Abstract
Background
Genome-wide association studies and candidate gene association studies have identified more than 180 genetic variants statistically associated with anthracycline-induced cardiotoxicity (AIC). However, the lack of functional validation has hindered the clinical translation of these findings.
Objectives
The aim of this study was to functionally validate all genes associated with AIC using human induced pluripotent stem cell–derived cardiomyocytes (hiPSC-CMs).
Methods
Through a systemic literature search, 80 genes containing variants significantly associated with AIC were identified. Additionally, 3 more genes with potential roles in AIC (GSTM1, CBR1, and ERBB2) were included. Of these, 38 genes exhibited expression in human fetal heart, adult heart, and hiPSC-CMs. Using clustered regularly interspaced short palindromic repeats/Cas9–based genome editing, each of these 38 genes was systematically knocked out in control hiPSC-CMs, and the resulting doxorubicin-induced cardiotoxicity (DIC) phenotype was assessed using hiPSC-CMs. Subsequently, functional assays were conducted for each gene knockout on the basis of hypothesized mechanistic implications in DIC.
Results
Knockout of 26 genes increased the susceptibility of hiPSC-CMs to DIC. Notable genes included efflux transporters (ABCC10, ABCC2, ABCB4, ABCC5, and ABCC9), well-established DIC-associated genes (CBR1, CBR3, and RAC2), and genome-wide association study–discovered genes (RARG and CELF4). Conversely, knockout of ATP2B1, HNMT, POR, CYBA, WDR4, and COL1A2 had no significant effect on the in vitro DIC phenotype of hiPSC-CMs. Furthermore, knockout of the uptake transporters (SLC28A3, SLC22A17, and SLC28A1) demonstrated a protective effect against DIC.
Conclusions
The present findings establish a comprehensive platform for the functional validation of DIC-associated genes, providing insights for future studies in DIC variant associations and potential mechanistic targets for the development of cardioprotective drugs.
Key Words: cardiomyocytes, doxorubicin, genomics, GWAS, human induced pluripotent stem cells
Central Illustration
Anthracyclines, primarily doxorubicin, constitute a key component in about 60% of cancer treatment regimens,1 with approximately 35% administered to patients with breast cancer. Despite their efficacy, doxorubicin-induced cardiotoxicity (DIC) occurs in a dose-dependent manner in about 9% of patients, and 98% of these cases emerge within the first year of treatment.2 DIC encompasses 4 major interrelated molecular mechanisms: 1) generation of reactive oxygen species (ROS); 2) mitochondrial dysfunction; 3) DNA damage involving TOP2B; and 4) calcium overload leading to sarcomere damage. These mechanisms collectively lead to cardiomyocyte death, clinically determined by troponin detection in peripheral blood and reduced left ventricular ejection fraction.
To date, 5 genome-wide association studies (GWAS)3, 4, 5, 6, 7 and 20 candidate gene association studies8 have identified 80 genes with single-nucleotide polymorphisms (SNPs) significantly linked to anthracycline-induced cardiotoxicity (AIC).8 Despite this, only 1 AIC-associated variant locus (in SLC28A3) has been independently replicated.9,10 Thus, the functional validation of AIC-associated variants remains a critical prerequisite before incorporating this information into clinical practice.
The AIC-associated genes can be categorized into 6 major groups on the basis of their hypothesized mechanistic function. 1) Genes associated with ROS production and handling: doxorubicin induces ROS generation predominantly by reduction in the mitochondria, producing semiquinone-producing superoxide (O2+) free radicals. 2) Genes related to DNA damage: doxorubicin inflicts DNA damage on cardiomyocytes either by direct intercalation with DNA or by disruption of DNA repair after cleavage by TOP2B. 3) Genes associated with iron uptake: doxorubicin, with a high affinity for iron, can alter iron metabolism through interactions with iron regulatory proteins, stabilizing transferrin transcripts and inhibiting the expression of iron-sequestering proteins.11 4) Genes associated with transporters controlling doxorubicin uptake and efflux: generally, variants in uptake transporters (ie, members of the solute carrier [SLC] family) are protective against AIC by reducing doxorubicin transport into cardiomyocytes.9,10,12,13 Conversely, variants in efflux transporters (adenosine triphosphate–binding cassette [ABC]) are linked to increased intracellular doxorubicin concentration and AIC.14 5) Genes involved in calcium handling: doxorubicin directly binds to ryanodine receptors, inducing calcium release from sarcoplasmic reticulum.15 It also enhances L-type calcium channel activity,16 leading to an increased level of intracellular calcium. 6) Genes related to altered electric currents in the cardiomyocytes: after doxorubicin treatment, this can result in impaired contractile function.17
Previously, we demonstrated that human induced pluripotent stem cell–derived cardiomyocytes (hiPSC-CMs) accurately recapitulate patient-specific cardiotoxic responses to doxorubicin.13,18,19 In the present study, we functionally validate the role of 38 genes associated with anthracycline cardiotoxicity in patients with cancer using clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9–based knockout (KO) in hiPSC-CMs, followed by an array of in vitro characterization assays. Our work confirms that 31 genes, identified in GWAS and candidate gene association studies, play mechanistic roles in DIC at the cellular level. This approach provides insights into the impact of each gene KO on DIC development and their potential mechanistic pathways.
Methods
Variant and gene candidate identification
Following a comprehensive PubMed search involving both original and review papers investigating genetic risk factors associated with DIC, we compiled a list of potential candidates. These candidates underwent testing for their expression levels in hiPSC-CMs, adult human heart, and fetal human heart19 (Supplemental Table 1).
Human induced pluripotent stem cell culture, CRISPR/Cas9-mediated KO generation, and differentiation to cardiomyocytes
For detailed information, refer to the Supplemental Methods. We used a control male hiPSC line with an exogenous TNNT2 promoter–driven phleomycin D1 resistance cassette for cardiomyocyte purification, as previously described.13 All protocols received approval from the Northwestern University Institutional Review Board. Pairs of CRISPR/Cas9 guide RNAs were designed with a separation of >50 bp to induce a large deletion within the earliest common exon of each gene. Supplemental Tables 2 to 4 display all used primers for single guide RNA expression vector generation, a list of potential off-targets, and sequencing primers. We selected 1 KO hiPSC line with the lowest expression of each gene, which was then differentiated into cardiomyocytes and assessed on day 30.
Statistical methods
Data are expressed as mean ± SEM. Comparisons were conducted using 1- or 2-way analysis of variance or unpaired 2-tailed Student’s t-tests, with significant differences defined as P < 0.05, P < 0.01, P < 0.001, and P < 0.0001.
For more detailed methods, refer to the Supplemental Appendix.
Results
DIC-associated gene prioritization, KO generation, and in vitro doxorubicin toxicity measurement
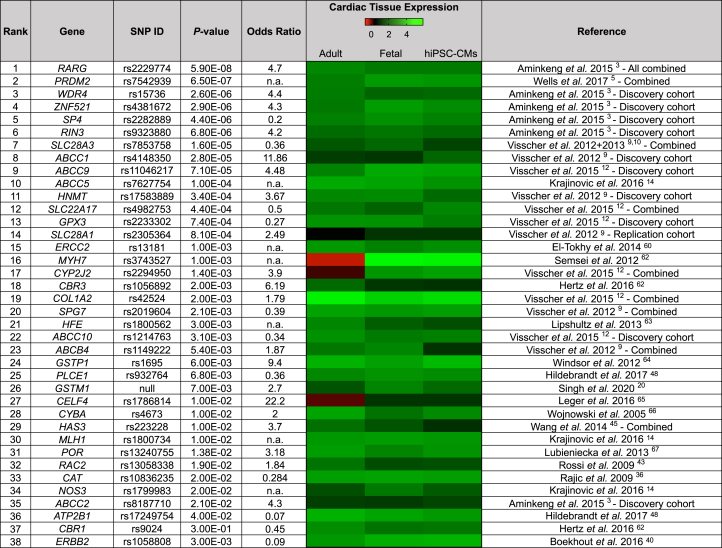
We compiled a table of 429 SNPs associated with DIC, with 180 being unique (Supplemental Table 1). From this meta-analysis, we identified 80 SNP-harboring genes significantly linked to AIC. Our bioinformatic analysis did not predict any gain of function resulting from these SNPs. Additionally, we included 3 other potential candidates: GSTM1, associated with DIC through deletion rather than a SNP;20 CBR1, considered one of the prototypical DIC-associated genes;21 and ERBB2, implicated in the cardiotoxicity of trastuzumab plus doxorubicin regimens.22 We assessed the expression of 83 genes using RNA sequencing in the fetal heart, adult human heart, and hiPSC-CMs. Subsequently, 38 genes consistently expressed (>10 transcripts per million for hiPSC-CMs) were chosen for KO generation (Figure 1, Supplemental Table 1).
