ABSTRACT
Multidrug-resistant Escherichia coli is a leading cause of global mortality. Transfer of plasmids carrying genes encoding beta-lactamases, carbapenamases, and colistin resistance between lineages is driving the rising rates of hard-to-treat nosocomial and community infections. Multidrug resistance (MDR) plasmid acquisition commonly causes transcriptional disruption, and while a number of studies have shown strain-specific fitness and transcriptional effects of an MDR plasmid across diverse bacterial lineages, fewer studies have compared the impacts of different MDR plasmids in a common bacterial host. As such, our ability to predict which MDR plasmids are the most likely to be maintained and spread in bacterial populations is limited. Here, we introduced eight diverse MDR plasmids encoding resistances against a range of clinically important antibiotics into E. coli K-12 MG1655 and measured their fitness costs and transcriptional impacts. The scale of the transcriptional responses varied substantially between plasmids, ranging from >650 to <20 chromosomal genes being differentially expressed. However, the scale of regulatory disruption did not correlate significantly with the magnitude of the plasmid fitness cost, which also varied between plasmids. The identities of differentially expressed genes differed between transconjugants, although the expression of certain metabolic genes and functions were convergently affected by multiple plasmids, including the downregulation of genes involved in L-methionine transport and metabolism. Our data show the complexity of the interaction between host genetic background and plasmid genetic background in determining the impact of MDR plasmid acquisition on E. coli.
IMPORTANCE
The increase in infections that are resistant to multiple classes of antibiotics, including those isolates that carry carbapenamases, beta-lactamases, and colistin resistance genes, is of global concern. Many of these resistances are spread by conjugative plasmids. Understanding more about how an isolate responds to an incoming plasmid that encodes antibiotic resistance will provide information that could be used to predict the emergence of MDR lineages. Here, the identification of metabolic networks as being particularly sensitive to incoming plasmids suggests the possible targets for reducing plasmid transfer.
KEYWORDS: multidrug resistance, plasmids, Escherichia coli, transcriptomics
INTRODUCTION
Escherichia coli is a leading cause of nosocomial antibiotic-resistant infections globally (1). Multidrug-resistant isolates of E. coli have been identified worldwide (2–7), with many resistance genes encoded on plasmids (8, 9). Understanding more about how these plasmids are disseminated therefore has the potential to reduce nosocomial and community spread of drug-resistant pathogens.
The E. coli species is hugely diverse both phenotypically and genotypically (10, 11), with MDR lineages found predominantly within phylogroups B2 (sequence types ST131 and ST1193), A (ST167, ST410), and F (ST648) (12–17). Within these lineages are clones that are defined in part by the MDR plasmids that they are known to carry. The H30Rx clone of ST131, for example, is proficient at acquiring multidrug resistance (MDR) plasmids and is differentiated from other ST131 lineages by the presence of an FII-type plasmid encoding the blaCTX-M-15β-lactamase (12, 17, 18). ST131-H30Rx has also been identified with plasmids encoding blaCTX-M-14 or blaCTX-M-27, albeit to a lesser extent (12). The carriage of this particular blaCTX-M-15-encoding plasmid is also a key feature of the B4/H24RxC clone of ST410 (15).
This raises the possibility that the nature of the plasmid may be influencing acquisition and therefore the emergence and spread of MDR E. coli lineages. The dissemination of blaCTX-M-14 in phylogroups A, B1, and D in Spain has been driven predominantly by K-type plasmids (19), and the spread of the β-lactamase blaCMY-2 in these phylogroups in China is via A/C-type plasmids (20). By contrast, blaCMY-2 spread in Brazil is not influenced entirely by A/C-type plasmids; there, the K-type and B/O-type were the predominant plasmid types (21). The carbapenemase blaNDM-1 has been identified on plasmid types including L/M, FII, N, A/C, and X3 (22–27), and OXA-type carbapenemases have been found on L/M-type (blaOXA-48, blaOXA-9), HI1B-type (blaOXA-1), and FII-type (blaOXA-1, blaOXA-9) plasmids (26, 28, 29). MDR genes can also co-exist on plasmids, with known examples including the colistin resistance gene mcr-1 with blaOXA-48 (30), blaCTX-M-55 (5), or blaCTX-M-65 (31). MDR plasmids are therefore highly variable, and it is important to ask to what extent this influences plasmid dissemination.
There is evidence that the genetic background of the recipient strain plays a role in determining the success of MDR plasmid transfer. This is somewhat evident in the lineage-specific nature of resistant E. coli strains, with ST131 clade C (12), ST648 (32–34), ST1193 (6, 35, 36), and ST410 (15, 37, 38) particularly prone to acquiring resistance. More definitively, recent work by Dunn et al. introduced an identical blaCTX-M-15 FII-type plasmid into a diverse set of E. coli strains from across the species phylogeny (39). This work showed that the transcriptional response to plasmid acquisition was largely strain dependent, with even closely related clade C strains exhibiting very different levels of transcriptional response to acquiring the plasmid.
The effect of plasmids on gene expression has also been investigated in other genera. Plasmids have been shown to alter gene expression in Pseudomonas aeruginosa PAO1, most notably of metabolic genes (40). In a multidrug-resistant strain of Acinetobacter baumannii, the presence of a conjugative plasmid resulted in the downregulation of genes involved in biofilm formation and an upregulation of genes related to glutamate/aspartate transport and iron uptake (41). When carrying a plasmid encoding a carbapenemase, carbohydrate metabolism and multidrug efflux systems genes were upregulated in hypervirulent Klebsiella pneumoniae, and genes including virulence factors were downregulated (42). These data suggest that metabolic networks may be particularly sensitive to altered expression following plasmid acquisition but whether the nature of the plasmid dictates this when the host background is controlled is not clear.
Here, we determine the extent to which plasmid genetic background can affect the transcriptional response of E. coli acquiring MDR plasmids. We use the term “MDR” here as a proxy for the presence of an extended-spectrum beta-lactamase (ESBL), a carbapenemase, or a colistin resistance gene as these generally co-occur with genes encoding resistance to other antibiotics (43, 44), although we recognize that this is not always the case. We introduced eight different MDR plasmids into E. coli K-12 MG1655 and measured the transcriptional response using RNA sequencing. The plasmids represent different replicons, different MDR gene types on similar plasmid types, and plasmids with multiple AMR gene types. We found plasmid-specific differential gene expression in response to plasmid acquisition in E. coli and that the level of response did not directly correlate to impacts on fitness. We show that metabolic gene expression is widely affected by MDR plasmid acquisition, with parallelisms in certain pathways and plasmid-specific effects in others, highlighting an important commonality in how MDR plasmids influence transcription in E. coli.
MATERIALS AND METHODS
Bacterial strains and plasmids
The recipient strain was E. coli K-12 MG1655. Eight different plasmid bearers (E. coli, Klebsiella pneumoniae) were used as donors to transfer different MDR plasmids by conjugation, including E. coli strains LA84 and LA232 isolated from travelers to Laos (45), and clinical K. pneumoniae isolates from patients in China (LL*). Donor strains and their corresponding MDR plasmids are detailed in Table 1. K. pneumoniae Ecl8 was used as an intermediate when the donor was E. coli. Here, we use “MDR plasmid” as a proxy for plasmids that carry an ESBL, carbapenemase, or colistin resistance gene.
TABLE 1.
The conjugative plasmids and resulting differential gene expression, including plasmid ID used throughout, the contig on which the plasmid is predicted to be located, the donor species and sequence type (ST), the predicted size [base pairs (bp)] of the plasmid, the plasmid replicon(s), MDR gene(s), other predicted resistance genes, and the total number of genes upregulated and downregulated in the transconjugants relative to the plasmid-free strain after a false discovery rate threshold of P > 0.05 and an absolute log fold change of at least 1 was applieda
| Plasmid | Contig | Donor species | Donor ST | Donor ref | Size (bp) | Replicon(s) | MDR gene(s) | Other resistance genes | Upregulated | Downregulated |
|---|---|---|---|---|---|---|---|---|---|---|
| p7E2_2 | 2 | K. pneumoniae | Not typable | 1,60,631 | A/C | blaCMY-2 | floR, tet(A), aph(6)-Id, aph(3″)-Ib, sul2, aac(3)-IIa, erm(B), mph(A) | 279 | 388 | |
| p7E2_3 | 3 | K. pneumoniae | As above | 1,51,377 | FIIK | None | sul1, aadA2, dfrA12 | |||
| p13F2 | 2 | K. pneumoniae | Not typable | 1,40,169 | FIIK,Q1 | blaCTX-M-3 | catB4, sul2, aph(3″)-Ib, aph(6)-Id, aac(3)-IId, aph(3′)-Ia, mph(A), sul1, qnrB2, aadA16, dfrA27, ARR-3, aac(6′)Ib-cr, tet(A), floR, blaTEM-1B, qnrS1 | 78 | 111 | |
| pLA84 | 2 | E. coli | 69 | 45 | 67,433 | FII | blaCTX-M-27 | None | 224 | 30 |
| pLA232 | 2 | E. coli | 101 | 45 | 98,197 | FII | blaCTX-M-55, mcr-3.4 | qnrS1, aac(3)-IId, catA2 | 173 | 107 |
| pLL19 | 2 | K. pneumoniae | Not typable | 1,17,199 | FIIK | blaCTX-M-15 | qnrS1, blaTEM-1B, aac(6′)Ib-cr, ARR-3, dfrA27, aadA16, sul1, mph(A) | 18 | 19 | |
| pLL34 | 2 | K. pneumoniae | 273 | PRJNA353728 | 57,456 | N | blaNDM-1 | dfrA14, qnrS1 | 190 | 139 |
| pLL35 | 2 | K. pneumoniae | 45 | 39 | 1,06,404 | FIIK | blaCTX-M-15′ blaOXA-9 | qnrS1, aac(6′)-Ib, ant(3″)-Ia, aac(3)-IId | 12 | 12 |
| pLL70 | 6 | K. pneumoniae | Not typable | 74,382 | I1 | blaCTX-M-55 | None | 77 | 93 | |
| p7E2_2 | 2 | K. pneumoniae | Not typable | 1,60,631 | A/C | blaCMY-2 | floR, tet(A), aph(6)-Id, aph(3″)-Ib, sul2, aac(3)-IIa, erm(B), mph(A) | 78 | 111 | |
| p7E2_3 | 3 | K. pneumoniae | As above | 1,51,377 | FIIK | None | sul1, aadA2, dfrA12 | 224 | 30 |
Resistance genes were considered present and reported here if they were scored by ABRicate as over 80% coverage and over 95% identity. All plasmid replicons reported by ABRicate using default cut-offs are reported here after confirmation using PubMLST. Publication references or BioProject numbers are given for donors where possible.
