Abstract
Signal peptidase cleavage at the C-prM junction in the flavivirus structural polyprotein is inefficient in the absence of the cytoplasmic viral protease, which catalyzes cleavage at the COOH terminus of the C protein. The signal peptidase cleavage occurs efficiently in circumstances where the C protein is deleted or if the viral protease complex is present. In this study, we used cDNA of Murray Valley encephalitis virus (MVE) to examine features of the structural polyprotein which allow this regulation of a luminal cleavage by a cytoplasmic protease. We found that the inefficiency of signal peptidase cleavage in the absence of the viral protease is not attributable solely to features of the C protein. Inhibition of cleavage still occurred when charged residues in C were mutated to uncharged residues or when an unrelated protein sequence (that of ubiquitin) was substituted for C. Also, fusion of the C protein did not inhibit processing of an alternative adjacent signal sequence. The cleavage region of the flavivirus prM translocation signal is unusually hydrophobic, and we established that altering this characteristic by making three point mutations near the signal peptidase cleavage site in MVE prM dramatically increased the extent of cleavage without requiring removal of the C protein. In addition, we demonstrated that luminal sequences downstream from the signal peptidase cleavage site contributed to the inefficiency of cleavage.
Polyprotein processing is important in the regulation of gene expression of many plus-strand RNA viruses (16, 19, 29, 41). The production from a polyprotein of precursor and mature proteins, which may have different functional activities, can be quantitatively and temporally modulated (9, 22, 43). This involves predominantly the alteration of cleavage specificities of virus-encoded cytoplasmic proteases. The regulation of a signal peptidase cleavage in the lumen of the endoplasmic reticulum (ER) by a cytoplasmic viral protease has been described for the processing of the structural polyprotein region of several flaviviruses (1, 23, 42). This is intriguing since signal peptidase cleavages are generally assumed to take place rapidly, during protein translocation across the ER membrane (4).
Flaviviruses are enveloped, positive-strand RNA viruses. The genome encodes a single polyprotein which is approximately 3,500 amino acids long and traverses the ER membrane multiple times (reviewed in reference 31). This polyprotein is cleaved to produce three structural and seven nonstructural proteins, and all but two of the necessary cleavages are catalyzed by the virus-encoded NS2B-3 protease in the cytoplasm or by signal peptidase at the luminal side of the ER membrane. The flavivirus structural proteins are encoded in the 5′ quarter of the genome. The capsid (C) protein, at the NH2 terminus of the polyprotein, is separated from the prM (precursor to membrane) protein by a signal sequence directing the translocation of prM. The NS2B-3 protease complex catalyzes cleavage at the COOH terminus of the C protein on the cytoplasmic side of the ER membrane. This is the only site in the structural polyprotein region which is cleaved by this enzyme. The type I transmembrane protein prM is anchored in the lipid bilayer by a COOH-terminal membrane anchor, which is immediately followed by the signal sequence for translocation of the E (envelope) protein, also a type I transmembrane protein. Thus the NH2 termini of the prM and E proteins are generated by signal peptidase cleavages. However, it has been noted for a number of flaviviruses that when the entire structural polyprotein region is expressed from cDNA, the signal peptidase-mediated cleavage at the NH2 terminus of prM does not occur efficiently, in contrast to that at the NH2 terminus of the E protein (23, 33, 36, 42). This inefficient production of prM is reflected in the deficiency of secretion of the prM-E heterodimer and, in turn, the lack of immunogenicity often observed when such constructs are used for vaccination (see, for example, references 10, 11, 18, 30, and 34).
Signal peptidase cleavage at the C-prM junction is greatly enhanced in the presence of the viral NS2B-3 protease (1, 23, 42) or when prM is expressed by using constructs which do not include the C protein-coding region (23, 42). Furthermore, cleavage at the NH2 terminus of prM by signal peptidase can be induced to occur posttranslationally following trypsin cleavage of the cytoplasmic C region of the C-prM precursor in crude microsomes in vitro (36). One of us has proposed that the covalent linkage of C to prM results in the positioning of the signal sequence of prM in the ER membrane such that the signal peptidase cleavage site is maintained in a cryptic conformation (23). In the present study we have investigated elements in the structural polyprotein region of a flavivirus, Murray Valley encephalitis virus (MVE), which allow the control of signal peptidase cleavage of prM by the viral protease.
MATERIALS AND METHODS
Site-directed mutagenesis.
In vitro site-directed mutagenesis was performed by the method of Kunkel et al. (20). DNA encompassing the target region was subcloned into M13mp19, and the recombinant phage was grown in Escherichia coli CJ236 in the presence of 0.25 μg of uridine per ml to give uracil-containing single-stranded template DNA. Mutagenic oligonucleotides were phosphorylated and annealed to template DNA before second-strand synthesis with Sequenase (U.S. Biochemicals) in the presence of T4 DNA ligase as previously described (37). E. coli TG1 cells were transformed with the reaction products, and phage progeny were screened for the desired mutation by sequence or restriction enzyme analysis. Sequences of mutagenic oligonucleotides are provided below, with pertinent restriction enzyme recognition sites in italics and mismatching nucleotides underlined.
Eukaryotic expression plasmids.
MVE cDNA encompassing most of the 5′ untranslated region and sequence encoding the NH2-terminal region of the polyprotein (MVE residues 1 to 1380, comprising C-prM-E-NS1-NS2A-6%NS2B) followed by 11 vector-specified amino acids is contained in pSTR (previously called pcDNA-STR [23]). Sequence coding for most of the C protein (residues 5 to 104) is deleted from this in pSTR.ΔC (formerly pcDNAΔC [23]). Plasmid pNS3/T (previously pcDNA-NS3/T [24]) contains MVE cDNA encoding the COOH-terminal region of the polyprotein (residues 1302 to 3434, equivalent to 31%NS2A-NS2B-NS3-NS4A-NS4B-NS5) with an amber termination codon at the NS3-4A junction. Schematic diagrams showing the expected topology in the ER membrane of translation products from plasmids containing MVE structural protein-coding regions are provided in Fig. 2B to 4B and 6B to 8B.
FIG. 2.
Substitution of basic residues in the C protein does not influence signal peptidase cleavage at the C-prM junction. (A) COS cells were transfected with pSTR (lanes 1 and 5), pSTR.Cmut2aa (lanes 2 and 6), pSTR.Cmut4aa (lanes 3 and 7), or pSTR.Cmut11aa (lanes 4 and 8), with (+) or without (−) pNS3/T. Lane 9 shows the results of transfection with pNS3/T only. Two days after transfection, the monolayers were metabolically labelled for 30 min and cell lysates were subjected to immunoprecipitation with anti-MVE mouse ascitic fluid. Polypeptides were separated by SDS-PAGE (12% polyacrylamide). Bands corresponding to MVE E, NS1, prM, and putative C-prM glycoproteins are labelled, and the numbers indicate the sizes (in kilodaltons) of marker proteins in lane M. (B) Schematic diagrams showing the expected topology at the ER membrane of polyproteins encoded by pSTR variants in the presence and absence of NS2B-3. M denotes the ER membrane, L denotes the ER lumen, and Cy denotes the cytoplasmic side of the membrane. Sites of efficient cleavage by signal peptidase are indicated by arrowheads, the proteolytic cleavage catalyzed by the viral NS2B-3 complex is denoted by an arrow, and regions of the MVE C protein which were altered in the various pSTR.Cmut constructs are indicated by circles in parentheses.
FIG. 4.
