Abstract
Clear cell renal cell carcinoma (ccRCC) is known as the most common type of renal cancer. Recently, a series of advances have been made in targeted therapy for ccRCC. To combat this highly metastatic tumor, novel therapeutic targets still need to be developed. C-type lectins (CLECs) contain a characteristic C-type lectin-like domain and affect several physiological functions. The effects of C-type lectin 2D (CLEC2D) on cancer progression have been revealed in several types of cancers; however, its expression in ccRCC tissues, and the possible effects on the progression and metastasis of ccRCC, are still unclear. Herein, we found the high mRNA and protein levels of CLEC2D in ccRCC tissues. We further found that CLEC2D expression was correlated with the prognosis of ccRCC patients and correlated with the tumor size (p = 0.019*) of patients. In addition, CLEC2D affected tumor immune infiltration, confirmed by the further analysis. CLEC2D knockdown suppressed the proliferation of ccRCC cells in vitro and restrained ccRCC tumor growth and immune infiltration in mice. Therefore, we believe that CLEC2D has the potential to serve as a promising ccRCC therapeutic target.
Keywords: Clear cell renal cell carcinoma (ccRCC), C-type lectin 2D (CLEC2D), Tumor size, TCGA database, Prognosis, Immune
1. Introduction
Clear cell renal cell carcinoma (ccRCC), accounting for approximately 2% of malignant cancers, is known as the most common renal cancer type [1]. Notably, the morbidity of ccRCC has increased obviously in recent years [2]. Worse, 20% of ccRCC patients had metastasized when diagnosed, and 6% of patients had metastasis-related symptoms [3]. Currently, the main treatment options for renal cell carcinoma include radical nephrectomy or nephron sparing surgery, including partial nephrectomy or thermal ablation techniques. Given the indolent biological behavior of tumors, early detection of ccpRCC can help patients avoid more invasive medical procedures [3]. The clinical characteristics of ccRCC mean that traditional surgical treatment is difficult to effectively remove, leading to tumor recurrence, while chemoradiotherapy may lead to poor therapeutic effects due to obvious side effects [4]. Recently, targeted therapy for ccRCC has made some progress and has good prospects in its treatment [5]. At present, the US FDA has approved more than 10 kinds of drugs for the first-line treatment of advanced ccRCC, including angiogenesis inhibitors, rapamycin target egg (mTOR) inhibitors and immune checkpoint inhibitors [5]. To combat this highly metastatic tumor, novel therapeutic targets still need to be developed.
D-type lectins (CLECs) contain a characteristic C-type lectin-like domain and affect several physiological functions, such as angiogenesis, inflammation, and development, due to the capacity to identify endogenous ligands [6]. CLEC2D could regulate B lymphocyte proteins during viral infection [7]. Viral infection also promoted CLEC2D expression in lymphocytes, led to the inhibition of the cytotoxic response and stimulated IFN-g release [8]. CLEC2D encodes LLT1, which can bind to the CD161 receptor, therefore mediating T-cell functions [9].
The effects of CLEC2D on tumor development have been revealed, such as in prostate cancer and cutaneous squamous cell carcinoma [9,10]. CLEC2D expression also correlated with the clinical features and prognosis of patients with head and neck squamous cell carcinoma [10]. Additionally, the CLEC2D-CD161 interaction promoted the natural killer cell-mediated lysis of triple-negative breast cancer cells [9,10]. In lung cancer, the expression of CLEC2D and its receptor CD161 correlated with patient prognosis [10]. However, its expression in ccRCC tissues, or the possible effects on the progression and metastasis of ccRCC, is still unclear.
In this study, we found obviously high mRNA and protein levels of CLEC2D in human ccRCC tissues and correlated them with the prognosis and clinical pathological features of ccRCC patients. We therefore thought that CLEC2D had the potential to serve as a target of ccRCC.
2. Materials and methods
2.1. Bioinformatic analysis
We conducted bioinformatics analysis through GEPIA (http://gepia.cancer-pku.cn/) to analyze expression levels and survival in The Cancer Genome Atlas (TCGA) database. The coexpressed genes of CLEC2D were calculated in cBioPortal (https://www.cbioportal.org/, Firehose Legacy cohort, and PanCancer Atlas cohort), and enrichment analysis of coexpressed genes was performed via DAVID (https://david.ncifcrf.gov/). Immune association analysis was carried out using TIMER 2.0 (http://timer.cistrome.org/).
2.2. Human tissue samples and clinical pathological analysis
Human ccRCC tissues and adjacent normal tissues were collected from 70 patients who received surgical therapy at our hospital. This study was reviewed and approved by the clinical ethics committee (CEC) of our hospital. Seventy patients signed informed consent forms. The clinical features were recorded and are listed in Table 1.
Table 1.
Relationships of CLEC2D and clinicopathological characteristics in 70 patients with clear cell renal cell carcinoma.
| Feature | All n = 70 | CLEC2D expression |
χ2 |
p |
|
|---|---|---|---|---|---|
| Low |
High |
||||
| n = 36 | n = 34 | ||||
| Age (year) | 3.225 | 0.073 | |||
| <55 | 44 | 19 | 25 | ||
| ≥55 | 26 | 17 | 9 | ||
| Gender | 1.377 | 0.241 | |||
| Male | 40 | 23 | 17 | ||
| Female | 30 | 13 | 17 | ||
| Tumor grade | 2.979 | 0.084 | |||
| Low | 30 | 19 | 11 | ||
| High | 40 | 17 | 23 | ||
| Tumor size | 5.505 | 0.019* | |||
| <4 cm | 24 | 17 | 7 | ||
| ≥4 cm | 46 | 19 | 27 | ||
*p < 0.05.
