Abstract
PURPOSE
About a third of patients with relapsed or refractory classic Hodgkin lymphoma (r/r CHL) succumb to their disease after high-dose chemotherapy followed by autologous stem-cell transplantation (HDC/ASCT). Here, we aimed to describe spatially resolved tumor microenvironment (TME) ecosystems to establish novel biomarkers associated with treatment failure in r/r CHL.
PATIENTS AND METHODS
We performed imaging mass cytometry (IMC) on 71 paired primary diagnostic and relapse biopsies using a marker panel specific to CHL biology. For each cell type in the TME, we calculated a spatial score measuring the distance of nearest neighbor cells to the malignant Hodgkin Reed Sternberg cells within the close interaction range. Spatial scores were used as features in prognostic model development for post-ASCT outcomes.
RESULTS
Highly multiplexed IMC data revealed shared TME patterns in paired diagnostic and early r/r CHL samples, whereas TME patterns were more divergent in pairs of diagnostic and late relapse samples. Integrated analysis of IMC and single-cell RNA sequencing data identified unique architecture defined by CXCR5+ Hodgkin and Reed Sternberg (HRS) cells and their strong spatial relationship with CXCL13+ macrophages in the TME. We developed a prognostic assay (RHL4S) using four spatially resolved parameters, CXCR5+ HRS cells, PD1+CD4+ T cells, CD68+ tumor-associated macrophages, and CXCR5+ B cells, which effectively separated patients into high-risk versus low-risk groups with significantly different post-ASCT outcomes. The RHL4S assay was validated in an independent r/r CHL cohort using a multicolor immunofluorescence assay.
CONCLUSION
We identified the interaction of CXCR5+ HRS cells with ligand-expressing CXCL13+ macrophages as a prominent crosstalk axis in relapsed CHL. Harnessing this TME biology, we developed a novel prognostic model applicable to r/r CHL biopsies, RHL4S, opening new avenues for spatial biomarker development.
INTRODUCTION
Microenvironment biology has been extensively explored in various cancers, including lymphomas, and many studies have revealed the pathologic importance of reactive immune cells in the tumor microenvironment (TME).1-3 Classic Hodgkin Lymphoma (CHL) is unique among virtually all cancers as the malignant Hodgkin and Reed Sternberg (HRS) cells are greatly outnumbered by reactive, non-neoplastic cells in the TME.4,5 Typically, the malignant HRS cells represent <1% of cells in an individual tumor. Despite progress in elucidating TME biology and the extensive cellular crosstalk by cytokines/chemokines in CHL pathogenesis,6,7 the molecular determinants of treatment failure remain mostly unknown.
CONTEXT
Key Objective
To determine the composition and spatial architecture of the tumor microenvironment (TME) in relapsed and refractory Hodgkin lymphoma associated with treatment failure to second-line chemotherapy and autologous stem-cell transplantation.
Knowledge Generated
Our study highlights the contrast between relatively persistent early relapse biology and more divergent late relapse biology with dynamic changes in the TME. Our data shed light on the unique and targetable spatial interaction between CXCR5+ Hodgkin and Reed Sternberg cells with CXCL13+ macrophages as a characteristic feature of relapsed/refractory classic Hodgkin lymphoma. To translate these biologic findings into the clinic, we developed and validated a spatially resolved biomarker assay (RHL4S).
Relevance (J.W. Friedberg)
-
This work extends previous studies demonstrating the importance of the TME and specifically the macrophage in the clinical behavior of Hodgkin lymphoma; future studies should leverage the assay to determine whether this represents a predictive biomarker in the therapeutic era of checkpoint blockade.*
*Relevance section written by JCO Editor-in-Chief Jonathan W. Friedberg, MD.
Despite recent treatment advances, about a third of patients with relapsed and refractory (r/r) CHL succumb to their disease after high-dose chemotherapy followed by autologous stem-cell transplantation (HDC/ASCT).8,9 Recent studies have reported that gene expression signatures representing non-neoplastic cells of the TME are associated with outcomes after therapy.7,10,11 Our group previously developed and validated a clinically applicable prognostic assay (RHL30), which identifies a subset of patients at high risk of treatment failure after HDC/ASCT in r/r CHL.10 The study also established that relapse biopsies are superior to diagnostic biopsies to develop biomarker assays for predicting post-ASCT outcomes. However, RHL30 does not incorporate important information about cellular interactions, specific expression features of HRS cells, and the spatial architecture of the TME. Recent progress with multiplex imaging techniques has enabled comprehensive spatial characterization of TME biology, suitable for formalin-fixed paraffin-embedded (FFPE) tissue. This was demonstrated in our recent publication using imaging mass cytometry (IMC) in primary CHL where novel interactions between tumor-specific immunosuppressive cell populations and malignant HRS cells were described.12 We therefore hypothesized that the more detailed description of spatially resolved ecosystems in samples at the timepoint of clinical relapse would lead to the development of refined biomarkers with the potential to improve risk stratification for postsecondary treatment outcomes.
Here, we performed IMC analysis of 71 paired pretreatment/relapse biopsies and nonrelapse biopsies (n = 22) to describe the unique changes in microenvironment architecture linked to relapse on a per-patient basis. Our data shed light on the unique interactions between cancer cells and the TME, highlighting the spatial interaction between CXCR5+ HRS cells with CXCL13+ macrophages as a characteristic feature of r/r CHL. Consequently, we developed a novel spatially resolved prognostic biomarker assay, RHL4S, that was also translated into a multicolor immunofluorescence (MC-IF) assay suitable for future implementation in routine pathology workflows.
PATIENTS AND METHODS
We analyzed IMC data using a customized marker panel for CHL from 164 CHL samples, including 71 patients with paired primary diagnostic and relapse specimens and 22 diagnostic control samples without relapse, termed the discovery cohort (Data Supplement, Appendix Fig A1 and Appendix Tables A1-A3 [online only]).10 Biopsies of these CHL cases, treated at BC Cancer between 1985 and 2011, are part of a tissue microarray (TMA) that was previously reported.10 Patients were classified as having early relapse if their CHL progressed within 12 months after initial diagnosis or was refractory to first-line treatment.
To develop a prognostic model for r/r CHL, we first calculated a spatial score for each cell type using IMC and MC-IF data, defined as the term (1 – average distance of HRS cells to the five nearest neighbor cells of that type capped at 50 μm) to distinguish relationships between HRS cells and clusters of interacting cells. This strategy enabled us to quantify the spatially resolved cellular architecture in CHL (Data Supplement, Appendix Fig A2). Then, we applied our new cross-format Least Absolute Shrinkage and Selection Operator (LASSO) plus algorithm on the combination of two separate lists of standardized spatial scores and standardized traditional protein-based cellular abundance percentages. Using the spatial scores of the four selected variables associated with post-ASCT failure-free survival (FFS), we developed a risk classification prediction model, RHL4S, on the basis of our new XGpred algorithm in cvmpv R package, which distinguished high- and low-risk groups of patients with respect to post-ASCT FFS.
