ABSTRACT
There is growing interest in engineering Pseudomonas putida KT2440 as a microbial chassis for the conversion of renewable and waste-based feedstocks, and metabolic engineering of P. putida relies on the understanding of the functional relationships between genes. In this work, independent component analysis (ICA) was applied to a compendium of existing fitness data from randomly barcoded transposon insertion sequencing (RB-TnSeq) of P. putida KT2440 grown in 179 unique experimental conditions. ICA identified 84 independent groups of genes, which we call fModules (“functional modules”), where gene members displayed shared functional influence in a specific cellular process. This machine learning-based approach both successfully recapitulated previously characterized functional relationships and established hitherto unknown associations between genes. Selected gene members from fModules for hydroxycinnamate metabolism and stress resistance, acetyl coenzyme A assimilation, and nitrogen metabolism were validated with engineered mutants of P. putida. Additionally, functional gene clusters from ICA of RB-TnSeq data sets were compared with regulatory gene clusters from prior ICA of RNAseq data sets to draw connections between gene regulation and function. Because ICA profiles the functional role of several distinct gene networks simultaneously, it can reduce the time required to annotate gene function relative to manual curation of RB-TnSeq data sets.
IMPORTANCE
This study demonstrates a rapid, automated approach for elucidating functional modules within complex genetic networks. While Pseudomonas putida randomly barcoded transposon insertion sequencing data were used as a proof of concept, this approach is applicable to any organism with existing functional genomics data sets and may serve as a useful tool for many valuable applications, such as guiding metabolic engineering efforts in other microbes or understanding functional relationships between virulence-associated genes in pathogenic microbes. Furthermore, this work demonstrates that comparison of data obtained from independent component analysis of transcriptomics and gene fitness datasets can elucidate regulatory-functional relationships between genes, which may have utility in a variety of applications, such as metabolic modeling, strain engineering, or identification of antimicrobial drug targets.
KEYWORDS: transposon insertion sequencing, RB-TnSeq, independent component analysis, machine learning, Pseudomonas putida, aromatic catabolism, amino acid metabolism, functional genomics
INTRODUCTION
Pseudomonas putida KT2440 (hereafter, P. putida) has garnered much interest as a host for the valorization of heterogeneous chemical streams, such as biomass- and plastic-derived feedstocks, owing to its ability to adapt to several, often toxic, environments and funnel heterogeneous substrates toward a single product (1–9). However, a deep understanding of the metabolic and stress tolerance capabilities of P. putida is essential for its use as an industrial biocatalyst (10–15).
For all organisms, including P. putida, activities of multiple gene products must be coordinated to form complex functional networks that permit survival and growth within the environments experienced by the cell, and metabolic engineering efforts do not always account for the complex, inter-related nature of these gene networks (16). One approach to illuminate these relationships is transposon insertion sequencing (TnSeq), a powerful functional genomics tool that combines high-density transposon mutagenesis with next-generation sequencing to simultaneously characterize bacterial gene essentiality and fitness (17, 18). By screening a pooled transposon library against various growth conditions, a TnSeq practitioner can establish the relative essentiality of each gene across the conditions tested, thereby generating a genotype-phenotype link that can help to elucidate the function of each feature. A variation on the TnSeq method, randomly barcoded transposon insertion sequencing (RB-TnSeq), uses unique barcode sequences encoded within transposons to reduce the sequencing burden of traditional TnSeq approaches (19). To date, RB-TnSeq has aided in the elucidation of the catabolism of aromatic acids, alcohols, fatty acids, lysine, and various nitrogen sources in addition to probing gene involvement during ionic, aromatic, and aliphatic acid stress in P. putida (20–24).
Nonetheless, while RB-TnSeq can aid in the assignment of gene function, this technique is restricted to the range of conditions studied, and identification of functionally related gene groups is often assessed through manual curation of the data sets. In contrast to principle component analysis (PCA), which uses dimensionality reduction to compress multivariate information, independent component analysis (ICA) is an unsupervised, multivariate signal separation algorithm used to decompose mixed signals into their individual parts (25). This approach performs best among many algorithms to identify sets of co-expressed genes (26) and has been successfully applied to microarray and RNAseq transcriptomics data (27–32).
In this work, ICA was applied to gene fitness data obtained from a set of diverse, previously conducted RB-TnSeq experiments, enabling rapid deconvolution of the complex genetic network of P. putida into groups of functionally independent genes. These functional groups, named fModules (for “functional modules”), represent sets of co-functioning genes that show correlated fitness performance across all conditions in the RB-TnSeq data set. The function of several genes was then examined in vivo to validate fModule membership. These included genes involved in hydroxycinnamate metabolism and tolerance, nitrogen assimilation from amino acid substrates, and acetyl-coenzyme A (CoA) utilization. The functional clustering data obtained from ICA of RB-TnSeq data sets were also compared with prior gene regulatory clustering data to establish regulation-function relationships between sets of genes in P. putida (32).
RESULTS AND DISCUSSION
ICA of multivariate gene fitness data separates genes into fModules
The appropriateness of applying ICA to gene fitness data was assessed through review of the required assumptions for the approach. ICA assumes that (i) independent components are statistically independent and (ii) independent components have a non-Gaussian distribution. For the first criterion, the statistical independence of functional gene networks seems counterintuitive given that the metabolic network consists of sets of intersecting anabolic, catabolic, and energy transfer reactions (33). However, empirical results have shown that ICA estimation of meaningful components is robust against some violation of the independence assumption (34). In practice, this means that ICA allows for variable membership to multiple independent components (as with metabolic intersection) but requires that interactions among members of an independent component be stronger than interactions between components to be effective. Therefore, the modularity of functional processes, like those within the metabolic network satisfies the requirement for independence (33, 35). This is similar to the successful application of ICA to expression data based upon the modularity of regulatory networks, where large sets of genes are transcriptionally controlled by relatively independent sets of global regulators that, in some cases, can also control the expression of other global regulators (regulatory intersection) (30).
The second requirement for non-Gaussian distribution of the independent components is also satisfied by the modularity of physiological processes, where the fitness distribution of a mutant strain is driven to non-Gaussian behavior, based upon the functional role of the gene product in each growth condition. For example, if a gene plays a functional role in efflux of a toxic compound, fitness outcomes from disrupting that gene would be skewed negative during growth in the presence of the toxic compound but neutral in conditions where the toxin is absent. If several genes are required for growth in the presence of the toxin, then they will exhibit similar, non-Gaussian distributions in the data set and be grouped together into a single fModule.
With the applicability of ICA of gene fitness data established, a broad panel of publicly available RB-TnSeq data from P. putida was compiled into a set of gene fitness measurements, covering 4,732 of 5,564 protein-coding genes in P. putida (36) from 332 samples grown in 179 unique conditions (Fig. 1A; see “Data Availability”). Gene fitness values were derived from a variety of selection conditions, including growth on single carbon sources, growth on single nitrogen sources, metabolite and osmotic stress, and volumetric scales ranging from microtiter plates to 2-L bioreactors (20–23, 37, 38). This data set was then subjected to matrix decomposition by ICA to obtain individual groups of genes unified by shared functional influence upon specific cellular processes.
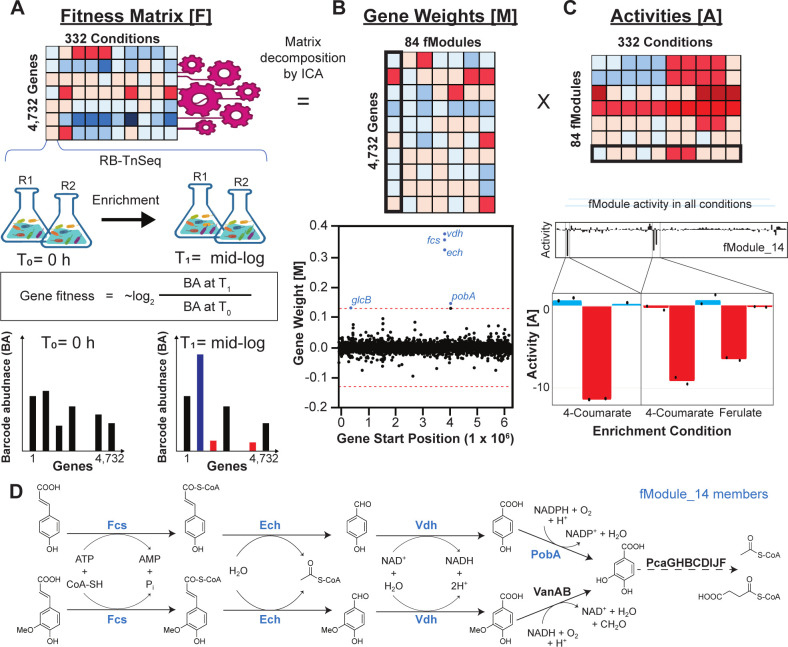
Fig 1.
Decomposition of RB-TnSeq multivariate gene fitness profiles using ICA. (A) Schematic of the general RB-TnSeq approach. A randomly barcoded transposon insertion library in P. putida is cultivated in a baseline (“time zero”, T0) condition and then enriched in a selective condition to mid-log (T1). Normalized barcode counts, which are indicative of specific mutant frequency in the population, are enumerated before and after enrichment to calculate a fitness value for each gene disruption. Genes with larger fitness magnitudes are expected to play a more important functional role for a given condition. In ICA, (A) fitness values, , from RB-TnSeq experiments were decomposed into a matrix of (B) gene weights, , and a matrix of (C) condition-specific activities, . In , genes with outlier weight values were grouped together into a single functional group, termed a “functional module”, which we abbreviate as “fModule”. In , the activity profile of an fModule characterizes the related biological functions of its member genes in each RB-TnSeq enrichment condition. Data from fModule_14 is provided as an example of ICA fModule gene weight and fModule activity outputs. (D) fModule_14 contains several genes involved in hydroxycinnamate metabolism. The negative activity of fModule_14 for conditions with 4-coumarate or ferulate as a sole carbon source in panel C is consistent with the essential role of its member gene products in catabolism of these substrates.
When ICA was applied to the P. putida RB-TnSeq data set, , gene fitness data were decomposed into a matrix of individual gene weight coefficients, , for 84 underlying fitness profiles (fModules) (Fig. 1B), and a matrix of condition-specific activities, , for each fModule (Fig. 1C). Most gene weight coefficients in an fModule were near zero, indicating that the underlying functional signal corresponding to each fModule affected a small number of significant genes. Genes with weight coefficients outside a predetermined threshold (see “Materials and Methods”) were removed, resulting in a set of outlier genes, which were identified as “member genes” for each fModule. Member genes with negative or positive gene weights displayed fitness profiles negatively or positively correlated with fModule activity, respectively. For example, if an fModule displayed negative activity for a particular condition, transposon-mediated disruption of its positively weighted member genes would be detrimental to growth in that condition. In this way, underlying fitness values were decomposed into a matrix of activities, , which reflected the concerted changes in fitness displayed by all member genes for each condition. In this analysis, only fModule_80 failed to contain a gene that satisfied the predetermined weight cutoff, resulting in an fModule with zero members. The 84 fModules contained 543 of the 4,732 unique genes used as input [11% of input, 9% of all P. putida open reading frames (ORFs)] with a median of 7 genes per fModule. In total, 83% of gene fitness variance within the RB-TnSeq data set was explained by the 84 fModules, and the proportion of total RB-TnSeq data set fitness variance explained by each fModule ranged from 0.14% to 8.47% (Table 1).
