Abstract
We have explored the requirements for host proteins in the integration of Moloney murine leukemia virus (MoMuLV) cDNA in vitro. Following infection, it is possible to lyse cells and obtain preintegration complexes (PICs) capable of integrating the MoMuLV cDNA into an added target DNA in vitro (intermolecular integration). PICs can be stripped of required proteins by gel filtration in high-salt buffers (600 mM KCl), allowing the nature of the removed factors to be investigated by in vitro reconstitution. In a previous study of human immunodeficiency virus type 1 (HIV-1) PICs, the host protein HMG I(Y) was found to be able to restore activity to salt-stripped PICs. In contrast, salt stripping and reconstitution of MoMuLV PICs led to the proposal that a host factor is important for a different activity, blocking integration into the cDNA itself (autointegration). In this report, we investigated reconstitution of salt-stripped MoMuLV PICs and found that addition of cellular extract from uninfected NIH 3T3 cells could block autointegration and also restore intermolecular integration. Isolation of the intermolecular integration-complementing activity yielded HMG I(Y), as in the HIV-1 case. However, HMG I(Y) could not block autointegration, implicating a different host factor in this process. Additionally, when MoMuLV PICs were partially purified but not salt stripped, the intermolecular integration activity was reduced but could be stimulated by the addition of any of several purified DNA binding proteins. In summary, three activities were detected: (i) the intermolecular integration cofactor HMG I(Y), (ii) an autointegration barrier protein, and (iii) stimulatory DNA binding proteins.
An essential step in retroviral replication is the integration of viral cDNA into host cell DNA (11, 20, 54). In vivo, the linear viral cDNA is assembled into large nucleoprotein complexes called preintegration complexes (PICs) (2, 26, 44). PICs can be isolated from newly infected cells and are capable of integrating the viral cDNA into an added target DNA in vitro (3, 16, 21). We have used such PIC-based assays to investigate host factors important for integration of Moloney murine leukemia virus (MoMuLV) cDNA.
One of the components of MoMuLV PICs is inferred to be integrase. Though the protein has not yet been detected physically in isolated MoMuLV PICs, integrase is expected to be present, since the enzymatic activity is essential for integration in vivo (4, 14, 50). For human immunodeficiency virus type 1 (HIV-1), integrase has been shown to be associated with PICs (5, 18, 23). Recombinant integrase proteins from ASLV (avian sarcoma-leukosis virus), MoMuLV, HIV-1, and other retroviruses have been shown to be capable of removing 2 nucleotides from 3′ ends of model long terminal repeat (LTR) DNAs (12, 30, 38, 51) and joining the resulting recessed 3′ hydroxyl group to the target DNA in vitro (8, 12, 15, 35, 37).
Purified integrase proteins differ in their abilities to carry out coordinated joining of pairs of LTR ends, thereby generating integration intermediates with a defined spacing between points of joining at each end of the cDNA (called coupled joining) (Fig. 1). In the case of HIV-1 integrase, only single LTR analogs generally become joined to target DNA, mimicking events at one cDNA end only (7, 8). More-efficient coupled joining is seen when lysed HIV-1 virions are used as a source of integrase (29). Purified MoMuLV and ASLV integrases yield coupled products more efficiently than does integrase of HIV (1, 12, 24, 37, 55).
FIG. 1.
Inferred pathway for intermolecular integration and autointegration. MoMuLV cDNA (thin lines) and target DNA (thick lines) are indicated. The 5′ ends (small solid circles) and 3′ ends (arrows) are indicated. II, integration intermediate.
Unlike purified integrase, reactions with PICs yield coupled integration products as the only detectable forms, producing the integration intermediate (II) illustrated in Fig. 1 (3, 16, 21). Integration is thought to be completed in vivo by host DNA repair enzymes which connect the remaining unjoined strands in the integration intermediate and complete proviral synthesis.
Several previous studies have outlined the organization and composition of PICs. In the case of MoMuLV, PICs are known to be large particles (around 160S) potentially containing several viral proteins (2). In the case of HIV-1, the linear viral cDNA is packaged into a large particle with a diameter of 54 nm as estimated by gel filtration (for comparison, the diameter of a virion is about 120 nm) (44). The ends of the cDNA in PICs are protected from exonuclease digestion by bound proteins (44). The two ends of viral cDNA appear to be held together by a protein bridge, since the cDNA can be cleaved internally without disrupting coordinated integration of the two cDNA ends (44). Of the viral proteins, integrase, reverse transcriptase (RT), matrix (MA) and Vpr proteins have been proposed to be the components of PICs and NC protein has been detected in some studies (5, 23, 28, 32, 44). In addition, a host protein, HMG I(Y) was found to be present in HIV-1 PICs and required for function in vitro (18). In that study, a salt-stripping procedure was used to remove a subset of proteins from HIV-1 PICs, which abolished intermolecular integration activity. Human HMG I(Y) protein was found to restore the activity (18).
A salt-stripping method was used previously to identify a host activity involved in the function of MoMuLV PICs (41). Salt-stripped PICs of MoMuLV not only lose normal intermolecular integration activity but also carry out autointegration, a suicidal intramolecular integration reaction in which the PIC uses its own cDNA as an integration target (41). As a result of autointegration, the linear viral cDNA becomes circularized and biologically inactivated (22, 41) (Fig. 1, autointegration). It was found that an extract from uninfected host cells could block autointegration and restore intermolecular integration activity, implicating an as yet unidentified host factor or factors (41).
Here we have isolated a factor that stimulates intermolecular integration by salt-stripped MoMuLV PICs and found the factor to be murine HMG I(Y). We have determined that murine HMG I(Y) is not itself the autointegration barrier factor. Furthermore, we investigated stimulation by additional DNA binding proteins in different experimental settings. These data identify several activities important for function of MoMuLV PICs in vitro.
MATERIALS AND METHODS
Materials.
Proteins were obtained from the following sources: Escherichia coli single-stranded DNA binding protein (SSB), Stratagene; T4 gene 32 protein, Boehringer Mannheim; and RNase A, Qiagen. HIV-1MN p7 was obtained through the AIDS Research and Reference Reagent Program, National Institute of Allergy and Infectious Diseases, National Institutes of Health from Louis Henderson (39, 52).
PIC preparation.
