Summary
Background
Chemoresistance is a critical factor contributing to poor prognosis in clinical patients with cancer undergoing postoperative adjuvant chemotherapy. The role of gut microbiota in mediating resistance to tumour chemotherapy remains to be investigated.
Methods
Patients with CRC were categorised into clinical benefit responders (CBR) and no clinical benefit responders (NCB) based on chemotherapy efficacy. Differential bacterial analysis using 16S rRNA sequencing revealed Desulfovibrio as a distinct microbe between the two groups. Employing a syngeneic transplantation model, we assessed the effect of Desulfovibrio on chemotherapy by measuring tumour burden, weight, and Ki-67 expression. We further explored the mechanisms underlying the compromised chemotherapeutic efficacy of Desulfovibrio using metabolomics, western blotting, colony formation, and cell apoptosis assays.
Findings
In comparison, Desulfovibrio was more abundant in the NCB group. In vivo experiments revealed that Desulfovibrio colonisation in the gut weakened the efficacy of FOLFOX. Treatment with Desulfovibrio desulfuricans elevates serum S-adenosylmethionine (SAM) levels. Interestingly, SAM reduced the sensitivity of CRC cells to FOLFOX, thereby promoting the growth of CRC tumours. These experiments suggest that SAM promotes the growth and metastasis of CRC by driving the expression of methyltransferase-like 3 (METTL3).
Interpretation
A high abundance of Desulfovibrio in the intestines indicates poor therapeutic outcomes for postoperative neoadjuvant FOLFOX chemotherapy in CRC. Desulfovibrio drives the manifestation of METTL3 in CRC, promoting resistance to FOLFOX chemotherapy by increasing the concentration of SAM.
Funding
This study is supported by Wuxi City Social Development Science and Technology Demonstration Project (N20201005).
Keywords: Colorectal cancer, FOLFOX, Desulfovibrio, S-adenosylmethionine, Methyltransferase-like 3
Research in context.
Evidence before this study
Chemoresistance is a significant issue leading to reduced treatment efficacy in clinical patients with CRC. Increasing evidence indicates that the gut microbiota affects the efficacy of patients with CRC. For example, Fusobacterium nucleatum confers resistance to oxaliplatin and 5-FU by inducing protective autophagy, thereby preventing cell apoptosis.1, 2, 3 Fusobacterium nucleatum has been demonstrated to promote the activation of fatty acid oxidation and Notch signalling to enhance the self-renewal of colon cancer stem cells.4 Bacteroides vulgatus mediates the attenuation of nucleotide synthesis in response to radiation and chemotherapy in rectal cancer.5 The FOLFOX regimen is the preferred choice for postoperative adjuvant chemotherapy in clinical patients with cancer. Desulfovibrio exacerbates the progression of colitis, thereby promoting the development of CRC.6, 7, 8 However, the relationship between Desulfovibrio and postoperative FOLFOX adjuvant chemotherapy remains unclear.
Added value of this study
We observed an increased abundance of Desulfovibrio in the NCB group, which was higher than that observed in the CBR group. Desulfovibrio reduces the efficacy of FOLFOX chemotherapy. The concentration of SAM can be elevated by Desulfovibrio; indeed, SAM induces a decrease in the reactivity of CRC cells to chemotherapeutic agents through the increased expression of METTL3, leading to treatment resistance. Knockdown expression of METTL3 diminishes the growth of CRC cells and enhanced their responsiveness to FOLFOX.
Implications of all the available evidence
This study provides evidence that Desulfovibrio is a detrimental factor in CRC chemotherapy. METTL3 is involved in the attenuation of FOLFOX chemotherapy efficacy by Desulfovibrio. The knockdown of METTL3 expression offers a new therapeutic approach for postoperative adjuvant chemotherapy.
Introduction
Colorectal cancer is one of the most prevalent malignant tumours of the gastrointestinal tract. The FOLFOX regimen is the classic chemotherapy protocol for advanced CRC or postoperative adjuvant chemotherapy for CRC. Therefore, we chose FOLFOX therapy for our study.9,10 Existing data indicate that the gut microbiota affects the efficacy of cancer treatment in patients with CRC. Our goal is to compare the different treatment outcomes among colorectal cancer patients receiving the same chemotherapy regimen, thereby elucidating the factors within the gut microbiota that interfere with chemotherapy effectiveness. By addressing this interference, we aim to benefit a larger number of patients from chemotherapy.
Desulfovibrio is a sulphate-reducing bacterium. In some cases, they may also be associated with diseases and health issues, especially as their presence within the gut microbiota can impact the host's health. Some studies have suggested that Desulfovibrio may be linked to gastrointestinal diseases, metabolic disorders, and other health conditions.6, 7, 8
Additionally, some studies have explored the association between Desulfovibrio and CRC, demonstrating that a high-fat diet increases the abundance of Desulfovibrio and enhances the liver metastatic potential of CRC.11 Importantly, as a member of the gut microbiota, Desulfovibrio is significantly enriched in the intestines and tumour sites of patients with CRC. This suggests that Desulfovibrio is likely to play a crucial role in the development, progression, and treatment of colorectal cancer. However, the specific mechanisms and functions require further investigation in research studies.12,13
Methods
Chemicals and reagents
Oxaliplatin was purchased from Hengrui (Jiangsu, China), 5-fluorouracil injection was obtained from Jinyao (Tianjin, China), and leucovorin calcium from MedChem Express (New Jersey, USA). FOLFOX injections were formulated according to the clinical guidelines and established research protocols. Vancomycin (V2002), ampicillin (A1593), neomycin sulphate (N6386), and metronidazole (M1547) were purchased from Aldrich (St. Aldrich, St. Louis, Missouri, USA).
Human faeces sample collection and preparation
A total of 61 faecal samples were collected from patients who underwent postoperative chemotherapy with the FOLFOX regimen at the Jiangnan University Affiliated Hospital (Wuxi, China) between September 2020 and February 2022. Among these, 49 samples were categorised into the clinical benefit group (CBR), which included patients with clinical computed tomography (CT) assessments classified as complete response (CR), partial response (PR), or stable disease (SD), and 12 samples were categorised into the non-clinical benefit group (NCB), which included patients with clinical CT assessments indicating disease progression (PD). We gathered faecal specimens and promptly stored them in a freezer at −80 °C. The exclusion criteria included diarrhoea or constipation, distinctive dietary behaviours, recent use of antibiotics, and invasive medical interventions in the past 3 months. This study was approved by the Ethics Committee of the Jiangnan University Affiliated Hospital (LS2020058).
Formalin-fixed paraffin-embedded (FFPE) sample collection
Tumour samples from 65 patients diagnosed with CRC between January 2016 and January 2017 were included in the analysis. Samples were formalin-fixed and paraffin-embedded (FFPE). All procedures were conducted in accordance with the ethical guidelines of the World Medical Association (Declaration of Helsinki) and adhered to the guidelines of the Institutional Review Board of Jiangnan University Affiliated Hospital (LS2020058). The clinical and pathological features of the patients were summarised, including age, sex, clinical stage, treatment response, degree of differentiation, and Desulfovibrio levels.
Desulfovibrio desulfuricans culture and detection
Desulfovibrio desulfuricans was purchased from the American Type Culture Collection (catalogue No. ATCC 29577; Manassas, Virginia, USA) and revived according to the manufacturer's instructions. D. desulfuricans was cultured in a 37 °C anaerobic workstation. After centrifugation at 3500g for 10 min, the strain was washed twice with physiological saline and resuspended in sterile phosphate buffer solution to a final concentration of 6 × 109 CFU/mL. PCR was performed to identify the presence of other bacterial species in the inoculum. To detect D. desulfuricans colonisation in the mouse colon, DNA was extracted from mouse faeces using the QIAamp DNA Stool Mini Kit (Qiagen, Hilden, Germany). Primers targeting D. desulfuricans were used for real-time PCR to assess faecal microbiota, Dsv691-F: CCGTAGATATCTGGAGGAACATCAG, Dsv826-R: ACATCTAGCATCCATCGTTTACACAGC. QPCR was used to compare the CT values of Desulfovibrio before and after inoculation with the aim of determining the colonisation of the target bacterial genus.
16S rRNA gene sequencing
The V4 region of the 16S rRNA gene was enlarged using the universal Bacteriological primers 27F (5′-GTGCCAGCMGCCGCGGTAA-3′) and 1492R (5′-GGACTACHVGGGTWTCTAAT-3′). The primers used for amplification included converters and single-end barcodes for the HiSeq platform, allowing sequence merging and demultiplexing of the PCR products. Qualified PCR products were sequenced using a 2 × 150 bp paired-end sequencing strategy on a HiSeq platform (Illumina, Inc., San Diego, CA, USA).
Animal experiments
Male C57BL/6J mice, aged 5–6 weeks, were provided by Gempharmatech Co., Ltd. The mice were kept in a temperature-regulated setting (24 ± 2 °C) on a 12h light/dark cycle. This study was conducted in compliance with the “Guidelines for the Care and Use of Laboratory Animals,” and was approved by the Institutional Animal Ethics Committee of Jiangnan University (Approval No: JN. No 20211215c0400516).
For the intestinal clearance procedure, mice were orally dosed with a wide-ranging antibiotic mixture of ABX containing vancomycin (100 mg/kg), neomycin sulphate (200 mg/kg), metronidazole (200 mg/kg), and ampicillin (200 mg/kg) daily for 7 consecutive days (from days −16 to −9) to exhaust the intestinal flora. Following antibiotic clearance, a 2-day recovery period was allowed. Subsequently, the mice were treated via oral gavage every other day with D. desulfuricans at a dose of 1 × 109 colony-forming units (CFU)/mouse throughout the study (from day −7 to day 22). Stool samples were collected on days −7 and 0 for D. desulfuricans gut colonization assessment. On day −7, 1 × 106 MC38 cells were subcutaneously inoculated into the flank of the mice. When the tumour volumes reached approximately 100 mm3 (day 0), the mice were randomly assigned to the following groups: control (n = 6), ABX (n = 6), Dsv (n = 6), ABX-Dsv (n = 6), FOLFOX (n = 6), ABX-FOLFOX (n = 6), FOLFOX-Dsv (n = 6), and ABX-FOLFOX-Dsv (n = 6). According to previous studies, FOLFOX (oxaliplatin 6 mg/kg, 2 h after 5-FU 50 mg/kg and Calcium Folinate 90 mg/kg treatment) was administered via intraperitoneal injection once weekly (on day 2, day 9 and day 16). The control group received injections of a solvent equivalent to the drug volume, the ABX group received equivalent volumes of antibiotics, and the ABX-FOLFOX and ABX-FOLFOX-Dsv groups received antibiotics and solvent volumes equivalent to the drug volume. Throughout the experiment, the tumour volume was monitored using callipers. Upon termination of the experiment (day 22), all mice were sacrificed, tumours were harvested, and preparations were prepared for immunohistochemical analysis.
Measurement of serum metabolites concentration by liquid chromatography mass spectrometry
For the sample preparation prior to liquid chromatography-mass spectrometry (LC-MS) analysis, 400 μL methanol was added and vortexed for 1 min, before subjecting to low-temperature centrifugation, concentrating and drying. The sample was resuspended in a solution of 2-chloro-1-phenylalanine (4 ppm) prepared with 80% methanol–water, before filtering the supernatant through a 0.22-μm membrane, and transferring it to a detection vial for LC-MS analysis. Mass spectrometric detection of metabolites was performed using an Orbitrap Exploris 120 (Thermo Fisher Scientific, USA).
Haematoxylin and eosin staining (H&E)
Mouse tumours were fixed in 4% formaldehyde for 48 h and embedded in paraffin. Blocks were sectioned into 4-μm slices and tinted with H&E reagents (BASO, Zhuhai, China). The pathologists were unbiased regarding the treatment-assessed images.
