Abstract
The capsid (CA) and nucleocapsid domains of the human immunodeficiency virus type 1 Gag polyprotein are separated by the p2 spacer peptide, which is essential for virus replication. Previous studies have revealed that p2 has an important role in virus morphogenesis. In this paper, we show that a crucial assembly determinant maps to the highly conserved N terminus of p2, which is predicted to form part of an α-helix that begins in CA. A mutational analysis indicates that the ability of the N terminus of p2 to adopt an α-helical structure is essential for its function during virus assembly. To prevent CA-p2 processing, it was necessary to mutate both the CA-p2 cleavage site and an internal cleavage site within p2. Virions produced by the double mutant lacked a conical core shell and instead contained a thin electron-dense shell about 10 nm underneath the virion membrane. These results suggest that p2 is transiently required for proper assembly, but needs to be removed from the C terminus of CA to weaken CA-CA interactions and allow the rearrangement of the virion core shell during virus maturation.
The internal structural proteins of the human immunodeficiency virus type 1 (HIV-1) virion are synthesized in the form of a polyprotein (Pr55gag) which can efficiently form enveloped virus-like particles even when expressed alone (17). Pr55gag is modified by N-terminal myristylation, which is required for its stable association with the inner leaflet of the plasma membrane, where virus assembly occurs (4, 21). During or after the release of an immature particle from the plasma membrane, Pr55gag is cleaved by the viral protease. The major Gag cleavage products are matrix (MA), capsid (CA), nucleocapsid (NC), and p6 (25, 34). MA, which has a crucial role in the incorporation of the viral surface glycoproteins (10, 52), remains associated with the host cell-derived lipid envelope of the virion (16). CA forms the shell of the characteristic cone-shaped core of the mature virion which encloses the viral genomic RNA (16, 27). NC is essential for the encapsidation of the viral genome and is believed to coat the viral RNA within the core of the virion (2, 19, 30). The C-terminal p6 domain of Pr55gag facilitates the release of assembled viral particles from the cell surface (20) and is also needed for the incorporation of the regulatory viral protein Vpr (31, 39).
Within the context of Pr55gag, two spacer peptides, p2 and p1, are located between CA and NC and between NC and p6, respectively (24, 25). Cleavage between CA and p2 is much slower than that between p2 and NC or between MA and CA (41). As a consequence, a CA-p2 protein (p25) accumulates in virus-producing cells (34). However, CA-p2 is normally found only in trace amounts in virions. In addition to p2, which comprises 14 amino acids (Ala-363 through Met-376) of the HIV-1HXB2 Gag precursor, a 10-amino-acid p2 fragment which extends from Ser-367 through Met-376 has been isolated from HIV-1 virions, indicating that the viral protease can also cleave within p2 (24, 25).
Genetic analyses indicate that the region surrounding the CA-p2 boundary has an important role in particle assembly (21, 28, 50). Within CA, the N-terminal two-thirds forms a domain which appears dispensable for particle assembly but is required for the formation of the cone-shaped core of the mature virion (8, 44, 51). Recent structure determinations have revealed that the N-terminal HIV-1 CA domain is largely α-helical (18, 35). An exposed loop region between two α-helices interacts with the prolyl isomerase cyclophilin A (14), which leads to the incorporation of the cellular enzyme into virions (13, 48). The C-terminal third of CA forms a distinct domain which is essential for Gag oligomerization and particle assembly (8, 12, 44). While genetic and structural studies indicate that the N-terminal boundary of the CA assembly domain coincides with a uniquely conserved sequence, termed the major homology region (8, 15, 18, 32), its C-terminal boundary remains less well defined.
The replacement of the scissile dipeptide Leu-Ala at the CA-p2 boundary with Ser-Arg in a mutant designated SVC-C2 led to the formation of grossly distorted capsid structures and caused a significant reduction in particle yield, indicating that the very C terminus of CA and/or p2 is crucial for HIV-1 morphogenesis (21). The possibility that the CA assembly domain extends into p2 is also suggested by the finding that the precise deletion of p2 from Pr55gag markedly reduced particle production (28). Electron microscopy revealed an accumulation of large electron-dense plaques underneath the plasma membrane in the absence of p2 (28), a phenotype which is similar to that observed for the SVC-C2 cleavage site mutant (21). However, the role of p2 in virus assembly remains controversial, because its removal appeared to have no effect on particle release in another study (41).
In the present study, we focused on the N-terminal portion of p2, since it is considerably more conserved than the C terminus and because it is predicted to be part of an α-helix which begins in CA. The analysis of a panel of single-amino-acid changes shows that the conserved N terminus of p2 is essential for virus replication and indicates that its predicted α-helical conformation is crucial for virus assembly. In contrast, a deletion which removed 5 out of 10 amino acids between a previously reported cleavage site within p2 and NC delayed but did not abolish virus replication, demonstrating that this relatively variable region of p2 has no essential function in the viral life cycle. We also show that processing of CA-p2 can be essentially prevented by disrupting both the CA-p2 cleavage site and the reported Met-Ser site (25) within p2. Interestingly, the mutant particles often contained a prominent circular structure underneath the viral membrane, indicating that the presence of p2 at the C terminus of CA prevented the rearrangement of the core into a conical tube.
MATERIALS AND METHODS
Proviral DNA constructs.