TABLE 1.
fModule annotation and explained variancea
| fModule | Description | # of genes | Explained variance | fModule | Description | # of genes | Explained variance |
|---|---|---|---|---|---|---|---|
| 1 | Fatty Acid Metabolism | 3 | 0.21% | 43 | Phenylalanine Catabolism | 11 | 0.18% |
| 2 | Nitrogen metabolism | 6 | 2.63% | 44 | Beta-alanine catabolism | 10 | 0.31% |
| 3 | RidA | 1 | 0.98% | 45 | Spermidine and propandiamine catabolism | 17 | 0.27% |
| 4 | Transcriptional regulation | 13 | 0.32% | 46 | Butyrolactam catabolism | 5 | 0.15% |
| 5 | Benzoate catabolism | 13 | 0.23% | 47 | Transcriptional regulation and cell:surface adhesion | 6 | 0.65% |
| 6 | Glycine, serine, and threonine metabolism | 9 | 0.14% | 48 | Acetic acid stress | 7 | 0.57% |
| 7 | Choline, betaine, and carnitine catabolism | 22 | 0.22% | 49 | 4-Hydroxyvalerate catabolism | 2 | 0.45% |
| 8 | Propanediamine catabolism | 22 | 0.61% | 50 | 1,4-Butanediol catabolism | 7 | 0.34% |
| 9 | Maintenance of lipid asymmetry (Mla) system | 14 | 0.50% | 51 | Phenolic acid stress | 19 | 0.24% |
| 10 | Bioreactor growth | 3 | 0.40% | 52 | Flagellar biosynthesis | 25 | 0.39% |
| 11 | Levulinic acid catabolism | 16 | 0.52% | 53 | Fatty acid metabolism | 11 | 0.88% |
| 12 | Biotin biosynthesis and pyruvate carboxylase | 9 | 1.24% | 54 | ParB | 1 | 0.29% |
| 13 | Molybdopterin biosynthesis and benzaldehyde tolerance | 20 | 0.42% | 55 | Lactic acid stress | 14 | 0.78% |
| 14 | 4-Coumarate and ferulate catabolism | 5 | 0.15% | 56 | Thymine degradation | 6 | 0.20% |
| 15 | Glutamate metabolism | 3 | 1.09% | 57 | Branched-chain Amino acid biosynthesis | 5 | 7.48% |
| 16 | Benzoate, 4-coumarate, and ferulate catabolism | 16 | 1.96% | 58 | Fatty acid catabolism | 4 | 0.64% |
| 17 | Pyrroloquinoline biosynthesis and short-chain alcohol catabolism | 20 | 0.41% | 59 | Thiamine diphosphate biosynthesis | 6 | 2.45% |
| 18 | Phenylacetate catabolism | 16 | 0.30% | 60 | 4-Aminobutyric acid catabolism | 7 | 0.18% |
| 19 | Phosphoenolpyruvate:sugar phosphotransferase system | 2 | 0.37% | 61 | Nitrogen metabolism | 6 | 0.20% |
| 20 | 4-Coumarate, ferulate, and catabolic intermediate stress tolerance | 6 | 0.34% | 62 | Galacturonic acid and glucuronic acid catabolism | 11 | 0.26% |
| 21 | Arginine metabolism | 14 | 3.12% | 63 | Methionine biosynthesis | 5 | 3.80% |
| 22 | Cobalamin biosynthesis and ethanolamine catabolism | 22 | 0.50% | 64 | NadC | 1 | 2.51% |
| 23 | Uncharacterized | 4 | 0.71% | 65 | Glutamate transport | 12 | 0.36% |
| 24 | Nitrogen stress sensor and glycogen biosynthesis | 3 | 0.43% | 66 | Proline metabolism | 2 | 2.05% |
| 25 | Bioreactor growth | 14 | 0.68% | 67 | Valine biosynthesis | 3 | 3.71% |
| 26 | Uncharacterized | 11 | 0.27% | 68 | Lysine metabolism | 18 | 0.60% |
| 27 | Membrane to surface adhesion | 5 | 0.96% | 69 | Histidine biosynthesis | 2 | 3.82% |
| 28 | Ferulate and catabolic intermediate catabolism | 9 | 0.32% | 70 | Butyrate catabolism | 9 | 0.34% |
| 29 | Transport of butylamine-containing compounds | 6 | 0.16% | 71 | HisA | 1 | 0.50% |
| 30 | Histidine biosynthesis | 11 | 6.89% | 72 | Lysine metabolism | 5 | 0.27% |
| 31 | Protocatechuate stress | 8 | 0.58% | 73 | L-Lysine catabolism | 3 | 0.24% |
| 32 | Nitrogen assimilation | 4 | 0.95% | 74 | Tryptophan biosynthesis | 9 | 8.47% |
| 33 | Biotin biosynthesis and isopentanol/isoprenol catabolism | 12 | 0.35% | 75 | Uncharacterized | 6 | 1.86% |
| 34 | Improved fitness factors | 7 | 1.91% | 76 | Sodium tolerance | 21 | 0.58% |
| 35 | Uncharacterized | 6 | 1.12% | 77 | Fatty acid metabolism | 2 | 0.15% |
| 36 | Possible Tween-20 catabolism | 12 | 0.24% | 78 | Possible butyrate metabolism | 7 | 0.18% |
| 37 | Lipopolysaccharide biosynthesis | 12 | 0.27% | 79 | Fatty ester metabolism | 7 | 0.32% |
| 38 | RuvC | 1 | 0.37% | 80 | No gene | 0 | 0.21% |
| 39 | PP_1410 | 1 | 0.18% | 81 | SerA | 1 | 1.32% |
| 40 | Acetate catabolism | 3 | 0.23% | 82 | ProI | 1 | 0.44% |
| 41 | Hexanoate and valerate catabolism | 10 | 0.20% | 83 | Beta-ketoadipate and levulinic acid stress | 4 | 0.22% |
| 42 | NADH:ubiquinone oxidoreductase I | 14 | 1.68% | 84 | Nitrate metabolism | 3 | 0.15% |
Functional annotations for each fModule, the number of genes in each fModule, and the proportion of the RB-TnSeq data set variance explained by each fModule. 543 unique genes are contained within the 84 fModules. Full details for each fModule are available at https://fmodules.github.io/putida.
The functional role of each fModule was initially classified using the Database for Annotation, Visualization, and Integrated Discovery (DAVID) database functional annotation clustering tool (39–41). The fModules were then manually curated using Kyoto Encyclopedia of Genes and Genomes (KEGG) orthology designations and fModule activity patterns to assign a putative functional annotation for all fModules (42). An example of this process is provided for fModule_14, which contained five genes (Fig. 1B). DAVID was unable to assign a functional clustering annotation, but fModule_14 activity was negative for growth enrichments containing 4-coumarate or ferulate as a sole carbon source (Fig. 1C), and GO terms revealed that 4/5 genes played known roles in hydroxycinnamate catabolism (Fig. 1D). These findings led to fModule_14 being provided with the functional annotation of “4-coumarate and ferulate catabolism” (File S1). Table 1 provides a list of all 84 fModules with putative functional annotations and the explained variance for each fModule. Most fModules could be assigned a functional annotation, but four fModules were left uncharacterized, due to ambiguity associated with member genes and corresponding fitness profiles (Table 1). Overall, over 57% of variance was explained by fModules annotated as amino acid metabolism, carbon metabolism, and nitrogen metabolism (Fig. S2). This is consistent with the fact that most conditions in the RB-TnSeq data set were designed to screen against various sole nitrogen sources, carbon sources, or amino acid dropout media compositions. Full details for each fModule, including gene weights, , and fModule activities, , are available at https://fmodules.github.io/putida.
Previous work applying ICA to E. coli transcriptomics data sets revealed that increasing the number and diversity of samples analyzed by ICA leads to the identification of a greater number of gene modules, and these modules are of higher quality (31). Since the application of ICA to gene fitness data sets does not mathematically differ from its application to transcriptomics data sets, the number and quality of identified fModules characterized are also expected to increase as sample size of the input data set increases. Therefore, future studies appending additional conditions to the data set may further refine P. putida fModule abundance and quality described in this work.
Also of note, the mariner transposon used in the KT2440 library does not contain an outward-facing promoter, making polar mutations even more likely in situations where a disrupted gene is encoded within an operon. This is a potential pitfall of transposon mutagenesis, in which affected genes may display strong fitness scores despite a lack of true involvement in a biological process. fModules may therefore include a small number of physiologically irrelevant genes, underscoring the need for careful consideration of gene context in any reverse-engineering campaign based on ICA assignments.
ICA successfully identifies several well-established metabolic pathways as fModules
Many fModules contained genes with well-characterized shared functional roles, underscoring the utility of ICA for grouping genes according to common function. For example, fModule_21 contained genes with described roles in L-arginine biosynthesis, transport, catabolism, and regulation (File S1) (43). The arginine biosynthesis genes displayed positive gene weights, while all other genes displayed negative gene weights. Accordingly, fModule_21 exhibited negative activity in all conditions lacking L-arginine supplementation (Fig. S3). In another example, fModule_68 and fModule_73, which exhibited negative activity when L-lysine, D-lysine (D-Lys), or catabolic intermediates were used as carbon or nitrogen sources (Fig. S4), contained genes with recently described roles in lysine regulation, transport, and catabolism (File S1) (20). In an example unrelated to amino acid metabolism, fModule_5 contained genes with characterized roles in benzoate catabolism (File S1) and displayed negative activity in conditions where benzoate was a sole carbon source (Fig. S5). Interestingly, negative weight gene members of fModule_5 were involved in the catabolism of protocatechuate toward the same product from benzoate catabolism, β-ketoadipate enol-lactone (44).
An additional, notable example of ICA successfully grouping enzymatically distinct but functionally related genes was demonstrated by fModule_28 (File S1). fModule_28 exhibited strongly negative activity for growth on the O-methylated aromatic compounds ferulate, vanillate, and vanillin (Fig. S6). Accordingly, this fModule contained vanAB, encoding the vanillate O-demethylase essential for growth in these conditions (45, 46). Formaldehyde is liberated as a product of O-demethylation by the native Rieske non-heme iron monooxygenase system, VanAB (46–48), and fModule_28 also includes frmA (encoding a glutathione-dependent formaldehyde dehydrogenase). Consistent with the glutathione and zinc cofactor requirements of FrmA, gshB (PP_4993, glutathione synthetase) and znuB1 (PP_0117, inner membrane pore of the zinc ABC transporter (49)) were also members of fModule_28. Disruption of znuB1 presented complexity due to its in-frame arrangement with znuC1 (PP_0118), but disruption of the transporter binding protein, znuA1 (PP_0120), led to poor growth with vanillate, relative to the wild-type, in the presence of zinc (Fig. S7).
Overall, the examples from arginine, lysine, benzoate, and catabolism of O-methylated aromatic substrates above demonstrate that ICA applied to fitness data can identify sets of genes with known functional relationships. Additionally, while the gene annotations of member genes within fModule_21, fModule_68 and fModule_73 together, fModule_5, and fModule_28 could be used to assign functional roles for these fModules in arginine metabolism, lysine catabolism, benzoate catabolism, and O-methylated aromatic catabolism, respectively, the activity profiles for these fModules were consistent with their functional annotations (Fig. S3 to S6), underscoring the power in using fModule activity profiles to assign function.