Plasmid analysis
Plasmid replicons and plasmid-encoded resistance genes were predicted using ABRicate (v0.8) to query the PlasmidFinder and ResFinder databases, respectively, from the long-read (LA232, LL19, LL35) and hybrid (13F2, 7E2, LA84, LL34, LL70) assemblies. Plasmid types were confirmed using PubMLST (45). This information, alongside plots generated using Bandage (v0.8.1), was used to identify the contigs that contained the plasmids. These were separated from the rest of the assembly using TigSPLIT (https://github.com/stevenjdunn/TigSPLIT) and annotated using Prokka (46) (v1.14.6). Plasmid maps were generated using SnapGene (v6.1.0). A pangenome of the plasmids was constructed using Panaroo (47) (v1.2.10) with default parameters and a sensitive clean mode. A phylogeny of the plasmid sequences was generated using MashTree (v1.2.0), and this plus the gene_presence_absence_roary output from Panaroo was visualized using Phandango (48).
Assigning function to plasmid-encoded genes
Functions were assigned to genes present in the contigs containing the plasmids using the method described in reference (49). Briefly, the pangenome reference file produced by Panaroo was translated from nucleotide into peptide sequence using the CDSToAminoAcids.py custom Python script from https://github.com/C-Connor/GeneralTools. Functional annotation was performed using eggNOG-mapper (v2.1.9) (50) based on eggNOG orthology data (51) to assign COG categories to each gene. Sequence searches were performed using DIAMOND (v2.0.15). Genes that could not be assigned a functional category were designated “-.” For visualization purposes, genes that were assigned a mixed function category (NU, for example) were grouped under “Other.”
Gene ontology enrichment analysis
To assess which functions were enriched in the differentially expressed (DE) genes, gene ontology (GO) enrichment analysis was performed on the significant genes as a whole and for each transconjugant individually using the PANTHER 17.0 classification system, with Fisher’s Exact test and corrected using false discovery rate (52, 53). Hypothetical genes or those without an assigned identification were excluded.
Plasmid conjugation
Liquid broth conjugations from LL19, LL34, 13F2, and LL35 (as in (54)) donors were performed as per methods detailed in reference (39). Briefly, an overnight culture was grown in 5 mL LB (E. coli MG1655 recipient) or 5 mL LB (E & O Laboratories Ltd) +4 µg/mL cefotaxime (Alfa Aesar) (donors) at 37°C from a single colony. The 5 mL overnight culture was centrifuged at 8,000 rpm (Eppendorf MiniSpin F-4512–11) for 3 minutes, the pellet was resuspended in 1 mL phosphate-buffered saline (PBS, VWR), and this washing step was repeated twice before resuspending in a final volume of 500 µL PBS. A sample of 20 µL donor and 80 µL recipient was mixed by brief vortexing before transfer into 5 mL brain-heart infusion (BHI, Sigma) broth. The suspension was inverted to mix and incubated for 18 hours at 37°C without agitation. Cultures were then serially diluted in PBS to single colonies and spread onto UTI ChromoSelect chromogenic agar plates (Millipore) containing 4 µg/mL cefotaxime before overnight incubation at 37°C. Transconjugants were identified by color and were then plated onto fresh chromogenic agar plates with 4 µg/mL cefotaxime to check purity. Liquid conjugation from the LA84 donor was performed as described above, with the additional step of conjugation via a K. pneumoniae Ecl8 intermediate.
Filter mating conjugations from 7E2, LA232, and LL70 donors were performed by growing an overnight culture in 5 mL LB (E. coli MG1655 recipient) or 5 mL LB + 4 µg/mL cefotaxime (donors) at 37°C from a single colony. A 1 mL sample of overnight suspension was centrifuged for 5 minutes at 8,000 rpm (Eppendorf MiniSpin F45-12-11), the pellet was resuspended in 1 mL PBS, and this washing step was then repeated twice before resuspending in a final volume of 500 µL PBS. Sterile filter paper discs were placed on LB agar plates before 10 µL donor and 10 µL recipient added to each. Plates were inverted and incubated for 7 hours at 37°C. Filter discs were removed with sterile forceps and vortexed thoroughly in 1 mL PBS, serially diluted in PBS to single colonies, and 100 µL spread onto chromogenic agar plates containing 4 µg/mL cefotaxime. Agar plates were incubated overnight at 37°C before the presence of pure transconjugants was assessed as for previous conjugations. Filter conjugation from the LA232 donor was performed as described above, with the additional step of conjugation via a K. pneumoniae Ecl8 intermediate. All conjugations were performed in biological triplicate.
Genome sequencing
Long-read sequencing of the transconjugants containing plasmids from the LA232, LL19, and LL35 donors was performed using MinION sequencing (Oxford Nanopore Technologies, UK). Genomic DNA was extracted from overnight cultures using the Monarch Genomic DNA Purification Kit (New England Biolabs). DNA was quantified using a Qubit 4 fluorometer (Invitrogen) and accompanying broad-range double-stranded DNA assay kit (Invitrogen). Sequencing libraries were prepared using the SQK-LSK109 ligation sequencing kit and EXP-NBD104 native barcode expansion (Oxford Nanopore Technologies, UK), as per the manufacturer’s instructions. Long-read sequencing was performed on a MinION sequencer using an R9 flow cell (Oxford Nanopore Technologies, UK). Sequences were base called using Guppy (v6.0.1). Reads were filtered using Filtlong (v0.2.1) using a cut-off of 600,000,000 target bases, demultiplexed using qcat (v1.1.0), and assembled using Flye (v2.8.3-b1695). Hybrid assemblies of the transconjugants containing plasmids from the 7E2, 13F2, LA84, LL34, and LL70 donors were generated by MicrobesNG using their enhanced sequencing service. A hybrid assembly for the plasmid-free parent strain has been published previously (55). Variants were called against the parent strain using breseq (v0.38.1) with a consensus minimum variant coverage of 10, minimum mapping quality of 20, and consensus frequency cutoff of 0.9, as in reference (54).
Transcriptome sequencing
RNA sequencing was performed on biological triplicates of the plasmid-free and the transconjugant strains. For sample preparation, a single colony for each replicate was picked following overnight growth on LB agar and added to 5 mL of LB broth (Sigma-Aldrich, United Kingdom). LB was supplemented with 4 µg/mL cefotaxime for transconjugant culture. A 50 µL suspension of each overnight culture was then transferred into 5 mL fresh LB/LB + cefotaxime media and incubated at 37°C with agitation until an optical density at 600 nm (OD600) of approximately 0.85. A 1.5 mL sample was centrifuged for 5 minutes at 8,000 rpm (Eppendorf MiniSpin F-45–12-11), resuspended in 1 mL PBS, and this wash step was repeated. The supernatant was aspirated and the pellet frozen prior to processing and RNA sequencing by GENEWIZ from Azenta Life Sciences (Frankfurt, Germany) using their standard RNA sequencing service. Differential gene expression was quantified using Kallisto (v0.48.0). The long-read assembly, annotated using Prokka (v1.14.6), of the plasmid-free strain was used as a reference. The annotated assembly was processed using genbank_to_kallisto.py (https://github.com/AnnaSyme/genbank_to_kallisto.py). GNU parallel (56) was used for job parallelization. Differential gene expression was analyzed using the Voom/Limma method in Degust (v4.1.1) with a false discovery rate threshold of P < 0.05 and an absolute log fold change of at least one. EcoCyc (57), Uniprot (58), and KEGG (59) databases were used to identify gene function and associated biochemical pathways.
