This study delves into the complex landscape of CRC metastasis, revealing distinct molecular profiles of metastatic seeds within primary tumors. The research identifies genetic alterations and communication capabilities driving metastasis, offering insights into clinical prognosis and potential anti-metastatic therapeutic targets for CRC patients.
Abstract
Metastasis of primary tumors remains a challenge for early diagnosis and prevention. The cellular properties and molecular drivers of metastatically competent clones within primary tumors remain unclear. Here, we generated 10–16 single cell–derived lines from each of three colorectal cancer (CRC) tumors to identify and characterize metastatic seeds. We found that intrinsic factors conferred clones with distinct metastatic potential and cellular communication capabilities, determining organ-specific metastasis. Poorly differentiated or highly metastatic clones, rather than drug-resistant clones, exhibited poor clinical prognostic impact. Personalized genetic alterations, instead of mutation burden, determined the occurrence of metastatic potential during clonal evolution. Additionally, we developed a gene signature for capturing metastatic potential of primary CRC tumors and demonstrated a strategy for identifying metastatic drivers using isogenic clones with distinct metastatic potential in primary tumors. This study provides insight into the origin and mechanisms of metastasis and will help develop potential anti-metastatic therapeutic targets for CRC patients.
Graphical Abstract
Introduction
Despite encouraging progress in cancer treatment, such as advances in surgical techniques and targeted therapies, metastatic disease remains the leading cause of cancer-associated deaths due to the lack of effective metastasis-specific therapies (Lambert et al., 2017; Turajlic and Swanton, 2016). Traditionally, metastasis is considered to be a product of evolution that appears in the late or end stages of tumor development (Birkbak and McGranahan, 2020). Emerging evidence suggests that the initiation of metastasis may be attributed to select subpopulations within primary tumors that possess unique characteristics. For example, in colorectal cancer (CRC) tumors, residual EMP1+ cells (Cañellas-Socias et al., 2022), and TP53 wild-type cancer cells with a fetal gene signature (Solé et al., 2022), as well as primary tumor clusters with plakoglobin-dependent intercellular adhesion in breast cancer (Aceto et al., 2014). The metastatic potential of cancer cells is an appropriate parameter for designing optimal strategies to prevent metastasis early and specifically targeting metastatic clones. However, it remains a methodological challenge to capture, identify, and characterize these metastatically competent clones within human primary tumors (Lawson et al., 2018; Turajlic and Swanton, 2016).
Currently, our understanding of the intrinsic properties of potential metastatic cells within primary tumors is limited. It is unclear whether specific mechanisms exist by which metastatic seeds within the primary tumors colonize specific organ soils, known as metastatic organotropism. The intrinsic molecular properties that predispose clones with the ability to adapt and colonize specific organ environments remain largely unexplored. Additionally, metastatic cells are characterized by stem cell traits and undergo epithelial–mesenchymal transition (EMT) (Brabletz et al., 2005; Cheung et al., 2020; Gkountela et al., 2019; Lambert and Weinberg, 2021), but it is uncertain to what extent differentiation programs predispose to metastatic potential in individual primary tumors (Brabletz, 2012; Lambert et al., 2017). There is also controversy surrounding whether metastases arise from primary tumor cells that are resistant to chemotherapy (Lambert et al., 2017; Oskarsson et al., 2014). Since each primary tumor consists of genetically and functionally diverse cancer cells (Greaves, 2015; Kreso et al., 2013; Marusyk et al., 2020; McGranahan and Swanton, 2015), it is essential to characterize the extent of inter- and intratumoral heterogeneity in the characteristics of metastatic cells. Therefore, the challenge is to move from observational studies toward deeper functional studies and to integrate the genotypes, phenotypes, and functions into a single cell (Lawson et al., 2018; Marusyk et al., 2020).
Questions regarding the evolution of clones with metastatic potential in human primary tumors, and whether there are metastasis-specific genes and genetic alterations, have long been debated and remain unanswered (Hunter et al., 2018; Oskarsson et al., 2014). As a result, the success of precision oncology, based on molecular-driven cancer treatment, has yet to be applied to guide therapies against metastasis. Large-scale cohort analysis has found little evidence of recurrent or universal driver mutations in specific “metastasis genes” (Birkbak and McGranahan, 2020; Brannon et al., 2014; Robinson et al., 2017; Zehir et al., 2017). This information implicates that metastatic evolution may depend on the heterogeneous progression of individual tumors (Kim et al., 2015). Comparative genetic studies of primary tumors and metastases have revealed certain alterations that are enriched at metastatic lesions (Armenia et al., 2018; Bertucci et al., 2019; Goswami et al., 2015; Turajlic et al., 2018; Xie et al., 2014; Yates et al., 2017). Nevertheless, these genetic differences between primary tumors and matched metastases do not accurately indicate metastatic drivers due to the continuous genomic evolution of metastasis. Additionally, bulk tumor analysis may potentially mask metastatic divergences, particularly when subclonal driver mutations are present at a low frequency. Therefore, the challenge remains to accurately decipher the natural occurrence of metastasis and explore specific genes and mutations that engender metastatic potential in human primary tumors.
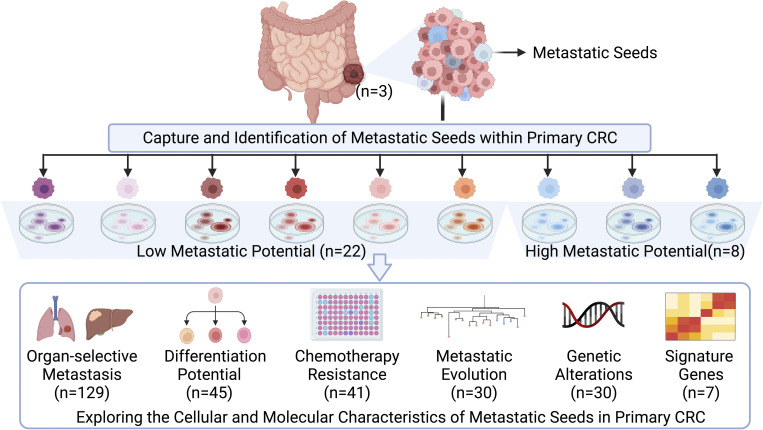
In this study, we captured, identified, and characterized metastatic seeds within primary CRC by generating 10–16 single cell–derived clonal cell lines from each tumor of three patients. We found that these metastatic seeds exhibit inter- and intratumoral heterogeneity, characterized by their organ-selective metastatic potential, differentiation potential, chemoresponse, and clonal evolutionary patterns. We analyzed that clonal cell lines with poor differentiation or high metastatic potential are significantly associated with clinical prognosis, rather than drug-resistant clones. We also investigated the molecular mechanisms behind metastatic organotropism and explored genetic alterations and signature genes associated with metastasis to capture the metastatic potential of primary CRC tumors. Our insights into the cellular and molecular characteristics of metastatic seeds within primary CRC tumors advance our multifaceted understanding of the natural occurrence and mechanisms of metastatic potential in primary tumors and provide potential therapeutic antimetastatic targets for CRC patients.
Results
Generating single cell–derived clonal cell lines from individuals
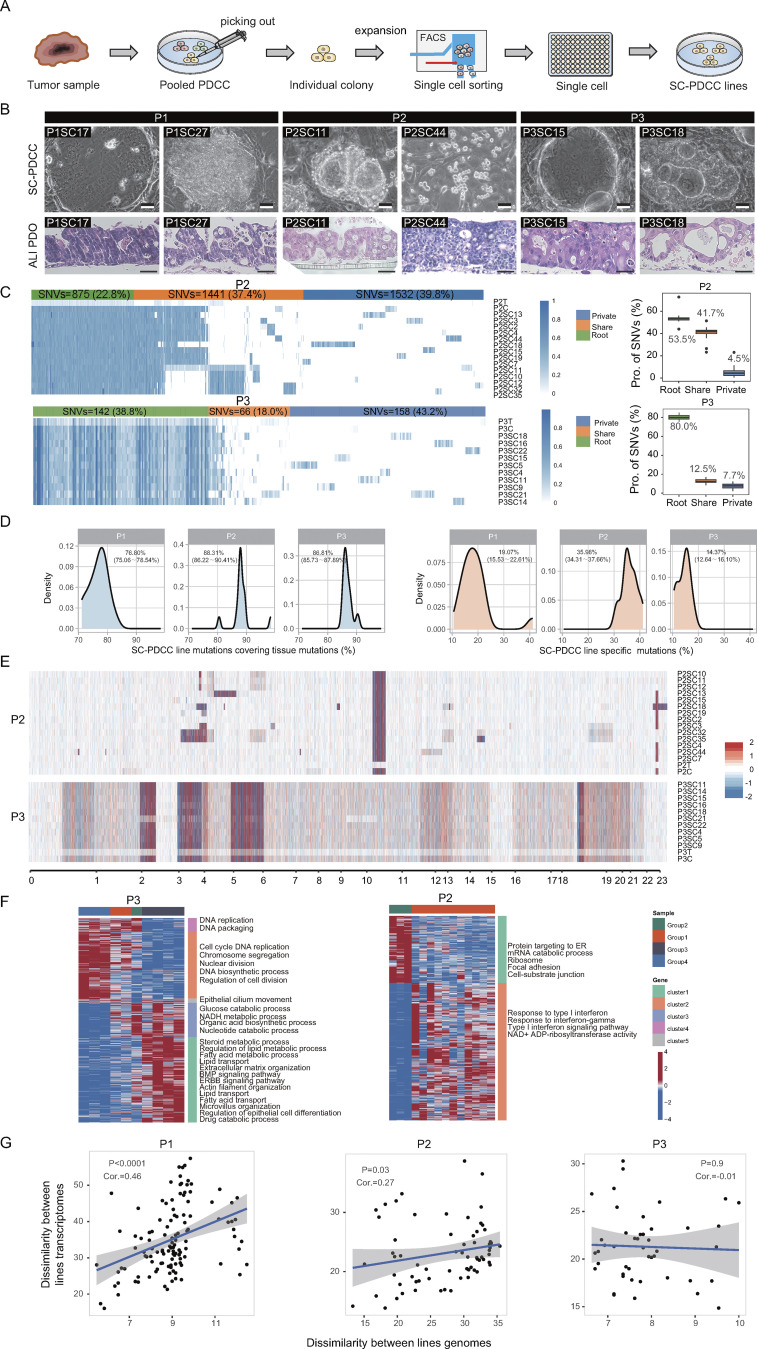
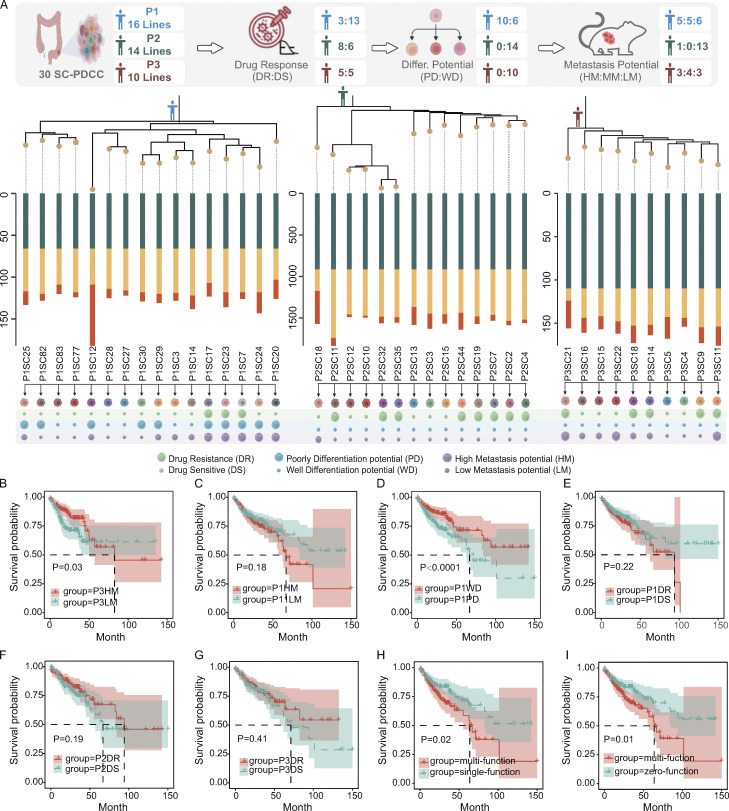
To investigate which cellular and molecular features confer metastasis potential to clones in the widely diverse clonal lineages within human primary CRC tumors, we cultured patient-derived cancer cells (PDCCs) from six patients (P1–P6; Table S1). We applied a previously established 2D and 3D model system for culturing PDCCs and air–liquid interface organoids (ALI PDOs) to capture inter- and intratumor heterogeneity of CRC tumors (Zhao et al., 2022). Briefly, each tumor tissue was minced and divided into three parts for clonal derivation, genomic analysis, and transcriptomic analysis, respectively. Primary cancer cells formed pooled colonies on the 3T3-J2 feeder layer. We established 10–16 single cell–derived clonal cell lines (SC-PDCC lines) from pooled colonies of each primary CRC tumor of three untreated patients without clinically overt metastatic lesions (P1, P2, and P3, see Materials and methods; Fig. S1 A). These 2D SC-PDCC lines from the same individual tumor showed heterogeneous morphologies and 3D ALI PDOs showed heterogeneous histological architecture (Fig. S1 B). The SC-PDCC lines represent 10–16 individual cancer cells within each patient’s primary tumor that should, in theory, allow us to study the cellular and molecular features that may contribute to metastasis potential.
Figure S1.
SC-PDCC lines recapitulate the intratumoral heterogeneity of the histology, genomic, and transcriptional landscape, related to Fig. 1. (A) Schematic procedure of establishing SC-PDCC lines from CRC patient. Colonies were generated after processing tissues, different morphological colonies were picked and expanded, then sorted using EPCAM+ to obtain single cell per well. SC-PDCC lines were established after expansion. (B) Representative bright field microscopy and H&E staining images of SC-PDCC colonies and matched 3D ALI PDOs from three patients. Scale bar, 50 µm. (C) In the left panel, a heatmap illustrating the allele frequency of SNVs in the parental tumor, pooled PDCCs, and SC-PDCC lines of P2 (up, n = 16) and P3 (down, n = 12). Variations are classified as: root (present in all samples), share (present in multiple samples but not all), and private (present in specific samples). In the right panel, the boxplot shows the distribution of each SNV types among samples, and the labeled numbers indicate the median proportion of mutations. (D) Distribution of mutations in SC-PDCC lines, including retained parental tumor mutations (left) and SC-PDCC line-specific mutations (right). (E) Heatmap shows CNAs in parental tumor, pooled PDCCs, and different SC-PDCC lines of P2 (up, n = 16) and P3 (down, n = 12). Red denotes copy number gains, and blue denotes copy number loss. (F) GOEA of consensus clustered genes in P2 (right, n = 14) and P3 (left, n = 10). Each column represents a SC-PDCC sample, and samples were grouped based on the top 1,000 highly variable genes. Rows represent DEGs (padj < 0.05 and |log2fold-change| > 0.5), which was the result of comparing each sample group with other samples. (G) Effects of mutations in exons on the transcriptome. Each data point represents the genomic dissimilarity (x-axis) and transcriptomic dissimilarity (y-axis) of a cell line pair. Consistency was assessed using the Spearman correlation coefficient.
Intratumoral molecular heterogeneity of single cell–derived clonal cell lines
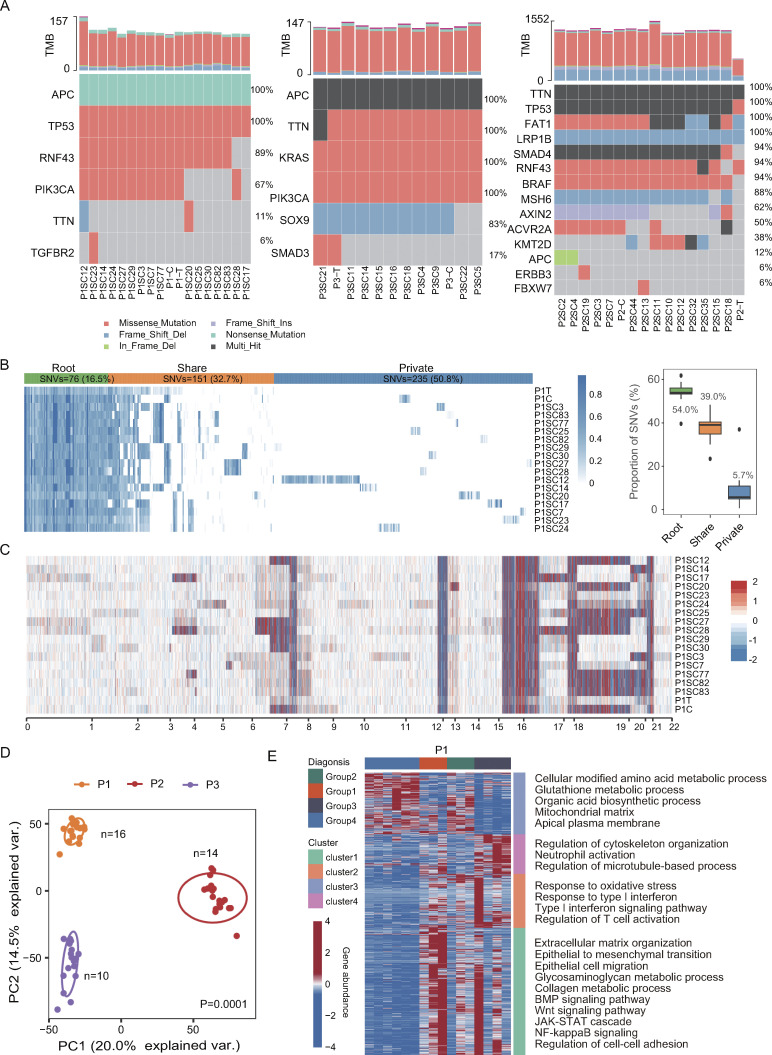
We verified whether SC-PDCC lines can capture the intratumor heterogeneity at molecular level. First, to assess the genomic diversification of SC-PDCC lines, we performed exome sequencing analysis on patient-matched samples from three CRC patients (P1, P2, and P3, Table S2), including three pooled PDCCs, 40 SC-PDCC lines, three primary tumor tissues, and three corresponding adjacent normal biopsies (as references to distinguish germline mutations). Analysis of somatic single nucleotide variants (SNVs) revealed that the majority of CRC-associated somatic mutations were presented in pooled PDCCs and SC-PDCC lines (Fig. 1 A). In these three patients, the root SNVs were 54.0%, 53.5%, and 80.0%; the shared SNVs were 39.0%, 41.7%, and 12.5%; and the private SNVs were 5.7%, 4.5%, and 7.7%, respectively (Fig. 1 B and Fig. S1 C). On average, 76.80%, 88.31%, and 86.81% of SNVs detected in parental tumors were retained in the SC-PDCC lines of P1, P2, and P3, respectively. However, 19.07%, 35.98%, and 14.37% of SNVs in the SC-PDCC lines of P1, P2, and P3, respectively, were undetectable in the corresponding parental tumors (Fig. S1 D). This may be due to the limited biomass of cells carrying these mutations in tissues, resulting in the failure of tissue-based analyses to detect these mutations. The majority of copy number alterations (CNAs) in parental tumors were well preserved in pooled PDCCs and SC-PDCC lines. However, distinct somatic mutations, amplifications, and deletions occurred in each SC-PDCC line (Fig. 1 C and Fig. S1 E), which represent intrinsic molecular features of each SC-PDCC line and their intratumoral heterogeneity.
