Extended Data Fig. 5 |. Geneformer boosted predictions in multiclass cell type annotation.
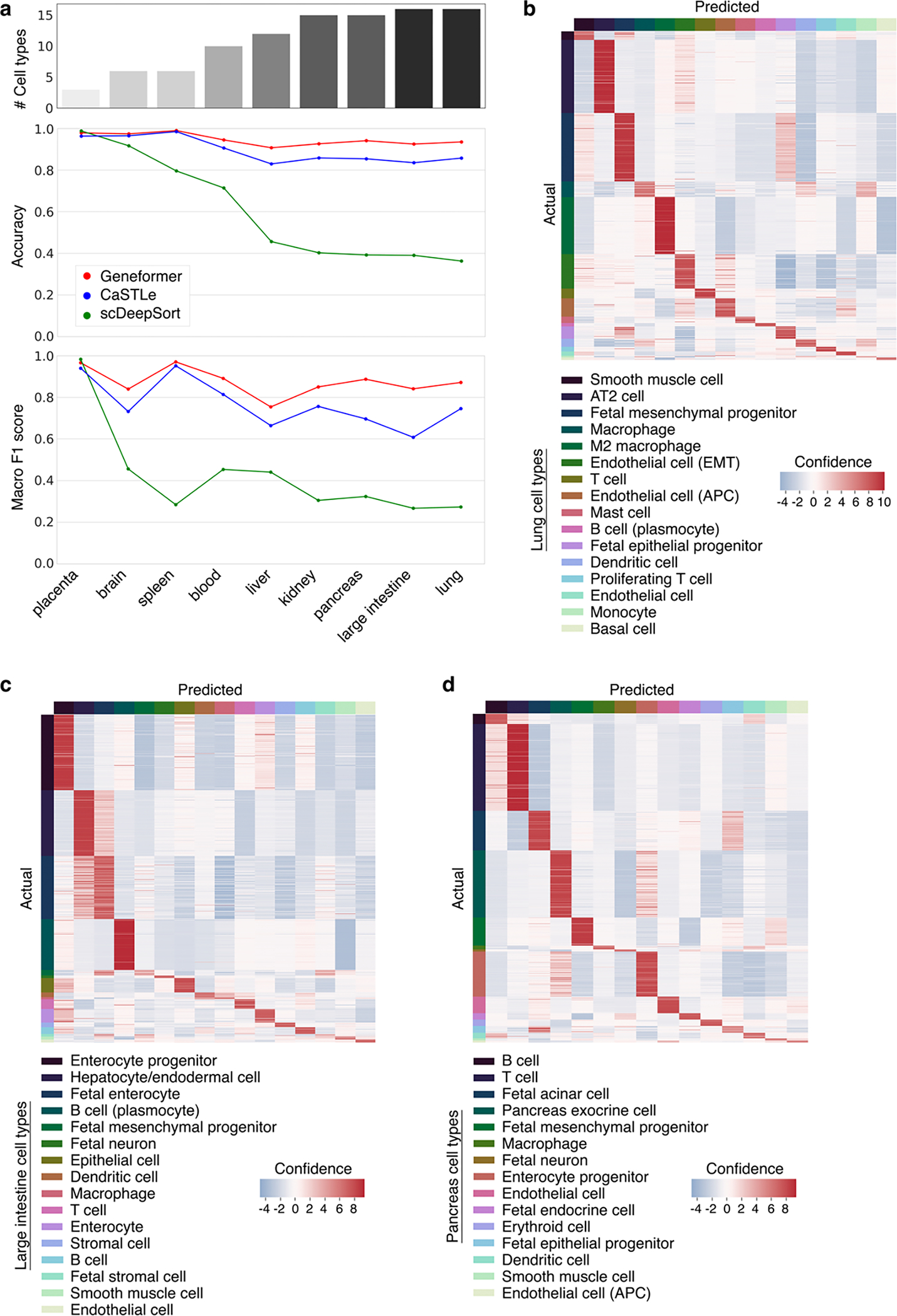
a, Predictive potential (as measured by accuracy and macro F1 score) of Geneformer fine-tuned for cell type annotation in the indicated human tissues as compared to XGBoost (CaSTLe) and deep neural network-based (scDeepSort) methods. The top bar graph indicates the number of cell type classes for each tissue; the gap in performance of Geneformer compared to alternatives increased as the number of cell type classes increased, indicating that Geneformer was robust in even increasingly complex multiclass prediction applications. b, Lung, c, large intestine, or d, pancreas out of sample predictions by Geneformer fine-tuned to distinguish cell types in each tissue (training on 80% of cells, predictions on held-out 20% of cells).