Relative fitness assay
We conducted pair-wise competitions to assess the relative fitness of the plasmid bearers compared to GFP-labeled plasmid-free MG1655 (60). Overnight cultures were created from LB agar plates in 10 mL LB (plasmid-free) and 5 LB with 4 µg/mL cefotaxime (transconjugants) from single colonies of the plasmid-free strain and the biological triplicates of the transconjugants. All cultures were diluted to approximately 1 × 109 cells/mL in PBS. Transconjugant cultures were then centrifuged for 5 minutes at 10,000 rpm (Eppendorf MiniSpin F-45–12-11), resuspended in 1 mL PBS, centrifuged for 5 minutes at 10,000 rpm, and resuspended in 1 mL LB. Plasmid-free cultures were then centrifuged for 10 minutes at 3,600 rpm (Thermo Scientific Megafuge 40R TX-1000), resuspended in 28 mL PBS, centrifuged for 10 minutes at 3,600 rpm, and resuspended in 28 mL LB. All cultures were then serially diluted to approximately 1 × 105 cells/mL before 50 µL plasmid-free and 50 µL transconjugant were mixed in 5 mL LB and incubated for 24 hours at 37°C. Samples of 0-hour and 24-hour mixed populations were stored at −80°C until flow cytometry analysis.
Flow cytometry
Plasmid-free and plasmid-bearers pre- and post-competitions were quantified using the Attune NxT flow cytometer and its associated software package. Prior to analysis, 2 mL of the 0 hour and 1 mL of the 24-hour mixed populations were thawed and washed three times for 5 minutes at 12,000 rpm in 1 mL sterile, filtered HEPES buffer solution (Gibco). The whole populations were then stained using 10 µM SYTO 84 (Invitrogen) as per reference (61), thus resulting in single (SYTO 84) stained transconjugants and double (SYTO 84 plus GFP) stained plasmid-free cells. SYTO 84 was visualized using a 561 nm yellow laser and 585/16 nm emission filter (YL1 channel) (61). GFP was visualized with a 488 nm blue laser and 530/30 nm emission filter (BL1 channel). Example gating is given in Fig. S1, and raw .fcs files are available at 10.6084 /m9.figshare.24476680. A competition index (CI) was calculated as per the Miles and Misra method (62) as transconjugant/plasmid-free, whereby a CI larger than one represented competition in favor of the transconjugant. The Pearson correlation coefficient to measure the relationship between a total number of DE genes and CI was calculated using pearsonr from the SciPy package.
RESULTS
Collating a panel of diverse conjugative plasmids encoding MDR genes
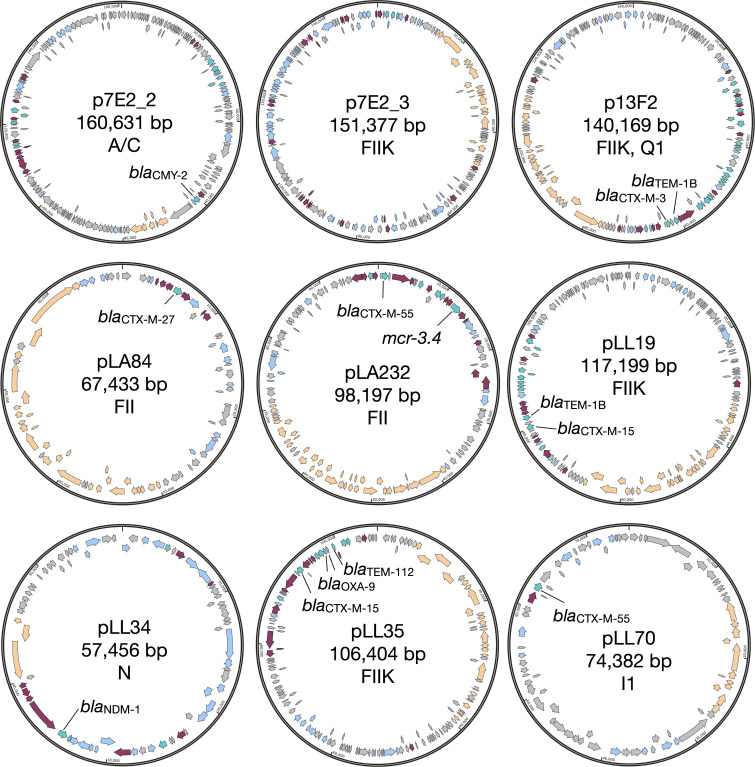
A panel of clinical E. coli and K. pneumoniae donors carrying conjugative MDR plasmids was selected. The plasmids were selected based on their ability to conjugate into a common E. coli host and to cover a wide range of different sizes, replicons and resistance genes to produce as diverse a test set as possible. The plasmid sizes range from 160,631 bp (p7E2_2) to 57,456 bp (pLL34), encode MDR genes including blaNDM-1 (pLL34), blaOXA-9 (pLL35), mcr3.4 (pLA232), and various blaCTX-M genes, and possess replicons including FII, N, and I (Table 1; Fig. 1). Eight unique transconjugants were generated in biological triplicate using E. coli K-12 MG1655 as a recipient. One transconjugant (7E2) was found to have taken up two plasmids, only one of which was classified as an MDR plasmid (p7E2_2) (Fig. 1).
Fig 1.
Maps for the nine plasmids conjugated into E. coli MG1655 Turquoise = resistance genes, including ones not identified by ResFinder. Orange = conjugal transfer genes, including potential gene/gene fragments not identified by Prokka but found using manual BLAST queries. Maroon = transposase. Grey = hypothetical protein. Blue = annotated gene of other function.
The remaining plasmid-encoded genes are also variable, with many having unknown function (Fig. S2). Of those to which a COG functional category could be assigned, the largest proportion of each plasmid is involved in replication, recombination, and repair (Fig. S3). The only exception to this is the plasmid encoded by the LL70 transconjugant, for which the largest proportion of known genes has functions that straddle several COG categories. Genes involved in inorganic ion transport and metabolism are particularly abundant on p7E2_3 (n = 14) in comparison to the other plasmids (n = 0 for pLL70, n = 1 for pLA232, pLL34, pLL35). Other genes relating to transport and metabolism are scarce on all plasmids. Overall, while some plasmids do share some genes, the majority of genes appear to be specific to the individual plasmid (Fig. S3). These plasmids therefore provide a suitably diverse panel from which to examine the immediate transcriptional response upon their acquisition.
Scale of transcriptional response does not correlate to plasmid fitness cost
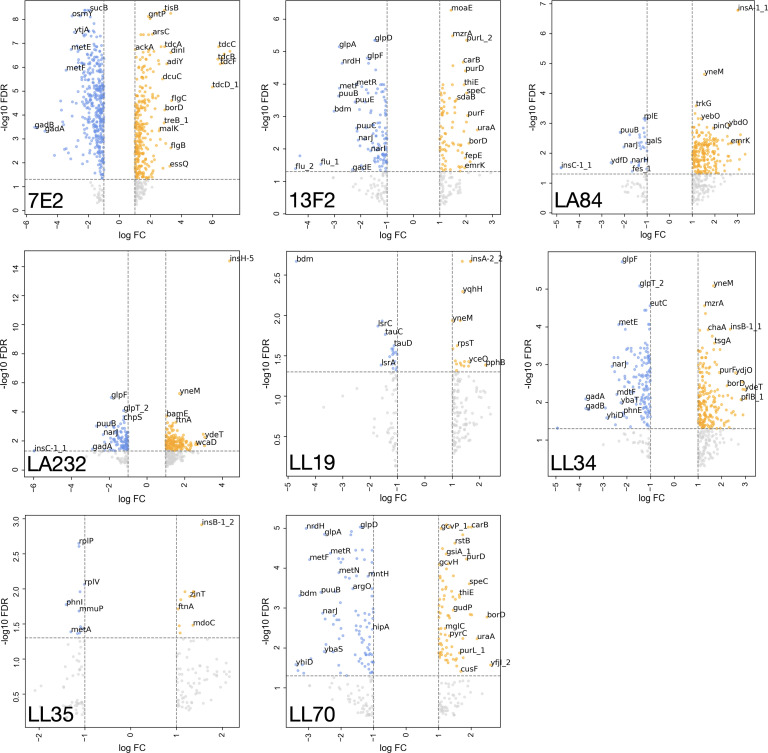
Gene expression of the eight different transconjugants was analyzed by RNA sequencing to assess the immediate transcriptomic impact of MDR plasmid acquisition. We uncovered large variability in the number of differentially expressed (DE) genes in response to plasmid acquisition (Fig. 2). For example, acquisition of the N-type pLL34 (blaNDM-1) and the FII-type pLA232 (blaCTX-M-55, mcr-3.4) resulted in relatively high numbers of upregulated genes (190 and 173, respectively), and the largest transcriptional response was observed on acquisition of p7E2, an A/FII plasmid encoding blaCMY-2, both with regard to upregulated (n = 279) and downregulated (n = 388) genes. There was no consistent pattern between the transconjugants as to whether there were more upregulated genes than downregulated, or vice versa (Table 1).
Fig 2.
Genes significantly upregulated (orange) and downregulated (blue) FDR threshold of P < 0.05 and an absolute log fold change (FC) of at least one] in the transconjugants in comparison to the plasmid-free control. Genes that had an absolute log FC of at least one but did not reach the FDR threshold are shown in gray and are considered to not be significantly differentially expressed. Plots are labeled by the donor with respect to Table 1. Select genes are labeled.
