Abstract
Objective
Adipose tissue mass is maintained by a balance between lipolysis and lipid storage. The contribution of adipose tissue lipogenesis to fat mass, especially in the setting of high-fat feeding, is considered minor. Here we investigated the effect of adipose-specific inactivation of the peroxisomal lipid synthetic protein PexRAP on fatty acid synthase (FASN)-mediated lipogenesis and its impact on adiposity and metabolic homeostasis.
Methods
To explore the role of PexRAP in adipose tissue, we metabolically phenotyped mice with adipose-specific knockout of PexRAP. Bulk RNA sequencing was used to determine transcriptomic responses to PexRAP deletion and 14C-malonyl CoA allowed us to measure de novo lipogenic activity in adipose tissue of these mice. In vitro cell culture models were used to elucidate the mechanism of cellular responses to PexRAP deletion.
Results
Adipose-specific PexRAP deletion promoted diet-induced obesity and insulin resistance through activation of de novo lipogenesis. Mechanistically, PexRAP inactivation inhibited the flux of carbons to ethanolamine plasmalogens. This increased the nuclear PC/PE ratio and promoted cholesterol mislocalization, resulting in activation of liver X receptor (LXR), a nuclear receptor known to be activated by increased intracellular cholesterol. LXR activation led to increased expression of the phospholipid remodeling enzyme LPCAT3 and induced FASN-mediated lipogenesis, which promoted diet-induced obesity and insulin resistance.
Conclusions
These studies reveal an unexpected role for peroxisome-derived lipids in regulating LXR-dependent lipogenesis and suggest that activation of lipogenesis, combined with dietary lipid overload, exacerbates obesity and metabolic dysregulation.
Keywords: Adipose tissue, Cholesterol, De novo lipogenesis, Diabetes, Plasmalogen, Liver X receptor
Highlights
-
•
Adipose-specific knockout of PexRAP promotes diet-induced obesity and insulin resistance.
-
•
PexRAP inactivation decreases ethanolamine plasmalogens, peroxisome-derived phospholipids.
-
•
PexRAP deletion alters adipocyte membrane phospholipid composition, causing cholesterol mislocalization and LXR activation.
-
•
LXR activation induces FASN-mediated lipogenesis, leading to diet-induced obesity and metabolic dysregulation.
1. Introduction
Obesity continues to rise globally, increasing risk and prevalence of diabetes, hypertension, and non-alcoholic fatty liver disease [1]. Unhealthy expansion of adipose tissue in obesity promotes disease through complex and multifactorial mechanisms. Adipose tissue mass is maintained through a balance between lipolysis and lipid storage. Disruption of this balance can lead to storage of lipids in other tissues, such as liver and skeletal muscle, impairing their functions via lipotoxicity [2]. The modern western diet contains a high proportion of calories from energy-rich fats, and in the setting of positive energy balance, these lipids are stored in adipose tissue, increasing susceptibility to obesity. In such a state of lipid excess, the role of lipogenesis, the endogenous synthesis of lipids from simple carbon precursors, is not obvious.
Increasing evidence implicates peroxisomes, organelles specialized for lipid metabolism, including lipid synthesis, in metabolic regulation [3]. Peroxisomes are versatile metabolic organelles involved in ether lipid biosynthesis, fatty acid oxidation, bile acid synthesis, and reactive oxygen species production and neutralization [4]. The importance of peroxisomes to mammalian physiology is demonstrated by the deleterious phenotypes associated with Zellweger spectrum peroxisome biogenesis disorders [5]. Ether lipids, such as plasmalogens, have recently been shown to regulate ferroptosis susceptibility, extend Caenorhabditis elegans lifespan, and function as cellular antioxidants [[6], [7], [8]]. Plasmalogens are also important for assembly of lipid raft microdomains, cholesterol-rich membrane regions involved in cellular signaling [9]. Plasmalogen deficiency leads to disruption of lipid rafts and cholesterol mislocalization to a perinuclear compartment [10]. However, the broader physiological roles of these peroxisome-derived lipids are unclear. Of note, how peroxisomal lipid synthesis affects metabolic homeostasis remains unknown.
Peroxisomes use the glycolytic intermediate dihydroxyacetone phosphate (DHAP) as a substrate for lipid biosynthesis. When conditions favor the routing of carbon away from glycolysis and towards peroxisomal ether lipid synthesis, peroxisomal membrane protein 2 (encoded by Pxmp2) shuttles DHAP into peroxisomes for entry into the acyl DHAP pathway, localized in peroxisomes and the endoplasmic reticulum (ER), for production of ether lipids and other phospholipid species. In the peroxisomal component of this pathway, DHAP is converted into acyl-DHAP, which may subsequently have its acyl group exchanged for an alkyl group to yield alkyl-DHAP. In the final peroxisome-localized step of this pathway, the sn-1 carbonyl of acyl- or alkyl-DHAP is reduced by acyl/alkyl DHAP reductase [4]. We previously reported that this protein is encoded by Dhrs7b and renamed the protein PexRAP (Peroxisomal Reductase Activating PPARγ) due to its role in generating partial agonists for the nuclear receptor PPARγ [11].
PexRAP-catalyzed reduction of acyl- or alkyl-DHAP yields lysophosphatidic acid (LPA), a diacyl phospholipid precursor, or 1-O-Alkyl-G3P (AGP), an ether lipid precursor, respectively. PexRAP is localized to both ER and peroxisomes and appears to have distinct, location-dependent preferences for acyl-versus alkyl-DHAP in HeLa cells, though this effect has not been explored in other contexts [12]. LPA, like ether lipids, is highly bioactive and functions as a cellular signaling molecule, which can agonize PPARγ and activate five known G protein-coupled receptors, in addition to serving as a precursor for downstream diacyl phospholipid synthesis [13,14]. As a key enzyme in the conversion of DHAP to LPA, AGP, and their downstream diacyl phospholipids and ether lipids, PexRAP represents a central node in peroxisomal lipid biosynthesis. We previously reported that global knockout of PexRAP in mice is lethal, but acutely induced whole-body knockout of PexRAP increases adipocyte browning through the ability of PexRAP to compete with the thermogenic transcriptional co-regulator PRDM16 for binding to PPARγ [15]. To further explore the role of PexRAP in metabolic homeostasis, we generated an adipose-specific PexRAP knockout mouse model (PexRAP-AKO).
Here, we show that PexRAP-AKO mice unexpectedly have increased diet-induced obesity and impaired glucose homeostasis despite elevated expression of thermogenic genes, such as UCP1, in subcutaneous white adipose tissue (WAT). Mechanistically, PexRAP inactivation in adipocytes alters nuclear membrane phospholipid composition, decreasing ether lipid analogs of phosphatidylethanolamine (PE) and increasing certain species of conventional diacyl phosphatidylcholine (PC), leading to increased PC/PE ratio and cholesterol mislocalization. Cholesterol mislocalization to the nucleus activates the sterol regulated nuclear receptor liver X receptor (LXR), which promotes phospholipid remodeling and increases de novo lipogenesis (DNL). In adipose tissue, the PexRAP inactivation-induced increase in lipogenesis is sufficient to drive a systemic metabolic phenotype characterized by increased diet induced obesity, insulin resistance, and glucose intolerance. This deleterious phenotype is notably absent in a brown adipose specific PexRAP knockout mouse model (PexRAP-BKO), suggesting that the PexRAP-AKO phenotype is driven by processes in white adipose tissue. At the cellular level, these results uncover a novel relationship between peroxisome-derived phospholipids and DNL. Systemically, our findings imply that while adipose tissue DNL is generally considered to be metabolically beneficial, aberrant adipose tissue DNL is deleterious and may be induced in obese states characterized by adipose tissue inflammation and insulin resistance. Together, these studies suggest that peroxisomal lipid synthesis mediated by PexRAP regulates metabolic homeostasis by balancing adipocyte browning with lipogenesis activation and the associated metabolic dysregulation.
2. Materials and methods
2.1. Experimental animals
Production of PexRAPLox/Lox mice on the C57BL/6J genetic background has been described previously [16]. To generate mice with adipose-specific knockout of PexRAP (PexRAP-AKO), PexRAP floxed animals were crossed with adiponectin-Cre mice obtained from the Jackson Laboratory (Stock No. 028020). To generate mice with brown adipose-specific knockout of PexRAP (PexRAP-BKO), PexRAP floxed animals were crossed with Ucp1-Cre mice obtained from the Jackson Laboratory (Stock No. 024670). PexRAPLox/Lox mice without Cre were used as a control for PexRAP-AKO and PexRAP-BKO mice. Genotyping was performed using previously described primer sets [11,16]. All experiments used male mice unless otherwise stated. All mice were kept on a 12-h light/dark cycle and provided ad libitum access to food (Purina 5053) and water. For high fat diet feeding experiments, animals were maintained on chow diet until 8 weeks of age, at which time they were given high fat diet (Research Diets D12492, 60% kcal from fat). For the experiment in Figure S1B a high fat diet containing 42% kcal from fat (Envigo TD.88137) was used. All animal experiments were performed in accordance with procedures approved by the Institutional Animal Care and Use Committee at Washington University School of Medicine.
2.2. Cell lines
Stromal vascular fraction (SVF) cells from adult iWAT were isolated from wild-type C57BL/6J mice by collagenase and dispase digestion, immortalized, and differentiated into adipocytes as previously reported [15]. 3T3-L1 cells were maintained in DMEM with 10% newborn calf serum. To differentiate 3T3-L1 cells to white adipocytes, they were first allowed to grow confluent for two days, and then treated with DMEM with 10% fetal bovine serum, 1 μg/ml insulin, 0.25 μM dexamethasone, and 0.5 mM methyl isobutylxanthine for three days. Next, DMEM with 10% fetal bovine serum and 1 μg/ml insulin was added for two days. The then-differentiated 3T3-L1 adipocytes were maintained in DMEM with 10% fetal bovine serum. Human embryonic kidney 293T (HEK293T) cells were maintained in DMEM supplemented with 10% FBS.
2.3. Plasmid constructs
Plasmids encoding lentivirus shRNA for mouse PexRAP (TRCN0000181732), SREBP1 (TRCN0000055325), LXRα (TRCN0000026011), and LXRβ (TRCN0000026048) were obtained from Sigma-Aldrich (St. Louis, MO). Packaging vector psPAX2 (12260), envelope vector pMD2.G (12259), and scrambled shRNA plasmid (1864) were obtained from Addgene. The LXRE-EGFP-fLuc lentiviral reporter plasmid was obtained from System Biosciences (TR102PA-P).