Immunohistochemistry (IHC)
Tumour sections from both humans and mice were deparaffinised with xylene and rehydrated using a gradient of ethanol and water. Antigen retrieval was carried out using ethylenediaminetetraacetic acid (EDTA)-sodium citrate buffer with microwave heating, followed by blocking of endogenous peroxidase activity with H2O2. Sections were then incubated overnight at 4 °C with Ki-67 antibody (1:400, Cell Signalling Technology, D3B5) and METTL3 antibody (1:500, Cell Signalling Technology, E3F2A). Subsequently, the sections were incubated for 1 h with horseradish peroxidase (HRP)-conjugated secondary antibodies. Finally, ImageJ software (Media Cybernetics, USA) was used to assess the staining intensity of the tumour tissues.
Western blot analysis
Tumour tissues and cell samples were lysed with protein lysis buffer and centrifuged at 4 °C. Protein lysates were divided by 10% SDS-PAGE and transferred onto a PVDF membrane. The membrane was blocked with 5% skim milk at room temperature, followed by overnight incubation at 4 °C on a shaker with METTL3 antibody (1:1000, Cell Signalling Technology, E3F2A), JAK2 antibody (1:1000, Cell Signalling Technology, D2E12), STAT3 antibody (1:1000, Cell Signalling Technology, D3Z2G), p-STAT3 antibody (1:1000, Cell Signalling Technology, D3A7). After washing with TBST, the membranes were incubated with secondary antibodies at room temperature. SuperSignal West Pico chemiluminescent substrate (Thermo Scientific, 34094) was used for detection. β-actin (1:2000, Cell Signalling Technology, 8H10D10) was applied as an internal reference. Protein bands were scanned and quantified using ImageJ software via densitometry analysis.
Quantitative real-time PCR
Total RNA was extracted from the cells using TRIzol reagent (Invitrogen). For transcription, 1000 ng RNA was transcribed into cDNA using a Hifair III RT Kit (Yeasen). QRT-PCR was performed using the indicated primers and Hieff qPCR SYBR Green Master Mix (Yeasen) on a CFX-96 Quantitative PCR system (Bio-Rad, Hercules, California, USA). The primer sequences are listed in Table 1. The relative levels were established relative to GAPDH using the 2−ΔΔCt method.
Table 1.
Primer and sequences.
| Primers for PCR and real time PCR | ||
|---|---|---|
| Primers | Sequences-F | Sequences-R |
| Dsv | 5′-CCGTAGATATCTGGAGGAACATCAG -3′ | 5′-ACATCTAGCATCCATCGTTTACACAGC -3′ |
| GAPDH-homo | 5′-AATGGGCAGCCGTTAGGAAA-3′ | 5′-GCGCCCAATACGACCAAATC-3′ |
| GAPDH-Mus | 5′-AGGTCGGTGTGAACGGATTTG-3′ | 5′-TGTAGACCATGTAGTTGAGGTCA-3′ |
| c-Myc-homo | 5′-GCAATGCGTTGCTGGGTTAT -3′ | 5′-TCCCTCCGTTCTTTTTCCCG-3′ |
| c-Myc-Mus | 5′-CACAGAACCCGATCCTTTCC-3′ | 5′-GTCGTTTCCGCAACAAGGA-3′ |
| CycD-homo | 5′-GCTGCGAAGTGGAAACCAGA-3′ | 5′-CCTCCTTCTGCACACATTTGA-3′ |
| CycD-Mus | 5′-CGTGGCCTCTAAGATGAAGG-3′ | 5′-GCTGGTAGTAGGACAGGAAGG-3′ |
| METTL3-hom | 5′-TCAGCATCGGAACCAGCAAA-3′ | 5′-TGGGGATTTCCTTTGACACCA-3′ |
| METTL3-Mus | 5′-CTGGGCACTTGGATTTAAGGA-3′ | 5′-TGAGAGGTGGTGTAGCAACTT-3′ |
| Sequences for siRNAs | ||
| si-METTL3#1 | GCUCAACAUACCCGUACUATT | UAGUACGGGUAUGUUGAGCTT |
| si-METTL3#2 | GCUACCUGGACGUCAGUAUTT | UAGUGAACAUACUUGCAGGTT |
| si-METTL3#3 | GGUUGGUGUCAAAGGAAAUTT | AUUUCCUUUGACACCAACCTT |
| si-METTL3#4 | CCUGCAAGUAUGUUCACUATT | UAGUGAACAUACUUGCAGGTT |
| si-METTL3##1 | GGCACUUGGAUUUAAGGAATT | UUCCUUAAAUCCAAGUGCCTT |
| si-METTL3##2 | CCUCCAAGAUGAUGCACAUTT | AUGUGCAUCAUCUUGGAGGTT |
| si-METTL3##3 | CCAAGGAAGAGUGCAUGAATT | UUCAUGCACUCUUCCUUGGTT |
| si-METTL3##4 | CCUGCAAAUAUGUUCACUATT | UAGUGAACAUAUUUGCAGGTT |
Small interfering RNA transfection
Transfection of 80% of overgrown cell cultures was performed using a Lipofectamine 3000 kit (Invitrogen, Carlsbad, CA, USA) with 55 nM siRNA. The four siRNA sequences for METTL3 were as follows: #1: GCUCAACAUACCCGUACUATT, #2: GCUACCUGGACGUCAGUAUTT, #3: GGUUGGUGUCAAAGGAAAUTT, and #4: CCUGCAAGUAUGUUCACUATT. Non-targeting NC siRNA (GenePharma, A06001) served as a control.
Cell proliferation, colony formation, and EdU incorporation assay
For cell proliferation, the cells were plated at a concentration of 8 × 103 cells/well in 96-well plates and allowed to attach overnight. Three replicates were performed for each experiment. CRC cells were exposed to FOLFOX (50 μM 5-Fu + 6 μM OXA + 90 μM CF) with or without 2 μM SAM for 24 h. Subsequently, 10 μL of CCK-8 solution (A311-01/02, Vazyme) was added, and the cells were incubated for 2 h. Cell viability was determined at OD 450 nm using a microplate reader (Thermo Fisher Scientific).
For the colony formation assay, CRC cells (600 cells/well) were seeded in 6-well plates and allowed to adhere overnight. Subsequently, cells were administered FOLFOX (50 μM 5-Fu + 6 μM OXA + 90 μM CF) with or without 2 μM SAM for 48 h. The medium was then changed, and the cells were maintained for approximately 14 days. The colonies were preserved in 4% paraformaldehyde (PFA) and stained with 0.5% (w/v) crystal violet. Subsequently, the colonies were imaged and counted.
For the EdU assay, 8 × 103 cells were seeded into each well of a 96-well plate and allowed to adhere overnight. Three replicates were performed for each experiment. CRC cells were treated with FOLFOX (50 μM 5-Fu + 6 μM OXA + 90 μM CF) with or without 2 μM SAM for 24 h. Cell proliferation was detected using an EdU-Apollo DNA Proliferation Assay Kit (C10310-1, RiboBio, China), according to the manufacturer's instructions. Hoechst staining was used for nuclear staining.
Migration and invasion assay
As for cell migration assay, 3 × 105 cells in 200 μL serum-free medium were introduced into the inserts of 8.0-μm Transwell chambers (354578, Corning). The cells were then incubated for 24 h and preserved in 4% PFA. The cells were stained with 0.5% crystal violet solution, followed by three PBS washes. Microscopic examination was conducted on the cells that displayed positive staining.
Matrigel-coated inserts (354483, Corning) were used for cell invasion assays. The cells were cultured for 48 h and the remaining steps were similar to those of the migration assay.
Multiplex immunofluorescence analyses
Multiplex immunofluorescence staining with a four-color IHC kit (Absin, China) was performed on tumour samples from CBR group and NCB group patients for counting immune cells infiltrating different tumours. The slides were deparaffinized in xylene, rehydrated, and washed in ethylenediaminetetraacetic acid (EDTA)-sodium citrate buffer for microwave antigen retrieval. Endogenous peroxidase activity was blocked using an antibody diluent/blocking agent (PerkinElmer, USA). Primary antibodies CD3 (1:500, Abcam, ab699), CD8 (1:500, Abcam, ab101500), and CD68 (1:500, Cell Signalling Technology, E3O7V) were incubated at room temperature for 1 h. Polymer HRP Ms + Rb was incubated for 10 min at 37 °C. Subsequently, the slides were incubated at room temperature for 10 min with TSA fluorescent dyes (TSA520, TSA570, and TSA650) diluted in signal amplification solution. Antigen-antibody complexes were stripped by microwave treatment using 0.05% Tris–EDTA buffer. TSA single-stained slides were counterstained with DAPI for 5 min and then coverslipped. Three observers, blinded to the experimental design of each sample, evaluated random fields of view at 100x magnification and calculated the number of immune cells in each field. Images were acquired using a confocal laser-scanning microscope (Zeiss Microscopy, USA).
m6A dot blot assay
RNA samples were spotted onto Amersham Hybond-N+ membrane (GE Healthcare) and subsequently cross-linked with ultraviolet (UV) light. After air-drying, the membrane was blocked for 1 h and then incubated overnight at 4 °C with a specific anti-m6A antibody (1:1000, Cell Signalling Technology, D9D9W). Subsequently, the membrane was incubated at room temperature for 1 h with HRP-conjugated anti-rabbit immunoglobulin G (1:3000, Abcam). Finally, the membrane was detected using a chemiluminescent HRP substrate kit (Millipore, USA) to capture images. Staining with 0.02% methylene blue (Sigma–Aldrich, USA) was performed and photographed to visualize total RNA content.
Immunofluorescence (IF)
8 × 103 CRC cells) were seeded on coverslips and allowed to attach overnight. The cells were plated in triplicate. CRC cells were given FOLFOX (50 μM 5-Fu + 6 μM OXA + 90 μM CF) with or without 2 μM SAM for 24 h. After fixation, the cells were rinsed in PBS containing 0.2% BSA, followed by permeabilization with PBS containing 0.1% Triton X-100 for 10 min. After blocking with 0.5% BSA, then cultivated with anti-METTL3 antibody (1:300) at 4 °C overnight, later subjected to incubation with Alexa Fluor® 555-conjugated secondary antibody (1:500) for 1 h. After three washes with PBST, DAPI staining of cells was performed for 5 min. Images were captured using a Nikon inverted microscope equipped with a 200 × objective lens.
Apoptosis analysis
CRC cells were treated with FOLFOX (50 μM 5-Fu + 6 μM OXA + 90 μM CF) with or without 2 μM SAM for 48 h. For apoptosis analysis, then reconstituted in 100 μL Binding Buffer. Next, 5 μL Annexin V-FITC and 5 μL PI were joined, followed by incubation in the dark for 15 min. Finally, 400 μL of Binding Buffer was mixed in and mixed thoroughly, and cell apoptosis was assessed via flow cytometry (FACSAriaⅢ, BD).
Fluorescence in situ hybridization (FISH)
The paraffin-embedded tissue blocks that were gathered were sliced into sections measuring 4-μm in thickness., and performed FISH on the sections according to the previously described protocol. Sections were de-paraffinized, hydrated, and incubated with Triton X-100 at 37 °C for 15 min. The slides were washed twice with PBS and incubated with lysozyme for 10 min, followed by hybridisation with a D. desulfuricans-specific probe and DAPI staining. The sequence of the D. desulfuricans-specific probe (Desu458, Cy3-labeled) was 5′-GGTCGCCCCCCGACACCT-3'. Five random fields of view at 200× magnification were evaluated by three observers who were blinded to the experimental design of each sample, and the average bacterial count per field was calculated. Images were acquired using a confocal laser-scanning microscope (Zeiss Microscopy, USA).
