Abstract
The discovery of non-coding RNAs (ncRNAs) has unveiled a wide range of transcripts that do not encode proteins but play key roles in several cellular and molecular processes. Long noncoding RNAs (lncRNAs) are specific class of ncRNAs that are longer than 200 nucleotides and have gained significant attention due to their diverse mechanisms of action and potential involvement in various pathological conditions. In the current review, the authors focus on the role of lncRNAs, specifically highlighting the Myocardial Infarction Associated Transcript (MIAT), in non-oncological context. MIAT is a nuclear lncRNA that has been directly linked to myocardial infarction and is reported to control post-transcriptional processes as a competitive endogenous RNA (ceRNA) molecule. It interacts with microRNAs (miRNAs), thereby limiting the translation and expression of their respective target messenger RNA (mRNA) and regulating protein expression. Yet, MIAT has been implicated in other numerous pathological conditions such as other cardiovascular diseases, autoimmune disease, neurodegenerative diseases, metabolic diseases, and many others. In this review, the authors emphasize that MIAT exhibits distinct expression patterns and functions across different pathological conditions and is emerging as potential diagnostic, prognostic, and therapeutic agent. Additionally, the authors highlight the regulatory role of MIAT and shed light on the involvement of lncRNAs and specifically MIAT in various non-oncological pathological conditions.
Keywords: LncRNAs, MIAT, Non-oncological conditions, Biomarkers, Therapeutic targets, Disease pathogenesis
1. Introduction
Through the advancement of transcript mapping and RNA sequencing technologies, it has been discovered that although the human genome is extensively transcribed, only a minor percentage of RNAs (around 2%) are responsible for encoding proteins and a significant portion of the remaining RNAs exhibit noncoding functions [1]. The latest developments in next-generation technologies and RNA-based techniques have contributed to the identification of non-coding RNAs (ncRNAs), a class of transcripts that do not code for proteins [[2], [3], [4], [5]].
These ncRNAs are often classified as housekeeping transcripts or regulatory transcripts. The length of housekeeping ncRNAs is typically from 50 to 500 nucleotides [[6], [7], [8]], and regulatory ncRNAs are classified into two broad categories: long non-coding RNAs (lncRNAs), that include more than 200 nucleotides in length [[9], [10], [11]], and small non-coding RNAs (sncRNAs), that include less than 200 nucleotides in length [[12], [13], [14], [15]].
MicroRNAs (miRNAs) have been extensively studied as potential biomarkers and therapeutic targets for several diseases [3,[16], [17], [18], [19], [20], [21]], however, the focus has now shifted to lncRNAs [[22], [23], [24], [25], [26], [27]]. Genomes undergo extensive transcription, resulting in the generation of lncRNAs [28]. These lncRNAs are classified based on their localization, which is typically either nuclear or cytoplasmic [29]. LncRNAs are still considered novel class of ncRNAs [30]. LncRNAs are produced from two genomic regions: the opposite strand of protein-coding genes, such as natural antisense transcripts, or from intergenic regions of the genome, such as large intergenic ncRNAs [31]. LncRNAs control gene expression by binding to chromatin regulators and interfering RNAs, which can influence the response of cells [32,33]. Aberrant expression of lncRNAs has been correlated to numerous oncological and non-oncological pathological conditions [[34], [35], [36]].
Myocardial Infarction Associated Transcript (MIAT) is a nuclear lncRNA. MIAT was originally linked to an increased risk of myocardial infarction (MI) in genome-wide association studies [37,38]. MIAT has been reported to control post-transcriptional processes, particularly as a competitive endogenous RNA (ceRNA) molecule [11,39]. Indeed, growing research suggests that a real battle between mRNAs and MIAT for the binding of distinct miRNAs. Overall, MIAT either directly binds to miRNAs sponging their effect on target mRNAs or directly bind to the 3′ untranslated regions (3′ UTRs) of the mRNA. This, in turn, limits translation and the production of end product, hence controlling protein expression [40,41].
MIAT has been implicated in a variety of cellular processes and pathological conditions [42]. MIAT can influence different types of oncological conditions such as gastric cancer, hepatocellular carcinoma, colorectal cancer, esophageal cancer, pancreatic cancer, cholangiocarcinoma, ovarian cancer, breast cancer, cervical cancer, renal cell carcinoma, prostate cancer, lung cancer, and thyroid cancer [43]. Apart from cancer, MIAT plays a crucial role in other non-oncological conditions, including MI [44,45], paranoid schizophrenia [46], microvascular dysfunction [47], diabetes mellitus [48], multiple sclerosis [49], neurogenic commitment [50], ischemic stroke [51], osteoarthritis [52], and nuclear body formation [53]. In this review article, the authors will mainly focus on the modulatory vital roles of lncRNAs with a specific emphasis on the role of MIAT in non-oncological conditions.
2. What are LncRNAs and how they work?
LncRNAs are produced from two genomic regions: the opposite strand of protein-coding genes, such as natural antisense transcripts, or from intergenic regions of the genome, such as large intergenic ncRNAs [31]. While lncRNAs do not encode proteins, they share certain similarities with mRNAs in terms of their biology. Similar to mRNAs, the majority of lncRNAs are transcribed by RNA polymerase II (Pol II) and undergo processes such as capping and polyadenylation [54]. It was believed at first that lncRNAs are not stable, however, this is the case for only a small portion of lncRNAs [55]. The majority of lncRNAs find stability through polyadenylation [55,56], whereas non-polyadenylated lncRNAs can be made stable by secondary structures like triple-helical structures at their 3’ ends [57].
These 3’ sequence features not only play a stabilizing role but also aid in an efficient nuclear export [58]. Due to extensive alternative splicing, the number of potential isoforms for the vast majority of lncRNAs dramatically increased [59]. Different studies have shown that although lncRNA splicing efficiency is lower than that of mRNAs [60], lncRNAs are considerably more alternatively spliced than mRNAs [61,62].
LncRNA expression levels are generally lower than those of mRNAs [63]. However, they exhibit stronger tissue-specific expression patterns, indicating their crucial involvement in cell type-specific processes [60,64]. Unlike mRNAs, lncRNAs are not highly conserved in terms of sequence among species. Although this makes it more challenging to assess lncRNA function, it can provide valuable insights into the roles that lncRNAs have evolved to play in different species. Nonetheless, some subsets of lncRNAs exhibit sequence or genomic position conservation and may function similarly across species [65,66].
A recent investigation by Guo et al. [67] found that some lncRNAs exhibit sequence and positional conservation between human and mouse embryonic stem cells. However, these lncRNAs are processed differently, leading to their localization in different subcellular compartments and ultimately serving distinct functions in mouse and human cells. In contrast, conserved mRNAs showed similar localization patterns in both species. Hence, lncRNA sequence conservation does not always indicate conserved functional roles, and lncRNA processing and binding partners significantly impact subcellular distribution and function. A study by Chen L.L [68]. brought attention to the importance and impact of lncRNA localization on their function.
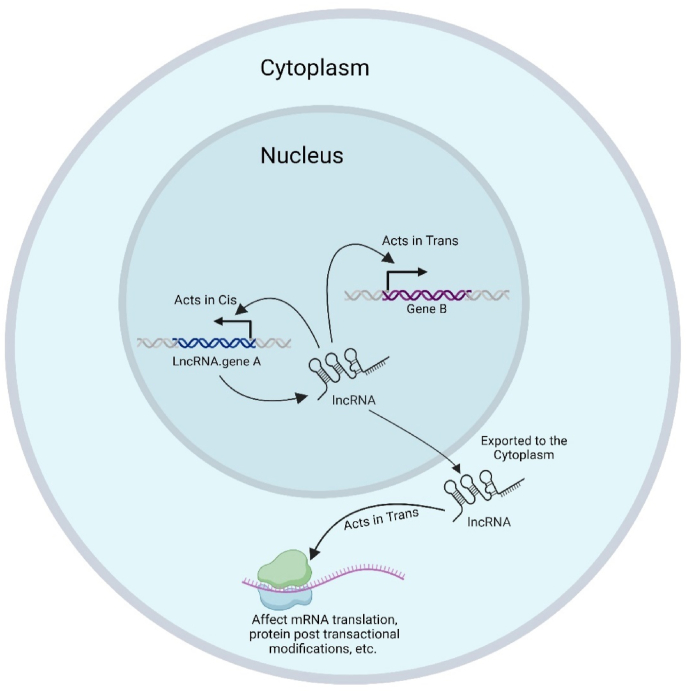
Certain lncRNAs are recognized as important regulators of nuclear functions and exhibit unique patterns of nuclear localization [69,70], while others must be transported to the cytoplasm to carry out their regulatory function. To better understand the relationship between lncRNA localization and function, lncRNAs can be classified based on their subcellular localization. These classifications include those that are strictly localized in the nucleus in cis, those that are primarily localized in the nucleus and function in trans, and those that are predominantly localized and function in the cytoplasm (Fig. 1) [68].
Fig. 1.
LncRNAs modes of Actions. LncRNAs mode of action; they can be strictly localized in the nucleus in cis or localized in the nucleus and function in trans. Additionally, they can be localized and function in the cytoplasm.
The past decade has witnessed extensive research focusing on lncRNAs due to their intriguing nature and diverse mechanisms of action [30]. These RNA molecules have been found to exert their functions through various means [71]. For instance, they can modulate transcription, epigenetic modifications, protein and RNA stability, translation, and post-translational modifications by interacting with DNA [[72], [73], [74]], RNAs [[75], [76], [77], [78]], and proteins as previously reviewed [30,[79], [80], [81]]. Recently, it was also discovered that lncRNAs can directly interact with signaling receptors [82].
An increasing number of lncRNAs have been discovered and have gained recognition for their pivotal involvement in various cellular processes such as cell cycle [8,83], metabolism [84,85], differentiation [[86], [87], [88]], and their association with several diseases [[89], [90], [91]] including malignant and non-malignant disorders [92]. In this review, the authors will focus on the role of lncRNAs in multiple non-oncological illnesses.
3. LncRNAs as central players in different pathological conditions
NcRNAs play a crucial role in several physiological and pathological processes [[93], [94], [95], [96]]. A database of experimentally verified lncRNA-related pathological conditions has been created by compiling data from over 500 publications. This database has listed 321 lncRNAs that are associated with 221 diseases. It is worth noting that lncRNAs have been identified to have a robust link with diverse types of pathological conditions where cardiovascular diseases and neurodegenerative disorders were ranked as highest two categories of diseases associated with lncRNAs after cancer [97].
In oncology, 6 lncRNAs have been identified in prostate cancer, with three being highly specific to the prostate: prostate cancer-associated lncRNA transcript 1 (PCAT1), prostate cancer gene expression marker 1 (PCGEM1), and prostate cancer antigen 3 (PCA3) [98]. HOX antisense intergenic RNA (HOTAIR) lncRNA is implicated in multiple cancers, such as breast cancer, gastric adenocarcinoma, and colorectal cancer. The colon cancer associated transcript 1 (CCAT1) lncRNA has been found to enhance long-range chromatin looping between the myelocytomatosis oncogene (MYC) promoter and its enhancers. Additionally, an increasing number of cancer-associated lncRNAs are being identified [99].
In cognitive abilities, lncRNAs play a significant role as well, given that the brain is the second most prevalent organ that expresses many lncRNAs. Moreover, several lncRNAs are exclusive to primates and mammals, indicating their potential importance in cognitive functions [100,101]. When the natural antisense lncRNA β-site amyloid precursor protein cleaving enzyme 1 (BACE1)-antisense transcript (BACE1-AS) is upregulated, it enhances the stability of BACE1, resulting in sustained high levels of the BACE1 enzyme. This, in turn, could potentially contribute to the pathophysiology of Alzheimer's disease [102].
In immune response processes, lncRNAs have been found to play a keystone role. For instance, the long noncoding-interleukin-7 receptor α-subunit gene (lnc-IL7R) overlaps with the 3′-UTR region of the interleukin-7 receptor α-subunit gene. When this lncRNA is repressed, it leads to a reduction in trimethylation of H3K27 at the proximal promoter regions of inflammatory mediators, thereby decreasing lipopolysaccharide (LPS)-induced inflammatory responses [103]. Additionally, lncRNAs can bind to the signal transducer and activator of transcription 3 (STAT3) in the cytoplasm and promote STAT3 phosphorylation, which is crucial for dendritic cell differentiation and T cell activation [104].
In growth and development medical conditions, the lncRNA IPW, imprinted gene in the region of the Prader-Willi syndrome, is typically transcribed from chromosome 15 specifically from the paternal allele. It interacts with histone lysine methyltransferase to preserve the trimethylation of Histone H3 at Lysine 4 (H3K9me3) state at the DLK1-DIO3 region on chromosome 14, which helps to suppress maternally expressed genes (MEGs) [105]. The abnormal upregulation of MEGs could potentially contribute to the manifestation of Prader-Willi phenotypes.
4. LncRNAs as potential theranostic agents (diagnostic, prognostic, and therapeutic agents)
The discovery of ncRNAs has sparked a new field in biomarker research [106]. This is mainly attributed to the prominent abundance of ncRNAs in various cellular compartments, participating in multiple cellular functions [18]. They have been detected in cells, tissues, or biological fluids [4]. LncRNAs exhibit distinctive expression patterns and functional versatility across a range of diseases, making them valuable in the fields of theranostics (diagnosis, prognosis, and therapy) for multiple pathological conditions [107].
HOXA Cluster Antisense RNA 3 (HOXA-AS3) is implicated in several non-oncological conditions such as atherosclerosis, pulmonary arterial hypertension, and mesenchymal stem cell (MSC) lineage commitment. In atherosclerosis, HOXA-AS3 contributes to endothelial inflammation and influences angiogenesis progression. In pulmonary arterial hypertension, HOXA-AS3 promotes cell proliferation and migration while suppressing apoptosis. Regarding MSC lineage commitment, HOXA-AS3 plays a role in the differentiation of MSCs into adipocytes and osteoblasts. These findings emphasize the significance of HOXA-AS3 in the pathogenesis and advancement of non-cancerous diseases. HOXA-AS3 abnormal expression and functional roles also position it as a potential biomarker and therapeutic target for both cancers and non-cancerous diseases [108].
In cardiovascular diseases (CVDs), several lncRNAs show promise as diagnostic and prognostic markers. For instance, LncRNA LIPCAR (long intergenic noncoding RNA predicting cardiac remodeling) is a potential biomarker for atherosclerosis. Furthermore, Antisense Noncoding RNA in the INK4 Locus (ANRIL) is linked to atherosclerosis susceptibility, in-stent restenosis, and ventricular dysfunction post-MI. Other lncRNAs such as growth arrest specific 5 (GAS5), non-coding repressor of NFAT (NRON), myosin heavy chain associated RNA Transcripts (MHRT), MIAT, uc004cov.4, and uc022bqu.1 are also being explored as potential biomarkers in several pathological conditions [109]. Furthermore, the deregulation of lncRNAs in CVDs indicated that they might be therapeutic targets. Modulating lncRNA expression levels using recombinant plasmids, viruses, small nucleotide molecules, and gene-editing techniques has shown potential in animal models, improving cardiac dysfunction and related gene expression [109].
LncRNAs play a crucial role in autophagy regulation and are associated with neurological diseases. While some lncRNAs contribute to the development of neurological diseases, others can slow their progression. Understanding the involvement of lncRNAs in autophagy regulation holds promise for identifying therapeutic targets and biomarkers for neurological diseases. LncRNAs function through diverse mechanisms, such as interacting with miRNAs, influencing autophagy-related signaling pathways, and modulating the expression of autophagy-related genes [110]. Notably, in specific neurological disorders such as Alzheimer's disease, lncRNA 17A enhances autophagy by increasing the expression of the Gamma-Aminobutyric Acid Type B Receptor Subunit 2 (GABABR2) gene, leading to elevated secretion of β-amyloid peptides [111]. In Parkinson's disease, lncRNA nuclear enriched abundant transcript 1 (NEAT1) stabilizes the PINK1 protein, thereby promoting autophagy and safeguarding dopaminergic neurons [112]. In ischemic stroke, lncRNA H19 activates autophagy by regulating the dual-specificity phosphatase 5 (DUSP5)- extracellular signal-regulated kinase 1/2 (ERK1/2) axis, potentially worsening cerebral ischemia-reperfusion injury [113]. In epilepsy, lncRNA Metastasis Associated Lung Adenocarcinoma Transcript 1 (MALAT1) inhibits the phosphoinositide 3-kinase (PI3K)/protein kinase B (AKT)/mammalian (or mechanistic) target of rapamycin (mTOR) signaling pathway, triggering autophagy and contributing to the degeneration of hippocampal neurons [114]. In glioma, lncRNA Maternally Expressed Gene 3 (MEG3) suppresses the PI3K/AKT/mTOR signaling pathway, activating autophagy and suppressing cell proliferation and migration [115].
