Abstract
Hepatitis delta virus (HDV) encodes two isoforms of its principal gene product, hepatitis delta antigen (HDAg). These two forms play distinctive and complementary roles in viral replication. Here we report that the large (LHDAg), but not the small (SHDAg), isoform of HDAg has the capacity to activate the expression of cotransfected genes driven by a variety of promoters, including the pre-S, S, and C promoters of hepatitis B virus. Mutational analysis of the C-terminal 19 amino acids unique to LHDAg shows that changing prolines to alanines in the two PXXP motifs in this region specifically ablates the activation function without abolishing another activity of LHDAg, namely, its ability to inhibit HDV RNA synthesis. However, C-terminal truncations that also disrupt these PXXP motifs only slightly diminished the activation function, indicating that the proline mutations were not acting by inactivating potential SH3 interactions that could be mediated by these motifs. Mutation of the isoprenylated cysteine to serine decreases but does not abolish the activation activity, and overexpression of SHDAg does not interfere with the transactivation function of LHDAg. Although the mechanism and biological significance of this activity of LHDAg remain unknown, the presence of this activity serves as yet another marker that functionally distinguishes this protein from the closely related isoform SHDAg.
Hepatitis delta virus (HDV) is an RNA virus that requires coinfection with hepatitis B virus (HBV) to complete its life cycle. The helper function supplied by HBV is limited to the provision of envelope proteins (hepatitis B surface antigens) for the completion of HDV assembly (28, 29, 31). HDV RNA replication is independent of its HBV helper (19). In fact, the presence of HDV suppresses HBV replication in vivo (30, 39). Nonetheless, clinical studies have shown that HDV infection can be associated with more severe hepatitis than HBV alone and is often implicated in cases of fulminant hepatitis (4, 32).
The genome of HDV is a circular, single-stranded RNA of about 1,700 nucleotides (nt), of which approximately 70% are self-complementary (for a review, see references 20 and 21). This self-complemetarity allows the genome to form an unbranched rod-like structure. A unique functional protein, hepatitis delta antigen (HDAg), is encoded by the genome (3, 38), and two isoforms of this protein are produced during infection. The canonical small form of HDAg (SHDAg) is 195 amino acids (aa) long; it harbors an N-terminal coiled-coil domain responsible for oligomerization (37), a central domain responsible for binding to the RNA genome (7, 23), a nuclear localization signal (2, 7), and a C-terminal glycine- and proline-rich region with an uncertain function. This form of HDAg is essential for viral RNA replication, although it is not itself a polymerase. Host RNA polymerase II is thought to supply the polymerase function for replication (15, 26). During viral replication, an RNA editing event occurs at the UAG termination codon of SHDAg, allowing readthrough of another 19 aa (Fig. 1) to generate the large isoform of the protein, LHDAg (25). Since LHDAg contains all of the domains of SHDAg, it too can form multimers with itself and with the SHDAg isoform, bind HDV RNA (as a homo- or heteromultimer), and be localized to the nucleus.
FIG. 1.
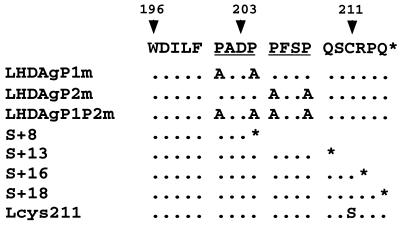
Sequence of the 19 aa unique to the C terminus of LHDAg. The PXXP motifs are underlined. Below are shown the amino acid changes present in the mutants employed in this study. The positions of the termination codons introduced into the truncation mutants are indicated by asterisks.
Despite these similarities, the two HDAgs have very distinct functions (22) and play complementary roles in HDV replication, which takes place largely in the nuclei of infected cells (34). While SHDAg activates HDV RNA replication, LHDAg is a trans-dominant inhibitor of this process (8). By contrast, LHDAg, but not SHDAg, is capable of interacting with the HBV envelope proteins to mediate envelopment of the HDV ribonucleoprotein in viral assembly (6). This interaction has been shown to require farnesylation of a cysteine residue found in the C-terminal 19 aa unique to LHDAg (27, 16). Furthermore, it has been shown recently that only LHDAg is phosphorylated in cells (1).
In this report, we describe yet another activity of LHDAg that further differentiates it from the related isoform SHDAg, i.e., the ability to activate gene expression in trans.
MATERIALS AND METHODS
Plasmids.
All wild-type (WT) and mutant HDAg coding regions were cloned into the KpnI and XbaI sites of pcDNA3 (Invitrogen). The complete LHDAg open reading frame was first cloned into pBluescript II SK(−) (Stratagene) to generate pHDL. All LHDAg mutants were prepared by PCR with pHDL as the template.
The plasmids expressing LHDAgP1m, LHDAgP2m, and LHDAgP1P2m were constructed by two rounds of PCR. In the first round, two independent PCRs were performed with two pairs of primers. The primers were designed such that the 3′-end primer of one pair has the complementary sequence of the 5′-end primer of another pair, and desired mutations were introduced into these two primers. For LHDAgP1m, the 5′-end primer HD1 (5′-GACTTCTAGAGGATCCCCCGCTTTATTCACTGG-3′ [nt 944 to 960; the sequence and numbering are according to reference 19]) was paired with the 3′-end primer 5′-GATATACTCTTCGCAGCCGATGCGCCCTTTTCTC-3′, corresponding to nt 1010 to 977 (underlined nucleotides represent mutations), and the 5′-end primer 5′-GAGAAAAGGGCGCATCGGCTGCGAAGAGTATATC-3′ (nt 977 to 1010) was paired with the 3′-end primer HD2 (5′-TTCGTCCCCAATCTGCAGGGAGTC-3′ [nt 1100 to 1077]). For LHDAgP2m, primer HD1 was paired with 3′-end primer 5′-CAGCCGATCCGGCCTTTTCTGCCCAGAGTTGTC-3′ (nt 997 to 965) and 5′-end primer 5′-GACAACTCTGGGCAGAAAAGGCCGGATCGGCTG-3′ (nt 965 to 997) was paired with primer HD2. For LHDAgP1P2m, primer HD1 was paired with the 3′-end primer 5′-GATATACTCTTCGCAGCCGATGCGGCCTTTTC-3′ (nt 1010 to 979) and primer 5′-GAAAAGGCCGCATCGGCTGCGAAGAGTATATC-3′ (nt 979 to 1010) was paired with primer HD2. pHDL was used as the template in all PCRs save in that for LHDAgP1P2m, in which LHDAgP2m was used as the template. After gel purification, the products from two PCRs were pooled. The presence of the complementary sequence in the primers allows annealing of part of the sequence between the PCR products. The resulting molecules were amplified in the second-round PCR by using primers HD1 and HD2. The final PCR products were then gel purified, digested with PstI and XbaI, and cloned into pHDL cut with PstI and XbaI. The resulting plasmids were digested with KpnI and XbaI and cloned into pcDNA3.