ICA predicts genes with complementary roles in otherwise well-characterized functional groups
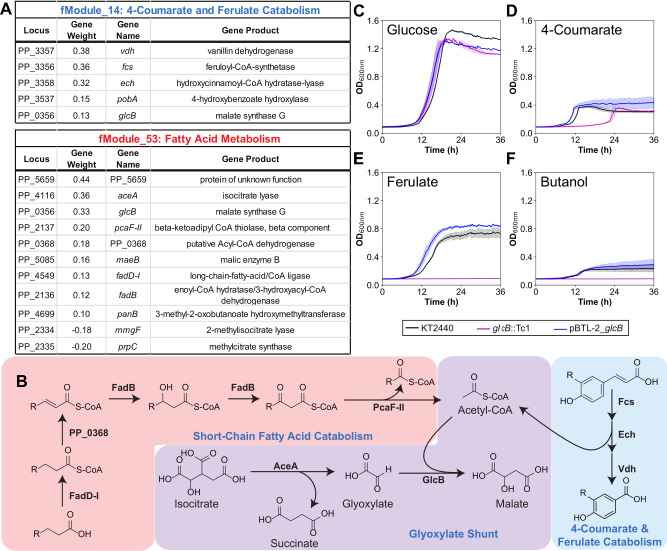
In several cases, the function of an fModule could be assigned based upon established roles for its constituent genes in a well-characterized pathway, but it also contained one or more genes with unintuitive roles in the pathway. One example of this observation is the inclusion of glcB (PP_0356), annotated as a malate synthase, in the well-characterized functional gene groups for fatty acid metabolism (fModule_53) and 4-coumarate and ferulate catabolism (fModule_14) (Fig. 2A; File S1). fModule_53 also included aceA, which together with glcB forms the glyoxylate shunt pathway to assimilate acetyl-CoA into the tricarboxylic acid (TCA) cycle (50). In conditions where catabolism of substrates results in the production of acetyl-CoA, such as with butanol and acetate, organisms are required to divert flux away from the oxidative steps of the TCA cycle and toward the anaplerotic steps of the glyoxylate shunt, conserving carbon for gluconeogenesis and subsequent biomass production (51). Accordingly, the abundance of glyoxylate shunt enzymes has been shown to increase in response to butanol (52), helping to explain the presence of glcB and aceA within fModule_53 for fatty acid metabolism.
Fig 2.
GlcB plays an important functional role within short-chain fatty acid metabolism and 4-coumarate and ferulate catabolism. (A) Membership and associated gene weights for fModule_14 and fModule_53. (B) Metabolic pathways for P. putida short-chain fatty acid metabolism, the glyoxylate shunt, and 4-coumarate (R=H) and ferulate (R=OCH3) catabolism. Growth profiles of a functional knockout of glcB (glcB::Tc1) and an overexpression mutant (pBTL-2_glcB) were compared with wild-type (KT2440) in M9 minimal medium supplemented with (C) 20 mM glucose, (D) 10 mM 4-coumarate, (E) 10 mM ferulate, and (F) 10 mM butanol. Error shading indicates the standard deviation from the mean for three biological replicates.
Upregulation of the glyoxylate shunt genes has also been demonstrated during benzoate degradation, likely as a means to assimilate the acetyl-CoA generated by PcaF (53). Acetyl-CoA is also a product of the Fcs:Ech:Vdh pathway for 4-coumarate and ferulate catabolism (Fig. 2B), and the genes for these enzymes are members of fModule_14, along with glcB (Fig. 2A). The membership of glcB in both fModule_53 and fModule_14 was bolstered by growth experiments, where the transposon disruption mutant of glcB grew no different from wild-type P. putida in minimal medium supplemented with glucose (Fig. 2C), but its growth was severely inhibited on 4-coumarate (Fig. 2D) and ferulate (Fig. 2E; Fig. S8) and entirely abolished on butanol (Fig. 2F). Expression of additional copies of glcB from a plasmid (strain ACB329; Table S1) led to modest growth improvements on 4-coumarate and ferulate but did not affect growth on glucose or butanol, relative to the wild-type (Fig. 2C through F, blue lines). The poor growth of the functional glcB knockout strain on 4-coumarate and ferulate indicates a critical role for the glyoxylate shunt in facilitating the anaplerotic assimilation of acetyl-CoA generated during 4-coumarate and ferulate catabolism. Importantly, since catabolism of 4-coumarate or ferulate produces both acetyl-CoA and succinyl-CoA products (53), the glyoxylate shunt is not essential to the same degree as with the catabolism of butanol and acetate, where acetyl-CoA is the sole product. Nonetheless, engineering strategies that leverage glcB may offer an underutilized, broadly applicable approach to improve growth outcomes for processes that rely upon the catabolism of feedstocks containing ferulate or 4-coumarate.
ICA aids in reannotation of gene function
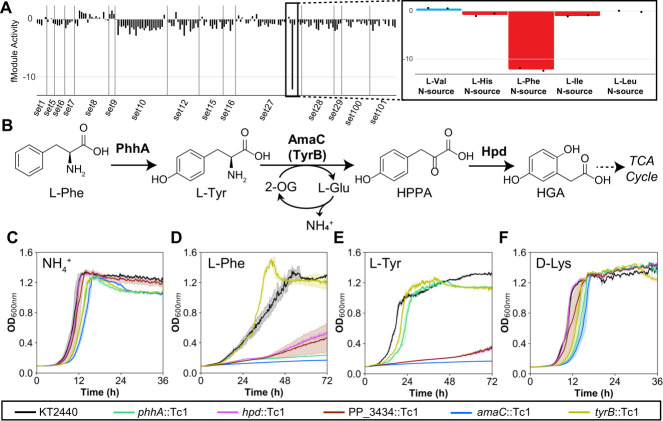
Occasionally, gene annotation seemed to be in conflict with a functional annotation derived from ICA. The membership of amaC (PP_3590, annotated as a D-lysine aminotransferase), in the phenylalanine catabolism functional module (fModule_43) serves as one example. fModule_43 exhibited a strongly negative activity during growth with L-phenylalanine (L-Phe) as the nitrogen source, so it was putatively annotated as a functional gene module for phenylalanine catabolism (Fig. 3A; Table 1). L-Phe catabolism proceeds through L-tyrosine (L-Tyr) in P. putida (Fig. 3B), but L-Tyr was not examined as a nitrogen source in the RB-TnSeq data set. To validate the role of genes included in fModule_43 (file S1) in L-Phe catabolism, single transposon disruption mutants of member genes phhA, amaC, and hpd were cultivated in M9 minimal medium with 20 mM glucose as a carbon source and either ammonium, L-glutamate (L-Glu), L-Phe, L-Tyr, or D-Lys as the sole nitrogen source (Fig. 3C through F). As expected, all mutants grew similarly to the wild-type strain when ammonium was used as the nitrogen source (Fig. 3C), and only a slight increase in lag time was observed for all mutants, except hpd when L-glutamate, a product of L-tyrosine aminotransferase, was used as the nitrogen source (Fig. S9). A substantial growth defect was observed when functional knockouts of phhA, hpd, and PP_3434 were grown with L-Phe as the sole source of nitrogen (Fig. 3D). The PP_3434 gene lies immediately upstream of hpd and presumably elicits a polar effect on the expression of hpd, but this was not explored further in the current work. While Hpd acts downstream of nitrogen assimilation within the L-Phe and L-Tyr catabolic pathway, it is possible that 4-hydroxyphenylpyruvic acid accumulation inhibits the activity of the upstream L-Tyr aminotransferase. This is bolstered by prior work, where an hpd knockout was used to increase the L-tyrosine concentration in P. putida (54). In accordance with the function of PhhA as a phenylalanine-4-hydroxylase, disruption of phhA did not substantially inhibit growth on L-Tyr (Fig. 3E). Surprisingly, disruption of tyrB (PP_1972, annotated as an L-Tyr aminotransferase) did not inhibit growth with L-Phe or L-Tyr, but disruption of amaC [PP_3590, sometimes called tyrB2 (55)] completely abrogated growth with L-Phe and L-Tyr (Fig. 3D and E) and did not substantially inhibit growth on D-Lys (Fig. 3F), inconsistent with its annotation as a D-lysine aminotransferase. Given these results and findings from previous work suggesting that AmaD, not AmaC, is the predominant D-lysine aminotransferase in P. putida (20, 55), we propose re-annotation of AmaC as an L-Tyr aminotransferase.
Fig 3.
ICA suggests reannotation of AmaC from a D-lysine aminotransferase to an L-tyrosine aminotransferase. (A) Activity profile of fModule_43 across all conditions in the RB-TnSeq data set, with the inset showing strongly negative activity during growth with L-Phe as a nitrogen source. (B) Proposed catabolic pathway for L-Phe and L-Tyr in P. putida. The growth of individual transposon disruption mutants was compared with wild-type (KT2440), using M9 minimal medium supplemented with 20 mM glucose as a carbon source and either (C) 5 mM ammonium, (D) 5 mM L-Phe, (E) 5 mM L-Tyr, or (F) 5 mM D-Lys as the sole nitrogen source. Error shading indicates the standard deviation from the mean of three biological replicates. 2-OG, 2-oxoglutarate; HPPA, 4-hydroxyphenylpyruvic acid; HGA, homogentisic acid.
fModules uncover previously uncharacterized functional relationships between genes
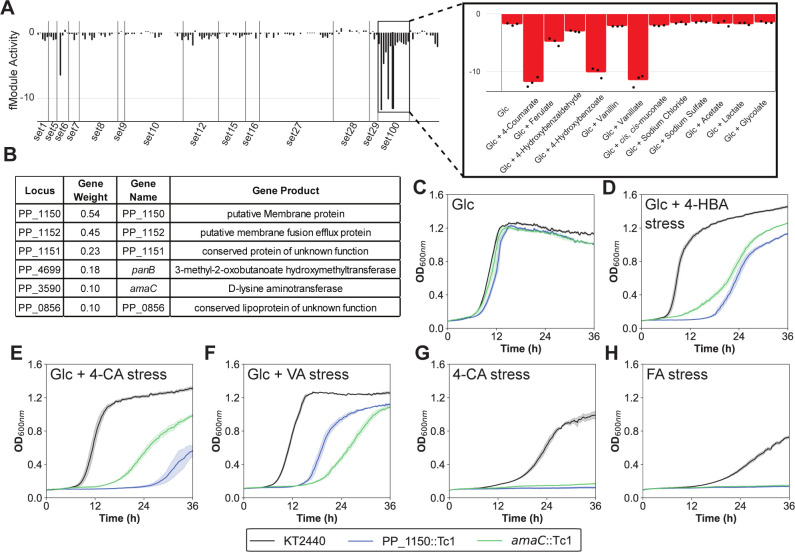
Activity and gene membership of fModules also helped define pathways for tolerance to hydroxycinnamate stress. The activity of fModule_20 was strongly negative during growth on glucose with stressful concentrations of hydroxycinnamic acids such as 4-coumarate, 4-hydroxybenzoate, and vanillate (Fig. 4A). This module contained only six genes: PP_1150–1152, amaC (PP_3590), panB (PP_4699), and PP_0856 (Fig. 4B). Transposon disruption mutants of these genes were utilized for growth assays to probe the function assigned by the fModule (Fig. 4C through H). PanB is involved in the biosynthesis of pantothenate, a precursor to CoA (56, 57); therefore, this mutant was unable to grow in M9 minimal medium without supplementation of pantothenate. PP_0856 was also excluded from analysis since no transposon disruption mutants were isolated for this gene. Nonetheless, growth assays with functional knockouts of PP_1150 and amaC effectively recapitulated the activity trends observed for fModule_20, where both mutants grew similarly to the wild-type strain with glucose as a sole carbon source (Fig. 4C), but both exhibited growth defects during growth on glucose with high concentrations of hydroxycinnamates (Fig. 4D through F) or during growth with high concentrations of 4-coumarate or ferulate as the sole carbon and energy source (Fig. 4G and H). PP_1150–1152 constitute a membrane protein complex, so these genes may be responsible for osmotic stress mitigation. Additionally, the operon is at least partially regulated by FleQ (32, 58), and previous reports have shown that overexpression of PP_1150–1152 enables enhanced growth on high concentrations of ferulate and 4-coumarate, relative to wild-type P. putida (22). The amaC gene, which was also included in fModule_43 for its role in L-Phe catabolism, has not been previously identified as a fitness contributor for hydroxycinnamate tolerance, so we engineered P. putida to overexpress amaC (strains ACB272 and ACB287; Table S3). Unfortunately, amaC overexpression failed to improve growth with 4-coumarate, ferulate, or protocatechuate, relative to the wild-type (Fig. S10), perhaps due to regulatory mechanisms or an as-yet misunderstood contribution of this gene to the function of fModule_20.
