Abstract
Cullin (CUL)-RING (Really Interesting New Gene) E3 ubiquitin (Ub) ligases (CRLs) are the largest E3 family. The E3 CRL core ligase is a subcomplex formed by the CUL C-terminal domain bound with the ROC1/RBX1 RING finger protein, which acts as a hub that mediates and organizes multiple interactions with E2, Ub, Nedd8, and the ARIH family protein, thereby resulting in Ub transfer to the E3-bound substrate. This report describes the modulation of CRL-dependent ubiquitination by small molecule compounds including KH-4-43, #33, and suramin, which target the CRL core ligases. We show that both KH-4-43 and #33 inhibit the ubiquitination of CK1α by CRL4CRBN. However, either compound’s inhibitory effect on this reaction is significantly reduced when a neddylated form of CRL4CRBN is used. On the other hand, both #33 and KH-4-43 inhibit the ubiquitination of β-catenin by CRL1β-TrCP and Nedd8-CRL1β-TrCP almost equally. Thus, neddylation of CRL1β-TrCP does not negatively impact the sensitivity to inhibition by #33 and KH-4-43. These findings suggest that the effects of neddylation to alter the sensitivity of CRL inhibition by KH-4-43/#33 is dependent upon the specific CRL type. Suramin, a compound that targets CUL’s basic canyon, can effectively inhibit CRL1/4-dependent ubiquitination regardless of neddylation status, in contrast to the results observed with KH-4-43/#33. This observed differential drug sensitivity of KH-4-43/#33 appears to echo CUL-specific Nedd8 effects on CRLs as revealed by recent high-resolution structural biology efforts. The highly diversified CRL core ligase structures may provide opportunities for specific targeting by small molecule modulators.
Keywords: ubiquitination, cullin-RING E3 ubiquitin ligase, Nedd8 modification, CRL core ligase, small molecule modulators
Cullin (CUL) -RING (Really Interesting New Gene) E3 ubiquitin (Ub) ligases (CRLs) are the largest E3 family defined by a signature Cullin-RING heterodimeric complex (1, 2). CUL proteins are molecular scaffolds that function to organize the CRL complex. Six canonical CULs, CUL1, CUL2/CUL5, CUL3 and CUL4A/CUL4B, assemble four major CRL classes designated as CRL1, CRL2/5, CRL3 and CRL4A/4B. Each CRL class is distinguished by the ability of a specific CUL type to utilize its distinct N-terminus to anchor an unique protein family of substrate receptor. These include CUL1 for the F-box protein family, CUL2/5 for the VHL-Box/SOCS-box family, CUL3 for the BTB (Bric-a-brac, Tramtrack, Broad-complex) family, and CUL4A/4B for the DCAF (DDB1 and CUL4 associated factor) family, respectively. On the other hand, the CUL’s C-terminal domains (CTDs) contain highly conserved cullin consensus sequence and possess a common globular structure that binds a RING finger protein, ROC1/RBX1 for CUL1 to 4 or ROC2 for CUL5, to form a core ligase complex.
The CRL core ligase is responsible for orchestrating the transfer of Ub to a substrate that is anchored by CRL’s receptor subunit, such as CRL1’s F-box protein or CRL4’s DCAF. The CRL core ligase activity is activated by the conjugation of Nedd8, an ubiquitin-like protein, to a CUL CTD’s conserved lysine residue (3). As revealed by a series of high-resolution structural and biochemical studies (4, 5, 6), the CRL core ligase is shown to act as a hub that mediates and organizes multiple interactions with E2, Nedd8, Ub, and in some cases, the ARIH family proteins. This CRL core ligase-centralized protein network is defined as a catalytic module (5). One distinct E3-E2 interaction is the interface between CUL CTD’s conserved basic canyon and E2 Cdc34’s acidic C-terminus (7, 8). The biochemical functions of the CRL core ligase-organized catalytic module are to coordinate the priming and extending reactions (9, 10) that lead to initial transfer of an Ub to the CRL-bound substrate (priming), followed by Ub chain elongation of the substrate-linked, initiator Ub (extending). Priming requires interactions between a CRL and an Ub carrying enzyme (6) that includes the E2 UbcH5 (UBE2D) family (9, 10) and the RBR E3 ARIH family members (4). On the other hand, Ub chain elongation is executed by the E2 Cdc34 (UBE2R) family members (9, 10) and UBE2G1 (11, 12).
Neddylation was initially thought to cause significant conformational changes that disrupt the autoinhibitory interactions between CUL CTD and ROC1/RBX1, which liberates the RING finger protein for productive interactions with an E2 enzyme to catalyze Ub transfer (13, 14). A series of subsequent high resolution structural studies by the Schulman group illustrates the details of mechanism by which Nedd8 activates E3 CRL and also reveals some different effects of the Nedd8 effects on different CULs. In the reported structures of Nedd8-CRL1-UBE2D (or ARIH1) (5, 6), Nedd8 engages in non-covalent interactions with the Nedd8-conjugated-CUL1, which allosterically stabilizes a specific form of Nedd8 conformation that enables interactions with priming enzyme UBE2D or ARIH1. In addition, Nedd8 causes CUL1 CTD conformational changes that allow positional flexibility of the neddylated domain, relative to the rest of CUL1. In the structure of Nedd8-CRL5-ARIH2 (15); however, there is no direct contact between Nedd8 and ARIH2. Instead, Nedd8 interacts with CUL5 at multiple domains to induce conformational changes, which result in new surfaces for binding to the priming enzyme ARIH2.
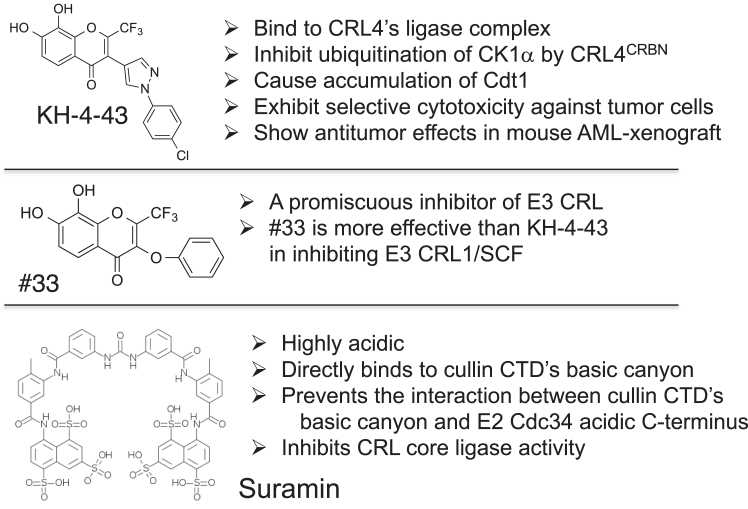
A selective small-molecule modulator of E3 CRL’s function allows us to address mechanistic and phenotypic questions about its targets in biochemical, cell-based, and animal studies. To this end, we have recently identified through a high throughput screen compound #33 (Fig. 1) that acts as a promiscuous inhibitor of all E3 CRLs (16). Medicinal chemistry structure-activity relationship (SAR) studies led to the discovery of compound KH-4-43 as a lead small molecule that selectively targets and inhibits the core ligase component of E3 CRL4 with potential antitumor therapeutical effects in cells and animal models (Fig. 1) (16).
Figure 1.
Properties of E3 CRL inhibitors KH-4-43, #33 and suramin.
The unrelated compound suramin is an anti-trypanosomal drug with the structure of highly acidic polysulphonated naphthylurea (Fig. 1). Previously, we reported suramin’s ability to directly bind E3 SCF’s core ligase module (the ROC1-CUL1 CTD subcomplex) by utilizing the compound’s highly acidic groups, most likely at the CUL1’s conserved basic canyon (17). Consequently, suramin was found to disrupt the electrostatic interactions between ROC1-CUL1 CTD and E2 Cdc34’s acidic C-terminus and also inhibit ubiquitination in vitro (17). While suramin may be limited with respect to its development into a more selective inhibitor of E3 CRL by medicinal chemistry optimization, its ability to block charged E3/E2 interactions can be exploited in mechanistic studies.
Using reconstituted ubiquitination of CK1α by E3 CRL4CRBN, and β-catenin by CRL1βTrCP, as described in the accompanying manuscript, we have analyzed the relative effects of #33, KH-4-43, and suramin, which provide insights into pharmacological perturbation of the CRL core ligase.
