Abstract
Renal fibrosis is a typical pathological change from chronic kidney disease (CKD) to end‐stage renal failure, which presents significant challenges in prevention and treatment. The progression of renal fibrosis is closely associated with the “gut‐kidney axis,” therefore, although clinical intervention to modulate the “gut‐kidney axis” imbalance associated with renal fibrosis brings hope for its treatment. In this study, we first identified the close relationship between renal fibrosis development and the intestinal microenvironment through fecal microtransplantation and non‐absorbable antibiotics experiments. Then, we analyzed the specific connection between the intestinal microenvironment and renal fibrosis using microbiomics and metabolomics, screening for the differential intestinal metabolite. Potential metabolite action targets were initially identified through network simulation of molecular docking and further verified by molecular biology experiment. We used flow cytometry, TUNEL apoptosis staining, immunohistochemistry, and Western blotting to assess renal injury and fibrosis extent, exploring the potential role of gut microbial metabolite in renal fibrosis development. We discovered that CKD‐triggered alterations in the intestinal microenvironment exacerbate renal injury and fibrosis. When metabolomic analysis was combined with experiments in vivo, we found that the differential metabolite xylitol delays renal injury and fibrosis development. We further validated this hypothesis at the cellular level. Mechanically, bromodomain‐containing protein 4 (BRD4) protein exhibits strong binding with xylitol, and xylitol alleviates renal fibrosis by inhibiting BRD4 and its downstream transforming growth factor‐β (TGF‐β) pathway. In summary, our findings suggest that the natural intestinal metabolite xylitol mitigates renal fibrosis by inhibiting the BRD4‐regulated TGF‐β pathway.
Study Highlights.
WHAT IS THE CURRENT KNOWLEDGE ON THE TOPIC?
Chronic kidney disease (CKD) has a global prevalence of ~8% to 16%, greatly affecting the quality of life of patients. Therefore, delaying renal fibrosis and saving renal functional units as much as possible are of great importance for patients with CKD.
WHAT QUESTION DID THIS STUDY ADDRESS?
We demonstrated that xylitol, a natural bacterial metabolite, significantly delayed renal fibrosis.
WHAT DOES THIS STUDY ADD TO OUR KNOWLEDGE?
The main mechanism of xylitol antifibrosis is inhibition of bromodomain protein 4 (BRD4)‐regulated transforming growth factor‐β pathway.
HOW MIGHT THIS CHANGE CLINICAL PHARMACOLOGY OR TRANSLATIONAL SCIENCE?
These findings evoke a novel therapeutic concept for renal fibrosis, the “xylitol‐BRD4‐renal fibrosis” axis may be exploited as a therapeutic target for protection against renal fibrosis.
INTRODUCTION
Chronic kidney disease (CKD) has a global prevalence of 8%–16% and its incidence is highly age‐dependent. CKD is often accompanied by increased mortality from cardiovascular and cerebrovascular diseases, which greatly affects the quality of life of patients. 1 Currently, CKD treatment consists of renal replacement therapy and symptomatic treatment. The continuing increase in global CKD mortality suggests that current treatments still have significant limitations and that the cost of treatment can significantly reduce the quality of life. 2 Therefore, preventing CKD, delaying renal fibrosis, and saving renal functional units as much as possible are of great importance for patients with CKD. The main features of CKD are irreversible damage to the functional units of the kidneys, impaired renal microcirculation, oxidative stress, and inflammation, leading to excessive deposition of the extracellular matrix (ECM) and eventually fibrosis. 3 , 4 Renal fibrosis includes glomerulosclerosis, tubulointerstitial fibrosis, and perivascular sclerosis. Fibrosis in these functional compartments is the progressive replacement of parenchymal tissue by the ECM, accompanied by irreversible damage. 5 Transforming growth factor‐β (TGF‐β) plays an important role in tissue fibrosis by inducing matrix protein synthesis and inhibiting matrix protein degradation in the process of renal fibrosis. 6
Recently, researchers have proposed the “gut‐renal axis” theory, in which one of the main ways in which gut microbes interact with their hosts is through metabolites, mainly from dietary components and their derivatives (such as short‐chain fatty acids [SCFAs] and indole derivatives), intermediate metabolites (such as lactic acid and succinic acid), and from bacterially modified microbial‐host co‐metabolites (secondary bile acids). 7 Wang et al. 8 found that the modulation of intestinal flora with the probiotic Bifidobacterium bifidum significantly reduced uremic toxin levels and kidney damage. Therefore, targeting the control and management of intestinal metabolites may be a potential strategy for slowing CKD progression.
In the present study, we found that mice that underwent fecal microbiota transplantation from patients with CKD also showed the same manifestation of renal fibrosis. Metabolomics of the intestinal contents revealed that xylitol, a natural intestinal metabolite, was reduced in mice with renal fibrosis following unilateral ureteral obstruction (UUO) surgery. However, there have been no reports on the role of xylitol in CKD. Therefore, we used a UUO mouse model to assess the effect of xylitol on the progression of renal fibrosis and elucidate the mechanisms of xylitol renal protection using untargeted and targeted metabolomics and proteomics.
METHODS
Recruitment of patients with CKD and healthy controls
The patient study was conducted in accordance with the Declaration of Helsinki and approved by the Ethics Committee of the First Affiliated Hospital of Guangzhou Medical University (No. 2020163). All volunteers were informed about the use of their feces, and informed consent was obtained from all participants. Stool samples were collected from 28 patients with CKD (obstructive nephropathy caused by urinary stones) and 41 healthy adults for flora transplantation and metabolite analysis. The diagnosis of CKD was performed based on the Kidney Disease Improving Global Outcomes Clinical Practice Guideline for the Evaluation and Management of Chronic Kidney Disease. Individuals who received probiotic or antibiotic therapy within the last 4 weeks were excluded. The detailed clinical information of the enrolled patients is shown in Table 1.
TABLE 1.
Baseline characteristics of the enrolled subjects.
| Total (n = 69) | Control (n = 41) | CKD (n = 28) | p value | |
|---|---|---|---|---|
| Age, year | 50 (44, 57) | 50 (44, 56) | 53 (46, 59) | 0.467 |
| Male, no. (%) | 33 (48%) | 19 (46%) | 14 (50%) | 0.765 |
| BMI, kg/m2 | 25.6 (22.0, 30.1) | 27.1 (22.6, 30.1) | 25.2 (21.8, 27.3) | 0.308 |
| Hypertension, no. (%) | 23 (33%) | 13 (32%) | 10 (36%) | 0.729 |
| Diabetes mellitus, no. (%) | 16 (23%) | 9 (22%) | 7 (25%) | 0.768 |
| Serum creatinine, μmol/L | 62 (51, 73) | 55 (48, 62) | 82 (66, 106) | <0.001 |
| BUN, mmol/L | 4.50 (3.20, 5.80) | 3.90 (2.30, 4.90) | 5.55 (4.50, 7.08) | <0.001 |
Note: Values for categorical variables are given as count (percentage); values for continuous variables are given as median (interquartile range).
Abbreviations: BMI, body mass index; BUN, blood urea nitrogen; CKD, chronic kidney disease.
