Abstract
The chemokine receptor CXCR4 is implicated in multiple diseases including inflammatory disorders, cancer growth and metastasis, and HIV/AIDS. CXCR4 targeting has been evaluated in treating cancer metastasis and therapy resistance. Cyclam derivatives, most notably AMD3100 (Plerixafor™), are a common motif in small molecule CXCR4 antagonists. However, AMD3100 has not been shown to be effective in cancer treatment as an individual agent. Configurational restriction and transition metal complex formation increases receptor binding affinity and residence time. In the present study, we have synthesized novel trans-IV locked cyclam-based CXCR4 inhibitors, a previously unexploited configuration, and demonstrated their higher affinity for CXCR4 binding and CXCL12-mediated signaling inhibition compared to AMD3100. These results pave the way for even more potent CXCR4 inhibitors that may provide significant efficacy in cancer therapy.
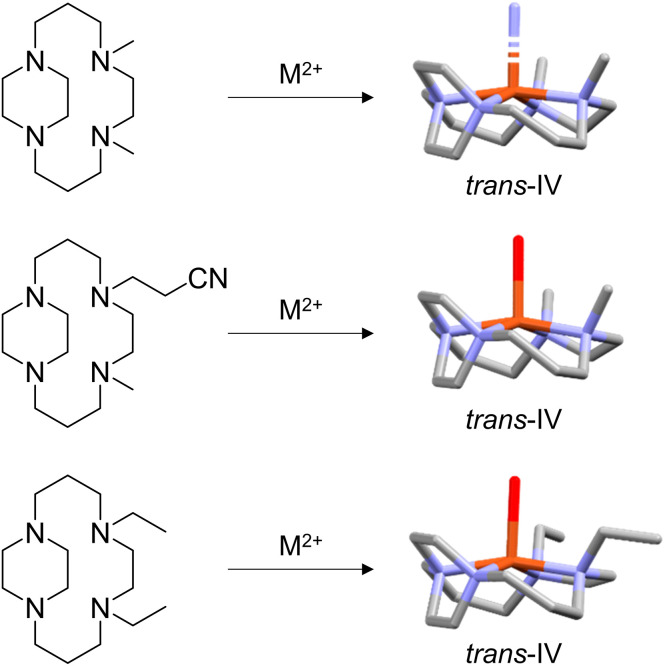
Alkylation of secondary amines on side-bridged cyclam derivatives locks the resulting metal complex in the trans-IV configuration. These derivatives offer new perspectives for the design and development of CXCR4-targeted therapeutic agents.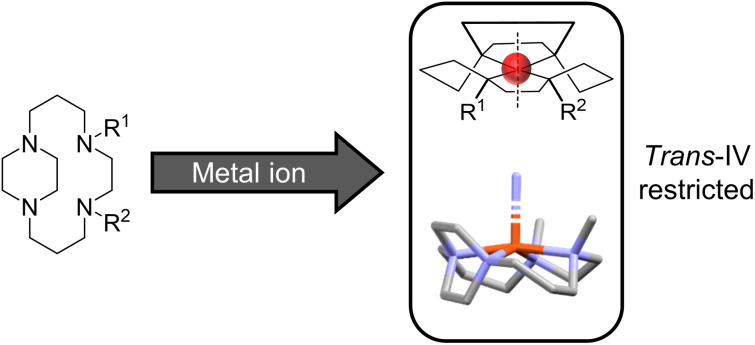
Introduction
Chemokine C–X–C motif receptor 4 (CXCR4) is a seven transmembrane helix G protein-coupled receptor which is, by interaction with its natural ligand CXCL12 (SDF-1α), essential for both organism development and normal physiological function.1 CXCR4 has been implicated in many diseases, including stroke, myocardial infarction and HIV infection.2–4 CXCR4 is overexpressed in multiple cancers; promoting tumour growth, metastatic spread to organs with high concentrations of CXCL12 (e.g. lung and liver), and resistance to standard chemo/radio/immuno-therapy.5–8 This results in particular relevance for clinical molecular imaging of cancers.9–12
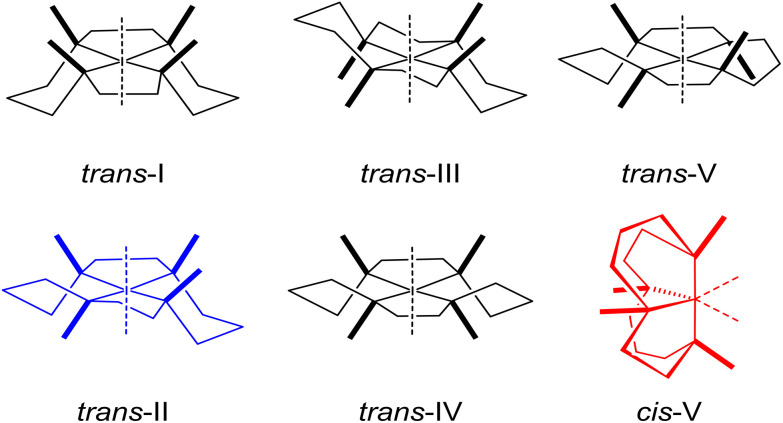
There is current interest in high affinity antagonists which bind to the CXCR4 receptor to prevent HIV entry and/or abrogate CXCL12 signalling.13 Preventing CXCL12-initiated CXCR4 signalling has resulted in a reduction in tumour growth, metastasis inhibition and increase in sensitivity to standard therapies in breast cancer, pancreatic cancer and gliomas.14–16 AMD3100 (Plerixafor™) is a cyclam-based CXCR4 antagonist which is used clinically, in combination with G-CSF, as a hematopoietic stem cell mobilization agent. However, AMD3100 lacks efficacy in cancer treatment and more potent CXCR4 inhibitors may lead to more effective receptor-mediated cancer therapy.13 The cyclam macrocycles in AMD3100 have the ideal cavity size for complex formation with transition metals, and some AMD3100 transition metal complexes have increased CXCR4 binding affinity in comparison to the metal-free compounds.17 Upon metal ion binding, flexible cyclam units can have six possible configurations in solution (Fig. 1), with trans-III the most stable due to its lower strain energy,18 leading to a stochastic overall affinity reflective of the equilibrium between configurations. In addition, transition metal cyclam complexes exhibit low kinetic and thermodynamic stabilities, precluding their use in vivo.19,20
Fig. 1. Six possible configurations of 1st row transition metal cyclam complexes.
