Abstract
A novel aerobic methanotrophic bacterium, designated as strain IN45T, was isolated from in situ colonisation systems deployed at the Iheya North deep-sea hydrothermal field in the mid-Okinawa Trough. IN45T was a moderately thermophilic obligate methanotroph that grew only on methane or methanol at temperatures between 25 and 56 °C (optimum 45–50 °C). It was an oval-shaped, Gram-reaction-negative, motile bacterium with a single polar flagellum and an intracytoplasmic membrane system. It required 1.5–4.0 % (w/v) NaCl (optimum 2–3 %) for growth. The major phospholipid fatty acids were C16 : 1ω7c, C16 : 0 and C18 : 1ω7c. The major isoprenoid quinone was Q-8. The 16S rRNA gene sequence comparison revealed 99.1 % sequence identity with Methylomarinovum caldicuralii IT-9T, the only species of the genus Methylomarinovum with a validly published name within the family Methylothermaceae. The complete genome sequence of IN45T consisted of a 2.42-Mbp chromosome (DNA G+C content, 64.1 mol%) and a 20.5-kbp plasmid. The genome encodes genes for particulate methane monooxygenase and two types of methanol dehydrogenase (mxaFI and xoxF). Genes involved in the ribulose monophosphate pathway for carbon assimilation are encoded, but the transaldolase gene was not found. The genome indicated that IN45T performs partial denitrification of nitrate to N2O, and its occurrence was indirectly confirmed by N2O production in cultures grown with nitrate. Genomic relatedness indices between the complete genome sequences of IN45T and M. caldicuralii IT-9T, such as digital DNA–DNA hybridisation (51.2 %), average nucleotide identity (92.94 %) and average amino acid identity (93.21 %), indicated that these two methanotrophs should be separated at the species level. On the basis of these results, strain IN45T represents a novel species, for which we propose the name Methylomarinovum tepidoasis sp. nov. with IN45T (=JCM 35101T =DSM 113422T) as the type strain.
Keywords: deep-sea, hydrothermal, methane oxidation, methanotroph, Okinawa Trough , thermophilic
Introduction
Methane sources can be found in a variety of marine environments [1], among which deep-sea hydrothermal vent fields are attractive due to their ecological uniqueness [2]. The results of molecular studies have indicated that several deep-sea hydrothermal fields harbour diverse aerobic methanotrophic bacteria [3,7], whereas no methanotrophic isolates from deep-sea hydrothermal environments have been described. In contrast, from shallow submarine hydrothermal systems, moderately thermophilic or thermotolerant marine methanotrophs have been isolated and described. These are Methylomarinovum caldicuralii IT-9T (growing at temperatures up to 55 °C) of the family Methylothermaceae from a water depth of 23 m [8] and Methylocaldum marinum S8T (growing at temperatures up to 47 °C) of the family Methylococcaceae from a water depth of 161 m [9], both members of the order Methylococcales within the class Gammaproteobacteria.
The family Methylothermaceae currently consists of three genera (Methylothermus, Methylohalobius and Methylomarinovum) and four methanotrophic species, including two species of the genus Methylothermus isolated from terrestrial hot springs [10,11], one species of the genus Methylohalobius from a hypersaline lake [12] and one species of the genus Methylomarinovum (i.e., M. caldicuralii IT-9T) from a shallow submarine hydrothermal system in a coral reef area [8]. All four species are moderate thermophiles and/or slight or moderate halophiles; these physiological characteristics are typical of members of the family Methylothermaceae. As molecular signatures of members of the family Methylothermaceae have been detected in deep-sea hydrothermal fields, such as the particulate methane monooxygenase gene pmoA from the 13 °N East Pacific Rise [6] and the Rainbow field in the Mid-Atlantic Ridge [6], and a near-complete genome sequence from the southern Lau Basin [7], it is likely that deep-sea hydrothermal fields are also habitats for methanotrophs of the family Methylothermaceae.
We have studied methanotrophic bacteria at the Original site in the Iheya North deep-sea hydrothermal field in the mid-Okinawa Trough, Japan, and recently reported the presence of various methanotrophs of the order Methylococcales [5]. In a series of studies in this field, we successfully isolated a moderately thermophilic methanotroph, designated as strain IN45T, from deep-sea habitats at a depth of approximately 1000 m. Rush et al. [13] have analysed IN45T previously for bacteriohopanepolyols, but it has not been described taxonomically. Herein, we characterised IN45T as representing a novel species of the genus Methylomarinovum within the family Methylothermaceae.
Methods
Enrichment and isolation
To collect microorganisms from the Original site in the Iheya North deep-sea hydrothermal field, in situ colonisation systems (ISCSs) were deployed in chemosynthetic animal colonies for 2 months from November 2013 (cruise NT13-22) to January 2014 (cruise KY14-01) using the remotely operated vehicle Hyper-Dolphin. ISCS-1 and ISCS-4 were used in this cultivation experiment. These two ISCSs were deployed in colonies of the galatheoid crab Shinkaia crosnieri in hydrothermal diffuse-flow areas at depths of 1058 m (27 ° 47′ 25″ N, 126 ° 54′ 02″ E) and 986 m (27 ° 47′ 27″ N, 126 ° 53′ 48″ E), respectively. The deployment and recovery of ISCSs and a detailed description of the site have been reported previously [5].
A methane-fed continuous flowthrough cultivation system [5] was used to enrich methanotrophs from a mixture of ceramic particles in ISCS-1 and ISCS-4. This system provides methanotrophs with a constant supply of low concentrations of growth substrates and avoids the accumulation of excreted metabolites. Compared with a batch cultivation system, it may provide conditions more similar to natural habitats and may help deep-sea methanotrophs to acclimate to laboratory conditions. The cultivation system consisted of a medium bottle connected to a bag containing a mixture of 83.8 % N2, 15 % CH4 and 1.2 % O2, a peristaltic pump (Masterflex L/S 7550–50; Cole-Parmer) controlled by an electronic on–off timer, and a cultivation column. The culture medium was periodically supplied at a rate of 20 ml min−1 for 1 min, followed by a 1 h pause. The cultivation column was heated to 45 °C in the incubator. The medium (pH 6.8) was prepared using REI-SEA Marine seawater (IWAKI) supplemented with (per litre) 0.1 g NaHCO3, 29 mg NH4NO3, 5 mg Na2HPO4, 2 mg KH2PO4, 0.025 mg CuSO4·5H2O, and 1.6 ml DSMZ 141 trace element solution.
Cultivation was initiated in February 2014 using the methane-fed continuous flowthrough system, and an enrichment culture of methanotrophic bacteria was obtained in April 2014 (5 weeks after the start of cultivation). For isolation, a portion of the enrichment culture was transferred to 3 ml of MJmet medium [8]. The medium was prepared in a 15 ml glass test tube, and the final pH was adjusted to 6.4 and 6.8 with gas phases of 28 % CH4, 13 % CO2, 6 % O2 and 28 % CH4, 5 % CO2, 6 % O2 (N2 balance, 150 kPa), respectively. The inoculated test tubes were incubated at 45 °C with shaking at 120 r.p.m., and cell growth was observed at pH 6.4 after 1 week of incubation. The culture was purified by repeating the serial dilution-to-extinction technique at least four times, as described previously [10]. As a result, a methanotrophic bacterium, designated as IN45T, was isolated. Heterotrophic contamination was tested using a medium containing 0.1–1 % (w/v) yeast extract instead of methane, but no such contamination was detected. The purity of the isolate was confirmed by successful direct sequencing of the partial 16S rRNA gene at least three times in independent cultures. Unless otherwise specified, IN45T was subsequently analysed by culturing under the same conditions as used for isolation.
