Abstract
The human papillomavirus type 18 (HPV-18) upstream regulatory region (URR) controls cell type-specific expression of viral oncoproteins E6 and E7. The HPV-18 URR is highly active in HeLa cells, but its activity is virtually undetectable in HepG2 cells. Previous work has shown that YY1 plays an important role in activation of the HPV-18 URR in HeLa cells, and this activating activity is dependent on its physical interaction with C/EBPβ, which binds to the switch region adjacent to the YY1 site in the URR. Overexpression of C/EBPβ in HepG2 cells restores C/EBPβ-YY1 interaction, resulting in strong activation of the HPV-18 URR activity. In this report, we show that, in contrast to the effect in HepG2 cells, overexpression of C/EBPβ represses the HPV-18 URR in HeLa cells. This C/EBPβ-induced repression of the HPV-18 URR in HeLa cells is binding site independent. It is also promoter specific, since it activates the albumin promoter under conditions in which it represses the URR in the same cells. Biochemical analysis shows that overexpression of C/EBPβ in HeLa cells specifically interferes with binding of TATA-binding protein to the TATA box of the HPV-18 URR, but its overexpression in HepG2 cells leads to activation of the HPV-18 URR. These results suggest that a molecular mechanism underlies the ability of C/EBPβ to regulate transcription in a cell type-specific manner and indicate the potential of using C/EBPβ to manipulate the activity of the HPV-18 URR in cervical carcinoma cells.
The human papillomavirus (HPV) family includes at present 80 distinct genotypes. HPV type 16 (HPV-16), HPV-18, and several additional anogenital types have been identified as high-risk viruses associated with the development of about 95% of cancers of the cervix and more than 50% of other anogenital cancers (reviewed in references 100 to 103). The regulation of viral gene expression is complex and is controlled by cellular and viral transcription factors. In cervical carcinoma cells, HPV DNA is usually integrated into the host genome, often disrupting the E1 and E2 genes, resulting in deregulated expression of the major oncoproteins E6 and E7 (2, 20, 71, 76). The immortalizing activities of the HPV E6 and E7 oncoproteins (27, 53) are correlated with their ability to stimulate cell proliferation by activating cyclins E and A (as shown for E7 [86, 99]) and by interfering with the functions of the tumor suppressor proteins p53 (as shown for E6 [69, 96]) and retinoblastoma protein (Rb) (as shown for E7 [21, 28, 54rsqb;) (for reviews, see references 100 to 103).
Transcription of the HPV-18 E6 and E7 genes initiates at the P105 promoter (named after the nucleotide at which transcription starts) (70, 84) and is regulated by the upstream regulatory region (URR). For the integrated HPV-18 genome, expression of the E6 and E7 genes is under the control of cellular transcription factors. A number of cellular transcription factors, including AP-1, C/EBPβ, SP1, YY1, and KRF-1, contribute either positively or negatively to the regulation of HPV-18 URR activity in a cell type-specific manner (3–5, 15, 24, 29, 34, 49, 50, 59, 68, 85).
YY1 (72) (also known as NF-E1 [62], δ [26], and UCRBP [23]) is a multifunctional transcription factor which activates or represses transcription of many cellular as well as viral genes (for reviews, see references 73 and 75). Its functional versatility may be attributable to its multiple transcriptional domains (9, 45, 46, 72). YY1 has been shown to be involved in transcription in both HPV-16 and HPV-18 (3–5, 51, 58). In the latter case, YY1 was first described to be a repressor of the proximal promoter of HPV-18 in HeLa cells (3). Further analyses revealed that, in the context of the complete HPV-18 URR, YY1 acts as an activator of the HPV-18 URR in HeLa cells (4), and this activating activity is dependent on its physical interaction with C/EBPβ, which binds to the switch region located upstream of the proximal-promoter YY1 binding site OL13 (5). The functional interplay between YY1 and C/EBPβ plays a critical role in regulating HPV-18 URR activity in a cell type-specific manner. For instance, the HPV-18 URR is virtually inactive in HepG2 cells. This is believed to be due to the lack of C/EBPβ-YY1 interaction which ensures that YY1 functions as an activator of the HPV-18 URR, as is the case in HeLa cells. Introduction of C/EBPβ into HepG2 cells can restore the C/EBPβ-YY1-switch region interaction and therefore activate the HPV-18 URR (5). These results strongly suggest that C/EBPβ-YY1 is a positive regulator of HPV-18 which contributes to cell type-specific HPV-18 URR activity (5).
C/EBPβ was first characterized as a protein whose mRNA synthesis is regulated by interleukin-6 (IL-6) and other cytokines (1). C/EBPβ binds to promoters of many cytokine genes, including that of IL-6 genes as well as viral promoters, implying an important role for this protein in the regulation of expression of cytokine and virus genes (1). C/EBPβ belongs to the family of CCAAT/enhancer-binding proteins (C/EBP), a highly conserved family of leucine zipper-type (bZIP) DNA-binding proteins currently including C/EBPα, C/EBPβ (also known as NF-IL6 and CRP2), C/EBPδ (also known as NF-IL6β and CRP3), C/EBPγ, CRP1, Ig-C/EBP, and GADD153 (also known as CHOP) (1, 10, 11, 16, 33, 35, 41, 63, 65, 67, 97). Different C/EBP isoforms are generally characterized by a high degree of sequence homology in the leucine zipper and basic regions but much less conserved N-terminal regulatory and transactivation domains (10, 32, 40). Additional isoforms can be generated by translation from internal AUGs (17, 30, 61). C/EBPs have the potential to form homo- and heterodimers with other C/EBP family members (22, 36, 42, 43, 63, 90, 93, 97), with other bZIP proteins (31, 37, 89, 92), and with non-leucine zipper-containing proteins (5, 13, 19, 44, 47, 48, 56, 57, 81–83).
In the present study, we performed experiments to further evaluate the functional role of C/EBPβ in regulating HPV-18 transcription in HeLa cells, a cervical carcinoma cell line. We showed that a slight increase in C/EBPβ level results in strong repression of URR activity in HeLa cells. In contrast to the switch region-dependent C/EBPβ-induced activation of the HPV-18 URR (5) in C/EBPβ-negative HepG2 cells (5, 47, 87, 91), repression of the HPV-18 URR in HeLa cells by overexpression of C/EBPβ is independent of the switch region. Within a given concentration range, we found that C/EBPβ specifically repressed the HPV-18 URR but activated the albumin promoter in the same cells. However, a further increase in C/EBPβ also resulted in repression of the albumin promoter in the same cells, indicating that both promoters are regulated by C/EBPβ positively or negatively, depending on the level of C/EBPβ in the cell. To understand the mechanisms that underlie the repressive effects of C/EBPβ on URR activity, we showed that overexpression of C/EBPβ could disrupt the interaction of TATA-binding protein (TPB) and the HPV-18 URR TATA box in HeLa cells. Consistent with the finding that C/EBPβ activates HPV-18 URR in HepG2 cells, the same amount of transfected C/EBPβ did not interfere with TBP-TATA box complex formation in HepG2 cells. Taken together, these results suggest that disruption of TBP binding may be an important mechanism that underlies the ability of overexpressed C/EBPβ to repress HPV-18 URR-mediated transcription in HeLa cells.
MATERIALS AND METHODS
Cell cultures, transfections, and CAT assays.
HeLa and HepG2 cells were grown in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal calf serum and were transfected by the calcium phosphate coprecipitation method (12). Transfections were performed as previously described (3) and were done with 3 to 10 μg of the various chloramphenicol acetyltransferase (CAT) gene-containing reporter constructs, up to 2 μg of various cytomegalovirus (CMV)-driven expression plasmids (the amounts of DNA are given in the figure legends), and 0.6 μg of a Rous sarcoma virus-luciferase gene construct (RSV/L) (18) as an internal control. CAT assays were performed on extracts with equivalent luciferase counts. To circumvent a possible effect of the effector plasmids on the cotransfected RSV-luciferase construct, CAT assays were also performed in parallel, using the same protein concentrations. All transfections were repeated at least five times. CAT assays were quantified by cutting spots from thin-layer chromatography plates and determining the radioactivity by liquid scintillation counting. The results for individual transfections varied by less than 15%.
RNA isolation and Northern blot analysis.
To obtain RNA from HeLa cells transfected with various amounts of C/EBPβ or the empty vector pcDNAI, transfections were done together with 0.6 μg of RSV/L (18) as recently described (5). In parallel, as a control for transfections with pcDNAI, cells were transfected with 0.6 μg of RSV/L only. In addition, nontransfected cells were kept and harvested under the same conditions. Briefly, to monitor for transfection efficiency, 1/10 of the cells were assayed for luciferase activity. Total RNA was isolated by standard procedures with acid guanidinium isothiocyanate as described previously (14). Five micrograms of total RNA was analyzed by electrophoresis on a 1.2% agarose formaldehyde gel followed by transfer to a Hybond N membrane (Amersham). The filter was hybridized with HPV-18 DNA probes labeled by random priming at 65°C and then washed at 65°C in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–0.1% sodium dodecyl sulfate.
Plasmids and their construction.
The cloning of various HPV-18 URR CAT plasmids has been previously described (3, 4). Plasmid p18URR contains the complete HPV-18 URR (nucleotides 6930 to 103 of the HPV-18 genome) fused to the CAT gene of pBLCAT3 (3). In plasmid p18URR-22M1, mutation OL22M1 (resulting in a mutated switch sequence) was introduced by PCR (4). The sequence for OL22M1, with mutated nucleotides shown in lowercase letters, is 5′-AGTTTGTTTccgcggAAGCTA-3′. The mutated TATA box sequence of HPV-18 (5′-AACGGTGTccgcggAAGATGTGAG-3′) was introduced into plasmids p18URR and p18URR-22M1 by PCR amplification as follows. To construct p18URR-TATAM1 and p18URR-22M1-TATAM1, each containing the mutated TATAM1 box sequence of the HPV-18 genome, plasmids p18URR and p18URR-22M1, respectively, and internal primers PTATAM1-A (5′-GAAAACGGTGTccgcggAAGATGTGAGAAACACAC-3′; sense strand) and PTATAM1-B (5′-GTTTCTCACATCTTccgcggACACCGTTTTCGGTCCCGAC-3′; antisense strand) were mixed with primers CAT2 (5′-GCTCCTGAAAATCTCGCCAAGCTC-3′, antisense strand), which binds in the CAT gene, and C1 (5′-GTAACGCCAGGGTTTTCCCAGTCAC-3′; sense strand), which binds to vector sequences 5′ of the HPV-18 sequences, respectively. Amplification was performed first at 94°C for 3 min and then for 35 cycles of 94, 50, and 72°C for 2 min at each temperature. The purified products were mixed, and amplification was repeated with primers C1 and CAT2, except that annealing of the primers was performed, after one 3-min 94°C precycle, at 94°C for 1.5 min, 45°C for 2 min, and 72°C for 3 min. The resulting products were digested with HindIII and BamHI, purified, and inserted into the corresponding sites of pBLCAT3. Plasmids p18URR-TATAM1 and p18URR-22M1-TATAM1 were sequenced across the internal mutations.
Plasmid pAlbCAT (constructed by U. Schibler) was used as a reporter construct of the albumin promoter (25). The following C/EBP expression plasmids were used: CMV-C/EBPβ (containing the full-length [rat] cDNA of C/EBPβ; a gift of C. Nerlov), CMV-LAP and CMV-LIP (17), CMV-C/EBPα (pCMVαWT [55]), CMV–NF-IL6 (1), and CMV-trunc.NF-IL6 [pcmNF-IL6(spl) 89].