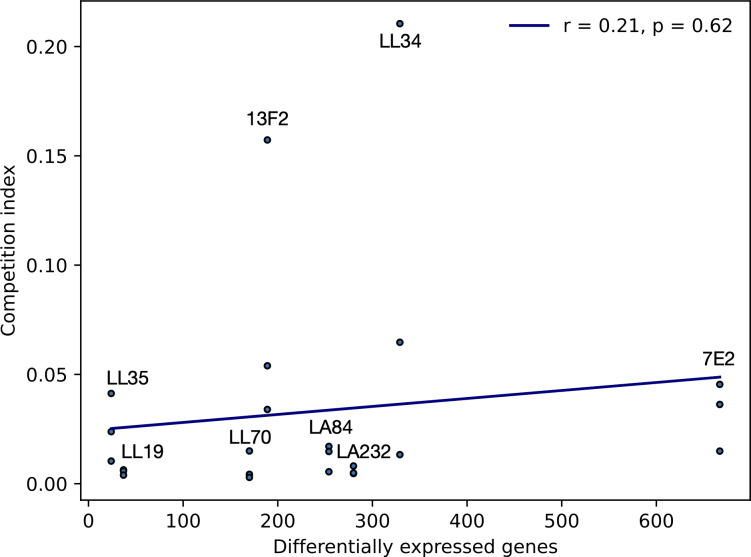
No significant correlation was observed between the fitness cost of the plasmid and the total number of DE genes (Fig. 3, Pearson correlation coefficient, P = 0.62). We also found minimal evidence for mutation accumulation during plasmid acquisition, with only a small number of changes identified (Table S2). Three transconjugants (LL19, LL34, and LL35) had no significant changes. One base pair deletions were found in ynaE (an uncharacterized protein) in 7E2, and in the intergenic region between yehA_2 (an uncharacterized fimbrial-like protein) and rcnB (involved in nickel/cobalt homeostasis, possibly transport) in LL70. In addition, plasmid size did not affect the scale of differential gene expression. The pLL34 plasmid was the smallest but triggered one of the highest number of DE genes (n = 329), whereas LA232 also has a high number of DE genes (n = 280) after acquiring a much larger plasmid (Table 1). The transconjugant that acquired two plasmids (160,631 bp and 151,377 bp), 7E2, did however show substantially more DE genes (n = 667). Together, these data suggest that transcriptional response to MDR plasmid acquisition in MG1655 is likely influenced by multiple factors, including the presence of another plasmid-encoded gene, rather than one primary driver.
Fig 3.
Correlation (Pearson) between a total number of differentially expressed genes and competition index (CI) of the transconjugants against a plasmid-free strain (mean of biological triplicate), whereby a value less than one indicates the plasmid-free is fitter in the competition condition. Individual CI values are plotted.
Majority of differentially expressed genes are plasmid specific
We then analyzed the significant DE genes to identify commonalities across the transconjugants. Only one gene (rpsT, encoding the 30S ribosomal subunit protein S20) was significantly upregulated in all eight transconjugants (Fig. S4). In all, 34 genes were upregulated in parallel in LA84, LA232, and LL34, including argACF (L-arginine biosynthesis from L-glutamate via L-ornithine), holE (DNA polymerase III subunit), and ydiM (inner membrane transport protein). In total, 21 genes were significantly DE in both LA84 and LA232, including crcB (fluoride-specific ion channel, citC (citrate lyase synthetase), and genes encoding uncharacterized proteins (ybbD, ybcK). The number of genes that were only upregulated in a single transconjugant varied between the recipients (7E2, n = 131; 13F2, n = 3; LA84, n = 59; LA232, n = 11; LL19, n = 3; LL34, n = 17; LL35, n = 0; LL70, n = 16). This implies that the response to certain plasmids, for example, pLL35, may produce a more general response than others, given the evidence that all upregulated genes in that transconjugant were also significantly upregulated in response to at least one other plasmid. There were no genes that were significantly downregulated in all eight of the transconjugants. Together, these data suggest that transcriptional response to MDR plasmid acquisition is specific to the incoming plasmid and that the response to certain plasmids (p7E2) is larger and more plasmid-specific than for others (pLL35).
Differentially expressed genes enriched in metabolic and transport functions
We performed GO enrichment analysis on the collection of all DE genes across the set of transconjugants to establish an overall pattern. Of the 23 significantly enriched GO terms, nine were related directly to transport (including metal ions) and a further three to the membrane (Fig. 4; Table S3). In addition, three GO terms related metabolic processes cover a total of 136 DE genes across the data set. This suggests that metabolic processes, including transport, are significantly impacted by the acquisition of MDR plasmids.
Fig 4.
GO terms significantly enriched in the combined set of all transconjugants. A value greater than one indicates more genes observed than expected. Terms are grouped by their role in biological (orange box), molecular (pink), or cellular (blue) function. GO IDs and P values given in Table S3.
Convergence observed in the downregulation of L-methionine transport and metabolism genes
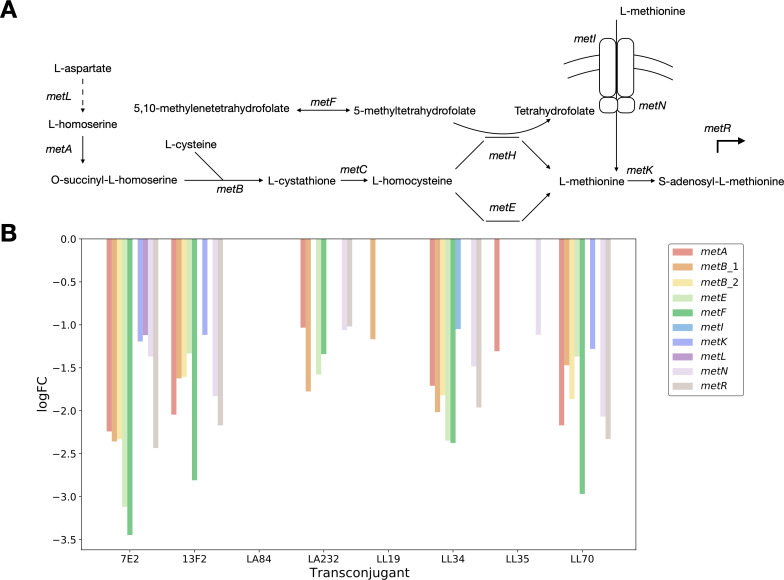
GO enrichment analysis of the whole transconjugant set identified transport and metabolism as key functions of DE genes. When the GO terms of the DE genes were analyzed for each transconjugant individually, five of the eight transconjugants showed enrichment in amino acid metabolism (Data Set S1). This includes aspartate (7E2), methionine (13F2, LL70), arginine (LA232, LL34), glutamate (LL34), and glutamine (LL34, LL70) metabolic pathways, as well as amino acid biosynthesis pathways more generally (7E2, 13F2, LL34, LL70).
The specific patterns of up- or downregulation of genes in these pathways were then examined individually. The 13F2 and LL70 transconjugants showed enrichment in genes involved in L-methionine biosynthetic (13F2, LL70) and metabolic (13F2) processes. Indeed, widespread downregulation in L-methionine transport and metabolism was observed across the transconjugants. The metA and metB genes that convert L-homoserine to L-cystathionine via O-succinyl-L-homoserine in the production of L-methionine from L-aspartate were downregulated in six transconjugants (Fig. 5). The ATP binding subunit of the MetNI ABC transporter was also significantly downregulated in five transconjugants, whereas the membrane subunit of the same transporter was only downregulated in LL34 (encoding blaNDM-1). The LA84 transconjugant (blaCTX-M-27) had no significant DE genes in this pathway, and only metB_1 is downregulated in LL19 (blaCTX-M-15). The convergence observed across this pathway provides evidence toward a hypothesis that the downregulation of L-methionine uptake and metabolism may be important in MDR plasmid acquisition and stable integration.
Fig 5.
(A) Simplified L-methionine biosynthetic pathway, with key genes highlighted and (B) the logFC values for the genes downregulated on the acquisition of an MDR plasmid, colored by gene involved in L-methionine transporter and biosynthesis.
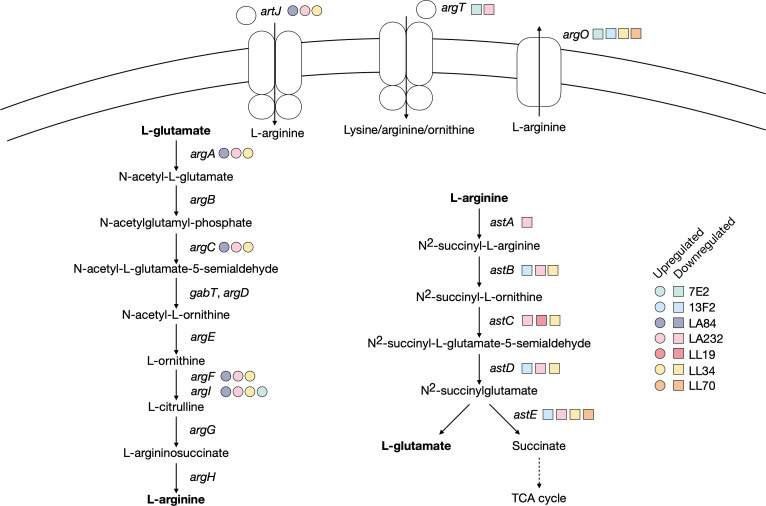
A proposed requirement for L-arginine following plasmid acquisition
GO analysis further highlighted enrichment in L-arginine and L-glutamate metabolic pathways, specifically in LA232 (arginine catabolic processes to glutamate and succinate, arginine metabolic process) and LL34 (arginine and glutamate metabolic processes).