Figure 1.
Prioritization of Doxorubicin-Induced Cardiotoxicity–Associated Loci
Table showing the 38 doxorubicin-induced cardiotoxicity–associated loci ranked on the basis of the single-nucleotide polymorphism (SNP) with highest P value from their respective publications.23, 24, 25, 26, 27, 28, 29, 30 The heat map of cardiac tissue expression shows the expression of anthracycline-induced cardiotoxicity–associated genes in adult human heart (n = 2), in fetal human heart (n = 2), and in human induced pluripotent stem cell–derived cardiomyocytes (hiPSC-CMs) (n = 7) by RNA sequencing. n = number of distinct patient-specific samples.
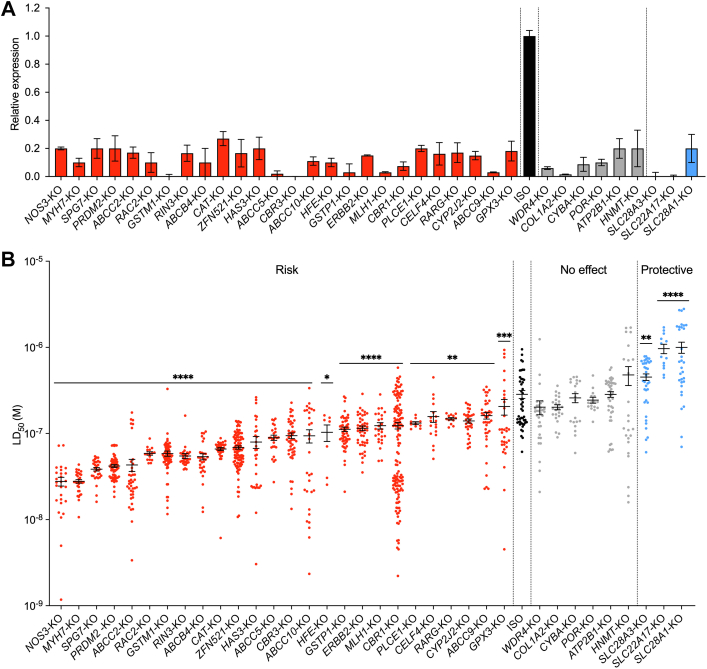
We generated KO hiPSC-CM lines for each of the 38 genes, with all guide RNA sequences listed in Supplemental Table 2. All KOs were validated using Sanger sequencing (Supplemental Figures 1 to 4, Supplemental Tables 4 and 5). The absence of the knocked-out protein was confirmed using western blot analysis (Supplemental Figures 5 and 6, Supplemental Tables 6 and 7). Additionally, attenuation of the KO gene in the KO lines was further confirmed using quantitative reverse transcriptase polymerase chain reaction (Figure 2A, Supplemental Table 8). Relevant SNPs contained in the control human induced pluripotent stem cells are detailed in Supplemental Table 9. Of the 38 KOs attempted, KO of 2 genes (ERCC2 and ABCC1) proved incompatible with hiPSC survival, and 1 gene (SP4) was incompatible with cardiac differentiation (Supplemental Table 1).
Figure 2.
Validation of KOs of 36 Genes Significantly Associated With Doxorubicin-Induced Cardiotoxicity
(A) Validation of successful clustered regularly interspaced short palindromic repeats/Cas9–based gene knockout (KO) in human induced pluripotent stem cell–derived cardiomyocytes (hiPSC-CMs) using quantitative reverse transcriptase polymerase chain reaction. The data presented here are the relative expression of each studied gene in its own KO lines relative to isogenic control (ISO). (B) Effect of gene KOs on hiPSC-CM viability after doxorubicin treatments (72 hours). Each data point represents 1 median lethal dose (LD50) calculation on the basis of an individual experimental replicate derived from a 5-log doxorubicin dosing. Central bar represents mean. Error bars represent ±SEM. P values (∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001, and ∗∗∗∗P < 0.0001) are derived from differences between gene KO and ISO (Mann-Whitney U test). Raw dose-response curves from which these LD50 values are derived are provided in Supplemental Figures 4 to 6.
Next, the remaining 35 KO hiPSC lines were differentiated into cardiomyocytes and exposed to 5-log doses of doxorubicin (10−8 to 10−4 M) for 72 hours, followed by viability assessment to establish the dose required to kill 50% of the cells (median lethal dose [LD50]) (Figure 2B). KO of the uptake drug transporters SLC28A3, SCL22A17, and SLC28A1 increased viability after treatments with doxorubicin, with LD50 values between 6.35 and 11.2 μM compared with 3.78 μM in isogenic control hiPSC-CMs (Figure 2B, Supplemental Figure 7). KO of ATP2B1, HMNT, POR, CYBA, WDR4, and COL1A2 did not alter viability after doxorubicin treatments (Figure 2B, Supplemental Figure 8). KO of the rest of the genes (n = 26) increased the hiPSC-CMs’ sensitivity to doxorubicin (Figure 2B, Supplemental Figures 9 and 10). LD50 values for KOs with increased sensitivity to doxorubicin ranged between 0.32 μM (NOS3-KO and MYH7-KO cardiomyocytes; P < 0.0001) and 2 μM (GSTM1-KO cardiomyocytes; P = 0.0018) compared with 3.78 μM for isogenic control hiPSC-CMs.