Figure 1.
Establishment of SC-PDCCs with molecular heterogeneity. (A) Somatic mutations related to CRC were detected in all samples from P1, P2, and P3. C, pooled PDCCs; T, parental tumor; P1SC12, SC-PDCC lines #12 from P1, and so on. n = 46. (B) In the left panel, a heatmap illustrating the allele frequency of SNVs in the parental tumor, pooled PDCCs, and 16 SC-PDCC lines of P1. Variations are classified as root (present in all samples), share (present in multiple samples but not all), and private (present in specific samples). In the right panel, the boxplot shows the distribution of each SNV type among samples, and the labeled numbers indicate the median proportion of mutations. n = 18. (C) Heatmap shows CNAs in parental tumor, pooled PDCCs, and different SC-PDCC lines of P1. Red denotes copy number gains, and blue denotes copy number loss. n = 18. (D) PCA plot of transcriptome variation of SC-PDCC lines within the individual tumors. Different SC-PDCC lines in each patient are dispersed. Significance was determined by PERMANOVA test, P = 0.0001. P1, n = 16; P2, n = 14; P3, n = 10. (E) GOEA of consensus clustered genes. Each column represents a SC-PDCC sample of P1, and samples were grouped based on the top 1,000 highly variable genes. Rows represent DEGs (Padj < 0.05 and |log2fold-change| > 0.5), which was the result of comparing each sample group with other samples. n = 16.
Next, to determine the diversification of SC-PDCC lines on gene expression profiling, we performed RNA sequencing (RNA-seq) on patient-matched samples (Table S2). Principal component analysis (PCA) and hierarchical clustering showed that SC-PDCC lines derived from the same individuals displayed distinct subgroups and exhibited intratumor transcriptomic variations (Fig. 1, D and E; and Fig. S1 F). Analysis of RNA and exome sequencing data demonstrated a significant correlation between genomic and transcriptional heterogeneity in SC-PDCC lines from P1 and P2 (Fig. S1 G). However, this correlation was not observed in P3, possibly due to the relatively low degree of intratumoral heterogeneity in the SC-PDCC lines derived from P3 (Fig. 1 B and Fig. S1 C). These data demonstrate that we have captured a variety of SC-PDCC lines from the primary CRC tumors that exhibit both genomic and transcriptional heterogeneity, despite the fact that our culture conditions and expansion processes may favor certain cell types and/or may cause some changes in molecular program and cellular phenotype.
Identifying metastatic seeds within primary CRC tumors
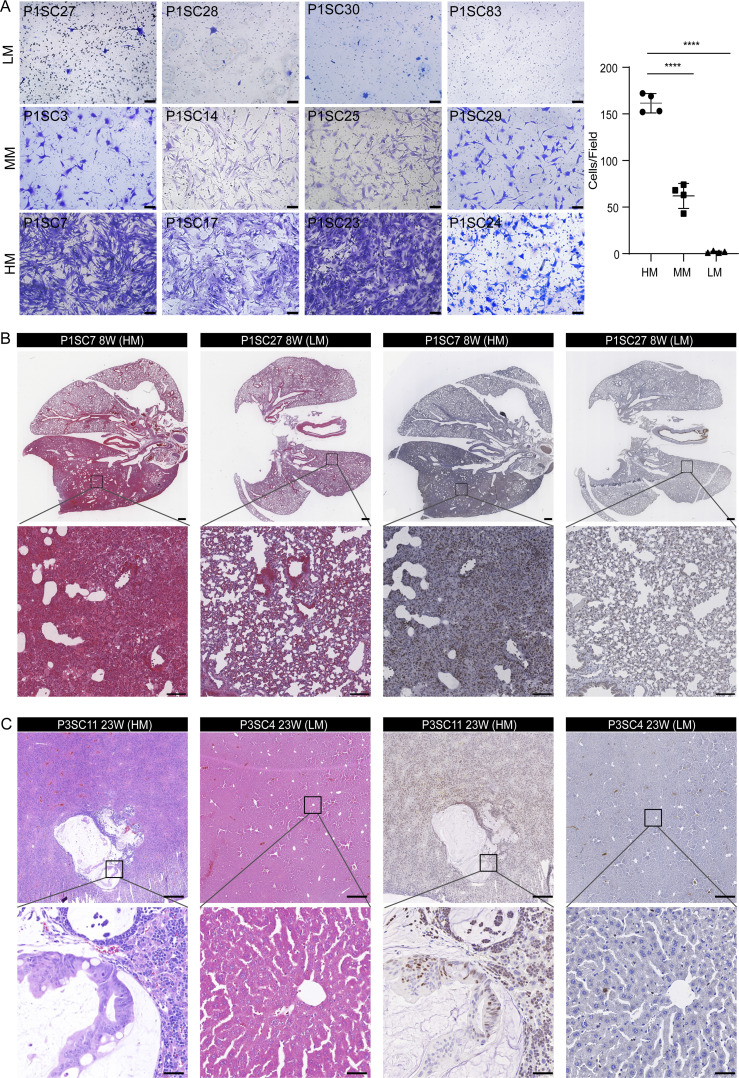
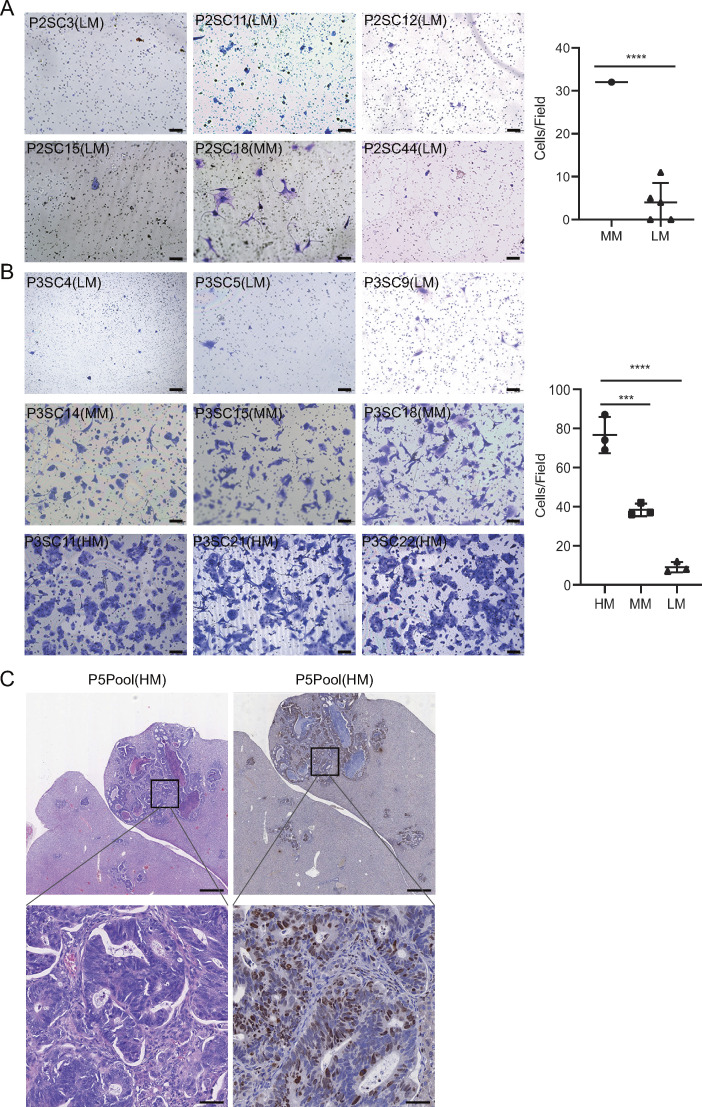
Given the high intratumor heterogeneity of primary CRC tumors at the molecular level (Punt et al., 2017), we investigated whether SC-PDCC lines have varying degrees of metastatic potential. We first measured the invasive capability of SC-PDCC lines using the Transwell cell invasion assay (Li and Hanahan, 2013). We found varying degrees of invasive potential among the SC-PDCC lines from P1, P2, and P3, with high (>75 cells/field), moderate (25–75 cells/field), and low (<25 cells/field) invasive ability. Among the SC-PDCC lines of P1, five, six, and five lines showed high, low, and intermediate invasive capability, respectively (Fig. 2 A and Table S2). In P2, all except one of the 14 SC-PDCC lines showed low invasive capability (Fig. S2 A and Table S2). In P3, three highly invasive, four moderately invasive, and three low invasive lines were obtained, respectively (Fig. S2 B and Table S2).
Figure 2.
Identification of SC-PDCC lines with distinct metastatic potential from the same primary CRC tumor. (A) In vitro invasion assay of 12 SC-PDCC lines in P1. Cells were seeded in a Matrigel-coated Transwell and were cultured in serum-free media. The bar graphs represent the relative numbers of invaded cells. The invaded cells were counted in five randomly chosen areas (repeat, n = 3). Error bars represent SD of the mean. ****, P < 0.0001; by two-tailed unpaired Student’s t test. n = 4 for each group. Scale bar, 100 µm. HM, high metastatic potential; MM, moderate metastatic potential; LM, low metastatic potential. (B) Representative H&E staining and KI67 staining images of lung metastases derived from P1 SC-PDCC lines with high and low metastatic potential. P1SC7, n = 8; P1SC27, n = 15. Scale bar: up, 500 µm; down, 100 µm. (C) Representative H&E staining and KI67 staining images of liver metastases derived from P3 SC-PDCC lines with high and low metastatic potential. P3SC11, n = 4; P3SC4, n = 9. Scale bar: up, 500 µm; down, 50 µm.
Figure S2.
Evaluation of metastatic potential of SC-PDCC lines and pooled PDCCs, related to Fig. 2. (A) In vitro invasion assay of six SC-PDCC lines in P2. The invaded cells were counted in five randomly chosen areas. Moderate metastatic potential (MM) compared with low metastatic potential (LM) group; ****, P < 0.0001; t test, two-tailed; error bars represent SD of the mean. Scale bar, 100 µm; n = 6. (B) In vitro invasion assay of nine SC-PDCC lines in P3. ***, P < 0.001; ****, P < 0.0001; t test, two-tailed. Error bars represent SD of the mean. Scale bar, 100 µm; n = 9. HM, high metastatic potential. (C) Representative H&E staining and KI67 staining images of liver metastases derived from pooled PDCC lines of P5 with diagnosed liver metastasis. Scale bar: up, 500 µm; down, 50 µm.
Next, we performed a tail vein injection assay for lung metastasis and an intrasplenic injection assay for liver metastasis using immunodeficient NSG (NOD.Cg-Prkdcscid Il2rgtm1) mice (Bouvet et al., 2006; Golovko et al., 2015; Khanna and Hunter, 2005; Sonoda et al., 2006) to evaluate the metastatic potential of the high and low invasive SC-PDCC lines from P1 and P3. We did not evaluate the metastatic potential in vivo of the P2 clonal cell lines because only one of them showed moderate invasive capability in vitro, whereas all other lines showed low invasive capability. After 8–13 wk, we observed a much higher rate of lung metastasis in mice injected with high invasive SC-PDCC lines of P1 (75–80%) than in mice injected with low invasive SC-PDCC lines of P1 (0–7%) in the tail vein injection mouse model (Fig. 2 B and Table S3). However, neither the high nor the low invasive SC-PDCC lines of P3 yielded lung metastasis even 16–20 wk after transplantation (Table S3). We observed that moderate and high invasive SC-PDCC lines from P3 yielded a much higher rate of liver metastasis (25–40%) than the low invasive SC-PDCC line (0%) 23–26 wk after intrasplenic injection (Fig. 2 C and Table S3). However, no liver metastasis was observed in mice even 16–32 wk after splenic injection of SC-PDCC lines of P1 (Table S3). KI67 staining patterns showed a high proportion of proliferating cancer cells in the metastases (Fig. 2, B and C). These results demonstrate a strong correlation between the invasive potential of SC-PDCC lines and their metastatic potential in vivo. Consequently, we classified all SC-PDCC lines into three categories based on their invasive ability: high (>75 cells/field), moderate (25–75 cells/field), and low (<25 cells/field) metastatic potential.
We then evaluated whether the metastasis potential of PDCCs in mice correlates with clinical metastasis in patients from whom they were derived. NSG mice were intrasplenically injected with pooled PDCCs derived from primary CRC of P1, P3, and P4 (CRC patients without diagnosed liver metastases at the time of sample collection), and of P5 and P6 (CRC patients with diagnosed liver metastasis at the time of sample collection). We observed liver metastasis in mice 15–32 wk after splenic injection of pooled PDCCs of P5 (metastasis rate 80%) and P6 (metastasis rate 100%), while no liver metastasis was observed from mice with transplantation of pooled PDCCs from P1, P3, and P4 (Fig. S2 C and Table S4). This result shows that liver metastasis potential of pooled PDCCs in mice correlates with clinical liver metastasis in patients.
Taken together, we have successfully captured and identified SC-PDCC lines with varying degrees of metastatic potential within primary CRC tumors. Interestingly, the high metastatic SC-PDCC lines (serve as metastatic seeds) derived from different patients exhibited organ-selective metastatic potential, colonizing specific organs, such as the lung or liver. Such clones, with intrinsically different metastatic potential and organotropsim, provide a valuable resource to understand the cellular and molecular features and mechanisms that confer metastatic potential to metastatic seeds within primary CRC tumors.
Genetic alterations of metastatic potential
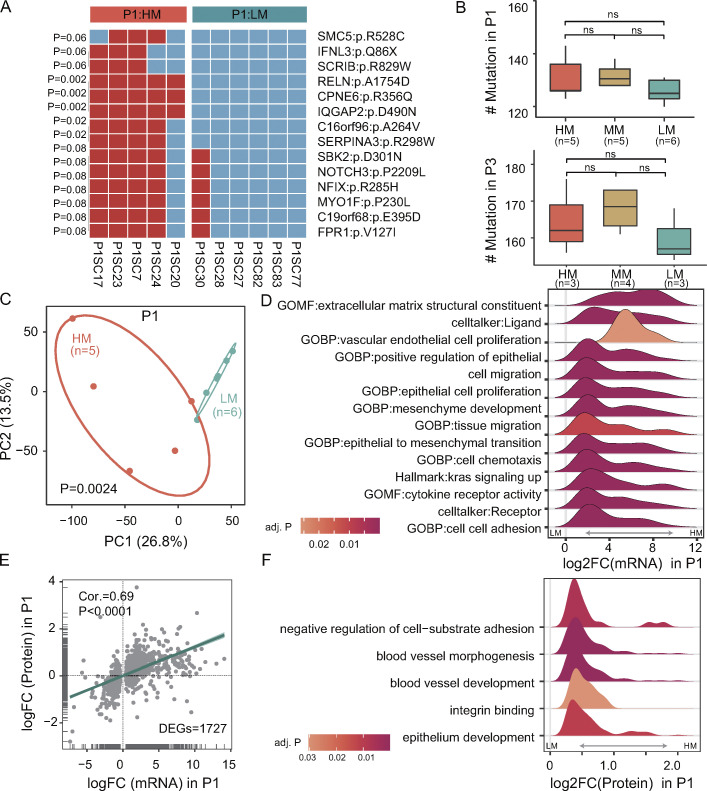
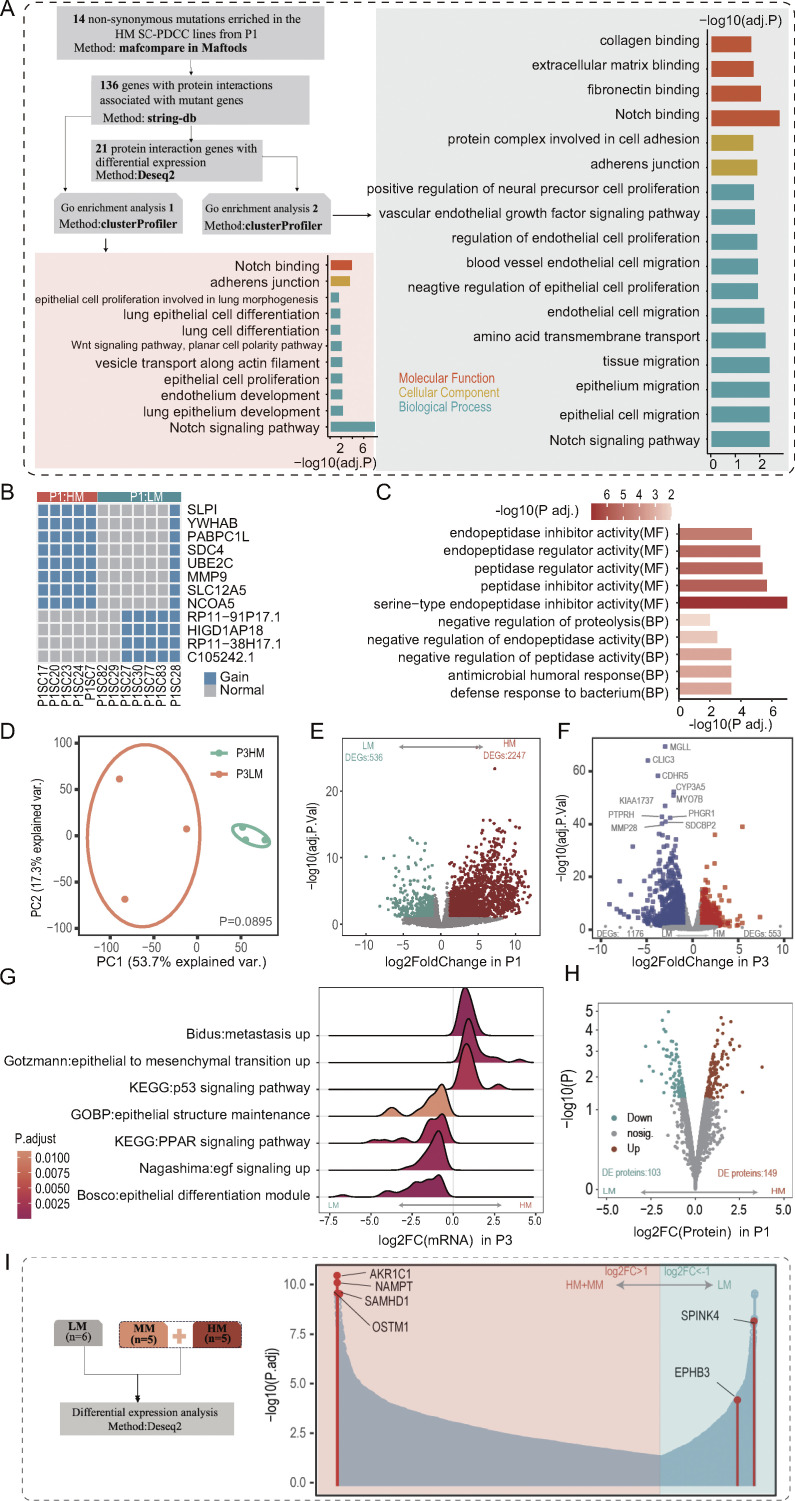
The existence of specific metastasis-driver alterations has long been a contentious and unresolved issue (Hunter et al., 2018; Oskarsson et al., 2014; Turajlic and Swanton, 2016). However, the genetic divergence found in SC-PDCC lines with distinct metastatic potential from the same individual may contribute to the metastatic potential, making them an ideal resource for exploring genetic alterations and genes associated with metastasis. Exome sequencing data of each SC-PDCC line of P1 revealed 222–261 somatic SNVs, with each line harboring 4–72 unique SNVs (Table S2 and Fig. 1 B). We identified 14 shared non-synonymous mutant genes in highly metastatic SC-PDCC lines from P1 (Fig. 3 A). Specifically, SCRIB, SERPINA3, NOTCH3, and FPR1 have been previously associated with CRC metastasis (Cao et al., 2018; Li et al., 2017; Shen et al., 2021; Sugiura et al., 2023). Interestingly, the protein interaction genes of these mutant genes are primarily involved in the development of lung epithelium and endothelium (Fig. S3 A), which is consistent with their potential for specific lung metastasis (Fig. 2 B and Table S3). Additionally, the protein-interacting genes that showed significant transcriptional alterations (Table S5) were found to be enriched in the pathway of migration and proliferation of epithelial and endothelial cells, as well as cell adhesion (Fig. S3 A). Additionally, SC-PDCC lines with high metastatic potential in P1 had a unique CNAs signature that enriched the copy number amplifications of specific genes (Fig. S3 B and Data S1), some of which (e.g., SLP1 [Wei et al., 2020], SERINC3 [Haan et al., 2014], TTPAL [Haan et al., 2014], PABPC1L [Wu et al., 2019], SDC4 [Jechorek et al., 2021], and UBE2C [Wang et al., 2017]), were previously reported as associate with metastasis, while other genes such as R3HDML, FITM2, and RIMS4 were newly found. The CNAs signature was particularly enriched with genes involved in serine-type endopeptidase inhibitor activity (Fig. S3 C).