16S rRNA gene analysis
Genomic DNA was extracted from IN45T using a DNeasy UltraClean Microbial Kit (Qiagen). The 16S rRNA gene was amplified via PCR using Bac27F and U1492R primers [14], and the purified PCR products were directly Sanger-sequenced. The resulting sequence (1466 bp) was subjected to a blast search (https://blast.ncbi.nlm.nih.gov/Blast.cgi), and sequence similarity was further analysed using GENETYX-MAC version 21.0.1 (GENETYX). The 16S rRNA gene sequences of IN45T and reference strains were aligned using sina [15] on the Silva website (https://www.arb-silva.de/). The alignment was corrected manually where necessary and ambiguous regions were deleted. A maximum likelihood phylogenetic tree was reconstructed using the IQ-TREE web server [16] with the substitution model of Tamura and Nei with empirical base frequencies, a proportion of invariable sites and the discrete gamma model with four rate categories (TN+F+I+G4) selected by ModelFinder [17] and ultrafast bootstrap analysis of 1000 replicates [18].
The 16S rRNA gene sequence has been deposited at DDBJ/EMBL/GenBank (accession number LC770110).
Whole-genome analysis
The whole-genome sequences of IN45T and M. caldicuralii IT-9T were determined. A culture of M. caldicuralii IT-9T, which has been maintained in our laboratory since its isolation, was used for the analysis. Genomic DNA was extracted using a NucleoSpin Tissue kit (Macherey-Nagel). Sequencing was performed on the MiSeq (Illumina) and Sequel (PacBio) platforms. PacBio subreads were assembled using the HGAP4 pipeline [19] from the PacBio SMRT toolkit (SMRT Link v6.0.0). Subsequently, the contigs were extended and combined with Illumina reads using PRICE version 1.0 [20]. Finally, the assemblies were error-corrected using Pilon version 1.18 [21]. Protein-coding genes in both genomes were predicted using Prodigal version 2.6.3 [22]. Their functional annotations were assigned using the Kyoto Encyclopedia of Genes and Genomes (KEGG) orthology database as a reference. Noncoding RNA genes, such as rRNAs and tRNAs, were predicted using INFERNAL version 1.1.4 [23] and tRNAscan-SE version 1.3.1 [24].
To infer the taxonomic position of the isolate, overall genomic relatedness was assessed. Digital DNA–DNA hybridisation (dDDH) values were estimated using the Genome-to-Genome Distance Calculator 3.0 (GGDC; http://ggdc.dsmz.de/ggdc.php#) [25]. Average nucleotide identity (ANI) and average amino acid identity (AAI) were estimated using OrthoANI [26] and AAI calculator (http://enve-omics.ce.gatech.edu/aai/), respectively. A genome-based phylogenetic tree was reconstructed using a concatenated amino acid alignment of 96 single-copy marker genes identified using the Genome Taxonomy Database Toolkit [27]. A maximum likelihood phylogenomic tree was inferred using RAxML [28] with the Le and Gascuel four-matrix model fused with free-rate heterogeneity and gamma rate heterogeneity (LG4X+G) and 300 bootstrap replicates.
For phylogenetic analysis of PmoA, the complete PmoA sequence (254 amino acid positions) deduced from the genome of IN45T was aligned with reference PmoA sequences using muscle [29]. A maximum likelihood phylogenetic tree was reconstructed using the IQ-TREE web server [16] with the substitution model Le and Gascuel with empirical base frequencies and the discrete gamma model with four rate categories (LG+F+G4) selected by ModelFinder [17] and ultrafast bootstrap analysis of 1000 replicates [18].
The genome sequences have been deposited at DDBJ/EMBL/GenBank under accession numbers AP024718 and AP024719 for the chromosome and plasmid of IN45T, respectively, and AP024714 for the genome of M. caldicuralii IT-9T.
Morphological analysis
Cells were routinely observed by phase-contrast microscopy using a BX51 microscope (Olympus). The Gram reaction of cells was determined using the KOH method [30]. The presence of flagella was determined by negative staining of cells with 1 % (w/v) neutral phosphotungstic acid. Ultrathin sections of cells were prepared by modifying a method reported previously [31]. Briefly, cells at the exponential growth phase were fixed with 2.5 % (w/v) glutaraldehyde and then subjected to high-pressure freezing (EM-PACT2, Leica) and freeze substitution. To enhance the contrast of intracellular organelles, the samples were en bloc stained via sequential treatment with osmium tetroxide and thiocarbohydrazide (OTO staining) [32] prior to dehydration and then embedded in epoxy resin. Transmission electron microscopy was performed under a Tecnai 20 electron microscope (FEI/Thermo Fisher Scientific) operating at 120 kV.
Physiological analysis
Growth conditions were examined using MJmet medium, which was modified as necessary. For the pH range test, media with different pH values were prepared by adding HCl or NaOH. Growth on solid media was assessed at 37 °C using MJmet medium solidified at a slant with 0.8 % gellan gum (Nacalai Tesque) or 1.5 % Noble agar (Difco) in a 100 ml vial with a butyl rubber cap. Before solidification, each solidifying agent was autoclaved in distilled water and then combined with an appropriate concentration of MJmet medium. The final pH of the solidified media was adjusted from 6.0 to 6.3. The gas phase of the vial was prepared as 16–23 % CH4, 5–10 % CO2 and 5–6 % O2 (N2 balance, 120–130 kPa).
Growth on carbon substrates other than methane was examined by replacing methane with N2. Methanol was evaluated at a concentration range of 0.1–6 % (v/v). The following carbon substrates were also tested: 0.05 % (w/v) formate, acetate, citrate, succinate, glucose, fructose, ribose, mannitol, methylamine, dimethylamine and ethanol as well as 0.1 % (w/v) yeast extract and casamino acids. Nitrogen sources for growth were examined using MJmet medium prepared without nitrogen compounds as the base medium. The following substrates were tested: 0.05 and 0.1 % (w/v) casamino acids and 0.05 % (w/v) NH4Cl, NaNO3, NaNO2, urea, Tris, methylamine, dimethylamine and l-aspartate.
N2O and O2 were analysed using cultures (3 ml of medium in a 15 ml tube) grown under low-oxygen (1 % O2, 140 kPa) or normal-oxygen (7 % O2, 140 kPa) conditions. The headspace gas (0.2 ml for N2O and 0.3 ml for O2) in the culture tubes was analysed using a 7890A GC system (Agilent Technologies) equipped with a thermal conductivity detector. The analysis of N2O was performed on a ShinCarbon ST micropacked column (100/120 mesh, 2 m×1 mm internal diameter, Restek) with a helium carrier at a flow rate of 7.0 ml min−1 and an oven temperature programme as follows: 100 °C for 2 min, ramp at 15 °C min−1 to 300 °C and hold for 6 min. The analysis of O2 was performed on a Molesieve 5A micropacked column (80/100 mesh, 2 m×1 mm internal diameter, Restek) with an argon carrier at a flow rate of 7.0 ml min−1 and an oven temperature programme as follows: 30 °C for 6.5 min, ramp at 60 °C min−1 to 120 °C and hold for 2 min.
Chemotaxonomic analysis
Isoprenoid quinones, polar lipids and whole-cell fatty acids were analysed using exponentially grown cells. Isoprenoid quinones and polar lipids were extracted from lyophilised cells using methods described previously [33]. Isoprenoid quinones were purified by thin-layer chromatography and analysed using HPLC [34]. Polar lipids were determined using two-dimensional thin-layer chromatography [33,34]. Fatty acid methyl esters were prepared following the Sherlock Microbial Identification System protocol [35]. To determine the double bond positions, a portion of the fatty acid methyl esters was derivatised with dimethyl disulphide [36]. Fatty acid methyl esters were analysed on a JMS-Q1500GC GC-MS system (JEOL) using two capillary columns of different polarity. The first analysis was performed on a Supelco SP-2560 column (100 m×0.25 mm internal diameter, 0.20 µm film thickness) at a helium carrier flow rate of 1.0 ml min−1 with an oven temperature programme of 160–240 °C (2 °C min−1) and hold time of 15 min. The second analysis was performed on an Agilent DB-5MS column (30 m×0.25 mm internal diameter, 0.25 µm film thickness) at a helium flow rate of 1.5 ml min−1 with an oven temperature programme of 120–280 °C (320 °C for dimethyl disulphide derivatives, 4 °C min−1) and hold time of 5 min. All data were combined, and the fatty acid composition was calculated.
