ABSTRACT
Rotavirus (RV) NSP2 is a multifunctional RNA chaperone that exhibits numerous activities that are essential for replication and viral genome packaging. We performed an in silico analysis that highlighted a distant relationship of NSP2 from rotavirus B (RVB) to proteins from other human RVs. We solved a cryo-electron microscopy structure of RVB NSP2 that shows structural differences with corresponding proteins from other human RVs. Based on the structure, we identified amino acid residues that are involved in RNA interactions. Anisotropy titration experiments showed that these residues are important for nucleic acid binding. We also identified structural motifs that are conserved in all RV species. Collectively, our data complete the structural characterization of rotaviral NSP2 protein and demonstrate its structural diversity among RV species.
IMPORTANCE
Rotavirus B (RVB), also known as adult diarrhea rotavirus, has caused epidemics of severe diarrhea in China, India, and Bangladesh. Thousands of people are infected in a single RVB epidemic. However, information on this group of rotaviruses remains limited. As NSP2 is an essential protein in the viral life cycle, including its role in the formation of replication factories, it may be a target for future antiviral strategy against viruses with similar mechanisms.
KEYWORDS: double‐stranded RNA virus, rotavirus, NSP2, protein structure
INTRODUCTION
Rotaviruses (RVs) are the most common cause of diarrheal disease. They belong to the Sedoreoviridae family, a group of double-stranded RNA (dsRNA) viruses. Ten serologically defined species were initially recognized by the International Committee on Taxonomy of Viruses (ICTV) (rotaviruses A–J; RVA–RVJ), but due to the lack of any additional virus isolates or sequencing data, RVE was withdrawn as a species by the ICTV in 2019 (1). In addition, the existence of two other species [rotavirus K (RVK) and rotavirus L (RVL)] has recently been reported (2, 3). Three species (RVA, RVB, and RVC) have been shown to be the most common infectious agents in humans (4, 5). RVA causes diarrhea in infants and has been studied extensively. RVB, also called adult diarrhea RV, was the cause of several outbreaks of diarrheal disease (4–8). The first reported RVB epidemics occurred in China in 1982, affecting more than one million people. Smaller outbreaks were later reported in India and Bangladesh (8). Antibodies to RVB have also been detected in human serum samples from other parts of the world (8–11). Phylogenetic analyses of RVB sequences suggest that viruses infecting humans are distinct from viruses infecting other animals (12, 13). Hoxie and Dennehy reported evidence of the reassortment of most segments in RVA (including segment 8, encoding NSP2) and emphasized that it may be an important driver of RV evolution and may affect the diversity of RV species (14, 15). Despite its importance for human health and epidemic potential, information on RVB species remains limited.
The RV genome consists of 11 dsRNA molecules that encode six structural proteins (VPs) and five or six nonstructural proteins (NSPs) in different strains. Each of the 11 RNA molecules must be replicated and incorporated into nascent virions during the viral life cycle. These challenging and complex processes are orchestrated by the cooperation of viral proteins (16–19). One key factor is NSP2, an octameric, multifunctional enzyme. NSP2 has been shown to bind RNA in a largely sequence-independent manner. A sequence-dependent NSP2–RNA interaction has also been proposed (20–23). Crystallographic studies revealed that the 5′ GG of the bound oligoribonucleotide interacts extensively with highly conserved residues in the RVA NSP2 enzymatic cleft. While these residues provide GG-specific interactions, however, surface plasmon resonance studies implicate the C-terminal helix (CTH) and other basic residues outside the enzymatic cleft in the sequence-independent RNA binding of NSP2 (20). NSP2 exhibits diverse RNA-related activities, such as RNA 5′-triphosphatase (RTPase) activity, RNA annealing, and adenosine triphosphate-independent dsRNA helix destabilization (23–29). NSP2 has been reported to be essential for rotaviral genome replication. It interacts with NSP5 to form viroplasms, also known as “viral factories,” cytoplasmic liquid-liquid phase separation inclusions where components that are required for genome replication are assembled (30–34). Moreover, NSP2 interacts directly with other proteins that are involved in viral replication, including RNA-dependent RNA polymerase (RdRP/VP1) and inner capsid protein (VP2) (32, 35). Given that NSP2 possesses nucleoside triphosphatase (NTPase) activity, undergoes conformational changes upon different ligand binding, and promotes inter-segment RNA–RNA interactions, it has been suggested to act as an RNA chaperone for RNA packaging into nascent virions (22, 23, 29, 36, 37).
An in silico analysis of amino acid sequences of NSP2 proteins from various species of RVs showed that RVB NSP2 belongs to a clade that is distinct from those from RVA and RVC. This suggests that NSP2 from RVB species may possess a different tertiary structure. Several NSP2 structures have been determined, including RVA and RVC NSP2s (20, 27, 38–40). However, no structural information is currently available for the RVB counterpart or any other species from the RVB clade. Here, we describe a cryo-electron microscopy (cryo-EM) structure of RVB NSP2 and present a comparison with its orthologs from other human RVs, revealing important structural differences and similarities among NSP2 proteins. To better understand the role of RVB NSP2 in RNA chaperone activity, we identified amino acid residues that are essential for RNA binding.
RESULTS
In silico analysis of rotaviral NSP2 sequences
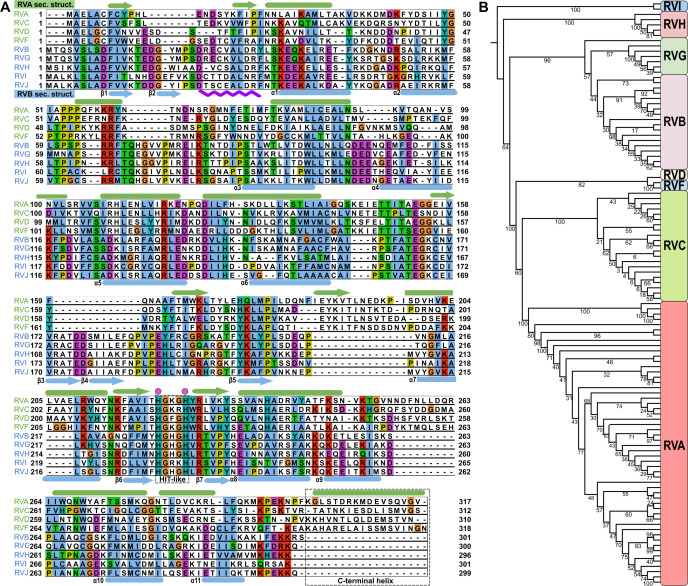
At least three species of RVs (RVA, RVB, and RVC) can infect humans and cause severe diarrhea, with RVA being the most common pathogen. Amino acid sequence analysis revealed limited similarity and identity (31.9% and 17.8%, respectively) between RVB and RVA NSP2s (Fig. 1A). We performed a phylogenetic analysis based on NSP2 amino acid sequences, which led to subdivision of the Rotavirus genus into two major clades, one that comprises RVs from RVA, RVC, RVD, and RVF species and another that comprises RVs from RVB, RVG, RVH, and RVI species. We refer to these species as RVA and RVB clades, respectively. This analysis also indicated that RVA and RVC are more closely related to each other than to RVB (Fig. 1B).