ELISA
The mouse SAM enzyme-linked immunosorbent assay (ELISA) kit (Yanqi, Shanghai, China) was used to measure SAM levels in mouse faecal extracts, caecal contents, tumour tissues, and serum samples.
Cell lines and culture
Human CRC cell lines, HCT116 and DLD1, were obtained from the American Type Culture Collection (Manassas, VA, USA) in 2015. Normal colon epithelial cell line NCM460 and mouse-derived CRC cell line MC38 were purchased from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China) in 2016. HCT116, DLD1, and NCM460 cells were cultured in DMEM (Gibco, Carlsbad, CA, USA) supplemented with 10% foetal bovine serum (Gibco, Carlsbad, CA, USA) and 1% penicillin-streptomycin at 37 °C in a humidified atmosphere containing 5% CO2. The MC38 cells were cultured in RPMI-1640 medium (Gibco, Carlsbad, CA, USA).
Statistical analysis
In the processing and analysis of microbiome data obtained from 16S rRNA sequencing, we utilized qiime2 software for data processing and microbiome analysis. Simultaneously, for statistical analysis and data visualization, we employed the R language (version 3.6.3). The data are expressed as the mean value ± the standard error of the mean (SEM). In this study, one- and two-way analyses of variance (ANOVA), Kaplan–Meier analysis, log-rank test, survival analysis, Tukey's multiple comparison test, Wilcoxon test, and Student's t-test were used as appropriate. Differences were considered statistically significant at p < 0.05. Statistical analyses were performed using GraphPad Prism 9.0 (GraphPad Software, La Jolla, CA, USA). p-values are represented as ∗p < 0.05, ∗∗p < 0.01 and ∗∗∗p < 0.001.
Role of funders
This work was supported by Wuxi City Social Development Science and Technology Demonstration Project (N20201005). The funders played no role in the study design, data collection, data analysis, interpretation, or writing of the report.
Results
Gut microbiota composition disparities between colorectal cancer patients in the CBR and NCB groups
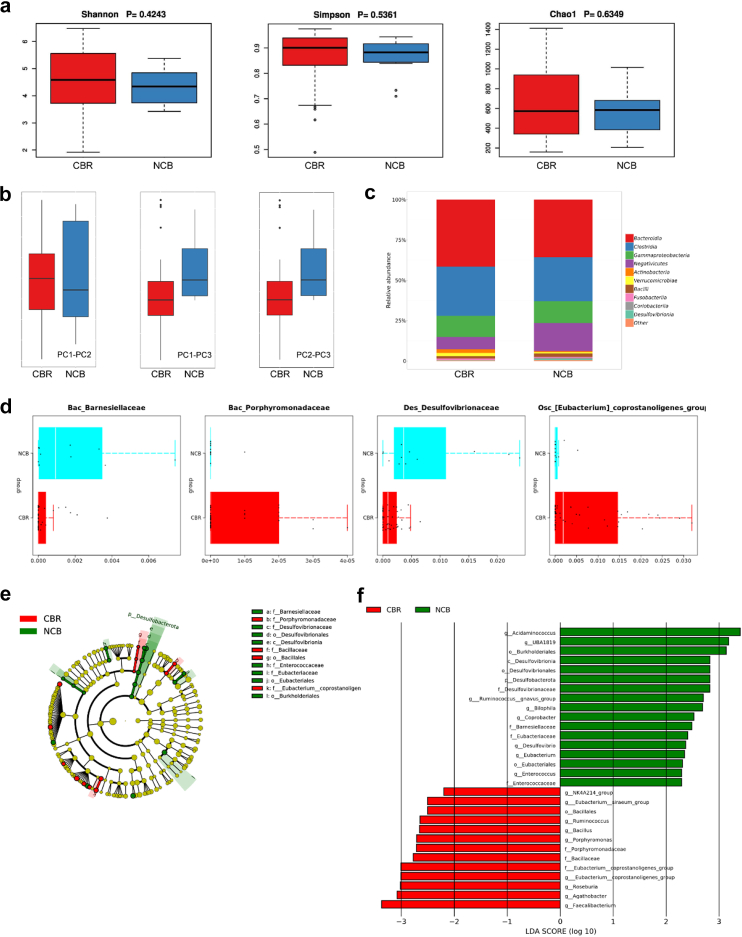
We recruited chemotherapy patients who were either preparing to undergo the FOLFOX chemotherapy regimen or were already receiving it. The collection of stool samples included both before and during the chemotherapy treatment. During the stool collection period, the patients were not subjected to any interference from other chemotherapy regimens. We collected 61 faecal samples from patients with CRC. During this period, a total of 49 samples were categorized into the clinical benefit group (CBR), with therapeutic assessments indicating complete response (CR), partial response (PR), and stable disease (SD); there were 12 samples categorized into the non-clinical benefit group (NCB), with therapeutic assessments indicating disease progression (PD). To investigate the impact of gut microbiota on chemotherapy capability in patients undergoing curative surgery, we performed 16S rRNA sequencing (n = 61) to assess the distribution of the gut microbiota composition of patients in the CBR (n = 49) and NCB groups (n = 12). The α-diversity of faecal samples, including Shannon, Simpson, and Chao1 indices, exhibited no notable distinctions among the CBR and NCB groups (Fig. 1a). However, compared with the CBR group, the NCB group exhibited increased levels of β-diversity. Bray–Curtis distance-based principal coordinate analysis showed evident inter-individual separations between faecal samples in the NCB group, as opposed to the CBR group (Fig. 1b, PERMANOVA: R2 = 0.0263, PPC1-PC2 = 0.052, PPC1-PC3 = 0.035, PPC2-PC3 = 0.042). We assessed the relative prevalence of the gut microbiota in both groups. The results revealed that The NCB group exhibited an elevated abundance of Bacilli, Desulfovibrionia, and Coriobacteriia and a decreased abundance of Actinobacteria and Verrucomicrobiae (Fig. 1c). Furthermore, we compared differences in microbial distribution between the two groups at the family level. The results showed that in the NCB group, the relative abundance of Bac_Barnesiellaceae and Des_Desulfovibrionaceae elevated, while in the CBR group, the relative abundance of Bac_Porphyromonadaceae and Osc_[Eubacterium] coprostanoligenes_group increased (Fig. 1d, Wilcoxon test, Bac_Barnesiellaceae: p = 0.0116; Des_Desulfovibrionaceae: p = 0.0017; Bac_Porphyromonadaceae: p = 0.0151; Osc [Eubacterium]_coprostanoligenes_group: p = 0.0120). To identify the microbial taxa influencing the differential efficacy of the FOLFOX chemotherapy regimen, we employed linear discriminant analysis. In the LEfSe evolutionary branch plot, a pronounced increase in Desulfovibrionaceae was observed in the NCB group (Fig. 1e). By setting a logarithmic linear discriminant analysis score >2.0, we confirmed the presence of differentially abundant microbial taxa in the CBR and NCB groups (Fig. 1f). Notably, Desulfovibrio in the NCB group showed enrichment across various taxonomic levels, including phylum, class, order, family, and genus. This suggests a likely correlation between the enrichment of Desulfovibrio in the gut and the diminished efficacy of postoperative adjuvant FOLFOX chemotherapy.
Fig. 1.
Relationship between postoperative adjuvant chemotherapy efficacy of FOLFOX regimen and differential distribution of gut microbiota. (a) 16S rRNA sequencing analysis of faecal samples from the CBR group (n = 49) and NCB group (n = 12) revealed no significant differences in α-diversity. (b) PCA indicates the differences in Bray–Curtis distances between individuals in the two groups of samples. PPC1-PC2 = 0.052, PPC1-PC3 = 0.035, PPC2-PC3 = 0.042. (c) Bar chart depicting the composition of microbial communities within the phylum category in faecal samples from the CBR (n = 49) and NCB groups (n = 12). (d) At the class level, variations were observed in the abundance distribution of the gut microbiota when comparing the CBR (n = 49) and NCB groups (n = 12). The Wilcoxon test is used for statistical analysis. (e, f) Evolutionary branch diagram and bar chart of LDA values generated by LEfSe illustrate the enriched microbial genera in each group, with red indicating enrichment in the CBR group (n = 49) and green indicating enrichment in the NCB group (n = 12). Statistical significance was set at LDA scores >2 and p-values <0.05 were seen as indicative of significance.
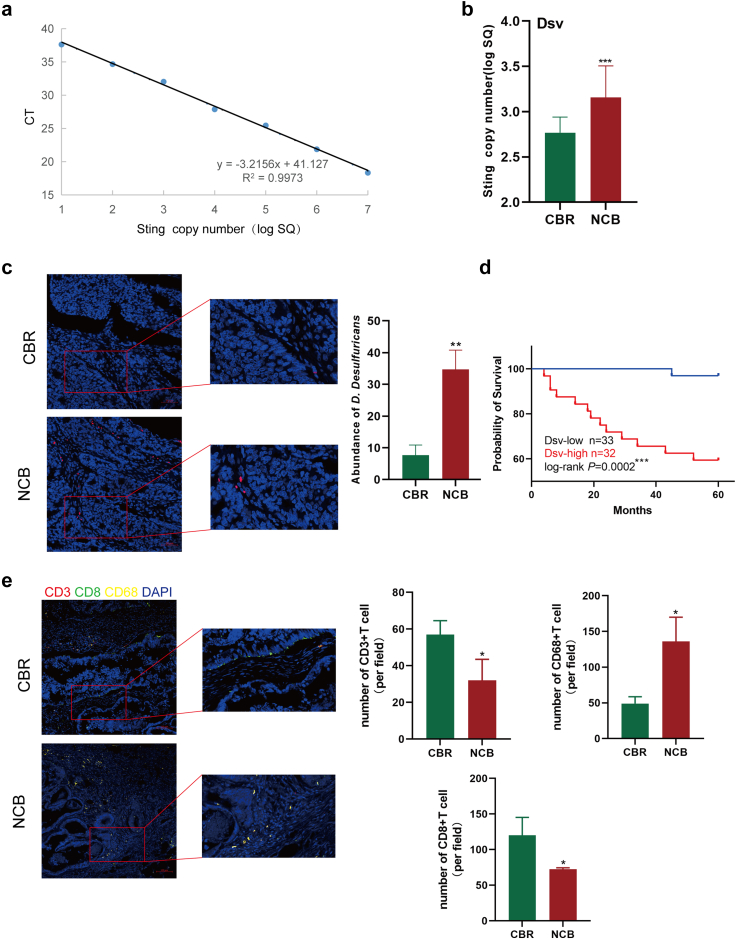
D. desulfuricans showed a higher degree of colonization in the tumour tissue of the NCB group in comparison to the CBR group
To further investigate the colonisation of D. desulfuricans at tumour sites, we obtained paraffin-embedded tissue. In this cohort of 65 patients, 58% were female, aged between 33 and 82 years with a median age of 67, and had a BMI ranging from 16.7 to 32.7, with a median of 23.8, indicating good nutritional status. We prepared a standard curve for the absolute quantification of D. desulfuricans using qPCR (Fig. 2a).9 Next, we extracted DNA from 30 paraffin samples and measured the expression levels of D. desulfuricans in each tissue sample. Absolute quantification by qPCR demonstrated that the presence of D. desulfuricans was higher in tumour samples from the NCB group than in those from the CBR group (Fig. 2b). Specific in situ hybridisation of D. desulfuricans in the samples yielded the same results as before (Fig. 2c). The results show that in CRC, Desulfovibrio colonizes areas in contact with tumour cells. This suggests that Desulfovibrio may influence the occurrence and development of CRC either through direct contact with tumour cells or through its metabolic products. Setting the median value of D. desulfuricans copy numbers as the threshold, all samples were separated into Dsv-low and Dsv-high groups. We analysed the relationship between D. desulfuricans levels and patient survival and found that patients with high colonization of D. desulfuricans in the gut had a worse prognosis (Fig. 2d). Furthermore, we also investigated the immune cell infiltration in tumour tissues of patients from the CBR group and NCB group, using CD3 to label T cells, CD8 to mark CD8+ T cells, and CD68 to identify macrophages. As shown in the figure, the infiltration of T cells and CD8+ T cells was significantly higher in the CBR group compared to the NCB group, while macrophage infiltration was higher in the NCB group (Fig. 2e). It can be observed that the type and quantity of immune cell infiltration in the tumour microenvironment have a complex relationship with tumour progression. By combining clinical factor analysis, we observed that D. desulfuricans levels were independent prognostic factors for poor outcomes in CRC (Supplementary Table S1). These findings indicate a close association between the high abundance of D. desulfuricans in the intestines of patients and poor response to FOLFOX chemotherapy, and unfavourable prognosis.
