Abstract
We have determined the complete nucleotide sequence of a replication-competent clone of bovine foamy virus (BFV) and have quantitated the amount of splice pol mRNA processed early in infection. The 544-amino-acid Gag protein precursor has little sequence similarity with its primate foamy virus homologs, but the putative nucleocapsid (NC) protein, like the primate NCs, contains the three glycine-arginine-rich regions that are postulated to bind genomic RNA during virion assembly. The BFV gag and pol open reading frames overlap, with pro and pol in the same translational frame. As with the human foamy virus (HFV) and feline foamy virus, we have detected a spliced pol mRNA by PCR. Quantitatively, this mRNA approximates the level of full-length genomic RNA early in infection. The integrase (IN) domain of reverse transcriptase does not contain the canonical HH-CC zinc finger motif present in all characterized retroviral INs, but it does contain a nearby histidine residue that could conceivably participate as a member of the zinc finger. The env gene encodes a protein that is over 40% identical in sequence to the HFV Env. By comparison, the Gag precursor of BFV is predicted to be only 28% identical to the HFV protein.
Spumaviruses (foamy viruses) were first described in 1954 by Enders and Peebles as cytopathogenic agents of primary monkey kidney cells (7). Since that time, these viruses have been isolated from other species including humans, felines, and bovines (12, 23, 27). Based on serology, foamy virus infections appear to be quite common in monkeys (simian foamy viruses [SFVs]) and cattle (bovine foamy viruses [BFVs]) (1, 2, 24, 31). Despite their early identification and widespread presence, the foamy viruses remain the least well characterized among the retroviruses, probably because they have not been associated with disease. It is interesting that while these viruses cause a pronounced cytopathic effect in a wide variety of tissue culture cells, they appear to be benign in vivo. This lack of apparent pathogenicity has led to the idea that the foamy viruses may be suitable delivery vectors for gene therapy (40, 43). However, it has been suggested that their presence in the host may contribute to diseases caused by other pathogens (25). Also, transgenic mice expressing human foamy virus (HFV) proteins have been reported to present a progressive encephalopathy and myopathy, suggesting that foamy viruses have pathogenic potential (5, 45).
The HFV and isolates of SFV type 1 (SFV-1) and SFV-3 have been molecularly cloned, sequenced, and characterized previously (9, 12, 17, 26, 35). The most prominent structural feature of these viruses is the presence of two accessory genes, one of which (bel1 or taf) encodes a transcriptional transactivator (19, 34). The primate foamy viruses are unique in their lack of Cys-His motifs in the nucleocapsid (NC) domain of the Gag protein and the major homology region in the capsid (CA) domain of Gag. The foamy viruses contain an internal promoter that directs transcription of their 3′ accessory genes early in infection (6, 20–22, 29, 34). Further, the Pol polyprotein in primate foamy viruses is synthesized from a spliced mRNA, rather than by frameshifting or termination suppression mechanisms (4, 8, 14, 46). Finally, in contrast to the other retroviruses, cells infected with HFV produce a large proportion of virions that contain genomic-length DNA. These several properties suggest that the foamy viruses may be related to the pararetroviruses and hepadnaviruses (39, 46).
The relationship of primate foamy viruses and nonprimate foamy viruses has not been clearly established. To determine if the unique aspects reported for the primate foamy viruses represent general hallmarks of foamy viruses, we have extended the reported sequence analysis of the long terminal repeat (LTR) and 3′ portion of BFV (36–38) to include the entire genome of an infectious DNA clone. In addition, we have carried out experiments to measure the levels of spliced pol mRNA. The results imply that BFV is similar in all respects to the primate foamy viruses, sharing with them an internal promoter in env, a spliced pol mRNA, and an NC protein that contains Gly-Arg-rich motifs rather than Cys-His motifs. The levels of BFV spliced pol mRNA, which has not been quantitated in other foamy virus studies, are notably high. Surprisingly, the predicted Env proteins of the foamy viruses are considerably more closely related to each other than to the Gag proteins, suggesting that different foamy viruses may use a highly conserved receptor.
