Abstract
In order to analyze bonding contacts that stabilize the virion or promote capsid assembly, bovine papillomavirus (BPV) virions were subjected to buffer conditions known to disrupt polyomavirus virions. At physiologic ionic strength, incubation with dithiothreitol (DTT), EGTA, or DTT plus EGTA did not disrupt BPV virions as determined by electron microscopy. However, incubation of virions with DTT rendered the BPV L1 protein susceptible to trypsin cleavage at its carboxy terminus and rendered the genome susceptible to digestion with DNase I. When DTT-treated BPV virions were analyzed by analytical ultracentrifugation, they sedimented at 230S compared with 273S for untreated virions, suggesting a capsid shell expansion. Incubation with EGTA had no effect on trypsin or DNase I sensitivity and only a small effect upon the virion S value. A single cysteine residue conserved among BPV and human papillomavirus (HPV) L1 proteins resides within the trypsin-sensitive carboxy terminus of L1, which is required for capsid assembly. A recombinant HPV type 11 L1 protein, which was purified after expression in Escherichia coli and which has a Cys-to-Gly change at this position (Cys424), formed pentamers; however, unlike the wild-type protein, these mutant pentamers could no longer assemble in vitro into capsid-like structures. These results indicate an important role for interpentamer disulfide bonds in papillomavirus capsid assembly and disassembly and suggest a mechanism of virus uncoating in the reducing environment of the cytoplasm.
Papovaviruses are small nonenveloped DNA viruses which include murine polyomavirus, simian virus 40 (SV40), and the papillomaviruses. These viruses have a common capsid structure composed of 72 capsomeric subunits arranged in a T=7d icosahedral lattice. For polyomaviruses, the capsomeres are pentamers of the VP1 structural protein, while for the papillomaviruses the capsomeres are pentamers of the L1 protein (1, 15). Although atomic-resolution structures for murine polyomavirus and SV40 (14, 24, 25) and high-resolution cryoelectron microscopic structures for several papillomaviruses (1, 2, 4, 26) are now known, much less is known about the biology of virion assembly and disassembly.
For polyomaviruses, virion disruption experiments initially indicated the importance of disulfide bonds and bound calcium in capsid stability (5–7, 28). In agreement with these observations, the in vitro self-assembly of the recombinant polyomavirus VP1 protein supported the importance of calcium and ionic bonds in the formation of capsids (19, 20). Most recently, crystallographic data have explicitly identified the calcium binding sites, disulfide-bonded residues, and other interactions which link capsomeres in virions (14, 24, 25). A crystallographic structure is not yet available for any papillomavirus. Virus-like particles (VLPs) generated by expressing the L1 protein in insect cells with recombinant baculovirus vectors (12, 17, 27) or in mammalian cells with vaccinia virus vectors (11, 29) have been useful for both cryoelectron microscopic structural analysis and to assess the assembly and disassembly properties for L1-containing capsids. For example, Sapp et al. (21) determined that human papillomavirus type 33 (HPV33) VLPs were disassembled by dithiothreitol (DTT) treatment, suggesting disulfide bonds between a subset of capsomeres in agreement with the observations of Doorbar and Gallimore with native HPV1 virions (8).
The recombinant expression of HPV11 L1 in Escherichia coli (13) now permits a similar analysis of capsid assembly for papillomaviruses as previously carried out for polyomaviruses (19, 20, 23). In order to assess their biological significance, in vitro assembly conditions for the recombinant L1 protein require correlation with conditions that affect the stability of native papillomavirus virions. However, although VLP disruption experiments have been performed, a systematic analysis of papillomavirus virion disruption similar to that carried out for polyomavirus has not been described. Therefore, we have used purified bovine papillomavirus type 1 (BPV) virions to test conditions affecting virion stability. Based on these results, disulfide bonds were found to be critical for virion integrity, and their reduction led to a substantial conformational change in the capsid which may be a precursor state for disassembly in vivo.
MATERIALS AND METHODS
Virion purification.
BPV virions were isolated from cow warts. A 100-ml volume of buffer H (20 mM HEPES [pH 8.0], 100 mM NaCl, 0.1 mM CaCl2) was added to 100 g of minced tissue, and the suspension was homogenized (Brinkmann Kinematica) at setting 4 to 5 for 5 to 10 min. The homogenate was then centrifuged at 14,000 × g for 15 min at 4°C. The supernatant was saved. The pellet was rehomogenized in 50 ml of buffer H with 1 M NaCl and centrifuged as before. The supernatant was saved, and the pellet was rehomogenized with buffer H with 0.05% sodium deoxycholic acid. A 25-ml volume of 1,1,2-trichlorotrifluroethane (Freon) was added, and the mixture was rehomogenized and centrifuged as described above. All supernatants were combined, sodium deoxycholic acid was added to a final concentration of 0.05%, and the mixture was centrifuged for 30 min at 20,000 × g and at 4°C. The clarified supernatant was then centrifuged in an SW28 rotor for 4 h at 25,000 rpm and 20°C. The supernatant was discarded, and the pellets were gently resuspended with 1 ml of buffer R (20 mM HEPES [pH 8.0], 1 mM CaCl2) with agitation at 4°C for 24 to 48 h. CsCl (≈100 mg) was added to each resuspended pellet before Dounce homogenization with a B pestle (Wheaton). The homogenate was brought to a final volume of 10 ml with a stock solution containing 1.33 g of CsCl per ml and 20 mM HEPES (pH 8.0). The density was adjusted to 1.33 g g/ml, as determined by refractive index, by the addition of solid CsCl. Samples were then centrifuged in an SW50.1 rotor at 35,000 rpm for 18 h at 20°C. Gradients were manually fractionated from the top into 200- to 500-μl aliquots with a pipetman. Full virions banded at ≈1.34 g/ml, while empty virions banded at ≈1.29 g/ml. Fractions were monitored for purity by electron microscopy (see below). Virion fractions were typically banded twice in CsCl before use. Polyomavirus virions were purified as previously described (18).