2.4. Immunohistochemistry and immunofluorescence imaging
For iWAT SVF cell immunofluorescence imaging, cells were fixed with 10% formalin and permeabilized with 0.1% Triton X-100 in PBS (PBS-T). Cells were blocked with PBS with 3% BSA for 1 h and subsequently incubated overnight with both rabbit polyclonal PexRAP at 1:100 dilution and PMP70-atto-488 (Sigma) in PBS with 1% BSA overnight. Custom-made PexRAP antibody obtained from Cocalico Biologicals was produced by injecting rabbit with GST-PexRAP and then affinity purified. Alexa Fluor conjugated secondary antibody (Invitrogen A-11008) was used at a dilution of 1:500 in PBS with 1% BSA at room temperature for 1 h. Cells were mounted with Vectashield mounting medium (H-1000) and imaged with a 20× oil objective and 3× zoom on a Nikon A1R confocal microscope. For the Filipin III stain in Figure 6B, differentiated 3T3-L1 adipocytes were stained according to the Abcam Cholesterol Assay Kit protocol (ab133116), with the only modification being that after the final wash step the cells were incubated with Nuclear-ID dye (Enzo Life Sciences, ENZ-52406) according to the manufacturer's protocol. Cells were finally imaged in Vectashield mounting medium (H-1000). For IHC detection of F4/80, formalin-fixed, paraffin-embedded adipose tissue sections were de-paraffinized with a series of xylene and ethanol solutions, incubated with anti-F4/80 antibody (Cell Signaling Technology), stained with the Vectastain Universal Elite ABC kit (Vector Labs) and imaged with a Leica DMI4000B microscope.
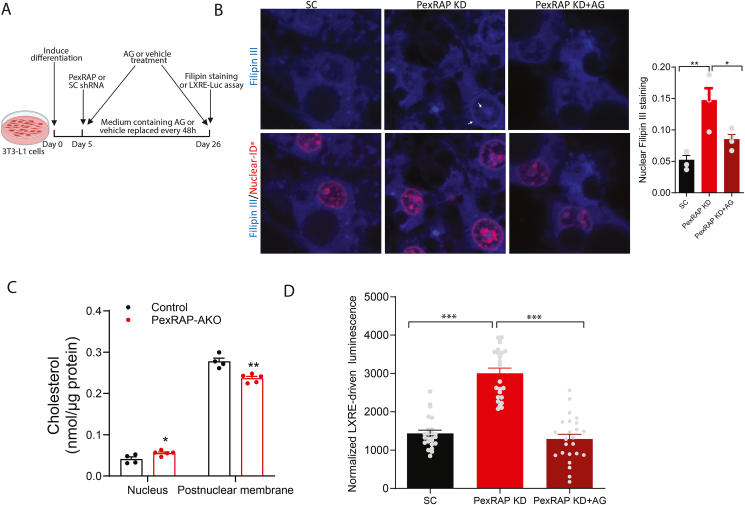
Figure 6.
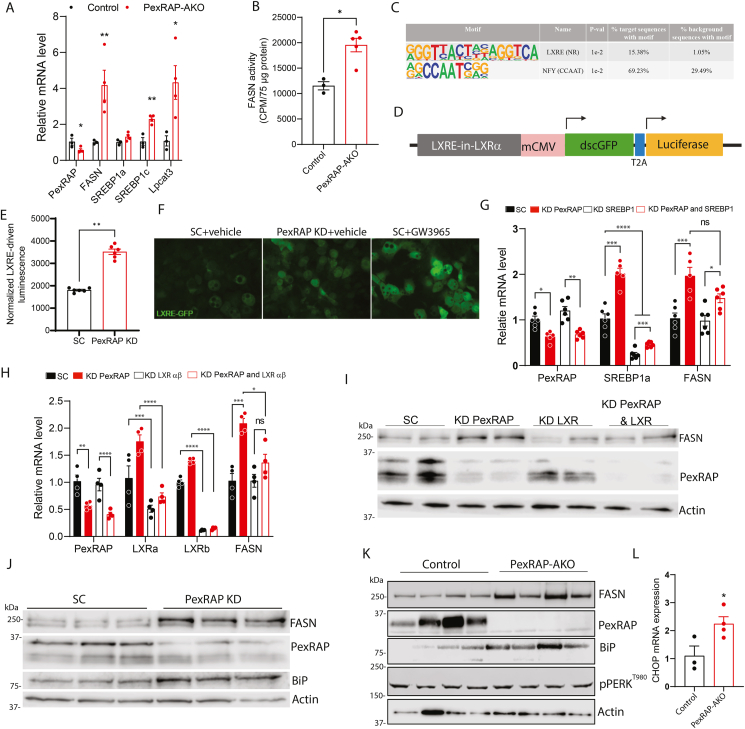
PexRAP depletion is associated with cholesterol mislocalization to the nucleus and LXR activation in an alkyl glycerol-dependent manner.A) Schematic depicting timeline of alkyl glycerol (AG) rescue in PexRAP-KD or control 3T3-L1 adipocytes. B) Fluorescence microscopy of cholesterol (Filipin III, blue) and nuclei (Nuclear-ID, red) in differentiated 3T3-L1 adipocytes. Nuclear localization of cholesterol is indicated by white arrows. Quantification of mean fluorescence of nuclear Filipin III staining is shown on the right (n = 4). C) GC–MS quantification of total cholesterol in nuclear and postnuclear membrane fractions of iWAT tissue from HFD-fed control or PexRAP-AKO mice (n = 4–5). D) Normalized luminescence of 3T3-L1 cells stably transfected with a plasmid expressing GFP and firefly luciferase under control of a LXR response element (LXRE), differentiated to white adipocytes, treated with scrambled (SC) or PexRAP shRNA and finally treated with AG (n = 24). Data are shown as mean ± SEM. Comparisons between groups were made with a two-tailed unpaired Student's t-test (B,C,D).
2.5. LXRE reporter assay
3T3-L1 cells were transduced with LXRE-GFP-fLuc lentivirus for two days and subsequently selected with puromycin for four days. Cells were then differentiated as described above. On day 5 of differentiation, cells were treated with lentivirus for scrambled or PexRAP shRNA for 48 h. Cells were maintained in DMEM with 10% FBS for another 48 h prior to being harvested. Luminescence was measured using a Promega Luciferase Assay kit (E1500) and normalized to cellular protein as measured with Pierce BCA Assay kit (Thermo Fisher 23227). For analysis of GFP signal, another plate of cells was treated equivalently, however instead of being harvested these cells were visualized with a Leica DMI4000B microscope and Leica CTR4000 laser system.
2.6. RT-qPCR
Total RNA was isolated using TRIzol reagent (Invitrogen 15596026). 2 μg of isolated RNA was reverse transcribed using High-Capacity cDNA Reverse Transcription kit (Applied Biosystems 4368814). Quantitative PCR (qPCR) was performed with a StepOnePlus Real-Time PCR System (Applied Biosystems) using PowerUP SYBR Green Master Mix (Applied Biosystems A25742). Assays were performed in duplicate, normalized to ribosomal protein L32, and analyzed with the ΔΔCt method.
2.7. Ex vivo lipolysis assay
An ex vivo lipolysis assay was performed based on the protocol presented in [17]. Briefly, 50–100 mg chunks of gWAT tissue were harvested from control and HFD-fed PexRAP-AKO mice and immediately transferred to prewarmed (37 °C) KRBH buffer in 12-well plates. Once all tissues were harvested, the buffer was aspirated and replaced with a prewarmed (37 °C) KRBH-BSA buffer containing either vehicle or 10 μM isoproterenol. Tissues were incubated for 2 h at 37 °C, after which 200 μL aliquots of media were taken and analyzed with a Lipolysis Colorimetric Assay Kit (Sigma MAK211) according to the manufacturer's instructions. Measurements were normalized to exact tissue weight, which was recorded during harvest of tissues.
2.8. RNA-sequencing and analysis
iWAT was collected from 8-week-old male control or PexRAP-AKO mice. RNA was harvested using RNeasy Lipid Tissue Mini Kit (Qiagen). 1 μg of total RNA was used for the construction of sequencing libraries. Samples were then submitted to the Genome Technology Access Center (GTAC) at Washington University School of Medicine in St. Louis for RNA sequencing and analysis. Samples were prepared according to library kit manufacturer's protocol, indexed, pooled, and sequenced on an Illumina HiSeq. Reads were aligned to Mus musculus reference build Ensembl_R76 with STAR v2.0.4b, after which gene counts were obtained using the Subread featureCount program (v1.4.5). Data are deposited to NCBI GEO under GEO Accession GSE227912.
Differential expression analysis was performed from gene-level counts using the R/Bioconductor package edgeR. Genes with Benjamini–Hochberg false-discovery rate less than or equal to 0.05 were considered differentially expressed. Gene ontology-based pathway enrichment analysis was performed with the R/Bioconductor package limma using functions goana and topGO. Gene set enrichment analysis was conducted on a gene list ranked by log(fold-change) times −log10(FDR) using the R/Bioconductor package clusterProfiler. RNA-sequencing and pathway enrichment results were visualized with the R packages ggplot2 and ComplexHeatmap.
Motif enrichment near the transcription start sites of differentially expressed genes was analyzed using hypergeometric optimization of motif enrichment (HOMER). Briefly, the findMotifs.pl function was called on all differentially expressed genes (excluding PexRAP itself) to search for motifs of length 8, 10, or 12 from −400 to +100 relative to the transcription start site using the mouse (M. musculus) promoter set. Motifs with p-value less than or equal to 0.05 were considered significantly enriched.
2.9. 14C-malonyl CoA de novo lipogenesis assay
De novo lipogenesis was assayed in whole tissue lysate using a protocol adapted from previously described methods [18,19]. Briefly, either fresh or snap-frozen iWAT tissue was homogenized using a Dounce tissue homogenizer in 3× volume of 0.25 M sucrose (Sigma Aldrich), 2 mM EDTA, 0.1 M KPO4, pH 7 buffer containing protease and phosphatase inhibitors (MilliporeSigma). Lysates were spun at 1,000 × g for 10 min at 4 °C and supernatant was moved to a clean tube. Protein concentration was measured with Pierce BCA Assay kit (Thermo Fisher 23227) and 75 μg of total protein from each lysate was moved to a clean tube and brought to a volume of 147 μL with the buffer. Each lysate was prewarmed at 37 °C for exactly 5 min before 103 μL of a prewarmed cocktail was added such that each reaction had 0.1 M KPO4 (pH 7), 20 nmol acetyl CoA (Cayman 16160), 20 nmol 12C-malonyl CoA (Cayman 16455), 0.5 mM NADPH, (Cayman 9000743) 12 mM DTT (Sigma Aldrich), 12 mM EDTA, and 0.1 μCi 14C-malonyl CoA (PerkinElmer). A no-NADPH control was included in each assay to ensure specificity of 14C incorporation to lipid fraction. The complete reactions were incubated at 37 °C for 15 min and quenched with 60% perchloric acid. The lipid fraction was extracted using 1:3 ethanol:petroleum ether. The petroleum ether fraction was dried overnight at room temperature. 3 mL of Ecoscinct XR (National Diagnostics) was added to each vial and samples were counted for 5 min each in a Beckman Coulter LS6500 liquid scintillation counter.
2.10. Subcellular fractionation
Inguinal WAT tissues were cut into small pieces and homogenized in 1× peroxisome extraction buffer (from Sigma Peroxisome Isolation Kit) using a glass dounce homogenizer. The homogenates were centrifuged at 800 × g for 10 min at 4 °C. The supernatants (membrane fraction) were transferred to a new tube and the pellets (nuclear fraction and tissue) were resuspended and totally homogenized using a dounce homogenizer. The clean nuclear fraction was pelleted by centrifuging at 400 × g for 10 min at 4 °C. For membrane fractions, the supernatants were centrifuged at 22,000 × g for 15 min at 4 °C and the pellets were collected.