Figure 3.
Molecular characteristics of metastatic SC-PDCC lines. (A) 14 non-synonymous mutations enriched in the high metastatic potential (HM) SC-PDCC lines of P1. Fisher test, two-tailed; HM, n = 5; LM, n = 6. (B) Comparison of the numbers of SNVs between low metastatic potential (LM), moderate metastatic potential (MM), and HM in P1 and P3. P > 0.1; the Mann–Whitney test, two-tailed, was used. For P1, HM, n = 5; LM, n = 6. For P3, HM, n = 3; LM, n = 3. (C) PCA showing the essential difference between HM (red, n = 5) and LM (blue, n = 6) SC-PDCC lines of P1. Significance determined by PERMANOVA test, P = 0.0024. (D) GSEA analysis of differential enrichment of molecular pathways at the mRNA level in the HM (n = 5) and LM (n = 6) SC-PDCC lines from P1. The gene sets were collected from the Molecular Signatures Database. (E) Correlation between mRNA and protein levels of 1,727 DEGs (P < 0.1) in HM (n = 4) and LM (n = 2) SC-PDCC lines from P1. Evaluated using Spearmans correlation coefficient. (F) GSEA analysis of pathways differentially enriched at the protein level in HM (n = 4) and LM (n = 2) SC-PDCC lines from P1. FC, fold change.
Figure S3.
Molecular divergence of metastatic SC-PDCC lines from individual tumors, related to Fig. 3. (A) 14 non-synonymous mutations were enriched in HM SC-PDCC lines of P1. GOEA of genes with protein interactions with these mutations, as well as signaling pathways enriched for DEGs associated with these mutant genes. (B) Specific amplified genes present in HM SC-PDCC lines from P1. Fisher’s test was used to analyze different CNAs; HM, n = 5; LM, n = 7. (C) GO terms enriched by the differential CNAs between HM SC-PDCC lines and LM SC-PDCC lines in P1. BP, biological process; MF, molecular function. (D) PCA showing the difference between HM SC-PDCC lines (n = 3) and LM SC-PDCC lines (n = 3) in P3. The P value was determined by PERMANOVA test, P = 0.089. (E and F) Volcano plot of genes that were differentially expressed between HM SC-PDCCs and LM SC-PDCCs in P1 (C, HM, n = 5, LM, n = 6) and P3 (D, HM, n = 3, LM, n = 3). (G) GSEA showing significantly enriched pathway in HM and LM SC-PDCC lines of P3, respectively, P values are shown. GOBP, GO terms of biological process. (H) Volcano plot showing the differentially expressed proteins between HM SC-PDCCs (n = 4) and LM SC-PDCCs (n = 2) in P1. (I) Six genes were selected for functional verification. Differential analysis was performed based on SC-PDCC lines from P1, the number (n) is indicated. HM, high metastatic potential; MM, moderate metastatic potential; LM, low metastatic potential.
Previous studies suggested that higher burden of somatic mutations and CNAs are usually characteristics of metastatic tumors compared with cohorts of primary tumors (Armenia et al., 2018; Birkbak and McGranahan, 2020; Robinson et al., 2017). We, therefore, compared the genetic features of SC-PDCC lines with high and low metastatic potential and found that in the same primary tumor (P1 and P3, Table S2), high metastatic SC-PDCC lines did not exhibit an increased somatic mutation burden compared with low metastatic SC-PDCC lines (Fig. 3 B). Taken together, these results suggest that, at least in some cases, personalized genetic mutations and CNAs, rather than mutational burden, determine the occurrence of metastatic potential during clonal evolution. The use of SC-PDCC lines showed to be a valuable resource for identifying newly metastasis-associated genetic alterations and investigating the genetic mechanisms underlying metastasis.
Molecular properties of metastatic potential
We next assessed whether metastatic seeds have unique molecular features at the transcriptional and protein levels that endow them with metastatic potential in human primary tumors. PCA plots clearly separated high and low metastatic SC-PDCC lines from P1 and P3 into two groups, respectively (Fig. 3 C and Fig. S3 D). We identified 2,783 differentially expressed genes (DEGs) between high and low metastatic SC-PDCC lines of P1, including 2,247 upregulated genes and 536 downregulated genes in high metastatic SC-PDCC lines (Fig. S3 E). Gene set enrichment analysis (GSEA) identified significant enrichment of the following biological processes in high metastatic SC-PDCC lines of P1: EMT, cell migration, tissue migration, ligand, receptor, vascular endothelial cell (EC) proliferation, epithelial cell proliferation, mesenchyme development, cell chemotaxis, KRAS signaling, cell adhesion, and extracellular matrix structural constituent (Fig. 3 D). In high metastatic SC-PDCC lines of P3, we observed 553 upregulated and 1,176 downregulated genes compared with low metastatic SC-PDCC lines (Fig. S3 F). GSEA analysis revealed significant enrichments in EMT, metastasis, and P53 signaling pathways in the high metastatic SC-PDCC lines of P3. In contrast, GSEA analysis showed significant enrichments in epithelial structure maintenance, peroxisome proliferator-activated receptor signaling pathway, epidermal growth factor (EGF) signaling, and the epithelial differentiation module in the low metastatic SC-PDCC lines of P3 (Fig. S3 G).
We performed a global proteomics analysis of P1 SC-PDCC lines using isobaric tandem mass tags (TMT) to validate the protein-level expression of genes related to metastasis (Table S2). We investigated whether mRNA with differential expression trends (P < 0.1) in SC-PDCC lines with high and low metastatic potential of P1 also exhibited consistent changes at the protein level. The analysis revealed a significantly positive correlation (Fig. 3 E). We identified 149 upregulated proteins and 103 downregulated proteins in the P1 high metastatic SC-PDCC lines compared to the P1 low metastatic SC-PDCC lines (Fig. S3 H). GSEA indicated significant enrichment of pathways associated with angiogenesis, integrin binding, epithelial development, and negative regulation of cell-substrate adhesion in high metastatic SC-PDCC lines of P1 (Fig. 3 F). These data show that high metastatic SC-PDCC lines have a unique gene expression profile characterized by pathways related to metastasis.
The metastatic signature evaluates the metastatic potential of primary CRC tumors
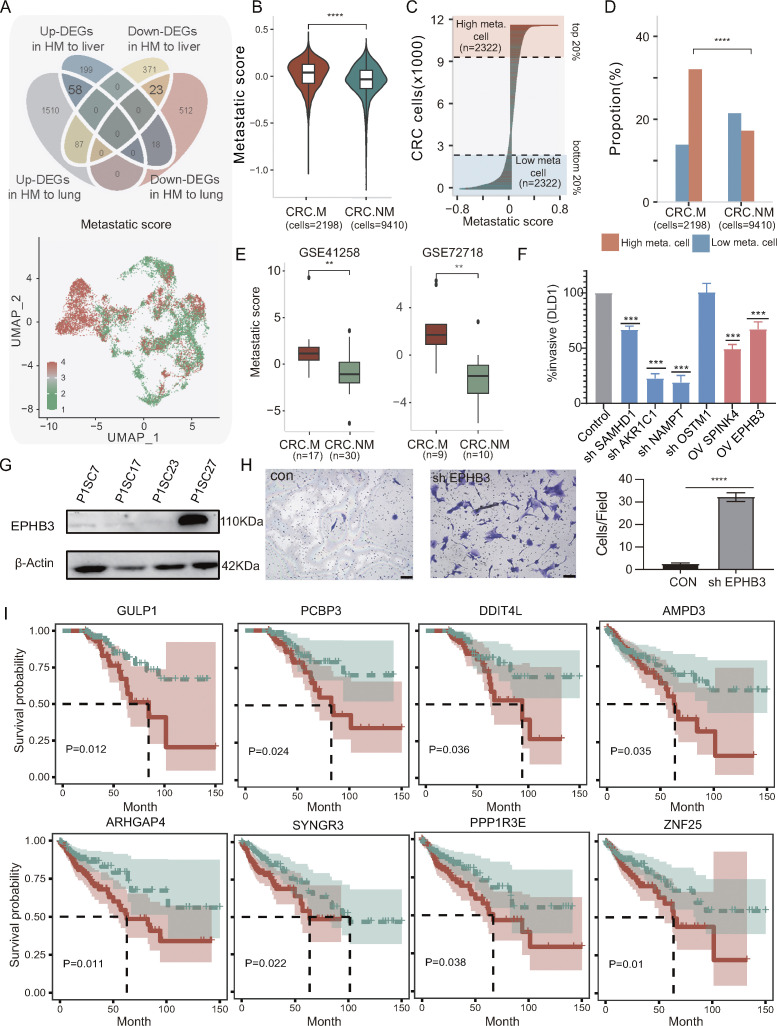
We then investigated whether a metastatic signature could be identified from these SC-PDCC lines with distinct metastatic potential to capture the metastatic potential of primary CRC tumors. To confirm the signature’s validity in vivo, only SC-PDCC lines validated by the tail vein injection mouse model were analyzed. By analyzing the overlapping DEGs between high and low metastatic SC-PDCC lines from P1 and P3 (Table S2), we identified a metastatic gene signature consisting of 58 upregulated genes and 23 downregulated genes that were shared among high metastatic SC-PDCC lines of both patients (Fig. 4 A and Data S2). Subsequently, we evaluated this metastatic signature in cancer cells using published single-cell RNA-seq (scRNA-seq) data (Xu et al., 2022) obtained from seven primary CRC tumors without observed metastasis (CRC.NM) and six primary CRC tumors with preoperative or intraoperative metastasis (CRC.M) (Fig. S4, A and B). Uniform Approximation and Projection (UMAP) plots demonstrated consistent distribution of upregulated and downregulated gene signatures in 11,608 cancer cells (Fig. 4 A). The metastatic signature was significantly higher in CRC.M cells than CRC.NM cells (P < 0.0001, Wilcoxon test) (Fig. 4 B). High-scoring cells were significantly enriched in CRC.M (P < 0.0001, Fisher test) (Fig. 4, C and D; and Table S6). Furthermore, our analysis of publicly available bulk RNA-seq data (GSE41258 and GSE72718) confirmed a significantly higher signature in primary CRC with metastasis than in those without metastasis (P < 0.05, Wilcoxon test) (Fig. 4 E). The findings suggest the promising utility of this gene signature in evaluating the metastatic potential of primary CRC tumors.
Figure 4.
Identification of metastatic signature and driver genes in primary CRC tumors. (A) Top: Venn diagram illustrating the metastatic signature derived from the overlapping sets of DEGs between high and low metastatic SC-PDCC lines from P1 and P3. For P1, high metastasis, n = 2; low metastasis, n = 2; for P3, high metastasis, n = 2; low metastasis, n = 1. Bottom: Distribution of high metastatic potential (HM) and low metastatic potential (LM) signature in cancer cells. Each dot represents a cell (Xu et al. [2022] dataset PRJNA748525). The HM score and LM signature were calculated based on shared significantly upregulated and downregulated genes in high metastatic SC-PDCC lines from P1 and P3. (B) Comparison of the difference in metastatic score between the CRC.M and CRC.NM groups (from Xu et al. [2022] dataset PRJNA748525). Metastatic score was calculated by subtracting the LM signature from the HM signature. The P value was measured by Wilcoxon test; the number (n) is indicated. (C) Distribution of metastatic score among cells; the number (n) is indicated. (D) Proportion comparison of different cell sources between risk groups. P values were calculated by Chisq test. ****, P < 0.0001. (E) Comparison of metastatic scores between primary CRC with and without liver metastasis. P value calculated by Wilcoxon test, with the number of samples (n) indicated. Data obtained from GSE41258 and GSE72718 datasets. **, P < 0.01. (F) Quantification of invasive assays following knockdown or overexpression of target gene in DLD1, respectively. Knockdown assays were performed using two independent shRNAs per gene (repeat, n = 3). Differences in invasion phenotype relative to control shRNAs (control) were significant by two-tailed t test, error bars show SD across three reduplicates, ***, P < 0.001. (G) Western blot analysis showing elevated expression of EPHB3 in LM SC-PDCC line (P1SC27), repeat, n = 3. (H) Knockdown of EPHB3 increases invasion of LM SC-PDCCs. The invaded cells were counted in five randomly chosen areas (n = 3; ****, P < 0.0001). Scale bar, 50 µm. (I) The Kaplan–Meier curves of patient outcomes were plotted for the TCGA-COAD cohort (cases = 458) based on indicated genes. The red and blue lines represent patients with upregulated and downregulated genes in HM SC-PDCC lines, respectively. The significance of differences between the two groups was assessed using a Cox test. Dotted line represents median survival. The plus signs represent the censored cases. Source data are available for this figure: SourceData F4.
Figure S4.
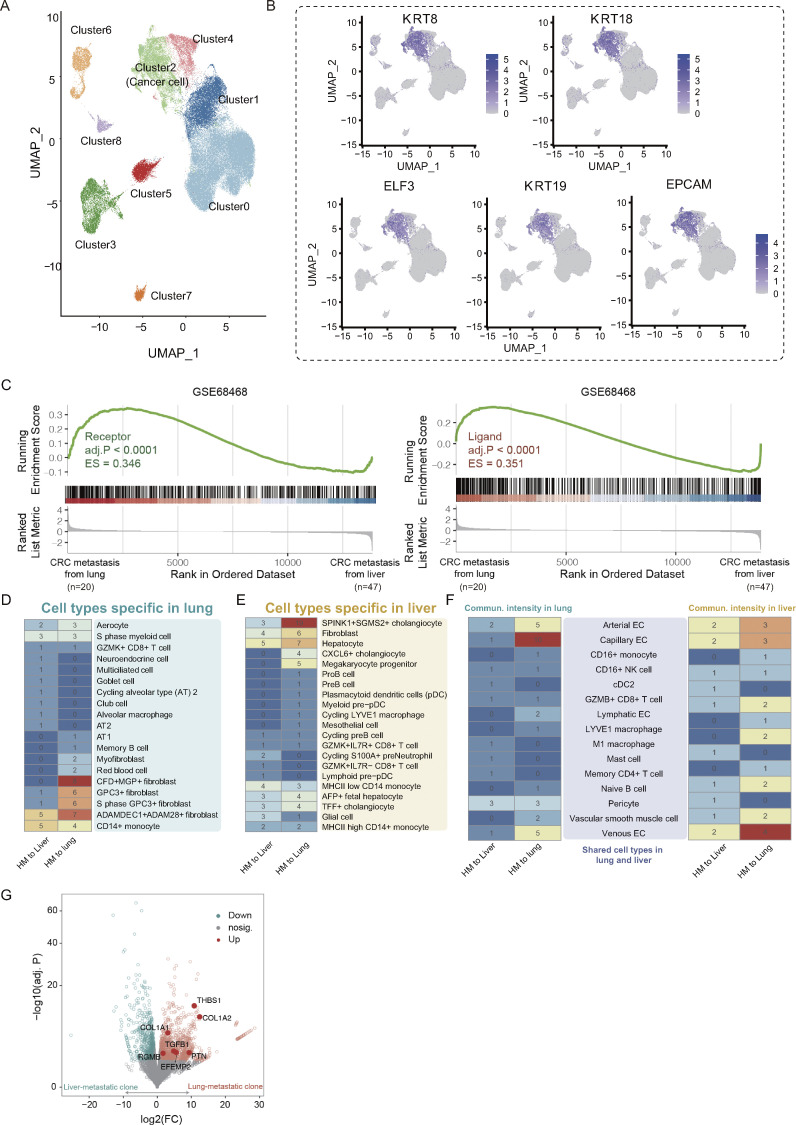
Analysis of primary CRC tumor scRNA-seq data and cellular communication capacity of SC-PDCC lines, related to Figs. 4 and 5. (A) An UMAP plot shows all cells from the public dataset PRJNA748525 (Xu et al., 2022), which retained a total of 87,717 cells after quality control and filtration. Cluster 2 was identified as a cluster of cancer cells based on the expression of five typical cancer cell markers. (B) The expression of indicated cancer cell markers in the cells of the public dataset PRJNA748525 (Xu et al., 2022). (C) GSEA analysis of publicly available data (the GSE68468 dataset) revealed a higher expression of receptors and ligands in CRC metastatic tissues from the lung (n = 20) compared to CRC metastatic tissues from the liver (n = 47). (D) Analysis of communication strength between P1 lung-metastatic lines and P3 liver-metastatic lines with lung-specific cells. (E) Analysis of communication strength between P1 lung-metastatic lines and P3 liver-metastatic lines with liver-specific cells. (F) Analysis of communication strength between P1 lung-metastatic lines and P3 liver-metastatic lines with shared cells for liver and lung. The numbers represent the intensity of cellular communication, determined by the number of receptor-ligand pairs. (G) Volcano plot of genes that were differentially expressed between lung-metastatic lines in P1(n = 2) and liver-metastatic lines in P3 (n = 2). P value < 0.05, |log2(fold-change)| > 1. HM, high metastatic potential.