PICs were prepared as described previously (3, 27) with some modifications. Plasmid pMoMuLV-K, which contains a complete genome of MoMuLV, was obtained from A. Dusty Miller. The plasmid was transfected into NIH 3T3 cells, and MoMuLV was harvested 2 days later. Fresh NIH 3T3 cells were infected by the virus and passaged to establish a chronically infected cell line. The infected cells (2.4 × 106 cells) were plated together with uninfected NIH 3T3 cells (9.6 × 106) in a 150-mm-diameter dish. Cells were harvested after 16 h of coculture by trypsinization. The trypsinized cells were washed twice with buffer A (20 mM HEPES-NaOH [pH 7.4], 150 mM KCl, 5 mM MgCl2, 1 mM dithiothreitol) and then lysed with 450 μl of buffer A containing 0.025% digitonin (Calbiochem) and 2 μg of aprotinin (Calbiochem) per ml. The lysate containing PICs was clarified by centrifugation as described previously (27). The crude extract, which contains approximately 10 ng of viral cDNA per ml and 6 mg of protein per ml was frozen in liquid nitrogen and stored at −70°C. All the cells were grown in Dulbecco modified Eagle’s medium with 10% fetal calf serum (Hyclone).
Extract containing the PICs used in Fig. 7 was prepared by a modification of the method described in reference 13. Newly infected cells were harvested by trypsinization and washed with ice-cold phosphate-buffered saline with 5 mM MgCl2. Cells were then incubated with 4 packed cell volumes of buffer B (10 mM Tris-HCl [pH 7.9], 10 mM KCl, 1.5 mM MgCl2, 1 mM dithiothreitol) on ice for 20 min and homogenized with 20 strokes (with a B pestle) in a Dounce homogenizer at 4°C. The cell lysate was clarified by low-speed centrifugation (1,000 × g, 3 min). The supernatant containing cytoplasmic PICs was further clarified by high-speed centrifugation (8,000 × g, 3 min), and the salt concentration was adjusted to 150 mM. The supernatant containing cytoplasmic PICs was frozen in liquid nitrogen and stored at −70°C.
FIG. 7.
Stimulation of the intermolecular integration activity of partially purified but not salt-stripped PICs by purified DNA binding proteins. The PICs were purified by hypotonic lysis of cells followed by gel filtration through Sepharose CL-4B in 150 mM KCl. The PICs were then mixed with the purified protein at the concentrations indicated. All the reactions were performed under standard reaction conditions in a total volume of 110 μl. BSA, bovine serum albumin.
Salt-stripping of PICs.
One milliliter of crude PICs was partially purified by low-ionic-strength precipitation as described previously (19), except that 8,000 × g instead of 2,000 × g was used to pellet PICs. After low-ionic-strength precipitation, 80 to 90% of PICs could be recovered. After the PIC pellet was suspended in buffer A, 30% of the PICs could carry out intermolecular integration and no autointegration was detected. The protein concentration of the partially purified PICs was ∼0.18 mg/ml, and the concentration of viral DNA was ∼8 ng/ml.
To carry out high salt depletion, the pelleted PICs were suspended in 680 μl of buffer A with 600 mM KCl and incubated at room temperature for 10 min. The PICs were then subjected to gel filtration through Sepharose CL-4B spin columns equilibrated in buffer A with 600 mM KCl. The resulting salt-stripped PICs were concentrated by a Centricon-100 to a final volume of 50 to 100 μl. The concentrated salt-stripped PICs were then diluted to the desired volume with buffer A containing 600 mM KCl. Upon dilution to 150 mM KCl, the salt-stripped PICs lacked intermolecular integration activity and 10 to 15% of the PICs had autointegration activity.
Integration assay.
The integration assay was carried out in buffer A with 30% glycerol and 10 μg of linearized φX174 RFI per ml. The reaction mixture was incubated at 37°C for 30 min. In the assay for complementing activity, the cellular extract or purified protein was incubated with salt-stripped PICs at room temperature for 10 min. The reaction buffer was then adjusted to the condition described above and further incubated at room temperature for 10 min. Linearized φX174 RFI was added last, and the reaction mixture was incubated at 37°C for 30 min.
PICs containing about 500 pg of viral cDNA were used in each reaction mixture. The total reaction volume was adjusted to 110 μl in all experiments. The DNA was purified and analyzed by Southern blotting with a 32P-labeled probe containing an LTR sequence. The intermolecular integration and autointegration activities were quantitated by PhosphorImager analysis (see Fig. 3A and 7). The intermolecular integration activity is defined as the percentage of viral DNA integrated into added target DNA. The autointegration activity is defined as the percentage of viral DNA converted to the inverted circular form (autointegration into the opposite strand in Fig. 1) times two. The measured value is doubled, since integration into the same cDNA strand yields two circular species with different sizes from molecule to molecule. Hence, these circles do not migrate as a discrete form in gels and are not detected.
FIG. 3.
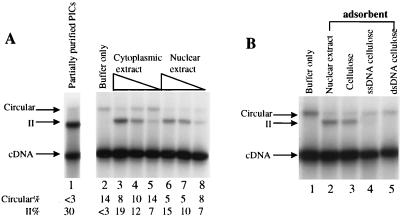
Integration-complementing activity and autointegration barrier activity of extracts of NIH 3T3 cells and DNA binding by the integration-complementing activity. (A) Reconstitution of salt-stripped MoMuLV PICs with cytoplasmic or nuclear extract. Lane 1 shows the activity of PICs purified by low-ionic-strength precipitation. The partially purified PICs were salt stripped and reconstituted with 10 μl of buffer alone (lane 2), cytoplasmic extract (10, 2, and 0.4 μl [lanes 3 to 5, respectively]) or nuclear extract (10, 2, and 0.4 μl [lanes 6 to 8, respectively]). Each reaction mixture was adjusted to standard solution conditions prior to addition of target DNA and incubated at 37°C for 30 min. The percentages of intermolecular integration activity (II%) and autointegration activity (circular%) were measured with a Molecular Dynamics PhosphorImager. (B) DNA binding by the integration-complementing activity. Salt-stripped PICs were reconstituted with 10 μl of buffer only (lane 1), 10 μl of nuclear extract (lane 2), 10 μl of extract depleted by cellulose (lane 3), single-stranded DNA-cellulose (lane 4), or double-stranded DNA-cellulose (lane 5).