The parental HIV-1 proviral construct used in this study was HXBH10/R+ (9), a vpu+ and vpr+ variant of the infectious HXB2 proviral clone. For site-directed mutagenesis, single-stranded DNA was prepared from plasmid pGEMgag-pol (21) and used as a template for the annealing of oligonucleotides and primer extension with T4 DNA polymerase as described previously (29). To regenerate full-length proviral clones after mutagenesis, 0.5-kb SpeI-ApaI fragments (nucleotides 1510 to 2009) carrying the desired mutations were inserted into HXBH10/R+ in exchange for the wild-type fragment. The following oligonucleotides were used to obtain mutant clones: Δ2 (5′-AGAGTTTTGGCCGCGATGAGCCAAGTA-3′), Δ6–10 (5′-GAAGCAATGAGCGCTACCATAATG-3′), Δ5–14 (5′-GAGTTTTGGCTGAAGCAATGATGCAGAGAGGCAATTTTAGG-3′), E2Q (5′-AGAGTTTTGGCGCAAGCAATGAGC-3′), E2A (5′-AGAGTTTTGGCCGCGGCAATGAGC-3′), E2G (5′-AGAGTTTTGGCCGGCGCAATGAGC-3′), E2P (5′-AGAGTTTTGGCGCCAGCAATGAGC-3′), CA1 (5′-GGCAAGAGTTATCGCGGAAGCAATGAG-3′), M4I (5′-GTTTTGGCTGAAGCGATATCCCAAGTAACAA-3′), and CA1/M4I (5′-AAGGCAAGAGTTATCGCGGAAGCGATATCCCAAGTAACAA-3′). The presence of the mutations in the final proviral constructs was confirmed by restriction enzyme digestion and DNA sequence analysis.
Cell culture and transfections.
HeLa cells were grown in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal calf serum. Jurkat cells were maintained in RPMI 1640 medium with 10% fetal calf serum. HeLa cells (106) were seeded into 80-cm2 tissue culture flasks 24 h prior to transfection. The cultures were transfected with proviral plasmid DNA by a calcium phosphate precipitation technique (7). Jurkat cells were transfected by the DEAE-dextran method (43).
Viral protein analysis.
HeLa cells were metabolically labeled with [35S]methionine (50 μCi/ml) from 48 to 60 h posttransfection. Viral particles released during the labeling period were pelleted through 20% sucrose cushions (in phosphate-buffered saline) for 90 min at 4°C and at 27,000 rpm in a Beckman SW28 rotor. Pelleted virions were lysed in radioimmunoprecipitation assay (RIPA) buffer (140 mM NaCl, 8 mM Na2HPO4, 2 mM NaH2PO4, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.05% sodium dodecyl sulfate [SDS]), and viral proteins were analyzed directly by SDS-polyacrylamide gel electrophoresis (PAGE).
Electron microscopy.
Transfected HeLa cell cultures were fixed in fresh 2.5% glutaraldehyde in phosphate-buffered saline and postfixed in 1% osmium tetroxide. The cells were embedded in Epon, and sections were made approximately 60 to 80 nm thick to accommodate the volume of the core structure parallel to the section plane. The sections were stained with 1% uranyl acetate, and specimens were analyzed with a Zeiss CEM 902 electron microscope at an accelerating voltage of 80 kV. A liquid nitrogen cooling trap was used to reduce beam damage.
RESULTS
A critical assembly determinant in p2 maps to its conserved N terminus.
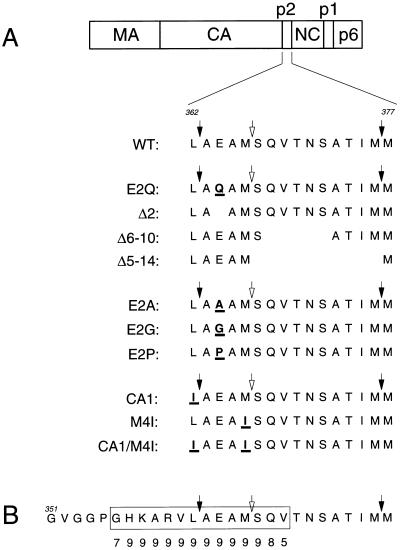
Sequence alignment of different isolates of HIV-1 and other primate lentiviruses shows a high degree of conservation of the four N-terminal residues of p2 (37). In contrast, the rest of p2 is poorly conserved among primate lentiviruses (37). The four conserved residues (Ala-Glu-Ala-Met in HIV-1HXB2) separate the C terminus of CA from a reported internal cleavage site in p2 (25) (Fig. 1). To examine the biological significance of this highly conserved region, Glu-2 of p2 was either conservatively replaced by Gln (mutant E2Q) or deleted (mutant Δ2). To investigate the role of the less-conserved region between the putative internal cleavage site in p2 and the N terminus of NC, we created a 10-amino-acid deletion which precisely removed this region (mutant Δ5–14). Additionally, in an attempt to preserve the p2/NC cleavage site we created the Δ6–10 mutant, which harbors a five-amino-acid deletion between the putative internal cleavage site in p2 and the N terminus of NC (Fig. 1).
FIG. 1.
(A) Location of mutations. The domain organization of the HIV-1 Gag precursor Pr55gag is illustrated at the top. The amino acid sequences of wild-type and mutant p2 together with N- and C-terminal flanking residues are shown below. Substitutions are underlined, and blank spaces represent deletions. Numbers refer to the positions of residues counting from the N terminus of Pr55gag. The CA-p2 and p2-NC cleavage sites are indicated by solid arrows, and a reported internal cleavage site in p2 (25) is indicated by open arrows. (B) Secondary structure analysis with the PHD program (45–47). Residues 351 to 377 of Pr55gag are shown, and amino acids predicted to form an α-helix are boxed. A reliability index for the secondary structure prediction with values from 0 (lowest reliability) to 9 (highest reliability) is given for each of the boxed residues.