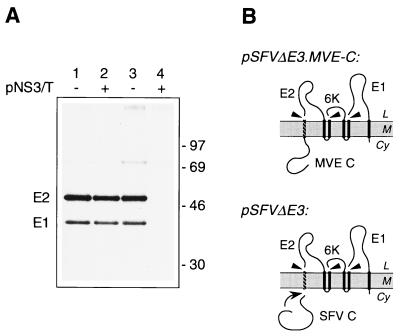
NH2-terminal linkage of the MVE C protein to an idealized signal sequence does not inhibit cleavage by signal peptidase. (A) COS cells transfected with pSFVΔE3.MVE-C (lane 1), pSFVΔE3 (lane 3), or pNS3/T (lane 4) or cotransfected with pSFVΔE3.MVE-C and pNS3/T (lane 2) were metabolically labelled for 3 h. The products immunoprecipitated by an anti-SFV mouse serum were separated by SDS-PAGE (10% polyacrylamide) under nonreducing conditions to separate E2 and E1. Bands corresponding to the SFV E2 and E1 glycoproteins are shown, and the numbers indicate the positions and sizes (in kilodaltons) of marker proteins. (B) Schematic diagrams showing the expected topology at the ER membrane of polyproteins encoded by pSFVΔE3.MVE-C and pSFVΔE3. Abbreviations are as listed in the legend to Fig. 2. Autocatalytic cleavage of the unmutated SFV C protein is indicated by a curved arrow.
FIG. 6.
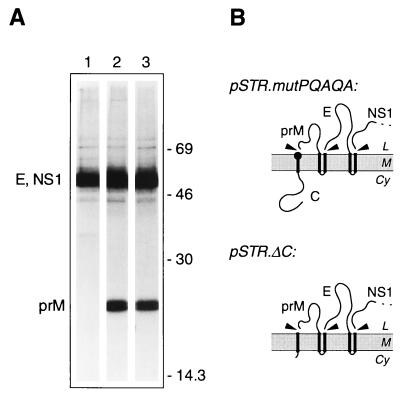
Mutations in the signal sequence of prM induce efficient signal peptidase cleavage independent of the presence of the NS2B-3 protease. (A) COS cells transiently transfected with pSTR (lane 1), pSTR.mutPQAQA (lane 2), or pSTR.ΔC (lane 3) were metabolically labelled for 3 h. Polypeptides immunoprecipitated from cell lysates with anti-MVE mouse ascitic fluid were resolved by SDS-PAGE (12% polyacrylamide). Bands corresponding to MVE E, NS1, and prM glycoproteins are labelled, and the numbers indicate the positions and sizes (in kilodaltons) of marker proteins. (B) Schematic diagrams showing the expected topology at the ER membrane of polyproteins encoded by pSTR.mutPQAQA and pSTRΔC. Abbreviations are as listed in the legend to Fig. 2. The location of the altered amino acid residues encoded by pSTR.mutPQAQA is represented by a solid circle.
FIG. 8.
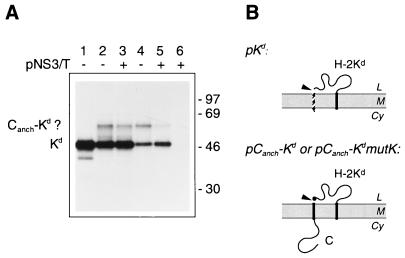
The production of H-2Kd from a Canch-Kd fusion protein is not dependent on the viral protease. (A) COS cells were transiently transfected with pKd (lane 1), pCanch-Kd (lanes 2 and 3), or pCanch-KdmutK (lanes 4 and 5), with (+) or without (−) pNS3/T. Lane 6 shows the results of transfection with pNS3/T only. Cell monolayers were pulse-labelled for 30 min, and the products immunoprecipitated from cell lysates with an anti-Kd monoclonal antibody (Hb159) were subjected to SDS-PAGE (12% polyacrylamide). Bands corresponding to the Kd and possible Canch-Kd fusion proteins are labelled, and the numbers indicate the positions and sizes (in kilodaltons) of marker proteins. (B) Schematic diagrams showing the expected topology at the ER membrane of polyproteins encoded by pKd, pCanch-Kd and pCanch-KdmutK. Abbreviations are as listed in the legend to Fig. 2, and the Pro→Lys substitution encoded in pCanch-KdmutK is represented by a solid circle.
(i) pSTR charge variants.
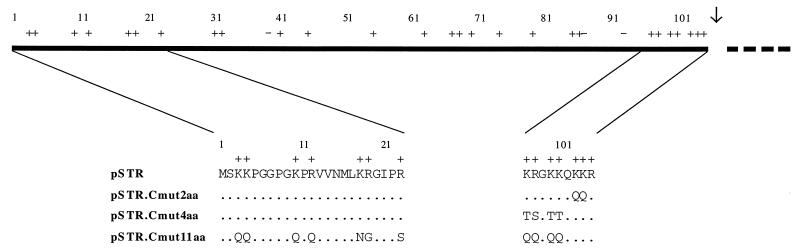
M13.Canch, a construct for use as a mutagenesis template, was created by subcloning from pSTR into M13mp19 a 521-bp HindIII fragment containing MVE cDNA encoding Canch (anchored C, composed of the C protein and the adjacent prM translocation signal). Site-directed mutagenesis was performed to alter codons for charged residues in the C protein. In some cases, recombinant phage carrying introduced mutations were grown in E. coli CJ236 in the presence of uridine to generate template DNA for further mutagenesis. Fragments containing appropriate combinations of mutations were substituted for the original coding sequence in pSTR. Oligonucleotide 203 (5′-C GAC CTG GGG CTG CCC G-3′) introduced the changes Lys10→Gln and Arg12→Gln; 204 (5′-T TTG TTG TTG GCC CTG TTG GTT CAC C-3′) gave Lys97→Gln, Arg98→Gln, Lys100→Gln, and Lys101→Gln; 205 (5′-CC TCC TGG TTG TTG AGA CAT TTG AA-3′) gave Lys3→Gln and Lys4→Gln; 206 (5′-GAA TAC GCT GGG TAT GCC GCC ATT TAG CAT-3′) gave Lys18→Asn, Arg19→Gly, and Arg23→Ser; 207 (5′-CC ACC TCT TTG TTG TTG TTT TTT GC-3′) gave Lys103→Gln and Lys104→Gln; and 208 (5′-TTT CTT TTG TGT TGT GCC ACT TGT GTT CAC CAC-3′) gave Lys97→Thr, Arg98→Ser, Lys100→Thr, and Lys101→Thr. Combinations of changes incorporated into particular constructs are illustrated in Fig. 1.
FIG. 1.
Schematic diagram showing the locations of alterations to charged residues in derivatives of the MVE C protein. Plasmid pSTR encodes the C-prM-E-NS1-NS2A-6%NS2B region of the MVE polyprotein. The C protein is represented by a solid line, and the beginning of the prM translocation signal is indicated by a dashed line. Positions of charged residues are indicated by + for Arg or Lys and − for Asp or Glu. Residues 1 to 23 and 97 to 105 of the C protein encoded by this construct are shown in single-letter amino acid code. Residues in these regions which are unchanged in the charge mutant derivatives of pSTR are denoted by dots. Sequences outside the marked regions were identical in all constructs. The site of cleavage by the viral NS2B-3 protease is marked with an arrow.
(ii) pSTR.Ub and pSTR.UbmutV.