Fig 1.
Amino acid sequence comparison of different RV species. (A) Amino acid sequence alignment of NSP2 from nine different RV species. Species that belong to the RVA clade (green) are at the top of the alignment. Species that belong to the RVB clade (blue) are at the bottom. Representations of RVA NSP2 secondary structures (green) are shown above the alignment panels. Representations of RVB NSP2 secondary structures (blue) are shown below the alignment panels. Arrows indicate β-strands. Cylinders indicate α-helices. The HIT-like motif and C-terminal helices are indicated by a gray rectangle. Conserved catalytic histidine residues are indicated by a pink dots above the alignment. The purple wavy line below the alignment represents a 10-amino-acid insertion in RVB NSP2. The following amino acid sequences were used: rotavirus A (PDB ID: 2R7C) (RVA), rotavirus B (AAF72868.1) (RVB), rotavirus C (AKH39851.1) (RVC), rotavirus D (AXL64569.1) (RVD), rotavirus F (AXL64586.1) (RVF), rotavirus G (AXF38053.1) (RVG), rotavirus H (QKY66970.1) (RVH), rotavirus I (AYH64828.1) (RVI), and rotavirus J (YP_010086027.1) (RVJ). (B) Phylogenetic analysis of NSP2 based on amino acid sequences from all nine RV species. Bootstrap values are indicated.
RVB NSP2 forms octamers
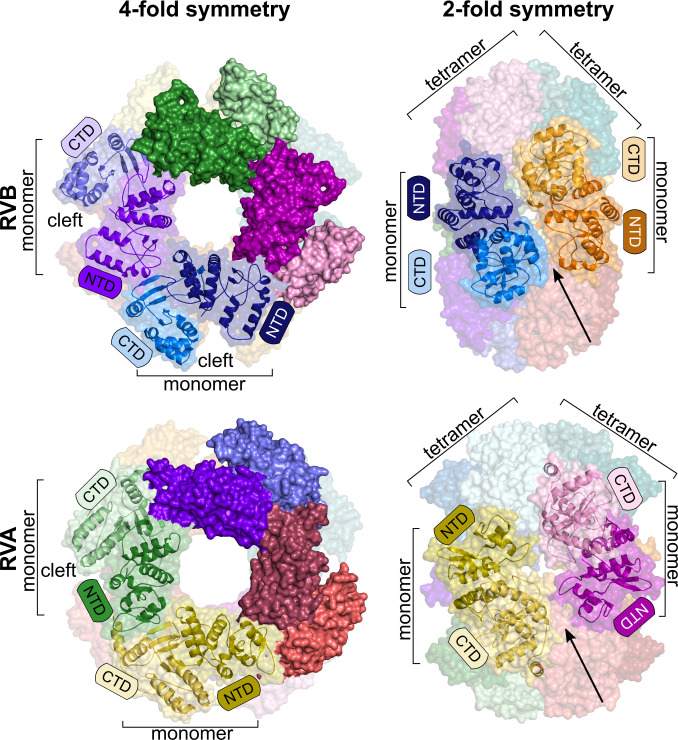
In contrast to RVA and RVC NSP2s, no structural information is available for RVB NSP2. Considering the distant relationship of RVB NSP2 to other human RV orthologs, we found that RVB NSP2 is an interesting target for structural studies. We determined the cryo-EM structure of RVB NSP2 at a global resolution of 3.8 Å. The overall shape of the RVB NSP2 structure is similar to orthologs from RVA and RVC. The proteins form an octameric ring (D4 symmetry) with a central cavity. The size of the RVB NSP2 octamer is similar to RVA NSP2 [Protein Data Bank (PDB) ID: 7PKO], except that the width along the 2-fold symmetry axis of the octamer is larger for the RVA protein (Fig. 2). Another common feature of rotaviral NSP2 is a catalytic cleft that is located between the N-terminal domain (NTD) and the C-terminal domain (CTD) of the single chain, which is essential for NTPase and RTPase activities (Fig. 3A and B). NSP2 tetramers are formed through the head-to-tail arrangement of subunits, and octamers are formed by stacking the same sides of the two tetrameric rings (Fig. 2). An important feature of the structure is the four grooves that run diagonally across the tetramer–tetramer interface (Fig. 2).
Fig 2.
Comparison of octamers formed by RVB NSP2 (present study, top panel) and RVA NSP2 (PDB ID: 7PKO, bottom panel). Structures are shown along the 4-fold symmetry axis (left) or 2-fold symmetry axis (right). The darker shade of each color represents the N-terminal domain, and the lighter shade corresponds to the C-terminal domain of each subunit. Surface and cartoon representations show the subunit of each octamer. Arrows indicate basic grooves between two tetramers. Catalytic clefts are indicated.
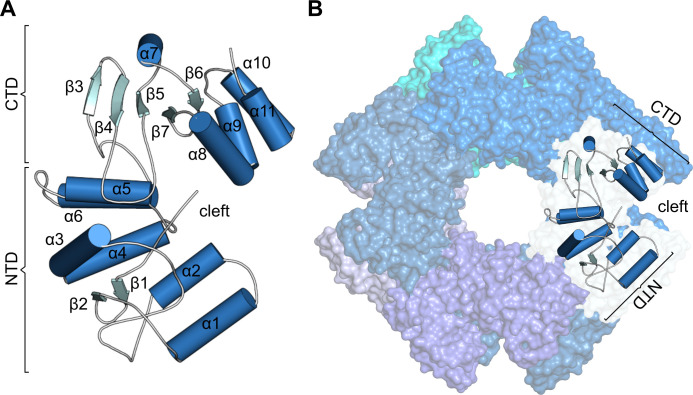
Fig 3.
Structure of RVB NSP2. (A) Single subunit of NSP2. (B) Surface representation of the NSP2 octamer with a cartoon representation of the single chain. NTD, N-terminal domain; CTD, C-terminal domain.
Structures of RVA and RVB NSP2s are different
RVB NSP2 comprises two domains. The NTD is formed by two antiparallel β-strands, followed by six α-helices (α1–α6). It is separated from the CTD by the deep catalytic cleft. The CTD comprises one β-sheet of three strands and an α-helix (α7), followed by two antiparallel β-strands and the last four α-helices (α8–α11; Fig. 3A).