5. Myocardial Infarction Associated Transcript (MIAT)
5.1. MIAT discovery, structure and chromosomal location
LncRNA MIAT is the main focus of the current review due to its recent involvement in several malignant and non-malignant conditions and their prominent role in modulating several signaling cascades simultaneously. MIAT is a nuclear lncRNA with four splicing variants. In 2000, MIAT was discovered [116] and then found to be located at 22q12.1 with a length of 30,051 base pairs. A susceptible locus for MIAT has been revealed through a massive case-control association research study using 52,608 haplotype-based single nucleotide polymorphism (SNP) markers. Within this locus, scientists extracted a complete cDNA of a novel gene, which they named MIAT [44]. MIAT is also known as Gomafu in humans or Rncr2 in mice [44,117,118].
MIAT gene is composed of five exons and all its splice junctions are believed to adhere to the fundamental GT/AG rule [119,120]. Despite the finding that typical sequence alignment searches identified no orthologous MIAT genes in non-mammals [118], a search indicated that putative MIAT was orthologous to Xenopus tropicalis and chick [121]. Remarkably, every orthologous gene's largest exon had many clustered sequence repetitions of the sequence (ACUAACC), with five to eight repeats detected within each transcript [122]. This repeat sequence contains the (ACUAAY) consensus recognition site for the RNA binding protein Quaking that controls RNA subcellular stability and distribution [123,124]. MIAT, compared to additional nuclear-retained lncRNAs such as antisense of IGF2R non-protein coding RNA (AIRN) and X-inactive specific transcript (XIST), is strongly linked with the nuclear matrix rather than chromatin [118].
MIAT is a potentially important functional lncRNA. It influences cellular processes such as apoptosis, proliferation, and invasion across multiple medical conditions and its abnormal expression has a strong association in several diseases’ progression. MIAT expression is altered in various diseases as it is upregulated in some conditions such as MI, cardiomyopathy, cataract, ischemic stroke, non-small-cell lung cancer, and prostate cancer. In contrast, it is downregulated in other pathological conditions such as bone diseases, schizophrenia, and diabetic nephropathy [121]. In this review, the authors will shed the light on such paradoxical expression profile of MIAT in several non-oncological pathological conditions such as neurological conditions, cardiovascular-related diseases and metabolic disorders and linking it with respective molecular signaling pathways tuned by MIAT.
5.1.1. MIAT regulatory networks for non-oncological disorders
Several ncRNA databases have been reported to store curated data across multiple domains including genome annotation, molecular interaction, expression profiles, as well as therapeutic insights for related diseases [125]. RNAcentral and miRbase are versatile ncRNA database that various tissues and species, as well as serve as primary databases that harness huge data collection from other resources for up-to-date ncRNA specifications [126,127].
Moreover, LncTarD and LncRNA2Target are two lncRNA databases that include potential lncRNA interactions across oncological and non-oncological samples collected from all published lncRNA studies [128,129]. Cytoscape v.3.10 was used to construct lncRNA regulatory networks using mentioned data.
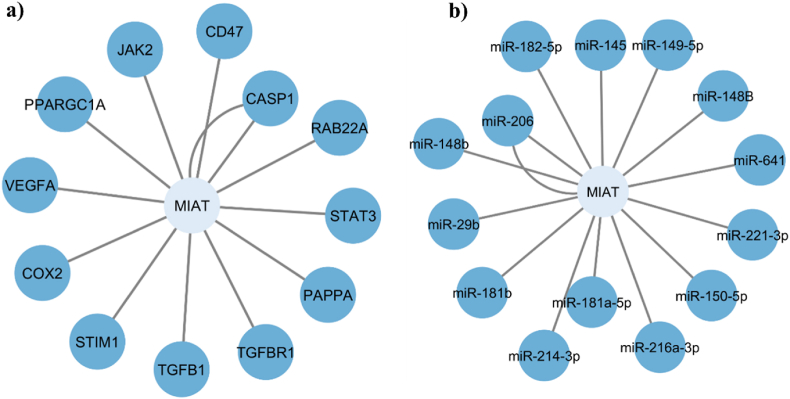
It is worth noting that similar to most of lncRNAs, MIAT has been associated with competing endogenous RNA (ceRNA) hypothesis that suggests that ncRNAs and mRNAs communicate by competing for binding to shared miRNAs, which accordingly modulate targets’ gene expression [130]. Following screening of MIAT lncRNA on both databases, our results revealed 60 MIAT-mRNA-miRNA interactions, where non-oncological disorders showed 12 MIAT-mRNA interactions, as well as 14 MIAT-miRNA ones across different tissues and disease states.
Our in-silico results showed that MIAT-mRNA regulatory network revealed that STAT3, RAB22A, CASP1, CD47, JAK2, PPARGC1A, VEGFA, COX2, STIM1, TGFB1, TGFBR1, and PAPPA genes have significant interaction with MIAT, while in MIAT-miRNA regulatory network, miR-182-5p, miR-145, miR-149-5p, miR-148B, miR-641, miR-221-3p, miR-150-5p, miR-216A-3p, miR-181a-5p, miR-214-3p, miR-181b, miR-29b, miR-148b, and miR-206 were regulated by MIAT as shown in Fig. 2.
Fig. 2.
MIAT-mRNA and MIAT-miRNA regulatory networks in non-oncological disorders. MIAT-mRNA and MIAT-miRNA regulatory networks in non-oncological disorders generated from LncTarD and LncRNA2Target databases. (a) Network plot for MIAT encompassing 12 lncRNA-mRNA interactions. (b) Network plot for MIAT encompassing 14 lncRNA-miRNA interactions.
Experimentally, it has been also validated that MIAT is involved in several pathological conditions through the ceRNA regulatory network. Investigations have reported that MIAT acts as a ceRNA for miR-150-5p [47,131], miR-24 [132], miR-22-3p [133], and miR-150 [134]. Moreover, MIAT can also activate a variety of signaling pathways through regulating various proteins such as Nrf2 (nuclear factor erythroid 2-related factor 2), which is critical in high glucose-induced renal tubular epithelial injury [135], and SRSF1(serine/arginine-rich splicing factor 1), which contributes to the pathophysiology of schizophrenia [136]. MIAT also creates a positive feedback regulatory loop with its transcriptional regulator octamer-binding transcription factor 4 (OCT4) in malignant mature B cells to enhance cellular proliferation [137].
5.2. Neurological diseases
5.2.1. Multiple sclerosis (MS)
MS stands as the most predominant inflammatory and demyelinating disease impacting the central nervous system (CNS), with a higher prevalence among young adults, especially women. According to the Atlas of MS, approximately 2.9 million individuals are affected globally, and its incidence has been on the rise since 2013 [138]. It can be categorized under neuropathological condition as well as autoimmune diseases [139].
Immunotherapies have shown promise in halting the progression and relapse of MS, but their efficacy appears to be more pronounced in the early stages of the disease compared to the later, progressive stages [140,141]. The complex pathophysiology of MS involves demyelination, neurodegeneration, immune-mediated processes, and inflammation. Although the exact causative factors remain unidentified, research indicates that genetic, environmental, infectious, nutritional, and epigenetic components play a role in the development of MS [[142], [143], [144]]. The onset of MS is believed to be linked to autoimmune responses involving adaptive T- and B-lymphocytes targeting the CNS [145]. Therefore, there is a crucial need to comprehend the underlying mechanisms driving MS and develop effective therapies to combat this neuropathological condition.
The dysregulation of lncRNAs has been implicated in the causes of various neurological diseases including MS [146]. LncRNAs play a role in regulating gene expression and controlling immune system responses [147,148]. Several lncRNAs, including MIAT, have been associated with the pathogenesis, prognosis, and diagnosis of MS [[147], [148], [149], [150]]. MIAT has also been linked to the induction of inflammatory responses. Further research into the role of MIAT and other lncRNAs may provide valuable insights into the molecular mechanisms underlying MS and potential therapeutic targets.
5.2.1.1. Exploring the role of MIAT in MS: expression profile, signaling pathways and potential implications
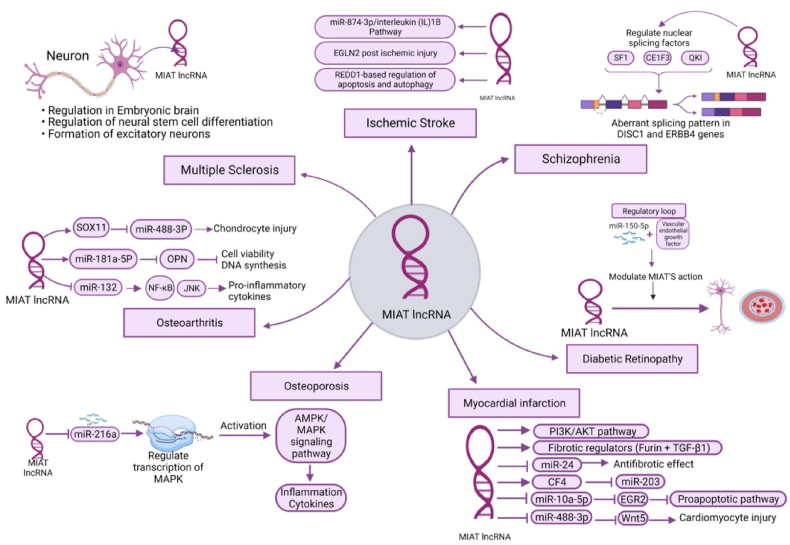
MIAT is predominantly expressed in neuronal cells, particularly in the nucleus of post-mitotic neurons [151,152]. The physiological functions of MIAT in CNS have been extensively discussed in the literature. It is known that MIAT is involved in the regulation of several processes in the embryonic brain, such as the regulation of neural stem cell differentiation and the formation of excitatory neurons as shown in Fig. 3 [153,154]. MIAT-deficient mice did not show developmental abnormalities but resulted in hyperactive behavior [151]. These findings highlight the significance of MIAT in neuronal development and functioning. Further research is needed to fully elucidate its precise mechanisms of action in the CNS and its potential implications for neurological disorders.
Fig. 3.
Role of lncRNA MIAT in several pathological diseases.
In recent studies, MIAT expression in the peripheral blood of MS patients has been investigated, revealing controversial findings. One study reported a significantly higher expression of MIAT in MS patients compared to controls [49]. In contrast, another study found downregulation of MIAT in peripheral blood mononuclear cells (PBMCs) of MS patients compared to controls; however, validation on a different cohort did not show significant differences in MIAT expression between the two groups [155]. This could be attributed to age differences between cohorts and different ethnic groups.
Given the conflicting results, further research is warranted to fully understand the role of MIAT in the context of MS. It is crucial to determine whether MIAT expression correlates with MS prognosis, diagnosis, or the potential for therapeutic targeting. These investigations hold promise for advancing our understanding of MS pathogenesis and may have implications for the development of targeted interventions to improve patient outcomes.
5.2.2. Schizophrenia (SZ)
Schizophrenia (SZ) is a complex psychiatric condition that affects around 24 million individuals worldwide, particularly young adults [156]. It exerts a profound impact on normal life functioning and has a high mortality and suicidal rates [157]. The development of SZ is believed to be influenced by a combination of environmental, genetic, and epigenetic factors, ultimately resulting in neural and glial cells abnormalities [[158], [159], [160]]. However, the precise mechanisms through which genetic and epigenetic factors contribute to the pathophysiology of the disease remain unclear.
5.2.2.1. Exploring the role of MIAT in SZ: expression profile, signaling pathways and potential implications
Alternative splicing, the process that generates multiple protein isoforms from a single gene, has been implicated as a significant contributor to genetic dysregulation in SZ [161,162]. Among the genes affected are disrupted in schizophrenia 1 (DISC1) and v-Erb A erythroblastic leukemia viral oncogene homolog 4 (ErbB4), which show aberrant splicing patterns in SZ [161,162]. Notably, lncRNAs have been found to influence gene transcription by modulating the alternative splicing of their target genes [163]. In this context, MIAT has been shown to regulate nuclear splicing factors like Splicing Factor 1 (SF1), EBF Transcription Factor 3 (EBF3), and QKI, KH Domain Containing RNA Binding (QKI) as shown in Fig. 3, which, in turn, alter the splicing patterns of DISC1 and ErbB4 in a manner similar to that observed in SZ [[164], [165], [166]].
In the pursuit of understanding the potential link between MIAT dysregulation and SZ, researchers conducted a study on the Chinese Han population to identify specific SNPs in SZ patients compared to controls [167]. Among the 485 SNPs examined within MIAT, only one SNP was found to be associated with paranoid SZ [167]. This intriguing finding suggests that certain SNPs within the MIAT gene might contribute to the vulnerability to SZ, providing valuable insights for further research into the genetic underpinnings of this complex psychiatric disorder.
Recent studies have shed light on the expression of MIAT in the brain tissues of SZ patients. One investigation found that MIAT expression in the brains of SZ patients was lower compared to controls [166]. Additionally, data from the PsychEncode Consortium revealed that MIAT was among the downregulated genes in a SZ cohort that included postmortem brain samples from patients with various psychiatric conditions and controls [168,169]. These intriguing findings suggest the potential involvement of MIAT in the pathophysiology of SZ and provide valuable insights into its role in susceptibility to the disorder.
The lack of blood-based biomarkers and the high heterogeneity of the disease among patients present significant hurdles to SZ diagnosis and treatment efficacy [170]. Due to the predominant presence of lncRNAs in PBMCs and their high transcriptomic resemblance to lncRNAs expressed in the brain, researchers are actively investigating the potential of lncRNAs as diagnostic non-invasive biomarkers for SZ [171,172]. In a small study conducted on a Chinese Han cohort, researchers examined the expression of MIAT in PBMCs and observed that MIAT was overexpressed in SZ patients compared to controls [173]. These findings were inconsistent with previous results obtained from post-mortem brain cohorts. The authors discussed potential reasons for such contradictory outcomes. Furthermore, the study indicated that treatment with anti-psychotics led to a further increase in MIAT expression; however, this increase did not correlate with improved disease progression for SZ patients [173].
Another study, involving a small cohort of 50 SZ patients and 50 normal individuals, found no statistically significant differences in MIAT expression levels between the two groups [174]. However, the study reported that the diagnostic accuracy of the MIAT gene to differentiate between SZ patients and controls was 68% [174]. On the other hand, transcriptome profiling of amygdala tissues from SZ patients and controls revealed no differences in MIAT expression between both groups [175]. Furthermore, another study using mouse models of SZ showed significantly lower MIAT expression in the brains of SZ-induced mice, but MIAT was not detected in peripheral blood samples taken from the same mice [176].
Another study revealed that MIAT expression was reduced in the brains of fear-conditioned mice.
Furthermore, knocking down MIAT in mice led to increased anxiety-like behavior and higher expression of Crystallin Beta B1 (Crybb1), a gene associated with SZ. The mechanism behind the upregulation of Crybb1 was also found to occur through the interaction of MIAT with BMI1 Proto-Oncogene, Polycomb Ring Finger (BMI1), a component of the polycomb repressive complex. Under normal circumstances, this interaction would repress Crybb1 expression. However, when MIAT was repressed through knocking down experiments or due to fear conditioning in mice, the interaction between MIAT and BMI1 was disrupted, leading to the de-repression of Crybb1 [177].
The controversy surrounding MIAT's expression in SZ patients and its potential as a possible diagnostic and prognostic non-invasive biomarker emphasizes the necessity for further research on its expression in the blood of SZ patients. These investigations are crucial in determining its validity as a potential biomarker in liquid biopsies of SZ patients. It is also worth noting that to ensure consistency between different studies, it is essential to establish standardized protocols for quantifying MIAT from blood samples, thus minimizing discrepancies.
5.2.3. Ischemic stroke (IS)
Ischemic strokes (IS) are a widespread global health concern, affecting 15 million people annually, according to the WHO [178]. They are associated with high morbidity and mortality rates, with approximately 67% of stroke sufferers experiencing death or permanent disability [178]. As strokes are more prevalent in individuals older than 40 years, the absolute number of cases continues to rise due to the increasing aging population [178].
High blood pressure and smoking are the main causative factors of stroke. IS is the most common type, accounting for 62.4% of all strokes, as estimated in 2019 [179]. IS occurs due to insufficient blood supply to the brain, leading to neuronal death and severe neurovascular damage [180]. Treatment options for IS include plasminogen activating therapy or mechanical thrombectomy. Plasminogen activating therapy is currently the only FDA-approved treatment for IS and is most effective when administered within the first few hours after the onset of the stroke [181,182].
In terms of diagnostics, there is an urgent need for improvement as the current methods rely heavily on clinical examination and imaging techniques, making the detection of small infarcts challenging and lengthy in terms of duration [183]. Therefore, the discovery of new targets and biomarkers for IS remains crucial to combating the high mortality rate associated with such disease.