Results and discussion
Genetic identification of IN45T
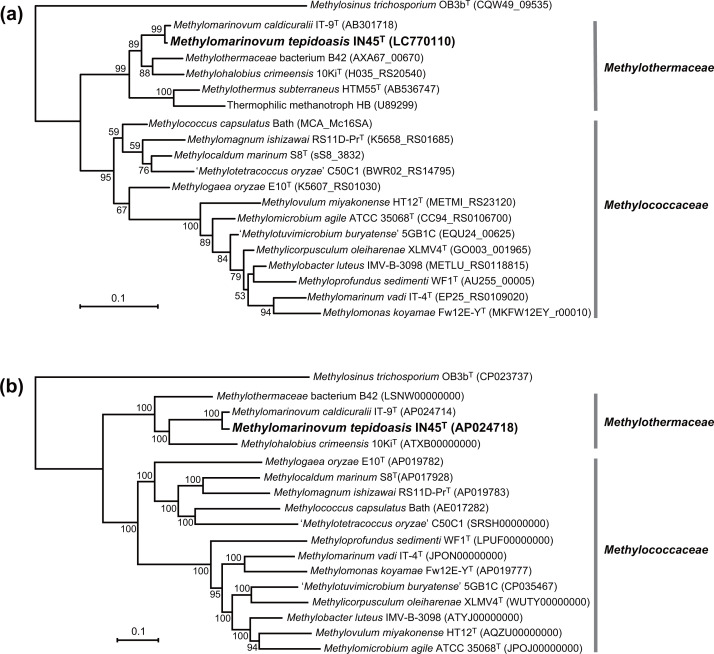
A methanotrophic enrichment culture was obtained from ISCSs deployed in chemosynthetic animal colonies in the Iheya North deep-sea hydrothermal field, and from which a methanotrophic bacterium, designated as IN45T, was isolated at 45 °C. The 16S rRNA gene sequence (1466 bp) was obtained for IN45T via PCR amplification and direct sequencing. Sequence comparison revealed that IN45T was closely related to M. caldicuralii IT-9T within the family Methylothermaceae, with a sequence identity of 99.1 %. This value was above the proposed species boundary cut-off (98.65 %) [37]. IN45T was moderately related to other members of the family Methylothermaceae, including Methylohalobius crimeensis 10KiT (94.7 % identity), species of the genus Methylothermus (91.4–91.6 %) and the uncultured bacterium B42 identified from a deep-sea hydrothermal vent (94.6 %) [7] The phylogenetic tree of 16S rRNA gene sequences is shown in Fig. 1a.
Fig. 1. Phylogenetic position of strain IN45T among reference species of the families Methylothermaceae and Methylococcaceae. (a) Maximum likelihood phylogenetic tree based on 16S rRNA gene sequences (1393 nucleotide positions). Bootstrap values >50 % are indicated at nodes. (b) Phylogenomic tree based on the concatenated amino acid sequences of 96 single-copy marker genes. The reference strains with available high-quality genomes were used in the analysis. Accession numbers or gene locus tags are shown in parentheses. Methylosinus trichosporium OB3bT was used as an outgroup species.
Owing to the high degree of similarity between the 16S rRNA gene sequences of IN45T and M. caldicuralii IT-9T, it was necessary to compare their genomes in order to assign a taxonomic position to IN45T. Accordingly, we performed whole-genome sequencing of IN45T. In addition, the reference bacterium M. caldicuralii IT-9T, for which no genome sequence was available, was analysed. As a result, the complete genome sequences of IN45T and M. caldicuralii IT-9T were obtained. The genome of IN45T was reconstructed as a 2.42 Mbp chromosome and a 20.5 kbp plasmid, and the chromosomal DNA G+C content was 64.1 mol%. The M. caldicuralii IT-9T genome was 2.69 Mbp in size, with a DNA G+C content of 64.6 mol%. The genome features are summarised in Table 1.
Table 1. Summary of genomic and phenotypic features of strain IN45T and a related species of the genus Methylomarinovum.
Strains: 1, IN45T (data from this study); 2, Methylomarinovum caldicuralii IT-9T (data from this study and from 8); +, Present or positive; −, absent or negative; pMMO, particulate methane monooxygenase; sMMO, soluble methane monooxygenase; RuMP, ribulose monophosphate; RuBisCO, ribulose-1,5-bisphosphate carboxylase; PS, phosphatidylserine; APL, unknown aminophospholipid; PL, unknown phospholipid; Q-8, ubiquinone 8.
| Characteristic | 1 | 2 |
| Genome information: | ||
| Genome accession numbers | AP024718, AP024719 | AP024714 |
| Assembly status | Complete | Complete |
| Number of contigs | 2 (Chromosome, 1; Plasmid, 1) | 1 |
| Chromosome size (bp) | 2 421 873 | 2 694 844 |
| Plasmid size (bp) | 20 514 | − |
| Chromosome DNA G+C content (mol%) | 64.1 | 64.6 |
| Plasmid DNA G+C content (mol%) | 58.7 | − |
| Genome coverage | 1058× | 870× |
| Number of protein coding genes | 2380 | 2614 |
| Number of rRNA genes (16S, 23S, 5S) | 2, 2, 2 | 2, 2, 2 |
| Number of tRNA genes | 46 | 47 |
| Presence of genes for: | ||
| pMMO | + | + |
| sMMO | − | − |
| MxaFI methanol dehydrogenase | + | + |
| XoxF methanol dehydrogenase | + | + |
| Nitrogen fixation | − | − |
| Hydroxylamine dehydrogenase | + | + |
| Dissimilatory nitrate reductase | + | − |
| Assimilatory nitrate reductase | + | − |
| RuMP pathway | + | + |
| Serine pathway | − | − |
| RuBisCO | − | − |
| Hemerythrin | + | − |
| Growth conditions: | ||
| Temperature (optimum) (°C) | 25–56 (45–50) | 30–55 (45–50) |
| pH (optimum) | 5.2–6.9 (5.9–6.4) | 5.3–6.9 (6.0–6.4) |
| NaCl concentration (optimum) (%, w/v) | 1.5–4 (2–3) | 1–5 (3) |
| Nitrogen sources | Ammonium, Nitrate | Ammonium, Urea |
| Vitamin requirement | + | − |
| Cell morphology (cell size, µm) | Ovoids (1.0–3.0×0.8–1.5) | Ovoids or Cocci (0.9–1.5×0.6–1.3) |
| Motility | + (polar flagellum) | + (polar flagellum) |
| Major fatty acids (>10 %) | C16 : 1ω7c, C16 : 0, C18 : 1ω7c | C16 : 0, C18 : 1ω7c |
| Major polar lipids | PS | APL, PL |
| Major quinone | Q-8 | Q-8 |
Overall genomic relatedness based on dDDH, ANI and AAI was determined between IN45T and M. caldicuralii IT-9T. The dDDH value was 51.2 % and the probability of a dDDH value ≥70 % was 22.5 % (formula 2). The ANI and AAI values were 92.94 and 93.21 %, respectively. All these values were below the proposed thresholds for species delineation (70 % for dDDH, 95–96 % for ANI, and 95 % for AAI) [25,38,40], indicating that IN45T is distinct from M. caldicuralii IT-9T at the species level. In the genome-based phylogenetic tree, IN45T was found to cluster with M. caldicuralii IT-9T within the family Methylothermaceae (Fig. 1b). The phylogenetic tree of deduced PmoA sequences also showed a clustering of these two strains within this family (Fig. S1, available in the online version of this article).