Functional validation of DIC-associated genes
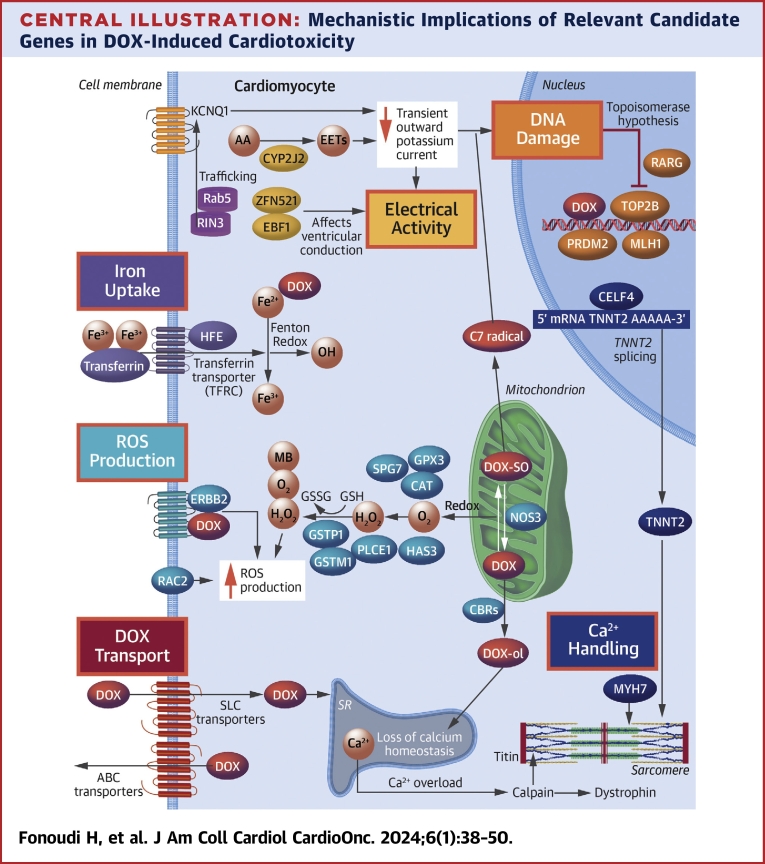
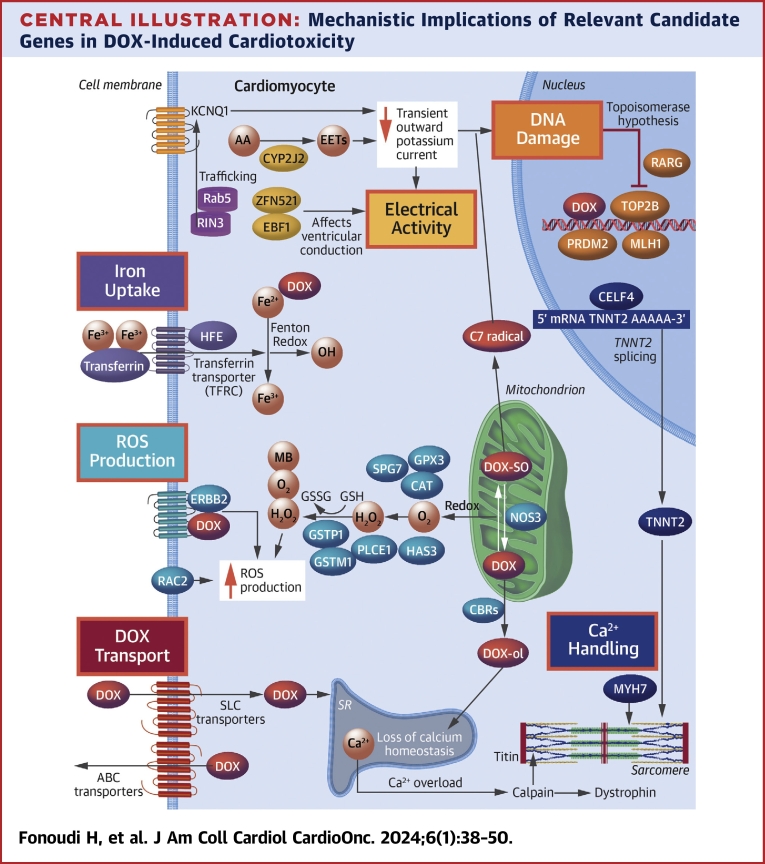
Next, for each gene KO, we performed a functional study to investigate their potential mechanistic implications for DIC. We categorized the genes into 6 functional groups (Table 1) and adopted a functional assay relevant to their mechanisms of action (Central Illustration).
Table 1.
Mechanistic Implications of Genes Associated With DIC
| Group | Gene | First Author (Year) Ref# | Mechanistic Implication |
|---|---|---|---|
| 1 | CAT | Rajic et al (2009)31 | ROS generation/handling32 |
| CBR1 | Armenian et al (2013)33 | ROS generation/handling34 | |
| CBR3 | Visscher et al (2012),9 Armenian (2013)33 | ROS generation/handling34 | |
| ERBB2 | Boekhout et al (2016)35 | ROS generation/handling36 | |
| GPX3 | Visscher et al (2015)12 | ROS generation/handling37 | |
| GSTM1 | Singh et al (2020)20 | ROS generation/handling | |
| GSTP | Visscher et al (2012),9 Rossi et al (2009)38 | ROS generation/handling39 | |
| HAS3 | Wang et al (2014)40 | ROS generation/handling41 | |
| NOS3 | Krajinovic et al (2016)14 | ROS generation/handling42 | |
| PLCE1 | Hildebrandt et al (2017)43 | ROS generation/handling44 | |
| RAC2 | Armenian et al (2013),33 Rossi et al (2009)38 | ROS generation/handling45 | |
| SPG7 | Visscher et al (2013)10 | ROS generation/handling46 | |
| 2 | PRDM2 | Wells et al (2017)5 | DNA damage47 |
| MLH1 | Krajinovic et al (2016)14 | DNA damage48 | |
| RARG | Aminkeng et al (2015)3 | DNA damage3 | |
| 3 | HFE | Armenian et al (2013)33 | Iron uptake and homeostasis49 |
| 4 | SLC22A17 | Visscher et al (2015)12 | DOX uptake13 |
| SLC28A1 | Visscher et al (2012)9 | DOX uptake50 | |
| SLC28A3 | Visscher et al (2012),9 Visscher et al (2013)10 | DOX uptake19 | |
| ABCB4 | Visscher et al (2012)9 | DOX efflux51 | |
| ABCC2 | Aminkeng et al (2015)3 | DOX efflux52 | |
| ABCC5 | Krajinovic et al (2016)14 | DOX efflux14 | |
| ABCC9 | Visscher et al (2015)12 | DOX efflux53 | |
| ABCC10 | Visscher et al (2015)12 | DOX efflux53 | |
| 5 | CELF4 | Wang et al (2016)6 | Calcium handling54 |
| MYH7 | Wasielewski et al (2014)55 | Calcium handling56 | |
| 6 | CYP2J2 | Visscher et al (2015)12 | Cardiac electrical activity57 |
| RIN3 | Aminkeng et al (2015)3 | Cardiac electrical activity58 | |
| ZFN521 | Aminkeng et al (2015)3 | Cardiac electrical activity59 |
DIC = doxorubicin-induced cardiotoxicity; DOX = doxorubicin; ROS = reactive oxygen species.
Central Illustration.
Mechanistic Implications of Relevant Candidate Genes in DOX-Induced Cardiotoxicity
The different mechanisms of cardiotoxicity after exposure to doxorubicin (DOX) are highlighted in colored boxes. AA = arachidonic acid; ABC = adenosine triphosphate–binding cassette; EET = epoxyeicosatrienoic acid; mRNA = messenger RNA; ROS = reactive oxygen species; SLC = solute carrier; SR = sarcoplasmic reticulum.
Genes involved in ROS production and handling
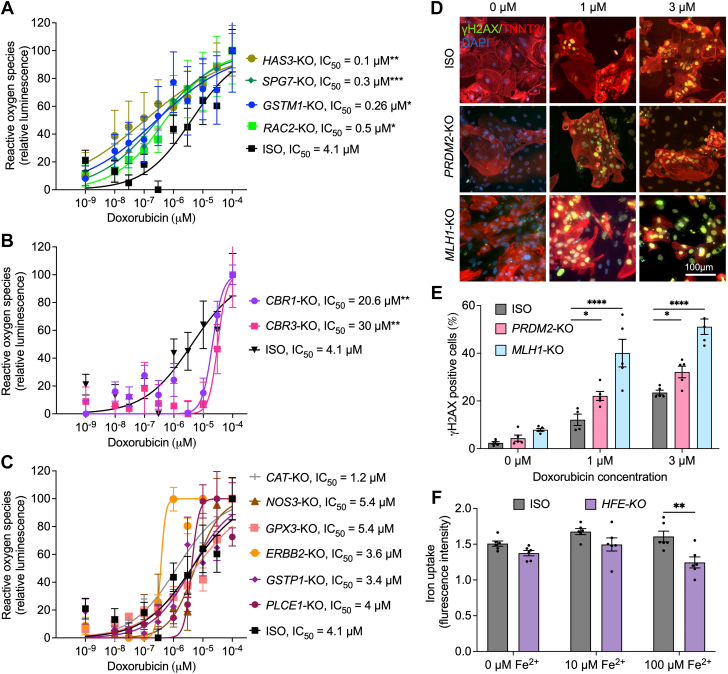
The largest group of genes in our list (12 of 35) is associated with ROS production and handling (Table 1). We assessed the H2O2 levels in hiPSC-CMs after 24 hours of doxorubicin treatment (Figures 3A to 3C). After doxorubicin treatment, our analysis indicated that ROS levels were lower in CBR1-KO (half maximal inhibitory concentration [IC50] = 20.6 mM; P = 0.0024) and CBR3-KO (IC50 = 31 mM; P = 0.0069) hiPSC-CMs compared with isogenic control hiPSC-CMs (IC50 = 4.1 mM) (Figure 3A). However, after doxorubicin treatment, ROS levels were increased in SPG7-KO (IC50 = 0.3 mM; P = 0.0006), HAS3-KO (IC50 = 0.1 mM; P = 0.0023), GSTM1-KO (IC50 = 0.26 mM; P = 0.039), and RAC2-KO (IC50 = 0.5 mM; P = 0.04) hiPSC-CMs (Figure 3B). No significant differences in ROS production were detected in PLCE1-KO, ERBB2-KO, CAT-KO, GSTP1-KO, NOS3-KO, and GPX3-KO hiPSC-CMs (Figure 3C).
