Visual Abstract

Key Words: alternative splicing, atrial cardiomyopathy, atrial fibrillation, genetics, RBM20
Highlights
-
•
Loss of function variants in the key cardiac splicing regulator RBM20 were associated with atrial fibrillation in 2 independent cohorts.
-
•
Loss of RBM20 facilitated structural and transcriptional changes in the atria and affected mitochondrial function.
-
•
Alternative splicing may act as a novel proarrhythmic mechanism in the atria through atrial remodeling and altered mitochondrial energetics.
Abstract
We showed an association between atrial fibrillation and rare loss-of-function (LOF) variants in the cardiac splicing regulator RBM20 in 2 independent cohorts. In a rat model with loss of RBM20, we demonstrated altered splicing of sarcomere genes (NEXN, TTN, TPM1, MYOM1, and LDB3), and differential expression in key cardiac genes. We identified altered sarcomere and mitochondrial structure on electron microscopy imaging and found compromised mitochondrial function. Finally, we demonstrated that 3 novel LOF variants in RBM20, identified in patients with atrial fibrillation, lead to significantly reduced splicing activity. Our results implicate alternative splicing as a novel proarrhythmic mechanism in the atria.
Atrial fibrillation (AF) is the most common sustained cardiac arrhythmia1 and conveys a substantially increased risk of stroke, heart failure, and premature death.2 Although AF represents a substantial and increasing health care burden,3 current treatment regimens have limited efficacy and have significant adverse effects.4
In the last 2 decades, some progress has been made in the understanding of the genetic architecture of AF. To date, large-scale genome-wide association studies (GWAS) have associated more than 130 unique genetic loci with AF.5,6 However, GWAS are limited in their coverage of rare genetic variation; thus, the vast majority of identified genetic variants are relatively common and only modestly affect disease risk. Therefore, recent studies have focused on examining rare loss-of-function (LOF) variants that often confer a substantially increased risk of disease. This approach may serve to complement large-scale GWAS and uncover novel insights into pathophysiologic mechanisms.
Recently, rare titin truncating variants were associated with a considerable risk of early-onset AF,7,8 indicating that an atrial myopathy may be involved in the development of AF in this patient group. Titin, encoded by the gene TTN, is a large protein that plays a crucial role in sarcomere structure and function. Several different isoforms exist, the expression of which are closely regulated by the splicing regulator RNA binding motif protein 20 (RBM20).9 The protein is encoded by the gene RBM20 and acts as a splicing regulator of several key cardiac genes related to sarcomere structure and intracellular calcium handling.9,10 Changes in isoform expression have been reported to play an important role in ventricular form and function,11 but its role in the atria has yet to be studied in detail.
In the present study, we examined the impact of rare variations in RBM20 on AF risk using a broad range of methodologic approaches. Our study showed several key findings: We identified an association between rare LOF variants in RBM20 and AF in a Scandinavian cohort of early-onset AF patients and replicated the association in whole-exome sequencing data from the UK Biobank. We identified altered expression of numerous cardiac genes in atrial tissue from an RBM20-deficient rat model using long-read nanopore sequencing. In the same rat model, we demonstrated compromised sarcomere structure and altered mitochondrial function through electron microscopy imaging and studies of mitochondrial function. Finally, we observed reduced splicing activity in HEK293-cell models with variants identified in the early-onset AF cohort.
Methods
Early-onset AF cohort
We recruited Scandinavian patients with early-onset AF and no other cardiovascular disease. Early-onset AF was defined as onset of AF at <50 years of age. Danish patients were identified and recruited through the Danish National Patient Registry, with AF defined by International Classification of Diseases, 10th Revision code I48. The validity of the AF diagnosis in the Danish National Health Registry has previously been shown to be approximately 92.5%.12 Individuals fulfilling the inclusion criteria were given written information and were offered the opportunity to participate. We collected clinical information and peripheral blood samples. The study was approved by the scientific ethics committee of the Capital Region of Denmark (protocol number: H-20048862).
Norwegian patients were recruited from the region of Oslo and Vestre Viken, Norway. Patients were identified through clinical practice and the AF registries at Vestre Viken Hospital Trust and Oslo University Hospital. AF was defined as AF for more than 30 seconds on an electrocardiogram. The study was approved by the South-Eastern Regional Ethics Committee (Project no.: 240149) in Norway.
Patients with diabetes, hypertension, hyperthyroidism, or other cardiac disease (congenital heart defects, valvular disease, congestive heart failure, ischemic heart disease) at inclusion were excluded. All participants gave written informed consent.
We extracted genomic DNA from leukocytes in peripheral blood samples from early-onset AF patients. Endonucleases were used to fragment the DNA, and gene-specific probes from the Illumina TruSight Cardio (Illumina Sequencing) sequencing kit were used to create hybridized molecules. We then captured the hybridized molecules using magnetic beads, and following polymerase chain reaction amplification, performed sequencing using Illumina HiSeq 2500 and NextSeq technology. The sequenced reads were trimmed from adapter sequences, filtered for low-quality reads, aligned to the Human Reference Genome (build GRCh37/bg19) using Burrows-Wheeler Aligner software,13 and postprocessed according to Genome Analysis Toolkit best practice guidelines.14 Our sequencing methods and bioinformatics pipeline have previously been described in detail by Ahlberg et al.7
Referent populations
We compared the burden of variants predicted to cause LOF in RBM20 in our data with available genetic data from 2 large population-based datasets: the SweGen cohort containing genetic data on 1,000 participants, representing a cross section of the Swedish population,15 and the Genome Aggregation Database (gnomAD) version 3, containing genetic information on 76,156 unrelated individuals.16
Categorical data are presented as counts and percentages and compared between the early-onset AF and control cohorts using the Fisher exact test because of small sample sizes. R version 4.1.2 was used for analysis.17 A P value of <0.05 was considered significant.
UK Biobank exomes
The UK Biobank is a large population-based cohort study, comprising almost 500,000 individuals aged 40 to 69 years at inclusion. The biobank includes genetic and clinical information, including health records, representative of the general population. All participants in the UK Biobank gave written informed consent. The study protocol and data collection have been described in detail by Bycroft et al.18
We examined the burden of rare LOF variants in RBM20 in whole-exome sequencing data from the UK Biobank.19 We accessed the whole-exome sequencing dataset, of which 404,012 participants were of European ancestry. Detailed information on exome sequencing methodology, quality control, alignment, variant calling, and annotation in the dataset has been described in detail previously.20 Only variant sites where at least 90% of the genotypes had a read depth of >10 were included in the subsequent analyses. We filtered samples based on heterozygosity, missing rates, excess relativeness, and kinship inference, based on UK Biobank resource 531, and excluded samples with disagreement between reported sex and genetically determined sex.
AF was defined by UK Biobank data field 131351, corresponding to International Classification of Diseases, 10th Revision code I48. The AF diagnosis in the UK Biobank was based on in-patient hospital records, death records, primary care records, and self-reports from interviews conducted by qualified nurses. We then conducted rare variant association tests for LOF variants with a minor allele frequency of <0.01 in RBM20. Association tests were conducted using the logistic regression model integrated in REGENIE,21 adjusted for sex, age at inclusion, and 10 first principal components. We tested for association across all rare LOF variants collapsed into a single gene-based test as well as for each rare LOF variant individually. We applied Firth logistic correction to account for potential bias in maximum likelihood estimates caused by case-control imbalance and small sample size. Results from the logistic regressions are presented as ORs with 95% CIs.
Finally, we tested for association between rare LOF variants in RBM20 and cardiac structure and function in a subset of UK Biobank participants who had undergone cardiac magnetic resonance imaging. Imaging annotation has previously been described in detail for right heart structure,22 left ventricular structure,23 and left atrial structure.24 Genetic association tests were conducted in REGENIE and adjusted for sex, age at imaging, body surface area at imaging, imaging center, and 10 first principal components.