A SacII site was introduced at the 5′ end of DNA corresponding to the MVE prM translocation signal by using oligonucleotide 218 (5′-TC ACT GCC ACC GCGGCG CTT TTG TTT TTT GC-3′) for site-directed mutagenesis of M13.Canch. Sequence containing the ubiquitin gene was obtained as a 0.2-kb EcoRI-SacII fragment from pRB269 (3). This fragment was inserted in place of the MVE C coding region in the M13 mutagenesis intermediate. The resulting ubiquitin-prM signal fusion sequence was transferred into pSTR as a HindIII fragment (replacing the 521-bp fragment containing the MVE C gene), giving pSTR.Ub. This construct encodes ubiquitin (76 amino acids) fused to the MVE prM-E-NS1-NS2A-6%NS2B sequence, with Gly residues at positions 75 and 76 doubling as the last two residues of the ubiquitin sequence and the first two residues of the prM translocation signal. A related construct, pSTR.UbmutV, was obtained by using oligonucleotide 219 (5′-GGA TGT TTC ACT GAC ACC GCG GAG-3′) to change the codon corresponding to Gly76 of ubiquitin to Val before exchanging the HindIII fragment into pSTR.
(iii) pSFVΔE3.MVE-C.
The MVE C gene was amplified by PCR from plasmid 2/1/22 (8) with oligonucleotide 211 (5′-ATT AGA TCT GCG TGA GCT TCC-3′), which contains a BglII site and sequence in the MVE 5′ untranslated region, and oligonucleotide 212 (5′-CT GGATCC TCT TTT CTT TTG T-3′), which is composed of a BamHI site and sequence complementary to the 3′ end of the MVE C gene. The PCR product was cut with BglII and BamHI and ligated into the BamHI site in the eukaryotic expression vector pcDNA1 (Invitrogen). DNA encoding the structural proteins of Semliki Forest virus (SFV) with the E3 protein-coding sequence replaced by an artificial cleavable translocation signal (25) was digested with BamHI, and a fragment corresponding to the artificial translocation signal-E2-6K-E1 was ligated into the BamHI site downstream of the MVE C protein gene. After failure to detect E1 protein of the expected size among proteins expressed from this construct, the HindIII-EcoRI fragment encoding the MVE C/SFV E2 region was used to replace the corresponding sequence (encoding SFV C-artificial signal-E2) in an alternative SFVΔE3 simian virus 40-based expression plasmid (25) (referred to here as pSFVΔE3), generating pSFVΔE3.MVE-C. This plasmid thus contains coding sequence for the 105 amino acids of the MVE C protein, fused in frame to sequence encoding 20 residues of an artificial cleavable signal sequence followed by the E2-6K-E1 polyprotein of SFV.
(iv) pSTR.mutPQAQA.
With M13.Canch as a template, oligonucleotide 213 (5′-G CTT TAA GGC TTG GGC TTG TGG AAT CAG CAT GAA-3′) was used to alter the region coding for the COOH-terminal amino acids of the prM translocation signal so that the sequence Gly-Phe-Ala-Ala-Ala was replaced with Pro-Gln-Ala-Gln-Ala. The mutated HindIII fragment was ligated in place of the corresponding region in pSTR to give pSTR.mutPQAQA.
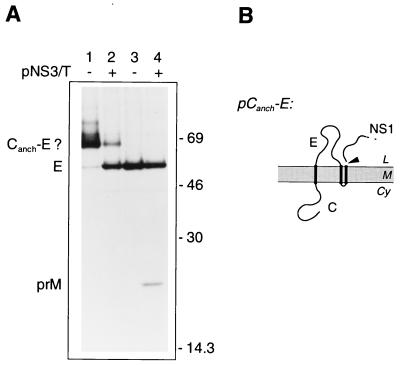
(v) pCanch-E.
A 1,975-bp region of pSTR encompassing the cDNA encoding MVE C, prM, and the NH2-terminal two-thirds of E was transferred as a PstI fragment into M13mp19. This construct was mutagenized with oligonucleotide 220 (5′-G CTT TAA GGC CGC GGC AAA TCC-3′) to introduce a silent SacII recognition site at the COOH-terminal end of the translocation signal for prM and oligonucleotide 221 (5′-CAG ACA GTT AAA AGC CGC GGC AGG AGC-3′) to introduce a SacII site and accompanying codon changes Tyr-Ser→Ala-Ala immediately before the E gene. The 488-bp PstI-SacII fragment (containing coding sequence for Canch) and the 968-bp SacII-PstI fragment (encompassing the 5′ two-thirds of the E gene) were then substituted for the region between the corresponding PstI sites in pSTR to yield pCanch-E. This construct thus encodes the MVE C protein and translocation signal for prM (MVE residues 1 to 125) fused directly to the MVE E-NS1-NS2A-6%NS2B sequence (MVE residues 293 to 1380) at the first residue of the MVE E protein.
(vi) pKd, pCanch-Kd, and pCanch-KdmutK.
The pcDNA3 (Invitrogen)-based expression plasmid pKd contains cDNA encoding the 368-amino-acid mouse H-2Kd major histocompatibility complex (MHC) antigen (21). To generate a Canch-Kd fusion construct, an ApaI site (and accompanying codon changes Leu-Lys→Gly-Pro) was introduced 3′ of the sequence encoding the prM translocation signal by mutagenesis of M13.Canch with oligonucleotide 214 (5′-G GCA TGC AAG CGG GCC CGC AGC GGC AA-3′). DNA encoding H-2Kd without the NH2-terminal signal sequence was then subcloned as a 1,096-bp ApaI fragment into the M13 mutagenesis intermediate to create M13.C-Kd. The Canch-Kd coding sequence was subsequently transferred as a 1,573-bp BamHI-XbaI fragment into pcDNA3 to obtain pCanch-Kd, which encodes the MVE C protein and translocation signal for prM (MVE residues 1 to 125) fused directly to the H-2Kd molecule (residues 22 to 368 of the H-2Kd preprotein). Oligonucleotide 216 (5′-CCT CAG CGA ATG TTT GCC CGC AGC GG-3′) was used to change the second codon in the H-2Kd sequence from Pro to Lys in the M13 mutagenesis intermediate, and the mutated sequence was transferred as a 1,573-bp BamHI-XbaI fragment into pcDNA3 to produce pCanch-KdmutK.
Transient expression in COS-7 cells.
COS-7 cells were transfected with 1 to 2 μg of eukaryotic expression vector DNA (or this amount of each plasmid in cotransfections) as previously described (24).
Metabolic labelling, lysis, immunoprecipitation, electrophoresis, and fluorography.
Two days after transfection, cell monolayers were washed twice with phosphate-buffered saline and starved in methionine- and cysteine-free medium for 0.5 to 1 h. The cells were metabolically labelled by incubation for 0.5 to 3 h with 0.25 to 1 ml of the above medium containing a mixture of l-[35S]methionine and l-[35S]cysteine (Amersham or Du Pont) at 100 to 500 μCi/ml.
Dishes of labelled cells were placed on ice and washed with phosphate-buffered saline. Monolayers were solubilized by incubation on ice for 10 to 30 min in 0.5 ml of NP-40 lysis buffer (1% Nonidet P-40, 50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 2 mM EDTA) containing 20 μg of phenylmethylsulfonyl fluoride per ml. Nuclei were removed by centrifugation, and the lysates were precleared by incubation for 2 to 3 h with protein A–Sepharose CL-4B (Pharmacia). The lysates were subjected to immunoprecipitation by incubation (usually overnight) with antibodies as detailed in the figure legends, and immune complexes were collected with protein A–Sepharose CL-4B. After being washed twice with buffer A (0.2% Nonidet P-40, 10 mM Tris-HCl [pH 7.5], 150 mM NaCl, 2 mM EDTA), twice with buffer B (0.2% Nonidet P-40, 10 mM Tris-HCl [pH 7.5], 500 mM NaCl, 2 mM EDTA), and once with buffer C (10 mM Tris-HCl [pH 7.5]), the immunoprecipitated proteins were denatured in SDS sample buffer (2% sodium dodecyl sulfate [SDS], 100 mM Tris-HCl [pH 8.8], 10% glycerol, 2.5 mM EDTA, 0.01% bromophenol blue) which contained 5% β-mercaptoethanol unless otherwise indicated in the figure legends. The samples were heated at 90°C for 2 to 3 min and subjected to SDS-polyacrylamide gel electrophoresis (PAGE) through discontinuous polyacrylamide gels. After fixation in 20% acetic acid, the gels were rinsed and then impregnated for fluorography by being soaked in 1 M sodium salicylate. Dried gels were exposed to Kodak X-Omat AR film at −70°C.