Identifying metastatic drivers using isogenic SC-PDCC lines with distinct metastatic potential
Deciphering the driver genes that confer the metastatic potential of cancer cells is challenging. This is because genetic differences between primary tumors and metastases do not necessarily indicate metastatic drivers, and subclonal driver genes at low frequencies may be masked by bulk tumor analysis. To address this challenge, we provided a demonstration of the feasibility of identifying metastatic drivers at the single-cell level using SC-PDCC lines with distinct metastatic potential within the primary tumor of the same patient origin. Within an individual's primary tumor, clones with high and low metastatic potential share similar genetic backgrounds. However, their genetic divergences can provide valuable information for exploring metastatic driver genes. We selected the top four upregulated genes (AKR1C1, NAMPT, SAMHD1, and OSTM1) and two downregulated genes (SPINK4 and EPHB3) in the high and moderate metastatic potential SC-PDCC lines of P1 (Fig. S3 I and Table S2), and performed functional verification in the colorectal cell line DLD1. We found that knocking down SAMHD1, AKR1C1, and NAMPT or overexpressing SPINK4 and EPHB3 significantly reduced invasive capability as tested by the Transwell cell invasion assay (Fig. 4 F). However, changing the expression of OSTM1 had no effect on the metastatic potential. In addition, previous studies using colorectal cell lines have shown a suppressive role for EPHB3 in CRC metastasis (Chiu et al., 2009). Western blot analysis clearly indicates that the EPHB3 protein was highly expressed in the low metastatic SC-PDCC line (P1SC27) but not in the three high metastatic SC-PDCC lines (Fig. 4 G). We then verified the function of EPHB3 in this low metastatic SC-PDCC line and found that knocking down EPHB3 significantly enhanced its metastatic potential (Fig. 4 H). Moreover, we also found several new potential metastasis-associated genes (i.e., GULP1, PCBP3, DDIT4L, AMPD3, ARHGAP4, SYNGR3, PPP1R3E, and ZNF25) from the upregulated and downregulated genes in the high metastatic potential lines of P1. These genes have a significant impact on poor clinical prognosis (Fig. 4 I). In summary, we innovatively explored metastatic driver genes from the intrinsically distinct metastatic potential of single cell–derived lines within human primary CRC. This research provides valuable resources and information for understanding the origin and mechanism of metastasis and will help develop antimetastatic targets within primary CRC tumors.
Intrinsic cellular communication capabilities endow metastatic seeds with soil preselection
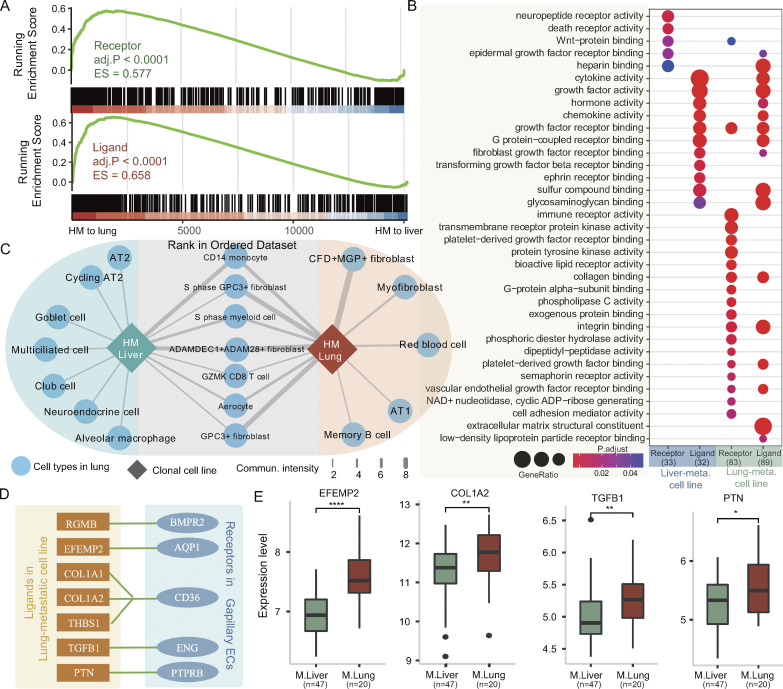
Elucidating the cellular and molecular factors of metastatic organotropism may help identify therapeutic targets for organ-specific metastasis. However, our understanding of the mechanisms underlying metastatic organotropism is still limited. To investigate this, we analyzed ligand–receptor mediated multicellular signaling (Ramilowski et al., 2016) using RNA-seq data from P1 high metastatic SC-PDCC lines and P3 high metastatic SC-PDCC lines (Tables S2 and S3). Our analysis included 708 ligands, 691 receptors, and 2,557 ligand–receptor pairs (Ramilowski et al., 2015). GSEA analysis revealed that P1 high metastatic SC-PDCC lines with a specific inclination for lung metastasis (lung-metastatic lines, SC-PDCC 7# and 23#) showed significantly higher expression of both receptor and ligand genes than P3 high metastatic SC-PDCC lines with a specific inclination for liver metastasis (liver-metastatic lines, SC-PDCC 11# and 22#) (Fig. 5 A). Our findings were further supported by publicly available data (GSE68468) (Fig. S4 C), which revealed significantly increased expression of receptor and ligand genes in lung metastases compared with liver metastases in CRC patients.
Figure 5.
Intrinsic cellular properties and communication capabilities endow metastatic seeds with soil preselection. (A) GSEA shows higher expression of receptor and ligand genes in lung-metastatic lines (n = 2) compared with liver-metastatic lines (n = 2). ES, enrichment score. (B) Functional enrichment analysis of receptor and ligand genes in liver-metastatic and lung-metastatic lines. (C) The thickness of the lines represents the intensity of cellular communication between P1 lung-metastatic lines and P3 liver-metastatic lines with specific cell types in human lung tissue. (D) Lung-metastatic lines exhibited significant overexpression of ligand genes whose corresponding receptors were expressed on the cell surface of capillary ECs. (E) Comparison of ligand genes between CRC lung- and liver-metastatic tissues. P values were calculated using the R package limma. The y-axis represents the signal intensity after log2 RNA signal transformation. M.lung, lung metastases from CRC; M.liver, liver metastases from CRC. *, P < 0.05; **, P < 0.01; ****, P < 0.0001.
Functional enrichment analysis of receptors and ligands expressed in lung-metastatic lines and liver-metastatic lines revealed that although they have distinct gene expression profiles, there was a high degree of sharing of pathways involved in their ligand genes, whereas the pathways involved in their receptor genes showed significant differences (Fig. 5 B). Both kinds of lines were significantly enriched ligand genes involved in pathways associated with fibroblast growth factor receptor binding, chemokine activity, growth factor receptor binding, glycosaminoglycan binding, G protein−coupled receptor binding, and heparin binding, suggesting an implicated mechanism in the general metastasis process (Fig. 5 B). In contrast, integrin binding, immune receptor activity, cell adhesion mediator activity, and extracellular matrix structural constituent were significantly enriched in lung-metastatic lines, but not in liver-metastatic lines, suggesting that a specific molecular mechanism for lung tropism exists in the metastatic cells of P1 (Fig. 5 B).
To further analyze the genetic basis of metastatic organotropism in these clonal cell lines, we constructed a map of cell–cell communication using RNA-seq data of SC-PDCC lines from P1 and P3 (Tables S2 and S3), as well as scRNA-seq data of 64 primary cell types in the human lung and 73 primary cell types in the human liver from the DISCO database (Li et al., 2022). We first quantified the strength of cellular communication by analyzing the ligand–receptor binding potential of each SC-PDCC line with cell types in the human lung and liver. Interestingly, we found that P1 lung-metastatic lines had a higher communication potential than P3 liver-metastatic lines (Fig. S4, D–F). Specifically, lung-metastatic lines communicate better with cell types in lung tissue than liver-metastatic lines (Fig. 5 C and Fig. S4, D–F). Lung-metastatic lines show high potential for communication with fibroblasts that are specific to the lung (CFD+MGP+ fibroblast, myofibroblast, ADAMDEC1+ADAM28+ fibroblast, GPC3+ fibroblast, S phase GPC3+ fibroblast), as well as with vascular-associated cells present in both liver and lung tissues (capillary ECs, arterial ECs, and venous ECs). Compared with liver-metastatic lines, lung-metastatic lines show significantly higher expression of some ligand genes, such as RGMB, EFEMP2, COL1A1, COL1A2, THBS1, TGFB1, and PTN (Fig. S4 G), all of which can activate the receptors BMPR2, AQP1, CD36, ENG, and PTPRB expressed on the cell surface of capillary ECs (Fig. 5 D). Moreover, through analysis of a publicly available dataset (GSE68468), we have validated that EFEMP2, COL1A2, TGFB1, and PTN, among the ligand genes, show significantly higher expression in CRC lung metastases compared with CRC liver metastases (Fig. 5 E). These mechanisms may be required for P1 lung metastases because, unlike the fenestrated endothelial layer of the liver sinusoid, the endothelial layer in the lung has tight junctions between ECs and an intact basement membrane. P1 metastatic seeds therefore need to interact more with capillary ECs to facilitate extravasation. Notably, CD36 on capillary ECs can bind to the ligands COL1A1, COL1A2, and THBS1 of lung-metastatic lines. It has been reported that low expression of CD36 on vascular ECs can inhibit angiogenesis (Bou Khzam et al., 2020). Therefore, the activation of CD36 on vascular ECs by ligands of P1 metastatic seeds may stimulate angiogenesis, aiding in metastatic colonization in the lung. The specific molecular mechanisms involved need to be confirmed by further experimental validation.
Taken together, our findings suggest that unique intrinsic molecular properties, as well as specific and robust cellular communication capabilities, confer the potential for metastatic seeds in P1 and P3 primary CRC tumors to metastasize in their preselected organ soil.
Distinct differentiation potential of metastatic seeds
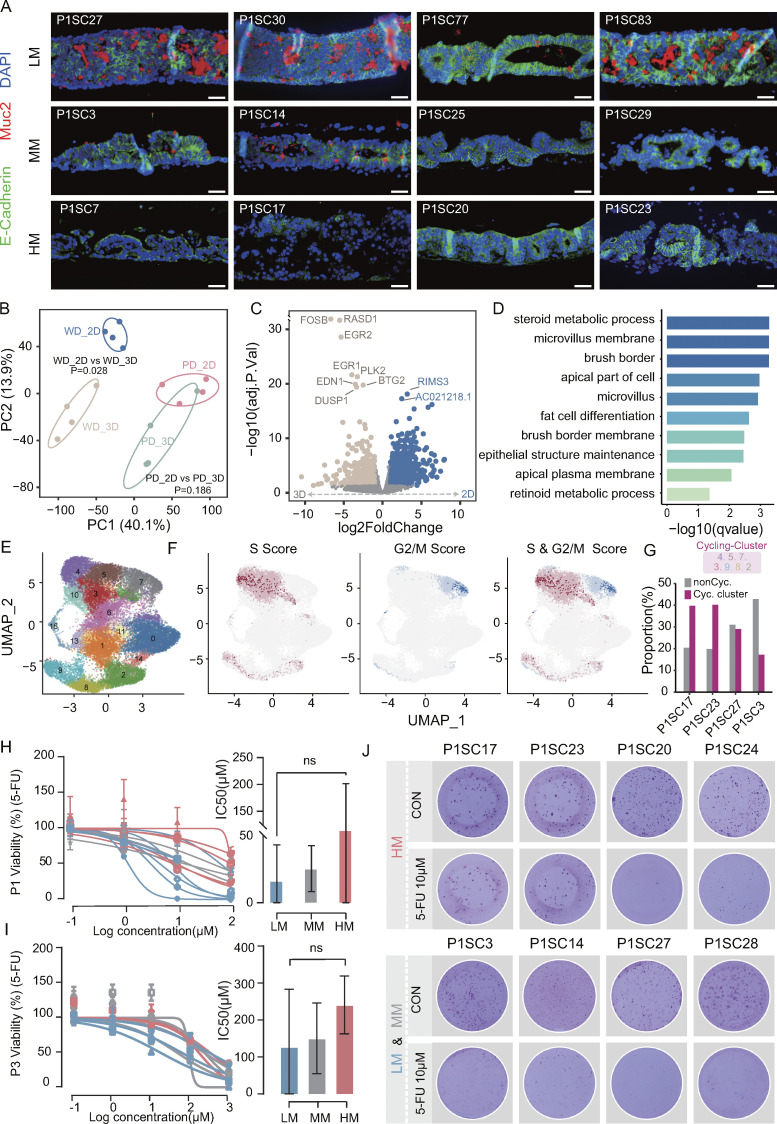
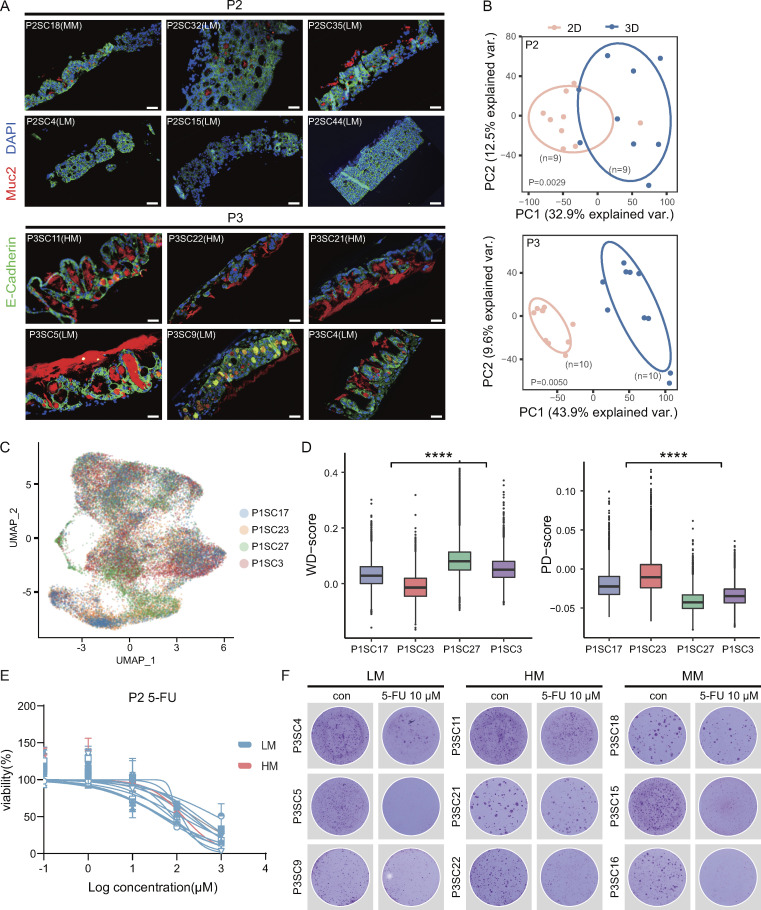
The differentiation status of tumor cells has been a major aspect of histopathological grading, and poorly differentiated clusters are currently considered a major adverse prognostic factor in CRC (Barresi et al., 2015; Jögi et al., 2012). Previous studies suggested that metastatic cancer cells of CRC have stem cell characteristics (de Sousa e Melo et al., 2017; Dieter et al., 2011); however, it remains unclear to what extent the differentiation program predisposes to metastatic potential (Brabletz, 2012; Lambert et al., 2017). To explore this issue, we evaluated the differentiation capability of SC-PDCC lines with distinct metastatic potential using 3D ALI PDOs. We classified the phenotypes as either well-differentiated or poorly differentiated by combining histopathological evaluation with MUC2 (a goblet cell–specific marker to indicate the differentiation status) positivity or negativity (Fig. 6 A and Fig. S5 A; and Table S2). The results showed that high metastatic SC-PDCC lines showed a poorly differentiated phenotype in P1, while low and moderate metastatic lines exhibited both well and poorly differentiated phenotypes (Fig. 6 A and Table S2). Moderate and low metastatic SC-PDCC lines in P2 had a well-differentiated phenotype, and all SC-PDCC lines in P3 exhibited well-differentiation potential regardless of metastatic potential (Fig. S5 A and Table S2).
Figure 6.