We then assembled a second, independent cohort for validation, termed the validation cohort (n = 44) used for MC-IF using a simplified panel (Data Supplement, Appendix Tables A4 and A5). This panel contains the multiparametric and spatial information of the four variables from the IMC-based model to establish a prognostic model that can be generalized and is applicable to routine pathology procedures. For the validation cohort, we selected patients with r/r CHL treated at BC Cancer between 2012 and 2021 with available FFPE relapse biopsies according to the same selection criteria as the discovery cohort. Patient characteristics of the validation cohort are summarized in the Data Supplement (Appendix Table A1 and Appendix Fig A1). To confirm the technical concordance between spatial scores derived from IMC versus MC-IF, we applied the MC-IF panel to a subset of the discovery cohort (n = 19). To adjust RHL4S scores between IMC and MC-IF methodologies, calibration was performed between the two techniques correcting scores by a calibration value defined as the mean difference between RHL4S IMC and MC-IF scores (Data Supplement, Appendix Figs A3 and A4).
Single-cell RNA sequencing (scRNA-seq) was performed as previously described12 using sorted enriched HRS cells (Data Supplement, Appendix FigA5 and Appendix Table A6) from cell suspensions of CHL.13,14 To boost transcriptome information for cell-to-cell interaction analyses, hybridization capture of marker genes (Data Supplement, Appendix Table A7) was performed as described previously15 and run on Illumina Nextseq550. Further details are provided in the Data Supplement.
Written informed consent or consent waivers were obtained from all patients. This study was reviewed and approved by the University of British Columbia-BC Cancer Agency Research Ethics Board (H14-02304), in accordance with the Declaration of Helsinki.
RESULTS
The Spatially Resolved TME of r/r CHL
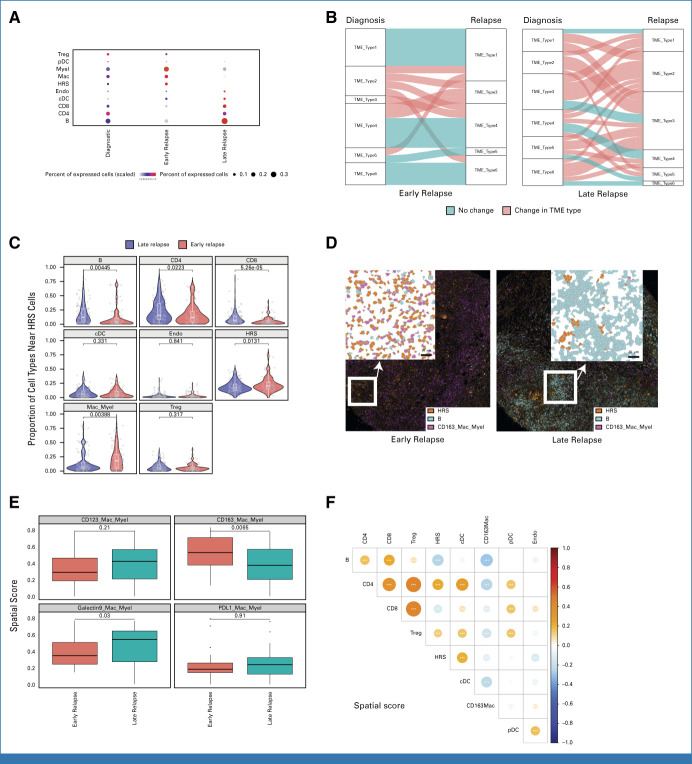
Using a customized marker panel specific to CHL biology (Data Supplement, Appendix Table A2), we obtained highly multiplexed images for a total of 7,146,042 cells at a resolution of 1 μm2. To define the spatial architecture and related cellular interactions, we first investigated the differences in TME characteristics according to disease status (initial diagnosis v early relapse v late relapse), showing differential abundance of each immune cell type (Fig 1A). While diagnostic samples demonstrated known CD4 T-cell enrichment patterns, relapse samples showed a unique immune cell abundance profile. Since high-resolution characterization beyond cellular abundance is needed to delineate the complexity of the cellular ecosystem in the TME,16 we defined comprehensive TME ecotypes using unsupervised clustering by leveraging a multitude of TME features (Data Supplement, Appendix Fig A6). When determining TME ecotypes in individual diagnostic and relapsed biopsy pairs per patient, we observed significant differences in TME dynamics between early relapse and late relapse samples, with a significantly increased number of TME type transitions in late relapse (P < .01; Fig 1B). Late relapse samples were characterized by an abundance of nonmalignant B cells (P < .01) along with CD4 (P < .05) and CD8 T-cell enrichment (P < .01; Figs 1A, 1C and 1D). Conversely, CHL biopsies from patients with early relapse demonstrated more similar TME patterns between diagnostic and relapse samples (Fig 1B). In particular, macrophage/myeloid cell–enriched TME ecotypes 1 and 6 were constant over time between diagnostic and early relapse biopsies (Figs 1B and 1C). Further analyses revealed that a CD163+ macrophage population,17 indicative of M2 polarization, was significantly enriched in early relapse samples (Figs 1D and 1E). Consistent with these findings, spatial analyses of relapse samples revealed an inverse correlation of CD163+ macrophages and B cells in cellular neighborhoods (Fig 1F).
FIG 1.
Distinct spatially resolved TME features according to relapse status. (A) Proportion for the indicated immune cell population by IMC-based cluster assignment. (B) The alluvial plot shows the TME types and their dynamic change between diagnostic samples and relapse samples according to relapse status. Horizontal ribbons represent individual cases and can be followed from left to right. Blue color of the ribbons indicates that there is no TME type change between diagnostic and relapse samples, whereas red-colored samples indicate the change in TME type. (C) Violin plot indicating the spatial score for the indicated cell types near HRS cells according to relapse status. (D) IMC analysis from FFPE sections of CHL shows localization of immune cells according to relapse status. (Left) A representative case with early relapse CHL case shows numerous CD163+ macrophage/myeloid cells and rare B cells. By contrast, (right) a representative late relapse HL case shows few CD163+ macrophage/myeloid cells and abundant B cells. (E) Box plot indicating the spatial scores of macrophage/myeloid cell subtypes near HRS cells according to relapse status. (F) Dot plot showing correlation of spatial scores of major immune cell markers by IMC. Dot size and color summarize Pearson correlation values, with positive correlations represented in red and negative correlations represented in blue. Asterisks represent associated P values (*P < .05; **P < .01; ***P < .001). cDC, conventional dendritic cell; CHL, classic Hodgkin lymphoma; FFPE, formalin-fixed paraffin-embedded; HRS, Hodgkin and Reed Sternberg; IMC, imaging mass cytometry; pDC, plasmacytoid dendritic cell; TME, tumor microenvironment; Treg, T regulatory cell.