The argACFI genes were significantly upregulated in the LA232, LA84, and LL34 transconjugants (Fig. 6). The argA gene encodes N-acetyl-glutamate synthase that converts L-glutamate into N-acetyl-L-glutamate in the ornithine biosynthesis pathway for the eventual production of L-ornithine from L-glutamate. The argC gene (N-acetylglutamylphosphate reductase) functions in the same pathway, and argF and argI (ornithine carbamoyltransferase chains F and I) catalyze the first step in the conversion of L-ornithine to L-arginine. The periplasmic binding protein of the L-arginine ABC transporter artJ is also upregulated in these three transconjugants.
Fig 6.
Genes upregulated (circle) and downregulated (square) across all transconjugants that relate to L-arginine biosynthetic pathways. Selected metabolites in each reaction are shown for simplicity, with key metabolites or reactions highlighted in bold.
By contrast, astABCDE in the arginine succinyltransferase pathway were downregulated after the acquisition of pLA232, and all but astA were downregulated in LL34. These genes encode the five enzymes in the pathway that converts L-arginine into L-glutamate and succinate for the TCA cycle. The periplasmic binding protein of the L-ornithine/L-arginine/L-lysine ABC transporter argT was also downregulated in LA232 and 7E2, and the L-arginine exporter argO was downregulated in LL34, 13F2, 7E2, and LL70. This pattern is not observed in, for example, 7E2, where argI is the only common upregulated gene, or in LL35 where none are differently expressed. This suggests two points. First, the exact metabolic requirement following MDR plasmid acquisition is plasmid specific. Second, in response to some MDR plasmids (pLA232, pLL34), there may be a preferential need for L-arginine or a diversion of resources away from the TCA cycle.
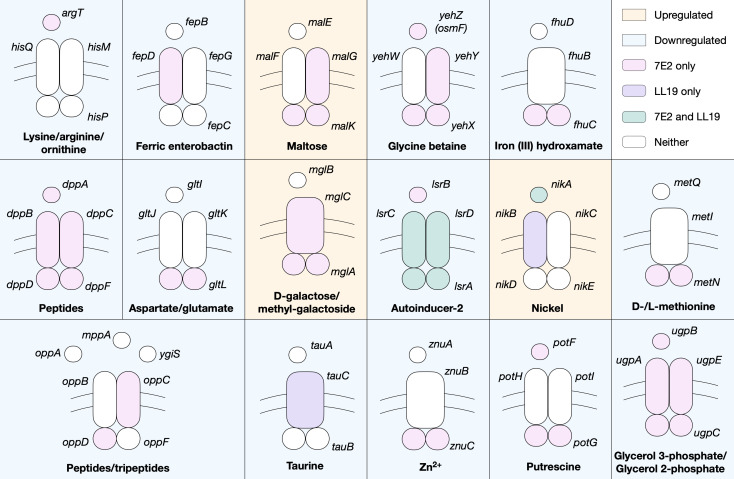
Plasmid-specific enrichment in ATP-binding cassette transporters
Furthermore, GO enrichment analysis identified significant differential expression in genes relating to transport across five of the eight transconjugants (except LL34 and LL35). The nature of the transporters, both in terms of specificity and type, varies between the transconjugants with some evidence of commonalities.
7E2 and 13F2, for example, show significant differential expression of inorganic ion and cation transmembrane transport, with changes also in transition metal and iron transport in the former (Supplementary Data Sheet). GO terms relating to ATP-binding cassette (ABC) transporters are also enriched in 7E2 and LL19. The majority of these genes are downregulated, including the entire dppABCDF and ugpABCE transporters in 7E2 (responsible for the transport of peptides and glycerol 3/2-phosphate, respectively) (Fig. 7). Components of the lysine/arginine/ornithine, ferric enterobactin, and iron (III) hydroxamate, ABC transporters, among others, are also downregulated in 7E2, and lsrACD is downregulated in both 7E2 and LL19 upon plasmid acquisition. There were fewer examples of ABC transporter genes being upregulated but it was noted in components of the maltose, nickel, and D-galactose/methyl-galactoside systems.
Fig 7.
Components of ABC transporters that are significantly upregulated (orange background) or downregulated (blue background) in 7E2 (pink), LL19 (purple), both (green), or neither (white). Simplified schematics of the periplasmic binding, membrane, and ATP-binding proteins are shown. Transporters with significant DE genes with unknown or putative specificity are not included but are detailed in the Supplementary Data Sheet.
A point of note is the significant downregulation of potF and potG, encoding components of the putrescine ABC transporter, in 7E2, whereas plaP, encoding a putrescine/H + symporter, is upregulated. The puuABC genes from the putrescine degradation pathway are also downregulated (Data Set S1). The low affinity of PlaP coupled with evidence that it has functions relating to type 1 pili (63) suggests that putrescine itself is not a desired metabolite under these conditions.
Together, this suggests a diversion of resources away from high-affinity transporters in a plasmid-dependent manner as the cell responds to the incoming plasmid.
DISCUSSION
The spread of MDR is of increasing concern, particularly within clinical settings. While the mechanisms by which MDR plasmids spread are well established, less is understood about the role that the recipient plays in plasmid acquisition; specifically, whether the transcriptional response is host or plasmid specific. To begin to unravel this, we introduced eight plasmids from clinical isolates into a laboratory strain of E. coli via conjugation and measured the resulting transcriptional response. These plasmids encoded a variety of MDR genes, including different blaCTX-M genes, and were of different plasmid types. We did not find convincing evidence of mutation accumulation in the transconjugants during plasmid acquisition. This is likely due to the lack of passage post-transfer, although this result should be taken with the caveat that only one of the three independent biological replicates per condition was sequenced.
We uncovered extensive plasmid-dependent differences in transcriptional response to MDR plasmid acquisition, both in terms of the absolute number of genes and the nature of the genes themselves. Genes relating to transport and metabolic processes were particularly DE across the transconjugants. Our work supports existing data from other species; in P. aeruginosa, plasmids have been shown to trigger differential expression of metabolic genes (40), indicating metabolic networks are particularly sensitive to the effects of incoming plasmids across genera.
We identified transconjugant-specific upregulation of genes involved in the conversion of L-glutamate to L-arginine alongside the downregulation of genes responsible for converting L-arginine into succinate, mostly notably in LA232 although with some specific gene examples in other transconjugants. This suggests that L-arginine might be important immediately following the acquisition of certain MDR plasmids. A link between L-arginine and MDR plasmids has been reported previously; specifically, strains carrying an MDR plasmid were serially passaged in the presence and absence of antibiotic selection and the artP gene encoding the L-arginine ABC transporter ATP binding protein was found to be the most highly mutated gene (54). While we did not find this particular gene to be differentially regulated to the same extent, we did find significant upregulation of artJ, encoding the periplasmic binding protein of the same transporter. There is existing evidence of amino acid transport and metabolism genes being consistently upregulated in A. baumannii in the presence of antibiotics (64), so an interesting extension of this work may be to measure gene expression immediately following plasmid acquisition in the presence versus absence of antibiotics.
The differential regulation of the pathways relating to L-arginine biosynthesis was particularly notable in the LA232 transconjugant but the downregulation of components of the L-methionine biosynthesis pathway was common to all but LA84. While the current understanding of the link between specific metabolic pathways and MDR is not complete, there are examples of other plasmids affecting the expression of genes relating to L-methionine biosynthesis in E. coli. Specifically, the E. coli plasmid pEX18Gm has been shown to increase L-methionine biosynthesis via the upregulation of genes including metH (20). MetH is involved in the cobalamin-dependent conversion of L-homocysteine, and while we did not find significant differential expression of metH we did identify a downregulation of metE, encoding the cobalamin-independent reaction. This suggests that the presence or absence of methionine may be important immediately following plasmid acquisition in a plasmid and/or strain-specific manner.
Among the significant DE genes, ABC transporters were also notably enriched in two transconjugants (7E2 and LL19), suggesting their importance may be plasmid specific. The majority of these genes were down- rather than upregulated, with the implication that metabolite import may not be a priority after plasmid acquisition. Components of the putrescine ABC transporter were downregulated in 7E2. Putrescine is the major polyamine in E. coli (65), and polyamines are known to decrease the permeability of the outer membrane in E. coli under stress conditions (66). It is possible that the genes were differentially expressed as part of a stress response or, when considered alongside the downregulation of genes in the putrescine degradation pathway, a diversion of resources away from its use in metabolic pathways. While there is not enough information here to establish the underlying cause, it presents an interesting avenue for future research.
It is already known that plasmids can manipulate host gene expression by plasmid-encoded transcriptional regulators. A plasmid transcriptional regulator has been shown to have wide-reaching effects on a strain of Pseudomonas fluorescens via extensive remodeling of the host proteome, including influencing metabolite uptake (67), and other plasmids have been shown to repress expression of the type VI secretion system to aid successful conjugation (41, 68). This manipulation of host behavior is hypothesized to be an adaptation of the plasmid to increase its fitness and likelihood of transmission, either by enhancing host competitiveness in a given environment (vertical transmission) or by promoting conjugation (horizontal transmission) (69). Viewed from this perspective, it is possible that the significant DE of metabolic genes across the transconjugants might play a role in increasing the competitiveness of the host outside of a laboratory environment.
