Abstract
Background
Circadian rhythms play a key role in the failing heart, but the exact molecular mechanisms linking changes in the expression of circadian rhythm-related genes to heart failure (HF) remain unclear.
Methods
By intersecting differentially expressed genes (DEGs) between normal and HF samples in the Gene Expression Omnibus (GEO) database with circadian rhythm-related genes (CRGs), differentially expressed circadian rhythm-related genes (DE-CRGs) were obtained. Machine learning algorithms were used to screen for feature genes, and diagnostic models were constructed based on these feature genes. Subsequently, consensus clustering algorithms and non-negative matrix factorization (NMF) algorithms were used for clustering analysis of HF samples. On this basis, immune infiltration analysis was used to score the immune infiltration status between HF and normal samples as well as among different subclusters. Gene Set Variation Analysis (GSVA) evaluated the biological functional differences among subclusters.
Results
13 CRGs showed differential expression between HF patients and normal samples. Nine feature genes were obtained through cross-referencing results from four distinct machine learning algorithms. Multivariate LASSO regression and external dataset validation were performed to select five key genes with diagnostic value, including NAMPT, SERPINA3, MAPK10, NPPA, and SLC2A1. Moreover, consensus clustering analysis could divide HF patients into two distinct clusters, which exhibited different biological functions and immune characteristics. Additionally, two subgroups were distinguished using the NMF algorithm based on circadian rhythm associated differentially expressed genes. Studies on immune infiltration showed marked variances in levels of immune infiltration between these subgroups. Subgroup A had higher immune scores and more widespread immune infiltration. Finally, the Weighted Gene Co-expression Network Analysis (WGCNA) method was utilized to discern the modules that had the closest association with the two observed subgroups, and hub genes were pinpointed via protein-protein interaction (PPI) networks. GRIN2A, DLG1, ERBB4, LRRC7, and NRG1 were circadian rhythm-related hub genes closely associated with HF.
Conclusion
This study provides valuable references for further elucidating the pathogenesis of HF and offers beneficial insights for targeting circadian rhythm mechanisms to regulate immune responses and energy metabolism in HF treatment. Five genes identified by us as diagnostic features could be potential targets for therapy for HF.
Keywords: Heart failure, Circadian rhythm, Machine learning, Immune infiltration, Unsupervised clustering, Bioinformatics
1. Introduction
Heart failure (HF) represents a complex clinical syndrome and is among the primary causes of mortality globally [1]. At present, there are limited treatment options for heart failure (HF), resulting in a poor prognosis and a 5-year survival rate of approximately 50% [2,3]. From a clinical viewpoint, heart failure represents the final outcome of numerous cardiac disorders in which the heart fails to pump blood efficiently enough to match the oxygen and nutrient requirements of tissues and organs, ultimately leading to impaired bodily function [4].While some progress has been made in HF research in recent decades, many areas regarding the causes and molecular mechanisms of heart failure remain unknown. The causes of heart failure are diverse, encompassing coronary artery disease, cardiomyopathy, cardiac surgery, substance abuse, ageing, and numerous other factors. Moreover, recent scholarship has indicated a possible link between the onset of heart failure and variations in genes that regulate circadian rhythms [4]. Recent evidence indicates that circadian rhythms play a role in the development of multiple cardiovascular diseases, such as myocardial infarction, ischemia-reperfusion injury post-myocardial infarction, and HF [5]. The cardiovascular system is particularly sensitive to circadian rhythm regulation, impacting various cardiovascular processes, including heart rate, signal transduction, contractility, metabolism and vascular tone [6,7]. Thus, comprehending circadian rhythm pattern alterations in patients with HF could better elucidate the relationship between circadian rhythm and HF.
Circadian rhythms are regulated by the molecular clock mechanism across various organs and tissues [8,9]. Mouse models have directly demonstrated significant cardiac phenotypes resulting from the key functions of peripheral core clocks in the heart. These core transcription factors, BMAL1 and CLOCK, regulate the circadian rhythms in the heart through transcription and translation [10]. Gene knockouts that disrupt the cardiac clock have been shown to result in various cardiomyopathies, including dilated cardiomyopathy and time-dependent cardiomyopathy [11,12]. Song et al. found that alterations in REVERB/BMAL1 rhythms are linked with the severity of dilated cardiomyopathy [13]. The deficiency of Bmal1 protein disrupts the rhythmic expression of genes encoding for Pik3r1, Akt, and Gsk3β proteins, ultimately leading to impaired cardiac contractility [14]. Moreover, the disturbance of circadian rhythms in the heart impacts mitochondrial function, leading to elevated levels of oxidative stress and the activation of pathways related to apoptosis, autophagy, inflammation, and remodeling [15]. Disruption of circadian rhythms also impacts the rhythm of cardiac metabolic activities, including fatty acid oxidation and glucose metabolism [16,17]. Moreover, the circadian clock can regulate immune and inflammatory responses, with the circadian/metabolic axis potentially being a key driver of immune function rhythms [18]. It is now known that circadian rhythm disturbances are risk factors for inflammatory bowel disease [19]. However, research on the interrelationships among circadian rhythms, immune responses, and metabolism in HF is still lacking.
The rapid advances in microarray and high-throughput sequencing technologies have expanded our comprehension of the genome, transcriptome, and epigenetics, resulting in a broader knowledge base of these fields [20,21]. A increasing number of researchers are willing to share their sequencing data through the Gene Expression Omnibus (GEO) database. This presents a prospect to unveil comprehensive insights into gene function, disease mechanisms and biological processes [22]. However, microarray or high-throughput sequencing data comprises a vast number of features at every level, which augments data dimensionality, redundancy, and noise. Subsequently, data analysis, visualization, and interpretation pose complex challenges. To tackle these issues, machine learning algorithms are significantly applied for feature selection and creating classification models, thereby enhancing the reliability of feature selection [23,24]. Iain S. Forrest and his co-authors used the Boruta machine learning algorithm to measure biomarkers associated with atherosclerosis and mortality risk using continuous spectra, without invasive procedures [25]. Additionally, LASSO models, SVM models, and Random Forest (RF) have also been employed for constructing risk models for ischemic stroke [26]. Based on this, the cross-combination of machine learning feature selection algorithms, when they converge on certain genes, indicates a stronger consensus regarding the importance of these genes. Therefore, we employ multiple machine learning algorithms to select more robust candidate genes.
This study systematically evaluates changes in circadian rhythm patterns in HF patients. Firstly, DEGs screened based on the publicly available microarray dataset GSE57338 and intersected with CRGs to obtain DE-CRGs. Subsequently, four machine learning algorithms were employed to screen for genes that could be used as features in disease onset prediction. Nomogram models were constructed, and the correlation between five risk genes and infiltration of immune cells was discussed. Moreover, samples from patients with HF were clustered based on the expression profiles of CRGs, and two different types of HF with distinct biological functions were identified. The variation in immune cell populations between these two distinct groups was further analyzed. Finally, using NMF clustering methods based on DEGs among subclusters, HF patients were divided into two subgroups with distinct molecular and clinical features, and hub genes related to the phenotype were screened through WGCNA. Through these studies, it was found that CRGs have a significant impact on the immune microenvironment and metabolic processes of HF patients.
2. Methods
2.1. Data collection and processing
Microarray data of heart failure patients analyzed in this study were retrieved from the GEO database (www.ncbi.nlm.nih.gov/geo/). Four datasets were included: GSE57338 [27],GSE79962 [28],GSE26887 [29] and GSE5406 [30]. Table 1 shows the detailed information of these datasets. To obtain symbols for genes corresponding to each probe, download the corresponding platform files and construct a matrix of gene expression. All microarray datasets included in this study were normalized with the "Limma" R package [31] and transformed to a log2 scale for subsequent analysis. Furthermore, CRGs included in this study were derived from GeneCard (https://www.genecards.org/).
Table 1.
Details of the dataset used in this study, including number, sequencing platform, number of samples, etc.
2.2. Identification and functional analysis of DE-CRGs
Dataset GSE57338 contains 177 HF patient samples and 136 control samples. Using the R package "Limma", differential gene analysis was performed and DEGs were identified by applying cutoff criteria of |logFC| > 0.5 and adjusted P-value <0.05. By intersecting DEGs with CRGs, we obtained DE-CRGs. The R package "ClusterProfiler" was utilized to conduct enrichment analysis and provide insights into the biological functions of DEGs [32].
2.3. Establishment and validation of CRGs diagnostic signature
The prediction of both the onset and progression risk of HF relies on the identification of stable and sensitive diagnostic markers. Firstly, four machine learning algorithms (namely, Boruta [33], Random Forest(RF) [34], Support Vector Machine_Recursive Feature Elimination (SVM_RFE) [35],and XGBoost [36]) were utilized to filter potential genes for HF diagnosis based on the expression profiles of differentially expressed genes related to circadian rhythms. We performed gene selection of features using the default parameters. Boruta, RF, and SVM_RFE machine learning algorithms demonstrated superior classification performance, whilst the XGBoost model displayed below-par performance and limited ability to generalize. Hence, employing disease classification as the dependent variable and circadian related DEGs as the explanatory variables, we randomly divided the entire GSE57338 dataset into a training set (70%) and an internal validation set (30%) to search for the best hyperparameter combination. In the training dataset, we employed grid search techniques to adjust the parameters and identify the optimal combination. The adjusted parameters include eta, max_depth, min_child_weight, gamma, subsample, colsample_bytree, alpha and lambda. Next, we evaluated the model's performance on the validation dataset. Finally, we utilized the optimized XGBoost predictive model to select feature genes and gauge variables' impact on the onset and progression of HF. We measured these impacts through Shapley Additive Explanation (SHAP) values. Ultimately, we analyzed the genes that the four algorithms identified at the intersection in greater depth.