Uninfected NIH 3T3 cell extract.
The cytoplasmic and nuclear extracts of NIH 3T3 cells were prepared as described previously (13). The protein concentration of each extract was adjusted to ∼4 μg/μl.
Purification of HMG proteins.
Uninfected NIH 3T3 cells (2 × 109) cells were scraped from 150-mm-diameter dishes, and HMG proteins were purified as described previously (18). The His-tagged HMG I(Y) was purified as described previously (18). HMG I and HMG Y are transcribed from the same gene but differ by an internal splice that removes 11 amino acids from HMG I to yield HMG Y (25). The two proteins usually cofractionate and function identically. Therefore, they are referred to together as HMG I(Y). The His-tagged variant described above actually encodes HMG I only.
For purification of HMG-1 [note that HMG I(Y) and HMG-1 are unrelated in sequence and function despite the similarity in names], pET11d-His-HMG-1 (a kind gift of Hui Ge) was transformed into HMS174(DE3) (Novagen). The bacteria containing pET11d-His-HMG-1 were grown to mid-log phase and induced by isopropyl-β-d-thiogalactopyranoside (IPTG) at a final concentration of 1 mM. After 2 h of growth at 37°C, the bacteria were harvested and His-tagged HMG-1 was purified as described by the manufacturer (Novagen).
RESULTS
Depletion of required factors from MoMuLV PICs with high concentrations of salt.
To investigate host activities involved in integration, a salt-stripping protocol was tested to determine whether it removed factors required for integration by MoMuLV PICs, as had previously been observed for HIV-1 PICs (18). MoMuLV PICs were prepared from newly infected cells as described previously (3, 27) and partially purified by precipitation in the presence of low-ionic-strength buffers (75 mM KCl). Thirty percent of the partially purified PICs could integrate their viral cDNA into added φX174 target DNA, and no autointegration activity was detected (Fig. 2B, lane 2; see Fig. 3A, lane 1, also). To deplete the partially purified PICs of protein factors, low-salt-precipitated PICs were suspended in high-salt buffer (600 mM KCl) and subjected to gel filtration through Sepharose CL-4B in 600 mM KCl. Complexes such as PICs with a molecular mass greater than 2 × 107 Da passed through the column but free proteins were retained.
FIG. 2.
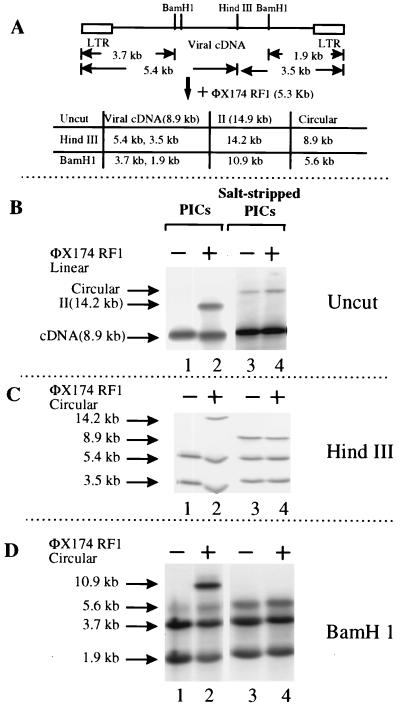
Analysis of intermolecular integration and autointegration products by restriction enzyme digestion. (A) Map of the MoMuLV genome and sizes of DNA fragments from substrates (viral cDNA and φX174 RFI), integration product (II), and autointegration products (circular) expected to be detected by the LTR probe. (B to D) Southern blot analyses of integration products generated by PICs partially purified by low-ionic-strength precipitation (lanes 1 and 2) or salt-stripped PICs (lanes 3 and 4) in the absence (−) or presence (+) of linearized target DNA (B) or circular target DNA (C and D). DNAs were digested with HindIII (C) or BamHI (D) prior to electrophoresis.
As previously reported (41), the resulting salt-stripped PICs had autointegration activity, as indicated by the formation of a circular form of the viral cDNA, but lacked intermolecular integration activity, as indicated by a disappearance of form II (Fig. 2B, lane 4). Note that the production of the circular autointegration product was independent of the presence or absence of added φX174 target DNA (Fig. 2B, compare lanes 3 and 4). These results indicated that the salt-stripping process removed factors required both for the protection of viral cDNA from autointegration and for efficient intermolecular integration.
Analysis of reaction products by cleavage with restriction enzymes.
To verify that the DNA products were those illustrated in Fig. 1, the structures of the reaction products were probed by cleavage with restriction enzymes (Fig. 2C and D). HindIII cleaves MoMuLV cDNA once but does not cut the φX174 target DNA. BamHI cleaves MoMuLV cDNA three times but does not cleave the target. Reaction products made with (Fig. 2, lanes 2 and 4) or without target DNA (Fig. 2, lanes 1 and 3) were digested with HindIII (Fig. 2C) or BamHI (Fig. 2D). The sizes of fragments expected to be detected by the LTR probe are shown in Fig. 2A, and sizes detected on Southern blots are shown beside the autoradiograms. Note that the expected autointegration product described is the inverted-circle form. A second autointegration product, pairs of DNA circles, are not detected by this method (see Materials and Methods and reference 22 for details). All the DNA sizes observed in Fig. 2B to D are as expected by the model in Fig. 1 and Fig. 2A, indicating that the observed DNA bands are likely the expected intermolecular integration and autointegration products. A similar analysis of MoMuLV autointegration by cleavage with BamHI was published previously (41).
Reconstitution of salt-stripped PICs with host cell extract.
The nature of the factor(s) required to restore intermolecular integration and block autointegration was then investigated. Cytoplasmic or nuclear extract from NIH 3T3 cells was used to reconstitute salt-stripped PICs (Fig. 3A, lanes 3 to 8). Each extract was able to restore intermolecular integration (indicated by the arrow labeled II) and showed some ability to block autointegration.
The intermolecular integration-complementing activity binds DNA.