Differentiation potential and drug response of SC-PDCC lines with different metastatic potential. (A) MUC2 (red) and E-cadherin (green) staining of ALI PDOs derived from different SC-PDCC lines with different metastatic potential in P1. Scale bar, 50 µm. HM, high metastatic potential; MM, moderate metastatic potential; LM, low metastatic potential; HM, n = 4; MM, n = 4; LM, n = 4. (B) PCA showing high similarity between the SC-PDCC lines (PD_2D) and poorly differentiated ALI PDOs (PD_3D) (P = 0.186), and great difference between the SC-PDCC lines (WD_2D) and moderately differentiated ALI PDOs (WD_3D) (P = 0.028). Significance was determined by PERMANOVA test (all SC-PDCC lines from P1; n = 4 for each group). WD, well differentiation; PD, poor differentiation. (C) Volcano plot of genes that were differentially expressed between WD SC-PDCCs and their derived ALI PDOs. DEGs were determined based on adjusted (P < 0.05) and log2 fold-change (absolute value >1). n = 4 for both the WD_2D group and the WD_3D group. (D) GOEA of DEGs between WD SC-PDCCs and their derived ALI PDOs in P1. (E) UMAP plot of single-cell RNA expression from two HM SC-PDCC lines (P1SC17 and P1SC23) and one MM SC-PDCC lines (P1SC3) and one LM SC-PDCC lines (P1SC27). Color code for cell type assignment. (F) Cell cycle scores for each cell based on the expression of S and G2/M phase genes. (G) Relative fractions of cycling and noncycling cells across four SC-PDCC lines in P1. (H and I) Dose-response curves of SC-PDCC lines with different metastatic potential from P1 and P3 after 6 days treatment with 5-FU. Error bars represent SEM of three independent experiments; HM, n = 5; MM, n = 3; LM, n = 6. (J) The drug response indicated by SC-PDCC lines from P1 treated with 10 µM 5-FU. Cells were fixed, rhodamine stained, and photographed after 6 days of treatment. HM, n = 4; MM, n = 2; LM, n = 2. Three technical replicates for each SC-PDCC line.
Figure S5.
Assessment of differentiation potential and drug response of SC-PDCC lines with different metastatic potential, related to Fig. 6. (A) MUC2 (red) and E-cadherin (green) staining of ALI PDOs derived from different SC-PDCC lines in P2 (n = 6) and P3 (n = 6), respectively. Scale bar, 50 µm. (B) PCA showing large difference between the SC-PDCC lines (2D) and corresponding ALI PDOs (3D) in P2 (n = 9) and P3 (n = 9). Significance was determined by PERMANOVA test; P2, P = 0.0029; P3, P = 0.005. (C) UMAP plot of single-cell RNA expression from two PD SC-PDCC lines (n = 2; P1SC17 and P1SC23) and two MD SC-PDCC lines (n = 2; P1SC3 and P1SC27) in P1. Color code for different samples. MD, moderate differentiation; PD, poor differentiation. (D) Differentiation score and undifferentiated score of single cells in four SC-PDCC lines. These scores were evaluated by the expression of DEGs from bulk RNA data of corresponding WD SC-PDCCs and PD SC-PDCCs. Two WD SC-PDCC lines (P1SC3 and P1SC27) showed higher WD scores (left), and two PD SC-PDCC lines (P1SC17 and P1SC23) showed higher PD scores (right). The Mann–Whitney test, two-tailed, was used. ****, P < 0.0001. (E) Dose-response curves of SC-PDCC lines (n = 14) with different metastatic potential from P2 after 6 days treatment with 5-FU (repeat, n = 3). Error bars represent the SEM of three independent experiments. (F) The functional phenotypes of drug response indicated by SC-PDCC lines from P3 treated with 10 µM 5-FU (n = 9). Cells were fixed, rhodamine stained, and photographed after 6 days of treatment. Three technical replicates for each SC-PDCC line. HM, high metastatic potential; MM, moderate metastatic potential; LM, low metastatic potential.
We analyzed gene expression levels to assess differentiation status. Poorly differentiated 3D ALI PDO samples clustered with their original 2D SC-PDCC lines in P1, while well-differentiated groups for P1, P2, and P3 had distinct clusters for 3D and 2D samples (Fig. 6 B and Fig. S5 B). This suggests that the poorly differentiated group has a similar gene expression profile to their 2D SC-PDCC lines, while the well-differentiated group has a distinct profile. DEGs in the well-differentiated group were enriched in signaling pathways related to cell differentiation (Fig. 6, C and D).
Finally, we evaluated the differentiation status by analyzing cell cycle phases at the single-cell level using scRNA-seq on 30,304 cells isolated from poorly differentiated ALI PDOs (P1SC17 and P1SC23) and well-differentiated ALI PDOs (P1SC27 and P1SC3) (Fig. 6 E and Fig. S5 C). We identified seven clusters representing cycling cells and eight clusters containing noncycling cells (Fig. 6 E and Fig. S5 C). The proportion of cycling cells was higher in the poorly differentiated ALI PDOs, while the proportion of noncycling cells was higher in the well-differentiated ALI PDOs (Fig. 6, F and G). The differentiation score of well-differentiated ALI PDOs was also higher than that of poorly differentiated ALI PDOs (Fig. S5 D). Taken together, these results show intratumoral heterogeneity in the differentiation potential of metastatic seeds.
Distinct chemoresponse of metastatic seeds
It is unclear whether metastatic cells are inherently more resistant to chemotherapy than primary cancer cells (Lambert et al., 2017; Oskarsson et al., 2014). To investigate this, we compared the chemoresponse of high and low metastatic SC-PDCC lines in primary CRC using 5-fluorouracil (5-FU), a frequently used chemotherapeutic agent (Vodenkova et al., 2020). Chemoresponse was determined by assessing the half-maximal inhibitory concentration (IC50) and by using dose-response curves (Broutier et al., 2017; van de Wetering et al., 2015). We found that SC-PDCC lines derived from P1, P2, and P3 exhibited striking differences in response to 5-FU (Table S2; Fig. 6, H and I; and Fig. S5 E). To validate the response to 5-FU, we performed PDCC colony counting, and the results were consistent with the dose-response curves (Fig. 6 J and Fig. S5 F). In summary, this result demonstrates intratumor heterogeneity in response to 5-FU in metastatic seeds of primary CRC.
Personalized evolution of metastatic seeds within primary CRC
It is still unclear how clones with metastatic potential evolve within human primary tumors, and whether the acquisition of metastatic potential is contingent upon the evolution of genetic mutations during tumor progression (Birkbak and McGranahan, 2020; Lambert et al., 2017). We therefore explored the clonal evolution of metastatic seeds within the primary tumor of P1, P2, and P3 based on SNV data from SC-PDCC lines (Fig. 7 A). These phylogenetic trees integrated intratumoral heterogeneities for the genetic landscape and for metastatic potential, revealing both genomic and metastatic evolution trajectories of SC-PDCC lines in each primary tumor. In the clonal evolution of P1, a branch of clonal clusters with high metastatic potential was evident, while this pattern was not evident for high metastatic SC-PDCC lines of P3. Although only one SC-PDCC line in P2 was identified as having moderate metastatic potential, it was clearly separated into a distinct branch from the low metastatic SC-PDCC lines. These results suggest that the natural occurrence of metastatic potential in primary CRC is the result of genetic evolution in some individuals, which is contingent upon the individualized evolution of the tumor. In addition, poorly differentiated SC-PDCC lines in P1, as well as chemoresistant SC-PDCC lines in three patients, were widely scattered throughout the phylogenetic tree, revealing distinct evolution trajectories compared with that of metastatic potential (Fig. 7 A). These phylogenetic trees reveal diverse evolutionary patterns of metastatic seeds in different primary CRC tumors and provide informative clues to understand the mechanisms by which specific metastatic seeds arise during personalized tumor evolution.
Figure 7.
Metastasis evolution and the impact of clinical prognosis of metastatic potential. (A) Phylogenetic trees were constructed using SNVs from SC-PDCC lines of three individuals, integrating functional heterogeneity. Branch lengths indicate mutation numbers. Colored labels correspond to each function. The size represents the strength of each function. (B and C) Kaplan–Meier curves of patients according to DEGs between HM and LM SC-PDCC lines from P3 (B) and P1 (C) in the TCGA-CAOD cohort (cases = 458). Red and blue lines denote the patients with genes highly expressed in HM and LM SC-PDCC lines, respectively. Significance of differences between the two groups was assessed by Cox test (B, P = 0.03; C, P = 0.18). HM, high metastatic potential; LM, low metastatic potential. Dotted line represents median survival. The plus signs represent the censored cases. (D) Kaplan–Meier curves of patients according to DEGs between WD and PD SC-PDCC lines from P1 in the TCGA-CAOD cohort (cases = 458). Red and blue lines denote the WD and PD SC-PDCC lines, respectively (P < 0.0001). WD, well differentiation potential; PD, poor differentiation potential. (E–G) Kaplan–Meier curves of patients according to DEGs between DR and DS SC-PDCC lines from P1 (E), P2 (F), and P3 (G) in the TCGA-CAOD cohort (cases = 458). Red and blue lines denote the DR and DS SC-PDCC lines, respectively (E, P = 0.22; F, P = 0.19; G, P = 0.41). DR, drug resistant; DS, drug sensitive. (H and I) Kaplan–Meier curves of patients according to DEGs between multitask and single-task SC-PDCC lines (H) and between multitask and zero-task SC-PDCC lines (I) from P1 in the TCGA-CAOD cohort (cases = 458). Red line denotes the mutitask SC-PDCCs, blue line denotes the single-task (H) or zero-task (I) SC-PDCC lines (H, P = 0.02; I, P = 0.01).
Significant impact of metastatic seeds on clinical prognosis
The studies mentioned above indicate that SC-PDCC lines derived from primary CRC tumors exhibit significant heterogeneity both at the molecular and functional levels (Fig. 7 A), which poses a challenge in determining which clones should be prioritized as therapeutic targets to achieve optimal clinical outcomes. Therefore, we predicted the relationship between different phenotypic SC-PDCC lines and clinical prognosis based on their gene expression profiles in P1, P2, and P3 (Table S2). The Kaplan–Meier curves of patients based on DEGs between SC-PDCC groups with distinct functions were used to characterize the prognostic impact of SC-PDCC lines. Through bioinformatics prediction, we found for the first time that different clones in a single tumor showed varying degrees of clinical impact. In P3, the survival probability of high metastatic SC-PDCC lines was significantly lower than that of low metastatic lines (P = 0.03; Fig. 7 B), although this result in P1 was obvious but not significant (P = 0.18; Fig. 7 C). In P1, poorly differentiated SC-PDCC lines showed significantly lower survival probability compared with well-differentiated SC-PDCC lines (P < 0.0001; Fig. 7 D). There was no significant difference in the survival probability between 5-FU resistant SC-PDCC lines and 5-FU sensitive lines derived from the three patients (P = 0.22, 0.19, 0.41, respectively; Fig. 7, E–G). Especially in P1, the survival probability of multifunction SC-PDCC lines was significantly lower than that of both single-function lines (P = 0.02; Fig. 7 H) and zero-function lines (P = 0.01; Fig. 7 I).
In conclusion, the prognostic impact of clones largely depends on whether the clone has the functional property of poor differentiation or high metastatic potential, and whether the clone has multiple functions. It is worth noting that for these three patients, drug-resistant clones may not lead to poor prognosis. To improve clinical outcomes, we should develop strategies to specifically target clones with poor differentiation or high metastatic potential, especially these multitasking clones that play a more dangerous role in clinical prognosis.
Discussion
Metastasis remains the major challenge to clinical treatment and experimental investigation (Birkbak and McGranahan, 2020; Steeg, 2016). It has been noted that metastatic dissemination of malignant CRC tumors occurs far earlier than clinical diagnosis (Hu et al., 2019), and that disseminated tumor cells and numerous micrometastatic lesions can spread throughout the body (Marusyk et al., 2012). Therefore, to achieve substantial improvements in therapeutic outcomes against metastasis, we must target and eliminate these metastatic seeds before they sprout and develop into clinically overt metastatic lesions. Sequencing technology provides valuable insights into the phylogenetic relationship of metastases and primary tumors at the genetic level (Hunter et al., 2018; Sylvester and Vakiani, 2015), while how to assign the metastatic potential to specific clones within the primary tumor remains a challenge (Lawson et al., 2018). To address this, our study cultured and identified multiple single cell–derived clonal cell lines with different degrees of metastatic potential in the human primary CRC through in vitro and in vivo experimental validation. Those clones with high metastatic potential may serve as seeds for the origin of future metastases. Our insights on the cellular and molecular characteristics of these clones with different degrees of metastatic potential provide potential therapeutic targets for personalized primary CRC.
Moreover, we have identified the clinically relevant characteristics of clones with metastatic potential in terms of organ-selective metastatic potential, differentiation potential, and chemoresponse. Interestingly, we have found that metastatic seeds exhibit intertumoral heterogeneity in their ability to selectively colonize certain organs. This suggests that there may be an inherent mechanism within metastasis seeds for selecting specific soil for colonization. Our pioneering analysis with organ-specific metastatic seeds from primary CRC tumors shows that specific and robust cellular communication capabilities confer the potential for metastatic seeds to metastasize in a lung-specific manner. This result suggests that robust cellular communication capabilities play a crucial role in lung-specific metastasis, shedding light on potential therapeutic targets.
Genetically distinct clones arise through branching evolution in primary tumors (Burrell et al., 2013; Greaves and Maley, 2012; McGranahan and Swanton, 2017), but whether genetic or non-genetic factors dictate the metastatic potential remains poorly understood. Therefore, to determine the genetic divergence of metastatic clones within the primary tumor at the single-cell level, the ideal strategy is to integrate genotypes and metastatic capacity into a single cell. Recently, single cell–derived organoids have accomplished the integration of molecular features and drug response (Roerink et al., 2018). Using this strategy, we have achieved the integration of genotype–phenotype at the single-cell level, as well as the integration of mutational phylogenetic trees and metastatic evolution patterns in individual primary tumors. We have used this integration to develop a gene signature that shows promising potential for predicting the risk of metastasis in primary CRC tumors. It implies that, despite the heterogeneity in the evolution of metastatic potential in individual tumors, there may be common genetic factors or pathways that consistently contribute to facilitating metastasis across a broader spectrum of cases. This gene signature likely represents a subset of genes that are consistently associated with metastatic potential in CRC, even if the precise mechanisms and mutations leading to metastasis differ between individual tumors. Nevertheless, it is important to note that while this signature may be effective in predicting metastatic risk in a general sense, it may not capture all the intricacies of the diverse pathways to metastasis seen in individual tumors. We anticipate that more precise and comprehensive prediction tools, potentially tailored to specific tumor subtypes, may emerge in the future as we gather data from a larger and more diverse patient cohort.
In summary, we have constructed a more comprehensive and diverse landscape of metastatic seeds in primary tumors. Our findings have highlighted the inter- and intratumoral heterogeneity of metastatic seeds, characterized by their metastatic organs, differentiation potential, chemoresponse, and evolutionary trajectory. We have also identified genetic and molecular characteristics associated with metastatic potential, as well as a metastatic signature that can be used to capture the metastatic potential of primary CRC tumors. Our study provides valuable insights into the mechanisms underlying metastasis and may contribute to the development of personalized treatment strategies for CRC patients by identifying and targeting metastatic seeds and their specific signature genes.
Materials and methods
Study design
This study aimed to identify and characterize single cell–derived clonal cell lines with varying degrees of metastatic potential within primary CRC tumors. 10–16 single-cell clonal cell lines were established from each of five primary CRC tumors to evaluate their metastatic potential both in vitro and in vivo. The clone’s differentiation potential, chemoresponse, and genetic and molecular characteristics associated with metastatic potential were investigated to profile these metastatic seeds in primary CRC. A metastatic signature was then identified and used to capture the metastatic potential of 13 primary CRC tumors with or without metastasis. Finally, some of the metastatic signature genes were validated through functional experiments in a colorectal cell line.
Human samples
The study was approved by the ethics committee of institution review board of the Tsinghua University (#20170019 and #20190303), National Cancer Center/Cancer Hospital, Chinese Academy of Medical Sciences, and Peking Union Medical College (#19/172-1956). Normal and CRC tissue samples were obtained from patients who were diagnosed with CRC and underwent surgical resection at the hospital. All patients gave written informed consent. Information on cancer and non-cancer tissue specimens is shown in Table S1.
SC-PDCC culture
CRC tissue was excised after surgery, stored, and transported in wash buffer: F12 (Gibco), 5% FBS (Hyclone), 1% penicillin/streptomycin (Gibco), 0.1% Amphotericin B (Gibco), 0.25% Gentamicin (Gibco), 1% HEPES (Gibco), and 5 μM Rock inhibitor (Calbiochem) at 4°C. Tumor tissues were cut into small pieces and incubated in 1 mg/ml collagenase type XI buffer (Gibco) at 37°C for 10–15 min. The digested cell solution was filtered through a 70-μm cell strainer (Falcon) and washed four times with wash buffer. Isolated cells were resuspended in stem cell medium (SCM): advanced DMEM/F12 (Hyclone) supplemented with 10% FBS, 1% penicillin/streptomycin, 1% L-Glutamine (Hyclone), 0.1% Amphotericin B, 0.5% Gentamicin, 0.18 mM Adenine (Sigma-Aldrich), 5 μg/ml insulin (Sigma-Aldrich), 2 nM T3 (Sigma-Aldrich), 200 ng/ml hydrocortisone (Sigma-Aldrich), 125 ng/ml R-Spondin 1 (R&D), 100 ng/ml Noggin (Peprotech), 2.5 μM Rock inhibitor (Calbiochem), 2 μM SB431542 (Cayman chemical), 10 mM Nicotinamide (Sigma-Aldrich), and 10 ng/ml EGF (Upstate Biotechnology). After resuspension, the cells were seeded onto irradiated 3T3-J2 feeder cells that were paved 1 day in advance and cultured at 37°C in 7.5% CO2. The primary cancer cells formed numerous colonies on the feeder layer, collectively referred to as pooled PDCCs. From each individual primary tumor of P1, P2, and P3, 10–16 individual colonies from the primary pooled PDCC culture without passage were selected and expanded separately. Subsequently, single cells were obtained by performing flow sorting on the disaggregated single-cell suspension, and each single cell–derived line (SC-PDCC line) was generated from a single cancer cell of each expanded colony. The culture medium was replaced every 2 days. We propagated PDCCs into the medium without R-Spondin-1 to remove the contamination of normal intestinal stem cell clones.