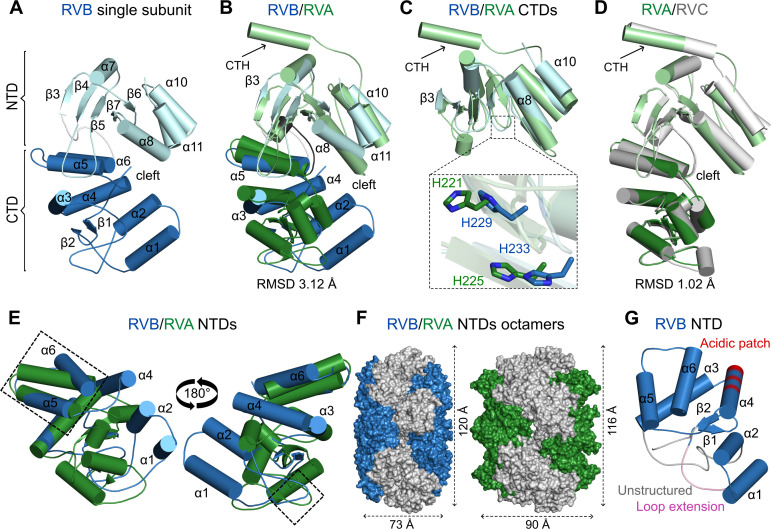
The overall structure of the single chain of NSP2 is similar for all three species of RVs (Fig. 4A through D). Catalytic clefts that are formed between the NTD and the CTD are a common feature of RV NSP2s. The main difference between structures of RVB and RVA (PDB ID: 2R7C) orthologs is observed in the N-terminal portion of the structures. In both cases, the N-terminus is predominantly α-helical. However, when structures of the NTDs are superposed, the three α-helices (α1, α2, and α4 in the case of RVB NSP2) of the two proteins adopt nearly perpendicular positions (Fig. 4B and E). In particular, these RVB NSP2 α-helices follow the loop with the 10-amino-acid insertion (residues 21–30), which is absent in RVA NSP2 (Fig. 1A and 4G). The presence of this loop results in a different arrangement of the α-helices. Additionally, one of these α-helices, α4 (residues 102–116), contains an acidic patch that is typical for species from the RVB clade (Fig. 1A and 4F). There is also a region of the NTD that is structurally similar between RVA and RVB proteins. It consists of two α-helices (RVB NSP2 residues 124–134 and 145–156; RVA NSP2 residues 107–119 and 124–149) and an intervening loop (residues 135–144 for RVB NSP2 and residues 120–123 in the case of the RVA counterpart; Fig. 1A and 4E). This similarity may stem from the fact that this region is responsible for intramolecular interactions of subunits that are involved in octamer formation. RVA and RVC NSP2s are almost identical (40). When their monomeric structures are superposed, the resulting root mean square deviation is 1.02 Å for 227 pairs of Cα atoms (Fig. 4D). A small difference between the two structures is observed in the loop that is formed by residues 170–185 (40).
Fig 4.
Comparison of individual subunits of NSP2 from RVA, RVB, and RVC species of RVs. (A) RVB NSP2 subunit (present study). (B) Superposition of RVA NSP2 (PDB ID: 2R7C, green) on RVB NSP2 (present study, blue). (C) Superposition of CTDs of RVB and RVA NSP2s. A close-up view of catalytic histidine residues is shown in the inset. (D) Superposition of RVC NSP2 (PDB ID: 2GU0, gray) and RVA NSP2 (PDB ID: 2R7C, green). (E) Superposition of NTDs of RVB and RVA NSP2s (similar regions are indicated by rectangles). (F) View along the 2-fold symmetry axis of the NSP2 octamer with the NTD in blue (RVB) and green (RVA). (G) N-terminal domain of RVB NSP2 with unique features of the RVB clade marked as the following: unstructured region of RVB NSP2 in gray, conserved loop extension in pink, and acidic patch present in the RVB clade in red. In all panels, particular domains are indicated by a darker shade (NTDs) and lighter shade (CTDs) of each color. The C-terminal helix (CTH) is indicated by arrows.
The loop that contains catalytic residues and is located between two β-strands, β6 and β7 (residues 225–237), is longer in RVB NSP2. This results in slight differences in the position of catalytic histidine residues essential for NTPase activity (H229 and H233 in RVB NSP2 and H221 and H225 in RVA NSP2; Fig. 4C). In the CTDs of NSP2 proteins, the RVB ortholog has an additional α-helix that comprises residues 254–262. The corresponding regions in RVA and RVC proteins lack secondary structures. A prominent difference between the NSP2 orthologs is the presence of a CTH in RVA and RVC proteins that is absent in RVB NSP2 (Fig. 1A and 4A through D). In RVA and RVC NSP2s, this α-helix is connected through a long, flexible linker that allows NSP2 to switch between open and closed conformations (20).
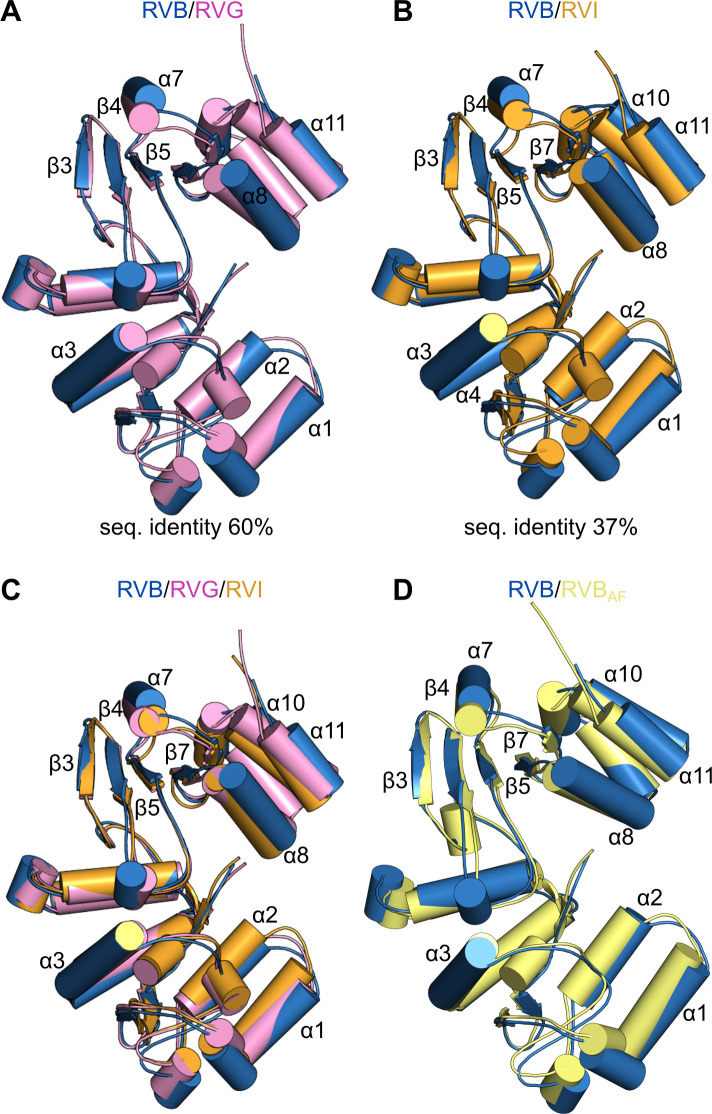
Structure of NSP2 is conserved among species within the RVB clade
The phylogenetic analysis of NSP2 divided the Rotavirus genus into two major clades: RVA and RVB clades. The amino acid sequences of NSP2 differ even within the same clade (Fig. 1A). Therefore, we compared the NSP2 structure of different species within the RVB clade. RVG NSP2 appears to have the closest evolutionary relationship with RVB NSP2, in which they share high sequence similarity and identity (78% and 60%, respectively). In contrast, RVI NSP2 is the most distant, with limited sequence similarity and identity with RVB NSP2 (60% and 37%, respectively). We generated predictions of NSP2 structures from these species using AlphaFold2, an artificial intelligence system that predicts three-dimensional structures of proteins (41), and compared them with our RVB NSP2 cryo-EM structure. There are only minor differences when RVG and RVB NSP2s are superposed (Fig. 5A). Slight changes in arrangements of α-helices in the C-terminal region of the two proteins are observed (Fig. 5A). Surprisingly, although the amino acid sequence identity between NSP2 from species B and I is limited, the overall structure of the two proteins is almost identical. In this case, we can also see modest differences in C-terminal regions of the proteins (Fig. 5B). Despite the diverse phylogenetic relationships, the superposition of all three structures shows that they adopt a nearly identical structure (Fig. 5C). The high accuracy of AlphaFold2 modeling is demonstrated by superposing the cryo-EM-solved RVB NSP2 subunit and the AlphaFold2-generated model of RVB NSP2 (Fig. 5D).
