 ubiquitination modification is considered one of the major signals for degradation by the proteasome pathway (34). We found that Nsp1β undergoes mainly K48-linked ubiquitination modifications (Fig. 6A). Further experiments showed that USP1 treatment was significantly reduced by ML323 treatment. Importantly, Nsp1β expression was remarkably downregulated, and K48 ubiquitination of Nsp1β was significantly enhanced (Fig. 6B). Similarly, USP1 expression reduced the K48 ubiquitination modification of Nsp1β (Fig. 6C). These results suggest that USP1 stabilizes Nsp1β protein by cleaving the K48 ubiquitination modification of Nsp1β.
Fig 6.
Nsp1β K48-linked ubiquitination is formed at lysine position 45. (A) HEK293T cells were cotransfected with vector, WT-Ub-HA, K48-Ub-HA, and K63-Ub-HA plasmids and Nsp1β-Flag, and the immunoprecipitation detection assay was performed 24 h later. (B) HEK293T cells were pretreated with DMSO and ML323, transfected with Nsp1β-Flag and K48-Ub-HA plasmids for 24 h, and then detected by Co-IP assay. (C) HEK293T cells were transfected with the indicated plasmids for 24 h and then detected by Co-IP.
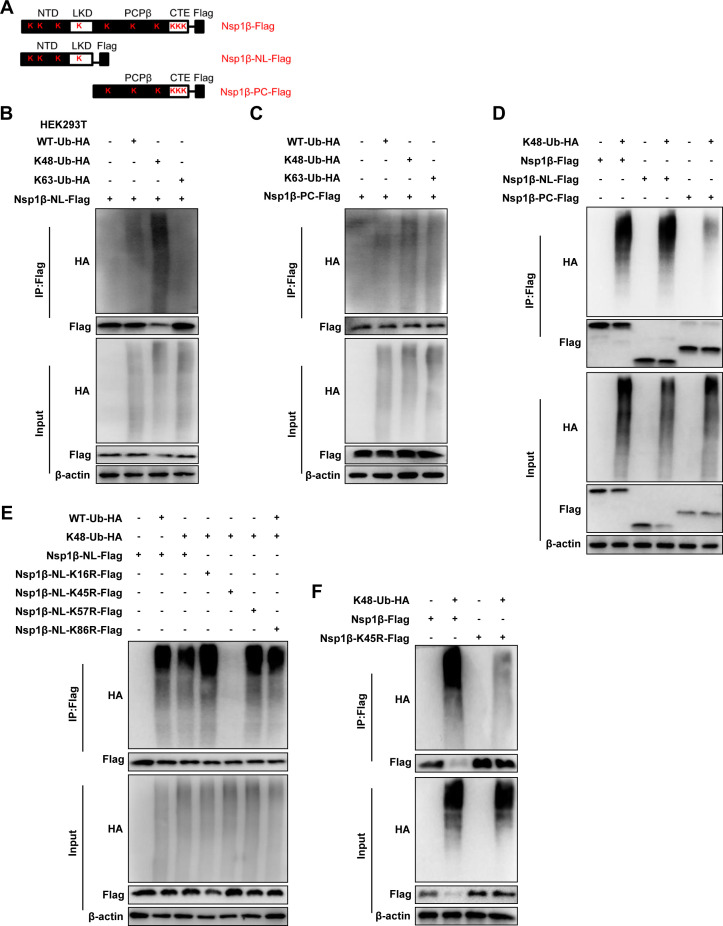
We mapped the domains of Nsp1β and constructed the corresponding truncated body, which consists of amino acids (aa) 1 to 111 (Nsp1β-NL-Flag) and aa 112 to 229 (Nsp1β-PC-Flag) (Fig. 7A). Co-IP results showed that the K48 ubiquitination modification was mainly formed at the Nsp1β-NL, whereas the Nsp1β-PC mutant had no obvious ubiquitination modification (Fig. 7B and C). Further results showed that both wild-type Nsp1β and Nsp1β-NL mutants had significant K48 ubiquitination compared with Nsp1β-PC (Fig. 7D). With the aforementioned results, we focused the Nsp1β ubiquitination sites in the 1 to 111 domain. Subsequently, to identify the ubiquitination sites of Nsp1β, we constructed Nsp1β-NL mutants (K16R, K45R, K57R, and K86R). Co-IP results showed that the Nsp1β-NL K45R mutant did not form ubiquitination, indicating that K45 may be the ubiquitination site of Nsp1β (Fig. 7E). The Nsp1β K45R mutant was further constructed, and the results showed that Nsp1β does not undergo K48 ubiquitination after silencing of the 45 lysine of Nsp1β (Fig. 7F). These results indicate that USP1 stabilizes Nsp1β protein by eliminating K48 ubiquitination of the lysine at position 45.
Fig 7.
USP1 modulates Nsp1β stabilization by deubiquitinating Nsp1β. (A) Diagram showing Nsp1β and its lysine-mutant variants. (B) Nsp1β-NL-Flag was cotransfected with the K48-Ub-HA plasmid in HEK293T cells, and a Co-IP assay was performed 24 h later. (C) Nsp1β-PC-Flag was cotransfected with the K48-Ub-HA plasmid in HEK293T cells, and a Co-IP assay was performed 24 h later. (D) Nsp1β-Flag, Nsp1β-NL-Flag, Nsp1β-PC-Flag, and K48-Ub-HA were cotransfected into HEK293T cells, and a Co-IP assay was performed after 24 h. (E) The Nsp1β-NL-terminal mutant plasmid was cotransfected into HEK293T cells and K48-Ub-HA. (F) Nsp1β-K45R-Flag was cotransfected into HEK293T cells with Nsp1β-Flag and K48-Ub-HA, and Co-IP assay was performed 24 h later for the Co-IP assay.