MATERIALS AND METHODS
DNA sequencing.
The sequence of the BFV genome was determined from p11-1 and p11-10 subclones derived from the infectious bovine syncytial virus type 11 (BSV-11) λ clone (38). Plasmids were prepared by alkaline minipreparation and were manually sequenced with Sequenase (U.S. Biochemical). Sequence data from small random subclones was derived from BSV-11 by shearing to approximately 250 bp or obtained by processively walking through p11-1 and p11-10 with sequence-specific primers. The BFV genome was assembled with AssemblyLIGN and analyzed with MacVector, DNAStar, and the Wisconsin Genetics Computer Group package. The EcoRI junction point of subclones 11-1 and 11-10 was sequenced from a PCR-generated clone that spanned the internal EcoRI site of BSV-11. Additionally, the PCR products spanning the gag-pol junction of six independent isolates obtained from the Cornell Veterinary School Diagnostic Laboratory were sequenced. Isolate 489770 was collected in Vermont in 1989, 422809 was from Ohio in 1988, 792152 was collected in New York in 1994, 336848 was collected in New York in 1987, 334862 was collected also in New York in 1987, and 438340 was collected in Connecticut in 1988.
Identification of a spliced pol mRNA.
Total RNA was isolated from Cf2Th (ATCC CRL-1430) tissue culture cells productively infected with BSV with the Genosys RNA isolation kit. The RNA was reverse transcribed with random hexamers and murine leukemia virus reverse transcriptase (RT) (New England Biolabs) according to the manufacturer’s specifications. The cDNA from these reactions was amplified with a forward primer from nucleotides (nt) 20 to 39 that contained an added KpnI site and two reverse primers at positions 2120 to 2145 and 2420 to 2445. The PCR products were gel purified with a Qiagen kit; cut with KpnI and NaeI, which occurs at position 2020; and cloned into pBluescriptSK− cut with KpnI and EcoRV. These products were sequenced as described above.
Relative quantitation of the spliced pol transcript.
Total RNA was isolated from Cf2Th cells and BFV-infected cells as described above. BFV-infected Cf2Th cells, ca. 80% confluent with approximately 10% of the cells displaying syncytia, were trypsinized and seeded at a 1:10 dilution onto uninfected Cf2Th cells (100-mm plates) that had been trypsinized and seeded (1:10) the previous day. At 2, 3, and 4 days postinoculation, total RNA was isolated from three separate plates for a total of nine samples. RNAs were isolated from three uninfected Cf2Th cultures to serve as controls. All RNA samples were treated for 4 h at 37°C with RNase-free DNase (Boehringer) in reverse transcription buffer, followed by phenol-chloroform extraction and ethanol precipitation. The samples were dissolved in 25 μl of water, and 0.5 μg was used in a 25-μl reverse transcription reaction as described above. The resultant cDNAs were diluted to 50 μl with water and used as templates for competitive PCR.
Competitive PCR.