Configurational restriction, to lock tetraazamacrocycle derivatives into one configuration rather than being in equilibrium, can be used to optimize amino acid side-chain based receptor coordinate bond interactions and is a route to formation of high affinity antagonists with longer receptor residence times and increased metal complex stabilities.21–28
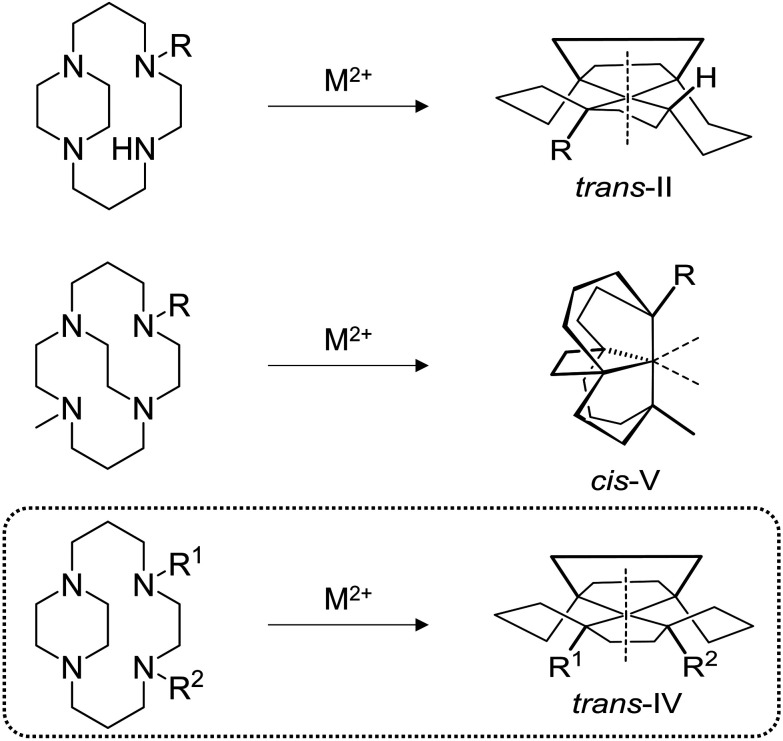
Linking adjacent cyclam nitrogen atoms with an ethylene bridge to form a piperazine ring can be carried out to form “side-bridged” derivatives, which, upon transition metal complex formation, are locked in the trans-II configuration (Fig. 2). Side-bridged (SB) trans-II restricted copper(ii) and zinc(ii) complexes demonstrate superior CXCR4-mediated anti-HIV properties compared with their respective AMD3100 analogues.21–25 Linking of non-adjacent nitrogen atoms with an ethylene linker to form “cross-bridged” (CB) derivatives, and transition metal complex formation, provides macrocycles which are exclusively locked in the cis-V configuration (Fig. 2). The copper(ii) complexes of CB-Bicyclam have multiple advantages over AMD3100 and Cu2AMD3100, including increased affinity, longer receptor residence times and increased complex stability.26,29
Fig. 2. N-Functionalization patterns of the side-bridged and cross-bridged cyclam macrocycles and their corresponding configurations for 1st row transition metal complexes. R = alkyl, R1, R2 ≠ H; M = Cu, Ni, Zn.
In the present work, we demonstrate a route to the formation of a third generation of configurationally restricted CXCR4 antagonists, employing strategies developed by Kaden and co-workers to exclusively form the trans-IV configuration (Fig. 2).30,31 The general synthetic methodology is presented, followed by three significantly different novel examples to show how this methodology can be applied. Biological evaluation of the novel transition metal complexes was carried out in multiple assays to assess inhibitory properties of molecules with this configuration against the CXCR4 receptor.
Results and discussion
Synthesis of trans-IV configurationally restricted compounds
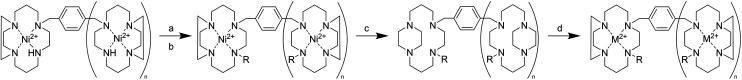
The formation of novel trans-IV derivatives follows a general synthetic pathway (Scheme 1). First, the nickel(ii) trans-II complex is alkylated in a two-step in situ process. n-BuLi is used to deprotonate the secondary amine to form the nickel(ii) stabilized amido complex, which can subsequently be alkylated using an alkyl halide. After formation of the nickel(ii) trans-IV species, the free ligand can be synthesized by decomposing the nickel(ii) complex using NaCN, which can subsequently be reacted with transition metal salts to form complexes with other metal centres.
Scheme 1. General scheme for the formation of trans-IV cyclam complexes; (a) n-BuLi, DMSO, (b) MeI, (c) NaCN, MeCN/H2O, (d) M(OAc)2, MeOH (n = 0 or 1).
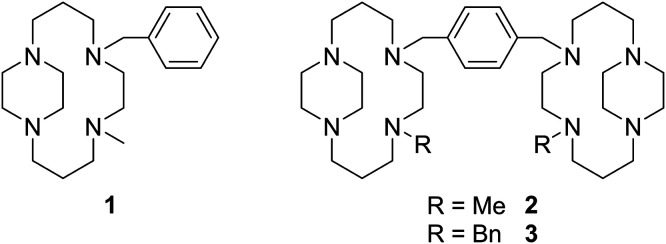
To develop and optimise the methodology, the synthesis of a mono-cyclam derivative 1 was first attempted (Fig. 3). The side-bridged NH precursor was synthesized in three steps following our previously described protocols.25,27,32 Formation of the trans-II nickel(ii) complex was carried out using nickel(ii) nitrate in dry methanol, using methodologies we have described previously.27 Alkylation of the secondary amine to form the novel trans-IV structures was carried out by a method analogous to that first described by Kaden and co-workers.30 The secondary amine nickel(ii) complex was deprotonated using n-BuLi in dry DMSO. Successful deprotonation can be clearly observed by noting the colour change from orange of the starting material to blue of the amido derivative, which is stable in aprotic conditions. The amido complex can subsequently be alkylated using methyl iodide, with observation of an orange colour indicating product formation. The complex [Ni1]2+ can then be isolated as the hexafluorophosphate salt. The alkylated trans-IV nickel(ii) complex can be decomposed to form the free ligand 1 using sodium cyanide, in a method similar to that used for nickel(ii) templated cyclam synthesis,33 with minor synthetic modifications introduced due to the lower solubility of the hexafluorophosphate complex. Complex formation reactions with zinc(ii) and copper(ii) acetate were then be carried out following our previously described methodology to synthesise trans-II complexes,21,22,26 with the trans-IV complexes forming in good yields.
Fig. 3. Novel ligands synthesized following the described general synthetic pathway.
In our previous studies we have shown that either mechanical restriction (cis-V) or sterics (trans-II) can fix the configuration of the cyclam complex on addition of an ethylene bridge between ring nitrogen donor atoms. Examination of previously determined X-ray crystal structures of alkylated side-bridge cyclam complexes demonstrates that the trans-IV configuration is adopted, although this was not highlighted at the time of publication. A search of the CCDC found a total of three deposited single crystal X-ray structures of transition metal complexes where a side-bridged cyclam chelator had been alkylated in a similar synthetic procedure to give related compounds. Despite variations in substituents on the cyclam nitrogens (methyl, ethylcyano or ethyl), all three structures clearly adopt the trans-IV configuration (Fig. 4). In contrast to the trans-II complexes, this steric restriction favours the two non-bridged nitrogen methyl groups oriented on the same side of the macrocyclic ring plane with the side bridge piperazine carbons oriented to the other side of the plane. These complexes offer the opportunity to probe the biological impact of this fixed configuration on coordination interactions with the CXCR4 chemokine receptor and utilising biological assays to determine binding potency and inhibitory activity.