The Unique Spatial Architecture Associated With CXCR5+ HRS Cells in Relapse Biopsies
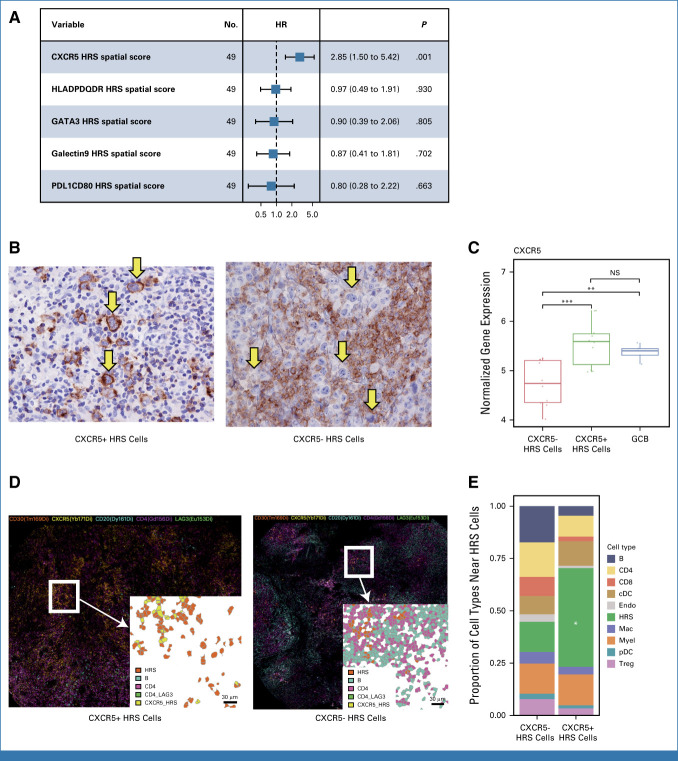
In previous studies, the most prominent obstacle for biomarker development in CHL was the scarcity of the malignant HRS cells and the heterogeneity of TME composition within individual tumor biopsies. Our IMC panel was designed to simultaneously quantify protein expression on HRS cells and the TME, including known variably expressed markers on HRS cells, such as CD30, PD-L1, and major histocompatibility classes I and II (MHC-I and MHC-II). Unsupervised clustering identified several new subsets within the HRS cellular compartment, defined by, for example, high GATA3 and CXCR5 expression. These phenotypic HRS cell definitions are additive to other known subsets, such as HRS cells with high PD-L1 or CD123 expression18,19 (Data Supplement, Appendix Fig A7). Next, we evaluated the prognostic impact of these HRS features in the spatial context of r/r CHL. LASSO analysis identified CXCR5+ HRS cells as the most significant HRS cell phenotype correlated with post-ASCT FFS, in contrast to the absence of an outcome that correlated with all HRS cells, not further specified (Fig 2A, Data Supplement, Appendix Fig A8). To confirm CXCR5 expression patterns on HRS cells by both protein and RNA levels, we assessed the expression of CXCR5 using immunohistochemistry (IHC) and reanalyzed published Affymetrix gene expression data generated from microdissected HRS cells of primary HL samples.20 These analyses confirmed that variable CXCR5 surface protein expression correlated well with mRNA expression and CXCR5 was highly expressed in a subset of CHL tumors (Fig 2B, Data Supplement, Appendix Fig A8) comparable with expression levels found in CD77+ germinal center B cells (Fig 2C). Interestingly, CXCR5+ HRS cells were spatially arranged together with other CXCR5+ HRS cells forming cell clusters as an architectural feature (P < .05). CXCR5+ HRS cell clusters were also characterized by a lower abundance of nonmalignant immune cells, including CD4+ T reg cells and CD20+ B cells when compared with CXCR5− HRS cells (Figs 2D-2F), suggesting distinct TME characteristics associated with CXCR5+ HRS cells.
FIG 2.
Characteristics of the tumor microenvironment of CHL associated with CXCR5 positivity on HRS cells. (A) Forest plots summarize the prognostic factors in relapsed CHL treated with high-dose chemotherapy/autologous stem-cell transplantation according to HRS cells' features by IMC. (B) IHC staining for CXCR5 in representative cases with either (left) positive or (right) negative HRS cells (×400). (C) Expression of CXCR5 in microdissected HRS cells from primary CHL samples (separated by CXCR5 status evaluated by IHC) and germinal center cells from reactive tonsil tissue (GCB; t test; NS: P > .05; *P ≤ .05; **P ≤ .01 ; ***P ≤ .001). (D) IMC image for selected immune subsets in representative cases with either (left) CXCR5-positive or (right) CXCR5-negative HRS cells. (E) Relative proportion of cell subtypes near either negative (left) or positive (right) HRS cells. *P < .05. cDC, conventional dendritic cell; CHL, classic Hodgkin lymphoma; GCB, germinal center B cell; HR, hazard ratio; HRS, Hodgkin and Reed Sternberg; IHC, immunohistochemistry; IMC, imaging mass cytometry; NS, nodular sclerosis; pDC, plasmacytoid dendritic cell; Treg, T regulatory cell.
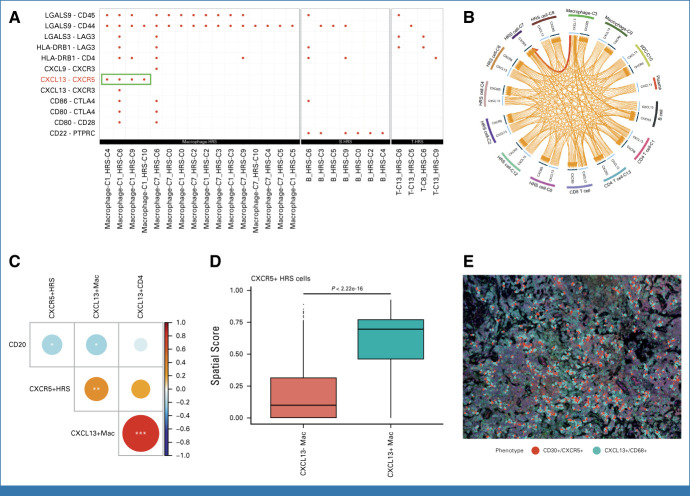
We next sought to understand the cellular interactions between CXCR5+ HRS cells and other immune cell populations. Although we observed fewer TME components surrounding CXCR5+ HRS cells in the IMC data (Fig 2E), we hypothesized that previously unknown immune cell populations, which cannot be defined with the current IMC panel, might interact with CXCR5+ HRS cells. Therefore, we additionally performed scRNA-seq on CHL samples with enrichment of the HRS cell population by cell sorting (Data Supplement, Appendix Figs A5 and A9). Using the cell-to-cell communication tool, Cell Chat,21 we predicted CXCR5-CXCL13 interaction between CXCR5+ HRS cells and CXCL13+ macrophages (Fig 3A). This significant interaction was also validated by the alternative iTALK method22 (Fig 3B). CXCL13 is a well-described cell attractant via the CXCL13/CXCR5 axis. Intriguingly, CXCL13+ macrophages mostly (>99%) did not coexpress M2 macrophage markers, such as CD163 or CD20611 (Data Supplement, Appendix Fig A10), indicating a distinct profile of this population. Since CXCL13 was not part of the original IMC panel, we were not able to study its expression in the IMC data. Therefore, to further describe the spatial relationship between CXCR5+ HRS cells and CXCL13+ macrophages, we applied MC-IF to the discovery cohort TMA that was used for IMC. Consistent with the results of the scRNAseq-based cell-to-cell interaction prediction, we observed a strong positive correlation of the abundance of CXCR5+ HRS cells with CXCL13+ macrophages in the MC-IF data (Fig 3C). Furthermore, CXCL13+ macrophages were located in close proximity to CXCR5+ HRS cells (Figs 3D and 3E), supporting the importance of the CXCR5/CXCL13 axis in CHL.
FIG 3.