2.3. Immunohistochemical (IHC) assays
To further investigate the possible link between CLEC2D expression and ccRCC progression, immunohistochemical (IHC) assays were performed. Sections were fixed with 4% PFA and blocked with 2% BSA in PBS for 25 min. Slides were then incubated with anti-CLEC2D antibody (1:100 dilution, ab239222, Abcam, Cambridge, UK) at 4 °C overnight. Subsequently, the sections were incubated with a secondary antibody conjugated to horseradish peroxidase (HRP). Then, the staining reaction was detected by an ECL kit. Two experienced pathologists independently observed the staining results.
2.4. Staining index
According to the staining results, CLEC2D was mainly located in the cytoplasm of ccRCC tissues. The score method is briefly described as follows. The percentage of positively stained cells was divided as follows: 0, negative cells; 1, 10–50% positive cells and 2, over 50% positive cells. The staining intensity was evaluated on a score of 0 (no staining), 1 (moderate staining) or 2 (strong staining). The CLEC2D expression level was calculated based on the staining intensity score + positive cell score. A score of 0–2 was considered low expression, whereas a score of 3 or 4 was considered high CLEC2D expression.
2.5. Cell culture
Clear cell renal cell carcinoma cell lines, 786-O and Caki-2, were obtained from the Committee of Type Culture Collection of the Chinese Academy of Sciences (Shang Hai, China). The cells were maintained in RPMI-1640 (G40618 Yuan Pei Biotechnology Corporation, Shanghai, China) supplemented with 10% fetal bovine serum (FBS, Gibco) at 37 °C in humidified 5% CO2.
2.6. Transfection
The CLEC2D short hairpin RNA oligonucleotide sequence (shRNA) was inserted into a lentiviral vector (GV115) to construct 4 predesigned shRNAs in the pAV-U6-GFP vector + 1 negative control with scrambled sequences. Four different CLEC2D-RNAi lentiviral vectors and one common lentiviral vector were incubated with 786-O and Caki-2 cells, respectively. The silencing effects of the two cell lines were detected by quantitative PCR and immunoblotting. The biological behavior of sh CLEC2D cells transfected with CLEC2D-RNAi lentiviral vector-transfected sh CLEC2D cells and cells transfected with control lentiviral vector were compared.
2.7. Real-time quantitative PCR
Total mRNA was extracted using TRIzol reagent (Thermo, USA) according to the manufacturer's instructions. RNA was reverse transcribed into cDNA using a cDNA reverse transcription kit (Thermo, USA), including 5 μL of template RNA, 1 μL of random primer, 6 μL of 4 μL of nuclease water, incubation at 65 °C for 5 min, 1 μL of RNase inhibitor, 2 μL of 10 μM dNTP mix, 4 μL of 5x reaction buffer, and a total of 1 μL of Aid total 20 μL. Quantitative PCR was performed using SGExcelFastSYBR high ROX mix from Thermo Scientific, USA. The 2 −ΔΔCt method was used to quantify the results. CLEC2D primer: 5′-GGGAAAGGCAAATGGAAAAC-3′, 5′-CCCAAGGCCATACAAGTGTT-3'. GAPDH primers: 5′-GAGTCAACGGATTTGGTCGT-3′ and 5′-TTGATTTTGGAGGGATCTCG-3' (Sheng Gong, Shanghai, China).
2.8. Western blot
Proteins were extracted using RIPA buffer (R0020 Solaibio Beijing China), and the concentration was measured by the BCA method. SDS‒PAGE (10%) was performed for 2 h, and then the proteins were transferred onto PVDF membranes. After blocking, the membranes were incubated with primary antibodies for 8 h. The secondary antibodies were incubated for 1 h. Proteins were visualized using an enhanced chemiluminescence detection reagent (Pierce; Thermo Fisher Scientific, Inc.) and were analyzed using ImageJ9.0 software.
2.9. Colony formation assay
786-O and Caki-2 cells were seeded in 6-well plates and incubated for 14 days. The cells were then washed with PBS, fixed with absolute ethanol for 15 min, and stained with 0.2% crystal violet for 25 min. Finally, colony formation was observed with the naked eye or a microscope (low magnification). The number of cell colonies was calculated.
2.10. Cell proliferation assay
786-O and Caki-2 cells were seeded in a 96-well plate at a density of 5000 cells/well, and the proliferation capacity of the two cell lines was measured by CCK-8 assay. The proliferation curve was described after 5 days, and the differences between the groups were measured at 450 nm wavelength. There were 5 replicate wells in each group.
2.11. In vivo tumor growth and immune infiltration detection assay
Animal experiments were performed with the approval of the Animal Care and Use Committee. (License No.: TMUaMEC2015008). A total of eight female nude mice (age, 8 weeks;weight, 22–24 g; n = 4 mice/group) were purchased from Beijing Vital River Laboratory Animal Technology Co., Ltd. The mice were sacrificed by cervical dislocation, and the lack of heartbeat was confirmed to validate death. The mice were monitored once a day before the experiment began and twice a day until the experiment ended.The stable cell line 786-O shRNA transfected or control cells were injected subcutaneously into the left axilla (3 × 106 cells). Twenty-nine days after subcutaneous implantation, the tumors were isolated and measured.
To isolate the CD8+T cells, we performed FCM assays. Flow cytometry was performed on Attune NxT Flow Cytometer and analyzed using FlowJo software. Cell-sorting experiments were performed on BD Aria II. Antibodies used for staining were: antimouse IFN-γ (Thermo Fisher Scientific Cat#12- 7311- 81), and antimouse CD8 (BD Biosci-ences Cat# 552877). The degree of CD8+ T cells and INF-γ levels were measured.
2.12. Statistics
Statistically significant correlation between CLEC2D expression level and survival rate in ccRCC patients were analyzed in TCGA database (TCGA; https://cancergenome.nih.gov/). The correlations between the clinical features of ccRCC patients and CLEC2D expression were analyzed through χ2 analysis. Student's t-test was used for statistical analysis. p < 0.05 was considered statistically significant.