The deletion mutants were prepared by a PCR in which the 5′- end primers were 5′-GACTCTAGACCCGCTTTATTCAATCGGCTGGGAAG-3′ (nt 990 to 1002) for the plasmid expressing S+8, 5′-GACTCTAGACCCGCTTTATTCAGGGAGAAAAGGGC-3′ (nt 975 to 987) for the construct S+13, 5′-GACTCTAGACCCGCTTTATTCAACAACTCTGGGGA-3′ (nt 966 to 978) for S+16, and 5′-GACTCTAGACCCGCTTTATTCAGGGTCGACAACTC-3′ (nt 959 to 972) for S+18, and the 3′-end primer was HD2. Lcys211 was also generated by a PCR in which the 5′-end primer 5′-GACTCTAGAGGATCCTATTCACTGGGGTCGAGAACTCTGGGGAG-3′ (nt 951 to 979) was paired with primer HD2. pHDL was used as the template. The PCR products were gel purified, digested with PstI and XbaI, and cloned into pHDL cut with PstI and XbaI. The inserts were prepared by digestion with KpnI and XbaI and cloned into pcDNA3.
N-myc140CAT was constructed by a PCR in which primer N-myc5′ (5′-CAGTCTCGAGTTCCCAGCAGTCGCCTG-3′) was paired with primer N-myc3′ (5′-TCGCCTGCCCGCC-3′) (13). N-myc84CAT was also constructed by a PCR in which the primer 5′-CCCTAGGGAAAGGAAGCAC-3′ was paired with N-myc3′. N-myc2-CAT was used as the template (35). N-myc140Sp1(−)CAT was constructed by two PCR rounds. In the first round, N-myc5′ paired with primer 5′-GTGTGCGGAGAGGGGGGACAATAGCCA-3′ and primer 5′-TGGCTATTGTCCCCCCTCTCCGCACAC-3′ paired with N-myc3′ were used in two independent PCRs in which N-myc2-CAT was the template. The PCR products were then pooled. The second PCR round was performed with N-myc5′ and N-myc3′ as the primer. The final PCR products were cloned into the XhoI and XbaI sites of pCAT, which was described previously (36). 3Sp1CAT was generated by annealing the sense oligonucleotide CAG CTC GAG ATT GCC CCC GCC CTC ATT GCC CCC GCC CTC ATT GCC CCC GCC CTC TAG ATA C to the antisense oligonucleotide GTA TCT AGA GGG CGG GGG CAA TGA GGG CGG GGG CAA TGA GGG CGG GGG CAA TCT CGA GCT G, which contains three Sp1 binding sites. The double-stranded oligonucleotide was cut with XhoI and XbaI and cloned into pCAT. 3Sp1mCAT was constructed in the same way with the sense oligonucleotide CAG CTC GAG ATT GTC CCC CCT CTC ATT GTC CCC CCT CTC ATT GTC CCC CCT CTC TAG ATA C and the antisense oligonucleotide GTA TCT AGA GAG GGG GGA CAA TGA GAG GGG GGA CAA TGA GAG GGG GGA CAA TCT CGA GCT G, which contain three mutated Sp1 binding sites.
The nucleotide sequence of the fragments from PCR was confirmed by conventional dideoxy sequencing.
Plasmid pDL452 was a gift from D. Lazinski (Tufts University) (unpublished data). The promoter constructs (hsp70 TATA series and simian virus 40 [SV40] early series) were the gift of D. Lukac and J. Alwine (University of Pennsylvania) and R. Kingston (Harvard Medical School) and have been described elsewhere (24, 33). pPreS1CAT, pSCAT, and pUCAT9 were the gift of T. S. B. Yen (University of California, San Francisco). pPreS1CAT and pUCAT9 have been described previously (17, 40). pSCAT was obtained by cloning the HBV fragment spanning nt 3125 to 33 in pCAT. Luciferase reporter vector pGL2-promoter (SV40 promoter) was obtained from Promega.
Cell transfection and CAT and luciferase assays.
HepG2, 293, and CCL13 (Chang liver) cells were cultured in Dulbecco modified Eagle medium (DMEM) supplemented with 10% bovine serum and antibiotics. Cells were transfected by calcium phosphate coprecipitation using 1 μg of reporter plasmid and 4 μg of transactivator plasmids. (For the Western immunoblot assay, 10 μg of DNA was transfected.) At 12 h after transfection, cells were washed with phosphate-buffered saline and cultured for another 24 h, at which time cell extracts were assayed for chloramphenicol acetyltransferase (CAT) activity or HDAg (by immunoblotting with anti-HDAg). For HDV RNA replication analyses, cells were harvested 4 to 6 days after transfection. The CAT assay method used is described in detail elsewhere (36). For the luciferase assay, cells were harvested 48 h after transfection and lysed in lysis buffer containing 25 mM Tris-HCl (pH 8.0), 8 mM MgCl2, 1% Triton X-100, 1% bovine serum albumin, 15% glycerol, and 1 mM dithiothreitol. Cellular extract was measured for luciferase activity in the lysis buffer in the presence of 0.4 mM ATP and 1 mM luciferin.
Northern blot analysis.
Total RNA was isolated from cells with RNAzol B (TEL-TEST), electrophoresed through a standard 1% agarose–2.2 M formaldehyde gel, and transferred to a positively charged nylon membrane (Boehringer Mannheim) in 10 × SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate). Membranes were hybridized at 72°C with an HDV antigenome-specific riboprobe uniformly labeled with digoxigenin (5). After hybridization, blots were washed at 72°C with 0.1% sodium dodecyl sulfate and 2×, 0.5×, and 0.1× SSC, successively. The probe was detected as previously described (12).
Western immunoblot analysis.