Fig 4.
Functional knockouts of genes in fModule_35 reflect its strongly negative activity during hydroxycinnamate stress. (A) Activity profile of fModule_20 across all conditions in the data set, with the inset demonstrating negative activity during hydroxycinnamate catabolism and stress. (B) Membership and associated gene weights for fModule_20. The growth of individual transposon disruption mutants of P. putida (PP_1150::Tc1 and amaC::Tc1) was compared with wild-type (KT2440), using M9 minimal medium supplemented (C) 20 mM glucose as the carbon source, (D) 20 mM glucose with 60 mM 4-hydroxybenzoate stress, (E) 20 mM glucose with 60 mM 4-coumarate stress, (F) 20 mM glucose with 60 mM vanillate stress, (G) 100 mM 4-coumarate as the carbon source, or (H) 160 mM ferulate as the carbon source. Error shading indicates the standard deviation from the mean of three biological replicates. Glc, glucose; 4-HBA, 4-hydroxybenzoate; 4-CA, 4-coumarate; VA, vanillate; FA, ferulate.
Inclusion of genes in multiple fModules reveals pathway integration
Notably, multiple genes were members of two fModules (70 genes), three fModules (22 genes), four fModules (nine genes), five fModules (four genes), and six fModules (two genes). Given the promiscuity of many proteins and the integrated nature of genetic networks, it is perhaps unsurprising to see several genes playing a role in multiple functionally distinct fModules. For example, cbrB (PP_4696) and cysB (PP_2327) were the two genes hat were each present in six different fModules. CbrB is a σ54 response regulator known to regulate central carbon metabolism and amino acid uptake (59, 60), while CysB is a LysR-type regulator that controls sulfate metabolism in P. putida (61). Consequently, both genes play far-reaching roles across metabolism, explaining their membership to several fModules. Overall, the ability for ICA to assign multiple functional groups to a gene better approximates the complex nature of biological systems and distinguishes this approach from other unsupervised machine learning clustering algorithms that cannot place genes into more than one group, such as k-means and agglomerative clustering (62, 63).
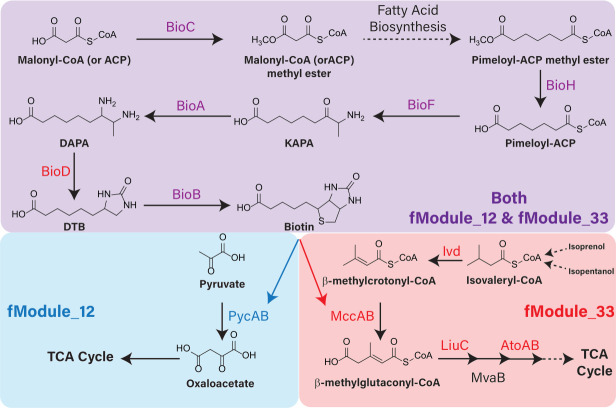
In six instances, cofactor biosynthesis genes were placed in the same fModule as genes for metabolic pathways requiring that cofactor. These are fModule_12 (biotin biosynthesis and pyruvate carboxylate) (64), fModule_13 (molybdopterin biosynthesis and benzaldehyde tolerance) (65), fModule_17 (pyrroloquinoline biosynthesis and short-chain alcohol catabolism) (23, 66), fModule_22 (cobalamin biosynthesis and ethanolamine catabolism) (67), fModule_28 (glutathione biosynthesis and vanillin catabolism/formaldehyde detoxification), and fModule_33 (biotin biosynthesis and isopentanol/isoprenol catabolism) (23).
fModule_28 is described above, but another notable example of cofactors being grouped with enzymes dependent on those cofactors involves bioBFHC and the independently transcribed bioA gene, which encode biotin biosynthesis enzymes (68) and are members to both fModule_12 and fModule_33 (Fig. 5; File S1). fModule_12 included genes encoding the two subunits of the biotin-dependent pyruvate carboxylase, PycAB (PP_5346–5347) (64), and its regulator (PP_5348). fModule_33 included the ivd:mccB:liuC:mccA operon, where MccA and MccB form a biotin-binding enzyme complex (69). The ivd:mccB:liuC:mccA operon and the two remaining members of fModule_33, atoAB, are critical for the catabolism of isopentanol and isoprenol, consistent with negative activity of this fModule when isopentanol or isoprenol were used as sole carbon sources (23).
Fig 5.
Biotin biosynthesis genes are members of both fModule_12 and fModule_33, in which each also contains genes encoding biotin-dependent carboxylases.
Inclusion of biotin biosynthetic pathway genes in the same fModules as those containing biotin-dependent carboxylase genes provides a powerful illustration of the physiological relationships predicted by ICA. Additionally, the presence of biotin biosynthesis genes in multiple fModules helps illustrate the potential of ICA for capturing the complex nature of biological systems, where genes and pathways can have several distinct functional relationships with other gene sets. Overall, ICA of RB-TnSeq fitness data may help identify uncharacterized links between metabolic processes and cofactor requirements.
Single-gene fModules often contain genes with unique, far-reaching physiological roles
Interestingly, eight fModules contained only a single gene (fModules 3, 38, 39, 54, 64, 71, 81, and 82; File S1). All the single-gene fModules, excluding fModule_39, displayed negative activity across most of the conditions tested. fModule_64 contained nadC (PP_0787), encoding a phosphoribosyltransferase that catalyzes the third step in NAD+ biosynthesis from L-aspartate (70). fModule_81, contained serA (PP_5155), which plays a critical role in serine biosynthesis and NAD+/NADH recycling (71). fModule_03 contained ridA (PP_5303), a conserved deaminase that controls accumulation of reactive and toxic enamine and imine intermediates generated by several metabolic processes (72). fModule_71 contained hisA (PP_0292), a gene with a role in histidine and purine biosynthesis (73). fModule_82 contained proI (PP_5095), which is involved in the final step of proline biosynthesis. Surprisingly, fModule_82 also displayed negative gene fitness in conditions containing supplemented proline, suggesting that proI may play a role beyond proline biosynthesis (Files S1 and S2). Interestingly, fModule_38 and fModule_54 contained ruvC (PP_1215) and parB (PP_001), respectively. These genes are involved in DNA processing and repair, where ParB is a chromosome-partitioning protein and RuvC is a crossover junction endodeoxyribonuclease (74, 75). Curiously, activities for fModules 38 and 54, while generally negative across all conditions, were positive for conditions where P. putida was grown in a bioreactor. Overall, most single-gene fModules contained genes with known far-reaching and pleiotropic metabolic roles. Therefore, identification of single-gene fModules during the application of ICA to TnSeq data sets from other organisms may be a useful tool for identifying important genes with several connections to distinct metabolic and physiological processes.
Comparing fModule data with iModulon data uncovers regulatory control of functional elements
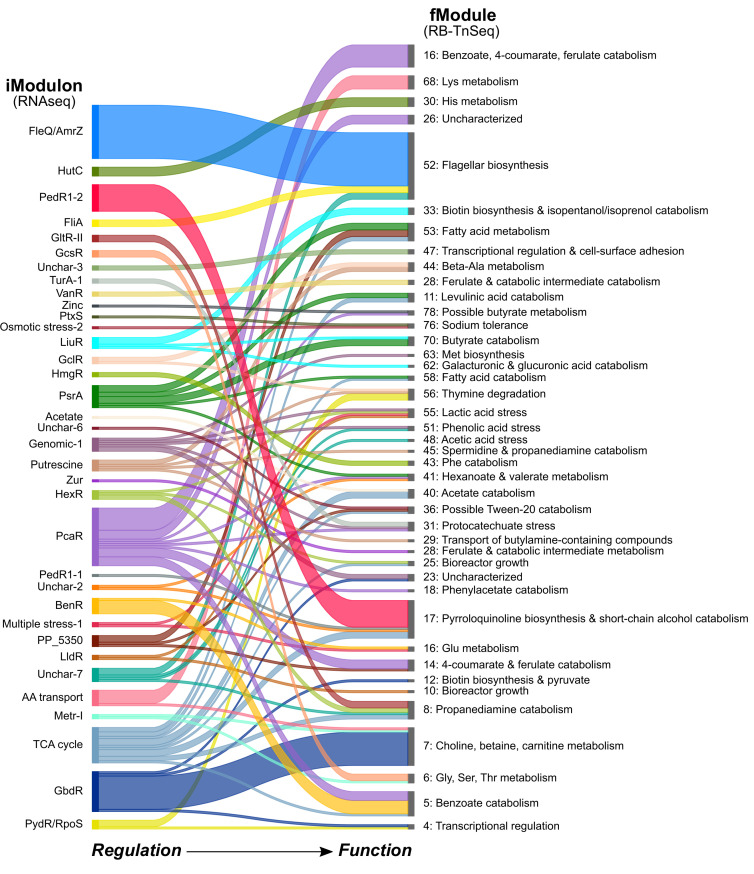
In a previous study, ICA was applied to transcriptomics data from P. putida for revealing co-regulated sets of genes, termed “iModulons” (32). The fModules described in this work are distinct from iModulons because fModules delineate groups of genes with shared function but not necessarily those with shared regulation. Nevertheless, synchronizing the expression of genes with shared physiological functions is critical to cell survival and proliferation (76). Therefore, the extent to which genes within a single fModule were colocalized within a single iModulon was explored (Fig. 6; File S2). In total, 176 genes that were grouped into an fModule were also found in one or more iModulons, and the extent to which groupings were conserved between the two analyses varied.
Fig 6.
ICA assigns genes to different groups, depending on the input data set. Some KT2440 genes with membership in fModules (ICA of RB-TnSeq data, this study) were also members of iModulons (ICA of transcriptomics data, a separate study [32]). Genes that were members of the same iModulon (i.e., co-regulated) were not necessarily members of the same fModule (i.e., co-functioning). The thickness of each line corresponds to the number of genes, and labels on left and right indicate iModulon and fModule annotations, respectively. Numerical data for this figure are provided in File S2. Code for the Sankey diagram plotting function was adapted from https://github.com/anazalea/pySankey/blob/master/pysankey/sankey.py and is provided in File S3.