Fig. 4. Examples of N-functionalization of the side-bridged cyclam resulting in trans-IV configuration of the M2+ metal complex and associated crystal structures.
Mono-ring configurationally restricted macrocyclic complexes are not ideal as CXCR4 antagonists as they can interact with only one receptor amino acid side-chain via a coordination interaction.28 Bis-macrocyclic complexes have significantly higher affinities as the metal ions can form coordination interactions with both Asp171 and Asp262.34,35 The methodologies developed for the synthesis of 1 were successfully translated to the bis-macrocyclic analogue 2 without modification (Fig. 3). The three-step ligand-to-ligand reactions from the secondary amine to the tetra-alkylated amine was carried out in an overall yield of 76% using gram amounts. Identity and purity were confirmed by analysis using 1H NMR, mass spectrometry and elemental analysis.
It is of interest to ascertain if the methodology described to synthesize trans-IV bis-macrocyclic CXCR4 antagonists is limited to highly reactive alkylating agents such as methyl iodide or has wider utility. To assess this, methyl iodide used in the synthesis of 2 was replaced with benzyl bromide, containing a less reactive halogen leaving group and bulkier substituent, to form 3 (Fig. 3). Reaction conditions did not need to be modified and 3 could be synthesized as easily as 2 in a 69% three-step ligand-to-ligand overall yield. Versatility in substitution indicates the potential for formation of a wider range of derivatives of bis-macrocyclic trans-IV CXCR4 antagonists, allowing for variables such as lipophilicity, π–π stacking ability and steric effects to be evaluated for binding effects. Alkylation with a protected functional group may also allow for further functionalization.24,27,32
In vitro evaluation of the CXCR4 affinity of the novel trans-IV metal complexes
The synthesised trans-IV complexes were screened for their CXCR4 binding properties using three in vitro assays: anti-HIV replication, CXCL12 competition binding and CXCL12-induced calcium signalling. CXCR4 is one of the two main co-receptors involved in the infection process of HIV-1.36,37 The effect of the compounds was tested on the viral infection of the CXCR4-using HIV-1 strain NL4-3 (×4). Binding competition with the natural chemokine (CXCL12) binding was directly determined using AlexaFluor647-labelled CXCL12.38,39 As a measure of abrogation of the natural CXCR4/CXCL12 signalling pathway, a functional downstream assay can be carried out to measure Ca2+ ions released from the endoplasmic reticulum.40,41 All assays were carried out over a concentration range and are presented as IC50 values (Table 1).
CXCR4-dependent effects and cellular toxicity of trans-IV metal complexes.
| CXCR4-mediated effects, IC50 (nM) | CC50 (μM) | |||
|---|---|---|---|---|
| Anti-HIV activitya | CXCL12 bindingbx | Ca2+ signallingc | Cellular toxicityd | |
| 1 | >5000 | >1000 | >10 000 | >100 |
| [Ni1]2+ | >5000 | >3000 | >2000 | 26.0 ± 6.4 |
| [Cu1]2+ | >5000 | >10 000 | >2000 | 68.2 ± 3.4 |
| [Zn1]2+ | >5000 | >1000 | >2000 | >100 |
| 2 | >5000 | >1000 | >10 000 | >100 |
| [Ni22]4+ | >5000 | >1000 | >2000 | 11.7 ± 2.8 |
| [Cu22]4+ | >5000 | >1000 | >2000 | 37.9 ± 7.1 |
| [Zn22]4+ | 642.0 ± 193.7 | 5.5 ± 0.7 | 67.5 ± 38.9 | 46.7 ± 4.0 |
| 3 | >5000 | >1000 | >10 000 | >50 |
| [Ni23]4+ | >5000 | 668.0 ± 106.1 | >2000 | 10.6 ± 0.7 |
| [Cu23]4+ | 332.5 ± 6.4 | 25.5 ± 3.5 | 108.0 ± 11.3 | 32.7 ± 0.3 |
| [Zn23]4+ | 327.0 ± 25.5 | 4.5 ± 0.7 | 72.5 ± 38.9 | 40.7 ± 0.2 |
| AMD3100 | 20.0 ± 1.4 | 18.0 ± 4.2 | 203.5 ± 19.429 | >100 |
| [Ni2AMD3100]4+ | 7.942,e | ND | 2.042,e | >10042,e |
| [Cu2AMD3100]4+ | 22.442,e | ND | 48.742,e | >10042,e |
| [Zn2AMD3100]4+ | 6.842,e | ND | 2.942,e | >10042,e |
Concentration required to inhibit the replication of HIV-1 strain NL4-3 (×4) by 50% in MT-4 cells.
Concentration required to inhibit CXCL12 binding by 50%.
Concentration required to reduce the level of Ca2+ ions observed during a normal signalling process by 50% in U87.CD4.CXCR4 cells.
Concentration required to reduce cell viability by 50% in MT-4 cells.
Concentration values published in μg mL−1 converted to nM.
AMD3100 binds to CXCR4 via hydrogen bonding to aspartate residues 171 and 262.34 Tetra-alkylated macrocyclic ligands have very low hydrogen bonding potential and therefore demonstrate no measurable affinity towards the CXCR4 receptor (Table 1). Mono-macrocyclic transition metal complexes are known to weakly bind to the receptor,27,28 relative to our high affinity bis-macrocycles. In this study, mono-macrocyclic complexes [Ni1]2+, [Cu1]2+ and [Zn1]2+ demonstrated micromolar level binding in the CXCL12 binding assay and no measurable activity up to 5 μM or 2 μM for anti-HIV activity and calcium signalling, respectively. Hence, they are characterized as relatively low affinity binding agents. For the methyl substituted bis-macrocyclic trans-IV complexes (2), the zinc(ii) complex is the only compound to demonstrate any significant affinity/activity; with the copper(ii) and nickel(ii) complexes showing similar activity to their mono-ring counterparts.
Modification of the methyl substituent to a benzyl (3) on either of the nickel(ii) or zinc(ii) complex does not significantly alter the activity in these assays, however an increase in potency is observed for the copper(ii) complex, giving it a similar profile to that of [Zn23]4+. [Zn22]4+ and [Zn23]4+ are more effective at blocking CXCL12 interactions with the receptor than AMD3100 in both the signalling and ligand binding assays but are less potent in the anti-HIV assay. All compounds were assessed in cellular toxicity assays and have CC50 values >10 μM.