A point of note was the relatively small transcriptional response observed in the two transconjugants that acquired the plasmids encoding blaCTX-M-15 (LL19, LL35). This was notably distinct from those encoding alternative blaCTX-M genes (13F2, LA84, LL70). The multidrug-resistant lineage of E. coli ST131 that harbors blaCTX-M-15 is arguably the most successful clone of this species in terms of global dissemination (12, 70). This gene is also found in E. coli lineages including ST410 (15, 38) and ST1193 (71, 72). We could therefore speculate that the prevalence of plasmid-encoded blaCTX-M-15 could potentially be due to the small transcriptional response that its acquisition triggers in the recipient. The pLL19 and pLL35 plasmids are among the larger in the set, suggesting the small transcriptional disruption is not due entirely to plasmid size. The plasmids are also distinct in terms of gene content, depicted through the construction of the plasmid phylogeny and accompanying gene presence/absence data (Fig. S3), and relatedness does not appear to inform the scale of transcriptional disruption. Overall, we speculate that the scale of transcriptional disruption is likely due to a combination of multiple factors and that pathways in metabolism are the most highly affected.
We now know that there are strain- and plasmid-specific responses to the acquisition of MDR plasmids. We have shown here and in two previous studies that differential expression of genes involved in metabolism is common during MDR plasmid acquisition (39, 54), but that the exact metabolic pathways affected and to what extent is plasmid dependent. Recent studies have also shown the importance of metabolic genes in the evolution of AMR in clinical E. coli (73). This study therefore contributes to an understanding of the intrinsic link between metabolism and MDR and should be explored further in existing and emerging pandemic lineages.
ACKNOWLEDGMENTS
Enhanced genome sequencing was provided by MicrobesNG (http://www.microbesng.com). Standard RNA sequencing was performed by GENEWIZ from Azenta Life Sciences. GFP-labeled E. coli MG1655 was provided by Michael J Bottery. The Flow Cytometry Platform at the University of Birmingham provided support for flow cytometry experiments. R.J.H. was supported by the University of Birmingham College of Medical and Dental Sciences Research Development Fund award, and by a NERC grant (NE/T01301X/1) awarded to AM.
R.J.H.: methodology, formal analysis, investigation, data curation, writing—original draft, and visualization. A.E.S.: investigation and data curation. M.T.: visualization. M.A.B.: conceptualization. A.M.: conceptualization, methodology, supervision, and funding acquisition. All authors: writing—review & editing.
The authors declare no competing financial interests in relation to the work described.
Contributor Information
Alan McNally, Email: a.mcnally.1@bham.ac.uk.
Alejandra Rodríguez-Verdugo, University of California, Irvine, USA.
DATA AVAILABILITY
The DNA and RNA datasets generated and analyzed during the current study are available from the NCBI BioProject with accession PRJNA1034827
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/msystems.01193-23.
Significantly enriched GO terms.
Significantly expressed genes.
Supplemental figures and tables.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Murray CJL, Ikuta KS, Sharara F, Swetschinski L, Robles Aguilar G, Gray A, Han C, Bisignano C, Rao P, Wool E. 2022. Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet 399:629–655. doi: 10.1016/S0140-6736(21)02724-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gong L, Tang N, Chen D, Sun K, Lan R, Zhang W, Zhou H, Yuan M, Chen X, Zhao X, Che J, Bai X, Zhang Y, Xu H, Walsh TR, Lu J, Xu J, Li J, Feng J. 2020. A nosocomial respiratory infection outbreak of carbapenem-resistant Escherichia coli ST131 with multiple transmissible blaKPC–2 carrying plasmids. Front. Microbiol 11:2068. doi: 10.3389/fmicb.2020.02068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hornsey M, Phee L, Wareham DW. 2011. A novel variant, NDM-5, of the New Delhi metallo--lactamase in a multidrug-resistant Escherichia coli ST648 isolate recovered from a patient in the United Kingdom. Antimicrob Agents Chemother 55:5952–5954. doi: 10.1128/AAC.05108-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Johnson JR, Tchesnokova V, Johnston B, Clabots C, Roberts PL, Billig M, Riddell K, Rogers P, Qin X, Butler-Wu S, et al. 2013. Abrupt emergence of a single dominant multidrug-resistant strain of Escherichia coli. J Infect Dis 207:919–928. doi: 10.1093/infdis/jis933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Xu L, Wang P, Cheng J, Qin S, Xie W. 2019. Characterization of a novel blaNDM-5 -harboring incFII plasmid and an mcr-1-bearing Inci2 plasmid in a single Escherichia coli ST167 clinical isolate. Infect Drug Resist 12:5119. doi: 10.2147/IDR.S192998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Johnson JR, Johnston BD, Porter SB, Clabots C, Bender TL, Thuras P, Trott DJ, Cobbold R, Mollinger J, Ferrieri P, Drawz S, Banerjee R, Diekema DJ. 2019. Rapid emergence, subsidence, and molecular detection of Escherichia coli sequence type 1193-FimH64, a new disseminated multidrug-resistant commensal and extraintestinal pathogen. J Clin Microbiol 57:e01664-18. doi: 10.1128/JCM.01664-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Banerjee R, Johnson JR. 2014. A new clone sweeps clean: the enigmatic emergence of Escherichia coli sequence type 131. Antimicrob Agents Chemother 58:4997–5004. doi: 10.1128/AAC.02824-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Schaufler K, Semmler T, Wieler LH, Wöhrmann M, Baddam R, Ahmed N, Müller K, Kola A, Fruth A, Ewers C, Guenther S. 2016. Clonal spread and interspecies transmission of clinically relevant ESBLproducing Escherichia coli of ST10-another successful pandemic clone?. FEMS Microbiol Ecol 92:fiv155. doi: 10.1093/femsec/fiv155 [DOI] [PubMed] [Google Scholar]
- 9. Wink PL, Almeida EK, Crispim MN, de Lima-Morales D, Zavascki AP, Barth AL. 2020. First report of IMP-1 in a clinical isolate of Escherichia coli in Latin America. Infect Control Hosp Epidemiol 41:997–998. doi: 10.1017/ice.2020.44 [DOI] [PubMed] [Google Scholar]
- 10. Chaudhuri RR, Henderson IR. 2012. The evolution of the Escherichia coli phylogeny. Infect Genet Evol 12:214–226. doi: 10.1016/j.meegid.2012.01.005 [DOI] [PubMed] [Google Scholar]
- 11. Erb A, Stürmer T, Marre R, Brenner H. 2007. Prevalence of antibiotic resistance in Escherichia coli: overview of geographical, temporal, and methodological variations. Eur J Clin Microbiol Infect Dis 26:83–90. doi: 10.1007/s10096-006-0248-2 [DOI] [PubMed] [Google Scholar]
- 12. Stoesser N, Sheppard AE, Pankhurst L, De Maio N, Moore CE, Sebra R, Turner P, Anson LW, Kasarskis A, Batty EM, Kos V, Wilson DJ, Phetsouvanh R, Wyllie D, Sokurenko E, Manges AR, Johnson TJ, Price LB, Peto TEA, Johnson JR, Didelot X, Walker AS, Crook DW, Modernizing Medical Microbiology Informatics Group (MMMIG) . 2016. Evolutionary history of the global emergence of the Escherichia coli epidemic clone ST131. mBio 7:e02162. doi: 10.1128/mBio.02162-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Platell JL, Trott DJ, Johnson JR, Heisig P, Heisig A, Clabots CR, Johnston B, Cobbold RN. 2012. Prominence of an O75 clonal group (clonal complex 14) among non-ST131 fluoroquinolone-resistant Escherichia coli causing extraintestinal infections in humans and dogs in Australia. Antimicrob Agents Chemother 56:3898–3904. doi: 10.1128/AAC.06120-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zong Z, Fenn S, Connor C, Feng Y, McNally A. 2018. Complete genomic characterization of two Escherichia coli lineages responsible for a cluster of carbapenemresistant infections in a Chinese hospital. J Antimicrob Chemother 73:2340–2346. doi: 10.1093/jac/dky210 [DOI] [PubMed] [Google Scholar]
- 15. Roer L, Overballe-Petersen S, Hansen F, Schønning K, Wang M, Røder BL, Hansen DS, Justesen US, Andersen LP, Fulgsang-Damgaard D, Hopkins KL, Woodford N, Falgenhauer L, Chakraborty T, Samuelsen Ø, Sjöström K, Johannesen TB, Ng K, Nielsen J, Ethelberg S, Stegger M, Hammerum AM, Hasman H. 2018. Escherichia coli sequence type 410 is causing new International highrisk clones. mSphere 3:e00337-18. doi: 10.1128/mSphere.00337-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Schaufler K, Semmler T, Wieler LH, Trott DJ, Pitout J, Peirano G, Bonnedahl J, Dolejska M, Literak I, Fuchs S, Ahmed N, Grobbel M, Torres C, McNally A, Pickard D, Ewers C, Croucher NJ, Corander J, Guenther S. 2019. Genomic and functional analysis of emerging virulent and multidrug-resistant Escherichia coli lineage sequence type 648 . Antimicrob Agents Chemother 63. doi: 10.1128/AAC.00243-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cummins EA, Snaith AE, McNally A, Hall RJ. 2021. The role of potentiating mutations in the evolution of pandemic Escherichia coli clones. Eur J Clin Microbiol Infect Dis 2021. doi: 10.1007/s10096-021-04359-3 [DOI] [PubMed] [Google Scholar]