Figure 3.
Reactive Oxygen Species Production, DNA Damage, and Iron Uptake
(A-C) Hydrogen peroxide levels measured by ROS-Glo assay (luminescence) in hiPSC-CMs after doxorubicin treatments (24 hours). (D) Representative images for γH2AX immunofluorescent staining in hiPSC-CMs after treatments with doxorubicin (24 hours, 1 and 3 μM). (E) Quantification of DNA damage on the basis of γH2AX staining in hiPSC-CMs using flow cytometry (ISO, n = 5; PRDM2-KO, n = 5; and MLH1-KO, n = 5). (F) Effect of HFE knockout on hiPSC-CM iron uptake (ISO, n = 6; and HFE-KO, n = 6) measured using calcein staining. Error bars represent ±SEM. n = full independent experimental replicates. ∗P < 0.05, ∗∗P ≤ 0.01, ∗∗∗P < 0.001, and ∗∗∗∗P < 0.0001 by Mann-Whitney U test (A-C) and 2-way analysis of variance (E and F). DAPI = 4′,6-diamidino-2-phenylindole; IC50 = half maximal inhibitory concentration; other abbreviations as in Figure 2.
Genes involved in DNA damage
RARG, PRDM2, and MLH1 were linked to DNA damage response3,19,47, 48, 60 (Table 1). Given our comprehensive study of the role of RARG in DIC,19 we directed our focus to PRDM2 and MLH1 to quantify double-stranded DNA breaks in hiPSC-CMs after doxorubicin treatment. Our immunofluorescence analysis suggests increased γH2AX staining in PRDM2-KO and MLH1-KO compared with isogenic hiPSC-CMs (Figure 3D). The elevated levels of γH2AX+ cells were further confirmed using flow cytometry after 1- and 3-μM doxorubicin treatments (Figure 3E). PRDM2 (22.1% ± 1.88% [P = 0.0283] and 32.16% ± 2.43% [P = 0.047]) and MLH1 (40.07% ± 5.79% [P < 0.0001] and 51.11% ± 3.2% [P < 0.0001]) KOs expressed higher levels of γH2AX after 1- and 3-μM doxorubicin treatments compared with isogenic control hiPSC-CMs (12.1% ± 2.37% and 23.52% ± 0.98%, respectively).
Genes involved in iron uptake and homeostasis
HFE encodes the homeostatic iron regulator protein, which controls iron transport and metabolism. The intracellular iron disposition in cardiomyocytes was examined by measuring iron-calcein quenching. No significant differences in iron uptake were observed in the presence of 0 and 10 μM Fe2+. However, HFE-KO hiPSC-CMs exposed to 100 μM of Fe2+ exhibited significantly reduced iron uptake (Figure 3F).
Genes involved in doxorubicin uptake and efflux
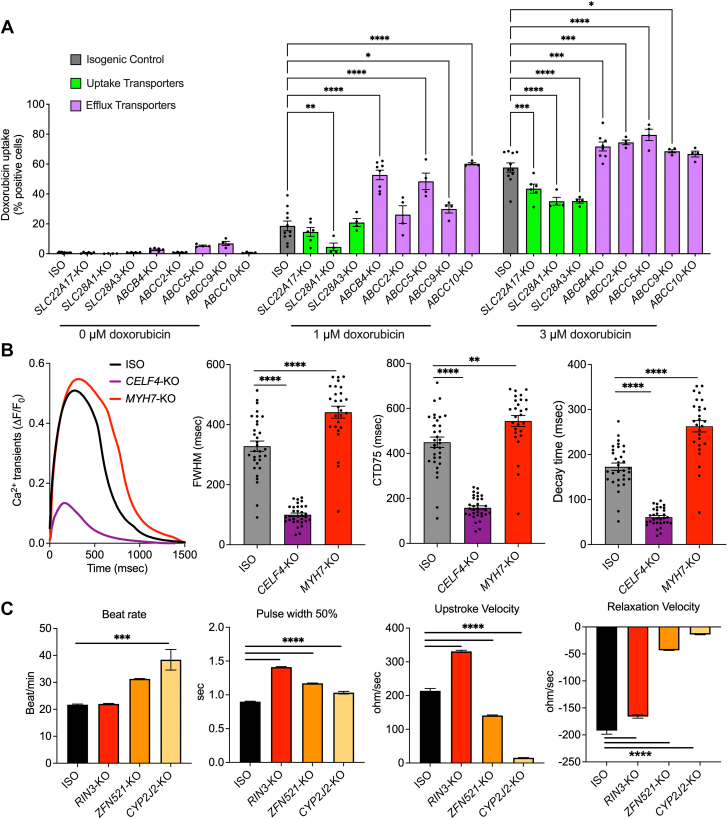
Doxorubicin uptake was assessed by measuring intracellular doxorubicin autofluorescence using flow cytometry after drug treatments.13 After the 1-μM doxorubicin treatment, SLC28A1-KO cardiomyocytes exhibited significantly reduced doxorubicin uptake (4.6% ± 2.5%; P = 0.0039) compared with isogenic control hiPSC-CMs (18.63% ± 3.24%) (Figure 4A). Conversely, doxorubicin uptake was significantly higher in ABCB4-KO (52.73% ± 3.2%; P < 0.0001), ABCC5-KO (48.45% ± 5.5%; P < 0.0001), ABCC9-KO (29.98% ± 2.7%; P = 0.0327), and ABCC10-KO hiPSC-CMs (60.18% ± 2.6%; P < 0.0001) (Figure 4A). After 1-μM treatments, no significant differences in doxorubicin uptake were observed in SLC22A17-KO, SLC28A3-KO, and ABCC2-KO hiPSC-CMs (Figure 4A).
Figure 4.
Doxorubicin Uptake, Calcium Handling, and Contractility
(A) Doxorubicin uptake in hiPSC-CMs with knockouts of SLC and ABC transporters (isotype, n = 11; SLC28A3-KO, n = 4; SLC22A17-KO, n = 6; SLC28A1-KO, n = 4; ABCC2-KO, n = 4; ABCB4-KO, n = 7; ABCC5-KO, n = 4; ABCC9-KO, n = 4; and ABCC10-KO, n = 4). n = full independent experimental replicates. (B) Effect of knocking out MYH7 and CELF4 on calcium transients. Left: representative calcium transients. Middle left: full width at half maximum (FWHM). Middle right: calcium transient duration 75% (CTD75). Right: decay time. (C) Contractility analysis using impedance measurement in RIN3-KO, ZFN521-KO, and CYP2J2-KO hiPSC-CMs (ISO, n = 41; RIN3-KO, n = 48; ZFN521-KO, n = 48; and CYP2J2-KO, n = 89). n = number of assessed wells of 96-well plate derived from at least 3 independent rounds of differentiation (B and C). Error bars represent ±SEM. ∗P < 0.05, ∗∗P ≤ 0.01, ∗∗∗P < 0.001, and ∗∗∗∗P < 0.0001 by 2-way analysis of variance (A) and 1-way analysis of variance (B and C). Abbreviations as in Figure 2.