USP1 does not promote the replication of PRRSV rSD16-K45R
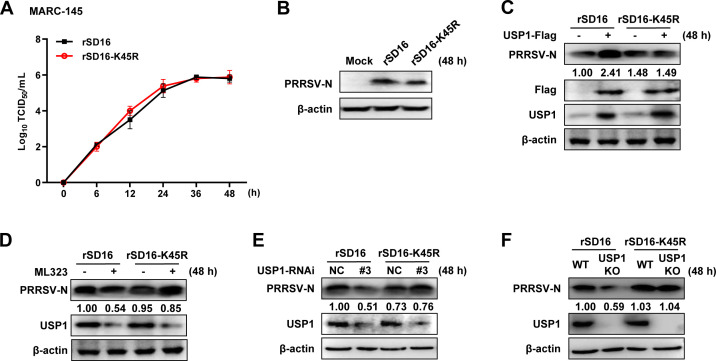
Next, we investigated the effect of K48 ubiquitination of Nsp1β on viral replication. We constructed a mutant PRRSV rSD16-K45R virus and compared the mutant virus with a wild-type virus. The findings demonstrated that both viruses’ titers peaked at 48 h, and the mutant virus rSD16-K45R’s growth kinetics were identical to those of rSD16 (Fig. 8A). Western blot showed that after PRRSV infected the cells, the N protein expression of the two viruses was comparable (Fig. 8B). These findings suggested that the rSD16-K45R virus was capable of in vitro stable replication. We also looked at how USP1 affected the propagation of the rSD16-K45R virus. WT virus was used as a control, and rSD16-K45R N protein expression did not change with USP1 expression (Fig. 8C). Upon ML323 treatment, the endogenous USP1 protein level was reduced, but rSD16-K45R N protein was not changed, and WT virus was used as a control (Fig. 8D). Consistently, rSD16-K45R N protein was not degraded after knockdown or knockout of USP1 (Fig. 8E and F). These results indicated that lysine at position 45 of Nsp1β is crucial for viral replication and that rSD16-K45R viruses are more stable.
Fig 8.
In vitro replication of the PRRSV rSD16-K45R virus. (A) MARC-145 was infected with rSD16 and rSD16-K45R (MOI = 10) at different time points (0, 6, 12, 24, 36, and 48 h), and the viral life cycle was determined by the viral titer. (B) Western blot was used to detect the expression of N protein in MARC-145 cells infected with rSD16 and rSD16-K45R (MOI = 10) for 48 h. (C) MARC-145 cells transfected with the empty plasmid and USP1-Flag plasmid were infected with rSD16-K45R and rSD16 (MOI = 10) for 48 h, and the change of PRRSV N was detected by Western blot. The ratio of PRRSV-GFP to β-actin was analyzed by ImageJ. (D) MARC-145 cells transfected with ML323 were infected with rSD16-K45R and rSD16 (MOI = 10) for 48 h, and the expression of PRRSV N was detected by Western blot. (E) rSD16-K45R and rSD16 were infected with siUSP1#3-transfected MARC-145 cells (MOI = 10), and PRRSV N protein expression was found 48 h later. (F) The N protein expression of rSD16-K45R and rSD16 (MOI = 10) in USP1 KO MARC-145 cells was detected by Western blot.
DISCUSSION
The first line of host defense against viral infections is innate immunity (35). PRRSV prolongs the acute and chronic phases of infection by using various strategies to subvert the host’s antiviral defenses and foster favorable conditions for its survival (36). Because of abrupt pandemic outbreaks and fast-changing PRRSV strains in swine herds, the present PRRSV prevention strategy, which relies on vaccines, is continually tested (37). As a result, there is a tremendous need to create small-molecule medications that can modulate host cellular components and signaling pathways essential for PRRSV infection. Although the intricate interaction between the PRRSV and the host cell is still not fully understood, our study may serve as a theoretical foundation for developing new biotherapeutics to manage PRRS. Hence, our study provides new PRRSV prevention and control ideas by screening therapeutic small-molecule drugs for PRRSV infection and further exploring its mechanism.