Fig 5.
Comparison of NSP2 structures from RVs in RVB clade. (A) Superposition of RVB NSP2 subunit (blue) and AlphaFold2-generated model of RVG NSP2 (pink). (B) Superposition of RVB NSP2 subunit (blue) and AlphaFold2-generated model of RVI NSP2 (orange). (C) Superposition of three structures of NSP2 from RVB (blue), RVG (pink), and RVI (orange). (D) Superposition of RVB NSP2 subunit solved by cryo-EM (blue) and AlphaFold2-generated model of RVB NSP2 (yellow).
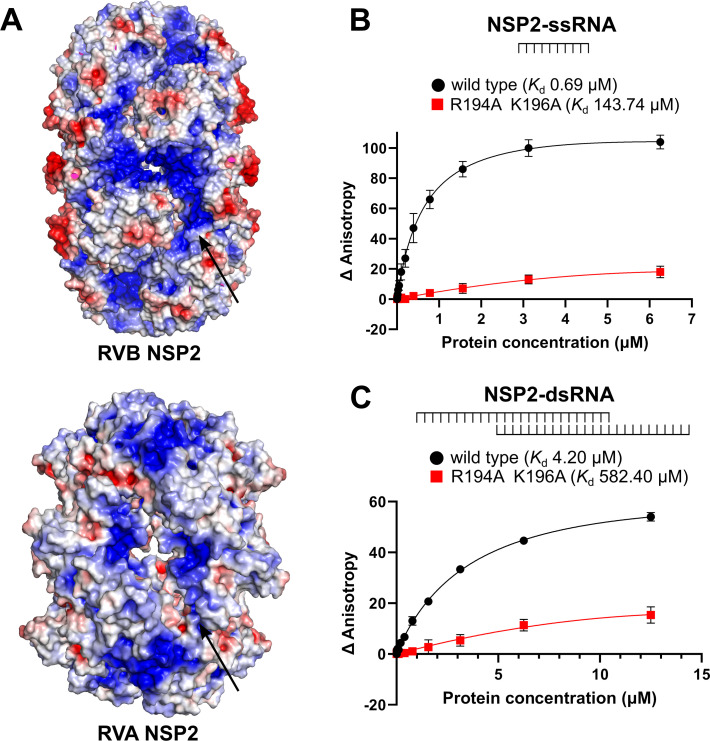
NSP2 interacts with RNA via basic grooves
RVs are dsRNA viruses. Their capsid contains 11 dsRNA segments whose expression must be coordinated to ensure proper viral replication. NSP2 is thought to be an RNA chaperone with many RNA-related functions. Thus, RNA binding is a crucial ability to fulfill its complex role in the RV life cycle. To identify NSP2 regions that interact with nucleic acids, we calculated electrostatic surface potential of the protein. This revealed the presence of basic grooves in the octamer, which could be RNA binding sites (Fig. 6A). A similar pattern is observed for the RVA and RVC NSP2 octamers. The difference is that RVC NSP2 generally appears to be less basic (40). To test whether these were indeed RNA binding sites, we purified a variant of RVB NSP2, NSP2mut (R194A/K196A), with two amino acid substitutions of basic residues in the groove (Fig. 6A). These residues were chosen because they both have a positive charge, are located in the groove, and are conserved within the RVB clade.
Fig 6.
RNA binding by RVB NSP2. (A) Electrostatic surface potential is shown in a view along the 2-fold symmetry axis of the RVB and RVA NSP2 octamers. Red color indicates negative potential, and blue color indicates positive potential. The arrows indicate putative nucleic acid binding sites and electropositive residues that are predicted to be involved in the NSP2–RNA interaction. (B) Fluorescence anisotropy titration of fluorescently labeled ssRNA with increasing concentrations of NSP2 (wild type and mutant protein). (C) Fluorescence anisotropy titration of fluorescently labeled, partially dsRNA with increasing concentrations of NSP2 (wild type and mutant protein). Changes in fluorescence anisotropy are plotted against NSP2 concentration. Three independent measurements are plotted for each concentration. Error bars indicate the standard deviation.
We performed binding assays using fluorescence anisotropy titrations to quantitatively analyze RNA binding ability. We used single-stranded RNA (ssRNA) and dsRNA with 3′ and 5′ ssRNA overhangs (see Materials and Methods). Compared to wild-type NSP2, the affinity for ssRNA was lower for mutant NSP2 (Kd = 0.69 µM and Kd = 143.7 µM, respectively; Fig. 6B). The affinity for dsRNA was significantly lower than that for the ssRNA substrate (Fig. 6B and C). For dsRNA, we also observed a lower affinity for the NSP2 mutant (Kd = 582.4 µM) compared to the wild-type counterpart (Kd = 4.2 µM; Fig. 6C). In conclusion, the affinity for both ssRNA and dsRNA was decreased in the NSP2 mutant (Fig. 6B and C). The fact that the mutations in the grooves disrupt the interaction between NSP2 and RNA, confirmed by fluorescence anisotropy experiments, suggests that the grooves are RNA binding sites.
DISCUSSION
At least 12 species of RV have been reported to date, but the ICTV has so far recognized nine species of RV. We performed a phylogenetic analysis of amino acid sequences of NSP2 from all recognized by ICTV RV species, resulting in the classification of RV species into two major clades. The first clade contains RV species RVA, RVC, RVD, and RVF. The second clade includes species RVB, RVG, RVH, RVI, and RVJ. Similar divisions have been observed in phylogenetic analyses based on VP1, VP6, and NSP1 proteins (13, 42, 43).
The most common species in the RVB clade that is able to infect humans is RVB. However, in contrast to RVA and RVC, RVB NSP2 has been poorly studied both functionally and structurally. Our cryo-EM structure of RVB NSP2 in the present study shows that it adopts an octameric form, similar to RVA and RVC proteins. The fact that all published NSP2 structures remain octameric despite clear differences in amino acid sequences underscores the importance of this oligomeric state. One role of octamerization of the protein may be to form grooves at the tetramer–tetramer interface that serve as platforms for RNA interactions. The importance of these grooves is supported by our RNA binding assays, which showed lower affinity for RNA of NSP2 with substitutions of arginine and lysine residues in the grooves. This suggests that the octameric form of NSP2 is conserved because it is required for its sequence-independent RNA binding and consequently for its RNA-related activities. We attempted to determine the structure of the NSP2–RNA complex. In our cryo-EM reconstruction, we observed an additional density that may correspond to RNA (Supplemental Fig. S2). This is supported by the fact that it is located in the grooves. However, the resolution of this density is too low to allow us to build an RNA model. This suggests the transient nature of nucleic acid binding or RNA flexibility. The basic grooves may also serve as NSP5 binding platforms, which has been shown to condense in the presence of polyarginine (33). The NSP2–NSP5 interaction results in the formation of viroplasms, liquid–liquid phase separation inclusion bodies where viral genome replication occurs. This is consistent with previous reports that NSP5 shares the same binding site with RNA within the groove in the NSP2 octamer (44). The mechanism of NSP5 binding to NSP2 would be important to establish because the NSP2–NSP5 interaction could be a target for the development of antiviral strategies (33, 34).