5.2.3.1. Exploring the role of MIAT in IS: expression profile, signaling pathways and potential implications
LncRNAs have been implicated in the pathophysiology of IS and are currently under investigation as potential diagnostic biomarkers. Among these lncRNAs, MIAT comes on top of the list. MIAT has been found to be upregulated in the blood of IS patients and showed positive correlations with the National Institutes of Health Stroke Scale (NIHSS), high-sensitivity C-reactive protein, and infarct volume [184]. Notably, MIAT was identified as a potential prognostic marker for IS, as patients with higher MIAT expression exhibited increased mortality rates [184]. Multivariate analysis demonstrated that MIAT independently predicted the outcome and death in IS patients [184]. Additionally, MIAT was observed to possess diagnostic value for IS patients [184]. To fully realize the promising potential of MIAT as a diagnostic and prognostic biomarker for IS, further clinical trials on larger cohorts are imperative to validate these findings. By advancing our understanding of MIAT's role in IS, we may pave the way for improved patient management and targeted therapeutic interventions in the future.
A comprehensive investigation was conducted to examine the potential involvement of MIAT in the pathogenesis of IS through the miR-874-3p/IL-1β pathway as shown in Fig. 3 [185]. Zhang et al. observed that the expression of MIAT and IL-1β in serum was upregulated in IS patients, while miR-874-3p was downregulated [185]. To further explore this pathway, the researchers performed in-vivo experiments, demonstrating that knockdown of either MIAT or IL-1β reduced neuronal apoptosis and inflammatory factors in the brains of mice, resulting in alleviated brain damage [185]. Remarkably, the mice exhibited improved neurological functions and behaviors [185].
Another study utilizing middle cerebral artery occlusion models discovered that MIAT increased the stability of Egl-9 Family Hypoxia Inducible Factor 2 (EGLN2) post-ischemic injury [186]. This increased stability reduced EGLN2 degradation, which had a detrimental effect on the volume of the infarct and neuronal apoptosis [186].
Additionally, an interaction between MIAT and Regulated in Development and DNA Damage Responses 1 (REDD1) was revealed in another study as shown in Fig. 3 [187]. REDD1 plays crucial regulatory roles in autophagy and apoptosis in stressed neurons [187]. The study observed increased MIAT and REDD1 levels in the brains of IS-induced mice, along with augmented infarct volumes [187]. Intriguingly, knocking down of MIAT resulted in the downregulation of REDD1, decreased apoptotic markers, and reduced infarct volumes [187]. The interaction between MIAT and REDD1 was further confirmed through RNA-pulldown and RIP assays [187].
These findings collectively suggest that MIAT plays a pivotal role in the progression of IS and hold promising potential as a therapeutic target as well as promising diagnostic and prognostic biomarker for IS. The in-depth exploration of the MIAT-mediated molecular pathways in IS opens new avenues for potential interventions to combat the detrimental effects of such neuropathological condition.
5.3. Cardiovascular diseases
5.3.1. Myocardial infarction (MI)
Cardiovascular diseases are the leading cause of global mortality, with coronary heart disease (CHD) accounting for the largest proportion [188]. Among CHD complications, MI is the predominant one, and is more prevalent in the elderly (>60 years) [188]. MI is an ischemic heart disease resulting from inadequate blood flow, leading to reduced oxygen supply, cell death in the ischemic region, and activation and proliferation of cardiac fibroblasts, contributing to cardiac fibrosis [[189], [190], [191]]. The deterioration of functional cardiomyocytes with fibrotic tissue worsens cardiac function, especially in the left ventricle (LV), ultimately leading to irreversible heart failure [192]. Cardiomyocytes are terminally differentiated cells lacking the ability to divide, limiting the heart's regenerative capacity in adult mammals [193]. As a result, the replacement of deceased cardiomyocytes with new functional ones becomes a formidable challenge, significantly impacting the severity of MI and its potential long-term effects on cardiac function [193]. Despite improved clinical outcomes for MI, CHD remains a leading cause of death worldwide. This emphasizes the urgent need to identify new therapeutic targets and potential stable biomarkers to reduce the chances of myocardial injuries in MI.
5.3.1.1. Exploring the role of MIAT in MI: expression profile, signaling pathways and potential implications
In the 2000s, researchers identified a SNP within MIAT that was highly prevalent in MI patients, suggesting its potential involvement in MI pathogenesis [194]. This finding was further validated in a larger cohort in 2006 [195]. In a subsequent clinical trial focusing on the expression of specific lncRNAs in MI, Vausort et al. observed moderate expression of MIAT in PBMCs of MI patients compared to controls [196].
Interestingly, when analyzing the two subtypes of MI, namely ST-segment–elevation MI (STEMI) and non–ST-segment–elevation MI (NSTEMI), Vausort et al. found that MIAT levels were lower in STEMI patients relative to NSTEMI patients [196]. Moreover, MIAT expression was negatively correlated with cardiac biomarkers such as troponin T [196]. Another study focusing on acute MI also reported similar findings, with MIAT expression being lower in STEMI compared to NSTEMI [197]. Furthermore, Vausort et al. found that MIAT expression showed positive correlations with cardiovascular risk factors, including hypertension and smoking [196]. Additionally, MIAT levels were positively correlated with the percentage of lymphocytes and negatively correlated with neutrophils and platelets [196]. The study also indicated that MIAT was a weak predictor of LV dysfunction, a condition commonly associated with MI [196].
In another study, researchers investigated whether MIAT's expression could serve as a diagnostic biomarker for MI. They found that plasma levels of MIAT in MI patients were significantly higher than in controls [198]. Furthermore, the expression of MIAT showed a positive correlation with cardiac markers, including creatine kinase-MB and troponin T [198]. Similar investigations in animal models also demonstrated a similar trend, with increased MIAT levels in MI-induced rats compared to controls. Knocking down experiments further revealed MIAT's involvement in cardiomyocyte apoptosis [199]. Similar findings were also reported by Wang et al., where MIAT was found to be more highly expressed in MI patients, although its expression was not positively correlated with cardiac biomarkers [200]. Additionally, a more recent clinical study conducted on a Chinese population also observed higher MIAT expression in MI patients compared to normal individuals [201]. These collective findings provide valuable insights into the potential utility of MIAT as a diagnostic biomarker in the context of MI.
In mouse models of MI, MIAT expression in MI hearts was significantly higher compared to controls. Interestingly, MIAT knockdown was found to prevent cardiac fibrosis, likely achieved by inhibiting MIAT's effects on collagen production, leading to improved cardiac function [202]. MIAT's involvement in heart failure models has been linked to the activation of the PI3K/AKT/mTOR signaling pathway, both in in-vivo and in-vitro settings, and its role in collagen production was once again validated, indicating its potential contribution to promote cardiac fibrosis [203].
Additionally, MIAT upregulation was associated with increased expression of fibrotic regulators, including furin and transforming growth factor (TGF-β1), and downregulation of miR-24 that is known for its role in mediating anti-fibrotic effects [202]. This suggests that the interplay between MIAT, miR-24, and fibrotic regulators contributes to the pathogenesis of MI as shown in Fig. 3.
It is well-established that exercise can significantly improve the quality of life for MI patients and reduce mortality [204]. In light of this, a recent study sought to investigate the impact of exercise on the expression of lncRNAs in rats [205]. The researchers conducted experiments in rats to demonstrate that moderate exercise indeed had positive effects on the progression of MI [205]. They observed improvements in LV function and a reduction in fibrosis and apoptosis of cardiomyocytes. Interestingly, the expression of MIAT was found to be downregulated in mice after the exercise regimen [205]. This intriguing finding, combined with the previously mentioned pro-fibrotic effects of MIAT, suggests that the reduction of fibrosis post-exercise may be attributed to the repression of MIAT expression levels. However, further investigations are required to validate the potential of MIAT as a therapeutic target for managing MI. These findings offer valuable insights into the potential therapeutic benefits of exercise in the context of MI and highlight the need for continued research to better understand the underlying mechanisms involved.
Another study demonstrated an interplay between miR-203, MIAT, and mitochondrial coupling factor 6 (CF6) in MI, where MIAT and CF6 were upregulated but miR-203 was downregulated in mouse myocardial tissues following MI [206]. CF6 is known to aggravate MI, and knocking down of MIAT led to downregulation of CF6 expression, suggesting MIAT and CF6 as downstream players of miR-203 in MI pathogenesis [206].
Furthermore, MIAT was found to interact with miR-10a-5p, downregulating its expression [207]. This repression of miR-10a-5p had repressive effects on Early Growth Response 2 (EGR2) expression, a factor associated with activating pro-apoptotic pathways [207]. The study demonstrated that MIAT silencing contributed to cardio protection in MI by modulating the miR-10a-5p/EGR2 axis as shown in Fig. 3 [207].
In another study, a distinct role of MIAT in MI was uncovered, involving its interaction with a mitochondrial membrane protein in cardiac cells known as Translocator protein (TSPO) [208]. TSPO plays a crucial role in regulating mitochondrial membrane potential and is involved in various signaling cascades, including apoptosis [208]. The study revealed that the interaction between MIAT and TSPO had significant consequences on mitochondrial membrane integrity in the infarct zone [208]. This interaction appeared to have detrimental effects on mitochondrial function and energy production within cardiac cells, leading to impaired contractile function of the heart [208]. Knocking down of MIAT not only represses TSPO levels but also rescues mitochondrial damage and reduced apoptosis [208]. To investigate further, the researchers initially examined the expression of MIAT in the hearts of MI mouse models and cultured neonatal mouse ventricular cells (NMVCs) after being subjected to hypoxic insult [208]. They observed that MIAT was significantly more highly expressed in both the induced-MI models and the NMVCs compared to their respective controls [208]. These findings provide valuable insights into the molecular mechanisms underlying MI-induced cardiac damage and could potentially pave the way for future therapeutic strategies aimed at targeting MIAT or TSPO to mitigate the detrimental effects of MI on the heart's contractile function. Such interventions may hold promise for improving the prognosis and quality of life for individuals affected by this devastating condition.
In a recent model of hypoxia-induced cardiomyocyte injury, another regulatory network governed by MIAT was unveiled. MIAT was found to suppress miR-488-3p, a miRNA known for its cardioprotective effects on cardiomyocyte injury, particularly through the downregulation of Wnt family member 5A (Wnt5A) [209]. The Wnt-β-catenin signaling pathway has been extensively discussed in the pathogenesis of MI [209]. This suggests that the MIAT-miR-488-3p-Wnt5 axis may also play a role in the pathogenesis of MI, highlighting the importance of normalizing the expression of MIAT in MI patients.
In another study exploring the cardioprotective properties of the phytochemical lutein in a mouse model of isoprenaline-induced MI, researchers found that treatment with lutein led to the suppression of MIAT expression [210]. These findings suggest that targeting MIAT could potentially offer cardiovascular benefits to MI patients.
These findings add to the growing body of evidence supporting the multifaceted role of MIAT in cardiac pathophysiology, particularly in heart failure and MI. The identification of these regulatory networks provides valuable insights into potential therapeutic targets for managing cardiac fibrosis and improving outcomes in patients with heart failure and MI. Further research is warranted to fully elucidate the mechanisms underlying MIAT's molecular pathways and explore its potential as a theranostic agent in cardiovascular diseases.
5.3.2. Coronary artery disease (CAD)
Coronary artery disease (CAD) is a prevalent condition stemming from atherosclerotic processes occurring in the coronary arteries. This disorder may exhibit no symptoms or lead to life-threatening situations. Notably, CAD stands as the primary cause of mortality in CHD [211]. Atherosclerosis is recognized as a gradually advancing inflammatory state wherein oxidative, hemodynamic, and biochemical factors impair the functionality of endothelial cells [212]. Subsequent changes in endothelial cell permeability, the accumulation of macrophages, the release of inflammatory substances, and the activation of smooth muscle cells represent additional stages in the progression of atherosclerosis [213,214].
5.3.2.1. Exploring the role of MIAT in CAD: expression profile, signaling pathways and potential implications
Tan et al. [215] conducted a study with the objective of uncovering the clinical significance of MIAT expression in predicting CAD. In pursuit of this goal, the researchers evaluated the serum levels of MIAT, tumor necrosis factor-α (TNF-α), and interleukin-6 (IL-6) in 102 CAD patients and 89 healthy controls. The findings indicated that the expression of MIAT was approximately three times higher in CAD patients compared to healthy donors. Corroborating this observation, linear correlation analysis demonstrated a positive association between MIAT, IL-6, and TNF-α in CAD. Furthermore, the results of multivariate analysis highlighted that, apart from hypertension, diabetes, and HDL-C, serum MIAT stood out as an independent risk factor for CAD. Considering these outcomes, the authors concluded that serum MIAT may serve as a potential biomarker for both diagnosing and prognosticating CAD. However, it is crucial to note that this study did not delve into the potential correlation between serum MIAT and coronary risk factors, such as hypertension, hyperlipidemia, diabetes, sex, and age.
Toraih et al. [216] further supported and expanded upon this concept by examining the circulating levels of MIAT in 110 stable CAD patients and 117 control volunteers using qPCR. They then correlated the expression of MIAT with both clinical and laboratory data. The results revealed a striking 12-fold increase in the median MIAT level in patients compared to controls. These compelling findings led the authors to suggest that MIAT holds diagnostic significance in CAD patients. However, the regulatory role of MIAT in the pathogenesis of atherosclerosis of coronary arteries is still needs further investigations.
5.3.3. Cardiac hypertrophy (CH)
Cardiac hypertrophy (CH) primarily arises as a response to heightened preload or afterload, aiming to maintain cardiac output. Infrequently, it may result from genetic mutations or exposure to growth factors. When confronted with increased workload, CH leads to enhanced contractility, initially reduces LV wall stress through increased wall thickness, accordance with Laplace's law, and induces alterations in gene expression. This, in turn, brings about modifications in heart metabolism, contractility, and the survival of cardiomyocytes. The classification of CH includes physiological and pathological forms. Despite their differences, both conditions involve the enlargement of cardiomyocytes and represent adaptive responses to cardiac stress [217].
5.3.3.1. Exploring the role of MIAT in CH: expression profile, signaling pathways and potential implications
Multiple studies employing various animal models have demonstrated the role of MIAT in the progression of CH. The findings from these investigations unanimously converge on the conclusion that MIAT functions as a growth stimulator, promoting hypertrophic changes in the heart. The initial study, conducted by Zhu et al. [218] investigated a mouse model of CH induced by Angiotensin II (AngII) and utilized an in-vitro model with rat heart-derived H9c2 cells. In both the in-vivo and in-vitro models, a substantial 5–6-fold increase in MIAT was observed in CH. Upon silencing of MIAT levels, it markedly mitigated the upregulation of hypertrophic biomarkers including atrial natriuretic peptide (ANP), B-type natriuretic peptide (BNP), and beta-myosin heavy chain (β-MHC) in H9c2 cells. Additionally, it significantly reduced the increase in cell surface area and protein synthesis induced by AngII. In a ceRNA manner, overexpression of MIAT in H9c2 cells resulted in the downregulation of miR-150 expression, while knockdown of MIAT upregulated it. Furthermore, forced expression of miR-150 inhibited the expression of hypertrophic marker genes and suppressed hypertrophic phenotypes, aligning with other studies that demonstrate the anti-hypertrophic action of this miRNA [219].
Later, another study was conducted to unravel the underlying mechanism through which MIAT regulates the development of CH. In their findings, MIAT and miR-93 exhibited contrasting expression patterns in AngII-treated cardiomyocytes; MIAT showed high expression, while miR-93 displayed low expression levels. These reciprocal changes prompted the researchers to establish the targeting relationship between MIAT and miR-93, employing a combination of qPCR, luciferase assays, and RNA immunoprecipitation (RIP) techniques. The results confirmed MIAT as a ceRNA of miR-93, subsequently regulating Toll-like receptor 4 (TLR4) expression. Consequently, their data suggest that MIAT knockdown exerts inhibitory effects on CH through the miR-93/TLR4 pathway [220].
In another experimental work, researchers affirmed the pro-hypertrophic nature of MIAT in both a mouse model of CH induced by intraperitoneal isoproterenol (ISO) and a cellular model of cardiomyocyte hypertrophy involving neonatal rat ventricular myocytes (NRVMs) [221]. Their findings demonstrated a significant 4.5-fold increase in MIAT transcripts upon ISO stimulation, observed both in-vivo and in-vitro. Again this study showed that knocking down of MIAT resulted in decreased expression levels of hypertrophic marker genes and a reduction in cell surface area. Furthermore, they presented evidence supporting MIAT's capability to inhibit miR-150 and extended their investigation to identify P300, an acetyltransferase and transcriptional coactivator known to promote cardiac growth and drive CH [222], as a target gene for miR-150. Subsequently, they established that MIAT reduces P300 expression by sequestering miR-150.
Yet another research team detailed their investigation into the impact of berberine, an effective natural supplement compound derived from various plants, on CH. They conducted experiments using a rat model with abdominal aorta constriction and H9C2 cells stimulated with AngII to induce cardiomyocyte hypertrophy [223]. Their findings indicated that berberine significantly alleviates CH, reduces cardiomyocyte enlargement, and diminishes the expression of hypertrophy marker proteins. Notably, berberine also suppresses the expression of MIAT in CH. Additionally, berberine downregulates phosphorylated mTOR, phosphorylated adenosine monophosphate-activated protein kinase (AMPK), and microtubule-associated proteins 1A/1B light chain 3B (LC3), while simultaneously upregulating autophagy-related genes. Despite concluding that berberine exerts beneficial effects on CH by decreasing MIAT expression and enhancing autophagy, the authors did not establish a connection between MIAT and autophagy, nor did they clarify the mechanism through which berberine suppresses the expression of MIAT [224].