Nsp1 is an important antagonist to inhibit IFNβ production (38). Nsp1α and Nsp1β are involved in suppressing IFNβ luciferase activity. Nsp1β decreases the phosphorylation of IRF3 (Ser 396) and inhibits nuclear translocation of IRF3 (39). Nsp1β inhibits phosphorylation of STAT1 and nuclear translocation of ISGF3 (40). These reports suggest an important role for Nsp1β in PRRSV immune escape. Recent studies have found that Nsp1β mediates stress granules (SG) from PRK restriction and suppresses the inflammatory response (41). Studies on the involvement of ubiquitination modifications in PRRSV replication have also shown new advances, with claims that hypoxia-inducible factor 1 was further stimulated by von Hippel–Lindau tumor suppressor (pVHL)-stabilized Nsp1β via K63-linked ubiquitination. These findings mainly focus on how the host protein pVHL maintained Nsp1β via K63-linked ubiquitination, maintaining Nsp1β protein production (42). Based on these results, we also found that Nsp1β undergoes ubiquitination modifications, mainly K48 ubiquitination. Among them, Nsp1β has five lysine residues, and we found that the lysine at position 45 is the essential site for K48 ubiquitination, which establishes that ubiquitination is involved in PRRSV replication, but studies on the role of deubiquitinating enzymes in PRRSV replication need to be supplemented. PRRSV-1 has frequent gene recombination and high genetic diversity and is more prevalent in Europe and Africa. Correspondingly, the PRRSV-2 genome is more stable and is prevalent mainly in North America and East Asia (43). We analyzed the Nsp1β genes of PRRSV-1 and PRRSV-2, and the lysine residue at position 45 was not conserved. In this study, we focused on PRRSV-2 and labeled the virus type in the material methods. We performed a preliminary screen of deubiquitinating enzyme inhibitors for this purpose, and the USP1 inhibitor, ML323, produced a significant inhibitory effect on PRRSV invasion. These studies revealed that the host degrades Nsp1β by ubiquitinating it through post-translational regulation, but PRRSV uses the deubiquitinating enzyme USP1 to evade the host immune response. However, we demonstrated the K48 ubiquitination modification of Nsp1β and the role of USP1 in its deubiquitination in our study. However, the ubiquitinating enzymes involved in PRRSV ubiquitination require further research. All the aforementioned phenomena indicate that ubiquitinating and deubiquitinating enzyme lines are indeed involved in PRRSV replication, which shows that the virus’ strategy of immune escape by using host proteins is complicated, giving new significance to our study.
UPS plays a double-edged role in viral infections (44). On the one hand, many viruses hijack UPS proteins to maintain normal function (45). On the other hand, UPS proteins are involved in host defense to eliminate viruses (46). Viruses can also use UPS proteins to degrade or eliminate host defense factors (47). Importantly, UPS proteins can degrade proteins that impede viral replication and are involved in the replication and proliferation of plus-strand RNA viruses (45). It has been reported that UPS proteins are involved in viral pathogenesis and have antiviral effects. A viral limiting factor in the innate immune response of MARC-145 cells and alveolar macrophages is the porcine ubiquitin-specific peptidase 18. CH25H of host cells degrades the nonstructural protein Nsp1α of PRRSV through the ubiquitin–proteasome pathway (48). Nsp1α uses the ubiquitin–proteasome system to degrade the host cell’s CBP and SLA-I proteins. Deubiquitinating enzymes function during viral infections and are involved in cancer, inflammation, and autoimmune diseases, so developing small-molecule inhibitors against deubiquitinate has become a hot topic of research (49). Many of them have good performance in antiviral mechanisms (50, 51). USP1 has a regulatory function on PRRSV replication, and USP1 may become a new antiviral target, while the inhibitor ML323 may provide new possibilities for preventing and treating PRRS.
Our study found that the nonstructural protein Nsp1β of PRRSV forms mainly K48 ubiquitination modifications. We hypothesized that this is a host strategy to inhibit viral infection by degrading viral proteins. However, PRRSV may escape host defenses in other ways. In our study, we screened the USP1 inhibitor ML323 to antagonize PRRSV. We then focused on the role of USP1 in PRRSV infection. PRRSV replication was significantly slowed by inhibitor treatment and upon deletion of USP1. We further investigated the type and site of ubiquitination of the PRRSV nonstructural protein Nsp1β and found that the lysine at position 45 is critical for the stability of Nsp1β. This work offers proof that PRRSV infection depends on ubiquitin alterations. In vitro testing of the particular USP1 inhibitor ML323 revealed notable antiviral activity. This study provides evidence that ubiquitin modification is important for PRRSV infection.
MATERIALS AND METHODS
Cells and viruses
MARC-145 cells, HEK293T cells, and PAM-Tang cells were obtained from the American Type Culture Collection (VA, USA). MARC-145 and HEK293T cells were cultured separately in Dulbecco’s modified Eagle medium (DMEM; Gibco, Grand Island, NY, USA), which also contained 10% heat-inactivated fetal bovine serum (FBS) and 100 U/mL penicillin and 100 µg/mL streptomycin sulfate (FBS; Gibco). The PRRSV-sensitive cell line PAM-Tang was grown in RPMI-1640 media obtained from Corning Inc. Fetal bovine serum, obtained from Gibco with 10% (vol/vol) fetal bovine serum, was used. All cells were cultured at 37°C and 5% CO2.
PRRSV BJ-4 strain (PRRSV-2) is a typical North American strain reported in China in 1996 (GenBank: AF331831) and labeled as BJ-4 in Fig. 1 to 4. Based on the genetic makeup of the SD16 (PRRSV-2) strain that expressed enhanced green fluorescent protein (EGFP) as an additional ORF, Professor En-min Zhou of Northwest A&F University created EGFP-PRRSV (rHP-PRRSV/SD16/EGFP) (52), which is given as rSD16 in the figure. The virus was reproduced, examined in MARC-145 cells, and stored at 80°C.
Antibodies and reagents
The following are the sources and antibodies used: Proteintech Group Inc. provided the following antibodies: anti-USP1 (1:1,000, 66069-1-Ig), anti-Flag (1:1,000, 80010-1-RR), and anti-actin (1:2,000, 66009-1-lg). Monoclonal anti-Flag-HRP antibody (1:1,000, A8592), anti-HA-HRP antibody (1:1,000, H6533), and anti-Flag M2 beads were purchased from Sigma-Aldrich. ML323 (HY-17543), PR-619 (HY-13814), USP7-IN-1 (HY-16709), P22077 (HY-13865), P005091 (HY-15667), HBX-19818 (HY-17540), USP7-USP47 inhibitor (HY-13487), DUBs-IN-1 (HY-50736), DUBs-IN-2 (HY-50737A), and DUBs-IN-3 (HY-50737) were purchased from MedChemExpress. We bought protease inhibitors from Roche. Phenyl methane sulfonyl fluoride (PMSF) was purchased from Solarbio Life Sciences, Beijing, China.