Comparisons of individual chains of NSP2 proteins from different RV species show the main differences in N-terminal regions. NSP2s from RVA and RVC are almost identical, whereas the NTD of RVB NSP2 differs significantly. A major difference is the arrangement of α-helices (formed by residues 34–35, 50–59, and 103–116 in RVB NSP2 and residues 2–6, 23–32, 54–61, and 96–106 in RVA NSP2). Likely because of this difference, the NTD in RVA NSP2 is directed toward the proximal part of the octamer in the 4-fold symmetry axis view. This results in a larger width of the octamer along its 2-fold symmetry axis (Fig. 4F). Interestingly, despite differences in the structure of NTDs, there are also structural features that are conserved across RV species. Two α-helices (RVB NSP2 residues 124–139 and 135–146 vs RVA NSP2 residues 107–119 and 124–149) and the loop between them (residues 135–144 for RVB NSP2 and residues 120–123 for the RVA counterpart) have a similar arrangement in all structures. These regions contain amino acid residues that are responsible for subunit-subunit interactions to form tetramers of NSP2 and are consequently essential for protein oligomerization.
When comparing the CTDs, one of the most striking differences between RVB NSP2 and its counterparts from RVA and RVC species of RVs is the lack of the CTH. Interestingly, this feature is characteristic of all NSP2 proteins from RVs clustered together with RVB NSP2 (Fig. 1A). The function of the CTH in RVA NSP2 has been extensively studied. It allows NSP2 to switch between open and closed conformations (20). It may also play a role in RNA binding, but this hypothesis requires further investigation. Bravo et al. investigated the CTH in the context of different NSP2 functions in the viral life cycle (38). They described the acidic patch in the helix, suggesting its role in RNA dissociation from NSP2. Perhaps, in the case of RVB NSP2, this function is fulfilled by the acidic patch in the different α-helix (residues 103–116), which is conserved in all species of the RVB clade (Fig. 1A and 4G). It has also been shown that the CTH is important for the regulation of the binding and release of RNA from NSP2, demonstrating its role in nucleic acid binding (20). Point mutations in the CTH abrogated viral replication, underlying its importance (38). The fact that the helix is absent in RVB NSP2 suggests that its role is complex. Nevertheless, the overall fold of the CTDs of RVA, RVB, and RVC is similar. This is likely because of the fact that it resembles the ubiquitous cellular histidine triad (HIT) group of nucleotidyl hydrolases. It contains the HIT-like motif, implying that the active site for NTP hydrolysis is located in the cleft, with one of the histidine residues acting as a key catalytic residue (20).
The cryo-EM structure of NSP2 that is presented herein is the first structure for species from the RVB clade. The amino acid sequence analysis of NSP2 showed that they differ even between RV species of the same clade. We used AlphaFold2 to generate prediction models of NSP2 from species with the highest and lowest levels of amino acid sequence identity and similarity (RVG and RVI, respectively) (41). Surprisingly, despite differences in the amino acid sequences of NSP2 from three RV species, the structures of the single subunits are almost identical. This applies to comparisons of RVB/RVG NSP2 (amino acid sequence identity of 60%) and RVB/RVI NSP2 (limited amino acid sequence identity of 37%). It strongly suggests that the overall three-dimensional structure of NSP2 from RVs that belong to the RVB clade is evolutionarily conserved, regardless of distinct levels of their phylogenetic relationships.
Despite key differences in NSP2 structures between species of human RVs, we also identified structural similarities that might be conserved in all species of RVs. These similarities include an octameric form of the proteins, the presence of basic grooves, and the arrangement of regions that are responsible for oligomerization and activity. We also reported the conservation of NSP2 structures among RV species of the RVB clade. The unique features of NSP2 are considered to affect the viral life cycle, although the exact mechanism is not fully understood. It has been previously reported that differences in homologous proteins from RVA and RVB impact viral pathogenicity. Diller et al. demonstrated that RVB NSP1-1 mediates syncytium formation in cultured human cells, a mechanism that is most likely unique for species from the RVB clade (45). Both similarities and differences in NSP2 appear to be factors that might be used to discriminate between members of different species and unclassified viruses. To date, because of its strong conservation, VP6 has been the major target of diagnostic assays for RVs (1, 46). However, unique features of NSP2 might be another molecular indicator that is useful for the classification of new RV species.
In conclusion, our research fills a gap in the structural knowledge of NSP2 proteins from RVs that infect humans. Our comparison of structures of NSP2s from three different RV species showed that the main differences are in the arrangement of the N-terminal α-helices and the absence of the α-helix at the C-terminus. We also identified structural elements of NSP2 that are conserved across RV species and important for protein oligomerization. Further studies will explain how RVB NSP2 functions without this α-helix or why NTDs exhibit such pronounced differences. In particular, it will be interesting to study how differences in NSP2 structures are related to the viral life cycle, pathogenesis, and host interactions.
MATERIALS AND METHODS
In silico analysis
RVB NSP2 (Q86197) was used as a query for a PSI-BLAST search. The search was conducted against non-redundant protein sequences (nr). It was restricted to the sequences in the database that correspond to RVs (taxid:10912). Sequences with low information content [models (sequences with accession number prefix XM_ or XP_), uncultured/environmental sample sequences, and non-redundant RefSeq proteins (sequences with accession number prefix WP)] were excluded. After three iterations, 2,577 sequences were found. Partial sequences were removed from the obtained set of sequences, and only RVA, RVB, and RVC sequences were used for further analysis. Finally, redundancy of this set of sequences was reduced to 95% by the CD-HIT software (47). This resulted in 90 sequences that were subjected to multi-sequence alignment with the MUSCLE algorithm (https://www.ebi.ac.uk/Tools/msa/muscle/).
The resulting alignments were further subjected to phylogenetic analysis with the Simple Phylogeny software (https://www.ebi.ac.uk/Tools/phylogeny/simple_phylogeny/) using UPGMA method, and the obtained phylogenetic tree was midpoint rooted and visualized with the Interactive Tree Of Life (iTOL) software (48).
Protein expression and purification
The synthetic gene that encodes wild-type NSP2 was purchased from Biomatik and subcloned into a pET28 expression vector that carried an N-terminal His6-SUMO tag that is removable by SUMO protease. The NSP2 variant with point substitutions was obtained according to Stratagene's QuikChange protocol.