RESULTS
Influence of basic residues in the C protein on signal peptidase cleavage of prM.
We have proposed that a structural feature of the flavivirus C protein, possibly its large positive charge, might influence the position of the attached prM signal sequence within the ER membrane and hence maintain the cleavage site of the signal peptide in a position that is inaccessible to the active site of signal peptidase (23). The MVE C protein contains 27 positively charged (Arg or Lys) and only 3 negatively charged (Asp or Glu) residues (8). To test the influence of the highly charged nature of the C protein on processing at the prM signal peptidase site, we replaced up to 11 basic residues in the NH2- and COOH-terminal ends of the C protein with uncharged polar amino acids (Fig. 1).
C protein charge variants were expressed in the context of the MVE structural polyprotein region by transient transfection of COS-7 cells. Transfected cells were pulse-labelled, and the MVE proteins were immunoprecipitated and analyzed by SDS-PAGE and fluorography to assess the efficiency of processing at the C-prM junction (Fig. 2). As is the case for transfection with unmutated MVE cDNA (pSTR; Fig. 2A, lane 1), the production of prM was very inefficient following transfection with each of the mutated C protein constructs (lanes 2 to 4). In each instance, and as described previously for pSTR (23), production of prM was greatly enhanced when the viral NS2B-3 protease was provided in trans by cotransfection with pNS3/T (lanes 5 to 8) to induce cleavage at the COOH terminus of the C protein. The control experiment (lane 9) ensured that no protein of similar size was detectable as a direct result of pNS3/T transfection. Although authentic signal peptidase cleavage of prM generated from the C protein charge variant constructs was not verified by NH2-terminal sequence analysis of prM, this protein had SDS-PAGE mobility identical to that of prM from virus-infected cells or recombinant prM (36), which has previously been isolated and subjected to NH2-terminal sequencing (data not shown). We have observed that after removal of the N-linked glycans with endoglycosidase H, this cleaved prM migrates faster during SDS-PAGE (12% polyacrylamide) than does a signal sequence-containing form of prM generated by in vitro translation (data not shown). Therefore, we conclude that the observed prM in Fig. 2 (and subsequent figures) no longer retains the 20-residue translocation signal.
A putative C-prM precursor was seen during expression of the MVE structural polyprotein region, with or without the C protein charge mutations, which disappeared when the viral NS2B-3 protease was coexpressed (Fig. 2). Consistent with its identification as uncleaved C-prM, this polypeptide had an estimated size of about 34 kDa and was glycosylated (data not shown). Also, by using antiserum raised against a fusion peptide including the NH2-terminal 16 amino acids of C, this band was observed among proteins immunoprecipitated from cells transfected with pSTR, pSTR.Cmut2aa, and pSTR.Cmut4aa but not pSTR.Cmut11aa, which contains 4 amino acid substitutions in the region encompassed by the peptide (data not shown).
These results demonstrate that significant changes can be made to the charge profile of the C protein (and thus presumably also to its conformation) without reducing its inhibitory influence on signal peptidase cleavage at the C-prM junction in the absence of NS2B-3.
Influence of cytoplasmic NH2-terminal sequences on signal peptidase cleavage of prM.
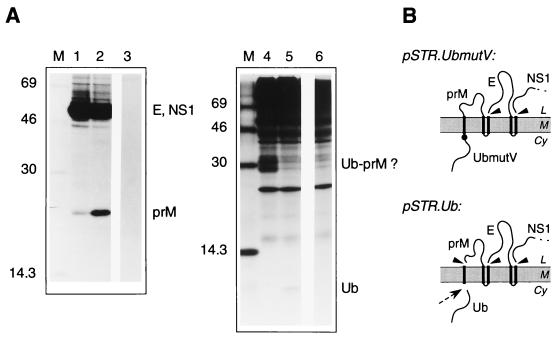
The upstream presence of the MVE C protein inhibits signal peptidase cleavage of the adjacent prM translocation signal (1, 23, 36, 42). To examine whether this inhibition is due to sequence-specific features of the C protein, we engineered cDNAs encoding ubiquitin fused to the NH2 terminus of the prM signal sequence (Fig. 3). Ubiquitin is a small cytoplasmic protein whose COOH terminus contains a site which is cleaved in eukaryotic cells by ubiquitin-specific proteases (2). This site was left intact in plasmid pSTR.Ub and was destroyed in pSTR.UbmutV by mutating the COOH-terminal Gly76 of the ubiquitin sequence to Val so as to inhibit cleavage by the cellular enzymes (17). Transient expression of the two ubiquitin fusion constructs in COS cells revealed that the foreign polypeptide covalently attached upstream of the signal sequence of prM could, like the C protein, significantly inhibit signal peptidase cleavage of prM (Fig. 3). Very little prM was immunoprecipitated from cells transfected with pSTR.UbmutV (Fig. 3A, lane 1), whereas prM was readily detected in cells transfected with pSTR.Ub, where ubiquitin is expected to be rapidly removed from the NH2-terminus of the prM signal sequence by the ubiquitin-specific proteases (lane 2). Detection (in shorter exposures of the fluorograph) of similar amounts of MVE E protein in each case confirmed that the observed difference was not due to reduced polyprotein synthesis from the mutant construct. Ubiquitin (predicted size, about 8 kDa) was detected by immunoprecipitation with ubiquitin-specific antiserum from cells transfected with pSTR.Ub (lane 5), whereas higher-molecular-weight products which probably correspond to uncleaved ubiquitin(Val76)-prM were seen in cells transfected with pSTR.UbmutV (lane 4). Lanes 3 and 6 show that none of these bands were immunoprecipitated by either the MVE- or the ubiquitin-specific antisera from cells transfected with a control plasmid. It is interesting that uncleaved ubiquitin-prM was not observed among the products of immunoprecipitation with anti-MVE mouse ascitic fluid (lane 1) when the same antibody preparation does seem to precipitate a C-prM precursor (Fig. 2A, lanes 1 to 4). This may be due to a difference between these fusion proteins in complex formation with the E protein.
FIG. 3.
Replacement of the C protein with ubiquitin at the NH2 terminus of the prM signal sequence does not alter the efficiency of prM cleavage by signal peptidase. (A) COS cells were transiently transfected with the ubiquitin fusion construct pSTR.UbmutV (lanes 1 and 4) or pSTR.Ub (lanes 2 and 5), in which the cleavage site for ubiquitin-specific proteases was destroyed or left intact, respectively, or with pNS3/T (lanes 3 and 6). The cells were metabolically labelled for 30 min, and polypeptides immunoprecipitated from lysates with anti-MVE mouse ascitic fluid (lanes 1 to 3) or antiserum against a ubiquitin–glutathione S-transferase fusion protein (lanes 4 to 6) were separated by SDS-PAGE (12% polyacrylamide) (lanes 1 to 3) or SDS-PAGE (15% polyacrylamide) (lanes 4 to 6). Bands corresponding to MVE E, NS1, and prM glycoproteins, as well as ubiquitin (Ub) and putative Ub-prM fusion products, are labelled, and the numbers indicate the sizes (in kilodaltons) of marker proteins in lane M. (B) Schematic diagrams showing the expected topology at the ER membrane of polyproteins encoded by pSTR.UbmutV and pSTR.Ub. Abbreviations are as listed in the legend to Fig. 2. Ubiquitin-specific protease cleavage is indicated by a dotted arrow, and a solid circle shows the approximate position of the Gly76→Val substitution mutation.