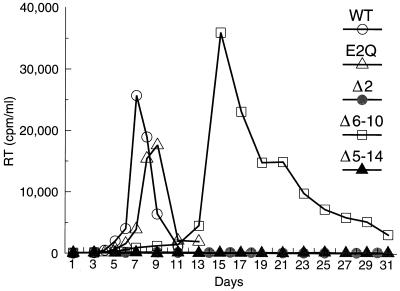
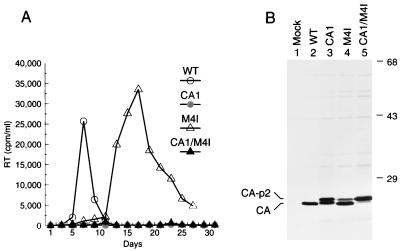
Full-length proviruses were then constructed which differ from the parental HXBH10/R+ proviral clone only by the mutations in the p2 coding region. To determine the ability of the mutants to initiate a productive infection, the parental HXBH10/R+ provirus and the mutant DNAs were transfected into the permissive cell line Jurkat. Virus replication was monitored by measuring particle-associated reverse transcriptase (RT) activity in the culture supernatants. This analysis showed that the E2Q mutant spread rapidly and replicated with only slightly delayed kinetics relative to wild-type HIV-1, indicating that the negative charge of Glu-2 is only of minor importance (Fig. 2). In contrast, the deletion of Glu-2 prevented virus replication (Fig. 2). The more extensive Δ6–10 deletion still allowed virus replication after a delay of about 1 week relative to the parental virus (Fig. 2). However, transfection of the Δ5–14 mutant did not result in a productive infection (Fig. 2).
FIG. 2.
Effects of alterations in different regions of p2 on virus replication. Jurkat cells were transfected with the parental proviral construct HXBH10/R+ (wild type [WT]) or with the indicated p2 mutants, and virus replication was monitored by measuring RT activity in the culture supernatants.
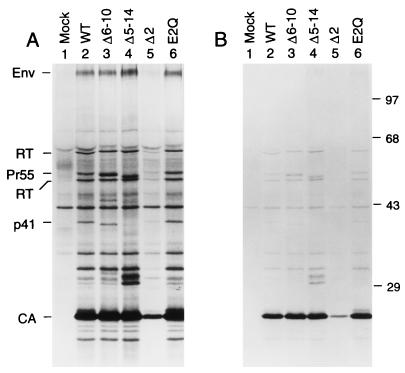
To determine whether the alterations in p2 affected the ability of the mutants to form viral particles, the wild-type and mutant proviruses were transfected into HeLa cells, which do not support virus replication because they lack the CD4 receptor. After metabolic labeling with [35S]methionine, viral particles released into the supernatant were pelleted through 20% sucrose cushions. Pelleted virions were then lysed in RIPA buffer, and their protein content was directly analyzed by SDS-PAGE. The E2Q, Δ6–10, and Δ5–14 mutants produced normal amounts of particles, as judged from the levels of CA in the pelletable fractions (Fig. 3). CA was expected to yield the most prominent viral protein band due to the presence of a large number of methionine residues. As anticipated, the Gag polyproteins produced by the Δ6–10 and Δ5–14 mutants migrated slightly faster in SDS-PAGE than wild-type Pr55gag. Additionally, the p41 Gag cleavage intermediate migrated slightly faster following the deletion of five amino acids from p2, indicating that this product, which corresponds to MA-CA, also includes p2 (Fig. 3, lane 3). In case of the Δ5–14 mutant, two novel bands with sizes of about 30 and 31 kDa were visible (Fig. 3, lane 4). The novel bands presumably correspond to CA-NC and CA-NC-p1. In contrast to the more extensive Δ6–10 and Δ5–14 deletions, the Δ2 deletion caused a significant defect in viral particle production, and the amount of pelletable CA protein was reduced by at least 10-fold (Fig. 3, lane 5). However, the CA protein which was present in the particulate fraction was mostly fully processed, indicating that cleavage at the CA-p2 site was not significantly affected.
FIG. 3.
A critical assembly determinant maps to the N terminus of p2. HeLa cells were transfected with wild-type proviral DNA (WT) or with the indicated mutants and metabolically labeled from 48 to 60 h posttransfection. Viral particles released during the labeling period were pelleted through 20% sucrose cushions, disrupted in RIPA buffer, and directly analyzed by SDS-PAGE. Two different exposures of the same gel are shown. The positions of specific viral proteins are indicated on the left. The positions of migration of molecular mass markers (in kilodaltons) are indicated on the right. Mock, mock transfection.
Role of a predicted α-helical structure at the CA-p2 boundary.
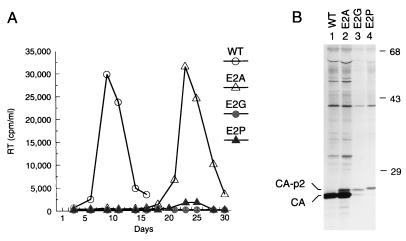
Secondary structure analysis with the PHD program (45–47) predicts that within the context of Pr55gag the conserved N terminus of p2 forms an α-helix which begins in CA (Fig. 1B). Since the PHD program correctly predicted the location of six out of seven α-helices in the N-terminal domain of CA (1), we introduced single-amino-acid substitutions into p2 which were designed either to preserve or to disrupt the predicted α-helical structure at the CA-p2 boundary. Glu-2 of p2, which is located near the center of the predicted α-helical region, was replaced by Ala (mutant E2A), Gly (mutant E2G), or Pro (mutant E2P). Full-length proviruses carrying these mutations were transfected into Jurkat cells, and virus replication was monitored by measuring RT activity in the culture supernatants. Three weeks posttransfection, RT activity began to rise rapidly in a culture transfected with the E2A mutant (Fig. 4A). However, in another culture transfected with the same mutant, RT activity rose only slowly during a 4-week observation period (data not shown), suggesting that a primary- or secondary-site reversion had occurred in the first culture. No evidence for virus replication was detected for up to 30 days in cultures transfected with the E2G and E2P mutants (Fig. 4A).
FIG. 4.