To test whether the activity required to reconstitute salt-stripped PICs was a DNA binding protein, depletion tests were carried out. Nuclear extract from uninfected cells was incubated with cellulose, single-stranded DNA-cellulose, or double-stranded DNA-cellulose. The cellulose was then removed by centrifugation, and the remaining supernatant was used to complement salt-stripped PICs. The nuclear extract depleted by cellulose could complement intermolecular integration (Fig. 3B, lane 3). However, extract depleted by single- or double-stranded DNA-cellulose failed to complement (Fig. 3B, lanes 4 and 5). Evidently the complementing activity is contributed at least in part by a DNA binding protein, particularly one that can bind both single- and double-stranded DNA.
Analysis of the target of the complementing activity by order-of-addition experiments.
To investigate whether the complementing factor acts on the salt-stripped PIC or the target DNA, an order-of-addition experiment was performed. The salt-stripped PICs were incubated with nuclear extract either before or after addition of target DNA. Note that the φX174 target DNA was present in great excess over viral DNA (10 μg/ml versus 8 ng/ml), so the φX174 DNA can act as a sink for added DNA-binding factors as well as an integration target. Premixing target DNA with complementing nuclear extract substantially reduced intermolecular integration upon addition of salt-stripped PICs (Fig. 4, compare lanes 2 to 4 with lanes 5 to 7). These findings support a model in which the complementing activity acts on PICs and not on target DNA.
FIG. 4.
Analysis of the site of action of nuclear extract by order of addition. Two orders of addition were compared: (i) addition of nuclear extract to salt-stripped PICs and then addition of target DNA (lanes 2 to 4) or (ii) addition of nuclear extract to target DNA and then addition of salt-stripped PICs (lanes 5 to 7). The amounts of nuclear extract added per reaction mixture are indicated above the autoradiogram. Lane 1 (Buffer) contained no added protein. The amounts of reactants were as follows: φX174 DNA, 2.6 × 10−9 M; HMG I(Y) in 10 μl of nuclear extract, 1.1 × 10−8 M; and cDNA in PICs, 8.3 × 10−13 M. The number of binding sites for HMG I(Y) in φX174 DNA is not known but is likely to be large.
Fractionation of HMG proteins in NIH 3T3 extract identified HMG I(Y) as the intermolecular integration-complementing factor.
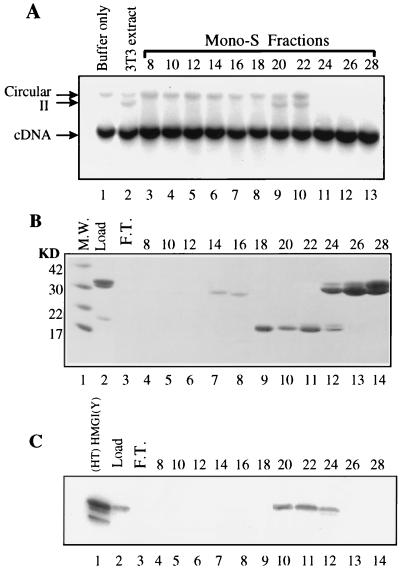
Since the factor restoring activity to salt-stripped PICs of HIV-1 was also a DNA binding protein and proved to be an HMG protein (18), HMG proteins were tested for the ability to complement salt-stripped MoMuLV PICs. Total HMG proteins were extracted from NIH 3T3 cells with 5% perchloric acid and loaded onto a Mono-S ion-exchange column. The column was eluted with a NaCl concentration gradient. Fractions were analyzed for complementing activity (Fig. 5A) and for protein content by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis followed by staining with Coomassie brilliant blue (Fig. 5B). Fractions 20 and 22 complemented intermolecular integration activity of salt-stripped PICs (Fig. 5A, lanes 10 and 11).
FIG. 5.
Fractionation of HMG proteins of NIH 3T3 cells identified HMG I(Y) as the intermolecular integration-complementing activity. (A) Analysis of the activity of HMG fractions eluted from a Mono-S column. The HMG proteins were extracted from uninfected NIH 3T3 cells (3T3 extract) with 5% perchloric acid and loaded onto a Mono-S column. Proteins were then eluted with a NaCl concentration gradient. Fractions 8 to 28 were collected and assayed for complementing activity. Salt-stripped PICs were incubated with buffer only (lane 1), 2 μl of nuclear extract (lane 2), or fractions 8 to 28 (lanes 3 to 13). (B) Analysis of the Mono-S fractions by SDS–15% polyacrylamide gel electrophoresis. The gel was stained with Coomassie brilliant blue. Molecular mass (M.W.) standards (in kilodaltons) are shown in lane 1. (C) Western blot analysis of Mono-S fractions with an anti-HMG I(Y) antibody.
Proteins in fractions 20 and 22 had the mobility on SDS-polyacrylamide gels and the fractionation profile on the Mono-S ion-exchange column expected for HMG I(Y) (18). Western blot analysis using an anti-HMG I(Y) antibody established that these fractions contained HMG I(Y) (Fig. 5C).
In addition to fractions 20 and 22, fraction 24 also contained HMG I(Y) but lacked complementing activity (Fig. 5A, lane 11, and Fig. 5C, lane 12). However, this fraction also contained histone H1 (Fig. 5B, lanes 12 to 14, band at about 30 kDa) which was previously found to inhibit intermolecular integration (18, 18a). Histone H1 also inhibited autointegration (Fig. 5A, lanes 11 to 13). Therefore, with the exception of the fractions which contained inhibitory histone H1, all the fractions containing HMG I(Y) were able to complement intermolecular integration by salt-stripped PICs.
As described above, the intermolecular integration-complementing activity was present in both cytoplasmic and nuclear fractions, as is HMG I(Y) (18). The relative amounts of activity in each did not correlate with the abundance of HMG I(Y) in a simple way since each fraction is a mixture of stimulatory HMG I(Y) and inhibitory factors such as histone H1 (unpublished data). In addition, the complementing activity could bind to both single- and double-stranded DNA-cellulose, as is known to be the case for HMG I(Y) (18). These observations further support the attribution of the intermolecular integration-complementing activity to HMG I(Y).
Complementation with purified proteins.