EMSAs.
Electrophoretic mobility shift assays (EMSAs) were performed in a total volume of 25 μl with nuclear extracts from HeLa and HepG2 cells, which were incubated in a reaction mixture containing 10 mM Tris-HCl (pH 7.5), 100 mM NaCl, 4 mM dithiothreitol, 4% glycerol, 0.5 μg of poly(dI-dC) (as a nonspecific competitor), and an end-labeled oligonucleotide probe at 25°C for 10 min. EMSAs using bacterially expressed human recombinant TBP (rTBP) (usually 0.5 μl), purchased from Upstate Biotechnology Inc., were performed in the same buffer, but without poly(dI-dC), and under the same conditions. For competition experiments, the reaction mixtures were incubated together with the unlabeled oligonucleotides at a 10- to 100-fold molar excess, as indicated in the figure legends. Usually 1 μl of the same concentrated antibodies was used in EMSAs and antibody disruption or antibody shift experiments. Reactions were carried out for 15 min at 25°C, and the DNA-protein complexes were resolved on a 5% polyacrylamide gel in 0.25× Tris-borate-EDTA at 25°C unless otherwise noted. Anti-TBP antibodies were a kind gift from M. Meisterernst, and anti-rTBP antibodies were purchased from Upstate Biotechnology Inc. Anti-C/EBPβ antibodies were a kind gift from E. Ziff and C. Nerlov.
Preparations of nuclear extracts.
Crude nuclear extracts were prepared essentially as described earlier (4, 5). To obtain nuclear extracts from HeLa or HepG2 cells transfected with various amounts of C/EBPβ or the empty vector pcDNAI, transfections and extract preparation were done as recently described (5). In parallel, as a second control (in addition to using pcDNAI in transfections), nontransfected cells were kept and harvested under the same conditions. Briefly, to monitor for transfection efficiency, cells were cotransfected with 0.6 μg of RSV/L (18), and 1/10 of the cells were assayed for luciferase activity. Nuclear extracts were prepared by mixing pelleted cells with a lysis buffer (5); after incubation on ice for 10 min, cytosol was separated by low-speed centrifugation and nuclei were resuspended in a 420 mM NaCl-containing extraction buffer (5). After 30 min of incubation on ice, nuclear extracts were cleared by centrifugation, aliquoted, and frozen in liquid nitrogen.
Oligonucleotides used in EMSAs.
The sequences of the oligonucleotide double-stranded portions are as follows, with mutations underlined: OL22 (the switch sequence [4] of the HPV-18 URR), 5′-AGTTTGTTTTTACTTAAGCTA-3′; 18TATA (the TATA box sequence of the HPV-18 URR), 5′-AACGGTGTATATAAAAGATGTGAG-3′; 18TATA-M1 (a mutated TATA box sequence of the HPV-18 URR), 5′-AACGGTGTCCGCGGAAGATGTGAG-3′; 18TATAS (a shorter TATA box sequence of the HPV-18 URR), 5′-TGTATATAAAAGATGT-3′; 18TATAS-M1 (a shorter mutated TATA box sequence of the HPV-18 URR), 5′-TGTCCGCGGAAGATGT; 16TATA (the TATA box sequence of the HPV-16 URR), 5′-AACGGTTAGTATAAAAGCAGACAT-3′; 16TATA-M1 (a mutated TATA box sequence of the HPV-16 URR), 5′-AACGGTTACCGCGGAAGCAGACAT-3′; AlbTATA (the TATA box sequence of the mouse albumin promoter), 5′-TAAAGAAGTATATTAGAGCGAGTCT-3′; and AlbTATA-M1 (a mutated TATA box sequence of the mouse albumin promoter), 5′-TAAAGAAGTCCGCGGGAGCGAGTCT-3′.
RESULTS
Overexpression of C/EBPs in HeLa cells represses HPV-18 URR activity.
The high level of HPV-18 URR transcriptional activity in HeLa cells can in part be attributed to the C/EBPβ-YY1 complex formed on switch region OL22 of the HPV-18 URR (Fig. 1). In HepG2 cells, in which the HPV-18 URR is virtually inactive, C/EBPβ and the C/EBPβ-YY1 complex are not detectable. Overexpression of C/EBPβ restores C/EBPβ-YY1 formation on OL22 in HepG2 cells and results in activation of the HPV-18 URR, suggesting a strong correlation between the presence of the C/EBPβ-YY1 complex and the high-level activity of the HPV-18 URR in HeLa cells (5).
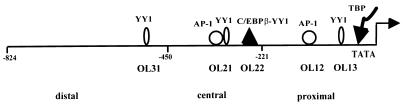
FIG. 1.
Schematic representation of the HPV-18 URR. The 824-bp HPV-18 URR is located within the 1,049-bp BamHI fragment of the HPV-18 genome. The URR consists of 5′ distal (positions −824 to −450), central (positions −450 to −221), and 3′ proximal (positions −221 to −1) fragments. The transcription initiation site is indicated by a crooked arrow. The relative positions of the distal YY1 site, the central overlapping AP-1 and YY1 sites, the C/EBPβ-YY1 site, the promoter-proximal AP-1 and YY1 sites, and the TBP-TATA box binding site are indicated.
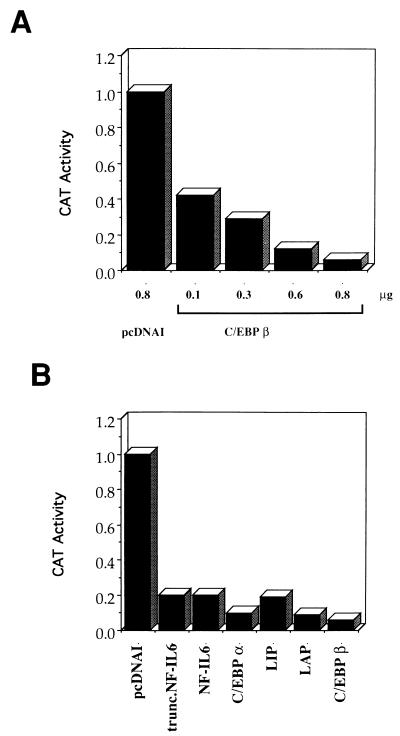
To further analyze the effect of C/EBPβ on HPV-18 URR activity in HeLa cells, p18URR (which contains the wild-type HPV-18 URR cloned into pBLCAT3 upstream of the CAT gene [4]) was cotransfected into HeLa cells with CMV-C/EBPβ. As shown in Fig. 2A, in contrast to what was observed in HepG2 cells (5), overexpression of C/EBPβ reduced HPV-18 URR activity in a dose-dependent manner. Cotransfection of 0.8 μg of CMV-C/EBPβ with 3 μg of p18URR almost completely abrogated the URR activity (Fig. 2A). Previously we showed that C/EBPβ, but not C/EBPα, can activate the HPV-18 URR in HepG2 cells (5). To determine whether other isoforms of C/EBP also repress the HPV-18 URR, p18URR was cotransfected with expression plasmids encoding C/EBPα, liver-enriched transcriptional activator protein (LAP), liver-enriched inhibitory protein (LIP), NF-IL6 (the human homolog of rat C/EBPβ), and a truncated NF-IL6 that contains mainly the C-terminal bZIP region, similar to LIP (89). As shown in Fig. 2B, all of these C/EBP variants repressed the HPV-18 URR to similar extents. These results suggest that the bZIP region may be critical for C/EBP-mediated repression of URR activity in HeLa cells.
FIG. 2.
Overexpression of the C/EBP family of transcription factors represses HPV-18 URR activity in HeLa cells. (A) C/EBPβ represses HPV-18 URR activity in HeLa cells in a dose-dependent manner. HeLa cells were transfected with 3 μg of p18URR together with increasing amounts of CMV-C/EBPβ (0.1 to 0.8 μg) or 0.8 μg of pcDNAI, as indicated at the bottom of the figure, and 0.6 μg of RSV/L (18) as an internal control. Relative CAT activities were quantified relative to the activity obtained with p18URR cotransfected with 0.8 μg of pcDNAI, which was set at 1. The results are shown as bar graphs, with relative CAT activities for p18URR as follows: 0.8 μg of pcDNAI, 1.0; 0.1 μg of CMV-C/EBPβ, 0.41; 0.3 μg of CMV-C/EBPβ, 0.29; 0.6 μg of CMV-C/EBPβ, 0.12; and 0.8 μg of CMV-C/EBPβ, 0.06. (B) Different members of the C/EBP family of transcription factors repress the HPV-18 URR in HeLa cells. HeLa cells were transfected with 3 μg of p18URR together with 0.8 μg of C/EBP expression plasmids and 0.6 μg of RSV/L (18) as an internal control. CAT activities were quantified relative to the activity obtained with p18URR cotransfected with 0.8 μg of pcDNAI, which was set at 1, and the results are shown as bar graphs. The relative CAT activities for p18URR transfected with the different expression plasmids were as follows: control vector pcDNAI, 1.0; truncated NF-IL6, 0.196; NF-IL6, 0.197; C/EBPα, 0.095; LIP, 0.189; LAP, 0.086; and C/EBPβ, 0.06.
Overexpression of C/EBPβ or LIP in HeLa cells disrupts C/EBPβ-YY1 complex formation on OL22.
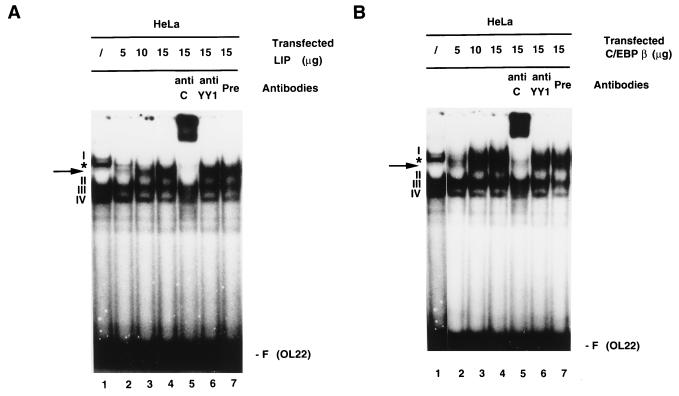
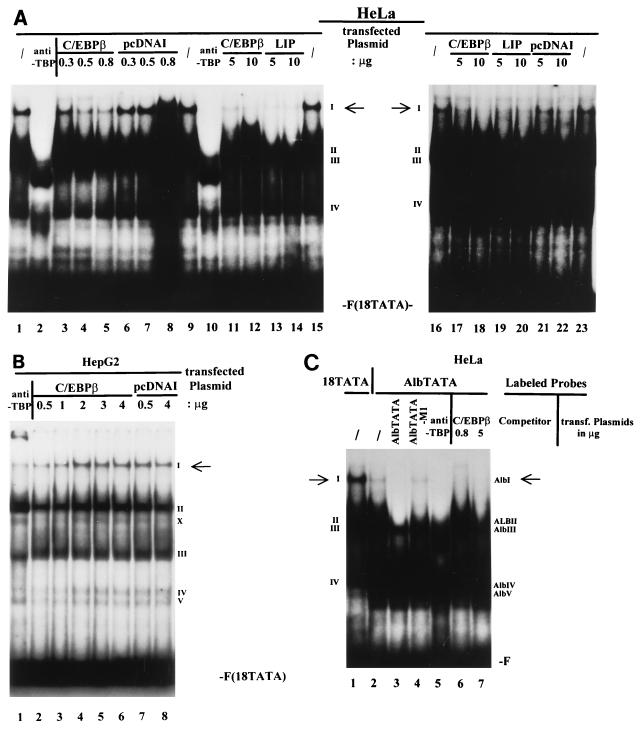
To understand the mechanisms underlying C/EBP-mediated repression of the HPV-18 URR in HeLa cells, the possibility that the C/EBPβ-YY1 complex is affected by overexpression of C/EBPs was investigated. Increasing amounts of plasmid DNAs encoding either LIP or C/EBPβ were transfected into HeLa cells, and EMSAs were performed with extracts prepared from these cells. As shown in Fig. 3A, increasing the amount of LIP caused disruption of complex I, formed on OL22, in a dose-responsive manner (lanes 1 to 4). Complex I was previously shown to be the specific complex formed between C/EBPβ and YY1 binding to OL22 in HeLa cells (5). Concomitant with the disappearance of complex I, a new complex was formed (marked by an arrow) which was specifically supershifted by anti-C/EBPβ but not by anti-YY1 or preimmune antibodies (Fig. 3A, lanes 5 to 7). Although the exact nature of this new complex is currently unknown, it is likely that it contains LIP but not YY1.