Animal models
Male C57BL/6J mice (18–22 g), 6–8 weeks of age, were purchased from Beijing Yancheng Bioscience Co., Ltd. (Beijing, China). All animal studies were performed according to the National Institutes of Health Guidelines for the Care and Use of Laboratory Animals and approved by the Animal Ethics Committee of the First Affiliated Hospital of Guangzhou Medical University. Mice were housed in pathogen‐free and ventilated cages in a 12‐h light/dark cycle with a room temperature of 25 ± 2°C and humidity between 40% and 60%, and all animals had free access to water and food for the duration of the experiment. A mouse renal fibrosis model was established by a known UUO procedure. 9 Under general anesthesia, complete UUO was performed by double ligation of the left ureter with a 4–0 silk wire. The ureter of sham‐surgery mice was exposed but not ligated.
Fecal microbiota transplantation experiment
For the fecal microbiota transplantation (FMT) study, stool samples were randomly collected from eight patients with CKD and eight healthy individuals with matched age, gender, and body mass index. Feces from patients with CKD and healthy individuals of the same wet weight were mixed and suspended in sterile phosphate‐buffered saline at a concentration of 0.125 g/mL. Next, the fecal suspension from CKD or healthy individuals was shaken and centrifuged at 94 g and 4°C for 5 min, after which the supernatant was collected. The supernatant was mixed with a volume of 20% sterile glycerol and stored at −80°C until transplantation.
Experiment design
First, male mice were divided into two groups: the FMT‐control group (receiving fecal transplants from healthy individuals, n = 10) and the FMT‐CKD group (receiving fecal transplants from patients with CKD, n = 10). The mice were treated with a mixture of ABX (100 mg/kg vancomycin, 200 mg/kg neomycin sulfate, 200 mg/kg metronidazole, and 200 mg/kg ampicillin) by oral gavage for 3 days to deplete the intestinal microbiota. Then, 200 μL of the fecal supernatant from CKD or healthy controls was gavaged to mice once daily for 5 days. Next, UUO surgery was performed, and the fecal supernatant was administered to mice every two days. Finally, the blood and kidneys of mice were collected on the seventh day after UUO surgery for the next analysis.
In a subsequent study, male mice were divided into four groups: the sham group (n = 10), the sham + xylitol group (n = 10), the UUO group (n = 10), and the UUO + xylitol group (n = 10). The mice were euthanized 14 days after UUO. For xylitol intervention, mice were given xylitol (2 g/kg) by gavage for 14 consecutive days after UUO surgery. The stool, colonic tissue, colonic contents, and kidney tissues were collected.
16S rRNA sequence
Feces were collected for fecal bacteria DNA which was extracted using a commercial DNA extraction kit (Mabio, Guangzhou, China) and the DNA concentration of all samples was quantified as 10 ng/μL. Primers V4F (5′‐GTGTGTCAGCMGCCGGTAA‐3′) and V4R (5′‐CCGGACTACNVGGGTWTCTAAT‐3′) were used to perform polymerase chain reaction (PCR) on variable region 4 (V4) of bacterial 16S rRNA. Novozymes Biologicals (Beijing) was commissioned to analyze the raw data to identify the gut microbial profile.
Metabolomic analysis
For metabolomic analysis, feces were obtained from patients with CKD (n = 10) or healthy individuals (n = 10). The metabolomics sequencing was commercially processed by Beijing Genomics Institution (China). Raw data were imported into Compound Discoverer 3.3 (Thermo Fisher Scientific, USA) and combined with the BGI Metabolome Database, mzCloud database, and ChemSpider online database for analysis.
Quantitative real‐time PCR
RNA was extracted using TRIzol reagent (Thermo Scientific, USA), and the extracted RNA was reverse‐transcribed using the ReverTra‐Ace qPCR RT kit (Toyobo, Shanghai, China). Quantitative real‐time PCR (qRT‐PCR) was performed using the 7500 Real‐Time PCR System (Applied Biosystems, USA). Relative gene expression levels were normalized to 18S ribosomal RNA.
Xylitol quantification by high‐performance liquid chromatography
We used an Agilent C18 column (4.6 × 150 mm, 5 μm) with a mobile phase of acetonitrile: water = 60:40 (v:v). The pH was adjusted to 2.95–2.98 with glacial acetic acid, and the injection volume was 20 μL with a flow rate of 1.0 mL/min. The column temperature was 25°C, the detection wavelength was 274 nm, and running time was 15 min.
Culture of HK‐2 cells
Human kidney proximal tubular epithelial cell (HK‐2; CH1168; NEWGAINBIO, China) were cultured in Dulbecco's Modified Eagle Medium (C11995500BT; Gibco) supplemented with 10% fetal bovine serum (FBS; 164210‐50, Procell) and 1% penicillin/streptomycin (P1400; Solarbio). Before experimental intervention, cells were given FBS‐free medium for 24 h. When the cells reached 40%–50% confluence, they were incubated for 24 h with 10 ng/mL of TGF‐β1. For xylitol intervention, cells were incubated with 0.5 mmol/L xylitol for 24 h. For bromodomain‐containing protein 4 (BRD4) inhibitor treatment, cells were first administered with 0.5 μM JQ‐1 for 1 h and then incubated with 10 ng/mL TGF‐β1 for 24 h.
Hematoxylin–eosin staining and Masson's trichrome staining
The left kidneys were collected and fixed in 4% paraformaldehyde (Leagene, Beijing, China) for 36 h. Paraffin sections with a thickness of 5 μm were subjected to hematoxylin & eosin (H&E) and Masson's trichrome staining (Solarbio, Beijing, China). At least six areas from each sample were randomly selected for observation.
Terminal deoxynucleotidyl transferase‐mediated nick end labeling assay
Paraffin‐embedded kidney tissue sections were stained for terminal deoxynucleotidyl transferase‐mediated nick end labeling (TUNEL) assay using a commercial assay kit (BL646C; Biosharp) according to the manufacturer's instructions. To determine the number of TUNEL‐positive cells, at least six areas per slide were randomly selected, and the images were analyzed for the apoptosis rate using ImageJ software.
Flow cytometry
To evaluate the extent of apoptosis in HK‐2 cells across different groups, we treated cell samples with the Annexin V‐FITC/PI apoptosis kit (MultiSciences Biotech, AP101), following the manufacturer's instructions. Subsequently, we analyzed the treated samples using a BD‐FACS flow cytometer, with the FITC detection channel for Annexin V‐FITC (Ex = 488 nm; Em = 530 nm) and the PI detection channel for propidium iodide (Ex = 535 nm; Em = 615 nm). Finally, we conducted post‐acquisition analysis using FlowJo software to assess the apoptotic levels in each group.
Immunohistochemistry
Paraffin‐embedded kidney tissues were cut into 5‐μm sections for immunohistochemistry with antibodies for α‐SMA (14395‐1‐AP; Proteintech), TGF‐β (21898‐1‐AP; Proteintech), connective tissue growth factor (C‐TGF; 23936‐1‐AP; Proteintech), or BRD4 (DF2905; Affinity). The positive area of immunohistochemistry staining was calculated with ImageJ software.
Immunofluorescence
HK‐2 cells were inoculated onto 24‐well plates and incubated with α‐SMA (1:1000, 14395‐1‐AP; Proteintech), TGF‐β (1:600, 21898‐1‐AP; Proteintech), C‐TGF (1:600, 23936‐1‐AP; Proteintech), or BRD4 (1:200, DF2905; Affinity) specific primary antibody, followed by incubation with horseradish peroxidase (HRP)‐conjugated goat anti‐rabbit IgG (1:200, GB23303; Servicebio) for 1 h at room temperature, and finally the nuclei were stained using 4′,6‐diamidino‐2‐phenylindole and six fields of view were randomly obtained. The images were analyzed for fluorescence staining intensity using the ImageJ software.