Plasmids
PrimeSTAR HS DNA Polymerase was used to amplify the USP1 (GenBank: NW_023666033.1) genes from MARC-145 cDNA before cloning them into the p3×Flag-CMV-14 or pEGFP-N1 expression vectors, referred to as USP1-Flag and USP1-GFP, respectively.
The Nsp1β gene was amplified from PRRSV BJ-4-infected Marc-145 cell cDNA using PrimeSTAR HS DNA Polymerase and cloned into p3×Flag-CMV-14 or pCAGGS-HA expression vectors, referred to as Nsp1β-Flag and Nsp1β-HA, respectively. The truncated mutant (amino acids 1–111 and 112–229) plasmids p3×Flag-CMV-Nsp1β-NL-Flag and p3×Flag-CMV-Nsp1β-PC-Flag were constructed using the Nsp1β-Flag vector as the template, referred to as Nsp1β-NL-Flag and Nsp1β-PC-Flag, respectively. The mutant plasmids of Nsp1β-NL ubiquitin sites (K16, K45, K57, and K86) and Nsp1β-K45 were constructed and cloned into the p3×Flag-CMV-14 plasmid.
According to the manufacturer’s instructions, polyethylenimine (PEI) (Polyscience, IL, USA) transfected WT-Ub-HA, K48-Ub-HA, or K63-Ub-HA plasmids from the International Joint Research Center of National Animal Immunology.
Construction of stable cell lines and lentivirus infection
Lentiviral particles were created in HEK293T cells transfected with the empty vector or LentiCRISPR v2-USP1 plasmid utilizing PEI and two packaging plasmids (psPAX2 and pMD2.G). The recombinant viruses were filtered after 48 h and added to MARC-145 and HEK293T cells with polybrene (8 µg/mL, Cat#H8641, Solarbio, China). Puromycin (Cat#IP1160, Solarbio) was used to select cells for 5 days at a final concentration of 8 μg/mL. The restricted dilution approach was applied to generate monoclonal cells in 96-well plates, and USP1 antibody immunoblotting was used to identify them. The following sequences were targeted for USP1 mRNA: human-USP1sgRNA: 5′-CACCGTCCAAGGAACACCAGTCAT-3′; monkey-USP1sgRNA: 5′-CACCGGTGGAGTGAGGAAGACCCTAG-3′.
RNA-mediated interference (RNAi)
We designed different siRNAs for monkey-derived USP1. Duplexes of short-interfering RNA were transfected into MARC-145 cells for transient gene silencing using the Lipofectamine 2000 reagent from Invitrogen following the recommended procedure. The siRNA sequences were as follows: siUSP1#1: 5′-GGAUUUCACAGAUUCUCAATT-3′; siUSP1#2: 5′-GCAGUUUACAGUCCUUAAUTT-3′; siUSP1#3: 5′-GGAUUUGAAUCUCCAGGAATT-3′.
Reverse transcription-quantitative PCR
Total RNA was isolated to measure the mRNA expression of the investigated genes using RT-qPCR. The gene expression was normalized with β-actin as the internal control. The primer sequences were as follows: monkey USP1-Fw: 5′-ACACAACTCATGCTTGTGGATT-3′; monkey USP1-Rv: 5′-ACTGGCACACTGATGTCCTGCTG-3′; monkey β-actin-Fw: 5′-CGTGGACATCCGTAAAGAC-3′; monkey β-actin-Rv: 5′-GGAAGGTGGACAGCGAGGC-3′; PRRSV ORF7-Fw: 5′-AGATCATCGCCCAACAAAAC-3′; PRRSV ORF7-Rv: 5′-ACACAATTGCCGCTCACTA-3′.
Immunoprecipitation and Western blot analysis
After 24 h of transfection, whole-cell extracts were taken for immunoprecipitation and lysed in M2 buffer containing MG132 (HY-13259, MCE), a protease inhibitor cocktail from Merck, 0.5% (vol/vol) Nonidet P-40, 20 mM Tris-HCl, 3 mM EDTA, and 3 mM EGTA. The cell supernatants were collected after centrifugation for 10 minutes at 12,000 g and mixed with 10 µL monoclonal anti-Flag M2 beads (SIGMA) for 3-h incubation. The incubated beads were washed five times with elution buffer containing 0.5% (vol/vol) Nonidet P-40, 20 mM Tris-HCl, 3 mM EDTA, 3 mM EGTA, and 500 mM NaCl. The immunoprecipitates were eluted by boiling in 1% (wt/vol) SDS sample buffer. Cell lysates were extracted with 2% (wt/vol) SDS for Western blot examination. As previously reported, the whole-cell lysates and the immunoprecipitates prepared before were submitted to SDS-PAGE, which was then transferred onto nitrocellulose membranes for blotting (53).