Animal model characteristics
We used atrial tissue from rats carrying a 95-kb deletion, resulting in the removal of exons 2 through 14 in RBM20. The rats were described by Guo et al9 and outcrossed to Sprague Dawley rats. We examined atrial tissue from 8 heterozygote RBM20+/– and 8 homozygote RBM20–/– animals. For controls, we used atrial tissue from 5 unrelated Sprague Dawley rats. The animal studies complied with all institutional and national requirements for the care and use of laboratory animals (Berlin State Office for Health and Social Affairs: Reg Y-9004/14).
Transmission electron microscopy
Atrial tissue from 3-month-old (n = 6) RBM20+/– siblings and unrelated 3-month-old WT (n = 5) rats was dissected out and separated into left and right atria. The samples were fixed in a 2% volume/volume glutaraldehyde in 0.05-mol/L sodium phosphate buffer (pH 7.4) for 24 hours. After a brief rinse in a sodium cacodylate buffer (0.15 mol/L, pH 7.4), a subsequent 2-hour postfixation step followed (1% weight/volume OsO4; 0.05-mol/L potassium ferricyanide in 0.12-mol/L sodium cacodylate buffer; pH 7.4). Samples were then dehydrated in ethanol and transferred to propylene oxide before being embedded in Epon. Samples were cut into sections of approximately 80 nm and collected on copper grids in Formvar supporting membranes. Samples were subsequently stained using uranyl acetate and lead citrate. Samples were imaged using transmission electron microscopy on a Philips CM 100 transmission electron microscope (Philips), operated at an accelerating voltage of 80 kV. Images were captured using an OSIS Veleta digital slow scan 2k × 2k charge-coupled device camera (Olympus).
Direct complementary DNA sequencing
We performed direct complementary DNA (cDNA) sequencing on atrial tissue from 3 RBM20+/– rats and 3 RBM20–/– rats and compared them with 3 WT controls. RNA was isolated from left and right atrial tissue and purified using TRIzol Reagent (Thermo Fisher Scientific) combined with the RNeasy Micro kit (Qiagen).
Reads were aligned to the Norwegian Brown reference transcriptome using fasta and gtf files provided by Ensembl (Rnor 7.2).25 We used the minimap2 software (version 2.24-r1112),26 with arguments -ax map-ont for direct cDNA nanopore sequencing data. Transcript abundance was then estimated using Salmon (version 1.8.0).27
Detailed methods for direct cDNA sequencing, read alignment, and transcript abundance quantification are summarized in the Supplemental Appendix.
Differential transcript usage analysis
For differential transcript usage (DTU), we based our approach on the one described in the pipeline-transcriptome-de pipeline. Based on expression data from the Protein Atlas 28 we focused on genes that were part of the Heart Specific Proteome. Using the gene symbols in the Ensembl database, we matched the 13,668 genes on this list with 11,936 orthologues in the rat reference genome. The remaining genes were excluded before DTU analysis. Raw counts were filtered (≥1 transcript count in ≥4 samples), and a data frame was constructed using the DRIMseq package (version 1.22.0)29 and analyzed using the DEXseq package (version 1.34.0).30 We controlled for multiple testing using the Benjamini-Hochberg false discovery rate (FDR) post hoc method. Associations with an FDR of <5% were considered statistically significant. All analyses were conducted in R version 4.1.2.17
Differential gene expression analysis
Before differential gene expression analysis, we performed prefiltering to ensure high quality of the data. Only genes with ≥10 counts in ≥4 samples and part of the heart-specific proteome (described earlier) were included in the analysis. Differential gene expression analysis was conducted in R with the DESeq2 package,31 using standard settings, and an FDR cutoff of <0.05 to account for multiple testing (using the Benjamini-Hochberg method). Following analysis, results were shrunk using the lfcshrink function in DEseq2, set to use the apeglm method.32 For visualization of data, data were normalized by the regularized logarithm transformation function in the DESeq2 package, and a principal component plot was constructed according to the instructions of the package vignette. Differentially expressed genes were visualized on volcano plots using the ggplot2 package in R.33 Finally, we conducted a pathway analysis of significantly enriched gene sets in RBM20–/– rats using gene set enrichment analysis,34,35 as described in detail in the Supplemental Appendix.
Mitochondrial function analysis
Samples for mitochondrial analyses were dissected out and were immediately placed in ice-cold Buffer X as described by Perry et al.36 Mitochondrial respiratory capacity was analyzed using high-resolution fluororespirometry by Oxygraph-2k technology (Oroboros Instruments) in accordance with that previously optimized for skeletal muscle.37 Our analyses were conducted with mitochondrial complex I– and II–linked and maximal oxidative phosphorylation (OXPHOS)–linked substrates (ie, glutamate, malate, succinate, and adenosine diphosphate). Mitochondrial respiration and hydrogen peroxide (H2O2) formation were measured in real time simultaneously. H2O2 levels were determined using Amplex UltraRed (Thermo Fisher Scientific) according to previously published methods.37 H2O2 is a stable form of reactive oxygen species (ROS) and will from here on be referred to as ROS. We performed statistical analyses in R (version 3.4.1).17 Residual and Q-Q plots were used to confirm homogeneity of covariance and normal distribution of the data. The main effect was analyzed using analysis of variance. Group differences were analyzed using a linear mixed model with the Tukey post hoc test and adjusted using the DFR. A value of P < 0.05 was considered statistically significant, and values are reported as mean ± SEM.
Identification of human RBM20 mutations with reduced activity
We used site-directed mutagenesis to examine the splicing activity of the included variants from the Scandinavian AF cohort. Mutagenesis was performed using the Q5 proofreading DNA polymerase (New England Biolabs) and a standard protocol. The primers used (Supplemental Table 1) were designed using the QuickChange Primer Design Program and have been described in detail elsewhere.38 Final constructs were sequence-verified. Detailed methods for analysis of RNA and protein expression are described in the Supplemental Appendix.
Results
Identification of rare variants in RBM20 in early-onset AF patients
We included 531 Scandinavian individuals with early-onset AF (age at AF onset: <50 years) Individuals with left ventricular ejection fraction of <55% or structural heart disease as determined by echocardiographic examination were excluded. The median age at AF onset was 30 years (IQR, 25th-75th percentile: 24-36 years), and there was a male predominance (83.4% men).
In the early-onset AF cohort, 3 individuals carried rare LOF variants (∼0.56%), all of which were novel. The clinical characteristics of variant carriers have been summarized in Supplemental Table 2. One individual (A-01) carried a variant resulting in a premature stop codon (p.Gln345∗) and developed AF at age 47 years without predisposing comorbidity. Another individual (A-11) carried a frameshift variant (p.Glu725Valfs∗34). This individual developed AF at age 26 years with no other comorbidities. At the time of inclusion, the individual had AF with monthly episodes. Both electric cardioversion and radiofrequency ablation had been attempted without success. A third individual (A-13) carried a variant resulting in a premature stop codon in RBM20 (p.Glu787∗). The individual developed AF at the age of 22 years with no other comorbidities.
LOF variants in RBM20 are associated with an increased risk of AF
We evaluated whether there was an association between an increased burden of rare LOF variants in RBM20 and AF. The number of variants in the Scandinavian early-onset AF cohort was compared to the number of RBM20 variants in the 2 population-based databases of normal variation, gnomAD16 and SweGen.15
The burden of LOF variants in RBM20 was significantly higher in patients with AF compared with the gnomAD cohort (0.5% vs 0.04%; Fisher exact test: P = 0.003). In a sensitivity analysis, excluding individuals of non-European ancestry from the gnomAD population, we saw consistent results (0.5% vs 0.06%; P = 0.007). We performed the same association test using the population-based, whole-genome sequencing data from SweGen representing a cross section of the Swedish population15 and found similar results (0.5% vs 0%; P = 0.041).