5.4. Metabolic disorders
5.4.1. Diabetes mellitus (DM)
Diabetes mellitus (DM) is a highly prevalent disease, affecting approximately half a billion people worldwide according to the International Diabetes Federation Atlas in 2021 [225]. It is a cluster of metabolic disorders characterized by chronic hyperglycemia stemming from defects in insulin secretion, insulin action, or both [[226], [227], [228]]. This condition leads to elevated blood sugar levels, resulting in severe complications that often cause dysfunction or impairment in various organs. The heart, eyes, nerves, kidneys, and blood vessels are particularly susceptible to damage or adverse effects due to chronic DM, contributing to the high morbidity and mortality associated with the disease [225].
DM is categorized into type 1 and type 2, both influenced by several genetic, epigenetic, and environmental factors [226]. Type I DM is an autoimmune disease affecting individuals younger than 30 years and accounts for only 5–10% of all DM cases. On the other hand, type II DM is the most prevalent, affecting individuals over the age of 40 and is often associated with obesity [225,226].
5.4.1.1. Exploring the role of MIAT in DM: expression profile, signaling pathways and potential implications
Numerous studies have highlighted the significant roles of lncRNAs in the pathogenesis, prognosis, and complications of DM. In this context, MIAT has drawn particular attention due to its potential implications for type 2 DM. Several investigations have demonstrated elevated MIAT expression in the blood of patients with type 2 DM compared to controls [229,230]. One study identified a positive correlation between MIAT expression and markers of cardiac remodeling, suggesting its potential as a prognostic or diagnostic factor for type 2 DM [230]. Another study focusing on PBMCs in type 2 DM also found upregulation of senescent and pro-inflammatory markers, along with a positive correlation between MIAT expression and poor glycemic control and insulin resistance [229]. Although direct links between MIAT expression and downstream effects on senescent and proinflammatory markers require further exploration, such investigations could yield valuable insights into the underlying mechanisms of MIAT's impact on type 2 DM [229].
Moreover, the intriguing VITA cohort study investigated lncRNA expression in non-diabetic older adults and conducted a follow-up after 7.5 years to correlate lncRNA expression with the incidence of type 2 DM [231]. Consistent with previous findings, MIAT was found to be upregulated in patients who developed type 2 DM during follow-up [231]. Furthermore, its expression was correlated with both baseline and follow-up glycated hemoglobin levels, indicating that MIAT's expression in the blood may predict the development of diabetes even before disease onset, suggesting a potential role for MIAT in the progressive loss of glucose homeostasis over time [231]. These studies highlight the importance of MIAT in the context of type 2 DM and warrant further research to elucidate its precise mechanisms and potential implications as a diagnostic or prognostic biomarker in DM.
In a recent study investigating the expression of ncRNAs in type 1 DM, MIAT was also identified as one of the upregulated differentially expressed genes in such metabolic disorder [232]. Notably, the differentially expressed genes also encompassed crucial pathways relevant to DM, including the phosphatidylinositol signaling system and the insulin signaling pathway [232]. While these findings indicate the potential involvement of MIAT in the pathogenesis of type 1 DM, further investigations are necessary to validate its role and significance in disease development and diagnosis.
The role of MIAT in the pathogenesis of DM has been extensively explored in a study conducted by Yan et al. In their project, they discovered that MIAT plays a significant role in microvascular complications that lead to diabetic retinopathy (DR) [233]. To gain deeper insights into the underlying mechanisms of MIAT's contribution to DM pathogenesis, the researchers uncovered a regulatory loop involving vascular endothelial growth factor (VEGF) and miR-150-5p [233]. This regulatory loop was found to modulate MIAT's actions on neural and vascular cell functions [233].
Moreover, Zhou et al. investigated the role of MIAT in hyperglycemia-induced renal tubular epithelial injury in diabetes-induced rats [234]. Their findings suggested that MIAT may exacerbate diabetic kidney dysfunction, potentially contributing to the progression of diabetic nephropathy [234].
These studies collectively shed light on the diverse roles of MIAT in the pathogenesis of DM and its related complications. Unraveling the underlying molecular mechanisms holds promise for potential therapeutic interventions by targeting MIAT to ameliorate the microvascular and organ-specific complications associated with DM. Further investigations are warranted to fully comprehend MIAT's multifaceted impact on DM pathogenesis and its potential as a therapeutic target.
5.4.1.2. Novel crosstalk between MIAT and diabetic retinopathy (DR)
A recent study studied the utility MIAT as a diagnostic marker for DR, a serious complication of DM that can lead to visual impairment and blindness [235]. However, no association was found between MIAT and DR disease progression in this study [235]. Yet, Biswas et al. conducted further investigations and confirmed that MIAT was upregulated in the serum of DR patients [236]. Moreover, another recent study linked the expression of a specific variant of MIAT to DR prognosis and its ability to predict a poor response to aflibercept treatment [237].
These findings collectively suggest the potential value of MIAT as a blood-based biomarker for DR. However, to establish its clinical utility, further validation studies on larger cohorts are required. Additionally, standardizing the protocol for MIAT quantification is essential to ensure consistent and reliable results as previously mentioned.
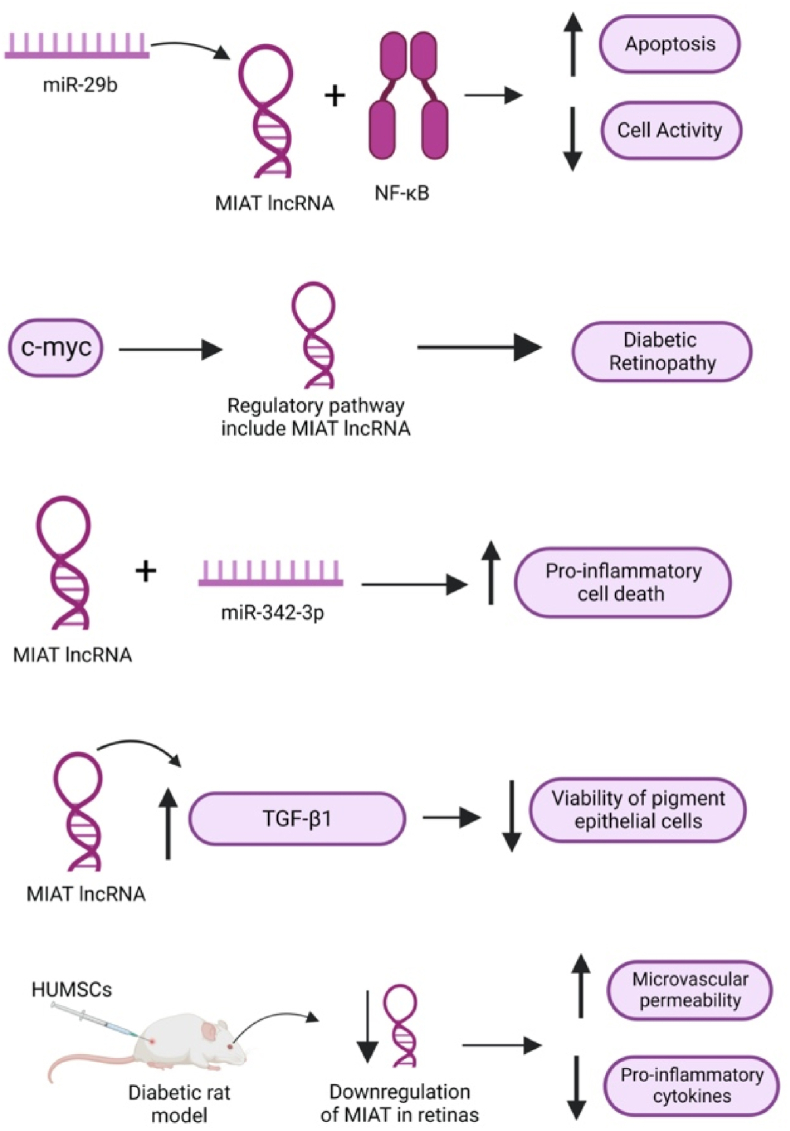
On both in-vitro and in-vivo levels, DR-induced rats and retinal Müller cells exhibited higher levels of MIAT expression compared to control groups [238]. Hyperglycemia induced an interaction between MIAT and nuclear factor-κB (NF-κB), leading to increased apoptosis and reduced cell activity, which were reversed upon knockdown of MIAT as shown in Fig. 4 [238]. In this regulatory network, miR-29b was also found to be involved in the regulation of MIAT [238]. A recent study indicated that MIAT is part of a pathway regulated by c-myc, which subsequently triggers the release of proinflammatory cytokines from retinal Müller cells, contributing to the pathogenesis of DR [239].
Fig. 4.
MIAT as a regulator of multiple pathways in diabetic retinopathy.
Furthermore, in another study using primary human retinal pericytes, MIAT was found to be upregulated and was observed to interact with miR-342-3p as shown in Fig. 4 [240]. This interaction was associated with increased proinflammatory cell death, providing additional insights into the role of MIAT in the inflammatory processes contributing to DR [240]. In adult retinal pigment epithelial cells, another study revealed that MIAT upregulates TGF-β1 signaling, leading to a reduction in the viability of pigment epithelial cells [241]. These findings offer further understanding of the diverse impact of MIAT in various cellular contexts related to the development and progression of DR.
In an interesting study, the impact of human umbilical-cord mesenchymal stem cells (HUMSCs) on diabetic rat models was investigated [242]. The study revealed that post-injection of HUMSCs, MIAT expression in the retinas of diabetic rats was downregulated [242]. This downregulation was associated with enhanced microvascular permeability and a reduction in the levels of pro-inflammatory cytokines known to play a role in DR [242]. The findings of this study established a correlation between MIAT expression and proinflammatory cytokines in DR, shedding the light onto the potential role of MIAT in regulating these cytokines (Fig. 4) [242]. These results underscore the importance of further investigations to explore the precise mechanisms by which MIAT influences the inflammatory processes in DR, potentially paving the way for innovative therapeutic interventions.
5.4.1.3. Novel crosstalk between MIAT and diabetic cardiomyopathy (DCM)
Diabetic cardiomyopathy (DCM) is a serious complication of DM that can lead to severe manifestations such as heart failure and arrhythmias [243]. However, treatment options for DCM are limited, highlighting the need to understand the underlying mechanisms driving this condition.
In the context of DCM, MIAT has emerged as a potential key player, as a study indicated its over-expression in DCM rats compared to controls. Notably, MIAT has been linked to the over-expression of IL-17, a proinflammatory cytokine with significant roles in cardiac fibrosis, thereby contributing to the pathogenesis of DCM [244]. This effect on IL-17 is mediated through the repressive impact of MIAT on miR-214-3p [244]. These findings underscore the importance of the MIAT/miR-214-3p/IL-17 regulatory pathway as a crucial contributor to cardiac fibrosis in DCM [244].
Another significant pathway involved in promoting pyroptosis of cardiomyocytes in rat models of DCM was reported, where MIAT was found to downregulate miR-214-3p and upregulate caspase 1 [245]. These findings strongly implicate MIAT in the pathogenesis of DCM and emphasize the need for further studies to investigate targeting this pathway and validate its role as an upstream regulator of cardiac fibrosis.
Despite these compelling findings, studies analyzing the levels of MIAT in the blood of DCM patients are still lacking. Correlating the expression of MIAT with pro-fibrotic and pro-apoptotic markers is also essential to unravel the true role of MIAT in DCM. Such investigations will provide critical insights into the potential use of MIAT as a diagnostic and/or prognostic biomarker for DCM and may shed light on its therapeutic potential in combating the detrimental effects of DCM.
5.4.2. Osteoporosis (OP)
Osteoporosis (OP) is a chronic metabolic disease characterized by an imbalance in bone formation and resorption, leading to impaired bone mass and an increased risk of fractures [246]. The impact of OP is substantial, with an alarming frequency of bone fractures worldwide, as estimated by the International Osteoporosis Foundation, where a bone breaks every 3 s due to this condition [247]. With an estimated 500 million affected individuals globally, OP poses a significant health burden, particularly in the aging population [247]. Among postmenopausal women, 30% experience post-menopausal OP, which remains challenging to manage due to limited treatment options [248,249]. Therefore, there is a pressing need to delve into research aimed at identifying therapeutic targets and gaining a deeper understanding of the mechanisms contributing to this debilitating condition. Such research endeavors hold the key to developing more effective treatments and interventions to combat the detrimental impact of OP on bone health and overall quality of life.
5.4.2.1. Exploring the role of MIAT in OP: expression profile, signaling pathways and potential implications
In a recent study, researchers investigated the levels of MIAT in the blood of postmenopausal OP patients [250]. The findings revealed that MIAT, along with IL-6, TNF-α, and p38 mitogen-activated protein kinases (p38MAPK) were upregulated in PBMCs of postmenopausal OP patients compared to healthy controls [250]. Furthermore, the study identified that MIAT acts as a repressor of miR-216a expression in PBMCs, thereby regulating the transcription of p38MAPK [250]. This regulation of p38MAPK is crucial for AMPK/MAPK signaling activation, which, in turn, influences the secretion of inflammatory cytokines [250]. These unique insights suggest that MIAT could serve as a promising predictive biomarker for postmenopausal OP and may be involved in the underlying pathogenesis of the condition. However, it is essential to conduct future studies on larger cohorts to validate these findings and fully elucidate the intricate pathways through which MIAT regulates OP. Such investigations may open up new avenues for understanding the mechanisms involved in OP development and potentially lead to the development of targeted therapeutic regimens for managing such debilitating condition.
Jin et al. investigated the role of MIAT in the osteogenic differentiation of human adipose-derived stem cells (hASCs) [251]. It is known that hASCs possess self-renewal capabilities, indicating their potential for bone tissue engineering [251]. Despite their promise, the precise mechanisms underlying hASCs' contribution to osteogenic differentiation remain poorly understood [251]. The study revealed a crucial regulatory role for MIAT in osteogenic differentiation [251]. During the process of osteogenic differentiation, MIAT expression was found to be downregulated [251]. Knockdown experiments demonstrated that reduced MIAT levels led to increased differentiation of osteoblasts [251]. Remarkably, MIAT knockdown effectively rescued the inhibition of osteogenesis induced by TNF α-mediated inflammation [251]. These findings propose a mechanism that contributes to the differentiation of hASCs, which holds particular significance for future clinical applications in tissue engineering. Understanding the role of MIAT in osteogenic differentiation may pave the way for improved strategies to harness the therapeutic potential of hASCs in bone regenerative medicine. Further investigations are warranted to fully elucidate the intricate molecular pathways involved in this process.
5.5. Bone diseases
5.5.1. Osteoarthritis (OA)
Based on the Global Burden of Disease systemic analysis in 2019, approximately 500 million people worldwide are afflicted by osteoarthritis (OA) [252]. OA is an autoimmune diseases that profoundly impacts daily life functions due to its association with joint pain and disability [253]. While it is more prevalent in the older population (over 55), it can also affect younger individuals [253]. The primary features of OA encompass the progressive depletion of cartilage and the remodeling of the subchondral bone. Treatment options for OA are quite limited, leaving patients with joint replacement surgery as their primary recourse [254]. Additionally, drug therapy primarily focuses on alleviating pain, as there is a scarcity of drugs available that effectively slow down or reverse joint damage in OA [255]. This emphasizes the importance of identifying biomarkers and diagnostic targets that can help mitigate joint damage in OA.
5.5.1.1. Exploring the role of MIAT in OA: expression profile, signaling pathways and potential implications
In a study involving 60 participants in China, the expression of MIAT in knee cartilage tissues was found to be significantly higher in individuals with OA compared to those with normal knee conditions [256]. Additionally, this study identified miR-488-3p and SRY-Box Transcription Factor 11 (SOX11) as targets for MIAT. Previous research has established a link between SOX11 and OA, as it plays a crucial role in the regulation of proinflammatory cytokines [256]. To investigate the relationship between MIAT expression, miR-488-3p, and SOX11, the researchers employed a cell model of OA in their study [256]. The results revealed a positive correlation between MIAT expression and SOX11 levels, which was attributed to the downregulation of miR-488-3p [256]. This correlation was also found to be associated with the potentiation of chondrocyte injury, indicating a potential mechanism by which MIAT impacts OA pathogenesis.
Previous studies have reported on osteopontin (OPN)'s function as a regulator of various factors related to OA, affecting chondrocyte proliferation and apoptosis [[257], [258], [259]]. In human chondrocytes isolated from OA cartilage, MIAT has been associated with its role as an upstream regulator of miR-181a-5p and OPN [260]. Notably, MIAT was found to be upregulated in OA cartilage tissue [260]. When targeted in chondrocytes, it led to increased expression of miR-181a-5p, which, in turn, inhibited OPN overexpression [260]. This resulted in reduced cell viability, decreased DNA synthesis, and promoted apoptosis in human chondrocytes [260]. Another study using a cell model of OA revealed that MIAT was involved in inducing apoptosis and the release of pro-inflammatory cytokines through the regulation of NF-κB and c-Jun N-terminal kinases (JNK) by inhibiting the expression of miR-132 [261]. These findings suggest that MIAT acts as an upstream regulator within several networks contributing to the pathogenesis of OA.