Upon increasing the doxorubicin concentration to 3 μM, all the cells, except for ABCC10-KO, exhibited significant differences in doxorubicin uptake compared with control. Doxorubicin uptake was significantly lower in SLC22A17-KO (43.55% ± 3.04%; P = 0.0005), SLC28A1-KO (35.11% ± 2.63%; P < 0.0001), and SLC28A3-KO (35.24% ± 1.36%; P < 0.0001) hiPSC-CMs compared with isogenic control hiPSC-CMs (57.68% ± 3.11%) (Figure 4A). Conversely, 3-μM doxorubicin treatment led to significantly higher doxorubicin uptake in ABCB4-KO (71.77% ± 3.02%; P = 0.0001), ABCC2-KO (74.52% ± 1.57%; P = 0.0003), ABCC5-KO (79.52% ± 3.79%; P < 0.0001), and ABCC9-KO (68.57% ± 1.02%; P = 0.04) hiPSC-CMs (Figure 4A).
Genes involved in calcium handling
CELF4 and MYH7 play crucial roles in controlling cardiomyocyte function by affecting calcium transients. KO of CELF4 resulted in a significant shortening of full width at half maximum (99.89% ± 5.67%; P < 0.0001), calcium transient duration at 75% (158.04% ± 8.79%; P < 0.0001), and decay time (60.83% ± 3.5%; P < 0.0001) of calcium transients compared with isogenic control hiPSC-CMs (328.15% ± 17.27%, 449.41% ± 23.04%, and 172.73% ± 8.27%, respectively) (Figure 4B). On the contrary, KO of MYH7 resulted in a significant prolongation in full width at half maximum (441.44% ± 18.21%; P < 0.0001), calcium transient duration at 75% (543.82% ± 22.4%; P < 0.0023), and decay time (262.82% ± 11.56%; P < 0.0001) compared with isogenic control hiPSC-CMs (Figure 4B).
Gene involved in cardiac electric activity
We identified 3 genes, RIN3, ZFN521, and CYP2J2, associated with cardiac electric activity57, 58, 59 (Table 1). Impedance measurement revealed that KO of RIN3 increased the pulse width at 50% (1.41% ± 0.007% vs 0.89% ± 0.006%; P < 0.0001) and upstroke velocity (331% ± 3.37% vs 214% ± 7.02%; P < 0.0001) and reduced the relaxation velocity (−166% ± 2.94% vs −192% ± 6.74%; P < 0.0001), compared with isogenic control hiPSC-CMs (Figure 4B). KO of ZFN521 resulted in hiPSC-CMs with increased pulse width at 50% (1.17% ± 0.005%; P < 0.0001) and upstroke velocity (141% ± 1.55%; P < 0.0001) and reduced relaxation velocity (−43.2% ± 0.32%; P < 0.0001). KO of CYP2J2 resulted in increased beat rate (38.39% ± 3.82% vs 21.7% ± 0.26%; P < 0.0004) and pulse width at 50% (1.03% ± 0.01%; P < 0.0001) and decreased upstroke velocity (15.53% ± 0.5%; P < 0.0001) and relaxation velocity (−13.98% ± 0.58%; P < 0.0001), compared with isogenic control hiPSC-CMs (Figure 4B).
Discussion
Inter-individual cardiotoxic response to doxorubicin is variable, indicating that genetics plays an important role in DIC response. The functional validation of genes and variants associated with cardiotoxicity risk is essential to improve the outcome of anthracycline regimens and to develop cardioprotective treatments.
Knocking out DIC-related genes alters the hiPSC-CM response to doxorubicin treatment
Of the 35 genes studied, 3 gene KOs were protective of DIC (SLC28A3, SLC22A17, and SLC28A1), 6 gene KOs did not have a significant effect on DIC (ATP2B1, HNMT, POR, CYBA, WDR4, and COL1A2), and 26 KOs increased DIC (ABCC10, ABCC2, ABCB4, ABCC5, ABCC9, CAT, CBR1, CBR3, CYP2J2, ERBB2, GPX3, GSTM1, GSTP1, HAS3, HFE, MLH1, MYH7, NOS3, PLCE1, PRDM2, RAC2, RARG, RIN3, SPG7, CELF4, and ZFN521).
ROS production is the major mechanism causing DIC
The largest functional group studied comprised genes related to ROS production. H2O2 production analysis in cardiomyocytes exposed to doxorubicin revealed reduced production in CBR1-KO and CBR3-KO hiPSC-CMs, indicating a potential link to cardiotoxicity via reduced metabolism of doxorubicin to doxorubicinol by carbonyl reductases. In contrast, HAS3-KO, SPG7-KO, GSTM1-KO, and RAC2-KO exhibited higher ROS production and increased sensitivity to doxorubicin, indicating a role of these genes in H2O2 handling. Notably, no significant differences in H2O2 production were observed in CAT-KO, NOS3-KO, PLCE1-KO, GPX3-KO, ERBB2-KO, and GSTP1-KO. This might suggest that the DIC observed in the KO of these genes is due to the defects in the handling and detoxification of additional ROS products and/or participation in other mechanisms related to doxorubicin toxicity besides ROS generation. In fact, it has been shown that CAT, glutathione S-transferases, and glutathione peroxidases have played crucial roles in detoxifying metabolites generated during oxidative stress.61
DNA damage response is elevated in PRDM2-KO and MHL1-KO
RARG inhibits TOP2B, which binds to DNA and stabilizes the intermediate TOP2B-mediated double-stranded DNA breaks. KOs of the RARG gene activate this DNA damage pathway, leading to increased cardiac cell death. Similar mechanisms are thought to exist in PRDM2 and MLH1 gene KOs. We detected higher levels of DNA damage after doxorubicin treatments PRDM2-KO and MHL1-KO, suggesting a protective role for these genes against doxorubicin-induced DNA damage.
HFE-KO cardiomyocytes exhibit reduced iron uptake
Our results indicate reduced iron uptake in HFE-KO cardiomyocytes compared with isogenic control, particularly evident at high Fe2+ concentrations (100 μM). Considering the important role of iron in mitochondrial function and the energy-demanding nature of cardiomyocytes, this impaired iron uptake may contribute to the heightened DIC observed in HFE-KO cardiomyocytes, as evidenced by the exacerbated risk for heart failure in the context of iron deficiency.62
KOs of SLC transporters reduce and ABC transporters KOs increase the doxorubicin uptake
KOs of SLC family members in hiPSC-CMs showed reduced doxorubicin uptake compared with isogenic controls. In addition, KO of ABC family members in hiPSC-CMs resulted in an elevation of doxorubicin uptake. Our results indicate that SLC transporters play a crucial role in facilitating the influx of doxorubicin into the cardiomyocytes, thereby contributing to an increased risk for DIC. Conversely, ABC transporters are implicated in the efflux of doxorubicin out of the cells, reducing intracellular doxorubicin levels and establishing them as potential cardioprotective gene targets.
CELF4-KO and MYH7-KO hiPSC-CMs demonstrate altered calcium transients
CELF4 is involved in regulatory splicing events essential for the proper functioning of cardiac troponin T, which in turn plays an essential role in proper calcium signaling in cardiomyocytes.63 MYH7, a gene encoding myosin heavy chain beta isoform, is associated with increased intracellular calcium levels in patients with hypertrophic cardiomyopathy.64 Our analysis revealed alterations in multiple aspects of calcium transients in both CELF4-KO and MYH7-KO, suggesting impaired calcium handling as one of the mechanisms underlying DIC.
Contractile properties of ZFN521-KO, RIN3-KO, and CYP2J2-KO hiPSC-CMs are affected
ZFN521 regulates the ventricular conduction system in the heart,65 while RIN3 interacts with and regulates RAB5, facilitating membrane trafficking of the voltage-gated potassium channel KCNQ1 and thereby regulating potassium currents.66 CYP2J2 KO has been shown to result in QT-interval prolongation on echocardiography.67 Impedance analysis of ZFN521-KO, RIN3-KO, and CYP2J2-KO hiPSC-CMs revealed significant differences in contractile properties, including beat rate, pulse width at 50%, upstroke velocity, and relaxation velocity, compared with the isogenic control.