FIG. 3.
Overexpression of LIP or C/EBPβ in HeLa cells disrupts YY1-C/EBPβ complex formation on the switch region OL22. (A) Expression of LIP disrupts complex I and results in the formation of a new complex on OL22. HeLa cells were transfected with increasing amounts of CMV-LIP (5, 10, or 15 μg). 32P-labeled oligonucleotide OL22 was used in EMSAs, and anti-C/EBPβ antibodies, anti-YY1 antibodies, or preimmune serum was included in the binding reaction mixtures. Lanes: 1, standard binding reaction with extracts from nontransfected cells; 2 to 4, binding reactions with nuclear extracts from HeLa cells transfected with increasing amounts of CMV-LIP (lane 2, 5 μg; lane 3, 10 μg; lane 4, 15 μg); 5 to 7, binding reactions with nuclear extracts from cells transfected with 15 μg of CMV-LIP, incubated with anti-C/EBPβ antibodies (anti C), (lane 5), anti-YY1 antibodies (anti YY1) (lane 6), or preimmune serum (Pre) (lane 7). Specific DNA-protein complexes are indicated by numbers (I to IV). A nonspecific DNA-protein complex is indicated by an asterisk. The new complex formed on OL22 through expression of transfected CMV-LIP is indicated by an arrow. F (OL22), free probe OL22. (B) Expression of C/EBPβ leads to the formation of new complexes on OL22. Essentially the same experiments as described for panel A were carried out, but a plasmid expressing C/EBPβ was used instead of a LIP-expressing plasmid. The EMSA products shown in panels A and B were resolved on a 6.5% polyacrylamide gel.
When HeLa cells transfected with CMV-C/EBPβ were analyzed for the formation of complex I on OL22, essentially the same results were obtained: increasing the amount of C/EBPβ resulted in the disruption of complex I, accompanied by the appearance of two new complexes. The faster-migrating complex, marked by an arrow in Fig. 3B, is similar to that seen in LIP-transfected HeLa cells. This is not surprising since CMV-C/EBPβ encodes both the LAP and the LIP proteins (54a). The other complex migrated to a position slightly above that of complex I. Both new complexes were supershifted by anti-C/EBPβ but not by anti-YY1 or preimmune antibodies (Fig. 3B). Taken together, these results suggest that disruption of the C/EBPβ-YY1 complex may be in part responsible for C/EBP-mediated repression of the HPV-18 URR in HeLa cells.
HPV-18 URR activity is strongly repressed by C/EBPβ.
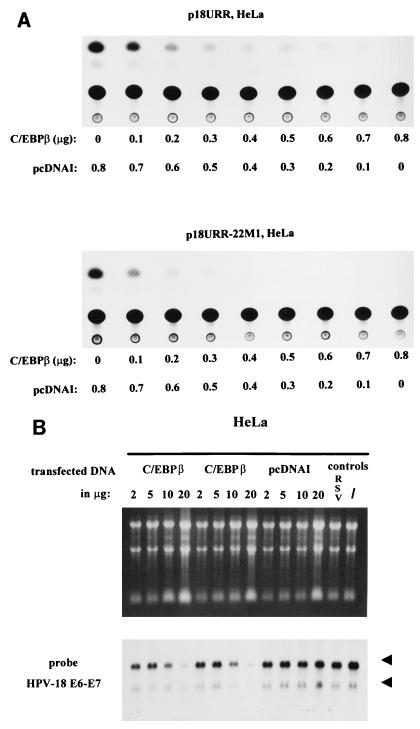
To further examine whether overexpression of C/EBPβ represses the HPV-18 URR activity via the switch region which binds C/EBPβ-YY1, we investigated whether wild-type URR is repressed better than the URR with the mutated switch region. As shown in Fig. 4A, wild-type HPV-18 URR (p18URR) and HPV-18 URR with a mutated switch region (p18URR-22M1) were repressed at the same level by overexpression of C/EBPβ in HeLa cells. This result suggests that unlike activation of the HPV-18 URR in HepG2 cells by C/EBPβ, which is dependent on the switch region (5), repression of HPV-18 URR activity by C/EBPβ in HeLa cells is by and large independent of the switch region. Therefore, although the C/EBPβ-YY1-switch region complex in HeLa cells overexpressing C/EBPβ is disrupted, elimination of this complex is unlikely to be the main cause of repression of the HPV-18 URR mediated by C/EBPβ. We also analyzed several p18URRs with mutations outside the switch region, such as in binding sites for the distal YY1 site OL31, the enhancer AP-1–YY1 site OL21, and the promoter-proximal YY1 site OL13. All of these URRs were repressed as efficiently as the wild-type URR by overexpression of C/EBPβ (data not shown). In addition, truncated URR, i.e., the promoter-proximal fragment (nucleotides −221 to −1 relative to the transcription start site [4]), which is the smallest 5′ deletion of the URR resulting in detectable CAT activity, is also completely repressed by C/EBPβ in HeLa cells (data not shown). Finally, and significantly, our analysis of HeLa cells transfected with CMV-C/EBPβ, but not the vector DNA, shows a strong downregulation of the endogenous E6-E7 mRNA expression level (Fig. 4B).
FIG. 4.
C/EBPβ represses HPV-18 activity. (A) C/EBPβ-induced repression of the HPV-18 URR is independent of the switch region OL22 in HeLa cells. CAT assays were carried out with extracts from HeLa cells transfected with a CAT plasmid containing the wild-type HPV-18 URR (p18URR) (upper panel) or with a construct containing mutations in the switch region of the HPV-18 URR (p18URR-22M1) (lower panel) (4). HeLa cells were transfected with 3 μg of p18URR or p18URR-22M1 together with increasing amounts of CMV-C/EBPβ (0.1 to 0.8 μg) and 0.6 μg RSV/L (18) as an internal control. Relative CAT activities were quantified relative to the activities obtained with p18URR and p18URR-22M1 cotransfected with 0.8 μg of pcDNAI, which were set at 1, and the results of representative CAT assays are shown. Relative CAT activities were as follows. Upper panel (p18URR): 0 μg of C/EBPβ, 1.0; 0.1 μg of C/EBPβ, 0.51; 0.2 μg of C/EBPβ, 0.36; 0.3 μg of C/EBPβ, 0.24; 0.4 μg of C/EBPβ, 0.20; 0.5 μg of C/EBPβ, 0.18; 0.6 μg of C/EBPβ, 0.12; 0.7 μg of C/EBPβ, 0.08; 0.8 μg of C/EBPβ, 0.06. Lower panel (p18URR-22M1): 0 μg of C/EBPβ, 1.0; 0.1 μg of C/EBPβ, 0.47; 0.2 μg of C/EBPβ, 0.25; 0.3 μg of C/EBPβ, 0.19; 0.4 μg of C/EBPβ, 0.15; 0.5 μg of C/EBPβ, 0.13; 0.6 μg of C/EBPβ, 0.09; 0.7 μg of C/EBPβ, 0.06; 0.8 μg of C/EBPβ, 0.05. (B) C/EBPβ represses E6-E7 mRNA expression in HeLa cells. Cells were transfected with increasing amounts of CMV-C/EBPβ or pcDNAI, as indicated in the figure, and 0.6 μg of RSV/L (18) to monitor for transfection efficiency. Cytoplasmic RNA was extracted and separated in a 1.2% agarose gel (upper panel). The filter was hybridized with a 32P-labeled HPV-18 E6-E7 DNA probe and was exposed to X-ray film for 3 days. Beside controls with transfected pcDNAI, RNA from HeLa cells transfected only with 0.6 μg of RSV/L only (18) (lane 13) and RNA from nontransfected HeLa cells (/) (lane 14) were used. The arrowheads indicate the positions of the 28S and 18S RNAs.
C/EBPβ activates the albumin promoter in HeLa cells.
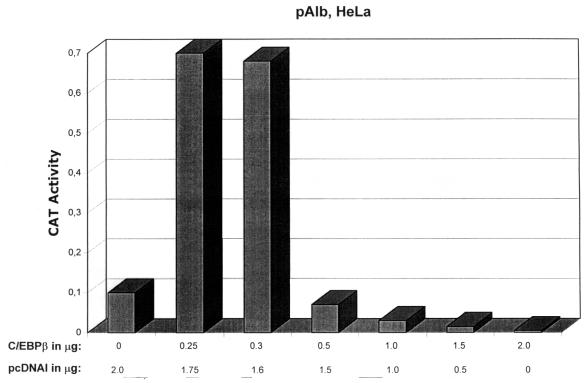
To determine whether repression caused by overexpressing C/EBPs in HeLa cells is specific to the HPV-18 URR, we analyzed the C/EBPβ-responsive mouse albumin promoter, pAlb (25). As shown in Fig. 5, overexpression of C/EBPβ results in activation of pAlb within a concentration range that efficiently inhibits p18URR activity almost completely (compare Fig. 5, 0.25 μg of C/EBPβ, with Fig. 4A, 0.2 μg of C/EBPβ). However, a further increase in C/EBPβ results in a decrease in pAlb activity (Fig. 5, 0.5 μg of C/EBPβ or more), consistent with previous reports (88, 97). This result indicates that (i) C/EBPβ can activate transcription in HeLa cells, which is in line with our previous finding showing that C/EBPβ is involved in activation of the HPV-18 URR in HeLa cells (4, 5); and (ii) C/EBPβ not only activates but also represses transcription, depending on its expression level (5) (Fig. 2A, 4, and 5).
FIG. 5.
C/EBPβ, depending on its level of expression, activates or represses the albumin promoter in HeLa cells. HeLa cells were transfected with 10 μg of DNA from plasmid pALB (containing the mouse albumin promoter upstream of the CAT gene [25]) together with increasing amounts of CMV-C/EBPβ (as indicated below the bar graphs) and 0.6 μg of RSV/L (18) as an internal control. CAT activities were quantified relative to the activity obtained with pAlb cotransfected with 2.0 μg of pcDNAI, which was set at 0.1. Relative CAT activities were as follows: 0 μg of C/EBPβ, 0.1; 0.25 μg of C/EBPβ, 0.7; 0.3 μg of C/EBPβ, 0.68; 0.5 μg of C/EBPβ, 0.07; 1.0 μg of C/EBPβ, 0.03; 1.5 μg of C/EBPβ, 0.015; and 2.0 μg of C/EBPβ, 0.005.