Genomic characteristics of IN45T
The genome sequence indicated that IN45T possesses key genes and pathways for methane metabolism (Table 1). Selected key genes are listed in Table S1. For methane oxidation, the genome contains two copies of the pmoCAB gene cluster and two orphan pmoC encoding particulate methane monooxygenase, but no soluble methane monooxygenase genes or pxmABC for another membrane-bound monooxygenase. For methanol oxidation, the genome contains one copy each of mxaFI and xoxF (clade 5), which encode different types of methanol dehydrogenases, and genes involved in the synthesis of the redox cofactor pyrroloquinoline quinone (pqqABCDE). The genes involved in the following pathways are also encoded: the tetrahydromethanopterin-mediated and tetrahydrofolate-mediated C1 transfer pathways, the ribulose monophosphate (RuMP) pathway for carbon assimilation, the Embden–Meyerhof–Parnas (EMP) glycolytic pathway, the tricarboxylic acid cycle and the aerobic respiratory chain. However, the serine pathway is incomplete, and the Calvin–Benson–Basham cycle and the Entner–Doudoroff glycolytic pathway are absent. Glycogen synthesis genes (glgABC) are encoded, indicating that the strain stores carbon as glycogen. A homologue of hemerythrin, a nonheme iron protein thought to transport oxygen, is encoded by the genome. Cytoplasmic hemerythrin has been reported to enhance the activity of particulate methane monooxygenase [41] and/or aerobic respiration [42] in methanotrophs, although the overall molecular mechanisms are not fully understood.
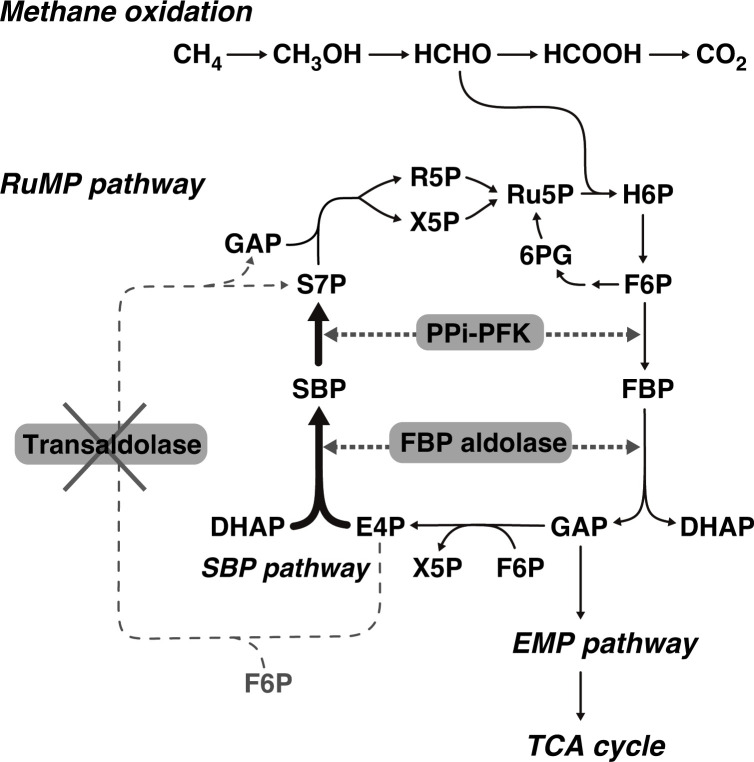
As described above, IN45T will assimilate carbon via the RuMP pathway. Notably, the transaldolase gene was not found in the IN45T genome. Transaldolase produces sedoheptulose 7-phosphate (S7P) as an obligate intermediate in the RuMP pathway and is also generally involved in the pentose phosphate pathway. However, despite the absence of the transaldolase gene, IN45T is likely to produce S7P via the sedoheptulose 1,7-bisphosphate (SBP) pathway [43], which is essentially identical to part of the Calvin–Benson–Basham cycle. Two canonical RuMP pathway enzymes, fructose-1,6-bisphosphate aldolase and pyrophosphate-dependent 6-phosphofructokinase, can catalyse two reactions in the SBP pathway (Fig. 2). The reactions are as follows: fructose-1,6-bisphosphate aldolase forms SBP by condensation of erythrose 4-phosphate and dihydroxyacetone phosphate, and then pyrophosphate-dependent 6-phosphofructokinase dephosphorylates SBP to form S7P (Fig. 2). In addition to the canonical reactions with fructose phosphates, these two enzymes have been shown to catalyse the above-mentioned reactions with sedoheptulose phosphates (regardless of the direction of the reactions) in different bacteria, including methanotrophs [43,49]. Furthermore, it has been reported that the SBP pathway is feasible or likely to have evolved as part of the RuMP or pentose phosphate pathways in several bacteria, including native [50] and synthetic [51,52] methylotrophs and bacteria assimilating pentose sugars without the transaldolase gene [43,44, 46].
Fig. 2. Simplified scheme of central carbon metabolism in IN45T. Thin dashed arrows indicate a reaction of transaldolase. In IN45T, transaldolase is absent and its absence can be compensated for by the sedoheptulose 1,7-bisphosphate (SBP) pathway (bold unbroken arrows). The SBP pathway can be catalysed by two canonical ribulose monophosphate (RuMP) pathway enzymes, fructose 1,6-bisphosphate (FBP) aldolase and pyrophosphate-dependent 6-phosphofructokinase (PPi-PFK). Abbreviations: Ru5P, ribulose 5-phosphate; H6P, 3-hexulose 6-phosphate; F6P, fructose 6-phosphate; 6 PG, 6-phosphogluconate; DHAP, dihydroxyacetone phosphate; GAP, glyceraldehyde 3-phosphate; E4P, erythrose 4-phosphate; X5P, xylulose 5-phosphate; S7P, sedoheptulose 7-phosphate; R5P, ribose 5-phosphate; EMP, Embden–Meyerhof–Parnas; TCA, tricarboxylic acid.
Regarding nitrogen metabolism, IN45T carries genes for ammonium assimilation (glnA and GDH2), nitrate assimilation (nasA and nirBD), partial denitrification of nitrate to N2O (narGHJI, nirK, and norBC) and hydroxylamine dehydrogenase (hao). This is consistent with the finding that IN45T used both ammonium and nitrate as the sole nitrogen source and that the strain produced N2O in the presence of nitrate, as described later.
Other genes of interest in IN45T are homologues of bcsABZC for cellulose synthesis. These genes have been found in some gammaproteobacterial methanotrophic strains, but are less common in methanotrophs [5]. Cellulose is believed to play a role in the survival strategies of some cellulose-producing bacteria in terms of adhesion, colonisation and self-protection [53,54]. Thus, the ability to produce cellulose could be advantageous for the survival of microorganisms in deep-sea hydrothermal fields, where physicochemical conditions are highly variable.
Genomic characteristics of M. caldicuralii IT-9T
In this study, the genome of the reference species M. caldicuralii IT-9T was analysed and its genomic features are briefly summarised in Table 1. A list of key genes is given in Table S1. The genome revealed a high similarity between M. caldicuralii IT-9T and IN45T with respect to central carbon metabolism. The striking similarity is the unusual absence of transaldolase in the RuMP pathway, indicating that the alternative SBP pathway functions similarly to that in IN45T (Fig. 2). In contrast, the nitrogen metabolism of the two strains differs significantly. M. caldicuralii IT-9T lacks all genes for nitrate assimilation and denitrification. This is consistent with its inability to grow on nitrate when used as the sole nitrogen source [8]. Another notable feature is the absence of a homologue of the oxygen carrier hemerythrin despite its prevalence in many methanotrophs, including IN45T. Therefore, the absence of hemerythrin combined with the absence of denitrification enzymes in M. caldicuralii IT-9T may reduce its metabolic activity under low-oxygen conditions. Intriguingly, similarly to IN45T, M. caldicuralii IT-9T possesses bcsABZC homologues; however, M. caldicuralii IT-9T most probably cannot synthesise cellulose because the bcsA homologue is disrupted by transposon insertion.