Replication in a population-based cohort
We then assessed whole-exome sequencing data from 404,012 unrelated individuals of European ancestry from the UK Biobank.18 The individuals in the cohort had a median age at inclusion of 58 years (IQR, 25th-75th percentile: 51-63 years), and 54.1% were of female sex. We identified 31,843 individuals diagnosed with AF and considered the remaining 372,169 individuals as controls. Characteristics of the cohort are summarized in Supplemental Table 3.
In this population-based cohort, we replicated the association between rare LOF variants in RBM20 and AF (OR: 2.43; 95% CI: 1.48-4.00; P < 0.001). We identified 5 unique LOF variants identified in the cohort, of which 3 were associated with an increased OR for AF (Supplemental Table 4). We examined cardiac structure and function in a subset of individuals who had undergone cardiac magnetic resonance imaging (44,123 and 47,005 individuals for atria and ventricles, respectively). We found that loss of RBM20 was associated with a significant increase in left atrial minimum volume (β = 0.77; P = 0.035). We also observed some suggestive, borderline-significant associations with increased left atrial maximum volume (β = 0.66; P = 0.067) and decreased left atrial total emptying fraction (β = –0.62; P = 0.089). Results are illustrated in Figure 1 and summarized in Supplemental Table 5.
Figure 1.
Associations Between Loss of RBM20 and Cardiac Structure and Function
Volcano plot of results from genetic association test between LOF variants in RBM20 and cardiac magnetic resonance imaging data. The figure shows an association with increased left atrial minimum volume. Associations with P < 0.10 are labeled with beta coefficients (β) and P values. LAMAX = left atrial maximum volume; LAMIN = left atrial minimum volume; LATEF = left atrial total emptying fraction; LOF = loss of function; LVEDV = left ventricular end-diastolic volume; LVESV = left ventricular end-systolic volume.
Differential isoform usage in RBM20-dependent transcripts
We investigated the transcriptional consequences of loss of RBM20 function using nanopore long-read direct cDNA sequencing on 9 atrial tissue samples from wild-type (WT) rats (n = 3), rats with heterozygous loss of RBM20 (RBM20+/–) (n = 3), and rats with homozygous loss of RBM20 (RBM20–/–) (n = 3). The mean number of quality control passed reads per sample was 2,105,938 (range: 1,398,064-3,708,577). Metrics of read lengths, quality, and mapping parameters are summarized in Supplemental Table 6. All findings were corrected for multiple testing using a Benjamini-Hochberg FDR of <5%.39
In an analysis of DTU, we observed a switch in isoform expression in several putative splicing targets of RBM20. In atria from RBM20+/– rats, we observed altered expression of transcripts in the TPM1 gene, encoding the actin-binding tropomyosin 1 protein and in LDB3, which encodes the important sarcomere-stabilizing LIM domain binding 3 protein (Table 1). In RBM20–/– rat atria, DTU was also identified in LDB3 and TPM1 and several additional genes spliced by RBM20, including TTN. We also identified DTU in NEXN, encoding the Z-disk protein nexilin; MYOM1, encoding the M-band protein myomesin 1; and KCNIP2, which encodes the ion-channel interacting protein Kv channel-interacting protein 2 (Table 2). Results from the DTU analysis are summarized in the Supplemental Appendix. All putative splicing targets of RBM20 are summarized in Supplemental Table 7.
Table 1.
Altered Isoform Expression in Splicing Targets (RBM20+/– Atria)
| Gene | Protein | Isoform | Transcript ID | Protein Size (aa) | Log2 Fold Change | P Value | Adjusted P Value (FDR) |
|---|---|---|---|---|---|---|---|
| LDB3 | LIM domain binding 3 | Ldb3-206 | ENSRNOT00000081631 | 330 | –0.0501 | 2.54 × 10–6 | 0.0003 |
| Ldb3-203 | ENSRNOT00000083623 | 729 | 4.0394 | 0.008 | 0.12 | ||
| Ldb3-205 | ENSRNOT00000086627 | 707 | –1.0161 | 0.12 | 0.47 | ||
| Ldb3-202 | ENSRNOT00000087521 | 771 | 0.7909 | 0.82 | 0.97 | ||
| Ldb3-201 | ENSRNOT00000085414 | 679 | 0.3489 | 0.88 | 0.98 | ||
| TPM1 | Tropomyosin 1 | Tpm1-208 | ENSRNOT00000024493 | 287 | 1.0371 | 1.02 × 10–6 | 1.76 × 10–4 |
| Tpm1-201 | ENSRNOT00000057641 | 326 | –2.6584 | 6.69 × 10–4 | 0.023 | ||
| Tpm1-202 | ENSRNOT00000024575 | 284 | 0.8738 | 0.010 | 0.13 | ||
| Tpm1-207 | ENSRNOT00000048044 | 284 | –0.3951 | 0.011 | 0.14 | ||
| Tpm1-209 | ENSRNOT00000090288 | 300 | 1.5946 | 0.067 | 0.37 | ||
| Tpm1-210 | ENSRNOT00000099012 | 267 | 2.2200 | 0.070 | 0.38 | ||
| Tpm1-204 | ENSRNOT00000040808 | 281 | –1.3919 | 0.10 | 0.45 | ||
| Tpm1-206 | ENSRNOT00000112475 | 257 | 0.7770 | 0.10 | 0.46 | ||
| Tpm1-203 | ENSRNOT00000085894 | 294 | 0.2243 | 0.13 | 0.49 | ||
| Tpm1-205 | ENSRNOT00000024617 | 248 | –1.4616 | 0.13 | 0.49 |
aa = amino acids; FDR = false discovery rate.
Table 2.
Altered Isoform Expression in Splicing Targets (RBM20–/– Atria)
| Gene | Protein | Isoform | Transcript ID | Protein Size (aa) | Log2 Fold Change | P Value | Adjusted P Value (FDR) |
|---|---|---|---|---|---|---|---|
| LDB3 | LIM domain binding 3 | Ldb3-205 | ENSRNOT00000086627 | 707 | 1.2314 | 0.002 | 0.03 |
| Ldb3-206 | ENSRNOT00000081631 | 330 | –0.1636 | 0.007 | 0.07 | ||
| Ldb3-201 | ENSRNOT00000085414 | 679 | –1.2065 | 0.15 | 0.45 | ||
| Ldb3-203 | ENSRNOT00000083623 | 729 | 3.8914 | 0.31 | 0.65 | ||
| Ldb3-202 | ENSRNOT00000087521 | 771 | –0.4304 | 0.44 | 0.76 | ||
| KCNIP2 | Potassium voltage-gated channel interacting protein 2 | Kcnip2-201 | ENSRNOT00000086164 | 225 | –2.0340 | 9.55 × 10–6 | 4.65 × 10–4 |
| Kcnip2-202 | ENSRNOT00000024709 | 270 | 1.5962 | 1.93 × 10–5 | 8.32 × 10–4 | ||
| Kcnip2-203 | ENSRNOT00000024750 | 220 | 2.1061 | 0.0174 | 0.13 | ||
| MYOM1 | Myomesin 1 | Myom1-203 | ENSRNOT00000095102 | 1,592 | –1.4593 | 0.002 | 0.030 |
| Myom1-202 | ENSRNOT00000112958 | 1,689 | 1.6058 | 0.007 | 0.072 | ||
| Myom1-201 | ENSRNOT00000085362 | 1,676 | 1.5243 | 0.046 | 0.23 | ||
| NEXN | Nexilin | Nexn-201 | ENSRNOT00000111795 | 669 | –1.4136 | 8.23 × 10–9 | 8.22 × 10–7 |
| Nexn-202 | ENSRNOT00000110679 | 592 | 2.2290 | 4.59 × 10–4 | 0.011 | ||
| Nexn-203 | ENSRNOT00000100634 | 535 | 0.2620 | 8.29 × 10–4 | 0.017 | ||
| Nexn-207 | ENSRNOT00000099274 | 656 | 2.6718 | 0.001 | 0.020 | ||
| Nexn-206 | ENSRNOT00000066780 | 670 | 0.7464 | 0.026 | 0.170 | ||
| TPM1 | Tropomyosin 1 | Tpm1-208 | ENSRNOT00000024493 | 287 | 1.0537 | 4.06 × 10–5 | 0.002 |
| Tpm1-202 | ENSRNOT00000024575 | 284 | 0.9882 | 0.021 | 0.15 | ||
| Tpm1-205 | ENSRNOT00000024617 | 248 | –1.9824 | 0.026 | 0.17 | ||
| Tpm1-204 | ENSRNOT00000040808 | 281 | –1.8077 | 0.030 | 0.18 | ||
| Tpm1-206 | ENSRNOT00000112475 | 257 | 1.0082 | 0.045 | 0.23 | ||
| Tpm1-201 | ENSRNOT00000057641 | 326 | –0.8091 | 0.080 | 0.32 | ||
| Tpm1-210 | ENSRNOT00000099012 | 267 | 2.2215 | 0.16 | 0.46 | ||
| Tpm1-207 | ENSRNOT00000048044 | 284 | –0.1326 | 0.21 | 0.54 | ||
| Tpm1-203 | ENSRNOT00000085894 | 294 | 0.04213 | 0.67 | 0.88 | ||
| Tpm1-209 | ENSRNOT00000090288 | 300 | 0.5827 | 0.74 | 0.92 | ||
| TTN | Titin | Ttn-202 | ENSRNOT00000101577 | 26,931 | –0.9696 | 0.001 | 0.021 |
| Ttn-201 | ENSRNOT00000114553 | 35,375 | 1.1073 | 0.002 | 0.028 | ||
| Ttn-203 | ENSRNOT00000108121 | 5475 | –0.0054 | 0.20 | 0.54 |
Significant P values adjusted by an FDR of <5% are marked in bold.