Comparison can also be made to the published assay values for the metal complexes of AMD3100 in related assays, showing a similar trend to AMD3100, with higher potency for [M2AMD3100]4+ in the anti-HIV assay compared to the trans-IV complexes. The calcium signalling assay shows that significant disruption of the binding interaction has occurred for the nickel(ii) complex [Ni23]4+ relative to [Ni2AMD3100]4+. Further comparison in the CXCL12 binding assay would be useful to determine any differences in the binding inhibition and residence time assays are needed to better understand the potential in vivo properties. The trans-IV configuration may confer additional advantages in terms of residence time and complex stability due to the structural rigidity.26
Experimental
General information
Bulk solvent was removed by evaporation under reduced pressure and trace solvent was removed using a Schlenk line. NMR spectra were recorded with a Jeol ECZ 400S spectrometer (400.2 MHz for 1H and 100.6 MHz for 13C). Chemical shifts (δ) are reported in ppm, using internal TMS or residual non-deuterated solvent signal as standards. High- and low-resolution mass spectra were recorded using an electrospray ion-trap LC-MS in positive mode. Elemental analysis was performed using a CHN analyser EA1108 (Carlo Erba). UV-vis spectra were obtained using an Agilent 800 diode spectrometer. Side-bridged secondary amine ligands were synthesised following our previously reported methods.21,25,27
Nickel(ii) 5-benzyl-1,5,8,12-tetraazabicyclo[10.2.2]hexadecane complex
5-Benzyl-1,5,8,12-tetraazabicyclo[10.2.2]hexadecane (1.10 g, 3.47 mmol) was dissolved in degassed anhydrous MeOH (25 mL). An anhydrous methanolic (10 mL) solution of nickel(ii) nitrate hexahydrate (1.11 g, 3.82 mmol) was added dropwise and the mixture was refluxed under argon for 12 h. Solvent was removed in vacuo and the macrocycle complex was purified via size exclusion chromatography (Sephadex LH20) in MeOH to yield bright orange crystals (1.51 g, 87%). Elemental analysis: calculated (%) for [C19H32N4Ni](NO3)2·0.1H2O·0.1MeOH: C, 45.50; H, 6.52; N, 16.67. Found (%): C, 45.51; H, 6.77; N, 16.65. MS (ESI): m/z = 436.19 ([[C19H32N4Ni](NO3)]+). HRMS (ESI): calculated for [C19H32N4Ni]2+, 187.0985; found 187.0981.
Di-nickel(ii) 1,4-bis(1,5,8,12-tetraazabicyclo[10.2.2]hexadecan-5-yl)xylene complex
First reported by Smith et al.27 Amounts used: 1,4-bis(1,5,8,12-tetraazabicyclo[10.2.2]hexadecan-5-yl)xylene (3.3 g, 5.95 mmol) in dry MeOH (50 mL), an anhydrous methanolic (20 mL) solution of nickel(ii) nitrate hexahydrate (3.80 g, 13.08 mmol). To yield bright orange crystals (4.91 g, 90%). Elemental analysis: calculated (%) for [C32H58N8Ni2](NO3)4·0.5H2O·2MeOH: C, 41.11; H, 6.80; N, 16.92. Found: C, 41.07; H, 6.77; N, 16.98. MS (ESI): m/z = 857.40 ([[C32H58N8Ni2](NO3)3]+). HRMS (ESI): calculated for [C32H58N8Ni2]4+, 167.5867; found 167.5864. UV-vis (H2O): λmax = 526 nm.
General procedure for N-alkylation of Ni(ii) complex to generate trans-IV structures
The nickel(ii) complex was dissolved in dry DMSO (15 mL) and stirred in vacuo for 1 h. The orange solution was purged with argon for 10 minutes. A solution of n-butyl lithium (2.5 M) in hexane was added dropwise over 30 min. A blue colour developed and the mixture was stirred at room temperature for 30 min. Alkyl halide was added dropwise and an orange solution re-appeared, which was stirred at room temperature for 1 h. The solution was stirred under vacuum again to remove hexane and alkyl halide. The DMSO solution was added dropwise to a saturated solution of ammonium hexafluorophosphate in ethanol. The orange precipitate was filtered and washed with diethyl ether (100 mL). The orange solid was dissolved in acetonitrile and evaporated in vacuo to yield an orange crystalline solid.
[Ni1]2+
Amounts used: Nickel(ii) 5-benzyl-1,5,8,12-tetraaza-bicyclo[10.2.2]hexadecane nitrate (1.2 g, 2.4 mmol), n-butyl lithium (2.5 M) in hexane (0.4 mL, 4.8 mmol, 2 equiv.), methyl iodide (1.36 g, 9.6 mmol, 4 equiv.), ammonium hexafluorophosphate (40 equiv.) in ethanol (40 mL). Yield (1.5 g, 92%). Elemental analysis: calculated (%) for [C20H34N4Ni](PF6)2·2.2H2O·0.15 MeCN: C, 33.63; H, 5.40; N, 8.02. Found (%): C, 33.71; H, 5.70; N, 8.01. MS (ESI): m/z = 533.19 ([[C20H34N4Ni](PF6)]+). HRMS (ESI): calculated for [[C20H34N4Ni](NO3)]+, 450.2010; found 450.1998.
[Ni22]4+
Amounts used: Di-nickel(ii) 1,4-bis(1,5,8,12-tetraazabicyclo[10.2.2]hexadecan-5-yl)xylene complex (1.5 g, 1.63 mmol), n-butyl lithium (2.5 M) in hexane (0.6 ml, 6.52 mmol, 4 equiv.), methyl iodide (1.85 g, 13.04 mmol, 8 equiv.), ammonium hexafluorophosphate (60 equiv.) in ethanol (60 mL). Yield (1.88 g, 90%). Elemental analysis: calculated (%) for [C34H62N8Ni2](PF6)4·2.4H2O: C, 30.86; H, 5.09; N, 8.47. Found: C, 30.80; H, 5.12; N, 8.61. MS (ESI): m/z = 991.08 ([[C34H62N8Ni2](H)(PF6)2]+). HRMS (ESI): calculated for [[C34H62N8Ni2](CH3COO)2]2+, 408.2030; found 408.2028.
[Ni23]4+
Amounts used: Di-nickel(ii) 1,4-bis(1,5,8,12-tetraazabicyclo[10.2.2]hexadecan-5-yl)xylene complex (1.2 g, 1.30 mmol), n-butyl lithium (2.5 M) in hexane (0.5 mL, 0.33 g, 5.22 mmol, 4 equiv.), benzyl bromide (1.78 g, 10.4 mmol, 8 equiv.), ammonium hexafluorophosphate (60 equiv.) in ethanol (60 mL). Yield (1.01 g, 85%). Elemental analysis: calculated (%) for [C46H70N8Ni2](PF6)4·1.4H2O: C, 37.91; H, 5.03; N, 7.69. Found: C, 37.88; H, 5.02; N, 7.60. MS (ESI): m/z = 570.19 ([[C46H70N8Ni2](PF6)2]2+). HRMS (ESI): calculated for [[C34H62N8Zn2](NO3)2]2+, 487.2088; found 487.2075.