Western blotting
For protein extraction and quantification, tissues or cells were lysed in RIPA buffer on ice and denatured by heating in sodium dodecyl sulfate (SDS) loading buffer for 5 min. Protein lysates were resolved using SDS‐polyacrylamide gel electrophoresis and transferred to a nitrocellulose membrane. The membranes were mixed with primary antibody against α‐SMA (1:5000, 14395‐1‐AP; Proteintech), TGF‐β (1:2000, 21898‐1‐AP; Proteintech), C‐TGF (1:2000, 23936‐1‐AP; Proteintech), or BRD4 (1:2000, DF2905; Affinity) primary antibody in a shaker at 4°C overnight. After incubation with HRP‐conjugated goat anti‐rabbit IgG (1:3000, GB23303; Servicebio), the protein bands on the membranes were detected using an enhanced chemiluminescence assay (LGSJ01‐402‐1; Biosharp). Specific bands indicating target proteins were analyzed using the ImageJ software.
Molecular docking
The structure of BRD4 protein (UniProtKB:O60885) was obtained from the AlphaFold Protein Structure Database (https://alphafold.ebi.ac.uk/), whereas the molecular structure of xylitol (PubChem CID:6912) was retrieved from the PubChem database (https://pubchem.ncbi.nlm.nih.gov/). To conduct the network simulation of molecular docking, we utilized the Schrodinger Suite (version 2017–1). Finally, for data visualization, we used the Discovery Studio Visualizer software (Accelrys, USA).
Statistical analysis
Statistical analysis was performed with IBM SPSS Statistics 26 (SPSS, IBM, USA) and GraphPad Prism 8.0 (GraphPad Software, USA). All tests were conducted at least three times. Chi‐squared tests were used to perform the rate comparisons between different groups. Two‐tailed Student's t‐test (parametric data) or Mann–Whitney U test (nonparametric data) was utilized to analyze the differences in statistics between both groups, whereas one‐way analysis of variance was utilized to analyze differences between over two groups, and then Tukey's multiple comparison assay was implemented. Any p < 0.05 was considered statistically significant.
RESULTS
Alterations in the intestinal microenvironment have important implications for renal fibrosis
To investigate the relationship between the human intestinal microenvironment and renal fibrosis, we collected 28 fecal samples from patients with confirmed CKD (Table 1) and transplanted their fecal bacteria into the intestines of normal 6–8 week old C57BL/6 mice whose intestinal flora were depleted by ABX. We found that the kidney tissues of mice transplanted with fecal bacteria from patients with CKD (the CKD group) revealed significant fibrous alterations compared to those of mice transplanted with normal human fecal bacteria (the control group; Figure 1a–c). The intestinal microenvironment of patients with CKD also aggravated kidney cell apoptosis in mice (Figure 1d,e). Furthermore, we found that the expression of inflammatory factors was increased in mice that received fecal microbiota transplantation from patients with CKD when compared to those of mice transplanted with normal human fecal bacteria (Figure 1f).
FIGURE 1.
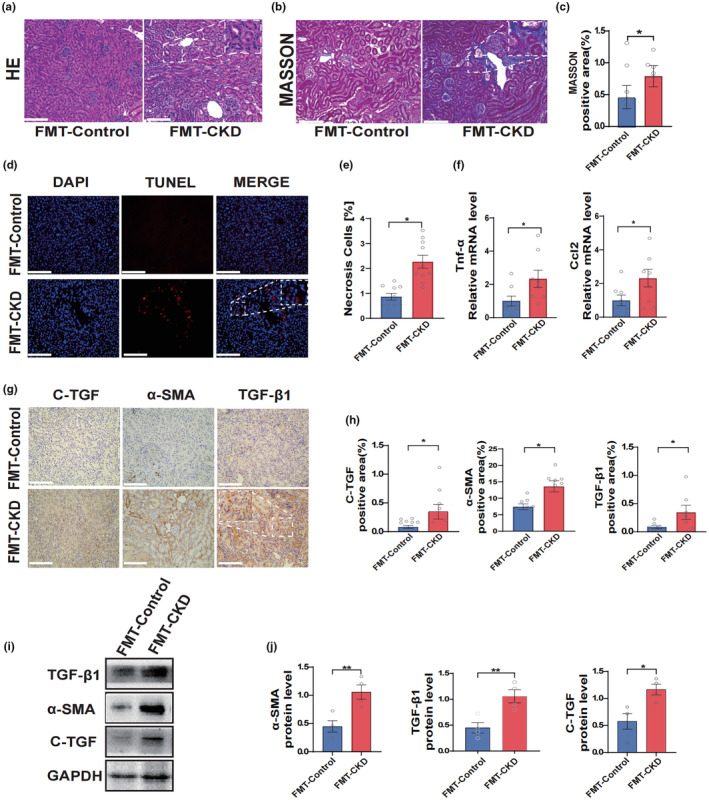
Renal fibrosis exacerbated by intestinal microenvironment in patients with CKD. (a, b) Representative microscopic photographs of H&E staining and Masson's staining of the left kidney of mice after fecal transplantation from patients with CKD and healthy adults (H&E and Masson's staining, scale bar, 100 μm, magnification, ×200). (c) Statistical analysis of Masson's staining for fiber positive area (n = 5). (d, e) Experimental and statistical analysis of renal apoptosis after fecal transplantation (TUNEL, scale bar, 100 μm, magnification, ×200, n = 10). (f) The qPCR experiments after reverse transcription of RNA from the kidneys after fecal transplantation (n = 8). (g, h) Immunohistochemistry experiments and statistical analysis of kidneys of mice in the CKD group and control group (IHC, scale bar, 100 μm, magnification, ×400, n = 6). (i, j) Protein blotting assay demonstrating renal fibronectin expression in the CKD and control groups (n = 4). Data are presented as mean ± SEM. *p < 0.05, **p < 0.01. CKD, chronic kidney disease; FMT, fecal microbiota transplantation; H&E, hematoxylin & eosin; IHC, immunohistochemical; qPCR, quantitative polymerase chain reaction; TUNEL, terminal deoxynucleotidyl transferase‐mediated nick end labeling.
As shown by immunohistochemistry and Western blot analyses, the intestinal microenvironment of patients with CKD significantly increased the levels of several profibrotic markers in mouse kidneys, including C‐TGF, α smooth muscle actin (α‐SMA) and TGF‐β1 (Figure 1g–j). With these data, together with the observation of H&E and Masson staining of renal microscopic sections, it is reasonable to assume that the human intestinal microenvironment is significantly correlated with chronic renal fibrosis and that alterations in the intestinal microenvironment may be an important factor in exacerbating renal fibrosis.
Altered intestinal microenvironment in patients with CKD
To investigate the potential mechanisms by which the intestinal microenvironment regulates chronic kidney fibrosis, we sequenced 16 s flora in the stools of patients clinically diagnosed with CKD (the CKD group) and healthy adults (the control group). Compared with the control group, patients in the CKD group showed significant changes in the total bacterial load and flora structure in the intestine (Figure 2c–e). Principal Co‐ordinate analysis (PCoA) analysis also revealed the separation of the two clusters in the CKD and control groups (Figure 2a,b).