Effects of single-amino-acid substitutions in p2 designed to preserve or to disrupt a predicted α-helical structure on virus replication and on particle formation. (A) RT activity in culture supernatants of Jurkat cells transfected with the indicated proviruses. (B) Direct SDS-PAGE analysis of [35S]methionine-labeled particulate material released from HeLa cells transfected with the indicated proviruses. WT, wild type.
To examine the effects of the single-amino-acid substitutions on viral particle formation, the mutants were transfected into HeLa cells. Virions produced during metabolic labeling with [35S]methionine were pelleted through 20% sucrose, and their protein composition was then directly analyzed by SDS-PAGE. The replacement of Glu-2 with Ala, a strongly helix-favoring amino acid (3, 33, 38), did not affect the efficiency of viral particle production (Fig. 4B, lane 2). However, while fully processed CA predominated by far, virions produced by the E2A mutant contained more CA-p2 than wild-type virions, indicating that processing at the CA-p2 site and/or the internal cleavage site in p2 was slightly affected. In contrast to the E2A mutant, the E2G and E2P mutants exhibited a severe defect in particle production which was at least as pronounced as that of the ΔE2 mutant (Fig. 4B, lanes 3 and 4). The E2G and E2P substitutions also markedly interfered with the conversion of CA-p2 to CA. In particles produced by the E2G mutant, the CA-p2 form was slightly more prominent than fully mature CA, while in E2P mutant particles, the CA-p2 form clearly predominated. Since Gly and Pro have a very low α-helix propensity (3, 5, 38), these results provide genetic evidence that an α-helical conformation of the CA-p2 boundary is crucial for the role of this region in HIV-1 particle production.
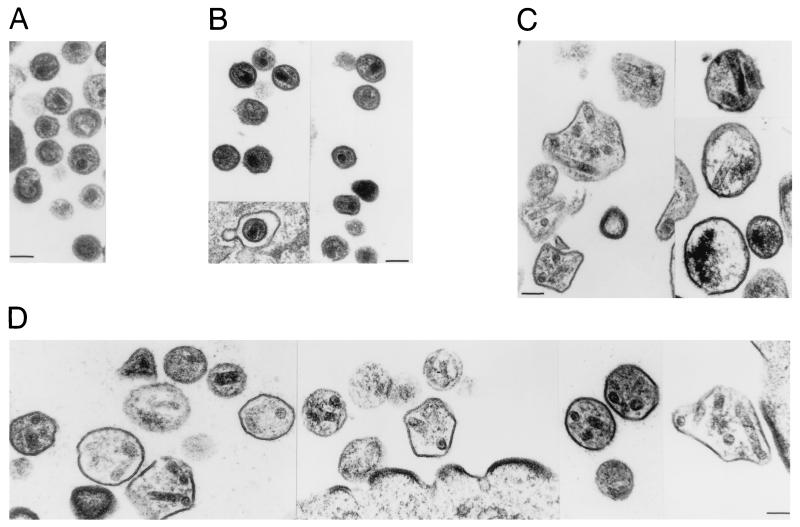
Transmission electron microscopy of HeLa cells transfected with the E2A mutant revealed the production of homogeneous particles (Fig. 5B) of a size comparable to that of wild-type virions (Fig. 5A). Mature virions frequently contained cone-shaped cores similar to those seen in wild-type virions. A remarkably different phenotype was observed for the E2G mutant (Fig. 5C), which did, however, resemble that obtained with the ΔE2 mutant (Fig. 5D). Electron-dense patches were seen at the cell membrane which only rarely assumed the uniform curvature of the budding structures produced by wild-type HIV-1. Extracellular viral particle-like structures were very heterogeneous in size and shape and on average were significantly larger than normal HIV-1 virions. The viral particle-like structures frequently contained one or more tube-shaped cores with a diameter of between 40 and 55 nm. These tubes, which varied in length, are reminiscent of the tubular structures that were recently obtained in vitro with purified HIV-1 CA (22).
FIG. 5.
Electron micrographs of HeLa cell cultures transfected with the parental HXBH10/R+ provirus (A) or with the p2 mutants E2A (B), E2G (C), and Δ2 (D). The magnification is the same for all micrographs (bars represent 100 nm).
CA-p2 processing is required for virion core rearrangement.
A previous study indicated that processing of CA-p2 occurred at an internal Met-Ser site when cleavage at the HIV-1 CA-p2 site was abolished by a point mutation termed CA1 (28). The CA1 mutation introduced Ile into the P1 position of the CA-p2 cleavage site (28), based on the observation that β-branched amino acids are excluded from the P1 position and can block cleavage when introduced by mutagenesis (42). In an attempt to prevent CA-p2 processing, we introduced the CA1 mutation into the HXBH10/R+ provirus in combination with a mutation termed M4I, which replaces the codon for Met-4 of p2 with a codon specifying Ile (Fig. 1). Each mutation was also individually introduced into HXBH10/R+.
In repeated experiments, virus replication was delayed by 1 to 2 weeks relative to that of the wild type after transfection of Jurkat cells with the M4I mutant (Fig. 6A and data not shown). However, when virus was harvested at peak virus production, normalized for RT activity, and used to infect fresh Jurkat cells, wild-type and mutant-derived virus stocks gave similar replication curves (data not shown). To determine whether the M4I mutant had reverted to the wild type, nuclear DNA was extracted from infected cells and used as a template for PCR amplification of the viral gag gene. DNA sequencing analysis of the PCR product revealed that residue 4 of p2 had reverted from Ile to Met, while adjacent silent mutations introduced to create a restriction site were retained (data not shown). In contrast to the M4I mutant, neither the CA1 mutant nor the CA1/M4I double mutant yielded any RT activity for up to 31 days after transfection into Jurkat cells (Fig. 6A).
FIG. 6.