All proteins (wild type and mutant) were produced in Escherichia coli BL21 Star (DE3) cells in Lysogen Broth medium and induced with 0.4 mM 1-thio-β-D-galactopyranoside at 18°C. The cells were then harvested and suspended in 25 mM HEPES (pH 7.5), 200 mM NaCl, 5% glycerol, 10 mM imidazole, and 10 mM β-mercaptoethanol and incubated on ice in the presence of 1 mg/mL lysozyme, protease inhibitor cocktail, and viscolase. Following sonication, the cleared lysate was applied to a HisTrap column (GE Healthcare) that was equilibrated with 25 mM HEPES (pH 7.5), 200 mM NaCl, 5% glycerol, 10 mM imidazole, and 5 mM β-mercaptoethanol. After a wash step with 60 mM imidazole, the protein was eluted with 300 mM imidazole. The eluted fraction was dialyzed overnight against 25 mM HEPES (pH 7.5), 200 mM NaCl, 5% glycerol, and 5 mM β-mercaptoethanol with SUMO protease to remove the SUMO-His tag and loaded onto a second HisTrap column (GE Healthcare) that was equilibrated with a buffer that contained 25 mM HEPES (pH 7.5), 200 mM NaCl, 5% glycerol, 10 mM imidazole, and 5 mM β-mercaptoethanol. Proteins were further purified on a Superdex 200 column (GE Healthcare) that was equilibrated with 25 mM HEPES (pH 7.5), 200 mM NaCl, and 1 mM dithiothreitol (DTT). Because of nucleic acid contamination (high A260/A280 ratio), proteins were further purified on a heparin column (GE Healthcare) that was equilibrated with the same buffer. Protein was eluted with a linear gradient of 200–1,000 mM NaCl. Selected fractions were concentrated, flash frozen in liquid nitrogen, and stored at −20°C.
Fluorescence anisotropy
The interaction between RNA and NSP2 or its mutated variant was studied using fluorescence anisotropy. The assay was conducted in 96-well, black, flat-bottom polystyrene NBS plates (Corning 3650) in a total reaction volume of 50 µL. Fluorescently (Cy5) labeled ssRNA (5′ ACACCACUCA 3′) and dsRNA oligonucleotide [with 5′ and 3′ single-stranded 10-nt overhangs produced by annealing of two RNAs: (5′ UUUUUUUUUUCAGCGUUACGAUAUGC 3′; 5′ UUUUUUUUUUGCAUAUCGUAACGCUG 3′)] were used at a concentration of 50 nM, and protein concentration ranged from 0 to 12.5 µM for dsRNA variant of experiment and from 0 to 6.25 for ssRNA variant. Dilutions of the oligonucleotides and protein were prepared in buffer A [50 mM HEPES (pH 7.0), 100 mM NaCl, 1 mM DTT, and 10 mM MgCl2]. The reactions were prepared in triplicate. Fluorescence anisotropy was measured using a Tecan Infinite M1000 microplate reader at excitation/emission wavelengths of 635/670 nm for Cy5-labeled oligonucleotide. The saturation curve was fitted using one-site-specific binding and GraphPad Prism software.
Cryo-EM sample preparation and data collection
The dsRNA oligonucleotide with 3′ single-stranded 10-nt overhang that was produced by annealing of two RNAs, 5′ GCAUAUCGUAACGCUGUUUUUUUUUU 3′ and 5′ CAGCGUUACGAUAUGC 3′, was mixed with RVB NSP2 in a 4:1 molar ratio (50 µM RNA and 12.5 µM NSP2). The 3 µL of mixture was applied to a glow-discharged C-flat-2/1 mesh 200 Cu grid and vitrified in liquid ethane with an FEI Vitrobot Mark IV (Thermo Fisher Scientific) at 4°C with 95% humidity and a 4 s blot time. Data collection was performed on a Titan Krios G3i electron microscope (Thermo Fisher Scientific) that operated at 300 kV and was equipped with a BioQuantum energy filter (with 20 eV energy slit) and K3 camera (Gatan) at the SOLARIS National Synchrotron Radiation Centre (Krakow, Poland). A total of 4,770 movies were recorded with 30° stage tilt in counting mode with a physical pixel size of 0.86 Å (nominal magnification of 105,000×), 50 µm C2 condenser aperture, and retracted objective aperture. The nominal defocus range was −2.5, −2.0, and 1.5 µm. The total dose (fractionated into 40 frames) was 40.92 e/Å2, and the dose rate was 16.70 e/pixel/s.
Cryo-EM data processing and refinement
Cryo-EM images were processed with RELION-3.1 (49) and cryoSPARC 3.2 (50). Raw movies were motion-corrected, and binned 2× using RELION’s implementation of MotionCor2 software (51). After motion correction, micrographs were imported to cryoSPARC, and the Patch CTF estimation was performed to fit the contrast transfer function. Manual picker, Template picker, and Inspect picker were used to pick 2,705,762 particles. After extraction (box size of 140 pixels; pixel size of 1.72 Å/pixel), particles were subjected to several rounds of two-dimensional classification (Supplemental Fig. S1). A total of 66,999 selected particles were used to generate the initial three-dimensional model (ab initio reconstruction with four classes) in cryoSPARC. Two selected classes (37,768 particles) were refined with imposed D4 symmetry, resulting in a 4.05 Å reconstruction (derived from the Fourier Shell Correlation between the half maps). Refined particles were re-imported into RELION with scripts from UCSF pyem, re-extracted with a pixel size of 1.72 Å/pixel, and subjected to Bayesian polishing (with re-extraction with an unbinned pixel size of 0.86 Å/pixel), followed by another round of two-dimensional classification (cryoSPARC), resulting in the selection of 37,667 particles that were again imported to RELION and subjected to a second round of Bayesian polishing. Afterward, an additional round of two-dimensional classification was performed, and a final number of 22,131 particles was selected. Non-uniform refinement in cryoSPARC resulted in 3.77 Å reconstruction. The final map was sharpened locally with the Local Filter tool in cryoSPARC (Supplemental Fig. S1). The initial model for structure refinement was generated by AlphaFold2 (41). It was manually edited in Coot (52) and refined using real-space refinement in Phenix (53).
ACKNOWLEDGMENTS
We would like to acknowledge Małgorzata Figiel, PhD, for her support in the initial stages of the project. We thank Michael Arends for proofreading of the manuscript.
Funding for open access charges was obtained from the IIMCB Statutory Funding.
We acknowledge SOLARIS Centre for the access to cryo-EM infrastructure, where the measurements were performed in collaboration with SOLARIS staff.
Contributor Information
Marcin Nowotny, Email: mnowotny@iimcb.gov.pl.
Christiane E. Wobus, University of Michigan Medical School, Ann Arbor, Michigan, USA
DATA AVAILABILITY
The cryo-EM structure of rotavirus B NSP2 has been deposited in the Protein Data Bank (https://www.wwpdb.org/) under the accession code 8P00. All other relevant data are available from the authors.
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/jvi.01660-23.