General procedure for free ligand formation
The nickel(ii) complex was dissolved in a mixture of water/MeCN (90 : 10) to form a bright orange solution. Sodium cyanide was added to the orange solution in one portion, the mixture became less intensely coloured and was heated at 80 °C for 24 h. The resulting mixture was cooled to room temperature, then in an ice bath. The nickel cyanide salt was removed by filtration and the filtrate was concentrated in vacuo to remove MeCN. KOH was added to the aqueous residue to adjust the pH to 14. The basic aqueous layer was extracted with DCM (5 × 40 mL). The combined organic layers were dried over MgSO4, filtered and concentrated in vacuo to yield a colourless oil, which was dried on a Schlenk line overnight to yield a white solid.
Ligand 1
Amounts used: [Ni1]2+ (1.4 g, 2.06 mmol) in 10 mL water/MeCN, sodium cyanide (400 mg, 8.24 mmol). Yield (622 mg, 91%). 1H NMR (CDCl3): δ 7.45–7.23 (m, 5H, Ar–H), 4.87 (s, 2H, CH2–Ar), 4.33 (s, 4H, CH2–N), 4.24–4.18 (m, 2H, CH2–N), 3.12–3.01 (m, 2H, CH2–N), 2.86 (s, 2H, CH2–N), 2.70–2.58 (m, 5H, CH2–N), 2.43–2.19 (m, 5H, CH2–N), 1.81 (s, 3H, CH3), 1.54 (m, 2H, CH2– –N), 1.48 (m, 2H, CH2– –N). 13C NMR (CDCl3): δ 23.51 (N–CH2), 25.62 (N–CH2), 44.88 (N–CH3), 48.19 (N–CH2), 50.21 (N–CH2), 55.32 (N–CH2), 55.71 (N–CH2), 56.46 (N–CH2), 56.91 (N–CH2), 61.11 (CH2–Ar), 124.32 (Ar–C), 129.51 (Ar–CH), 129.82 (Ar–CH), 143.22 (Ar–C). Elemental analysis: calculated (%) for C20H34N4·2.3H2O: C, 64.58; H, 10.46; N, 15.06. Found: C, 64.20; H, 10.37; N, 15.19. MS (ESI): m/z = 331.2 ([M + H]+). HRMS (ESI): calculated for [M + H]+, 331.2856; found 331.2849.
Ligand 2
Amounts used: [Ni22]4+ (1.8 g, 1.40 mmol) in 20 mL water/MeCN, sodium cyanide (700 mg, 14 mmol). Yield (770 mg, 94%). 1H NMR (CDCl3): δ 7.20 (s, 4H, CH–Ar), 3.63 (s, 4H, N–CH2–Ar), 3.22 (m, 4H, N–CH2), 3.01 (m, 4H, N–CH2), 2.82 (m, 4H, N–CH2), 2.61 (m, 4H, N–CH2), 2.52 (m, 8H, N–CH2), 2.44 (m, 12H, N–CH2), 2.14 (m, 4H, N–CH2), 2.01 (s, 6H, CH3), 1.75 (m, 4H, N–CH2), 1.66 (m, 4H, N–CH2). 13C NMR (CDCl3): δ 19.51 (N– –CH2), 20.45 (N– –CH2), 41.12 (N–CH3), 44.73 (N–CH2), 49.13 (N–CH2), 51.02 (N–CH2), 54.21 (N–CH2), 57.43 (N–CH2), 59.31 (N–CH2), 63.55 (N–CH2–Ar), 122.82 (Ar–CH), 141.22 (Ar–C). Elemental analysis: calculated (%) for C34H62N8·1.35H2O: C, 67.25; H, 10.74; N, 18.45. Found: C, 67.18; H, 10.59; N, 18.63. MS (ESI): m/z = 583.5 ([M + H]+). HRMS (ESI): calculated for [M + H]+, 583.5170; found 583.5167.
Ligand 3
Amounts used: [Ni23]4+ (1 g, 0.7 mmol) in 20 mL water/MeCN, sodium cyanide (350 mg, 7 mmol). Yield (465 mg, 90%). 1H NMR (CDCl3): δ 7.20–7.37 (s, 14H, C–H–Ar), 3.61 (s, 8H, N–CH2–Ar), 3.15 (m, 4H, N–CH2), 2.97 (m, 4H, N–CH2), 2.79 (m, 4H, N–CH2), 2.57 (m, 4H, N–CH2), 2.46 (m, 8H, N–CH2), 2.24 (m, 12H, N–CH2), 2.04 (m, 4H, N–CH2), 1.69 (m, 4H, N–CH2), 1.62 (m, 4H, N–CH2). 13C NMR (CDCl3): δ 22.53 (N– –CH2), 23.47 (N– –CH2), 47.12 (N–CH2), 49.11 (N–CH2), 57.73 (N–CH2), 59.28 (N–CH2), 66.12 (N–CH2–Ar), 67.19 (N–CH2–Ar), 118.44 (Ar–CH), 130.07 (Ar–CH), 131.94 (Ar–CH), 133.44 (Ar–CH), 141.22 (Ar–C), 144.18 (Ar–C). Elemental analysis: calculated (%) for C46H70N8·0.8H2O: C, 73.71; H, 9.63; N, 14.95. Found: C, 73.70; H, 9.52; N, 14.79. MS (ESI): m/z = 735.58 ([M + H]+).
General procedure for metal complex formation reaction
The macrocycle ligand was dissolved in degassed anhydrous MeOH. A solution of metal(ii) acetate in anhydrous MeOH was added dropwise to the mixture at 80 °C. The reaction was heated to reflux for 24 h. Solvent was removed in vacuo to 1 mL and the complex was purified via size exclusion chromatography (Sephadex LH20). Solvent was removed to yield the desired compound.
[Cu1]2+
Amounts used: ligand 1 (200 mg, 0.6 mmol) in methanol (10 mL), copper acetate monohydrate (132 mg, 0.66 mmol) in methanol (5 mL). Yield a green–blue solid (290 mg, 94%). Elemental analysis: calculated (%) for [C20H34N4Cu](CH3COO)2·2.6H2O·1MeOH: C, 50.80; H, 8.39; N, 9.48. Found (%): C, 50.92; H, 8.47; N, 9.18. MS (ESI): m/z = 452.22 ([[C20H34N4Cu](CH3COO)]+). HRMS (ESI): calculated for [C20H34N4Cu]2+, 196.6034; found 196.6030. UV-vis (H2O): λmax = 533 nm.
[Zn1]2+
Amounts used: ligand 1 (200 mg, 0.6 mmol) in methanol (10 mL), zinc acetate dihydrate (145 mg, 0.66 mmol) in methanol (5 mL). Yield a yellow solid (290 mg, 94%). Elemental analysis: calculated (%) for [C20H34N4Zn](CH3COO)2·2.2H2O·1.2MeOH: C, 51.12; H, 8.38; N, 9.46. Found (%): C, 50.99; H, 8.43; N, 9.52. MS (ESI): m/z = 453.22 ([[C20H34N4Zn](CH3COO)]+). HRMS (ESI): calculated for [[C20H34N4Zn](CH3COO)]+, 453.2202; found 453.2222. UV-vis (H2O): λmax = 526 nm.