FIGURE 2.
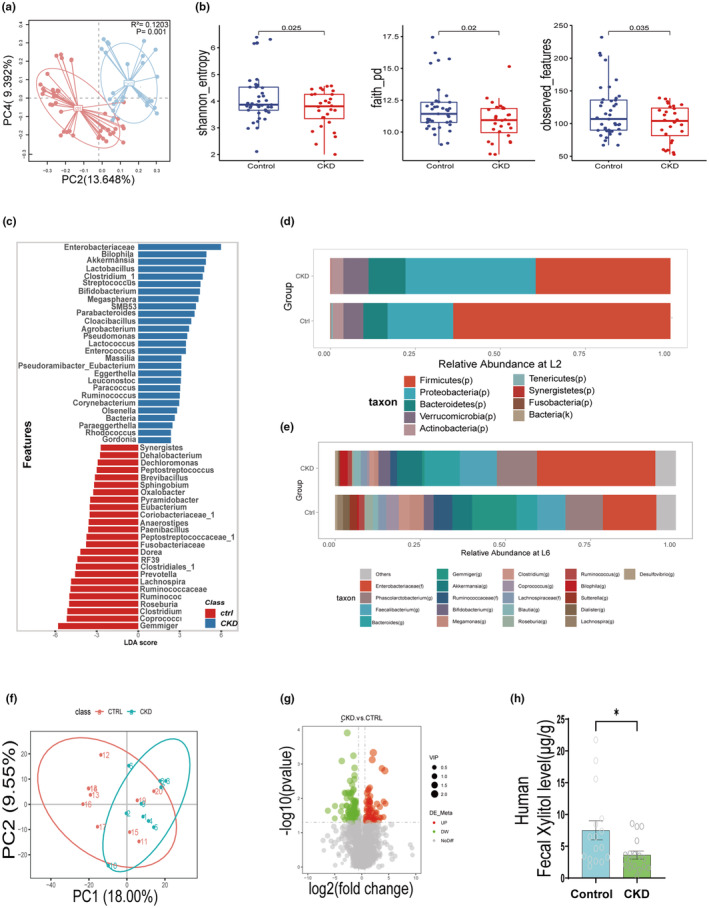
Changes in the intestinal microenvironment in patients with CKD. (a) PCoA of intestinal flora in patients with CKD (n = 28) with healthy adults (control, n = 41). (b) Comparison of intestinal flora abundance in patients with CKD (n = 28) with healthy adults (n = 41). (c) Graph of intestinal flora LEfse analysis in patients with CKD (n = 28) with healthy adults (n = 41). (d, e) Comparison of relative abundance of intestinal flora at L2 and L6 in patients with CKD (n = 28) with healthy adults (n = 41). (f) PCoA of intestinal bacterial metabolites in patients with CKD (n = 10) versus healthy adults (n = 10). (g) Volcano plot of intestinal flora metabolites in patients with CKD (n = 10) versus healthy adults (n = 10). (h) HPLC determination of xylitol content in the intestines of patients with CKD (n = 17) versus healthy adults (n = 16). Data are presented as mean ± SEM. *p < 0.05. CKD, chronic kidney disease; HPLC, high‐performance liquid chromatography; LDA, linear discriminant analysis; LEfSe, linear discriminant analysis effect size; PCoA, principal component analysis.
Next, we examined the intestinal bacterial metabolites in the CKD group versus the control group population using untargeted metabolomics analysis, which revealed that the intestinal metabolic profile was significantly altered in the CKD group, as expected based on the PCoA analysis (Figure 2f). In addition, we identified several metabolites with significantly lower levels in the CKD group, including xylitol (Figure 2g).
Further validation of xylitol content variation in mice through high‐performance liquid chromatography analysis and ABX intervention
To further confirm that xylitol is a differential gut flora metabolite in patients with CKD and healthy individuals, we examined feces from patients with CKD and normal adults using high‐performance liquid chromatography. We found that the intestinal xylitol content was significantly lower in patients with CKD than in healthy adults (Figure 2h). We obtained the same results when we examined the feces and serum of mice in the sham and UUO groups (Figure 3a,b). After depletion of the intestinal flora by ABX, it was found that the xylitol content in the feces and serum of mice in the ABX group was significantly lower compared to the control group (Figure 3c,d). These data strongly suggest that xylitol is a differential metabolite resulting from altered intestinal flora in CKD. Therefore, we hypothesized that the natural differential intestinal metabolite xylitol could delay the progression of renal fibrosis in CKD.
FIGURE 3.
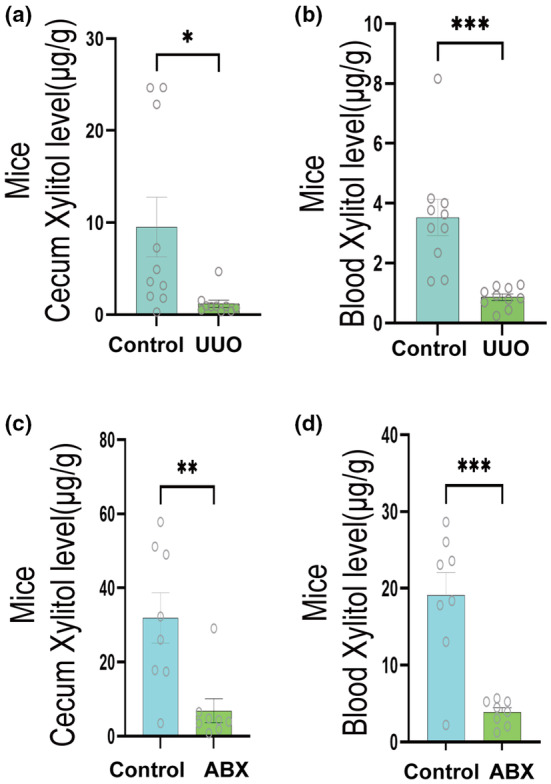
Further validation of xylitol content variation in mice through HPLC analysis and ABX intervention. (a, b) HPLC determination of xylitol content in the intestine and serum of mice in the UUO group versus controls (n = 10). (c, d) After ABX removal of intestinal flora, HPLC determination of xylitol content in the intestine and serum of mice in the ABX group versus controls (n = 8). Data are presented as mean ± SEM. *p < 0.05, **p < 0.01, and ***p < 0.001. ABX, 100 mg/kg vancomycin, 200 mg/kg neomycin sulfate, 200 mg/kg metronidazole, and 200 mg/kg ampicillin; HPLC, high‐performance liquid chromatography; UUO, unilateral ureteral obstruction.
Xylitol attenuates renal fibrosis in the UUO animal model
To test this hypothesis, we examined the nephroprotective effects of xylitol in UUO mice. The mice were randomly divided into four groups: sham, sham + xylitol, UUO, and UUO + xylitol. H&E and Masson staining results showed that xylitol administration significantly improved renal morphology in UUO model mice. Xylitol reduced glomerulosclerosis, atrophy, tubular dilation, tubulointerstitial fibrosis, and inflammatory cell infiltration (Figure 4a‐c). According to the TUNEL assay results, xylitol treatment reduced autophagy and apoptosis in renal cells (Figure 4d,e). In addition, xylitol treatment reduced the levels of several pro‐fibrotic markers, such as α‐SMA, TGF‐β1, and C‐TGF in immunohistochemistry (Figure 4f,g) and Western blot analyses (Figure 4h,i). Based on these experimental data, we concluded that xylitol significantly attenuates renal fibrosis in animals in vivo.