To confirm that HMG I(Y) was the intermolecular integration-complementing activity, recombinant HMG I(Y) protein was purified after overexpression in bacteria and tested. In this experiment, HMG I(Y) was attached to a hexahistidine tag for convenient purification [named (HT)HMG I(Y)]. In the presence of the purified (HT)HMG I(Y), the intermolecular integration activity of salt-stripped PICs was partially restored (Fig. 6, lanes 2 to 4). However, (HT)HMG I(Y) had consistently lower complementing activity than uninfected-cell nuclear extract. Ten microliters of nuclear extract, which contained only 15 ng of HMG I(Y) (Fig. 3A, lane 6), was sufficient for maximal reconstitution of intermolecular integration activity, while 1 μg of (HT)HMG I(Y) was required (Fig. 6, lane 3). In addition, the maximal achievable activity was lower with (HT)HMG I(Y).
FIG. 6.
Reconstitution of salt-stripped PICs with purified proteins. Salt-stripped PICs were reconstituted with the indicated purified protein at the concentrations indicated over the gels. All the reactions were performed under the standard reaction conditions in a total volume of 110 μl. BSA, bovine serum albumin.
Several other DNA binding proteins, including HMG-1 [a HMG protein unrelated to HMG I(Y) in sequence and function], HIV-1 nucleocapsid (NC), RNase A, E. coli single-stranded binding protein (Fig. 6) and T4 phage gene 32 protein (data not shown) were also tested for complementing activity. At all concentrations tested, none of these DNA binding proteins displayed complementing activity.
A separate requirement for blocking autointegration.
Several observations indicate that the activity that restores intermolecular integration is different from the previously reported factor (41) that blocks autointegration. (i) Although nuclear extract complemented salt-stripped PICs as well as cytoplasmic extract, nuclear extract had much higher autointegration blocking activity (2 μl of nuclear extract could reduce autointegration from 14 to 8%, while 2 μl of cytoplasmic extract had no detectable effect on autointegration [both extracts at 4 μg/μl] [Fig. 3A]). These results suggested that different factors were responsible for each activity. (ii) DNA cellulose depleted the integration-complementing activity more efficiently than the autointegration-blocking activity (Fig. 3B). (iii) Upon fractionation of HMG proteins (Fig. 5), although fractions capable of complementing intermolecular integration by salt-stripped PICs were found, no clear autointegration-blocking activity was detected apart from inhibition of both reactions by histone H1. (iv) (HT)HMG I(Y) from a bacterial expression system could restore the intermolecular integration activity but could not block autointegration (Fig. 6, lanes 2 to 4).
Stimulation of intermolecular integration by DNA binding proteins.
In addition, another assay method revealed a different response to added DNA binding proteins. The activity of partially purified PICs which were not salt stripped was found in some cases to be reduced. However, the activity of such complexes could be stimulated by addition of any of several different DNA binding proteins. This observation may parallel previous reports of many DNA binding proteins stimulating the activity of purified integrase proteins in vitro (1, 6, 8, 10, 26, 36, 43).
To test for stimulatory factors, PICs were obtained from a cytoplasmic fraction by hypotonic lysis and subjected to gel filtration through Sepharose CL-4B in 150 mM KCl. Such PICs displayed low intermolecular integration activity (roughly 7% integration compared to the usual 30%). However, when these complexes were incubated with selected purified DNA binding proteins, the intermolecular integration activity was increased (Fig. 7). In the case of HMG I(Y), 1 μg of the protein stimulated the reaction twofold (Fig. 7, lane 3). Addition of 5 μg of the protein was inhibitory (Fig. 7, lane 2). HMG-1 and HIV-1 NC displayed threefold stimulation (Fig. 7, lanes 5 to 12). Several single-stranded binding proteins (RNase A and E. coli SSB [Fig. 7, lanes 13 to 20] and phage T4 gene 32 protein [data not shown]) were also tested, since they were previously found to stimulate the activity of purified MoMuLV integrase protein in vitro (12a). At the highest concentration, the integration activity was increased by 1.5- to twofold (Fig. 7, lanes 13 to 20). Addition of bovine serum albumin did not stimulate the reaction (Fig. 7, lanes 21 to 24), indicating that simply increasing the bulk protein concentration did not account for the observed stimulation. Similar results were obtained (data not shown) when PICs were partially purified by a different method (digitonin lysis followed by low-ionic-strength precipitation and gel filtration through Sepharose CL-4B in 150 mM KCl). The observed diversity of DNA binding proteins that satisfy this requirement is in contrast to the strict requirement for HMG I(Y) in reconstituting intermolecular integration by salt-stripped PICs.
DISCUSSION
Here we report the isolation of a factor that restores intermolecular integration by salt-stripped MoMuLV PICs and its identification as murine HMG I(Y). A requirement for human HMG I(Y) was reported previously for HIV-1 PICs (18). The previously described autointegration barrier factor (41) was also detected in our experiments and found to be separate from HMG I(Y). We also identified another activity. When PICs were partially purified but were not salt stripped, they often had reduced intermolecular integration activity. This reduced activity permitted another approach to reconstitution, which revealed a requirement that could be satisfied by several different DNA binding proteins.
Possible roles of HMG I(Y).
The mechanism by which HMG I(Y) complements the intermolecular integration by salt-stripped PICs is unclear, although several models can be proposed based on previous work. HMG I(Y) binds A/T-rich DNA sequences and such sequences are present in the LTRs of MoMuLV and HIV-1 (17, 42, 48, 49, 53). Each HMG I(Y) monomer has three DNA binding motifs (48) and one HMG I(Y) molecule can bridge two DNAs by binding to sites on each (33), leading to the possibility that HMG I(Y) acts by bringing distant DNA domains into proximity.
One possible model is that one HMG I(Y) molecule binds both viral cDNA and target DNA, thus bridging the PIC and target. However, a study of 61 integration sites generated by HIV-1 infection did not show an enrichment for HMG I(Y) binding sites near integration sites (10a), which is evidence against this model. Furthermore, the order-of-addition experiment in Fig. 4 also indicates that HMG I(Y) probably acts on PICs and not on target DNA.
An attractive model holds that HMG I(Y) may serve as an architectural element for the formation of active PICs, helping shape the DNA for optimal function. Host DNA binding proteins have been shown to serve as architectural elements in the assembly of recombination complexes in several well-studied site-specific recombination and transposition systems (31, 40, 45). In addition, HMG I(Y) itself has been proposed to serve as an architectural element in the assembly of transcription complexes (for a review see reference 9).