Binding of TBP to the HPV-18 TATA box.
The results presented above suggest that repression of HPV-18 URR activity in HeLa cells by C/EBPβ is independent of the switch region. Furthermore, there is no evidence that this repression is dependent on any specific transcription factor binding sites. For example, the activity of the mutant URR with the smallest 5′ deletion was also completely repressed by C/EBPβ. This fragment contains one of the two AP-1 binding sites of the HPV-18 URR. It was reported that C/EBPβ can interfere with AP-1 (31). In our analysis of several AP-1-dependent promoters, we could not detect any major effects of overexpression of C/EBPβ on them (6). In addition, we failed to detect any differences in the responses to C/EBPβ of the wild-type URR and the URR constructs containing mutations in the AP-1 site. These results suggest that, in the case of the HPV-18 URR, C/EBPβ is unlikely to exert its effects by interacting with AP-1. Because the promoter-proximal fragment encompasses the TATA box and transcription start site of the URR, we asked if C/EBPβ repression is due to its ability to interfere with binding of TBP to the TATA box.
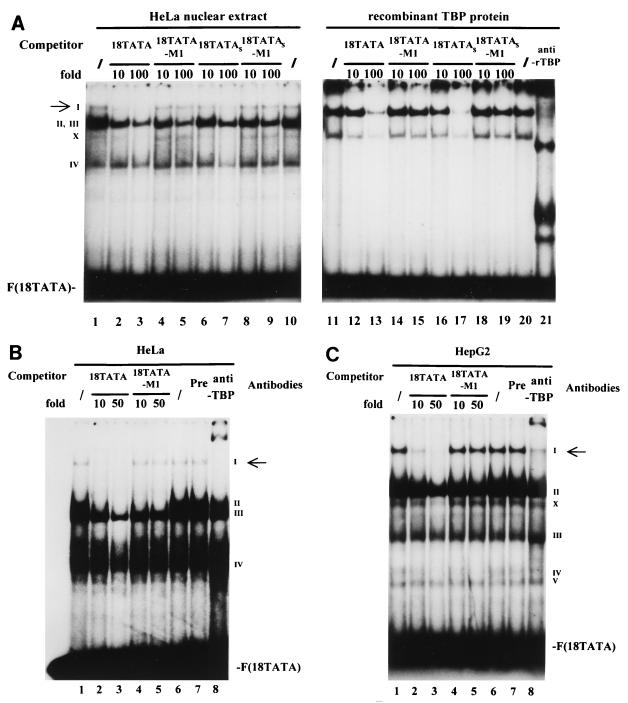
We first analyzed the HPV-18 TATA sequence-protein interaction by performing EMSAs. As shown in Fig. 6A, with HeLa cell nuclear extracts, a specific complex called complex I, indicated by an arrow, was abolished by addition of a molar excess of unlabeled 18TATA (lanes 2 and 3) but not by addition of a molar excess of the mutated 18TATA-M1 oligonucleotides (lanes 4 and 5). Complex I was also abolished by addition of an unlabeled, shorter oligonucleotide, 18TATAS (lanes 6 and 7), encompassing the TATA box and only four or five additional nucleotides on both sides, but not by addition of the mutant 18TATAS-M1 (lanes 8 and 9). In contrast, complexes II, III, and IV are not affected by an excess of TATA oligonucleotides. Therefore, complex I represents an interaction between TBP and the TATA box of the HPV-18 URR. When the same probe was incubated with bacterial rTBP (lanes 11 to 21), a DNA-protein complex was formed which comigrated slightly below that of complex I formed with HeLa nuclear extracts (lanes 1 to 10). This complex was also clearly abolished by addition of a 100-fold molar excess of the unlabeled 18TATA or its shorter form, 18TATAS (lanes 13 and 17), but not by molar excesses of the two mutated TATA oligonucleotides (18TATA-M1 [lane 15] and 18TATAS-M1 [lane 19]). A second, faster-migrating complex (labeled as X) was also formed with rTBP protein (as well as with HeLa extract) on 18TATA and was similarly abolished by wild-type but not mutant TATA oligonucleotides. A TBP-specific antibody (anti-rTBP) interacted with both complexes formed with rTBP protein in a supershift experiment (lane 21). Although the exact nature of this faster-migrating complex is unknown, it is likely caused by binding of partially degraded TBP to the probe.
FIG. 6.
Binding of TBP to the TATA box of the HPV-18 URR. (A) Formation of the specific TBP-TATA box complex I. 32P-labeled oligonucleotide 18TATA was used in EMSAs with HeLa nuclear extracts (lanes 1 to 10) or bacterial rTBP (lanes 11 to 21). Binding reactions were carried out with HeLa cell nuclear extracts in the absence (/) (lanes 1 and 10) or presence (lanes 2 to 9) of unlabeled competitor oligonucleotides. Unlabeled competitor oligonucleotides 18TATA, 18TATA-M1, 18TATAS, and 18TATAS-M1 were each used in 10- and 100-fold molar excesses, as indicated above the lanes. Binding reactions were carried out with bacterial rTBP in the absence (/) (lanes 11 and 20) or presence (lanes 12 to 19) of unlabeled competitor oligonucleotides. Unlabeled competitor oligonucleotides were used in the same manner as for the competition experiments with HeLa cell nuclear extracts. A specific antibody against bacterial rTBP (anti-rTBP) was included in the binding reaction shown in lane 21. A specific DNA-protein complex is indicated by a I (marked by an arrow); numerals II to IV indicate nonspecific DNA-protein complexes. The complex marked by an X is likely caused by binding of partially degraded TBP to the probe. F(18TATA), free probe 18TATA. EMSAs were resolved on a 6.5% polyacrylamide gel. (B) Anti-human TBP antibody specifically recognizes complex I in HeLa cells. Binding reactions with 32P-labeled 18TATA were carried out with HeLa cell nuclear extracts in the absence (/) (lanes 1 and 6) and presence (lanes 2 to 5) of unlabeled competitor. Unlabeled competitor oligonucleotides 18TATA and 18TATA-M1 were used in a 10- or 50-fold molar excess, as indicated above the lanes. A specific antibody against human TBP protein (1 μl) was included in the binding reaction shown in lane 8 (anti-TBP). In lane 7, the binding reaction mixture included 1 μl of preimmune serum (Pre). A specific DNA-protein complex is indicated by the I (marked by an arrow); numerals II to IV indicate nonspecific DNA-protein complexes. F(18TATA), free probe 18TATA. (C) Anti-human TBP antibody specifically recognizes complex I in HepG2 cells. Essentially the same binding reactions as described for panel B were carried out with nuclear extracts from HepG2 cells. A specific DNA-protein complex is indicated by the I (marked by an arrow); numerals II to V indicate nonspecific DNA-protein complexes. The complex marked by an X is likely caused by binding of partially degraded TBP to the probe. F(18TATA), free probe 18TATA.
To determine if complex I indeed contains TBP in HeLa cells, EMSA-antibody experiments were carried out. As shown in Fig. 6B, complex I, which was specifically abolished by unlabeled 18TATA (lanes 2 and 3) but not its mutated form (lanes 4 and 5), was supershifted by a specific antibody directed against human TBP (lane 8) (kindly provided by M. Meisterernst). In a similar experiment, nuclear extracts from HepG2 cells were incubated with 18TATA (Fig. 6C). In addition to several nonspecific bands (complexes II to V), complex I was formed on 18TATA, as seen with HeLa nuclear extracts (Fig. 6B), and the complex was specifically disrupted by addition of excess unlabeled 18TATA (lanes 2 and 3) as well as supershifted by the addition of the specific anti-TBP antibody (lane 8). In further control experiments, we found that the anti-TBP antibody itself did not bind the labeled 18TATA oligonucleotide (data not shown).
C/EBPβ interferes with TBP binding.
We next analyzed the effect of overexpression of C/EBPβ on the TATA complex (complex I) described above. EMSA experiments were performed with nuclear extracts prepared from HeLa cells transfected with increasing amounts of the CMV-C/EBPβ expression plasmid. As shown in Fig. 7A, increasing amounts of C/EBPβ disrupt complex I formed on 18TATA, in a dose-responsive manner (lanes 3 to 5). Transfection with the empty-vector plasmid pcDNAI had no effect on the formation of the TBP-TATA box complex I (lanes 6 to 8). As described earlier, we also observed strong repression of URR activity by LIP (Fig. 2). We therefore analyzed extracts of HeLa cells in which LIP was overexpressed. Similar to the results shown in Fig. 3, overexpression of LIP also completely disrupted the binding of TBP to the TATA box of the HPV-18 URR (Fig. 7A; compare lanes 11–12 and 17–18 with lanes 13–14 and 19–20). Control transfections with the empty-vector plasmid pcDNAI showed only a slight decrease in the formation of complex I with the highest amount of transfected pcDNAI (10 μg; lane 22). These results indicate that disruption of the TBP-TATA box interaction may, at least in part, be responsible for C/EBPβ-mediated repression of HPV-18 URR activity.
FIG. 7.
C/EBPβ interferes with TBP binding. (A) Overexpression of C/EBPβ or LIP disrupts formation of TBP-TATA complex I on oligonucleotide 18TATA in HeLa cells. HeLa cells were transfected with increasing amounts of CMV-C/EBPβ or CMV-LIP. Standard binding reactions were carried out with extracts from nontransfected cells (/) (lanes 1, 9, 15, 16, and 23) or with nuclear extracts from cells transfected with increasing amounts of CMV-C/EBPβ (lanes 3 to 5, 11, 12, 17, and 18), CMV-LIP (lanes 13, 14, 19, and 20), or pcDNAI (lanes 6 to 8, 21, and 22) as indicated above the lanes. In lanes 2 and 10, anti-human TBP antibodies were included in the binding reactions with nuclear extract from nontransfected cells. A specific DNA-protein complex is indicated by the I (marked by the arrows); numerals II to IV indicate nonspecific DNA-protein complexes. F(18TATA), free probe 18TATA. (B) Inability of C/EBPβ to disrupt TBP-TATA complex I formation in HepG2 cells. Standard binding reactions were carried out with nuclear extracts from HepG2 cells transfected with increasing amounts of CMV-C/EBPβ (lanes 2 to 6) or pcDNAI (lanes 7 and 8) as indicated above the lanes. Anti-human TBP antibodies were included in the binding reaction with nuclear extract from nontransfected HepG2 cells (lane 1). A specific DNA-protein complex is indicated by the I (marked by an arrow); numerals II to V indicate nonspecific DNA-protein complexes. The complex marked by an X is likely caused by binding of partially degraded TBP to the probe. F(18TATA), free probe 18TATA. (C) Overexpression of C/EBPβ disrupts TBP-TATA box complex formation on the albumin promoter in HeLa cells. Standard binding reactions with 32P-labeled oligonucleotide AlbTATA (encompassing the TATA box of the albumin promoter) were carried out with nuclear extracts from HeLa cells transfected (transf.) with increasing amounts of CMV-C/EBPβ as indicated above the lanes. Binding reactions with extracts from nontransfected cells were carried out in the absence (/) (lane 2) and presence (lanes 3 and 4) of unlabeled competitor oligocleotides. Unlabeled competitor oligonucleotides AlbTATA and AlbTATA-M1 were used in a 50-fold molar excess. Anti-human TBP antibodies were included in the binding reaction with nuclear extract from nontransfected HeLa cells (lane 5). In lane 1, a standard binding reaction was carried out with HeLa cell nuclear extract and 32P-labeled probe 18TATA. On the left side (for probe 18TATA), a specific DNA-protein complex is indicated by the I (marked by an arrow); numerals II to IV indicate nonspecific DNA-protein complexes. On the right side (for probe AlbTATA), a specific complex is indicated by AlbI (marked by an arrow); complexes AlbII to ALBV indicate nonspecific DNA-protein complexes. F, free probe 18TATA (lane 1) or AlbTATA (lanes 2 to 7).