The data described so far demonstrate the significance of the presence of a cytoplasmic, NH2-terminal polypeptide for control of prM cleavage by signal peptidase. However, a structurally distinct protein can replace the flavivirus C protein in this function.
Cytoplasmic fusion of the MVE C protein to an idealized signal sequence.
To examine whether the efficiency of cleavage of a different signal peptide would be altered by COOH-terminal fusion to the MVE C protein, we used plasmid pSFVΔE3.MVE-C. This construct encodes the MVE C protein upstream of an artificial signal sequence, which was designed to be cleavable by signal peptidase (25), followed by the envelope proteins E2 and E1 of SFV. SFV E2 and E1 were separated, as in the viral polyprotein, by the 6,000-molecular-weight (6K) region (Fig. 4B). Figure 4 shows that the NH2-terminal addition of the MVE C protein did not reduce the efficiency of cleavage of the idealized signal sequence and hence did not reduce the production of the adjacent E2 protein in transiently transfected COS cells. There was no detectable difference in the ratio of the SFV envelope proteins E1 and E2 produced between this construct (lane 1) and a control plasmid, pSFVΔE3 (25), encoding the autocatalytically removed SFV capsid protein upstream of the ideal signal sequence (lane 3). In both constructs, the NH2 terminus of E1 is generated by cleavage of the authentic E1 signal sequence. Coexpression of the MVE NS2B-3 protease did not increase the production of SFV E2 from pSFVΔE3.MVE-C (lane 2). E1 and E2 were not immunoprecipitated from lysates of cells transfected only with pNS3/T (lane 4). Although not contradicting a model involving C protein-mediated control of the accessibility of the prM signal peptidase site, this result demonstrates that the presence of the MVE C protein is not by itself sufficient to inhibit the processing of an adjacent signal sequence and that therefore some features of the signal sequence and/or prM must also contribute to the inefficiency of processing observed for MVE C-prM in the absence of the NS2B-3 protease.
The cleavage region of the prM signal sequence is important in the regulated production of prM.
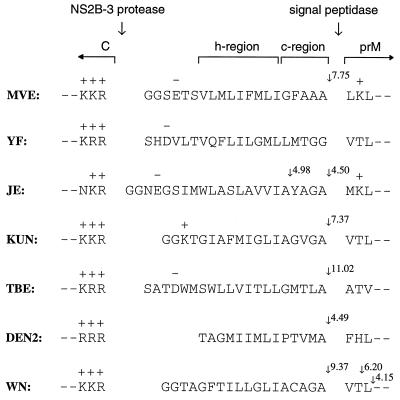
The MVE prM signal sequence (Fig. 5) conforms to the requirements predicted to be of importance for translocation and signal peptidase processing (reviewed in reference 40). It contains basic residues in the NH2-terminal region (n-region), which may determine the orientation of insertion of the peptide into the membrane, a hydrophobic core (h-region) composed of a stretch of hydrophobic residues uninterrupted by charged or polar amino acids, and a COOH-terminal cleavage domain (c-region), which, although relatively hydrophobic, is consistent with the “(−3,−1) rule,” which dictates that for signal peptidase recognition the residue in position −1 with respect to the cleavage site must be small and the residue in position −3 must not be aromatic, charged, or large and polar. Cleavage-potential scores determined by a computer program (written by Mantei [15]) based on the weight-matrix algorithm of von Heijne (39) show a maximum of 7.75 at the experimentally determined NH2 terminus of prM, which is within the range of scores characteristic of known signal sequences (typical scores at known cleavage sites are between 5 and 12, with only 2% of a sample of 161 signal sequences having a maximum score less than 3.5 [39]). Although all flavivirus prM signal sequences fit this general pattern, they show considerable heterogeneity in both the length and sequence of the h-region and in the sequence of the c-region (Fig. 5). One feature which is generally apparent, however, and which is not characteristic of “typical” signal sequences (40) is a lack of polar residues in the c-region.
FIG. 5.
Alignment of flavivirus amino acid sequences around the C-prM junction. The MVE prM translocation signal with flanking sequences encoded by pSTR is shown at the top, with the sequences we have considered to be the h- and c-regions indicated. Corresponding sequences are also shown for yellow fever virus (YF) (32), Japanese encephalitis virus (JE) (38), Kunjin virus (KUN) (7), tick-borne encephalitis virus (TBE) (26), dengue virus type 2 (DEN2) (13) and West Nile virus (WN) (6). Large arrows indicate sites of cleavage by the viral NS2B-3 protease complex and signal peptidase. Small arrows show sites at which cleavage potential scores based on the algorithm of von Heijne (39) are greater than an arbitrary value of 4. Positively (+) and negatively (−) charged amino acid residues are labelled.
To increase the polar nature of the MVE prM signal c-region, we replaced Gly, Phe, and Ala at positions −5, −4, and −2 (from the signal peptidase cleavage site) with Pro, Gln, and Gln, respectively. This mutation increased the theoretical cleavage score to 10.58 without introducing predicted alternative cleavage sites. Transient expression in COS cells of the MVE structural polyprotein region from a plasmid containing these point mutations (pSTR.mutPQAQA) (Fig. 6A, lane 2) resulted in the efficient production of prM (seen as a doublet due to carbohydrate modifications during the long labelling period [23]) at levels much higher than those observed from the unmutated construct (pSTR, lane 1) and comparable to those seen with construct pSTR.ΔC, in which the C protein-coding region is deleted (lane 3). Furthermore, coexpression of the flavivirus protease during transient transfections did not further enhance the production of prM from pSTR.mutPQAQA (data not shown).
Influence of the downstream protein on processing at the prM signal peptidase site.
To examine whether it was possible to use the C protein-prM signal sequence to impose regulation of signal peptidase cleavage on a protein other than prM, we engineered fusion constructs with Canch (C protein plus the signal sequence for prM) upstream of the NH2 termini of the MVE E protein or the mouse MHC class I Kd molecule (H-2Kd), replacing the natural signal peptides. Both E and H-2Kd are type I transmembrane proteins, and cleavage at their native signal peptidase sites is thought to occur cotranslationally.
We transfected COS cells with pCanch-E (encoding the Canch-E fusion protein) or pSTR (in which E is preceded by its native signal) and performed immunoprecipitations with a monoclonal antibody which recognizes the MVE E protein (M2-8E7 [14]) to avoid precipitating NS1, which migrates to a nearby position during SDS-PAGE. Figure 7 shows that during expression of the Canch-E fusion protein, the amount of E immunoprecipitated from lysates was significantly reduced in comparison with that seen in cells transfected with pSTR (Fig. 7A, compare lane 1 with lanes 3 and 4). Expression of the Canch-E fusion construct also resulted in the appearance of a band with an apparent size some 11 to 12 kDa greater than that of the E protein. This protein was also immunoprecipitated by antiserum raised against the NH2 terminus of the MVE C protein (data not shown) and is presumably a membrane-inserted but uncleaved Canch-E fusion. When the viral NS2B-3 protease was coexpressed with this construct, the amount of the putative fusion protein was greatly reduced and there was a parallel increase in the amount of E protein detected (lane 2).
FIG. 7.
The production of MVE E from a Canch-E fusion protein is dependent on the viral protease. (A) COS cells transiently transfected with pCanch-E (lanes 1 and 2) or pSTR (lanes 3 and 4), in the absence (−) or presence (+) of pNS3/T, were metabolically labelled for 30 min. Products immunoprecipitated from the lysates with anti-MVE E monoclonal antibody M2-8E7 (14) were examined by SDS-PAGE (12% polyacrylamide). Positions of E, prM (coprecipitated as part of a prM-E heterodimer), and putative Canch-E fusion glycoproteins are labelled, and the numbers indicate the positions and sizes (in kilodaltons) of marker proteins. (B) Schematic diagram showing the expected topology at the ER membrane of the polyprotein encoded by pCanch-E. Abbreviations are as listed in the legend to Fig. 2.