CXCL13/CXCR5 interaction in CHL. (A) The dot plot shows significant ligand and receptor interaction between HRS cells (receptor) and immune cell populations (ligand) using Cell Chat. (B) An interaction between CXCL13 and CXCR5 on immune cells and HRS cells in CHL samples was predicted using the iTALK tool. (C) Dot plot showing correlations of the proportions of selected immune cell subsets with emphasis on CXCL13/CXCR5 interaction (multicolor immunohistochemistry). Dot size and color summarize Pearson correlation values, with positive correlations represented in blue and negative correlations represented in red. Asterisks represent associated P values (*P < .05; **P < .01; ***P < .001). (D) Boxplot showing the spatial score of CXCL13+ and CXCL13− macrophages in the region surrounding CD30+ cells (HRS). (E) Membrane map depicting CD68+CXCL13+ macrophages (light blue) and CD30+CXCR5+ HRS cells (red). CHL, classic Hodgkin lymphoma; HRS, Hodgkin and Reed Sternberg.
Development of a Prognostic Model Leveraging Spatial TME Information
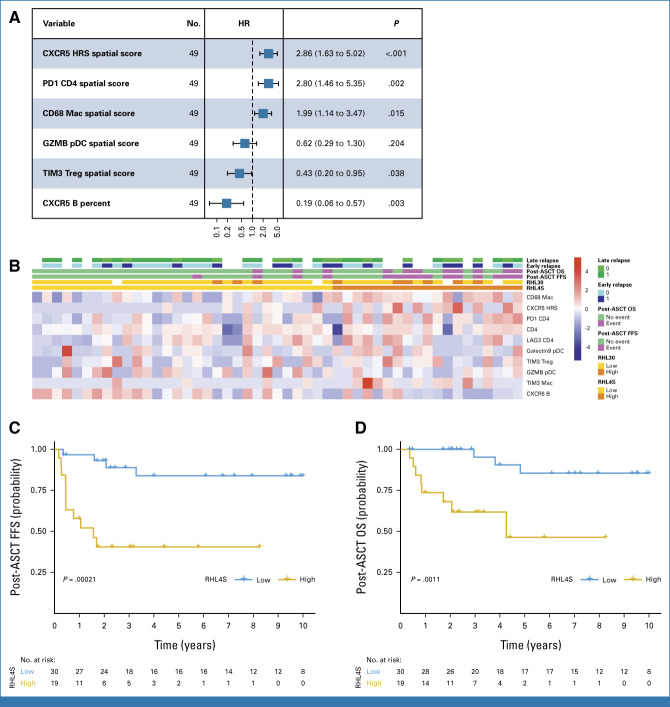
Next, we sought to construct a prognostic model for post-ASCT outcomes in patients with r/r CHL taking advantage of simultaneously capturing HRS cell and TME biology by IMC. Considering the previous demonstration of the superiority of relapse specimens for outcome prediction in r/r CHL,10 we focused our analyses on biomarker measurements in relapse samples. For feature selection, we first performed cross-format LASSO plus analysis on two separate sets of variables, namely, standardized spatial scores and conventional cellular abundance percentages (on the basis of protein expression markers; see the Data Supplement, Method, Appendix file). Strikingly, all top five variables, which were significantly associated with post-ASCT FFS P ≤ .05, were spatial scores (Fig 4A), confirming the importance of spatially informed parameters for outcome prediction. Consistent with the independent prognostic importance of these variables (Fig 4A), each patient sample showed distinct spatial patterns, which were linked to these cellular components (Fig 4B). Four variables, CXCR5+ HRS cells (hazard ratio [HR], 2.86 [95% CI, 1.63 to 5.02]), PD1+ CD4+ T cells (HR, 2.80 [95% CI, 1.46 to 5.35]), CD68+ macrophages (HR, 1.99 [95% CI, 1.14 to 3.47]), and CXCR5+ B cells (HR, 0.19 [95% CI, 0.06 to 0.57]), were identified as factors most significantly associated with post-ASCT FFS. By contrast, we could not observe any significant prognostic associations of the T follicular helper (TFH) subset or other macrophage subsets such as CD163+ macrophages and PD-L1+ macrophages (Data Supplement, Appendix Fig A11). The CD4+PD1+ T-cell population coexpressed other inhibitory receptors such as TIM3 and showed a significant positive correlation with PD-L1+ HRS cells (Data Supplement, Appendix Fig A12).
FIG 4.
Development of a novel prognostic model, RHL4S, which predicts FFS after ASCT. (A) Forest plots summarize the prognostic factors in relapsed classic Hodgkin lymphoma treated with high-dose chemotherapy/ASCT according to IMC. (B) Heatmap of the spatial scores in RHL4S according to IMC. Cases are ordered by the RHL4S model score. Kaplan-Meier curves of the high- versus low-risk groups for (C) post-ASCT FFS and (D) post-ASCT OS as identified by RHL4S. P values were calculated using a log-rank test. ASCT, autologous stem-cell transplantation; FFS, failure-free survival; HR, hazard ratio; HRS, Hodgkin and Reed Sternberg; IMC, imaging mass cytometry; OS, overall survival.
To leverage the multifactorial spatial biologic features that are linked to r/r biology, we developed a risk prediction model, RHL4S, using the spatial scores from these four variables. RHL4S identified high- and low-risk groups of patients with the high-risk group of patients having significantly inferior post-ASCT FFS (5-year post-ASCT FFS: 41% v 81%; P < .0001; Fig 4C) and inferior post-ASCT overall survival (OS; 5-year post-ASCT OS: high-risk, 46% v low-risk, 85%; P = .001; Fig 4D).
We then investigated the independent prognostic value of RHL4S with respect to previously reported prognostic factors of post-ASCT outcomes including RHL30 (Fig 4B), time to first relapse, chemoresistance, B symptoms at relapse, age ≥45 years at ASCT, and stage IV disease at initial diagnosis. Pairwise multivariable Cox regression analysis including RHL4S and these known prognostic variables demonstrated that the RHL4S risk group was statistically independent (P <.05; Data Supplement, Appendix Fig A13). In line with a previous study,10 RHL4S scores using expression measurements from the initial diagnostic biopsy could not predict post-ASCT outcomes, confirming the superiority of relapse biopsies for predicting post-ASCT outcomes in r/r CHL (Data Supplement, Appendix Fig A14).
Independent Validation of RHL4S
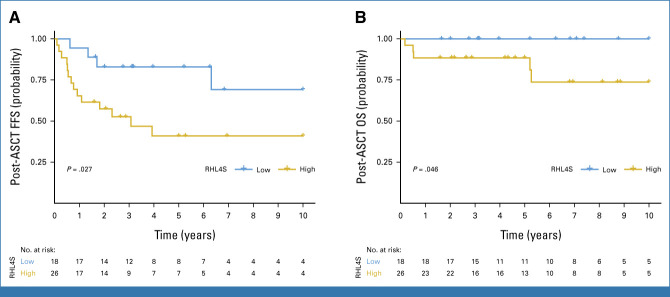
To establish a prognostic model that can be readily used for diagnostic procedures in clinical practice, we next translated the IMC-based RHL4S assay into MC-IF methodology. We built a simplified marker panel encompassing the four expression variables of the predictive model (Data Supplement, Appendix Table A3 and Appendix Fig A15) and calculated calibrated individual spatial scores from the MC-IF data in the independent validation cohort of relapse biopsies from 44 patients with r/r CHL uniformly treated with salvage treatment and consolidating ASCT (Data Supplement, Appendix Table A1). In the validation cohort, first-line brentuximab vedotin (BV) plus doxorubicin, vinblastine, and dacarbazine was used for only one patient and 10 patients received BV consolidation (Data Supplement, Appendix Tables A1 and A6-A7), whereas no patients in this study received the PD-1 blockade before ASCT. Consistent with the finding in the discovery cohort, high-risk patients (defined by RHL4S) displayed unfavorable post-ASCT FFS (5-year post-ASCT FFS: 41% v 83%; P = .027; Fig 5A) and post-ASCT OS (5-year post-ASCT OS: 88% v 100%; P = .046; Fig 5B) in the independent validation cohort. Finally, we investigated the independent prognostic value of RHL4S with respect to previously reported prognostic factors of post-ASCT outcomes in the validation cohort. Pairwise multivariable Cox regression analysis demonstrated the independence of RHL4S (P < .05) or trends toward independence against these markers (Data Supplement, Appendix Fig A16). Of note, although the mRNA gene expression–based RHL30 assay showed similar performance to RHL4S in the discovery cohort, RHL30 did not show a significant difference in post-ASCT FFS between the high- and low-risk groups in the validation cohort (Data Supplement, Appendix Fig A17), indicating superior performance of RHL4S.