We utilized the "rms" and "forestplot" R packages to construct nomograms and forest plots, respectively. Discriminative ability of the model was evaluated by analyzing receiver operating characteristic (ROC) curves, while calibration was assessed using calibration plots. The model's accuracy was further validated in both an internal training set and two external validation sets.
2.4. Evaluation of immune cell infiltration
The infiltration levels of 28 types of immune cells were calculated using the ssGSEA method. Furthermore, statistical analyses were conducted by performing T-tests to compare the levels of immune cell infiltration. The optimal immune cell variables were determined using the least absolute shrinkage and selection operator (LASSO) algorithm [37] and ten-fold cross-validation to screen for the best λ value. Additionally, the study utilized correlation analysis to determine how hub genes are related to immune cells and investigate their underlying connections.
To enhance comprehension of immune infiltration variations across different clustering patterns in heart failure patients, the ssGSEA method was employed to investigate the immune microenvironment in HF, and T-tests were executed to compare disparities in immune cell infiltration.
2.5. Unsupervised clustering analysis of DE-CRGs
Consensus clustering analysis was conducted using the R package "ConsensusClusterPlus" to recognize related patterns of different CRGs clustering in HF patients [38]. The optimum number of subtypes was based on evaluating the performance of cumulative distribution function (CDF) curves, relative changes in the area under the CDF curve, consensus matrix, and consistent cluster scores. Subsequently, the differences among the number of clustering samples were visualized using the "ggpubr" package.
2.6. GSEA and GSVA enrichment analysis
The "clusterprofiler" R package was used to perform GSEA enrichment analysis on the DEGs between the two clusters generated by the first clustering to explore biological processes associated with these genes.
To comprehend the biological functional differences within subgroups, we employed the "GSVA" [39] and "Limma" packages to investigate the underlying causes for these phenotypic differences. The background gene lists, Hall.v2023.1.Hs.symbols.gmt, C5.go.bp.v2023.1.Hs.symbols.gmt, and C2.cp.kegg.v2023.1.Hs.symbols.gmt, were downloaded from the Molecular Signatures Database (MSigDB) (https://www.gsea-msigdb.org/gsea/msigdb/).
2.7. Non-negative matrix factorization algorithm
The NMF algorithm extracts biologically relevant coefficients within the gene expression matrix and organizes genes and samples to capture the internal structural features of the data for sample grouping [40]. Using the empirical Bayesian method, DEGs between clusters A and B were acquired through the R package "Limma", with selection criteria of |logFC| > 0.5 and adjusted p-value <0.05. The "NMF" R package was subsequently employed to conduct a clustering analysis of all HF samples using the DEGs identified between the subclusters, with the aim of uncovering possible molecular subtypes. The "brunet" algo rithm was chosen with 100 iterations for each value within the range. The number of clusters was set from 2 to 10. The optimal number of clusters is determined by the combination of cophenetic, dispersion and silhouette.
2.8. Weighted gene Co-expression network
We used the median absolute deviation method to select the top 5000 genes with the largest differences between samples. First, low-quality data were removed after assessing the data quality of both samples and genes. Second, samples with obvious outliers were excluded after using the "hclust" function to perform hierarchical clustering on the samples. Using the "pickSoftThreshold" function in the "WGCNA" R package, an appropriate soft-thresholding power β was chosen according to the standard of a scale-free network, with the range varying from 1 to 20. Selection criteria were as follows: R2 > 0.85, slope approximately equal to −1, and choosing the value at the inflection point of the R2 and power relationship plot. Next, a WGCNA was constructed to identify gene modules, with a combined cutting height of 0.25 and a minimum number of module genes set to 30.
Additionally, we computed the interaction network of genes within the black module using the STRING online database (https://string-db.org/) and ranked the importance of these genes using the MCC algorithm in the CytoHubba plugin. Furthermore, we confirmed the association of these genes with HF through the Comparative Toxicogenomics Database (CTD) (http://ctdbase.org/).
3. Results
3.1. Expression landscape and functional enrichment analysis of CRGs
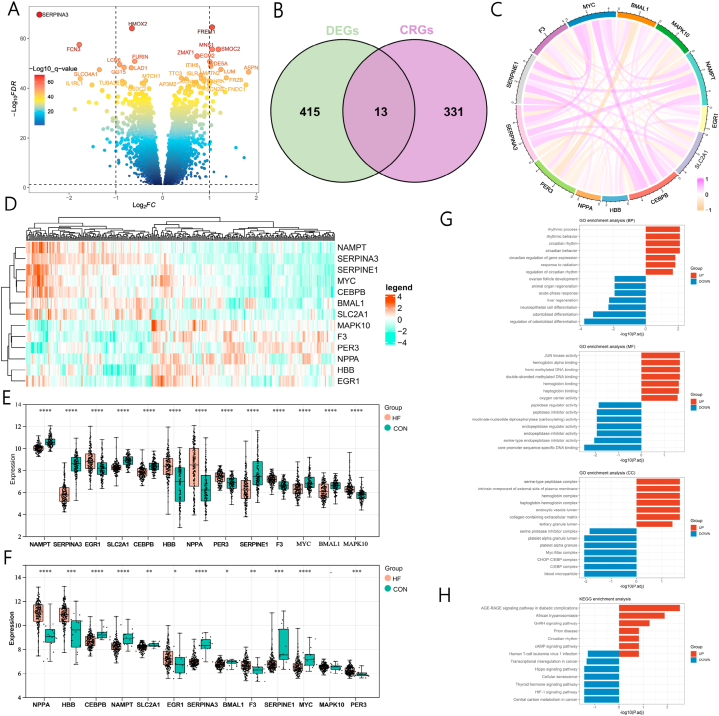
Fig. 1 shows the flowchart of this study. A differential analysis was performed on the expression profile data of 136 normal samples and 177 HF normalized samples, resulting in a total of 428 DEGs identified, including 234 significantly upregulated genes and 194 significantly downregulated genes. (Fig. 2A). Furthermore, by using correlation scores to examine the relationship between genes and circadian rhythms, we obtained a list of 344 circadian rhythm genes with correlation scores exceeding 3.5. To extract circadian rhythm genes that play a crucial role in regulating transcription and have potential research value in HF, we overlapped differentially expressed genes (DEGs) with circadian rhythm genes (CRGs). Eventually, this resulted in a set of circadian rhythm-related genes that are differentially expressed in HF patients. (Fig. 2B and Supplementary Table 1). Correlation analysis indicated that genes SERPINA3, CEBPB, MYC, and NAMPT were significantly positively correlated (Fig. 2C). Fig. 2D and E demonstrate the significant differences in the 13 CRGs between the control group and HF samples. To further verify the expression of the 13 CRGs, an independent HF cohort was used for validation. Fig. 2F revealed that a significant proportion of the CRGs displayed consistent expression levels, with notable differences.
Fig. 1.
Flowchart of this study.
Fig. 2.
Identification and functional enrichment analysis of DE-CRGs. (A) DEGs between HF and normal samples. (B) The common genes of DEGs and CRGs in HF. (C) Correlation chord diagram of DE-CRGs. Red represents positive correlations, and orange represents negative correlations. (D) Heatmap showing the expression levels of 13 CRGs in HF and control samples. (E, F) Boxplots and scatter plots displaying the expression differences of thirteen CRGs in GSE57338 and GSE5406. (G) GO enrichment analysis of DE-CRGs. (H) KEGG enrichment analysis of DE-CRGs. DE-CRGs, differentially expressed circadian rhythm-related genes; CRGs: circadian rhythm-related gene; DEGs: differentially expressed genes; HF: heart failure; GO: gene ontology; KEGG: Kyoto Encyclopedia of Genes and Genomes. *P < 0.05, **P < 0.01, ***P < 0.001. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
To more finely explore the potential roles of DEGs, we conducted a functional enrichment analysis on the DE-CRGs. GO analysis indicated significant enrichments in rhythmic process, rhythmic behavior, circadian rhythm, and circadian regulation of gene expression (Fig. 2G). Additionally, KEGG results revealed that upregulated genes were significantly enriched in pathways, including the AGE-RAGE signaling pathway in diabetic complications, African trypanosomiasis, GnRH signaling pathway, and Circadian rhythm, while downregulated genes were mainly enriched in the HIF-1 signaling pathway and Human T-cell leukemia virus 1 infection signal pathways (Fig. 2H).