HMG I(Y) protein is not the autointegration barrier activity.
Our data indicate that HMG I(Y) is not the autointegration barrier activity. Fractionation of murine HMG proteins yielded an activity that stimulated intermolecular integration by salt-stripped PICs, but no activity that redirected autointegration to intermolecular integration was seen. Purified HMG I(Y) could restore intermolecular integration activity of salt-stripped PICs but could not block autointegration. In addition, the relative amounts of the two activities differed in nuclear and cytoplasmic fractions of uninfected host cells, consistent with different factors accounting for each.
Stimulation of partially purified PICs by diverse DNA binding proteins.
We found that MoMuLV PICs that had been partially purified but not salt stripped were often diminished in intermolecular integration activity, and the activity could be boosted two- to fourfold by addition of any of several DNA binding proteins. RNase A, T4 gene 32 protein, E. coli SSB, HMG I(Y), HIV-1 NC, and HMG-1 all supported this activity. This stimulation is distinct from the reconstitution of intermolecular integration activity of salt-stripped PICs, since only HMG I(Y) could reconstitute in this setting. Of the proteins that stimulated PICs in the two assays, so far only HMG I(Y) has been found in association with PICs (in the HIV-1 system [18]). Similar composition studies have not yet been possible in the MoMuLV system.
It is unclear how these diverse DNA binding proteins promote integration in vitro. Previously many of these proteins have been found to stimulate reactions with purified integrase proteins. Purified MoMuLV integrase was stimulated 3 to 13-fold by E. coli SSB, RNase A, and T4 gene 32 protein (12a), purified ASLV integrase was stimulated two- to fourfold by HMG-1 and HU proteins (1), and purified HIV-1 integrase was stimulated very strongly by NC protein (10), two- to fourfold by RNase T1 (43), 10 to 20-fold by Ini-1 (36), and strongly by a cocktail of DNA binding proteins (8). The fact that so many proteins support this activity raises the possibility that the mechanism involves nonselective coating of DNA, perhaps by shielding negative charges on the DNA phosphates (though for a different view, see reference 1). It will be of interest to determine whether this activity is important for integration in vivo and, if so, what proteins are involved.
Possible further host activities important for the function of PICs.
HMG I(Y) purified after overexpression in E. coli was less active for complementation than extracts from uninfected NIH 3T3 cells, raising the possibility that still other factors are involved. Furthermore, in the HIV-1 case, purified HMG I(Y) also did not reconstitute activity as well as crude extracts did (18). The complementation experiments illustrated in Fig. 6 used human HMG I, not murine HMG I which differs from human HMG I at two amino acid positions (34). Therefore, it is also possible that this difference explains the discrepancy in activities. HMG I(Y) has been reported to be posttranslationally modified by glycosylation, phosphorylation, acetylation, methylation, and adenylation (46, 47). Possibly, the lack of posttranslational modifications in the purified HMG I(Y) accounts for the low complementing activity. Alternatively, further protein factors may be involved. The methods for stripping and reconstituting PICs described offer an approach to identifying possible additional factors.
ACKNOWLEDGMENTS
We thank members of the Bushman lab and Leonard Evans, Mike Januzeski, Paula Cannon, and Amy Lin for suggestions and comments on the manuscript; S. Goff, A. Dusty Miller, and Hung Fan for MoMuLV cDNA; and Hui Ge for the HMG-1 expression vector.
This work was supported by Genetic Therapy Inc./Novartis and by grants AI34786 and AI37489 to F.D.B. F.D.B. is a Scholar of the Leukemia Society of America.
REFERENCES
- 1.Aiyar A, Hindmarsh P, Skalka A M, Leis J. Concerted integration of linear retroviral DNA by the avian sarcoma virus integrase in vitro: dependence on both long terminal repeat termini. J Virol. 1996;70:3571–3580. doi: 10.1128/jvi.70.6.3571-3580.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bowerman B, Brown P O, Bishop J M, Varmus H E. A nucleoprotein complex mediates the integration of retroviral DNA. Genes Dev. 1989;3:469–478. doi: 10.1101/gad.3.4.469. [DOI] [PubMed] [Google Scholar]
- 3.Brown P O, Bowerman B, Varmus H E, Bishop J M. Correct integration of retroviral DNA in vitro. Cell. 1987;49:347–356. doi: 10.1016/0092-8674(87)90287-x. [DOI] [PubMed] [Google Scholar]
- 4.Brown P O, Bowerman B, Varmus H E, Bishop J M. Retroviral integration: structure of the initial covalent complex and its precursor, and a role for the viral IN protein. Proc Natl Acad Sci USA. 1989;86:2525–2529. doi: 10.1073/pnas.86.8.2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bukrinsky M I, Sharova N, McDonald T L, Pushkarskaya T, Tarpley G W, Stevenson M. Association of integrase, matrix, and reverse transcriptase antigens of human immunodeficiency virus type 1 with viral nucleic acids following acute infection. Proc Natl Acad Sci USA. 1993;90:6125–6129. doi: 10.1073/pnas.90.13.6125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bushman F D, Craigie R. Sequence requirements for integration of Moloney murine leukemia virus DNA in vitro. J Virol. 1990;64:5645–5648. doi: 10.1128/jvi.64.11.5645-5648.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bushman F D, Craigie R. Activities of human immunodeficiency virus (HIV) integration protein in vitro: specific cleavage and integration of HIV DNA. Proc Natl Acad Sci USA. 1991;88:1339–1343. doi: 10.1073/pnas.88.4.1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bushman F D, Fujiwara T, Craigie R. Retroviral DNA integration directed by HIV integration protein in vitro. Science. 1990;249:1555–1558. doi: 10.1126/science.2171144. [DOI] [PubMed] [Google Scholar]
- 9.Bustin M, Reeves R. High-mobility-group chromosomal proteins: architectural components that facilitate chromatin Function. Prog Nucleic Acid Res Mol Biol. 1996;••:35–100. doi: 10.1016/s0079-6603(08)60360-8. [DOI] [PubMed] [Google Scholar]
- 10.Carteau S, Batson S C, Poljak L, Mouscadet J-F, Rocquigny H, Darlix J-L, Roques B P, Kas E, Auclair C. Human immunodeficiency virus type 1 nucleocapsid protein specifically stimulates Mg2+-dependent DNA integration in vitro. J Virol. 1997;71:6225–6229. doi: 10.1128/jvi.71.8.6225-6229.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10a.Carteau, S., C. Hoffmann, and F. D. Bushman. Submitted for publication.