Morphology
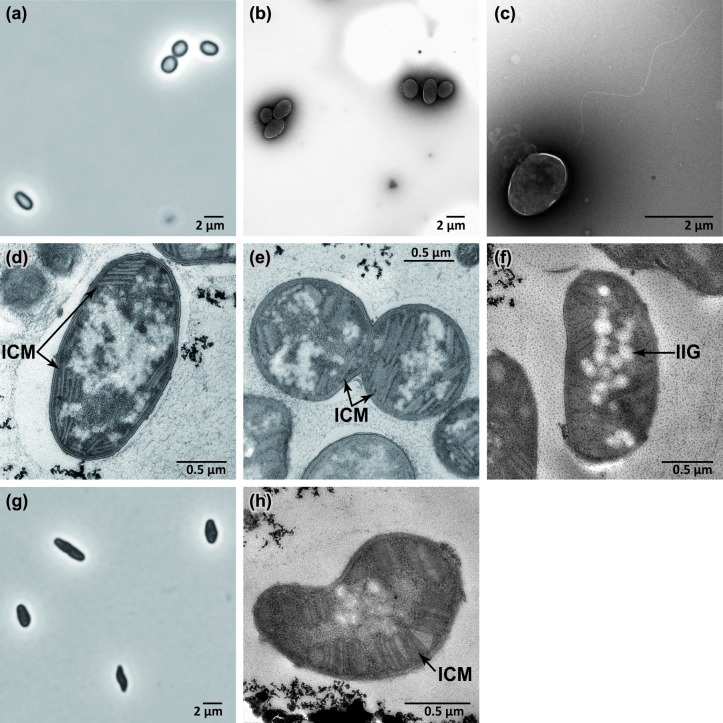
Cells of IN45T were Gram-reaction-negative and predominantly oval, partly coccoid or plump rod-shaped under optimum pH conditions (Fig. 3a–c). At pH 6.7 and above, most cells showed a deformed morphology (Fig. 3g, h). Cell size was approximately 1.0–3.0 µm long and 0.8–1.5 µm wide. Cells were motile with a single polar flagellum (Fig. 3c) and contained type I intracytoplasmic membranes and inclusion granules (Fig. 3d–f, h). In some cells of IN45T, the bundles of intracytoplasmic membranes showed a somewhat disordered or sparse arrangement (Fig. 3d), and a similar membrane arrangement has been reported in M. caldicuralii IT-9T [8]. No cyst-like cells were observed in the culture after 1 month of storage.
Fig. 3. Images of cells of IN45T obtained by phase-contrast microscopy (a, g) and transmission electron microscopy (b–f, h). Using transmission electron microscopy, negatively stained cells (b, c) and ultrathin sections of cells (d–f, h) were observed. Panels (a) – (f) show the cells grown at pH 6.1 (optimum pH for growth). Panels (g) and (h) show the cells grown at pH 6.7, where most of the cells were deformed. ICM, intracytoplasmic membranes; IIG, intracellular inclusions of granules.
Physiology
IN45T grew at temperatures between 25 and 56 °C (optimum 45–50 °C), but not at 23 °C or 57 °C. When a culture that had been stored for a few weeks or longer was used as inoculum, the two-step incubation, first at 37 °C for 1 day and then at 45 °C, appeared to shorten the growth lag. The strain grew at a pH range 5.2–6.9 (optimum pH 5.9–6.4) but not at pH 5.0 or 7.0. It seems strange that the strain could not grow at pH 7 or higher, since onboard measurements recorded pH 7.5 for fluid samples collected at the site of ISCS-4, one of the sources of isolation [5]. However, high-temperature hydrothermal fluids have been reported to be acidic (pH ≤5) [55], and mixing them with slightly alkaline cold seawater would create a gradient of environmental factors (temperature, pH and concentrations of substances, etc.) in the microbial habitats of this hydrothermal field. NaCl was required for growth at a concentration range 1.5–4 % (w/v) (optimum 2–3 %). Growth was not stable with 1 % NaCl and was not observed with 0.5 % or 5 % NaCl. At the time of isolation, Balch’s vitamin mixture [56] was routinely added to the medium, and the results of subsequent tests indicated that the strain required the vitamin mixture for growth. Under optimum conditions, IN45T exhibited a maximum specific growth rate of 0.13–0.18 h–1 (doubling time of 4.0–5.4 h, n=3) and a maximum cell density of approximately 1–2×108 cells ml–1.
Methanol supported the growth of IN45T instead of methane at concentrations of 0.1–5 % (v/v), but not at 6 %. Neither formate nor any of the multicarbon substrates tested supported its growth. In the nitrogen source test, active growth was observed on NH4Cl and NaNO3. Although weak growth on urea was observed, no urease genes were found in the genome, so the strain may not have actually used urea for growth. Decomposition of urea in aqueous solution, where the product is generally ammonia, has been reported to occur over a wide temperature range, not only at high temperatures [57], and the possibility that some of the urea decomposed during incubation cannot be excluded. No apparent growth was observed on NaNO2, Tris, methylamine, dimethylamine, l-aspartate or casamino acids at the concentrations tested. In addition, gaseous N2 did not support growth. The growth test on solid media was performed three times independently; however, the strain did not form visible colonies on solid media, gellan gum or Noble agar, during incubation for 3 weeks.
As stated earlier, the genome of IN45T encodes genes for partial denitrification of nitrate to N2O. We thus examined N2O production in batch cultures grown to stationary phase with or without NaNO3 supplementation (0.04 %, w/v). NH4Cl was added as nitrogen source. When IN45T was grown with NaNO3 under low-oxygen conditions (1 % of the initial O2 concentration, no gas exchange during cultivation), N2O was detected in the headspace gas at concentrations of 21–126 ppm (average 81 ppm, n=4). In contrast, N2O concentrations in cultures without NaNO3 were low and below the limit of quantification (<5 ppm, n=4). During cultivation, O2 decreased from 1 % to 0.1–0.2 %. The medium with NaNO3 but not inoculated with cells (negative control) contained no detectable amounts of N2O (<2 ppm, n=2). This indicates that most of the N2O was produced from nitrate via denitrification, with a small amount from ammonium. IN45T carries the hao gene, so the small amount of N2O was probably produced from ammonium via nitrifier denitrification, although this was not investigated in this study.
Under low-oxygen conditions, the final cell density of stationary-phase cultures was 2–3×107 cells ml–1, which was several to 10 times lower than the density under normal oxygen conditions (1–2×108 cells ml–1, 7 % O2). Even under normal-oxygen conditions, N2O was detected, probably due to the decreased oxygen levels (<2 % O2) in stationary-phase cultures; however, the N2O levels per cell were several times lower than those under low-oxygen conditions (data not shown). This is consistent with the general knowledge that denitrification is promoted under oxygen limitation [58]. The cell yield per ml oxygen was roughly estimated to be in the range of 3–4×108 cells ml−1 oxygen, irrespective of low- or normal-oxygen conditions.
Partial denitrification of nitrate and N2O production have been reported in methanotrophic strains, particularly in members of the genus Methylomonas, including Methylomonas methanica, Methylomonas koyamae and Methylomonas lenta [59]. Furthermore, ‘Methylomonas denitrificans’ FJG1 has been shown to couple partial denitrification of nitrate and methane oxidation and to increase intracellular ATP levels under oxygen limitation [60]. The genes required for partial denitrification of nitrate are also present in some other proteobacterial methanotrophs [5]. Whether IN45T can promote its growth or conserve cellular energy under oxygen limitation via denitrification remains to be investigated . If it can, the properties of this strain would be advantageous for survival in nature, as methane is generally abundant in anoxic environments [1].
Chemotaxonomy
The major isoprenoid quinone in strain IN45T was ubiquinone 8 (Q-8), representing 96.3 % of the quinone profile. The minor quinones detected were ubiquinone 7 (3.1 %), ubiquinone 6 (0.5 %) and ubiquinone 9 (0.1 %). Q-8 is the predominant quinone shared by members of the family Methylothermaceae, M. caldicuralii IT-9T [8] and 'Methylothermus thermalis' MYHT [11]. The major polar lipid in IN45T was phosphatidylserine, while phosphatidylethanolamine, diphosphatidylglycerol, phosphatidylglycerol and several unknown phospholipids were also detected (Fig. S2). The polar lipid profile of IN45T differed markedly from that of M. caldicuralii IT-9T, in which unknown phospholipids were detected as the major polar lipids [8].