3. Results
3.1. Bioinformatics analysis data showed that the CLEC2D mRNA level was upregulated in ccRCC tissues and correlated with prognosis
To assess CLEC2D expression in ccRCC tissues, we first investigated the mRNA levels of CLEC2D in human ccRCC and normal tissues from the TCGA database. A total of 523 ccRCC tissues and 100 normal tissues were included. We noticed high mRNA levels of CLEC2D in ccRCC tissues (Fig. 1A, p < 0.05). Additionally, we assessed the correlations between CLEC2D mRNA levels and the prognosis of ccRCC patients through the TCGA database. Kaplan‒Meier (KM) survival analysis was performed to assess whether CLEC2D expression affected ccRCC patient prognosis. Interestingly, we found that CLEC2D expression correlated with the overall survival (OS) rates (p = 0.0041) of ccRCC patients (Fig. 1B). Furthermore, another database (TCGA, Firehose Legacy cohort, 448 samples) also showed significant differences (p = 4.48e-06) in overall survival (Fig. 1C). In addition, we found that high expression of CLEC2D was associated with tumor grade and stage in ccRCC patients via the TCGA database (Fig. 1D). Therefore, we believe that CLEC2D is upregulated in human ccRCC tissues and correlated with prognosis.
Fig. 1.
CLEC2D mRNA was upregulated in ccRCC tissues and correlated with prognosis. (A). CLEC2D mRNA levels in 523 ccRCC tissues and 100 normal tissues according to the TCGA database. (B). CLEC2D mRNA levels were correlated with the survival rates (p < 0.05) of ccRCC patients based on the TCGA database (B) and based on another database (C) (TCGA, Firehose Legacy cohort, 448 samples). D. CLEC2D mRNA expression in independent clinical grade and stage.
3.2. CLEC2D protein expression was upregulated in human ccRCC tissues
Given the crucial capacity of CLEC2D in tumors, we examined the expression level in The Human Protein Atlas (Atlas, https://www.proteinatlas.org/), finding that high expression of CLEC2D was associated with poor OS in renal cancer (Fig. S1A). Otherwise, the Atlas IHC database showed that CLEC2D has a high expression in colorectal cancer, breast cancer, prostate cancer, lung cancer and liver cancer (Fig. S1B).
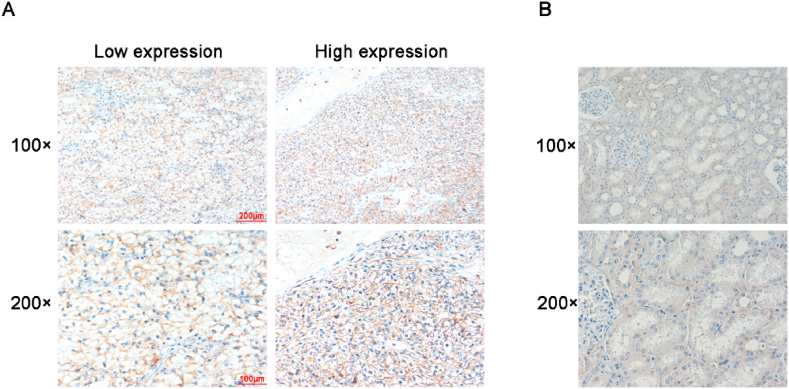
To further investigate the expression of CLEC2D in ccRCC tissues, IHC assays were conducted. The expression of ccRCC was detected in 70 tumor tissues and the corresponding normal tissues of patients in our hospital. Furthermore, the differential expression of CLEC2D was compared between tumor tissues and normal tissues. Importantly, we found that ccRCC tissues showed obvious higher CLEC2D expression than normal tissues (Fig. 2A and B). Therefore, CLEC2D protein expression was upregulated in human ccRCC tissues.
Fig. 2.
CLEC2D protein expression was enhanced in human ccRCC tissues. (A, B) IHC assays were performed to assess CLEC2D expression levels in human ccRCC tissues (A) and corresponding normal tissues (B), and representative photographs are shown (100 × and 200 × magnification, respectively).
3.3. CLEC2D expression was associated with the clinical features of ccRCC patients
We analyzed the correlation between CLEC2D expression and the clinical features of ccRCC patients. Patient age, sex, tumor grade, and tumor size were evaluated. The 70 patients were divided into low or high groups according to the expression of CLEC2D. We noticed that 36 patients showed low CLEC2D expression (51.4%, Table 1), and 34 patients (48.6%) exhibited high CLEC2D expression. According to the results, no significant correlation was found in clinical features, including patient age (p = 0.073), sex (p = 0.241), and tumor grade (p = 0.084), between the CLEC2D low and high-expression groups (Table 1). Importantly, we noticed that CLEC2D expression was obviously associated with the tumor size (p = 0.019) of ccRCC patients (Table 1). Therefore, we thought that CLEC2D expression was related to the tumor size of ccRCC patients.
3.4. CLEC2D co-expression gene enrichment analysis
To further investigate the potential biological function and mechanism of CLEC2D in ccRCC, gene co-expression analysis was performed via the cBioPortal dataset in the Firehose Legacy cohort and PanCancer Atlas cohort. As shown in Fig. 3A and 36 genes were merged in the top 50 genes of the two cohorts, including Firehose Legacy cohort and PanCancer Atlas cohort, suggesting that these genes could have correlations with CLEC2D (Table 2, Table 3). In detail, the significant association between the expression of CLEC2D and PVRIG, MIR155HG,TRAF5, which are reported to affect the progression of immune infiltration, is shown in Fig. 3B. GO and KEGG enrichment analysis of 36 coexpressed genes showed that CLEC2D may contribute to immune infiltration in ccRCC, affecting T-cell activation, immunodeficiency, Th1 and Th2 cell differentiation, and NF-κB pathway (Fig. 3C). Notably, these cellular processes and pathways are involved in the progression of immune infiltration. In conclusion, CLEC2D co-expression gene enrichment analysis confirmed that CLEC2D affected immune infiltration in ccRCC.