Results
Inhibition of E3 CRL4-dependent ubiquitination by KH-4-43 and effects of Nedd8
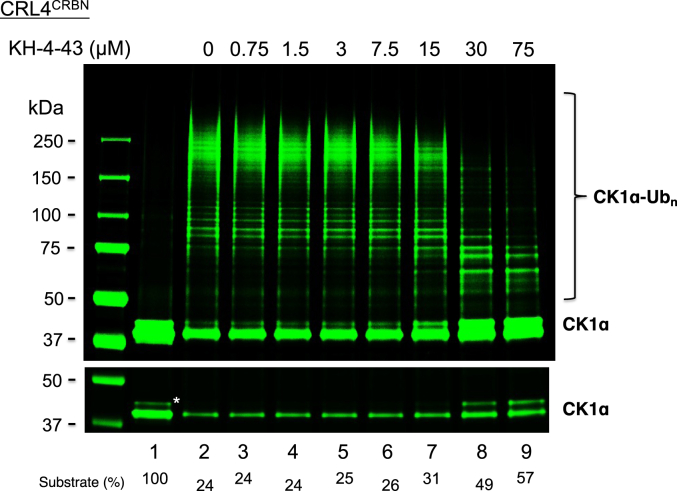
We previously reported inhibition of the CRL4CRBN-dependent ubiquitination of CK1α by KH-4-43 using the immunoprecipitation approach (16). We have now improved the CK1α ubiquitination assay reconstituted with all pure recombinant proteins in solution without the need for immune immobilization, which enables better activity measurement (accompanying manuscript). We tested the effects of KH-4-43 in the newly improved CK1α ubiquitination assay in solution (Fig. 2). We observed that at higher concentrations (30 and 75 μM), KH-4-43 blocked substrate utilization (lanes 8 and 9, lower panel; see quantification). At concentrations of 15 to 75 μM, KH-4-43 inhibited Ub chain elongation in a dose dependent manner (lanes 7–9, top panel). Thus KH-4-43 inhibits the ability of CRL4CRBN for both initiation and elongation reactions.
Figure 2.
Inhibition of CRL4CRBN-dependent CK1α ubiquitination by compound KH-4-43. Lenalidomide is required for interactions between substrate receptor CRBN and neo-substrates that include CK1α (25, 26). The detailed procedure is described under “Experimental Procedure.” Low-intensity image of the substrate is also shown to better indicate substrate utilization. Species∗ most likely represents auto-phosphorylated form of CK1α (27). The substrate levels after reactions were quantified and shown. Note that similar inhibition of the CRL4CRBN-dependent CK1α ubiquitination by KH-4-43 was observed in more than a dozen of independent experiments, including those shown in Figures 3C and 4, underscoring a highly reproducible effect.
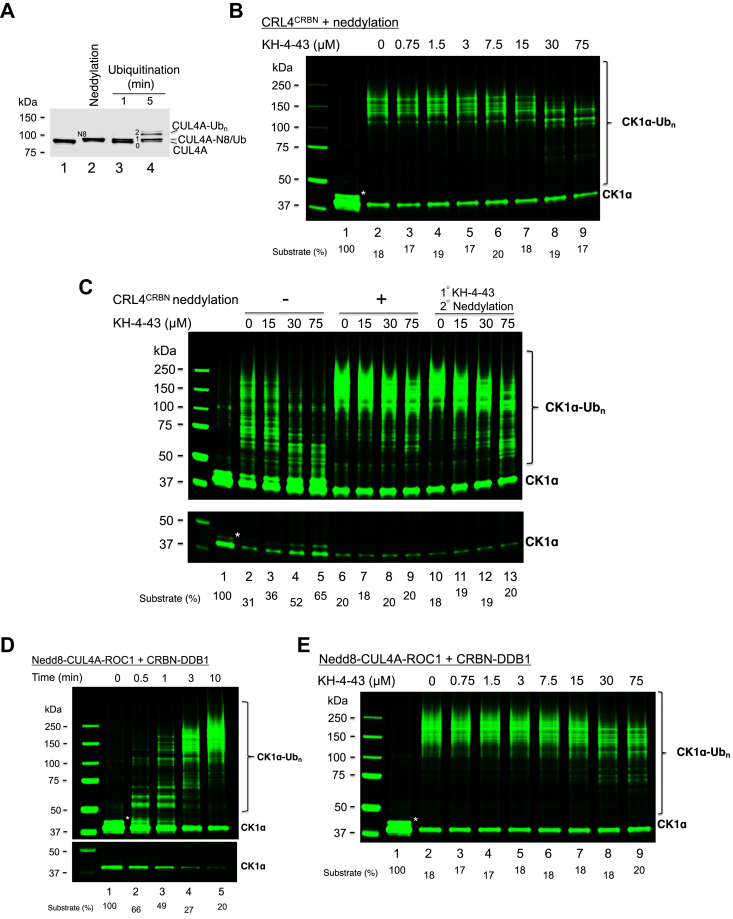
We examined the effects of neddylation on CK1α ubiquitination. First, immunoblot confirmed Nedd8 conjugation to CUL4A as a result of a neddylation reaction (Fig. 3A, compare lanes 1 and 2). As shown, the inhibitory effects of KH-4-43 in the ubiquitination of CK1α were significantly reduced with CRL4CRBN that had received prior treatment with neddylation reagents (Fig. 3B). Specifically, KH-4-43 was no longer able to block substrate utilization (compare Fig. 3B, lanes 8 and 9, with Fig. 2, lanes 8 and 9), suggesting that this compound loses its ability to inhibit Nedd8-CRL4CRBN/UbcH5c for priming substrate ubiquitination. In addition, while KH-4-43 still inhibited Ub chain elongation to some extent, it was evident that the inhibitory effects were much less pronounced in comparison to reactions with CRL4CRBN in the absence of neddylation (compare Fig. 3B, lanes 8 and 9, with Fig. 2, lanes 8 and 9). These results were confirmed by side-by-side comparisons of reactions with CRL4CRBN that received no treatment (Fig. 3C, lanes 2–5), or prior-treatment with neddylation agents (Fig. 3C, lanes 6–9). When CRL4CRBN was first mixed with KH-4-43 and then after with neddylation agents (Fig. 3C, lanes 10–13), no inhibition of substrate utilization was observed, indicating that prior formation of the E3-inhibtor complex could not restore the ability of KH-4-43 to inhibit UbcH5c-catalyzed priming reactions as seen with Nedd8-free CRL4CRBN (Fig. 2, lanes 8 and 9). However, prior formation of the E3-KH-4-43 complex appeared to slightly enhance the ability of the compound to inhibit Ub chain elongation (compare Fig. 3C, lanes 9 and 13).
Figure 3.
CRL4CRBNneddylation reduced sensitivity to KH-4-43 inhibition on CK1α ubiquitination.A, immunoblot confirmed CUL4A neddylation and ubiquitination. Neddylated or ubiquitinated CRL4CRBN was prepared using procedures as described under the section of “Immunoblot” in the “Experimental Procedure.” Lane 2 supports Figure 3, B and C by revealing that CUL4A is extensively neddylated. Lanes 3 and 4 provide support for Figure 4 by showing that incubation of CRL4CRBN with UbcH5c, E1 and ubiquitin prior to the addition of KH-4-43 and Cdc34b∼Ub yielded CUL4A in ubiquitin-modified forms. B, effects of increasing concentrations of KH-4-43 on CK1α ubiquitination by CRL4CRBN that received prior treatment with neddylation agents. C, side-by-side comparison of KH-4-43 effects on CK1α ubiquitination by CRL4CRBN that received no treatment with neddylation agents, prior treatment with neddylation agents, or prior treatment with KH-4-43 first and then neddylation agents. Low intensity image of the substrate is also shown to better indicate substrate utilization. D, CK1α ubiquitination kinetics with Nedd8-CRL4CRBN that was assembled by mixing Nedd8-CUL4A-ROC1 and CRBN-DDB1 (free of neddylation agents). Low intensity image of the substrate is also shown to better indicate substrate utilization. E, effects of increasing concentrations of KH-4-43 on CK1α ubiquitination by Nedd8-CRL4CRBN free of neddylation agents. The detailed procedure is described under “Experimental Procedure.” Species∗ most likely represent the auto-phosphorylated form of CK1α (27). The substrate levels after reactions were quantified and shown. The observation that Nedd8-CRL4CRBN is less sensitive to inhibition by KH-4-43 is established by three independent experiments shown in Figure 3, B, C and E.