These observations demonstrate that the control mediated by Canch over prM production could be transferred to a second type I transmembrane protein when it was COOH-terminally fused to Canch. However, this was not a general phenomenon. Expression of the Canch-Kd fusion construct in transfected COS cells revealed little inhibitory influence of Canch on the signal peptidase cleavage required to produce H-2Kd (Fig. 8). The mobility during SDS-PAGE of H-2Kd produced from pCanch-Kd (Fig. 8A, lane 2) was apparently identical to that of H-2Kd encoded by a control plasmid, pKd, which encoded the MHC class I molecule preceded by its authentic signal sequence (lane 1). A small amount of a protein with an apparent size 11 to 13 kDa greater than that of H-2Kd was also precipitated, and this may represent an uncleaved Canch-Kd fusion. If so, this indicates that the Canch domain may also have a slight inhibitory effect on signal peptidase cleavage when fused upstream of the H-2Kd luminal domain, resulting in a small proportion of molecules escaping cotranslational cleavage. However, cotransfection with pCanch-Kd of pNS3/T to provide viral NS2B-3 protease activity did not increase the production of H-2Kd or reduce the intensity of the putative precursor band (compare lane 3 with lane 2) and did not contribute any nonspecific immunoprecipitation products (lane 6).
By using the algorithm of von Heijne (39), an alternative site for signal peptidase cleavage was predicted to occur in the Canch-Kd fusion construct following a Pro 2 residues past the authentic cleavage site of H-2Kd. Additionally, studies with engineered mutants of pro-sucrase-isomaltase indicate that Pro at position +2 downstream of a signal peptidase cleavage site might be beneficial for cleavage (15). Therefore, we engineered a Pro→Lys substitution (Lys being the amino acid present in the corresponding position of the prM protein) at residue 2 of the H-2Kd sequence in the Canch-Kd fusion (pCanch-KdmutK). This mutated fusion protein was also subject to cleavage by signal peptidase (as evidenced by the appearance of a band corresponding to the H-2Kd molecule in Fig. 8A, lane 4), and the presence of the viral protease did not greatly enhance this cleavage (compare lane 5 with lane 4). Thus, the presence of the downstream Pro residue (and consequent potential for a second or improved cleavage site) is apparently not the only feature responsible for the high efficiency of cleavage of H-2Kd fused to the prM translocation signal. As with the unmutated construct, a small amount of putative uncleaved Canch-KdmutK fusion was apparent in each lysate.
To establish that the similarity in the amounts of H-2Kd detected from the lysates in Fig. 8 in the presence and absence of the viral protease was not due to saturating amounts of antigen, we performed a second round of immunoprecipitation. Ratios of the amounts of protein collected from each of the lysates were similar to those seen in the initial immunoprecipitation, and for each lysate the radioactivity precipitated in the second round was only 30 to 40% of that collected in the first round (data not shown). We also used the addition of further protein A-Sepharose beads after the initial immunoprecipitation to confirm that the quantity of beads used for the collection of immune complexes was not limiting the amount of H-2Kd recovered. This suggests that similar amounts of H-2Kd were indeed produced from these constructs in the presence and absence of pNS3/T.
These results demonstrate that in addition to the requirement for covalently linked NH2-terminal cytoplasmic polypeptide and the structure of the c-region of the prM signal sequence, luminal sequences downstream of the signal peptidase cleavage site also play a role in maintaining the dependence of efficient cleavage at this site on the presence of the viral protease.
DISCUSSION
Signal peptidase processing at the NH2 terminus of prM during expression of the structural region of the MVE polyprotein is greatly increased in the presence of the viral protease NS2B-3, which cleaves at the COOH terminus of the C protein (1, 23, 42). The conversion of a signal peptidase cleavage site from a cryptic to an accessible conformation during the synthesis of a viral protein is a previously unrecognized mechanism for polyprotein processing-mediated regulation of viral gene expression. Here we have investigated elements in the structural polyprotein region of MVE which are important for maintenance of the control mediated by the cytoplasmic viral protease over luminal cleavage at the C-prM junction.
The finding that removal of the C protein from the signal sequence of prM by enzymatic cleavage or deletion mutagenesis greatly enhanced the production of prM (1, 23, 36, 42) was consistent with a model proposing a pronounced influence of the C protein on the maintenance of the signal peptidase site of prM in a cryptic conformation. Here we have demonstrated that this effect is not a consequence of the large number of positively charged amino acids or other sequence-specific features of the flavivirus C protein but that it is dependent only on the linkage of an NH2-terminal cytoplasmic polypeptide to the signal sequence of prM. We observed that replacement of as many as 11 of 27 positively charged amino acids in the MVE C protein with uncharged residues did not influence the dependence of efficient prM cleavage on the presence of the viral protease complex. Furthermore, replacement of the C protein in the MVE structural polyprotein region with the small cytoplasmic protein ubiquitin (after modification of the COOH terminus of the protein to inhibit ubiquitin-specific protease cleavage) did not markedly improve the efficiency of signal peptidase processing of prM. Finally, we could not transpose signal peptidase cleavage inhibition to an unrelated type I transmembrane protein, the E2 protein of SFV, by inserting the MVE C protein NH2 terminally to an idealized signal sequence.
The internal location of the prM signal sequence per se cannot account for the inefficiency of cleavage at the NH2 terminus of prM, since other internal signal sequences (preceding the E and NS1 proteins) in the flavivirus structural polyprotein are efficiently processed. Since we could rule out a general inhibitory effect of the MVE C protein on signal peptidase processing at sites located downstream of C, we predicted that structural elements in the prM signal sequence also contribute to the prevention of cotranslational processing. Cleavable signal sequences typically consist of three domains: a positively charged n-region, 1 to 5 amino acids in length; a membrane-spanning h-region, consisting of 7 to 15 hydrophobic amino acids; and a c-region, 3 to 7 residues long, characterized by the presence of residues conforming to the “(−3,−1) rule” for signal peptidase recognition and enriched in residues with polar side chains (40). The presence of amino acids with α-helix-breaking properties, such as proline or glycine, at the start of the c-region may also be important for proper cleavage (28). In cases where a potential cleavage site exists, the length of the h-region appears to be the dominant factor which determines the transition between a cleavable signal sequence and a signal peptide-membrane anchor (27). In the MVE prM signal sequence, this hydrophobic stretch preceding the c-region is about 9 amino acids long (Fig. 5), which conforms well with the characteristics of cleavable signal sequences. However, the c-region is also deficient in amino acids with polar side chains, resulting in a stretch of hydrophobic amino acids which is much longer than the core necessary for translocation (up to 15 amino acids if the Leu residue beyond the cleavage site is included and 17 amino acids if the uncharged but polar residues toward the NH2-terminal side of the membrane are also counted). One could therefore envisage that the h- and nonpolar c-regions of the MVE prM signal sequence are, in combination, of the critical length and hydrophobicity to function as a membrane anchor with the potential cleavage site buried in the membrane when the preceding C protein is covalently attached. Removal of the cytoplasmic domain would allow the signal sequence sufficient freedom of movement to permit the interaction of the cleavage site with the active site of signal peptidase. This proposition was supported experimentally by using a mutant in which the c-region of the MVE prM signal sequence was changed from Gly-Phe-Ala-Ala-Ala to Pro-Gln-Ala-Gln-Ala, a sequence which would not be readily accommodated in the membrane. This optimization of the c-region for signal peptidase processing resulted in greatly improved cleavage of the prM signal sequence. Importantly, processing of the C protein by the flavivirus NS2B-3 protease did not further enhance cleavage by signal peptidase of this mutated signal sequence, demonstrating that these three point mutations could overcome the dependence of the proteolytic processing of prM on the cytoplasmic protease.