Fig. 2.
Colonization of D. desulfuricans in the tumour sites of patients with CRC. (a) Standard qPCR curve of D. desulfuricans. (b) The expression levels of D. desulfuricans in 30 paraffin samples (n = 15/group). (c) Representative images of specific fluorescence in situ hybridization (FISH) for D. desulfuricans in tumour tissues of the NCB (n = 10) and CBR groups (n = 10). Scale bar, 50 μm. (d) Kaplan–Meier analysis was used to determine the overall survival of the 65 patients with CRC based on their Dsv levels, and the log-rank test was conducted to evaluate the discrepancies in survival between the Dsv-high group (n = 32) and Dsv-low groups (n = 33). (e) Immune cell infiltration was analysed through multiplex immunofluorescence in the tumour tissues of patients from the CBR group (n = 10) and NCB group (n = 10), including CD3+ T cells, CD8+ T cells, and CD68+ macrophages. Scale bar, 100 μm. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.005, ∗∗∗∗p < 0.001. Statistical significance was determined using one-way ANOVA and Tukey's test for multiple comparisons.
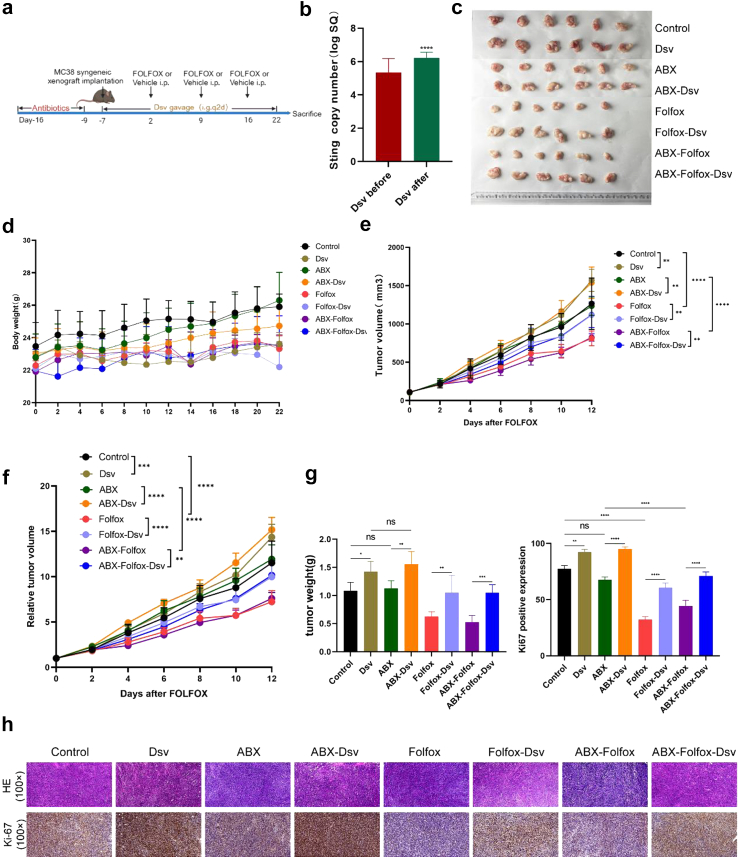
Colonization of D. desulfuricans led to a decrease in the effectiveness of FOLFOX treatment
To validate the influence of D. desulfuricans on the efficacy of the FOLFOX regimen, we established a syngeneic xenograft chemotherapy model of D. desulfuricans-colonised MC38 colon cancer cells (Fig. 3a).6, 7, 8,10,14,15 Stool samples from mice were collected before and one week after oral administration of D. desulfuricans to assess its colonization in the mouse gut, as shown in (Fig. 3b).16 The copy number of D. desulfuricans increased significantly after oral administration, indicating successful colonisation, and the inhibitory effect of FOLFOX chemotherapy on MC38 tumours was significantly weakened in the Folfox-Dsv and ABX-Folfox-Dsv groups (Fig. 3c). During the experiment, D. desulfuricans treatment had no effect on the body weight of the mice (Fig. 3d). However, in the groups in which FOLFOX chemotherapy was combined with D. desulfuricans administration (Folfox-Dsv and ABX-Folfox-Dsv groups), tumour growth advanced without hindrance, with an increase in both volume and weight (Fig. 3e, f, and g). Furthermore, in the D. desulfuricans-treated group, there was a substantial increase in Ki-67 expression in the tumour tissue, which further corroborated the above findings (Fig. 3h). In summary, in vivo animal experiments confirmed that the enrichment of D. desulfuricans in the gut leads to a reduction in the efficacy of FOLFOX chemotherapy. In conclusion, findings from in vivo animal experiments provide evidence that the enrichment of D. desulfuricans in the gut is associated with a diminishment in the therapeutic efficacy of FOLFOX chemotherapy.
Fig. 3.
Treatment with D. desulfuricans significantly diminished the anticancer efficacy of the FOLFOX regimen. (a) The in vivo experimental design of this study. 1 × 106 MC38 cells were subcutaneously injected into the lateral flank of the mice. D. Desulfuricans were delivered via gavage at a dose of 1 × 109 colony-forming units (CFU) per mouse every other day. FOLFOX chemotherapy was administered according to the dosages of oxaliplatin (6 mg/kg), 5-FU (50 mg/kg), and calcium folinate (90 mg/kg), with oxaliplatin administered 2 h after 5-FU once a week via intraperitoneal injection. (b) QPCR was used to compare the absolute abundance of D. desulfuricans in mouse faecal samples before and 7 days after oral administration (n = 24 mice/group). (c, e, and f) Comparison of tumour volumes among groups (n = 6 mice/group). (d) Changes in body weight of mice during the treatment period (n = 6 mice/group). (g) Comparison of tumour weights among different groups (n = 6 mice/group). (h) Histological images of tumour tissues stained with H&E and IHC staining for Ki-67 expression in various groups (scale bar, 100 μm). The results are presented as mean ± SEM. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.005, ∗∗∗∗p < 0.001. Statistical significance was determined using one-way ANOVA and Tukey's test for multiple comparisons.
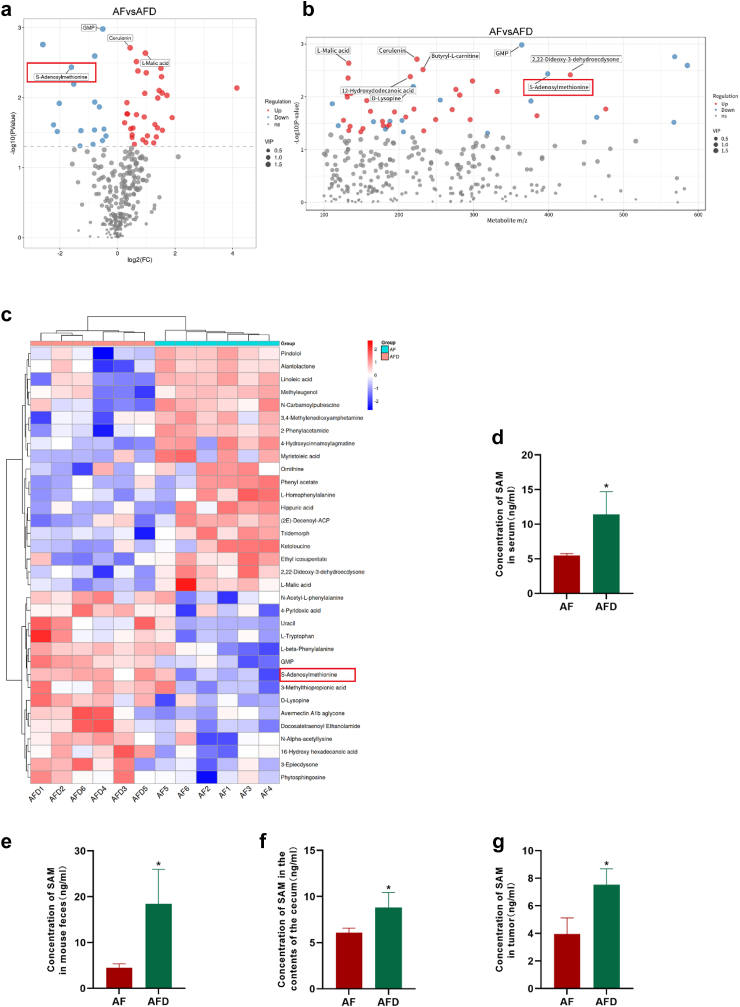
Concentration of S-adenosylmethionine in the serum metabolites of mice in the FOLFOX chemotherapy combined with D. desulfuricans administration group was increased
Based on these results, we developed a keen interest in gut-derived metabolites following D. desulfuricans administration. Given that we established a syngeneic xenograft model, we conducted a non-targeted metabolomic analysis using mouse serum samples. The analysis indicated increased concentrations of metabolites such as Cerulenin, l-Malic acid, and Butyryl-l-carnitine in the ABX-Folfox group (AF). In the ABX-Folfox-Dsv group (AFD), in addition to increased levels of nucleoside-like compounds such as GMP and Uracil, S-adenosylmethionine (SAM) exhibited a notable elevation in serum metabolites (Fig. 4a, b, and 4c). We re-evaluated serum SAM concentrations in both groups of mice to confirm the stability of its elevation in the AFD group (Fig. 4d). To investigate whether SAM was derived from the gut, we measured the SAM levels in mouse faeces and caecal contents (Fig. 4e and f). These results consistently indicate higher SAM concentrations in the AFD group than in the AF group, suggesting that SAM may originate from the gut. We also measured the concentration of SAM in tumour tissues. We found that the concentration of SAM in the tumour tissues of the AFD group was higher than that in the AF group, which is consistent with earlier research findings (Fig. 4g). These findings suggest that SAM may be a gut microbiota-derived metabolite following D. desulfuricans administration, which is absorbed into the bloodstream and circulates in the tumour tissue, exerting a role in chemotherapy resistance.
Fig. 4.
The levels of S-adenosylmethionine (SAM) were elevated in the D. desulfurican-treated chemotherapy model group. (a, b, and c) Comparison of serum metabolomic profiles between the ABX-Folfox and ABX-Folfox-Dsv groups in mice: (a) volcano plot, (b) mass-to-charge ratio vs. p-value scatter plot, and (c) heatmap of differential metabolites. Metabolomics analysis was conducted using non-targeted LC-MS/MS. years (n = 6 mice/group). ELISA was performed to measure the SAM concentration in the AF and AFD groups. (d) Concentration of SAM in serum; (e) Concentration of SAM in mouse faeces; (f) Concentration of SAM in mouse caecal contents; (g) Concentration of SAM in mouse tumour tissues. years (n = 6 mice/group). The results are presented as mean ± SEM. ∗p < 0.05. Statistical significance was determined using a one-way ANOVA.