Transfected cells were harvested and lysed in a solution containing 50 mM Tris-HCl (pH 7.5), 400 mM NaCl, 0.2% Nonidet P-40, and protease inhibitors. Lysates were fractionated by sodium dodecyl sulfate-polyacrylamide (12.5%) gel electrophoresis and transferred to Immobilon-P membranes (Millipore). The membranes were incubated with an anti-HDAg polyclonal antibody (1:10,000 dilution) for 1 h at room temperature. Immune complexes were detected with horseradish peroxidase-conjugated goat antibodies to rabbit immunoglobulin G (1:7,500 dilution) (Gibco BRL) and enhanced chemiluminescence (Amersham).
RESULTS
Evidence for transactivation activity of LHDAg.
The possibility that LHDAg harbors an activation activity was initially suggested by experiments aimed at identifying cellular factors that might interact with the protein in the yeast two-hybrid assay. In those experiments, a fusion protein containing the DNA binding domain of Gal4p fused to intact LHDAg was expressed in yeast cells. Surprisingly, this Gal4p-LHDAg fusion protein was able to strongly activate transcription of a lacZ reporter gene under the control of a promoter containing multiple Gal4p binding sites, even in the absence of any other interacting activator proteins. No comparable activation was seen when a similar fusion protein containing SHDAg was expressed in the same yeast strain (5). However, since many proteins that do not activate transcription under normal conditions can score in this assay, we decided to conduct further, more rigorous tests of gene activation by LHDAg.
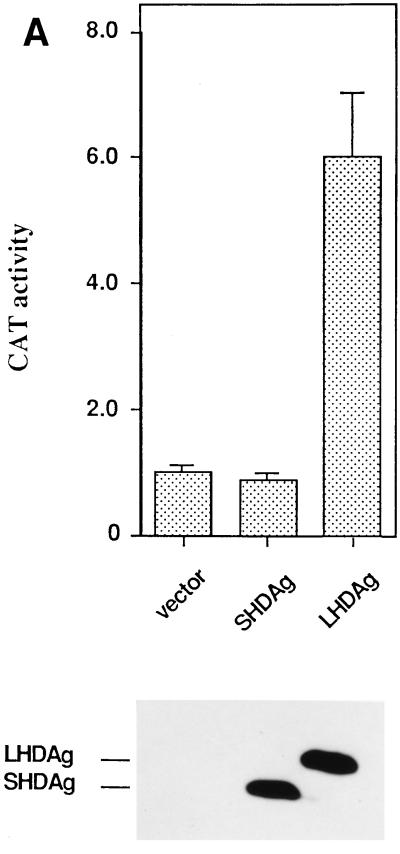
To examine the ability of LHDAg to activate transcription in mamalian cells, plasmids expressing LHDAg or SHDAg were cotransfected into HepG2 cells along with a CAT reporter gene driven by the cellular N-myc2 promoter. Forty-eight hours later, the cells were assayed for CAT activity; the results are expressed as fold CAT activity above that induced in the absence of LHDAg (each value represents the mean result of at least three independent experiments). A plasmid expressing human growth hormone was used as an internal control for transfection efficiency. When the CAT gene was under control of the N-myc2 promoter, CAT activity was increased more than sixfold in the presence of LHDAg (Fig. 2A). In contrast, no activation of the N-myc2 promoter by SHDAg was detected (Fig. 2A). Since immunoblotting with anti-HDAg confirmed the expression of both LHDAg and SHDAg in these experiments (Fig. 2A), the absence of transactivation activity of SHDAg is not due to the absence of the protein. By contrast, a CAT gene under the control of a minimal promoter (TATA box) from adenovirus E1B was not activated by either LHDAg or SHADg (data not shown), suggesting that activation by LHDAg likely operates through factors bound upstream of the TATA box (see also Fig. 2D).
FIG. 2.
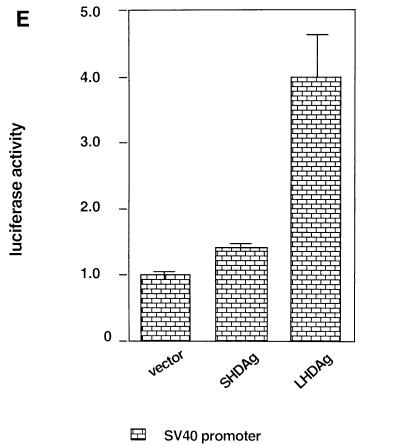
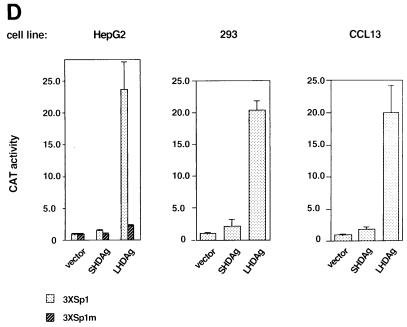
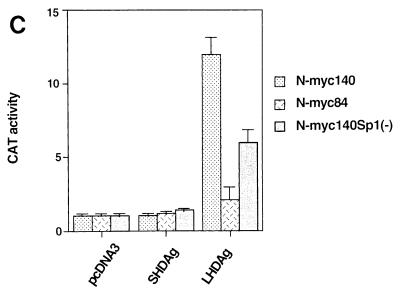
LHDAg has transactivation activity. (A) One microgram of a plasmid containing a CAT gene under the control of the woodchuck N-myc2 promoter was transfected with 4 μg of the pcDNA3 vector or a plasmid expressing SHDAg or LHDAg. CAT activity was measured, and that expressed in the cells transfected with the reporter plus the pcDNA3 vector was set arbitrarily at 1.0; other assay results were normalized to this value. At the bottom is a Western blot showing expression of SHDAg and LHDAg. (B) Part of the sequence of the woodchuck N-myc2 promoter. The TATA element is boxed. Sequences bearing putative binding sites for transcription factors are underlined. The transcription start site is designated +1 (13). Mutations introduced into the Sp1 site in 3×Sp1mCAT are shown separately below. (C) CAT assay of deletion mutants of the N-myc2 promoter. One microgram of a CAT reporter plasmid under the control of the N-myc promoter containing 140 bp (N-myc140), 84 bp (N-myc84), or 140 bp with a mutated Sp1 binding site [N-myc140Sp1(−)] regulatory sequence was transfected with 4 μg of the pcDNA3 vector or with a plasmid expressing SHDAg or LHDAg. A CAT assay was performed as described for A. (D) Activation of Sp1 by LHDAg in different cell lines. A CAT gene driven by the E1b TATA element with three WT or mutated Sp1 sites, as indicated, was cotransfected with either the pcDNA3 vector or a vector expressing SHDAg or LHDAg. Cells were assayed for CAT activity 48 h after transfection, and results were normalized to the level of CAT expressed from the pcDNA3 cotransfection, as described for A. (E) Detection of the activation activity of LHDAg by using the luciferase gene as a reporter. The luciferase gene driven by the SV40 early promoter was cotransfected with either the pcDNA3 vector or a vector expressing SHDAg or LHDAg, and its activity was measured 48 h after transfection. The error bars show the standard deviation from the mean of at least three independent experiments.