Virion incubations.
Purified virions were first dialyzed into either BPV buffer (200 mM NaCl, 10 mM HEPES [pH 7.4], 0.1 mM CaCl2) or low-ionic-strength buffer (10 mM Tris-HCl [pH 7.9]). In a typical incubation, 20 μl of virions (protein content, approximately 1.4 mg/ml as determined by optical density at 280 nm and the Bio-Rad protein assay) was used. To this volume, additions were made to a final volume of 25 μl (making the final NaCl concentration 160 mM for virions in BPV buffer). Stock solutions of 40,000 U of DNase I (Sigma) per ml, 1 M DTT (Boehringer-Mannheim), 0.5 M EGTA (pH 7.4), 100 mM phenylmethylsulfonyl fluoride (PMSF), 0.5 M EDTA (pH 7.2), 1 M MgCl2, and 0.5 mg of sequencing grade trypsin (Boehringer-Mannheim) per ml were used.
For trypsin digestion analysis, virion samples were first incubated with 10 mM DTT, 5 mM EGTA, or 10 mM DTT–5 mM EGTA for 15 min at 37°C. Trypsin was then added to a final concentration of 1 μg/25 μl unless otherwise indicated, and the mixture was incubated at 37°C for 15 min. The digestion was terminated by the addition of PMSF to a final concentration of 5 mM, and the samples were stored at 4°C until electron microscopy analysis.
For DNase I digestion analysis, virion samples were first incubated in 10 mM DTT or 5 mM EGTA for 15 min at 37°C. DNase I and MgCl2 were then added to final concentrations of 40 U and 5 mM per 25 μl, respectively, and the mixture was incubated for 15 min at 37°C. To terminate the digestion and to prepare the samples for extraction, stock solutions were added to final concentrations of 2.5 mM PMSF, 15 mM EDTA, and 1% sodium dodecyl sulfate (SDS), and the samples were incubated at 37°C for 15 min. The viral DNA was then extracted with phenol-chloroform, and the samples were resolved on a 1% agarose gel in TAE buffer (40 mM Tris-acetate, 1 mM EDTA [pH 8.5]). Ethidium bromide-stained bands were detected with a UV transilluminator and photographed.
Immunoblot analysis.
SDS-10% polyacrylamide gel electrophoresis gels were transferred to Immobilon polyvinylidine difluoride membranes (Millipore) with a dry blot apparatus (Hoefer). The membranes were blocked for 1 h in TBST (100 mM Tris-HCl [pH 8.0], 1.5 M NaCl, 0.05% Tween 20) with 2% bovine serum albumin and then were incubated with primary antibody in TBST for 1 h. The membranes were washed for 5 min three times with TBST and then incubated with secondary antibody in TBST for 1 h. The membranes were washed for 5 min three times with TBST and developed with goat anti-rabbit alkaline phosphatase secondary antibody (1:15,000) (Promega). Primary antibodies were rabbit anti-BPV L2 (from John Schiller) and rabbit anti-HPV11 L1 (1:100; R-2,399; from Robert Rose), which cross-reacts with BPV L1.
Analytical ultracentrifugation.
Velocity sedimentation experiments were performed with a Beckman model XLA analytical ultracentrifuge. All sedimentation experiments were performed at 20°C with a Beckman four-hole An-60Ti rotor by using 1.2-cm-path-length double-sector cells with quartz windows. Virions were dialyzed into 10 mM HEPES–200 mM NaCl–0.1 mM CaCl2 (pH 7.4). Sample channels contained virions diluted to a final volume of 400 μl at an optical density at 280 nm of 0.5 in buffer containing 10 mM DTT, 10 mM EGTA, or buffer alone. Samples were incubated for 15 min at 37°C and placed at 4°C until they were loaded into the sample cells. The blank channel contained no virions but contained the corresponding additions to the buffer. The movement of the boundary at 280 nm was monitored every 10 min for 8 h at 6,000 rpm. Sedimentation coefficients were determined by the transport method (3, 22), with software provided by Beckman.
L1 mutagenesis.
The wild-type HPV11 L1 DNA sequence obtained by PCR amplification from a baculovirus plasmid pVL11L1 has been described elsewhere (13). Plasmid pET-17b containing the HPV11 L1 gene was purified with the Plasmid Maxi kit (Qiagen) and used as the template for site-directed mutagenesis. Mutations were introduced into the HPV11 L1 coding region with the QuikChange Site-Directed Mutagenesis kit (Stratagene). The mutant L1 coding sequence with a Cys-to-Gly change at residue 424 was generated by PCR amplification with the primers GTCACAGGCCATTACCGGTCAGAAACCCACACCTG and CAGGTGTGGGTTTCTGACCGGTAATGGCCTGTGAC.
Purification of recombinant HPV11 L1 proteins.
The wild-type and mutant HPV11 L1 proteins were expressed from pET-17b-HPV11 L1 plasmids in the bacterial host BL21 (DE3). Cells were grown at 37°C in M9 medium with ampicillin (100 μg/ml). L1 protein expression was induced by addition of 1 mM isopropyl-β-d-thiogalactopyranoside for 6 h. L1 protein purification after expression in E. coli has been previously described (13).
Electron microscopy.