FIGURE 4.
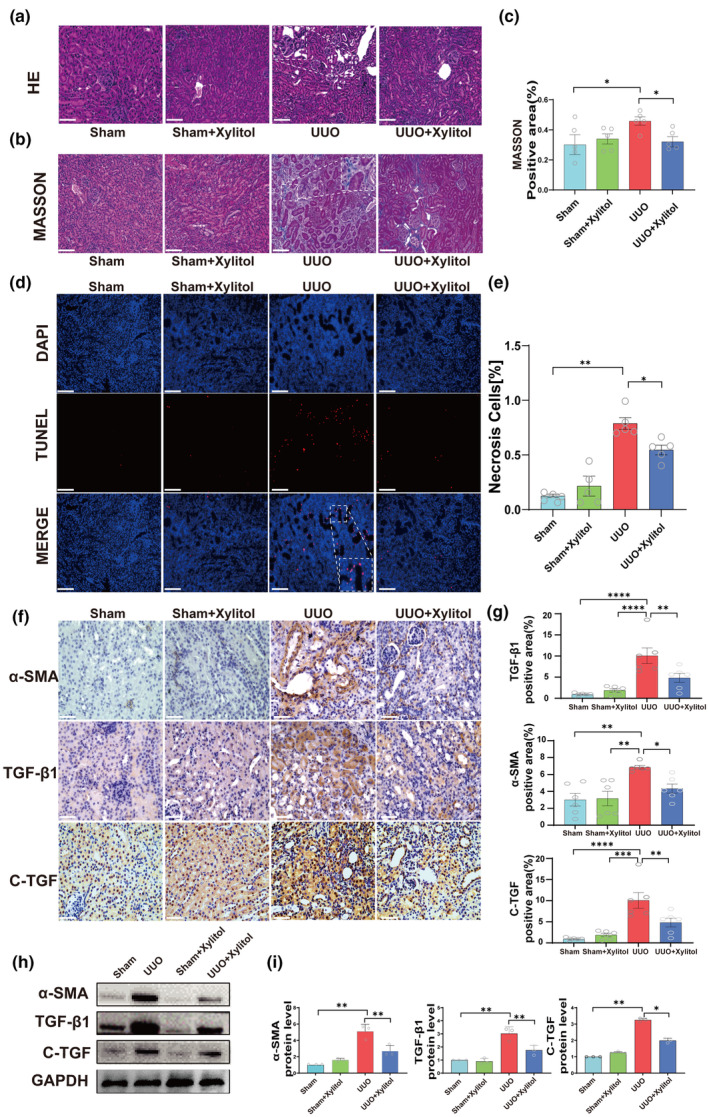
Xylitol attenuates renal fibrosis in the UUO animal model. (a, b) Representative microscopic photographs of H&E staining and Masson staining of the left kidney of mice in the sham group, sham + xylitol group, UUO group, and UUO + xylitol group (H&E and Masson's staining, scale bar, 100 μ m, magnification, ×200). (c) Statistical analysis of the positive area of Masson's staining fibers (n = 5). (d, e) Apoptosis experiments and statistical analysis of left kidney cells in the sham group, sham + xylitol group, UUO group, and UUO + xylitol group (TUNEL, scale bar, 100 μm, n = 5). (f, g) Immunohistochemical assay and statistical analysis of representative fibronectin in the left kidneys of four groups of mice (IHC, scale bar, 100 μm, magnification, ×400, n = 6). (h, i) Protein blotting experiments for fibronectin (n = 3). Data are presented as mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001. H&E, hematoxylin & eosin; IHC, immunohistochemical; TUNEL, terminal deoxynucleotidyl transferase‐mediated nick end labeling; UUO, unilateral ureteral obstruction.
Xylitol mitigates fibrosis marker levels in a TGF‐β1‐stimulated HK‐2 cell model
To further confirm the protective effect of xylitol on the kidneys at the cellular level, we used a TGF‐β1‐stimulated HK‐2 cell model. We evaluated the effect of xylitol on cellular activity using the CCK‐8 method and found that xylitol had no effect on HK‐2 cell viability when used at doses below 1 mmol/L (Figure 5a).
FIGURE 5.
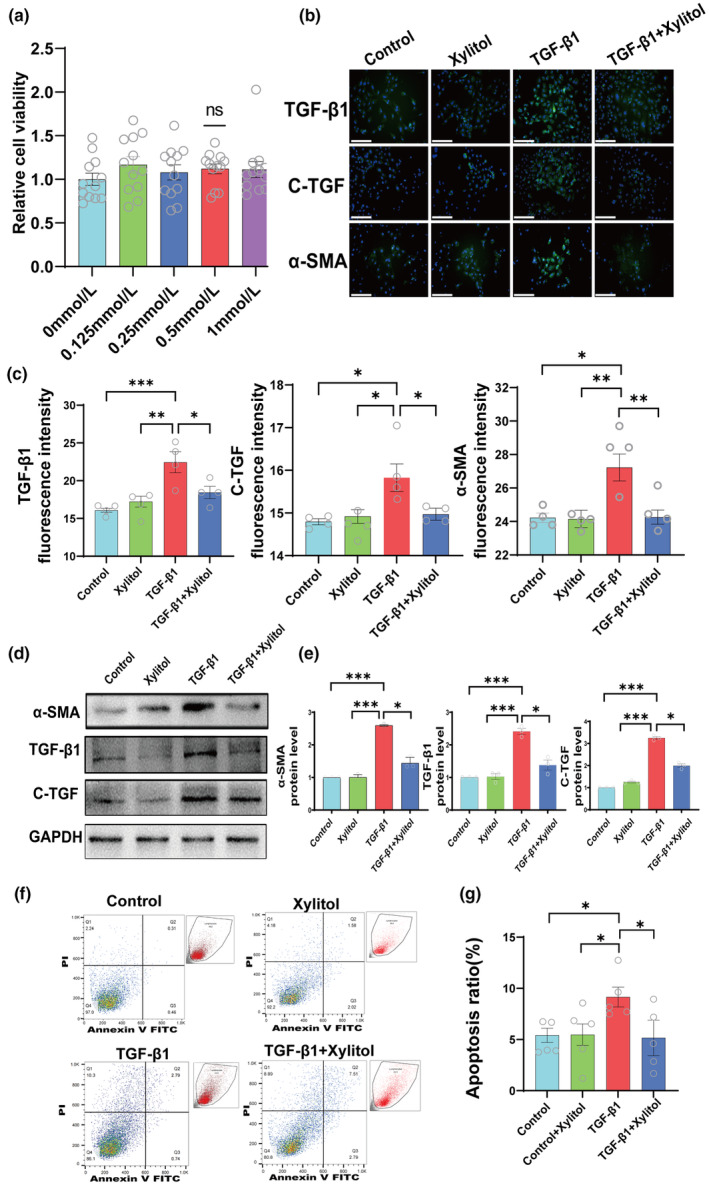
Xylitol mitigates fibrosis marker levels in a TGFβ1‐stimulated HK‐2 cell model. (a) Toxicity assay by CCK‐8 method after four doses of xylitol intervention on HK‐2 (n = 12). (b, c) Immunofluorescence experiments of HK‐2 cells in the control group, xylitol group, TGF‐β1 group, and TGF‐β1 + xylitol group against representative fibronectin (IF, scale bar, 100 μm, magnification, ×400, n = 4). (d, e) Protein blotting experiments for four groups against representative fibronectin (n = 3). (f, g) Flow apoptosis experiments for four groups of HK‐2 cells, (n = 5). All of the in vitro experiments were conducted at least three times. Data are presented as mean ± SEM. *p < 0.05, **p < 0.01, and ***p < 0.001. ns, not significant.