The N-myc2 gene (originally cloned from woodchuck liver) is a member of the myc gene family (14) whose promoter has been extensively studied in HepG2 cells (13). To map the sites of LHDAg responsiveness, we constructed several deletions of sequences in the N-myc2 regulatory region (cf. Fig. 2B and C). Deletions to −140 (relative to the cap site of the mRNA) maintained the transactivation activity of LHDAg, while deletions to −84 reduced activation by 5.7-fold (Fig. 2C). A computer search of the sequence from −140 to −84 revealed the presence of binding sites for Sp1 and for members of the GATA family of transcription factors (Fig. 2B). To determine if Sp1 might be a target of activation, we mutated the Sp1 site in this promoter and tested the ability of the mutant to be activated; this lesion reduced LHDAg induction twofold (Fig. 2C). From the fact that constructs with inactive Sp1 binding sites continue to show a residual level of activation (Fig. 2C), we infer that additional factors can also be up-regulated by LHDAg (see below).
To provide a direct test of the ability of Sp1 to be activated in trans (directly or indirectly) by LHDAg, we cotransfected LHDAg or SHDAg into HepG2 cells together with a vector containing three Sp1 binding sites upstream of the minimal E1B TATA box-driven CAT gene (Fig. 2D). CAT activity was measured 48 h after transfection. As expected, the presence of SHDAg had no effect on CAT activity, whereas LHDAg strongly activated the CAT gene linked to the three Sp1 sites (over 20-fold) (Fig. 2D). The specificity of Sp1 activation by LHDAg was further demonstrated by introducing mutations into the Sp1 sites. Control experiments with nuclear extracts showed that oligonucleotides bearing these mutant Sp1 sites do not bind Sp1 in vitro (data not shown). Correspondingly, transcription of a CAT gene linked to the mutant Sp1 sites was not up-regulated by LHDAg (Fig. 2D). This result confirms that Sp1 is one target of activation by LHDAg.
To explore the generality of this activation response, we examined if it can be observed in cell lines other than HepG2. Accordingly, we cotransfected the 3×Sp1E1B CAT reporter gene with the plasmids expressing SHDAg and LHDAg into 293 (human embryonal kindey) and CCL13 (human liver) cells and assayed for CAT activity. In both cell lines, LHDAg activated CAT gene expression as strongly as in HepG2 cells (Fig. 2D); as in HepG2 cells, SHDAg was inactive in this assay in both lines.
The fact that CAT genes with differing 5′ regions respond differently to LHDAg makes it unlikely that the prime target of this activation resides within CAT sequences. However, to exclude the possibility of reporter-specific artifacts, we also examined the ability of LHDAg to activate a luciferase reporter, in this case driven by an SV40 promoter. As shown in Fig. 2E, this reporter was reproducibly up-regulated fourfold, an amount similar to that observed with CAT reporters driven by promoters containing SV40 components (Fig. 3). No effect of SHDAg on luciferase expression was observed.
FIG. 3.
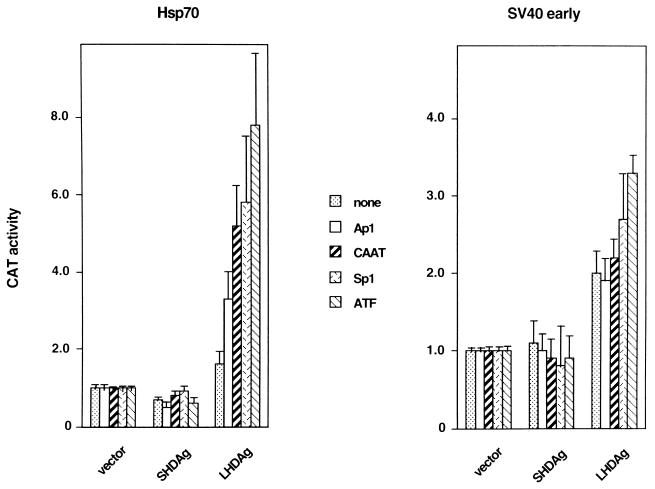
Activation of several upstream elements by LHDAg. CAT genes driven by various upstream stimulatory elements, including binding sites for ATF, CAAT, Sp1, and AP1, paired with either the hsp70 TATA element or the SV40 early promoter TATA element, were cotransfected into HepG2 cells with either the pcDNA3 vector or a vector expressing SHDAg or LHDAg. Cellular extracts were assayed for CAT activity, and the results were normalized as described in the legend to Fig. 2.
Effect of LHDAg on various promoters.
To assess the range of potential transcription factor targets of LHDAg, we tested two series of promoters in transient transfection assays. The hsp70 minimal promoter and the TATA box of the SV40 early promoter were each paired with a single copy of a variety of upstream stimulatory elements (USEs), including CAAT, Ap1, Sp1, ATF, and a nonsense sequence (none), all driving CAT reporter genes (for detailed sequences, see references 24 and 33). Each of these plasmids was cotransfected into HepG2 cells with constructs expressing either LHDAg or SHDAg, and the transfected cells were assayed for CAT activity. As shown in Fig. 3, the increase in CAT activity caused by LHDAg was higher with the promoters based on the hsp70 minimal promoter than those based on the SV40 early promoter for the same USE. When different USEs were linked to the same promoter and tested for response to LHDAg, there was a hierarchy of responses, in the following order: ATF > Sp1 = CAAT > AP1. These data affirm that Sp1 is not the only factor that can be activated by LHDAg expression. By contrast, preliminary experiments failed to show activation of Oct-1 and HNF4 (data not shown), suggesting that LHDAg is not an indiscriminate activator of all upstream activators.
Effect of LHDAg mutations on transactivation activity.