Enhancement of SAM increases CRC cells' resistance to FOLFOX
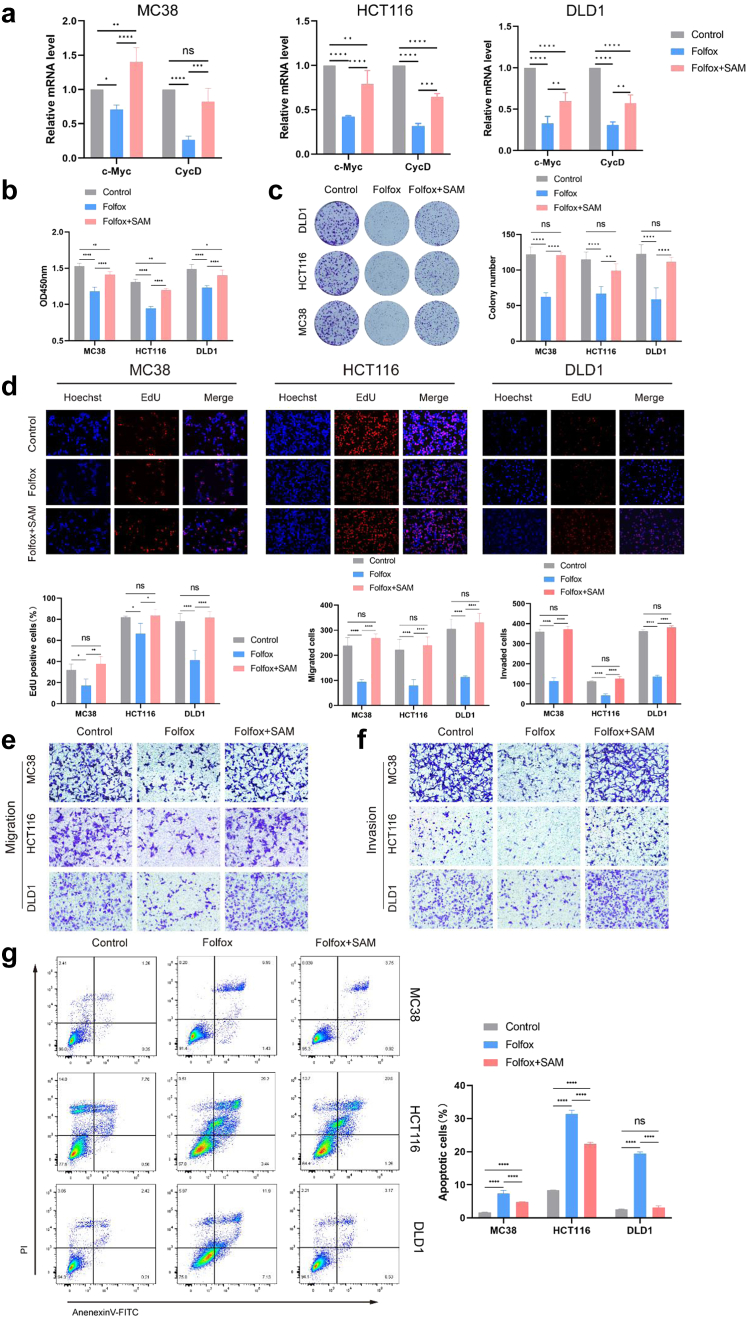
To investigate the potential effect of SAM on chemotherapy efficacy, we conducted experiments using three types of CRC cells: MC38, HCT116, and DLD1. We employed the concentrations of the three chemotherapy drugs in the FOLFOX regimen as follows: (50 μM 5-Fluorouracil (5-Fu) + 6 μM Oxaliplatin (OXA) + 90 μM Calcium Folinate (CF). Under this concentration ratio, the cell viability of CRC cells was approximately 50% of the control group (Supplementary Figure S1a); meanwhile, the concentration of 2 μM SAM exhibited a consistent effect in promoting tumour growth (Supplementary Figure S1b). Our findings revealed that regardless of whether it was combined with chemotherapy, SAM consistently promoted tumour cell proliferation (Supplementary Figure S1c and d). Considering that our study primarily focused on the relationship between metabolites and FOLFOX chemotherapy, we ultimately selected control, FOLFOX chemotherapy, and FOLFOX combined with SAM treatment groups for in vitro validation. After SAM combined with chemotherapy, we observed an increase in the mRNA expression levels of the proliferation genes c-Myc and CycD (Fig. 5a). Compared with cells subjected to FOLFOX chemotherapy alone, we observed increased cell viability when SAM was combined with chemotherapy (Fig. 5b). Exogenously added SAM decreased the sensitivity of tumour cells to FOLFOX chemotherapy, accompanied by enhanced proliferative capacity (Fig. 5c and d). Furthermore, after combined treatment with SAM and FOLFOX, CRC cells exhibited significantly increased migration and invasion capabilities compared to the FOLFOX only group (Fig. 5e and f). Additionally, we conducted apoptosis assays and found that the proportion of apoptotic cells in the FOLFOX + SAM group was lower than that in the FOLFOX group (Fig. 5g). In summary, these findings suggest that SAM can reduce the sensitivity of CRC cells to FOLFOX chemotherapy, leading to anticancer resistance, and ultimately weakening the therapeutic efficacy of chemotherapy, thereby promoting the growth of colorectal tumours.
Fig. 5.
SAM Impairs the efficacy of FOLFOX chemotherapy. (a) The mRNA expression levels of c-Cyc and CycD after SAM combined with chemotherapy treatment in CRC cells. (b) Compared to the FOLFOX group, the combination of SAM with FOLFOX enhanced the viability of MC38, HCT116, and DLD1 CRC cells. (c) Compared with the FOLFOX group, the combined use of SAM and FOLFOX enhanced colony formation in CRC cells. (d) Compared to the FOLFOX group, the combination of SAM and FOLFOX enhanced the proliferative ability of CRC cells. (e, f) In the Folfox + SAM group, both the migration (e) and invasion (f) abilities of the CRC cells were higher than those in the Folfox group. (g)The apoptosis rates of the three types of CRC cells in the Folfox + SAM group were lower than those in the Folfox group. Data were from one representative of three independent experiments. The results are presented as mean ± SEM. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.005, ∗∗∗∗p < 0.001. Statistical significance was determined using one-way or two-way analysis of variance (ANOVA).
SAM mediates chemoresistance in CRC cells through METTL3
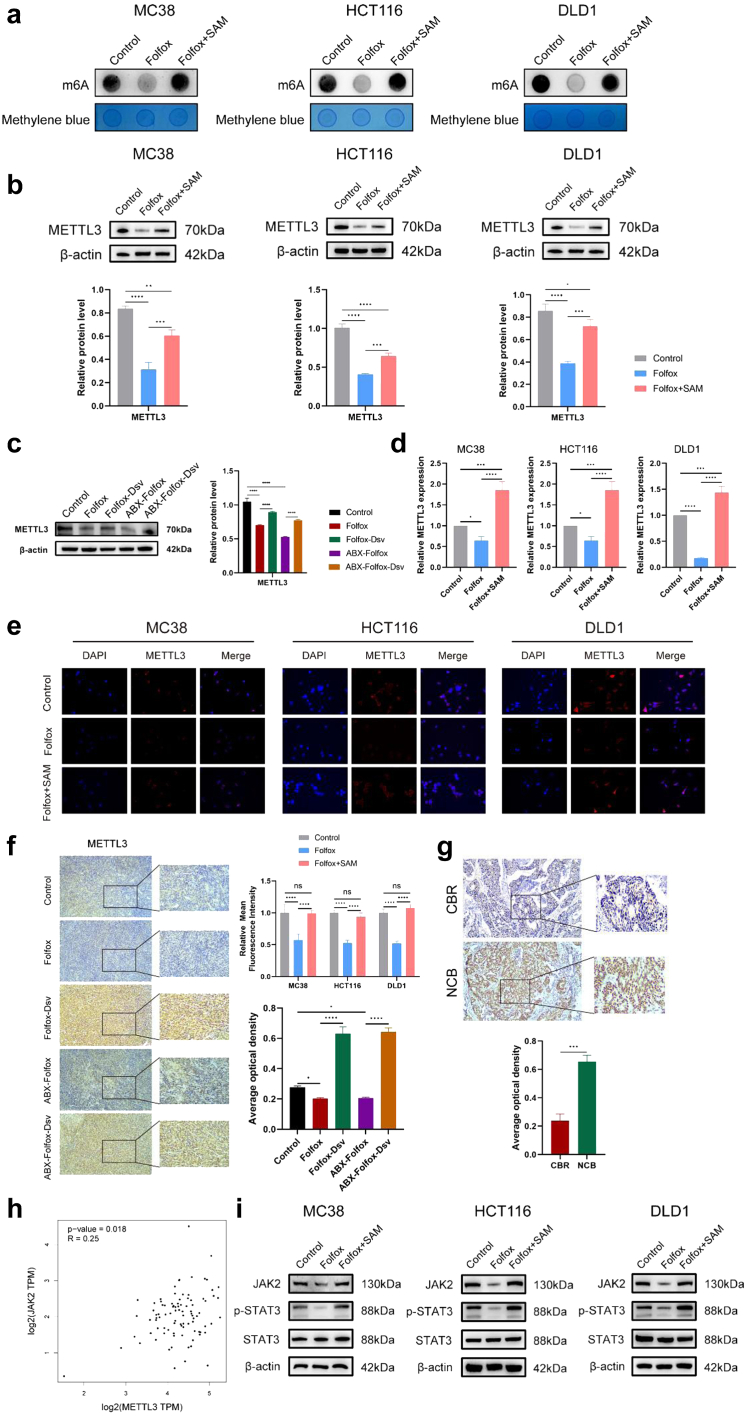
Several studies, SAM has demonstrated anticancer properties.17,18 However, SAM also serves as a crucial physiological molecule that participates in biochemical reactions within organisms through processes such as methylation, sulphur transfer, and transamination. SAM acts as a methyl donor during intracellular methylation reactions. Methyltransferase-like 3 (METTL3) serves as a crucial mediator of methylation and is also a SAM-dependent methyltransferase, containing a SAM-binding pocket. In contrast, METTL14 lacks a SAM-binding pocket. Dysregulation of METTL3 has been linked to various conditions, including cancer, cardiovascular diseases, and neurological conditions. Importantly, METTL3 is SAM-dependent and serves as a methyltransferase in methylation reactions. We examined the methylating effect of SAM in CRC cells, and the results showed that the m6A methylation levels in CRC cells were significantly higher when treated with SAM in combination with chemotherapy compared to the chemotherapy-only group (Fig. 6a). To elucidate the mechanism underlying the SAM-induced resistance of CRC cells to FOLFOX chemotherapy, we examined the expression of METTL3. We observed that the protein expression of METTL3 was higher in the CRC cell lines MC38, HCT116, and DLD1 treated with SAM in combination with chemotherapy than in the chemotherapy-only group (Fig. 6b). Similar results were obtained for the tumour tissues from mice (Fig. 6c). The combination of SAM and chemotherapy elevated the transcriptional level of METTL3 in CRC cells (Fig. 6d). Immunofluorescence analysis revealed increased METTL3 expression in SAM-treated CRC cells (Fig. 6e). Immunohistochemical results from mouse tumour tissues indicated elevated positivity for METTL3 after D. desulfuricans administration (Fig. 6f). In addition, in the earlier patient-derived tumour tissues, the expression of METTL3 was higher in the NCB group than in the CBR group, consistent with the aforementioned results (Fig. 6g). To further explore downstream targets regulated by METTL3 in chemotherapy resistance, we analysed a CRC RNA-seq dataset and found a positive correlation between METTL3 and JAK2 (Fig. 6h). As a result, we validated the JAK2-STAT3 signalling pathway in three CRC cell lines and found that SAM promotes chemotherapy resistance by activating the METTL3-mediated classical JAK2-STAT3 signalling pathway (Fig. 6i). These findings imply that the combination of SAM and FOLFOX chemotherapy may reduce the effectiveness of chemotherapy by upregulating the expression of METTL3. Previous studies have also indicated that elevated levels of METTL3 are commonly associated with poor prognosis in CRC.19,20
Fig. 6.