Abbreviations as in Table 1.
Long-read cDNA sequencing reveals differential gene expression
In RBM20+/– rat atria, we identified 444 differentially expressed genes compared with WT rats, (Figures 2E and 2F). The altered gene expression was more pronounced in RBM20–/– rat atria, where we observed 585 genes with differential expression compared with WT rats (Figures 3E and 3F). Among the most significant changes in expression were an up-regulation of the MYL4 gene (log2 fold change: 1.17; P = 3.27 × 10–21) and of RYR2 (log2 fold change: 0.95; P = 1.47 × 10–11) in RBM20+/– rat atria, with similar findings in RBM20–/– rat atria. Gene set enrichment analysis indicated enrichment of altered expression in genes that are involved in mitochondrial and sarcomere function. Among the 10 most up-regulated gene sets in both RBM20+/– and RBM20–/– atria were cytoplasmic translation and cellular respiration (Figures 2F and 3F), whereas gene sets related to cell migration such as actin-cytoskeleton reorganization were down-regulated (Figure 2F and 3F). Notably, we also observed the gene set sarcomere organization among the most up-regulated pathways in RBM20–/– atria (Figure 3F) (P = 0.001). Results from differential gene expression and pathway analyses are summarized in the Supplemental Appendix.
Figure 2.
Electron Microscopy Imaging and Differential Gene Expression in RBM20+/– Rat Atria
(A, B) TEM images of left atria of a 3-month-old WT rat. Sarcomeres are well organized, with clearly defined Z-disks and I-bands. (C, D) TEM images from left atria of a 3-month-old RBM20+/– rat. In contrast to WT, Z-disks appear thin and fuzzy, and I-bands and M-bands are blurry and poorly defined. Mitochondria are abundant and show compromised structure. (E) A volcano plot of differentially expressed genes in RBM20+/– rat atria compared with WT rat atria. Genes shown in red are significantly up-regulated, and genes shown in blue are significantly down-regulated. (F) The 10 most up- and down-regulated gene sets by biological process in RBM20+/– rat atria compared with WT rat atria. The x-axis denotes normalized enrichment score, and the y-axis lists gene ontology pathways. Data points are colored by adjusted P value. TEM = transmission electron microscope; WT = wild type.
Figure 3.
Electron Microscopy Imaging and Differential Gene Expression in RBM20–/– Rat Atria
(A, B) TEM images of left atria of 3-month-old WT rats. Sarcomeres are well organized, with clearly defined Z-disks and I-bands. (C, D) TEM images from left atria of 3-month-old RBM20–/– rats. The images show disorganized sarcomeres and compromised mitochondrial structure. (E) A volcano plot of differentially expressed genes in RBM20–/– rat atria compared with WT rat atria. Genes shown in red are significantly up-regulated, and genes shown in blue are significantly down-regulated. (F) The 10 most up- and down-regulated gene sets by biological process in RBM20–/– rat atria compared with WT rat atria. The x-axis denotes normalized enrichment score, and the y-axis lists gene ontology pathways. Data points are colored by adjusted P value. Abbreviations as in Figure 2.
Altered atrial tissue structure in an RBM20-deficient rat model
We imaged atrial tissue samples from RBM20+/–, RBM20–/–, and WT rats, using transmission electron microscopy (Figure 2). Compared with WT rats, the tissue from RBM20+/– rats showed an overall tissue degradation, with disorganized sarcomere structures showing poorly defined M- and I-bands and thin, fuzzy Z-discs (Figures 2C and 2D). Interestingly, mitochondria appeared to be more abundant in RBM20+/– rats; however, their structure was less well defined, with a high number of the present mitochondria showing altered morphology, appearing bloated with poorly defined cristae and degraded membranes. A similar phenotype was observed in atria from RBM20–/– rats (Figures 3C and 3D).
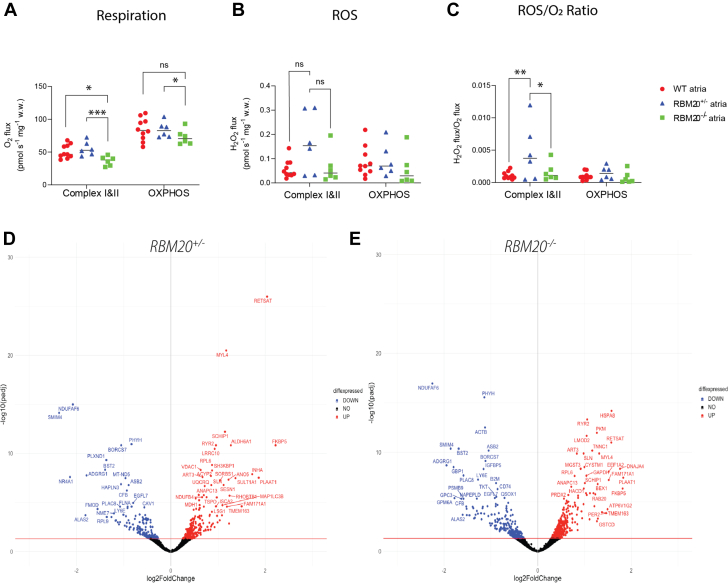
Impaired mitochondrial function in RBM20-deficient rats
Based on the transmission electron microscopy imaging, we followed up with analyses on mitochondrial function. Respiration and absolute formation of ROS (measured as hydrogen peroxide) were significantly affected in the RBM20-deficient rats compared with WT rats (group effect, respiration: P = 0.008; ROS: P = 0.01). The RBM20–/– atria showed significantly lower respiration in complexes I and II than the WT (P = 0.024) and the RBM20+/– atria (P < 0.001). Respiration with the maximal OXPHOS-linked substrates was also lower in the RBM20–/– atria compared with RBM20+/– atria (P = 0.042), although the difference was not statistically significant compared with WT atria (P = 0.075) (Figure 4A). We observed no difference in the absolute ROS level in RBM20+/– and RBM20–/– rats compared to WT (complexes I and II: 0.058 vs RBM20+/– 0.16 pmol·s–1·mg w/v–1 and vs RBM20–/– 0.064 pmol·s–1·mg w/v–1; P = 0.058 and P = 0.90, respectively. OXPHOS: RBM20+/– 0.093 and RBM20–/– 0.056 vs 0.086 pmol·s–1·mg w.w.–1; P = 0.90 and P = 0.70, respectively) (Figure 4B). However, when the formation was expressed relatively as ROS formation per oxygen used by the mitochondria, ROS/O2, RBM20+/– rats were found to have a significantly higher ROS/O2 ratio than both the WT and RBM20–/– rats with complexes I and II–linked substrates (0.0046 vs WT 0.0011 pmol·s–1·mg w.w.–1 and vs RBM20–/– 0.0016 pmol·s–1·mg w.w.–1; P = 0.001 and P = 0.011, respectively) (Figure 4C). This difference in ROS/O2 was no longer observed at maximal OXPHOS (RBM20+/– 0.0015 vs WT 0.00098 and RBM20–/– 0.00077 pmol·s–1·mg w.w.–1; both P = 0.90).