LHDAg and SHDAg are identical in sequence, save for the additional 19 aa present at the C terminus of LHDAg. Since SHDAg does not possess the transactivation activity displayed by LHDAg, it seemed likely that this activity was endowed by this 19-aa C-terminal extension. To explore this possibility, we constructed several classes of LHDAg mutants, including both truncations and amino acid substitutions. In a preliminary inspection of the sequence of this region, two particular features that pointed to the potential for protein-protein interactions attracted our attention. First, we noted the presence of two PXXP motifs (Fig. 1); in some proteins, such motifs have been implicated in interactions with other proteins harboring SH3 domains (9). The second and more well-established feature was the CXXX motif responsible for the known farnesylation of LHDAg at cysteine 211 (27) that is essential for interactions with HBsAg (16). Special attention was paid to these two features in the design of mutations in this region.
The mutations we constructed are summarized in Fig. 1. Three lesions were specifically designed to target the PXXP motifs. Mutations in LHDAgP1m and LHDAgP2m consist of replacement of prolines with alanines in the first and second PXXP motifs, respectively (Fig. 1), while LHDAgP1P2m bears mutations in both PXXP motifs (Fig. 1). Mutant Lcys211 changes Cys 211 to serine, thereby ablating isoprenylation. The remaining mutations introduce stop codons after codons 8, 13, 16, and 18 of this 19-codon region (Fig. 1) and are designated S+8, S+13, S+16, and S+18, respectively. Each mutant was transfected into HepG2 cells and tested for both transactivation of the 3×Sp1CAT reporter and for the expression of the mutant proteins (as assayed by immunoblotting with anti-HDAg). As shown in Fig. 4C, all of the mutants expressed similar levels of LHDAg proteins and had electrophoretic mobilities consonant with their predicted chain lengths.
FIG. 4.
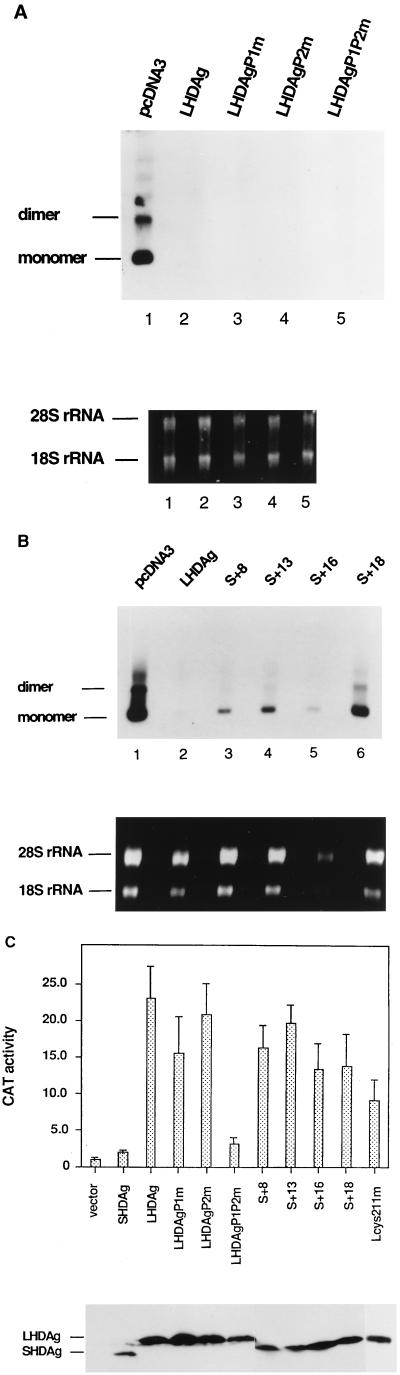
Effect of mutations on the replication inhibition and transactivation activities of LHDAg. (A) Inhibition of HDV RNA replication by PXXP motif mutants. Three micrograms of HDV plasmid pDL452 DNA, which is capable of initiating viral replication upon transfection, was transfected into 293 cells with 10 μg of plasmids expressing the mutant LHDAgs indicated above the gels (top). Six days posttransfection, total RNA was extracted and 3 μg of this RNA was used for Northern blot analysis. Membranes were hybridized with a specific antigenomic probe uniformly labeled with digoxigenin. At the bottom are the ethidium bromide-stained rRNAs used as a loading control. (B) Inhibition of HDV genome replication by truncation mutants. Three micrograms of pDL452 DNA was transfected with 10 μg of a plasmid expressing WT or truncated LHDAg into 293 cells. Northern blot analysis was performed as for B. The ethidium bromide-stained rRNAs shown were used as a loading control. (C) Effect of LHDAg mutations on transactivation activity (top). One microgram of plasmid 3×Sp1CAT was transfected with 4 μg of the indicated WT or mutant HDAg expression vector into HepG2 cells, CAT assays were performed, and the results were normalized as described in the legend to Fig. 2. The immunoblot at the bottom shows expression of the indicated WT or mutant HDAgs in transfected cells.
While mutations in the individual PXXP motifs did not significantly impair transactivation, the P1P2m double mutant virtually ablated this activity (Fig. 4C) and was, in fact, the only mutant in this series with this phenotype. Although this initially suggested that SH3 interaction motifs might be important for the activation activity, this simple interpretation is countermanded by the phenotype of mutant S+8. In this mutant, one PXXP motif is deleted and the other is severely crippled (Fig. 1), yet transactivation is preserved at nearly WT levels (Fig. 4A). Thus, it appears that the phenotype of the P1P2m mutant is due not to disruption of putative SH3 interaction but to effects on another aspect of the structure or conformation of this region. However, we note that the P1P2m lesion is highly specific and its phenotype is not the trivial result of a global disruption of protein structure. The protein stably accumulates to WT levels, remains immunoreactive, and continues to function as an inhibitor of SHDAg-mediated HDV RNA replication. The latter result is shown in the experiment of Fig. 4A, in which 10 μg of the WT or mutant LHDAg expression vector was cotransfected into 293 cells with 3 μg of replication-competent overlength HDV plasmid pDL452, which is designed to express HDV genomic RNA. HDV RNA of antigenomic polarity was then assayed by Northern blotting; such RNA can only be produced in these cells by authentic (SHDAg-dependent) viral RNA replication (19). As can be seen in Fig. 4A, all PXXP mutants, including P1P2m, retain the ability to dominantly inhibit replication, implying that all of them retain the ability to hetero-oligomerize with SHDAg. Thus, the P1P2m lesion selectively impairs the transactivation function of LHDAg.