The scarcity of amino acids with polar side chains in the c-region of the prM signal sequence is a conserved feature of flaviviruses (Fig. 5). We therefore suggest that the control over processing at the C-prM junction noted during expression of the structural polyprotein region of several flaviviruses is mediated by a common mechanism which involves the conserved hydrophobic nature of the c-region and the internal location of the prM signal sequence. In support of the generality of this mechanism for the regulation of flavivirus structural gene expression, we have recently observed that replacement of the c-region residues Leu-Leu-Met-Thr-Gly-Gly by Val-Pro-Gln-Ala-Gln-Ala in the yellow fever virus prM signal sequence also significantly enhanced signal peptidase cleavage (35a).
We also noted an influence of different type I transmembrane proteins on cleavage at the prM signal peptidase site when such proteins were fused downstream of MVE Canch (i.e., the C protein plus signal sequence for prM). Canch linked to the NH2 terminus of the MVE E protein was inefficiently cleaved by signal peptidase, and the extent of cleavage was increased by provision of the NS2B-3 protease complex, akin to the proteolytic processing events at the authentic C-prM junction. In contrast, replacement of the natural signal sequence of the MHC class I molecule H-2Kd by Canch resulted in little apparent reduction of signal peptidase cleavage or detectable effect from the presence of the viral protease. We did not investigate the characteristics of the two type I transmembrane proteins that could account for these differences. However, it is interesting that folding and assembly of MHC class I glycoproteins involves transient interaction with a host of ER resident proteins, including immunoglobulin binding protein, gp96, calnexin, the transporter associated with antigen processing, calreticulin, and tapasin (reviewed in reference 35). Similar to models which implicate luminal binding of molecular chaperones in unidirectional protein translocation (5, 12), interactions of the translocated MHC class I glycoprotein with ER-resident proteins could influence slippage of the signal peptide in the lipid bilayer and/or the accessibility of the signal peptidase site so that rapid cleavage occurs.
The conservation among flaviviruses of the controlled nature of the prM signal peptidase cleavage during proteolytic processing of the structural polyprotein region implies functional significance for this regulation in flavivirus replication. One of the aims of this work was to identify elements in the flavivirus C-prM region which could be subjected to mutagenesis in order to overcome the controlled order of cleavages at the C-prM junction. The finding that a combination of only three amino acid substitutions could override the influence of the C protein on the signal peptidase cleavage of prM is suggestive of selective pressure against such sequences. These small changes in the prM signal sequence are well suited to insertion into a full-length flavivirus cDNA clone, allowing investigation of the biological function of the control of signal peptidase processing of prM by a cytoplasmic protease.
ACKNOWLEDGMENTS
We thank Ron Weir for providing anti-MVE hyperimmune ascitic fluid, Arno Müllbacher for providing antiserum against SFV, Roy Hall for providing hybridoma culture M2-8E7, and Rohan Baker for providing ubiquitin cDNA and antibodies raised against ubiquitin. We are also grateful to N. Mantei for making his AnalyseSignalase2.03 program for the Macintosh publicly available.
REFERENCES
- 1.Amberg S M, Nestorowicz A, McCourt D W, Rice C M. NS2B-3 proteinase-mediated processing in the yellow fever virus structural region: in vitro and in vivo studies. J Virol. 1994;68:3794–3802. doi: 10.1128/jvi.68.6.3794-3802.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bachmair A, Finley D, Varshavsky A. In vivo half-life of a protein is a function of its amino-terminal residue. Science. 1986;234:179–186. doi: 10.1126/science.3018930. [DOI] [PubMed] [Google Scholar]
- 3.Baker R T, Smith S A, Marano R, McKee J, Board P G. Protein expression using cotranslational fusion and cleavage of ubiquitin. J Biol Chem. 1994;269:25381–25386. [PubMed] [Google Scholar]
- 4.Blobel G, Dobberstein B. Transfer of proteins across membranes. I. Presence of proteolytically processed and unprocessed nascent immunoglobulin light chains on membrane-bound ribosomes of murine myeloma. J Cell Biol. 1975;67:835–851. doi: 10.1083/jcb.67.3.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brodsky J L. Post-translational protein translocation: not all hsc70s are created equal. Trends Biochem Sci. 1996;21:122–126. [PubMed] [Google Scholar]
- 6.Castle E, Nowak T, Leidner U, Wengler G, Wengler G. Sequence analysis of the viral core protein and the membrane-associated proteins V1 and NV2 of the flavivirus West Nile virus and of the genome sequence for these proteins. Virology. 1985;145:227–236. doi: 10.1016/0042-6822(85)90156-4. [DOI] [PubMed] [Google Scholar]
- 7.Coia G, Parker M D, Speight G, Byrne M E, Westaway E G. Nucleotide and complete amino acid sequences of Kunjin virus: definitive gene order and characteristics of the virus-specified proteins. J Gen Virol. 1988;69:1–21. doi: 10.1099/0022-1317-69-1-1. [DOI] [PubMed] [Google Scholar]
- 8.Dalgarno L, Trent D W, Strauss J H, Rice C M. Partial nucleotide sequence of the Murray Valley encephalitis virus genome. Comparison of the encoded polypeptides with yellow fever virus structural and non-structural proteins. J Mol Biol. 1986;187:309–323. doi: 10.1016/0022-2836(86)90435-3. [DOI] [PubMed] [Google Scholar]
- 9.de Groot R J, Hardy W R, Shirako Y, Strauss J H. Cleavage-site preferences of Sindbis virus polyproteins containing the non-structural proteinase. Evidence for temporal regulation of polyprotein processing in vivo. EMBO J. 1990;9:2631–2638. doi: 10.1002/j.1460-2075.1990.tb07445.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deubel V, Kinney R M, Esposito J J, Cropp C B, Vorndam A V, Monath T P, Trent D W. Dengue 2 virus envelope protein expressed by a recombinant vaccinia virus fails to protect monkeys against dengue. J Gen Virol. 1988;69:1921–1929. doi: 10.1099/0022-1317-69-8-1921. [DOI] [PubMed] [Google Scholar]
- 11.Fonseca B A, Pincus S, Shope R E, Paoletti E, Mason P W. Recombinant vaccinia viruses co-expressing dengue-1 glycoproteins prM and E induce neutralizing antibodies in mice. Vaccine. 1994;12:279–285. doi: 10.1016/0264-410x(94)90206-2. [DOI] [PubMed] [Google Scholar]
- 12.Glick B S. Can Hsp70 proteins act as force-generating motors? Cell. 1995;80:11–14. doi: 10.1016/0092-8674(95)90444-1. [DOI] [PubMed] [Google Scholar]
- 13.Hahn Y S, Galler R, Hunkapiller T, Dalrymple J M, Strauss J H, Strauss E G. Nucleotide sequence of dengue 2 RNA and comparison of the encoded proteins with those of other flaviviruses. Virology. 1988;162:167–180. doi: 10.1016/0042-6822(88)90406-0. [DOI] [PubMed] [Google Scholar]