It was possible that neddylation agents (Nedd8, Nedd8 E1, Ubc12), rather than E3 neddylation, might in some way negatively impact KH-4-43’s inhibitory activity. To test this possibility, purified forms of Nedd8-CUL4A-ROC1 and CRBN-DDB1 were mixed to assemble Nedd8-CRL4CRBN free of neddylation agents. The assembled Nedd8-CRL4CRBN was highly active in supporting the ubiquitination of CK1α as revealed by the kinetics experiment (Fig. 3D). CK1α ubiquitination with Nedd8-CRL4CRBN (free of neddylation agents) still showed reduced inhibitory effects by KH-4-43 (Fig. 3E). Taken together, these findings suggest that E3 CRL4 neddylation causes significant reduction of sensitivity to KH-4-43 inhibition. While KH-4-43 appears to retain inhibitory activity on Ub chain elongation to some extent, it can no longer inhibit substrate utilization, suggesting a profound defect to antagonize the priming reaction catalyzed by UbcH5c.
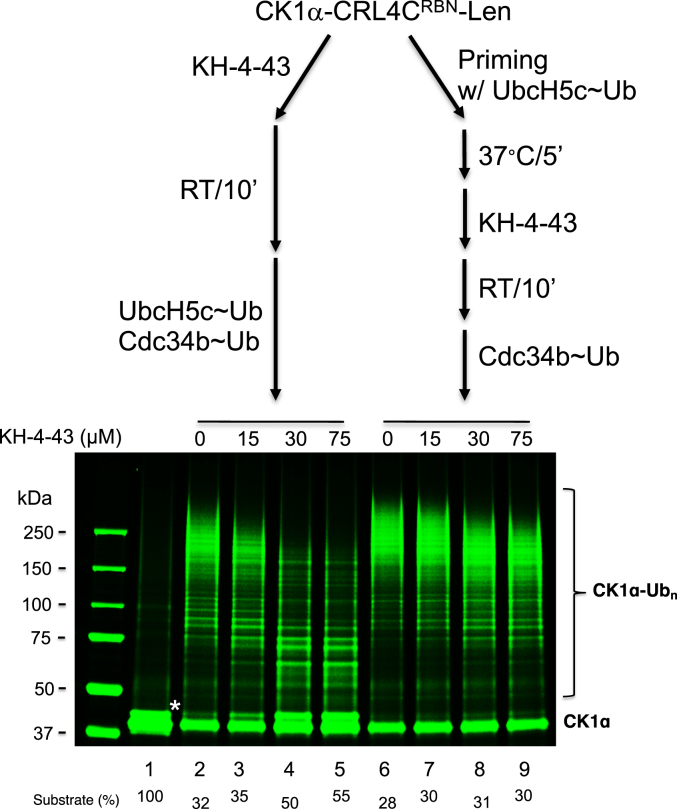
Prior priming reverses the inhibitory effects of KH-4-43 on CK1α ubiquitination
During the course of investigating the inhibitory effects of KH-4-43, we found that such effects can be influenced by the way in which the ubiquitination reaction was assembled. In Scheme 1 (Fig. 4, top left), the E3-substrate complex was formed by incubating CK1α, CRL4CRBN, and Lenolidomide. KH-4-43 was added to the CK1α/CRL4CRBN/Lenolidomide complex for a brief incubation, followed by the addition of a pre-charged mix that contained both UbcH5c∼Ub and Cdc34b∼Ub thiol-ester complexes. Under this condition, KH-4-43 was able to inhibit the ubiquitination of CK1α (Fig. 4, lanes 2–5), which was entirely consistent with previous observations (Fig. 2). In Scheme 2 (Fig. 4, top right), the preformed CK1α/CRL4CRBN/Lenolidomide complex was first mixed with the pre-charged UbcH5c∼Ub thiol-ester complex for a brief incubation. This was then followed by the addition of KH-4-43 along with pre-charged Cdc34b∼Ub thiol-ester. However, under the condition of Scheme 2, the inhibitory effects by KH-4-43 was significantly reduced (Fig. 4, lanes 6–9). Thus, the priming reaction with UbcH5c∼Ub appeared to prevent E3 CRL4CRBN being inhibited by KH-4-43. Note that these findings cannot be explained by the possibility that KH-4-43 inhibits the priming reaction selectively, because we have previously shown that KH-4-43 is able to inhibit the Ub chain elongation reaction by E3 CRL4CRBN and E2 Cdc34 (16).
Figure 4.
Priming with UbcH5c∼Ub reverses inhibitory effects by KH-4-43 on the ubiquitination of CK1α. The prior reaction of CK1α-CRL4CRBN-lenalidomide with UbcH5c∼Ub caused resistance to KH-4-43 inhibition. The reaction scheme is shown on the top. The detailed procedure is described under “Experimental Procedure.” Species∗ most likely represents the auto-phosphorylated form of CK1α (27). The substrate levels after reactions were quantified and shown. This experiment was repeated once.
The observed reduction in the inhibitory effects of CRL4CRBN by KH-4-43 as a result of the priming reaction (Fig. 4) appears to resemble the effects of neddylation in causing drug insensitivity (Fig. 3). These findings may be explained by CUL ubiquitination that has been reported previously by several groups (8, 13, 16, 18, 19). Indeed, as shown by immunoblot experiment, nearly 50% of CUL4A became mono-ubiquitinated after 1 min incubation of CRL4CRBN with pre-charged UbcH5c∼Ub (Fig. 3A, compare lanes 1 and 3), with longer incubation resulting in CUL4A receiving multiple Ub moieties (Fig. 3A, lane 4). In the light of previous observations suggesting that CUL mono-ubiquitination mimics the effects of neddylation (13), we postulate that mono-ubiquitination of CUL4 may resemble the effects of neddylation in decreasing the ability of CRL4CRBN to respond to KH-4-43 inhibition.
Effects of compound #33 on the ubiquitination of CK1α
Compound #33 is a high throughput hit molecule acting as a promiscuous inhibitor of all E3 CRLs tested (Fig. 1; (16)). Fig. 5A showed that #33 inhibited the ubiquitination of CK1α by CRL4CRBN in a manner similar to KH-4-43 (Fig. 3). Also similar to KH-4-43, #33’s inhibitory effects on CK1α ubiquitination was much reduced with prior neddylation of CRL4CRBN (Fig. 5B). Thus, #33 and KH-4-43 are highly similar in their ability to inhibit CRL4CRBN, and their inhibitory effects are significantly reduced by CRL4 neddylation.
Figure 5.
Effects of compound #33 on CRL4CRBN-dependent CK1α ubiquitination.A, reactions with CRL4CRBN. Low-intensity image of the substrate is also shown to better indicate substrate utilization. B, reactions with Nedd8-CRL4CRBN. The detailed procedure is described under “Experimental Procedure.” Species∗ most likely represents the auto-phosphorylated form of CK1α (27). The substrate levels after reactions were quantified and shown. These experiments were repeated once.
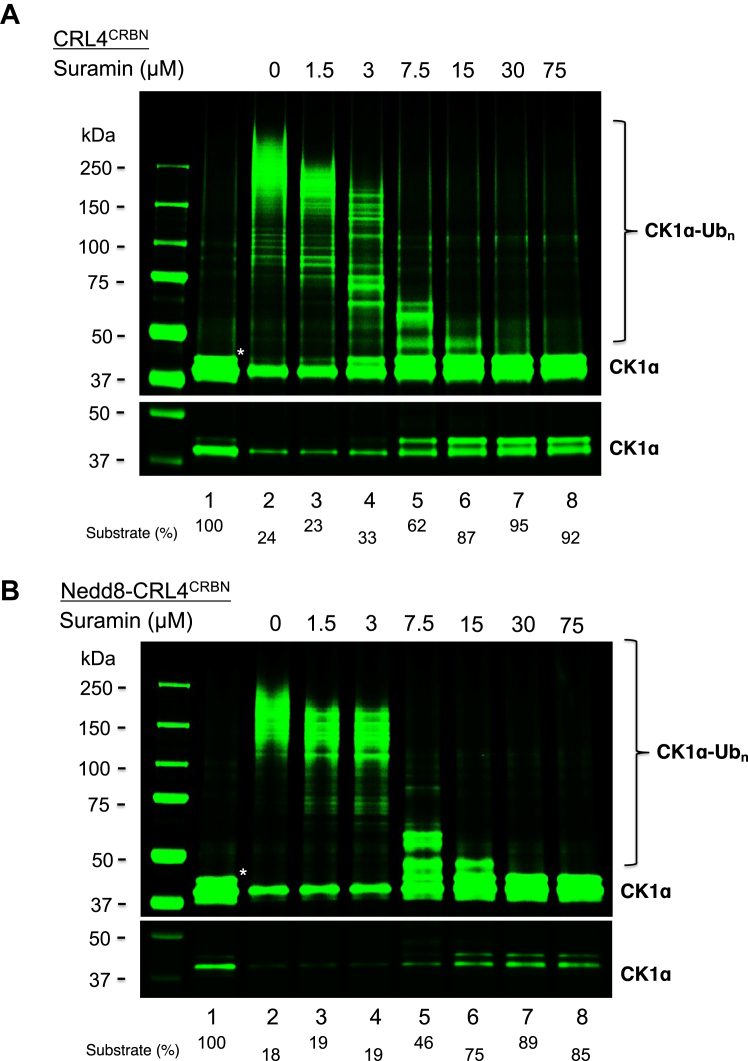
Effects of suramin on the ubiquitination of CK1α
Suramin is an anti-trypansomal drug with the structure of highly acidic polysulfonated naphthylurea (Fig. 1). Suramin inhibited the ubiquitination of CK1α by CRL4CRBN (Fig. 6A). Note that at low concentrations (1.5 and 3 μM), suramin primarily inhibited high molecular weight Ub chain growth, presumably through blocking Cdc34 recruitment (Fig. 6A, lanes 3 and 4). But at high concentrations (>7.5 μM), it completely blocked ubiquitination by inhibiting UbcH5c priming activity as well (Fig. 6A, lanes 5–8). These findings are consistent with our previously published observations (17). Suramin was still able to block the ubiquitination of CK1α by CRL4CRBN that was neddylated, albeit with slight reduction of inhibitory effects, most evidently at concentrations of 3 to 7.7 μM (Fig. 6B, lanes 4–5). Overall, in contrast to KH-4-43/#33, suramin largely retains its ability to inhibit CRL4 activity, even when it is neddylated.