[Cu22]4+
Amounts used: ligand 2 (200 mg, 0.34 mmol) in methanol (10 mL), copper acetate monohydrate (150 mg, 0.75 mmol) in methanol (5 mL). Yield a purple-blue solid (300 mg, 93%). Elemental analysis: calculated (%) for [C34H62N8Cu2](CH3COO)4·1.2H2O·1MeOH: C, 51.65; H, 8.11; N, 11.21. Found (%): C, 51.62; H, 8.17; N, 11.25. HRMS (ESI): calculated for [[C34H62N8Cu2](CH3COO)2]2+, 413.1972; found 413.1966. UV-vis (H2O): λmax = 518 nm.
[Zn22]4+
Amounts used: ligand 2 (200 mg, 0.34 mmol) in methanol (10 mL), zinc acetate dihydrate (137 mg, 0.75 mmol) in methanol (5 mL). Yield a yellow solid (270 mg, 92%). Elemental analysis: calculated (%) for [C34H62N8Zn2](CH3COO)4·2H2O·0.8MeOH: C, 50.82; H, 8.09; N, 11.08. Found (%): C, 50.89; H, 8.03; N, 11.18. MS (ESI): m/z = 415.20 ([[C34H62N8Zn2](CH3COO)2]2+). HRMS (ESI): calculated for [[C34H62N8Zn2](CH3COO)2]2+, 415.1956; found 415.1949. UV-vis (H2O): λmax = 536 nm.
[Cu23]4+
Amounts used: ligand 3 (100 mg, 0.14 mmol) in methanol (6 mL), copper acetate monohydrate (62 mg, 0.31 mmol) in methanol (2 mL). Yield a purple-blue solid (125 mg, 82%). Elemental analysis: calculated (%) for [C46H70N8Cu2](CH3COO)4·0.8H2O·0.7MeOH: C, 57.87; H, 7.67; N, 9.87. Found: C, 57.80; H, 7.65; N, 9.83. MS (ESI): m/z = 489.23 ([[C46H70N8Cu2](CH3COO)2]2+). HRMS (ESI): calculated for [[C46H70N8Cu2](CH3COO)2]2+, 489.2281; found 489.2274. UV-vis (H2O): λmax = 502 nm.
[Zn23]4+
Amounts used: ligand 2 (100 mg, 0.14 mmol) in methanol (6 mL), zinc acetate dihydrate (68 mg, 0.31 mmol) in methanol (2 mL). Yield a yellow solid (120 mg, 78%). Elemental analysis: calculated (%) for [C46H70N8Zn2](CH3COO)4·0.9H2O·0.7MeOH: C, 57.60; H, 7.65; N, 9.82. Found (%): C, 57.58; H, 7.62; N, 9.80. MS (ESI): m/z = 491.23 ([[C46H70N8Zn2](CH3COO)2]2+). HRMS (ESI): calculated for [[C46H70N8Zn2](CH3COO)2]2+, 492.2264; found 492.2247. UV-vis (H2O): λmax = 535 nm.
Biological evaluation
Cell lines and compounds
Peripheral blood mononuclear cells (PBMCs) activated with phytohemagglutinin (PHA; Sigma, Belgium) were used for CXCL12 binding studies. The glioblastoma astrocytoma U87.CD4 cell line transfected with CXCR4 was used for measurement of CXCL12-induced calcium mobilization. The MT-4 cell line was previously described.43 The CXCR4 inhibitor AMD3100 (Plerixafor™) was a gift from Dr Gary Bridger affiliated at that time with AnorMED (Langley, Canada). The HIV-1 NL4.3 (CXCR4-using, ×4) strain was obtained through the AIDS Research and Reference Reagent Program (Division of AIDS, NIAID, NIH).
HIV infection assay
MT-4 cells were seeded (1 × 104 cells per well) in transparent 96-well plates in DMEM (Dulbecco's Modified Eagle Medium; Life Technologies, Waltham, MA, USA) supplemented with 10% Foetal Bovine Serum (FBS) and 10 mM HEPES. Subsequently, compounds were added and the cell/compound mixture was incubated at 37 °C for 30 min. Thereafter, HIV-1 NL4.3 was added (100 pg p-24 per well) and incubated with the cells for 48 h. The HIV-1 induced cytopathogenic effect (CPE) was evaluated microscopically and cell viability was quantified using a tetrazolium-based colorimetric assay as described before.43
Chemokine (CXCL12) binding inhibition assay
Human peripheral blood lymphocytes (PBLs) were washed once with assay buffer (Hanks’ balanced salt solution with 20 mM HEPES buffer and 0.2% bovine serum albumin, pH 7.4) and then incubated for 15 min at room temperature with CXCR4 antagonists, at a range of concentrations, diluted in assay buffer. Subsequently, AlexaFluor647-labelled CXCL12 (CXCL12-AF647) (25 ng mL−1) was added (30 min, room temperature) to the compound-incubated cells. Thereafter, the cells were washed twice in assay buffer, fixed in paraformaldehyde (1%) in PBS and analysed on the FL4 channel of a FACSCalibur flow cytometer equipped with a 635 nm red diode laser (Becton Dickinson, San Jose, CA, USA). The percentage inhibition of CXCL12-AF647 binding was calculated according to the formula: [1 − ((MFI − MFINC)/(MFIPC − MFINC))] × 100 where MFI is the mean fluorescence intensity of the cells incubated with CXCL12-AF647 in the presence of the inhibitor, MFINC is the mean fluorescence intensity measured in the negative control (i.e., autofluorescence of unlabeled cells) and MFIPC is the mean fluorescence intensity of the positive control (i.e., cells exposed to CXCL12-AF647 alone).