Figure 4.
Mitochondrial Dysfunction in RBM20+/– and RBM20–/– Rat Atria
(A) Significantly decreased mitochondrial respiration in RBM20–/– rat atria compared with RBM20+/– and WT rat atria. (B) A tendency toward higher ROS formation in RBM20+/– rat atria compared with WT rat atria (not significant). (C) Increased formation of mitochondrial ROS/O2 in RBM20+/– rat atria compared with RBM20–/– and WT rat atria. (D, E) Volcano plots of differentially expressed genes related to cellular respiration in RBM20+/– and RBM20–/– rat atria, respectively. Significantly up-regulated genes are depicted in red, and down-regulated genes are depicted in blue. Group differences were analyzed using a linear mixed model with the Tukey post hoc test, and values are reported as mean ± SEM. ∗∗∗P < 0.001, ∗∗P < 0.01, ∗P < 0.05. NS (not significant) indicates P > 0.05. diffexpressed = differentially expressed; OXPHOS = maximal oxidative phosphorylation; ROS = reactive oxygen species; SWT = wild type.
Reduced splicing activity in human RBM20 variants
To examine the effects on TTN splicing in human RBM20 variants, we constructed a series of human RBM20 single nucleotide base exchange mutants (point mutants) based on the variants identified in our early-onset AF cohort. Here, we included all rare variants (minor allele frequency: <0.1%) predicted to be deleterious in in silico analyses (combined annotation-dependent depletion score: >20). (Figures 5A and 5B).
Figure 5.
Rare RBM20 Variants Identified in an Early-Onset AF Cohort Affect Splicing Activity
(A to D) RBM20 variants mediate splicing repression on titin-derived splicing reporter in cell culture. (A) Schematic representation of (human) RBM20 and the single nucleotide base exchange mutants (point mutants). The 2 zinc finger (ZnF) domains are depicted in blue, the RNA recognition motif (RRM) is in green, and the serine-rich domain-like domain is in orange. Unstructured regions are indicated as small gray boxes. RBM20 amino acid residues are indicated underneath the horizontal line in the scheme, and the vertical lines show changes in amino acid residue and their approximate position in the protein sequence. (B) Table of rare variants in RBM20 identified in our early-onset AF cohort. (C) Real-time reverse transcription polymerase chain reaction (SYBR Green, Thermo Fisher Scientific) analysis of HEK293 cells transfected with the TTN241-3 splicing reporter and the mutant RBM20 expression constructs; n = 3; ∗∗∗P < 0.001. (D) WesTM (ProteinSimple) analysis of RBM20 mutants, transfected into HEK293 cells, using anti-RBM20 antibody and anti-β-tubulin as the loading control. Group comparisons were analyzed by 1-way analysis of variance. AF = atrial fibrillation; HEK293 = human embryonic kidney 293 cells; NA = not available; refSNV (formerly SNP) = reference single-nucleotide variation.
Quantitative reverse transcription polymerase chain reaction (analysis showed that all 3 LOF variants (RBM20-E725Vfs∗34, RBM20-E787∗, and RBM20-Q345∗) led to significantly reduced splicing activity (Figure 5C). The missense variants did not show a significant impact on protein expression, except for the RBM20-P411L variant, which showed a modest reduction of RBM20 expression. The 4 variants that displayed reduced splicing activity also resulted in significantly reduced protein expression levels. The RBM20-Q345∗ variant was not detected because of a lack of expression of the C-terminal part of the RBM20 protein, which is required for the anti-RBM20 antibody to bind.
Discussion
In this study, we used targeted next-generation sequencing and whole-exome data to uncover a novel association between LOF in RBM20 and AF in 2 independent cohorts. In a cohort of highly selected early-onset AF patients, the prevalence of LOF variants in RBM20 was significantly higher in individuals with AF compared with population-based control individuals. In the UK Biobank, an independent population-based cohort consisting of approximately 400,000 exomes, AF was similarly associated with a higher burden of rare LOF variants in RBM20.
Studies have shown RBM20 to be an important regulator of splicing of numerous cardiac genes, including key sarcomere genes (eg, TTN, TPM1, LDB3) and genes handling intracellular Ca2+ dynamics (eg, CACNA1C, CAMK2D, RYR2).9,10 We hypothesized that altered splicing resulting from a loss of RBM20 function would affect atrial structure and function, resulting in an atrial cardiomyopathy phenotype. To examine this systematically, we conducted electron microscopy imaging and long-read cDNA sequencing of atrial tissue from a rat model with loss of RBM20. Electron microscopy imaging revealed substantial changes in tissue integrity, including sarcomere structure and altered mitochondrial morphology, while long-read cDNA sequencing revealed differential transcript use in several putative splice targets of RBM20.
In RBM20+/–, atria we observed altered splicing of TPM1 and LDB3. TPM1 encodes the protein tropomyosin 1, which is important in thin filament function and actin-myosin interaction in sarcomeres.40 Pathogenic variants in the gene have previously been associated with cardiomyopathy41 and reduced sarcomere content.42 Furthermore, alternative splicing of TPM1 isoforms has been observed in patients with heart failure and cardiomyopathy.43 LDB3, which encodes the LIM domain binding 3 protein, ensures the structural integrity and stability of the sarcomere Z-disks, as evidenced by the disruption of Z-disks seen in mice with LDB3 mutations.44 Specifically, RBM20 regulates the inclusion of the cardiac isoform–specific exon 4, whereas loss of RBM20 function leads to the inclusion of exons 5 and 6 instead, normally only expressed in LDB3 isoforms of skeletal muscle.9 In line with these results, our data showed a down-regulation of the short cardiac isoform of LDB3 in RBM20+/– rat atria (ENSRNOT00000081631).
We found more profound effects on splicing in RBM20–/– atria, with altered splicing of the sarcomere genes TPM1, LDB3, NEXN, TTN, MYOM1, and KCNIP2, which encodes an ion-channel interacting protein. Numerous isoforms were differentially expressed in NEXN, which encodes the crucial Z-disk protein nexilin. Mutations in the NEXN gene have previously been associated with cardiomyopathy.45 Myomesin 1, the protein product of MYOM1, is part of the M-band and is important for sarcomere integrity.46 Alternative splicing of MYOM1 is known to affect the stability of sarcomere M-bands,47 and an up-regulation of a fetal isoform of myomesin 1 has been linked with dilated cardiomyopathy (DCM).48
The key sarcomere protein titin, encoded by TTN, plays an essential role in the sarcomere contractile function. Changes in titin isoform composition modulate cardiac elasticity and stiffness,49 and protein-truncating variants in titin have been associated with early-onset AF.7 In RBM20–/– atria, we observed a down-regulation of the short isoform of titin (ENSRNOT00000101577) and up-regulation of the long isoform (ENSRNOT00000114553). A similar switch from short to long titin isoforms has previously been reported in human DCM hearts.47
At the gene expression level, we also found differential expression of several genes involved in sarcomere function, such as MYL4, TNNT2, TCAP, and SYNPO2L. We speculate that the changes in gene expression of these genes may be part of a compensatory mechanism. Collectively, these changes in gene expression and isoform composition may explain the sarcomere degradation found in electron microscopy imaging, where we observed unorganized thick and thin filaments, fuzzy Z-discs, and blurred M-lines.