Figure 6.
Effects of suramin on CRL4CRBN-dependent CK1α ubiquitination.A, reactions with CRL4CRBN. This experiment has been repeated four times. B, reactions with Nedd8-CRL4CRBN. This experiment was repeated once. The detailed procedure is described under “Experimental Procedure.” Low-intensity image of the substrate is also shown to better indicate substrate utilization. Species∗ most likely represents the auto-phosphorylated form of CK1α (27). The substrate levels after reactions were quantified and shown.
As suramin is postulated to target cullin CTD’s conserved basic canyon (17), the differential effects on the sensitivity between KH-4-43/#33 and suramin suggest that KH-4-43/#33 target cullin CTD at a site that is different from the basic canyon.
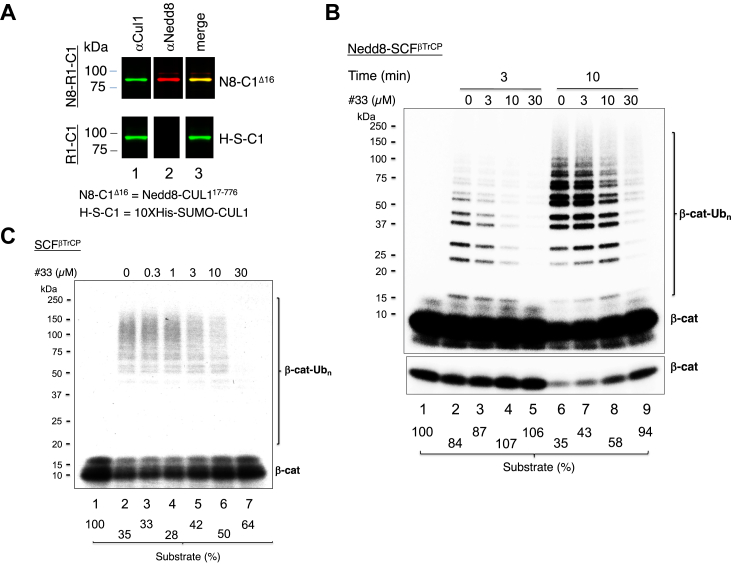
Compound #33 inhibits the ubiquitination of β-catenin
We recently reconstituted the ubiquitination of the β-catenin phosphor-degron peptide by CRL1β-TrCP with UbcH5c as priming E2 and Cdc34b as elongating E2 as described in the accompanying manuscript. Note that CRL1 was originally designated SCF (Skp1-CUL1-F box protein), in which Skp1 is an adaptor protein linking CUL1 to an F-box protein that binds to a substrate (1, 2). We thus use CRL1 and SCF interchangeably in this report. We tested the effects of #33 in β-catenin ubiquitination using both Nedd8-SCFβ-TrCP (Fig. 7B) and SCFβ-TrCP (Fig. 7C). Immunoblot confirmed extensive neddylation of CUL1 (Fig. 7A). Note the significantly lowered reaction efficiency without Nedd8 modification. The results show the ability of #33 to inhibit β-catenin ubiquitination with either Nedd8-SCFβ-TrCP or SCFβ-TrCP (compare Fig. 7, B and C). It is evident that #33 inhibited both initiation and Ub chain elongation, as it blocked substrate utilization (Fig. 7B, lanes 7–9; see quantification of percentage of remaining substrate), and caused accumulation of products of smaller size (Fig. 7B, lane 8). Quantification revealed that #33 exhibited inhibitory effects on substrate utilization with Nedd8-SCFβ-TrCP at levels more pronounced than those with SCFβ-TrCP (compare Fig. 7, B and C). These findings demonstrate that neddylation of SCFβ-TrCP does not negatively impact its sensitivity to #33.
Figure 7.
Effects of compound #33 on CRL1/SCFβ-TrCP-dependent β-catenin ubiquitination. A, immunoblot confirmed extensive neddylation of CUL1. 100 ng of Nedd8-ROC1/Rbx1-CUL1 (lanes 1 and 2, top; a gift of Ning Zheng of University of Washington, Seattle) or ROC1/Rbx1-CUL1 (lanes 1 and 2, bottom; purchased from R&D Systems) was subjected to immunoblot analysis using anti-CUL1 (lane 1) or anti-Nedd8 (lane 2) antibodies. Antibody-reactive signals were visualized by Li-COR Odyssey scanner. Nedd8-ROC1/Rbx1-CUL1, but not ROC1/Rbx1-CUL1, reacted to anti-Nedd8 antibody. Signal merge in lane 3 shows the complete overlap of anti-CUL1 and anti-Nedd8 reactive signals, indicating quantitative neddylation of CUL1. B, reactions with Nedd8-CRL1/SCFβ-TrCP. Incubation is 10 min at 37 °C. Low intensity image of the substrate is also shown to better indicate substrate utilization. Note that similar inhibitory effects of #33 on the Nedd8-SCFβ-TrCP-dependent β-catenin ubiquitination were observed in at least five independent experiments. C, reactions with CRL1/SCFβ-TrCP. Incubation is 20 min at 37 °C. This experiment was repeated once. The detailed procedure is described under “Experimental Procedure.” The substrate levels after reactions were quantified and shown.