5.6. Conclusion and future perspectives
In the current review, the authors have unveiled the significance of lncRNAs in various physiological and pathological conditions. LncRNAs have distinct expression patterns and versatile functions across different diseases. Accordingly, disease-associated lncRNAs can be utilized as diagnostic and prognostic biomarkers and/or therapeutic targets tailoring a personalized medicine approach for patients with rare complex diseases. This review was mainly concerned about the non-oncological pathological conditions where lncRNAs were found to be implicated in neurological diseases, cardiovascular diseases, metabolic disorders and some bone diseases. In particular, MIAT lncRNA was reported to be involved in most of previously mentioned categories of non-oncological pathological conditions. The authors cast the light upon the controversial studies regarding the expression profile, diagnostic and prognostic potential of MIAT in several diseases as summarized in (Table 1). Nonetheless, a snapshot of all validated signaling pathways drawn downstream MIAT lncRNA in non-oncological conditions have been highlighted in the current review.
Table 1.
Expression status of MIAT in multiple pathological diseases.
| Disease | Expression Profile | Validated Target Genes/Signaling pathway | References |
|---|---|---|---|
| Multiple Sclerosis | Controversial | SF1 CE1f3 QKI |
[49,155,166,168,169,176] |
| Schizophrenia | Controversial | BMI1 Crybb1 |
[174,177] |
| Ischemic stroke | Upregulated | miR-874-3p/IL-1β pathway | [185] |
| EGLN2 | [186] | ||
| REDD1 | [187] | ||
| Myocardial infarction | Upregulated | involvement in cardiomyocyte apoptosis | [189,198] |
| Activation of the PI3K/Akt signaling pathway | [203] | ||
| miR-24/furin/TGF-β1 | [202] | ||
| miR-203 | [206] | ||
| miR-10a-5p/EGR2 axis. | [207] | ||
| TSPO modulating mitochondrial membrane integrity in the infarct zone | [208] | ||
| Wnt-β-catenin signaling pathway | [209] | ||
| Coronary artery disease | Upregulated | chemokine signaling pathways NOD-like receptors |
[215,216] |
| Cardiac hypertrophy | Upregulated | ANP BNP β-MHC |
[218,219] |
| miR-93/TLR4 PI3K/Akt/mTOR pathway |
[220] | ||
| miR-150/P300 | [222] | ||
| Diabetes mellitus | Upregulated | N/A | [229,230] |
| Diabetic retinopathy | Upregulated | NF-κB | [238] |
| miR-342-3p | [240] | ||
| TGF-β1 signaling pathway | [241] | ||
| Diabetic cardiomyopathy | Upregulated | miR-214-3p/IL-17 | [244] |
| miR-214-3p-CASP1 axis in pyroptosis of cardiomyocytes | [245] | ||
| Osteoporosis | Upregulated | miR-216a/p38MAPK signaling pathway | [250] |
| Osteoarthritis | Upregulated | miR-488-3p/SOX11 | [256] |
| miR-181a-5p/OPN | |||
| miR-132/NF-κB |
It is also worth noting the MIAT lncRNA also show high therapeutic potential in several conditions highlighting a great promise in preclinical studies. However, further research is needed to fully elucidate the underlying molecular mechanisms and evaluate their effectiveness and safety in clinical settings. Future research should focus on comprehensive functional studies to uncover the specific roles of MIAT in different diseases. Exploring the interactions between lncRNAs and other ncRNAs, such as miRNAs, may reveal complex regulatory networks and therapeutic opportunities. Developing innovative technologies for accurate detection and quantification of lncRNAs will be crucial for their clinical use. In summary, lncRNAs have the potential to revolutionize the diagnosis, prognosis, and treatment of various non-oncological diseases. Continued research efforts and interdisciplinary collaborations are necessary to unlock the full potential of lncRNAs as diagnostic biomarkers, therapeutic targets, and interventions in the era of personalized medicine.
CRediT authorship contribution statement
Yousra Zeinelabdeen: Writing – original draft, Methodology, Investigation, Data curation. Tasneem Abaza: Writing – original draft, Validation, Methodology, Data curation. Montaser Bellah Yasser: Data curation, Formal analysis, Methodology, Software. Noha M. Elemam: Writing – review & editing, Validation, Methodology, Formal analysis. Rana A. Youness: Writing – review & editing, Writing – original draft, Validation, Supervision, Project administration, Methodology, Investigation, Data curation, Conceptualization.
Declaration of competing interest
The authors declare no conflict of interest.
References
- 1.Djebali S., et al. Landscape of transcription in human cells. Nature. 2012;489(7414):101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Salzberg S.L. Open questions: how many genes do we have? BMC Biol. 2018;16(1):1–3. doi: 10.1186/s12915-018-0564-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.El-Daly S.M., et al. Editorial: recent breakthroughs in the decoding of circulating nucleic acids and their applications to human diseases. Front. Mol. Biosci. 2023;10 doi: 10.3389/fmolb.2023.1203495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dawoud A., et al. Circular RNAs: new layer of complexity evading breast cancer heterogeneity. Noncoding RNA Res. 2023;8(1):60–74. doi: 10.1016/j.ncrna.2022.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.El-Aziz M.K.A., et al. Decoding hepatocarcinogenesis from a noncoding RNAs perspective. J. Cell. Physiol. 2023;238(9):1982–2009. doi: 10.1002/jcp.31076. [DOI] [PubMed] [Google Scholar]
- 6.Zhang P., et al. Non-coding RNAs and their integrated networks. J. Integr. Bioinform. 2019;16(3) doi: 10.1515/jib-2019-0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Awad A.R., et al. An acetylated derivative of vitexin halts MDA-MB-231 cellular progression and improves its immunogenic profile through tuning miR- 20a-MICA/B axis. Nat. Prod. Res. 2021;35(18):3126–3130. doi: 10.1080/14786419.2019.1686372. [DOI] [PubMed] [Google Scholar]
- 8.Ahmed Youness R., et al. A methoxylated quercetin glycoside harnesses HCC tumor progression in a TP53/miR-15/miR-16 dependent manner. Nat. Prod. Res. 2020;34(10):1475–1480. doi: 10.1080/14786419.2018.1509326. [DOI] [PubMed] [Google Scholar]
- 9.Soliman A.H., et al. Phytochemical-derived tumor-associated macrophage remodeling strategy using Phoenix dactylifera L. boosted photodynamic therapy in melanoma via H19/iNOS/PD-L1 axis. Photodiagnosis Photodyn. Ther. 2023;44 doi: 10.1016/j.pdpdt.2023.103792. [DOI] [PubMed] [Google Scholar]
- 10.Mekky R.Y., et al. MALAT-1: immunomodulatory lncRNA hampering the innate and the adaptive immune arms in triple negative breast cancer. Transl. Oncol. 2023;31 doi: 10.1016/j.tranon.2023.101653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Youness R.A., et al. The long noncoding RNA sONE represses triple-negative breast cancer aggressiveness through inducing the expression of miR-34a, miR-15a, miR-16, and let-7a. J. Cell. Physiol. 2019;234(11):20286–20297. doi: 10.1002/jcp.28629. [DOI] [PubMed] [Google Scholar]
- 12.Su Y., et al. Regulatory non-coding RNA: new instruments in the orchestration of cell death. Cell Death Dis. 2016;7(8) doi: 10.1038/cddis.2016.210. e2333-e2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Selem N.A., et al. Let-7a/cMyc/CCAT1/miR-17-5p circuit Re-sensitizes atezolizumab resistance in triple negative breast cancer through modulating PD-L1. Pathol. Res. Pract. 2023;248 doi: 10.1016/j.prp.2023.154579. [DOI] [PubMed] [Google Scholar]
- 14.Abdallah R.M., et al. Hindering the synchronization between miR-486-5p and H19 lncRNA by hesperetin halts breast cancer aggressiveness through tuning ICAM-1. Anti Cancer Agents Med. Chem. 2022;22(3):586–595. doi: 10.2174/1871520621666210419093652. [DOI] [PubMed] [Google Scholar]
- 15.Nafea H., et al. Dual targeting of H(2)S synthesizing enzymes; cystathionine β-synthase and cystathionine γ-lyase by miR-939-5p effectively curbs triple negative breast cancer. Heliyon. 2023;9(10) doi: 10.1016/j.heliyon.2023.e21063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.ZeinElAbdeen Y.A., AbdAlSeed A., Youness R.A. Decoding insulin-like growth factor signaling pathway from a non-coding RNAs perspective: a step towards precision oncology in breast cancer. J. Mammary Gland Biol. Neoplasia. 2022;27(1):79–99. doi: 10.1007/s10911-022-09511-z. [DOI] [PubMed] [Google Scholar]
- 17.Fahmy S.A., et al. Molecular engines, therapeutic targets, and challenges in pediatric brain tumors: a special emphasis on hydrogen sulfide and RNA-based nano-delivery. Cancers. 2022;14(21) doi: 10.3390/cancers14215244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Youssef S.S., et al. miR-516a-3P, a potential circulating biomarker in hepatocellular carcinoma, correlated with rs738409 polymorphism in PNPLA3. Pres. Méd. 2022;19(6):483–493. doi: 10.2217/pme-2022-0005. [DOI] [PubMed] [Google Scholar]
- 19.El Kilany F.H., et al. miR-744/eNOS/NO axis: a novel target to halt triple negative breast cancer progression. Breast Dis. 2021;40(3):161–169. doi: 10.3233/BD-200454. [DOI] [PubMed] [Google Scholar]
- 20.Youness R.A., et al. Targeting hydrogen sulphide signaling in breast cancer. J. Adv. Res. 2021;27:177–190. doi: 10.1016/j.jare.2020.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mekky R.Y., et al. Epigallocatechin gallate (EGCG) and miR-548m reduce HCV entry through repression of CD81 receptor in HCV cell models. Arch. Virol. 2019;164(6):1587–1595. doi: 10.1007/s00705-019-04232-x. [DOI] [PubMed] [Google Scholar]
- 22.Selem N.A., Youness R.A., Gad M.Z. What is beyond LncRNAs in breast cancer: a special focus on colon cancer-associated Transcript-1 (CCAT-1) Noncoding RNA Res. 2021;6(4):174–186. doi: 10.1016/j.ncrna.2021.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nafea H., et al. LncRNA HEIH/miR-939-5p interplay modulates triple-negative breast cancer progression through NOS2-induced nitric oxide production. J. Cell. Physiol. 2021;236(7):5362–5372. doi: 10.1002/jcp.30234. [DOI] [PubMed] [Google Scholar]
- 24.ElKhouly A.M., Youness R.A., Gad M.Z. MicroRNA-486-5p and microRNA-486-3p: multifaceted pleiotropic mediators in oncological and non-oncological conditions. Noncoding RNA Res. 2020;5(1):11–21. doi: 10.1016/j.ncrna.2020.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.El Din G.S., et al. miRNA-506-3p directly regulates rs10754339 (A/G) in the immune checkpoint protein B7-H4 in breast cancer. MicroRNA. 2020;9(5):346–353. doi: 10.2174/2211536609666201209152949. [DOI] [PubMed] [Google Scholar]
- 26.Youness R.A., Gad M.Z. Long non-coding RNAs: functional regulatory players in breast cancer. Noncoding RNA Res. 2019;4(1):36–44. doi: 10.1016/j.ncrna.2019.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Abaza T., et al. Emerging role of circular RNAs in hepatocellular carcinoma immunotherapy. Int. J. Mol. Sci. 2023;24(22) doi: 10.3390/ijms242216484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Statello L., et al. Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 2021;22(2):96–118. doi: 10.1038/s41580-020-00315-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gudenas B.L., Wang L. Prediction of LncRNA subcellular localization with deep learning from sequence features. Sci. Rep. 2018;8(1) doi: 10.1038/s41598-018-34708-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Abdel-Latif M., et al. MALAT-1/p53/miR-155/miR-146a ceRNA circuit tuned by methoxylated quercitrin glycoside alters immunogenic and oncogenic profiles of breast cancer. Mol. Cell. Biochem. 2022;477(4):1281–1293. doi: 10.1007/s11010-022-04378-4. [DOI] [PubMed] [Google Scholar]
- 31.Katayama S., et al. Antisense transcription in the mammalian transcriptome. Science. 2005;309(5740):1564–1566. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- 32.Rinn J.L., Chang H.Y. Genome regulation by long noncoding RNAs. Annu. Rev. Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Youness R.A., et al. A novel role of sONE/NOS3/NO signaling cascade in mediating hydrogen sulphide bilateral effects on triple negative breast cancer progression. Nitric Oxide. 2018;80:12–23. doi: 10.1016/j.niox.2018.07.004. [DOI] [PubMed] [Google Scholar]
- 34.Garzon R., et al. Expression and prognostic impact of lncRNAs in acute myeloid leukemia. Proc. Natl. Acad. Sci. USA. 2014;111(52):18679–18684. doi: 10.1073/pnas.1422050112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kumarswamy R., et al. Circulating long noncoding RNA, LIPCAR, predicts survival in patients with heart failure. Circ. Res. 2014;114(10):1569–1575. doi: 10.1161/CIRCRESAHA.114.303915. [DOI] [PubMed] [Google Scholar]
- 36.Takayama K.i., et al. Androgen‐responsive long noncoding RNA CTBP1‐AS promotes prostate cancer. EMBO J. 2013;32(12):1665–1680. doi: 10.1038/emboj.2013.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Boon R.A., et al. Long noncoding RNAs: from clinical genetics to therapeutic targets? J. Am. Coll. Cardiol. 2016;67(10):1214–1226. doi: 10.1016/j.jacc.2015.12.051. [DOI] [PubMed] [Google Scholar]
- 38.Liao J., et al. LncRNA MIAT: myocardial infarction associated and more. Gene. 2016;578(2):158–161. doi: 10.1016/j.gene.2015.12.032. [DOI] [PubMed] [Google Scholar]
- 39.Ghafouri-Fard S., Azimi T., Taheri M. Myocardial infarction associated transcript (MIAT): review of its impact in the tumorigenesis. Biomed. Pharmacother. 2021;133 doi: 10.1016/j.biopha.2020.111040. [DOI] [PubMed] [Google Scholar]
- 40.Jin H., et al. Long non-coding RNA MIAT competitively binds miR-150-5p to regulate ZEB1 expression in osteosarcoma. Oncol. Lett. 2019;17(1):1229–1236. doi: 10.3892/ol.2018.9671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jin S., et al. p53-targeted lincRNA-p21 acts as a tumor suppressor by inhibiting JAK2/STAT3 signaling pathways in head and neck squamous cell carcinoma. Mol. Cancer. 2019;18:1–18. doi: 10.1186/s12943-019-0993-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sun C., et al. Long non-coding RNA MIAT in development and disease: a new player in an old game. J. Biomed. Sci. 2018;25(1):23. doi: 10.1186/s12929-018-0427-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Da C.-m., et al. The role of long non-coding RNA MIAT in cancers. Biomed. Pharmacother. 2020;129 doi: 10.1016/j.biopha.2020.110359. [DOI] [PubMed] [Google Scholar]
- 44.Ishii N., et al. Identification of a novel non-coding RNA, MIAT, that confers risk of myocardial infarction. J. Hum. Genet. 2006;51(12):1087–1099. doi: 10.1007/s10038-006-0070-9. [DOI] [PubMed] [Google Scholar]
- 45.Vausort M., Wagner D.R., Devaux Y. Long noncoding RNAs in patients with acute myocardial infarction. Circ. Res. 2014;115(7):668–677. doi: 10.1161/CIRCRESAHA.115.303836. [DOI] [PubMed] [Google Scholar]