- 14.Hall R A, Kay B H, Burgess G W, Clancy P, Fanning I D. Epitope analysis of the envelope and non-structural glycoproteins of Murray Valley encephalitis virus. J Gen Virol. 1990;71:2923–2930. doi: 10.1099/0022-1317-71-12-2923. [DOI] [PubMed] [Google Scholar]
- 15.Hegner M, von Kieckebusch-Gück A, Falchetto R, James P, Semenza G, Mantei N. Single amino acid substitutions can convert the uncleaved signal-anchor of sucrase-isomaltase to a cleaved signal sequence. J Biol Chem. 1992;267:16928–16933. [PubMed] [Google Scholar]
- 16.Hellen C U T, Kräusslich H-G, Wimmer E. Proteolytic processing of polyproteins in the replication of RNA viruses. Biochemistry. 1989;28:9881–9890. doi: 10.1021/bi00452a001. [DOI] [PubMed] [Google Scholar]
- 17.Johnson E S, Bartel B, Seufert W, Varshavsky A. Ubiquitin as a degradation signal. EMBO J. 1992;11:497–505. doi: 10.1002/j.1460-2075.1992.tb05080.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Konishi E, Pincus S, Fonseca B A, Shope R E, Paoletti E, Mason P W. Comparison of protective immunity elicited by recombinant vaccinia viruses that synthesize E or NS1 of Japanese encephalitis virus. Virology. 1991;185:401–410. doi: 10.1016/0042-6822(91)90788-d. [DOI] [PubMed] [Google Scholar]
- 19.Kräusslich H-G, Wimmer E. Viral proteinases. Annu Rev Biochem. 1988;57:701–754. doi: 10.1146/annurev.bi.57.070188.003413. [DOI] [PubMed] [Google Scholar]
- 20.Kunkel T A, Roberts J D, Zakour R A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- 21.Lalanne J-L, Delarbre C, Gachelin G, Kourilsky P. A cDNA clone containing the entire coding sequence of a mouse H-2Kd histocompatibility antigen. Nucleic Acids Res. 1983;11:1567–1577. doi: 10.1093/nar/11.5.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lemm J A, Rümenapf T, Strauss E G, Strauss J H, Rice C M. Polypeptide requirements for assembly of functional Sindbis virus replication complexes: a model for the temporal regulation of minus- and plus-strand RNA synthesis. EMBO J. 1994;13:2925–2934. doi: 10.1002/j.1460-2075.1994.tb06587.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lobigs M. Flavivirus premembrane protein cleavage and spike heterodimer secretion require the function of the viral proteinase NS3. Proc Natl Acad Sci USA. 1993;90:6218–6222. doi: 10.1073/pnas.90.13.6218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lobigs M. Proteolytic processing of a Murray Valley encephalitis virus nonstructural polyprotein segment containing the viral proteinase: accumulation of a NS3-4A precursor which requires mature NS3 for efficient processing. J Gen Virol. 1992;73:2305–2312. doi: 10.1099/0022-1317-73-9-2305. [DOI] [PubMed] [Google Scholar]
- 25.Lobigs M, Zhao H, Garoff H. Function of Semliki Forest virus E3 peptide in virus assembly: replacement of E3 with an artificial signal peptide abolishes spike heterodimerization and surface expression of E1. J Virol. 1990;64:4346–4355. doi: 10.1128/jvi.64.9.4346-4355.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mandl C W, Heinz F X, Kunz C. Sequence of the structural proteins of tick-borne encephalitis virus (western subtype) and comparative analysis with other flaviviruses. Virology. 1988;166:197–205. doi: 10.1016/0042-6822(88)90161-4. [DOI] [PubMed] [Google Scholar]
- 27.Nilsson I, Whitley P, von Heijne G. The COOH-terminal ends of internal signal and signal-anchor sequences are positioned differently in the ER translocase. J Cell Biol. 1994;126:1127–1132. doi: 10.1083/jcb.126.5.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nothwehr S F, Gordon J I. Eukaryotic signal peptide structure/function relationships. J Biol Chem. 1989;264:3979–3987. [PubMed] [Google Scholar]
- 29.Palmenberg A C. Proteolytic processing of picornaviral polyprotein. Annu Rev Microbiol. 1990;44:603–623. doi: 10.1146/annurev.mi.44.100190.003131. [DOI] [PubMed] [Google Scholar]
- 30.Pincus S, Mason P W, Konishi E, Fonseca B A L, Shope R E, Rice C M, Paoletti E. Recombinant vaccinia virus producing the prM and E proteins of yellow fever virus protects mice from lethal yellow fever encephalitis. Virology. 1992;187:290–297. doi: 10.1016/0042-6822(92)90317-i. [DOI] [PubMed] [Google Scholar]
- 31.Rice C M. Flaviviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, Chanock R M, Melnick J L, Monath T P, Roizman B, Strauss S E, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 931–959. [Google Scholar]
- 32.Rice C M, Lenches E M, Eddy S R, Shin S J, Sheets R L, Strauss J H. Nucleotide sequence of yellow fever virus: implications for flavivirus gene expression and evolution. Science. 1985;229:726–733. doi: 10.1126/science.4023707. [DOI] [PubMed] [Google Scholar]
- 33.Ruiz-Linares A, Cahour A, Desprès P, Girard M, Bouloy M. Processing of yellow fever virus polyprotein: role of cellular proteases in maturation of the structural proteins. J Virol. 1989;63:4199–4209. doi: 10.1128/jvi.63.10.4199-4209.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sato T, Takamura C, Yasuda A, Miyamoto M, Kamogawa K, Yasui K. High-level expression of the Japanese encephalitis virus E protein by recombinant vaccinia virus and enhancement of its extracellular release by the NS3 gene product. Virology. 1993;192:483–490. doi: 10.1006/viro.1993.1064. [DOI] [PubMed] [Google Scholar]
- 35.Solheim J C, Carreno B M, Hansen T H. Are transporter associated with antigen processing (TAP) and tapasin class I MHC chaperones? J Immunol. 1997;158:541–543. [PubMed] [Google Scholar]
- 35a.Stocks, C., and M. Lobigs. Unpublished data.
- 36.Stocks C E, Lobigs M. Posttranslational signal peptidase cleavage at the flavivirus C-prM junction in vitro. J Virol. 1995;69:8123–8126. doi: 10.1128/jvi.69.12.8123-8126.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Su T-Z, El-Gewely M R. A multisite-directed mutagenesis using T7 DNA polymerase: application for reconstructing a mammalian gene. Gene. 1988;69:81–89. doi: 10.1016/0378-1119(88)90380-0. [DOI] [PubMed] [Google Scholar]
- 38.Sumiyoshi H, Mori C, Fuke I, Morita K, Kuhara S, Kondou J, Kikuchi Y, Nagamatu H, Igarashi A. Complete nucleotide sequence of the Japanese encephalitis virus genome RNA. Virology. 1987;161:497–510. doi: 10.1016/0042-6822(87)90144-9. [DOI] [PubMed] [Google Scholar]
- 39.von Heijne G. A new method for predicting signal sequence cleavage sites. Nucleic Acids Res. 1986;14:4683–4690. doi: 10.1093/nar/14.11.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.von Heijne G. The signal peptide. J Membr Biol. 1990;115:195–201. doi: 10.1007/BF01868635. [DOI] [PubMed] [Google Scholar]
- 41.Wellink J, van Kammen A. Proteases involved in the processing of viral polyproteins. Arch Virol. 1988;98:1–26. doi: 10.1007/BF01321002. [DOI] [PubMed] [Google Scholar]
- 42.Yamshchikov V F, Compans R W. Regulation of the late events in flavivirus protein processing and maturation. Virology. 1993;192:38–51. doi: 10.1006/viro.1993.1006. [DOI] [PubMed] [Google Scholar]
- 43.Ypma-Wong M F, Dewalt P G, Johnson V H, Lamb J G, Semler B L. Protein 3CD is the major poliovirus proteinase responsible for cleavage of the P1 capsid precursor. Virology. 1988;166:265–270. doi: 10.1016/0042-6822(88)90172-9. [DOI] [PubMed] [Google Scholar]