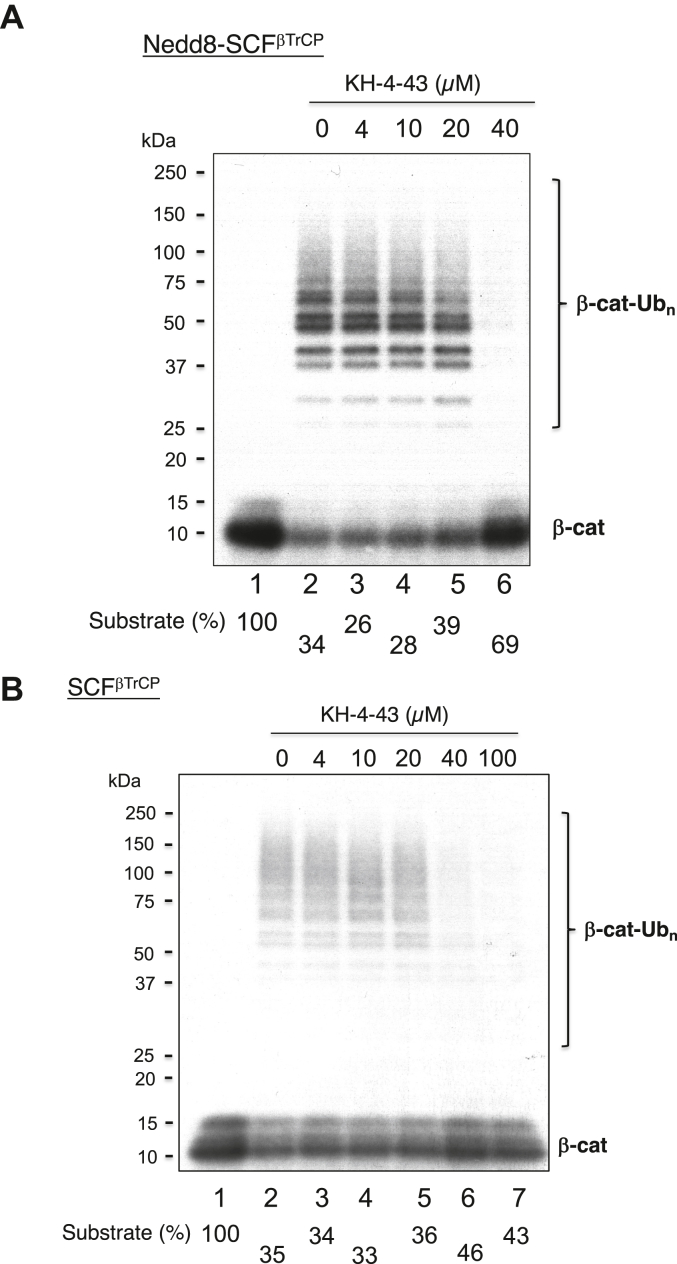
Effects of KH-4-43 on the ubiquitination of β-catenin
KH-4-43 at high levels inhibited the ubiquitination of the β-catenin by Nedd8-SCFβ-TrCP (Fig. 8A). Note that in comparison to #33, high levels of KH-4-43 are required for the inhibition, consistent with the notion that KH-4-43 is less inhibitory than #33 in inhibiting CRL1/SCF (16). The effects of KH-4-43 on the ubiquitination of the β-catenin by Nedd8-SCFβ-TrCP (Fig. 8A) were similar to those observed with the Nedd8-free E3 complex (Fig. 8B). Quantification revealed that KH-4-43 exhibited inhibitory effects on substrate utilization with Nedd8-SCFβ-TrCP at levels more pronounced than those with SCFβ-TrCP (compare Fig. 8, A and B). Thus, like #33, the sensitivity of SCFβ-TrCP to KH-4-43 is not negatively impacted by E3 neddylation. In all, it appears that the effects of neddylation to alter the sensitivity of CRL inhibition by KH-4-43/#33 is dependent upon the specific CRL type. Presumably, Nedd8-CRL1/SCF CTD and Nedd8-CRL4 CTD are substantially different such that the former, but not the latter, can be effectively targeted by KH-4-43/#33.
Figure 8.
Effects of compound KH-4-43 on CRL1/SCFβ-TrCP-dependent β-catenin ubiquitination.A, reactions with Nedd8-CRL1/SCFβ-TrCP. Incubation is 10 min at 37 °C. B, reactions with CRL1/SCFβ-TrCP. Incubation is 20 min at 37 °C. The detailed procedure is described under “Experimental Procedure.” The substrate levels after reactions were quantified and shown. These experiments were repeated once.
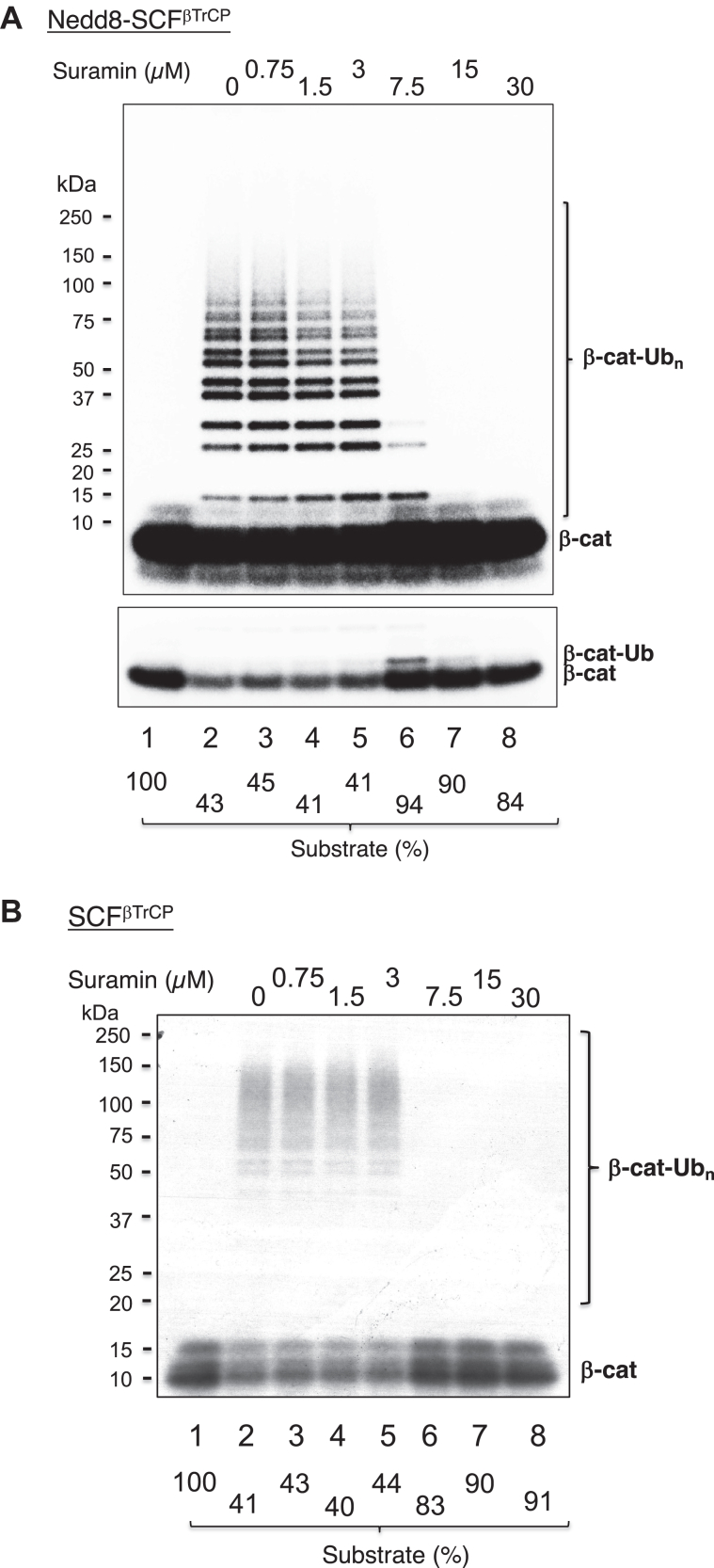
Synergistic effects of #33 and suramin on inhibition of the ubiquitination of β-catenin
Suramin inhibited the ubiquitination of β-catenin by Nedd8-SCFβ-TrCP (Fig. 9A) or SCFβTrCP (Fig. 9B) almost equally. At low concentrations (1.5 and 3 μM), suramin primarily inhibited high molecular weight Ub chain growth, presumably through blocking Cdc34 recruitment (Fig. 9A, lanes 4 and 5). However, at high concentrations (>7.5 μM), it completely blocked ubiquitination by inhibiting UbcH5c priming activity too (Fig. 9A, lanes 6–8).
Figure 9.
Effects of suramin on CRL1/SCFβ-TrCP-dependent β-catenin ubiquitination.A, reactions with Nedd8-CRL1/SCFβ-TrCP. Incubation is 10 min at 37 °C. Low-intensity image of the substrate is also shown to better indicate substrate utilization. B, reactions with CRL1/SCFβ-TrCP. Incubation is 20 min at 37 °C. The detailed procedure is described under “Experimental Procedure.” The substrate levels after reactions were quantified and shown. These experiments were repeated once.