Previously, we demonstrated that overexpression of C/EBPβ in HepG2 cells induces HPV-18 URR activity (5). We therefore analyzed the effect of C/EBPβ on TBP-TATA box complex formation in HepG2 cells. As shown in Fig. 7B, identical patterns of DNA-protein complexes were found for extracts prepared from C/EBPβ-transfected HepG2 cells (lanes 2 to 6) and those prepared from cells transfected with pcDNAI (Fig. 7B, lanes 7 and 8; see also Fig. 6C, lanes 1 and 6, for an example of nontransfected HepG2 cells). The inability of C/EBPβ (within the concentration range shown in Fig. 7B) to disrupt the TBP-TATA complex is consistent with our observation that C/EBPβ activates, rather than represses, the HPV-18 URR in HepG2 cells.
Since we also observed repression of the albumin promoter with higher amounts of overexpressed C/EBPβ, we were interested in determining if TBP-albumin TATA box complex formation is also affected by overexpressed C/EBPβ in HeLa cells. EMSAs were performed with nuclear extracts prepared from HeLa cells transfected with increasing amounts of the CMV-C/EBPβ expression plasmid. As shown in Fig. 7C, similar to complex I observed with 18TATA (lane 1), a specific complex, AlbI, is formed on the oligonucleotide AlbTATA encompassing the TATA box of the albumin promoter. Complex AlbI was specifically disrupted by a 50-fold molar excess of AlbTATA oligonucleotide (lane 3), but not by its mutated form AlbTATA-M1 (lane 4). Addition of the specific human antibody anti-TBP supershifted complex AlbI (lane 5). Increasing amounts of C/EBPβ disrupt complex AlbI formed on AlbTATA in a dose-responsive manner (lanes 6 and 7). These results indicate that disruption of the TBP-TATA box interaction may also be responsible for C/EBPβ-mediated repression of albumin promoter activity in HeLa cells. Taken together, these results suggest that different promoters may have different sensitivities to repression mediated by the overexpressed C/EBPβ. Further, they provide additional support for the hypothesis that disruption of TBP-TATA interaction is a common mechanism that underlies repression caused by overexpression of C/EBPβ.
Mutation of the TBP binding site strongly represses the HPV-18 URR.
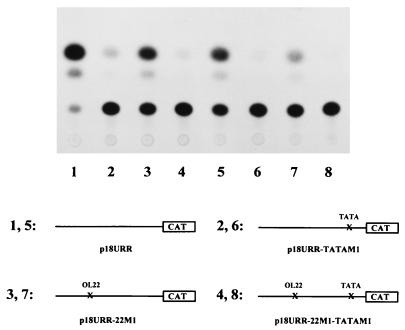
We reasoned that if overexpression of C/EBPβ in HeLa cells abolishes binding of TBP to the URR TATA box (Fig. 7A), mutation of the TATA box in the context of the URR would be expected to have the same effect, i.e., repression of URR activity. To determine the functional role of the TBP binding site TATA in the context of the authentic HPV-18 URR, mutation 18TATA-M1 was introduced by site-directed mutagenesis into the HPV-18 URR. In addition, mutations in the TATA box were created in an HPV-18 URR that also carries mutations in the switch region (p18URR-22M1 [4, 5]). Mutated URR fragments were cloned into plasmid pBLCAT3 upstream of the CAT gene to form constructs p18URR-TATAM1 and p18URR-22M1-TATAM1. These constructs, in parallel with their parental URR-CAT plasmids (wild-type plasmid p18URR as well as plasmid p18URR-22M1 containing a mutated switch region, respectively), were transiently transfected into HeLa cells, and extracts of transfected cells were used to determine CAT activity. As shown in Fig. 8, while mutation of the switch region resulted in about 50% loss of wild-type URR activity (compare lane 1 with lane 3 or lane 5 with lane 7), as previously reported (5), mutations in the TATA box strongly decreased URR activity in both constructs (p18URR-TATAM1 and p18URR-22M1-TATAM1). This suggests that binding of TBP to the TATA box plays a major role in determining HPV-18 URR activity and that no other elements in these promoter constructs could substitute for the loss of the TBP-TATA interactions.
FIG. 8.
Mutation of the TATA box strongly represses HPV-18 URR activity in HeLa cells. CAT assays were carried out with extracts prepared from HeLa cells transfected with plasmids p18URR (wild type), p18URR-22M1 (containing the mutated switch sequence OL22M1), p18URR-TATAM1 (containing the mutated TATA sequence TATA-M1), and p18URR-22M1-TATAM1 (containing the mutated switch sequence and a mutated TATA sequence). HeLa cells were transfected with 5 μg (lanes 1 to 4) or 3 μg (lanes 5 to 8) of DNA from each plasmid, as indicated in the figure, together with 0.6 μg of RSV/L (18) as an internal control. CAT activities were quantified relative to the activity obtained with p18URR (5 μg of transfected DNA) (lane 1), which was set at 1 (for 3 μg of transfected DNAs [lanes 5 to 8], the activities quantified relative to p18URR [set at 1] are shown in parentheses). The results of a representative CAT assay are shown. Relative CAT activities were as follows: p18URR (lane 1), 1.00; p18URR-TATAM1 (lane 2), 0.10; p18URR-22M1 (lane 3), 0.51; p18URR-22M1-TATAM1 (lane 4), 0.04; p18URR (lane 5), 0.447 (1); p18URR-TATAM1 (lane 6), 0.025 (0.056); p18URR-22M1 (lane 7), 0.15 (0.34); and p18URR-22M1-TATAM1 (lane 8), 0.014 (0.031).
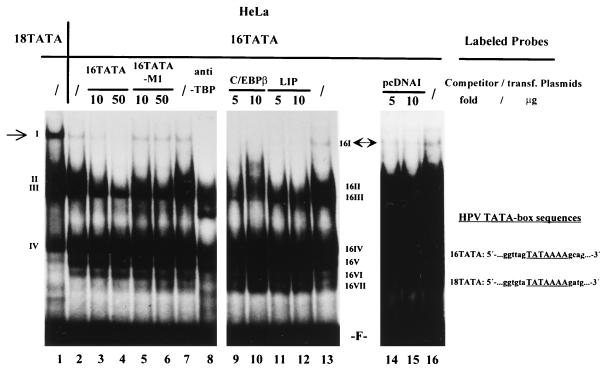
Like that of HPV-18, HPV-16 URR activity is also repressed by C/EBPβ (NF-IL6) (39). Comparison of the TATA boxes of HPV-16 and HPV-18 reveals a high degree of homology (Fig. 9). Therefore, we were interested in determining if both types of viruses are regulated by C/EBPβ through a similar mechanism. To test this possibility, we analyzed the HPV-16 TATA sequence-protein interactions by EMSAs. As shown in Fig. 9, a specific complex, 16I, was observed (lane 2). The mobility of complex 16I is identical to that of complex I formed with the HPV-18 TATA oligonucleotide probe 18TATA (compares lane 1 and 2). The formation of complex 16I on 16TATA was also abolished by addition of a molar excess of unlabeled 16TATA (lanes 3 and 4), but not by the mutant 16TATA-M1 (lanes 5 and 6). Furthermore, human TBP antibody specifically supershifted this complex 16I (lane 8), as it did complex I (Fig. 6B). This suggests that complex 16I represents TBP bound to the TATA box of HPV-16. We performed EMSAs to determine whether overexpression of C/EBPβ affects the formation of complex 16I. HeLa cells were transfected with increasing amounts of CMV-C/EBPβ as well as of CMV-LIP expression plasmids, and the extracts were used for EMSAs. As shown in Fig. 9, increasing amounts of both proteins disrupted the specific TBP complex 16I on the HPV-16 TATA box (lanes 9 to 12), whereas large amounts of pcDNAI only slightly affected the formation of TBP-TATA box complex I (lanes 14 and 15). This suggests that repression of both virus types by C/EBPβ may be mediated through disruption of TBP-TATA interactions.
FIG. 9.
Overexpression of C/EBPβ disrupts formation of the TBP-TATA complex on the HPV-16 URR in HeLa cells. 32P-labeled oligonucleotide 16TATA was used in EMSAs with HeLa cell nuclear extracts. Binding reactions were carried out with extracts from nontransfected cells in the absence (/) (lanes 2, 7, 13, and 16) and presence (lanes 3 to 6) of unlabeled competitor oligonucleotides. Unlabeled competitor oligonucleotides 16TATA and 16TATA-M1 were used in a 10- or 50-fold molar excess, as indicated above the lanes. Anti-human TBP antibodies were included in the binding reaction with nuclear extract from nontransfected HeLa cells (lane 8). Binding reactions were also carried out with nuclear extracts from HeLa cells transfected with increasing amounts of CMV-C/EBPβ (lanes 9 and 10), CMV-LIP (lanes 11 and 12), or pcDNAI (lanes 14 and 15) as indicated above the lanes. In lane 1, a standard binding reaction was carried out with HeLa cell nuclear extract and 32P-labeled probe 18TATA. On the left side (for probe 18TATA), a specific DNA-protein complex is indicated by the I (marked by an arrow); numerals II to IV indicate nonspecific DNA-protein complexes. On the right side (for probe 16TATA), a specific DNA-protein complex is indicated by 16I (marked by an arrow); 16II to 16VII indicate nonspecific DNA-protein complexes. F, free probe 18TATA (lane 1) or free probe 16TATA (lanes 2 to 16). trans., transfected.
DISCUSSION
The HPV-18 URR directs transcription of the E6 and E7 oncogenes. Expression of these oncoproteins is considered to be the first step in the process of HPV-induced carcinogenesis. It is therefore important to understand the molecular mechanisms underlying virus gene expression determined by the viral URR. In this report, we have provided evidence that overexpression of C/EBPβ represses HPV-18 URR activity in HeLa cells. This is in contrast to our previous observation that C/EBPβ activates the HPV-18 URR in HepG2 cells. Furthermore, repression by C/EBPβ in HeLa cells is independent of the switch region, which is essential for C/EBPβ-induced activation of the HPV-18 URR. Instead, we find that repression by overexpression of C/EBPβ is likely due to its ability to disrupt the binding of TBP to the TATA box.