2.11. Lipidomic analysis and carbon flux tracing
3T3-L1 cells were cultured and differentiated as described above. On day 5 of differentiation, cells were treated with lentivirus for scrambled or PexRAP shRNA for 48 h. Cells were then maintained in [U13C6]-glucose DMEM media with 10% FBS for 7 days. Cells were lysed in 1× peroxisome extraction buffer (from Sigma Peroxisome Isolation Kit) using a dounce homogenizer. Samples were normalized by total protein content (Thermo Fisher 23227) and lipids were extracted with 1:1 chloroform:methanol. The samples were dried under a stream of nitrogen and redissolved in methanol with 0.5% NH4+. Lipids were analyzed by ESI-MS on a Thermo Fisher LTQ Orbitrap Velos in both positive and negative ion modes scanning from 500 to 1000 m/z with resolution 100,000 (at m/z 400 Da). Data were processed by built-in Xcalibur software. PE and PC intensities were normalized to the spiked-in internal standards containing both PE (14:0/14:0) (Avanti) and PC (14:0/14:0) (Avanti) for normalization in negative and positive ion modes, respectively.
Nuclear and post-nuclear membrane fraction lipids were analyzed similarly. 3T3-L1 cells were cultured differentiated as described above. On day 5 of differentiation, cells were treated with lentivirus for scrambled or PexRAP shRNA for 48 h. Cells were then maintained in DMEM media with 10% FBS for 7 days prior to being harvested. Cells were lysed in 1× peroxisome extraction buffer (from Sigma Peroxisome Isolation Kit) using a dounce homogenizer. The nuclear fraction was pelleted by spinning the whole cell lysate at 1,000 × g for 10 min at 4 °C. The supernatant was subsequently spun at 21,000 × g for 25 min at 4 °C to pellet a post-nuclear membrane fraction. These fractions were normalized by total protein content. Lipid extraction and mass spectrometry analysis was the same as above.
2.12. Cholesterol measurement using GC–MS
For measurement of free cholesterol levels, lipids were extracted and dried as described above. The dried lipid residue was resuspended in 100 μL of a mixture of 1:0.8:2.2 (v/v/v) BSTFA with 1% TMCS:pyridine:acetonitrile, vortexed, and heated at 65 °C for 1 h.
Electron ionization (EI) GC–MS analysis was conducted on an Agilent (Santa Clara, CA USA) 7890A GC coupled with Agilent 5975C MSD using a 25-m Agilent J & W capillary column (DB-1; inner diameter, 0.25 mm; film thickness, 0.1 μm). An aliquot of 2 μL of the derivatized solution was injected in a 1/10 split mode onto the GC/MS system controlled by Agilent ChemStation software. The temperature program started at 200 °C and held for 3 min, ramped 10 °C/min until 300 °C, held at 300 °C for 7 min.
GC/MS analysis of cholesterol was acquired in selected ion mode. The dwelling times for ions selected for monitoring cholesterol (m/z 458.4, 368.3 and 329.3) and d7-cholesterol (m/z 465.4, 376.3, 336.3) were set at 100 ms. d7-Cholesterol and cholesterol peaks were eluted at 14.657 and 14.689 min, respectively, and the peak areas of the Indigenous cholesterol and the added d7-cholesterol internal standard peaks from samples were integrated and quantitation was achieved using the standard curve established by a series of injection of various cholesterol/d7-cholesterol ratio mixtures.
2.13. Statistical analysis
Data are presented as mean ± standard error of mean (SEM) unless stated otherwise. Comparisons between two groups were performed using an unpaired, two-tailed t test. When n > 19 comparisons were made, p values were adjusted for multiple comparisons with a Benjamini–Hochberg adjustment (referred to as false discovery rate, FDR). ANOVA with Bonferroni's or Tukey's multiple comparison test was used for comparisons between more than 2 groups. A p value (or adjusted p value, when relevant) less than 0.05 was considered significant. Statistical analysis and graphs were generated using either GraphPad Prism software or the R statistical programming language.
3. Results
3.1. Adipose-specific knockout of PexRAP promotes diet-induced obesity
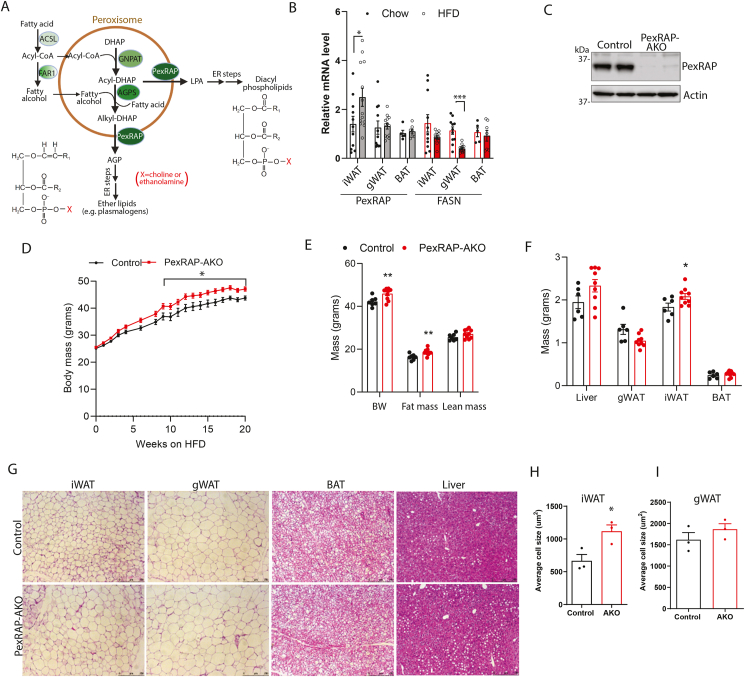
The acyl DHAP pathway of lipid synthesis is required for the production of ether-linked phospholipids, such as plasmalogens, and also contributes to the synthesis of conventional diacyl phospholipids [20]. The process begins in peroxisomes but is completed in the ER (Figure 1A). PexRAP, the terminal enzyme in the peroxisomal component of the pathway, reduces alkyl- or acyl-DHAP into the ether lipid precursor AGP or LPA, a precursor of conventional diacylphospholipids [11]. Immunofluorescence analysis confirmed that PexRAP is localized in peroxisomes in iWAT adipocytes (Figure S1A). We previously reported that inducible global knockout of PexRAP promotes robust browning of subcutaneous WAT and demonstrated that this effect is cell autonomous [15]. However, how PexRAP is related to diet-induced obesity has not been investigated. In wild-type C57BL/6J mice fed a high fat diet (HFD), PexRAP expression increased in iWAT, a subcutaneous adipose tissue depot which is prone to browning, while no change was detected in gonadal WAT (gWAT) or brown adipose tissue (BAT). Conversely, high fat feeding decreased the expression of the lipogenic enzyme fatty acid synthase (FASN) as expected, especially in gWAT (Figure 1B).
Figure 1.
PexRAP-AKO mice have increased diet induced obesity.A) Diagram of the peroxisomal pathway involved in ether lipid synthesis. The initial steps of ether lipid synthesis occur in peroxisomes, generating 1-O-alkyl glycerol-3-phosphate (AGP), a precursor of ether lipids, such as plasmalogens. The subsequent steps take place in the endoplasmic reticulum (ER). Peroxisomes can also generate lysophosphatidic acid (LPA), a precursor of conventional diacyl phospholipids. Structures of a diacyl phospholipid and a plasmalogen are shown, where X is usually a choline or an ethanolamine. Abbreviations: ACSL, acyl-CoA synthetase; AGPS, alkylglycerone phosphate synthase; DHAP, dihydroxyacetone phosphate; FAR1, fatty acyl-CoA reductase; GNAPT, glyceronephosphate O-acyltransferase; PexRAP, Peroxisomal Reductase activating PPARγ. B) qPCR analysis of PexRAP and FASN gene expression in wild-type C57BL/6J mice fed a chow or high-fat diet (60% kcal fat) for 22 weeks (n = 11–13 for iWAT and gWAT, 5–7 for BAT). C) Western blot analysis of iWAT tissue from control or PexRAP-AKO mice. D) Body weight of male control and PexRAP-AKO mice fed a high fat diet (HFD, 60% kcal fat) beginning at 8 weeks of age (n = 8–9). E) Body composition results from EchoMRI analysis of male control and PexRAP-AKO mice fed a HFD for 16 weeks (n = 7–9). F) Mass of tissues from male control and PexRAP-AKO mice fed a HFD for 16 weeks (n = 6–8). G) Hematoxylin and eosin (H&E) staining of formalin-fixed paraffin embedded (FFPE) tissues from male control and PexRAP-AKO mice fed a HFD for 19 weeks from which H) iWAT and I) gWAT cell area were quantified (n = 3 mice/group) using AdipoSoft/ImageJ [60]. Data are shown as mean ± SEM. Comparisons between groups were made with a two-tailed unpaired Student's t-test (B,D,E,F,H,I).
To investigate the role of PexRAP in diet-induced obesity and metabolic homeostasis, we studied mice with an adipose-specific knockout of PexRAP (PexRAP-AKO) that were generated by crossing adiponectin-Cre mice with PexRAP-floxed mice. PexRAP knockout in adipose tissue was validated by Western blot analysis (Figure 1C). Consistent with our previously published results [15], PexRAP-AKO mice fed an adjusted calories diet containing 42% kcal from fat (TD.88137) had no significant difference in body weight or body composition (Figure S1B). Surprisingly, high fat feeding (60 kcal% fat; Research Diets D12492) resulted in significantly higher body weight in PexRAP-AKO compared to control animals (Figure 1D). Body composition analysis using Echo-MRI revealed a specific elevation of fat mass with no change in lean mass in PexRAP-AKO mice (Figure 1E). The knockout mice had heavier iWAT fat depots, though masses of gWAT, liver, and BAT did not differ from control mice (Figure 1F). Histologic analysis of adipose tissue depots and liver revealed that the adipocytes in iWAT were markedly larger in PexRAP-AKO mice, while there were no major differences in the morphology of other tissues (Figure 1G). Quantification of adipocyte size confirmed that adipocytes in iWAT, but not gWAT, were significantly larger in the knockout animals (Figure 1H–I).
The increased adiposity in PexRAP-AKO was not associated with altered food intake, ambulatory movement, fecal output, or fecal fat content in HFD-fed mice, suggesting that the exacerbated diet induced obesity in these mice is not due to changes in caloric intake, nutrient absorption, or physical activity (Figure S1C–F). Resting energy expenditure measured after 12 weeks of high fat feeding, prior to emergence of a body weight difference between the genotypes, trended lower in PexRAP-AKO mice, but the effects did not reach statistical significance (Figure S1G–H), possibly because indirect calorimetry is not sufficiently sensitive to detect subtle changes in energy expenditure that impact body weight over a prolonged period [21,22]. Moreover, indirect calorimetry indicated respiratory exchange ratio was not altered in PexRAP-AKO mice (Figure S1I).