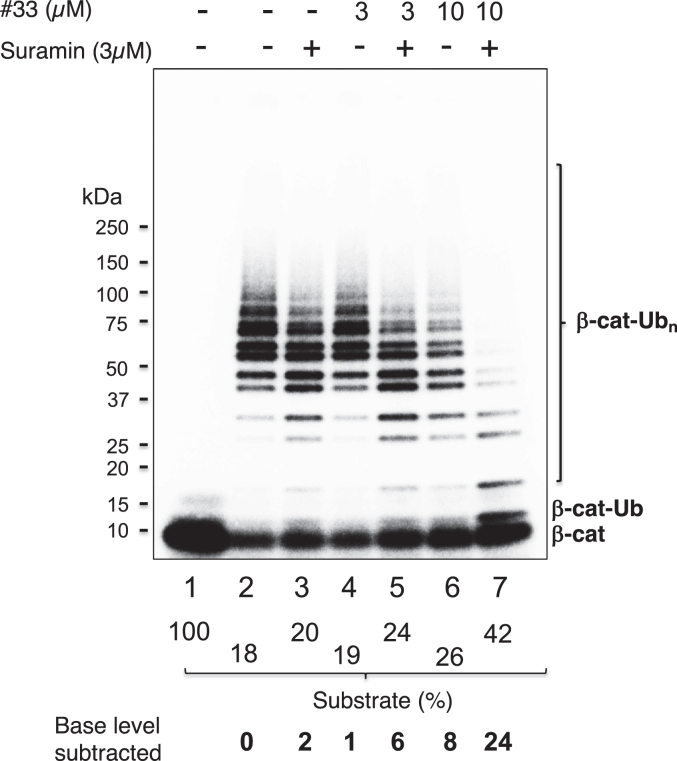
To probe for synergistic effects, the β-catenin ubiquitination model was chosen because it contains only one Ub acceptor lysine residue. At low levels of suramin (3 μM), only small inhibitory effects were observed (Fig. 10, lane 3). Compound #33 alone at 10 μM also caused low level of inhibition as well (Fig. 10, lane 6). However, combination of both compounds yielded inhibition at levels that were far greater than those expected for additive effects of each compound alone (Fig. 10, lane 7). Specifically, with base level subtracted, suramin (3 μM) or #33 (10 μM) alone resulted in inhibition of substrate utilization by 2 and 8%, respectively (Fig. 10, lanes 3 and 6). When combined, the reaction was inhibited by 24% (Fig. 10, lane 7), which was significantly larger than 10%, which would be expected for additive effects (2 + 8%). These results suggest that suramin and #33 act synergistically. It further supports the notion that #33 targets a site on ROC1-CUL1 CTD that is different from CUL1’s basic canyon.
Figure 10.
Synergistic effects of #33 and suramin on Nedd8-CRL1/SCFβ-TrCP-dependent β-catenin ubiquitination. #33 or suramin, alone or in combination as indicated, were added to the reaction. The detailed procedure is described under “Experimental Procedure.” The substrate levels after reactions were quantified and shown. In lane 2 without compound, the remaining substrate level is 18% of the input. “18%” was subtracted from the remaining substrate level in reactions with various combinations of the compounds, resulting in a set of “background-subtracted” numbers as shown. This experiment was repeated once.
Discussion
The E3 CRL core ligase is defined as a sub-complex formed by the CUL CTD bound with the ROC1/RBX1 RING finger protein (1, 2). The multifaced interactions between the CUL CTD, ROC1/RBX1, Nedd8, E2∼Ub, and the ARIH family protein are essential for positioning the thiol-ester-linked Ub optimally to enable its transfer to the CRL-bound substrate lysine, or the receptor lysine of an Ub located on the distal end of an Ub chain. Pharmacological perturbation of the CRL core ligase is of fundamental importance at both basic science and potential therapeutic settings. Suramin, an anti-trypansomal drug, contains highly acidic polysulfonated naphthylurea (Fig. 1), a substructure capable of direct binding to E3 SCF’s core ligase (the ROC1-CUL1 CTD subcomplex) (17). The inhibitory effects of suramin on CRL ubiquitination were revealed in this report using reconstituted ubiquitination systems with the CK1α (Fig. 6) and β-catenin (Fig. 9) substrates.
Our recently reported small molecule inhibitors #33 and KH-4-43 directly bind to CRL core ligases and inhibit CRL-dependent ubiquitination (Fig. 1; ref. (16)). The analyses of these compounds using improved reconstituted ubiquitination of CK1α by CRL4CRBN and β-catenin by SCFβ-TrCP have revealed the following new insights into pharmacological perturbation of the CRL core ligases. First, both KH-4-43 and #33 inhibit the ubiquitination of CK1α by CRL4CRBN (Figs. 2 and 5A). However, both compound’s inhibitory effects on this reaction are significantly reduced when a neddylated form of CRL4CRBN is used (Figs. 3 and 5B). Secondly, while KH-4-43 and #33 appear to retain inhibitory activity on Ub chain elongation to some extent, they can no longer inhibit substrate utilization (Figs. 3 and 5B), suggesting a profound defect to block the priming reaction catalyzed by UbcH5c. Thirdly, both #33 and KH-4-43 inhibit the ubiquitination of β-catenin by CRL1/SCFβ-TrCP and Nedd8-CRL1/SCFβ-TrCP almost equally (Figs. 7–8). Thus, neddylation of CRL1/SCFβ-TrCP does not negatively impact its sensitivity to inhibition by #33 and KH-4-43, suggesting that the effects of neddylation to alter the sensitivity of CRL inhibition by KH-4-43/#33 is dependent upon the specific CRL type. Fourth, suramin inhibits the ubiquitination of CK1α by CRL4CRBN (Fig. 6A). This inhibitory effect is reduced only slightly with Nedd8-CRL4CRBN (Fig. 6B). Suramin inhibit the ubiquitination of β-catenin by SCFβ-TrCP irrespectively of the E3 neddylation status (Fig. 9). Thus, in contrast to KH-4-43/#33, suramin appears largely insensitive to CRL neddylation.
What are the possible molecular bases for the differential responses of KH-4-43/#33 to Nedd8-CRL4 and Nedd8-CRL1? Presumably, the structures of Nedd8-CRL1 and Nedd8-CRL4 are substantially different, such that the former, but not the latter, can be effectively inhibited by KH-4-43/#33. In this regard, previous studies have noted that despite a common globular CTD domain adopted by CUL1-5, different CUL CTDs have divergent folds (13, 20, 21, 22), and different total areas of interface with ROC1/Rbx1, resulting in significant divergent orientation of the ROC1/Rbx1 RING domain among CRLs (22). Of particular interest are the CUL-specific Nedd8 effects that were revealed by recent high resolution structural work by the Schulman group (23). In Nedd8-CRL1/SCF (5, 6), Nedd8 acquires a new conformation that allows direct contacts with E2 UBE2D (UbcH5) or E3 ARIH1 to facilitate priming. However, in Nedd8-CRL5 (15), there is no direct contact between Nedd8 and ARIH2. Instead, new surfaces created by Nedd8-induced CUL5 conformational changes enhance the binding to priming partner ARIH2. Thus, there are clear differences between Nedd8-CRL1 and Nedd8-CRL5 interactions with enzyme partners critical for priming. If Nedd8-CRL1 and Nedd8-CRL4 were to show substantial differences in engaging in priming enzymes, it could provide an explanation for our observed differential drug sensitivities. While Nedd8-CRL1’s surface for priming enzyme is still sensitive to targeting by KH-4-43/#33, its counterpart in Nedd8-CRL4 may be sufficiently different such that it could no longer be targetable to these inhibitors.
Clearly, future work is required to resolve the structures of Nedd8-CRL1 and Nedd8-CRL4 in comparison with the structures of these E3s in complexes with KH-4-43/#33. Intriguingly, the CRL4CRBN-neddylation-caused insensitivity to inhibition by KH-4-43/#33 can be re-created by subjecting the E3 complex to ubiquitination with UbcH5c∼Ub prior to mixing with the inhibitors (Fig. 4). This finding supports previous observations suggesting that CUL mono-ubiquitination mimics the effects of neddylation (13). However, at present time, it is unclear whether CUL mono-ubiquitination is simply a result of in vitro side-reaction, or bears any biological significance.
It should be noted that previously we reported KH-4-43’s ability to inhibit substrate-independent ubiquitination that is mediated by the Nedd8-ROC1-CUL4A complex (16). The differential effects of KH-4-43 on substrate-dependent (CK1α; Fig. 3) and substrate-independent (16) ubiquitination underscore the mechanistic differences between these two reactions. The catalytic architecture for substrate ubiquitination is far more complex than that for substrate-free Ub-Ub ligation. Thus, a note of caution is that results obtained through substrate-independent ubiquitination reactions need to be validated by substrate-dependent reactions.
The specific binding sites of KH-4-43/#33 on CUL CTD remain to be determined. The observations that both compounds are impacted by CRL4CRBN neddylation (Figs. 3 and 5A) suggest that KH-4-43/#33 may target a CUL CTD in the vicinity of the conserved neddylation lysine receptor site. The results with suramin in this work support the notion that KH-4-43/#33 target a site different from suramin, which is known to bind to CUL CTD’s basic canyon. First, suramin appears largely insensitive to CRL1/CRL4 neddylation (Figs. 6 and 9). Second, suramin and #33 act synergistically to inhibit the ubiquitination of β-catenin by SCFβ-TrCP (Fig. 10).
It is intriguing that both previous high-resolution structural work (5, 6, 15) and drug sensitivity studies with CRL core ligase inhibitors presented in this report reveal CUL-specific Nedd8 effects. Nedd8-CRL core ligase-UbcH5∼Ub/ARIH∼Ub are highly diversified architectures, which may provide opportunities for specific targeting by small molecule modulators. The differential drug sensitivities demonstrated in this work provides proof-of-principle evidence of feasibility. Ongoing structure-activity relationship studies with KH-4-43/#33 may expand the CRL inhibitor tool compound repertoire with capabilities to target CRL core ligases and Nedd8-CRLs in a more highly specific manner.