- 18. McNally A, Kallonen T, Connor C, Abudahab K, Aanensen DM, Horner C, Peacock SJ, Parkhill J, Croucher NJ, Corander J. 2019. Diversification of colonization factors in a multidrug-resistant Escherichia coli lineage evolving under negative frequency- dependent selection. mBio 10:e00644-19. doi: 10.1128/mBio.00644-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Valverde A, Cantón R, Garcillán-Barcia MP, Novais A, Galán JC, Alvarado A, de la Cruz F, Baquero F, Coque TM. 2009. Spread of blaCTX-M-14 is driven mainly by IncK plasmids disseminated among Escherichia coli phylogroups A, B1, and D in Spain. Antimicrob Agents Chemother 53:5204–5212. doi: 10.1128/AAC.01706-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Guo Y-F, Zhang W-H, Ren S-Q, Yang L, Lü D-H, Zeng Z-L, Liu Y-H, Jiang H-X. 2014. IncA/C plasmidmediated spread of CMY-2 in multidrug-resistant Escherichia coli from food animals in China. PLoS ONE 9:e96738. doi: 10.1371/journal.pone.0096738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ferreira JC, Penha Filho RAC, Andrade LN, Berchieri Junior A, Darini ALC. 2017. Diversity of plasmids harboring blaCMY-2 in multidrug-resistant Escherichia coli isolated from poultry in Brazil. Diagn Microbiol Infect Dis 88:361–364. doi: 10.1016/j.diagmicrobio.2017.04.014 [DOI] [PubMed] [Google Scholar]
- 22. Ho PL, Lo WU, Yeung MK, Lin CH, Chow KH, Ang I, Tong AHY, Bao J-J, Lok S, Lo JYC. 2011. Complete sequencing of pNDM-HK encoding NDM-1 carbapenemase from a multidrug-resistant Escherichia coli strain isolated in Hong Kong. PLoS ONE 6:e17989. doi: 10.1371/journal.pone.0017989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rahman M, Mukhopadhyay C, Rai RP, Singh S, Gupta S, Singh A, Pathak A, Prasad KN. 2018. Novel variant NDM-11 and other NDM-1 variants in multidrug-resistant Escherichia coli from South India. J Glob Antimicrob Resist 14:154–157. doi: 10.1016/j.jgar.2018.04.001 [DOI] [PubMed] [Google Scholar]
- 24. Bonnin RA, Poirel L, Carattoli A, Nordmann P. 2012. Characterization of an IncfII plasmid encoding NDM-1 from Escherichia coli ST131. PLoS ONE 7:e34752. doi: 10.1371/journal.pone.0034752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Poirel L, Bonnin RA, Nordmann P. 2011. Analysis of the resistome of a multidrugresistant NDM-1-producing Escherichia coli strain by high-throughput genome sequencing. Antimicrob Agents Chemother 55:4224–4229. doi: 10.1128/AAC.00165-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Khajuria A, Praharaj AK, Kumar M, Grover N. 2014. Emergence of Escherichia coli, co-producing NDM-1 and OXA-48 carbapenemases, in urinary isolates, at a tertiary care centre at central India. J Clin Diagn Res 8:DC01–4. doi: 10.7860/JCDR/2014/7952.4413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Feng Y, Yang P, Xie Y, Wang X, McNally A, Zong Z. 2015. Escherichia coli of sequence type 3835 carrying blaNDM-1, blaCTX-M-15, blaCMY-42 and blaSHV12. Sci Rep 5:12275. doi: 10.1038/srep12275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pfeifer Y, Schlatterer K, Engelmann E, Schiller RA, Frangenberg HR, Stiewe D, Holfelder M, Witte W, Nordmann P, Poirel L. 2012. Emergence of OXA-48-type carbapenemase-producing enterobacteriaceae in German hospitals. Antimicrob Agents Chemother 56:2125–2128. doi: 10.1128/AAC.05315-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Dortet L, Poirel L, Al Yaqoubi F, Nordmann P. 2012. OXA-48 and OXA-181 carbapenemase-producing enterobacteriaceae in sultanate of Oman. Clin Microbiol Infect 18:E144–8. doi: 10.1111/j.1469-0691.2012.03796.x [DOI] [PubMed] [Google Scholar]
- 30. Muktan B, Thapa Shrestha U, Dhungel B, Mishra BC, Shrestha N, Adhikari N, Banjara MR, Adhikari B, Rijal KR, Ghimire P. 2020. Plasmid mediated colistin resistant mcr-1 and co-existence of OXA48 among Escherichia coli from clinical and poultry isolates: first report from Nepal. Gut Pathog 12:44. doi: 10.1186/s13099-020-00382-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Long H, Feng Y, Ma K, Liu L, McNally A, Zong Z. 2019. The co-transfer of plasmid-borne colistin-resistant genes mcr-1 and mcr-3.5, the carbapenemase gene blaNDM-5 and the 16S methylase gene rmtB from Escherichia coli. Sci Rep 9:696. doi: 10.1038/s41598-018-37125-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhang H, Seward CH, Wu Z, Ye H, Feng Y. 2016. Genomic insights into the ESBL and MCR-1-producing ST648 Escherichia coli with multi-drug resistance. Science Bulletin 61:875–878. doi: 10.1007/s11434-016-1086-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Paulshus E, Thorell K, Guzman-Otazo J, Joffre E, Colque P, Kühn I, Möllby R, Sørum H, Sjöling Å. 2019. Repeated isolation of extended-spectrum-Lactamase-positive Escherichia coli sequence types 648 and 131 from community wastewater indicates that sewage systems are important sources of emerging clones of antibiotic-resistant bacteria. Antimicrob Agents Chemother 63:e00823-19. doi: 10.1128/AAC.00823-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sherchan JB, Hayakawa K, Miyoshi-Akiyama T, Ohmagari N, Kirikae T, Nagamatsu M, Tojo M, Ohara H, Sherchand JB, Tandukar S. 2015. Clinical epidemiology and molecular analysis of extendedspectrum--lactamase-producing Escherichia coli in Nepal: characteristics of sequence types 131 and 648. Antimicrob Agents Chemother 59:3424–3432. doi: 10.1128/AAC.00270-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tchesnokova V, Radey M, Chattopadhyay S, Larson L, Weaver JL, Kisiela D, Sokurenko EV. 2019. Pandemic fluoroquinolone resistant Escherichia coli clone ST1193 emerged via simultaneous homologous recombinations in 11 gene Loci. Proc Natl Acad Sci USA 116:14740–14748. doi: 10.1073/pnas.1903002116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nguyen Q, Nguyen TTN, Pham P, Chau V, Nguyen LPH, Nguyen TD, Ha TT, Le NTQ, Vu DT, Baker S, Thwaites GE, Rabaa MA, Pham DT. 2021. Genomic insights into the circulation of pandemic fluoroquinolone-resistant extra-intestinal pathogenic Escherichia coli ST1193 in Vietnam. Microb Genom 7:000733. doi: 10.1099/mgen.0.000733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Nadimpalli ML, de Lauzanne A, Phe T, Borand L, Jacobs J, Fabre L, Naas T, Le Hello S, Stegger M. 2019. Escherichia coli ST410 among humans and the environment in Southeast Asia. Int J Antimicrob Agents 54:228–232. doi: 10.1016/j.ijantimicag.2019.05.024 [DOI] [PubMed] [Google Scholar]
- 38. Feng Y, Liu L, McNally A, Zong Z. 2019. Coexistence of three blaKPC-2 genes on an Incf/Incr plasmid in ST11 Klebsiella pneumoniae. J Glob Antimicrob Resist 17:90–93. doi: 10.1016/j.jgar.2018.11.017 [DOI] [PubMed] [Google Scholar]
- 39. Dunn S, Carrilero L, Brockhurst M, McNally A. 2021. Limited and strain-specific transcriptional and growth responses to acquisition of a multidrug resistance plasmid in genetically diverse Escherichia coli lineages. mSystems 6:e00083-21. doi: 10.1128/mSystems.00083-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. San Millan A, Toll-Riera M, Qi Q, Betts A, Hopkinson RJ, McCullagh J, MacLean RC. 2018. Integrative analysis of fitness and metabolic effects of plasmids in Pseudomonas aeruginosa PAO1. ISME J 12:3014–3024. doi: 10.1038/s41396-018-0224-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Di Venanzio G, Flores-Mireles AL, Calix JJ, Haurat MF, Scott NE, Palmer LD, Potter RF, Hibbing ME, Friedman L, Wang B, Dantas G, Skaar EP, Hultgren SJ, Feldman MF. 2019. Urinary tract colonization is enhanced by a plasmid that regulates uropathogenic Acinetobacter baumannii chromosomal genes. Nat Commun 10:2763. doi: 10.1038/s41467-019-10706-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Long D, Zhu L-L, Du F-L, Xiang T-X, Wan L-G, Wei D-D, Zhang W, Liu Y. 2019. Phenotypical profile and global transcriptomic profile of hypervirulent Klebsiella pneumoniae due to carbapenemase-encoding plasmid acquisition. BMC Genomics 20:480. doi: 10.1186/s12864-019-5705-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pitout JDD, Laupland KB. 2008. Extended-spectrum -lactamase-producing enterobacteriaceae: an emerging public-health concern. Lancet Infect Dis 8:159–166. doi: 10.1016/S1473-3099(08)70041-0 [DOI] [PubMed] [Google Scholar]