3.2. Development and selection of diagnostic markers
To screen for genes related to diagnosis from the 13 intersecting genes, three machine learning algorithms with feature selection were employed. The importance of the 13 features was ranked using the RF algorithm, and the top ten contributing genes were selected for subsequent analysis (Fig. 3A and B). According to the results of the SVM-REF algorithm, the classifier error was minimal when the number of features was 13 (Fig. 3C). Boruta algorithm results showed that the maximum Z-score of shadow features clearly distinguished important and unimportant features. The Z-Score values of all variables in a single random forest run were higher than the maximum Z-Score among shadow attributes (MZSA) in the random variable shadow features (Fig. 3D and E). Therefore, all 13 variables were confirmed as important features. To determine the machine learning model for predicting HF, the dataset consisting of 313 samples was randomly grouped into training and test cohorts, and an XGBoost machine learning model was established to predict the outcomes. Fig. 3G demonstrates that the XGBoost model exhibited perfect accuracy (AUC = 0.988). The impact of each feature variable on the predictive model based on the XGBoost model was elucidated using SHAP values in the training set. Based on the SHAP summary plot, the feature variable importance ranking indicated that SERPINA3 was the variable with the highest contribution to the XGBoost model (Fig. 3F). Ultimately, the intersection of the results of the four machine learning algorithms yielded the final nine diagnostic markers (Fig. 3H and Supplementary Table 2). The fold changes of the nine biomarkers are presented in Fig. 3I.
Fig. 3.
Multiple machine learning methods for screening circadian rhythm-related diagnostic biomarkers. (A) Random forest tree. (B) Gini importance measure. (C) SVM-RFE algorithm selects feature genes. N = 13 represents the optimal number of selectable variables. (D) Z-score changes during Boruta operation. Green represents confirmed features, and blue represents the importance of minimum, average, and maximum shadow features, respectively. (E) MZSA distinguishes important and non-important features. Yellow is determined to be an important feature. (F) XGBoost ranks the importance of genes. (G) ROC curve evaluating the accuracy of the XGBoost model. (H) Intersection of genes selected by the four machine learning algorithms. (I) Fold change of genes with potential diagnostic value. MZSA, maximum Z score among shadow attributes. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.3. Assessment and validation of diagnostic features of CRGs for HF
While complex machine learning models can model the features of the data at a finer level, in practice, simple linear regression models are interpretable, highly predictive, and easier to translate into actionable models for clinical practice. Thus, a logistic regression model was constructed to explore the diagnostic value of characterized genes. The expression of nine markers was independently associated with HF, as revealed by multivariate logistic regression analysis shown in the forest plot (Fig. 4A). The nomogram assigns a score point to each feature variable value, and the total risk score for developing HF is obtained by adding up the scores of all feature variables (Fig. 4B). The accuracy of the nomogram in diagnosing HF was confirmed by the calibration curve (Fig. 4C). To better illustrate the diagnostic ability of the feature genes for HF, we further verified their diagnostic capacity in two external datasets. The results indicate that there were significant differences in the expression levels of NPPA, NAMPT, SLC2A1, MAKP10, and SERPINA3 in both datasets (Fig. 4D and E). Subsequently, ROC analysis was performed in the training cohort and the two external cohorts, and AUC values were calculated to assess the accuracy of each diagnostic gene. Our findings indicated that the five diagnostic genes had high predictive values, particularly SERPINA3 (Fig. 4F–H).
Fig. 4.
Development and validation of CRGs as diagnostic features for HF. (A) Forest plot of multivariable logistic regression analysis. (B) Nomogram showing the probability of HF occurrence based on nine genes. (C) Calibration curve validation of the Nomogram. (D, E) Expression levels of nine diagnostic markers in two external datasets. *P < 0.05 **P < 0.01 ***P < 0.001. (F) Receiver operating characteristic (ROC) curves of diagnostic markers that showed expression differences in both external datasets. (G) ROC curves showing the diagnostic value of five feature genes in the GSE26887 dataset. (H) ROC curves of the five feature genes in the GSE79962 dataset.
3.4. Immune infiltration analysis between normal and HF samples
As shown in Fig. 5A and B, significant differences in the abundance of 20 immune infiltrating cells were observed between HF and normal samples, including activated CD4 T cells, activated dendritic cells, CD56bright natural killer cells, CD56dim natural killer cells, central memory CD4 T cells, central memory CD8 T cells, effector memory CD8 T cells, effector memory CD4 T cells, immature dendritic cells, natural killer T cells, regulatory T cells, type 2 T helper cells, type 17 T helper cells, natural killer cells, macrophages, MDSCs, monocytes, natural killer cells, neutrophils, and plasmacytoid dendritic cells (Fig. 5A and B). Furthermore, we employed the LASSO algorithm to identify characteristic immune cells associated with HF progression. Ultimately, nine optimal variables with non-zero coefficients were identified from the immune cell subgroups mentioned above: activated CD8 T cells, CD56bright natural killer cells, CD56dim natural killer cells, central memory CD4 T cells, central memory CD8 T cells, effector memory CD4 T cells, effector memory CD8 T cells, type 17 T helper cells, and type 2 T helper cells (Fig. 5C and D).
Fig. 5.
SsGSEA analysis of HF and normal samples. (A) Heatmap demonstrating the difference in immune cell infiltration in HF and normal samples (B) Infiltration scores of 28 immune cells in cardiac tissue from HF patients and normal. ns = not significant, *p < 0.05, **p < 0.01, ***p < 0.001 (C, D) The best variable with a non-zero coefficient in the immune cell subpopulation was obtained by LASSO regression. ssGSEA ,Single Sample Gene Set Enrichment Analysis.
The correlation analysis between feature genes and immune cells revealed that SERPINA3 and SLC2A1 were strongly positively correlated with central memory CD8 T cells and negatively correlated with type 2 T helper cells (Fig. 6A and E). NPPA was significantly negatively correlated with type 17 T helper cells (Fig. 6C). MAPK10 was significantly positively correlated with type 2 T helper cells (Fig. 6D). Interestingly, NAMPT exhibited significant correlations with all immune cells (Fig. 6B). Taken together, these results suggest the presence of immune dysregulation in HF, influenced by changes in the expression of circadian rhythm genes.
Fig. 6.
(A–E) Correlation between immune cells and SERPINA3, NAMPT, NPPA, MAPKI0 and SLC2A1.
3.5. Consensus clustering analysis of circadian rhythm gene sets
Using the R package "ConsensusClusterPlus," we conducted consensus clustering analysis on 177 HF samples based on 13 DE-CRGs. The results of the clustering demonstrated that a perfect cluster number appeared to be k = 2, which accurately dispersed HF patients into two clusters: Cluster A (n = 99) and Cluster B (n = 78) (Fig. 7A–D). Clear differences between Cluster A and Cluster B were revealed through PCA analysis (Fig. 7E). Furthermore, several CRGs exhibited significant heterogeneity between the two clusters (Fig. 7F), specifically MYC, NPPA, SERPINA3, and SERPINE1, which displayed higher expression in Cluster A than in Cluster B.
Fig. 7.
Unsupervised clustering analysis based on DE-CRGs expression spectrum. (A) Consensus clustering matrix for k = 2, defining two different subtypes of circadian expression patterns. (B) Cumulative distribution function (CDF) curves for k = 1–9. (C) CDF delta area curves. (D) Consensus clustering scores when k is 2–9. (E) Principal component analysis (PCA) visualization of the sample distribution of the two clusters. (F) Expression of the 13 CRGs in the two clusters. (G) Gene set variance analysis (GSVA) based on the HALLMARK gene set. DE-CRGs, differentially expressed circadian rhythm-related genes.
3.6. Identification of distinct circadian rhythm patterns of immune microenvironment and biological function features
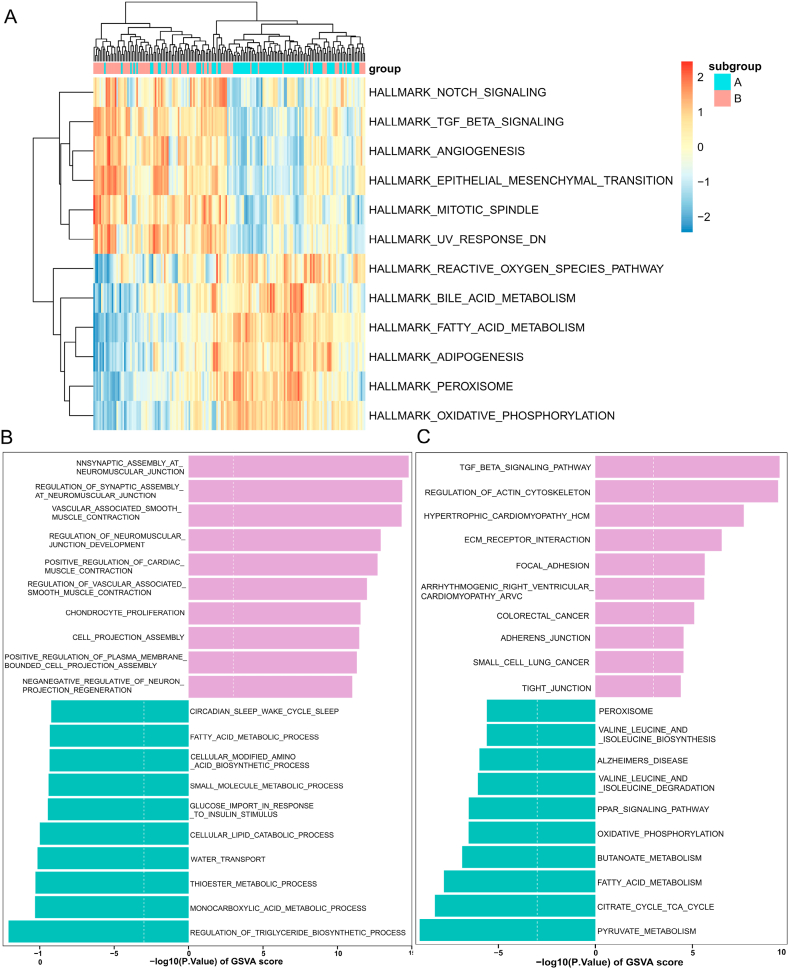
The HALLMARK gene set was used to perform GSVA analysis, which revealed significant enrichment of TGF-β signaling, angiogenesis, NFκB-mediated signaling, TNF-α signaling, coagulation function, epithelial-mesenchymal transition, and IL6-JAK-STAT3 signaling pathways in Cluster A. In contrast, Cluster B showed primary enrichment in metabolic-related pathways such as fatty acid metabolism and oxidative phosphorylation (Fig. 7G). Moreover, we conducted further analysis of the biological function and pathway enrichment differences between Cluster A and Cluster B. Regulation of insulin biosynthesis and protein acylation were significantly enriched in Cluster B, while positive regulation of cardiac muscle contraction, negative regulation of platelet activation, and phagosome maturation mediated by regulatory particles were significantly enriched in Cluster A (Supplementary Fig. 1A). Based on KEGG, GSVA results indicated significant enrichment of TGF-β signaling, ECM receptor interactions, and regulation of actin cytoskeleton-related pathways in Cluster A. Conversely, metabolic pathways such as lipid metabolism, valine and isoleucine degradation, and butanoate metabolism were enriched in Cluster B (Supplementary Fig. 1B). Furthermore, we compared changes in infiltrating immune cell abundance between the two clusters, observing increased T follicular helper cell infiltration in Cluster B, as shown in Supplementary Figs. 2A and B. Overall, the study revealed that there are two distinct circadian rhythm patterns which correlate with different immune features and pathways. This finding suggests a critical role of circadian rhythm in the regulation of immune microenvironment and metabolic processes in HF.