Immunofluorescence assay
For the immunofluorescence assay, Nsp1β-Flag-transfected MARC-145 cells were grown for 24 h at 37°C before being fixed for 10 minutes with 4% buffered paraformaldehyde. Cells were then permeabilized in 0.1% Triton X-100 for 5 minutes at room temperature. Before being washed three times with phosphate-buffered saline (PBS), cells were exposed to anti-Flag pAb (1:500) and anti-USP1 protein mAb (1:500) at room temperature for 1 h. Goat anti-rabbit IgG (H&L) (1:500) and goat anti-mouse IgG (H&L) (1:500) staining procedures were then performed on the cells for an hour at room temperature and in the dark. 4',6-Diamidino-2-phenylindole (DAPI) (1:5,000) was used as a counterstain to stain nuclei, which were detected by a fluorescence microscope from the Olympus Corporation (Tokyo, Japan). Immunofluorescence was observed, and mock-infected cells were used for the background staining.
Flow cytometry
For viral proliferation assays, cells were infected with recombinant viruses for 24–48 h and detached from cell plates by trypsin–EDTA digestion. Next, cells were collected by centrifugation and re-suspended in PBS before the flow cytometry assay. The percentage of GFP-positive cells was calculated using flow cytometry and a BD AccuriC6 instrument. The CytExpert software was used to analyze all the data.
Viral titration
Viral titration assays were applied to determine viral infectivity, including the TCID50 assay. On day 0, a cell density of 1 × 104 cells per well was used to seed cells into 96-well plates. On day 1, the viruses were applied to the cells, serially diluted 10 times, and then infected. The cells were then incubated at 37°C for 2 h. After incubation, the extra viral inoculum was removed by washing with PBS. Then, each well received 100 µL of maintenance media (2% FBS/DMEM), and cells were maintained for an additional 5–7 days. The TCID50 value was determined using the Reed–Muench method, and the cytotoxic effects were monitored daily.
Reverse genetics-based mutagenesis of PRRSV
Using the targeted mutagenesis primer (5′-GCCGAAGGGAGAATCTCCTGG-3′), the aa residue at position 45 of Nsp1β was altered. Following Kas I and Pml I digestion, cDNA fragments containing the required mutations were ligated to rSD16, and constructions were later verified by DNA sequencing. According to the manufacturer’s instructions, Attractene Transfection Reagent (Qiagen, Hilden, Germany) was used to transfect rSD16 and constructs with mutated Nsp1β sites into MARC-145 cells to rescue mutated viruses. Three days after transfection, cell culture supernatants were collected, and Western blot tests were carried out to verify the replication of recombinant viruses. In MARC-145 cells, the cell culture supernatants were serially passaged three times. Whole-genome sequencing was then performed to confirm the virus rescue. The designation for recovered viruses was rSD16-K45R.
The identification of site-specific modification on Nsp1β
First, vector plasmid, WT-Ub-HA, K48-Ub-HA, or K63-Ub-HA, and Nsp1β-Flag plasmid were cotransfected in HEK293T cells. After 24-h transfection, the cell lysates were precipitated with anti-Flag M2 beads and further detected by a Western blot with anti-Flag MAb and anti-HA antibodies, respectively. To learn more about the mechanism of action of USP1, HEK293T cells were pretreated with 10 µM ML323 for 4 h before being transfected with the appropriate plasmid and having cell lysates prepared 24 h later. A Flag antibody was used for immunoprecipitation, and a Western blot with an anti-HA antibody was used to confirm that the protein had been ubiquitinated.
To target specific modification, the vector encoding Nsp1β-NL-Flag or Nsp1β-PC-Flag or Nsp1β mutants (shown in Fig. 7), WT-Ub-HA, K48-Ub-HA, or K63-Ub-HA were cotransfected in HEK293T cells for 24 h. Immunoprecipitates were probed with Flag and HA antibodies, and the targets were detected with the whole-cell lysate by Western blot.
Statistical analysis
The experiments were run in triplicate. The means and standard errors of the data were shown. All statistical analyses were performed using one-way analysis of variance. The significant differences of the corresponding controls are set at *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
ACKNOWLEDGMENTS
We acknowledge Jiang Wang (Henan Agricultural University) for providing mutant PRRSV and helpful discussions.
This study was supported by grants from the National Natural Science Foundation of China (31490601 and 32272987). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Contributor Information
Zhengjie Kong, Email: kongzhengjie@pku.edu.cn.
Bo Wan, Email: wanboyi2000@163.com.
Martin Schwemmle, University Medical Center Freiburg, Freiburg, Germany.
DATA AVAILABILITY
All data generated or analyzed during this study are included in the article.