In some cases, genes that were members of a single iModulon were also present within a single fModule. One example of this includes the relationship between the “FleQ/AmrZ” iModulon and fModule_52 (flagellar biosynthesis), where all gene members from the “FleQ/AmrZ” iModulon with membership to an fModule belong to fModule_52 (File S2). Another example includes the “HutC” iModulon and its strong relationship to fModule_30 (His metabolism). These instances exemplify the occurrence of specific transcriptional circuits, where a transcriptional regulator, such as FleQ or HutC, controls the expression of genes all involved in a specific physiological function, such as flagellar motility or histidine metabolism, respectively (58, 77).
In other cases, gene members of a single iModulon were dispersed across several distinct, but related, fModules. As an example, several genes within fModules relevant to catabolism of specific fatty acids were members of the iModulon for PsrA, a transcriptional repressor of the β-oxidation pathway for catabolism of all fatty acids in P. putida (78). The “TCA cycle” iModulon offered a variation on this theme, where member genes were dispersed across several catabolic fModules that are all expected to funnel carbon toward the TCA cycle. These cases are indicative of instances where a single global transcriptional circuit may coordinate expression of related, interconnected functions.
Finally, there were instances where fModules contained genes present in two or more iModulons. Examples include fModule_5 (benzoate catabolism) and fModule_55 (lactic acid stress). In the case of fModule_5, six genes involved in the catabolism of benzoate to β-ketoadipate were included as part of the “BenR” iModulon, while four genes involved in the parallel pathway for catabolism of protocatechuate toward β-ketoadipate were members of the “PcaR” iModulon (File S2). Coordinated expression of benzoate and protocatechuate catabolic genes has been observed previously in P. putida, and coordination of these peripheral pathways is believed to be important for hierarchical assimilation of related metabolites that share a common downstream pathway (44, 79). Generally, these are examples of integrated transcriptional circuits, where a common cellular function is subject to multiple points of transcriptional control.
In isolation, analysis of an fModule or iModulon data set can provide high-throughput functional or regulatory information, respectively. However, comparison of the two data sets can characterize whether functional gene sets are subject to regulatory control by specific or global transcriptional circuits and determine the extent to which these circuits are integrated. Furthermore, this comparative analysis illustrates that concerted changes in fitness are not necessarily driven by concerted changes in transcriptional regulation.
In some cases, transcriptomic data may inform the function of poorly annotated fModules and vice versa. For example, fModule_26 was annotated as “Uncharacterized,” but one of its member genes belongs to the “PcaR” iModulon (File S2), which primarily contains genes required for the catabolism of aromatic compounds. Similarly, iModulon “Unchar-3” contains two genes within the “Transcriptional regulation & cell-surface adhesion” fModule (fModule_46) (File S2). Together, these data suggest that fModule_26 and the iModulon “Unchar-3”may play roles in aromatic catabolism and cell-to-surface adhesion, respectively.
Of the appropriately sized RB-TnSeq data sets that exist for other organisms, a companion iModulon data set already exists for Escherichia coli BW25113 (30), opening the possibility of comparing future fModule data from E. coli BW25113 with existing iModulon data for characterizing regulatory control of functional elements in this organism. However, for both P. putida and E. coli BW25113, the publicly available RB-TnSeq and transcriptomics data were not collected following growth in a shared set of conditions. While this did not preclude the extraction of meaningful regulatory information from the P. putida data sets, future studies, where gene fitness and expression data are obtained from a shared set of growth conditions, may yield stronger relationships between functional sets of genes than those achieved here.
Conclusions
This work describes the application of ICA to a P. putida RB-TnSeq data set for elucidating functional relationships between genes. In total, this approach successfully (i) identified well-characterized functional relationships between sets of genes, (ii) uncovered new gene members with overlooked roles in otherwise well-characterized functional gene groups, (iii) was used to reannotate one gene’s functional role, (iv) uncovered otherwise overlooked functional gene relationships, (v) revealed instances of pathway integration, (vi) highlighted genes with far-reaching pleotropic functional roles, and (vii) was analyzed alongside iModulon data to uncover the transcriptional control mechanisms governing expression of various functional gene sets.
Overall, the technique presented in this work represents a rapid and automated approach to characterize functional modules within complex genetic networks and elucidate how an organism coordinates the expression of these modules. The approach also represents an opportunity to extract newfound value from previously generated data sets. ICA has successfully been applied to transcriptomics data sets containing as few as 23 unique conditions (80). Publicly available RB-TnSeq data sets containing at least 23 unique screening conditions exist for 37 additional organisms on the Fitness Browser (https://fit.genomics.lbl.gov/cgi-bin/myFrontPage.cgi), so it is possible that the use of these RB-TnSeq compendia might also reveal informative functional relationships between genes. Furthermore, the approach is applicable to functional genomics data sets from any organism, regardless of whether they were obtained specifically through the RB-TnSeq method. This opens ICA to a wide range of industrially, medically, or environmentally important organisms, where its ability to simultaneously identify several functional groups of genes can expedite the annotation of relatively uncharacterized organisms, as compared with classical genetic and biochemical approaches or stepwise analysis of individual data sets.
MATERIALS AND METHODS
Generation of an RB-TnSeq fitness data compendium for P. putida KT2440
An initial RB-TnSeq fitness data compendium was generated by collecting 332 publicly available P. putida RB-TnSeq data sets collected between 2017 and 2022, consisting of a mixture of duplicate or triplicate samples spanning 183 unique growth conditions. The Fitness Browser (https://fit.genomics.lbl.gov/cgi-bin/myFrontPage.cgi) was used to obtain data for 254/332 of the samples, and the remaining data were generated by the Beckham group, available through the NCBI Sequence Read Archive (SRA) with accession numbers PRJNA809672, PRJNA856070, and PRJNA1011287 (22, 38). In instances where gene fitness data for a particular gene did not exist across all 332 data sets, the gene was eliminated from analysis. Biological and technical replicates present in the data were not averaged or normalized in any other way prior to analysis. This resulted in a final data set, where 4,732/5,564 protein-coding genes from P. putida contained fitness data for the 332 samples (36). Unlike previous ICA experiments using RNAseq data (32), fitness values were not normalized by batch, since each fitness measurement was already normalized by transposon insertion counts in the baseline (“time zero”) condition. Gene fitness values, associated statistics, and metadata for each sample are available at https://github.com/beckham-lab/fModule.
ICA of the RB-TnSeq data set
ICA was performed for the final compendium containing fitness values for 4,732 genes profiled across 332 samples, using the FastICA algorithm of the scikit-learn package (v0.23.2), as described previously (30). This algorithm was executed 100 times with random seeds and a convergence tolerance of 10−7. The resulting independent components (ICs) were clustered and compared using DBSCAN (81) with an epsilon of 0.1 and minimum cluster size of 50, to identify robust ICs. To account for the occurrence identical ICs with opposite signs, a distance metric (d) was used to compute the distance matrix as follows:
where ρx,y is the Pearson correlation between components x and y. The final robust ICs were then defined as the centroids of the cluster.
Since the number of selected dimensions during ICA can alter the results of the analysis (82), the optimal dimensionality was determined by comparing the number of ICs with single genes to the number of ICs correlated (Pearson R > 0.7) with ICs in the largest dimension, using increasing dimension from 10 to 180 and a step size of 10. The optimal dimension was chosen to be 130, in which the final number of ICs (fModules) was greater than the number of multi-gene components, while minimizing the number of single-gene ICs (Fig. S1).
Using the optimal dimensionality of 130, the member genes for each fModule were determined by iteratively removing genes with the largest gene weight size in the fModule and computing the D’Agostino K2 test statistic (83) for the remaining genes. All genes that were removed prior to the test statistic dropping below a cutoff value were deemed member genes of the fModule. The threshold was determined as the value for K2, where non-removed genes were sufficiently normally distributed around 0, as described previously (30).
Characterization of fModules
Functional clustering annotations for each fModule were initially explored using the DAVID 2021 functional annotation clustering tool, with P. putida KT2440 set as the analysis background and classification stringency set to the lowest setting, keeping all other settings at default. The gene membership list for each fModule was used as input, and the DAVID functional annotation output is provided in File S1. These annotations were refined by KEGG and Cluster of Orthologous Groups (COG) information obtained using the EggNOG-mapper (84) together with fModule activity patterns across all test conditions to manually assign putative functional annotations for all fModules ( File S1). Known transcription factor associations and any iModulon membership(s) for each gene were taken from previous assignments in the study by Lim et al. (32).
The interactive P. putida fModuleDB page was generated by using the imodulondb_export function in the Pymodulon package, adapted from the use for the iModulonDB (85).
Bacterial strains, plasmids, and growth conditions
Plasmids used in this study are described in Table S1, primers are listed in Table S2, bacterial strains are described in Table S3, and synthetic DNA sequences are listed in Table S4. PCR reactions were performed with Q5 High-Fidelity 2X Master Mix (New England Biolabs), and the pACB131 plasmid was assembled by the method of Gibson using NEBuilder HiFi DNA Assembly Master Mix (New England Biolabs). Plasmid-bearing E. coli strains were grown at 37°C and 225 rpm. P. putida and mutants were maintained in lysogeny broth (LB) medium at 30°C unless otherwise indicated. For overexpression of PP_3590 (amaC) in P. putida strains ACB272 and ACB287, overnight cultures of the P. putida KT2440 wild-type were electroporated with 500 ng of pACB127 or pACB131, respectively, according to an established method (86). Plasmids contained 1,000-bp homology arms on either side of the Ptac:PP_3590 construct, which enabled recombination at the desired loci in the P. putida genome. Recombination was enabled with a previously established protocol in which transformants were selected twice on LB agar with 50 mg/L kanamycin (Km) and counter-selected twice on YT agar with 25% sucrose (87). The same procedure was used for knockout of PP_0120 (znuA1) in P. putida strain KDD007. For plasmid-based overexpression of glcB in P. putida strain ACB329, an overnight culture of the P. putida KT2440 wild-type was electroporated with 100 ng of pACB145, and plasmid-bearing mutants were selected on LB agar supplemented with 50 mg/L Km. All subsequent cultivations of ACB329 were performed in media supplemented with 50 mg/L Km.
Isolation of P. putida strains from an individually arrayed insertion mutant library
All RB-TnSeq data in the ICA data set were generated with a previously described, randomly barcoded transposon mutant library in P. putida KT2440 (Putida_ML5) (88). This pooled library was individually arrayed, and barcode assignments were determined for each transposon mutant as previously described (89). Individual transposon insertion mutants (Table S3) were withdrawn from the arrayed library by scraping a small amount of relevant glycerol stock into a round-bottom tube filled with 3 mL of LB medium with 50 mg/L Km. Each mutant culture was grown overnight at 30°C and 225 rpm, and permanent stocks were generated by combining overnight culture with sterile glycerol to 20% (vol/vol). Additionally, 1 µL of overnight culture was used as a template for a PCR reaction to verify the barcode sequence of each mutant. Each PCR reaction contained 12.5 µL of Q5 High-Fidelity 2× Master Mix (New England Biolabs), 1.25 µL each of the previously described BarSeq_P1 and BarSeq_P2 primers (Table S2), 0.5 µL of dimethyl sulfoxide (Sigma-Aldrich), 1 µL of overnight culture, and water to 25 µL. Thermal cycles were as follows: (i) 98°C for 4 min; (ii) 25 cycles of 98°C for 30 s, 55°C for 30 s, and 72°C for 30 s; and (iii) 72°C for 5 min. PCR products were Sanger sequenced with oACB441 (Table S2) to verify the barcode sequence of each mutant.