Samples were applied to glow-discharged, carbon-coated, 400-mesh grids and stained with 2% uranyl acetate. The images were photographed with a Philips CM 10 electron microscope at a nominal magnification of ×39,000 or ×73,000.
RESULTS
This study of BPV virion stability was based on buffer conditions that were previously determined to disrupt polyomavirus and SV40 virions (5–7, 28). In particular, calcium chelation with EGTA or EDTA, disulfide reduction with DTT, and variation in ionic strength (10 mM Tris-HCl versus 160 mM NaCl) were tested. The effects of each treatment on virion structure and stability were assayed by the sensitivity of the L1 capsid protein to trypsin digestion, the electron microscopic appearance of the virions, and the resistance of the viral genome to DNase I digestion. In all instances examined for BPV virions, no differences were observed between treatments with either EGTA or EDTA.
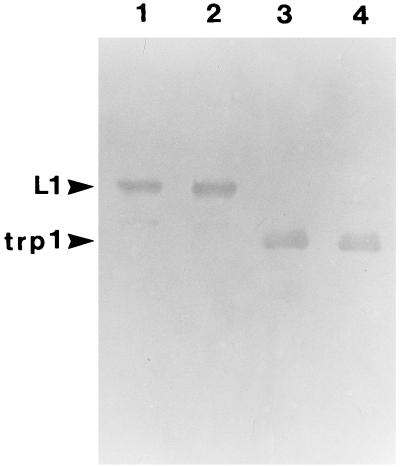
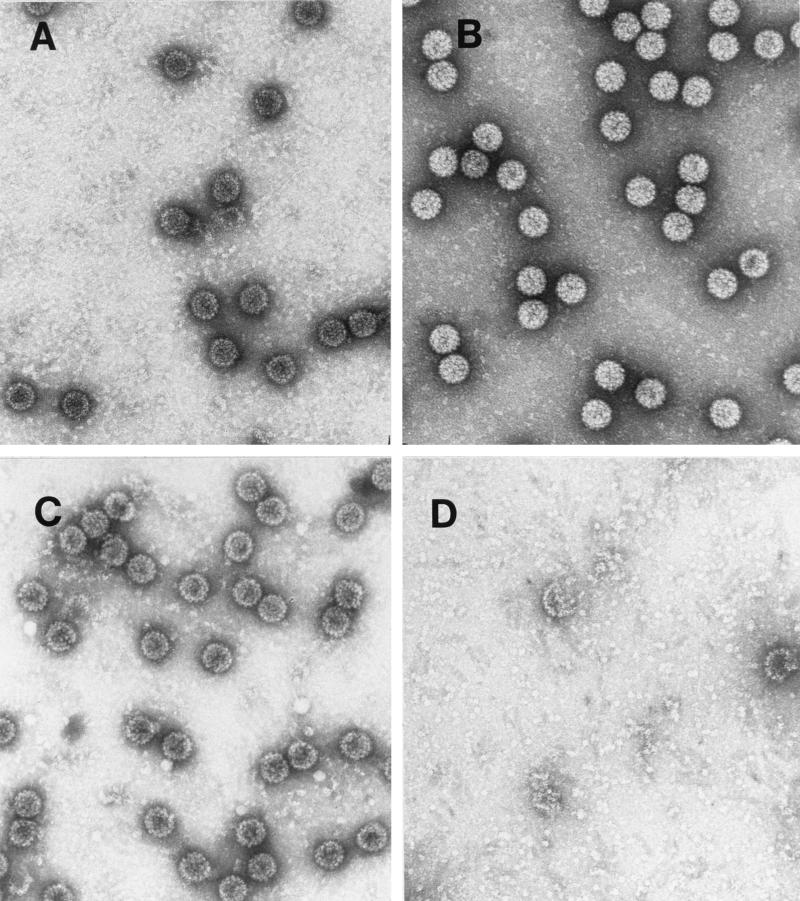
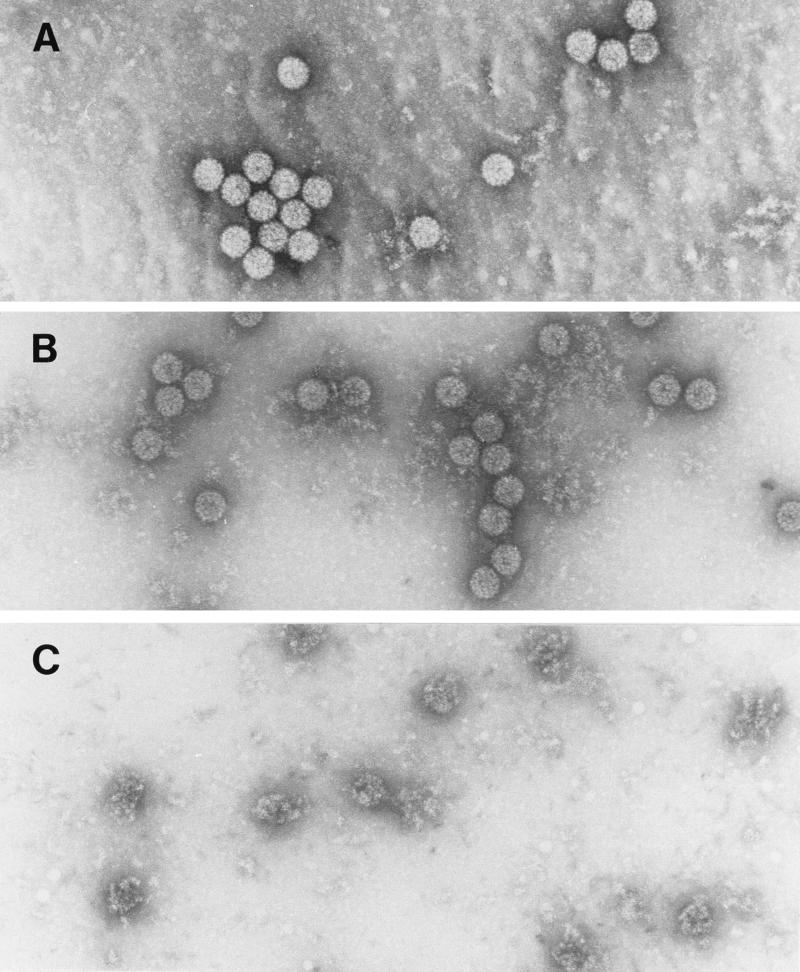
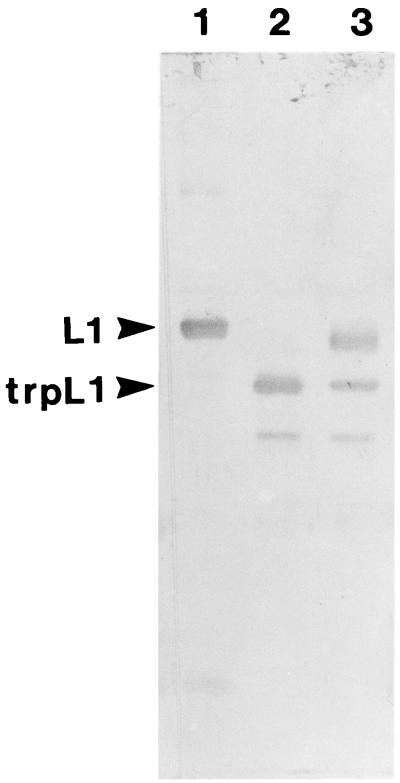
Trypsin was chosen as a probe for virion structure based on previous results with the recombinant HPV11 L1 protein purified after expression in E. coli. Digestion of the recombinant L1 protein with trypsin yielded two major cleavage products: trpL1 (≈42 kDa), resulting from cleavage at R415 (87 residues from the carboxy terminus); and an intermediate species termed L1a (≈48 kDa), likely generated from cleavage at R462 (13). BPV virions are resistant to trypsin digestion (16). BPV virions preincubated with EGTA or DTT in 160 mM NaCl buffer were digested with trypsin for 15 min at 37°C. As shown in Fig. 1, the L1 protein was susceptible to trypsin digestion only after the virions were treated with DTT, and a limit digest was obtained with an L1 protein having an electrophoretic mobility corresponding to that of trpL1. When these samples were analyzed by electron microscopy (Fig. 2), the virions appeared intact after incubation with either EGTA or DTT and