The PCR strategy using mutant competitors of the same size as the amplification products produced from native mRNAs has been described elsewhere (28). Briefly, competitors for specific transcripts were generated by PCR with one primer approximately 45 bases in length that spans a restriction site. This primer is used to incorporate an alternative restriction site in the same position as the restriction site in the native sequence. To quantitate the amount of the spliced pol mRNA, the primers homologous to nt 52 to 91 and 1941 to 1960 were used to amplify a 175-bp competitor from a plasmid clone that has a BamHI site substituted for the BglII site at nt 80 to 85. Competitors for transcripts containing gag sequences and Borf-2 sequences were similarly synthesized with primers homologous to nt 588 to 630 and 751 to 770 (gag) and 9573 to 9592 and 9711 to 9753 (Borf-2). The gag competitor has a BglII site substituted for the BamHI site at nt 619 to 624; the Borf-2 competitor also has a BglII site substituted for the BamHI site at nt 9717 to 9722. Following amplification of the respective competitors, each was purified from an agarose gel (Qiagen kit) and quantitated spectrophotometrically. Experimentally, a dilution series of each competitor was used in PCRs containing 1 μl of the reverse transcription reaction mixtures described above. One sample from each RNA time point was processed in triplicate with each competitor series to determine the reproducibility of the PCRs. In addition, all samples were run individually to control for variability at the level of RNA isolated at different time points and the efficiency of cDNA synthesis. A cocktail containing buffer, Mg2+, deoxynucleoside triphosphates, and primers was dispensed, and the cDNA and competitors were added. The primer pairs used in these amplifications were homologous to nt 52 to 71 and 1941 to 1960 (pol), 588 to 607 and 751 to 770 (gag), and 9573 to 9592 and 9734 to 9711 (Borf-2). The samples were heated at 96°C for 5 min and immediately cooled in ice, and Taq polymerase was added. The samples were amplified for 35 cycles of 94°C for 30 s, 50°C for 30 s, and 72°C for 40 s, followed by a 72°C extension for 10 min. Five microliters from each sample was digested with BamHI or BglII to analyze whether the PCR product was made predominantly from the competitor or from native cDNA. When the levels of digestion products generated from the competitor and native cDNA were equal, as seen by ethidium bromide staining, the amount of viral mRNA from the infected cells above was inferred. These results are not quantitative in an absolute sense, but relative amounts of specific mRNAs can be deduced.
Nucleotide sequence accession number.
The sequence of the BFV proviral genome has been deposited in the GenBank database (accession no. U94514).
RESULTS
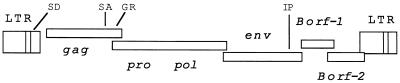
The genomic organization of the BFV provirus is shown in Fig. 1. The sequences of the BFV LTR, Borf-1, and Borf-2 have been reported elsewhere (36, 37). The results reported here use the putative transcriptional start at nt 982 in the BFV LTR (36) as position 1.
FIG. 1.
Genomic organization of BFV. The open reading frames for BFV were determined from the DNA sequence of λ BSV-11 described by Renshaw et al. (38). The approximate positions of the viral splice donor (SD), splice acceptor site for pol mRNA (SA), the Gly-Arg-rich region in Gag (GR), and the internal promoter (IP) are shown.
gag gene.
The Gag polyproteins of primate foamy viruses range in size from 646 amino acid (aa) residues in SFV-3 to 673 aa residues in SFVcpz (19). The sequence of BFV type 11 (BFV-11) shows that the gag open reading frame (nt 433 to 2064) encodes a Gag polyprotein of 544 aa residues. Protein alignments with the DNAStar package showed that the BFV Gag polyprotein has no amino acid sequence similarity with retroviruses outside the Spumavirinae and only limited identity with the primate Gag proteins, i.e., 24% identity with SFV-3 and 29% identity with SFV-1 (Table 1). Despite the primary sequence divergence among the foamy virus Gag proteins, their hydrophilicity profiles are quite similar and each has a hydrophilic C terminus (data not shown). The proteolytic products of foamy virus Gag polyproteins have not been purified and thus are not clearly defined. While there is no evidence that the spumavirus Gag polyprotein is cleaved during a budding-associated maturation process, as seen with other retroviruses, putative cleavage has been identified in lysates of HFV-infected BHK-21 cells by Western blot analysis (30). Recently, it was reported that the HFV Gag polyprotein is cleaved early after infection, producing a matrix (MA) protein of 26 kDa, a CA protein of 32 kDa, and an NC protein of 18 kDa (10). The BFV Gag polypeptide has a very proline-rich segment of approximately 48 residues starting at aa 157 that is reminiscent of the segment seen between MA and CA in other retroviruses, e.g., the murine leukemia virus and the avian sarcoma and leukosis viruses. By analogy with other Gag polyproteins, the BFV MA protein is likely encoded at the amino-terminal end of Gag and is comprised of approximately 155 to 200 aa residues. Starting at ca. position 200 in the predicted BFV Gag protein is a region of amino acid conservation that aligns with ca. position 300 to about 320 in the primate Gag proteins. We suggest that this is a region within CA and that the difference in overall lengths of the BFV and primate gag-encoded proteins is in the region connecting MA and CA, as is found in other retroviruses.