Figures S1 and S2; Tables S1 and S2.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Walker PJ, Siddell SG, Lefkowitz EJ, Mushegian AR, Adriaenssens EM, Alfenas-Zerbini P, Dempsey DM, Dutilh BE, García ML, Curtis Hendrickson R, et al. 2022. Recent changes to virus taxonomy ratified by the international committee on taxonomy of viruses (2022). Arch Virol 167:2429–2440. doi: 10.1007/s00705-022-05516-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Johne R, Schilling-Loeffler K, Ulrich RG, Tausch SH. 2022. Whole genome sequence analysis of a prototype strain of the novel putative rotavirus species L. Viruses 14:462. doi: 10.3390/v14030462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Johne R, Tausch SH, Ulrich RG, Schilling-Loeffler K. 2023. Genome analysis of the novel putative rotavirus species K. Virus Res 334:199171. doi: 10.1016/j.virusres.2023.199171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chen CM, Hung T, Bridger JC, McCrae MA. 1985. Chinese adult rotavirus is a group B rotavirus. Lancet 2:1123–1124. doi: 10.1016/s0140-6736(85)90710-x [DOI] [PubMed] [Google Scholar]
- 5. Chitambar SD, Lahon A, Tatte VS, Maniya NH, Tambe GU, Khatri KI, Desai HS, Ugare MR, Kulkarni SV, Waghmare AP. 2011. Occurrence of group B rotavirus infections in the outbreaks of acute gastroenteritis from western India. Indian J Med Res 134:399–400. [PMC free article] [PubMed] [Google Scholar]
- 6. Hung T, Chen GM, Wang CG, Yao HL, Fang ZY, Chao TX, Chou ZY, Ye W, Chang XJ, Den SS. 1984. Waterborne outbreak of rotavirus diarrhoea in adults in China caused by a novel rotavirus. Lancet 1:1139–1142. [PubMed] [Google Scholar]
- 7. Joshi MS, Ganorkar NN, Ranshing SS, Basu A, Chavan NA, Gopalkrishna V. 2017. Identification of group B rotavirus as an etiological agent in the gastroenteritis outbreak in Maharashtra, India. J Med Virol 89:2244–2248. doi: 10.1002/jmv.24901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sanekata T, Ahmed MU, Kader A, Taniguchi K, Kobayashi N. 2003. Human group B rotavirus infections cause severe diarrhea in children and adults in Bangladesh. J Clin Microbiol 41:2187–2190. doi: 10.1128/JCM.41.5.2187-2190.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. 1995. Infections of the gastrointestinal tract. Raven Press, New York. [Google Scholar]
- 10. Sen A, Kobayashi N, Das S, Krishnan T, Bhattacharya SK, Naik TN. 2001. The evolution of human group B rotaviruses. Lancet 357:198–199. doi: 10.1016/S0140-6736(00)03596-0 [DOI] [PubMed] [Google Scholar]
- 11. Christensen ML. 1989. Human viral gastroenteritis. Clin Microbiol Rev 2:51–89. doi: 10.1128/CMR.2.1.51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Diller JR, Parrington HM, Patton JT, Ogden KM, López S. 2019. Rotavirus species B encodes a functional fusion-associated small transmembrane protein. J Virol 93:e00813-19. doi: 10.1128/JVI.00813-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Suzuki T, Kuga K, Miyazaki A, Tsunemitsu H. 2011. Genetic divergence and classification of non-structural protein 1 among porcine rotaviruses of species B. J Gen Virol 92:2922–2929. doi: 10.1099/vir.0.036426-0 [DOI] [PubMed] [Google Scholar]
- 14. Hoxie I, Dennehy JJ. 2020. Intragenic recombination influences rotavirus diversity and evolution. Virus Evol 6:vez059. doi: 10.1093/ve/vez059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jere KC, Mlera L, Page NA, van Dijk AA, O’Neill HG. 2011. Whole genome analysis of multiple rotavirus strains from a single stool specimen using sequence-independent amplification and 454 pyrosequencing reveals evidence of intergenotype genome segment recombination. Infect Genet Evol 11:2072–2082. doi: 10.1016/j.meegid.2011.09.023 [DOI] [PubMed] [Google Scholar]
- 16. Viral gastroenteritis. 1996. Springer Vienna, Vienna. Available from: http://link.springer.com/10.1007/978-3-7091-6553-9. Retrieved 23 Nov 2022. [Google Scholar]
- 17. Jayaram H, Estes MK, Prasad BVV. 2004. Emerging themes in rotavirus cell entry, genome organization, transcription and replication. Virus Res. 101:67–81. doi: 10.1016/j.virusres.2003.12.007 [DOI] [PubMed] [Google Scholar]
- 18. Pesavento JB, Crawford SE, Estes MK, Prasad BVV. 2006. Rotavirus proteins: structure and assembly. Curr Top Microbiol Immunol 309:189–219. doi: 10.1007/3-540-30773-7_7 [DOI] [PubMed] [Google Scholar]
- 19. 2006. Reoviruses: entry, assembly and morphogenesis: with 4 tables. Springer, Berlin Heidelberg. [Google Scholar]
- 20. Hu L, Chow D-C, Patton JT, Palzkill T, Estes MK, Prasad BVV. 2012. Crystallographic analysis of rotavirus NSP2-RNA complex reveals specific recognition of 5′ GG sequence for RTPase activity. J Virol 86:10547–10557. doi: 10.1128/JVI.01201-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kattoura MD, Clapp LL, Patton JT. 1992. The rotavirus nonstructural protein, NS35, possesses RNA-binding activity in vitro and in vivo . Virology 191:698–708. doi: 10.1016/0042-6822(92)90245-k [DOI] [PubMed] [Google Scholar]
- 22. Schuck P, Taraporewala Z, McPhie P, Patton JT. 2001. Rotavirus nonstructural protein NSP2 self-assembles into octamers that undergo ligand-induced conformational changes. J Biol Chem 276:9679–9687. doi: 10.1074/jbc.M009398200 [DOI] [PubMed] [Google Scholar]
- 23. Taraporewala Z, Chen D, Patton JT. 1999. Multimers formed by the rotavirus nonstructural protein NSP2 bind to RNA and have nucleoside triphosphatase activity. J Virol 73:9934–9943. doi: 10.1128/JVI.73.12.9934-9943.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Aponte C, Poncet D, Cohen J. 1996. Recovery and characterization of a replicase complex in rotavirus-infected cells by using a monoclonal antibody against NSP2. J Virol 70:985–991. doi: 10.1128/JVI.70.2.985-991.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gallegos CO, Patton JT. 1989. Characterization of rotavirus replication intermediates: a model for the assembly of single-shelled particles. Virology 172:616–627. doi: 10.1016/0042-6822(89)90204-3 [DOI] [PubMed] [Google Scholar]
- 26. Helmberger-Jones M, Patton JT. 1986. Characterization of subviral particles in cells infected with simian rotavirus SA11. Virology 155:655–665. doi: 10.1016/0042-6822(86)90225-4 [DOI] [PubMed] [Google Scholar]