after trypsin digestion of EGTA-treated virions. However, trypsin digestion of the DTT-treated virions resulted in complete virion disruption (Fig. 2D). As seen in the electron micrograph (Fig. 2D), the disruption products included donut-like structures resembling individual capsomeric subunits. These structures are similar to those seen after trypsin digestion of the recombinant L1 protein (13). BPV virions treated with both DTT and EGTA appeared identical to the DTT-treated virions by electron microscopy (data not shown) and were similarly sensitive to trypsin (Fig. 1, lane 4).
FIG. 1.
Trypsin digestion of BPV virions. Immunoblot of BPV L1 after virion digestion with 1 μg of trypsin for 15 min at 37°C (see Materials and Methods). Lanes: 1, no pretreatment; 2, 5 mM EGTA; 3, 10 mM DTT; 4, 5 mM EGTA plus 10 mM DTT.
FIG. 2.
Electron micrographs of BPV virions digested with trypsin. Samples were incubated for 2 h at 20°C in dialysis buffer with 10 mM EGTA (A), 10 mM EGTA plus trypsin (B), 10 mM DTT (C), or 10 mM DTT plus trypsin (D).
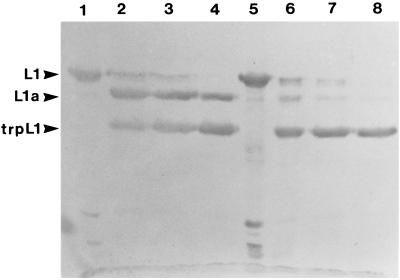
The extent of virion disruption by trypsin digestion could be varied with increasing concentrations of trypsin, and a progressive increase in the trpL1 species seen by immunoblotting (Fig. 3) corresponded to increased virion disruption as determined by electron microscopy (data not shown). In comparing the immunoblot with the electron microscopy data, it appears that generation of trpL1 is sufficient for virion disruption in the presence of DTT, although the digestion may proceed further, yielding small amounts of a faster-migrating species (Fig. 3, lane 2). In all cases in which L1 was digested by trypsin, the L2 protein was also completely digested, as determined by immunoblot analysis (data not shown).
FIG. 3.
Trypsin digestion of BPV virions. BPV virions were preincubated with 10 mM DTT and then digested with trypsin at the indicated concentrations for 15 min at 37°C. Lanes: 1, no trypsin; 2 to 5, 1, 0.1, 0.01, and 0.001 μg of trypsin, respectively.