Experimental procedures
Proteins
The following reagents were prepared using established protocols or purchased. β-TrCP-Skp1, Ub E1, UbcH5c, Cdc34a, and Ubc12 were prepared using methods described in Wu et al. (24). E3 CRL4CRBN, Nedd8-ROC1/Rbx1-CUL4A, ROC1/Rbx1-CUL1, CRBN-DDB1, Cdc34b, UBE2G1, Nedd8, Nedd8 E1, and human Ub were purchased from R&D Systems. Nedd8-ROC1-CUL1 was a gift from Ning Zheng of University of Washington, Seattle.
Chemicals
Lenalidomide and suramin were purchased from Selleckchem and Sigma, respectively. KH-4-43 and #33 were prepared as previously described (16).
Effects of compounds on substrate ubiquitination
Reactions with Nedd8-free CRL
For ubiquitination experiments described in Figures 2 and 3C (lanes 2–5), Figures 5A, 6A, 7C, 8B and 9B, the reaction was assembled as follows. To assemble the E3-substrate complex, a mixture (3 μl) containing E3 (100 nM), fluorescently or radioactively labeled substrate (CK1α, 250 nM; β-catenin, 240 nM), lenalidomide (10 μM), 50 mM Tris-HCl, pH 7.4, 5 mM MgCl2, 2 mM ATP, 0.5 mM DTT, 0.1 mg/ml BSA, was incubated for 10 min at room temperature. For reactions with β-catenin (Figs. 7C, 8B and 9B), 70 mM Na-glutamate was added. Compounds in concentrations as specified, were added to the above E3-substrate mix. The volume was adjusted to 5 μl with a buffer that contained 50 mM Tris-HCl, pH 7.4, 5 mM MgCl2, 2 mM ATP, 0.5 mM DTT, 0.1 mg/ml BSA. The mixture was incubated for 10 min at room temperature.
The E2 charging reaction was assembled in a separate tube in a mixture (5 μl) that contained 50 mM Tris-HCl, pH 7.4, 5 mM MgCl2, 2 mM ATP, 0.5 mM DTT, 0.1 mg/ml BSA, Ub (20 μM), E1 (100 nM), UbcH5c (1 μM) and Cdc34 (1 μM). The reaction was incubated for 5 min at room temperature.
To initiate ubiquitination, the above two reaction mixtures were combined (in a final volume of 10 μl) and incubated at 37 °C for times as indicated. After separation by 4 to 20% SDS-PAGE the reaction products were visualized and quantified by fluorescent or phosphor imaging (Typhoon).
Reactions with Nedd8-CRL
To assemble Nedd8-CRL-substrate complex as described in Figure 3, B and C (lanes 6–9)Figures 5B and 6B, E3 CRL4CRBN was first neddylated. For this purpose, the E3 complex (100 nM) was incubated in a reaction (3 μl) containing 50 mM Tris-HCl, pH 7.4, 5 mM MgCl2, 2 mM ATP, 0.5 mM DTT, 0.1 mg/ml BSA, Nedd8 (10 μM), Nedd8 E1 (7.5 nM) and Ubc12 (1.7 μM). The reaction was incubated at room temperature for 10 min. Fluorescently labeled CK1α 250 nM) and lenalidomide (10 μM) were added, followed by incubation at room temperature for 10 min. Compounds in concentrations as specified, were added to the above E3-substrate mix. The volume was adjusted to 5 μl with a buffer that contained 50 mM Tris-HCl, pH 7.4, 5 mM MgCl2, 2 mM ATP, 0.5 mM DTT, 0.1 mg/ml BSA. The mixture was incubated for 10 min at room temperature.
To assemble Nedd8-CRL-substrate complex as described in Figures 3, D and E, 7B, 8A, 9A and 10, Nedd8-ROC1-CUL (100 nM) and substrate receptor – adaptor complex (100 nM; CRBN-DDB1 in Fig. 3, D and E; and β-TrCP-Skp1 in Figs. 7B, 8A, 9A and 10) were mixed and then incubated at room temperature for 10 min. Fluorescently or radioactively labeled substrate (CK1α, 250 nM; β-catenin, 240 nM) and lenalidomide (10 μM; Fig. 3, D and E) were added, followed by incubation at room temperature for 10 min. For reactions with β-catenin (Figs. 7B, 8A, 9A and 10), 70 mM Na-glutamate was added. Compounds in concentrations as specified, were added to the above E3-substrate mix. The volume was adjusted to 5 μl with a buffer that contained 50 mM Tris-HCl, pH 7.4, 5 mM MgCl2, 2 mM ATP, 0.5 mM DTT, 0.1 mg/ml BSA. The mixture was incubated for 10 min at room temperature.
The assembly of E2-Ub charge mix, ubiquitination initiation and incubation, as well as analysis of reaction products, are the same as described earlier.
Reactions with prior formation of E3-inhibitor complex
For ubiquitination experiment described in Fig. 3C (lanes 10–13), the reaction was assembled as follows. E3 was first mixed with the inhibitor in a mixture (2 μl) containing 50 mM Tris-HCl, pH 7.4, 5 mM MgCl2, 2 mM ATP, 0.5 mM DTT, 0.1 mg/ml BSA, CRL4CRBN (100 nM), and KH-4-43 in concentrations as indicated. The mixture was incubated for 10 min at room temperature. Fluorescent CK1α (250 nM) and lenalidomide (10 μM) were added and incubation continued at room temperature for 10 min. Ubiquitination was initiated by the addition of the preformed E2-Ub charge mix, assembled as described above. The reaction was incubated for 10 min at 37 °C. The reaction products were analyzed as described above.
Priming reverses the inhibitory effects of KH-4-43
The E3-substrate complex was formed by mixing CRL4CRBN (100 nM), fluorescent CK1α (250 nM), lenalidomide (10 μM), 50 mM Tris-HCl, pH 7.4, 5 mM MgCl2, 2 mM ATP, 0.5 mM DTT, 0.1 mg/ml BSA in a volume of 4.7 μl (Fig. 4). The mixture was incubated for 10 min at room temperature.
In two additional separate tubes, two E2-Ub charge mixes were prepared. UbcH5c∼Ub mixture (2.5 μl) contained 50 mM Tris-HCl, pH 7.4, 5 mM MgCl2, 2 mM ATP, 0.5 mM DTT, 0.1 mg/ml BSA, Ub (20 μM), E1 (100 nM), and UbcH5c (1 μM). Cdc34b∼Ub mixture was identical to the above mix except that Cdc34 (1 μM) replaces UbcH5c. The reaction was incubated for 5 min at room temperature.
For reactions shown in Figure 4, lanes 2 to 5, KH-4-43 (0.3 μl) in concentrations as indicated was added to the E3-substrate mix, followed by incubation for 10 min at room temperature. This was then followed by the addition of both UbcH5c∼Ub and Cdc34b∼Ub. The reaction was incubated for 20 min at 37 °C.
For reactions shown in Figure 4, lanes 6 to 9, the UbcH5c∼Ub mix was added to the E3-substrate mix, The reaction was incubated for 5 min at 37 °C. KH-4-43 (0.3 μl) in concentrations as indicated was added to the E3-substrate mix, followed by incubation for 10 min at room temperature. This was then followed by the addition of Cdc34b∼Ub. The reaction was incubated for 20 min at 37 °C.
All reaction products were analyzed as described above.
Immunoblot
The CUL4 blots shown in Figure 3A were performed as follows. For CRL4CRBN neddylation, the E3 complex (1pmol) was incubated in a reaction (3 μl) containing 50 mM Tris-HCl, pH 7.4, 5 mM MgCl2, 2 mM ATP, 0.5 mM DTT, 0.1 mg/ml BSA, Nedd8 (10 μM), Nedd8 E1 (7.5 nM) and Ubc12 (1.7 μM). The reaction was incubated at room temperature for 10 min. For mono-ubiquitination of CRL4CRBN, the E3 complex (1pmol) was incubated with 50 mM Tris-HCl, pH 7.4, 5 mM MgCl2, 2 mM ATP, 0.5 mM DTT, 0.1 mg/ml BSA, Ub (20 μM), E1 (100 nM), and UbcH5c (1 μM), at 37 °C for 1 or 5 min. Untreated CRL4CRBN (1pmol) was used as a control. These materials were separated by 4 to 20% SDS-PAGE and transferred to membrane blot. CUL4A was visualized by immunoblot analysis using anti-CUL4A antibodies and Odyssey scanner.