Mutant Lcys211, in which cysteine 211 was changed to a serine, has previously been shown to be unprenylated (16), as well as to be somewhat less active as an inhibitor of replication (18). As shown in Fig. 4C, the Lcys211 mutation reproducibly diminished the activation activity (ca. twofold) but did not abolish it. We conclude that farnesylation of LHDAg is not essential for this activity. The transactivation results obtained with the truncation mutants are also shown in Fig. 4C. Clearly, all of the C-terminal truncations tested were stably expressed and were able to activate expression of the reporter, with only modest and variable reductions in activity. Figure 4B shows that all were also still able to inhibit HDV RNA replication, although there was clear variation in their potency in this regard. There was little apparent correlation between this inhibitory activity and the strength of transactivation.
Effect of SHDAg on transactivation activity of LHDAg.
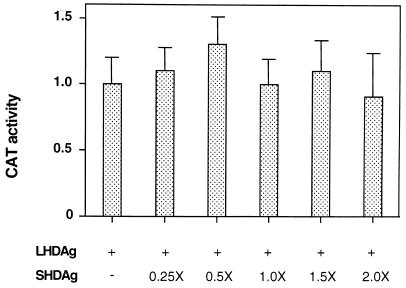
During viral replication, SHDAg and LHDAg coexist in cells and their relative stoichiometries are important in determining function. For example, the inhibitory effects of LHDAg on viral RNA replication can be overcome by overexpression of the SHDAg isoform. To determine if expression of SHDAg can influence the transactivation activity of LHDAg, HepG2 cells were transfected with a fixed quantity of the LHDAg expression plasmid and increasing amounts of an SHDAg-expressing construct, and the ability to activate CAT expression from a cotransfected reporter gene was assayed. Over an eightfold range of relative SHDAg-to-LHDAg plasmid concentrations, no significant effect on the level of transactivation was observed (Fig. 5).
FIG. 5.
Expression of SHDAg does not affect the transactivation activity of LHDAg. One microgram of 3×Sp1CAT DNA was transfected with a fixed amount (4 μg) of a plasmid expressing LHDAg and increasing amounts of a plasmid expressing SHDAg into HepG2 cells. The molar ratio of SHDAg over LHDAg DNA is indicated; pcDNA3 plasmid DNA was added to each transfection to bring the total amount of DNA used in each transfection to a constant level. At 48 h posttransfection, CAT activity was determined and normalized to the level produced in the presence of LHDAg alone, which was set at 1.0.
Effect of LHDAg on HBV promoters.
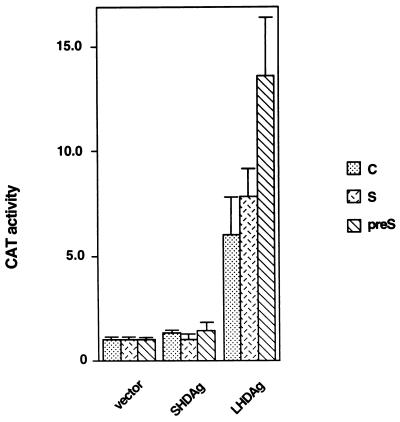
Since HBV is the natural helper virus of HDV, we wanted to know what effect expression of LHDAg would have on HBV promoters. pPreS1CAT, pSCAT, and pUCAT9 are plasmids in which the CAT gene is under the control of the HBV pre-S, S, and C promoters, respectively (17, 40). Each of these plasmids was cotransfected with the LHDAg expression vector into HepG2 cells, and extracts were assayed for CAT activity. As shown in Fig. 6, all of the HBV promoters tested were activated by LHDAg; again, SHDAg expression was without effect.
FIG. 6.
Activation of HBV promoters by LHDAg. One microgram of the indicated HBV-CAT reporter construct was cotransfected into HepG2 cells with 4 μg of pcDNA3 (vector) or derivatives encoding SHDAg or LHDAg. CAT activity was measured 48 h after transfection, and the results for each reporter were normalized to the level produced in the presence of the pcDNA3 vector alone, which was set at 1.0.
DISCUSSION
These studies document that, at least in acutely transfected cell cultures, LHDAg, but not SHDAg, displays a novel activity, the ability to activate gene expression in trans. Activation is independent of the reporter function used and occurs in several cell types and on a wide variety of natural and synthetic promoters but is not observed on minimal (basal) promoter elements, suggesting that it operates principally on accessory rather than basal transcription factors.
We know little of the mechanism by which this activation occurs. The yeast two-hybrid result that initially drew our attention to this phenomenon could be interpreted to mean that LHDAg can interact directly or indirectly with the basal transcriptional machinery once targeted to DNA, and certainly binding to accessory transcription factors might be one way to achieve such targeting in vivo. However, since many proteins with no transcriptional roles in vivo can score in two-hybrid assays, we are reluctant to place too much emphasis on this result as a clue to the mechanism. Many alternative mechanisms can be envisioned, including much more indirect ones. For example, LHDAg might function from a cytoplasmic location (e.g., the cytosolic face of the endoplasmic reticulum) to initiate a signal transduction cascade that ultimately converges on nuclear transcription factors. Such a proposal would be formally similar to certain models proposed for the activation of gene expression by the HBV X protein (11). Additional in vivo and in vitro studies are required to address the mechanistic issues raised here.
The potential biological significance of this activity is likewise a matter of conjecture. For example, we do not know if activation occurs at levels of LHDAg achieved during infection in vivo. Attempts to address this question in transfected cells have been impeded by technical difficulties. Starting from cloned HDV DNA, it takes days in cell culture before RNA editing generates substantial LHDAg; by this time, most of the input reporter CAT plasmid has disappeared. We have been unable to examine rigorously whether native LHDAg expressed from edited RNA can activate a reporter gene, although we think this highly likely. The larger question is what, if any, functional consequences such transactivation might have. It seems unlikely that such an activity would be required for HDV RNA synthesis, since the latter is dependent upon SHDAg and is achieved prior to peak LHDAg levels. By contrast, induction of HBV envelope protein expression by LHDAg could well serve to enhance HDV assembly, and, as shown in Fig. 6, the HBV pre-S and S promoters are clearly responsive to LHDAg expression.