When the colonies were generated after processing tissue, individual colonies were picked and cultured in 48-well plates for expanding. For each primary tumor, 10–16 individual colonies were picked from pooled PDCC cultures (not passaged) and expanded separately. Then, cells were digested in a 0.25% trypsin-EDTA solution (Gibco) for 5–8 min at 37°C and cell suspensions were passed through 30-µm filters (Miltenyi Biotec). 106 cells were blocked with 0.1% FBS at 4°C for 30 min and then incubated with CD326 Monoclonal Antibody (1:50, 53-9326-42; Thermo Fisher Scientific) at 4°C for 30 min. Samples were collected and sorted with an Aria SORP Cell Sorter (BD). Single cells were seeded into 96-well plates coated with feeder cells. Thus, single cells were obtained by performing flow sorting on the disaggregated single-cell suspensions, and a single cell–derived clonal cell line (SC-PDCC) was generated from each expanded colony.
3D ALI-PDO culture
ALI PDO culture was performed as described (Wang et al., 2015). Initially, each Transwell insert (Corning) was coated with 20% Matrigel (growth factor reduced, BD Biosciences) and incubated at 37°C for 30 min for polymerization. Then, 200,000 feeder cells were seeded into each Transwell insert and incubated overnight at 37°C in 7.5% CO2. PDCCs were digested in a 0.25% trypsin-EDTA solution at 37°C for 8 to10 min and passed through 30 µm filters (Miltenyi Biotec) to obtain single cells. The PDCCs pellets were then resuspended in 80 μl F12 and 20 μl mouse feeder removing MicroBeads (130-095-531; Miltenyi Biotec), followed by an incubation at 4°C for 15 min in low-light conditions. The columns were placed in the magnetic MACS Separator and rinsed with 3 ml F12 buffer. Then 400 μl F12 buffer was added to the 100 μl PDCCs solution incubated with MicroBeads, gently mixed and added to the column to separate the PDCCs from mouse feeder cells. After counting, between 200,000–300,000 PDCCs were seeded onto each Transwell insert and cultured with SCM medium. After PDCCs' growth reached confluence (3–7 days), the medium on the Transwell's apical side was carefully removed by pipetting, leaving only the medium (SCM minus nicotinamide, named SCM-6) on the bottom side of the Transwell. The PDCCs were further cultured for 6–12 days in SCM-6 medium prior to analysis. The medium was refreshed daily.
Liver and lung metastasis in mice
All animal procedures were conducted in accordance with the guidelines for the welfare of animals in cancer research provided by the Institutional Review Board of the Chinese Academy of Medical Sciences Cancer Institute and Tsinghua University Animal Ethics Committee guidelines. NSG mice (6–12 wk old) were obtained from HFK Bioscience Co., Ltd. To induce forced liver metastases, PDCCs were harvested and resuspended in 50% Matrigel (Corning) and 50% PBS media. Then, 1 × 106 cells in 25 μl of PDCC suspension were injected into spleen of male NOD/SCID mice. The livers were isolated 15–32 wk after transplantation. Forced lung metastases were induced by injecting 100 μl of PBS containing 1 × 106 PDCC cells into the tail vein of the mice. The lungs were isolated 10–32 weeks after transplantation. The number of mice used for each SC-PDCC line is shown in Table S3. Finally, all samples were fixed in 4% paraformaldehyde for subsequent immunohistochemical analysis.
RNA-seq analysis
RNA was isolated from PDCCs and tissues with Trizol Reagent (Invitrogen) and with RNeasy mini kit (QIAGEN). 2 µg of prepared input RNA was sequenced on the Illumina platform generating 150 bp paired-end reads. Gene abundance was quantified using HISAT2 (Kim et al., 2019) (version 2.0.5) and StringTie (Kovaka et al., 2019) (version 1.3.4) pipeline (Pertea et al., 2016). DESeq2 (version 1.24.0) (Love et al., 2014) was used to perform differential expression analysis of high abundance genes (mean reads >10). DEGs were detected at a strict threshold of adjusted P < 0.01 and |log2(fold-change)| > 1.
GSEA and Gene Ontology term enrichment analysis (GOEA) were performed with R package clusterProfiler (version 3.12.0) (Yu et al., 2012). PCA was performed based on the top 10,000 most variable genes by prcomp function in R and was visualized with ggbioplot (version 0.55) (https://github.com/vqv/ggbiplot). Multivariate Analysis of Variance Using Distance Matrices (PERMANOVA) was performed using the adonis function in the vegan package (https://github.com/vegandevs/vegan).
Whole-exome sequencing (WES) analysis
Genomic DNA was extracted with DNeasy Blood & Tissue kit (Qiagen). 1–3 µg DNA was used for WES. Single base mutations, insertions, and deletions were identified by Genome Analysis Toolkits (version 4.1.0.0) pipeline (McKenna et al., 2010). First, potentially contaminating DNA of mouse cells from feeder layer was filtered by mapping reads to mouse reference genome (GRCm38) by Bowtie2 (version 2.3.2) (Langmead and Salzberg, 2012). Then, filtered reads were quality-filtered and aligned to the human reference genome (hg19) by TrimGalore (https://github.com/FelixKrueger/TrimGalore) and BWA (version 0.7.15) (Li and Durbin, 2009), respectively. Next, Picard (version 2.18.27) was used to mark duplicate reads and Mutect2 was used to call somatic mutations in tumor samples by comparison to their matched adjacent normal tissues. In detail, the parameters (1) --af-of-alleles-not-in-resource was set to 0.0000025 to filter germline variant, and (2) –annotation was set as UniqueAltReadCount. Subsequently, GATK FilterMutectCalls (--unique-alt-read-count 5) and FilterByOrientationBias were used to perform second filtration with default parameters. Variants were annotated to the functional consequence using ANNOVAR (version Apr 2018) based on the human genome (Wang et al., 2010). CNAs were called using FACETS (version 0.5.14), which are integer copy number calls that correct for tumor purity, ploidy, and clonal heterogeneity (Shen and Seshan, 2016). Fisher’s test was used to analyze differences in the frequency of mutations in samples with different phenotypes.
Correlation analysis between genomic and transcriptional heterogeneity
To assess the potential impact of genomic mutations on the transcriptome, we generated a binary matrix that encoded mutations within the exonic regions. Subsequently, the Euclidean distance was applied to quantify the genomic dissimilarity across diverse SC-PDCC lines. For transcriptome analysis, we identified the top 10,000 genes with the highest coefficient of variation across distinct SC-PDCC lines. Following a log2+1 transformation of transcripts per million values, we computed the Euclidean distance to assess transcriptomic dissimilarity across different cell lines. Lastly, we utilized Spearman’s correlation coefficient to evaluate the overall consistency of the SC-PDCC lines.
scRNA-seq
Single-cell suspensions were achieved as described above and then resuspended into 1 × PBS for 10× genomics processing. The aimed target cell recovery for each library was 8,000 and the libraries were performed on an Illumina HiSeq X Ten platform. Data produced by scRNA-seq were processed, aligned, and summarized for unique molecular identifier (UMI) counts against the human (hg19) and mouse (mm10) reference genome by Cellranger software (version 3.1.0) (Zheng et al., 2017) with default parameters. The raw, unfiltered count matrices were imported into R for further processing by Seurat (version 3.2.0) (Stuart et al., 2019). Quality control was performed to discard genes detected <200 counts, genes detected in mouse more than in human, as well as those with a mitochondrial transcription ratio >30%. The function FindIntegrationAnchors from Seurat was then used to integrate different samples and remove batch effects and biological covariates (Tran et al., 2020).
Dimensionality reduction was performed using PCA. Cells were clustered in the reduced dimension space using the Seurat package (resolution = 0.8) and were visualized using UMAP plots. We used the function AddModuleScore from Seurat and the list of G2M-associated genes to calculate a cell cycle score for each cell (Scialdone et al., 2015). To analyze the well-differentiation (WD) score and poor-differentiation (PD) score in each cell, 237 PD marker genes and 122 WD marker genes were detected by DEG analysis of bulk RNA-seq data from P1. Then the function AddModuleScore was used to calculate the WD and PD scores in each cell. The P value was calculated based on wilcox.test in R.
We found 15 cell clusters including seven cycling clusters and eight noncycling clusters. Cluster marker genes were detected with the FindAllMarkers in Seurat package using the wilcox test. scCATCH (version 2.1) (Shao et al., 2020) and SCSA (version) (Cao et al., 2020) were used to preliminary annotate cell clusters. They were further confirmed manually. Cluster marker genes with adjusted P < 0.05 and log2(fold-change) > 5 were selected to perform GOEA by Enrichr (Kuleshov et al., 2016; Xie et al., 2021).
The somatic mutation-based phylogenetic trees
Phylogenies were constructed based on a binary matrix of mutations present or absent in each sample (Roerink et al., 2018). The program pipline of seqboot, Mix, and Consense in Phylip (version 3.695) (https://phylipweb.github.io/phylip/install.html) were used with the same parameters as published (Roerink et al., 2018). In short, all private mutations were excluded because they are uninformative and misleading. The seqboot program was used to generate bootstrap replicates, the program Mix (with Wagner method) reconstructed each bootstrap replicate by maximum parsimony, and a consensus of all trees was built by the program Consense with the majority rule (extended) option. The tree was visualized by R package ggtree (version 1.16.6) (Yu, 2020).
Processing scRNA-seq data, cell clustering, and metastatic score
Publicly available single-cell sequence data (PRJNA748525) of 13 CRC samples were downloaded and reanalyzed. In detail, the raw reads were downloaded and aligned against GRCh38 human reference genome provided by Cell Ranger (version 5.0.0, 10× genomics). Then, the R package Seurat (version 4.1.1) was used to remove genes expressed in fewer than three cells, cells with fewer than 500 genes, and cells with high percentages of mitochondrial genes (more than 15%). After quality control, the UMI counts were log-normalized using a scale of 10,000. The top 1,000 variable genes were selected with the ‘‘FindVariableGenes’’ function of Seural. Next, “var.to.regress” option UMI’s and percent mitochondrial content were used to regress out unwanted sources of variation. Next, Harmony (version 0.1.0) was used to correct for batch effects and biological covariates. The “FindClusters” function was used to detect clusters, which employs an optimization algorithm of nearest-neighbor modularity implemented in Seurat. The identified clusters were then visualized using the UMAP algorithm. The annotation of each cell cluster was confirmed by the expression of canonical marker genes. The canonical marker genes KRT8, KRT18, EPCAM, ELF3, and KRT19 were used to identify the cluster of cancer cells (Che et al., 2021), and a total of 11,608 tumor cells were identified.
Next, all cancer cells were extracted and reanalyzed according to the above procedure to identify cell subcluster. Significantly high and low expression of genes in high metastatic SC-PDCC lines were used to calculate metastatic and non-metastatic scores for each cell by the function “AddModuleScore.” The combined metastatic risk score was obtained using the metastatic score minus the non-metastatic scores. According to the metastatic risk scores, we identified cells with the top 20% and bottom 20% scores as high metastatic risk cells and low metastatic risk cells, respectively.
Cell communication analysis
Cell communication between different cell types and tissues primarily relies on the interaction between ligands secreted by cells and cell surface receptors (Ramilowski et al., 2016). To investigate the strength of cell communication among different SC-PDCC lines and between lung and liver, we performed an analysis using the DISCO database (Li et al., 2022), which integrates over 18 million single-cell omics data from 4,593 samples. From this database, we obtained and curated the significantly upregulated genes in each cell type of lung or liver tissues. These genes were defined as exhibiting significantly higher (adjusted P value <0.01) expression in the respective cell type compared with other cells in the tissue. We identified 53 liver-specific cell types, 44 lung-specific cell types, and 20 cell types shared between the two tissues. DEG analysis of significantly upregulated genes between different SC-PDCC lines was performed using the R package DESeq2 (Love et al., 2014). For the significantly upregulated genes between SC-PDCC lines with high metastatic to liver and high metastatic to lung, we applied the thresholds of adjusted P value <0.01 and |log2fold-change| > 1. For SC-PDCC lines with low metastatic to lung and high metastatic to lung, the thresholds used were adjusted P value <0.05 and |log2fold-change| > 1. For SC-PDCC lines with low metastatic to liver and high metastatic to liver, the thresholds were P value <0.05 and |log2fold-change| > 1.
Information on pair of ligands and receptors was obtained from the built-in dataset “ramilowski_pairs” in the R package celltalker (Cillo et al., 2020), which includes 691 receptors and 708 ligands, forming a total of 2,557 receptor–ligand pairs (Ramilowski et al., 2015). To calculate the communication density between SC-PDCC lines, we counted the pairs of highly expressed receptor genes in SC-PDCC lines and highly expressed ligand genes in each cell type, as well as the pairs of highly expressed ligand genes in SC-PDCC lines and highly expressed receptor genes in each cell type. By determining the number of receptor–ligand pairs between them, we defined the communication density between SC-PDCC lines and different cell types of each tissue.
Enrichment analysis of receptors and ligands in SC-PDCC lines
The GSEA analysis was performed to analyze DEGs in SC-PDCC lines with different phenotypes. The gene lists of receptors and ligands were obtained from the R package celltalker (Cillo et al., 2020). The GSEA function from the R package clusterProfiler (Wu et al., 2021) was used for enrichment analysis of the receptor gene set and ligand gene set.
For GO analysis, first, we identified DEGs between SC-PDCC lines associated with the ability of high metastatic to liver and that with the ability of high metastatic to lung with the thresholds adjusted P value <0.05 and |log2fold-change| > 1. In total, we identified 789 upregulated DEGs in high metastatic to liver, with 33 receptor genes and 83 ligand genes. In SC-PDCC lines with high metastatic to lung, 1,381 DEGs were upregulated, including 32 receptor genes and 89 ligand genes. Subsequently, the obtained receptor and ligand genes were subjected to GO enrichment analysis using the compareCluster function from the R package clusterProfiler (Wu et al., 2021).
Processing of public data
GSE68468 contains expression profiles of 20 CRC metastatic tissue with lung metastasis and 47 samples of metastatic tissue with CRC liver metastasis (Gavert et al., 2007). GSE72718 contains expression profiles of nine samples of primary CRC with liver metastasis and 10 samples of primary CRC without liver metastasis (Gao et al., 2016). GSE41258 contains expression profiles of 17 ascending colon samples of primary CRC with metastasis and 30 ascending colon samples of primary CRC without liver metastasis (Martin et al., 2018)). The expression profiles for these three datasets were obtained using the “GEOquery” package, available at https://github.com/seandavi/GEOquery. Probes were converted to gene symbols using an annotation file provided by the manufacturer. The metastatic signature for GSE72718 and GSE41258 was calculated using the “gsva” function with the method set to “zscore” in the R package GSVA (Hänzelmann et al., 2013). The differential gene expression analysis for GSE68468 was performed using the limma package (Ritchie et al., 2015).
Drug response test
Five concentrations (0, 0.1, 1, 10, and 100 μM) of chemotherapeutic agent 5-FU were prepared and assayed. PDCCs were gently disrupted into single cells and feeder cells were removed with magnetic beads. 10,000 cells were plated on 96-well plates coated with 10% Matrigel. Drug was added individually after overnight incubation, and cell viability was measured using CellTiter-Glo reagent (Promega) after 6 days. Each SC-PDCC was performed with three technical replicates and two biological replicates with different passages. The results were normalized to controls and expressed as percent cell viability. The determination of IC50 values was conducted using Graph Pad Prism9. For 2D PDCCs, the drug was added to SCM medium at final concentration of 10 µM on the second day of PDCC culture. 0.1% DMSO was used as control. Following 6 days of treatment, the cells were washed twice with PBS, fixed with 4% paraformaldehyde for 20 min, and stained with 10% rhodamine staining solution. Surviving cells were counted under a bright-field microscope. PDCCs with survival rate >50% were defined as drug resistant, and PDCCs with survival rate <50% were defined as drug sensitive.
Proteome analysis
SC-PDCCs from P1 were homogenized in 200 μl lysis buffer. The lysis buffer consisted of 1 mM PMSF (Sigma-Aldrich). Lysates were centrifuged at 20,000 g for 10 min and protein concentrations of the clarified lysates were measured by BCA assay (Pierce). For each sample, 200 μg peptides were prepared by vacuum centrifugation dryness for the following TMT labeling experiment. The isobaric labeling experiment was conducted according to the TMT kit instructions. For each set of TMT 11-plex labeling experiment, the mixed peptides were labeled with channel 126 as the internal reference, and three low metastatic SC-PDCC samples and four high metastatic SC-PDCC samples were labeled with the other seven channels (low metastatic labeled with 127N, 127C, 129N; high metastatic labeled with 129C, 130N, 130C, and 131).
Data were normalized using the median centering method across total proteins to correct sample loading differences. In normalized samples, these proteins should have a log TMT ratio value centered at zero. Normalized proteins/phosphorylation sites with SwissProt ID were converted to Human Genome Nomenclature Committee’s (HGNC) HUGO symbols provided by HGNC (https://www.genenames.org). First, Limma (version 3.40.6) was used to detect significantly changed proteins. Then, GSEA was performed in clusterProfiler (version 3.12.0) to analyze the pathways of significant changes. Pearson correlation coefficients were used to detect the consistency of transcriptome and proteome changes.
Survival analysis
Transcriptomic data were collected from the The Cancer Genome Atlas Colon Adenocarcinoma Collection (TCGA-COAD) dataset. The Fragments Per Kilobase Million (FPKM) normalized expression data were downloaded by R package GEOquery (version 2.52.0) (Davis and Meltzer, 2007) and TCGAbiolinks (version 2.12.6) (Colaprico et al., 2016). For single genes, the top 40 and bottom 40% of samples (based on FPKM) were aligned to the high and low abundance groups. A list of phenotypic-related genes with adjusted P < 0.001 and |log2(fold-change)| > 2 were detected by DESeq2 (with samples classified based on the z-score transformed method). In detail for each sample, the sum of the z-score was used as the final overall score to classify the samples to the high (top 40%) and low (lower 40%) groups. Kaplan–Meier survival curves were generated using the ggsurvplot function from the survminer package (version 0.4.8) (https://github.com/kassambara/survminer). P-values of survival differences were calculated with age and gender covariates in the coxph function of the survival (version 2.44.1.1) package.