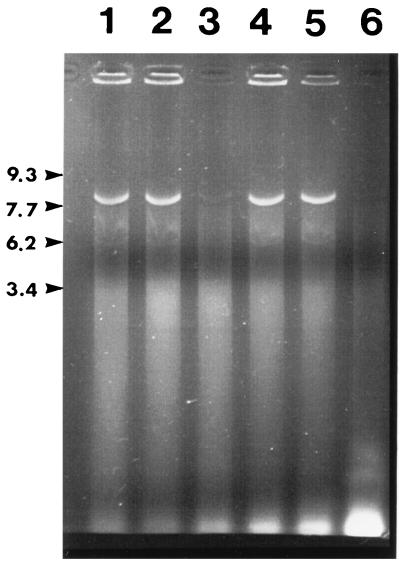
As another probe of virion integrity, DTT- and EGTA-treated virions were digested with DNase I. As shown in Fig. 4, the viral genome was resistant to digestion, except when DTT was included in the virion pretreatment buffer (lanes 3 and 6). The electron microscopic appearance of the reaction products showed that the DTT-treated, DNase I-digested virions had many empty capsids present, although complete capsid disruption was also noted (data not shown). In contrast, polyomavirus virions were not susceptible to either trypsin or DNase I unless both EGTA and DTT were used in the preincubation. Under these conditions, the virions were completed disrupted by the preincubation (data not shown), as previously described (5, 6).
FIG. 4.
BPV virions digested with DNase I; agarose gel electrophoresis of viral DNA extracted after digestion of BPV virions with DNase I. Virions were preincubated in BPV buffer (see Materials and Methods) with 15 mM MgCl2 (no DNase I) (lane 1), 5 mM EGTA (followed by DNase I) (lane 2), 5 mM EGTA plus 10 mM DTT (followed by DNase I) (lane 3), 5 mM MgCl2 (no DNase I) (lane 4), 5 mM MgCl2 (followed by DNase I) (lane 5), and 10 mM DTT (followed by DNase I) (lane 6).
The effects of EGTA and DTT on BPV virion stability were distinctly different when the treatments were performed at low (10 mM Tris-HCl) ionic strength. As shown in Fig. 5, although EGTA treatment had no effect on gross virion morphology, DTT treatment alone resulted in complete virion disruption. However, under these conditions of low ionic strength, L1 was susceptible to trypsin digestion after pretreatment with EGTA (Fig. 6), suggesting a difference in the effects of chelating agents at low versus physiologic ionic strength. A faster-migrating L1 cleavage product was also detected under these conditions.
FIG. 5.
Electron micrographs of BPV virions disrupted at low ionic strength. (A) Virions in buffer containing 10 mM Tris-HCl (pH 7.9); (B and C) the same buffer incubated for 15 min at 37°C with 1 mM EGTA and 10 mM DTT, respectively.
FIG. 6.
Trypsin digestion of BPV virions in low-ionic-strength buffer. Immunoblot of BPV virions (lane 1) incubated (as in Fig. 5) with DTT (lane 2) or EGTA (lane 3) and then digested with trypsin for 15 min at 37°C.
Analytical ultracentrifuge analysis.
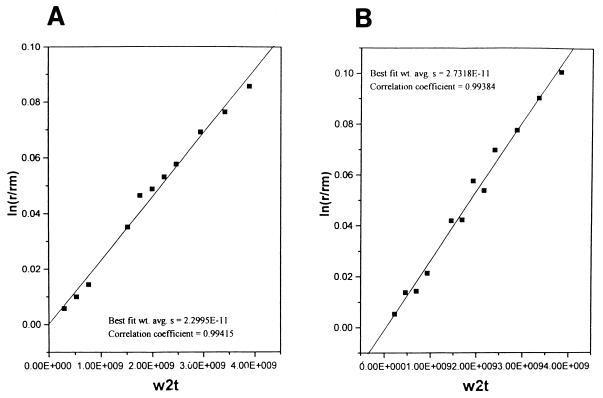
The susceptibility of L1 and the viral genome to enzymatic digestion after virion treatment with DTT suggested that a conformational change allowing access of the enzymes to the substrates may have occurred. In order to evaluate such a conformational change, virions incubated with either EDTA or DTT (in 160 mM NaCl) were analyzed by analytical ultracentrifugation. Untreated virions sedimented at 273S, EDTA-treated virions sedimented at 256S (not shown), and DTT-treated virions sedimented at 230S (Fig. 7). The substantial decrease in the S value of the DTT-treated virions, given their intact appearance by electron microscopy, suggests capsid swelling. Virions treated with DTT, which were determined to be 230S and sensitive to trypsin, were then redialyzed into buffer without DTT. These virions regained resistance to trypsin digestion (data not shown), demonstrating that the conformational change was reversible. Polyomavirus virions treated with either EGTA or DTT did not change their S values (data not shown).
FIG. 7.
Sedimentation analysis of BPV virions. BPV virions in BPV buffer plus 10 mM DTT (A) or buffer alone (B) were analyzed by analytical ultracentrifugation (see Materials and Methods). The DTT-treated virions sedimented at 230S, compared with 273S for untreated virions. w2t, product of the square of the angular velocity and time; r, radius of boundary; rm, radius of the meniscus; wt. avg., weight average.
L1 Cys424Gly mutant protein does not assemble into capsids.