Effects of cleavage site mutations on virus replication and CA processing. (A) RT activity in culture supernatants of Jurkat cells transfected with HXBH10/R+ (WT) or the indicated cleavage site mutants. (B) Direct SDS-PAGE analysis of [35S]methionine-labeled particulate material released from HeLa cells transfected with the parental provirus or the indicated cleavage site mutants. Molecular mass markers (in kilodaltons) are indicated on the right. Mock, mock transfection.
Transfection into HeLa cells revealed that the M4I, CA1, and CA1/M4I mutants each retained the ability to produce viral particles as efficiently as the wild-type construct (Fig. 6B). Particles produced by the M4I mutant contained CA and CA-p2 at about a 10:1 molar ratio as determined by densitometry (Fig. 6B, lane 4). As previously reported (28), CA1 particles did not contain fully processed CA protein. Rather, about equal amounts of CA-p2 and of a novel CA species which migrated slightly slower than fully mature CA were seen (Fig. 6B, lane 3). It was previously suggested that the latter species results from the use of the Met-Ser site within p2 (28). This view is supported by the effect of the M4I mutation, which, when combined with the CA1 mutation, almost completely prevented CA-p2 processing (Fig. 6B, lane 5).
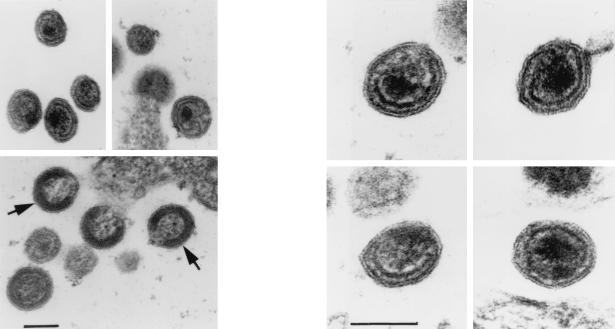
To examine whether the removal of p2 from the C terminus of CA has a role in HIV-1 morphogenesis, HeLa cells transfected with the CA1/M4I double mutant were examined by electron microscopy. Numerous virus particles of normal size were seen, but no cone- or rod-shaped cores were observed in the mutant particles. Immature particles exhibited a ring- or crescent-shaped distribution of electron-dense material assembled underneath the viral envelope (Fig. 7), as is typically seen in an infection with wild type HIV-1. Interestingly, a thin ring- or crescent-shaped submembrane layer persisted in CA1/M4I virions with an otherwise mature appearance (Fig. 7). These distinct electron-dense structures, which frequently appeared to encircle condensed core material, in general were separated from the viral envelope by an electron-lucent space of about 10 nm (Fig. 7).
FIG. 7.
Electron micrographs of HeLa cells transfected with the CA1/M4I double mutant. Arrows point to examples of immature viral particles. Note the electron-dense layer approximately 10 nm below the viral lipid membrane visible in several particles with a mature appearance. Bars represent 100 nm.
DISCUSSION
The presence of a spacer peptide which separates the CA and NC domains in the Gag precursor is a conserved feature among lentiviruses as well as among avian retroviruses (11, 23, 26, 40, 49). Previous studies have shown that the spacer peptides of both HIV-1 and Rous sarcoma virus (RSV) are essential for virus replication (6, 28, 40, 41). Deletions in RSV which removed part or all of the spacer peptide allowed efficient particle assembly, but increased the sensitivity of the virion core to detergent (6, 40). In the case of HIV-1, deletion of the p2 spacer peptide had no apparent effect on the efficiency of virus assembly in one study (41), but had drastic effects on particle production and morphology in another (28).
In the present report, we confirm that p2 harbors a determinant required for efficient particle production and show that this determinant is confined exclusively to the conserved N terminus of p2. Its disruption by point mutations resulted in the production of highly aberrant particles reminiscent of those previously seen with a mutant which lacked p2 entirely (28) and with a mutant which harbored a Ser-Arg substitution at the CA-p2 cleavage site (21). The primary defect of these mutants appears to be in the assembly of uniformly curved buds rather than in the lateral aggregation of Gag precursor molecules per se. The location of the critical determinant in p2 suggests that it may form part of a larger assembly domain which is primarily provided by CA. Recent studies revealed that CA has two domains which have different roles in virus morphogenesis (8, 15, 18, 44). The N-terminal domain of CA is essential for the formation of the characteristic cone-shaped core of the mature virion, but insertions or deletions in this domain generally had little effect on particle assembly (8, 44, 51). In contrast, mutations in the C-terminal third of CA, which forms a distinct domain (15), frequently blocked or severely impaired the ability of Pr55gag to form viral particles (8, 44, 50). An extension of the C-terminal CA assembly domain into p2 may allow its modification through proteolytic processing during the course of virus maturation.
Nuclear magnetic resonance and crystal structures show that the N-terminal domain of CA is largely α-helical (18, 35). The PHD program (45–47) predicts that the C-terminal assembly domain of CA contains four α-helices (1), in excellent agreement with the very recently reported three-dimensional structure (15). Interestingly, when the secondary structure of the CA assembly domain is analyzed in the context of the Gag precursor, the PHD program predicts a fifth α-helix which overlaps the CA-p2 boundary (Fig. 1B). Based on this prediction, we tested the hypothesis that the function of the N terminus of p2 may depend on its ability to adopt an α-helical structure. Consistent with this hypothesis, particle production and virion morphology were not noticeably affected when p2 residue Glu-2 near the center of the predicted helix was replaced by Gln or Ala, but were profoundly impaired when Glu-2 was replaced by Gly or Pro. While the difference between the −CH3 side group in Ala and the −H side group in Gly appears relatively minor, the helical propensities of these residues differ considerably, with Ala being strongly favored and Gly being strongly disfavored in α-helices (3, 5, 33, 38). Pro, which is even more disfavored than Gly near the center of α-helices (36), had an effect comparable to that of Gly on particle production, but a more pronounced effect on CA-p2 processing.