Fig. 3.
CLEC2D co-expression gene enrichment analysis. (A) CLEC2D gene co-expression analysis (TOP50 genes) was performed via the cBioPortal dataset in the Firehose Legacy cohort and PanCancer Atlas cohort. (B) Representative pictures of CLEC2D co-expression gene analysis. (C) GO and KEGG enrichment analysis of CLEC2D coexpressed genes.
Table 2.
CLEC2D co-expressed genes in the Firehose Legacy cohort (top 50).
| Correlated Gene | Cytoband | Spearman's Correlation | p Value |
|---|---|---|---|
| MIR155HG | 21q21.3 | 0.705 | 3.54E-68 |
| TRAF5 | 1q32.3 | 0.652 | 2.15E-55 |
| PARP15 | 3q21.1 | 0.644 | 1.46E-53 |
| ZAP70 | 2q11.2 | 0.642 | 3.81E-53 |
| ARRDC5 | 19p13.3 | 0.635 | 8.71E-52 |
| SEPTIN1 | 16p11.2 | 0.63 | 1.10E-50 |
| IRF1-AS1 | 5q31.1 | 0.625 | 9.42E-50 |
| PVRIG | 7q22.1 | 0.624 | 1.33E-49 |
| TFAP2E | 1p34.3 | 0.62 | 8.21E-49 |
| SAMD3 | 6q23.1 | 0.618 | 2.10E-48 |
| XAF1 | 17p13.2 | 0.618 | 2.25E-48 |
| ACAP1 | 17p13.1 | 0.618 | 2.30E-48 |
| CARMIL2 | 16q22.1 | 0.617 | 4.04E-48 |
| KLRK1 | 12p13.2 | 0.617 | 4.38E-48 |
| LINC00426 | 13q12.3 | 0.612 | 3.86E-47 |
| STAT4 | 2q32.2 | 0.608 | 1.93E-46 |
| TMC8 | 17q25.3 | 0.606 | 4.21E-46 |
| MIAT | 22q12.1 | 0.606 | 5.41E-46 |
| DDB1 | 11q12.2 | −0.602 | 3.04E-45 |
| LAT | 16q13 | 0.601 | 3.41E-45 |
| KLRA1P | 12p13.2 | 0.6 | 4.91E-45 |
| APOBEC3D | 22q13.1 | 0.598 | 1.60E-44 |
| PARVG | 22q13.31 | 0.596 | 3.07E-44 |
| ARHGAP9 | 12q13.3 | 0.595 | 4.45E-44 |
| APOBEC3H | 22q13.1 | 0.594 | 7.06E-44 |
| GSDMB | 17q21.1 | 0.593 | 9.95E-44 |
| CTRL | 16q22.1 | 0.589 | 4.57E-43 |
| BCL11B | 14q32.2 | 0.589 | 5.15E-43 |
| TOGARAM2 | 2p23.2 | 0.589 | 6.49E-43 |
| SCML4 | 6q21 | 0.587 | 1.11E-42 |
| INTS6L | Xq26.3 | 0.587 | 1.20E-42 |
| HEATR9 | 17q12 | 0.587 | 1.23E-42 |
| ZNF700 | 19p13.2 | 0.587 | 1.40E-42 |
| C5ORF58 | 5q35.1 | 0.587 | 1.45E-42 |
| UNC13D | 17q25.3 | 0.586 | 1.55E-42 |
| CXORF65 | Xq13.1 | 0.586 | 2.12E-42 |
| TTC24 | 1q22 | 0.585 | 2.25E-42 |
| SYTL1 | 1p36.11 | 0.585 | 2.47E-42 |
| ZNF80 | 3q13.31 | 0.585 | 2.75E-42 |
| GFI1 | 1p22.1 | 0.585 | 2.96E-42 |
| APOBEC3G | 22q13.1 | 0.582 | 9.70E-42 |
| FAM111A | 11q12.1 | 0.582 | 1.02E-41 |
| VAMP1 | 12p13.31 | 0.581 | 1.15E-41 |
| TCERG1 | 5q32 | 0.579 | 2.42E-41 |
| HCG27 | 6p21.33 | 0.579 | 2.55E-41 |
| TSPAN32 | 11p15.5 | 0.578 | 3.47E-41 |
| ERMN | 2q24.1 | 0.578 | 3.54E-41 |
| RUFY4 | 2q35 | 0.575 | 1.13E-40 |
| CLK4 | 5q35.3 | 0.575 | 1.34E-40 |
| TBC1D10C | 11q13.2 | 0.574 | 1.84E-40 |
Table 3.
CLEC2D co-expressed genes in the PanCancer Atlas cohort (top 50).