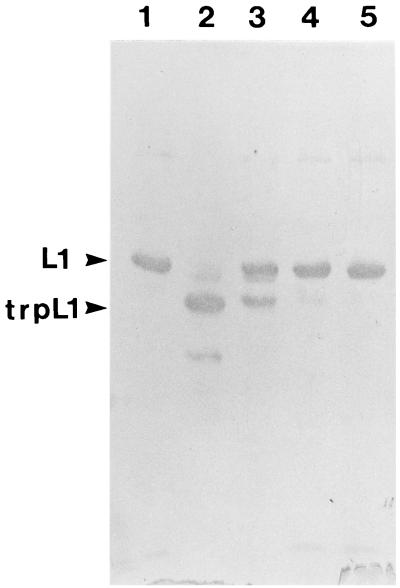
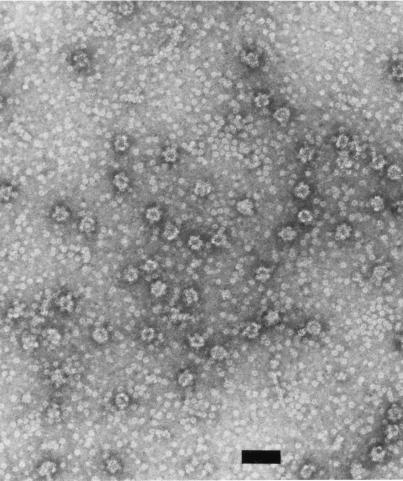
In comparing the L1 proteins of all sequenced papillomaviruses, there is only one cysteine residue in the carboxy-terminal domain that has been identified as critical for capsid formation (13). For HPV11 L1, this cysteine is residue 424. In order to test whether a recombinant mutant L1 protein with a substitution for this cysteine was capable of in vitro self-assembly, an L1 protein containing a Cys-to-Gly change at residue 424 (Cys424Gly) was generated, expressed in E. coli, and purified. Under conditions in which the wild-type protein assembled into capsid-like structures, the mutant protein formed capsomeres, but these capsomeres did not form capsids. As shown in Fig. 8, small aggregates, perhaps five or six capsomeres around one, can be seen, but larger structures were not observed. The formation of capsid-like structures in vitro may inhibit the ability of trypsin to digest L1 to trpL1 (13). As a further indication that the Cys424Gly mutant protein is defective in assembly, Fig. 9 shows the kinetics of trypsin digestion of the mutant versus wild-type protein. As seen in lanes 6 to 8, the digestion of the nonassembling mutant protein proceeds more rapidly than that of the wild type (lanes 2 to 4), presumably because the site of trpL1 digestion, R415, is not sterically shielded within capsid-like structures. Thus, Cys424 may be involved in interpentamer bonding.
FIG. 8.
Electron micrograph of L1 mutant protein Cys424Gly. The Cys424Gly mutant L1 protein was purified after expression in E. coli and incubated under assembly conditions with no reducing agent and 0.5 M NaCl (13). No capsid-like aggregates are seen, although the capsomeres cluster into small aggregates of five to six capsomeres around one.
FIG. 9.
Trypsin digestion of the Cys424Gly recombinant protein; immunoblot of HPV11 L1. Wild-type (lanes 1 to 4) or Cys424Gly (lanes 5 to 8) purified recombinant proteins were digested with trypsin (weight ratio, 10:1) at 20°C for 0 (lanes 1 and 5), 30 (lanes 2 and 6), 60 (lanes 3 and 7), and 90 (lanes 4 and 8) min.
DISCUSSION
BPV virions were found to undergo a structural change upon thiol reduction, leading to trypsin sensitivity of the L1 and L2 structural proteins, DNase I sensitivity of the viral genome, and a significant change in their sedimentation rates. This structural change did not affect the gross morphology of the virions as assessed by electron microscopy. BPV virions were more unstable at low ionic strength, at which thiol reduction resulted in complete disruption of the capsids, as determined by electron microscopy, and EGTA or EDTA treatment conferred trypsin sensitivity upon the virions, which was not observed at physiologic ionic strength. The stability of BPV virions, therefore, contrasts with that observed for polyomavirus virions, for which both thiol reduction and calcium chelation lead to complete virion disruption, and neither agent alone has a measurable effect.
Belnap et al. (4) have described an open structure for cottontail rabbit papillomavirus (CRPV) capsids, as determined by cryoelectron microscopic image reconstruction of capsids visualized at low ionic strength with chelating agents. These open structures have a 7% increase in diameter from the closed virion form. The structural change is accompanied by the appearance of holes in the capsid shell between the hexavalent capsomeric subunits. Such a structural transition would explain our BPV data. Although Belnap et al. did not examine DTT-treated virions at physiologic ionic strength, EGTA or EDTA affects the trypsin accessibility of L1 at low ionic strength, i.e., under the conditions in which CRPV was imaged. The holes could provide access for trypsin and DNase I, and the 7% increase in shell diameter could account for the change in the sedimentation coefficient of DTT-treated BPV virions. The data of Trus et al. (26) for BPV virions at a resolution of 9 Å show two cross-bridging densities between adjacent hexavalent capsomeres. It is possible that these cross-bridges contain the interpentamer thiol bond.
Capsid shell expansion allowing protease and nuclease attack has been well described for plant viruses after calcium chelation or pH changes. For example, Southern bean mosaic virus undergoes a 7% change in diameter after chelation (115S to 100S), and the virion becomes sensitive to nuclease and protease digestion. The conformational change is reversible upon readdition of divalent metal ions, and the protease digestion is nonrandom, yielding specific cleavage fragments. Cowpea chlorotic mottle virus is similarly affected (23), and high-resolution structures of the swollen form show that expansion occurs at the quasi-threefold axis of the virion, much like that seen for hole formation in the CRPV images. Thus, a structurally distinct swollen capsid conformation, which is characterized by a detectable change in S value and increased accessibility to proteases and nucleases, may be a general feature of virus assembly-disassembly pathways. For papillomavirus virions at physiologic ionic strength, thiol bonds appear to be critical, but the chemistry of the conformation switch may be dependent on the biology of the particular virus.