Immunohistochemistry and immunofluorescence
Sections of formalin-fixed, paraffin-embedded tissues, xenografts, and ALI PDOs were stained by standard hematoxylin and eosin (H&E) staining. For immunohistochemistry and immunofluorescence staining, slides were subjected to antigen retrieval in citrate buffer (pH 6.0; Sigma-Aldrich) at 95°C for 20 min, and a blocking procedure was performed overnight with 5% BSA (Sigma-Aldrich) and 0.05% Triton X-100 (Sigma-Aldrich) in Dulbecco's Phosphate-Buffered Saline(−) (Gibco) at 4°C. Primary antibodies used in this study included antibodies against MUC2 (1:200; Santa Cruz), KI67 (1:500; Thermo Fisher Scientific), CK20 (1:200; Dako), and E-Cadherin (1:200; R&D Systems). Secondary antibodies were either Alexa Fluor-488 or Alexa Fluor-594 Donkey anti-goat/mouse/rabbit IgG (Thermo Fisher Scientific). Images were acquired by Olympus IX73 microscopy.
Cell invasion assay
100,000 cells suspended in 100 μl serum-free culture medium were seeded in the upper chambers coated with 20% Matrigel of Transwells (Corning, pore size 8 µm) and the lower chambers were loaded with 600 μl culture medium with 10% FBS. Following incubation for 24 h, cells were fixed in ethanol overnight and stained with crystal violet. At least five fields were randomly selected and counted under a bright-field microscope. Assays were performed in duplicate or triplicate.
Quantitative PCR (qPCR)
Total RNA was extracted from PDCCs, ALI PDOs, and tissues using the RNeasy mini kit (QIAGEN), and cDNAs were synthesized using the high-capacity cDNA reverse transcription Kit (Thermo Fisher Scientific). Expression levels were measured with iTaq Universal SYBR Green Supermix (Biorad) and normalized to GAPDH. All experiments were carried out in triplicate.
Western blot
PDCCs were collected in radioimmunoprecipitation buffer (Thermo Fisher Scientific) containing protease inhibitor cocktail (Bimake) and phosphatase inhibitor cocktail (1:100; Bimake). Protein was quantified with BCA reagent (Thermo Fisher Scientific). The extracts were resolved by SDS-PAGE on a 10% gradient gel, transferred onto polyvinylidene difluoride membranes (GE healthcare life sciences), and incubated with primary antibodies EPHB3 (1:1,000; Abnova), and β-ACTIN (1:1,000; Cell signaling) overnight at 4°C. Incubation with secondary antibody (Anti-mouse IgG, HRP-linked Antibody; Cell signaling) was performed for 2 h at room temperature. After detection using an ECL Western blot Substrates (Thermo Fisher Scientific), images were acquired using an ImageQuant LAS 4000.
Cell line culture and transfection
Human colon cancer cell line DLD1 was obtained from ATCC and cultured in DMEM supplemented with 10% FBS at 37°C in 5% CO2. For stable knockdown of AKR1C1, NAMPT, SAMHD1, OSTM1, and for stable overexpression of EPHB3 and SPINK4, cells were infected with lentiviruses expression of two different shRNAs (Table S7).
For PDCCs, cells were dissociated into single cells as mentioned above. Then cells were resuspended in SCM medium and lentiviruses at a 1:1 ratio (1 ml each), with the addition of 8 µg/ml polybrene. The cell solution was transferred to a 6-well plate and incubated overnight at 37°C in 7.5% CO2. After 24 h, the medium was replaced with fresh SCM medium. 2 days after injection, PDCCs were cultured in SCM medium added with 4 µg/ml puromycin for 2 wk. The effect of knockdown or overexpression was confirmed by qPCR or Western blot.
Statistical analysis
Analysis procedures of genome and transcriptome data are provided in the relevant sections of Materials and methods. Statistical analysis was performed with GraphPad Prism and presented as mean values ± SD. Unpaired two-tailed Student’s t test was used to calculate P values between two groups. The ‘‘n’’ numbers for each experiment were provided in the text and figures. Corresponding statistical significance was denoted with *P < 0.05; **P < 0.01; ***P < 0.001 and ****P < 0.0001 in the figures and figures legends.
Online supplemental material
Fig. S1 illustrates how SC-PDCC lines can recapitulate the intratumoral heterogeneity of histology, genomic, and transcriptional landscape. Fig. S2 offers additional information on evaluating the metastatic potential of SC-PDCC lines. Fig. S3 demonstrates the molecular divergence of metastatic SC-PDCC lines from individual tumors. Fig. S4 includes the analysis of primary CRC tumor scRNA-seq data and the cellular communication capacity of SC-PDCC lines. Fig. S5 explains the differentiation potential and drug response of SC-PDCC lines with varying metastatic potential. Table S1 shows clinicopathological data of CRC patients. Table S2 shows SC-PDCC information and test results for various analyses. Table S3 shows liver and lung metastasis of SC-PDCCs in mice, related to Figs. 2 and S3. Table S4 shows the metastasis potential of pooled PDCC in mice, related to Figs. 2 and S3. Table S5 shows a total of 21 protein interaction genes with differential expression in high metastatic SC-PDCC lines from P1. Table S6 shows the proportion of cancer cells with metastatic signatures in each individual primary CRC tumor (from Xu et al. [2022] dataset PRJNA748525). Table S7 shows the shRNA information, related to Fig. 7 D. Data S1 shows differential CNAs between high and low metastatic SC-PDCC lines in P1. Data S2 shows the metastatic signature.
Supplementary Material
shows clinicopathological data of CRC patients.
shows SC-PDCC information and test results for various analyses.
shows liver and lung metastasis of SC-PDCCs in mice, related to Figs. 2 and S3.
shows the metastasis potential of pooled PDCC in mice, related to Figs. 2 and S3.
shows a total of 21 protein interaction genes with differential expression in HM SC-PDCC lines from P1.
shows the proportion of cancer cells with metastatic signature in each individual primary CRC tumor (from Xu et al. [2022] dataset PRJNA748525).
shows the shRNA information, related to Fig. 7 D.
shows differential CNAs between high and low metastatic SC-PDCC lines in P1.
shows the metastatic signature.
is the source file for Fig. 4.
Acknowledgments
We thank all patients who generously donated their tissues for this study. We thank Guoliang Li for his help in bioinformatic training for students. We thank Hans Clevers, Weiwei Zhai, Yeguang Chen, Wei Wu, Wei Xie, and Huili Hu for comments and suggestions. Some of the graphic icons were created with https://BioRender.com.
This work was supported by grants from the National Key Research and Development Program of China (2021YFA1100103 to X. Wang); the National Natural Science Foundation of China (32070796 to X. Wang); Institute for Intelligent Healthcare, Tsinghua University; Chinese Academy of Medical Sciences Innovation Fund for Medical Sciences (2022-I2M-1-009, 2021-I2M-1-067); National Nature Science Foundation of China (22193034 to H. Wang); Tsinghua University Initiative Scientific Research Program; and Tsinghua-Peking Center for Life Sciences (100084).
Author contributions: Y. Zhao generated and cultured all of the PDCCs and ALI PDOs, performed most of the biological experiments and data analysis, and prepared figures, figure legends, and methods. B. Zhang analyzed all sequencing data and prepared figures, figure legends, and methods. Y. Ma performed metastasis assay in vivo. F. Zhao and J. Chen provided patient specimens and maintained clinical records. B. Wang performed histopathological analysis. M. Guo, H. Jin, F. Zhao, J. Guan, and Q. Zhao participated in additional data collection and analyses. H. Wang supervised tumor tissue collection and metastasis assay in vivo. Q. Liu supervised tumor tissue collection and clinical records. F. Zhou supervised sequencing data analysis. X. Wang supervised project design and wrote the manuscript.
Footnotes
X. Wang is the lead contact.
Data availability
The raw sequence data reported in this study have been deposited in the Genome Sequence Archive at the National Genomics Data Center, China National Center for Bioinformation/Beijing Institute of Genomics, Chinese Academy of Sciences under the Bioproject accession codes PRJCA006448 and PRJCA023187. Specifically, the WES data and single-cell sequencing data have been deposited under the accession code HRA001280 (https://ngdc.cncb.ac.cn/gsa-human/browse/HRA001280). The RNA-seq data have been deposited under the accession code HRA006587 (https://ngdc.cncb.ac.cn/gsa-human/browse/HRA006587). All additional data supporting the findings of this study are located within the article, in the supplemental information files, or can be obtained from the corresponding author upon request.
Data availability
The raw sequence data reported in this study have been deposited in the Genome Sequence Archive at the National Genomics Data Center, China National Center for Bioinformation/Beijing Institute of Genomics, Chinese Academy of Sciences under the Bioproject accession codes PRJCA006448 and PRJCA023187. Specifically, the WES data and single-cell sequencing data have been deposited under the accession code HRA001280 (https://ngdc.cncb.ac.cn/gsa-human/browse/HRA001280). The RNA-seq data have been deposited under the accession code HRA006587 (https://ngdc.cncb.ac.cn/gsa-human/browse/HRA006587). All additional data supporting the findings of this study are located within the article, in the supplemental information files, or can be obtained from the corresponding author upon request.
References
- Aceto, N., Bardia A., Miyamoto D.T., Donaldson M.C., Wittner B.S., Spencer J.A., Yu M., Pely A., Engstrom A., Zhu H., et al. 2014. Circulating tumor cell clusters are oligoclonal precursors of breast cancer metastasis. Cell. 158:1110–1122. 10.1016/j.cell.2014.07.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armenia, J., Wankowicz S.A.M., Liu D., Gao J., Kundra R., Reznik E., Chatila W.K., Chakravarty D., Han G.C., Coleman I., et al. 2018. The long tail of oncogenic drivers in prostate cancer. Nat. Genet. 50:645–651. 10.1038/s41588-018-0078-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barresi, V., Reggiani Bonetti L., Ieni A., Caruso R.A., and Tuccari G.. 2015. Histological grading in colorectal cancer: New insights and perspectives. Histol. Histopathol. 30:1059–1067. 10.14670/HH-11-633 [DOI] [PubMed] [Google Scholar]
- Bertucci, F., Ng C.K.Y., Patsouris A., Droin N., Piscuoglio S., Carbuccia N., Soria J.C., Dien A.T., Adnani Y., Kamal M., et al. 2019. Genomic characterization of metastatic breast cancers. Nature. 569:560–564. 10.1038/s41586-019-1056-z [DOI] [PubMed] [Google Scholar]
- Birkbak, N.J., and McGranahan N.. 2020. Cancer genome evolutionary trajectories in metastasis. Cancer Cell. 37:8–19. 10.1016/j.ccell.2019.12.004 [DOI] [PubMed] [Google Scholar]
- Bou Khzam, L., Son N.H., Mullick A.E., Abumrad N.A., and Goldberg I.J.. 2020. Endothelial cell CD36 deficiency prevents normal angiogenesis and vascular repair. Am. J. Transl. Res. 12:7737–7761. [PMC free article] [PubMed] [Google Scholar]
- Bouvet, M., Tsuji K., Yang M., Jiang P., Moossa A.R., and Hoffman R.M.. 2006. In vivo color-coded imaging of the interaction of colon cancer cells and splenocytes in the formation of liver metastases. Cancer Res. 66:11293–11297. 10.1158/0008-5472.CAN-06-2662 [DOI] [PubMed] [Google Scholar]
- Brabletz, T. 2012. To differentiate or not--routes towards metastasis. Nat. Rev. Cancer. 12:425–436. 10.1038/nrc3265 [DOI] [PubMed] [Google Scholar]
- Brabletz, T., Jung A., Spaderna S., Hlubek F., and Kirchner T.. 2005. Opinion: Migrating cancer stem cells - an integrated concept of malignant tumour progression. Nat. Rev. Cancer. 5:744–749. 10.1038/nrc1694 [DOI] [PubMed] [Google Scholar]
- Brannon, A.R., Vakiani E., Sylvester B.E., Scott S.N., McDermott G., Shah R.H., Kania K., Viale A., Oschwald D.M., Vacic V., et al. 2014. Comparative sequencing analysis reveals high genomic concordance between matched primary and metastatic colorectal cancer lesions. Genome Biol. 15:454. 10.1186/s13059-014-0454-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broutier, L., Mastrogiovanni G., Verstegen M.M., Francies H.E., Gavarró L.M., Bradshaw C.R., Allen G.E., Arnes-Benito R., Sidorova O., Gaspersz M.P., et al. 2017. Human primary liver cancer-derived organoid cultures for disease modeling and drug screening. Nat. Med. 23:1424–1435. 10.1038/nm.4438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrell, R.A., McGranahan N., Bartek J., and Swanton C.. 2013. The causes and consequences of genetic heterogeneity in cancer evolution. Nature. 501:338–345. 10.1038/nature12625 [DOI] [PubMed] [Google Scholar]
- Cañellas-Socias, A., Cortina C., Hernando-Momblona X., Palomo-Ponce S., Mulholland E.J., Turon G., Mateo L., Conti S., Roman O., Sevillano M., et al. 2022. Metastatic recurrence in colorectal cancer arises from residual EMP1+ cells. Nature. 611:603–613. 10.1038/s41586-022-05402-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, L.L., Pei X.F., Qiao X., Yu J., Ye H., Xi C.L., Wang P.Y., and Gong Z.L.. 2018. SERPINA3 silencing inhibits the migration, invasion, and liver metastasis of colon cancer cells. Dig. Dis. Sci. 63:2309–2319. 10.1007/s10620-018-5137-x [DOI] [PubMed] [Google Scholar]
- Cao, Y., Wang X., and Peng G.. 2020. SCSA: A Cell Type Annotation Tool for Single-Cell RNA-seq Data. Front Genet. 11:490. 10.3389/fgene.2020.00490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Che, L., Liu J., Huo J., Luo R., Xu R., He C., Li Y., Zhou A., Huang P., Chen Y., et al. 2021. A single-cell atlas of liver metastases of colorectal cancer reveals reprogramming of the tumor microenvironment in response to preoperative chemotherapy. Cell Discov. 7:80. 10.1038/s41421-021-00312-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung, P., Xiol J., Dill M.T., Yuan W.C., Panero R., Roper J., Osorio F.G., Maglic D., Li Q., Gurung B., et al. 2020. Regenerative reprogramming of the intestinal stem cell state via hippo signaling suppresses metastatic colorectal cancer. Cell Stem Cell. 27:590–604.e9. 10.1016/j.stem.2020.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu, S.T., Chang K.J., Ting C.H., Shen H.C., Li H., and Hsieh F.J.. 2009. Over-expression of EphB3 enhances cell-cell contacts and suppresses tumor growth in HT-29 human colon cancer cells. Carcinogenesis. 30:1475–1486. 10.1093/carcin/bgp133 [DOI] [PubMed] [Google Scholar]
- de Sousa e Melo, F., Kurtova A.V., Harnoss J.M., Kljavin N., Hoeck J.D., Hung J., Anderson J.E., Storm E.E., Modrusan Z., Koeppen H., et al. 2017. A distinct role for Lgr5+ stem cells in primary and metastatic colon cancer. Nature. 543:676–680. 10.1038/nature21713 [DOI] [PubMed] [Google Scholar]
- Cillo, A.R., Kürten C.L., Tabib T., Qi Z., Onkar S., Wang T., Liu A., Duvvuri U., Kim S., Soose R.J., et al. 2020. Immune Landscape of Viral- and Carcinogen-Driven Head and Neck Cancer. Immunity. 52:183–199.e9. 10.1016/j.immuni.2019.11.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colaprico, A., Silva T.C., Olsen C., Garofano L., Cava C., Garolini D., Sabedot T.S., Malta T.M., Pagnotta S.M., Castiglioni I., et al. 2016. TCGAbiolinks: an R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res. 44:e71. 10.1093/nar/gkv1507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis, S., and Meltzer P.S.. 2007. GEOquery: a bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinformatics. 23:1846–1847. 10.1093/bioinformatics/btm254 [DOI] [PubMed] [Google Scholar]
- Dieter, S.M., Ball C.R., Hoffmann C.M., Nowrouzi A., Herbst F., Zavidij O., Abel U., Arens A., Weichert W., Brand K., et al. 2011. Distinct types of tumor-initiating cells form human colon cancer tumors and metastases. Cell Stem Cell. 9:357–365. 10.1016/j.stem.2011.08.010 [DOI] [PubMed] [Google Scholar]
- Gao, B., Shao Q., Choudhry H., Marcus V., Dong K., Ragoussis J., and Gao Z.. 2016. Weighted gene co-expression network analysis of colorectal cancer liver metastasis genome sequencing data and screening of anti-metastasis drugs. Int. J. Oncol. 49:1108–1118. 10.3892/ijo.2016.3591 [DOI] [PubMed] [Google Scholar]
- Gavert, N., Sheffer M., Raveh S., Spaderna S., Shtutman M., Brabletz T., Barany F., Paty P., Notterman D., Domany E., et al. 2007. Expression of L1-CAM and ADAM10 in human colon cancer cells induces metastasis. Cancer Res. 67:7703–7712. 10.1158/0008-5472.CAN-07-0991 [DOI] [PubMed] [Google Scholar]
- Gkountela, S., Castro-Giner F., Szczerba B.M., Vetter M., Landin J., Scherrer R., Krol I., Scheidmann M.C., Beisel C., Stirnimann C.U., et al. 2019. Circulating tumor cell clustering shapes DNA methylation to enable metastasis seeding. Cell. 176:98–112.e14. 10.1016/j.cell.2018.11.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golovko, D., Kedrin D., Yilmaz O.H., and Roper J.. 2015. Colorectal cancer models for novel drug discovery. Expert Opin. Drug Discov. 10:1217–1229. 10.1517/17460441.2015.1079618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goswami, R.S., Patel K.P., Singh R.R., Meric-Bernstam F., Kopetz E.S., Subbiah V., Alvarez R.H., Davies M.A., Jabbar K.J., Roy-Chowdhuri S., et al. 2015. Hotspot mutation panel testing reveals clonal evolution in a study of 265 paired primary and metastatic tumors. Clin. Cancer Res. 21:2644–2651. 10.1158/1078-0432.CCR-14-2391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greaves, M. 2015. Evolutionary determinants of cancer. Cancer Discov. 5:806–820. 10.1158/2159-8290.CD-15-0439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greaves, M., and Maley C.C.. 2012. Clonal evolution in cancer. Nature. 481:306–313. 10.1038/nature10762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haan, J.C., Labots M., Rausch C., Koopman M., Tol J., Mekenkamp L.J., van de Wiel M.A., Israeli D., van Essen H.F., van Grieken N.C., et al. 2014. Genomic landscape of metastatic colorectal cancer. Nat. Commun. 5:5457. 10.1038/ncomms6457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hänzelmann, S., Castelo R., and Guinney J.. 2013. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics. 14:7. 10.1186/1471-2105-14-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, Z., Ding J., Ma Z., Sun R., Seoane J.A., Scott Shaffer J., Suarez C.J., Berghoff A.S., Cremolini C., Falcone A., et al. 2019. Quantitative evidence for early metastatic seeding in colorectal cancer. Nat. Genet. 51:1113–1122. 10.1038/s41588-019-0423-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter, K.W., Amin R., Deasy S., Ha N.H., and Wakefield L.. 2018. Genetic insights into the morass of metastatic heterogeneity. Nat. Rev. Cancer. 18:211–223. 10.1038/nrc.2017.126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jechorek, D., Haeusler-Pliske I., Meyer F., and Roessner A.. 2021. Diagnostic value of syndecan-4 protein expression in colorectal cancer. Pathol. Res. Pract. 222:153431. 10.1016/j.prp.2021.153431 [DOI] [PubMed] [Google Scholar]