Next, we divided the cells into the control group, control + xylitol group, TGF‐β1 group, and TGF‐β1 + xylitol group, where the concentration of xylitol was 0.5 mmol/L. The expression of these pro‐fibrotic markers, α‐SMA, TGF‐β1, and C‐TGF, were detected by immunofluorescence, and the results revealed that the pro‐fibrotic markers in TGF‐β1 group expression was significantly increased and there was a significant decrease in the expression of the TGF‐β1 + xylitol group (Figure 5b,c).
To further confirm the role of xylitol in fibrosis progression at the cellular level, we extracted HK‐2 cellular proteins and performed Western blot quantification of these pro‐fibrotic markers. The results were the same as those of the immunofluorescence assay, and the levels of pro‐fibrotic markers were significantly decreased in the TGF‐β1 + xylitol group compared with those in the TGF‐β1 group (Figure 5d,e). Additionally, we performed an apoptosis assay using flow cytometry, and the same conclusion as in the animal experiments was drawn: xylitol reduced the apoptosis of HK‐2 cells in the kidneys (Figure 5f,g).
Xylitol mitigates renal fibrosis in vivo and in vitro by inhibiting BRD4 protein expression
Based on the experiment mentioned above, we concluded that xylitol significantly attenuates renal fibrosis in vivo and in vitro. However, the mechanism of action of xylitol is unclear, and we explored the targets of xylitol action.
Recently, investigators have found that the inhibition of BRD4 not only attenuates inflammatory factor expression in the UUO model but also delays glomerular structural damage. 10 In addition, in a UUO kidney injury model, the application of a specific BET inhibitor (JQ1) effectively attenuated renal fibrosis through a mechanism mainly related to the prevention of renal fibrosis by inhibiting BRD4, thereby blocking the TGF‐β pathway. 11 , 12 Therefore, we make the reasonable hypothesis that xylitol prevents renal fibrosis by inhibiting the expression of the BRD4 protein, thus blocking the downstream fibrosis pathway.
To test our hypothesis, we first performed qPCR on RNA from mouse kidney tissues after reverse transcription into DNA to detect BRD4 expression at the gene level. The results revealed that the mRNA expression of BRD4 was reduced in mice administered xylitol compared to that in the UUO group (Figure 6a). Subsequently, we confirmed the connection between xylitol and BRD4 activity utilizing molecular docking (Figure 6b). Next, we measured the BRD4 protein expression levels. We used TGF‐β1‐stimulated HK‐2 cell model and divided the cells into the control group, control + xylitol group, TGF‐β1 group, and TGF‐β1 + xylitol group, where the concentration of xylitol was 0.5 mmol/L. BRD4 protein was detected by immunofluorescence, and the results revealed that BRD4 protein level was decreased in HK‐2 cells following the administration of xylitol (Figure 6c,d). In addition, we used the renal tubular epithelial cell marker protein cytokeratin 18 (CK18), a member of the intermediate filament protein family that is widely expressed in epithelial and endothelial cells. 13 We used CK18 to target the renal tubular epithelial cells (RTECs) and probe the location of BRD4 expression at the cellular level. The results showed that BRD4 protein expression was significantly decreased in RTECs following xylitol administration (Figure 6e,f).
FIGURE 6.
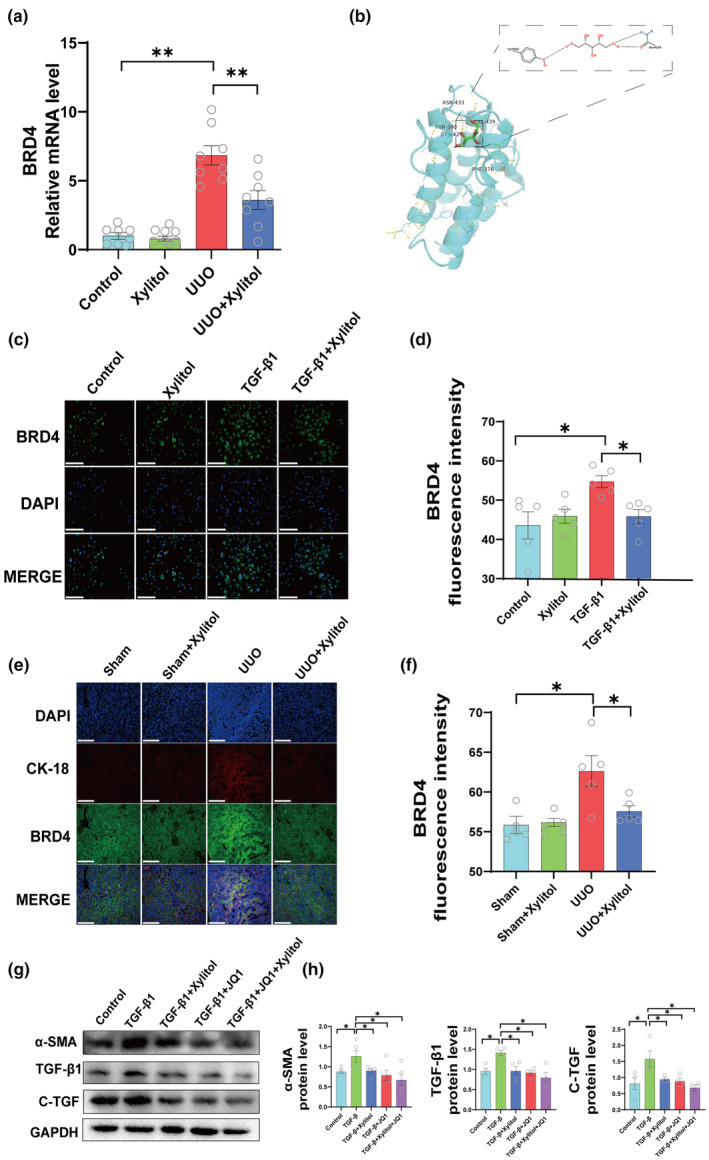
Xylitol attenuates renal fibrosis in vivo and in vitro by inhibiting BRD4 protein. (a) Left kidney qPCR experiments in mice of the sham group, sham + Xylitol group, UUO group, and UUO + Xylitol group. (n = 8). (b) Molecular docking to evaluate the interaction between xylitol and BRD4. (c, d) Immunofluorescence experiments against BRD4 in the control group, Xylitol group, TGF‐β1 group, TGF‐β1 + Xylitol group (IF, scale bar, 100 μm, magnification, ×400, n = 5). (e, f) Immunofluorescence experiments against BRD4 in the sham group, sham + Xylitol group, UUO group, and UUO + Xylitol group of mice left kidneys for BRD4 protein expression in tubular epithelial cells (IF, scale bar, 100 μm, magnification, ×400, n = 5). (g, h) Control group, TGF‐β1 group, TGF‐β1 + xylitol TGF‐β1 + JQ1, and TGF‐β1 + JQ1 + Xylitol groups for protein imprinting experiments of representative fibronectin by HK‐2 cells (n = 4). All of the in vitro experiments were conducted at least three times. Data are presented as mean ± SEM. *p < 0.05, and **p < 0.01. qPCR, quantitative polymerase chain reaction; UUO, unilateral ureteral obstruction.