The mitochondrial disarray and differential expression of numerous genes involved in cellular respiration and OXPHOS prompted us to perform analyses of mitochondrial respiration and ROS formation. Interestingly, we identified an increased formation of mitochondrial ROS/O2 in RBM20+/– rats, probably because of altered proton or electron leak and/or impaired antioxidant defense. In RBM20–/– rats, we observed similar evidence of compromised mitochondrial function, indicated by significantly lowered mitochondrial respiration. In a recent study on RNA sequencing data from cardiac tissue in humans with DCM, titin-truncating variants were reported to alter mitochondrial energetics.50 We propose that altered TTN isoform composition could also influence mitochondrial function in loss of RBM20.
The mitochondrial genes found to be differentially expressed in both RBM20+/– and RBM20–/– atria include the gene that codes for mitochondrial complex I (MT-ND5). Complex I, the largest complex in the respiratory chain, is the first enzyme in the respiratory chain and plays a major role in mitochondrial metabolism.51 Mutations in MT-ND5 have been shown to lead to lower complex I activity, consequently resulting in elevated ROS.52 A similar elevated ROS formation has been seen in a cancer model with a mutation in the MT-ND3 gene,53 which was also found to be differentially expressed in the RBM20+/– atria. Collectively, these factors could account for the significantly elevated ROS/O2 ratio found in the RBM20+/– atria and not in the RBM20–/– atria.
The additional differentially expressed mitochondrial gene found in RBM20–/– atria, COX7A1, could contribute to the observed worsened mitochondrial function compared to RBM20+/– atria. The COX7A1 gene codes for the cytochrome c oxidase (complex IV), the terminal component of the mitochondrial respiratory chain, and is generally accepted as the major regulator for OXPHOS.54 Thus, our observed differential expression of mitochondrial genes would have a definite impact on the electron transfer processes and the electron transport chain integrity, resulting in proton and/or electron leak, and may explain the subsequent overwhelming of the mitochondrial antioxidant defense system. Recent studies have implicated mitochondrial dysfunction and oxidative stress in the pathogenesis of heart disease and AF55,56 and found that increased oxidative stress may affect cardiomyocyte function through ROS-mediated modifications of sarcomere proteins, compromising cardiac contractility.57
One of the most significant changes in gene expression we observed was an up-regulation of RYR2 expression in both RBM20+/– and RBM20–/– rats. The RYR2 gene encodes the ryanodine receptor 2 channels (RYR2 channels). We speculated that this may be a compensatory mechanism in response to altered Ca2+ dynamics in dysfunctional sarcomeres. However, this could also be caused by increased oxidation of RYR2 channels by ROS. Previous studies have reported that RYR2 channels are susceptible to oxidation, inducing a conformational change that leads to a higher open probability of the channel and, thus, increased Ca2+ leak.58 Such Ca2+ leak and altered intracellular Ca2+ dynamics have been implicated in the pathogenesis of early afterdepolarizations,59 an important mechanism in the initiation of AF and other arrhythmias. Our results indicated differential expression of several genes involved in intracellular calcium handling (Figures 6C and 6D). Calcium sparks and calcium waves initiated by disturbed Ca2+ handling may facilitate early afterdepolarizations,60 mainly through an increase in the sodium-calcium exchanger current (INCX), which acts as a depolarizing current that can facilitate arrhythmia60 (Figure 6E). This could be compounded by the alternative splicing observed in KCNIP2, because the protein encoded by the gene has been reported to play an important role in cardiac electrical stability and arrhythmogenesis.61
Figure 6.
Potential Mechanisms of Arrhythmia
(A, B) Volcano plots of differentially expressed genes related to sarcomere function in RBM20+/– and RBM20–/– rat atria, respectively. (C, D) Volcano plots of differentially expressed genes related to intracellular calcium handling in RBM20+/– and RBM20–/– rat atria, respectively. Significantly up-regulated genes are depicted in red, and down-regulated genes are depicted in blue. (E) Hypothesized pathophysiologic mechanisms of arrhythmia in loss of RBM20. Electron microscopy, mitochondrial function analyses, and complementary DNA sequencing revealed sarcomere dysfunction and increased oxidative stress in a loss-of-RBM20 model. Ryanodine receptor 2 channels (RYR2) in the sarcoplasmic reticulum membrane play an essential role in intracellular Ca2+ dynamics. Oxidation of these channels leads to a Ca2+ leak and altered intracellular calcium dynamics. This increases the depolarizing current of the sodium calcium exchanger (NCX), facilitating AF through early afterdepolarizations. AF = atrial fibrillation; diffexpressed = differentially expressed.
Finally, we explored the effects of splicing in all potentially deleterious RBM20 variants detected in AF patients. Using point mutants in cell models, we observed that all LOF variants significantly reduced splicing activity. Most missense variants, except RBM20-P411L, did not seem to significantly affect splicing. Based on our findings, we hypothesize that rare LOF variants in RBM20 may lead to changes in the splicing of key sarcomere genes, resulting in a dysfunctional contractile function of cardiomyocytes. This may, in turn, increase stress on the metabolic systems of the cardiomyocyte such as the mitochondria, as indicated by the increased formation of ROS/O2. Finally, altered intracellular Ca2+ dynamics, facilitated by altered calcium handling and leak through oxidized and up-regulated RYR2 channels, may induce a proarrhythmic condition, which, in turn, may lead to a vicious, self-reinforcing cycle of AF and atrial dysfunction. Our findings are in line with previous studies that report a higher incidence of atrial arrhythmias in human patients and mouse models with DCM caused by missense variants in RBM20.62,63
Study limitations
Whereas our study provided novel insights into plausible mechanisms of arrhythmia and atrial cardiomyopathy in individuals with rare variants in RBM20, our approach also had some limitations.
First, our results may have limited generalizability across ancestry groups because of the inclusion of individuals of European ancestry only. Second, we do not have phenotypic information on the referent populations from gnomAD and SweGen. Thus, some of the control individuals may have AF, although this would lead to bias toward no association. The in-house cohort of early-onset AF patients, although highly selected and well defined, was also of medium size and not representative of the general population. Moreover, the number of individuals identified in the in-house cohort with RBM20 LOF mutations is small. Replicating our findings in the UK Biobank may have broadened the implications of our findings. Furthermore, because RBM20 is a known splicing regulator, we performed long-read sequencing of cDNA to enable the study of the entire transcriptome, which also affected read depth compared with short-read sequencing. In turn, this could negatively influence our power to detect differentially expressed genes and transcripts, and our results are therefore not exhaustive. Finally, imaging, cDNA sequencing, pathway analyses, and mitochondrial analyses were conducted on a rat model, in which we had only a very limited number of animals available, which may also have negatively affected power.
Conclusions
To our knowledge, our results provide the first comprehensive evidence of a link between rare LOF variants in the cardiac splicing regulator RBM20 and AF. Our experimental data suggest that loss of RBM20 function in the atria may lead to sarcomere and mitochondrial dysfunction as mechanisms underlying the development of atrial arrhythmia in individuals with such variants. Our findings contribute to a more complete understanding of the complex genetics of AF and offer new insights into the role of alternative splicing in cardiac arrhythmia.
Perspectives.
COMPETENCY IN MEDICAL KNOWLEDGE: AF is a complex phenotype with multifaceted pathophysiologic mechanisms. Our results link the RBM20 gene with AF and highlight that atrial cardiomyopathy may be an underlying driver of arrhythmia in some individuals presenting with AF. These findings expand our knowledge of the genetic component of AF and may contribute to possible future genetic risk stratification in individuals with early onset of AF.