Indeed, interference with TBP binding may be a common mechanism that underlies repression of other HPV serotypes by overexpression of C/EBPβ. Kyo et al. reported that HPV-16 is strongly repressed by C/EBPβ (NF-IL6) (39). Our analysis of HPV-16, revealing that HPV-16 URR repression by C/EBPβ is also correlated with disruption of TBP-TATA box interactions, is consistent with this possibility. In addition, it has been reported that HPV-11 (95) and bovine papillomavirus type 4 (52) are repressed by C/EBPβ. It thus appears that C/EBPβ may play an important role in the transcriptional regulation of different papillomavirus types and that one of the cellular defense mechanisms used to control HPV infection may involve stimulation of overexpression of C/EBPβ, which disrupts important DNA-protein interactions, resulting in repression of transcription from the viral promoters. Significantly, we have found that overexpression of C/EBPβ also results in a strong reduction in the quantity of endogenous HPV-18 E6-E7 mRNA in HeLa cells. Finally, in support of the importance of the TBP-TATA interaction in regulating URR activity, we found that mutating the TATA box of HPV-18 resulted in a complete repression of HPV-18 URR activity in HeLa cells, similar to the effect of overexpression of C/EBPβ, which also leads to the disruption of the TBP-TATA interaction. From these data, taken together, we conclude that different levels of C/EBPβ may differentially regulate HPV-18 URR activity and that it may be possible to modulate HPV-18 URR activity by altering the intracellular C/EBPβ level.
When we compared the URR activities of HPV-16 and HPV-18, we observed that the HPV-18 URR is highly active, compared to the HPV-16 URR, in HeLa cells (data not shown; see also reference 66). It is interesting that binding of TBP to the HPV-18 TATA box is at least 100-fold more effective (Fig. 9; compare lane 1 [HPV-18 TATA] with lane 2 [HPV-16 TATA]). Therefore, it is possible that binding of TBP to the TATA box determines the promoter activities of different serotypes. It will be interesting to analyze the binding of TBP to TATA boxes of different HPV types and to compare it with their URR activities (7). Comparison of the TATA box nucleotide compositions of HPV-18 and HPV-16 shows that the HPV-18 TATA box is a perfect match of thymidine/adenine nucleotide repeats. The HPV-18 sequence is 5′…ggtgtaTATAAAAga…3′, and the HPV-16 sequence is 5′…ggttagTATAAAAgc…3′ (Fig. 9). Similarly, binding of TBP to the albumin promoter TATA sequence was also weak compared to the interaction between TBP and 18TATA (compare lanes 1 and 2 of Fig. 7C). The albumin TATA box sequence is 5′…aagaagTATATTAga…3′, with a 5′ neighboring base (g) similar to that of HPV-16, suggesting that the nucleotide at this position, at least for HPV-18 and HPV-16, for which all other nucleotides of the TATA box sequence are identical, could be important in determining the affinity of TBP for the TATA box. This is consistent with previous reports that TFIID (TBP) binding is also influenced by bases immediately flanking the core TATA sequence (79, 80).
Our analysis of C/EBPβ regulation of HPV-18 URR activity showed that the shorter LIP protein strongly represses HPV-18 URR activity in HeLa cells (Fig. 2). Like C/EBPβ, overexpression of LIP also disrupts TBP-TATA box formation in the HPV-18 URR (Fig. 7A). Since the common element between C/EBPβ and LIP is the bZIP region, these observations suggest that the bZIP region may be responsible for the disruption of the TBP-TATA complex. Consistent with these results, it has been shown that repression of the HPV-16 URR by C/EBPβ is also dependent on the bZIP region of C/EBPβ (39). The truncated LIP protein is able to dimerize and bind DNA by its bZIP region and can act, when it is overexpressed, dominant negatively, presumably by competing with endogenous bZIP proteins for its dimerization partners and/or for the promoter DNA binding site. To elucidate the exact mechanism by which C/EBPβ or LIP disrupts TBP-TATA complex formation, we are currently studying the effects of so-called dominant-negative systems on C/EBP (8, 38, 60, 94).
As we have shown here and in previous reports (3–5), regulation of HPV-18 URR activity seems to be dependent, at least in part, on YY1 and C/EBPβ. The transcriptional activity of YY1 can be altered by the adenovirus E1A oncoproteins via binding of E1A to the coactivator p300 in a p300-YY1 complex (45). In this regard, it is interesting that C/EBPβ, which can alter the activity of YY1, has been suggested to be a cellular E1A-like activity that can substitute for the biological functions of E1A (77, 78). Since HPV E7 has many of the same biological and biological functions as E1A (98), it will be interesting to determine if E7 can modulate the activity of YY1 and C/EBPβ (74). Both C/EBPβ and E7 can bind Rb (13, 53, 54). It will be interesting to investigate whether HPV E7 affects the activity of YY1 and C/EBPβ via the Rb connection.
Finally, expression of high-risk HPV E6-E7 has been suggested to be a prerequisite for continued growth stimulation, and upregulation of E6-E7 expression in the course of progression toward invasive growth suggests a correlation between the quantity of the viral oncogene products and the severity of the lesion (103). In this context, perhaps it is important to note our finding that overexpression of C/EBPβ can cause significant downregulation of the HPV-18 E6-E7 mRNA level in HeLa cells. This suggests that molecular analysis of repression of the HPV-18 URR may be directly relevant to our objective of finding a means of regulating viral gene expression, which directly affects the oncogenic state of the cells. We are in the process of analyzing the growth status of cells in which HPV-18 E6-E7 mRNA is downregulated by overexpression of C/EBPβ.
In summary, we have provided evidence that overexpression of C/EBPβ can efficiently suppress HPV-18 URR activity during transient transfection as well as endogenous HPV-18 transcription in HeLa cells, possibly by disrupting the interaction of TBP with the TATA box of the HPV-18 URR. HPV-18 and HPV-16 together are responsible for about 70% of cervical carcinomas; the fact that both types of viruses can be repressed by overexpression of C/EBPβ suggests a potential use of C/EBPβ in the regulation of these oncogenic HPVs. Since C/EBPβ (NF-IL6) is strongly induced by the cytokine IL-6 (1), it will be interesting to analyze the effect of IL-6 on URR activities in precancerous lesions as well as in different cervical carcinoma cell lines or HPV-derived cervical tumors (64) and to determine whether these IL-6 effects are mediated by the stimulation of C/EBPβ expression. Finally, since C/EBPβ can be strongly induced by IL-6, the possibility that we will be able to modulate the C/EBPβ level by using IL-6, rather than by transfection, in order to affect the URR activity of HPV is appealing.
ACKNOWLEDGMENTS
We thank especially Harald zur Hausen for his great support. We thank Michael Meisterernst (Genzentrum München, Munich, Germany), Claus Nerlov (EMBL, Heidelberg, Germany), and Ed Ziff (New York University Medical Center, New York, N.Y.) for generous gifts of antibodies and expression plasmids, Georg Pougialis and Frank Herweck for technical assistance, and Hajo Delius and his group for performing oligonucleotide synthesis and DNA sequencing.
This work was supported in part by a grant to Y.S. from the National Institutes of Health (GM53874). Y.S. was a recipient of an American Cancer Society Junior Faculty Research Award during the period in which this research was conducted.
REFERENCES
- 1.Akira S, Isshiki H, Sugita T, Tanabe O, Kinoshita S, Nishio Y, Nakajima T, Hirano T, Kishimoto T. A nuclear factor for IL-6 expression (NF-IL6) is a member of a C/EBP family. EMBO J. 1990;9:1897–1906. doi: 10.1002/j.1460-2075.1990.tb08316.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baker C C, Phelps W C, Lindgren V, Braun M J, Gonda M A, Howley P M. Structural and transcriptional analysis of human papillomavirus type 16 sequences in cervical carcinoma cell lines. J Virol. 1987;61:962–971. doi: 10.1128/jvi.61.4.962-971.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bauknecht T, Angel P, Royer H-D, zur Hausen H. Identification of a negative regulatory domain in the human papillomavirus type 18 promoter: interaction with the transcriptional repressor YY1. EMBO J. 1992;11:4607–4617. doi: 10.1002/j.1460-2075.1992.tb05563.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bauknecht T, Jundt F, Herr I, Oehler T, Delius H, Shi Y, Angel P, zur Hausen H. A switch region determines the cell type-specific positive or negative action of YY1 on the activity of the human papillomavirus type 18 promoter. J Virol. 1995;69:1–12. doi: 10.1128/jvi.69.1.1-12.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bauknecht T, See R H, Shi Y. A novel C/EBP β-YY1 complex controls the cell-type-specific activity of the human papillomavirus type 18 upstream regulatory region. J Virol. 1996;70:7695–7705. doi: 10.1128/jvi.70.11.7695-7705.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bauknecht, T., and P. Angel. Unpublished data.
- 7.Bauknecht, T., H. Delius, and Y. Shi. Unpublished data.
- 8.Bauknecht, T., Y. Shi, and C. Vinson. Unpublished data.