TABLE 1.
Similarities of proteins predicted from the major open reading frames of BFV with those predicted from the sequences of the primate viruses
| Virus | % Similarity with protein encoded by:
|
||||
|---|---|---|---|---|---|
| gag | pol | env | taf (orf-1) | orf-2 | |
| BFV | |||||
| HFV | 28 | 57 | 41 | 17 | 15 |
| SFV-1 | 29 | 57 | 39 | 17 | 13 |
| SFVcpz | 27 | 47 | 36 | 13 | 11 |
| SFV-3 | 24 | 47 | 32 | 14 | 13 |
The NC proteins of the spumaviruses do not contain the zinc finger domain(s) of other retroviruses (reviewed in reference 18). They do, however, contain three glycine-arginine-rich regions (GR boxes 1, 2, and 3) that are postulated to bind genomic RNA during virion assembly and have recently been shown to direct transport of the HFV Gag precursor to the nuclei of infected cells (42). While there is no discernible amino acid sequence conservation of GR box 1 among any of the spumaviruses, within the 23-aa GR box 2, which is necessary for transport of Gag to the nucleus, there are 10 conserved positions that possibly constitute the functional core of this element (Fig. 2). Also, within the 34-aa stretch identified as GR box 3 there is a region where six of eight amino acid residues are identical to those in the primate spumavirus NC proteins. There is no obvious similarity in the carboxy-terminal regions of the spumavirus Gag proteins beyond GR box 3. As with the primate foamy viruses, the BFV gag and pol open reading frames are overlapping. We have confirmed this overlap by sequence analysis of PCR products from six BFV samples from the Cornell Veterinary Diagnostic Laboratory (data not shown).
FIG. 2.
Alignment of glycine-arginine-rich regions (GR boxes 1 to 3) within the putative NC proteins of spumaviruses (38). Amino acid residues that are conserved in the foamy virus NC proteins are in boldface and underlined. The amino acid positions of the GR boxes within the respective Gag polyproteins flank the amino acid sequences.
pro and pol genes.
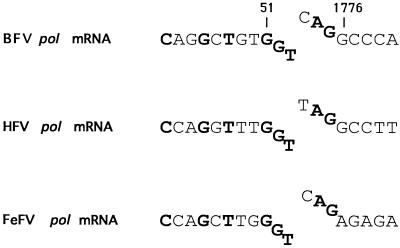
As in the genomes of the primate spumaviruses, the pro and pol genes of BFV are contiguous in the same open reading frame. In contrast to the other retroviruses that translate pol from full-length mRNA as part of a gag-pol polyprotein, it has been shown that the primate spumaviruses express pol gene products from a spliced mRNA (14, 46). Recently, it has also been reported that the feline foamy virus produces a spliced pol mRNA (4). BFV sequence similarity with the HFV splice donor and splice acceptor sites allowed us to design PCR primers that amplify across the putative pol mRNA splice junction. Figure 3 shows the splice junction of the pol mRNA obtained by RT-PCR. The junction is from nt 51 in the BFV leader sequence to nt 1776 in the BFV genome. Like that in the primate foamy viruses, the BFV pol mRNA is predicted to contain a long untranslated region, in this case one of 242 nt, prior to the first ATG codon of the pol open reading frame at nt 2018. We suggest that the translation of the pol open reading frame is likely to begin at this ATG codon, which encodes the first amino acid of protease. We were led to this conclusion based on the observation that the conserved PR sequence DSGA begins at aa 21, similar to the position (aa 25) of the DTGA sequence in the mature human immunodeficiency virus PR (33). The mature PR of primate spumaviruses is approximately 10 kDa, but the size of the BFV PR is yet to be determined. Within RT, the first recognizable motif is ENQV. By analogy with the other retroviral Pol proteins, we estimate that the amino terminus of RT is approximately 40 residues upstream of this sequence, consistent with a PR comprising approximately 100 aa residues.