- 27. Kumar M, Jayaram H, Vasquez-Del Carpio R, Jiang X, Taraporewala ZF, Jacobson RH, Patton JT, Prasad BVV. 2007. Crystallographic and biochemical analysis of rotavirus NSP2 with nucleotides reveals a nucleoside diphosphate kinase-like activity. J Virol 81:12272–12284. doi: 10.1128/JVI.00984-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Taraporewala ZF, Patton JT. 2001. Identification and characterization of the helix-destabilizing activity of rotavirus nonstructural protein NSP2. J Virol 75:4519–4527. doi: 10.1128/JVI.75.10.4519-4527.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Vasquez-Del Carpio R, Gonzalez-Nilo FD, Riadi G, Taraporewala ZF, Patton JT. 2006. Histidine triad-like motif of the rotavirus NSP2 octamer mediates both RTPase and NTPase activities. J Mol Biol 362:539–554. doi: 10.1016/j.jmb.2006.07.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Petrie BL, Greenberg HB, Graham DY, Estes MK. 1984. Ultrastructural localization of rotavirus antigens using colloidal gold. Virus Res 1:133–152. doi: 10.1016/0168-1702(84)90069-8 [DOI] [PubMed] [Google Scholar]
- 31. Fabbretti E, Afrikanova I, Vascotto F, Burrone OR. 1999. Two non-structural rotavirus proteins, NSP2 and NSP5, form viroplasm-like structures in vivo. J Gen Virol 80:333–339. doi: 10.1099/0022-1317-80-2-333 [DOI] [PubMed] [Google Scholar]
- 32. Criglar JM, Hu L, Crawford SE, Hyser JM, Broughman JR, Prasad BVV, Estes MK. 2014. A novel form of rotavirus NSP2 and phosphorylation-dependent NSP2-NSP5 interactions are associated with viroplasm assembly. J Virol 88:786–798. doi: 10.1128/JVI.03022-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Geiger F, Acker J, Papa G, Wang X, Arter WE, Saar KL, Erkamp NA, Qi R, Bravo JP, Strauss S, Krainer G, Burrone OR, Jungmann R, Knowles TP, Engelke H, Borodavka A. 2021. Liquid–liquid phase separation underpins the formation of replication factories in rotaviruses. EMBO J 40:e107711. doi: 10.15252/embj.2021107711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Papa G, Borodavka A, Desselberger U. 2021. Viroplasms: assembly and functions of rotavirus replication factories. Viruses 13:1349. doi: 10.3390/v13071349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Vende P, Tortorici MA, Taraporewala ZF, Patton JT. 2003. Rotavirus NSP2 interferes with the core lattice protein VP2 in initiation of minus-strand synthesis. Virology 313:261–273. doi: 10.1016/s0042-6822(03)00302-7 [DOI] [PubMed] [Google Scholar]
- 36. Borodavka A, Desselberger U, Patton JT. 2018. Genome packaging in multi-segmented dsRNA viruses: distinct mechanisms with similar outcomes. Curr Opin Virol 33:106–112. doi: 10.1016/j.coviro.2018.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Borodavka A, Dykeman EC, Schrimpf W, Lamb DC. 2017. Protein-mediated RNA folding governs sequence-specific interactions between rotavirus genome segments. Elife 6:e27453. doi: 10.7554/eLife.27453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bravo JPK, Bartnik K, Venditti L, Acker J, Gail EH, Colyer A, Davidovich C, Lamb DC, Tuma R, Calabrese AN, Borodavka A. 2021. Structural basis of rotavirus RNA chaperone displacement and RNA annealing. Proc Natl Acad Sci U S A 118:e2100198118. doi: 10.1073/pnas.2100198118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Jayaram H, Taraporewala Z, Patton JT, Prasad BVV. 2002. Rotavirus protein involved in genome replication and packaging exhibits a HIT-like fold. Nature 417:311–315. doi: 10.1038/417311a [DOI] [PubMed] [Google Scholar]
- 40. Taraporewala ZF, Jiang X, Vasquez-Del Carpio R, Jayaram H, Prasad BVV, Patton JT. 2006. Structure-function analysis of rotavirus NSP2 octamer by using a novel complementation system. J Virol 80:7984–7994. doi: 10.1128/JVI.00172-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, Tunyasuvunakool K, Bates R, Žídek A, Potapenko A, et al. 2021. Highly accurate protein structure prediction with AlphaFold. Nature 596:583–589. doi: 10.1038/s41586-021-03819-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Matthijnssens J, Otto PH, Ciarlet M, Desselberger U, Van Ranst M, Johne R. 2012. VP6-sequence-based cutoff values as a criterion for rotavirus species demarcation. Arch Virol 157:1177–1182. doi: 10.1007/s00705-012-1273-3 [DOI] [PubMed] [Google Scholar]
- 43. Ogden KM, Johne R, Patton JT. 2012. Rotavirus RNA polymerases resolve into two phylogenetically distinct classes that differ in their mechanism of template recognition. Virology 431:50–57. doi: 10.1016/j.virol.2012.05.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Jiang X, Jayaram H, Kumar M, Ludtke SJ, Estes MK, Prasad BVV. 2006. Cryoelectron microscopy structures of rotavirus NSP2-NSP5 and NSP2-RNA complexes: implications for genome replication. J Virol 80:10829–10835. doi: 10.1128/JVI.01347-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Diller JR, Parrington HM, Patton JT, Ogden KM, López S. 2019. Rotavirus species B encodes a functional fusion-associated small transmembrane protein. J Virol 93:e00813-19. doi: 10.1128/JVI.00813-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Matthijnssens J, Otto PH, Ciarlet M, Desselberger U, Van Ranst M, Johne R. 2012. VP6-sequence-based cutoff values as a criterion for rotavirus species demarcation. Arch Virol 157:1177–1182. doi: 10.1007/s00705-012-1273-3 [DOI] [PubMed] [Google Scholar]
- 47. Li W, Godzik A. 2006. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22:1658–1659. doi: 10.1093/bioinformatics/btl158 [DOI] [PubMed] [Google Scholar]
- 48. Letunic I, Bork P. 2021. Interactive tree of life (iTOL) V5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res 49:W293–W296. doi: 10.1093/nar/gkab301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zivanov J, Nakane T, Forsberg BO, Kimanius D, Hagen WJ, Lindahl E, Scheres SH. 2018. New tools for automated high-resolution cryo-EM structure determination in RELION-3. Elife 7:e42166. doi: 10.7554/eLife.42166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Punjani A, Rubinstein JL, Fleet DJ, Brubaker MA. 2017. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat Methods 14:290–296. doi: 10.1038/nmeth.4169 [DOI] [PubMed] [Google Scholar]
- 51. Zheng SQ, Palovcak E, Armache J-P, Verba KA, Cheng Y, Agard DA. 2017. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods 14:331–332. doi: 10.1038/nmeth.4193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60:2126–2132. doi: 10.1107/S0907444904019158 [DOI] [PubMed] [Google Scholar]
- 53. Liebschner D, Afonine PV, Baker ML, Bunkóczi G, Chen VB, Croll TI, Hintze B, Hung LW, Jain S, McCoy AJ, Moriarty NW, Oeffner RD, Poon BK, Prisant MG, Read RJ, Richardson JS, Richardson DC, Sammito MD, Sobolev OV, Stockwell DH, Terwilliger TC, Urzhumtsev AG, Videau LL, Williams CJ, Adams PD. 2019. Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr D Struct Biol 75:861–877. doi: 10.1107/S2059798319011471 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figures S1 and S2; Tables S1 and S2.
Data Availability Statement
The cryo-EM structure of rotavirus B NSP2 has been deposited in the Protein Data Bank (https://www.wwpdb.org/) under the accession code 8P00. All other relevant data are available from the authors.