- 11.Coffin J M. Retroviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley R M, editors. Virology. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 1767–1848. [Google Scholar]
- 12.Craigie R, Fujiwara T, Bushman F. The IN protein of Moloney murine leukemia virus processes the viral DNA ends and accomplishes their integration in vitro. Cell. 1990;62:829–837. doi: 10.1016/0092-8674(90)90126-y. [DOI] [PubMed] [Google Scholar]
- 12a.Craigie, R., and F. D. Bushman. Unpublished data.
- 13.Dignam J D, Martin P L, Shastry B S, Roeder R G. Eukaryotic gene transcription with purified components. Methods Enzymol. 1983;101:582–598. doi: 10.1016/0076-6879(83)01039-3. [DOI] [PubMed] [Google Scholar]
- 14.Donehower L A, Varmus H E. A mutant murine leukemia virus with a single missense codon in pol is defective in a function affecting integration. Proc Natl Acad Sci USA. 1984;81:6461–6465. doi: 10.1073/pnas.81.20.6461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dotan I, Scottoline B P, Heuer T S, Brown P T. Characterization of recombinant murine leukemia virus integrase. J Virol. 1995;69:456–468. doi: 10.1128/jvi.69.1.456-468.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ellison V H, Abrams H, Roe T, Lifson J, Brown P O. Human immunodeficiency virus integration in a cell-free system. J Virol. 1990;64:2711–2715. doi: 10.1128/jvi.64.6.2711-2715.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Elton T S, Nissen M S, Reeves R. Specific A · T sequence binding of RP-HPLC purified HMG-I. Biochem Biophys Res Commun. 1987;143:260–265. doi: 10.1016/0006-291x(87)90659-0. [DOI] [PubMed] [Google Scholar]
- 18.Farnet C, Bushman F D. HIV-1 cDNA integration: requirement of HMG I(Y) protein for function of preintegration complexes in vitro. Cell. 1997;88:1–20. doi: 10.1016/s0092-8674(00)81888-7. [DOI] [PubMed] [Google Scholar]
- 18a.Farnet, C., and F. D. Bushman. Unpublished data.
- 19.Farnet C, Lipford R, Wang B, Bushman F D. Differential inhibition of HIV-1 preintegration complexes and purified integrase protein by small molecules. Proc Natl Acad Sci USA. 1996;93:9742–9747. doi: 10.1073/pnas.93.18.9742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Farnet, C. M., and F. D. Bushman. 1996. HIV cDNA integration: molecular biology and inhibitor development. AIDS 10(Suppl. A):3–11. [PubMed]
- 21.Farnet C M, Haseltine W A. Integration of human immunodeficiency virus type 1 DNA in vitro. Proc Natl Acad Sci USA. 1990;87:4164–4168. doi: 10.1073/pnas.87.11.4164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Farnet C M, Haseltine W A. Circularization of human immunodeficiency virus type 1 DNA in vitro. J Virol. 1991;65:6942–6952. doi: 10.1128/jvi.65.12.6942-6952.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Farnet C M, Haseltine W A. Determination of viral proteins present in the human immunodeficiency virus type 1 preintegration complex. J Virol. 1991;65:1910–1915. doi: 10.1128/jvi.65.4.1910-1915.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fitzgerald M L, Vora A C, Zeh W G, Grandgenett D P. Concerted integration of viral DNA termini by purified avian myeloblastosis virus integrase. J Virol. 1992;66:6257–6263. doi: 10.1128/jvi.66.11.6257-6263.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Friedmann M, Holth L T, Zoghbi H, Reeves R. Organization, inducible-expression, and chromosome localization of the human HMG I(Y) nonhistone protein gene. Nucleic Acids Res. 1993;21:4259–4267. doi: 10.1093/nar/21.18.4259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fujiwara T, Craigie R. Integration of mini-retroviral DNA: a cell-free reaction for biochemical analysis of retroviral integration. Proc Natl Acad Sci USA. 1989;86:3065–3069. doi: 10.1073/pnas.86.9.3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fujiwara T, Mizuuchi K. Retroviral DNA integration: structure of an integration intermediate. Cell. 1988;54:497–504. doi: 10.1016/0092-8674(88)90071-2. [DOI] [PubMed] [Google Scholar]
- 28.Gallay P, Swingler S, Song J, Bushman F, Trono D. HIV nuclear import is governed by the phosphotyrosine-mediated binding of matrix to the core domain of integrase. Cell. 1995;17:569–576. doi: 10.1016/0092-8674(95)90097-7. [DOI] [PubMed] [Google Scholar]
- 29.Goodarzi G, Im G-J, Brackmann K, Grandgenett D. Concerted integration of retrovirus-like DNA by human immunodeficiency virus type 1 integrase. J Virol. 1995;69:6090–6097. doi: 10.1128/jvi.69.10.6090-6097.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grandgenett D P, Vora A C, Schiff R D. A 32,000-dalton nucleic acid-binding protein from avian retrovirus cores possesses DNA endonuclease activity. Virology. 1978;89:119–132. doi: 10.1016/0042-6822(78)90046-6. [DOI] [PubMed] [Google Scholar]
- 31.Heichman K A, Johnson R C. The Hin invertosome: protein-mediated joining of distant recombination sites at the enhancer. Science. 1990;249:511–517. doi: 10.1126/science.2166334. [DOI] [PubMed] [Google Scholar]
- 32.Heinzinger N K, Bukrinsky M I, Haggerty S A, Ragland A M, Kewalramani V, Lee M-A, Gendelman H E, Ratner L, Stevenson M, Emerman M. The Vpr protein of human immunodeficiency virus type 1 influences nuclear localization of viral nucleic acids in nondividing host cells. Proc Natl Acad Sci USA. 1994;91:7311–7315. doi: 10.1073/pnas.91.15.7311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huth J R, Bewley C A, Nissen M S, Evans J N S, Reeves R, Gronenborn A M, Clore G M. The solution structure of an HMG I(Y)-DNA complex defines a new architectural minor groove binding motif. Nature Struct Biol. 1997;4:657–665. doi: 10.1038/nsb0897-657. [DOI] [PubMed] [Google Scholar]