Irrespective of its role in HDV replication, the activation activity might influence other aspects of the infection, e.g., pathogenesis. Induction of host gene expression by LHDAg could play a role in enhancing pathogenesis; for example, if induction included class I major histocompatibility complex genes, elevated liver cell cytotoxicity might result from enhanced cytotoxic T-lymphocyte action. Induction of HBV gene expression (Fig. 6) could have similar effects in sensitized infected hosts. This activity could contribute to the clear association of HDV with fulminant hepatitis, which is thought to result largely from exaggerated cytotoxic responses to viral antigens. In this formulation, the reduction in HBV markers typically observed in HDV coinfection (30) would be the result of enhanced immune clearance (rather than direct repression of HBV genes by HDV gene products). Alternatively, it is possible that suppression of HBV in HDV-infected cells is mediated by host factors induced by LHDAg or that the inductive effect of LHDAg observed here is countered by a repressive effect of another viral gene product, e.g., SHDAg (39).
Finally, the transactivation function described here might have no function in present-day HDV replication but may simply represent a vestige of the evolutionary history of LHDAg. We recently identified a host gene, termed dipA, whose product interacts with HDAg (5). Characterization of this gene showed that it is distantly related to that for LHDAg, suggesting that present-day HDV may have been derived from the capture of a cellular gene by a primitive, viroid-like replicon. Of note is the fact that dipA also shares limited homology with members of the fra-encoded family of transcription factors (10), and the dipA gene is directly adjacent to fra-1 in the mouse genome (5a), again suggesting a relationship. If these tantalizing evolutionary connections to cellular transcription factors are correct, then the activation function of LHDAg may simply be the vestigial remnant of an activity that was once its central mission but which now is only a secondary or accessory function.
ACKNOWLEDGMENTS
We are grateful to Rob Brazas for many insightful discussions and for helpful comments on the manuscript. We thank David Lazinski for plasmid pDL452; David Lukac, James Alwine, and Robert Kingston for the promoter constructs based on hsp70 and SV40 early promoters; and T.-S. Benedict Yen for pPreS1CAT, pSCAT, and pUCAT9. We thank F. Bergametti, D. Sitterlin, and C. Transy for help with the luciferase assay.
REFERENCES
- 1.Bichko V, Barik S, Taylor J. Phosphorylation of the hepatitis delta virus antigens. J Virol. 1997;71:512–518. doi: 10.1128/jvi.71.1.512-518.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bichko V V, Taylor J M. Redistribution of the delta antigens in cells replicating the genome of hepatitis delta virus. J Virol. 1996;70:8064–8070. doi: 10.1128/jvi.70.11.8064-8070.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bonino F, Heermann K H, Rizzetto M, Gerlich W H. Hepatitis delta virus: protein composition of delta antigen and its hepatitis B virus-derived envelope. J Virol. 1986;58:945–950. doi: 10.1128/jvi.58.3.945-950.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bonino F, Negro F, Baldi M, Brunetto M R, Chiaberge E, Capalbo M, Maran E, Lavarini C, Rocca N, Rocca G. The natural history of chronic delta hepatitis. 234. Alan R. New York, N.Y: Liss; 1987. [PubMed] [Google Scholar]
- 5.Brazas R, Ganem D. A cellular homolog of hepatitis delta antigen: implications for viral replication and evolution. Science. 1996;274:90–94. doi: 10.1126/science.274.5284.90. [DOI] [PubMed] [Google Scholar]
- 5a.Brazas, R., and D. Ganem. Unpublished data.
- 6.Chang F L, Chen S J, Tu S J, Chiu M N, Wang C J, Chen D S. The large form of hepatitis delta antigen is crucial for the assembly of hepatitis delta virus. Proc Natl Acad Sci USA. 1991;88:8490–8494. doi: 10.1073/pnas.88.19.8490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang M F, Baker S C, Soe L H, Kamahora T, Keck J G, Makino S, Govindajajan S, Lai M M C. Human hepatitis delta antigen is a nuclear phosphoprotein with RNA-binding activity. J Virol. 1988;62:2403–2410. doi: 10.1128/jvi.62.7.2403-2410.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chao M, Hsieh S-Y, Taylor J. Role of two forms of the hepatitis delta virus antigen: evidence for a mechanism of self-limiting genome replication. J Virol. 1990;64:5066–5069. doi: 10.1128/jvi.64.10.5066-5069.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cicchetti P, Mayer B J, Thiel G, Baltimore B. Identification of a protein that binds to the SH3 region of abl and is similar to Bcr and GAP-rho. Science. 1992;257:803–806. doi: 10.1126/science.1379745. [DOI] [PubMed] [Google Scholar]
- 10.Cohen D R, Curran T. fra-1, a serum-inducible, cellular immediate-early gene that encodes a Fos-related antigen. Mol Cell Biol. 1988;8:2063–2069. doi: 10.1128/mcb.8.5.2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Doria M, Klein N, Lucito R, Schneider R J. The hepatitis B virus HBx protein is a dual specificity cytoplasmic activator of Ras and nuclear activator of transcription factors. EMBO J. 1995;14:4747–4757. doi: 10.1002/j.1460-2075.1995.tb00156.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Engler-Blum G, Meier M, Frank J, Müller G A. Reduction of background problems in nonradioactive Northern and Southern blot analyses enables higher sensitivity than 32P-based hybridizations. Anal Biochem. 1993;210:235–244. doi: 10.1006/abio.1993.1189. [DOI] [PubMed] [Google Scholar]
- 13.Fourel G, Transy C, Tennant B C, Buendia M A. Expression of the woodchuck N-myc2 retroposon in brain and in liver tumors is driven by a cryptic N-myc promoter. Mol Cell Biol. 1992;12:5336–5344. doi: 10.1128/mcb.12.12.5336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fourel G, Trépo C, Bougueleret L, Henglein B, Ponzetto A, Tiollais P, Buendia M A. Frequent activation of N-myc genes by hepadnavirus insertion in woodchuck liver tumours. Nature. 1990;347:294–298. doi: 10.1038/347294a0. [DOI] [PubMed] [Google Scholar]
- 15.Fu T-B, Taylor J. The RNAs of hepatitis delta virus are copied by RNA polymerase II in nuclear homogenates. J Virol. 1993;67:6965–6972. doi: 10.1128/jvi.67.12.6965-6972.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Glenn J S, Watson J A, Havel C M, White J M. Identification of a prenylation site in delta virus large antigen. Science. 1992;256:1331–1333. doi: 10.1126/science.1598578. [DOI] [PubMed] [Google Scholar]