| Correlated Gene | Cytoband | Spearman's Correlation | p Value |
|---|---|---|---|
| MIR155HG | 21q21.3 | 0.711 | 1.75E-55 |
| TFAP2E | 1p34.3 | 0.659 | 3.00E-45 |
| KLRK1 | 12p13.2 | 0.655 | 1.71E-44 |
| ARRDC5 | 19p13.3 | 0.645 | 9.73E-43 |
| XAF1 | 17p13.2 | 0.643 | 1.85E-42 |
| TRAF5 | 1q32.3 | 0.64 | 5.61E-42 |
| ZAP70 | 2q11.2 | 0.638 | 1.26E-41 |
| PARP15 | 3q21.1 | 0.633 | 7.33E-41 |
| SEPTIN1 | 16p11.2 | 0.627 | 8.44E-40 |
| STAT4 | 2q32.2-q | 0.627 | 8.83E-40 |
| IRF1-AS1 | 5q31.1 | 0.626 | 9.68E-40 |
| GSDMB | 17q21.1 | 0.623 | 2.72E-39 |
| ACAP1 | 17p13.1 | 0.616 | 3.34E-38 |
| SAMD3 | 6q23.1 | 0.614 | 8.62E-38 |
| KLRA1P | 12p13.2 | 0.613 | 9.86E-38 |
| CARMIL2 | 16q22.1 | 0.61 | 3.06E-37 |
| DDB1 | 11q12.2 | −0.608 | 6.24E-37 |
| SPDYA | 2p23.2 | 0.607 | 7.15E-37 |
| LINC00426 | 13q12.3 | 0.605 | 1.67E-36 |
| ARHGAP9 | 12q13.3 | 0.604 | 2.47E-36 |
| MIAT | 22q12.1 | 0.604 | 2.49E-36 |
| BCL11B | 14q32.2 | 0.604 | 2.51E-36 |
| HSPB11 | 1p32.3 | 0.599 | 1.08E-35 |
| IDO2 | 8p11.21 | 0.599 | 1.32E-35 |
| TMC8 | 17q25.3 | 0.598 | 1.43E-35 |
| CLK4 | 5q35.3 | 0.597 | 2.19E-35 |
| PVRIG | 7q22.1 | 0.594 | 7.04E-35 |
| TCERG1 | 5q32 | 0.593 | 7.22E-35 |
| TBX19 | 1q24.2 | 0.593 | 7.67E-35 |
| HCG27 | 6p21.33 | 0.593 | 8.00E-35 |
| SNHG12 | 1p35.3 | 0.593 | 8.89E-35 |
| LAT | 16q13 | 0.592 | 9.81E-35 |
| CTRL | 16q22.1 | 0.592 | 1.15E-34 |
| ZNF700 | 19p13.2 | 0.59 | 2.13E-34 |
| LINC00158 | 21q21.2 | 0.589 | 2.86E-34 |
| ZNF80 | 3q13.31 | 0.588 | 4.29E-34 |
| ZNF83 | 19q13.41 | 0.587 | 4.97E-34 |
| ANKLE1 | 19p13.11 | 0.584 | 1.30E-33 |
| FAM186A | 12q13.12 | 0.583 | 2.14E-33 |
| GCNA | Xq13.1 | 0.582 | 2.34E-33 |
| APOBEC3H | 22q13.1 | 0.58 | 4.71E-33 |
| N4BP2L2 | 13q13.1 | 0.58 | 4.86E-33 |
| FAM111A | 11q12.1 | 0.58 | 5.45E-33 |
| ARHGEF39 | 9p13.3 | 0.58 | 5.70E-33 |
| SYTL1 | 1p36.11 | 0.578 | 8.24E-33 |
| FAM13A | 4q22.1 | 0.578 | 9.48E-33 |
| TOGARAM2 | 2p23.2 | 0.577 | 1.42E-32 |
| PARVG | 22q13.31 | 0.576 | 1.55E-32 |
| FNBP4 | 11p11.2 | 0.576 | 1.82E-32 |
| INTS6L | Xq26.3 | 0.576 | 1.86E-32 |
3.5. Immune Analysis of CLEC2D in ccRCC
The above data suggest that CLEC2D may regulate tumor immunity in ccRCC. Next, the correlation between tumor immune infiltration and the CLEC2D expression level was analyzed via TIMER2.0. The results show that CLEC2D closely correlated with immune subtypes of ccRCC (Fig. 4A). Then, we further investigated the correlation between CLEC2D expression and tumor-infiltrating lymphocytes (TILs) and immuno-inhibitors. As shown in Fig. 4B and C, the top three TILs and immuno-inhibitors with a Spearman correlation test rho greater than 0.4 with TK1 expression are shown.
Fig. 4.
Immune Analysis of CLEC2D in ccRCC. (A) Relationships between CLEC2D mRNA expression and immune subtype in the TCGA ccRCC dataset. (B) Correlation between CLEC2D mRNA expression and tumor-infiltrating lymphocytes (TILs). (C) Correlation between CLEC2D mRNA expression and tumor-infiltrating lymphocytes (TILs) and immuno-inhibitors (C).
3.6. CLEC2D was depleted by shRNA plasmids in ccRCC cell lines
The total mRNAs of the 786-O and Caki-2 cell line experimental groups (CLEC2D shRNA transfection group, shCLEC2D) and the control group (control shRNA transfection group, shControl) were extracted. The qPCR assays showed that the mRNA expression of CLEC2D in the experimental group was lower than that in the control group (p < 0.01) (Fig. 5A). Through immunoblot assays, the protein levels of CLEC2D in control or CLEC2D-depleted 786-O and Caki-2 cells were extracted, and the protein expression of CLEC2D in the experimental group (shCLEC2D) was lower than that in the control group (p < 0.01) (Fig. 5B).
Fig. 5.
CLEC2D was effectively knocked down after transfection of its shRNAs in ccRCC cells. mRNA expression (qPCR) (A) and protein expression (WB) (B) of CLEC2D after transfection in 786-O and Caki-2 cells with shRNA and empty vector, respectively, were detected. Colony formation assays (C) and CCK-8 proliferation assays (D) were performed to detect the effects of CLEC2D on ccRCC cell proliferation.
3.7. Knockdown of CLEC2D inhibited cell proliferation in 786-O and Caki-2 cells
786-O and Caki-2 cells were transfected with CLEC2D shRNA and empty vector, respectively. After 9 days, the number of cells formed by the control clones was significantly higher than that of the CLEC2D depletion group (p < 0.05). Both 786-O and Caki-2 cells showed consistent results (Fig. 5C). The CCK-8 proliferation assay showed that the proliferation capacity of 786-O and Caki-2 cells was decreased after CLEC2D depletion compared to that of the control group (p < 0.01) (Fig. 5D).