3.7. Determination of HF gene subgroups based on DEGs related to circadian rhythm subtypes
To further investigate potential biological behaviors of different circadian rhythm-related phenotypes, we utilized the R package "Limma" to identify 72 DEGs associated with circadian rhythm phenotypes (Supplementary Fig. 2C). A heatmap displayed expression differences of the 72 DEGs between the two clusters (Fig. 8A). Subsequently, we conducted GO and KEGG analyses to determine biological functions (Supplementary Fig. 2E and Supplementary Table 3). Moreover, significant enrichment for leukocyte chemotaxis, regulation of blood pressure, aerobic electron transport chain, cellular response to lipopolysaccharide, and regulation of calcium ion transmembrane transporter activity was revealed by GSEA enrichment analysis (Fig. 8B). Thus, CRGs may affect HF by modulating cardiac material and energy metabolism, as well as mediating immune-related processes.
Fig. 8.
Identification of HF gene subgroups based on DEGs associated with CRG subtypes. (A) Heatmap of differential expression of DEGs between the two CRG subtypes. (B) GSEA enrichment analysis of DEGs between two CRG subtypes. (C) Heatmap of NMF clustering at k = 2. (D) Distribution of cophenetic, residuals, RSS, silhouette, Var and dispersion with a rank of 2–10. (E) PCA plots showing differences in sample clustering between two HF gene subgroups. (F) Variation in gene subgroups for two different clustering methods. (G) Expression box plots and scatter plots of 13 CRGs. (H) Immune cell infiltration scores in 28 between the two subgroups. NMF, non-negative matrix decomposition; GSEA, gene set enrichment analysis.
In addition, we performed cluster analysis based on the expression patterns of 72 DEGs in Clusters A and B using the NMF algorithm. The optimal cluster number, k = 2, was determined based on cophenetic, dispersion, and silhouette metrics (Fig. 8C and D). The HF samples were divided into two subgroups, Subgroup A (n = 88) and Subgroup B (n = 89), according to the distinct expression patterns of the 72 DEGs. PCA demonstrated significant expression differences between these subgroups (Fig. 8E). A dendrogram showed a similar grouping tendency for the two clusters and gene subgroups (Fig. 8F). The expression of 13 CRGs among the two subgroups is depicted in Fig. 8G and Supplementary Fig. 2D, with genes NPPA, NAMPT, and SERPINE1 exhibiting notable differences between the subgroups.
3.8. Identification of immune microenvironment and biological function features in different subgroups
Firstly, we conducted an analysis of the differences in immune microenvironments between Subgroup A and Subgroup B. In comparison to Subgroup A, Subgroup B exhibited higher levels of infiltration by monocyte immune cells as demonstrated by the results. Conversely, In Subgroup A, higher infiltration levels were observed in activated dendritic cells and type 17 T helper cells (Fig. 8H). Next, we performed GSVA between the different subgroups. Enrichment analysis of the HALLMARK gene set showed that Subgroup B was enriched in Notch signaling pathways, TNF-β signaling pathways, angiogenesis, and epithelial-mesenchymal transition. In contrast, cardiac metabolic processes such as fatty acid metabolism, oxidative phosphorylation, peroxidation products, and lipid metabolism were significantly enriched in Subgroup A (Fig. 9A). Biological processes upregulated in Subgroup B included Tnfsf11-mediated signaling pathways, positive regulation of the epithelial-to-mesenchymal transition during endocardial formation, and regulatory mechanisms for vascular-associated smooth muscle contraction (Fig. 9B). In Subgroup B, the expression levels of cardiomyopathy, arrhythmogenic right ventricular cardiomyopathy, and regulation of the actin cytoskeleton were significantly upregulated, whereas in Subgroup A, there was a significant enrichment of metabolic-related pathways such as fatty acid metabolism and butyrate metabolism (Fig. 9C). These findings suggest that patients with HF in Subgroup A have higher metabolic levels, whereas those in Subgroup B have higher levels of immune infiltration.
Fig. 9.
GSVA analysis to assess biological function and pathway differences between the two subgroups. (A) GSVA analysis based on the HALLMARK gene set. (C) Biological process analysis based on the GO gene set. (C) Enrichment pathway based on the KEGG pathway. GSVA, genomic variation analysis.
3.9. Identification and functional enrichment analysis of hub genes between two subgroups
The clustering results of the 177 samples are shown in Supplementary Fig. 3. Two outlier samples (GSM379815 and GSM380018) were excluded, and 175 samples were included. At a soft threshold of β = 6, the scale-free R2 value reached 0.9, resulting in a greater average connectivity (Fig. 10A). Hierarchical clustering identified a total of eleven modules (Fig. 10B). Among them, the black and green modules showed positive correlations (Fig. 10C). From the module-trait correlation heatmap, the black module was positively correlated with Subgroup A (R2 = 0.77) (Fig. 10D and E). Therefore, we constructed a PPI network by utilizing genes extracted from the black module and screened for hub genes using the MCC algorithm in cytoHubba plugin (Fig. 10F). Additionally, we performed biological function annotation for the top 50 ranked genes. The hub genes were found to be primarily associated with cardiac muscle tissue development, heart contraction, calcium ion transmembrane transport, and regulation of heart contraction according to GO analysis (Fig. 10G). KEGG analysis revealed significant enrichment of axon guidance, calcium signaling pathway, cAMP signaling pathway, and circadian entrainment pathways (Fig. 10H). We confirmed through the CTD that most hub genes participate in the occurrence and development of HF or other cardiovascular diseases (Fig. 11A–J).
Fig. 10.
Identification of hub genes between the two subgroups by the WGCNA method. (A) The network constructed when power index 6 was chosen as the appropriate soft threshold was more consistent with a scale-free topology. (B) Co-expression networks were constructed based on the optimal soft threshold to divide the genes into 11 different modules. (C) Cluster tree and correlation heat map between modules. (D) Correlation and significance between modules and features. the highest correlation was found between MEblack and features. (E) Scatter plot of module feature genes in the black module. mm and gs were positively correlated. (F) Protein-protein interaction network (PPI) ranking of gene importance. (G, H) GO and KEGG enrichment analysis of the top 50 significant genes in the PPI network. mm, module membership; GS, gene significance; GO, gene ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes.
Fig. 11.
Association of hub genes with cardiovascular disease, evidence from the CTD database. (A–J) Relationship between expression levels of AQP4, KALRN, LRCC7, DLG1, MAP2, NPPA, ERBB4, NRG1, NTN1, and GRIN2A and inferred scores of cardiovascular disease. CTD, Comparative Toxicogenomics Database.
4. Discussion
Circadian rhythms are a manifestation of the body's biological clock, regulating many physiological processes, including metabolism, immunity, and cell death [15,41,42]. Existing research suggests that there is a close relationship between circadian rhythms and HF, but the exact mechanisms of molecular clocks in these phenomena are yet to be fully understood. Therefore, we conducted bioinformatics approaches to investigate the role of CRGs in HF and reveal their association with immune and metabolic processes. In this study, we employed various machine learning algorithms to screen five diagnostic biomarkers that demonstrated excellent diagnostic ability for HF, yielding good predictive results in the validation set. Secondly, we performed unsupervised clustering of HF samples based on CRGs expression profiles for the first time, identifying two subclusters with different expression patterns. GSVA revealed that Cluster A was mainly associated with immune processes while Cluster B was mainly associated with metabolic processes. Finally, based on the DEGs between the two clusters from the first clustering, we further identified two gene subgroups with unique immune and metabolic features and revealed hub genes related to circadian rhythm changes through WGCNA.