Study limitations
Although we investigated the KO of gene candidates from GWAS and candidate gene association studies, these studies identified SNPs linked to AIC. Because not all the SNPs cause loss of function, studying individual SNP corrections and/or patient-specific hiPSC-CMs can shed further light on this matter. Additionally, we generated KOs of selected candidate genes on the basis of their potential link to AIC and expression in hiPSC-CMs. Although not all the generated KOs showed significant differences in LD50 after doxorubicin treatment, repeating this process using KO of a gene with no proven function in AIC could further validate these findings. Additionally, the application of more mature cardiomyocytes could assist to harness the differences in a more physiologically relevant system. The present study was focused only on the effect of KOs on cardiomyocytes, but cardiotoxicity might stem from malfunction in other cell types, including endothelial cells and cardiac fibroblasts. Finally, for each KO, only 1 functional study was performed; additional functional studies can provide a more detailed understanding of each gene contribution to the formation of DIC.
Conclusions
This study demonstrates that genomic analysis, coupled with human induced pluripotent stem cell modeling, is an efficient platform for assessing the mechanisms of DIC. Through high-throughput assays, we validate the influence of DIC genes on cell viability, ROS production, DNA damage, doxorubicin uptake, iron uptake, calcium handling, and electric activity in response to doxorubicin. Our results confirm that more than 55% of the DIC genes identified to date in association studies are expressed in cardiac cells, reliably recapitulating alterations in DIC phenotype in the hiPSC-CM model. Furthermore, for each gene, we provide a functional assay to validate its role in DIC.
Perspectives.
COMPETENCY IN MEDICAL KNOWLEDGE: In this study, we have functionally validated all current DIC-associated genes that are expressed in cardiomyocytes. This comprehensive validation represents the first step in translating functionally validated genotype-phenotype correlations into clinical tests. Once the relevance of a gene in DIC is identified, and its mechanism of action is known, the next step involves validating the role of genetic variants in modulating the response to doxorubicin. This information will serve as a unique platform for designing polygenic risk scores that can benefit patients undergoing doxorubicin treatment. In addition, a precise understanding of the genetic basis of DIC is crucial for identifying druggable targets and discovering innovative cardioprotective therapies.
TRANSLATIONAL OUTLOOK: One major barrier in the clinical application of this study lies in determining the genetic contribution to DIC risk for each gene for all sexes and ancestries. Currently, there is a scarcity of studies exploring the effects of sex and diverse genetic backgrounds on the mechanisms of DIC. Establishing patient-specific risk factors is a crucial prerequisite before the clinical implementation of the findings from this study can be applied.
Funding Support and Author Disclosures
This work was supported by National Institutes of Health grants R01 CA220002 and R01 CA261898, American Heart Association Transformational Project Award 18TPA34230105, and the Leducq Foundation (to Dr Burridge). The authors have reported that they have no relationships relevant to the contents of this paper to disclose.
Footnotes
The authors attest they are in compliance with human studies committees and animal welfare regulations of the authors’ institutions and Food and Drug Administration guidelines, including patient consent where appropriate. For more information, visit the Author Center.
Appendix
For supplemental methods, tables, and figures, please see the online version of this paper.
Appendix
References
- 1.Bhatia S. Genetics of anthracycline cardiomyopathy in cancer survivors: JACC: CardioOncology state-of-the-art review. J Am Coll Cardiol CardioOnc. 2020;2(4):539–552. doi: 10.1016/j.jaccao.2020.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cardinale D., Colombo A., Bacchiani G., et al. Early detection of anthracycline cardiotoxicity and improvement with heart failure therapy. Circulation. 2015;131(22):1981–1988. doi: 10.1161/CIRCULATIONAHA.114.013777. [DOI] [PubMed] [Google Scholar]
- 3.Aminkeng F., Bhavsar A.P., Visscher H., et al. A coding variant in RARG confers susceptibility to anthracycline-induced cardiotoxicity in childhood cancer. Nat Genet. 2015;47(9):1079–1084. doi: 10.1038/ng.3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schneider B.P., Shen F., Gardner L., et al. Genome-wide association study for anthracycline-induced congestive heart failure. Clin Cancer Res. 2017;23(1):43–51. doi: 10.1158/1078-0432.CCR-16-0908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wells Q.S., Veatch O.J., Fessel J.P., et al. Genome-wide association and pathway analysis of left ventricular function after anthracycline exposure in adults. Pharmacogenet Genomics. 2017;27(7):247–254. doi: 10.1097/FPC.0000000000000284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang X., Sun C.L., Quinones-Lombrana A., et al. CELF4 variant and anthracycline-related cardiomyopathy: a Children’s Oncology Group genome-wide association study. J Clin Oncol. 2016;34(8):863–870. doi: 10.1200/JCO.2015.63.4550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Park B., Sim S.H., Lee K.S., Kim H.J., Park I.H. Genome-wide association study of genetic variants related to anthracycline-induced cardiotoxicity in early breast cancer. Cancer Sci. 2020;111(7):2579–2587. doi: 10.1111/cas.14446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Magdy T., Burridge P.W. Use of hiPSC to explicate genomic predisposition to anthracycline-induced cardiotoxicity. Pharmacogenomics. 2021;22(1):41–54. doi: 10.2217/pgs-2020-0104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Visscher H., Ross C.J., Rassekh S.R., et al. Pharmacogenomic prediction of anthracycline-induced cardiotoxicity in children. J Clin Oncol. 2012;30(13):1422–1428. doi: 10.1200/JCO.2010.34.3467. [DOI] [PubMed] [Google Scholar]
- 10.Visscher H., Ross C.J., Rassekh S.R., et al. Validation of variants in SLC28A3 and UGT1A6 as genetic markers predictive of anthracycline-induced cardiotoxicity in children. Pediatr Blood Cancer. 2013;60(8):1375–1381. doi: 10.1002/pbc.24505. [DOI] [PubMed] [Google Scholar]
- 11.Minotti G., Recalcati S., Mordente A., et al. The secondary alcohol metabolite of doxorubicin irreversibly inactivates aconitase/iron regulatory protein-1 in cytosolic fractions from human myocardium. FASEB J. 1998;12(7):541–552. doi: 10.1096/fasebj.12.7.541. [DOI] [PubMed] [Google Scholar]
- 12.Visscher H., Rassekh S.R., Sandor G.S., et al. Genetic variants in SLC22A17 and SLC22A7 are associated with anthracycline-induced cardiotoxicity in children. Pharmacogenomics. 2015;16(10):1065–1076. doi: 10.2217/pgs.15.61. [DOI] [PubMed] [Google Scholar]