Measurement of intracellular CXCL12-induced calcium mobilization
CXCR4-positive U87 glioblastoma/astrocytoma cells were loaded (45 min, room temperature) with the fluorescent calcium indicator Fluo-3 acetoxymethyl (Molecular Probes) (4 μM) in assay buffer (Hanks’ balanced salt solution with 20 mM HEPES buffer and 0.2% bovine serum albumin, pH 7.4). After thorough washing with assay buffer, cells were pre-incubated (10 min, 37 °C) in the same buffer with the various CXCR4 antagonists at a range of concentrations. The intracellular calcium mobilization in response to CXCL12 addition was then measured by monitoring the fluorescence as a function of time in all the wells simultaneously by using a Fluorometric Imaging Plate Reader (FLIPR; Molecular Devices, Sunnyvale, CA, USA).40,41
Determination of cellular toxicity
Cytotoxicity measurements in MT-4 cells were based on the viability of the cells that had been exposed to various concentrations of the test compound.44 After the cells were allowed to proliferate for 5 days, the number of viable cells was quantified by a tetrazolium-based colorimetric method, as originally described by Pauwels et al.45
Conclusions
Some conclusions can be drawn about the potential for trans-IV configuration cyclam complexes as high affinity CXCR4 binding agents, for example, in comparison with the structurally similar trans-II configuration. Whilst nickel(ii), copper(ii) and zinc(ii) complexes as trans-II structures are all highly active (in the order zinc > nickel > copper),21,25,27 transition metal ion selection has a significant effect on the binding of trans-IV complexes. Neither of the nickel(ii) trans-IV compounds developed in this study demonstrated low nanomolar affinity for the CXCR4 receptor. Of the three fixed configurations studied to date (trans-II, cis-V and trans-IV), trans-IV is the most variable in CXCR4 binding properties across the range of transition metals tested, with copper(ii) and zinc(ii) complexes showing superiority to AMD3100 in some assays. Targeting CXCL12 binding, and subsequent CXCR4 receptor activation, is the key step in developing potent CXCR4 inhibition agents for cancer therapy. A further useful feature is the facile functionalization of the trans-IV structure, using the methodologies presented in this study. Variation in substituents can have a major impact on CXCR4 binding, as observed for the copper(ii) complex. This is consistent with the sensitivity of the binding affinity to the coordination environment for copper(ii) due to its coordination properties.
Overall, the high affinity and inhibitory action of zinc(ii) trans-IV structures (compared to AMD3100) coupled with the versatility in substitution to tune the molecular properties, opens the door to a new sub-class of compounds that may exhibit increased drug efficacy and improved properties as potential CXCR4-targeted anti-cancer agents.
Author contributions
Conceptualisation: S. O. A., G. Mc., D. S., S. J. A.; Methodology: G. Mc., B. P. B., T. J. H., D. S., S. J. A.; Validation: B. P. B., I. R.; Investigation: S. O. J., G. Mc., A. K., T. D'H, T. V. L., A. N. W., I. R.; Writing – original draft: S. O. A., G. Mc., T. D'H., T. V. L., I. R., D. S., B. P. B., S. J. A.; Writing – review & editing: A. K., I. R., T. J. H., D. S., B. P. B., S. J. A.; Supervision: T. J. H., D. S., S. J. A.; Funding acquisition: D. S., S. J. A.
Conflicts of interest
There are no conflicts to declare.
Supplementary Material
Acknowledgments
We gratefully acknowledge the Daisy Appeal Charity (Grant: DAhul0211 and fellowship funding for BPB), Yorkshire Cancer Research (HEND376), the KU Leuven (Grant: C22/17/008), and the University of Hull (UoH) for PET infrastructure support. We thank Dr Assem Allam and his family for their generous donation to help found the PET Research Centre at the UoH. Mass spectrometry data was acquired at the UK National Mass Spectrometry Facility (NMSF) at Swansea University. This work was supported by the Medical Research Council (grant number MC_PC_18049).
References
- Zou Y. R. Kottmann A. H. Kuroda M. Taniuchi I. Littman D. R. Nature. 1998;393:595–599. doi: 10.1038/31269. [DOI] [PubMed] [Google Scholar]
- Hummel S. Van Aken H. Zarbock A. Curr. Opin. Hematol. 2014;21:29–36. doi: 10.1097/MOH.0000000000000002. [DOI] [PubMed] [Google Scholar]
- Ruscher K. Kuric E. Liu Y. Walter H. L. Issazadeh-Navikas S. Englund E. Wieloch T. J. Cereb. Blood Flow Metab. 2013;33:1225–1234. doi: 10.1038/jcbfm.2013.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleul C. C. Wu L. Hoxie J. A. Springer T. A. Mackay C. R. Proc. Natl. Acad. Sci. U. S. A. 1997;94:1925–1930. doi: 10.1073/pnas.94.5.1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cojoc M. Peitzsch C. Trautmann F. Polishchuk L. Telegeev G. D. Dubrovska A. OncoTargets Ther. 2013;6:1347–1361. doi: 10.2147/OTT.S36109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duda D. G. Kozin S. V. Kirkpatrick N. D. Xu L. Fukumura D. Jain R. K. Clin. Cancer Res. 2011;17:2074–2080. doi: 10.1158/1078-0432.CCR-10-2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domanska U. M. Kruizinga R. C. Nagengast W. B. Timmer-Bosscha H. Huls G. de Vries E. G. Walenkamp A. M. Eur. J. Cancer. 2013;49:219–230. doi: 10.1016/j.ejca.2012.05.005. [DOI] [PubMed] [Google Scholar]
- Trautmann F. Cojoc M. Kurth I. Melin N. Bouchez L. C. Dubrovska A. Peitzsch C. Int. J. Radiat. Biol. 2014;90:687–699. doi: 10.3109/09553002.2014.906766. [DOI] [PubMed] [Google Scholar]
- Renard I. Domarkas J. Poty S. Burke B. P. Roberts D. P. Goze C. Denat F. Cawthorne C. J. Archibald S. J. Nucl. Med. Biol. 2023;120–121:108335. doi: 10.1016/j.nucmedbio.2023.108335. [DOI] [PubMed] [Google Scholar]
- Baghdadi N. E. Burke B. P. Alresheedi T. Nigam S. Saeed A. Almutairi F. Domarkas J. Khan A. Archibald S. J. Dalton Trans. 2021;50:1599–1603. doi: 10.1039/D0DT02626C. [DOI] [PubMed] [Google Scholar]
- Renard I. and Archibald S. J., in Medicinal Chemistry, ed. P. J. Sadler and R. VanEldik, 2020, vol. 75, pp. 447–476 [Google Scholar]