Growth analysis of the P. putida wild-type and mutants. Growth media were prepared by mixing equal volumes of 2× M9 medium and 2× carbon source solution to achieve a final concentration of 1× M9 medium (6.78 g/L Na2HPO4, 3 g/L KH2PO4, 0.5 g/L NaCl, 1 g/L NH4Cl, 2 mM MgSO4, 100 µM CaCl2, and 18 µM FeSO4) with the desired final concentration of each carbon source. For zinc experiments with strain KDD007, 100 nM ZnSO4 was added to 1× M9 medium. Carbon sources were glucose, 4-hydroxybenzoate, 4-coumarate, ferulate, vanillate, protocatechuate, vanillin, benzoate, and butanol. Where indicated, M9 medium omitted NH4Cl and instead utilized an alternative nitrogen source: 5 mM of L-Glu, L-Phe, L-Tyr, or D-Lys. Aromatic compounds were titrated with base (4 M NaOH) to solubilize the compound in aqueous solution prior to sterile filtration and addition to media. Amino acid nitrogen sources were titrated with acid (1 M H2SO4, for L-Tyr) or base (4 M NaOH, for D-Lys and L-Phe) to solubilize the compound in aqueous solution prior to sterile filtration and addition to media. The pH of solubilized L-Tyr was 3.8, but upon addition of this solution to M9 medium, the pH remained neutral (pH = 7.1), as with all other carbon and nitrogen sources. No precipitation of carbon or nitrogen sources was observed during growth. All chemicals for media preparation were obtained from Sigma-Aldrich, except for protocatechuate (Acros Organics) and D-Lys (Ambeed Inc.).
To assess the growth of the P. putida wild-type and all mutants, besides ΔznuA1, biological triplicate cultures of each strain were inoculated from single colonies into 4 mL of LB medium in round-bottom tubes and incubated overnight at 30°C and 225 rpm. For each condition, 2 µL of overnight culture was directly inoculated into 200 µL of medium (1:100 dilution) in a Honeycomb plate (Growth Curves Ltd.). Plates were incubated at 30°C and maximum shaking speed in a BioscreenC Pro instrument (Growth Curves Ltd.), and the optical density at 600 nm (OD600) was measured every 15 min.
For growth experiments with zinc added to the medium, biological triplicate cultures of the P. putida wild type and KDD007 were inoculated from single colonies into 4 mL of LB medium in round-bottom tubes and incubated overnight at 30°C and 225 rpm. The next day, overnight cultures were used to inoculate secondary seed cultures in 5 mL M9 medium + 30 mM glucose. Seed cultures were grown to mid-log phase (~4 h at 30°C and 225 rpm), and then, each culture was washed twice in 1× M9 salts and diluted to an OD600 of 3. Next, 10 µL of each cell suspension was directly inoculated into wells of a Nunc Edge 2.0 96-well plate (Thermo Scientific) containing 200 µL of M9 medium with 30 mM glucose or 20–80 mM vanillate as the carbon source. In half of the samples, media were amended to include 100 nM ZnSO4. Plates were incubated at 30°C and 500 rpm in a LogPhase 600 instrument (Agilent), and the OD600 was measured every 20 min. Background correction for each sample was performed by subtracting the average optical density of the appropriate negative controls.
ACKNOWLEDGMENTS
This work was partially authored by the Alliance for Sustainable Energy, LLC, the manager, and operator of the National Renewable Energy Laboratory for the U.S. Department of Energy (DOE), under Contract No. DE-AC36-08GO28308. A.J.B., A.C.B., Z.A.K., and G.T.B. were funded by The Center for Bioenergy Innovation, a U.S. DOE Bioenergy Research Center supported by the Office of Biological and Environmental Research in the DOE Office of Science. Work from H.G.L., K.R., and B.O.P. was conducted as part of the Joint BioEnergy Institute, supported by the Office of Science, Office of Biological and Environmental Research, of the U.S. DOE under Contract No. DE-AC02-05CH11231. Funding for A.C.B., K.D.D., T.L.H., and G.T.B. was partially provided by the U.S. Department of Energy Office of Energy Efficiency and Renewable Energy Bioenergy Technologies office (BETO) for the Agile BioFoundry. The views expressed herein do not necessarily represent the views of the DOE or the U.S. Government.
The authors thank Rex R. Malmstrom and Adam M. Deutschbauer for the donation of the individually arrayed P. putida transposon insertion mutant library and Kristian M. Eschenburg for discussions of statistical analysis.
A.J.B. did the conceptualization, data curation, data analysis and visualization, design of experiments, and writing – original draft, review, and editing. A.C.B. did data curation, data analysis and visualization, design and performance of experiments, and writing – original draft, review, and editing. H.G.L. did data curation, data analysis and visualization, software development, and writing – review and editing. K.R. did data analysis and software development. K.D.D. did plasmid and strain construction and performance of experiments. Z.A.K. did plasmid and strain construction and performance of experiments. T.L.H. did plasmid and strain construction and performance of experiments. B.O.P. did project administration, supervision, funding acquisition, and writing – review and editing. G.T.B. did project administration, supervision, funding acquisition, and writing – review and editing.
Contributor Information
Bernhard O. Palsson, Email: palsson@ucsd.edu.
Gregg T. Beckham, Email: gregg.beckham@nrel.gov.
Steven J. Hallam, University of British Columbia, Vancouver, British Columbia, Canada
DATA AVAILABILITY
Sequence data from RB-TnSeq data sets 100 and 101 are available at the NCBI SRA, with accession numbers PRJNA809672, PRJNA856070, and PRJNA1011287 (22, 38). Perl and Python scripts used for analysis of data sets 100 and 101 are accessible from https://github.com/beckham-lab/RB-TnSeq.git. All remaining fitness data are available at https://fit.genomics.lbl.gov/. Source code for fModule analysis and figures may be found at https://github.com/fModules/putida-code, which contains the Jupyter notebook files used for analysis as well as raw input and output data. The interactive website, containing activity and gene information for each fModule, can be found at: https://fmodules.github.io/putida. Gene fitness values, associated statistics, and metadata for each sample are available at https://github.com/beckham-lab/fModule. Python code for the Sankey diagram plotting function was adapted from https://github.com/anazalea/pySankey/blob/master/pysankey/sankey.py and is provided in File S3.
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/msystems.00942-23.
Functional annotation clustering for fModules.
Gene names and weights in fModules.
Code for generating Sankey diagram in Figure 6.
Figures S1-S10; Tables S1-S4.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Linger JG, Vardon DR, Guarnieri MT, Karp EM, Hunsinger GB, Franden MA, Johnson CW, Chupka G, Strathmann TJ, Pienkos PT, Beckham GT. 2014. Lignin valorization through integrated biological funneling and chemical catalysis. Proc Natl Acad Sci USA 111:12013–12018. doi: 10.1073/pnas.1410657111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Francois JM, Alkim C, Morin N. 2020. Engineering microbial pathways for production of bio-based chemicals from lignocellulosic sugars: current status and perspectives. Biotechnol Biofuels 13:118. doi: 10.1186/s13068-020-01744-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mezzina MP, Manoli MT, Prieto MA, Nikel PI. 2021. Engineering native and synthetic pathways in Pseudomonas putida for the production of tailored polyhydroxyalkanoates. Biotechnol J 16:e2000165. doi: 10.1002/biot.202000165 [DOI] [PubMed] [Google Scholar]
- 4. Werner AZ, Clare R, Mand TD, Pardo I, Ramirez KJ, Haugen SJ, Bratti F, Dexter GN, Elmore JR, Huenemann JD, Peabody GL, Johnson CW, Rorrer NA, Salvachúa D, Guss AM, Beckham GT. 2021. Tandem chemical deconstruction and biological upcycling of poly(ethylene terephthalate) to β-ketoadipic acid by Pseudomonas putida KT2440. Metab Eng 67:250–261. doi: 10.1016/j.ymben.2021.07.005 [DOI] [PubMed] [Google Scholar]
- 5. Kohlstedt M, Weimer A, Weiland F, Stolzenberger J, Selzer M, Sanz M, Kramps L, Wittmann C. 2022. Biobased PET from lignin using an engineered cis,cis-muconate-producing Pseudomonas putida strain with superior robustness, energy and redox properties. Metab Eng 72:337–352. doi: 10.1016/j.ymben.2022.05.001 [DOI] [PubMed] [Google Scholar]
- 6. Ling C, Peabody GL, Salvachúa D, Kim Y-M, Kneucker CM, Calvey CH, Monninger MA, Munoz NM, Poirier BC, Ramirez KJ, St. John PC, Woodworth SP, Magnuson JK, Burnum-Johnson KE, Guss AM, Johnson CW, Beckham GT. 2022. Muconic acid production from glucose and xylose in Pseudomonas putida via evolution and metabolic engineering. Nat Commun 13:4925. doi: 10.1038/s41467-022-32296-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sullivan KP, Werner AZ, Ramirez KJ, Ellis LD, Bussard JR, Black BA, Brandner DG, Bratti F, Buss BL, Dong X, Haugen SJ, Ingraham MA, Konev MO, Michener WE, Miscall J, Pardo I, Woodworth SP, Guss AM, Román-Leshkov Y, Stahl SS, Beckham GT. 2022. Mixed plastics waste valorization through tandem chemical oxidation and biological funneling. Science 378:207–211. doi: 10.1126/science.abo4626 [DOI] [PubMed] [Google Scholar]
- 8. Borchert AJ, Wilson AN, Michener WE, Roback J, Henson WR, Ramirez KJ, Beckham GT. 2023. Biological conversion of cyclic ketones from catalytic fast pyrolysis with Pseudomonas putida KT2440. Green Chem 25:3278–3291. doi: 10.1039/D3GC00084B [DOI] [Google Scholar]
- 9. Bujdoš D, Popelářová B, Volke DC, Nikel PI, Sonnenschein N, Dvořák P. 2023. Engineering of Pseudomonas putida for accelerated co-utilization of glucose and cellobiose yields aerobic overproduction of pyruvate explained by an upgraded metabolic model. Metab Eng 75:29–46. doi: 10.1016/j.ymben.2022.10.011 [DOI] [PubMed] [Google Scholar]
- 10. Beckham GT, Johnson CW, Karp EM, Salvachúa D, Vardon DR. 2016. Opportunities and challenges in biological lignin valorization. Curr Opin Biotechnol 42:40–53. doi: 10.1016/j.copbio.2016.02.030 [DOI] [PubMed] [Google Scholar]
- 11. Kim J, Salvador M, Saunders E, González J, Avignone-Rossa C, Jiménez JI. 2016. Properties of alternative microbial hosts used in synthetic biology: towards the design of a modular chassis. Essays Biochem 60:303–313. doi: 10.1042/EBC20160015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Calero P, Jensen SI, Bojanovič K, Lennen RM, Koza A, Nielsen AT. 2018. Genome-wide identification of tolerance mechanisms toward p-coumaric acid in Pseudomonas putida. Biotechnol Bioeng 115:762–774. doi: 10.1002/bit.26495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nikel PI, de Lorenzo V. 2018. Pseudomonas putida as a functional chassis for industrial biocatalysis: from native biochemistry to trans-metabolism. Metab Eng 50:142–155. doi: 10.1016/j.ymben.2018.05.005 [DOI] [PubMed] [Google Scholar]