FIG 5.
Validation of RHL4S in the independent cohort of relapsed and refractory classic Hodgkin lymphoma. Kaplan-Meier curves of the high- versus low-risk groups for (A) post-ASCT FFS and (B) post-ASCT OS as identified by RHL4S in the independent validation cohort, respectively. P values were calculated using a log-rank test. ASCT, autologous stem-cell transplantation; FFS, failure-free survival; OS, overall survival.
DISCUSSION
Here, we developed and validated a novel prognostic model, RHL4S, on the basis of spatial TME biology, which can predict outcome after ASCT in patients with r/r CHL. Our study establishes a paradigm for the strong prognostic value of spatially resolved biomarkers superior to traditional expression-based biomarkers in lymphoma and suggests the biologic importance of tumor architectural patterns for biomarker development and discovery of immunotherapeutic targets in other cancer types. Importantly, RHL4S was associated with post-ASCT outcomes regardless of relapse status (early relapse or late relapse).
Currently, RHL4S necessitates the use of the MC-IF platform. While we anticipate that MC-IF will remain feasible in many situations, especially in clinical trials, because of the ability to obtain only one unstained slide to apply MC-IF and the affordability of the technique, further steps will be required to make RHL4S readily applicable to routine clinical practice on widely available platforms, such as IHC. We anticipate that our resources will be valuable for future validation studies. Encouragingly, digital pathology systems have already been implemented for primary diagnostic purposes in clinical practice under high-quality standards such as the Clinical Laboratory Improvement Amendments program.23,24 Looking ahead, the application of artificial intelligence (AI)–based solutions and machine learning approaches might help overcome the current limitations of MC-IF.25 AI and machine learning can enhance applicability and accuracy, including cell segmentation, and potentially enable the calculation of spatial information from cancer cells using hematoxylin and eosin or single-stain IHC.
RHL4S includes four spatially resolved variables, including macrophages that were identified as a prognostic biomarker in CHL on the basis of raw cellular abundance in the TME in multiple studies.7,26-28 We now also identified a phenotypically defined subset of HRS cells (CD30+CXCR5+) that was associated with outcome, and intriguingly, we observed enrichment of CXCL13+ macrophages in regions surrounding CXCR5+ HRS cells. We also recently found that CXCL13+PD1+ TFH-like cells are enriched in a specific subtype of CHL (lymphocyte-rich CHL) and associated with poor clinical outcome in this rare subtype.6 These two studies indicate the importance of ligand (CXCL13) and receptor (CXCR5) interaction and their association with treatment failure in CHL. Importantly, CXCL13/CXCR5 targeting agents are currently under investigation in hematologic malignancies and in autoimmune disease.29-31 Therefore, additional investigations into the CXCR5/CXCL13 biology including migration phenotypes of CXCR5+ HRS cells are warranted to determine the therapeutic benefit in CHL.
Our study also highlights the contrast between early relapse and late relapse biology. We observed relatively similar TME features between diagnostic and relapse biopsies in early relapse cases, whereas differences were more pronounced in late relapse cases. This finding raises the hypothesis that the establishment of malignant cellular ecosystems, including HRS cell–driven shaping of the TME, is persistent over time after first-line treatment in early relapse CHL, and by contrast, the biology of late relapses is indicative of more divergent disease and more dynamic changes in the TME. The latter scenario might also be supported by reports of clonally independent malignant HRS cells between diagnostic and relapse time points.32
The multiple cellular components associated with treatment outcomes strongly suggest that the mechanism underlying relapsed disease might not be uniform (Fig 6). Nevertheless, delineating specific and targetable biology underlying r/r disease, such as the PD1-PDL1 and CXCR5-CXCL13 axes, might lead to a more effective delivery of precision oncology. In addition, coexpression patterns of multiple coinhibitory receptors including TIM3 and LAG3 in PD1+ CD4+ T-cell subsets might provide a rationale to consider multiple targeted treatments as investigated in clinical trials (eg, ClinicalTrials.gov identifiers: NCT05216835 and NCT03598608).33 The development of biology-driven biomarkers in CHL might also help pave the way for rational treatment selection, an approach that is currently emerging in diffuse large B-cell lymphoma on the basis of gene expression and mutational profiling.34-36
FIG 6.
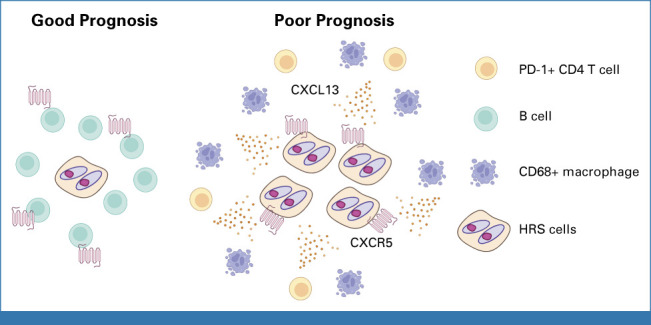
Graphical summary of findings in r/r CHL. r/r CHL with poor prognosis is characterized by CXCR5 positivity on HRS cells. CXCL13+ macrophages surround CXCR5+ HRS cells, and PD1+ CD4+ T cells were also present in the tumor microenvironment. By contrast, CXCR5+ B cells were enriched in r/r CHL with good prognosis. CHL, classic Hodgkin lymphoma; HRS, Hodgkin and Reed Sternberg; r/r, relapsed or refractory.
While RHL4S shows promise in guiding treatment selection and might complement response-adapted treatment algorithms, further studies are necessary to validate the clinical utility of RHL4S in patients with r/r CHL who are treated with novel therapies such as BV and/or PD-1 blockade in both first-line and second-line treatments.9,37-43 The underlying biology of r/r disease after these treatments may differ. Considering the mechanism of action, the PD-1 blockade might overcome treatment resistance linked to certain parameters such as the abundance of and spatial patterns involving CD4+PD1+ T cells. Using samples from completed or ongoing clinical trial cohorts could significantly contribute to addressing these unresolved questions. Indeed, our immunofluorescence-based RHL4S assay can be readily used for FFPE tissues collected according to trial correlative protocols and inform on critical TME components in relapse biopsies of patients treated with BV and/or checkpoint inhibitors. Moving forward, we anticipate that the value of spatially resolved biomarkers will be tested in additional lymphoma subtypes44 and other cancers45 with a biologically important TME component.