In addition to CA-p2, a novel CA species was present in virions when cleavage at the CA-p2 site was prevented. As previously suggested (28), the novel species presumably resulted from cleavage at an internal Met-Ser site in p2. In RSV virions, an equivalent CA species known as CA3, which has a three-amino-acid C-terminal extension, accounts for about one-third of the total CA in mature virions (40). The CA3 species results from the use of an internal cleavage site in the spacer peptide which separates the CA and NC domains of RSV (40). It has been noted that HIV-1 p2 shows a limited degree of sequence similarity to the corresponding spacer peptide in RSV (40). The similarity is particularly apparent in the sequence which separates CA from the internal cleavage site in each of the two spacer peptides (Ala-Glu-Ala-Met for HIV-1 and Ala-Ala-Met for RSV). In view of these similarities, the question arises whether HIV-1 virions contain a CA species equivalent to the CA3 species of RSV. Attempts to detect such a species in wild-type HIV-1 virions by SDS-PAGE were unsuccessful (1). However, it remains possible that the resolution achieved was insufficient to detect a minor variant with a small mass difference relative to that of fully mature CA.
CA-p2 processing was essentially abolished when both the CA-p2 site and the internal Met-Ser site in p2 were mutated at the P1 position. Remarkably, particles produced by the double mutant lacked a conical core shell but retained a thin ring- or crescent-shaped shell about 10 nm underneath the virion membrane, indicating that the collapse of the CA shell into a cone was blocked. Nevertheless, condensed material which presumably represented the nucleoprotein complex was seen near the center of mutant virions with a mature appearance. This observation is consistent with our previous finding that the viral nucleoprotein complex condenses into a circular structure in the absence of a surrounding core shell (21).
Our results raise the possibility that a major role of p2 is to increase the helix-forming tendency of the C terminus of CA during virus assembly. During virus maturation, cleavage at the CA-p2 site would disrupt the C-terminal helix and thereby weaken CA-CA interactions to permit the rearrangement of the virion core. This in turn may help to convert a relatively stable structure into a more flexible one primed for uncoating.
ACKNOWLEDGMENTS
We thank Paul Werner for help with electron microscopy studies.
M.A.A. was supported by National Cancer Institute training grant T32 CA09141. This work was supported by National Institutes of Health grants AI29873, AI28691 (Center for AIDS Research), and CA06516 (Cancer Center) and by a gift from the G. Harold and Leila Y. Mathers Charitable Foundation.
REFERENCES
- 1.Accola, M. A., and H. G. Göttlinger. Unpublished observation.
- 2.Aldovini A, Young R A. Mutations of RNA and protein sequences involved in human immunodeficiency virus type 1 packaging result in production of noninfectious virus. J Virol. 1990;64:1920–1926. doi: 10.1128/jvi.64.5.1920-1926.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blaber M, Zhang X-J, Matthews B W. Structural basis of amino acid α helix propensity. Science. 1993;260:1637–1640. doi: 10.1126/science.8503008. [DOI] [PubMed] [Google Scholar]
- 4.Bryant M, Ratner L. Myristoylation-dependent replication and assembly of human immunodeficiency virus 1. Proc Natl Acad Sci USA. 1990;87:523–527. doi: 10.1073/pnas.87.2.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bryson J W, Betz S F, Lu H S, Suich D J, Zhou H X, O’Neil K T, DeGrado W F. Protein design: a hierarchic approach. Science. 1995;270:935–941. doi: 10.1126/science.270.5238.935. [DOI] [PubMed] [Google Scholar]
- 6.Craven R C, Leure-duPree A E, Erdie C R, Wilson C B, Wills J W. Necessity of the spacer peptide between CA and NC in the Rous sarcoma virus Gag protein. J Virol. 1993;67:6246–6252. doi: 10.1128/jvi.67.10.6246-6252.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cullen B R. Use of eukaryotic expression technology in the functional analysis of cloned genes. Methods Enzymol. 1987;152:684–704. doi: 10.1016/0076-6879(87)52074-2. [DOI] [PubMed] [Google Scholar]
- 8.Dorfman T, Bukovsky A, Öhagen Å, Höglund S, Göttlinger H G. Functional domains of the capsid protein of human immunodeficiency virus type 1. J Virol. 1994;68:8180–8187. doi: 10.1128/jvi.68.12.8180-8187.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dorfman T, Göttlinger H G. The human immunodeficiency virus type 1 capsid p2 domain confers sensitivity to the cyclophilin-binding drug SDZ NIM 811. J Virol. 1996;70:5751–5757. doi: 10.1128/jvi.70.9.5751-5757.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dorfman T, Mammano F, Haseltine W A, Göttlinger H G. Role of the matrix protein in the virion association of the human immunodeficiency virus type 1 envelope glycoprotein. J Virol. 1994;68:1689–1696. doi: 10.1128/jvi.68.3.1689-1696.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elder J H, Schnölzer M, Hasselkus-Light C S, Henson M, Lerner D A, Phillips T R, Wagaman P C, Kent S B H. Identification of proteolytic processing sites within the Gag and Pol polyproteins of feline immunodeficiency virus. J Virol. 1993;67:1869–1876. doi: 10.1128/jvi.67.4.1869-1876.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Franke E K, Yuan H E H, Bossolt K L, Goff S P, Luban J. Specificity and sequence requirements for interactions between various retroviral Gag proteins. J Virol. 1994;68:5300–5305. doi: 10.1128/jvi.68.8.5300-5305.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Franke E K, Yuan H E H, Luban J. Specific incorporation of cyclophilin A into HIV-1 virions. Nature. 1994;372:359–362. doi: 10.1038/372359a0. [DOI] [PubMed] [Google Scholar]