3.2. PexRAP-AKO mice have disrupted glucose homeostasis, impaired insulin signaling, and increased adipose tissue inflammation
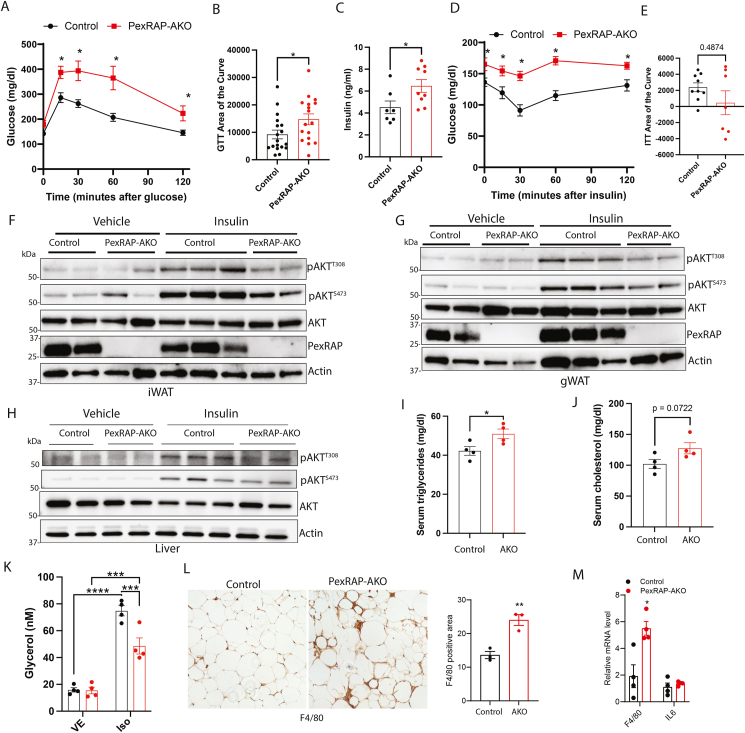
While excessive adiposity is typically associated with increased risk of developing diabetes, obesity does not necessarily lead to metabolic dysfunction [23,24]. We thus sought to characterize the metabolic phenotype of HFD fed PexRAP-AKO mice to determine where these mice fall on the disease spectrum. Under high fat feeding, PexRAP-AKO mice exhibited impaired glucose tolerance (Figure 2A–B) in the context of increased insulin secretion (Figure 2C), suggesting that the knockout mice are insulin resistant. Analysis of the area of the curve (AOC), which most accurately reflects glucose tolerance or insulin responsiveness independent of alterations in basal blood glucose [25], confirmed that adipose tissue PexRAP deficiency significantly increases glucose intolerance (Figure 2B). Insulin tolerance testing suggested that HFD-fed PexRAP-AKO mice have decreased insulin sensitivity, though the results were confounded by mildly increased basal glucose levels in the knockout mice (Figure 2D). Subtracting the baseline area (AOC) indicated a trend towards insulin resistance in PexRAP-AKO mice (Figure 2E). Elevated basal glucose levels despite increased insulin secretion suggested hepatic insulin resistance. However, there was no alteration in pyruvate tolerance in PexRAP-AKO mice (Figure S2A), suggesting that increased gluconeogenesis is not involved in the observed metabolic phenotype. Western blot analysis of insulin-stimulated AKT phosphorylation in iWAT (Figure 2F), gWAT (Figure 2G), liver (Figure 2H), BAT (Figure S2B) and skeletal muscle (Figure S2C) revealed impaired insulin signaling in these metabolic tissues and suggested that PexRAP-AKO mice have systemic insulin resistance.
Figure 2.
PexRAP-AKO mice fed a high fat diet (60% kcal fat) have impaired glucose homeostasis, decreased insulin signaling, hyperlipidemia, and increased immune cell infiltration of adipose tissue.A) Glucose tolerance test and B) area of the curve measurements of male control and PexRAP-AKO mice fed a HFD for 12 weeks (n = 17–18). C) Glucose-stimulated insulin secretion of male control and PexRAP-AKO mice fed a HFD (n = 7–9). D) Insulin tolerance test and E) area of the curve measurements of male control and PexRAP-AKO mice fed a HFD for 12 weeks (n = 7–9). F–H) Western blot analysis in iWAT (F), gWAT (G), and liver (H) of male HFD-fed control or PexRAP-AKO mice injected intraperitoneally with vehicle or insulin. Tissues were harvested 10 min after insulin injection. AKT phosphorylation was analyzed at Thr308 and Ser473. I) Serum triglyceride and J) cholesterol levels of ad-libitum fed male control or PexRAP-AKO mice following 12 weeks of HFD feeding (n = 4). K) Media glycerol levels (normalized to tissue weight) of fresh iWAT explants from HFD-fed control or PexRAP-AKO mice incubated in media containing vehicle or isoproterenol for 2 h (n = 4). L) F4/80 staining and quantification in gWAT tissue from male control and PexRAP-AKO mice fed a HFD for 16 weeks. M) qPCR analysis of gene expression of gWAT tissue from male control and PexRAP-AKO mice fed a HFD for 16 weeks. Data are shown as mean ± SEM. Comparisons between groups were made with a two-tailed unpaired Student's t-test (A,B,C,D,E,I,J,L,M) or two-way ANOVA with Bonferroni's multiple comparison test (K).
Serum analysis in ad-libitum fed PexRAP-AKO mice revealed that, although free fatty acid levels were unaffected (Figure S2D), triglycerides were significantly increased (Figure 2I) and cholesterol was almost significantly increased (Figure 2J). Assaying lipolysis ex-vivo showed that PexRAP inactivation does not alter basal lipolysis in iWAT of HFD-fed mice but decreases lipolytic capacity following stimulation with isoproterenol (Figure 2K), suggesting that the observed hyperlipidemia is not due to increased lipolysis and likely reflects increased adipose tissue lipid synthesis or elevated hepatic very-low density lipoprotein secretion associated with reduced insulin action in the liver. Further supporting a lipid-mediated origin of metabolic disruption, levels of circulating leptin and resistin, two key metabolic hormones, were unaltered in PexRAP-AKO mice (Figure S2E–F).
Finally, obesity is associated with adipose tissue inflammation, which contributes to insulin resistance and NAFLD [26]. Since obesity-associated inflammation is more intense in visceral WAT as compared to subcutaneous WAT [27], we assessed recruitment of macrophages in gWAT, a visceral adipose depot. Immunohistochemistry analysis demonstrated that PexRAP-AKO mice exhibit increased F4/80+ macrophage infiltration into gWAT with characteristic crown-like structures (Figure 2L), a result corroborated by quantitative real-time PCR (qPCR) analysis of F4/80 mRNA levels (Figure 2M). Together, these findings reveal that HFD-fed PexRAP-AKO mice have multiple hallmarks of metabolic syndrome and obesity-associated diabetes including insulin resistance, glucose intolerance, hyperlipidemia, and adipose tissue inflammation.
3.3. BAT-specific knockout of PexRAP is not sufficient to affect adiposity or metabolism in HFD-fed mice
To determine if deletion of PexRAP in brown fat alone is sufficient to affect adiposity and metabolism, we generated mice with BAT-specific knockout of PexRAP (PexRAP-BKO) by crossing PexRAP floxed mice with UCP1-Cre mice. Gene expression analysis confirmed the selective knockout of PexRAP in BAT with no change in PexRAP expression in iWAT (Figure S3A). Metabolic phenotyping of HFD fed mice revealed that PexRAP-BKO did not alter diet induced obesity (Figure S3B), body composition (Figure S3C), or systemic glucose homeostasis, including glucose tolerance (Figure S3D) or insulin sensitivity (Figure S3E). Gene expression analysis (Figure S3F) and cold tolerance testing (Figure S3G) indicated that the thermogenic function of BAT was also not affected by BAT-specific knockout of PexRAP. Together, these results suggest that metabolic dysfunction associated with pan-adipose knockout of PexRAP is likely mediated by disruption of peroxisomal lipid synthesis in WAT or both WAT and BAT.
3.4. PexRAP-AKO mice exhibit increased expression of genes involved in de novo lipogenesis and cholesterol synthesis in iWAT
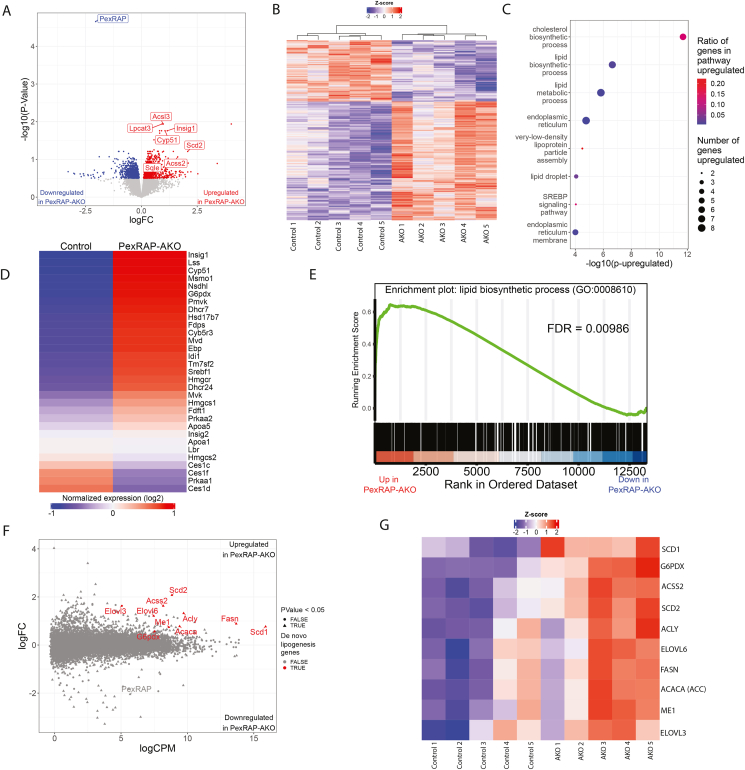
While the metabolic phenotype of HFD-fed PexRAP-AKO mice is systemic, we hypothesized that the underlying mechanism is adipose tissue-autonomous. To explore the primary effects of PexRAP inactivation in adipose tissue independent of secondary effects of the observed increases in diet-induced obesity and glucose homeostasis, we conducted bulk RNA sequencing (RNA-seq) on iWAT from chow-fed PexRAP-AKO and control mice. Differential expression analysis revealed that adipose-specific PexRAP knockout significantly alters the abundance of many mRNAs and confirmed effective PexRAP ablation (Figure 3A–B). In response to PexRAP knockout, the two most significantly upregulated genes are lysophosphatidylcholine acyltransferase 3 (Lpcat3) and acyl-CoA synthetase long chain family member 3 (Acsl3) (Figure 3A). Lpcat3 is a phospholipid remodeling enzyme that plays a key role in ameliorating lipotoxic ER stress in the liver [28,29], and Acsl3 activates long-chain fatty acids into fatty Acyl-CoA esters and is induced in response to ER stress in the liver [30,31], together suggesting that the predominant effects of PexRAP-AKO relate to its role in lipid metabolism and that ER stress might be one feature of lifelong PexRAP deletion.