3.8. Depletion of CLEC2D blocked the growth and immune infiltration of tumors in mice
To study the effects of CLEC2D on tumor growth, 3 × 106 shCLEC2D cells or shControl cells were implanted into the left ankle of BALB/C nude mice. After 29 days, the mice were dissected for tumor formation. The results showed that the tumor weight in the CLEC2D depletion group was significantly lower than that in the control group (p < 0.05), suggesting that downregulation of CLEC2D inhibited tumor growth in vivo (Fig. 6A). Then, immunohistochemistry (IHC) assays were performed to detect the expression of CLEC2D in tumor tissue from the two groups. Notably, the expression of CLEC2D in the experimental group (shCLEC2D) was lower than that in the control group (p < 0.01) (Fig. 6B). To further confirm the effects on immune infiltration, we isolated the CD8+ T cells from tumor tissues, and found the decreased levels of CD8+ T cells in CLEC2D depletion group (Fig. 6C). We further detected the IFN-γ in isolated CD8+ T cells, we confirmed that IFN-γlevels were decreased in CD8+ T cells from CLEC2D depletion group (Fig. 6D). Therefore, CLEC2D depletion suppressed immune infiltration of tumors.
Fig. 6.
CLEC2D promotes tumor growth and immune infiltration of ccRCC cells in vivo. CLEC2D shRNA and empty vector were stably transfected into 786-O cells, which were then implanted in nude mice to form tumors. Representative images and tumor growth curves are shown (A). Immunohistochemistry was performed to detect the expression of CLEC2D in tumor tissues from the indicated groups (B). (C, D). Proportions of intratumoral CD8+ T cells (C) and INF-γ. (D) produced was quantified from tumor tissues in shControl and shCLEC2D mice.
4. Discussion
ccRCC is thought to be a highly metastatic tumor [11]. Chemotherapy and immunotherapy usually extend survival by approximately one year [12]. The main clinical symptoms of ccRCC are the triad of renal carcinoma, namely, hematuria, pain, and lumbago [13]. In view of the lack of obvious symptoms in the early stage of ccRCC, it is often in the advanced stage at the time of clinical diagnosis, which aggravates the patient's condition [14]. Targeted therapy may be the most promising treatment for ccRCC [15]. Several drugs targeting ccRCC are currently in clinical trials to improve patient outcomes [16]. To further improve patient survival, more effective treatments need to be developed. Importantly, here, we found that a C-type lectin, CLEC2D, was obviously highly expressed in human ccRCC tissues. According to the TCGA database, we further found that CLEC2D expression correlated with the prognosis of ccRCC patients. Our data therefore suggest that CLEC2D could serve as a prognostic predictor of ccRCC.
Through clinical pathological analysis, we found that the expression of CLEC2D was significantly correlated with the tumor size of ccRCC patients. Therefore, we speculated that CLEC2D could regulate the proliferation of ccRCC cells and further affect the growth of ccRCC. The next step is to perform a series of in vitro assays and animal assays. We should detect the effects of CLEC2D on ccRCC cell proliferation and motility in ccRCC cells in vitro and tumor growth in mice. Collectively, our data suggest that CLEC2D could serve as a promising target.
Notably, the role of CLEC2D in tumorigenesis has been well reported [9,10,17]. Docetaxel could suppress the immunotherapy efficacy of natural killer cells toward castration-resistant prostate cancer cells through targeting CLEC2D (LLT1) [9,17,18]. Additionally, CLEC2D expression was correlated with clinical features, such as nodal metastasis, in patients with head and neck squamous cell carcinoma [19]. Moreover, CLEC2D is a marker of germinal center-derived B-cell non-Hodgkin's lymphomas [7,20]. These studies confirmed that CLEC2D has the potential to serve as a cancer target.
As a protein expressed on the surface of cells, CLEC2D could also affect immune responses [9,21,22]. In this study, we noticed high expression in human ccRCC tissues. CLEC2D-mediated activation of IFN-gamma production also involves the ERK pathway in natural killer cells [10,23]. We should next assess whether CLEC2D could promote the progression of ccRCC through the regulation of cell immune responses.
In a previous study, the CLEC2D reporter responded to lysates from necrotic cells [7]. Through biochemical purification, they identified histones as CLEC2D ligands [7]. CLEC2D knockout mice showed reduced proinflammatory responses to injected histones and further improved survival in a hepatotoxic injury model [24,25]. Additionally, another study indicated the contrasting expression of CLEC2D in HIV infection [7]. These studies all imply the important role of CLEC2D in immunity [9]. In this study, we found that abnormal expression of CLEC2D may be involved in the development of ccRCC. Further research is needed to confirm whether CLEC2D can influence the progression of ccRCC through immunomodulatory effects.
Importantly, C-type lectin receptors (CLRs) are known as carbohydrate binding pattern recognition receptors (PRRs), which could play a role in host recognition of pathogenic microorganisms [26]. Through CLRs, signaling shown on antigen-presenting cells dictates key adaptive immune responses [27]. Whether CLEC2D can thus affect tumor progression remains to be further investigated.
Notably, CLEC2D receptor for KLRB1 that protects target cells against natural killer cell-mediated lysis, inhibits osteoclast formation, inhibits bone resorption and modulates the release of interferon-gamma. It is also a ligand for the human NKR-P1A (CD161) receptor, present on NK cells and T cells [7,8]. WInterestingly, we here also revealed that CLEC2D affected the immune infiltration of ccRCC, and we guessed that it affected the progression of immune infiltration via mediating NK cells and T cells. We believed the effects of this protein on NK cells and T cells affected the immune infiltration of ccRCC. In addition, CLEC2D could form homodimers as well as heterodimers with TLR2 to negatively regulate IRF5-mediated antifungal immunity, suggesting the important role of CLEC2D in immunity [20]. Herein, we confirmed CLEC2D promotes tumor growth and immune infiltration.