- 9.Bushmeyer S, Park K, Atchison M L. Characterization of functional domains within the multifunctional transcription factor, YY1. J Biol Chem. 1995;270:30213–30220. doi: 10.1074/jbc.270.50.30213. [DOI] [PubMed] [Google Scholar]
- 10.Cao Z, Umek R M, McKnight S L. Regulated expression of three C/EBP isoforms during adipose conversion of 3T3-L1 cells. Genes Dev. 1991;5:1538–1552. doi: 10.1101/gad.5.9.1538. [DOI] [PubMed] [Google Scholar]
- 11.Chang C-J, Chen T-T, Lei H-Y, Chen D-S, Lee S-C. Molecular cloning of a transcription factor, AGP/EBP, that belongs to members of the C/EBP family. Mol Cell Biol. 1990;10:6642–6653. doi: 10.1128/mcb.10.12.6642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen C, Okayama H. High-efficiency transformation of mammalian cells by plasmid DNA. Mol Cell Biol. 1987;7:2745–2752. doi: 10.1128/mcb.7.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen P-L, Riley D J, Chen-Kiang S, Lee W-H. Retinoblastoma protein directly interacts with and activates the transcription factor NF-IL6. Proc Natl Acad Sci USA. 1996;93:465–469. doi: 10.1073/pnas.93.1.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 15.Demeret C, Yaniv M, Thierry F. The E2 transcriptional repressor can compensate for SP1 activation of the human papillomavirus type 18 early promoter. J Virol. 1994;68:7075–7082. doi: 10.1128/jvi.68.11.7075-7082.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Descombes P, Chojkier M, Lichtsteiner S, Falvey E, Schibler U. LAP, a novel member of the C/EBP gene family, encodes a liver-enriched transcriptional activator protein. Genes Dev. 1990;4:1541–1551. doi: 10.1101/gad.4.9.1541. [DOI] [PubMed] [Google Scholar]
- 17.Descombes P, Schibler U. A liver-enriched transcriptional activator protein, LAP, and a transcriptional inhibitory protein, LIP, are translated from the same mRNA. Cell. 1991;67:569–579. doi: 10.1016/0092-8674(91)90531-3. [DOI] [PubMed] [Google Scholar]
- 18.de Wet J R, Wood K V, DeLuca M, Helinski D R, Subramani S. Firefly luciferase gene: structure and expression in mammalian cells. Mol Cell Biol. 1987;7:725–737. doi: 10.1128/mcb.7.2.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Diehl J A, Hannink M. Identification of a C/EBP-Rel complex in avian lymphoid cells. Mol Cell Biol. 1994;14:6635–6646. doi: 10.1128/mcb.14.10.6635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dürst M, Gissmann L, Ikenberg H, zur Hausen H. A new papillomavirus DNA from a cervical carcinoma and its prevalence in cancer biopsy samples from different geographic regions. Proc Natl Acad Sci USA. 1983;80:3812–3815. doi: 10.1073/pnas.80.12.3812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dyson N, Howley P M, Münger K, Harlow E. The human papillomavirus-16 E7 oncoprotein is able to bind to the retinoblastoma gene product. Science. 1989;243:934–937. doi: 10.1126/science.2537532. [DOI] [PubMed] [Google Scholar]
- 22.Fawcett T W, Eastman H B, Martindale J L, Holbrook N J. Physical and functional association between GADD153 and CCAAT/enhancer-binding protein β during cellular stress. J Biol Chem. 1996;271:14285–14289. doi: 10.1074/jbc.271.24.14285. [DOI] [PubMed] [Google Scholar]
- 23.Flanagan J R, Becker K G, Ennist D L, Gleason S L, Driggers P H, Levi B-Z, Appella E, Ozato K. Cloning of a negative transcription factor that binds to the upstream conserved region of Moloney murine leukemia virus. Mol Cell Biol. 1992;12:38–44. doi: 10.1128/mcb.12.1.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gloss B, Bernard H-U. The E6/E7 promoter of human papillomavirus type 16 is activated in the absence of E2 proteins by a sequence-aberrant Sp1 distal element. J Virol. 1990;64:5577–5584. doi: 10.1128/jvi.64.11.5577-5584.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gorski K, Carneiro M, Schibler U. Tissue-specific in vitro transcription from the mouse albumin promoter. Cell. 1986;47:767–776. doi: 10.1016/0092-8674(86)90519-2. [DOI] [PubMed] [Google Scholar]
- 26.Hariharan N, Kelley D E, Perry R P. δ, a transcription factor that binds to downstream elements in several polymerase II promoters, is a functionally versatile zinc finger protein. Proc Natl Acad Sci USA. 1991;88:9799–9803. doi: 10.1073/pnas.88.21.9799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hawley-Nelson P, Vousden K H, Hubbert N L, Lowy D R, Schiller J T. HPV16 E6 and E7 proteins cooperate to immortalize human foreskin keratinocytes. EMBO J. 1989;8:3905–3910. doi: 10.1002/j.1460-2075.1989.tb08570.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heck D V, Yee C L, Howley P M, Münger K. Efficiency of binding the retinoblastoma protein correlates with the transforming capacity of the E7 oncoproteins of the human papillomaviruses. Proc Natl Acad Sci USA. 1992;89:4442–4446. doi: 10.1073/pnas.89.10.4442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hoppe-Seyler F, Butz K. Activation of human papillomavirus type 18 E6-E7 oncogene expression by transcription factor SP1. Nucleic Acids Res. 1992;20:6701–6706. doi: 10.1093/nar/20.24.6701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hsu W, Chen-Kiang S. Convergent regulation of NF-IL6 and Oct-1 synthesis by interleukin-6 and retinoic acid signaling in embryonal carcinoma cells. Mol Cell Biol. 1993;13:2515–2523. doi: 10.1128/mcb.13.4.2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hsu W, Kerppola T K, Chen P-L, Curran T, Chen-Kiang S. Fos and Jun repress transcription activation by NF-IL6 through association at the basic zipper region. Mol Cell Biol. 1994;14:268–276. doi: 10.1128/mcb.14.1.268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hurst H C. bZIP proteins. Protein Profile. 1994;1:123–168. [PubMed] [Google Scholar]
- 33.Johnson P F, Landschulz W H, Graver B J, McKnight S L. Identification of a rat liver nuclear protein that binds to the enhancer core element of three animal viruses. Genes Dev. 1987;1:133–146. doi: 10.1101/gad.1.2.133. [DOI] [PubMed] [Google Scholar]
- 34.Jundt F, Herr I, Angel P, zur Hausen H, Bauknecht T. Transcriptional control of human papillomavirus type 18 oncogene expression in different cell lines: role of transcription factor YY1. Virus Genes. 1995;11:53–58. doi: 10.1007/BF01701662. [DOI] [PubMed] [Google Scholar]
- 35.Katz S, Kowenz-Leutz E, Müller C, Meese K, Ness S A, Leutz A. The NF-M transcription factor is related to C/EBPβ and plays a role in signal transduction, differentiation and leukemogenesis of avian myelomonocytic cells. EMBO J. 1993;12:1321–1332. doi: 10.1002/j.1460-2075.1993.tb05777.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kinoshita S, Akira S, Kishimoto T. A member of the C/EBP family, NF-IL6β, forms a heterodimer and transcriptionally synergizes with NF-IL6. Proc Natl Acad Sci USA. 1992;89:1473–1476. doi: 10.1073/pnas.89.4.1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Klampfer L, Lee T H, Hsu W, Vilc̆ek J, Chen-Kiang S. NF-IL6 and AP-1 cooperatively modulate the activation of the TSG-6 gene by tumor necrosis factor alpha and interleukin-1. Mol Cell Biol. 1994;14:6561–6569. doi: 10.1128/mcb.14.10.6561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Krylov D, Olive M, Vinson C. Extending dimerization interfaces: the bZIP basic region can form a coiled coil. EMBO J. 1995;14:5329–5337. doi: 10.1002/j.1460-2075.1995.tb00217.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kyo S, Inoue M, Nishio Y, Nakanishi K, Akira S, Inoue H, Yutsudo M, Tanizawa O, Hakura A. NF-IL6 represses early gene expression of human papillomavirus type 16 through binding to the noncoding region. J Virol. 1993;67:1058–1066. doi: 10.1128/jvi.67.2.1058-1066.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lamb P, McKnight S L. Diversity and specificity in transcriptional regulation: the benefits of heterotypic dimerization. Trends Biochem Sci. 1991;16:417–422. doi: 10.1016/0968-0004(91)90167-t. [DOI] [PubMed] [Google Scholar]
- 41.Landschulz W H, Johnson P F, Adashi E Y, Graves B J, McKnight S L. Isolation of a recombinant copy of the gene encoding C/EBP. Genes Dev. 1988;2:786–800. doi: 10.1101/gad.2.7.786. [DOI] [PubMed] [Google Scholar]
- 42.Landschulz W H, Johnson P F, McKnight S L. The leucine zipper: a hypothetical structure common to a new class of DNA binding proteins. Science. 1988;240:1759–1764. doi: 10.1126/science.3289117. [DOI] [PubMed] [Google Scholar]
- 43.Landschulz W H, Johnson P F, McKnight S L. The DNA binding domain of the rat liver nuclear protein C/EBP is bipartite. Science. 1989;243:1681–1688. doi: 10.1126/science.2494700. [DOI] [PubMed] [Google Scholar]
- 44.LeClair K P, Blanar M A, Sharp P A. The p50 subunit of NF-κB associates with NF-IL6 transcription factor. Proc Natl Acad Sci USA. 1992;89:8145–8149. doi: 10.1073/pnas.89.17.8145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee J-S, Galvin K M, See R H, Eckner R, Livingston D, Moran E, Shi Y. Relief of YY1 transcriptional repression by adenovirus E1A is mediated by E1A-associated protein p300. Genes Dev. 1995;9:1188–1198. doi: 10.1101/gad.9.10.1188. [DOI] [PubMed] [Google Scholar]
- 46.Lee J-S, See R H, Galvin K M, Wang J, Shi Y. Functional interactions between YY1 and adenovirus E1A. Nucleic Acids Res. 1995;23:925–931. doi: 10.1093/nar/23.6.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee Y-H, Yano M, Liu S-Y, Matsunaga E, Johnson P F, Gonzalez F J. A novel cis-acting element controlling the rat CYP2D5 gene and requiring cooperativity between C/EBPβ and an Sp1 factor. Mol Cell Biol. 1994;14:1383–1394. doi: 10.1128/mcb.14.2.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee Y-H, Williams S C, Baer M, Sterneck E, Gonzalez F J, Johnson P F. The ability of C/EBPβ but not C/EBPα to synergize with an Sp1 protein is specified by the leucine zipper and activation domain. Mol Cell Biol. 1997;17:2038–2047. doi: 10.1128/mcb.17.4.2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lichy J H, Majidi M, Elbaum J, Tsai M M. Differential expression of the human ST5 gene in HeLa-fibroblast hybrid cell lines mediated by YY1: evidence that YY1 plays a part in tumor suppression. Nucleic Acids Res. 1996;24:4700–4708. doi: 10.1093/nar/24.23.4700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mack D H, Laimins L A. A keratinocyte-specific transcription factor, KRF-1, interacts with AP1 to activate expression of human papillomavirus type 18 in squamous epithelial cells. Proc Natl Acad Sci USA. 1991;88:9102–9106. doi: 10.1073/pnas.88.20.9102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.May M, Dong X-P, Beyer-Finkler E, Stubenrauch F, Fuchs P G, Pfister H. The E6/E7 promoter of extrachromosomal HPV16 DNA in cervical cancers escapes from cellular repression by mutation of target sequences for YY1. EMBO J. 1994;13:1460–1466. doi: 10.1002/j.1460-2075.1994.tb06400.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McCaffery R E, Jackson M E. An element binding a C/EBP-related transcription factor contributes to negative regulation of the bovine papillomavirus type 4 long control region. J Gen Virol. 1994;75:3047–3056. doi: 10.1099/0022-1317-75-11-3047. [DOI] [PubMed] [Google Scholar]
- 53.Münger K, Phelps W C, Bubb V, Howley P M, Schlegel R. The E6 and E7 genes of human papillomavirus type 16 together are necessary and sufficient for transformation of primary human keratinocytes. J Virol. 1989;63:4417–4421. doi: 10.1128/jvi.63.10.4417-4421.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Münger K, Werness B A, Dyson N, Phelps W C, Harlow E, Howley P M. Complex formation of human papillomavirus E7 proteins with the retinoblastoma tumor suppressor gene product. EMBO J. 1989;8:4099–4105. doi: 10.1002/j.1460-2075.1989.tb08594.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54a.Nerlov, C. Personal communication.
- 55.Nerlov C, Ziff E B. Three levels of functional interaction determine the activity of CCAAT/enhancer binding protein-α on the serum albumin promoter. Genes Dev. 1994;8:350–362. doi: 10.1101/gad.8.3.350. [DOI] [PubMed] [Google Scholar]
- 56.Nerlov C, Ziff E B. CCAAT/enhancer binding protein-α amino acid motifs with dual TBP and TFIIB binding ability co-operate to activate transcription in both yeast and mammalian cells. EMBO J. 1995;14:4318–4328. doi: 10.1002/j.1460-2075.1995.tb00106.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nishio Y, Isshiki H, Kishimoto T, Akira S. A nuclear factor for interleukin-6 expression (NF-IL6) and the glucocorticoid receptor synergistically activate transcription of the rat α1-acid glycoprotein gene via direct protein-protein interaction. Mol Cell Biol. 1993;13:1854–1862. doi: 10.1128/mcb.13.3.1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.O’Connor M J, Tan S-H, Tan C-H, Bernard H-U. YY1 represses human papillomavirus type 16 transcription by quenching AP-1 activity. J Virol. 1996;70:6529–6539. doi: 10.1128/jvi.70.10.6529-6539.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Offord E A, Beard P. A member of the activator protein 1 family found in keratinocytes but not in fibroblasts required for transcription from a human papillomavirus type 18 promoter. J Virol. 1990;64:4792–4798. doi: 10.1128/jvi.64.10.4792-4798.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Olive M, Williams S C, Dezan C, Johnson P F, Vinson C. Design of a C/EBP-specific, dominant-negative bZIP protein with both inhibitory and gain-of-function properties. J Biol Chem. 1996;271:2040–2047. doi: 10.1074/jbc.271.4.2040. [DOI] [PubMed] [Google Scholar]
- 61.Ossipow V, Descombes P, Schibler U. CCAAT/enhancer-binding protein mRNA is translated into multiple proteins with different transcription activation potentials. Proc Natl Acad Sci USA. 1993;90:8219–8223. doi: 10.1073/pnas.90.17.8219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Park K, Atchison M L. Isolation of a candidate repressor/activator, NF-E1 (YY-1, δ), that binds to the immunoglobulin κ 3′ enhancer and the immunoglobulin heavy-chain enhancer. Proc Natl Acad Sci USA. 1991;88:9804–9808. doi: 10.1073/pnas.88.21.9804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Poli V, Mancini F P, Cortese R. IL-6DBP, a nuclear protein involved in interleukin-6 signal transduction, defines a new family of leucine zipper proteins related to C/EBP. Cell. 1990;63:643–653. doi: 10.1016/0092-8674(90)90459-r. [DOI] [PubMed] [Google Scholar]
- 64.Randelzhofer, B., T. Bauknecht, and T. Bauknecht. Unpublished data.