Figure 3.
PexRAP-AKO increases transcription of genes involved in de novo lipogenesis and cholesterol synthesis in iWAT.A) Volcano plot of differentially expressed genes from bulk RNA-sequencing analysis of iWAT from PexRAP and control mice. Several genes that were very significantly increased are highlighted. B) Heatmap of logCPM value-derived Z-scores. C) Gene ontology (GO) pathway enrichment analysis of genes differentially expressed (FDR ≤ 0.05) in PexRAP-AKO mice. D) Heatmap of all genes in the cholesterol biosynthetic process GO term (GO:0006695). E) Enrichment plot of GO:0008610 (lipid biosynthetic process), a top result of an unbiased GSEA analysis. F) MA plot (logFC vs. logCPM) of results of differential expression analysis of RNA-seq data from control and PexRAP-AKO mice (n = 5/group for all). G) Heatmap of key lipid synthetic genes.
Paradoxically, despite the increased diet-induced obesity, PexRAP inactivation also resulted in increased expression of multiple genes involved in UCP1-dependent thermogenesis (Figure S4A), consistent with our previous observation that acute PexRAP deletion promotes adipocyte browning [15]. Significantly increased (p < 0.05) genes included the thermogenic transcription factors PPARα and Ppargc1a, and Cox8b, a subunit of the cytochrome c oxidase complex. The genes that trended higher included UCP1, Cidea and Dio2. However, genes involved in UCP1-independent thermogenic processes were minimally affected (Figure S4A). qPCR analysis confirmed that knockdown of PexRAP in iWAT-derived adipocytes using shRNA results in significant increases in the expression of genes involved in the classical thermogenesis pathway, including UCP1 and Cidea (Figure S4B). To determine the effect of PexRAP inactivation on thermogenesis, PexRAP-AKO and control mice were subjected to a 6-h cold challenge. Body temperature was not different between the genotypes (Figure S4C), indicating that the modestly increased adipocyte browning in PexRAP-AKO mice does not translate to increased thermogenesis. In conjunction with the differential expression RNA-seq analysis results (Figure 3A), these data suggest that while chronic PexRAP ablation increases thermogenic gene expression, the increases are modest, and the transcriptomic response to PexRAP-AKO is instead predominated by changes in lipid metabolic genes. We also measured lipogenic gene expression in iWAT of mice housed at thermoneutrality (30°C) for 14 days. FASN was still significantly increased in PexRAP-AKO mice compared to control animals (Figure S4D), further suggesting that the elevated lipogenic gene expression is unrelated to thermogenesis.
For an unbiased exploration into shared functions of differentially expressed genes, which we defined as those with false discovery rate (FDR) < 0.05, we next performed gene ontology (GO) based pathway-enrichment analysis. This analysis revealed a significant enrichment of differentially expressed genes involved in both cholesterol and lipid biosynthesis, sterol regulatory element binding protein (SREBP) signaling, and very-low density lipoprotein particle assembly (Figure 3C). Further exploration of cholesterol biosynthetic process (GO:0006695), the top pathway from the pathway-enrichment analysis, revealed that nearly all genes in this pathway were upregulated, and those downregulated genes include only various isoforms of Ces1 (Carboxylesterase 1), a lipase [32], and Prkaa1 (Protein Kinase AMP-Activated Catalytic Subunit Alpha 1), which inhibits lipid biosynthesis through its role as a catalytic subunit of AMPK (Figure 3D) [33]. Gene set enrichment analysis (GSEA) suggested that PexRAP-AKO increases abundance of mRNAs encoding lipid biosynthetic enzymes by returning the lipid biosynthetic process GO term (GO:0008610) as the most significantly enriched pathway (Figure 3E). Key genes involved in de novo lipogenesis, including stearoyl-CoA desaturase 1 and 2 (Scd1 and Scd2), acyl-CoA synthetase (Acss2), acetyl-CoA carboxylase (Acc1) and FASN were significantly upregulated (p < 0.05) despite being among the most abundantly expressed genes in the dataset (Figure 3F). Further analysis revealed that nearly all genes involved in de novo lipogenesis were elevated in iWAT of PexRAP-AKO mice (Figure 3G) in a manner we hypothesized to be physiologically relevant due to their high abundance (Figure 3F).
3.5. PexRAP ablation induces lipogenesis through activation of LXR
Since PexRAP-AKO mice exhibit increased diet-induced obesity and insulin resistance, we determined whether PexRAP deficiency also results in increased lipogenic gene expression in the context of high fat feeding. qPCR analysis demonstrated that the mRNA levels of lipogenic genes significantly increase in iWAT of HFD-fed PexRAP-AKO mice, with FASN being upregulated nearly 5-fold (Figure 4A). To determine if the elevated gene expression translates to increased FASN activity, we measured incorporation of 14C-malonyl-CoA into fatty acids in iWAT lysates from HFD-fed PexRAP-AKO and control mice. PexRAP knockout significantly increased FASN-mediated lipogenesis (Figure 4B).
Figure 4.
PexRAP inactivation induces lipogenesis through activation of LXR.A) qPCR analysis of lipogenic genes in iWAT from male control or PexRAP-AKO mice fed a HFD (60% kcal fat) for 14 weeks (n = 3–4). B) Counts per minute results of 14C-Malonyl CoA de-novo lipogenesis assay performed on iWAT from male control or PexRAP-AKO mice fed a HFD for 14 weeks (n = 3–4). C) Top results from hypergeometric motif enrichment analysis (HOMER) of genes differentially expressed (FDR ≤ 0.05) in PexRAP-AKO mice according to RNA-seq analysis (n = 5). D) Schematic of LXR reporter construct. E) Normalized luminescence and F) green fluorescence microscopy from 3T3-L1 cells stably transfected with a plasmid expressing GFP and firefly luciferase under control of a LXR response element (LXRE), differentiated to white adipocytes, and then treated with scrambled (SC) or PexRAP shRNA and GW3965 or vehicle (DMSO) (n = 6). G) qPCR analysis of lipogenic genes in 3T3-L1 cells differentiated to white adipocytes and then treated with scrambled (SC) or SREBP1 shRNA followed by SC or PexRAP shRNA (n = 5–6). H) qPCR analysis and I) Western blot analysis of 3T3-L1 cells differentiated to white adipocytes and then treated with scrambled (SC) or LXRαβ shRNA followed by SC or PexRAP shRNA (n = 4). J) Western blot analysis of 3T3-L1 cells differentiated to white adipocytes and then treated with scrambled (SC) or PexRAP shRNA. K) Western blot analysis of iWAT from male control or PexRAP-AKO mice fed a HFD for 14 weeks. L) CHOP mRNA expression in iWAT of PexRAP-AKO (N = 4) and control (N = 3) mice fed a HFD for 19 weeks. Data are shown as mean ± SEM. Comparisons between groups were made with a two-tailed unpaired Student's t-test (A,B,E, L) or two-way ANOVA with Tukey's multiple comparisons test (G,H).
We next sought to understand the molecular mechanism underlying the increased lipogenic gene expression associated with PexRAP inactivation. Based on the high enrichment of upregulated genes in GO terms concerning lipogenesis, cholesterol synthesis (Figure 3C), and SREBP regulation, we hypothesized that PexRAP-AKO increases SREBP-driven transcription of lipogenic genes. Transcription factor enrichment analysis with HOMER, however, revealed LXR and NFY as the only two transcription factors with known motifs significantly enriched near the transcription start sites (TSS) of differentially expressed genes in the RNA-seq data (Figure 4C) [34]. NFY is a highly conserved and ubiquitously expressed transcription factor [35] that can cooperate with SREBP1 and SP1 to coordinate transcription of various lipogenic gene programs [36]. LXR is a nuclear receptor that is activated in response to increased cellular levels of cholesterol and promotes cholesterol homeostasis and lipogenesis using oxysterols as endogenous ligands [37,38]. Oxysterols are formed by enzymatic or non-enzymatic oxidation of cholesterol [39]. Cyp51, a cytochrome p450 protein that promotes oxysterol production through its role in cholesterol synthesis [40], was significantly upregulated in the PexRAP-AKO iWAT RNA-seq dataset (Figure 3A). Moreover, HOMER analysis identified known LXR binding motifs near the TSS of the two most significantly upregulated genes in the PexRAP-AKO mice, Acsl3 and LPCAT3 (Figure 3A), which are both known to be direct LXR target genes with functional LXR response elements (LXRE) across multiple mammalian species [29,41]. To determine if PexRAP inactivation promotes LXR activation in adipocytes, we used an LXR-dependent reporter that expresses luciferase and GFP under the control of LXRE paired with a minimal CMV promoter (mCMV) (Figure 4D). PexRAP knockdown in 3T3-L1 adipocytes markedly increased LXRE-driven expression of luciferase and GFP, indicating LXR activation (Figure 4E–F). The validity of the LXRE-driven luciferase assay was verified through treatment with LXR agonist GW3965 [42], which elicited an increase in GFP fluorescence in differentiated 3T3-L1 cells (Figure 4F).
Next, we determined whether LXR is required for PexRAP-inactivation associated increases in lipogenic gene expression. Although SREBP1 KD did not eliminate the PexRAP-KD induced increase in FASN expression (Figure 4G), simultaneous knockdown of PexRAP, LXRα, and LXRβ (LXRab KD) in 3T3-L1 adipocytes demonstrated that LXR is a required mediator of increased FASN mRNA and protein levels in response to PexRAP KD (Figure 4H–I).
Given the increased expression of the LXR transcriptional target Lpcat3 (Figure 3, Figure 4A), a phospholipid remodeling enzyme that regulates ER stress response [29], we hypothesized that PexRAP knockout induces ER stress. Analysis of publicly available RNA-seq data (GSE129442) identified a striking similarity between the transcriptomic responses to PexRAP-AKO in iWAT and lipotoxic ER stress in 3T3-L1 adipocytes. Every GO term significantly upregulated in iWAT of PexRAP-AKO mice was also upregulated in palmitate-treated 3T3-L1 cells (Figure S5). Western blot analysis showed that PexRAP knockdown (KD) in differentiated 3T3-L1 adipocytes increases the protein levels of FASN and Grp78/BiP (Figure 4J), which activates the ER stress response [43]. Furthermore, HFD-fed PexRAP-AKO mice exhibited increased FASN and Grp78/BiP protein levels in iWAT (Figure 4K). We also assessed other downstream markers of ER stress, including PERK phosphorylation and CHOP levels. Although PERK phosphorylation was not affected (Figure 4K), CHOP mRNA expression was significantly increased in iWAT of HFD-fed mice (Figure 4L). Together, these data demonstrate that PexRAP inactivation increases adipocyte lipogenesis through LXR activation and initiates ER stress response that is partially attenuated, presumably because LXR activation mitigates ER stress [29].