The limitation of this study is that it failed to deeply reveal the molecular mechanism of CLEC2D's influence on the progression of ccRCC, failed to find downstream key proteins and signaling pathways, and failed to elucidate the cause of the influence on immune infiltration. We found CLEC2D promotes tumor growth and immune infiltration in mice, and the mechanism needs further study Therefore, in the following studies, we will try to use multidimensional omics to screen downstream key proteins and pathways, and verify them through various biochemical means. As for the effect of immune infiltration, we will try to construct more reasonable animal models to confirm the relevant hypothesis.
In conclusion, we confirmed that CLEC2D was obviously highly expressed in human ccRCC tissues and that the expression of CLEC2D correlated with ccRCC patient prognosis. In addition, we found that the expression of CLEC2D correlated with the tumor size of patients with ccRCC. Collectively, we found high expression of CLEC2D and thought that CLEC2D could serve as a promising target for ccRCC treatment.
Funding
No funding was received.
Ethical statement
The study methodology conformed to the standard set by the Declaration of Helsinki and was approved (2022-03-B163) by the Ethics Committee of the First Affiliated Hospital of Henan University of Science and Technology. All patients had signed inform consent forms.
Data availability statement
The data generated during the current study are available from the corresponding author.
CRediT authorship contribution statement
Huibing Li: Writing – review & editing, Writing – original draft, Visualization, Validation, Supervision, Software, Resources, Project administration, Methodology, Investigation, Funding acquisition, Formal analysis, Data curation, Conceptualization. Pengyi Zheng: Investigation, Formal analysis, Data curation. Zhijun Li: Software, Resources, Methodology. Qingjiang Han: Visualization, Validation, Software. Bisheng Zhou: Writing – original draft, Visualization. Kaixuan Wang: Resources, Methodology, Data curation.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Contributor Information
Huibing Li, Email: researcherlhb@163.com.
Pengyi Zheng, Email: zpydoctor@126.com.
References
- 1.Souto Filho Jtd, Silveira A.L.O., Lacerda A.P., Pires A.Z., Sales L.R., Dias Y.P. Erythropoietin-producing clear cell renal cell carcinoma associated with secondary polycythemia. Hematol Transfus Cell Ther. 2021;43(2):226–227. doi: 10.1016/j.htct.2020.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang K., Wu T., Chen Y., Song G., Chen Z. Prognostic effect of Preoperative Apolipoprotein B level in surgical patients with clear cell renal cell carcinoma. Oncol. Res. Treat. 2020;43:340–345. doi: 10.1159/000507964. [DOI] [PubMed] [Google Scholar]
- 3.Xie Y., Chen L., Gao Y., Ma X., He W., Zhang Y., Zhang F., Fan Y., Gu L., Li P., et al. miR-363 suppresses the proliferation, migration and invasion of clear cell renal cell carcinoma by downregulating S1PR1. Cancer Cell Int. 2020;20:227. doi: 10.1186/s12935-020-01313-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Che Z., Fan J., Zhou Z., Li Q., Ma Z., Hu Z., Wu Y., Jin Y., Su Y., Liang P., et al. Activation-induced Cytidine deaminase expression facilitates the malignant phenotype and epithelial-to-mesenchymal transition in clear cell renal cell carcinoma. DNA Cell Biol. 2020;39(7):1–14. doi: 10.1089/dna.2019.5119. [DOI] [PubMed] [Google Scholar]
- 5.Kostrzewa M., Zyla M., Wladzinski J., Stetkiewicz T., Stachowiak G., Wilczynski J.R. Metastases of renal clear cell carcinoma to ovary--case report and review of the literature. Eur. J. Gynaecol. Oncol. 2015;36(2):219–222. [PubMed] [Google Scholar]
- 6.Cummings R.D., McEver R.P., Type Lectins C. In: Essentials of Glycobiology. rd Varki A., Cummings R.D., Esko J.D., Stanley P., Hart G.W., Aebi M., Darvill A.G., Kinoshita T., Packer N.H., et al., editors. Cold Spring Harbor; NY: 2015. pp. 435–452. [Google Scholar]
- 7.Varaden D., Moodley J., Onyangunga O.A., Naicker T. Morphometric image analysis of placental C-type lectin domain family 2, member D (CLEC2D) immuno-expression in HIV associated pre-eclampsia. Eur. J. Obstet. Gynecol. Reprod. Biol. X. 2019;3 doi: 10.1016/j.eurox.2019.100039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bouzid D., Fourati H., Amouri A., Marques I., Abida O., Haddouk S., Ben Ayed M., Tahri N., Penha-Goncalves C., Masmoudi H. Association of ZAP70 and PTPN6, but Not BANK1 or CLEC2D, with inflammatory bowel disease in the Tunisian population. Genet. Test. Mol. Biomarkers. 2013;17(4):321–326. doi: 10.1089/gtmb.2012.0372. [DOI] [PubMed] [Google Scholar]
- 9.Mathew S.O., Chaudhary P., Powers S.B., Vishwanatha J.K., Mathew P.A. Overexpression of LLT1 (OCIL, CLEC2D) on prostate cancer cells inhibits NK cell-mediated killing through LLT1-NKRP1A (CD161) interaction. Oncotarget. 2016;7(42):68650–68661. doi: 10.18632/oncotarget.11896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.He X., Qian Y., Wu C., Feng J., Sun X., Zheng Q., Li X., Shen J. Entropy-Mediated High-Entropy MXenes Nanotherapeutics: NIR-II-Enhanced Intrinsic Oxidase Mimic Activity to Combat Methicillin Resistant Staphylococcus Aureus Infection. Adv. Mater. 2023;35(26):2211432. doi: 10.1002/adma.202211432. [DOI] [PubMed] [Google Scholar]