- 13.Magdy T., Jouni M., Kuo H.H., et al. Identification of drug transporter genomic variants and inhibitors that protect against doxorubicin-induced cardiotoxicity. Circulation. 2022;145(4):279–294. doi: 10.1161/CIRCULATIONAHA.121.055801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krajinovic M., Elbared J., Drouin S., et al. Polymorphisms of ABCC5 and NOS3 genes influence doxorubicin cardiotoxicity in survivors of childhood acute lymphoblastic leukemia. Pharmacogenomics J. 2016;16(6):530–535. doi: 10.1038/tpj.2015.63. [DOI] [PubMed] [Google Scholar]
- 15.Saeki K., Obi I., Ogiku N., Shigekawa M., Imagawa T., Matsumoto T. Doxorubicin directly binds to the cardiac-type ryanodine receptor. Life Sci. 2002;70(20):2377–2389. doi: 10.1016/s0024-3205(02)01524-2. [DOI] [PubMed] [Google Scholar]
- 16.Keung E.C., Toll L., Ellis M., Jensen R.A. L-type cardiac calcium channels in doxorubicin cardiomyopathy in rats morphological, biochemical, and functional correlations. J Clin Invest. 1991;87(6):2108–2113. doi: 10.1172/JCI115241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Benjanuwattra J., Siri-Angkul N., Chattipakorn S.C., Chattipakorn N. Doxorubicin and its proarrhythmic effects: a comprehensive review of the evidence from experimental and clinical studies. Pharmacol Res. 2020;151 doi: 10.1016/j.phrs.2019.104542. [DOI] [PubMed] [Google Scholar]
- 18.Burridge P.W., Li Y.F., Matsa E., et al. Human induced pluripotent stem cell-derived cardiomyocytes recapitulate the predilection of breast cancer patients to doxorubicin-induced cardiotoxicity. Nat Med. 2016;22(5):547–556. doi: 10.1038/nm.4087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Magdy T., Jiang Z., Jouni M., et al. RARG variant predictive of doxorubicin-induced cardiotoxicity identifies a cardioprotective therapy. Cell Stem Cell. 2021;28(12):2076–2089.e7. doi: 10.1016/j.stem.2021.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Singh P., Wang X., Hageman L., et al. Association of GSTM1 null variant with anthracycline-related cardiomyopathy after childhood cancer—a Children’s Oncology Group ALTE03N1 report. Cancer. 2020;126(17):4051–4058. doi: 10.1002/cncr.32948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Blanco J.G., Sun C.L., Landier W., et al. Anthracycline-related cardiomyopathy after childhood cancer: role of polymorphisms in carbonyl reductase genes—a report from the Children’s Oncology Group. J Clin Oncol. 2012;30(13):1415–1421. doi: 10.1200/JCO.2011.34.8987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gabrielson K., Bedja D., Pin S., et al. Heat shock protein 90 and ErbB2 in the cardiac response to doxorubicin injury. Cancer Res. 2007;67(4):1436–1441. doi: 10.1158/0008-5472.CAN-06-3721. [DOI] [PubMed] [Google Scholar]
- 23.El-Tokhy M.A., Hussein N.A., Bedewy A.M.L., Barakat M.R. XPD gene polymorphisms and the effects of induction chemotherapy in cytogenetically normal de novo acute myeloid leukemia patients. Hematology. 2014;19(7):397–403. doi: 10.1179/1607845413Y.0000000144. [DOI] [PubMed] [Google Scholar]
- 24.Semsei A.F., Erdelyi D.J., Ungvari I., et al. ABCC1 polymorphisms in anthracycline-induced cardiotoxicity in childhood acute lymphoblastic leukaemia. Cell Biol Int. 2012;36(1):79–86. doi: 10.1042/CBI20110264. [DOI] [PubMed] [Google Scholar]
- 25.Hertz D.L., Caram M.V., Kidwell K.M., et al. Evidence for association of SNPs in ABCB1 and CBR3, but not RAC2, NCF4, SLC28A3 or TOP2B, with chronic cardiotoxicity in a cohort of breast cancer patients treated with anthracyclines. Pharmacogenomics. 2016;17(3):231–240. doi: 10.2217/pgs.15.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lipshultz S.E., Cochran T.R., Franco V.I., Miller T.L. Treatment-related cardiotoxicity in survivors of childhood cancer. Nat Rev Clin Oncol. 2013;10(12):697–710. doi: 10.1038/nrclinonc.2013.195. [DOI] [PubMed] [Google Scholar]
- 27.Windsor R.E., Strauss S.J., Kallis C., Wood N.E., Whelan J.S. Germline genetic polymorphisms may influence chemotherapy response and disease outcome in osteosarcoma: a pilot study. Cancer. 2012;118(7):1856–1867. doi: 10.1002/cncr.26472. [DOI] [PubMed] [Google Scholar]
- 28.Leger K.J., Cushing-Haugen K., Hansen J.A., et al. Clinical and genetic determinants of cardiomyopathy risk among hematopoietic cell transplantation survivors. Biol Blood Marrow Transplant. 2016;22(6):1094–1101. doi: 10.1016/j.bbmt.2016.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wojnowski L., Kulle B., Schirmer M., et al. NAD(P)H oxidase and multidrug resistance protein genetic polymorphisms are associated with doxorubicin-induced cardiotoxicity. Circulation. 2005;112(24):3754–3762. doi: 10.1161/CIRCULATIONAHA.105.576850. [DOI] [PubMed] [Google Scholar]
- 30.Lubieniecka J.M., Liu J., Graham J., et al. Single-nucleotide polymorphisms in reductase genes are not associated with response to daunorubicin-based remission induction. Cancer Epidemiol Biomarkers Prev. 2013;22(10):1918–1920. doi: 10.1158/1055-9965.EPI-13-0671. [DOI] [PubMed] [Google Scholar]
- 31.Rajic V., Aplenc R., Debeljak M., et al. Influence of the polymorphism in candidate genes on late cardiac damage in patients treated due to acute leukemia in childhood. Leuk Lymphoma. 2009;50(10):1693–1698. doi: 10.1080/10428190903177212. [DOI] [PubMed] [Google Scholar]
- 32.Kang Y.J., Sun X., Chen Y., Zhou Z. Inhibition of doxorubicin chronic toxicity in catalase-overexpressing transgenic mouse hearts. Chem Res Toxicol. 2002;15(1):1–6. doi: 10.1021/tx015532n. [DOI] [PubMed] [Google Scholar]
- 33.Armenian S.H., Ding Y., Mills G., et al. Genetic susceptibility to anthracycline-related congestive heart failure in survivors of haematopoietic cell transplantation. Br J Haematol. 2013;163(2):205–213. doi: 10.1111/bjh.12516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oppermann U. Carbonyl reductases: the complex relationships of mammalian carbonyl- and quinone-reducing enzymes and their role in physiology. Annu Rev Pharmacol Toxicol. 2007;47:293–322. doi: 10.1146/annurev.pharmtox.47.120505.105316. [DOI] [PubMed] [Google Scholar]
- 35.Boekhout A.H., Gietema J.A., Milojkovic Kerklaan B., et al. Angiotensin II-receptor inhibition with candesartan to prevent trastuzumab-related cardiotoxic effects in patients with early breast cancer: a randomized clinical trial. JAMA Oncol. 2016;2(8):1030–1037. doi: 10.1001/jamaoncol.2016.1726. [DOI] [PubMed] [Google Scholar]
- 36.Belmonte F., Das S., Sysa-Shah P., et al. ErbB2 overexpression upregulates antioxidant enzymes, reduces basal levels of reactive oxygen species, and protects against doxorubicin cardiotoxicity. Am J Physiol Heart Circ Physiol. 2015;309(8):H1271–H1280. doi: 10.1152/ajpheart.00517.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Doroshow J.H., Esworthy R.S., Chu F.F. Control of doxorubicin-induced, reactive oxygen-related apoptosis by glutathione peroxidase 1 in cardiac fibroblasts. Biochem Biophys Rep. 2020;21 doi: 10.1016/j.bbrep.2019.100709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rossi D., Rasi S., Franceschetti S., et al. Analysis of the host pharmacogenetic background for prediction of outcome and toxicity in diffuse large B-cell lymphoma treated with R-CHOP21. Leukemia. 2009;23(6):1118–1126. doi: 10.1038/leu.2008.398. [DOI] [PubMed] [Google Scholar]
- 39.Yin Z., Ivanov V.N., Habelhah H., Tew K., Ronai Z. Glutathione S-transferase p elicits protection against H2O2-induced cell death via coordinated regulation of stress kinases. Cancer Res. 2000;60(15):4053–4057. [PubMed] [Google Scholar]