In recent years, the impact of CRG rhythmic expression changes on cardiac phenotypes has been demonstrated in knockout mouse models. Mice without BMAL1 exhibit disrupted circadian rhythms in blood pressure and heart rate, accompanied by cardiac electrophysiological and metabolic abnormalities [14,43]. Moreover, REV-ERBα agonists and antagonists have been shown to provide cardiac protection in mouse models, preventing heart failure [44,45]. Therefore, the exploration of novel biomarkers may bring unexpected benefits for HF treatment. Machine learning can perfectly solve data feature dimensionality and redundancy issues, enabling more accurate identification of diagnostically valuable genes [46]. Based on this, we combined four machine learning algorithms (including RF, Boruta, SVM-REF, and XGBoost) to obtain nine diagnostic biomarkers. In addition, we further validated the nine diagnostic biomarkers using multivariate logistic regression analysis and two external validation datasets, finding that only NAMPT, SERPINA3, MAPK10, NPPA, and SLC2A1 could accurately predict HF.
Current evidence indicates that immune and inflammatory activation have a significant impact on the development of HF, and immune responses are strictly controlled by circadian rhythm mechanisms [47,48]. Therefore, we conducted an analysis to investigate the correlation between gene expression and immune cell infiltration. The correlation analysis revealed that SERPINA3 and SLC2A1 were strongly positively correlated with Central memory CD8 T cells and negatively correlated with Type 2 T helper cells. SERPINA3 mainly activates the immune process by regulating neutrophil cathepsin G and leukocyte elastase [49]. Proteomic results also highlighted the role of SERPINA3 in the epicardial adipose tissue of HF patients, where regulation of SERPINA3 metabolism promotes epicardial adipose inflammation associated with HF [50]. Furthermore, SERPINA3 can improve prognostic stratification for patients with newly diagnosed or worsening heart failure [51]. Consistent with previous research, dysregulated or imbalanced SERPINA3 gene expression is associated with the progression of HF patients. SLC2A1 is an insulin-independent glucose transporter that predominantly facilitates basal cardiac glucose uptake in quiescent muscle cells [52]. Reduced SLC2A1 expression in the human heart indicates that reduced SLC2A1-mediated glucose uptake and utilization could contribute to the development of HF [53]. Notably, glucose availability is a key factor in regulating innate and adaptive immune responses as well as cancer. It has been shown that SLC2A1 is the rate-limiting glucose transporter on pro-inflammatory polarized macrophages. SLC2A1 overexpression results in increased glucose uptake and metabolism, exacerbating the inflammatory response [54]. A significant negative correlation was observed between NPPA and Type 17 T helper cells. Atrial natriuretic peptide (ANP), which is encoded by NPPA, is a member of the natriuretic peptide family and has been utilized as a diagnostic biomarker for HF [55], further confirming the accuracy of this study. MAPK10 was significantly positively correlated with Type 2 T helper cells. The gene is primarily expressed in the heart, brain, and testes and is a member of the JNK family [56]. MAPK10 knockdown alleviates diabetes-induced atrial fibrillation by inhibiting inflammation, fibrosis, electrical dysfunction, and apoptosis [57]. Specific knockdown in mice can lead to myocardial dysfunction, including diastolic dysfunction and cell apoptosis [58,59]. Furthermore, inhibiting MAPK10 phosphorylation can effectively alleviate synaptic dysfunction, inflammatory responses, and various neurodegenerative pathologies associated with Alzheimer's disease [60]. However, few reports exist on its role in HF patients. Based on the above analysis, MAPK10 may regulate the occurrence of heart failure by modulating inflammatory responses, myocardial cell apoptosis, and electrical signal conduction. Interestingly, NAMPT is significantly correlated with all immune cells. Various types of immune cells regulate NAMPT through diverse inflammatory signals. Inflammatory cytokines released by inflammatory cells in response to chronic inflammation can suppress the expression of NAMPT [61]. TNF-α inhibits the activation of clock genes induced by the CLOCK-BMAL1 complex, thereby disrupting the function of clock genes [62]. In addition, knocking down or overexpressing NAMPT both result in reduced protein acetylation, suppressed metabolic genes, and mitochondrial dysfunction [63]. Moreover, the expression of NAMPT is controlled by the transcription factor Kruppel-like factor 15 (KLF15) [64]. Research suggests that KLF15 regulates catabolic metabolism and oxidative stress responses in the heart through transcriptional activation of NAD-producing enzyme NAMPT [65]. In our study, we also observed an increase in NAMPT expression levels in metabolic subtype A. This may indicate a disruption of NAD homeostasis in HF patients. Consequently, NAMPT emerges as a crucial hub gene enabling regulation of cardiac metabolism and active oxygen scavenging. Integrating this information, we believe that disrupted normal CRGs expression may disrupt the cardiac energy balance and immune function imbalance, thereby exacerbating HF development.
To further explore the distinctions in immune features between normal and HF samples, as well as the relationship between gene features connected to circadian rhythm and immune cells, we performed quantification analysis of 28 immune cell infiltration scores utilizing the single-sample gene set enrichment analysis (ssGSEA) algorithm. Our findings indicate that the quantity of a majority of immune cells varies in HF and healthy control samples, comprising neutrophils, macrophages, and most lymphocytes. Research indicates that circadian rhythm fluctuations can convey information to the immune system, mediating both innate and adaptive immune responses [66,67]. The fluctuations of the circadian rhythm in neutrophils may aid their migration to various tissues during their active phase, whilst safeguarding blood vessels against the effect of these extremely active cells during their resting phase [68]. Macrophages release the inflammatory cytokines IL-1β, IL-6 and TNF-α, dependent on the molecular clock phase. Most genes expressed in a circadian rhythm in macrophages play a role in immune response at varying levels, including Toll-like receptor (TLR) expression [69]. In our investigation, we discovered that there were varying degrees of macrophage infiltration between two groups, and that the Toll-like receptor signaling pathway was significantly enriched. As a result, we hypothesize that circadian rhythms impact the regulation of macrophage expression in the context of HF. Research has also shown that CD8 T cells lacking Bmal1 produce reduced levels of IL-2, IFN-γ and TNF-α when co-cultured with wild-type cells [70]. Additionally, circadian rhythm genes regulate the periodic migration of T cells to lymph nodes and the timing of lymph node transitions [71]. Chronic disruption of circadian rhythms during HF could significantly disrupt various immune responses, perhaps by regulating immune cell cytokine release rhythmically. Although our study offers fresh insight into the link between circadian rhythms and immunity in HF, further research is necessary to determine the specific mechanisms.
To better understand the heterogeneity among HF patients, we carried out unsupervised clustering analysis based on CRGs. Cluster A was significantly enriched in TGF-β signaling pathway, angiogenesis, NFκB-mediated signal transduction, TNF-α signaling pathway, blood coagulation function, epithelial and stromal cell transition, and IL6-JAK-STAT3 signaling pathway. Cluster B mainly focused on metabolism-related pathways, such as fatty acid metabolism and oxidative phosphorylation. Additionally, NMF clustering based on differentially expressed genes also effectively classified HF patients into two subgroups. The Notch signaling pathway, TNF-β signaling pathway, angiogenesis, and epithelial and stromal cell transition were significantly enriched in Subgroup B, while cardiac metabolic processes such as fatty acid metabolism, lipid metabolism, reactive oxygen species, and oxidative phosphorylation were enriched in Subgroup A. Prior research has established that numerous genes related to metabolism, particularly those involved in fatty acid metabolism, display circadian oscillations [13]. Research has also shown that intramyocardial lipid content is higher in the early morning [72]. The immune system also participates in the process of heart failure. Hemodynamic changes in HF patients trigger cardiac immune responses [73]. Immune cell differentiation and cytokine release also exhibit time dependence [74]. Existing research confirms that targeting circadian rhythm mechanisms inhibits the formation of cardiac NLRP3 inflammasomes and subsequent cascade reactions [75]. Therefore, CRG-based subtyping can better help us understand the influence of different biological processes on heart failure, thereby adopting appropriate intervention measures to slow down heart failure development.
However, we must acknowledge the limitations of our study. Firstly, our results have not been validated in vivo or in vitro due to the absence of experimental conditions for follow-up trials. Second, the dataset was obtained from a public database, and we did not obtain clinical information related to patients and therefore could not perform a prognostic analysis. Lastly, transcriptomic features may vary at different stages of heart failure, and individual differences must be considered when including samples.
5. Conclusion
The machine learning based diagnostic model has higher accuracy and can perfectly discriminate HF patients from normal patients. In addition, we investigated the relationship between CRGs and immune infiltration, as well as the differences in biological function and immune cell infiltration between different clusters and subgroups. The subclusters of HF patients showed different immune and metabolic patterns among themselves. In summary, our study provides comprehensive analysis for the first time on the role of CRGs in HF, which offers valuable references for targeted inhibition of immune response and energy metabolism through regulating circadian rhythm mechanisms in the treatment of HF.
Source of funding
This work was supported by the Shanghai Shuguang Project [No. 21SG37] and the National Natural Science Foundation of China [No. 81770244].