The fatty acid compositions of IN45T and other members of the family Methylothermaceae are shown in Table 2. The major fatty acids of IN45T were C16 : 1ω7c, C16 : 0 and C18 : 1ω7c, which accounted for 94 % of the total fatty acids. The double bond positions of C16 : 1ω7c and C18 : 1ω7c were determined by analysis of dimethyl disulphide derivatives of these methyl esters. The GC–MS chromatogram and mass spectra of the dimethyl disulphide derivatives are shown in Fig. S3. The fatty acid profile of IN45T was similar to that of M. caldicuralii IT-9T; however, the abundance of C16 : 1ω7c in IN45T allowed the two strains to be distinguished. A relatively high abundance of C16 : 1ω7c (14.2–19.6 %) has also been reported in Methylohalobius crimeensis, another member of the family Methylothermaceae [12]. Currently, a common feature of the family members is the relatively high abundance of C18 : 1 species (28.6–60.5 %).
Table 2. Cellular fatty acid compositions of IN45T and reference strains of members of the family Methylothermaceae.
Strains: 1, IN45T (data from this study); 2, Methylomarinovum caldicuralii IT-9T (data from 8); 3, Methylohalobius crimeensis 10KiT and 4Kr (data from 12); 4, 'Methylothermus thermalis' MYHT (data from 11); 5, Methylothermus subterraneus HTM55T (data from 10).
| Fatty acid | 1 | 2 | 3 | 4 | 5 |
| C10 : 0 | – | 0.3 | – | – | – |
| C12 : 0 | – | 0.4 | 0.2 | – | 0.5 |
| C14 : 0 | 5.2 | 1.6 | 1.4–2.5 | 1.2 | 0.8 |
| C15 : 0 | 0.4 | 0.3 | 0.3–0.5 | 2.1 | 0.6 |
| C16 : 1ω9c | – | 0.2 | – | – | – |
| C16 : 1ω7c | 32.4 | 8.0 | 14.2–19.6 | 3.5 | 2.2 |
| C16 : 1ω7t+ω5c | 0.4 | 0.6 | – | – | – |
| C16 : 0 | 32.9 | 43.0 | 22.8–23.0 | 37.2 | 52.0 |
| C16 : 0 2-OH | – | 4.8 | – | 8.4 | – |
| C17 : 0cyclo | – | – | 0.7 | 4.7 | 1.7 |
| C17 : 0 | – | 0.3 | 0.3 | 2.5 | 1.0 |
| C18 : 1ω9c | – | – | – | 35.2 | – |
| C18 : 1ω7c | 28.6 | 39.1 | 51.9–60.5 | 0.4 | 34.8 |
| C18 : 1ω7t | – | 0.5 | – | – | – |
| C18 : 0 | 0.1 | 1.0 | 0.5–0.6 | 1.7 | 4.0 |
| C19 : 0cyclo | – | – | – | 2.4 | – |
| C19 : 1 | – | – | – | – | 2.5 |
Proposal of a novel species of the genus Methylomarinovum
On the basis of its morphological, physiological, chemotaxonomic and phylogenetic characteristics, IN45T represents a member of the genus Methylomarinovum within the family Methylothermaceae. The only species in this genus with a validly published name is M. caldicuralii IT-9T. The results indicate a high degree of similarity between IN45T and M. caldicuralii IT-9T in key taxonomic characteristics (Table 1), including growth conditions (temperature, pH and NaCl concentration), major fatty acid species, chromosomal DNA G+C content and 16S rRNA gene sequence. Furthermore, their basic carbon metabolism is similar, as indicated by the results of genome analysis.
However, a number of other characteristics distinguish IN45T from M. caldicuralii IT-9T (Table 1). Their major polar lipids were clearly different. Only IN45T assimilated nitrate and required vitamins for growth. Only IN45T carries genes for partial denitrification of nitrate, and the observed N2O production indicates that IN45T can perform partial denitrification under oxygen limitation. A hemerythrin homologue was found in IN45T but not in M. caldicuralii IT-9T. Furthermore, the overall genomic relatedness indices (dDDH, ANI and AAI) indicated that these two methanotrophs should be separated at the species level. In conclusion, strain IN45T represents a novel species of the genus Methylomarinovum, for which we propose the name Methylomarinovum tepidoasis sp. nov.
Description of Methylomarinovum tepidoasis sp. nov
Methylomarinovum tepidoasis (te.pid.o’a.sis. L. masc. adj. tepidus, moderately warm; L. fem. n. oasis, oasis; N.L. gen. fem. n. tepidoasis, of a warm oasis, as the type strain was isolated from warm sites in a deep-sea hydrothermal vent field, often likened to an oasis in a deep-sea desert).
Gram-reaction-negative, motile, oval cells; sometimes appearing as cocci or plump rods. Cell size approximately 1.0–3.0 µm long and 0.8–1.5 µm wide. Possesses a single polar flagellum and a type I intracytoplasmic membrane system. Reproduces by normal cell division. Does not form cysts. Moderately thermophilic, growing at temperatures of 25–56 °C (optimum 45–50 °C) and at pH 5.2–6.9 (optimum pH 5.9–6.4). Requires 1.5–4 % (w/v) NaCl for growth (optimum 2–3 %). Grows aerobically on methane or methanol. Possesses particulate methane monooxygenase but no soluble methane monooxygenase. Assimilates C1 compounds via the RuMP pathway. Uses ammonium or nitrate as a nitrogen source. Does not fix atmospheric nitrogen for growth. Requires vitamins for growth. The major fatty acids are C16 : 1ω7c, C16 : 0 and C18 : 1ω7c. The major polar lipid is phosphatidylserine. The major isoprenoid quinone is Q-8.
The type strain is IN45T (JCM 35101T =DSM 113422T), which was isolated from in situ colonisation systems deployed at the Original site in the Iheya North deep-sea hydrothermal field in the mid-Okinawa Trough, Japan. The genome of the type strain consists of a 2.42-Mbp chromosome and a 20.5-kbp plasmid. The G+C content of the chromosomal DNA is 64.1 mol%.
The GenBank/EMBL/DDBJ accession numbers are LC770110 for the 16S rRNA gene sequence and AP024718 and AP024719 for the chromosome and plasmid sequence.
supplementary material
Acknowledgements
We thank the captains and crews of R/V Natsushima and R/V Kaiyo, the ROV operations team, Drs Junichi Miyazaki and Hiroyuki Yamamoto for their support during the cruises. We thank Miwako Tsuda and Keiko Tanaka for their assistance with the experiments. We also thank Professors Aharon Oren and Bernhard Schink for their help with the nomenclature.
Footnotes
Funding: This work was partly supported by funding from the JSPS KAKENHI (grant no. 16K07498) to H.H. and Y.T.
Author contributions: H.H.: conceptualisation, investigation, formal analysis, data curation, visualisation, writing – original draft, funding acquisition. Y.T.: investigation, formal analysis, data curation, writing – original draft, funding acquisition. M.A.: investigation. M.M.: investigation, formal analysis, writing – original draft. K.U.: investigation, writing – original draft. Y.M.: investigation, formal analysis, writing – original draft. K.T.: writing – review and editing, supervision, funding acquisition.
Accession No: The GenBank/EMBL/DDBJ accession numbers for the sequences obtained are LC770110 for the 16S rRNA gene of Methylomarinovum tepidoasis IN45T, AP024718 and AP024719 for the chromosome and plasmid, respectively, of M. tepidoasis IN45T and AP024714 for the genome of Methylomarinovum caldicuralii IT-9T.
Contributor Information
Hisako Hirayama, Email: hirayamah@jamstec.go.jp.
Yoshihiro Takaki, Email: takakiy@jamstec.go.jp.
Mariko Abe, Email: m-abe@jamstec.go.jp.