REFERENCES
- 1. Lunney JK, Benfield DA, Rowland RRR. 2010. Porcine reproductive and respiratory syndrome virus: an update on an emerging and re-emerging viral disease of swine. Virus Res. 154:1–6. doi: 10.1016/j.virusres.2010.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Neumann EJ, Kliebenstein JB, Johnson CD, Mabry JW, Bush EJ, Seitzinger AH, Green AL, Zimmerman JJ. 2005. Assessment of the economic impact of porcine reproductive and respiratory syndrome on swine production in the United States. J Am Vet Med Assoc 227:385–392. doi: 10.2460/javma.2005.227.385 [DOI] [PubMed] [Google Scholar]
- 3. Meulenberg JJ, de Meijer EJ, Moormann RJ. 1993. Subgenomic RNAs of lelystad virus contain a conserved leader-body junction sequence. J Gen Virol 74:1697–1701. doi: 10.1099/0022-1317-74-8-1697 [DOI] [PubMed] [Google Scholar]
- 4. Wensvoort G, Terpstra C, Pol JM, ter Laak EA, Bloemraad M, de Kluyver EP, Kragten C, van Buiten L, den Besten A, Wagenaar F. 1991. Mystery swine disease in the Netherlands: the isolation of lelystad virus. Vet Q 13:121–130. doi: 10.1080/01652176.1991.9694296 [DOI] [PubMed] [Google Scholar]
- 5. Benfield DA, Nelson E, Collins JE, Harris L, Goyal SM, Robison D, Christianson WT, Morrison RB, Gorcyca D, Chladek D. 1992. Characterization of swine infertility and respiratory syndrome (SIRS) virus (isolate ATCC VR-2332). J Vet Diagn Invest 4:127–133. doi: 10.1177/104063879200400202 [DOI] [PubMed] [Google Scholar]
- 6. Kappes MA, Faaberg KS. 2015. PRRSV structure, replication and recombination: origin of phenotype and genotype diversity. Virology 479–480:475–486. doi: 10.1016/j.virol.2015.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tian K, Yu X, Zhao T, Feng Y, Cao Z, Wang C, Hu Y, Chen X, Hu D, Tian X, et al. 2007. Emergence of fatal PRRSV variants: unparalleled outbreaks of atypical PRRS in China and molecular dissection of the unique hallmark. PLoS One 2:e526. doi: 10.1371/journal.pone.0000526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhao K, Ye C, Chang XB, Jiang CG, Wang SJ, Cai XH, Tong GZ, Tian ZJ, Shi M, An TQ. 2015. Importation and recombination are responsible for the latest emergence of highly pathogenic porcine reproductive and respiratory syndrome virus in China. J Virol 89:10712–10716. doi: 10.1128/JVI.01446-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhou L, Wang Z, Ding Y, Ge X, Guo X, Yang H. 2015. NADC30-like strain of porcine reproductive and respiratory syndrome virus, China. Emerg Infect Dis 21:2256–2257. doi: 10.3201/eid2112.150360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Liu J, Wei C, Lin Z, Fan J, Xia W, Dai A, Yang X. 2019. Recombination in lineage 1, 3, 5 and 8 of porcine reproductive and respiratory syndrome viruses in China. Infect Genet Evol 68:119–126. doi: 10.1016/j.meegid.2018.12.006 [DOI] [PubMed] [Google Scholar]
- 11. Lager KM, Mengeling WL, Brockmeier SL. 1999. Evaluation of protective immunity in gilts Inoculated with the NADC-8 isolate of porcine reproductive and respiratory syndrome virus (PRRSV) and challenge-exposed with an antigenically distinct PRRSV isolate. Am J Vet Res 60:1022–1027. [PubMed] [Google Scholar]
- 12. Meng XJ. 2000. Heterogeneity of porcine reproductive and respiratory syndrome virus: implications for current vaccine efficacy and future vaccine development. Vet Microbiol 74:309–329. doi: 10.1016/s0378-1135(00)00196-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lunney JK, Fang Y, Ladinig A, Chen N, Li Y, Rowland B, Renukaradhya GJ. 2016. Porcine reproductive and respiratory syndrome virus (PRRSV): pathogenesis and interaction with the immune system. Annu Rev Anim Biosci 4:129–154. doi: 10.1146/annurev-animal-022114-111025 [DOI] [PubMed] [Google Scholar]
- 14. Ahlquist P, Noueiry AO, Lee WM, Kushner DB, Dye BT. 2003. Host factors in positive-strand RNA virus genome replication. J Virol 77:8181–8186. doi: 10.1128/JVI.77.15.8181-8186.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chasman D, Gancarz B, Hao L, Ferris M, Ahlquist P, Craven M. 2014. Inferring host gene subnetworks involved in viral replication. PLoS Comput Biol 10:e1003626. doi: 10.1371/journal.pcbi.1003626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Krishnan MN, Ng A, Sukumaran B, Gilfoy FD, Uchil PD, Sultana H, Brass AL, Adametz R, Tsui M, Qian F, Montgomery RR, Lev S, Mason PW, Koski RA, Elledge SJ, Xavier RJ, Agaisse H, Fikrig E. 2008. RNA interference screen for human genes associated with West Nile virus infection. Nature 455:242–245. doi: 10.1038/nature07207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hao L, Sakurai A, Watanabe T, Sorensen E, Nidom CA, Newton MA, Ahlquist P, Kawaoka Y. 2008. Drosophila RNAi screen identifies host genes important for influenza virus replication. Nature 454:890–893. doi: 10.1038/nature07151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yoo D, Song C, Sun Y, Du Y, Kim O, Liu HC. 2010. Modulation of host cell responses and evasion strategies for porcine reproductive and respiratory syndrome virus. Virus Res 154:48–60. doi: 10.1016/j.virusres.2010.07.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sun Y, Han MY, Kim C, Calvert JG, Yoo D. 2012. Interplay between interferon-mediated innate immunity and porcine reproductive and respiratory syndrome virus. Viruses 4:424–446. doi: 10.3390/v4040424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Costers S, Lefebvre DJ, Delputte PL, Nauwynck HJ. 2008. Porcine reproductive and respiratory syndrome virus modulates apoptosis during replication in alveolar macrophages. Arch Virol 153:1453–1465. doi: 10.1007/s00705-008-0135-5 [DOI] [PubMed] [Google Scholar]