Next, we used JQ1 to verify the effect of BRD4 on fibrosis. The cells were divided into the control, TGF‐β1, TGF‐β1 + xylitol, TGF‐β1 + JQ1, and TGF‐β1+ JQ1 + xylitol groups. Cellular proteins were extracted to detect the expression of pro‐fibrotic factors. As shown in the figure, the expression content of pro‐fibrotic markers (α‐SMA, TGF‐β1, and C‐TGF) was significantly decreased in the TGF‐β1 + xylitol group, TGF‐β1 + JQ1 group, and TGF‐β1 + JQ1 + xylitol group compared to the TGF‐β1 group of the fiber model group (Figure 6g,h).
Based on our experimental data, we demonstrate that xylitol prevents renal fibrosis by inhibiting the expression of the BRD4 protein to reduce the expression of pro‐fibrotic markers (Figure 7).
FIGURE 7.
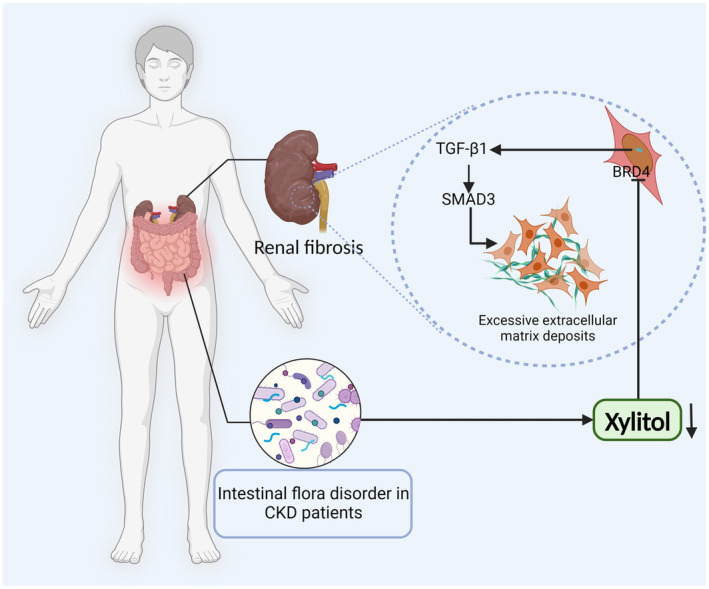
Schematic illustration of xylitol mitigates renal fibrosis by inhibiting the BRD4‐regulated TGF‐β pathway. CKD, chronic kidney disease.
DISCUSSION
The pathological factors in CKD may be epigenetic modifications or alterations, the most widely studied of which are DNA methylation, histone modifications, and changes in miRNA levels. 14 It is generally accepted that there is a complex interplay between the kidneys and the intestines. 15 For example, in patients with CKD, toxic substances, such as urotoxins and uric acid, accumulate in the body owing to the reduced excretory function of the kidneys, changing the microenvironment in the intestines. 16 , 17 , 18 A growing body of evidence suggests that gut flora has great potential for studying delayed renal fibrosis. 8 As an example, Vaziri et al. found that the administration of a fermentable dietary fiber diet reduced kidney injury and improved kidney function in a rat model of CKD. 19 A potential reason for this may be that dietary fiber increases the content of SCFAs in the intestines and improves intestinal barrier function. 19
Xylitol, an intestinal metabolite, is a 5‐carbon sugar alcohol compound that occurs naturally in various edible plants. 20 Xylitol is often added to food as an artificial sweetener to prevent dental caries. 21 Recent studies have demonstrated that xylitol has good inflammatory inhibitory effects, including the inhibition of cellular inflammatory factor expression and angiogenesis. 20 Besides, there are scholars concerned about the safety and efficacy of xylitol in pulmonary cystic fibrosis. 22 What is more, Xylitol is a low‐calorie sugar with strong antioxidant potential and can be used to treat diabetes and prevent cardiovascular disease. 23
In this study, we found that following transplantation of feces from patients with CKD into mice, the mice also developed renal fibrosis. This may be attributed to the expression of renal inflammatory factors. Next, we verified the changes in the intestinal microenvironment in mice with renal fibrosis, in which the xylitol content was significantly reduced. Based on our data, we show that xylitol not only attenuated chronic fibrosis in the animal model but also exhibited an equally significant effect at the cellular level. Xylitol decreased the expression of pro‐fibrotic factors and reduced the rate of apoptotic cell death.
BRD4 is a novel epigenetic target that regulates various renal diseases, including chronic nephritis, diabetic nephropathy, and experimental renal damage. 10 , 24 Recently, Sakamaki et al. defined BRD4 as a potential inhibitor of autophagy. 25 BRD4 inhibits the transcriptional process of lysosomal genes by binding to the acetylated histone H4 lysine 16 site (H4K16ac) in the promoter region of lysosomal genes. 25 In addition, BRD4 is involved in regulating the NF‐κB pathway. BRD4 inhibition blocks the transcriptional activation of NF‐κB in the kidney and decreased NF‐κB‐controlled genes, including CCL‐2 and IL‐17A. 10 BRD4 has a significant effect on renal fibrosis in addition to regulating autophagy and inflammation. It has been demonstrated that the BRD4 inhibitor JQ1 can prevent renal fibrosis. 11 , 12 The underlying mechanism may be related to the fact that JQ1 reduces α‐SMA expression in the kidney and inhibits TGF‐β1‐induced activation of renal fibroblasts. In another study using a nephrotoxic cisplatin‐induced acute kidney injury model, JQ1 restored certain renal function parameters, such as serum creatinine and urea nitrogen levels, and reduced renal injury. Therefore, we propose a reasonable hypothesis for the target of action of xylitol in this study, which acts on BRD4 protein and thus attenuates renal fibrosis.
Next, we explored this hypothesis. At the genetic level, we found that the expression of BRD4 decreased following administration of xylitol in an animal model using renal qPCR experiments. Immunofluorescence experiments revealed that BRD4 expression in renal tubular epithelial cells decreased following xylitol administration. At the cellular level, we found that the expression of profibrogenic factors in HK‐2 cells decreased following BRD4 inhibitor administration. Therefore, we conclude that the intestinal metabolite xylitol reduces TGF‐β1 expression by inhibiting BRD4 protein to attenuate chronic renal fibrosis. These findings evoke a novel therapeutic concept for renal fibrosis, the “xylitol‐BRD4‐renal fibrosis” axis may be exploited as a therapeutic target for protection against renal fibrosis.
However, our study has several shortcomings. We did not explore or demonstrate the potential mechanism of the “xylitol‐BRD4‐renal fibrosis” axis. There may be other types of cells in patients with CKD that express BRD4; however, in this study, we only demonstrated that RTECs significantly expressed BRD4 protein. The number of samples in our animal model was very small, and further research is needed to investigate this direction in depth.