- 11.Farhadi F., Nikpanah M., Paschall A.K., Shafiei A., Tadayoni A., Ball M.W., Linehan W.M., Jones E.C., Malayeri A.A. Clear cell renal cell carcinoma growth correlates with baseline diffusion-weighted MRI in Von Hippel‒Lindau disease. Radiology. 2020;295(3):E10. doi: 10.1148/radiol.2020204010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gao X., Yang J., Chen Y. Identification of a four immune-related genes signature based on an immunogenomic landscape analysis of clear cell renal cell carcinoma. J. Cell. Physiol. 2020;235(12):9834–9850. doi: 10.1002/jcp.29796. [DOI] [PubMed] [Google Scholar]
- 13.Ged Y., Chaim J.L., DiNatale R.G., Knezevic A., Kotecha R.R., Carlo M.I., Lee C.H., Foster A., Feldman D.R., Teo M.Y., et al. DNA damage repair pathway alterations in metastatic clear cell renal cell carcinoma and implications on systemic therapy. J Immunother Cancer. 2020;8(1) doi: 10.1136/jitc-2019-000230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Padala S.A., Kallam A. StatPearls. Treasure. 2020. Cancer, clear cell renal carcinoma. Island (FL) [Google Scholar]
- 15.Bergmann L., Grunwald V., Maute L., Grimm M.O., Weikert S., Schleicher J., Klotz T., Greiner J., Florcken A., Hartmann A., et al. A randomized phase IIa trial with temsirolimus versus sunitinib in advanced non-clear cell renal cell carcinoma: an intergroup study of the CESAR central European society for anticancer drug research-EWIV and the interdisciplinary working group on renal cell cancer (IAGN) of the German cancer society. Oncol. Res. Treat. 2020;43:333–338. doi: 10.1159/000508450. [DOI] [PubMed] [Google Scholar]
- 16.Bersanelli M., Brunelli M., Gnetti L., Maestroni U., Buti S. Pazopanib as a possible option for the treatment of metastatic non-clear cell renal carcinoma patients: a systematic review. Ther Adv Med Oncol. 2020;12 doi: 10.1177/1758835920915303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.He X., Hou J.T., Sun X., Jangili P., An J., Qian Y., Kim J.S., Shen J. NIR-II Photo-Amplified Sonodynamic Therapy Using Sodium Molybdenum Bronze Nanoplatform against Subcutaneous Staphylococcus Aureus Infection. Adv. Funct. Mater. 2022;32(38):2203964. [Google Scholar]
- 18.Germain C., Bihl F., Zahn S., Poupon G., Dumaurier M.J., Rampanarivo H.H., Padkjaer S.B., Spee P., Braud V.M. Characterization of alternatively spliced transcript variants of CLEC2D gene. J. Biol. Chem. 2010;285(46):36207–36215. doi: 10.1074/jbc.M110.179622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lai J.J., Cruz F.M., Rock K.L. Immune sensing of cell death through recognition of histone sequences by C-type lectin-receptor-2d causes inflammation and tissue injury. Immunity. 2020;52(1):123–135 e126. doi: 10.1016/j.immuni.2019.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Germain C., Guillaudeux T., Galsgaard E.D., Hervouet C., Tekaya N., Gallouet A.S., Fassy J., Bihl F., Poupon G., Lazzari A., et al. Lectin-like transcript 1 is a marker of germinal center-derived B-cell non-Hodgkin's lymphomas dampening natural killer cell functions. OncoImmunology. 2015;4(8) doi: 10.1080/2162402X.2015.1026503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He X., Koo S., Obeng E., Sharma A., Shen J., Kim J.S. Emerging 2D MXenes for antibacterial applications: Current status, challenges, and prospects. Coordin. Chem. Rev. 2023;492:215275. [Google Scholar]
- 22.Roth P., Mittelbronn M., Wick W., Meyermann R., Tatagiba M., Weller M. Malignant glioma cells counteract antitumor immune responses through expression of lectin-like transcript-1. Cancer Res. 2007;67(8):3540–3544. doi: 10.1158/0008-5472.CAN-06-4783. [DOI] [PubMed] [Google Scholar]
- 23.Germain C., Meier A., Jensen T., Knapnougel P., Poupon G., Lazzari A., Neisig A., Hakansson K., Dong T., Wagtmann N., et al. Induction of lectin-like transcript 1 (LLT1) protein cell surface expression by pathogens and interferon-gamma contributes to modulate immune responses. J. Biol. Chem. 2011;286(44):37964–37975. doi: 10.1074/jbc.M111.285312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Del Fresno C., Sancho D. Clec2d joins the cell death sensor ranks. Immunity. 2020;52(1):6–8. doi: 10.1016/j.immuni.2019.12.015. [DOI] [PubMed] [Google Scholar]
- 25.Williams K.J., Eaton H.E., Jones L., Rengan S., Burshtyn D.N. Vaccinia virus Western Reserve induces rapid surface expression of a host molecule detected by the antibody 4C7 that is distinct from CLEC2D. Microbiol. Immunol. 2016;60(11):754–769. doi: 10.1111/1348-0421.12451. [DOI] [PubMed] [Google Scholar]
- 26.Shimojima M., Takenouchi A., Shimoda H., Kimura N., Maeda K. Distinct usage of three C-type lectins by Japanese encephalitis virus: DC-SIGN, DC-SIGNR, and LSECtin. Arch. Virol. 2014;159(8):2023–2031. doi: 10.1007/s00705-014-2042-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schulke S., Vieths S. Dendritic cell targeting with C-type lectins for improvement of allergen immunotherapy. J Allergy Clin. 2016;138(2):568–570. doi: 10.1016/j.jaci.2016.06.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data generated during the current study are available from the corresponding author.