In the SAM combined with the FOLFOX chemotherapy group, the expression level of METTL3 is increased. (a) After SAM combined with chemotherapy, CRC cells exhibited an increase in m6A methylation levels. (b) The protein expression of METTL3 in MC38, HCT116, and DLD1 cells 48 h after SAM and FOLFOX treatment. (c) The protein expression of METTL3 in mouse tumour tissues. (d)The mRNA levels of METTL3 in MC38, HCT116, and DLD1 cells after 48 h of treatment with SAM combined with FOLFOX. (e) Immunofluorescence analysis of METTL3 levels in the three cell lines (MC38, HCT116, and DLD1) after 48 h of SAM and FOLFOX treatment. (f) Determination of METTL3 expression in mouse tumour tissues by IHC. Scale bar, 100 μm. (g) Immunohistochemical analysis of METTL3 expression in tumour tissues of patients with CRC. Scale bar, 100 μm. (h) RNA-seq sequencing analysis of CRC dataset revealed a positive correlation between METTL3 and JAK2. (i) After SAM combined with chemotherapy, the JAK2-STAT3 signalling pathway is activated in CRC cells, leading to an increase in protein expression. Data were from one representative of three independent experiments. The results are presented as mean ± SEM. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.005, ∗∗∗∗p < 0.001. Each experiment was repeated thrice. Statistical significance was determined using one-way or two-way analysis of variance (ANOVA).
Knocking down METTL3 can counteract the attenuating effect of SAM on chemotherapy
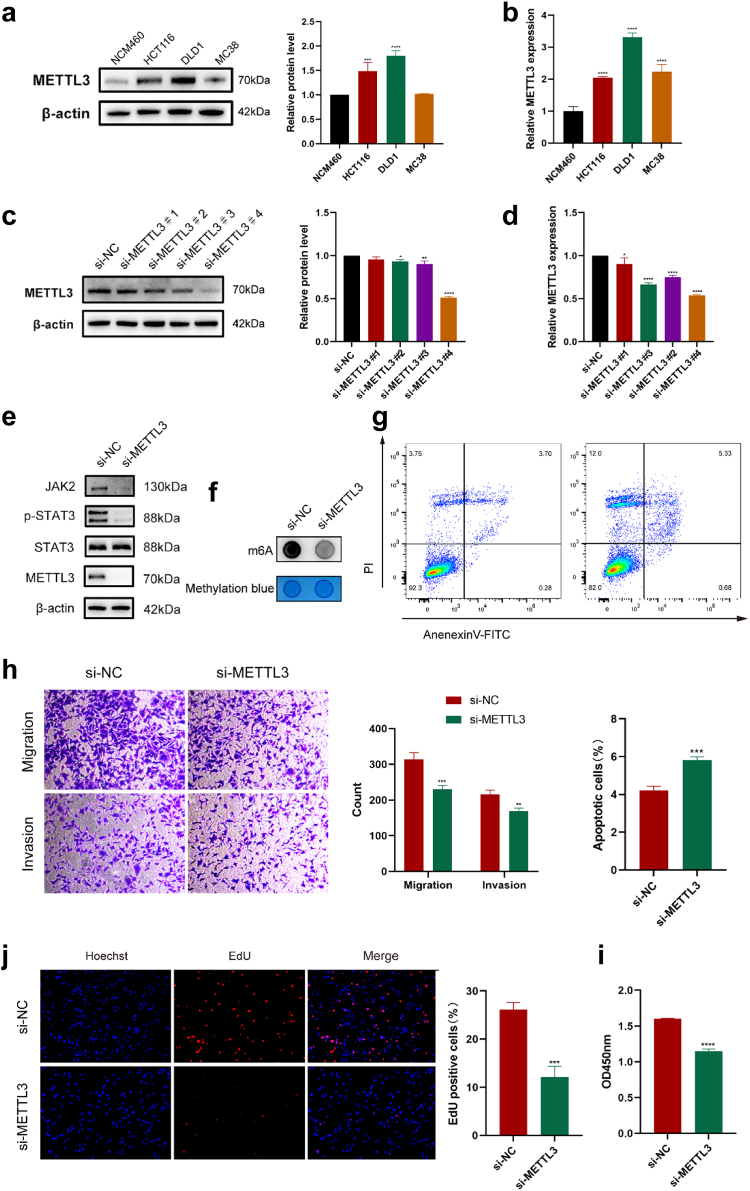
First, we compared the protein expression of METTL3 in four cell lines: NCM460 (human normal colonic epithelial cells), MC38, HCT116, and DLD1. NCM460 was used as a control and DLD1 exhibited the highest METTL3 expression (Fig. 7a). We also compared the transcription levels of METTL3 in these four cell lines, and the results were consistent with the earlier findings (Fig. 7b). Next, we selected DLD1 cells with high expression of METTL3 for knockdown experiments and designed four siRNAs to silence METTL3 in DLD1 cells; si-METTL3#4 showed the highest knockdown efficiency at both the protein and transcriptional levels (Fig. 7c and d). Subsequently, we validated the expression of proteins in the METTL3-mediated JAK2-STAT3 pathway. As shown in the figure, the expression of JAK2 and pSTAT3 significantly decreased after METTL3 silencing. This once again confirms that METTL3 exerts its role in chemotherapy resistance by mediating the JAK2-STAT3 pathway (Fig. 7e and Supplementary Figure S2a). After silencing METTL3, the m6A methylation levels in CRC cells significantly decreased (Fig. 7f). Therefore, we selected si-METTL3#4 for the knockdown experiments in DLD1 cells for further cell-based experiments. After knocking down METTL3, the apoptotic rate of DLD1 cells increased and their migration and invasion abilities decreased (Fig. 7g and h). Silencing of METTL3 also decreased post-chemotherapy cell viability and diminished the proliferative capacity of DLD1 cells (Fig. 7i and j). MC38 and HCT116 cells also exhibited the same results (Supplementary Figure S2b). These results indicate that METTL3 plays a crucial role in SAM-mediated chemoresistance in CRC cells. By reducing the manifestation of METTL3, the sensitivity of CRC cells to chemotherapeutic agents can be enhanced, thereby restoring the efficacy of FOLFOX chemotherapy (Fig. 8).
Fig. 7.
Silencing METTL3 enhances the efficacy of chemotherapy. (a) Protein expression of METTL3 in NCM460, MC38, HCT116, and DLD1 cells. (b) Transcript levels of METTL3 in NCM460, MC38, HCT116, and DLD1 cells. (c, d) Efficiency of the four siRNAs for knocking down METTL3 in DLD1 cells, (c) protein level, and (d) mRNA level. (e) Knocking down METTL3 resulted in a decrease in the protein expression of the JAK2-STAT3 signalling pathway. (f) Knocking down METTL3 resulted in a decrease in m6A methylation levels in DLD1 cells. (g) Flow cytometry analysis of the apoptotic ratio in METTL3-silenced DLD1 cells after chemotherapy. (h)Migration and invasion of METTL3-silenced DLD1 cells post chemotherapy. (i, j) Cell viability (i) and proliferation capacity (j) of METTL3-silenced DLD1 cells post chemotherapy. The results are presented as mean ± SEM. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.005, ∗∗∗∗p < 0.001. Each experiment was repeated three times, and statistical significance was determined using one-way or two-way ANOVA.
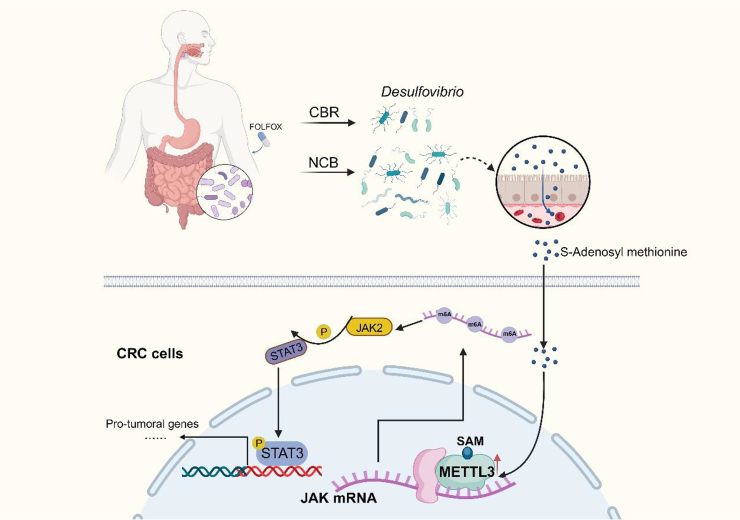
Fig. 8.
A schematic diagram illustrating this study.
Discussion
The efficacy of chemotherapy for CRC is closely correlated with the overall survival prognosis of patients. Patients with resected CRC often opt for adjuvant chemotherapy using the FOLFOX regimen to reduce the risk of recurrence. However, a subset of patients still experiences disease progression owing to chemotherapy resistance. After collecting faecal samples from patients undergoing FOLFOX chemotherapy, we observed a substantial increase in Desulfovibrio in the gut of patients who experienced disease progression during the course of chemotherapy. D. desulfuricans reportedly promotes progression and onset of CRC.13,21 In this study, we observed that D. desulfuricans diminished the efficacy of FOLFOX chemotherapy and facilitated the progression of CRC. The gut microbiota-derived metabolite, SAM, plays a crucial role in promoting chemoresistance. Mechanistically, SAM interacts with METTL3 to enhance its expression, thereby contributing to chemotherapeutic resistance. Silencing METTL3 can counteract the resistance effect, thereby restoring chemotherapy efficacy. These results indicate that the gut microbiome and its metabolic by-products may be key factors in postoperative adjuvant chemotherapy for CRC. Further research is warranted to elucidate their impact on treatment efficacy and the underlying mechanisms.
The effectiveness of postoperative adjuvant chemotherapy in CRC directly affects patient survival and quality of life. However, the ineffectiveness of CRC treatments is largely attributed to drug resistance. In this study, we retrieved faecal samples from patients who underwent postoperative adjuvant chemotherapy with FOLFOX. Through 16S rRNA sequencing of stool samples, we discovered a higher abundance of Desulfovibrio in the gut of patients who did not benefit from chemotherapy after treatment. Furthermore, D. desulfuricans was more prevalent at the tumour sites of non-benefiting patients than in benefiting patients. According to previous reports, there is a remarkable increase in the types and abundance of D. desulfuricans in the faeces of patients with ulcerative colitis, which may be related to the toxic effects of H2S produced by Desulfovibrio on intestinal epithelial cells.22 Compared to healthy volunteers, Desulfovibrio was enriched in the faeces and biopsies of patients with CRC.13,23 Moreover, Desulfovibrio is also significantly enriched in the intestines of CRC liver metastasis patients.11 This suggests a close association between D. desulfuricans and the appearance and advancement of colorectal adenomas, offering a conceptual foundation for this study. D. desulfuricans has been linked to various aspects of human health and disease. It exacerbates atherosclerosis by increasing circulating LPS levels and exerts protective effects in mice displaying Parkinson's disease symptoms through the microbiota-gut-brain axis.24,25 Desulfovibrio fairfieldensis has been implicated in damaging the intestinal epithelial barrier, inducing macrophage inflammation and apoptosis.26 Reports also suggest that D. desulfuricans may contribute to obesity and non-alcoholic fatty liver by increasing gut permeability and affecting lipid absorption.18,27 In summary, D. desulfuricans to a certain extent affects human health and disease development. Our results suggest that D. desulfuricans play a detrimental role in postoperative adjuvant chemotherapy for CRC. In other words, the colonisation of D. desulfuricans in the intestines of postoperative patients may serve as a predictive indicator of poor prognosis after FOLFOX chemotherapy.