Masayuki Miyazaki, Email: miyazakim@jamstec.go.jp.
Katsuyuki Uematsu, Email: uematsuk@jamstec.go.jp.
Yohei Matsui, Email: ymatsui@jamstec.go.jp.
Ken Takai, Email: kent@jamstec.go.jp.
References
- 1.Schubert CJ. Methane, origin. Encycl Geobiol. 2011:578–586. doi: 10.1007/978-1-4020-9212-1. [DOI] [Google Scholar]
- 2.Dubilier N, Bergin C, Lott C. Symbiotic diversity in marine animals: the art of harnessing chemosynthesis. Nat Rev Microbiol. 2008;6:725–740. doi: 10.1038/nrmicro1992. [DOI] [PubMed] [Google Scholar]
- 3.Dick GJ, Tebo BM. Microbial diversity and biogeochemistry of the Guaymas basin deep-sea hydrothermal plume. Environ Microbiol. 2010;12:1334–1347. doi: 10.1111/j.1462-2920.2010.02177.x. [DOI] [PubMed] [Google Scholar]
- 4.Distel DL, Lee HK, Cavanaugh CM. Intracellular coexistence of methano- and thioautotrophic bacteria in a hydrothermal vent mussel. Proc Natl Acad Sci U S A. 1995;92:9598–9602. doi: 10.1073/pnas.92.21.9598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hirayama H, Takaki Y, Abe M, Imachi H, Ikuta T, et al. Multispecies populations of methanotrophic Methyloprofundus and cultivation of a likely dominant species from the Iheya North deep-sea hydrothermal field. Appl Environ Microbiol. 2022;88:e0075821. doi: 10.1128/AEM.00758-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nercessian O, Bienvenu N, Moreira D, Prieur D, Jeanthon C. Diversity of functional genes of methanogens, methanotrophs and sulfate reducers in deep-sea hydrothermal environments. Environ Microbiol. 2005;7:118–132. doi: 10.1111/j.1462-2920.2004.00672.x. [DOI] [PubMed] [Google Scholar]
- 7.Skennerton CT, Ward LM, Michel A, Metcalfe K, Valiente C, et al. Genomic reconstruction of an uncultured hydrothermal vent gammaproteobacterial methanotroph (family Methylothermaceae) indicates multiple adaptations to oxygen limitation. Front Microbiol. 2015;6:1425. doi: 10.3389/fmicb.2015.01425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hirayama H, Abe M, Miyazaki M, Nunoura T, Furushima Y, et al. Methylomarinovum caldicuralii gen. nov., sp. nov., a moderately thermophilic methanotroph isolated from a shallow submarine hydrothermal system, and proposal of the family Methylothermaceae fam. Int J Syst Evol Microbiol. 2014;64:989–999. doi: 10.1099/ijs.0.058172-0. [DOI] [PubMed] [Google Scholar]
- 9.Takeuchi M, Kamagata Y, Oshima K, Hanada S, Tamaki H, et al. Methylocaldum marinum sp. nov., a thermotolerant, methane-oxidizing bacterium isolated from marine sediments, and emended description of the genus Methylocaldum. Int J Syst Evol Microbiol. 2014;64:3240–3246. doi: 10.1099/ijs.0.063503-0. [DOI] [PubMed] [Google Scholar]
- 10.Hirayama H, Suzuki Y, Abe M, Miyazaki M, Makita H, et al. Methylothermus subterraneus sp. nov., a moderately thermophilic methanotroph isolated from a terrestrial subsurface hot aquifer. Int J Syst Evol Microbiol. 2011;61:2646–2653. doi: 10.1099/ijs.0.028092-0. [DOI] [PubMed] [Google Scholar]
- 11.Tsubota J, Eshinimaev BT, Khmelenina VN, Trotsenko YA. Methylothermus thermalis gen. nov., sp. nov., a novel moderately thermophilic obligate methanotroph from a hot spring in Japan. Int J Syst Evol Microbiol. 2005;55:1877–1884. doi: 10.1099/ijs.0.63691-0. [DOI] [PubMed] [Google Scholar]
- 12.Heyer J, Berger U, Hardt M, Dunfield PF. Methylohalobius crimeensis gen. nov., sp. nov., a moderately halophilic, methanotrophic bacterium isolated from hypersaline lakes of Crimea. Int J Syst Evol Microbiol. 2005;55:1817–1826. doi: 10.1099/ijs.0.63213-0. [DOI] [PubMed] [Google Scholar]
- 13.Rush D, Osborne KA, Birgel D, Kappler A, Hirayama H, et al. The bacteriohopanepolyol inventory of novel aerobic methane oxidising bacteria reveals new biomarker signatures of aerobic methanotrophy in marine systems. PLoS One. 2016;11:e0165635. doi: 10.1371/journal.pone.0165635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lane DJ. 16S/23S Sequencing. New York: John Wiley & Sons; 1991. [Google Scholar]
- 15.Pruesse E, Peplies J, Glöckner FO. SINA: accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics. 2012;28:1823–1829. doi: 10.1093/bioinformatics/bts252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Trifinopoulos J, Nguyen L-T, von Haeseler A, Minh BQ. W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 2016;44:W232–5. doi: 10.1093/nar/gkw256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 2017;14:587–589. doi: 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoang DT, Chernomor O, von Haeseler A, Minh BQ, Vinh LS. UFBoot2: improving the ultrafast bootstrap approximation. Mol Biol Evol. 2018;35:518–522. doi: 10.1093/molbev/msx281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chin C-S, Alexander DH, Marks P, Klammer AA, Drake J, et al. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat Methods. 2013;10:563–569. doi: 10.1038/nmeth.2474. [DOI] [PubMed] [Google Scholar]
- 20.Ruby JG, Bellare P, Derisi JL. PRICE: software for the targeted assembly of components of (meta) genomic sequence data. G3. 2013;3:865–880. doi: 10.1534/g3.113.005967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, et al. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One. 2014;9:e112963. doi: 10.1371/journal.pone.0112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hyatt D, LoCascio PF, Hauser LJ, Uberbacher EC. Gene and translation initiation site prediction in metagenomic sequences. Bioinformatics. 2012;28:2223–2230. doi: 10.1093/bioinformatics/bts429. [DOI] [PubMed] [Google Scholar]
- 23.Nawrocki EP, Eddy SR. Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics. 2013;29:2933–2935. doi: 10.1093/bioinformatics/btt509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lowe TM, Chan PP. tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 2016;44:W54–7. doi: 10.1093/nar/gkw413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meier-Kolthoff JP, Auch AF, Klenk H-P, Göker M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinformatics. 2013;14:60. doi: 10.1186/1471-2105-14-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee I, Ouk Kim Y, Park S-C, Chun J. OrthoANI: an improved algorithm and software for calculating average nucleotide identity. Int J Syst Evol Microbiol. 2016;66:1100–1103. doi: 10.1099/ijsem.0.000760. [DOI] [PubMed] [Google Scholar]
- 27.Chaumeil P-A, Mussig AJ, Hugenholtz P, Parks DH. GTDB-Tk v2: memory friendly classification with the genome taxonomy database. Bioinformatics. 2022;38:5315–5316. doi: 10.1093/bioinformatics/btac672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Buck JD. Nonstaining (KOH) method for determination of Gram reactions of marine bacteria. Appl Environ Microbiol. 1982;44:992–993. doi: 10.1128/aem.44.4.992-993.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Steyer AM, Ruhwedel T, Möbius W. Biological sample preparation by high-pressure freezing, microwave-assisted contrast enhancement, and minimal resin embedding for volume imaging. J Vis Exp. 2019;2019:145. doi: 10.3791/59156. [DOI] [PubMed] [Google Scholar]
- 32.Deerinck TJ, Bushong EA, Thor A, Ellisman MH. NCMIR Methods for 3D EM: A New Protocol for Preparation of Biological Specimens for Serial Block Face Scanning Electron Microscopy. 2010. [Google Scholar]