TRANSLATIONAL OUTLOOK: Alternative splicing has been reported to play an important role in the form and function of the ventricles. Our study illustrates how changes in isoform expression may also act as a proarrhythmic mechanism in the atria. Future studies into RBM20 and other splicing regulators may identify novel therapeutic targets in atrial arrhythmia and remodeling.
Funding Support and Author Disclosures
This work was supported by the Novo Nordisk Foundation, John and Birthe Meyer Foundation, Research Foundation of the Heart Centre Rigshospitalet, Villadsen Family Foundation, Arvid Nilsson Foundation, Danish Council for Independent Research, Research Foundation at Rigshospitalet, Skibsreder Per Henriksens R., og hustrus fond, and Department of Clinical Medicine (University of Copenhagen). Dr Ahlberg has received a grant from the Novo Nordisk Foundation, BRIDGE–Translational Excellence Programme. Dr Gotthardt was supported by the Deutsche Forschungsgemeinschaft and the Leducq Foundation. Dr Svendsen has received a fee for participating in the Advisory Board (Medtronic); and speaker fees from Medtronic not related to this work. Dr Gotthardt has a consultancy agreement with River BioMedics; and has received speaker honoraria from Bayer not related to this work. Dr Olesen has received speaker fees from Biosense Webster not related to this work. All other authors have reported that they have no relationships relevant to the contents of this paper to disclose.
Acknowledgments
This research has been conducted using the UK Biobank resource under application number 43247. The authors thank all individuals for their participation in this study. The Visual Abstract and Figure 6E were created using BioRender.
Footnotes
The authors attest they are in compliance with human studies committees and animal welfare regulations of the authors’ institutions and Food and Drug Administration guidelines, including patient consent where appropriate. For more information, visit the Author Center.
Appendix
For supplemental material and tables, please see the online version of this paper.
Appendix
References
- 1.Kornej J., Börschel C.S., Benjamin E.J., Schnabel R.B. Epidemiology of atrial fibrillation in the 21st century. Circ Res. 2020;127:4–20. doi: 10.1161/CIRCRESAHA.120.316340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahlsson A., Manolis A.S., Casadei B., et al. 2016 ESC guidelines for the management of atrial fibrillation developed in collaboration with EACTS. Eur Heart J. 2016;37:2893–2962. doi: 10.1093/eurheartj/ehw210. [DOI] [PubMed] [Google Scholar]
- 3.Stewart S., Murphy N.F., Murphy N., Walker A., McGuire A., McMurray J.J.V. Cost of an emerging epidemic: an economic analysis of atrial fibrillation in the UK. Heart. 2004;90:286–292. doi: 10.1136/hrt.2002.008748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dobrev D., Nattel S. New antiarrhythmic drugs for treatment of atrial fibrillation. Lancet. 2010;375:1212–1223. doi: 10.1016/S0140-6736(10)60096-7. [DOI] [PubMed] [Google Scholar]
- 5.Roselli C., Chaffin M.D., Weng L.-C., et al. Multi-ethnic genome-wide association study for atrial fibrillation. Nat Genet. 2018;50:1225–1233. doi: 10.1038/s41588-018-0133-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nielsen J.B., Thorolfsdottir R.B., Fritsche L.G., et al. Biobank-driven genomic discovery yields new insight into atrial fibrillation biology. Nat Genet. 2018;50:1234–1239. doi: 10.1038/s41588-018-0171-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ahlberg G., Refsgaard L., Lundegaard P.R., et al. Rare truncating variants in the sarcomeric protein titin associate with familial and early-onset atrial fibrillation. Nat Commun. 2018;9:4316. doi: 10.1038/s41467-018-06618-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choi S.H., Weng L.-C., Roselli C., et al. Association between titin loss-of-function variants and early-onset atrial fibrillation. JAMA. 2018;320:2354–2364. doi: 10.1001/jama.2018.18179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guo W., Schafer S., Greaser M.L., et al. RBM20, a gene for hereditary cardiomyopathy, regulates titin splicing. Nat Med. 2012;18:766–773. doi: 10.1038/nm.2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maatz H., Jens M., Liss M., et al. RNA-binding protein RBM20 represses splicing to orchestrate cardiac pre-mRNA processing. J Clin Invest. 2014;124:3419–3430. doi: 10.1172/JCI74523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Makarenko I., Opitz C.A., Leake M.C., et al. Passive stiffness changes caused by upregulation of compliant titin isoforms in human dilated cardiomyopathy hearts. Circ Res. 2004;95:708–716. doi: 10.1161/01.RES.0000143901.37063.2f. [DOI] [PubMed] [Google Scholar]
- 12.Rix T.A., Riahi S., Overvad K., Lundbye-Christensen S., Schmidt E.B., Joensen A.M. Validity of the diagnoses atrial fibrillation and atrial flutter in a Danish patient registry. Scand Cardiovasc J. 2012;46:149–153. doi: 10.3109/14017431.2012.673728. [DOI] [PubMed] [Google Scholar]
- 13.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Van der Auwera G.A., Carneiro M.O., Hartl C., et al. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics. 2013;43:11.10.1–11.10.33. doi: 10.1002/0471250953.bi1110s43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ameur A., Dahlberg J., Olason P., et al. SweGen: a whole-genome data resource of genetic variability in a cross-section of the Swedish population. Eur J Hum Genet. 2017;25:1253–1260. doi: 10.1038/ejhg.2017.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Collins R.L., Brand H., Karczewski K.J., et al. A structural variation reference for medical and population genetics. Nature. 2020;581:444–451. doi: 10.1038/s41586-020-2287-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.R Core Team. R A language and environment for statistical computing. R Foundation for Statistical Computing. Accessed July 11, 2023. https://www.R-project.org/
- 18.Bycroft C., Freeman C., Petkova D., et al. The UK Biobank resource with deep phenotyping and genomic data. Nature. 2018;562:203–209. doi: 10.1038/s41586-018-0579-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karczewski K.J., Solomonson M., Chao K.R., et al. Systematic single-variant and gene-based association testing of thousands of phenotypes in 394,841 UK Biobank exomes. Cell Genom. 2022;2(9) doi: 10.1016/j.xgen.2022.100168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Backman J.D., Li A.H., Marcketta A., et al. Exome sequencing and analysis of 454,787 UK Biobank participants. Nature. 2021;599:628–634. doi: 10.1038/s41586-021-04103-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mbatchou J., Barnard L., Backman J., et al. Computationally efficient whole-genome regression for quantitative and binary traits. Nat Genet. 2021;53:1097–1103. doi: 10.1038/s41588-021-00870-7. [DOI] [PubMed] [Google Scholar]
- 22.Pirruccello J.P., Di Achille P., Nauffal V., et al. Genetic analysis of right heart structure and function in 40,000 people. Nat Genet. 2022;54:792–803. doi: 10.1038/s41588-022-01090-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pirruccello J.P., Bick A., Wang M., et al. Analysis of cardiac magnetic resonance imaging in 36,000 individuals yields genetic insights into dilated cardiomyopathy. Nat Commun. 2020;11:2254. doi: 10.1038/s41467-020-15823-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ahlberg G., Andreasen L., Ghouse J., et al. Genome-wide association study identifies 18 novel loci associated with left atrial volume and function. Eur Heart J. 2021;42:4523–4534. doi: 10.1093/eurheartj/ehab466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cunningham F., Allen J.E., Allen J., et al. Ensembl 2022. Nucleic Acids Res. 2022;50:D988–D995. doi: 10.1093/nar/gkab1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics. 2018;34:3094–3100. doi: 10.1093/bioinformatics/bty191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Patro R., Duggal G., Love M.I., Irizarry R.A., Kingsford C. Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods. 2017;14:417–419. doi: 10.1038/nmeth.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Uhlén M., Fagerberg L., Hallström B.M., et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347 doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 29.Nowicka M., Robinson M.D. DRIMSeq: a Dirichlet-multinomial framework for multivariate count outcomes in genomics. F1000Res. 2016;5:1356. doi: 10.12688/f1000research.8900.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Anders S., Reyes A., Huber W. Detecting differential usage of exons from RNA-seq data. Genome Res. 2012;22:2008–2017. doi: 10.1101/gr.133744.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhu A., Ibrahim J.G., Love M.I. Heavy-tailed prior distributions for sequence count data: removing the noise and preserving large differences. Bioinformatics. 2019;35:2084–2092. doi: 10.1093/bioinformatics/bty895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wickham H. Springer; 2016. ggplot2: Elegant Graphics for Data Analysis. [Google Scholar]