For CUL1 blots shown in Figure 7A, 100 ng of Nedd8-ROC1/Rbx1-CUL1 (a gift of Ning Zheng of University of Washington) or ROC1/Rbx1-CUL1 (purchased from R&D Systems) was subjected to immunoblot analysis using anti-CUL1 or anti-Nedd8 antibodies. Antibody-reactive signals were visualized by Li-COR Odyssey scanner.
Data availability
All data are contained within the manuscript.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We thank I. Lu for technical assistance, and N. Zheng for reagents.
Author contributions
K. W. and Z.-Q. P. methodology; K. W. and Z.-Q. P. investigation; K. W. and Z.-Q. P. formal analysis; R. J. D resources; Z.-Q. P. writing–original draft.
Funding and additional information
This work was supported by NIH grant CA251425 (to Z.-Q. P and R. J. D). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Reviewed by members of the JBC Editorial Board. Edited by George DeMartino
References
- 1.Petroski M.D., Deshaies R.J. Function and regulation of cullin-RING ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 2005;6:9–20. doi: 10.1038/nrm1547. [DOI] [PubMed] [Google Scholar]
- 2.Sarikas A., Hartmann T., Pan Z.Q. The cullin protein family. Genome Biol. 2011;12:220. doi: 10.1186/gb-2011-12-4-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pan Z.Q., Kentsis A., Dias D.C., Yamoah K., Wu K. Nedd8 on cullin: building an expressway to protein destruction. Oncogene. 2004;23:1985–1997. doi: 10.1038/sj.onc.1207414. [DOI] [PubMed] [Google Scholar]
- 4.Scott D.C., Rhee D.Y., Duda D.M., Kelsall I.R., Olszewski J.L., Paulo J.A., et al. Two distinct types of E3 ligases work in unison to regulate substrate ubiquitylation. Cell. 2016;166:1198–1214.e24. doi: 10.1016/j.cell.2016.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baek K., Krist D.T., Prabu J.R., Hill S., Klügel M., Neumaier L.M., et al. NEDD8 nucleates a multivalent cullin-RING-UBE2D ubiquitin ligation assembly. Nature. 2020;578:461–466. doi: 10.1038/s41586-020-2000-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Horn-Ghetko D., Krist D.T., Prabu J.R., Baek K., Mulder M.P.C., Klügel M., et al. Ubiquitin ligation to F-box protein targets by SCF-RBR E3-E3 super-assembly. Nature. 2021;590:671–676. doi: 10.1038/s41586-021-03197-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kleiger G., Saha A., Lewis S., Kuhlman B., Deshaies R.J. Rapid E2-E3 assembly and disassembly enable processive ubiquitylation of cullin-RING ubiquitin ligase substrates. Cell. 2009;139:957–968. doi: 10.1016/j.cell.2009.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sandoval D., Hill S., Ziemba A., Lewis S., Kuhlman B., Kleiger G. Ubiquitin-conjugating enzyme Cdc34 and ubiquitin ligase Skp1-cullin-F-box ligase (SCF) interact through multiple conformations. J. Biol. Chem. 2015;290:1106–1118. doi: 10.1074/jbc.M114.615559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu K., Kovacev J., Pan Z.Q. Priming and extending: an UbcH5/Cdc34 E2 handoff mechanism for polyubiquitination on a SCF substrate. Mol. Cell. 2010;37:784–796. doi: 10.1016/j.molcel.2010.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kovacev J., Wu K., Spratt D.E., Chong R.A., Lee C., Nayak J., et al. A snapshot at ubiquitin chain elongation: lysine 48-tetra-ubiquitin slows down ubiquitination. J. Biol. Chem. 2014;289:7068–7081. doi: 10.1074/jbc.M113.530576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lu G., Weng S., Matyskiela M., Zheng X., Fang W., Wood S., et al. UBE2G1 governs the destruction of cereblon neomorphic substrates. Elife. 2018;7 doi: 10.7554/eLife.40958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sievers Q.L., Gasser J.A., Cowley G.S., Fischer E.S., Ebert B.L. Genome-wide screen identifies cullin-RING ligase machinery required for lenalidomide-dependent CRL4CRBN activity. Blood. 2018;132:1293–1303. doi: 10.1182/blood-2018-01-821769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duda D.M., Borg L.A., Scott D.C., Hunt H.W., Hammel M., Schulman B.A. Structural insights into NEDD8 activation of cullin-RING ligases: conformational control of conjugation. Cell. 2008;134:995–1006. doi: 10.1016/j.cell.2008.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yamoah K., Oashi T., Sarikas A., Gazdoiu S., Osman R., Pan Z.Q. Autoinhibitory regulation of SCF-mediated ubiquitination by human cullin 1's C-terminal tail. Proc. Natl. Acad. Sci. U. S. A. 2008;105:12230–12235. doi: 10.1073/pnas.0806155105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kostrhon S., Prabu J.R., Baek K., Horn-Ghetko D., von Gronau S., Klügel M., et al. CUL5-ARIH2 E3-E3 ubiquitin ligase structure reveals cullin-specific NEDD8 activation. Nat. Chem. Biol. 2021;17:1075–1083. doi: 10.1038/s41589-021-00858-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu K., Huynh K.Q., Lu I., Moustakim M., Miao H., Yu C., et al. Inhibitors of cullin-RING E3 ubiquitin ligase 4 with antitumor potential. Proc. Natl. Acad. Sci. U. S. A. 2021;118 doi: 10.1073/pnas.2007328118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu K., Chong R.A., Yu Q., Bai J., Spratt D.E., Ching K., et al. Suramin inhibits cullin-RING E3 ubiquitin ligases. Proc. Natl. Acad. Sci. U. S. A. 2016;113:E2011–2018. doi: 10.1073/pnas.1601089113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu G., Xu G., Schulman B.A., Jeffrey P.D., Harper J.W., Pavletich N.P. Structure of a beta-TrCP1-Skp1-beta-catenin complex: destruction motif binding and lysine specificity of the SCF(beta-TrCP1) ubiquitin ligase. Mol. Cell. 2003;11:1445–1456. doi: 10.1016/s1097-2765(03)00234-x. [DOI] [PubMed] [Google Scholar]
- 19.Laplaza J.M., Bostick M., Scholes D.T., Curcio M.J., Callis J. Saccharomyces cerevisiae ubiquitin-like protein Rub1 conjugates to cullin proteins Rtt101 and Cul3 in vivo. Biochem. J. 2004;377:459–467. doi: 10.1042/BJ20030755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Angers S., Li T., Yi X., MacCoss M.J., Moon R.T., Zheng N. Molecular architecture and assembly of the DDB1-CUL4A ubiquitin ligase machinery. Nature. 2006;443:590–593. doi: 10.1038/nature05175. [DOI] [PubMed] [Google Scholar]
- 21.Zheng N., Schulman B.A., Song L., Miller J.J., Jeffrey P.D., Wang P., et al. Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex. Nature. 2002;416:703–709. doi: 10.1038/416703a. [DOI] [PubMed] [Google Scholar]
- 22.Cardote T.A.F., Gadd M.S., Ciulli A. Crystal structure of the Cul2-Rbx1-EloBC- VHL ubiquitin ligase complex. Structure. 2017;25:901–911. doi: 10.1016/j.str.2017.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dikic I., Schulman B.A. An expanded lexicon for the ubiquitin code. Nat. Rev. Mol. Cell Biol. 2023;24:273–287. doi: 10.1038/s41580-022-00543-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu K., Ching K., Chong R.A., Pan Z.Q. A new Forster resonance energy transfer-based platform to track substrate ubiquitination by fluorescence. J. Biol. Chem. 2020;296 doi: 10.1074/jbc.RA120.016858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krönke J., Fink E.C., Hollenbach P.W., MacBeth K.J., Hurst S.N., Udeshi N.D., et al. Lenalidomide induces ubiquitination and degradation of CK1α indel(5q) MDS. Nature. 2015;523:183–188. doi: 10.1038/nature14610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Petzold G., Fischer E.S., Thomä N.H. Structural basis of lenalidomide-induced CK1α degradation by the CRL4(CRBN) ubiquitin ligase. Nature. 2016;532:127–130. doi: 10.1038/nature16979. [DOI] [PubMed] [Google Scholar]
- 27.Budini M., Jacob G., Jedlicki A., Pérez C., Allende C.C., Allende J.E. Autophosphorylation of carboxy-terminal residues inhibits the activity of protein kinase CK1alpha. J. Cell Biochem. 2009;106:399–408. doi: 10.1002/jcb.22019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data are contained within the manuscript.