- 14.Gamble T R, Vajdos F F, Yoo S, Worthylake D K, Houseweart M, Sundquist W I, Hill C P. Crystal structure of human cyclophilin A bound to the amino-terminal domain of HIV-1 capsid. Cell. 1996;87:1285–1294. doi: 10.1016/s0092-8674(00)81823-1. [DOI] [PubMed] [Google Scholar]
- 15.Gamble T R, Yoo S, Vajdos F F, von Schwedler U K, Worthylake D K, Wang H, McCutcheon J P, Sundquist W I, Hill C P. Structure of the carboxy-terminal dimerization domain of the HIV-1 capsid protein. Science. 1997;278:849–853. doi: 10.1126/science.278.5339.849. [DOI] [PubMed] [Google Scholar]
- 16.Gelderblom H R, Hausmann E H, Özel M, Pauli G, Koch M A. Fine structure of human immunodeficiency virus (HIV) and immunolocalization of structural proteins. Virology. 1987;156:171–176. doi: 10.1016/0042-6822(87)90449-1. [DOI] [PubMed] [Google Scholar]
- 17.Gheysen D, Jacobs E, de Foresta F, Thiriart C, Francotte M, Thines D, De Wilde M. Assembly and release of HIV-1 precursor Pr55gag virus-like particles from recombinant baculovirus-infected insect cells. Cell. 1989;59:103–112. doi: 10.1016/0092-8674(89)90873-8. [DOI] [PubMed] [Google Scholar]
- 18.Gitti R K, Lee B M, Walker J, Summers M F, Yoo S, Sundquist W I. Structure of the amino-terminal core domain of the HIV-1 capsid protein. Science. 1996;273:231–235. doi: 10.1126/science.273.5272.231. [DOI] [PubMed] [Google Scholar]
- 19.Gorelick R J, Nigida S M, Jr, Bess J W, Jr, Arthur L O, Henderson L E, Rein A. Noninfectious human immunodeficiency virus type 1 mutants deficient in genomic RNA. J Virol. 1990;64:3207–3211. doi: 10.1128/jvi.64.7.3207-3211.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Göttlinger H G, Dorfman T, Sodroski J G, Haseltine W A. Effect of mutations affecting the p6 gag protein on human immunodeficiency virus particle release. Proc Natl Acad Sci USA. 1991;88:3195–3199. doi: 10.1073/pnas.88.8.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Göttlinger H G, Sodroski J G, Haseltine W A. Role of capsid precursor processing and myristoylation in morphogenesis and infectivity of human immunodeficiency virus type 1. Proc Natl Acad Sci USA. 1989;86:5781–5785. doi: 10.1073/pnas.86.15.5781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gross I, Hohenberg H, Kräusslich H G. In vitro assembly properties of purified bacterially expressed capsid proteins of human immunodeficiency virus. Eur J Biochem. 1997;249:592–600. doi: 10.1111/j.1432-1033.1997.t01-1-00592.x. [DOI] [PubMed] [Google Scholar]
- 23.Henderson L E, Benveniste R E, Sowder R, Copeland T D, Schultz A M, Oroszlan S. Molecular characterization of gag proteins from simian immunodeficiency virus (SIVMne) J Virol. 1988;62:2587–2595. doi: 10.1128/jvi.62.8.2587-2595.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Henderson L E, Bowers M A, Sowder II R C, Serabyn S A, Johnson D G, Bess J W, Jr, Arthur L O, Bryant D K, Fenselau C. Gag proteins of the highly replicative MN strain of human immunodeficiency virus type 1: posttranslational modifications, proteolytic processings, and complete amino acid sequences. J Virol. 1992;66:1856–1865. doi: 10.1128/jvi.66.4.1856-1865.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Henderson L E, Copeland T D, Sowder R C, Schultz A M, Oroszlan S. Analysis of proteins and peptides purified from sucrose gradient banded HTLV-III. In: Bolognesi D, editor. Human retroviruses, cancer, and AIDS: approaches to prevention and therapy. New York, N.Y: Alan R. Liss, Inc.; 1988. pp. 135–147. [Google Scholar]
- 26.Henderson L E, Sowder R C, Smythers G W, Oroszlan S. Chemical and immunological characterizations of equine infectious anemia virus gag-encoded proteins. J Virol. 1987;61:1116–1124. doi: 10.1128/jvi.61.4.1116-1124.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Höglund S, Öfverstedt L-G, Nilsson Å, Lundquist P, Gelderblom H, Özel M, Skoglund U. Spatial visualization of the maturing HIV-1 core and its linkage to the envelope. AIDS Res Hum Retroviruses. 1992;8:1–7. doi: 10.1089/aid.1992.8.1. [DOI] [PubMed] [Google Scholar]
- 28.Kräusslich H-G, Fäcke M, Heuser A-M, Konvalinka J, Zentgraf H. The spacer peptide between human immunodeficiency virus capsid and nucleocapsid proteins is essential for ordered assembly and viral infectivity. J Virol. 1995;69:3407–3419. doi: 10.1128/jvi.69.6.3407-3419.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kunkel T A, Roberts J D, Zakour R A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- 30.Linial M L, Miller A D. Retroviral RNA packaging: sequence requirements and implications. Curr Top Microbiol Immunol. 1990;157:125–152. doi: 10.1007/978-3-642-75218-6_5. [DOI] [PubMed] [Google Scholar]