- 65.Roman C, Platero J S, Shuman J, Calame K. Ig/EBP-1: a ubiquitously expressed immunoglobulin enhancer binding protein that is similar to C/EBP and heterodimerizes with C/EBP. Genes Dev. 1990;4:1404–1415. doi: 10.1101/gad.4.8.1404. [DOI] [PubMed] [Google Scholar]
- 66.Romanczuk H, Villa L L, Schlegel R, Howley P M. The viral transcriptional regulatory region upstream of the E6 and E7 genes is a major determinant of the differential immortalization activities of human papillomavirus types 16 and 18. J Virol. 1991;65:2739–2744. doi: 10.1128/jvi.65.5.2739-2744.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ron D, Habener J F. CHOP, a novel developmentally regulated nuclear protein that dimerizes with transcription factors C/EBP and LAP and functions as a dominant-negative inhibitor of gene transcription. Genes Dev. 1992;6:439–453. doi: 10.1101/gad.6.3.439. [DOI] [PubMed] [Google Scholar]
- 68.Rösl F, Das B C, Lengert M, Geletneky K, zur Hausen H. Antioxidant-induced changes of the AP-1 transcription complex are paralleled by a selective suppression of human papillomavirus transcription. J Virol. 1997;71:362–370. doi: 10.1128/jvi.71.1.362-370.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Scheffner M, Werness B A, Huibregtse J M, Levine A J, Howley P M. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell. 1990;63:1129–1136. doi: 10.1016/0092-8674(90)90409-8. [DOI] [PubMed] [Google Scholar]
- 70.Schneider-Gädicke A, Schwarz E. Different human cervical carcinoma cell lines show similar transcription patterns of human papillomavirus type 18 early genes. EMBO J. 1986;5:2285–2292. doi: 10.1002/j.1460-2075.1986.tb04496.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schwarz E, Freese U K, Gissmann L, Mayer W, Roggenbuck B, Stremlau A, zur Hausen H. Structure and transcription of human papillomavirus sequences in cervical carcinoma cells. Nature. 1985;314:111–114. doi: 10.1038/314111a0. [DOI] [PubMed] [Google Scholar]
- 72.Shi Y, Seto E, Chang L-S, Shenk T. Transcriptional repression by YY1, a human GLI-Krüppel related protein, and relief of repression by adenovirus E1A protein. Cell. 1991;67:377–388. doi: 10.1016/0092-8674(91)90189-6. [DOI] [PubMed] [Google Scholar]
- 73.Shi Y, Lee J-S, Galvin K M. Everything you have ever wanted to know about Yin Yang 1…. Biochim Biophys Acta. 1997;1332:F25–F48. doi: 10.1016/s0304-419x(96)00044-3. [DOI] [PubMed] [Google Scholar]
- 74.Shi, Y., M. Tommasino, and T. Bauknecht. Unpublished data.
- 75.Shrivastava A, Calame K. An analysis of genes regulated by the multi-functional transcriptional regulator Yin Yang-1. Nucleic Acids Res. 1994;22:5151–5155. doi: 10.1093/nar/22.24.5151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Smotkin D, Wettstein F O. Transcription of human papillomavirus type 16 early genes in a cervical cancer and a cancer-derived cell line and identification of the E7 protein. Proc Natl Acad Sci USA. 1986;83:4680–4684. doi: 10.1073/pnas.83.13.4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Spergel J M, Chen-Kiang S. Interleukin 6 enhances a cellular activity that functionally substitutes for E1A protein in transcription. Proc Natl Acad Sci USA. 1991;88:6472–6476. doi: 10.1073/pnas.88.15.6472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Spergel J M, Hsu W, Akira S, Thimmappaya B, Kishimoto T, Chen-Kiang S. NF-IL6, a member of the C/EBP family, regulates E1A-responsive promoters in the absence of E1A. J Virol. 1992;66:1021–1030. doi: 10.1128/jvi.66.2.1021-1030.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Starr D B, Hawley D K. TFIID binds in the minor groove of the TATA box. Cell. 1991;67:1231–1240. doi: 10.1016/0092-8674(91)90299-e. [DOI] [PubMed] [Google Scholar]
- 80.Starr D B, Hoopes B C, Hawley D K. DNA bending is an important component of site-specific recognition by the TATA binding protein. J Mol Biol. 1995;250:434–446. doi: 10.1006/jmbi.1995.0388. [DOI] [PubMed] [Google Scholar]
- 81.Stein B, Baldwin A S., Jr Distinct mechanisms for regulation of the interleukin-8 gene involve synergism and cooperativity between C/EBP and NF-κB. Mol Cell Biol. 1993;13:7191–7198. doi: 10.1128/mcb.13.11.7191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Stein B, Cogswell P C, Baldwin A S., Jr Functional and physical associations between NF-κB and C/EBP family members: a Rel domain-bZIP interaction. Mol Cell Biol. 1993;13:3964–3974. doi: 10.1128/mcb.13.7.3964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Stein B, Yang M X. Repression of the interleukin-6 promoter by estrogen receptor is mediated by NF-κB and C/EBPβ. Mol Cell Biol. 1995;15:4971–4979. doi: 10.1128/mcb.15.9.4971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Thierry F, Garcia-Carranca A, Yaniv M. Elements that control the transcription of genital papillomavirus type 18. Cancer Cells. 1987;5:23–32. [Google Scholar]
- 85.Thierry F, Spyrou G, Yaniv M, Howley P M. Two AP1 sites binding JunB are essential for human papillomavirus type 18 transcription in keratinocytes. J Virol. 1992;66:3740–3748. doi: 10.1128/jvi.66.6.3740-3748.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tommasino M, Adamczewski J P, Carlotti F, Barth C F, Manetti R, Contorni M, Cavalieri F, Hunt T, Crawford L. HPV16 E7 protein associates with the protein kinase p33CDK2 and cyclin A. Oncogene. 1993;8:195–202. [PubMed] [Google Scholar]
- 87.Trautwein C, Caelles C, van der Geer P, Hunter T, Karin M, Chojkier M. Transactivation by NF-IL6/LAP is enhanced by phosphorylation of its activation domain. Nature. 1993;364:544–547. doi: 10.1038/364544a0. [DOI] [PubMed] [Google Scholar]
- 88.Trautwein C, Rakemann T, Pietrangelo A, Plümpe J, Mantosi G, Manns M P. C/EBP-β/LAP controls down-regulation of albumin gene transcription during liver regeneration. J Biol Chem. 1996;271:22262–22270. doi: 10.1074/jbc.271.36.22262. [DOI] [PubMed] [Google Scholar]
- 89.Tsukada J, Saito K, Waterman W R, Webb A C, Auron P E. Transcription factors NF-IL6 and CREB recognize a common essential site in the human prointerleukin 1β gene. Mol Cell Biol. 1994;14:7285–7297. doi: 10.1128/mcb.14.11.7285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ubeda M, Wang X-Z, Zinszner H, Wu I, Habener J F, Ron D. Stress-induced binding of the transcription factor CHOP to a novel DNA control element. Mol Cell Biol. 1996;16:1479–1489. doi: 10.1128/mcb.16.4.1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Umek R M, Friedman A D, McKnight S L. CCAAT-enhancer binding protein: a component of a differentiation switch. Science. 1991;251:288–292. doi: 10.1126/science.1987644. [DOI] [PubMed] [Google Scholar]
- 92.Vallejo M, Ron D, Miller C P, Habener J F. C/ATF, a member of the activating transcription factor family of DNA-binding proteins, dimerizes with CAAT/enhancer-binding proteins and directs their binding to cAMP response elements. Proc Natl Acad Sci USA. 1993;90:4679–4683. doi: 10.1073/pnas.90.10.4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Vinson C R, Sigler P B, McKnight S L. Scissors-grip model for DNA recognition by a family of leucine zipper proteins. Science. 1989;246:911–916. doi: 10.1126/science.2683088. [DOI] [PubMed] [Google Scholar]
- 94.Vinson C R, Hai T, Boyd S M. Dimerization specificity of the leucine zipper-containing bZIP motif on DNA binding: prediction and rational design. Genes Dev. 1993;7:1047–1058. doi: 10.1101/gad.7.6.1047. [DOI] [PubMed] [Google Scholar]
- 95.Wang H, Liu K, Yuan F, Berdichevsky L, Taichman L B, Auborn K. C/EBPβ is a negative regulator of human papillomavirus type 11 in keratinocytes. J Virol. 1996;70:4839–4844. doi: 10.1128/jvi.70.7.4839-4844.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Werness B A, Levine A J, Howley P M. Association of human papillomavirus types 16 and 18 E6 proteins with p53. Science. 1990;248:76–79. doi: 10.1126/science.2157286. [DOI] [PubMed] [Google Scholar]
- 97.Williams S C, Cantwell C A, Johnson P F. A family of C/EBP-related proteins capable of forming covalently linked leucine zipper dimers in vitro. Genes Dev. 1991;5:1553–1567. doi: 10.1101/gad.5.9.1553. [DOI] [PubMed] [Google Scholar]
- 98.Wong H K, Ziff E B. The human papillomavirus type 16 E7 protein complements adenovirus type 5 E1A amino-terminus-dependent transactivation of adenovirus type 5 early genes and increases ATF and Oct-1 DNA binding activity. J Virol. 1996;70:332–340. doi: 10.1128/jvi.70.1.332-340.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zerfass K, Schulze A, Spitkovsky D, Friedman V, Henglein B, Jansen-Dürr P. Sequential activation of cyclin E and cyclin A gene expression by human papillomavirus type 16 E7 through sequences necessary for transformation. J Virol. 1995;69:6389–6399. doi: 10.1128/jvi.69.10.6389-6399.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.zur Hausen H. Papillomaviruses in anogenital cancer as a model to understand the role of viruses in human cancers. Cancer Res. 1989;49:4677–4681. [PubMed] [Google Scholar]
- 101.zur Hausen H. Viruses in human cancers. Science. 1991;254:1167–1173. doi: 10.1126/science.1659743. [DOI] [PubMed] [Google Scholar]
- 102.zur Hausen H. Human papillomaviruses in the pathogenesis of anogenital cancer. Virology. 1991;184:9–13. doi: 10.1016/0042-6822(91)90816-t. [DOI] [PubMed] [Google Scholar]
- 103.zur Hausen H. Papillomavirus infections—a major cause of human cancers. Biochim Biophys Acta. 1996;1288:F55–F78. doi: 10.1016/0304-419x(96)00020-0. [DOI] [PubMed] [Google Scholar]