To elucidate the mediators through which D. desulfuricans affects chemotherapy efficacy, we conducted LC-MS untargeted metabolomic analysis of mouse serum. We observed that the combination of D. desulfuricans and chemotherapy increased the serum concentrations of metabolites such as uracil, GMP, and SAM. Previous reports have demonstrated that nucleoside analogues, such as uracil and GMP, can assist CRC cells in resisting the effects of radiation and chemotherapy.5 Through in vitro experiments involving CCK8 assays, colony formation assays, EdU labelling, apoptosis analysis, and migration and invasion assays, we discovered that SAM plays a role in chemotherapy resistance when combined with FOLFOX. This observation is reminiscent of a previous study that indicated that SAM promotes CRC metastasis.28 Conversely, some studies have suggested that SAM inhibits P-glycoprotein expression and NF-κB activation, thereby increasing the sensitivity of CRC cells to 5-FU.29 SAM has also upregulated miRNAs to suppress CRC cell migration.30 These findings differ from those of our study. In light of these disparities, we suggest that the dosage of SAM administered to the colon cancer cells may have contributed to these discrepancies. The concentrations of SAM in the aforementioned reports were 500 μM, whereas our study employed a SAM concentration of 2 μM. We speculate that SAM might promote tumour progression at low doses, while exhibiting anti-tumour effects at high doses. At lower concentrations, SAM might accelerate tumour progression through the upregulation of METTL3 expression, whereas higher concentrations of SAM predominantly induce apoptosis and autophagy in tumour cells, exerting antitumour effects. Moreover, SAM combined with chemotherapy reportedly exerts synergistic anticancer effects.31,32 Other reports have suggested that SAM induces ER stress-triggered apoptosis in head and neck squamous cell carcinoma, enhancing sensitivity to cisplatin.33 Furthermore, a considerable amount of research has confirmed the safety and efficacy of SAM for the treatment of depression,34,35 osteoarthritis,36, 37, 38 and liver disease.39 The relationship between SAM and CRC treatment warrants further investigation.
As a crucial intracellular methyl donor, SAM interacts with METTL3, driving an increase in catalysed RNA methylation modification reactions. METTL3 dysregulation is associated with various diseases. In our study, we observed that in the SAM combined with chemotherapy group, the expression of METTL3 increased, along with enhanced cell proliferation. In clinical samples, we confirmed elevated levels of METTL3 expression in the tumour tissues of patients who experienced poor outcomes following postoperative adjuvant chemotherapy. This suggests that METTL3 negatively affects FOLFOX chemotherapy. Therefore, we silenced METTL3 in CRC cells and conducted further in vitro experiments, confirming that METTL3 knockdown in CRC cells yielded better outcomes when treated with SAM in combination with chemotherapy. This provides substantial evidence that METTL3 plays a critical role in chemotherapy resistance and offers potential new therapeutic targets for postoperative adjuvant chemotherapy in clinical patients with CRC. The interaction between SAM and METTL3 and their impact on chemotherapy resistance may have important implications for enhancing treatment outcomes.
Recent studies have extensively explored the functions of METTL3, particularly in CRC. METTL3, through m6A modifications, regulates proteins such as IGF2BP2, YTHDF2, and Hippo, thereby promoting the progression of cancer tumours.40, 41, 42, 43 METTL3 targets the m6A-BHLHE41-CXCL1/CXCR2 axis to inhibit antitumour immunity and facilitate the advancement of CRC.44 Furthermore, METTL3 has been found to activates glycolysis, which contributes to CRC progression.45 Research indicates that METTL3 is involved in the development of resistance, rendering CRC resistant to the chemotherapeutic drug 5-FU.46 These provide supportive evidence for the ongoing research.
In addition to CRC, METTL3 plays a carcinogenic role in other types of cancer. Reports indicate that METTL3 controls the distinction between regular haematopoietic and leukemic cells in the bone marrow, thereby sustaining myeloid leukaemia.47,48 Given METTL3's critical role in leukaemia development, studies suggest that small molecule inhibitors of METTL3 could be a strategy for treating myeloid leukaemia.49 METTL3 contributes to breast cancer progression by regulating Bcl-2 expression.50 In liver cancer, METTL3 enhances cancer advancement by targeting SOCS2, YTHDF1, and Snail.51, 52, 53 In bladder cancer, METTL3 facilitates progression through the AFF4/NF-κB/MYC signalling network.54 These reports underscore the indispensable role of METTL3 in various cancers, affecting tumour proliferation, response to chemotherapy, and immunotherapy. This strongly suggests the broad potential of METTL3 in cancer treatment. The findings of this study also highlighted the role of METTL3 in postoperative chemotherapy for CRC, implying that targeting METTL3 may enhance the prognosis of postoperative patients with CRC.
The relationship between gut microbiota and the occurrence and treatment of CRC has been found to have both positive and negative associations.2,55, 56, 57 Our research results, for the first time, suggest that Desulfovibrio can reduce the efficacy of postoperative adjuvant chemotherapy in patients with CRC. Through in vitro and in vivo experiments, we demonstrated that D. desulfuricans promoted chemotherapy resistance in CRC by driving METTL3 expression through SAM. However, our study has several limitations. In our study, the sample size of faecal samples from colorectal cancer patients was insufficient. To address this, we collected paraffin-embedded samples from colorectal cancer surgery patients and conducted tests for D. desulfuricans to provide supporting evidence. Secondly, during our experimental process, we did not use small-molecule inhibitors targeting METTL3. Our method primarily focused on knocking down METTL3. However, a treatment strategy combining small-molecule inhibitors with chemotherapy may offer greater clinical translational value. In subsequent research, this aspect requires further investigation. Despite the limitations of our study, we established that enrichment of D. desulfuricans in the gut of patients often indicates poor chemotherapy efficacy. Thus, METTL3 has the potential to be a novel target in CRC treatment. Our findings suggest that targeting METTL3 or combining it with chemotherapy could be a new therapeutic strategy. In patients with CRC with an enrichment of D. desulfuricans in the gut, the consumption of probiotics can disrupt the colonisation of D. desulfuricans and enhance the effectiveness of FOLFOX chemotherapy.
In conclusion, this study demonstrates that D. desulfuricans mediates CRC resistance to FOLFOX chemotherapy through SAM-driven upregulation of METTL3 expression. D. desulfuricans has emerged as a promising biomarker for predicting the prognosis of postoperative adjuvant chemotherapy in patients with CRC, and METTL3 has opened up new avenues and targets for CRC treatment strategies.
Contributors
Conceptualisation and Design: GF Li, Y Mao, F Wang. Data Acquisition: Y Mao, F Wang, GF Li, H Liu, YM Yu, Q Wang, C Yang. Data analysis and interpretation: GF Li, YM Yu, and H Liu, Y Yan. Validation of fundamental data: GF Li, F Wang, Y Yan, Q Wang, and C Yang. Research Guidance: Y Mao. Manuscript Drafting: GF Li. All the authors have read and approved the final version of the manuscript. GF Li, Y Yan, Q Wang, and C Yang accessed and validated the data and GF Li is responsible for the decision to submit the manuscript.
Data sharing statement
All primary data from this study will be made available upon request from the corresponding author in accordance with appropriate data-sharing agreements. The 16S-rRNA sequencing data were deposited in the National Genomic Data Center Gene Expression Omnibus under appropriate data-sharing protocols (PRJNA1013400).
Declaration of interests
No potential conflict of interest was reported by the authors.
Acknowledgements
These studies have received Wuxi City Social Development Science and Technology Demonstration Project (N20201005).
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.ebiom.2024.105041.
Contributor Information
Fang Wang, Email: wangfang_lukas@zju.edu.cn.
Yong Mao, Email: 9812015252@jiangnan.edu.cn.
Appendix A. Supplementary data
References
- 1.Ramos A., Hemann M.T. Drugs, bugs, and cancer: fusobacterium nucleatum promotes chemoresistance in colorectal cancer. Cell. 2017;170(3):411–413. doi: 10.1016/j.cell.2017.07.018. [DOI] [PubMed] [Google Scholar]
- 2.Wong C.C., Yu J. Gut microbiota in colorectal cancer development and therapy. Nat Rev Clin Oncol. 2023;20(7):429–452. doi: 10.1038/s41571-023-00766-x. [DOI] [PubMed] [Google Scholar]
- 3.Yu T., Guo F., Yu Y., et al. Fusobacterium nucleatum promotes chemoresistance to colorectal cancer by modulating autophagy. Cell. 2017;170(3):548–563.e16. doi: 10.1016/j.cell.2017.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu H., Du J., Chao S., et al. Fusobacterium nucleatum promotes colorectal cancer cell to acquire stem cell-like features by manipulating lipid droplet-mediated numb degradation. Adv Sci. 2022;9(12) doi: 10.1002/advs.202105222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Teng H., Wang Y., Sui X., et al. Gut microbiota-mediated nucleotide synthesis attenuates the response to neoadjuvant chemoradiotherapy in rectal cancer. Cancer Cell. 2023;41(1):124–138.e6. doi: 10.1016/j.ccell.2022.11.013. [DOI] [PubMed] [Google Scholar]
- 6.Iida N., Dzutsev A., Stewart C.A., et al. Commensal bacteria control cancer response to therapy by modulating the tumor microenvironment. Science. 2013;342(6161):967–970. doi: 10.1126/science.1240527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Limani P., Linecker M., Kachaylo E., et al. Antihypoxic potentiation of standard therapy for experimental colorectal liver metastasis through myo-inositol trispyrophosphate. Clin Cancer Res. 2016;22(23):5887–5897. doi: 10.1158/1078-0432.CCR-15-3112. [DOI] [PubMed] [Google Scholar]
- 8.Robinson S.M., Mann J., Vasilaki A., et al. Pathogenesis of FOLFOX induced sinusoidal obstruction syndrome in a murine chemotherapy model. J Hepatol. 2013;59(2):318–326. doi: 10.1016/j.jhep.2013.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fite A., Macfarlane G.T., Cummings J.H., et al. Identification and quantitation of mucosal and faecal desulfovibrios using real time polymerase chain reaction. Gut. 2004;53(4):523–529. doi: 10.1136/gut.2003.031245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gong S., Yan Z., Liu Z., et al. Intestinal microbiota mediates the susceptibility to polymicrobial sepsis-induced liver injury by granisetron generation in mice. Hepatology. 2019;69(4):1751–1767. doi: 10.1002/hep.30361. [DOI] [PubMed] [Google Scholar]