- 40.Wang X., Liu W., Sun C.L., et al. Hyaluronan synthase 3 variant and anthracycline-related cardiomyopathy: a report from the children’s oncology group. J Clin Oncol. 2014;32(7):647–653. doi: 10.1200/JCO.2013.50.3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Law C.H., Li J.M., Chou H.C., Chen Y.H., Chan H.L. Hyaluronic acid-dependent protection in H9C2 cardiomyocytes: a cell model of heart ischemia-reperfusion injury and treatment. Toxicology. 2013;303:54–71. doi: 10.1016/j.tox.2012.11.006. [DOI] [PubMed] [Google Scholar]
- 42.Neilan T.G., Blake S.L., Ichinose F., et al. Disruption of nitric oxide synthase 3 protects against the cardiac injury, dysfunction, and mortality induced by doxorubicin. Circulation. 2007;116(5):506–514. doi: 10.1161/CIRCULATIONAHA.106.652339. [DOI] [PubMed] [Google Scholar]
- 43.Hildebrandt M.A.T., Reyes M., Wu X., et al. Hypertension susceptibility loci are associated with anthracycline-related cardiotoxicity in long-term childhood cancer survivors. Sci Rep. 2017;7(1):9698. doi: 10.1038/s41598-017-09517-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tripaydonis A., Conyers R., Elliott D.A. Pediatric anthracycline-induced cardiotoxicity: mechanisms, pharmacogenomics, and pluripotent stem-cell modeling. Clin Pharmacol Ther. 2019;105(3):614–624. doi: 10.1002/cpt.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hordijk P.L. Regulation of NADPH oxidases: the role of Rac proteins. Circ Res. 2006;98(4):453–462. doi: 10.1161/01.RES.0000204727.46710.5e. [DOI] [PubMed] [Google Scholar]
- 46.Almontashiri N.A., Chen H.H., Mailloux R.J., et al. SPG7 variant escapes phosphorylation-regulated processing by AFG3L2, elevates mitochondrial ROS, and is associated with multiple clinical phenotypes. Cell Rep. 2014;7(3):834–847. doi: 10.1016/j.celrep.2014.03.051. [DOI] [PubMed] [Google Scholar]
- 47.Cho C., Jung-Ha H., Willis W.D., et al. Protamine 2 deficiency leads to sperm DNA damage and embryo death in mice. Biol Reprod. 2003;69(1):211–217. doi: 10.1095/biolreprod.102.015115. [DOI] [PubMed] [Google Scholar]
- 48.Romeo F., Falbo L., Di Sanzo M., et al. BRCA1 is required for hMLH1 stabilization following doxorubicin-induced DNA damage. Int J Biochem Cell Biol. 2011;43(12):1754–1763. doi: 10.1016/j.biocel.2011.08.011. [DOI] [PubMed] [Google Scholar]
- 49.Miranda C.J., Makui H., Soares R.J., et al. Hfe deficiency increases susceptibility to cardiotoxicity and exacerbates changes in iron metabolism induced by doxorubicin. Blood. 2003;102(7):2574–2580. doi: 10.1182/blood-2003-03-0869. [DOI] [PubMed] [Google Scholar]
- 50.Andreev E., Brosseau N., Carmona E., Mes-Masson A.M., Ramotar D. The human organic cation transporter OCT1 mediates high affinity uptake of the anticancer drug daunorubicin. Sci Rep. 2016;6 doi: 10.1038/srep20508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Smith A.J., van Helvoort A., van Meer G., et al. MDR3 P-glycoprotein, a phosphatidylcholine translocase, transports several cytotoxic drugs and directly interacts with drugs as judged by interference with nucleotide trapping. J Biol Chem. 2000;275(31):23530–23539. doi: 10.1074/jbc.M909002199. [DOI] [PubMed] [Google Scholar]
- 52.Lian G., Yuan J., Gao Y. In vitro transport ability of ABCC2 (G1249A) polymorphic variant towards anticancer drugs. Onco Targets Ther. 2020;13:1413–1419. doi: 10.2147/OTT.S207613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang J.Q., Wu Z.X., Yang Y., et al. ATP-binding cassette (ABC) transporters in cancer: a review of recent updates. J Evid Based Med. Sep. 2021;14(3):232–256. doi: 10.1111/jebm.12434. [DOI] [PubMed] [Google Scholar]
- 54.Ladd A.N., Stenberg M.G., Swanson M.S., Cooper T.A. Dynamic balance between activation and repression regulates pre-mRNA alternative splicing during heart development. Dev Dyn. 2005;233(3):783–793. doi: 10.1002/dvdy.20382. [DOI] [PubMed] [Google Scholar]
- 55.Wasielewski M., van Spaendonck-Zwarts K.Y., Westerink N.D., et al. Potential genetic predisposition for anthracycline-associated cardiomyopathy in families with dilated cardiomyopathy. Open Heart. 2014;1(1) doi: 10.1136/openhrt-2014-000116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lan F., Lee A.S., Liang P., et al. Abnormal calcium handling properties underlie familial hypertrophic cardiomyopathy pathology in patient-specific induced pluripotent stem cells. Cell Stem Cell. 2013;12(1):101–113. doi: 10.1016/j.stem.2012.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Arnold W.R., Baylon J.L., Tajkhorshid E., Das A. Arachidonic acid metabolism by human cardiovascular CYP2J2 is modulated by doxorubicin. Biochemistry. 2017;56(51):6700–6712. doi: 10.1021/acs.biochem.7b01025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Seebohm G., Strutz-Seebohm N., Birkin R., et al. Regulation of endocytic recycling of KCNQ1/KCNE1 potassium channels. Circ Res. 2007;100(5):686–692. doi: 10.1161/01.RES.0000260250.83824.8f. [DOI] [PubMed] [Google Scholar]
- 59.Singh A., Babyak M.A., Nolan D.K., et al. Gene by stress genome-wide interaction analysis and path analysis identify EBF1 as a cardiovascular and metabolic risk gene. Eur J Hum Genet. 2015;23(6):854–862. doi: 10.1038/ejhg.2014.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wu Q., Vasquez K.M. Human MLH1 protein participates in genomic damage checkpoint signaling in response to DNA interstrand crosslinks, while MSH2 functions in DNA repair. PLoS Genet. 2008;4(9) doi: 10.1371/journal.pgen.1000189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Allocati N., Masulli M., Di Ilio C., Federici L. Glutathione transferases: substrates, inhibitors and pro-drugs in cancer and neurodegenerative diseases. Oncogenesis. 2018;7(1):8. doi: 10.1038/s41389-017-0025-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang H., Zhabyeyev P., Wang S., Oudit G.Y. Role of iron metabolism in heart failure: From iron deficiency to iron overload. Biochim Biophys Acta Mol Basis Dis. 2019;1865(7):1925–1937. doi: 10.1016/j.bbadis.2018.08.030. [DOI] [PubMed] [Google Scholar]
- 63.Biesiadecki B.J., Elder B.D., Yu Z.B., Jin J.P. Cardiac troponin T variants produced by aberrant splicing of multiple exons in animals with high instances of dilated cardiomyopathy. J Biol Chem. 2002;277(52):50275–50285. doi: 10.1074/jbc.M206369200. [DOI] [PubMed] [Google Scholar]
- 64.Dainis A., Zaleta-Rivera K., Ribeiro A., et al. Silencing of MYH7 ameliorates disease phenotypes in human iPSC-cardiomyocytes. Physiol Genom. 2020;52(7):293–303. doi: 10.1152/physiolgenomics.00021.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Litvinukova M., Talavera-Lopez C., Maatz H., et al. Cells of the adult human heart. Nature. 2020;588(7838):466–472. doi: 10.1038/s41586-020-2797-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kajiho H., Sakurai K., Minoda T., et al. Characterization of RIN3 as a guanine nucleotide exchange factor for the Rab5 subfamily GTPase Rab31. J Biol Chem. 2011;286(27):24364–24373. doi: 10.1074/jbc.M110.172445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Solanki M., Pointon A., Jones B., Herbert K. Cytochrome P450 2J2: potential role in drug metabolism and cardiotoxicity. Drug Metab Dispos. 2018;46(8):1053–1065. doi: 10.1124/dmd.117.078964. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.