Data availability
The data relevant to this study (GSE57338, GSE79962, GSE26887, and GSE5406) have all been deposited in the publicly available repository, the GEO database (www.ncbi.nlm.nih.gov/geo/), as they originate from public databases. The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Summary
This study has shed light on the connection between circadian rhythms and heart failure (HF) pathogenesis. The study used machine learning algorithms to identify differentially expressed circadian rhythm-related genes (DE-CRGs) in HF patients, and diagnostic models were constructed based on these feature genes. Nine feature genes were obtained through cross-referencing results from four distinct machine learning algorithms, and multivariate LASSO regression and external dataset validation were performed to select five key genes with diagnostic value, including NAMPT, SERPINA3, MAPK10, NPPA, and SLC2A1. Consensus clustering analysis could divide HF patients into two distinct clusters, which exhibited different biological functions and immune characteristics. Additionally, two subgroups were distinguished using the non-negative matrix factorization (NMF) algorithm based on circadian rhythm associated differentially expressed genes. Studies on immune infiltration showed marked variances in levels of immune infiltration between these subgroups. Finally, the Weighted Gene Co-expression Network Analysis (WGCNA) method was utilized to discern the modules that had the closest association with the two observed subgroups, and hub genes were pinpointed via protein-protein interaction (PPI) networks. GRIN2A, DLG1, ERBB4, LRRC7, and NRG1 were circadian rhythm-related hub genes closely associated with HF.
The study provides valuable references for further elucidating the pathogenesis of HF, offering beneficial insights for targeting circadian rhythm mechanisms to regulate immune responses and energy metabolism in HF treatment. Five genes identified by the study as diagnostic features could be potential targets for therapy for HF. However, further research is needed to fully understand the relationship between circadian rhythms and heart failure.
CRediT authorship contribution statement
Xuefu Wang: Writing – original draft, Visualization, Software, Formal analysis, Data curation. Jin Rao: Methodology, Formal analysis, Data curation. Li Zhang: Validation, Supervision, Project administration, Methodology, Investigation. Xuwen Liu: Validation, Supervision, Software, Investigation. Yufeng Zhang: Writing – review & editing, Supervision, Project administration, Funding acquisition, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Many thanks to the uploaders of GEO data for providing us with transcriptome data that we can continue to explore.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e27049.
Appendix A. Supplementary data
The following is the supplementary data to this article:
References
- 1.McDonagh T.A., Metra M., Adamo M., Gardner R.S., Baumbach A., Böhm M., Burri H., Butler J., Čelutkienė J., Chioncel O., Cleland J.G.F., Coats A.J.S., Crespo-Leiro M.G., Farmakis D., Gilard M., Heymans S., Hoes A.W., Jaarsma T., Jankowska E.A., Lainscak M., Lam C.S.P., Lyon A.R., McMurray J.J.V., Mebazaa A., Mindham R., Muneretto C., Francesco Piepoli M., Price S., Rosano G.M.C., Ruschitzka F., Kathrine Skibelund A. 2021 ESC Guidelines for the diagnosis and treatment of acute and chronic heart failure. Eur. Heart J. 2021;42:3599–3726. doi: 10.1093/eurheartj/ehab368. [DOI] [PubMed] [Google Scholar]
- 2.Orso F., Fabbri G., Maggioni A.P. Epidemiology of heart failure. Handb. Exp. Pharmacol. 2017;243:15–33. doi: 10.1007/164_2016_74. [DOI] [PubMed] [Google Scholar]
- 3.Rossignol P., Hernandez A.F., Solomon S.D., Zannad F. Heart failure drug treatment. Lancet. 2019;393:1034–1044. doi: 10.1016/S0140-6736(18)31808-7. [DOI] [PubMed] [Google Scholar]
- 4.McDonagh T.A., Metra M., Adamo M., Gardner R.S., Baumbach A., Böhm M., Burri H., Butler J., Čelutkienė J., Chioncel O., Cleland J.G.F., Crespo-Leiro M.G., Farmakis D., Gilard M., Heymans S., Hoes A.W., Jaarsma T., Jankowska E.A., Lainscak M., Lam C.S.P., Lyon A.R., McMurray J.J.V., Mebazaa A., Mindham R., Muneretto C., Francesco Piepoli M., Price S., Rosano G.M.C., Ruschitzka F., Skibelund A.K. Eur Heart J; 2023. Focused Update of the 2021 ESC Guidelines for the Diagnosis and Treatment of Acute and Chronic Heart Failure. 2023. [DOI] [PubMed] [Google Scholar]
- 5.Tsao C.W., Aday A.W., Almarzooq Z.I., Anderson C.A.M., Arora P., Avery C.L., Baker-Smith C.M., Beaton A.Z., Boehme A.K., Buxton A.E., Commodore-Mensah Y., Elkind M.S.V., Evenson K.R., Eze-Nliam C., Fugar S., Generoso G., Heard D.G., Hiremath S., Ho J.E., Kalani R., Kazi D.S., Ko D., Levine D.A., Liu J., Ma J., Magnani J.W., Michos E.D., Mussolino M.E., Navaneethan S.D., Parikh N.I., Poudel R., Rezk-Hanna M., Roth G.A., Shah N.S., St-Onge M.P., Thacker E.L., Virani S.S., Voeks J.H., Wang N.Y., Wong N.D., Wong S.S., Yaffe K., Martin S.S. Heart disease and stroke statistics-2023 update: a report from the American heart association. Circulation. 2023;147:e93–e621. doi: 10.1161/CIR.0000000000001123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crnko S., Du Pré B.C., Sluijter J.P.G., Van Laake L.W. Circadian rhythms and the molecular clock in cardiovascular biology and disease. Nat. Rev. Cardiol. 2019;16:437–447. doi: 10.1038/s41569-019-0167-4. [DOI] [PubMed] [Google Scholar]
- 7.El Jamal N., Lordan R., Teegarden S.L., Grosser T., FitzGerald G. The circadian biology of heart failure. Circ. Res. 2023;132:223–237. doi: 10.1161/CIRCRESAHA.122.321369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Takahashi J.S. Transcriptional architecture of the mammalian circadian clock. Nat. Rev. Genet. 2017;18:164–179. doi: 10.1038/nrg.2016.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bass J., Lazar M.A. Circadian time signatures of fitness and disease. Science. 2016;354:994–999. doi: 10.1126/science.aah4965. [DOI] [PubMed] [Google Scholar]
- 10.Rabinovich-Nikitin I., Lieberman B., Martino T.A., Kirshenbaum L.A. Circadian-regulated cell death in cardiovascular diseases. Circulation. 2019;139:965–980. doi: 10.1161/CIRCULATIONAHA.118.036550. [DOI] [PubMed] [Google Scholar]
- 11.Alibhai F.J., Reitz C.J., Peppler W.T., Basu P., Sheppard P., Choleris E., Bakovic M., Martino T.A. Female ClockΔ19/Δ19 mice are protected from the development of age-dependent cardiomyopathy. Cardiovasc. Res. 2018;114:259–271. doi: 10.1093/cvr/cvx185. [DOI] [PubMed] [Google Scholar]
- 12.Martino T.A., Oudit G.Y., Herzenberg A.M., Tata N., Koletar M.M., Kabir G.M., Belsham D.D., Backx P.H., Ralph M.R., Sole M.J. Circadian rhythm disorganization produces profound cardiovascular and renal disease in hamsters. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2008;294:R1675–R1683. doi: 10.1152/ajpregu.00829.2007. [DOI] [PubMed] [Google Scholar]
- 13.Song S., Tien C.L., Cui H., Basil P., Zhu N., Gong Y., Li W., Li H., Fan Q., Min Choi J., Luo W., Xue Y., Cao R., Zhou W., Ortiz A.R., Stork B., Mundra V., Putluri N., York B., Chu M., Chang J., Yun Jung S., Xie L., Song J., Zhang L., Sun Z. Myocardial rev-erb-mediated diurnal metabolic rhythm and obesity paradox. Circulation. 2022;145:448–464. doi: 10.1161/CIRCULATIONAHA.121.056076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Young M.E., Brewer R.A., Peliciari-Garcia R.A., Collins H.E., He L., Birky T.L., Peden B.W., Thompson E.G., Ammons B.J., Bray M.S., Chatham J.C., Wende A.R., Yang Q., Chow C.W., Martino T.A., Gamble K.L. Cardiomyocyte-specific BMAL1 plays critical roles in metabolism, signaling, and maintenance of contractile function of the heart. J. Biol. Rhythm. 2014;29:257–276. doi: 10.1177/0748730414543141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang L., Jain M.K. Circadian regulation of cardiac metabolism. J. Clin. Invest. 2021;131 doi: 10.1172/JCI148276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Young M.E., Razeghi P., Cedars A.M., Guthrie P.H., Taegtmeyer H. Intrinsic diurnal variations in cardiac metabolism and contractile function. Circ. Res. 2001;89:1199–1208. doi: 10.1161/hh2401.100741. [DOI] [PubMed] [Google Scholar]
- 17.Xu W., Jain M.K., Zhang L. Molecular link between circadian clocks and cardiac function: a network of core clock, slave clock, and effectors. Curr. Opin. Pharmacol. 2021;57:28–40. doi: 10.1016/j.coph.2020.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cox S.L., O'Siorain J.R., Fagan L.E., Curtis A.M., Carroll R.G. Intertwining roles of circadian and metabolic regulation of the innate immune response. Semin. Immunopathol. 2022;44:225–237. doi: 10.1007/s00281-021-00905-5. [DOI] [PubMed] [Google Scholar]