- 14. Weimer A, Kohlstedt M, Volke DC, Nikel PI, Wittmann C. 2020. Industrial biotechnology of Pseudomonas putida: advances and prospects. Appl Microbiol Biotechnol 104:7745–7766. doi: 10.1007/s00253-020-10811-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Borchert AJ, Henson WR, Beckham GT. 2022. Challenges and opportunities in biological funneling of heterogeneous and toxic substrates beyond lignin. Curr Opin Biotechnol 73:1–13. doi: 10.1016/j.copbio.2021.06.007 [DOI] [PubMed] [Google Scholar]
- 16. Hartline CJ, Schmitz AC, Han Y, Zhang F. 2021. Dynamic control in metabolic engineering: theories, tools, and applications. Metab Eng 63:126–140. doi: 10.1016/j.ymben.2020.08.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. van Opijnen T, Camilli A. 2013. Transposon insertion sequencing: a new tool for systems-level analysis of microorganisms. Nat Rev Microbiol 11:435–442. doi: 10.1038/nrmicro3033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cain AK, Barquist L, Goodman AL, Paulsen IT, Parkhill J, van Opijnen T. 2020. A decade of advances in transposon-insertion sequencing. Nat Rev Genet 21:526–540. doi: 10.1038/s41576-020-0244-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wetmore KM, Price MN, Waters RJ, Lamson JS, He J, Hoover CA, Blow MJ, Bristow J, Butland G, Arkin AP, Deutschbauer A. 2015. Rapid quantification of mutant fitness in diverse bacteria by sequencing randomly bar-coded transposons. mBio 6:e00306-15. doi: 10.1128/mBio.00306-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Thompson MG, Blake-Hedges JM, Cruz-Morales P, Barajas JF, Curran SC, Eiben CB, Harris NC, Benites VT, Gin JW, Sharpless WA, Twigg FF, Skyrud W, Krishna RN, Pereira JH, Baidoo EEK, Petzold CJ, Adams PD, Arkin AP, Deutschbauer AM, Keasling JD, Lee SY. 2019. Massively parallel fitness profiling reveals multiple novel enzymes in Pseudomonas putida lysine metabolism. mBio 10:e02577-18. doi: 10.1128/mBio.02577-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Incha MR, Thompson MG, Blake-Hedges JM, Liu Y, Pearson AN, Schmidt M, Gin JW, Petzold CJ, Deutschbauer AM, Keasling JD. 2020. Leveraging host metabolism for bisdemethoxycurcumin production in Pseudomonas putida. Metab Eng Commun 10:e00119. doi: 10.1016/j.mec.2019.e00119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Borchert AJ, Bleem A, Beckham GT. 2023. RB-TnSeq identifies genetic targets for improved tolerance of Pseudomonas putida towards compounds relevant to lignin conversion. Metab Eng 77:208–218. doi: 10.1016/j.ymben.2023.04.007 [DOI] [PubMed] [Google Scholar]
- 23. Thompson MG, Incha MR, Pearson AN, Schmidt M, Sharpless WA, Eiben CB, Cruz-Morales P, Blake-Hedges JM, Liu Y, Adams CA, Haushalter RW, Krishna RN, Lichtner P, Blank LM, Mukhopadhyay A, Deutschbauer AM, Shih PM, Keasling JD, Zhou N-Y. 2020. Fatty acid and alcohol metabolism in Pseudomonas putida: functional analysis using random barcode transposon sequencing. Appl Environ Microbiol 86:e01665-20. doi: 10.1128/AEM.01665-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Schmidt M, Pearson AN, Incha MR, Thompson MG, Baidoo EEK, Kakumanu R, Mukhopadhyay A, Shih PM, Deutschbauer AM, Blank LM, Keasling JD, Zhou N-Y. 2022. Nitrogen metabolism in Pseudomonas putida: functional analysis using random barcode transposon sequencing. Appl Environ Microbiol 88:e0243021. doi: 10.1128/aem.02430-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Comon P. 1994. Independent component analysis, a new concept. Signal Processing 36:287–314. doi: 10.1016/0165-1684(94)90029-9 [DOI] [Google Scholar]
- 26. Saelens W, Cannoodt R, Saeys Y. 2018. A comprehensive evaluation of module detection methods for gene expression data. Nat Commun 9:1090. doi: 10.1038/s41467-018-03424-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Liebermeister W. 2002. Linear modes of gene expression determined by independent component analysis. Bioinformatics 18:51–60. doi: 10.1093/bioinformatics/18.1.51 [DOI] [PubMed] [Google Scholar]
- 28. Kong W, Vanderburg CR, Gunshin H, Rogers JT, Huang X. 2008. A review of independent component analysis application to microarray gene expression data. Biotechniques 45:501–520. doi: 10.2144/000112950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nascimento M, Silva FFE, Sáfadi T, Nascimento ACC, Ferreira TEM, Barroso LMA, Ferreira Azevedo C, Guimarães SEF, Serão NVL. 2017. Independent component analysis (ICA) based-clustering of temporal RNA-seq data. PLoS One 12:e0181195. doi: 10.1371/journal.pone.0181195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sastry AV, Gao Y, Szubin R, Hefner Y, Xu S, Kim D, Choudhary KS, Yang L, King ZA, Palsson BO. 2019. The Escherichia coli transcriptome mostly consists of independently regulated modules. Nat Commun 10:5536. doi: 10.1038/s41467-019-13483-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sastry AV, Hu A, Heckmann D, Poudel S, Kavvas E, Palsson BO. 2021. Independent component analysis recovers consistent regulatory signals from disparate datasets. PLoS Comput Biol 17:e1008647. doi: 10.1371/journal.pcbi.1008647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lim HG, Rychel K, Sastry AV, Bentley GJ, Mueller J, Schindel HS, Larsen PE, Laible PD, Guss AM, Niu W, Johnson CW, Beckham GT, Feist AM, Palsson BO. 2022. Machine-learning from Pseudomonas putida KT2440 transcriptomes reveals its transcriptional regulatory network. Metab Eng 72:297–310. doi: 10.1016/j.ymben.2022.04.004 [DOI] [PubMed] [Google Scholar]
- 33. Spirin V, Gelfand MS, Mironov AA, Mirny LA. 2006. A metabolic network in the evolutionary context: multiscale structure and modularity. Proc Natl Acad Sci USA 103:8774–8779. doi: 10.1073/pnas.0510258103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hyvärinen A. 2013. Independent component analysis: recent advances. Philos Trans A Math Phys Eng Sci 371:20110534. doi: 10.1098/rsta.2011.0534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yadav VG, De Mey M, Lim CG, Ajikumar PK, Stephanopoulos G. 2012. The future of metabolic engineering and synthetic biology: towards a systematic practice. Metab Eng 14:233–241. doi: 10.1016/j.ymben.2012.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Belda E, van Heck RGA, José Lopez-Sanchez M, Cruveiller S, Barbe V, Fraser C, Klenk H-P, Petersen J, Morgat A, Nikel PI, Vallenet D, Rouy Z, Sekowska A, Martins Dos Santos VAP, de Lorenzo V, Danchin A, Médigue C. 2016. The revisited genome of Pseudomonas putida KT2440 enlightens its value as a robust metabolic chassis. Environ Microbiol 18:3403–3424. doi: 10.1111/1462-2920.13230 [DOI] [PubMed] [Google Scholar]
- 37. Eng T, Banerjee D, Lau AK, Bowden E, Herbert RA, Trinh J, Prahl JP, Deutschbauer A, Tanjore D, Mukhopadhyay A. 2021. Engineering Pseudomonas putida for efficient aromatic conversion to bioproduct using high throughput screening in a bioreactor. Metab Eng 66:229–238. doi: 10.1016/j.ymben.2021.04.015 [DOI] [PubMed] [Google Scholar]
- 38. Borchert AJ, Bleem A, Beckham GT. 2022. Experimental and analytical approaches for improving the resolution of randomly barcoded transposon insertion sequencing (RB-TnSeq) studies. ACS Synth Biol 11:2015–2021. doi: 10.1021/acssynbio.2c00119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Dennis G, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. 2003. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol 4:1–11. [PubMed] [Google Scholar]
- 40. Huang DW, Sherman BT, Lempicki RA. 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4:44–57. doi: 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- 41. Huang DW, Sherman BT, Lempicki RA. 2009. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 37:1–13. doi: 10.1093/nar/gkn923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kanehisa M, Furumichi M, Sato Y, Kawashima M, Ishiguro-Watanabe M. 2023. KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Res 51:D587–D592. doi: 10.1093/nar/gkac963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Barrientos-Moreno L, Molina-Henares MA, Ramos-González MI, Espinosa-Urgel M, Kivisaar M. 2022. Role of the transcriptional regulator ArgR in the connection between arginine metabolism and c-di-GMP signaling in Pseudomonas putida. Appl Environ Microbiol 88:e0006422. doi: 10.1128/aem.00064-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Moreno R, Rojo F. 2008. The target for the Pseudomonas putida Crc global regulator in the benzoate degradation pathway is the BenR transcriptional regulator . J Bacteriol 190:1539–1545. doi: 10.1128/JB.01604-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Brunel F, Davison J. 1988. Cloning and sequencing of Pseudomonas genes encoding vanillate demethylase. J Bacteriol 170:4924–4930. doi: 10.1128/jb.170.10.4924-4930.1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Buswell JA, Ribbons DW. 1988. Vanillate O-demethylase from Pseudomonas species. Meth Enzymol:294–301. doi: 10.1016/0076-6879(88)61032-9 [DOI] [PubMed] [Google Scholar]
- 47. Hibi M, Sonoki T, Mori H. 2005. Functional coupling between vanillate O-demethylase and formaldehyde detoxification pathway. FEMS Microbiol Lett 253:237–242. doi: 10.1016/j.femsle.2005.09.036 [DOI] [PubMed] [Google Scholar]
- 48. Erickson E, Bleem A, Kuatsjah E, Werner AZ, DuBois JL, McGeehan JE, Eltis LD, Beckham GT. 2022. Critical enzyme reactions in aromatic catabolism for microbial lignin conversion. Nat Catal 5:86–98. doi: 10.1038/s41929-022-00747-w [DOI] [Google Scholar]
- 49. Cánovas D, Cases I, de Lorenzo V. 2003. Heavy metal tolerance and metal homeostasis in Pseudomonas putida as revealed by complete genome analysis. Environ Microbiol 5:1242–1256. doi: 10.1111/j.1462-2920.2003.00463.x [DOI] [PubMed] [Google Scholar]
- 50. Renilla S, Bernal V, Fuhrer T, Castaño-Cerezo S, Pastor JM, Iborra JL, Sauer U, Cánovas M. 2012. Acetate scavenging activity in Escherichia coli: interplay of acetyl–coa synthetase and the PEP–glyoxylate cycle in chemostat cultures. Appl Microbiol Biotechnol 93:2109–2124. doi: 10.1007/s00253-011-3536-4 [DOI] [PubMed] [Google Scholar]
- 51. Dolan SK, Welch M. 2018. The glyoxylate shunt, 60 years on. Annu Rev Microbiol 72:309–330. doi: 10.1146/annurev-micro-090817-062257 [DOI] [PubMed] [Google Scholar]
- 52. Simon O, Klebensberger J, Mükschel B, Klaiber I, Graf N, Altenbuchner J, Huber A, Hauer B, Pfannstiel J. 2015. Analysis of the molecular response of Pseudomonas putida KT2440 to the next-generation biofuel n-butanol. J Proteomics 122:11–25. doi: 10.1016/j.jprot.2015.03.022 [DOI] [PubMed] [Google Scholar]