- 46.Rao S.-Q., et al. Genetic variants in long non-coding RNA MIAT contribute to risk of paranoid schizophrenia in a Chinese Han population. Schizophr. Res. 2015;166(1–3):125–130. doi: 10.1016/j.schres.2015.04.032. [DOI] [PubMed] [Google Scholar]
- 47.Yan B., et al. lncRNA-MIAT regulates microvascular dysfunction by functioning as a competing endogenous RNA. Circ. Res. 2015;116(7):1143–1156. doi: 10.1161/CIRCRESAHA.116.305510. [DOI] [PubMed] [Google Scholar]
- 48.Chen Y., He Y., Zhou H. The potential role of lncRNAs in diabetes and diabetic microvascular complications. Endocr. J. 2020;67(7):659–668. doi: 10.1507/endocrj.EJ19-0574. [DOI] [PubMed] [Google Scholar]
- 49.Amiri M., et al. Expression and diagnostic values of MIAT, H19, and NRON long non-coding RNAs in multiple sclerosis patients. Egypt. J. Med. Hum. Genet. 2022;23(1):46. [Google Scholar]
- 50.Aprea J., et al. Transcriptome sequencing during mouse brain development identifies long non‐coding RNAs functionally involved in neurogenic commitment. EMBO J. 2013;32(24):3145–3160. doi: 10.1038/emboj.2013.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Guo X., et al. LncRNA-MIAT promotes neural cell autophagy and apoptosis in ischemic stroke by up-regulating REDD1. Brain Res. 2021;1763 doi: 10.1016/j.brainres.2021.147436. [DOI] [PubMed] [Google Scholar]
- 52.Wang J., et al. Roles of long non-coding RNA in osteoarthritis. Int. J. Mol. Med. 2021;48(1):1–8. doi: 10.3892/ijmm.2021.4966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ishizuka A., et al. Formation of nuclear bodies by the lnc RNA Gomafu‐associating proteins Celf3 and SF 1. Gene Cell. 2014;19(9):704–721. doi: 10.1111/gtc.12169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Derrien T., et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22(9):1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Clark M.B., et al. Genome-wide analysis of long noncoding RNA stability. Genome Res. 2012;22(5):885–898. doi: 10.1101/gr.131037.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Beaulieu Y.B., et al. Polyadenylation-dependent control of long noncoding RNA expression by the poly (A)-binding protein nuclear 1. PLoS Genet. 2012;8(11) doi: 10.1371/journal.pgen.1003078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Brown J.A., et al. Formation of triple-helical structures by the 3′-end sequences of MALAT1 and MENβ noncoding RNAs. Proc. Natl. Acad. Sci. USA. 2012;109(47):19202–19207. doi: 10.1073/pnas.1217338109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wilusz J.E., et al. A triple helix stabilizes the 3′ ends of long noncoding RNAs that lack poly (A) tails. Gene Dev. 2012;26(21):2392–2407. doi: 10.1101/gad.204438.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Deveson I.W., et al. Universal alternative splicing of noncoding exons. Cell Syst. 2018;6(2):245–255. e5. doi: 10.1016/j.cels.2017.12.005. [DOI] [PubMed] [Google Scholar]
- 60.Zuckerman B., Ulitsky I. Predictive models of subcellular localization of long RNAs. RNA. 2019;25(5):557–572. doi: 10.1261/rna.068288.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Melé M., et al. Chromatin environment, transcriptional regulation, and splicing distinguish lincRNAs and mRNAs. Genome Res. 2017;27(1):27–37. doi: 10.1101/gr.214205.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tilgner H., et al. Deep sequencing of subcellular RNA fractions shows splicing to be predominantly co-transcriptional in the human genome but inefficient for lncRNAs. Genome Res. 2012;22(9):1616–1625. doi: 10.1101/gr.134445.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mukherjee N., et al. Integrative classification of human coding and noncoding genes through RNA metabolism profiles. Nat. Struct. Mol. Biol. 2017;24(1):86–96. doi: 10.1038/nsmb.3325. [DOI] [PubMed] [Google Scholar]
- 64.Cabili M.N., et al. Localization and abundance analysis of human lncRNAs at single-cell and single-molecule resolution. Genome Biol. 2015;16:1–16. doi: 10.1186/s13059-015-0586-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hezroni H., et al. Principles of long noncoding RNA evolution derived from direct comparison of transcriptomes in 17 species. Cell Rep. 2015;11(7):1110–1122. doi: 10.1016/j.celrep.2015.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ulitsky I. Evolution to the rescue: using comparative genomics to understand long non-coding RNAs. Nat. Rev. Genet. 2016;17(10):601–614. doi: 10.1038/nrg.2016.85. [DOI] [PubMed] [Google Scholar]
- 67.Guo C.-J., et al. Distinct processing of lncRNAs contributes to non-conserved functions in stem cells. Cell. 2020;181(3):621–636. e22. doi: 10.1016/j.cell.2020.03.006. [DOI] [PubMed] [Google Scholar]
- 68.Chen L.-L. Linking long noncoding RNA localization and function. Trends Biochem. Sci. 2016;41(9):761–772. doi: 10.1016/j.tibs.2016.07.003. [DOI] [PubMed] [Google Scholar]
- 69.Batista P.J., Chang H.Y. Long noncoding RNAs: cellular address codes in development and disease. Cell. 2013;152(6):1298–1307. doi: 10.1016/j.cell.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chen L.-L., Carmichael G.G. Decoding the function of nuclear long non-coding RNAs. Curr. Opin. Cell Biol. 2010;22(3):357–364. doi: 10.1016/j.ceb.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mattick J.S., et al. Long non-coding RNAs: definitions, functions, challenges and recommendations. Nat. Rev. Mol. Cell Biol. 2023;24(6):430–447. doi: 10.1038/s41580-022-00566-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Arora R., et al. RNaseH 1 regulates TERRA-telomeric DNA hybrids and telomere maintenance in ALT tumour cells. Nat. Commun. 2014;5(1):5220. doi: 10.1038/ncomms6220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Clemson C.M., et al. XIST RNA paints the inactive X chromosome at interphase: evidence for a novel RNA involved in nuclear/chromosome structure. J. Cell Biol. 1996;132(3):259–275. doi: 10.1083/jcb.132.3.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Postepska-Igielska A., et al. LncRNA Khps1 regulates expression of the proto-oncogene SPHK1 via triplex-mediated changes in chromatin structure. Mol. Cell. 2015;60(4):626–636. doi: 10.1016/j.molcel.2015.10.001. [DOI] [PubMed] [Google Scholar]
- 75.Grelet S., et al. A regulated PNUTS mRNA to lncRNA splice switch mediates EMT and tumour progression. Nat. Cell Biol. 2017;19(9):1105–1115. doi: 10.1038/ncb3595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kleaveland B., et al. A network of noncoding regulatory RNAs acts in the mammalian brain. Cell. 2018;174(2):350–362. e17. doi: 10.1016/j.cell.2018.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zealy R.W., et al. Long noncoding RNA complementarity and target transcripts abundance. Biochim. Biophys. Acta - Gene Regul. Mech. 2018;1861(3):224–234. doi: 10.1016/j.bbagrm.2018.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ramzy A., et al. Drugless nanoparticles tune-up an array of intertwined pathways contributing to immune checkpoint signaling and metabolic reprogramming in triple-negative breast cancer. Biomed. Mater. 2022;18(1) doi: 10.1088/1748-605X/aca85d. [DOI] [PubMed] [Google Scholar]
- 79.Ahn J.-H., et al. nc886 is induced by TGF-β and suppresses the microRNA pathway in ovarian cancer. Nat. Commun. 2018;9(1):1166. doi: 10.1038/s41467-018-03556-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jiang L., et al. NEAT1 scaffolds RNA-binding proteins and the Microprocessor to globally enhance pri-miRNA processing. Nat. Struct. Mol. Biol. 2017;24(10):816–824. doi: 10.1038/nsmb.3455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yamazaki T., et al. Functional domains of NEAT1 architectural lncRNA induce paraspeckle assembly through phase separation. Mol. Cell. 2018;70(6):1038–1053. e7. doi: 10.1016/j.molcel.2018.05.019. [DOI] [PubMed] [Google Scholar]
- 82.Schmidt K., et al. Targeting the oncogenic long non-coding RNA SLNCR1 by blocking its sequence-specific binding to the androgen receptor. Cell Rep. 2020;30(2):541–554. e5. doi: 10.1016/j.celrep.2019.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kitagawa M., et al. Cell cycle regulation by long non-coding RNAs. Cell. Mol. Life Sci. 2013;70:4785–4794. doi: 10.1007/s00018-013-1423-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sirey T.M., et al. The long non-coding RNA Cerox1 is a post transcriptional regulator of mitochondrial complex I catalytic activity. Elife. 2019;8 doi: 10.7554/eLife.45051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sun X., Wong D. Long non-coding RNA-mediated regulation of glucose homeostasis and diabetes. Am. J. Cardiovasc. Dis. 2016;6(2):17. [PMC free article] [PubMed] [Google Scholar]
- 86.Ballarino M., et al. Non-coding RNAs in muscle differentiation and musculoskeletal disease. J. Clin. Invest. 2016;126(6):2021–2030. doi: 10.1172/JCI84419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Brazão T.F., et al. Long noncoding RNAs in B-cell development and activation. Blood. J. Am. Soc. Hematol. 2016;128(7):e10–e19. doi: 10.1182/blood-2015-11-680843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Delas M.J., et al. lncRNA requirements for mouse acute myeloid leukemia and normal differentiation. Elife. 2017;6 doi: 10.7554/eLife.25607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Esteller M. Non-coding RNAs in human disease. Nat. Rev. Genet. 2011;12(12):861–874. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 90.Wang Y., et al. Mammalian ncRNA-disease repository: a global view of ncRNA-mediated disease network. Cell Death Dis. 2013;4(8) doi: 10.1038/cddis.2013.292. e765-e765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yuan J.-h., et al. A long noncoding RNA activated by TGF-β promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell. 2014;25(5):666–681. doi: 10.1016/j.ccr.2014.03.010. [DOI] [PubMed] [Google Scholar]
- 92.Rahmoon M.A., et al. MiR-615-5p depresses natural killer cells cytotoxicity through repressing IGF-1R in hepatocellular carcinoma patients. Growth Factors. 2017;35(2–3):76–87. doi: 10.1080/08977194.2017.1354859. [DOI] [PubMed] [Google Scholar]
- 93.Youness R.A., et al. MicroRNA-486-5p enhances hepatocellular carcinoma tumor suppression through repression of IGF-1R and its downstream mTOR, STAT3 and c-Myc. Oncol. Lett. 2016;12(4):2567–2573. doi: 10.3892/ol.2016.4914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Youness R.A., et al. Contradicting interplay between insulin-like growth factor-1 and miR-486-5p in primary NK cells and hepatoma cell lines with a contemporary inhibitory impact on HCC tumor progression. Growth Factors. 2016;34(3–4):128–140. doi: 10.1080/08977194.2016.1200571. [DOI] [PubMed] [Google Scholar]
- 95.Aboelenein H.R., et al. Reduction of CD19 autoimmunity marker on B cells of paediatric SLE patients through repressing PU.1/TNF-α/BAFF axis pathway by miR-155. Growth Factors. 2017;35(2–3):49–60. doi: 10.1080/08977194.2017.1345900. [DOI] [PubMed] [Google Scholar]
- 96.Shaalan Y.M., et al. Destabilizing the interplay between miR-1275 and IGF2BPs by Tamarix articulata and quercetin in hepatocellular carcinoma. Nat. Prod. Res. 2018;32(18):2217–2220. doi: 10.1080/14786419.2017.1366478. [DOI] [PubMed] [Google Scholar]
- 97.Chen G., et al. LncRNADisease: a database for long-non-coding RNA-associated diseases. Nucleic Acids Res. 2012;41(D1):D983–D986. doi: 10.1093/nar/gks1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Walsh A.L., et al. Long noncoding RNAs and prostate carcinogenesis: the missing ‘linc’? Trends Mol. Med. 2014;20(8):428–436. doi: 10.1016/j.molmed.2014.03.005. [DOI] [PubMed] [Google Scholar]
- 99.Xiang J.-F., et al. Human colorectal cancer-specific CCAT1-L lncRNA regulates long-range chromatin interactions at the MYC locus. Cell Res. 2014;24(5):513–531. doi: 10.1038/cr.2014.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Morris K.V., Mattick J.S. The rise of regulatory RNA. Nat. Rev. Genet. 2014;15(6):423–437. doi: 10.1038/nrg3722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Necsulea A., et al. The evolution of lncRNA repertoires and expression patterns in tetrapods. Nature. 2014;505(7485):635–640. doi: 10.1038/nature12943. [DOI] [PubMed] [Google Scholar]
- 102.Faghihi M.A., et al. Expression of a noncoding RNA is elevated in Alzheimer's disease and drives rapid feed-forward regulation of β-secretase. Nat. Med. 2008;14(7):723–730. doi: 10.1038/nm1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Cui H., et al. The human long noncoding RNA lnc‐IL 7 R regulates the inflammatory response. Eur. J. Immunol. 2014;44(7):2085–2095. doi: 10.1002/eji.201344126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wang P., et al. The STAT3-binding long noncoding RNA lnc-DC controls human dendritic cell differentiation. Science. 2014;344(6181):310–313. doi: 10.1126/science.1251456. [DOI] [PubMed] [Google Scholar]
- 105.Stelzer Y., et al. The noncoding RNA IPW regulates the imprinted DLK1-DIO3 locus in an induced pluripotent stem cell model of Prader-Willi syndrome. Nat. Genet. 2014;46(6):551–557. doi: 10.1038/ng.2968. [DOI] [PubMed] [Google Scholar]
- 106.Youness R.A., et al. A snapshot of photoresponsive liposomes in cancer chemotherapy and immunotherapy: opportunities and challenges. ACS Omega. 2023;8(47):44424–44436. doi: 10.1021/acsomega.3c04134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Trzybulska D., Vergadi E., Tsatsanis C. miRNA and other non-coding RNAs as promising diagnostic markers. Ejifcc. 2018;29(3):221. [PMC free article] [PubMed] [Google Scholar]
- 108.Yao Q., et al. The integrated comprehension of lncRNA HOXA-AS3 implication on human diseases. Clin. Transl. Oncol. 2022;24(12):2342–2350. doi: 10.1007/s12094-022-02920-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Le L.T., Nhu C.X. The role of long non-coding RNAs in cardiovascular diseases. Int. J. Mol. Sci. 2023;24(18) doi: 10.3390/ijms241813805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Xu X., et al. Autophagy-associated lncRNAs: promising targets for neurological disease diagnosis and therapy. Neural Plast. 2020:2020. doi: 10.1155/2020/8881687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Massone S., et al. 17A, a novel non-coding RNA, regulates GABA B alternative splicing and signaling in response to inflammatory stimuli and in Alzheimer disease. Neurobiol. Dis. 2011;41(2):308–317. doi: 10.1016/j.nbd.2010.09.019. [DOI] [PubMed] [Google Scholar]
- 112.Yan W., et al. LncRNA NEAT1 promotes autophagy in MPTP-induced Parkinson's disease through stabilizing PINK1 protein. Biochem. Biophys. Res. Commun. 2018;496(4):1019–1024. doi: 10.1016/j.bbrc.2017.12.149. [DOI] [PubMed] [Google Scholar]