Shanee Chung
Honoraria: Astellas Pharma, Takeda, Novartis, Paladin, Pfizer
Consulting or Advisory Role: Takeda
Kayleigh Morris
Stock and Other Ownership Interests: Abbott Laboratories (I), Zoetis (I), Thermo Fisher Scientific (I), Johnson & Johnson/Janssen (I), Vertex (I), Danaher (I)
Lauren C. Chong
Employment: AbCellera
Stock and Other Ownership Interests: AbCellera
Monirath Hav
Employment: Macrogenics
Travel, Accommodations, Expenses: Macrogenics, Ionpath Inc
Anthony R. Colombo
Patents, Royalties, Other Intellectual Property: There was a patent filed with the Cedars-Sinai Cancer center that was derived from research findings from 2020/2021. The patent was a methodological approach to spatially identifying prognostic signatures in DLBCL that was derived from a published project
Maryse Power
Honoraria: Novartis Canada Pharmaceuticals Inc
Alina S. Gerrie
Honoraria: Janssen, AbbVie, AstraZeneca, BeiGene
Consulting or Advisory Role: Janssen, AbbVie, AstraZeneca, BeiGene
Research Funding: AbbVie (Inst), Roche Canada (Inst), Janssen (Inst), AstraZeneca (Inst)
Travel, Accommodations, Expenses: Janssen
Andrew P. Weng
Honoraria: BMS (I), Janssen (I), AbbVie (I), Seagen (I)
Consulting or Advisory Role: BMS (I), Seagen (I)
Research Funding: Roche (Inst), BMS (I)
Other Relationship: Beigene (I), Regeneron (I), AstraZeneca (I)
Aly Karsan
Honoraria: Jazz Pharmaceuticals
Research Funding: AstraZeneca, Pfizer
David W. Scott
Consulting or Advisory Role: Janssen, AbbVie, AstraZeneca, Incyte
Research Funding: Janssen, Roche/Genentech
Patents, Royalties, Other Intellectual Property: Named inventor on a pending patent describing gene expression profiling in prognostication in classical Hodgkin lymphoma, Named inventor on a pending patent describing using gene expression profiling to identify molecular subtypes of GCB-DLBCL, Named inventor on a patent on the use of gene expression profiling to assign cell-of-origin in diffuse large B-cell lymphoma, Named inventor on a pending patent on the use of gene expression profiling to determine the proliferation signature in mantle cell lymphoma
Kerry J. Savage
Honoraria: Bristol Myers Squibb, Janssen Oncology, AbbVie, Seagen
Consulting or Advisory Role: Bristol Myers Squibb, Seagen
Research Funding: Roche (Inst), Bristol Myers Squibb (Inst)
Uncompensated Relationships: Beigene, Regeneron, AstraZeneca
Brad H. Nelson
Leadership: Innovakine Therapeutics, Overture Therapeutics
Research Funding: Innovakine Therapeutics, Overture Therapeutics
Patents, Royalties, Other Intellectual Property: Microfluidic Devices And Methods for Use Thereof in Multicellular Assays of Secretion (provisional patent). United States. 61/806,329. Patent Status: Pending Funding Sources: University of British Columbia
Akil Merchant
Consulting or Advisory Role: Oncovalent Therapeutics
Christian Steidl
Consulting or Advisory Role: Bayer
Research Funding: Trillium Therapeutics (Inst), Epizyme (Inst)
Patents, Royalties, Other Intellectual Property: Holder of a patent “Method for Determining Lymphoma Type” using the Nanostring platform
Expert Testimony: Bayer
Travel, Accommodations, Expenses: Eisai
No other potential conflicts of interest were reported.
PRIOR PRESENTATION
Presented in part at the 64th ASH annual meeting; New Orleans, LA, December 10, 2022.
SUPPORT
Supported by Program Project Grant funding from the Terry Fox Research Institute (Grant No. 1061 and 1108); Large Scale Applied Research Project funding from Genome Canada (Grant No. 13124), Genome BC (Grant No. 271LYM), and CIHR (Grant No. GP1-155873); the Canadian Cancer Society Research Institute (Grant No. 705288); a Foundation grant from CIHR (Grant No. 148393); the BC Cancer Foundation; and the Paul G. Allen Frontiers Group (Distinguished Investigator award to C.S. Grant No. 12829). T.A. was supported by fellowships from the Japanese Society for The Promotion of Science, the Uehara Memorial Foundation, CIHR, and the Lymphoma Research Foundation. T.A. received research funding support from The Kanae Foundation for the Promotion of Medical Science. T.A. received research support as a Lymphoma Research Foundation Lymphoma Scientific Research Mentoring Program Scholar. S.R. was supported by the Gascoyne Lymphoma Research Fellowship (BC Cancer Foundation).
T.A., A.J., and A.X. contributed equally to this work. A.M. and C.S. contributed equally to this work as cosenior authors.
DATA SHARING STATEMENT
Single-cell RNA-seq counts (generated using Cell Ranger v2.1.0) and a merged SingleCellExperiment R object are available in the European Genome-phenome Archive (EGA; EGAD00001010892) via controlled access. IMC data are available at zenodo.org/deposit/7963681.46 Scripts used for data analysis are available upon request. Our in-house R packages, csmpv (https://github.com/ajiangsfu/csmpv), and RHL4S (https://github.com/ajiangsfu/RHL4S) are available on GitHub.