- 53. Sudarsan S, Dethlefsen S, Blank LM, Siemann-Herzberg M, Schmid A. 2014. The functional structure of central carbon metabolism in Pseudomonas putida KT2440. Appl Environ Microbiol 80:5292–5303. doi: 10.1128/AEM.01643-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Verhoef S, Ballerstedt H, Volkers RJM, de Winde JH, Ruijssenaars HJ. 2010. Comparative transcriptomics and proteomics of p-hydroxybenzoate producing Pseudomonas putida S12: novel responses and implications for strain improvement. Appl Microbiol Biotechnol 87:679–690. doi: 10.1007/s00253-010-2626-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Revelles O, Wittich R-M, Ramos JL. 2007. Identification of the initial steps in D-lysine catabolism in Pseudomonas putida. J Bacteriol 189:2787–2792. doi: 10.1128/JB.01538-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Cronan JE, Littel KJ, Jackowski S. 1982. Genetic and biochemical analyses of pantothenate biosynthesis in Escherichia coli and Salmonella typhimurium. J Bacteriol 149:916–922. doi: 10.1128/jb.149.3.916-922.1982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Rubio A, Downs DM. 2002. Elevated levels of ketopantoate hydroxymethyltransferase (PanB) lead to a physiologically significant coenzyme A elevation in Salmonella enterica serovar typhimurium. J Bacteriol 184:2827–2832. doi: 10.1128/JB.184.10.2827-2832.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Blanco-Romero E, Redondo-Nieto M, Martínez-Granero F, Garrido-Sanz D, Ramos-González MI, Martín M, Rivilla R. 2018. Genome-wide analysis of the FleQ direct regulon in Pseudomonas fluorescens F113 and Pseudomonas putida KT2440. Sci Rep 8:1–13. doi: 10.1038/s41598-018-31371-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Valentini M, García-Mauriño SM, Pérez-Martínez I, Santero E, Canosa I, Lapouge K. 2014. Hierarchical management of carbon sources is regulated similarly by the CbrA/B systems in Pseudomonas aeruginosa and Pseudomonas putida. Microbiology 160:2243–2252. doi: 10.1099/mic.0.078873-0 [DOI] [PubMed] [Google Scholar]
- 60. Monteagudo-Cascales E, García-Mauriño SM, Santero E, Canosa I. 2019. Unraveling the role of the CbrA histidine kinase in the signal transduction of the CbrAB two-component system in Pseudomonas putida. Sci Rep 9:9110. doi: 10.1038/s41598-019-45554-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kouzuma A, Endoh T, Omori T, Nojiri H, Yamane H, Habe H. 2008. Transcription factors CysB and SfnR constitute the hierarchical regulatory system for the sulfate starvation response in Pseudomonas putida. J Bacteriol 190:4521–4531. doi: 10.1128/JB.00217-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. MacQueen J. 1967. “Some methods for classification and analysis of multivariate observations” p 281–297 [Google Scholar]
- 63. Müllner D. 2011. Modern hierarchical, agglomerative clustering algorithms. Arxiv
- 64. Jitrapakdee S, Wallace JC. 1999. Structure, function and regulation of pyruvate carboxylase. Biochem J 340:1–16. doi: 10.1042/bj3400001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Luykx D, Duine JA, de Vries S. 1998. Molybdopterin radical in bacterial aldehyde dehydrogenases. Biochemistry 37:11366–11375. doi: 10.1021/bi972972y [DOI] [PubMed] [Google Scholar]
- 66. Mueller J, Willett H, Feist AM, Niu W. 2022. Engineering Pseudomonas putida for improved utilization of syringyl aromatics. Biotechnol Bioeng 119:2541–2550. doi: 10.1002/bit.28131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Costa FG, Escalante-Semerena JC. 2022. Localization and interaction studies of the Salmonella enterica ethanolamine ammonia‐lyase (EutBC), its reactivase (EutA), and the EutT corrinoid adenosyltransferase. Mol Microbiol 118:191–207. doi: 10.1111/mmi.14962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Molina-Henares MA, de la Torre J, García-Salamanca A, Molina-Henares AJ, Herrera MC, Ramos JL, Duque E. 2010. Identification of conditionally essential genes for growth of Pseudomonas putida KT2440 on minimal medium through the screening of a genome‐wide mutant library. Environ Microbiol 12:1468–1485. doi: 10.1111/j.1462-2920.2010.02166.x [DOI] [PubMed] [Google Scholar]
- 69. Förster-Fromme K, Höschle B, Mack C, Bott M, Armbruster W, Jendrossek D. 2006. Identification of genes and proteins necessary for catabolism of acyclic terpenes and leucine/isovalerate in Pseudomonas aeruginosa. Appl Environ Microbiol 72:4819–4828. doi: 10.1128/AEM.00853-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Bhatia R, Calvo KC. 1996. The sequencing, expression, purification, and steady-state kinetic analysis of quinolinate phosphoribosyl transferase from Escherichia coli. Arch Biochem Biophys 325:270–278. doi: 10.1006/abbi.1996.0034 [DOI] [PubMed] [Google Scholar]
- 71. Grant GA. 2018. Elucidation of a self-sustaining cycle in Escherichia coli L-serine biosynthesis that results in the conservation of the coenzyme, NAD+. Biochemistry 57:1798–1806. doi: 10.1021/acs.biochem.8b00074 [DOI] [PubMed] [Google Scholar]
- 72. Borchert AJ, Ernst DC, Downs DM. 2019. Reactive enamines and Imines in vivo: lessons from the RidA paradigm. Trends Biochem Sci 44:849–860. doi: 10.1016/j.tibs.2019.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Vázquez-Salazar A, Becerra A, Lazcano A. 2018. Evolutionary convergence in the biosyntheses of the imidazole moieties of histidine and purines. PLoS One 13:e0196349. doi: 10.1371/journal.pone.0196349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Iwasaki H, Takahagi M, Shiba T, Nakata A, Shinagawa H. 1991. Escherichia coli RuvC protein is an endonuclease that resolves the holliday structure. EMBO J 10:4381–4389. doi: 10.1002/j.1460-2075.1991.tb05016.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Funnell BE. 2016. ParB partition proteins: complex formation and spreading at bacterial and plasmid centromeres. Front Mol Biosci 3:44. doi: 10.3389/fmolb.2016.00044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Bervoets I, Charlier D. 2019. Diversity, versatility and complexity of bacterial gene regulation mechanisms: opportunities and drawbacks for applications in synthetic biology. FEMS Microbiol Rev 43:304–339. doi: 10.1093/femsre/fuz001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Naren N, Zhang X-X. 2020. Global regulatory roles of the histidine-responsive transcriptional repressor HutC in Pseudomonas fluorescens SBW25. J Bacteriol 202:00792–19. doi: 10.1128/JB.00792-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Fonseca P, de la Peña F, Prieto MA. 2014. A role for the regulator PsrA in the polyhydroxyalkanoate metabolism of Pseudomonas putida KT2440. Int J Biol Macromol 71:14–20. doi: 10.1016/j.ijbiomac.2014.04.014 [DOI] [PubMed] [Google Scholar]
- 79. Nichols NN, Harwood CS. 1995. Repression of 4-hydroxybenzoate transport and degradation by benzoate: a new layer of regulatory control in the Pseudomonas putida beta-ketoadipate pathway. J Bacteriol 177:7033–7040. doi: 10.1128/jb.177.24.7033-7040.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Hirose Y, Poudel S, Sastry AV, Rychel K, Lamoureux CR, Szubin R, Zielinski DC, Lim HG, Menon ND, Bergsten H, Uchiyama S, Hanada T, Kawabata S, Palsson BO, Nizet V, Zhang X-H. 2023. Elucidation of independently modulated genes in Streptococcus pyogenes reveals carbon sources that control its expression of hemolytic toxins. mSystems 8:e0024723. doi: 10.1128/msystems.00247-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Ester M, Kriegel H-P, Sander J, Xu X. 1996. A density-based algorithm for discovering clusters in large spatial databases with noise. In Abstr Proc ACM SIGKDD INT Portland. Oregon, AAAI Press. [Google Scholar]
- 82. McConn JL, Lamoureux CR, Poudel S, Palsson BO, Sastry AV. 2021. Optimal dimensionality selection for independent component analysis of transcriptomic data. BMC Bioinformatics 22:584. doi: 10.1186/s12859-021-04497-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. D’agostino RB, Belanger A, D’agostino RB. 1990. A suggestion for using powerful and informative tests of normality. Am Stat 44:316–321. doi: 10.1080/00031305.1990.10475751 [DOI] [Google Scholar]
- 84. Cantalapiedra CP, Hernández-Plaza A, Letunic I, Bork P, Huerta-Cepas J. 2021. eggNOG-mapper v2: functional annotation, orthology assignments, and domain prediction at the metagenomic scale. Mol Biol Evol 38:5825–5829. doi: 10.1093/molbev/msab293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Rychel K, Decker K, Sastry AV, Phaneuf PV, Poudel S, Palsson BO. 2021. iModulonDB: a knowledgebase of microbial transcriptional regulation derived from machine learning. Nucleic Acids Res 49:D112–D120. doi: 10.1093/nar/gkaa810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Choi K-H, Kumar A, Schweizer HP. 2006. A 10-min method for preparation of highly electrocompetent Pseudomonas aeruginosa cells: application for DNA fragment transfer between chromosomes and plasmid transformation. J Microbiol Methods 64:391–397. doi: 10.1016/j.mimet.2005.06.001 [DOI] [PubMed] [Google Scholar]
- 87. Johnson CW, Beckham GT. 2015. Aromatic catabolic pathway selection for optimal production of pyruvate and lactate from lignin. Metab Eng 28:240–247. doi: 10.1016/j.ymben.2015.01.005 [DOI] [PubMed] [Google Scholar]
- 88. Rand JM, Pisithkul T, Clark RL, Thiede JM, Mehrer CR, Agnew DE, Campbell CE, Markley AL, Price MN, Ray J, Wetmore KM, Suh Y, Arkin AP, Deutschbauer AM, Amador-Noguez D, Pfleger BF. 2017. A metabolic pathway for catabolizing levulinic acid in bacteria. Nat Microbiol 2:1624–1634. doi: 10.1038/s41564-017-0028-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Cole BJ, Feltcher ME, Waters RJ, Wetmore KM, Mucyn TS, Ryan EM, Wang G, Ul-Hasan S, McDonald M, Yoshikuni Y, Malmstrom RR, Deutschbauer AM, Dangl JL, Visel A, Dong X. 2017. Genome-wide identification of bacterial plant colonization genes. PLoS Biol 15:e2002860. doi: 10.1371/journal.pbio.2002860 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Functional annotation clustering for fModules.
Gene names and weights in fModules.
Code for generating Sankey diagram in Figure 6.
Figures S1-S10; Tables S1-S4.
Data Availability Statement
Sequence data from RB-TnSeq data sets 100 and 101 are available at the NCBI SRA, with accession numbers PRJNA809672, PRJNA856070, and PRJNA1011287 (22, 38). Perl and Python scripts used for analysis of data sets 100 and 101 are accessible from https://github.com/beckham-lab/RB-TnSeq.git. All remaining fitness data are available at https://fit.genomics.lbl.gov/. Source code for fModule analysis and figures may be found at https://github.com/fModules/putida-code, which contains the Jupyter notebook files used for analysis as well as raw input and output data. The interactive website, containing activity and gene information for each fModule, can be found at: https://fmodules.github.io/putida. Gene fitness values, associated statistics, and metadata for each sample are available at https://github.com/beckham-lab/fModule. Python code for the Sankey diagram plotting function was adapted from https://github.com/anazalea/pySankey/blob/master/pysankey/sankey.py and is provided in File S3.