- 34.Johnson K R, Lehn D A, Elton T S, Barr P J, Reeves R. Complete murine cDNA sequence, genomic structure, and tissue expression of the high mobility group protein HMG I(Y) J Biol Chem. 1988;263:18338–18342. [PubMed] [Google Scholar]
- 35.Jonsson C B, Donzella G A, Roth M J. Characterization of the forward and reverse integration reactions of the Moloney murine leukemia virus integrase protein purified from Escherichia coli. J Biol Chem. 1993;268:1–8. [PubMed] [Google Scholar]
- 36.Kalpana G V, Marmon S, Wang W, Crabtree G R, Goff S P. Binding and stimulation of HIV-1 integrase by a human homolog of yeast transcription factor SNF5. Science. 1994;266:2002–2006. doi: 10.1126/science.7801128. [DOI] [PubMed] [Google Scholar]
- 37.Katz R A, Merkel G, Kulkosky J, Leis J, Skalka A M. The avian retroviral IN protein is both necessary and sufficient for integrative recombination in vitro. Cell. 1990;63:87–95. doi: 10.1016/0092-8674(90)90290-u. [DOI] [PubMed] [Google Scholar]
- 38.Katzman M, Katz R A, Skalka A M, Leis J. The avian retroviral integration protein cleaves the terminal sequences of linear viral DNA at the in vivo sites of integration. J Virol. 1989;63:5319–5327. doi: 10.1128/jvi.63.12.5319-5327.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lam W-C, Maki A H, Casas-Finet J R, Erickson J W, Kane B P, Sowder R C, Henderson L E. Phosphorescence and optically detected magnetic resonance investigation of the binding of the nucleocapsid protein of the human immunodeficiency virus type 1 and related peptides to RNA. Biochemistry. 1994;33:10693–10700. doi: 10.1021/bi00201a017. [DOI] [PubMed] [Google Scholar]
- 40.Lavoie B D, Shaw G S, Millner A, Chaconas G. Anatomy of a Flexer-DNA complex inside a higher-order transposition intermediate. Cell. 1996;85:761–771. doi: 10.1016/s0092-8674(00)81241-6. [DOI] [PubMed] [Google Scholar]
- 41.Lee M S, Craigie R. Protection of retroviral DNA from autointegration: involvement of a cellular factor. Proc Natl Acad Sci USA. 1994;91:9823–9827. doi: 10.1073/pnas.91.21.9823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maher J F, Nathans D. Multivalent DNA-binding properties of the HMG-I proteins. Proc Natl Acad Sci USA. 1996;93:6716–6720. doi: 10.1073/pnas.93.13.6716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miller M, Bor Y-C, Bushman F D. Target DNA capture by HIV-1 integration complexes. Curr Biol. 1995;5:1047–1056. doi: 10.1016/s0960-9822(95)00209-0. [DOI] [PubMed] [Google Scholar]
- 44.Miller M D, Farnet C M, Bushman F D. Human immunodeficiency virus type 1 preintegration complexes: studies of organization and composition. J Virol. 1997;71:5382–5390. doi: 10.1128/jvi.71.7.5382-5390.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nash H A. Bending and supercoiling of DNA at the attachment site of bacteriophage λ. Trends Biochem Sci. 1990;15:222–227. doi: 10.1016/0968-0004(90)90034-9. [DOI] [PubMed] [Google Scholar]
- 46.Nissen M S, Langan T A, Reeves R. Phosphorylation by cdc2 kinase modulates DNA binding activity of high mobility group-I nonhistone chromatin protein. J Biol Chem. 1991;266:19945–19952. [PubMed] [Google Scholar]
- 47.Reeves R, Langan T A, Nissen M S. Phosphorylation of the DNA-binding domain of nonhistone high-mobility group I protein by cdc2 kinase: reduction of binding affinity. Proc Natl Acad Sci USA. 1991;88:1671–1675. doi: 10.1073/pnas.88.5.1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reeves R, Nissen M S. The A · T-binding domain of mammalian high mobility group I chromosomal proteins. J Biol Chem. 1990;265:8573–8582. [PubMed] [Google Scholar]
- 49.Reeves R, Wolffe A P. Substrate structure influences binding of the non-histone protein HMG I(Y) to free and nucleosomal DNA. Biochemistry. 1996;35:5063–5074. doi: 10.1021/bi952424p. [DOI] [PubMed] [Google Scholar]
- 50.Schwartzberg P, Colecilli J, Goff S P. Construction and analysis of deletion mutations in the pol gene of Moloney murine leukemia virus: a new viral function required for productive infection. Cell. 1984;37:1043–1052. doi: 10.1016/0092-8674(84)90439-2. [DOI] [PubMed] [Google Scholar]
- 51.Sherman P A, Fyfe J A. Human immunodeficiency virus integration protein expressed in Escherichia coli possesses selective DNA cleaving activity. Proc Natl Acad Sci USA. 1990;87:5119–5123. doi: 10.1073/pnas.87.13.5119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.South T L, Blake P R, Sowder R C, Arthur L O, Henderson L E, Summers M F. The nucleocapsid protein isolated from HIV-1 particles binds zinc and forms retroviral-type zinc fingers. Biochemistry. 1990;29:7786–7789. doi: 10.1021/bi00486a002. [DOI] [PubMed] [Google Scholar]
- 53.Strauss F, Varshavsky A. A protein binds to a satellite DNA repeat at three specific sites that would be brought into mutual proximity by DNA folding in the nucleosome. Cell. 1984;37:889–901. doi: 10.1016/0092-8674(84)90424-0. [DOI] [PubMed] [Google Scholar]
- 54.Varmus H E, Brown P O. Retroviruses. In: Berg D E, Howe M M, editors. Mobile DNA. Washington, D.C: American Society for Microbiology; 1989. pp. 53–108. [Google Scholar]
- 55.Vora A C, Grandgenett D P. Assembly and catalytic properties of retrovirus integrase-DNA complexes capable of efficiently performing concerted integration. J Virol. 1995;69:7483–7488. doi: 10.1128/jvi.69.12.7483-7488.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]