AUTHOR CONTRIBUTIONS
Z.T., Z.W., and G.K. wrote the manuscript. Z.T., Z.W., G.K., J.X., and H.Y. designed the research. Z.T., Z.W., G.K., Q.L.Z., X.L., Y.M.Z., S.L., Y.N.Z., C.J., and Z.H. performed the research. Z.T., Z.W., G.K., N.F., Q.Z., J.H., Y.M., and Z.Y.W. analyzed the data.
FUNDING INFORMATION
This study was supported by National Natural Science Foundation of China (No. 82060131, No. 82360154, and No. 81871551), Guangdong Basic and Applied Basic Research Foundation (No. 2023A1515012474), Sichuan Science and Technology Program (No. 2021YFS0159), and Guangzhou City Science and Technology Project (No. 2023A03J0344, No. 2023A03J0342, and No. 202201020508).
CONFLICT OF INTEREST STATEMENT
The authors declared no competing interests for this work.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
All experiments were approved by the Ethics Committee of the First Affiliated Hospital of Guangzhou Medical University.
ACKNOWLEDGMENTS
The authors thank Shenhai Gong for the excellent technical assistance.
Tan Z, Wang Z, Zeng Q, et al. Natural intestinal metabolite xylitol reduces BRD4 levels to mitigate renal fibrosis. Clin Transl Sci. 2024;17:e13770. doi: 10.1111/cts.13770
Zhouke Tan, Ze Wang, Qianglin Zeng, and Xiaoyou Liu contributed equally to this work as first authors.
Contributor Information
Jie Xiao, Email: 13600097893@163.com.
Hong Yang, Email: yhicu_1103@163.com.
Guibao Ke, Email: gbke@outlook.com.
DATA AVAILABILITY STATEMENT
All data supporting the findings of this study are available from the corresponding authors upon reasonable request.
REFERENCES
- 1. Stevens LA, Li S, Wang C, et al. Prevalence of CKD and comorbid illness in elderly patients in the United States: results from the kidney early evaluation program (KEEP). Am J Kidney Dis. 2010;3(Suppl 2):S23‐S33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Thomas B, Matsushita K, Abate KH, et al. Global cardiovascular and renal outcomes of reduced GFR. J Am Soc Nephrol. 2017;7:2167‐2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Li L, Fu H, Liu Y. The fibrogenic niche in kidney fibrosis: components and mechanisms. Nat Rev Nephrol. 2022;9:545‐557. [DOI] [PubMed] [Google Scholar]
- 4. Fontecha‐Barriuso M, Lopez‐Diaz AM, Guerrero‐Mauvecin J, et al. Tubular mitochondrial dysfunction, oxidative stress, and progression of chronic kidney disease. Antioxidants (Basel). 2022;7:1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Duffield JS. Cellular and molecular mechanisms in kidney fibrosis. J Clin Invest. 2014;6:2299‐2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Isaka Y. Targeting TGF‐β signaling in kidney fibrosis. Int J Mol Sci. 2018;9:2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Miller TL, Wolin MJ. Pathways of acetate, propionate, and butyrate formation by the human fecal microbial flora. Appl Environ Microbiol. 1996;5:1589‐1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wang X, Yang S, Li S, et al. Aberrant gut microbiota alters host metabolome and impacts renal failure in humans and rodents. Gut. 2020;12:2131‐2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fukasawa H, Yamamoto T, Togawa A, et al. Down‐regulation of Smad7 expression by ubiquitin‐dependent degradation contributes to renal fibrosis in obstructive nephropathy in mice. Proc Natl Acad Sci USA. 2004;23:8687‐8692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Suarez‐Alvarez B, Morgado‐Pascual JL, Rayego‐Mateos S, et al. Inhibition of Bromodomain and Extraterminal domain family proteins ameliorates experimental renal damage. J Am Soc Nephrol. 2017;2:504‐519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhou B, Mu J, Gong Y, et al. Brd4 inhibition attenuates unilateral ureteral obstruction‐induced fibrosis by blocking TGF‐β‐mediated Nox4 expression. Redox Biol. 2017;11:390‐402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Xiong C, Masucci MV, Zhou X, et al. Pharmacological targeting of BET proteins inhibits renal fibroblast activation and alleviates renal fibrosis. Oncotarget. 2016;43:69291‐69308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lebherz‐Eichinger D, Krenn CG, Roth GA. Keratin 18 and heat‐shock protein in chronic kidney disease. Adv Clin Chem. 2013;62:123‐149. [DOI] [PubMed] [Google Scholar]
- 14. Morgado‐Pascual JL, Marchant V, Rodrigues‐Diez R, et al. Epigenetic modification mechanisms involved in inflammation and fibrosis in renal pathology. Mediat Inflamm. 2018;2931049:1‐14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mocanu A, Bogos RA, Lazaruc TI, et al. Exploring a complex interplay: kidney‐gut Axis in pediatric chronic kidney disease. Nutrients. 2023;16:3609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Vaziri ND, Wong J, Pahl M, et al. Chronic kidney disease alters intestinal microbial flora. Kidney Int. 2013;2:308‐315. [DOI] [PubMed] [Google Scholar]
- 17. Yun Y, Yin H, Gao Z, et al. Intestinal tract is an important organ for lowering serum uric acid in rats. PLoS One. 2017;12:e0190194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Plata C, Cruz C, Cervantes LG, Ramírez V. The gut microbiota and its relationship with chronic kidney disease. Int Urol Nephrol. 2019;12:2209‐2226. [DOI] [PubMed] [Google Scholar]
- 19. Vaziri ND, Liu SM, Lau WL, et al. High amylose resistant starch diet ameliorates oxidative stress, inflammation, and progression of chronic kidney disease. PLoS One. 2014;12:e114881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yi EY, Kim YJ. Xylitol inhibits and angiogenesis by suppressing the NF‐κB and Akt signaling pathways. Int J Oncol. 2013;1:315‐320. [DOI] [PubMed] [Google Scholar]
- 21. Riley P, Moore D, Ahmed F, Sharif MO, Worthington HV. Xylitol‐containing products for preventing dental caries in children and adults. Cochrane Database Syst Rev. 2015;3:CD010743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Singh S, Hornick D, Fedler J, et al. Randomized controlled study of aerosolized hypertonic xylitol versus hypertonic saline in hospitalized patients with pulmonary exacerbation of cystic fibrosis. J Cyst Fibros. 2020;1:108‐113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chukwuma CI, Islam S. Xylitol improves anti‐oxidative defense SYSTEM in serum, liver, heart, kidney and pancreas of normal and type 2 diabetes model of rats. Acta Pol Pharm. 2017;3:817‐826. [PubMed] [Google Scholar]
- 24. Morgado‐Pascual JL, Rayego‐Mateos S, Tejedor L, Suarez‐Alvarez B, Ruiz‐Ortega M. Bromodomain and Extraterminal proteins as novel epigenetic targets for renal diseases. Front Pharmacol. 2019;10:1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sakamaki JI, Wilkinson S, Hahn M, et al. Bromodomain protein BRD4 is a transcriptional repressor of autophagy and lysosomal function. Mol Cell. 2017;4:517‐532. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data supporting the findings of this study are available from the corresponding authors upon reasonable request.