- 44. Pitout JDD, Nordmann P, Laupland KB, Poirel L. 2005. Emergence of enterobacteriaceae producing extended-spectrum beta-lactamases (ESBLs) in the community. J Antimicrob Chemother 56:52–59. doi: 10.1093/jac/dki166 [DOI] [PubMed] [Google Scholar]
- 45. Jolley KA, Bray JE, Maiden MCJ. 2018. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res 3:124. doi: 10.12688/wellcomeopenres.14826.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. doi: 10.1093/bioinformatics/btu153 [DOI] [PubMed] [Google Scholar]
- 47. Tonkin-Hill G, MacAlasdair N, Ruis C, Weimann A, Horesh G, Lees JA, Gladstone RA, Lo S, Beaudoin C, Floto RA, Frost SDW, Corander J, Bentley SD, Parkhill J. 2020. Producing polished prokaryotic pangenomes with the panaroo pipeline. Genome Biol 21:180. doi: 10.1186/s13059-020-02090-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hadfield J, Croucher NJ, Goater RJ, Abudahab K, Aanensen DM, Harris SR. 2018. Phandango: an interactive viewer for bacterial population genomics. Bioinformatics 34:292–293. doi: 10.1093/bioinformatics/btx610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Cummins EA, Hall RJ, Connor C, McInerney JO, McNally A. 2022. Distinct evolutionary trajectories in the Escherichia coli pangenome occur within sequence types. Microb Genom 8:11. doi: 10.1099/mgen.0.000903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Cantalapiedra CP, Hernández-Plaza A, Letunic I, Bork P, Huerta-Cepas J. 2021. eggNOG-mapper v2: functional annotation, orthology assignments, and domain prediction at the metagenomic scale. Mol Biol Evol 38:5825–5829. doi: 10.1093/molbev/msab293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Huerta-Cepas J, Szklarczyk D, Heller D, Hernández-Plaza A, Forslund SK, Cook H, Mende DR, Letunic I, Rattei T, Jensen LJ, von Mering C, Bork P. 2019. eggNOG 5.0: a hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res 47:D309–D314. doi: 10.1093/nar/gky1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. 2000. Gene ontology: tool for the unification of biology. Nat Genet 25:25–29. doi: 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. The Gene Ontology Consortium, Aleksander SA, Balhoff J, Carbon S, Cherry JM, Drabkin HJ, Ebert D, Feuermann M, Gaudet P, Harris NL, et al. 2023. The gene ontology knowledgebase in 2023. Genetics 224. doi: 10.1093/genetics/iyad031 [DOI] [Google Scholar]
- 54. Carrilero L, Dunn SJ, Moran RA, McNally A, Brockhurst MA. 2023. Evolutionary responses to acquiring a multidrug resistance plasmid are dominated by metabolic functions across diverse Escherichia coli lineages. mSystems 8:e0071322. doi: 10.1128/msystems.00713-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Hall RJ, Snaith AE, Element SJ, Moran RA, Smith H, Cummins EA, Bottery MJ, Chowdhury KF, Sareen D, Ahmad I, Blair JMA, Carter LJ, McNally A. 2023. Non-antibiotic pharmaceuticals exhibit toxicity against Escherichia coli at environmentally relevant concentrations with no evolution of cross-resistance to antibiotics. bioRxiv. doi: 10.1101/2023.08.21.554069 [DOI]
- 56. Tange O. 2018. In GNU parallel 2018 [Google Scholar]
- 57. Karp PD, Ong WK, Paley S, Billington R, Caspi R, Fulcher C, Kothari A, Krummenacker M, Latendresse M, Midford PE, Subhraveti P, Gama-Castro S, Muñiz-Rascado L, Bonavides-Martinez C, Santos-Zavaleta A, Mackie A, Collado-Vides J, Keseler IM, Paulsen I. 2018. The EcoCyc database. EcoSal Plus 8. doi: 10.1128/ecosalplus.ESP-0006-2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. The Uniprot consortium . 2019. UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res:D506–15. doi: 10.1093/nar/gky1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Kanehisa M, Goto S. 2000. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res 28:27–30. doi: 10.1093/nar/28.1.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Bottery MJ, Wood AJ, Brockhurst MA. 2017. Adaptive modulation of antibiotic resistance through Intragenomic coevolution. Nat Ecol Evol 1:1364–1369. doi: 10.1038/s41559-017-0242-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Whittle EE, Legood SW, Alav I, Dulyayangkul P, Overton TW, Blair JMA. 2019. Flow cytometric analysis of efflux by dye accumulation. Front Microbiol 10:2319. doi: 10.3389/fmicb.2019.02319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Miles AA, Misra SS, Irwin JO. 1938. The estimation of the bactericidal power of the blood. Epidemiology & Infection 38:732–749. doi: 10.1017/s002217240001158x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Kurihara S, Suzuki H, Oshida M, Benno Y. 2011. A novel putrescine importer required for type 1 Pili-driven surface motility induced by extracellular putrescine in Escherichia coli K-12. J Biol Chem 286:10185–10192. doi: 10.1074/jbc.M110.176032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Qin H, Lo N-S, Loo J, Lin X, Yim A-Y, Tsui S-W, Lau T-K, Ip M, Chan T-F. 2018. Comparative transcriptomics of multidrug-resistant Acinetobacter baumannii in response to antibiotic treatments. Sci Rep 8:3515. doi: 10.1038/s41598-018-21841-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Schneider BL, Reitzer L. 2012. Pathway and enzyme redundancy in putrescine catabolism in Escherichia coli. J Bacteriol 194:4080–4088. doi: 10.1128/JB.05063-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Tkachenko AG, Pozhidaeva ON, Shumkov MS. 2006. Role of polyamines in formation of multiple antibiotic resistance of Escherichia coli under stress conditions. Biochemistry (Moscow) 71:1042–1049. doi: 10.1134/s0006297906090148 [DOI] [PubMed] [Google Scholar]
- 67. Thompson CMA, Hall JPJ, Chandra G, Martins C, Saalbach G, Panturat S, Bird SM, Ford S, Little RH, Piazza A, Harrison E, Jackson RW, Brockhurst MA, Malone JG. 2023. Plasmids manipulate bacterial behaviour through translational regulatory crosstalk. PLOS Biol 21:e3001988. doi: 10.1371/journal.pbio.3001988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Weber BS, Ly PM, Irwin JN, Pukatzki S, Feldman MF. 2015. A multidrug resistance plasmid contains the molecular switch for type VI secretion in Acinetobacter baumannii. Proc Natl Acad Sci USA 112:9442–9447. doi: 10.1073/pnas.1502966112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Billane K, Harrison E, Cameron D, Brockhurst MA. 2021. Why do plasmids manipulate the expression of bacterial phenotypes?. Philos Trans R Soc Lond B Biol Sci 377:20200461. doi: 10.1098/rstb.2020.0461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Price LB, Johnson JR, Aziz M, Clabots C, Johnston B, Tchesnokova V, Nordstrom L, Billig M, Chattopadhyay S, Stegger M, Andersen PS, Pearson T, Riddell K, Rogers P, Scholes D, Kahl B, Keim P, Sokurenko EV, Parkhill J. 2013. The epidemic of extended-spectrum--lactamase-producing Escherichia coli ST131 is driven by a single highly pathogenic subclone, H30-Rx. mBio 4:e00377-13. doi: 10.1128/mBio.00377-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Tchesnokova VL, Rechkina E, Larson L, Ferrier K, Weaver JL, Schroeder DW, She R, Butler-Wu SM, Aguero-Rosenfeld ME, Zerr D, Fang FC, Ralston J, Riddell K, Scholes D, Weissman S, Parker K, Spellberg B, Johnson JR, Sokurenko EV. 2019. Rapid and extensive expansion in the United States of a new multidrugresistant Escherichia coli clonal group, sequence type 1193. Clin Infect Dis 68:334–337. doi: 10.1093/cid/ciy525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Ding Y, Zhang J, Yao K, Gao W, Wang Y. 2021. Molecular characteristics of the new emerging global clone ST1193 among clinical isolates of Escherichia coli from neonatal invasive infections in China. Eur J Clin Microbiol Infect Dis 40:833–840. doi: 10.1007/s10096-020-04079-0 [DOI] [PubMed] [Google Scholar]
- 73. Lopatkin AJ, Bening SC, Manson AL, Stokes JM, Kohanski MA, Badran AH, Earl AM, Cheney NJ, Yang JH, Collins JJ. 2021. Clinically relevant mutations in core metabolic genes confer antibiotic resistance. Science 371:eaba0862. doi: 10.1126/science.aba0862 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Significantly enriched GO terms.
Significantly expressed genes.
Supplemental figures and tables.
Data Availability Statement
The DNA and RNA datasets generated and analyzed during the current study are available from the NCBI BioProject with accession PRJNA1034827