- 31.Lu Y-L, Spearman P, Ratner L. Human immunodeficiency virus type 1 viral protein R localization in infected cells and virions. J Virol. 1993;67:6542–6550. doi: 10.1128/jvi.67.11.6542-6550.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mammano F, Öhagen Å, Höglund S, Göttlinger H G. Role of the major homology region of human immunodeficiency virus type 1 in virion morphogenesis. J Virol. 1994;68:4927–4936. doi: 10.1128/jvi.68.8.4927-4936.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marqusee S, Robbins V H, Baldwin R L. Unusually stable helix formation in short alanine-based peptides. Proc Natl Acad Sci USA. 1989;86:5286–5290. doi: 10.1073/pnas.86.14.5286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mervis R J, Ahmad N, Lillehoj E P, Raum M G, Salazar F H R, Chan H W, Venkatesan S. The gag gene products of human immunodeficiency virus type 1: alignment with the gag open reading frame, identification of posttranslational modifications, and evidence for alternative gag precursors. J Virol. 1988;62:3993–4002. doi: 10.1128/jvi.62.11.3993-4002.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Momany C, Kovari L C, Prongay A J, Keller W, Gitti R K, Lee B M, Gorbalenya A E, Tong L, McClure J, Ehrlich L S, Summers M F, Carter C, Rossmann M G. Crystal structure of dimeric HIV-1 capsid protein. Nat Struct Biol. 1996;3:763–770. doi: 10.1038/nsb0996-763. [DOI] [PubMed] [Google Scholar]
- 36.Muñoz V, Serrano L. Intrinsic secondary structure propensities of the amino acids, using statistical φ-ψ matrices: comparison with experimental scales. Proteins. 1994;20:301–311. doi: 10.1002/prot.340200403. [DOI] [PubMed] [Google Scholar]
- 37.Myers G, Korber B, Wain-Hobson S, Smith R F, Pavlakis G N. Human retroviruses and AIDS 1993. A compilation and analysis of nucleic acid and amino acid sequences. Los Alamos, N.Mex: Los Alamos National Laboratory; 1993. [Google Scholar]
- 38.Myers J K, Pace C N, Scholtz J M. A direct comparison of helix propensity in proteins and peptides. Proc Natl Acad Sci USA. 1997;94:2833–2837. doi: 10.1073/pnas.94.7.2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Paxton W, Connor R I, Landau N R. Incorporation of Vpr into human immunodeficiency virus type 1 virions: requirement for the p6 region of gag and mutational analysis. J Virol. 1993;67:7229–7237. doi: 10.1128/jvi.67.12.7229-7237.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pepinsky R B, Papayannopoulos I A, Chow E P, Krishna N K, Craven R C, Vogt V M. Differential proteolytic processing leads to multiple forms of the CA protein in avian sarcoma and leukemia viruses. J Virol. 1995;69:6430–6438. doi: 10.1128/jvi.69.10.6430-6438.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pettit S C, Moody M D, Wehbie R S, Kaplan A H, Nantermet P V, Klein C A, Swanstrom R. The p2 domain of human immunodeficiency virus type 1 Gag regulates sequential proteolytic processing and is required to produce fully infectious virions. J Virol. 1994;68:8017–8027. doi: 10.1128/jvi.68.12.8017-8027.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pettit S C, Simsic J, Loeb D D, Everitt L, Hutchison III C A, Swanstrom R. Analysis of retroviral protease cleavage sites reveals two types of cleavage sites and the structural requirements of the P1 amino acid. J Biol Chem. 1991;266:14539–14547. [PubMed] [Google Scholar]
- 43.Queen C, Baltimore D. Immunoglobulin gene transcription is activated by downstream sequence elements. Cell. 1983;33:741–748. doi: 10.1016/0092-8674(83)90016-8. [DOI] [PubMed] [Google Scholar]
- 44.Reicin A S, Paik S, Berkowitz R D, Luban J, Lowy I, Goff S P. Linker insertion mutations in the human immunodeficiency virus type 1 gag gene: effects on virion particle assembly, release, and infectivity. J Virol. 1995;69:642–650. doi: 10.1128/jvi.69.2.642-650.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rost B, Sander C. Prediction of protein secondary structure at better than 70% accuracy. J Mol Biol. 1993;232:584–599. doi: 10.1006/jmbi.1993.1413. [DOI] [PubMed] [Google Scholar]
- 46.Rost B, Sander C. Improved prediction of protein secondary structure by use of sequence profiles and neural networks. Proc Natl Acad Sci USA. 1993;90:7558–7562. doi: 10.1073/pnas.90.16.7558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rost B, Sander C. Combining evolutionary information and neural networks to predict protein secondary structure. Proteins. 1994;19:55–72. doi: 10.1002/prot.340190108. [DOI] [PubMed] [Google Scholar]
- 48.Thali M, Bukovsky A, Kondo E, Rosenwirth B, Walsh C, Sodroski J, Göttlinger H G. Functional association of cyclophilin A with HIV-1 virions. Nature. 1994;372:363–365. doi: 10.1038/372363a0. [DOI] [PubMed] [Google Scholar]
- 49.Tobin G J, Sowder II R C, Fabris D, Hu M Y, Battles J K, Fenselau C, Henderson L E, Gonda M A. Amino acid sequence analysis of the proteolytic cleavage products of the bovine immunodeficiency virus Gag precursor polypeptide. J Virol. 1994;68:7620–7627. doi: 10.1128/jvi.68.11.7620-7627.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Von Poblotzki A, Wagner R, Niedrig M, Wanner G, Wolf H, Modrow S. Identification of a region in the Pr55gag-polyprotein essential for HIV-1 particle formation. Virology. 1993;193:981–985. doi: 10.1006/viro.1993.1210. [DOI] [PubMed] [Google Scholar]
- 51.Wang C-T, Barklis E. Assembly, processing, and infectivity of human immunodeficiency virus type 1 Gag mutants. J Virol. 1993;67:4264–4273. doi: 10.1128/jvi.67.7.4264-4273.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yu X, Yuan X, Matsuda Z, Lee T-H, Essex M. The matrix protein of human immunodeficiency virus type 1 is required for incorporation of viral envelope protein into mature virions. J Virol. 1992;66:4966–4971. doi: 10.1128/jvi.66.8.4966-4971.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]