Disulfide bonds also contribute to the stability of SV40 and polyomavirus virions (24, 25). Crystallographic data have shown that in SV40, interpentamer disulfide bonds are formed (Cys104 to Cys104 between neighboring β,β′ VP1 monomers), while in polyomavirus an intrapentameric disulfide occurs between Cys114 (homologous to SV40 Cys104) and Cys19 near the amino terminus. Although the polyomavirus disulfides are intrapentameric, they may stabilize interpentamer bonding by contributing to a rigid conformation which fixes the invading carboxy-terminal arm from the neighboring pentamer (14). However, a Cys114Ala recombinant VP1 protein self-assembles in vitro into capsids similar to the wild-type protein, indicating that this contact may provide stability to the virion but is not necessary for capsid assembly (10a). For papillomaviruses, the Cys424 equivalent residue is the only cysteine residing within the carboxy-terminal domain essential for capsid formation. In analogy with both SV40 and polyomavirus, in which acidic residues from the invading carboxy terminus combine to form a calcium binding site with acidic residues in the body of a neighboring VP1 molecule, Cys424 likely bonds with a conserved cysteine in the body of a neighboring L1 molecule. The bond must be interpentameric, because thiol reduction leads to capsid disruption into capsomeres, not disruption of capsomeres, and the Cys424Gly mutant protein forms pentamers but does not assemble into capsids.
Divalent ion chelation by amino acids in L1 seems to play a minor role in papillomavirus stability, and its effect can be detected only at low ionic strength. Other bonding contacts must also be utilized, since the virion structure is grossly intact in the presence of both DTT and EGTA. The differences in stability seen at low versus physiologic ionic strength suggest that ionic or hydrophobic contacts play additional structural roles. Furthermore, the carboxy-terminal arm of L1 (or L2) appears critical for maintaining virion integrity, since once L1 is cleaved the capsid structure is completely disrupted. This region contains the major DNA binding domain of L1, and cleavage to trpL1 separates this domain from the pentamer. Thus, the bonds between the viral genome and capsid may also stabilize the structure when intercapsomeric thiol bonds are reduced.
Virions are necessarily metastable structures. They must be efficiently assembled, resistant to environmental degradation, and yet capable of rapid disassembly upon infection. In order to achieve this balance, conformational trigger mechanisms, likely with high-energy transition state intermediates, may be employed. For example, picornaviruses such as poliovirus (9, 10) change conformation upon receptor attachment, and, as described above, plant viruses may use calcium chelation or pH changes. Disulfide bonds take advantage of intra- versus extracellular oxidation conditions. Papillomavirus virion entry into the reducing environment of the cytoplasm would result in a change to become the 230S conformation, and subsequent proteolysis of the carboxy terminus of L1 would free the genome from the capsid to initiate infection. Since disulfide bond formation may not occur until virions exit the cell, open assembly intermediates may be present in the cell nucleus and may not be completely closed until oxidation after cell lysis. The biologic cues for assembly and disassembly are thus programmed by the chemistry of capsid formation.
ACKNOWLEDGMENTS
We thank John Schiller for L2 antiserum, Carl Olsen for cow warts, Robert Rose for the L1 antibody, and Roberta Thimmig for assistance with the analytical ultracentrifuge analysis.
This work was supported by grant CA37667 from the National Cancer Institute to R.L.G. P.B. was supported by Swiss Research against Cancer and the Swiss National Science Foundation.
REFERENCES
- 1.Baker T S, Drak J, Bina M. The capsid of small papova viruses contains 72 pentameric capsomeres: direct evidence from cryoelectron-microscopy of simian virus 40. Biophys J. 1989;55:243–253. doi: 10.1016/S0006-3495(89)82799-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baker T S, Newcomb W W, Olson N H, Cowsert L M, Olson C, Brown J C. Structures of bovine and human papillomaviruses: analysis by cryoelectron microscopy and three-dimensional image reconstruction. Biophys J. 1991;60:1445–1456. doi: 10.1016/S0006-3495(91)82181-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baldwin R L. Sedimentation coefficients of small molecules: methods of measurement based on the refractive-index gradient curve. The sedimentation coefficient of polyglucose A. Biochem J. 1953;55:644–648. doi: 10.1042/bj0550644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Belnap D M, Olson N H, Cladel N M, Newcomb W W, Brown J C, Kreider J W, Christensen N D, Baker T S. Conserved features in papillomavirus and polyomavirus capsids. J Mol Biol. 1996;259:249–263. doi: 10.1006/jmbi.1996.0317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brady J N, Winston V D, Consigli R A. Dissociation of polyomavirus by the chelation of calcium ions found associated with purified virions. J Virol. 1977;23:717–724. doi: 10.1128/jvi.23.3.717-724.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brady J N, Winston V D, Consigli R A. Characterization of a DNA-protein complex and capsomere subunits derived from polyomavirus by treatment with ethyleneglycol-bis-N,N′-tetraacetic acid and dithiothreitol. J Virol. 1978;27:193–204. doi: 10.1128/jvi.27.1.193-204.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Christiansen G, Landers T, Griffith J, Berg P. Characterization of components released by alkali disruption of simian virus 40. J Virol. 1977;21:1079–1084. doi: 10.1128/jvi.21.3.1079-1084.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Doorbar J, Gallimore P H. Identification of proteins encoded by the L1 and L2 open reading frames of human papillomavirus 1a. J Virol. 1987;61:2793–2799. doi: 10.1128/jvi.61.9.2793-2799.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Filman D J, Syed R, Chow M, Macadam A J, Minor P D, Hogel J M. Structural factors that control conformational transitions and serotype specificity in type 3 poliovirus. EMBO J. 1989;8:1567–1579. doi: 10.1002/j.1460-2075.1989.tb03541.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fricks C E, Hogle J M. Cell-induced conformational change in poliovirus: externalization of the amino terminus of VP1 is responsible for liposome binding. J Virol. 1990;64:1934–1945. doi: 10.1128/jvi.64.5.1934-1945.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10a.Garcea, R. Unpublished data.