- 21. Labarque G, Van Gucht S, Nauwynck H, Van Reeth K, Pensaert M. 2003. Apoptosis in the lungs of pigs infected with porcine reproductive and respiratory syndrome virus and associations with the production of apoptogenic cytokines. Vet Res 34:249–260. doi: 10.1051/vetres:2003001 [DOI] [PubMed] [Google Scholar]
- 22. Liu F, Du Y, Feng WH. 2017. New perspective of host microRNAs in the control of PRRSV infection. Vet Microbiol 209:48–56. doi: 10.1016/j.vetmic.2017.01.004 [DOI] [PubMed] [Google Scholar]
- 23. Van Reeth K, Labarque G, Nauwynck H, Pensaert M. 1999. Differential production of proinflammatory cytokines in the pig lung during different respiratory virus infections: correlations with pathogenicity. Res Vet Sci 67:47–52. doi: 10.1053/rvsc.1998.0277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. D’Arcy P, Wang X, Linder S. 2015. Deubiquitinase inhibition as a cancer therapeutic strategy. Pharmacol Ther 147:32–54. doi: 10.1016/j.pharmthera.2014.11.002 [DOI] [PubMed] [Google Scholar]
- 25. Nijman SMB, Huang TT, Dirac AMG, Brummelkamp TR, Kerkhoven RM, D’Andrea AD, Bernards R. 2005. The deubiquitinating enzyme USP1 regulates the Fanconi anemia pathway. Mol Cell 17:331–339. doi: 10.1016/j.molcel.2005.01.008 [DOI] [PubMed] [Google Scholar]
- 26. Gao W, Wang L, Ju X, Zhao S, Li Z, Su M, Xu J, Wang P, Ding Q, Lv G, Zhang W. 2022. The deubiquitinase USP29 promotes SARS-CoV-2 virulence by preventing proteasome degradation of ORF9b. mBio 13:e0130022. doi: 10.1128/mbio.01300-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Subramaniyan B, Larabee JL, Bodas M, Moore AR, Burgett AWG, Papin JF, Walters MS. 2023. Inhibition of the cellular deubiquitinase UCHL1 suppresses SARS-CoV-2 replication. Am J Respir Cell Mol Biol 69:367–370. doi: 10.1165/rcmb.2023-0076LE [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wang YC, Li Q, Hu DW, Gao DL, Wang WB, Wu KL, Wu JG. 2021. USP38 inhibits zika virus infection by removing envelope protein ubiquitination. Viruses 13:2029. doi: 10.3390/v13102029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chen H-Y, Tang R-C, Liang J-W, Zhao W, Yu S-S, Yao R-R, Xu R, Zhang A, Geng S, Sun X-Y, Ge Q, Zhang J. 2023. Deubiquitinase USP47 attenuates virus-induced type I interferon signaling. Int Immunopharmacol 118:110040. doi: 10.1016/j.intimp.2023.110040 [DOI] [PubMed] [Google Scholar]
- 30. Wang XM, Yang C, Zhao Y, Xu ZG, Yang W, Wang P, Lin DD, Xiong B, Fang JY, Dong C, Zhong B. 2020. The deubiquitinase USP25 supports colonic inflammation and bacterial infection and promotes colorectal cancer. Nat Cancer 1:811–825. doi: 10.1038/s43018-020-0089-4 [DOI] [PubMed] [Google Scholar]
- 31. Lin JY, Ai YX, Zhou HD, Lv Y, Wang MH, Xu JC, Yu C, Zhang HM, Wang MY. 2020. UL36 encoded by Marek’s disease virus exhibits linkage-specific deubiquitinase activity. IJMS 21:1783. doi: 10.3390/ijms21051783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Karlowitz R, Stanifer ML, Roedig J, Andrieux G, Bojkova D, Bechtel M, Smith S, Kowald L, Schubert R, Boerries M, Cinatl J, Boulant S, van Wijk SJL. 2022. USP22 controls type III interferon signaling and SARS-CoV-2 infection through activation of STING. Cell Death Dis 13:684. doi: 10.1038/s41419-022-05124-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Luo Y, Zhang X, Chen R, Li R, Liu Y, Zhang J, Liu Q, Si M, Liu J, Wu B, Wang X, Wu S, Zhang Y, Zhang X, Guo D, He X, Pan T, Zhang H. 2022. USP10 regulates B cell response to SARS-CoV-2 or HIV-1 nanoparticle vaccines through deubiquitinating AID. Signal Transduct Target Ther 7:7. doi: 10.1038/s41392-021-00858-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Martínez-Férriz A, Ferrando A, Fathinajafabadi A, Farràs R. 2022. Ubiquitin-mediated mechanisms of translational control. Semin Cell Dev Biol 132:146–154. doi: 10.1016/j.semcdb.2021.12.009 [DOI] [PubMed] [Google Scholar]
- 35. Chen XY, Liu SS, Goraya MU, Maarouf M, Huang SL, Chen JL. 2018. Host immune response to influenza A virus infection. Front Immunol 9:320. doi: 10.3389/fimmu.2018.00320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. An TQ, Li JN, Su CM, Yoo D. 2020. Molecular and cellular mechanisms for PRRSV pathogenesis and host response to infection. Virus Res 286:197980. doi: 10.1016/j.virusres.2020.197980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Thanawongnuwech R, Suradhat S. 2010. Taming PRRSV: revisiting the control strategies and vaccine design. Virus Res 154:133–140. doi: 10.1016/j.virusres.2010.09.003 [DOI] [PubMed] [Google Scholar]