AUTHOR CONTRIBUTIONS
Conception and design: Tomohiro Aoki, Aixiang Jiang, Alexander Xu, Yifan Yin, Maryse Power, Akil Merchant, Christian Steidl
Financial support: Akil Merchant, Christian Steidl
Administrative support: Monirath Hav, Akil Merchant, Christian Steidl
Provision of study materials or patients: Tomohiro Aoki, Monirath Hav, Merrill Boyle, Maryse Power, Alina S. Gerrie, Christian Steidl
Collection and assembly of data: Tomohiro Aoki, Aixiang Jiang, Alexander Xu, Yifan Yin, Alicia Gamboa, Katy Milne, Katsuyoshi Takata, Shanee Chung, Shinya Rai, Mary Warren, Talia Goodyear, Kayleigh Morris, Monirath Hav, Adele Telenius, Merrill Boyle, Andrew P. Weng, Pedro Farinha, David W. Scott, Kerry J. Savage, Brad H. Nelson, Akil Merchant, Christian Steidl
Data analysis and interpretation: Tomohiro Aoki, Aixiang Jiang, Alexander Xu, Yifan Yin, Alicia Gamboa, Katy Milne, Katsuyoshi Takata, Tomoko Miyata-Takata, Shaocheng Wu, Celia Strong, Talia Goodyear, Lauren C. Chong, Monirath Hav, Anthony R. Colombo, Susana Ben-Neriah, Maryse Power, Alina S. Gerrie, Aly Karsan, Andrew Roth, Pedro Farinha, Kerry J. Savage, Brad H. Nelson, Akil Merchant, Christian Steidl
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
Spatially Resolved Tumor Microenvironment Predicts Treatment Outcome in Relapsed/Refractory Hodgkin Lymphoma
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/jco/authors/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
Shanee Chung
Honoraria: Astellas Pharma, Takeda, Novartis, Paladin, Pfizer
Consulting or Advisory Role: Takeda
Kayleigh Morris
Stock and Other Ownership Interests: Abbott Laboratories (I), Zoetis (I), Thermo Fisher Scientific (I), Johnson & Johnson/Janssen (I), Vertex (I), Danaher (I)
Lauren C. Chong
Employment: AbCellera
Stock and Other Ownership Interests: AbCellera
Monirath Hav
Employment: Macrogenics
Travel, Accommodations, Expenses: Macrogenics, Ionpath Inc
Anthony R. Colombo
Patents, Royalties, Other Intellectual Property: There was a patent filed with the Cedars-Sinai Cancer center that was derived from research findings from 2020/2021. The patent was a methodological approach to spatially identifying prognostic signatures in DLBCL that was derived from a published project
Maryse Power
Honoraria: Novartis Canada Pharmaceuticals Inc
Alina S. Gerrie
Honoraria: Janssen, AbbVie, AstraZeneca, BeiGene
Consulting or Advisory Role: Janssen, AbbVie, AstraZeneca, BeiGene
Research Funding: AbbVie (Inst), Roche Canada (Inst), Janssen (Inst), AstraZeneca (Inst)
Travel, Accommodations, Expenses: Janssen
Andrew P. Weng
Honoraria: BMS (I), Janssen (I), AbbVie (I), Seagen (I)
Consulting or Advisory Role: BMS (I), Seagen (I)
Research Funding: Roche (Inst), BMS (I)
Other Relationship: Beigene (I), Regeneron (I), AstraZeneca (I)
Aly Karsan
Honoraria: Jazz Pharmaceuticals
Research Funding: AstraZeneca, Pfizer
David W. Scott
Consulting or Advisory Role: Janssen, AbbVie, AstraZeneca, Incyte
Research Funding: Janssen, Roche/Genentech
Patents, Royalties, Other Intellectual Property: Named inventor on a pending patent describing gene expression profiling in prognostication in classical Hodgkin lymphoma, Named inventor on a pending patent describing using gene expression profiling to identify molecular subtypes of GCB-DLBCL, Named inventor on a patent on the use of gene expression profiling to assign cell-of-origin in diffuse large B-cell lymphoma, Named inventor on a pending patent on the use of gene expression profiling to determine the proliferation signature in mantle cell lymphoma
Kerry J. Savage
Honoraria: Bristol Myers Squibb, Janssen Oncology, AbbVie, Seagen
Consulting or Advisory Role: Bristol Myers Squibb, Seagen
Research Funding: Roche (Inst), Bristol Myers Squibb (Inst)
Uncompensated Relationships: Beigene, Regeneron, AstraZeneca
Brad H. Nelson
Leadership: Innovakine Therapeutics, Overture Therapeutics
Research Funding: Innovakine Therapeutics, Overture Therapeutics
Patents, Royalties, Other Intellectual Property: Microfluidic Devices And Methods for Use Thereof in Multicellular Assays of Secretion (provisional patent). United States. 61/806,329. Patent Status: Pending Funding Sources: University of British Columbia
Akil Merchant
Consulting or Advisory Role: Oncovalent Therapeutics
Christian Steidl
Consulting or Advisory Role: Bayer
Research Funding: Trillium Therapeutics (Inst), Epizyme (Inst)
Patents, Royalties, Other Intellectual Property: Holder of a patent “Method for Determining Lymphoma Type” using the Nanostring platform
Expert Testimony: Bayer
Travel, Accommodations, Expenses: Eisai
No other potential conflicts of interest were reported.
REFERENCES
- 1.Scott DW, Gascoyne RD: The tumour microenvironment in B cell lymphomas. Nat Rev Cancer 14:517-534, 2014 [DOI] [PubMed] [Google Scholar]
- 2.Danenberg E, Bardwell H, Zanotelli VRT, et al. : Breast tumor microenvironment structures are associated with genomic features and clinical outcome. Nat Genet 54:660-669, 2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jain RK: Normalizing tumor microenvironment to treat cancer: Bench to bedside to biomarkers. J Clin Oncol 31:2205-2218, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Castellino SM, Geiger AM, Mertens AC, et al. : Morbidity and mortality in long-term survivors of Hodgkin lymphoma: A report from the Childhood Cancer Survivor Study. Blood 117:1806-1816, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kelly KM, Hodgson D, Appel B, et al. : Children's Oncology Group's 2013 blueprint for research: Hodgkin lymphoma. Pediatr Blood Cancer 60:972-978, 2013 [DOI] [PubMed] [Google Scholar]
- 6.Aoki T, Chong LC, Takata K, et al. : Single-cell profiling reveals the importance of CXCL13/CXCR5 axis biology in lymphocyte-rich classic Hodgkin lymphoma. Proc Natl Acad Sci USA 118:e2105822118, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Steidl C, Lee T, Shah SP, et al. : Tumor-associated macrophages and survival in classic Hodgkin's lymphoma. N Engl J Med 362:875-885, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gerrie AS, Power MM, Shepherd JD, et al. : Chemoresistance can be overcome with high-dose chemotherapy and autologous stem-cell transplantation for relapsed and refractory Hodgkin lymphoma. Ann Oncol 25:2218-2223, 2014 [DOI] [PubMed] [Google Scholar]
- 9.Spinner MA, Sica RA, Tamaresis JS, et al. : Improved outcomes for relapsed/refractory Hodgkin lymphoma after autologous transplantation in the era of novel agents. Blood 141:2727-2737, 2023 [DOI] [PubMed] [Google Scholar]
- 10.Chan FC, Mottok A, Gerrie AS, et al. : Prognostic model to predict post-autologous stem-cell transplantation outcomes in classical Hodgkin lymphoma. J Clin Oncol 35:3722-3733, 2017 [DOI] [PubMed] [Google Scholar]
- 11.Scott DW, Chan FC, Hong F, et al. : Gene expression-based model using formalin-fixed paraffin-embedded biopsies predicts overall survival in advanced-stage classical Hodgkin lymphoma. J Clin Oncol 31:692-700, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aoki T, Chong LC, Takata K, et al. : Single-cell transcriptome analysis reveals disease-defining T-cell subsets in the tumor microenvironment of classic Hodgkin lymphoma. Cancer Discov 10:406-421, 2020 [DOI] [PubMed] [Google Scholar]
- 13.Reichel J, Chadburn A, Rubinstein PG, et al. : Flow sorting and exome sequencing reveal the oncogenome of primary Hodgkin and Reed-Sternberg cells. Blood 125:1061-1072, 2015 [DOI] [PubMed] [Google Scholar]