- 33.Minnikin DE, O’Donnell AG, Goodfellow M, Alderson G, Athalye M, et al. An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J Microbiol Met. 1984;2:233–241. doi: 10.1016/0167-7012(84)90018-6. [DOI] [Google Scholar]
- 34.Komagata K, Suzuki K-I. In: Methods Microbiol. Colwell RR, Grigorova R, editors. 19: Academic Press; 1988. Lipid and cell-wall analysis in bacterial systematics; pp. 161–207. [Google Scholar]
- 35.Sasser M. MIDI Technical Note. Vol. 101. Newark, DE: MIDI, Inc; 1990. Bacterial identification by gas chromatographic analysis of fatty acid methyl esters (GC-FAME) vol. [Google Scholar]
- 36.Christie WW. In: Advances in Lipid Methodology - Four. Christie WW, editor. Dundee: Oily Press; 1997. Structural analysis of fatty acids; pp. 119–169. [Google Scholar]
- 37.Kim M, Oh H-S, Park S-C, Chun J. Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int J Syst Evol Microbiol. 2014;64:346–351. doi: 10.1099/ijs.0.059774-0. [DOI] [PubMed] [Google Scholar]
- 38.Goris J, Konstantinidis KT, Klappenbach JA, Coenye T, Vandamme P, et al. DNA–DNA hybridization values and their relationship to whole-genome sequence similarities. Int J Syst Evol Microbiol. 2007;57:81–91. doi: 10.1099/ijs.0.64483-0. [DOI] [PubMed] [Google Scholar]
- 39.Richter M, Rosselló-Móra R. Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci U S A. 2009;106:19126–19131. doi: 10.1073/pnas.0906412106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Konstantinidis KT, Rosselló-Móra R, Amann R. Uncultivated microbes in need of their own taxonomy. ISME J. 2017;11:2399–2406. doi: 10.1038/ismej.2017.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen KH-C, Wu H-H, Ke S-F, Rao Y-T, Tu C-M, et al. Bacteriohemerythrin bolsters the activity of the particulate methane monooxygenase (pMMO) in Methylococcus capsulatus (Bath) J Inorg Biochem. 2012;111:10–17. doi: 10.1016/j.jinorgbio.2012.02.019. [DOI] [PubMed] [Google Scholar]
- 42.Nariya S, Kalyuzhnaya MG. Hemerythrins enhance aerobic respiration in Methylomicrobium alcaliphilum 20ZR, a methane-consuming bacterium. FEMS Microbiol Lett. 2020;367:fnaa003. doi: 10.1093/femsle/fnaa003. [DOI] [PubMed] [Google Scholar]
- 43.Koendjbiharie JG, Hon S, Pabst M, Hooftman R, Stevenson DM, et al. The pentose phosphate pathway of cellulolytic clostridia relies on 6-phosphofructokinase instead of transaldolase. J Biol Chem. 2020;295:1867–1878. doi: 10.1074/jbc.RA119.011239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Garschagen LS, Franke T, Deppenmeier U. An alternative pentose phosphate pathway in human gut bacteria for the degradation of C5 sugars in dietary fibers. FEBS J. 2021;288:1839–1858. doi: 10.1111/febs.15511. [DOI] [PubMed] [Google Scholar]
- 45.Nakahara K, Yamamoto H, Miyake C, Yokota A. Purification and characterization of class-I and class-II fructose-1,6-bisphosphate aldolases from the cyanobacterium Synechocystis sp. PCC 6803. Plant Cell Physiol. 2003;44:326–333. doi: 10.1093/pcp/pcg044. [DOI] [PubMed] [Google Scholar]
- 46.Nakahigashi K, Toya Y, Ishii N, Soga T, Hasegawa M, et al. Systematic phenome analysis of Escherichia coli multiple-knockout mutants reveals hidden reactions in central carbon metabolism. Mol Syst Biol. 2009;5:306. doi: 10.1038/msb.2009.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reshetnikov AS, Rozova ON, Khmelenina VN, Mustakhimov II, Beschastny AP, et al. Characterization of the pyrophosphate-dependent 6-phosphofructokinase from Methylococcus capsulatus Bath. FEMS Microbiol Lett. 2008;288:202–210. doi: 10.1111/j.1574-6968.2008.01366.x. [DOI] [PubMed] [Google Scholar]
- 48.Rozova ON, Khmelenina VN, Mustakhimov II, Reshetnikov AS, Trotsenko YA. Characterization of recombinant fructose-1,6-bisphosphate aldolase from Methylococcus capsulatus bath. Biochemistry. 2010;75:892–898. doi: 10.1134/s0006297910070114. [DOI] [PubMed] [Google Scholar]
- 49.Rozova ON, Khmelenina VN, Trotsenko YA. Characterization of recombinant PPi-dependent 6-phosphofructokinases from Methylosinus trichosporium OB3b and Methylobacterium nodulans ORS 2060. Biochemistry. 2012;77:288–295. doi: 10.1134/S0006297912030078. [DOI] [PubMed] [Google Scholar]
- 50.Stolzenberger J, Lindner SN, Persicke M, Brautaset T, Wendisch VF. Characterization of fructose 1,6-bisphosphatase and sedoheptulose 1,7-bisphosphatase from the facultative ribulose monophosphate cycle methylotroph Bacillus methanolicus. J Bacteriol. 2013;195:5112–5122. doi: 10.1128/JB.00672-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fan L, Wang Y, Qian J, Gao N, Zhang Z, et al. Transcriptome analysis reveals the roles of nitrogen metabolism and sedoheptulose bisphosphatase pathway in methanol-dependent growth of Corynebacterium glutamicum. Microb Biotechnol. 2021;14:1797–1808. doi: 10.1111/1751-7915.13863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Woolston BM, King JR, Reiter M, Van Hove B, Stephanopoulos G. Improving formaldehyde consumption drives methanol assimilation in engineered E. coli. Nat Commun. 2018;9 doi: 10.1038/s41467-018-04795-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Augimeri RV, Varley AJ, Strap JL. Establishing a role for bacterial cellulose in environmental interactions: lessons learned from diverse biofilm-producing proteobacteria. Front Microbiol. 2015;6:1282. doi: 10.3389/fmicb.2015.01282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Römling U, Galperin MY. Bacterial cellulose biosynthesis: diversity of operons, subunits, products, and functions. Trends Microbiol. 2015;23:545–557. doi: 10.1016/j.tim.2015.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kawagucci S, Miyazaki J, Nakajima R, Nozaki T, Takaya Y, et al. Post‐drilling changes in fluid discharge pattern, mineral deposition, and fluid chemistry in the Iheya North hydrothermal field, Okinawa Trough. Geochem Geophys Geosyst. 2013;14:4774–4790. doi: 10.1002/2013GC004895. [DOI] [Google Scholar]
- 56.Balch WE, Fox GE, Magrum LJ, Woese CR, Wolfe RS. Methanogens: reevaluation of a unique biological group. Microbiol Rev. 1979;43:260–296. doi: 10.1128/mr.43.2.260-296.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kuwamoto K. Simple and quick quality confirmation method of aqueous urea solution for marine SCR system: evaluation of alkalinity of aqueous urea solution with Laquatwin pH meter. Readout. 2019;E52:28–31. [Google Scholar]
- 58.Skiba U. In: Encyclopedia of Ecology. Jørgensen SE, Fath BD, editors. Oxford: Academic Press; 2008. Denitrification; pp. 866–871. [Google Scholar]
- 59.Hoefman S, van der Ha D, Boon N, Vandamme P, De Vos P, et al. Niche differentiation in nitrogen metabolism among methanotrophs within an operational taxonomic unit. BMC Microbiol. 2014;14:83. doi: 10.1186/1471-2180-14-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kits KD, Klotz MG, Stein LY. Methane oxidation coupled to nitrate reduction under hypoxia by the Gammaproteobacterium Methylomonas denitrificans, sp. nov. type strain FJG1. Environ Microbiol. 2015;17:3219–3232. doi: 10.1111/1462-2920.12772. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.