- Jögi, A., Vaapil M., Johansson M., and Påhlman S.. 2012. Cancer cell differentiation heterogeneity and aggressive behavior in solid tumors. Ups. J. Med. Sci. 117:217–224. 10.3109/03009734.2012.659294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna, C., and Hunter K.. 2005. Modeling metastasis in vivo. Carcinogenesis. 26:513–523. 10.1093/carcin/bgh261 [DOI] [PubMed] [Google Scholar]
- Kim, D., Paggi J.M., Park C., Bennett C., and Salzberg S.L.. 2019. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 37:907–915. 10.1038/s41587-019-0201-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, T.M., Jung S.H., An C.H., Lee S.H., Baek I.P., Kim M.S., Park S.W., Rhee J.K., Lee S.H., and Chung Y.J.. 2015. Subclonal genomic architectures of primary and metastatic colorectal cancer based on intratumoral genetic heterogeneity. Clin. Cancer Res. 21:4461–4472. 10.1158/1078-0432.CCR-14-2413 [DOI] [PubMed] [Google Scholar]
- Kovaka, S., Zimin A.V., Pertea G.M., Razaghi R., Salzberg S.L., and Pertea M.. 2019. Transcriptome assembly from long-read RNA-seq alignments with StringTie2. Genome Biol. 20:278. 10.1186/s13059-019-1910-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreso, A., O’Brien C.A., van Galen P., Gan O.I., Notta F., Brown A.M., Ng K., Ma J., Wienholds E., Dunant C., et al. 2013. Variable clonal repopulation dynamics influence chemotherapy response in colorectal cancer. Science. 339:543–548. 10.1126/science.1227670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert, A.W., Pattabiraman D.R., and Weinberg R.A.. 2017. Emerging biological principles of metastasis. Cell. 168:670–691. 10.1016/j.cell.2016.11.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuleshov, M.V., Jones M.R., Rouillard A.D., Fernandez N.F., Duan Q., Wang Z., Koplev S., Jenkins S.L., Jagodnik K.M., Lachmann A., et al. 2016. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 44:W90–W97. 10.1093/nar/gkw377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert, A.W., and Weinberg R.A.. 2021. Linking EMT programmes to normal and neoplastic epithelial stem cells. Nat. Rev. Cancer. 21:325–338. 10.1038/s41568-021-00332-6 [DOI] [PubMed] [Google Scholar]
- Langmead, B., and Salzberg S.L.. 2012. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 9:357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson, D.A., Kessenbrock K., Davis R.T., Pervolarakis N., and Werb Z.. 2018. Tumour heterogeneity and metastasis at single-cell resolution. Nat. Cell Biol. 20:1349–1360. 10.1038/s41556-018-0236-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H., and Durbin R.. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 25:1754–1760. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, L., and Hanahan D.. 2013. Hijacking the neuronal NMDAR signaling circuit to promote tumor growth and invasion. Cell. 153:86–100. 10.1016/j.cell.2013.02.051 [DOI] [PubMed] [Google Scholar]
- Li, M., Zhang X., Ang K.S., Ling J., Sethi R., Lee N.Y.S., Ginhoux F., and Chen J.. 2022. Disco: A database of deeply integrated human single-cell omics data. Nucleic Acids Res. 50:D596–D602. 10.1093/nar/gkab1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, S.Q., Su N., Gong P., Zhang H.B., Liu J., Wang D., Sun Y.P., Zhang Y., Qian F., Zhao B., et al. 2017. The expression of formyl peptide receptor 1 is correlated with tumor invasion of human colorectal cancer. Sci. Rep. 7:5918. 10.1038/s41598-017-06368-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love, M.I., Huber W., and Anders S.. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, M.L., Zeng Z., Adileh M., Jacobo A., Li C., Vakiani E., Hua G., Zhang L., Haimovitz-Friedman A., Fuks Z., et al. 2018. Logarithmic expansion of LGR5+ cells in human colorectal cancer. Cell Signal. 42:97–105. 10.1016/j.cellsig.2017.09.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marusyk, A., Almendro V., and Polyak K.. 2012. Intra-tumour heterogeneity: A looking glass for cancer? Nat. Rev. Cancer. 12:323–334. 10.1038/nrc3261 [DOI] [PubMed] [Google Scholar]
- Marusyk, A., Janiszewska M., and Polyak K.. 2020. Intratumor heterogeneity: The rosetta stone of therapy resistance. Cancer Cell. 37:471–484. 10.1016/j.ccell.2020.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGranahan, N., and Swanton C.. 2015. Biological and therapeutic impact of intratumor heterogeneity in cancer evolution. Cancer Cell. 27:15–26. 10.1016/j.ccell.2014.12.001 [DOI] [PubMed] [Google Scholar]
- McGranahan, N., and Swanton C.. 2017. Clonal heterogeneity and tumor evolution: Past, present, and the future. Cell. 168:613–628. 10.1016/j.cell.2017.01.018 [DOI] [PubMed] [Google Scholar]
- McKenna, A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., et al. 2010. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20:1297–1303. 10.1101/gr.107524.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oskarsson, T., Batlle E., and Massagué J.. 2014. Metastatic stem cells: Sources, niches, and vital pathways. Cell Stem Cell. 14:306–321. 10.1016/j.stem.2014.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertea, M., Kim D., Pertea G.M., Leek J.T., and Salzberg S.L.. 2016. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 11:1650–1667. 10.1038/nprot.2016.095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punt, C.J., Koopman M., and Vermeulen L.. 2017. From tumour heterogeneity to advances in precision treatment of colorectal cancer. Nat. Rev. Clin. Oncol. 14:235–246. 10.1038/nrclinonc.2016.171 [DOI] [PubMed] [Google Scholar]
- Ramilowski, J.A., Goldberg T., Harshbarger J., Kloppmann E., Lizio M., Satagopam V.P., Itoh M., Kawaji H., Carninci P., Rost B., and Forrest A.R.. 2015. A draft network of ligand-receptor-mediated multicellular signalling in human. Nat. Commun. 6:7866. 10.1038/ncomms8866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramilowski, J.A., Goldberg T., Harshbarger J., Kloppmann E., Lizio M., Satagopam V.P., Itoh M., Kawaji H., Carninci P., Rost B., and Forrest A.R.R.. 2016. Corrigendum: A draft network of ligand-receptor-mediated multicellular signalling in human. Nat. Commun. 7:10706. 10.1038/ncomms10706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie, M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., and Smyth G.K.. 2015. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43:e47. 10.1093/nar/gkv007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson, D.R., Wu Y.M., Lonigro R.J., Vats P., Cobain E., Everett J., Cao X., Rabban E., Kumar-Sinha C., Raymond V., et al. 2017. Integrative clinical genomics of metastatic cancer. Nature. 548:297–303. 10.1038/nature23306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roerink, S.F., Sasaki N., Lee-Six H., Young M.D., Alexandrov L.B., Behjati S., Mitchell T.J., Grossmann S., Lightfoot H., Egan D.A., et al. 2018. Intra-tumour diversification in colorectal cancer at the single-cell level. Nature. 556:457–462. 10.1038/s41586-018-0024-3 [DOI] [PubMed] [Google Scholar]
- Scialdone, A., Natarajan K.N., Saraiva L.R., Proserpio V., Teichmann S.A., Stegle O., Marioni J.C., and Buettner F.. 2015. Computational assignment of cell-cycle stage from single-cell transcriptome data. Methods. 85:54–61. 10.1016/j.ymeth.2015.06.021 [DOI] [PubMed] [Google Scholar]
- Shao, X., Liao J., Lu X., Xue R., Ai N., and Fan X.. 2020. scCATCH: Automatic Annotation on Cell Types of Clusters from Single-Cell RNA Sequencing Data. iScience. 23:100882. 10.1016/j.isci.2020.100882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, R., and Seshan V.E.. 2016. FACETS: allele-specific copy number and clonal heterogeneity analysis tool for high-throughput DNA sequencing. Nucleic Acids Res. 44:e131. 10.1093/nar/gkw520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, H., Huang C., Wu J., Li J., Hu T., Wang Z., Zhang H., Shao Y., and Fu Z.. 2021. SCRIB promotes proliferation and metastasis by targeting hippo/YAP signalling in colorectal cancer. Front. Cell Dev. Biol. 9:656359. 10.3389/fcell.2021.656359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solé, L., Lobo-Jarne T., Álvarez-Villanueva D., Alonso-Marañón J., Guillén Y., Guix M., Sangrador I., Rozalén C., Vert A., Barbachano A., et al. 2022. p53 wild-type colorectal cancer cells that express a fetal gene signature are associated with metastasis and poor prognosis. Nat. Commun. 13:2866. 10.1038/s41467-022-30382-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonoda, K., Izumi K., Matsui Y., Inomata M., Shiraishi N., and Kitano S.. 2006. Decreased growth rate of lung metastatic lesions after splenectomy in mice. Eur. Surg. Res. 38:469–475. 10.1159/000095415 [DOI] [PubMed] [Google Scholar]
- Steeg, P.S. 2016. Targeting metastasis. Nat. Rev. Cancer. 16:201–218. 10.1038/nrc.2016.25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart, T., Butler A., Hoffman P., Hafemeister C., Papalexi E., W.M. Mauck, 3rd, Hao Y., Stoeckius M., Smibert P., and Satija R.. 2019. Comprehensive Integration of Single-Cell Data. Cell. 177:1888-1902 e1821. [DOI] [PMC free article] [PubMed]
- Sugiura, K., Masuike Y., Suzuki K., Shin A.E., Sakai N., Matsubara H., Otsuka M., Sims P.A., Lengner C.J., and Rustgi A.K.. 2023. LIN28B promotes cell invasion and colorectal cancer metastasis via CLDN1 and NOTCH3. JCI Insight. 8:e167310. 10.1172/jci.insight.167310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sylvester, B.E., and Vakiani E.. 2015. Tumor evolution and intratumor heterogeneity in colorectal carcinoma: Insights from comparative genomic profiling of primary tumors and matched metastases. J. Gastrointest. Oncol. 6:668–675. 10.3978/j.issn.2078-6891.2015.083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran, H.N., Ang K.S., Chevrier M., Zhang X., Lee N.S., Goh M., and Chen J.. 2020. A benchmark of batch-effect correction methods for single-cell RNA sequencing data. Genome Biol. 21:12. 10.1186/s13059-019-1850-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turajlic, S., and Swanton C.. 2016. Metastasis as an evolutionary process. Science. 352:169–175. 10.1126/science.aaf2784 [DOI] [PubMed] [Google Scholar]
- Turajlic, S., Xu H., Litchfield K., Rowan A., Chambers T., Lopez J.I., Nicol D., O’Brien T., Larkin J., Horswell S., et al. 2018. Tracking cancer evolution reveals constrained routes to metastases: TRACERx renal. Cell. 173:581–594.e12. 10.1016/j.cell.2018.03.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Wetering, M., Francies H.E., Francis J.M., Bounova G., Iorio F., Pronk A., van Houdt W., van Gorp J., Taylor-Weiner A., Kester L., et al. 2015. Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell. 161:933–945. 10.1016/j.cell.2015.03.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vodenkova, S., Buchler T., Cervena K., Veskrnova V., Vodicka P., and Vymetalkova V.. 2020. 5-fluorouracil and other fluoropyrimidines in colorectal cancer: Past, present and future. Pharmacol. Ther. 206:107447. 10.1016/j.pharmthera.2019.107447 [DOI] [PubMed] [Google Scholar]
- Wang, K., Li M., and Hakonarson H.. 2010. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38:e164. 10.1093/nar/gkq603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, R., Song Y., Liu X., Wang Q., Wang Y., Li L., Kang C., and Zhang Q.. 2017. UBE2C induces EMT through Wnt/β-catenin and PI3K/Akt signaling pathways by regulating phosphorylation levels of Aurora-A. Int. J. Oncol. 50:1116–1126. 10.3892/ijo.2017.3880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X., Yamamoto Y., Wilson L.H., Zhang T., Howitt B.E., Farrow M.A., Kern F., Ning G., Hong Y., Khor C.C., et al. 2015. Cloning and variation of ground state intestinal stem cells. Nature. 522:173–178. 10.1038/nature14484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, Z., Liu G., Jia R., Zhang W., Li L., Zhang Y., Wang Z., and Bai X.. 2020. Targeting secretory leukocyte protease inhibitor (SLPI) inhibits colorectal cancer cell growth, migration and invasion via downregulation of AKT. PeerJ. 8:e9400. 10.7717/peerj.9400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, T., Hu E., Xu S., Chen M., Guo P., Dai Z., Feng T., Zhou L., Tang W., Zhan L., et al. 2021. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation (Camb). 2:100141. 10.1016/j.xinn.2021.100141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, Y.Q., Ju C.L., Wang B.J., and Wang R.G.. 2019. PABPC1L depletion inhibits proliferation and migration via blockage of AKT pathway in human colorectal cancer cells. Oncol. Lett. 17:3439–3445. 10.3892/ol.2019.9999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, T., Cho Y.B., Wang K., Huang D., Hong H.K., Choi Y.L., Ko Y.H., Nam D.H., Jin J., Yang H., et al. 2014. Patterns of somatic alterations between matched primary and metastatic colorectal tumors characterized by whole-genome sequencing. Genomics. 104:234–241. 10.1016/j.ygeno.2014.07.012 [DOI] [PubMed] [Google Scholar]
- Xie, Z., Bailey A., Kuleshov M.V., Clarke D.B., Evangelista J.E., Jenkins S.L., Lachmann A., Wojciechowicz M.L., Kropiwnicki E., Jagodnik K.M., et al. 2021. Gene Set Knowledge Discovery with Enrichr. Curr. Protoc. 1:e90. 10.1002/cpz1.90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, Y., Wei Z., Feng M., Zhu D., Mei S., Wu Z., Feng Q., Chang W., Ji M., Liu C., et al. 2022. Tumor-infiltrated activated B cells suppress liver metastasis of colorectal cancers. Cell Rep. 40:111295. 10.1016/j.celrep.2022.111295 [DOI] [PubMed] [Google Scholar]
- Yates, L.R., Knappskog S., Wedge D., Farmery J.H.R., Gonzalez S., Martincorena I., Alexandrov L.B., Van Loo P., Haugland H.K., Lilleng P.K., et al. 2017. Genomic evolution of breast cancer metastasis and relapse. Cancer Cell. 32:169–184.e7. 10.1016/j.ccell.2017.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, G. 2020. Using ggtree to Visualize Data on Tree-Like Structures. Curr. Protoc. Bioinformatics. 69:e96. 10.1002/cpbi.96 [DOI] [PubMed] [Google Scholar]
- Yu, G., Wang L., Han Y., and He Q.. 2012. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 16:284–287. 10.1089/omi.2011.0118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zehir, A., Benayed R., Shah R.H., Syed A., Middha S., Kim H.R., Srinivasan P., Gao J., Chakravarty D., Devlin S.M., et al. 2017. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat. Med. 23:703–713. 10.1038/nm.4333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, Y., Zhang B., Ma Y., Zhao F., Chen J., Wang B., Jin H., Zhou F., Guan J., Zhao Q., et al. 2022. Colorectal cancer patient-derived 2D and 3D models efficiently recapitulate inter- and intratumoral heterogeneity. Adv. Sci. 9:e2201539. 10.1002/advs.202201539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, G.Y., Terry J.M., Belgrader P., Ryvkin P., Bent Z.W., Wilson R., Ziraldo S.B., Wheeler T.D., McDermott G.P., Zhu J., et al. 2017. Massively parallel digital transcriptional profiling of single cells. Nat. Commun. 8:14049. 10.1038/ncomms14049 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
shows clinicopathological data of CRC patients.
shows SC-PDCC information and test results for various analyses.
shows liver and lung metastasis of SC-PDCCs in mice, related to Figs. 2 and S3.
shows the metastasis potential of pooled PDCC in mice, related to Figs. 2 and S3.
shows a total of 21 protein interaction genes with differential expression in HM SC-PDCC lines from P1.
shows the proportion of cancer cells with metastatic signature in each individual primary CRC tumor (from Xu et al. [2022] dataset PRJNA748525).
shows the shRNA information, related to Fig. 7 D.
shows differential CNAs between high and low metastatic SC-PDCC lines in P1.
shows the metastatic signature.
is the source file for Fig. 4.
Data Availability Statement
The raw sequence data reported in this study have been deposited in the Genome Sequence Archive at the National Genomics Data Center, China National Center for Bioinformation/Beijing Institute of Genomics, Chinese Academy of Sciences under the Bioproject accession codes PRJCA006448 and PRJCA023187. Specifically, the WES data and single-cell sequencing data have been deposited under the accession code HRA001280 (https://ngdc.cncb.ac.cn/gsa-human/browse/HRA001280). The RNA-seq data have been deposited under the accession code HRA006587 (https://ngdc.cncb.ac.cn/gsa-human/browse/HRA006587). All additional data supporting the findings of this study are located within the article, in the supplemental information files, or can be obtained from the corresponding author upon request.
The raw sequence data reported in this study have been deposited in the Genome Sequence Archive at the National Genomics Data Center, China National Center for Bioinformation/Beijing Institute of Genomics, Chinese Academy of Sciences under the Bioproject accession codes PRJCA006448 and PRJCA023187. Specifically, the WES data and single-cell sequencing data have been deposited under the accession code HRA001280 (https://ngdc.cncb.ac.cn/gsa-human/browse/HRA001280). The RNA-seq data have been deposited under the accession code HRA006587 (https://ngdc.cncb.ac.cn/gsa-human/browse/HRA006587). All additional data supporting the findings of this study are located within the article, in the supplemental information files, or can be obtained from the corresponding author upon request.