3.6. PexRAP KO alters membrane phospholipid composition and results in cholesterol mislocalization
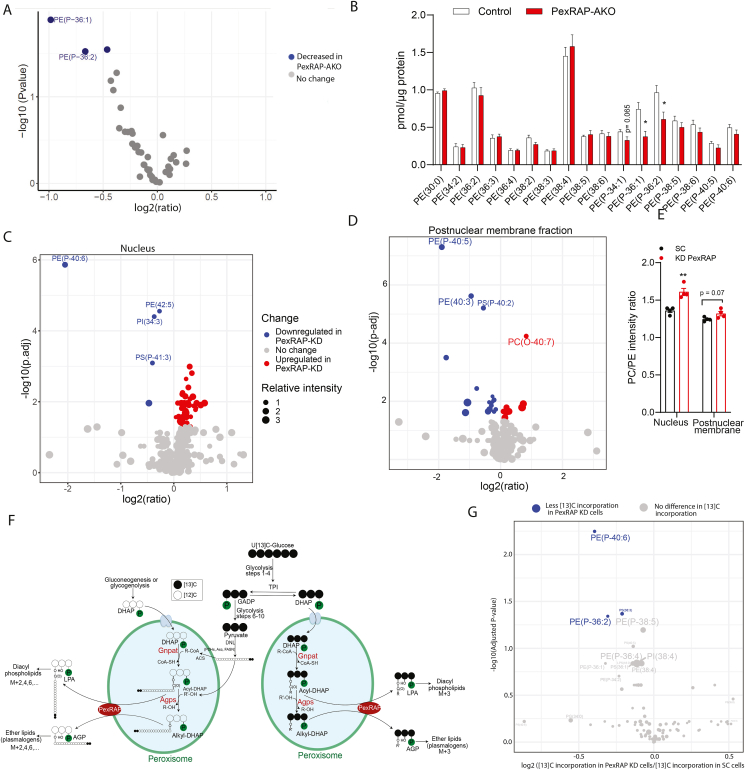
ER stress has been observed in adipose tissue of metabolically unhealthy humans and mice [44,45]. In liver, ER stress is known to dysregulate lipogenesis [46,47]. Aberrant lipogenesis can in turn further contribute to ER stress by increasing PC/PE ratio, forming a problematic positive feedback loop wherein ER stress induces lipogenesis, which in turn causes further ER stress [48]. PexRAP is involved in the synthesis of ether-linked phospholipids, but can also affect the levels of diacyl phospholipids, such as PC and PE. Global PexRAP knockout decreases the levels of ether lipid analogs of PC but results in compensatory increases in diacyl PC species in neutrophils [16]. To determine if adipose-specific PexRAP deletion affects phospholipid levels similarly, we conducted shotgun electrospray ionization mass spectrometry (ESI-MS) on adipose tissue of HFD-fed control or PexRAP-AKO mice. Analysis of PC and PE phospholipids revealed that PexRAP deletion markedly decreases levels of multiple PE plasmalogens in both nuclear (Figure 5A,B) and postnuclear membrane (Figure S6A and B) fractions. The purity of subcellular fractionation was confirmed via western blot against PexRAP alongside known peroxisomal, mitochondrial, and nuclear markers (Figure S6C). PexRAP knockdown in differentiated 3T3-L1 adipocytes elicited similar decreases in PE plasmalogens, suggesting that the lipodomic changes observed in vivo represent a cell-autonomous effect of PexRAP depletion (Figure 5C,D). More robust coverage of the lipidome in vitro allowed us to calculate PC/PE intensity ratios, which revealed that PexRAP knockdown significantly increased the PC/PE ratio in the nuclear fraction and trended towards increasing the ratio in the postnuclear membrane fraction (Figure 5E). Supplementary Table 1 and Supplementary Table 2 list all lipid species identified in the nuclear and postnuclear membrane fractions, respectively.
Figure 5.
PexRAP deletion is associated with decreased synthesis of PE plasmalogens and increased PC/PE ratio. Targeted ESI-MS analysis of PC and PE species from the nuclear fraction of iWAT from HFD fed male control and PexRAP-AKO mice, expressed as A) logFC values (PexRAP-AKO versus Control) and B) absolute concentrations. C) Shotgun ESI-MS analysis of PC and PE species from the nuclear fraction and D) post-nuclear membrane fraction of 3T3-L1 cells differentiated to white adipocytes and then treated with scrambled (SC) or PexRAP shRNA (species with adjusted p-value less than or equal to 0.01 are shown). E) PC/PE ratios based on lipid changes in C and D (n = 4). F) Schematic showing ways in which 13Carbon may be incorporated to phospholipids from glucose. G) Volcano plot of isotope incorporation in control cells versus PexRAP-KD cells from an ESI-MS-based 13Carbon-flux analysis of 3T3-L1 cells differentiated to white adipocytes, treated with scrambled (SC) or PexRAP shRNA, and then maintained in [U13C6]-glucose containing media for 5 days (n = 4, species with adjusted p-value less than or equal to 0.05 are shown). Data are shown as mean ± SEM (B, E). Comparisons between groups were made with a two-tailed unpaired Student's t-test adjusted for multiple comparisons using the Benjamini–Hochberg method (A,C,D,G) or a two-tailed unpaired Student's t-test (B, E).
Static levels of individual lipid species reflect the summed effect of synthesis, degradation, and remodeling processes. Given that PexRAP deletion alters levels of many key lipid synthesis and remodeling enzymes including FASN and Lpcat3, we sought to determine if the decreased levels of PE plasmalogens in PexRAP depleted cells reflects decreased synthesis of these lipids or instead results from increased activity of other lipid metabolic processes such as remodeling. To do this, we used ESI-MS to trace flux of 13C from [U13C6]-glucose into specific lipid species after knockdown of PexRAP in differentiated 3T3-L1 cells (Figure 5F). Isotope incorporation into an abundantly present plasmalogen species was detected well above the levels expected due to naturally occurring 13C alone in both control and KD samples, demonstrating the validity of our [U13C6]-glucose treatment dose and duration (Figure S6D). Furthermore, PexRAP knockdown resulted in a significant decrease in incorporation of 13C into PE-plasmalogen species relative to control cells (Figure 5G), suggesting that the decreased levels of PE plasmalogens in PexRAP depleted cells and tissue is a direct result of the decreased synthesis of these species.
Ether lipids, including plasmalogens, are required for the organization and stability of lipid rafts, cholesterol-rich membrane microdomains involved in cell signaling [49]. Previous studies show that plasmalogen-deficient GNPAT knockout mice exhibit disruption of lipid raft formation, resulting in mislocalization of cholesterol to a perinuclear compartment [10]. To test if PexRAP depletion has a similar effect, we knocked down PexRAP in differentiated 3T3-L1 adipocytes and imaged intracellular cholesterol using Filipin III. PexRAP-KD cells demonstrated an increase in nuclear cholesterol localization which was rescued with supplementation of the downstream ether lipid intermediate 1-O-alkyl glycerol (AG) (Figure 6A,B). Measuring total cholesterol levels in nuclear or postnuclear membrane fractions of iWAT from HFD fed mice using electrospray ionization gas chromatography mass spectrometry (GC–MS) confirmed this increase in nuclear cholesterol levels in vivo and revealed a concomitant decrease in postnuclear membrane cholesterol levels (Figure 6C). LXR, which is activated in response to PexRAP knockdown, is a nuclear receptor known to be activated endogenously by cholesterol derivatives called oxysterols [38]. Notably, we found that treating differentiated 3T3-L1 cells with AG, which rescues PexRAP-KD induced cholesterol mislocalization, also completely attenuates the PexRAP-KD induced activation of LXR (Figure 6D).
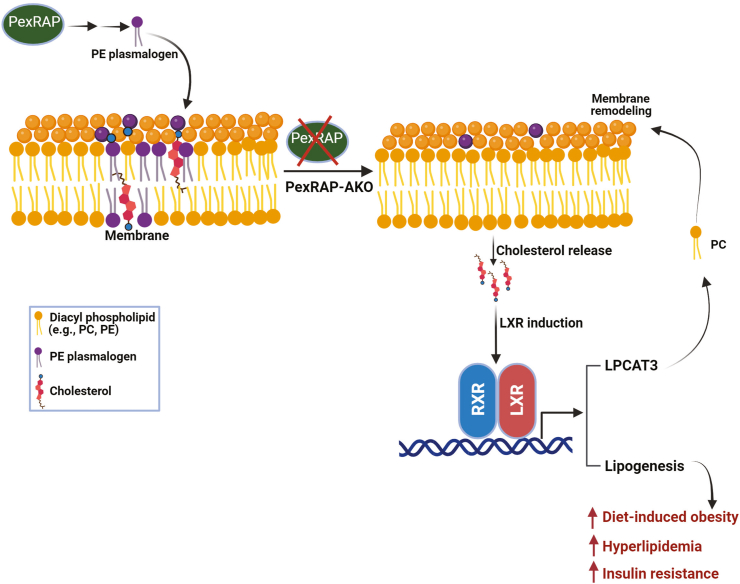
Together, these results suggest that the loss of PexRAP-derived PE plasmalogens disrupts cellular membrane phospholipid composition and thus induces cholesterol mislocalization and mild ER stress. Mislocalized cholesterol in turn activates LXR and its downstream de novo lipogenic gene program. In the context of HFD feeding, this aberrant activation of DNL is deleterious and promotes insulin resistance, glucose intolerance, and increased diet induced obesity (Figure 7).
Figure 7.
Proposed mechanism by which adipose-specific PexRAP deletion increases diet-induced obesity and insulin resistance. Loss of PexRAP decreases the amount of phosphatidylethanolamine (PE) plasmalogens present in cellular membranes. The absence of these key membrane lipids leads to release of membrane cholesterol, which is sensed by the sterol-regulated transcription factor Liver X receptor (LXR). LXR dimerizes with RXR to drive a transcriptional program characterized by increased expression of lysophosphatidylcholine acyltransferase 3 (Lpcat3), which remodels the PE-plasmalogen deficient membranes, and de novo lipogenesis genes. Aberrant activation of lipogenic genes in adipose tissue drives diet-induced obesity, hyperlipidemia and insulin resistance. Created with BioRender.
4. Discussion
Our previous studies indicate that PexRAP inhibits thermogenic gene expression by competing with PRDM16 for interaction with PPARγ [15]. Although PexRAP inactivation results in increased expression of brown adipocyte genes such as UCP1 and Cidea in subcutaneous WAT, here we report that adipose-specific knockout of PexRAP paradoxically promotes diet-induced obesity and insulin resistance through activation of LXR-dependent lipogenesis via an alternative mechanism. PexRAP ablation in adipocytes alters the nuclear membrane phospholipid composition through loss of certain species of PE plasmalogens, resulting in cholesterol mislocalization and activation of LXR, which increases expression of the membrane remodeling protein LPCAT3. LXR activation also promotes cholesterol homeostasis and FASN-mediated DNL. When challenged with a HFD, activation of adipose tissue DNL increases diet induced obesity and impairs glucose homeostasis. Together, these studies suggest that PexRAP regulates metabolic homeostasis by maintaining a balance between adipocyte browning and aberrant activation of lipogenesis. However, in the context of high fat feeding, the activation of lipogenesis has the dominant effect, resulting in metabolic dysregulation.