- 34.Subramanian A., Tamayo P., Mootha V.K., et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sergushichev A.A. 2016. An algorithm for fast preranked gene set enrichment analysis using cumulative statistic calculation. bioRxiv. [Google Scholar]
- 36.Perry C.G.R., Kane D.A., Lin C.-T., et al. Inhibiting myosin-ATPase reveals dynamic range of mitochondrial respiratory control in skeletal muscle. Biochem J. 2011;437:215–222. doi: 10.1042/BJ20110366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Collins M.M., Ahlberg G., Hansen C.V., et al. Early sarcomere and metabolic defects in a zebrafish pitx2c cardiac arrhythmia model. Proc Natl Acad Sci U S A. 2019;116:24115–24121. doi: 10.1073/pnas.1913905116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dauksaite V., Gotthardt M. Molecular basis of titin exon exclusion by RBM20 and the novel titin splice regulator PTB4. Nucleic Acids Res. 2018;46:5227–5238. doi: 10.1093/nar/gky165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol. 1995;57:289–300. [Google Scholar]
- 40.England J., Granados-Riveron J., Polo-Parada L., et al. Tropomyosin 1: multiple roles in the developing heart and in the formation of congenital heart defects. J Mol Cell Cardiol. 2017;106:1–13. doi: 10.1016/j.yjmcc.2017.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hershberger R.E., Givertz M.M., Ho C.Y., et al. Genetic evaluation of cardiomyopathy—a Heart Failure Society of America practice guideline. Jour Card Fail. 2018;24:281–302. doi: 10.1016/j.cardfail.2018.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dorsch L.M., Kuster D.W.D., Jongbloed J.D.H., et al. The effect of tropomyosin variants on cardiomyocyte function and structure that underlie different clinical cardiomyopathy phenotypes. Int J Cardiol. 2021;323:251–258. doi: 10.1016/j.ijcard.2020.08.101. [DOI] [PubMed] [Google Scholar]
- 43.Rajan S., Jagatheesan G., Karam C.N., et al. Molecular and functional characterization of a novel cardiac-specific human tropomyosin isoform. Circulation. 2010;121:410–418. doi: 10.1161/CIRCULATIONAHA.109.889725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pathak P., Blech-Hermoni Y., Subedi K., et al. Myopathy associated LDB3 mutation causes Z-disc disassembly and protein aggregation through PKCα and TSC2-mTOR downregulation. Commun Biol. 2021;4:1–15. doi: 10.1038/s42003-021-01864-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang H., Li Z., Wang J., et al. Mutations in NEXN, a Z-disc gene, are associated with hypertrophic cardiomyopathy. Am J Hum Genet. 2010;87:687–693. doi: 10.1016/j.ajhg.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lamber E.P., Guicheney P., Pinotsis N. The role of the M-band myomesin proteins in muscle integrity and cardiac disease. J Biomed Sci. 2022;29:18. doi: 10.1186/s12929-022-00801-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Weeland C.J., van den Hoogenhof M.M., Beqqali A., Creemers E.E. Insights into alternative splicing of sarcomeric genes in the heart. J Mol Cell Cardiol. 2015;81:107–113. doi: 10.1016/j.yjmcc.2015.02.008. [DOI] [PubMed] [Google Scholar]
- 48.Schoenauer R., Emmert M.Y., Felley A., et al. EH-myomesin splice isoform is a novel marker for dilated cardiomyopathy. Basic Res Cardiol. 2011;106:233–247. doi: 10.1007/s00395-010-0131-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu Y., Bell S.P., Trombitas K., et al. Changes in titin isoform expression in pacing-induced cardiac failure give rise to increased passive muscle stiffness. Circulation. 2002;106:1384–1389. doi: 10.1161/01.cir.0000029804.61510.02. [DOI] [PubMed] [Google Scholar]
- 50.Verdonschot Job A.J., Wang Ping, Derks Kasper W.J., et al. Clustering of cardiac transcriptome profiles reveals unique subgroups of dilated cardiomyopathy patients. J Am Coll Cardiol Basic Trans Science. 2023;8:406–418. doi: 10.1016/j.jacbts.2022.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sharma L.K., Lu J., Bai Y. Mitochondrial respiratory complex I: structure, function and implication in human diseases. Curr Med Chem. 2009;16:1266–1277. doi: 10.2174/092986709787846578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Singh R.K., Saini S., Verma D., Kalaiarasan P., Bamezai R.N.K. Mitochondrial ND5 mutation mediated elevated ROS regulates apoptotic pathway epigenetically in a P53 dependent manner for generating pro-cancerous phenotypes. Mitochondrion. 2017;35:35–43. doi: 10.1016/j.mito.2017.05.001. [DOI] [PubMed] [Google Scholar]
- 53.Singh R.K., Srivastava A., Kalaiarasan P., Manvati S., Chopra R., Bamezai R.N.K. mtDNA germ line variation mediated ROS generates retrograde signaling and induces pro-cancerous metabolic features. Sci Rep. 2014;4:6571. doi: 10.1038/srep06571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kadenbach B., Hüttemann M., Arnold S., Lee I., Bender E. Mitochondrial energy metabolism is regulated via nuclear-coded subunits of cytochrome c oxidase. Free Radic Biol Med. 2000;29:211–221. doi: 10.1016/s0891-5849(00)00305-1. [DOI] [PubMed] [Google Scholar]
- 55.Xie W., Santulli G., Reiken S.R., et al. Mitochondrial oxidative stress promotes atrial fibrillation. Sci Rep. 2015;5 doi: 10.1038/srep11427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Peoples J.N., Saraf A., Ghazal N., Pham T.T., Kwong J.Q. Mitochondrial dysfunction and oxidative stress in heart disease. Exp Mol Med. 2019;51:1–13. doi: 10.1038/s12276-019-0355-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Steinberg S.F. Oxidative stress and sarcomeric proteins. Circ Res. 2013;112:393–405. doi: 10.1161/CIRCRESAHA.111.300496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dridi H., Kushnir A., Zalk R., Yuan Q., Melville Z., Marks A.R. Intracellular calcium leak in heart failure and atrial fibrillation: a unifying mechanism and therapeutic target. Nat Rev Cardiol. 2020;17:732–747. doi: 10.1038/s41569-020-0394-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li N., Wehrens X.H.T. Extinguishing intracellular calcium leak—a promising anti-arrhythmic approach. Heart Rhythm. 2013;10:108–109. doi: 10.1016/j.hrthm.2012.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Qu Z., Xie L.-H., Olcese R., et al. Early afterdepolarizations in cardiac myocytes: beyond reduced repolarization reserve. Cardiovasc Res. 2013;99:6–15. doi: 10.1093/cvr/cvt104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nassal D.M., Wan X., Liu H., et al. KChIP2 is a core transcriptional regulator of cardiac excitability. Elife. 2017;6 doi: 10.7554/eLife.17304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Refaat M.M., Lubitz S.A., Makino S., et al. Genetic variation in the alternative splicing regulator RBM20 is associated with dilated cardiomyopathy. Heart Rhythm. 2012;9:390–396. doi: 10.1016/j.hrthm.2011.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ihara K., Sasano T., Hiraoka Y., et al. A missense mutation in the RSRSP stretch of Rbm20 causes dilated cardiomyopathy and atrial fibrillation in mice. Sci Rep. 2020;10 doi: 10.1038/s41598-020-74800-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.