- 113.Wang J., et al. Long non-coding RNA H19 induces cerebral ischemia reperfusion injury via activation of autophagy. Aging Dis. 2017;8(1):71. doi: 10.14336/AD.2016.0530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Wu Q., Yi X. Down-regulation of long noncoding RNA MALAT1 protects hippocampal neurons against excessive autophagy and apoptosis via the PI3K/Akt signaling pathway in rats with epilepsy. J. Mol. Neurosci. 2018;65(2):234–245. doi: 10.1007/s12031-018-1093-3. [DOI] [PubMed] [Google Scholar]
- 115.Xu D.H., et al. Wiley Online Library; 2019. Retracted: Long Noncoding RNA MEG3 Inhibits Proliferation and Migration but Induces Autophagy by Regulation of Sirt7 and PI3K/AKT/mTOR Pathway in Glioma Cells. [DOI] [PubMed] [Google Scholar]
- 116.Ohnishi Y., et al. Identification of 187 single nucleotide polymorphisms (SNPs) among 41 candidate genes for ischemic heart disease in the Japanese population. Hum. Genet. 2000;106:288–292. doi: 10.1007/s004390051039. [DOI] [PubMed] [Google Scholar]
- 117.Blackshaw S., et al. Genomic analysis of mouse retinal development. PLoS Biol. 2004;2(9):e247. doi: 10.1371/journal.pbio.0020247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Sone M., et al. The mRNA-like noncoding RNA Gomafu constitutes a novel nuclear domain in a subset of neurons. J. Cell Sci. 2007;120(15):2498–2506. doi: 10.1242/jcs.009357. [DOI] [PubMed] [Google Scholar]
- 119.Mount S.M. A catalogue of splice junction sequences. Nucleic Acids Res. 1982;10(2):459–472. doi: 10.1093/nar/10.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Shapiro M.B., Senapathy P. RNA splice junctions of different classes of eukaryotes: sequence statistics and functional implications in gene expression. Nucleic Acids Res. 1987;15(17):7155–7174. doi: 10.1093/nar/15.17.7155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Sun C., et al. Long non-coding RNA MIAT in development and disease: a new player in an old game. J. Biomed. Sci. 2018;25(1):1–7. doi: 10.1186/s12929-018-0427-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Rapicavoli N.A., Poth E.M., Blackshaw S. The long noncoding RNA RNCR2 directs mouse retinal cell specification. BMC Dev. Biol. 2010;10:1–10. doi: 10.1186/1471-213X-10-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Galarneau A., Richard S. Target RNA motif and target mRNAs of the Quaking STAR protein. Nat. Struct. Mol. Biol. 2005;12(8):691–698. doi: 10.1038/nsmb963. [DOI] [PubMed] [Google Scholar]
- 124.Ryder S.P., Williamson J.R. Specificity of the STAR/GSG domain protein Qk1: implications for the regulation of myelination. RNA. 2004;10(9):1449–1458. doi: 10.1261/rna.7780504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Balamurali D., Stoll M. Non-coding RNA databases in cardiovascular research. Noncoding RNA. 2020;6(3) doi: 10.3390/ncrna6030035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.The RNAcentral Consortium RNAcentral: a hub of information for non-coding RNA sequences. Nucleic Acids Res. 2018;47(D1):D221–D229. doi: 10.1093/nar/gky1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Kozomara A., Birgaoanu M., Griffiths-Jones S. miRBase: from microRNA sequences to function. Nucleic Acids Res. 2019;47(D1):D155–d162. doi: 10.1093/nar/gky1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Zhao H., et al. LncTarD 2.0: an updated comprehensive database for experimentally-supported functional lncRNA–target regulations in human diseases. Nucleic Acids Res. 2022;51(D1):D199–D207. doi: 10.1093/nar/gkac984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Cheng L., et al. LncRNA2Target v2.0: a comprehensive database for target genes of lncRNAs in human and mouse. Nucleic Acids Res. 2019;47(D1):D140–d144. doi: 10.1093/nar/gky1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Tay Y., Rinn J., Pandolfi P.P. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505(7483):344–352. doi: 10.1038/nature12986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Jiang Q., et al. Long non-coding RNA-MIAT promotes neurovascular remodeling in the eye and brain. Oncotarget. 2016;7(31) doi: 10.18632/oncotarget.10434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Zhu M., et al. Peripheral blood leukocyte expression of lncRNA MIAT and its diagnostic and prognostic value in ischemic stroke. J. Stroke Cerebrovasc. Dis. 2018;27(2):326–337. doi: 10.1016/j.jstrokecerebrovasdis.2017.09.009. [DOI] [PubMed] [Google Scholar]
- 133.Zhou X., et al. lncRNA MIAT functions as a competing endogenous RNA to upregulate DAPK2 by sponging miR-22-3p in diabetic cardiomyopathy. Cell Death Dis. 2017;8(7) doi: 10.1038/cddis.2017.321. e2929-e2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Zhang H.-Y., et al. The long non-coding RNA MIAT regulates zinc finger E-box binding homeobox 1 expression by sponging miR-150 and promoting cell invasion in non-small-cell lung cancer. Gene. 2017;633:61–65. doi: 10.1016/j.gene.2017.08.009. [DOI] [PubMed] [Google Scholar]
- 135.Zhou L., et al. Long non-coding MIAT mediates high glucose-induced renal tubular epithelial injury. Biochem. Biophys. Res. Commun. 2015;468(4):726–732. doi: 10.1016/j.bbrc.2015.11.023. [DOI] [PubMed] [Google Scholar]
- 136.Kim B.J., et al. Adipocytokines and ischemic stroke: differential associations between stroke subtypes. J. Neurol. Sci. 2012;312(1–2):117–122. doi: 10.1016/j.jns.2011.08.007. [DOI] [PubMed] [Google Scholar]
- 137.Sattari A., et al. Upregulation of long noncoding RNA MIAT in aggressive form of chronic lymphocytic leukemias. Oncotarget. 2016;7(34) doi: 10.18632/oncotarget.11099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Walton C., et al. Rising prevalence of multiple sclerosis worldwide: insights from the Atlas of MS. Mult. Scler. 2020;26(14):1816–1821. doi: 10.1177/1352458520970841. third edition. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Kiriacos C.J., et al. Prospective medicinal plants and their phytochemicals shielding autoimmune and cancer patients against the SARS-CoV-2 pandemic: a special focus on matcha. Front. Oncol. 2022;12 doi: 10.3389/fonc.2022.837408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Kappos L., et al. Contribution of relapse-independent progression vs relapse-associated worsening to overall confirmed disability accumulation in typical relapsing multiple sclerosis in a pooled analysis of 2 randomized clinical trials. JAMA Neurol. 2020;77(9):1132–1140. doi: 10.1001/jamaneurol.2020.1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Hoenes G., Hauser M., Pfleiderer G. Dynamic total fluorescence and anisotropy decay study of the dansyl fluorophore in model compounds and enzymes. Photochem. Photobiol. 1986;43(2):133–137. doi: 10.1111/j.1751-1097.1986.tb09504.x. [DOI] [PubMed] [Google Scholar]
- 142.Kuhlmann T., Antel J. Multiple sclerosis: 2023 update. Free Neuropathol. 2023;4 doi: 10.17879/freeneuropathology-2023-4675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Miclea A., et al. A brief review of the effects of vitamin D on multiple sclerosis. Front. Immunol. 2020;11:781. doi: 10.3389/fimmu.2020.00781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Ascherio A. Environmental factors in multiple sclerosis. Expert Rev. Neurother. 2013;13(12 Suppl):3–9. doi: 10.1586/14737175.2013.865866. [DOI] [PubMed] [Google Scholar]
- 145.Munz C., et al. Antiviral immune responses: triggers of or triggered by autoimmunity? Nat. Rev. Immunol. 2009;9(4):246–258. doi: 10.1038/nri2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Qureshi I.A., Mattick J.S., Mehler M.F. Long non-coding RNAs in nervous system function and disease. Brain Res. 2010;1338:20–35. doi: 10.1016/j.brainres.2010.03.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Yousuf A., Qurashi A. Non-coding RNAs in the pathogenesis of multiple sclerosis. Front. Genet. 2021;12 doi: 10.3389/fgene.2021.717922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Imamura K., et al. Long noncoding RNA NEAT1-dependent SFPQ relocation from promoter region to paraspeckle mediates IL8 expression upon immune stimuli. Mol. Cell. 2014;53(3):393–406. doi: 10.1016/j.molcel.2014.01.009. [DOI] [PubMed] [Google Scholar]
- 149.Sung T.L., Rice A.P. Effects of prostrating on Cyclin T1/P-TEFb function and the gene expression profile in primary resting CD4+ T cells. Retrovirology. 2006;3:66. doi: 10.1186/1742-4690-3-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Santoro M., et al. A pilot study of lncRNAs expression profile in serum of progressive multiple sclerosis patients. Eur. Rev. Med. Pharmacol. Sci. 2020;24(6):3267–3273. doi: 10.26355/eurrev_202003_20694. [DOI] [PubMed] [Google Scholar]
- 151.Sone M., et al. The mRNA-like noncoding RNA Gomafu constitutes a novel nuclear domain in a subset of neurons. J. Cell Sci. 2007;120(Pt 15):2498–2506. doi: 10.1242/jcs.009357. [DOI] [PubMed] [Google Scholar]
- 152.Ip J.Y., et al. Gomafu lncRNA knockout mice exhibit mild hyperactivity with enhanced responsiveness to the psychostimulant methamphetamine. Sci. Rep. 2016;6 doi: 10.1038/srep27204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Sheik Mohamed J., et al. Conserved long noncoding RNAs transcriptionally regulated by Oct4 and Nanog modulate pluripotency in mouse embryonic stem cells. RNA. 2010;16(2):324–337. doi: 10.1261/rna.1441510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Aprea J., et al. Transcriptome sequencing during mouse brain development identifies long non-coding RNAs functionally involved in neurogenic commitment. EMBO J. 2013;32(24):3145–3160. doi: 10.1038/emboj.2013.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Fenoglio C., et al. LncRNAs expression profile in peripheral blood mononuclear cells from multiple sclerosis patients. J. Neuroimmunol. 2018;324:129–135. doi: 10.1016/j.jneuroim.2018.08.008. [DOI] [PubMed] [Google Scholar]
- 156.Institute of health Metrics and Evaluation (IHME). Global Health Data Exchange (GHDx).
- 157.Correll C.U., et al. Mortality in people with schizophrenia: a systematic review and meta-analysis of relative risk and aggravating or attenuating factors. World Psychiatr. 2022;21(2):248–271. doi: 10.1002/wps.20994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Focking M., et al. Epigenetic factors in schizophrenia: mechanisms and experimental approaches. Mol. Neuropsychiatry. 2019;5(1):6–12. doi: 10.1159/000495063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Wahbeh M.H., Avramopoulos D. Gene-Environment interactions in schizophrenia: a literature review. Genes. 2021;12(12) doi: 10.3390/genes12121850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Stilo S.A., Murray R.M. Non-genetic factors in schizophrenia. Curr. Psychiatr. Rep. 2019;21(10):100. doi: 10.1007/s11920-019-1091-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Nakata K., et al. DISC1 splice variants are upregulated in schizophrenia and associated with risk polymorphisms. Proc. Natl. Acad. Sci. U. S. A. 2009;106(37):15873–15878. doi: 10.1073/pnas.0903413106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Zhang C.Y., et al. An alternative splicing hypothesis for neuropathology of schizophrenia: evidence from studies on historical candidate genes and multi-omics data. Mol. Psychiatr. 2022;27(1):95–112. doi: 10.1038/s41380-021-01037-w. [DOI] [PubMed] [Google Scholar]
- 163.Chen J., et al. Alternative splicing of lncRNAs in human diseases. Am. J. Cancer Res. 2021;11(3):624–639. [PMC free article] [PubMed] [Google Scholar]
- 164.Ishizuka A., et al. Formation of nuclear bodies by the lncRNA Gomafu-associating proteins Celf3 and SF1. Gene Cell. 2014;19(9):704–721. doi: 10.1111/gtc.12169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Tsuiji H., et al. Competition between a noncoding exon and introns: Gomafu contains tandem UACUAAC repeats and associates with splicing factor-1. Gene Cell. 2011;16(5):479–490. doi: 10.1111/j.1365-2443.2011.01502.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Barry G., et al. The long non-coding RNA Gomafu is acutely regulated in response to neuronal activation and involved in schizophrenia-associated alternative splicing. Mol. Psychiatr. 2014;19(4):486–494. doi: 10.1038/mp.2013.45. [DOI] [PubMed] [Google Scholar]
- 167.Rao S.Q., et al. Genetic variants in long non-coding RNA MIAT contribute to risk of paranoid schizophrenia in a Chinese Han population. Schizophr. Res. 2015;166(1–3):125–130. doi: 10.1016/j.schres.2015.04.032. [DOI] [PubMed] [Google Scholar]
- 168.Gandal M.J., et al. Transcriptome-wide isoform-level dysregulation in ASD, schizophrenia, and bipolar disorder. Science. 2018;362(6420) doi: 10.1126/science.aat8127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Psych E.C., et al. The PsychENCODE project. Nat. Neurosci. 2015;18(12):1707–1712. doi: 10.1038/nn.4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Lai C.Y., et al. Biomarkers in schizophrenia: a focus on blood based diagnostics and theranostics. World J. Psychiatr. 2016;6(1):102–117. doi: 10.5498/wjp.v6.i1.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Chen S., et al. Aberrant expression of long non-coding RNAs in schizophrenia patients. Med. Sci. Mon. Int. Med. J. Exp. Clin. Res. 2016;22:3340–3351. doi: 10.12659/MSM.896927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Ren Y., et al. A co-expression network analysis reveals lncRNA abnormalities in peripheral blood in early-onset schizophrenia. Prog. Neuro-Psychopharmacol. Biol. Psychiatry. 2015;63:1–5. doi: 10.1016/j.pnpbp.2015.05.002. [DOI] [PubMed] [Google Scholar]
- 173.Liu Y., et al. Changes in the level of Long Non-Coding RNA Gomafu gene expression in schizophrenia patients before and after antipsychotic medication. Schizophr. Res. 2018;195:318–319. doi: 10.1016/j.schres.2017.08.025. [DOI] [PubMed] [Google Scholar]
- 174.Badrlou E., et al. Expression of BDNF-associated lncRNAs in treatment-resistant schizophrenia patients. J. Mol. Neurosci. 2021;71(11):2249–2259. doi: 10.1007/s12031-020-01772-9. [DOI] [PubMed] [Google Scholar]
- 175.Tian T., et al. The long noncoding RNA landscape in amygdala tissues from schizophrenia patients. EBioMedicine. 2018;34:171–181. doi: 10.1016/j.ebiom.2018.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Jia J., et al. A preliminary analysis of LncRNA biomarkers for schizophrenia. Epigenomics. 2021;13(18):1443–1458. doi: 10.2217/epi-2021-0223. [DOI] [PubMed] [Google Scholar]
- 177.Spadaro P.A., et al. Long noncoding RNA-directed epigenetic regulation of gene expression is associated with anxiety-like behavior in mice. Biol. Psychiatr. 2015;78(12):848–859. doi: 10.1016/j.biopsych.2015.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.World Health Organization Stroke. Cerebrovascular Accident.