In the liver, the complex relationships between prolonged positive energy balance and DNL are well characterized. Excessive food intake activates hepatic DNL, increasing the cellular PC/PE ratio. The elevated PC/PE ratio in the ER membrane in turn impairs SERCA activity and thus Ca2+ transport, inducing ER stress and further promoting lipogenesis, constituting a positive feedback cycle [48]. In response to this lipotoxic ER stress, LXR is activated and induces Lpcat3 to promote phospholipid remodeling and incorporation of unsaturated phospholipids into the ER membrane [29]. Lpcat3-mediated incorporation of unsaturated phospholipids into the ER membrane has also been shown to promote post-translational processing of SREBP1, which is also regulated by LXR, demonstrating a close connection between the lipotoxic ER stress response and DNL since LXR simultaneously activates both processes [50]. LXR is activated by increased intracellular cholesterol [38]. Cholesterol is reported to be present in nuclear membranes, where it is implicated in promoting the formation of membrane microdomains [51,52]. Given the importance of plasmalogens in the assembly of microdomains [9,10], our studies indicate the existence of an alternative mechanism of LXR activation through regulation of cholesterol homeostasis by nuclear plasmalogens. Our results suggest that PexRAP deletion induces lipotoxic stress by disrupting the nuclear PC/PE ratio by decreasing PE plasmalogens, which releases cholesterol from nuclear membranes. This, in turn, results in LXR activation to increase lipogenesis.
These results shed light on the role of PexRAP-derived phospholipids in adipocytes, suggesting that they function to promote membrane homeostasis. Whether this reflects a general effect of PexRAP-derived lipids on membrane fluidity or simply that PexRAP contributes to the production of many unsaturated phospholipids remains unclear. Notably, the cellular effects of PexRAP-KO are reminiscent of Zellweger syndrome disease models such as Pex2−/− mice, which have hepatic increases in levels of Grp78/BiP, FASN, and Insig1 [53,54]. Loss of peroxisome-derived lipids might be an important link between peroxisome deficiency and disease, in which case supplementation of these lipids might partially correct the Zellweger phenotype.
The adipose-specific disruption of peroxisomal lipid synthesis in this study yields insight into the role of adipose tissue DNL in propagating the diabetic phenotype. Although chREBP-driven DNL in adipose tissue has emerged as a systemically beneficial insulin-sensitizing process [55], our results suggest that an LXR-driven program of adipose tissue lipogenesis impairs metabolic homeostasis, supporting earlier studies demonstrating that adipose-specific overexpression of SREBP1a promotes fatty liver and adipocyte hypertrophy [56]. Similarly, an inverse correlation between adipose tissue DNL and metabolic health has been observed in humans who have undergone bariatric surgery [57], further suggesting heterogeneity in the effects of DNL on systemic metabolism. The divergent effects of ChREBP- and LXR/SREBP-driven DNL might relate to their unique endogenous regulatory mechanisms. While ChREBP is activated by carbohydrates such as glucose [54], LXR and SREBP respond to intracellular or intramembrane sterol lipids [36]. The lipogenic activity of LXR limits the potential use of LXR agonists for treatment of atherosclerosis or Alzheimer disease [38].
Lipogenesis, by definition, is simply the conversion of sugars to lipids. Palmitate (C16:0), the primary product of DNL, is generally toxic but can be modified by various cellular elongases or desaturases to yield systemically-beneficial lipids such as palmitoleate or fatty acid–hydroxy–fatty acids [58]. It is thus not surprising that distinct lipogenic transcription factors might have opposing effects on physiology despite promoting expression of many of the same canonical lipogenesis genes. Furthermore, even within single lipogenic transcriptional programs, disruption of individual genes often has opposite, deleterious effects compared to perturbation of the master regulator such as ChREBP [59]. Characterizing these distinct lipogenic programs has the potential to identify strategies for modifying adipose tissue function to treat obesity and diabetes.
Credit authorship contribution statement
Brian Kleiboeker: Writing – review & editing, Writing – original draft, Methodology, Investigation, Formal analysis, Data curation, Conceptualization. Anyuan He: Methodology, Investigation, Data curation, Conceptualization. Min Tan: Investigation. Dongliang Lu: Writing – review & editing, Methodology, Investigation, Formal analysis. Donghua Hu: Writing – review & editing, Methodology, Investigation, Conceptualization. Xuejing Liu: Investigation. Parniyan Goodarzi: Writing – review & editing, Investigation. Fong-Fu Hsu: Methodology, Investigation. Babak Razani: Writing – review & editing, Resources, Conceptualization. Clay F. Semenkovich: Writing – review & editing, Resources, Conceptualization. Irfan J. Lodhi: Writing – review & editing, Writing – original draft, Resources, Funding acquisition, Formal analysis, Conceptualization.
Declaration of competing interest
The authors declare that no conflict of interest exists.
Acknowledgments
This work was supported by NIH grants DK115867, DK118333, DK132239, and T32DK007120. The core services of the Washington University Diabetes Research Center (DK020579), Mass Spectrometry Resource (GM103422), and the Nutrition Obesity Research Center (DK056341) also provided support for this work.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.molmet.2024.101913.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Data availability
Data will be made available on request.
References
- 1.Blüher M. Obesity: global epidemiology and pathogenesis. Nat Rev Endocrinol. 2019;15(5):288–298. doi: 10.1038/s41574-019-0176-8. [DOI] [PubMed] [Google Scholar]
- 2.Kusminski C.M., Bickel P.E., Scherer P.E. Targeting adipose tissue in the treatment of obesity-associated diabetes. Nat Rev Drug Discov. 2016;15(9):639–660. doi: 10.1038/nrd.2016.75. [DOI] [PubMed] [Google Scholar]
- 3.Kleiboeker B., Lodhi I.J. Peroxisomal regulation of energy homeostasis: effect on obesity and related metabolic disorders. Mol Metab. 2022;65 doi: 10.1016/j.molmet.2022.101577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wanders R.J.A., Baes M., Ribeiro D., Ferdinandusse S., Waterham H.R. The physiological functions of human peroxisomes. Physiol Rev. 2023;103(1):957–1024. doi: 10.1152/physrev.00051.2021. [DOI] [PubMed] [Google Scholar]
- 5.Klouwer F.C., Berendse K., Ferdinandusse S., Wanders R.J., Engelen M., Poll-The B.T. Zellweger spectrum disorders: clinical overview and management approach. Orphanet J Rare Dis. 2015;10:151. doi: 10.1186/s13023-015-0368-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cedillo L., Ahsan F.M., Li S., Stuhr N.L., Zhou Y., Zhang Y., et al. Ether lipid biosynthesis promotes lifespan extension and enables diverse pro-longevity paradigms in Caenorhabditis elegans. Elife. 2023;12 doi: 10.7554/eLife.82210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Engelmann B. Plasmalogens: targets for oxidants and major lipophilic antioxidants. Biochem Soc Trans. 2004;32(1):147–150. doi: 10.1042/bst0320147. [DOI] [PubMed] [Google Scholar]
- 8.Zou Y., Henry W.S., Ricq E.L., Graham E.T., Phadnis V.V., Maretich P., et al. Plasticity of ether lipids promotes ferroptosis susceptibility and evasion. Nature. 2020;585(7826):603–608. doi: 10.1038/s41586-020-2732-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pike L.J., Han X., Chung K.N., Gross R.W. Lipid rafts are enriched in arachidonic acid and plasmenylethanolamine and their composition is independent of caveolin-1 expression: a quantitative electrospray ionization/mass spectrometric analysis. Biochemistry. 2002;41(6):2075–2088. doi: 10.1021/bi0156557. [DOI] [PubMed] [Google Scholar]
- 10.Rodemer C., Thai T.P., Brugger B., Kaercher T., Werner H., Nave K.A., et al. Inactivation of ether lipid biosynthesis causes male infertility, defects in eye development and optic nerve hypoplasia in mice. Hum Mol Genet. 2003;12(15):1881–1895. doi: 10.1093/hmg/ddg191. [DOI] [PubMed] [Google Scholar]
- 11.Lodhi I.J., Yin L., Jensen-Urstad A.P., Funai K., Coleman T., Baird J.H., et al. Inhibiting adipose tissue lipogenesis reprograms thermogenesis and PPARγ activation to decrease diet-induced obesity. Cell Metab. 2012;16(2):189–201. doi: 10.1016/j.cmet.2012.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Honsho M., Tanaka M., Zoeller R.A., Fujiki Y. Distinct functions of acyl/alkyl dihydroxyacetonephosphate reductase in peroxisomes and endoplasmic reticulum. Front Cell Dev Biol. 2020;8:855. doi: 10.3389/fcell.2020.00855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin M.-E., Herr D.R., Chun J. Lysophosphatidic acid (LPA) receptors: signaling properties and disease relevance. Prostag Other Lipid Mediat. 2010;91(3–4):130–138. doi: 10.1016/j.prostaglandins.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McIntyre T.M., Pontsler A.V., Silva A.R., St Hilaire A., Xu Y., Hinshaw J.C., et al. Identification of an intracellular receptor for lysophosphatidic acid (LPA): LPA is a transcellular PPARgamma agonist. Proc Natl Acad Sci U S A. 2003;100(1):131–136. doi: 10.1073/pnas.0135855100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lodhi I.J., Dean J.M., He A., Park H., Tan M., Feng C., et al. PexRAP inhibits PRDM16-mediated thermogenic gene expression. Cell Rep. 2017;20(12):2766–2774. doi: 10.1016/j.celrep.2017.08.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lodhi I.J., Wei X., Yin L., Feng C., Adak S., Abou-Ezzi G., et al. Peroxisomal lipid synthesis regulates inflammation by sustaining neutrophil membrane phospholipid composition and viability. Cell Metab. 2015;21(1):51–64. doi: 10.1016/j.cmet.2014.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roy D., Myers J.M., Tedeschi A. Protocol for assessing ex vivo lipolysis of murine adipose tissue. STAR Protoc. 2022;3(3) doi: 10.1016/j.xpro.2022.101518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hsu R.Y., Wasson G., Porter J.W. The purification and properties of the fatty acid synthetase of pigeon liver. J Biol Chem. 1965;240(10):3736–3746. [PubMed] [Google Scholar]