- 17.Guo W, Chen M, Yen T S B, Ou J. Hepatocyte-specific expression of the hepatitis B virus core promoter depends on both positive and negative regulation. Mol Cell Biol. 1993;13:443–448. doi: 10.1128/mcb.13.1.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hwang S B, Lai M M C. Isoprenylation masks a conformational epitope and enhances trans-dominant inhibitory function of the large hepatitis delta antigen. J Virol. 1994;68:2958–2964. doi: 10.1128/jvi.68.5.2958-2964.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuo M Y-P, Chao M, Taylor J. Initiation of replication of the human hepatitis delta virus genome from cloned DNA: role of delta antigen. J Virol. 1989;63:1945–1950. doi: 10.1128/jvi.63.5.1945-1950.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lai M M C. The molecular biology of hepatitis delta virus. Annu Rev Biochem. 1995;64:259–286. doi: 10.1146/annurev.bi.64.070195.001355. [DOI] [PubMed] [Google Scholar]
- 21.Lazinski D W, Taylor J M. Recent developments in hepatitis delta virus research. Adv Virus Res. 1994;43:187–231. doi: 10.1016/s0065-3527(08)60049-4. [DOI] [PubMed] [Google Scholar]
- 22.Lazinski D W, Taylor J M. Relating structure to function in the hepatitis delta virus antigen. J Virol. 1993;67:2672–2680. doi: 10.1128/jvi.67.5.2672-2680.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin J-H, Chang M-F, Baker S C, Govindarajan S, Lai M M C. Characterization of hepatitis delta antigen: specific binding to hepatitis delta virus RNA. J Virol. 1990;64:4051–4058. doi: 10.1128/jvi.64.9.4051-4058.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lukac D M, Manuppello J R, Alwine J C. Transcriptional activation by the human cytomegalovirus immediate-early proteins: requirements for simple promoter structures and interactions with multiple components of the transcription complex. J Virol. 1994;68:5184–5193. doi: 10.1128/jvi.68.8.5184-5193.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luo G, Chao M, Hsieh S-Y, Sureau C, Nishikura K, Taylor J. A specific base transition occurs on replicating hepatitis delta virus RNA. J Virol. 1990;64:1021–1027. doi: 10.1128/jvi.64.3.1021-1027.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.MacNaughton T B, Gowans E J, McNamara S P, Burrell C J. Hepatitis delta antigen is necessary for access of hepatitis delta virus RNA to the cell transcriptional machinery but is not part of the transcriptional complex. Virology. 1991;184:387–390. doi: 10.1016/0042-6822(91)90855-6. [DOI] [PubMed] [Google Scholar]
- 27.Otto J C, Casey P J. The hepatitis delta virus large antigen is farnesylated both in vitro and in animal cells. J Biol Chem. 1996;27:4569–4572. doi: 10.1074/jbc.271.9.4569. [DOI] [PubMed] [Google Scholar]
- 28.Ponzetto A, Cote P J, Popper H, Boyer B H, London W T, Ford E C, Bonino F, Purcell R H, Gerin J L. Transmission of the hepatitis B-associated delta agent to the eastern woodchuck. Proc Natl Acad Sci USA. 1984;81:2208–2212. doi: 10.1073/pnas.81.7.2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rizzetto M, Canese M G, Arico J, Crivelli O, Bonino F, Trepo C G, Verme G. Immunofluorescence detection of a new antigen-antibody system associated to the hepatitis B virus in the liver and in the serum of HDsAg carriers. Gut. 1977;18:997–1003. doi: 10.1136/gut.18.12.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rizzetto M, Canese M G, Gerin J L, London W T, Sly D L, Purcell R H. Transmission of the hepatitis B virus-associated delta antigen to chimpanzees. J Infect Dis. 1980;141:590–602. doi: 10.1093/infdis/141.5.590. [DOI] [PubMed] [Google Scholar]
- 31.Rizzetto M, Hoyer B, Canese M G, Shih J W-K, Purcell R H, Gerin J L. Delta agent: association of delta antigen with hepatitis B surface antigen and RNA in serum of delta-infected chimpanzees. Proc Natl Acad Sci USA. 1980;77:6124–6128. doi: 10.1073/pnas.77.10.6124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saracco G, Macagno S, Rosina F, Rizzetto M. Serologic markers with fulminant hepatitis in persons positive for hepatitis B surface antigen. A worldwide epidemiologic and clinical survey. Ann Intern Med. 1988;108:380. doi: 10.7326/0003-4819-108-3-380. [DOI] [PubMed] [Google Scholar]
- 33.Taylor I C A, Kingston R E. Factor substitution in a human HSP70 gene promoter: TATA-dependent and TATA-independent interactions. Mol Cell Biol. 1990;10:165–175. doi: 10.1128/mcb.10.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Taylor J, Mason W, Summers J, Goldberg J, Aldrich C, Coates L, Gerin J, Gowans E. Replication of human hepatitis delta virus in primary cultures of woodchuck hepatocytes. J Virol. 1987;61:2891–2895. doi: 10.1128/jvi.61.9.2891-2895.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ueda K, Wei Y, Ganem D. Activation of N-myc2 gene expression by cis-acting elements of oncogenic hepadnaviral genomes: key role of enhancer II. Virology. 1996;217:413–417. doi: 10.1006/viro.1996.0133. [DOI] [PubMed] [Google Scholar]
- 36.Ueda K, Wei Y, Ganem D. Cellular factors controlling the activity of woodchuck hepatitis virus enhancer II. J Virol. 1996;70:4714–4723. doi: 10.1128/jvi.70.7.4714-4723.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang J-G, Lemon S M. Hepatitis delta virus antigen forms dimers and multimeric complexes in vivo. J Virol. 1993;67:446–454. doi: 10.1128/jvi.67.1.446-454.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weiner A J, Choo Q-L, Wang K-S, Govindarajan S, Redeker A G, Gerin J L, Houghton M. A single antigenomic open reading frame of the hepatitis delta virus encodes the epitope(s) of both hepatitis delta antigen polypeptides p24δ and p27δ. J Virol. 1988;62:594–599. doi: 10.1128/jvi.62.2.594-599.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu J-C, Chen P-J, Kuo M Y P, Lee S-D, Chen D-S, Ting L-P. Production of hepatitis delta virus and suppression of helper hepatitis B virus in a human hepatoma cell line. J Virol. 1991;65:1099–1104. doi: 10.1128/jvi.65.3.1099-1104.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou D, Yen T S B. Differential regulation of the hepatitis B virus surface gene promoters by a second viral enhancer. J Biol Chem. 1990;265:20731–20734. [PubMed] [Google Scholar]