- 14.Wienand K, Chapuy B, Stewart C, et al. : Genomic analyses of flow-sorted Hodgkin Reed-Sternberg cells reveal complementary mechanisms of immune evasion. Blood Adv 3:4065-4080, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deng Q, Han G, Puebla-Osorio N, et al. : Characteristics of anti-CD19 CAR T cell infusion products associated with efficacy and toxicity in patients with large B cell lymphomas. Nat Med 26:1878-1887, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Steen CB, Luca BA, Esfahani MS, et al. : The landscape of tumor cell states and ecosystems in diffuse large B cell lymphoma. Cancer Cell 39:1422-1437.e10, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scott DW, Steidl C: The Hlassical hodgkin lymphoma tumor microenvironment: Macrophages and gene expression-based modeling. Hematology Am Soc Hematol Educ Program 2014:144-150, 2014 [DOI] [PubMed] [Google Scholar]
- 18.Roemer MG, Advani RH, Ligon AH, et al. : PD-L1 and PD-L2 genetic alterations define classical Hodgkin lymphoma and predict outcome. J Clin Oncol 34:2690-2697, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ruella M, Klichinsky M, Kenderian SS, et al. : Overcoming the immunosuppressive tumor microenvironment of Hodgkin lymphoma using chimeric antigen receptor T cells. Cancer Discov 7:1154-1167, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Steidl C, Diepstra A, Lee T, et al. : Gene expression profiling of microdissected Hodgkin Reed-Sternberg cells correlates with treatment outcome in classical Hodgkin lymphoma. Blood 120:3530-3540, 2012 [DOI] [PubMed] [Google Scholar]
- 21.Jin S, Guerrero-Juarez CF, Zhang L, et al. : Inference and analysis of cell-cell communication using CellChat. Nat Commun 12:1088, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Y, Wang R, Zhang S, et al. : iTALK: an R Package to characterize and illustrate intercellular communication. bioRxiv 10.1101/507871 [DOI] [Google Scholar]
- 23.Hanna MG, Ardon O, Reuter VE, et al. : Integrating digital pathology into clinical practice. Mod Pathol 35:152-164, 2022 [DOI] [PubMed] [Google Scholar]
- 24.Zneimer SM, Hongo D: Preparing for Clinical Laboratory Improvement Amendments (CLIA) and College of American Pathologists (CAP) inspections. Curr Protoc 1:e324, 2021 [DOI] [PubMed] [Google Scholar]
- 25.Baxi V, Edwards R, Montalto M, et al. : Digital pathology and artificial intelligence in translational medicine and clinical practice. Mod Pathol 35:23-32, 2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Casulo C, Arcila M, Bohn OL, et al. : Tumor associated macrophages in relapsed and refractory Hodgkin lymphoma. Leuk Res 37:1178-1183, 2013 [DOI] [PubMed] [Google Scholar]
- 27.Kelley TW, Pohlman B, Elson P, et al. : The ratio of FOXP3+ regulatory T cells to granzyme B+ cytotoxic T/NK cells predicts prognosis in classical Hodgkin lymphoma and is independent of bcl-2 and MAL expression. Am J Clin Pathol 128:958-965, 2007 [DOI] [PubMed] [Google Scholar]
- 28.Tzankov A, Meier C, Hirschmann P, et al. : Correlation of high numbers of intratumoral FOXP3+ regulatory T cells with improved survival in germinal center-like diffuse large B-cell lymphoma, follicular lymphoma and classical Hodgkin's lymphoma. Haematologica 93:193-200, 2008 [DOI] [PubMed] [Google Scholar]
- 29.Bunse M, Pfeilschifter J, Bluhm J, et al. : CXCR5 CAR-T cells simultaneously target B cell non-Hodgkin's lymphoma and tumor-supportive follicular T helper cells. Nat Commun 12:240, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klimatcheva E, Pandina T, Reilly C, et al. : CXCL13 antibody for the treatment of autoimmune disorders. BMC Immunol 16:6, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zanetti C, Kumar R, Ender J, et al. : The age of the bone marrow microenvironment influences B-cell acute lymphoblastic leukemia progression via CXCR5-CXCL13. Blood 138:1870-1884, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Obermann EC, Mueller N, Rufle A, et al. : Clonal relationship of classical hodgkin lymphoma and its recurrences. Clin Cancer Res 17:5268-5274, 2011 [DOI] [PubMed] [Google Scholar]
- 33.Timmerman J, Lavie D, Johnson NA, et al. : Updated results from an open-label phase 1/2 study of favezelimab (anti-LAG-3) plus pembrolizumab in relapsed or refractory classical Hodgkin lymphoma after anti-PD-1 treatment. Blood 140:768-770, 2022. (suppl 1) [Google Scholar]
- 34.Wilson WH, Wright GW, Huang DW, et al. : Effect of ibrutinib with R-CHOP chemotherapy in genetic subtypes of DLBCL. Cancer Cell 39:1643-1653.e3, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chapuy B, Stewart C, Dunford AJ, et al. : Molecular subtypes of diffuse large B cell lymphoma are associated with distinct pathogenic mechanisms and outcomes. Nat Med 24:679-690, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schmitz R, Wright GW, Huang DW, et al. : Genetics and pathogenesis of diffuse large B-cell lymphoma. N Engl J Med 378:1396-1407, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ramchandren R, Domingo-Domenech E, Rueda A, et al. : Nivolumab for newly diagnosed advanced-stage classic Hodgkin lymphoma: Safety and efficacy in the phase II CheckMate 205 study. J Clin Oncol 37:1997-2007, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Advani RH, Moskowitz AJ, Bartlett NL, et al. : Brentuximab vedotin in combination with nivolumab in relapsed or refractory Hodgkin lymphoma: 3-Year study results. Blood 138:427-438, 2021 [DOI] [PubMed] [Google Scholar]
- 39.Ansell SM, Lesokhin AM, Borrello I, et al. : PD-1 blockade with nivolumab in relapsed or refractory Hodgkin's lymphoma. N Engl J Med 372:311-319, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brockelmann PJ, Buhnen I, Meissner J, et al. : Nivolumab and doxorubicin, vinblastine, and dacarbazine in early-stage unfavorable Hodgkin lymphoma: Final analysis of the randomized German Hodgkin Study Group phase II NIVAHL trial. J Clin Oncol 41:1193-1199, 2023 [DOI] [PubMed] [Google Scholar]
- 41.Mei MG, Lee HJ, Palmer JM, et al. : Response-adapted anti-PD-1-based salvage therapy for Hodgkin lymphoma with nivolumab alone or in combination with ICE. Blood 139:3605-3616, 2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moskowitz AJ, Shah G, Schoder H, et al. : Phase II trial of pembrolizumab plus gemcitabine, vinorelbine, and liposomal doxorubicin as second-line therapy for relapsed or refractory classical Hodgkin lymphoma. J Clin Oncol 39:3109-3117, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moskowitz CH, Nademanee A, Masszi T, et al. : Brentuximab vedotin as consolidation therapy after autologous stem-cell transplantation in patients with Hodgkin's lymphoma at risk of relapse or progression (AETHERA): A randomised, double-blind, placebo-controlled, phase 3 trial. Lancet 385:1853-1862, 2015 [DOI] [PubMed] [Google Scholar]
- 44.Phillips D, Matusiak M, Gutierrez BR, et al. : Immune cell topography predicts response to PD-1 blockade in cutaneous T cell lymphoma. Nat Commun 12:6726, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Galon J, Mlecnik B, Bindea G, et al. : Towards the introduction of the “Immunoscore” in the classification of malignant tumours. J Pathol 232:199-209, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xu AM, Jiang A, Aoki T, et al. : Single cell spatial analysis and biomarker discovery in Hodgkin lymphoma. bioRxiv 10.1101/2023.05.24.542195 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Single-cell RNA-seq counts (generated using Cell Ranger v2.1.0) and a merged SingleCellExperiment R object are available in the European Genome-phenome Archive (EGA; EGAD00001010892) via controlled access. IMC data are available at zenodo.org/deposit/7963681.46 Scripts used for data analysis are available upon request. Our in-house R packages, csmpv (https://github.com/ajiangsfu/csmpv), and RHL4S (https://github.com/ajiangsfu/RHL4S) are available on GitHub.