- 19.Rajagopal R., Zhang S., Wei X., Doggett T., Adak S., Enright J., et al. Retinal de novo lipogenesis coordinates neurotrophic signaling to maintain vision. JCI Insight. 2018;3(1) doi: 10.1172/jci.insight.97076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hajra A.K., Das A.K. Lipid biosynthesis in peroxisomes. Ann N Y Acad Sci. 1996;804:129–141. doi: 10.1111/j.1749-6632.1996.tb18613.x. [DOI] [PubMed] [Google Scholar]
- 21.Butler A.A., Kozak L.P. A recurring problem with the analysis of energy expenditure in genetic models expressing lean and obese phenotypes. Diabetes. 2010;59(2):323–329. doi: 10.2337/db09-1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cohen P., Levy J.D., Zhang Y., Frontini A., Kolodin D.P., Svensson K.J., et al. Ablation of PRDM16 and beige adipose causes metabolic dysfunction and a subcutaneous to visceral fat switch. Cell. 2014;156(1–2):304–316. doi: 10.1016/j.cell.2013.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Colditz G.A., Willett W.C., Rotnitzky A., Manson J.E. Weight gain as a risk factor for clinical diabetes mellitus in women. Ann Intern Med. 1995;122(7):481–486. doi: 10.7326/0003-4819-122-7-199504010-00001. [DOI] [PubMed] [Google Scholar]
- 24.Klein S., Wadden T., Sugerman H.J. AGA technical review on obesity. Gastroenterology. 2002;123(3):882–932. doi: 10.1053/gast.2002.35514. [DOI] [PubMed] [Google Scholar]
- 25.Virtue S., Vidal-Puig A. GTTs and ITTs in mice: simple tests, complex answers. Nat Metab. 2021;3(7):883–886. doi: 10.1038/s42255-021-00414-7. [DOI] [PubMed] [Google Scholar]
- 26.Reilly S.M., Saltiel A.R. Adapting to obesity with adipose tissue inflammation. Nat Rev Endocrinol. 2017;13(11):633–643. doi: 10.1038/nrendo.2017.90. [DOI] [PubMed] [Google Scholar]
- 27.Murano I., Barbatelli G., Parisani V., Latini C., Muzzonigro G., Castellucci M., et al. Dead adipocytes, detected as crown-like structures, are prevalent in visceral fat depots of genetically obese mice. J Lipid Res. 2008;49(7):1562–1568. doi: 10.1194/jlr.M800019-JLR200. [DOI] [PubMed] [Google Scholar]
- 28.Kazachkov M., Chen Q., Wang L., Zou J. Substrate preferences of a lysophosphatidylcholine acyltransferase highlight its role in phospholipid remodeling. Lipids. 2008;43(10):895–902. doi: 10.1007/s11745-008-3233-y. [DOI] [PubMed] [Google Scholar]
- 29.Rong X., Albert C.J., Hong C., Duerr M.A., Chamberlain B.T., Tarling E.J., et al. LXRs regulate ER stress and inflammation through dynamic modulation of membrane phospholipid composition. Cell Metab. 2013;18(5):685–697. doi: 10.1016/j.cmet.2013.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Magtanong L., Ko P.J., To M., Cao J.Y., Forcina G.C., Tarangelo A., et al. Exogenous monounsaturated fatty acids promote a ferroptosis-resistant cell state. Cell Chem Biol. 2019;26(3):420–432.e9. doi: 10.1016/j.chembiol.2018.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chang Y.S., Tsai C.T., Huangfu C.A., Huang W.Y., Lei H.Y., Lin C.F., et al. ACSL3 and GSK-3β are essential for lipid upregulation induced by endoplasmic reticulum stress in liver cells. J Cell Biochem. 2011;112(3):881–893. doi: 10.1002/jcb.22996. [DOI] [PubMed] [Google Scholar]
- 32.Riddles P.W., Richards L.J., Bowles M.R., Pond S.M. Cloning and analysis of a cDNA encoding a human liver carboxylesterase. Gene. 1991;108(2):289–292. doi: 10.1016/0378-1119(91)90448-k. [DOI] [PubMed] [Google Scholar]
- 33.Hardie D.G. AMP-activated/SNF1 protein kinases: conserved guardians of cellular energy. Nat Rev Mol Cell Biol. 2007;8(10):774–785. doi: 10.1038/nrm2249. [DOI] [PubMed] [Google Scholar]
- 34.Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38(4):576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maity S.N., de Crombrugghe B. Role of the CCAAT-binding protein CBF/NF-Y in transcription. Trends Biochem Sci. 1998;23(5):174–178. doi: 10.1016/s0968-0004(98)01201-8. [DOI] [PubMed] [Google Scholar]
- 36.Reed B.D., Charos A.E., Szekely A.M., Weissman S.M., Snyder M. Genome-wide occupancy of SREBP1 and its partners NFY and SP1 reveals novel functional roles and combinatorial regulation of distinct classes of genes. PLoS Genet. 2008;4(7) doi: 10.1371/journal.pgen.1000133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schultz J.R., Tu H., Luk A., Repa J.J., Medina J.C., Li L., et al. Role of LXRs in control of lipogenesis. Genes Dev. 2000;14(22):2831–2838. doi: 10.1101/gad.850400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang B., Tontonoz P. Liver X receptors in lipid signalling and membrane homeostasis. Nat Rev Endocrinol. 2018;14(8):452–463. doi: 10.1038/s41574-018-0037-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Samadi A., Sabuncuoglu S., Samadi M., Isikhan S.Y., Chirumbolo S., Peana M., et al. A comprehensive review on oxysterols and related diseases. Curr Med Chem. 2021;28(1):110–136. doi: 10.2174/0929867327666200316142659. [DOI] [PubMed] [Google Scholar]
- 40.Lorbek G., Lewinska M., Rozman D. Cytochrome P450s in the synthesis of cholesterol and bile acids – from mouse models to human diseases. FEBS J. 2012;279(9):1516–1533. doi: 10.1111/j.1742-4658.2011.08432.x. [DOI] [PubMed] [Google Scholar]
- 41.Weedon-Fekjaer M.S., Dalen K.T., Solaas K., Staff A.C., Duttaroy A.K., Nebb H.I. Activation of LXR increases acyl-CoA synthetase activity through direct regulation of ACSL3 in human placental trophoblast cells. J Lipid Res. 2010;51(7):1886–1896. doi: 10.1194/jlr.M004978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Collins J.L., Fivush A.M., Watson M.A., Galardi C.M., Lewis M.C., Moore L.B., et al. Identification of a nonsteroidal liver X receptor agonist through parallel array synthesis of tertiary amines. J Med Chem. 2002;45(10):1963–1966. doi: 10.1021/jm0255116. [DOI] [PubMed] [Google Scholar]
- 43.Ma Y., Hendershot L.M. The role of the unfolded protein response in tumour development: friend or foe? Nat Rev Cancer. 2004;4(12):966–977. doi: 10.1038/nrc1505. [DOI] [PubMed] [Google Scholar]
- 44.Boden G., Duan X., Homko C., Molina E.J., Song W., Perez O., et al. Increase in endoplasmic reticulum stress-related proteins and genes in adipose tissue of obese, insulin-resistant individuals. Diabetes. 2008;57(9):2438–2444. doi: 10.2337/db08-0604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ozcan U., Cao Q., Yilmaz E., Lee A.H., Iwakoshi N.N., Ozdelen E., et al. Endoplasmic reticulum stress links obesity, insulin action, and type 2 diabetes. Science. 2004;306(5695):457–461. doi: 10.1126/science.1103160. [DOI] [PubMed] [Google Scholar]
- 46.Werstuck G.H., Lentz S.R., Dayal S., Hossain G.S., Sood S.K., Shi Y.Y., et al. Homocysteine-induced endoplasmic reticulum stress causes dysregulation of the cholesterol and triglyceride biosynthetic pathways. J Clin Investig. 2001;107(10):1263–1273. doi: 10.1172/JCI11596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kammoun H.L., Chabanon H., Hainault I., Luquet S., Magnan C., Koike T., et al. GRP78 expression inhibits insulin and ER stress-induced SREBP-1c activation and reduces hepatic steatosis in mice. J Clin Investig. 2009;119(5):1201–1215. doi: 10.1172/JCI37007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fu S., Yang L., Li P., Hofmann O., Dicker L., Hide W., et al. Aberrant lipid metabolism disrupts calcium homeostasis causing liver endoplasmic reticulum stress in obesity. Nature. 2011;473(7348):528–531. doi: 10.1038/nature09968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gorgas K., Teigler A., Komljenovic D., Just W.W. The ether lipid-deficient mouse: tracking down plasmalogen functions. Biochim Biophys Acta. 2006;1763(12):1511–1526. doi: 10.1016/j.bbamcr.2006.08.038. [DOI] [PubMed] [Google Scholar]
- 50.Rong X., Wang B., Palladino E.N., de Aguiar Vallim, Ford D.A., Tontonoz P. ER phospholipid composition modulates lipogenesis during feeding and in obesity. J Clin Investig. 2017;127(10):3640–3651. doi: 10.1172/JCI93616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Codini M., Garcia-Gil M., Albi E. Cholesterol and sphingolipid enriched lipid rafts as therapeutic targets in cancer. Int J Mol Sci. 2021;22(2) doi: 10.3390/ijms22020726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dazzoni R., Grelard A., Morvan E., Bouter A., Applebee C.J., Loquet A., et al. The unprecedented membrane deformation of the human nuclear envelope, in a magnetic field, indicates formation of nuclear membrane invaginations. Sci Rep. 2020;10(1):5147. doi: 10.1038/s41598-020-61746-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kovacs W.J., Tape K.N., Shackelford J.E., Wikander T.M., Richards M.J., Fliesler S.J., et al. Peroxisome deficiency causes a complex phenotype because of hepatic SREBP/Insig dysregulation associated with endoplasmic reticulum stress. J Biol Chem. 2009;284(11):7232–7245. doi: 10.1074/jbc.M809064200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Faust P.L., Kovacs W.J. Cholesterol biosynthesis and ER stress in peroxisome deficiency. Biochimie. 2014;98:75–85. doi: 10.1016/j.biochi.2013.10.019. [DOI] [PubMed] [Google Scholar]
- 55.Herman M.A., Peroni O.D., Villoria J., Schön M.R., Abumrad N.A., Blüher M., et al. A novel ChREBP isoform in adipose tissue regulates systemic glucose metabolism. Nature. 2012;484(7394):333–338. doi: 10.1038/nature10986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Horton J.D., Shimomura I., Ikemoto S., Bashmakov Y., Hammer R.E. Overexpression of sterol regulatory element-binding protein-1a in mouse adipose tissue produces adipocyte hypertrophy, increased fatty acid secretion, and fatty liver. J Biol Chem. 2003;278(38):36652–36660. doi: 10.1074/jbc.M306540200. [DOI] [PubMed] [Google Scholar]
- 57.Garrido-Sanchez L., Vendrell J., Fernandez-Garcia D., Ceperuelo-Mallafre V., Chacon M.R., Ocana-Wilhelmi L., et al. De novo lipogenesis in adipose tissue is associated with course of morbid obesity after bariatric surgery. PLoS One. 2012;7(2) doi: 10.1371/journal.pone.0031280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yilmaz M., Claiborn K.C., Hotamisligil G.S. De novo lipogenesis products and endogenous lipokines. Diabetes. 2016;65(7):1800–1807. doi: 10.2337/db16-0251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wallace M., Metallo C.M. Tracing insights into de novo lipogenesis in liver and adipose tissues. Semin Cell Dev Biol. 2020;108:65–71. doi: 10.1016/j.semcdb.2020.02.012. [DOI] [PubMed] [Google Scholar]
- 60.Galarraga M., Campión J., Muñoz-Barrutia A., Boqué N., Moreno H., Martínez J.A., et al. Adiposoft: automated software for the analysis of white adipose tissue cellularity in histological sections. J Lipid Res. 2012;53(12):2791–2796. doi: 10.1194/jlr.D023788. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.