- 179.Collaborators G.B.D.S. Global, regional, and national burden of stroke and its risk factors, 1990-2019: a systematic analysis for the Global Burden of Disease Study 2019. Lancet Neurol. 2021;20(10):795–820. doi: 10.1016/S1474-4422(21)00252-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Ahad M.A., et al. Insights into the neuropathology of cerebral ischemia and its mechanisms. Rev. Neurosci. 2020;31(5):521–538. doi: 10.1515/revneuro-2019-0099. [DOI] [PubMed] [Google Scholar]
- 181.Marder V.J., et al. Thrombolysis with plasmin: implications for stroke treatment. Stroke. 2010;41(10 Suppl):S45–S49. doi: 10.1161/STROKEAHA.110.595157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Campbell B.C.V., et al. Ischaemic stroke. Nat. Rev. Dis. Prim. 2019;5(1):70. doi: 10.1038/s41572-019-0118-8. [DOI] [PubMed] [Google Scholar]
- 183.Zeng L., et al. MicroRNA-210 as a novel blood biomarker in acute cerebral ischemia. Front. Biosci. 2011;3(4):1265–1272. doi: 10.2741/e330. [DOI] [PubMed] [Google Scholar]
- 184.Zhu M., et al. Peripheral blood leukocyte expression of lncRNA MIAT and its diagnostic and prognostic value in ischemic stroke. J. Stroke Cerebrovasc. Dis. 2018;27(2):326–337. doi: 10.1016/j.jstrokecerebrovasdis.2017.09.009. [DOI] [PubMed] [Google Scholar]
- 185.Zhang S., et al. Long non-coding RNA MIAT impairs neurological function in ischemic stroke via up-regulating microRNA-874-3p-targeted IL1B. Brain Res. Bull. 2021;175:81–89. doi: 10.1016/j.brainresbull.2021.07.005. [DOI] [PubMed] [Google Scholar]
- 186.Li S., et al. LncRNA MIAT enhances cerebral ischaemia/reperfusion injury in rat model via interacting with EGLN2 and reduces its ubiquitin-mediated degradation. J. Cell Mol. Med. 2021;25(21):10140–10151. doi: 10.1111/jcmm.16950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Guo X., et al. LncRNA-MIAT promotes neural cell autophagy and apoptosis in ischemic stroke by up-regulating REDD1. Brain Res. 2021;1763 doi: 10.1016/j.brainres.2021.147436. [DOI] [PubMed] [Google Scholar]
- 188.Salari N., et al. The global prevalence of myocardial infarction: a systematic review and meta-analysis. BMC Cardiovasc. Disord. 2023;23(1):206. doi: 10.1186/s12872-023-03231-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Manabe I., Shindo T., Nagai R. Gene expression in fibroblasts and fibrosis: involvement in cardiac hypertrophy. Circ. Res. 2002;91(12):1103–1113. doi: 10.1161/01.res.0000046452.67724.b8. [DOI] [PubMed] [Google Scholar]
- 190.Dean R.G., et al. Connective tissue growth factor and cardiac fibrosis after myocardial infarction. J. Histochem. Cytochem. 2005;53(10):1245–1256. doi: 10.1369/jhc.4A6560.2005. [DOI] [PubMed] [Google Scholar]
- 191.Yu C.M., et al. Effects of combination of angiotensin-converting enzyme inhibitor and angiotensin receptor antagonist on inflammatory cellular infiltration and myocardial interstitial fibrosis after acute myocardial infarction. J. Am. Coll. Cardiol. 2001;38(4):1207–1215. doi: 10.1016/s0735-1097(01)01518-2. [DOI] [PubMed] [Google Scholar]
- 192.Garg P., et al. Left ventricular fibrosis and hypertrophy are associated with mortality in heart failure with preserved ejection fraction. Sci. Rep. 2021;11(1):617. doi: 10.1038/s41598-020-79729-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Kikuchi K., Poss K.D. Cardiac regenerative capacity and mechanisms. Annu. Rev. Cell Dev. Biol. 2012;28:719–741. doi: 10.1146/annurev-cellbio-101011-155739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Ohnishi Y., et al. Identification of 187 single nucleotide polymorphisms (SNPs) among 41 candidate genes for ischemic heart disease in the Japanese population. Hum. Genet. 2000;106(3):288–292. doi: 10.1007/s004390051039. [DOI] [PubMed] [Google Scholar]
- 195.Ishii N., et al. Identification of a novel non-coding RNA, MIAT, that confers risk of myocardial infarction. J. Hum. Genet. 2006;51(12):1087–1099. doi: 10.1007/s10038-006-0070-9. [DOI] [PubMed] [Google Scholar]
- 196.Vausort M., Wagner D.R., Devaux Y. Long noncoding RNAs in patients with acute myocardial infarction. Circ. Res. 2014;115(7):668–677. doi: 10.1161/CIRCRESAHA.115.303836. [DOI] [PubMed] [Google Scholar]
- 197.Eicher J.D., et al. Characterization of the platelet transcriptome by RNA sequencing in patients with acute myocardial infarction. Platelets. 2016;27(3):230–239. doi: 10.3109/09537104.2015.1083543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Azat M., et al. Long noncoding RNA MIAT: a potential role in the diagnosis and mediation of acute myocardial infarction. Mol. Med. Rep. 2019;20(6):5216–5222. doi: 10.3892/mmr.2019.10768. [DOI] [PubMed] [Google Scholar]
- 199.Chen L., et al. Targeting MIAT reduces apoptosis of cardiomyocytes after ischemia/reperfusion injury. Bioengineered. 2019;10(1):121–132. doi: 10.1080/21655979.2019.1605812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Wang X.M., et al. Corrigendum to "Long non-coding RNAs H19, MALAT1 and MIAT as potential novel biomarkers for diagnosis of acute myocardial infarction". Biomed. Pharmacother. 2019;118:109208. doi: 10.1016/j.biopha.2019.109208. Biomed Pharmacother, 2022. 151: p. 113206. [DOI] [PubMed] [Google Scholar]
- 201.Zeng H., et al. Expression of lncRNA APF in peripheral blood of patients with acute myocardial infarction caused by coronary heart disease and its clinical significance. Int. Heart J. 2022;63(4):742–748. doi: 10.1536/ihj.21-434. [DOI] [PubMed] [Google Scholar]
- 202.Qu X., et al. MIAT is a pro-fibrotic long non-coding RNA governing cardiac fibrosis in post-infarct myocardium. Sci. Rep. 2017;7 doi: 10.1038/srep42657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Zhao X., et al. The mechanism of myocardial fibrosis is ameliorated by myocardial infarction-associated transcript through the PI3K/Akt signaling pathway to relieve heart failure. J. Int. Med. Res. 2021;49(7) doi: 10.1177/03000605211031433. 3000605211031433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Xing Y., et al. The beneficial role of exercise training for myocardial infarction treatment in elderly. Front. Physiol. 2020;11:270. doi: 10.3389/fphys.2020.00270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Farsangi S.J., et al. Modulation of the expression of long non-coding RNAs H19, GAS5, and MIAT by endurance exercise in the hearts of rats with myocardial infarction. Cardiovasc. Toxicol. 2021;21(2):162–168. doi: 10.1007/s12012-020-09607-0. [DOI] [PubMed] [Google Scholar]
- 206.Wang F., et al. Overexpressing microRNA-203 alleviates myocardial infarction via interacting with long non-coding RNA MIAT and mitochondrial coupling factor 6. Arch Pharm. Res. (Seoul) 2021;44(5):525–535. doi: 10.1007/s12272-021-01324-8. [DOI] [PubMed] [Google Scholar]
- 207.Cao X., et al. Silencing long non-coding RNA MIAT ameliorates myocardial dysfunction induced by myocardial infarction via MIAT/miR-10a-5p/EGR2 axis. Aging (Albany NY) 2021;13(8):11188–11206. doi: 10.18632/aging.202785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Bai X., et al. LncRNA MIAT impairs cardiac contractile function by acting on mitochondrial translocator protein TSPO in a mouse model of myocardial infarction. Signal Transduct. Targeted Ther. 2021;6(1):172. doi: 10.1038/s41392-021-00538-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Zhang M., et al. Knockdown of long noncoding RNA MIAT attenuates hypoxia-induced cardiomyocyte injury by regulating the miR-488-3p/Wnt/beta-catenin pathway. Cell Biol. Int. 2023;47(1):63–74. doi: 10.1002/cbin.11945. [DOI] [PubMed] [Google Scholar]
- 210.Abdelmonem M., et al. Lutein exerts its cardioprotective effect against the experimental model of isoprenaline-induced myocardial infarction via MIAT/miR-200a/Nrf2/TXINP pathway. J. Biochem. Mol. Toxicol. 2021;35(11) doi: 10.1002/jbt.22899. [DOI] [PubMed] [Google Scholar]
- 211.Sanchis-Gomar F., et al. Epidemiology of coronary heart disease and acute coronary syndrome. Ann. Transl. Med. 2016;4(13) doi: 10.21037/atm.2016.06.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Matsuzawa Y., Lerman A. Endothelial dysfunction and coronary artery disease: assessment, prognosis and treatment. Coron. Artery Dis. 2014;25(8):713. doi: 10.1097/MCA.0000000000000178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Ding L., et al. Gene expressions underlying mishandled calcium clearance and elevated generation of reactive oxygen species in the coronary artery smooth muscle cells of chronic heart failure rats. Chinese Med J. 2017;130(4):460–469. doi: 10.4103/0366-6999.199825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Nurnberg S.T., et al. Coronary artery disease associated transcription factor TCF21 regulates smooth muscle precursor cells that contribute to the fibrous cap. PLoS Genet. 2015;11(5) doi: 10.1371/journal.pgen.1005155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Tan J., et al. LncRNA-MIAT increased in patients with coronary atherosclerotic heart disease. Cardiol. Res. Pract. 2019;2019 doi: 10.1155/2019/6280194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Toraih E.A., et al. Association of long non-coding RNA MIAT and MALAT1 expression profiles in peripheral blood of coronary artery disease patients with previous cardiac events. Genet. Mol. Biol. 2019;42:509–518. doi: 10.1590/1678-4685-GMB-2018-0185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Shimizu I., Minamino T. Physiological and pathological cardiac hypertrophy. J. Mol. Cell. Cardiol. 2016;97:245–262. doi: 10.1016/j.yjmcc.2016.06.001. [DOI] [PubMed] [Google Scholar]
- 218.Zhu X.-H., et al. LncRNA MIAT enhances cardiac hypertrophy partly through sponging miR-150. Eur. Rev. Med. Pharmacol. Sci. 2016;20(17) [PubMed] [Google Scholar]
- 219.Liu W., et al. MicroRNA‐150 protects against pressure overload‐induced cardiac hypertrophy. J. Cell. Biochem. 2015;116(10):2166–2176. doi: 10.1002/jcb.25057. [DOI] [PubMed] [Google Scholar]
- 220.Li Y., et al. LncRNA myocardial infarction-associated transcript (MIAT) contributed to cardiac hypertrophy by regulating TLR4 via miR-93. Eur. J. Pharmacol. 2018;818:508–517. doi: 10.1016/j.ejphar.2017.11.031. [DOI] [PubMed] [Google Scholar]
- 221.Li Z., et al. Long noncoding RNA myocardial infarction-associated transcript is associated with the microRNA-150-5p/P300 pathway in cardiac hypertrophy. Int. J. Mol. Med. 2018;42(3):1265–1272. doi: 10.3892/ijmm.2018.3700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Wei J.Q., et al. Quantitative control of adaptive cardiac hypertrophy by acetyltransferase p300. Circulation. 2008;118(9):934–946. doi: 10.1161/CIRCULATIONAHA.107.760488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Zeng Z., et al. Myocardial hypertrophy is improved with berberine treatment via long non-coding RNA MIAT-mediated autophagy. J. Pharm. Pharmacol. 2019;71(12):1822–1831. doi: 10.1111/jphp.13170. [DOI] [PubMed] [Google Scholar]
- 224.Yang C., Zhang Y., Yang B. MIAT, a potent CVD-promoting lncRNA. Cell. Mol. Life Sci. 2022;79(1):43. doi: 10.1007/s00018-021-04046-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Sun H., et al. IDF Diabetes Atlas: global, regional and country-level diabetes prevalence estimates for 2021 and projections for 2045. Diabetes Res. Clin. Pract. 2022;183 doi: 10.1016/j.diabres.2021.109119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.American Diabetes A. 2. Classification and diagnosis of diabetes: standards of medical care in diabetes-2019. Diabetes Care. 2019;42(Suppl 1):S13–S28. doi: 10.2337/dc19-S002. [DOI] [PubMed] [Google Scholar]
- 227.Zimmet P.Z., et al. Diabetes: a 21st century challenge. Lancet Diabetes Endocrinol. 2014;2(1):56–64. doi: 10.1016/S2213-8587(13)70112-8. [DOI] [PubMed] [Google Scholar]
- 228.Merovci A., et al. Dapagliflozin improves muscle insulin sensitivity but enhances endogenous glucose production. J. Clin. Invest. 2014;124(2):509–514. doi: 10.1172/JCI70704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Sathishkumar C., et al. Linking a role of lncRNAs (long non-coding RNAs) with insulin resistance, accelerated senescence, and inflammation in patients with type 2 diabetes. Hum. Genom. 2018;12(1):41. doi: 10.1186/s40246-018-0173-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.de Gonzalo-Calvo D., et al. Circulating long-non coding RNAs as biomarkers of left ventricular diastolic function and remodelling in patients with well-controlled type 2 diabetes. Sci. Rep. 2016;6 doi: 10.1038/srep37354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Wenzl F.A., et al. Circulating long noncoding RNA signatures associate with incident diabetes in older adults: a prospective analysis from the VITA cohort study. Diabetes Care. 2023;46(6):1239–1244. doi: 10.2337/dc23-0012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Shi Q., Yao H. Signature RNAS and related regulatory roles in type 1 diabetes mellitus based on competing endogenous RNA regulatory network analysis. BMC Med. Genom. 2021;14(1):133. doi: 10.1186/s12920-021-00931-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Yan B., et al. lncRNA-MIAT regulates microvascular dysfunction by functioning as a competing endogenous RNA. Circ. Res. 2015;116(7):1143–1156. doi: 10.1161/CIRCRESAHA.116.305510. [DOI] [PubMed] [Google Scholar]
- 234.Zhou L., et al. Long non-coding MIAT mediates high glucose-induced renal tubular epithelial injury. Biochem. Biophys. Res. Commun. 2015;468(4):726–732. doi: 10.1016/j.bbrc.2015.11.023. [DOI] [PubMed] [Google Scholar]
- 235.Toraih E.A., et al. Deciphering the role of circulating lncRNAs: RNCR2, NEAT2, CDKN2B-AS1, and PVT1 and the possible prediction of anti-VEGF treatment outcomes in diabetic retinopathy patients. Graefes Arch. Clin. Exp. Ophthalmol. 2019;257(9):1897–1913. doi: 10.1007/s00417-019-04409-9. [DOI] [PubMed] [Google Scholar]
- 236.Biswas S., et al. Expressions of serum lncRNAs in diabetic retinopathy - a potential diagnostic tool. Front. Endocrinol. 2022;13 doi: 10.3389/fendo.2022.851967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Mohammad H.M.F., et al. Long non-coding RNAs gene variants as molecular markers for diabetic retinopathy risk and response to anti-VEGF therapy. Pharmgenomics Pers. Med. 2021;14:997–1014. doi: 10.2147/PGPM.S322463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Zhang J., et al. Long non-coding RNA MIAT acts as a biomarker in diabetic retinopathy by absorbing miR-29b and regulating cell apoptosis. Biosci. Rep. 2017;37(2) doi: 10.1042/BSR20170036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Zhang J., et al. C-myc contributes to the release of Muller cells-derived proinflammatory cytokines by regulating lncRNA MIAT/XNIP pathway. Int. J. Biochem. Cell Biol. 2019;114 doi: 10.1016/j.biocel.2019.105574. [DOI] [PubMed] [Google Scholar]
- 240.Yu X., et al. Long noncoding RNA MIAT regulates primary human retinal pericyte pyroptosis by modulating miR-342-3p targeting of CASP1 in diabetic retinopathy. Exp. Eye Res. 2021;202 doi: 10.1016/j.exer.2020.108300. [DOI] [PubMed] [Google Scholar]
- 241.Li Q., et al. Long non-coding RNA of myocardial infarction associated transcript (LncRNA-MIAT) promotes diabetic retinopathy by upregulating transforming growth factor-beta1 (TGF-beta1) signaling. Med. Sci. Mon. Int. Med. J. Exp. Clin. Res. 2018;24:9497–9503. doi: 10.12659/MSM.911787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Yu C., et al. Downregulation of long noncoding RNA MIAT in the retina of diabetic rats with tail-vein injection of human umbilical-cord mesenchymal stem cells. Int. J. Med. Sci. 2020;17(5):591–598. doi: 10.7150/ijms.38078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Boudina S., Abel E.D. Diabetic cardiomyopathy, causes and effects. Rev. Endocr. Metab. Disord. 2010;11(1):31–39. doi: 10.1007/s11154-010-9131-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Qi Y., et al. LncRNA-MIAT-mediated miR-214-3p silencing is responsible for IL-17 production and cardiac fibrosis in diabetic cardiomyopathy. Front. Cell Dev. Biol. 2020;8:243. doi: 10.3389/fcell.2020.00243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Xiao W., et al. Long non-coding RNA MIAT is involved in the regulation of pyroptosis in diabetic cardiomyopathy via targeting miR-214-3p. iScience. 2021;24(12) doi: 10.1016/j.isci.2021.103518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Lane N.E. Epidemiology, etiology, and diagnosis of osteoporosis. Am. J. Obstet. Gynecol. 2006;194(2 Suppl):S3–S11. doi: 10.1016/j.ajog.2005.08.047. [DOI] [PubMed] [Google Scholar]
- 247.International Osteoporosis Foundation. Epidemiology of Osteoporosis and Fragility Fractures.
- 248.Zhao W., et al. The regulatory roles of long noncoding RNAs in osteoporosis. Am. J. Transl. Res. 2020;12(9):5882–5907. [PMC free article] [PubMed] [Google Scholar]
- 249.Jin C., et al. Long non-coding RNA MIAT knockdown promotes osteogenic differentiation of human adipose-derived stem cells. Cell Biol. Int. 2017;41(1):33–41. doi: 10.1002/cbin.10697. [DOI] [PubMed] [Google Scholar]
- 250.Li R., et al. Elevated lncRNA MIAT in peripheral blood mononuclear cells contributes to post-menopausal osteoporosis. Aging (Albany NY) 2022;14(7):3143–3154. doi: 10.18632/aging.204001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Rumman M., Dhawan J., Kassem M. Concise review: quiescence in adult stem cells: biological significance and relevance to tissue regeneration. Stem Cell. 2015;33(10):2903–2912. doi: 10.1002/stem.2056. [DOI] [PubMed] [Google Scholar]
- 252.Long H., et al. Prevalence trends of site-specific osteoarthritis from 1990 to 2019: findings from the global burden of disease study 2019. Arthritis Rheumatol. 2022;74(7):1172–1183. doi: 10.1002/art.42089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Sacitharan P.K. Ageing and osteoarthritis. Subcell. Biochem. 2019;91:123–159. doi: 10.1007/978-981-13-3681-2_6. [DOI] [PubMed] [Google Scholar]
- 254.Santaguida P.L., et al. Patient characteristics affecting the prognosis of total hip and knee joint arthroplasty: a systematic review. Can. J. Surg. 2008;51(6):428–436. [PMC free article] [PubMed] [Google Scholar]
- 255.Rodriguez-Merchan E.C. Topical therapies for knee osteoarthritis. Postgrad. Med. 2018;130(7):607–612. doi: 10.1080/00325481.2018.1505182. [DOI] [PubMed] [Google Scholar]
- 256.Pan W., et al. lncRNA myocardial infarction-associated transcript (MIAT) knockdown alleviates LPS-induced chondrocytes inflammatory injury via regulating miR-488-3p/sex determining region Y-related HMG-box 11 (SOX11) axis. Open Life Sci. 2021;16(1):511–522. doi: 10.1515/biol-2021-0023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Xu M., et al. Phosphorylation of osteopontin in osteoarthritis degenerative cartilage and its effect on matrix metalloprotease 13. Rheumatol. Int. 2013;33(5):1313–1319. doi: 10.1007/s00296-012-2548-4. [DOI] [PubMed] [Google Scholar]
- 258.Cheng C., et al. Osteopontin inhibits HIF-2 alpha mRNA expression in osteoarthritic chondrocytes. Exp. Ther. Med. 2015;9(6):2415–2419. doi: 10.3892/etm.2015.2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Zhang F.J., et al. Effect of osteopontin on TIMP-1 and TIMP-2 mRNA in chondrocytes of human knee osteoarthritis in vitro. Exp. Ther. Med. 2014;8(2):391–394. doi: 10.3892/etm.2014.1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Zeng S., Tu M. The lncRNA MIAT/miR-181a-5p axis regulates osteopontin (OPN)-mediated proliferation and apoptosis of human chondrocytes in osteoarthritis. J. Mol. Histol. 2022;53(2):285–296. doi: 10.1007/s10735-022-10067-9. [DOI] [PubMed] [Google Scholar]
- 261.Li C., et al. Silence of lncRNA MIAT protects ATDC5 cells against lipopolysaccharides challenge via up-regulating miR-132. Artif. Cells, Nanomed. Biotechnol. 2019;47(1):2521–2527. doi: 10.1080/21691401.2019.1626410. [DOI] [PubMed] [Google Scholar]