- Mayerhoefer M. E. Archibald S. J. Messiou C. Staudenherz A. Berzaczy D. Schoder H. J. Magn. Reson. Imaging. 2020;51:1325–1335. doi: 10.1002/jmri.26848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grande F. Giancotti G. Ioele G. Occhiuzzi M. A. Garofalo A. Eur. J. Med. Chem. 2017;139:519–530. doi: 10.1016/j.ejmech.2017.08.027. [DOI] [PubMed] [Google Scholar]
- Lefort S. Thuleau A. Kieffer Y. Sirven P. Bieche I. Marangoni E. Vincent-Salomon A. Mechta-Grigoriou F. Oncogene. 2017;36:1211–1222. doi: 10.1038/onc.2016.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S. Srivastava S. K. Bhardwaj A. Owen L. B. Singh A. P. Br. J. Cancer. 2010;103:1671–1679. doi: 10.1038/sj.bjc.6605968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurth R. Bajetto A. Harrison J. K. Barbieri F. Florio T. Front. Cell. Neurosci. 2014;8:144. doi: 10.3389/fncel.2014.00144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlach L. O. Jakobsen J. S. Jensen K. P. Rosenkilde M. R. Skerlj R. T. Ryde U. Bridger G. J. Schwartz T. W. Biochemistry. 2003;42:710–717. doi: 10.1021/bi0264770. [DOI] [PubMed] [Google Scholar]
- Bosnich B. Poon C. K. Tobe M. L. Inorg. Chem. 1965;4:1102–1108. doi: 10.1021/ic50030a003. [DOI] [Google Scholar]
- Nimmagadda S. Pullambhatla M. Stone K. Green G. Bhujwalla Z. M. Pomper M. G. Cancer Res. 2010;70:3935–3944. doi: 10.1158/0008-5472.CAN-09-4396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mewis R. E. Archibald S. J. Coord. Chem. Rev. 2010;254:1686–1712. doi: 10.1016/j.ccr.2010.02.025. [DOI] [Google Scholar]
- Valks G. C. McRobbie G. Lewis E. A. Hubin T. J. Hunter T. M. Sadler P. J. Pannecouque C. De Clercq E. Archibald S. J. J. Med. Chem. 2006;49:6162–6165. doi: 10.1021/jm0607810. [DOI] [PubMed] [Google Scholar]
- Silversides J. D. Allan C. C. Archibald S. J. Dalton Trans. 2007:971–978. doi: 10.1039/b615329a. doi: 10.1039/b615329a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan A. Greenman J. Archibald S. J. Curr. Med. Chem. 2007;14:2257–2277. doi: 10.2174/092986707781696618. [DOI] [PubMed] [Google Scholar]
- Khan A. Silversides J. D. Madden L. Greenman J. Archibald S. J. Chem. Commun. 2007:416–418. doi: 10.1039/b614557d. doi: 10.1039/b614557d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McRobbie G. Valks G. C. Empson C. J. Khan A. Silversides J. D. Pannecouque C. De Clercq E. Fiddy S. G. Bridgeman A. J. Young N. A. Archibald S. J. Dalton Trans. 2007:5008–5018. doi: 10.1039/b705800d. doi: 10.1039/b705800d. [DOI] [PubMed] [Google Scholar]
- Khan A. Nicholson G. Greenman J. Madden L. McRobbie G. Pannecouque C. De Clercq E. Ullom R. Maples D. L. Maples R. D. Silversides J. D. Hubin T. J. Archibald S. J. J. Am. Chem. Soc. 2009;131:3416–3417. doi: 10.1021/ja807921k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith R. Huskens D. Daelemans D. Mewis R. E. Garcia C. D. Cain A. N. Freeman T. N. Pannecouque C. De Clercq E. Schols D. Hubin T. J. Archibald S. J. Dalton Trans. 2012;41:11369–11377. doi: 10.1039/C2DT31137B. [DOI] [PubMed] [Google Scholar]
- Maples R. D. Cain A. N. Burke B. P. Silversides J. D. Mewis R. E. D'Huys T. Schols D. Linder D. P. Archibald S. J. Hubin T. J. Chemistry. 2016;22:12916–12930. doi: 10.1002/chem.201601468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke B. P. Miranda C. S. Lee R. E. Renard I. Nigam S. Clemente G. S. D'Huys T. Ruest T. Domarkas J. Thompson J. A. Hubin T. J. Schols D. Cawthorne C. J. Archibald S. J. J. Nucl. Med. 2020;61:123–128. doi: 10.2967/jnumed.118.218008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowallick R. Neuburger M. Zehnder M. Kaden T. A. Helv. Chim. Acta. 1997;80:948–959. doi: 10.1002/hlca.19970800326. [DOI] [Google Scholar]
- Siegfried L. Kowallick R. Kaden T. A. Supramol. Chem. 2001;13:357–367. doi: 10.1080/10610270108027490. [DOI] [Google Scholar]
- Abdulwahaab B. H. Burke B. P. Domarkas J. Silversides J. D. Prior T. J. Archibald S. J. J. Org. Chem. 2016;81:890–898. doi: 10.1021/acs.joc.5b02464. [DOI] [PubMed] [Google Scholar]
- Barefield E. K. Wagner F. Hodges K. D. Inorg. Chem. 1976;15:1370–1377. doi: 10.1021/ic50160a025. [DOI] [Google Scholar]
- Gerlach L. O. Skerlj R. T. Bridger G. J. Schwartz T. W. J. Biol. Chem. 2001;276:14153–14160. doi: 10.1074/jbc.M010429200. [DOI] [PubMed] [Google Scholar]
- Liang X. Parkinson J. A. Weishaupl M. Gould R. O. Paisey S. J. Park H. S. Hunter T. M. Blindauer C. A. Parsons S. Sadler P. J. J. Am. Chem. Soc. 2002;124:9105–9112. doi: 10.1021/ja0260723. [DOI] [PubMed] [Google Scholar]
- Schols D. Este J. A. Henson G. DeClercq E. Antiviral Res. 1997;35:147–156. doi: 10.1016/S0166-3542(97)00025-9. [DOI] [PubMed] [Google Scholar]
- Schols D. Struyf S. Van Damme J. Este J. A. Henson G. De Clercq E. J. Exp. Med. 1997;186:1383–1388. doi: 10.1084/jem.186.8.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatse S. Princen K. Liekens S. Vermeire K. De Clercq E. Schols D. Cytometry, Part A. 2004;61:178–188. doi: 10.1002/cyto.a.20070. [DOI] [PubMed] [Google Scholar]
- Schoofs G. Van Hout A. D’Huys T. Schols D. Van Loy T. J. Visualized Exp. 2018;133:57271. doi: 10.3791/57271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Princen K. Hatse S. Vermeire K. De Clercq E. Schols D. Cytometry, Part A. 2003;51:35–45. doi: 10.1002/cyto.a.10008. [DOI] [PubMed] [Google Scholar]
- Claes S. D’Huys T. Van Hout A. Schols D. Van Loy T. J. Visualized Exp. 2018;132:56780. doi: 10.3791/56780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esté J. A. Cabrera C. De Clercq E. Struyf S. Van Damme J. Bridger G. Skerlj R. T. Abrams M. J. Henson G. Gutierrez A. Clotet B. Schols D. Mol. Pharmacol. 1999;55:67–73. doi: 10.1124/mol.55.1.67. [DOI] [PubMed] [Google Scholar]
- Vermeire K. Princen K. Hatse S. De Clercq E. Dey K. Bell T. W. Schols D. AIDS. 2004;18:2115–2125. doi: 10.1097/00002030-200411050-00003. [DOI] [PubMed] [Google Scholar]
- Harada S. Koyanagi Y. Yamamoto N. Science. 1985;229:563–566. doi: 10.1126/science.2992081. [DOI] [PubMed] [Google Scholar]
- Pauwels R. Balzarini J. Baba M. Snoeck R. Schols D. Herdewijn P. Desmyter J. Declercq E. J. Virol. Methods. 1988;20:309–321. doi: 10.1016/0166-0934(88)90134-6. [DOI] [PubMed] [Google Scholar]