- 11.Hagensee M E, Olson N H, Baker T S, Galloway D A. Three-dimensional structure of vaccinia virus-produced human papillomavirus type 1 capsids. J Virol. 1994;68:4503–4505. doi: 10.1128/jvi.68.7.4503-4505.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kirnbauer R, Booy G, Cheng N, Lowy D R, Schiller J T. Papillomavirus L1 major capsid protein self-assembles into virus-like particles that are highly immunogenic. Proc Natl Acad Sci USA. 1992;89:12180–12184. doi: 10.1073/pnas.89.24.12180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li M, Cripe T P, Estes P A, Lyon M K, Rose R C, Garcea R L. Expression of the human papillomavirus type-11 L1 capsid protein in Escherichia coli: characterization of protein domains involved in DNA-binding and capsid assembly. J Virol. 1997;71:2988–2995. doi: 10.1128/jvi.71.4.2988-2995.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liddington R C, Yan Y, Moulai J, Sahli R, Benjamin T L, Harrison S C. Structure of simian virus 40 at 3.8-Å resolution. Nature. 1991;354:278–284. doi: 10.1038/354278a0. [DOI] [PubMed] [Google Scholar]
- 15.Rayment I, Baker T S, Caspar D L D, Murakami W T. Polyoma virus capsid structure at 22.5 Å resolution. Nature (London) 1982;295:110–115. doi: 10.1038/295110a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roden R B S, Hubbert N L, Kirnbauer R, Breitburd F, Lowy D R, Schiller J T. Papillomavirus L1 capsids agglutinate mouse erythrocytes through a proteinaceous receptor. J Virol. 1995;69:5147–5151. doi: 10.1128/jvi.69.8.5147-5151.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rose R C, Bonnez W, Reichman R C, Garcea R L. Expression of human papillomavirus type 11 L1 protein in insect cells: in vivo and in vitro assembly of viruslike particles. J Virol. 1993;67:1936–1944. doi: 10.1128/jvi.67.4.1936-1944.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sahli R, Freund R, Dubensky T, Garcea R, Bronson R, Benjamin T. Defect in entry and altered pathogenicity of a polyoma virus mutant blocked in VP2 myristylation. Virology. 1993;192:142–153. doi: 10.1006/viro.1993.1016. [DOI] [PubMed] [Google Scholar]
- 19.Salunke D M, Caspar D L D, Garcea R L. Self-assembly of purified polyomavirus capsid protein VP1. Cell. 1986;46:895–904. doi: 10.1016/0092-8674(86)90071-1. [DOI] [PubMed] [Google Scholar]
- 20.Salunke D M, Caspar D L D, Garcea R L. Polymorphism in the assembly of polyomavirus capsid protein VP1. Biophys J. 1989;56:887–900. doi: 10.1016/S0006-3495(89)82735-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sapp M, Volpers C, Müller M, Streeck R E. Organization of the major and minor capsid proteins in human papillomavirus type 33 virus-like particles. J Gen Virol. 1995;76:2407–2412. doi: 10.1099/0022-1317-76-9-2407. [DOI] [PubMed] [Google Scholar]
- 22.Schachman H K. Ultracentrifugation in biochemistry. New York, N.Y: Academic Press; 1959. [Google Scholar]
- 23.Speir J A, Munshi S, Wang G, Baker T S, Johnson J E. Structures of the native and swollen forms of cowpea chlorotic mottle virus determined by X-ray crystallography and cryo-electron microscopy. Structure. 1995;3:63–78. doi: 10.1016/s0969-2126(01)00135-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stehle T, Gamblin S J, Yan Y, Harrison S C. The structure of simian virus 40 refined at 3.1 Å resolution. Structure. 1996;4:165–182. doi: 10.1016/s0969-2126(96)00020-2. [DOI] [PubMed] [Google Scholar]
- 25.Stehle T, Harrison S C. Crystal structures of murine polyomavirus in complex with straight-chain and branched-chain sialyloligosaccharide receptor fragments. Structure. 1996;4:183–194. doi: 10.1016/s0969-2126(96)00021-4. [DOI] [PubMed] [Google Scholar]
- 26.Trus B L, Roden R B S, Greenstone H L, Vrhel M, Schiller J T, Booy F P. Novel structural features of bovine papillomavirus capsid revealed by a three-dimensional reconstruction to 9Å resolution. Nat Struct Biol. 1997;4:413–420. doi: 10.1038/nsb0597-413. [DOI] [PubMed] [Google Scholar]
- 27.Volpers C, Schirmacher P, Streeck R E, Sapp M. Assembly of the major and the minor capsid protein of human papillomavirus type 33 into virus-like particles and tubular structures in insect cells. Virology. 1994;200:504–512. doi: 10.1006/viro.1994.1213. [DOI] [PubMed] [Google Scholar]
- 28.Walter G, Deppert W. Intermolecular disulphide bonds: an important structural feature of the polyoma virus capsid. Cold Spring Harbor Symp Quant Biol. 1974;39:255–257. doi: 10.1101/sqb.1974.039.01.033. [DOI] [PubMed] [Google Scholar]
- 29.Zhou J, Stenzel D J, Sun X Y, Frazer I H. Synthesis and assembly of infectious bovine papillomavirus particles in vitro. J Gen Virol. 1993;74:763–768. doi: 10.1099/0022-1317-74-4-763. [DOI] [PubMed] [Google Scholar]