- 19.Galluzzi L., Vitale I., Aaronson S.A., Abrams J.M., Adam D., Agostinis P., Alnemri E.S., Altucci L., Amelio I., Andrews D.W., Annicchiarico-Petruzzelli M., Antonov A.V., Arama E., Baehrecke E.H., Barlev N.A., Bazan N.G., Bernassola F., Bertrand M.J.M., Bianchi K., Blagosklonny M.V., Blomgren K., Borner C., Boya P., Brenner C., Campanella M., Candi E., Carmona-Gutierrez D., Cecconi F., Chan F.K., Chandel N.S., Cheng E.H., Chipuk J.E., Cidlowski J.A., Ciechanover A., Cohen G.M., Conrad M., Cubillos-Ruiz J.R., Czabotar P.E., D'Angiolella V., Dawson T.M., Dawson V.L., De Laurenzi V., De Maria R., Debatin K.M., DeBerardinis R.J., Deshmukh M., Di Daniele N., Di Virgilio F., Dixit V.M., Dixon S.J., Duckett C.S., Dynlacht B.D., El-Deiry W.S., Elrod J.W., Fimia G.M., Fulda S., García-Sáez A.J., Garg A.D., Garrido C., Gavathiotis E., Golstein P., Gottlieb E., Green D.R., Greene L.A., Gronemeyer H., Gross A., Hajnoczky G., Hardwick J.M., Harris I.S., Hengartner M.O., Hetz C., Ichijo H., Jäättelä M., Joseph B., Jost P.J., Juin P.P., Kaiser W.J., Karin M., Kaufmann T., Kepp O., Kimchi A., Kitsis R.N., Klionsky D.J., Knight R.A., Kumar S., Lee S.W., Lemasters J.J., Levine B., Linkermann A., Lipton S.A., Lockshin R.A., López-Otín C., Lowe S.W., Luedde T., Lugli E., MacFarlane M., Madeo F., Malewicz M., Malorni W., Manic G., et al. Molecular mechanisms of cell death: recommendations of the nomenclature committee on cell death 2018. Cell Death Differ. 2018;25:486–541. doi: 10.1038/s41418-017-0012-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saeidian A.H., Youssefian L., Vahidnezhad H., Uitto J. Research techniques made simple: whole-transcriptome sequencing by RNA-seq for diagnosis of monogenic disorders. J. Invest. Dermatol. 2020;140:1117–1126.e1. doi: 10.1016/j.jid.2020.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hanauer D.A., Rhodes D.R., Sinha-Kumar C., Chinnaiyan A.M. Bioinformatics approaches in the study of cancer. Curr. Mol. Med. 2007;7:133–141. doi: 10.2174/156652407779940431. [DOI] [PubMed] [Google Scholar]
- 22.Joshi A., Rienks M., Theofilatos K., Mayr M. Systems biology in cardiovascular disease: a multiomics approach. Nat. Rev. Cardiol. 2021;18:313–330. doi: 10.1038/s41569-020-00477-1. [DOI] [PubMed] [Google Scholar]
- 23.Vijayasarveswari V., Andrew A.M., Jusoh M., Sabapathy T., Raof R.A.A., Yasin M.N.M., Ahmad R.B., Khatun S., Rahim H.A. Multi-stage feature selection (MSFS) algorithm for UWB-based early breast cancer size prediction. PLoS One. 2020;15 doi: 10.1371/journal.pone.0229367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li J., Xu Q., Wu M., Huang T., Wang Y. Pan-Cancer classification based on self-normalizing neural networks and feature selection. Front. Bioeng. Biotechnol. 2020;8:766. doi: 10.3389/fbioe.2020.00766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Forrest I.S., Petrazzini B.O., Duffy Á., Park J.K., Marquez-Luna C., Jordan D.M., Rocheleau G., Cho J.H., Rosenson R.S., Narula J., Nadkarni G.N., Do R. Machine learning-based marker for coronary artery disease: derivation and validation in two longitudinal cohorts. Lancet. 2023;401:215–225. doi: 10.1016/S0140-6736(22)02079-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qin X., Yi S., Rong J., Lu H., Ji B., Zhang W., Ding R., Wu L., Chen Z. Identification of anoikis-related genes classification patterns and immune infiltration characterization in ischemic stroke based on machine learning. Front. Aging Neurosci. 2023;15 doi: 10.3389/fnagi.2023.1142163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu Y., Morley M., Brandimarto J., Hannenhalli S., Hu Y., Ashley E.A., Tang W.H., Moravec C.S., Margulies K.B., Cappola T.P., Li M. RNA-Seq identifies novel myocardial gene expression signatures of heart failure. Genomics. 2015;105:83–89. doi: 10.1016/j.ygeno.2014.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matkovich S.J., Al Khiami B., Efimov I.R., Evans S., Vader J., Jain A., Brownstein B.H., Hotchkiss R.S., Mann D.L. Widespread down-regulation of cardiac mitochondrial and sarcomeric genes in patients with sepsis. Crit. Care Med. 2017;45:407–414. doi: 10.1097/CCM.0000000000002207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Greco S., Fasanaro P., Castelvecchio S., D'Alessandra Y., Arcelli D., Di Donato M., Malavazos A., Capogrossi M.C., Menicanti L., Martelli F. MicroRNA dysregulation in diabetic ischemic heart failure patients. Diabetes. 2012;61:1633–1641. doi: 10.2337/db11-0952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hannenhalli S., Putt M.E., Gilmore J.M., Wang J., Parmacek M.S., Epstein J.A., Morrisey E.E., Margulies K.B., Cappola T.P. Transcriptional genomics associates FOX transcription factors with human heart failure. Circulation. 2006;114:1269–1276. doi: 10.1161/CIRCULATIONAHA.106.632430. [DOI] [PubMed] [Google Scholar]
- 31.Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., Smyth G.K. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu G., Wang L.G., Han Y., He Q.Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kursa M.B., Rudnicki W.R. Feature selection with the Boruta package. J. Stat. Software. 2010;36:1–13. [Google Scholar]
- 34.Breiman L. Random forests. Mach. Learn. 2001;45:5–32. [Google Scholar]
- 35.Noble W.S. What is a support vector machine? Nat. Biotechnol. 2006;24:1565–1567. doi: 10.1038/nbt1206-1565. [DOI] [PubMed] [Google Scholar]
- 36.Chen T., Guestrin C. Proceedings of the 22nd Acm Sigkdd International Conference on Knowledge Discovery and Data Mining. 2016. Xgboost: a scalable tree boosting system; pp. 785–794. [Google Scholar]
- 37.Engebretsen S., Bohlin J. Statistical predictions with glmnet. Clin. Epigenet. 2019;11:123. doi: 10.1186/s13148-019-0730-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wilkerson M.D., Hayes D.N. ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinformatics. 2010;26:1572–1573. doi: 10.1093/bioinformatics/btq170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hänzelmann S., Castelo R., Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinf. 2013;14:7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gaujoux R., Seoighe C. A flexible R package for nonnegative matrix factorization. BMC Bioinf. 2010;11:367. doi: 10.1186/1471-2105-11-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang M., Pan W., Xu Y., Zhang J., Wan J., Jiang H. Microglia-mediated neuroinflammation: a potential target for the treatment of cardiovascular diseases. J. Inflamm. Res. 2022;15:3083–3094. doi: 10.2147/JIR.S350109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kohsaka A., Das P., Hashimoto I., Nakao T., Deguchi Y., Gouraud S.S., Waki H., Muragaki Y., Maeda M. The circadian clock maintains cardiac function by regulating mitochondrial metabolism in mice. PLoS One. 2014;9 doi: 10.1371/journal.pone.0112811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Curtis A.M., Cheng Y., Kapoor S., Reilly D., Price T.S., Fitzgerald G.A. Circadian variation of blood pressure and the vascular response to asynchronous stress. Proc. Natl. Acad. Sci. U. S. A. 2007;104:3450–3455. doi: 10.1073/pnas.0611680104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Solt L.A., Wang Y., Banerjee S., Hughes T., Kojetin D.J., Lundasen T., Shin Y., Liu J., Cameron M.D., Noel R., Yoo S.H., Takahashi J.S., Butler A.A., Kamenecka T.M., Burris T.P. Regulation of circadian behaviour and metabolism by synthetic REV-ERB agonists. Nature. 2012;485:62–68. doi: 10.1038/nature11030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang L., Zhang R., Tien C.L., Chan R.E., Sugi K., Fu C., Griffin A.C., Shen Y., Burris T.P., Liao X., Jain M.K. REV-ERBα ameliorates heart failure through transcription repression. JCI Insight. 2017;2 doi: 10.1172/jci.insight.95177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liang S., Ma A., Yang S., Wang Y., Ma Q. A review of matched-pairs feature selection methods for gene expression data analysis. Comput. Struct. Biotechnol. J. 2018;16:88–97. doi: 10.1016/j.csbj.2018.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Keller M., Mazuch J., Abraham U., Eom G.D., Herzog E.D., Volk H.D., Kramer A., Maier B. A circadian clock in macrophages controls inflammatory immune responses. Proc. Natl. Acad. Sci. U. S. A. 2009;106:21407–21412. doi: 10.1073/pnas.0906361106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Orozco-Solis R., Aguilar-Arnal L. Circadian regulation of immunity through epigenetic mechanisms. Front. Cell. Infect. Microbiol. 2020;10:96. doi: 10.3389/fcimb.2020.00096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sorokin V., Woo C.C. Role of Serpina3 in vascular biology. Int. J. Cardiol. 2020;304:154–155. doi: 10.1016/j.ijcard.2019.12.030. [DOI] [PubMed] [Google Scholar]