- 11.Yu Y., Cai Y., Yang B., et al. High-fat diet enhances the liver metastasis potential of colorectal cancer through microbiota dysbiosis. Cancers. 2022;14(11) doi: 10.3390/cancers14112573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van Liere E., de Boer N.K.H., Dekker E., van Leerdam M.E., de Meij T.G.J., Ramsoekh D. Systematic review: non-endoscopic surveillance for colorectal neoplasia in individuals with Lynch syndrome. Aliment Pharmacol Ther. 2022;55(7):778–788. doi: 10.1111/apt.16824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yan Y., Drew D.A., Markowitz A., et al. Structure of the mucosal and stool microbiome in lynch syndrome. Cell Host Microbe. 2020;27(4):585–600.e4. doi: 10.1016/j.chom.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hou X., Zhang P., Du H., et al. Akkermansia muciniphila potentiates the antitumor efficacy of FOLFOX in colon cancer. Front Pharmacol. 2021;12:725583. doi: 10.3389/fphar.2021.725583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang C., Zhou Q., Li M., et al. Upregulation of CYP2S1 by oxaliplatin is associated with p53 status in colorectal cancer cell lines. Sci Rep. 2016;6 doi: 10.1038/srep33078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hou X.Y., Zhang P., Du H.Z., et al. Prevotella contributes to individual response of FOLFOX in colon cancer. Clin Transl Med. 2021;11(9):e512. doi: 10.1002/ctm2.512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jensen-Pergakes K., Tatlock J., Maegley K.A., et al. SAM-competitive PRMT5 inhibitor PF-06939999 demonstrates antitumor activity in splicing dysregulated NSCLC with decreased liability of drug resistance. Mol Cancer Ther. 2022;21(1):3–15. doi: 10.1158/1535-7163.MCT-21-0620. [DOI] [PubMed] [Google Scholar]
- 18.Lin Y.C., Lin H.F., Wu C.C., Chen C.L., Ni Y.H. Pathogenic effects of Desulfovibrio in the gut on fatty liver in diet-induced obese mice and children with obesity. J Gastroenterol. 2022;57(11):913–925. doi: 10.1007/s00535-022-01909-0. [DOI] [PubMed] [Google Scholar]
- 19.Liu X., He H., Zhang F., et al. m6A methylated EphA2 and VEGFA through IGF2BP2/3 regulation promotes vasculogenic mimicry in colorectal cancer via PI3K/AKT and ERK1/2 signaling. Cell Death Dis. 2022;13(5):483. doi: 10.1038/s41419-022-04950-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peng W., Li J., Chen R., et al. Upregulated METTL3 promotes metastasis of colorectal Cancer via miR-1246/SPRED2/MAPK signaling pathway. J Exp Clin Cancer Res. 2019;38(1):393. doi: 10.1186/s13046-019-1408-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kushkevych I., Dordević D., Vítězová M. Possible synergy effect of hydrogen sulfide and acetate produced by sulfate-reducing bacteria on inflammatory bowel disease development. J Adv Res. 2021;27:71–78. doi: 10.1016/j.jare.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rowan F., Docherty N.G., Murphy M., Murphy B., Calvin Coffey J., O'Connell P.R. Desulfovibrio bacterial species are increased in ulcerative colitis. Dis Colon Rectum. 2010;53(11):1530–1536. doi: 10.1007/DCR.0b013e3181f1e620. [DOI] [PubMed] [Google Scholar]
- 23.Hale V.L., Chen J., Johnson S., et al. Shifts in the fecal microbiota associated with adenomatous polyps. Cancer Epidemiol Biomarkers Prev. 2017;26(1):85–94. doi: 10.1158/1055-9965.EPI-16-0337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang K., Qin X., Qiu J., et al. Desulfovibrio desulfuricans aggravates atherosclerosis by enhancing intestinal permeability and endothelial TLR4/NF-κB pathway in Apoe (-/-) mice. Genes Dis. 2023;10(1):239–253. doi: 10.1016/j.gendis.2021.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao Z., Ning J., Bao X.Q., Shang M., Ma J., et al. Fecal microbiota transplantation protects rotenone-induced Parkinson's disease mice via suppressing inflammation mediated by the lipopolysaccharide-TLR4 signaling pathway through the microbiota-gut-brain axis. Microbiome. 2021;9(1):226. doi: 10.1186/s40168-021-01107-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nie Y., Xie X.Q., Zhou L., et al. Desulfovibrio fairfieldensis-derived outer membrane vesicles damage epithelial barrier and induce inflammation and pyroptosis in macrophages. Cells. 2022;12(1) doi: 10.3390/cells12010089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Petersen C., Bell R., Klag K.A., et al. T cell-mediated regulation of the microbiota protects against obesity. Science. 2019;365(6451) doi: 10.1126/science.aat9351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang Y., Yu H., Zhang J., et al. Cul4A-DDB1-mediated monoubiquitination of phosphoglycerate dehydrogenase promotes colorectal cancer metastasis via increased S-adenosylmethionine. J Clin Invest. 2021;131(21) doi: 10.1172/JCI146187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mosca L., Pagano M., Borzacchiello L., et al. S-adenosylmethionine increases the sensitivity of human colorectal cancer cells to 5-fluorouracil by inhibiting P-glycoprotein expression and NF-κB activation. Int J Mol Sci. 2021;22(17) doi: 10.3390/ijms22179286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Borzacchiello L., Veglia Tranchese R., Grillo R., et al. S-adenosylmethionine inhibits colorectal cancer cell migration through mirna-mediated targeting of Notch signaling pathway. Int J Mol Sci. 2022;23(14) doi: 10.3390/ijms23147673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cave D.D., Desiderio V., Mosca L., et al. S-Adenosylmethionine-mediated apoptosis is potentiated by autophagy inhibition induced by chloroquine in human breast cancer cells. J Cell Physiol. 2018;233(2):1370–1383. doi: 10.1002/jcp.26015. [DOI] [PubMed] [Google Scholar]
- 32.Chik F., Machnes Z., Szyf M. Synergistic anti-breast cancer effect of a combined treatment with the methyl donor S-adenosyl methionine and the DNA methylation inhibitor 5-aza-2'-deoxycytidine. Carcinogenesis. 2014;35(1):138–144. doi: 10.1093/carcin/bgt284. [DOI] [PubMed] [Google Scholar]
- 33.Mosca L., Pagano M., Ilisso C.P., et al. AdoMet triggers apoptosis in head and neck squamous cancer by inducing ER stress and potentiates cell sensitivity to cisplatin. J Cell Physiol. 2019;234(8):13277–13291. doi: 10.1002/jcp.28000. [DOI] [PubMed] [Google Scholar]
- 34.Galizia I., Oldani L., Macritchie K., et al. S-adenosyl methionine (SAMe) for depression in adults. Cochrane Database Syst Rev. 2016;10(10):Cd011286. doi: 10.1002/14651858.CD011286.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ullah H., Khan A., Rengasamy K.R.R., Di Minno A., Sacchi R., Daglia M. The efficacy of S-adenosyl methionine and probiotic supplementation on depression: a synergistic approach. Nutrients. 2022;14(13) doi: 10.3390/nu14132751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aury-Landas J., Bazille C., Allas L., et al. Anti-inflammatory and chondroprotective effects of the S-adenosylhomocysteine hydrolase inhibitor 3-Deazaneplanocin A, in human articular chondrocytes. Sci Rep. 2017;7(1):6483. doi: 10.1038/s41598-017-06913-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.De Silva V., El-Metwally A., Ernst E., Lewith G., Macfarlane G.J. Evidence for the efficacy of complementary and alternative medicines in the management of osteoarthritis: a systematic review. Rheumatology. 2011;50(5):911–920. doi: 10.1093/rheumatology/keq379. [DOI] [PubMed] [Google Scholar]
- 38.Rutjes A.W., Nüesch E., Reichenbach S., Jüni P. S-Adenosylmethionine for osteoarthritis of the knee or hip. Cochrane Database Syst Rev. 2009;2009(4) doi: 10.1002/14651858.CD007321.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mato J.M., Lu S.C. Role of S-adenosyl-L-methionine in liver health and injury. Hepatology. 2007;45(5):1306–1312. doi: 10.1002/hep.21650. [DOI] [PubMed] [Google Scholar]
- 40.Chen H., Gao S., Liu W., et al. RNA N (6)-methyladenosine methyltransferase METTL3 facilitates colorectal cancer by activating the m (6)A-GLUT1-mTORC1 Axis and is a therapeutic target. Gastroenterology. 2021;160(4):1284–1300.e16. doi: 10.1053/j.gastro.2020.11.013. [DOI] [PubMed] [Google Scholar]
- 41.Li T., Hu P.S., Zuo Z., et al. METTL3 facilitates tumor progression via an m (6)A-IGF2BP2-dependent mechanism in colorectal carcinoma. Mol Cancer. 2019;18(1):112. doi: 10.1186/s12943-019-1038-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pan J., Liu F., Xiao X., et al. METTL3 promotes colorectal carcinoma progression by regulating the m6A-CRB3-Hippo axis. J Exp Clin Cancer Res. 2022;41(1):19. doi: 10.1186/s13046-021-02227-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou D., Tang W., Xu Y., et al. METTL3/YTHDF2 m6A axis accelerates colorectal carcinogenesis through epigenetically suppressing YPEL5. Mol Oncol. 2021;15(8):2172–2184. doi: 10.1002/1878-0261.12898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen H., Pan Y., Zhou Q., et al. METTL3 inhibits antitumor immunity by targeting m (6)A-BHLHE41-CXCL1/CXCR2 Axis to promote colorectal cancer. Gastroenterology. 2022;163(4):891–907. doi: 10.1053/j.gastro.2022.06.024. [DOI] [PubMed] [Google Scholar]
- 45.Shen C., Xuan B., Yan T., et al. m (6)A-dependent glycolysis enhances colorectal cancer progression. Mol Cancer. 2020;19(1):72. doi: 10.1186/s12943-020-01190-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li M., Xia M., Zhang Z., et al. METTL3 antagonizes 5-FU chemotherapy and confers drug resistance in colorectal carcinoma. Int J Oncol. 2022;61(3) doi: 10.3892/ijo.2022.5396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Barbieri I., Tzelepis K., Pandolfini L., et al. Promoter-bound METTL3 maintains myeloid leukaemia by m (6)A-dependent translation control. Nature. 2017;552(7683):126–131. doi: 10.1038/nature24678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vu L.P., Pickering B.F., Cheng Y., et al. The N (6)-methyladenosine (m (6)A)-forming enzyme METTL3 controls myeloid differentiation of normal hematopoietic and leukemia cells. Nat Med. 2017;23(11):1369–1376. doi: 10.1038/nm.4416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yankova E., Blackaby W., Albertella M., et al. Small-molecule inhibition of METTL3 as a strategy against myeloid leukaemia. Nature. 2021;593(7860):597–601. doi: 10.1038/s41586-021-03536-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang H., Xu B., Shi J. N6-methyladenosine METTL3 promotes the breast cancer progression via targeting Bcl-2. Gene. 2020;722 doi: 10.1016/j.gene.2019.144076. [DOI] [PubMed] [Google Scholar]
- 51.Chen M., Wei L., Law C.T., et al. RNA N6-methyladenosine methyltransferase-like 3 promotes liver cancer progression through YTHDF2-dependent posttranscriptional silencing of SOCS2. Hepatology. 2018;67(6):2254–2270. doi: 10.1002/hep.29683. [DOI] [PubMed] [Google Scholar]
- 52.Lin X., Chai G., Wu Y., et al. RNA m (6)A methylation regulates the epithelial mesenchymal transition of cancer cells and translation of Snail. Nat Commun. 2019;10(1):2065. doi: 10.1038/s41467-019-09865-9. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 53.Xu H., Wang H., Zhao W., et al. SUMO1 modification of methyltransferase-like 3 promotes tumor progression via regulating Snail mRNA homeostasis in hepatocellular carcinoma. Theranostics. 2020;10(13):5671–5686. doi: 10.7150/thno.42539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cheng M., Sheng L., Gao Q., et al. The m (6)A methyltransferase METTL3 promotes bladder cancer progression via AFF4/NF-κB/MYC signaling network. Oncogene. 2019;38(19):3667–3680. doi: 10.1038/s41388-019-0683-z. [DOI] [PubMed] [Google Scholar]
- 55.Sugimura N., Li Q., Chu E.S.H., et al. Lactobacillus gallinarum modulates the gut microbiota and produces anti-cancer metabolites to protect against colorectal tumourigenesis. Gut. 2021;71(10):2011–2021. doi: 10.1136/gutjnl-2020-323951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ternes D., Tsenkova M., Pozdeev V.I., et al. The gut microbial metabolite formate exacerbates colorectal cancer progression. Nat Metab. 2022;4(4):458–475. doi: 10.1038/s42255-022-00558-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yang J., Wei H., Zhou Y., et al. High-fat diet promotes colorectal tumorigenesis through modulating gut microbiota and metabolites. Gastroenterology. 2022;162(1):135–149.e2. doi: 10.1053/j.gastro.2021.08.041. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.