- 38. Shi XB, Zhang GP, Wang L, Li XW, Zhi YB, Wang FY, Fan JM, Deng RG. 2011. The nonstructural protein 1 papain-like cysteine protease was necessary for porcine reproductive and respiratory syndrome virus nonstructural protein 1 to inhibit interferon-β induction. DNA Cell Biol 30:355–362. doi: 10.1089/dna.2010.1188 [DOI] [PubMed] [Google Scholar]
- 39. Chen Z, Lawson S, Sun Z, Zhou X, Guan X, Christopher-Hennings J, Nelson EA, Fang Y. 2010. Identification of two auto-cleavage products of nonstructural protein 1 (nsp1) in porcine reproductive and respiratory syndrome virus infected cells: nsp1 function as interferon antagonist. Virology 398:87–97. doi: 10.1016/j.virol.2009.11.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Patel D, Nan YC, Shen MY, Ritthipichai K, Zhu XP, Zhang YJ. 2010. Porcine reproductive and respiratory syndrome virus inhibits type I interferon signaling by blocking STAT1/STAT2 nuclear translocation. J Virol 84:11045–11055. doi: 10.1128/JVI.00655-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gao P, Liu YY, Wang H, Chai Y, Weng WL, Zhang YN, Zhou L, Ge XN, Guo X, Han J, Yang HC. 2022. Viral evasion of PKR restriction by reprogramming cellular stress granules. Proc Natl Acad Sci U S A 119:e2201169119. doi: 10.1073/pnas.2201169119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Pang Y, Zhou YR, Wang YC, Sun Z, Liu J, Li CY, Xiao SB, Fang LR. 2022. Porcine reproductive and respiratory syndrome virus nsp1β stabilizes HIF-1α to enhance viral replication. Microbiol Spectr 10:e0317322. doi: 10.1128/spectrum.03173-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Chae H, Roh HS, Jo YM, Kim WG, Chae JB, Shin SU, Kang JW. 2023. Development of a one-step reverse transcription-quantitative polymerase chain reaction assay for the detection of porcine reproductive and respiratory syndrome virus. PLoS One 18:e0293042. doi: 10.1371/journal.pone.0293042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lange SM, Armstrong LA, Kulathu Y. 2022. Deubiquitinases: from mechanisms to their inhibition by small molecules. Mol Cell 82:15–29. doi: 10.1016/j.molcel.2021.10.027 [DOI] [PubMed] [Google Scholar]
- 45. Gao WY, Wang LL, Ju XH, Zhao SM, Li ZL, Su MM, Xu JC, Wang PH, Ding Q, Lv GY, Zhang WY. 2022. The deubiquitinase USP29 promotes SARS-CoV-2 virulence by preventing proteasome degradation of ORF9b. mBio 13. doi: 10.1128/mbio.01300-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Guo YY, Jiang F, Kong LL, Wu HF, Zhang HH, Chen XR, Zhao J, Cai BS, Li YQ, Ma CH, Yi F, Zhang L, Liu BY, Zheng Y, Zhang LQ, Gao CJ. 2021. OTUD5 promotes innate antiviral and antitumor immunity through deubiquitinating and stabilizing STING. Cell Mol Immunol 18:1945–1955. doi: 10.1038/s41423-020-00531-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sun H, Zhang Q, Jing YY, Zhang M, Wang HY, Cai Z, Liuyu TZ, Zhang ZD, Xiong TC, Wu Y, Zhu QY, Yao J, Shu HB, Lin DD, Zhong B. 2017. USP13 negatively regulates antiviral responses by deubiquitinating STING. Nat Commun 8:15534. doi: 10.1038/ncomms15534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ke WT, Fang LR, Jing HY, Tao R, Wang T, Li Y, Long SW, Wang D, Xiao SB. 2017. Cholesterol 25-hydroxylase inhibits porcine reproductive and respiratory syndrome virus replication through enzyme activity-dependent and -independent mechanisms. J Virol 91:e00827-17. doi: 10.1128/JVI.00827-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Schauer NJ, Magin RS, Liu X, Doherty LM, Buhrlage SJ. 2020. Advances in discovering deubiquitinating enzyme (DUB) inhibitors. J Med Chem 63:2731–2750. doi: 10.1021/acs.jmedchem.9b01138 [DOI] [PubMed] [Google Scholar]
- 50. Ming SL, Zhang S, Wang Q, Zeng L, Zhou LY, Wang MD, Ma YX, Han LQ, Zhong K, Zhu HS, Bai YL, Yang GY, Wang J, Chu BB. 2022. Inhibition of USP14 influences alphaherpesvirus proliferation by degrading viral VP16 protein via ER stress-triggered selective autophagy. Autophagy 18:1801–1821. doi: 10.1080/15548627.2021.2002101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Große M, Setz C, Rauch P, Auth J, Morokutti-Kurz M, Temchura V, Schubert U. 2022. Inhibitors of deubiquitinating enzymes interfere with the SARS-CoV-2 papain-like protease and block virus replication in vitro. Viruses 14:Viruses–Basel. doi: 10.3390/v14071404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wang CB, Huang BC, Kong N, Li QY, Ma YP, Li ZJ, Gao JM, Zhang C, Wang XP, Liang C, Dang L, Xiao SQ, Mu Y, Zhao Q, Sun Y, Almazan F, Enjuanes L, Zhou EM. 2013. A novel porcine reproductive and respiratory syndrome virus vector system that stably expresses enhanced green fluorescent protein as a separate transcription unit. Vet Res 44. doi: 10.1186/1297-9716-44-104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Xiong TC, Wei MC, Li FX, Shi M, Gan H, Tang Z, Dong HP, Liuyu TZ, Gao P, Zhong B, Zhang ZD, Lin DD. 2022. The E3 ubiquitin ligase ARIH1 promotes antiviral immunity and autoimmunity by inducing mono-ISGylation and oligomerization of cGAS. Nat Commun 13:5973. doi: 10.1038/s41467-022-33671-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in the article.