- 50.Zhao L., Guo Z., Wang P., Zheng M., Yang X., Liu Y., Ma Z., Chen M., Yang X. Proteomics of epicardial adipose tissue in patients with heart failure. J. Cell Mol. Med. 2020;24:511–520. doi: 10.1111/jcmm.14758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Delrue L., Vanderheyden M., Beles M., Paolisso P., Di Gioia G., Dierckx R., Verstreken S., Goethals M., Heggermont W., Bartunek J. Circulating SERPINA3 improves prognostic stratification in patients with a de novo or worsened heart failure. ESC Heart Fail. 2021;8:4780–4790. doi: 10.1002/ehf2.13659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mueckler M., Makepeace C. Transmembrane segment 6 of the Glut1 glucose transporter is an outer helix and contains amino acid side chains essential for transport activity. J. Biol. Chem. 2008;283:11550–11555. doi: 10.1074/jbc.M708896200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Luptak I., Yan J., Cui L., Jain M., Liao R., Tian R. Long-term effects of increased glucose entry on mouse hearts during normal aging and ischemic stress. Circulation. 2007;116:901–909. doi: 10.1161/CIRCULATIONAHA.107.691253. [DOI] [PubMed] [Google Scholar]
- 54.Freemerman A.J., Johnson A.R., Sacks G.N., Milner J.J., Kirk E.L., Troester M.A., Macintyre A.N., Goraksha-Hicks P., Rathmell J.C., Makowski L. Metabolic reprogramming of macrophages: glucose transporter 1 (GLUT1)-mediated glucose metabolism drives a proinflammatory phenotype. J. Biol. Chem. 2014;289:7884–7896. doi: 10.1074/jbc.M113.522037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Goetze J.P., Bruneau B.G., Ramos H.R., Ogawa T., de Bold M.K., de Bold A.J. Cardiac natriuretic peptides. Nat. Rev. Cardiol. 2020;17:698–717. doi: 10.1038/s41569-020-0381-0. [DOI] [PubMed] [Google Scholar]
- 56.Shvedova M., Anfinogenova Y., Atochina-Vasserman E.N., Schepetkin I.A., Atochin D.N. c-Jun N-terminal kinases (JNKs) in myocardial and cerebral ischemia/reperfusion injury. Front. Pharmacol. 2018;9:715. doi: 10.3389/fphar.2018.00715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu F., Deng Y., Zhao Y., Li Z., Gao J., Zhang Y., Yang X., Liu Y., Xia Y. Time series RNA-seq analysis identifies MAPK10 as a critical gene in diabetes mellitus-induced atrial fibrillation in mice. J. Mol. Cell. Cardiol. 2022;168:70–82. doi: 10.1016/j.yjmcc.2022.04.013. [DOI] [PubMed] [Google Scholar]
- 58.Urano F., Wang X., Bertolotti A., Zhang Y., Chung P., Harding H.P., Ron D. Coupling of stress in the ER to activation of JNK protein kinases by transmembrane protein kinase IRE1. Science. 2000;287:664–666. doi: 10.1126/science.287.5453.664. [DOI] [PubMed] [Google Scholar]
- 59.Deng Y.W., Liu F., Li Z.T., Gao J.H., Zhao Y., Yang X.L., Xia Y.L. Hyperglycemia promotes myocardial dysfunction via the ERS-MAPK10 signaling pathway in db/db mice. Lab. Invest. 2022;102:1192–1202. doi: 10.1038/s41374-022-00819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shen Q., Liu L., Gu X., Xing D. Photobiomodulation suppresses JNK3 by activation of ERK/MKP7 to attenuate AMPA receptor endocytosis in Alzheimer's disease. Aging Cell. 2021;20 doi: 10.1111/acel.13289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kralisch S., Klein J., Lossner U., Bluher M., Paschke R., Stumvoll M., Fasshauer M. Interleukin-6 is a negative regulator of visfatin gene expression in 3T3-L1 adipocytes. Am. J. Physiol. Endocrinol. Metab. 2005;289:E586–E590. doi: 10.1152/ajpendo.00090.2005. [DOI] [PubMed] [Google Scholar]
- 62.Cavadini G., Petrzilka S., Kohler P., Jud C., Tobler I., Birchler T., Fontana A. TNF-alpha suppresses the expression of clock genes by interfering with E-box-mediated transcription. Proc. Natl. Acad. Sci. U. S. A. 2007;104:12843–12848. doi: 10.1073/pnas.0701466104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Byun J., Oka S.I., Imai N., Huang C.Y., Ralda G., Zhai P., Ikeda Y., Ikeda S., Sadoshima J. Both gain and loss of Nampt function promote pressure overload-induced heart failure. Am. J. Physiol. Heart Circ. Physiol. 2019;317:H711–h725. doi: 10.1152/ajpheart.00222.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li L., Li H., Tien C.L., Jain M.K., Zhang L. Kruppel-like factor 15 regulates the circadian susceptibility to ischemia reperfusion injury in the heart. Circulation. 2020;141:1427–1429. doi: 10.1161/CIRCULATIONAHA.119.041664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li L., Xu W., Zhang L. KLF15 regulates oxidative stress response in cardiomyocytes through NAD( ) Metabolites. 2021;11 doi: 10.3390/metabo11090620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cano G., Sved A.F., Rinaman L., Rabin B.S., Card J.P. Characterization of the central nervous system innervation of the rat spleen using viral transneuronal tracing. J. Comp. Neurol. 2001;439:1–18. doi: 10.1002/cne.1331. [DOI] [PubMed] [Google Scholar]
- 67.Elenkov I.J., Wilder R.L., Chrousos G.P., Vizi E.S. The sympathetic nerve--an integrative interface between two supersystems: the brain and the immune system. Pharmacol. Rev. 2000;52:595–638. [PubMed] [Google Scholar]
- 68.Adrover J.M., Del Fresno C., Crainiciuc G., Cuartero M.I., Casanova-Acebes M., Weiss L.A., Huerga-Encabo H., Silvestre-Roig C., Rossaint J., Cossío I., Lechuga-Vieco A.V., García-Prieto J., Gómez-Parrizas M., Quintana J.A., Ballesteros I., Martin-Salamanca S., Aroca-Crevillen A., Chong S.Z., Evrard M., Balabanian K., López J., Bidzhekov K., Bachelerie F., Abad-Santos F., Muñoz-Calleja C., Zarbock A., Soehnlein O., Weber C., Ng L.G., Lopez-Rodriguez C., Sancho D., Moro M.A., Ibáñez B., Hidalgo A. A neutrophil timer coordinates immune defense and vascular protection. Immunity. 2019;50:390–402.e10. doi: 10.1016/j.immuni.2019.01.002. [DOI] [PubMed] [Google Scholar]
- 69.Logan R.W., Sarkar D.K. Circadian nature of immune function. Mol. Cell. Endocrinol. 2012;349:82–90. doi: 10.1016/j.mce.2011.06.039. [DOI] [PubMed] [Google Scholar]
- 70.Sutton C.E., Finlay C.M., Raverdeau M., Early J.O., DeCourcey J., Zaslona Z., O'Neill L.A.J., Mills K.H.G., Curtis A.M. Loss of the molecular clock in myeloid cells exacerbates T cell-mediated CNS autoimmune disease. Nat. Commun. 2017;8:1923. doi: 10.1038/s41467-017-02111-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Druzd D., Matveeva O., Ince L., Harrison U., He W., Schmal C., Herzel H., Tsang A.H., Kawakami N., Leliavski A., Uhl O., Yao L., Sander L.E., Chen C.S., Kraus K., de Juan A., Hergenhan S.M., Ehlers M., Koletzko B., Haas R., Solbach W., Oster H., Scheiermann C. Lymphocyte circadian clocks control lymph node trafficking and adaptive immune responses. Immunity. 2017;46:120–132. doi: 10.1016/j.immuni.2016.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ith M., Stettler C., Xu J., Boesch C., Kreis R. Cardiac lipid levels show diurnal changes and long-term variations in healthy human subjects. NMR Biomed. 2014;27:1285–1292. doi: 10.1002/nbm.3186. [DOI] [PubMed] [Google Scholar]
- 73.Adamo L., Rocha-Resende C., Prabhu S.D., Mann D.L. Reappraising the role of inflammation in heart failure. Nat. Rev. Cardiol. 2020;17:269–285. doi: 10.1038/s41569-019-0315-x. [DOI] [PubMed] [Google Scholar]
- 74.Haspel J.A., Anafi R., Brown M.K., Cermakian N., Depner C., Desplats P., Gelman A.E., Haack M., Jelic S., Kim B.S., Laposky A.D., Lee Y.C., Mongodin E., Prather A.A., Prendergast B.J., Reardon C., Shaw A.C., Sengupta S., Szentirmai É., Thakkar M., Walker W.E., Solt L.A. Perfect timing: circadian rhythms, sleep, and immunity - an NIH workshop summary. JCI Insight. 2020;5 doi: 10.1172/jci.insight.131487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Reitz C.J., Alibhai F.J., Khatua T.N., Rasouli M., Bridle B.W., Burris T.P., Martino T.A. SR9009 administered for one day after myocardial ischemia-reperfusion prevents heart failure in mice by targeting the cardiac inflammasome. Commun. Biol. 2019;2:353. doi: 10.1038/s42003-019-0595-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data relevant to this study (GSE57338, GSE79962, GSE26887, and GSE5406) have all been deposited in the publicly available repository, the GEO database (www.ncbi.nlm.nih.gov/geo/), as they originate from public databases. The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.