FIG. 3.
Alignment of the pol mRNA splice junctions of BFV, HFV, and feline foamy virus (FeFV). Conserved nucleotides are shown in boldface. The nucleotide positions of the BFV splice donor and acceptor are shown above the sequence.
The BFV pol gene encodes amino acid sequences typically seen in other retroviral enzymes. Since the amino terminus of RT is not known, we used the N-terminal methionine of PR as the reference for numbering residues of RT and integrase (IN). The BFV pol gene encodes the RT signature sequence YVDD at aa 309 to 312. Other conserved RT motifs that can be aligned with known retroviral RTs are found at aa 248 to 250 (LDL) and 281 to 284 (LPQG). The conserved sequences TDSY and TDS at positions 368 to 371 and 667 to 669, respectively, identify the RNase H domain of RT. Motifs typical of IN are seen at aa 885 to 888 (DYIG) and 991 to 994 (SDQG). Interestingly, the proline residue at position 819 disrupts the canonical HH-CC zinc finger motif present in all characterized retroviral INs (15, 32). Provocatively, the histidine residue at position 816 in BFV is not present in any other spumaviruses, suggesting the possibility that it may participate as a member of the Zn2+ finger. Alternatively, the subclone of BSV-11, used for the sequencing, has had a single base change, CAC to CCC, resulting in the substitution of Pro for His, which could result in an inactive virus. This seems unlikely, however, because the BSV-11 clone has been shown elsewhere to be infectious (38) and the sequences of the PCR products derived from two independent infectious isolates (489770 and 422809) also encode His and Pro at positions 816 and 819, respectively.
env gene.
The predicted BFV Env protein is 990 aa in length, which is comparable to the primate foamy virus Env proteins, which are 984 (SFV-1) to 1020 (SFVcpz) aa in length. Interestingly, alignment of the foamy virus Env proteins does not identify hypervariable regions that are typical of many retroviral Env proteins but rather a nearly uniform distribution of conserved amino acids (Fig. 4). This data suggests that the Env proteins of foamy viruses from genetically distant hosts may be functionally conserved. The amino acid identity of the BFV Env with members of this group ranges from 32.9% (SFV-3) to 41.3% (HFV) (Table 1). A putative hydrophobic leader peptide can be identified from aa 54 to 89. The sequence KRTRR is present in Env starting at aa position 568. This sequence is analogous to the SFV and HFV Env protein sequences suggested to be the recognition sequences for proteolysis leading to the production of SU and TM (19). Consistent with this hypothesis is the presence of 11 potential N-linked glycosylation sites in the putative BFV SU. Assuming that KRTRR is the SU-TM cleavage site, there is 25% amino acid sequence identity in SU among all the foamy virus isolates. The smaller TM has approximately 38% amino acid sequence identity with each of the foamy virus isolate TM proteins. The sequence xxGxxxxxAxxTLSxxS occurs near the amino terminus of TM. This sequence is conserved among the foamy viruses and is thought to be important for membrane fusion. Near the carboxy terminus of the BFV TM is a hydrophobic region that aligns with an analogous region in the other primate foamy viruses and probably represents the membrane anchor domain of TM. This domain is followed by a very short (11 aa in BFV), highly basic region that forms the cytoplasmic tail of TM. It has been suggested that this highly basic tail may be important to the interactions of TM with the gag-derived MA protein during virus assembly (19). The TM proteins of the primate foamy viruses have lysine residues at positions −5 and −3 from their respective carboxy termini (11). This motif is an endoplasmic retrieval signal. The BFV TM differs by having an Arg at position −5 from the carboxy terminus. It was previously suggested that the Arg residues at −4, −5, and −6 may compensate for the lack of a lysine at position −5 (13).
FIG. 4.
Alignment of foamy virus Env proteins. Identical amino acids conserved among the foamy viruses are shown in bold. Putative functional domains are shown; the putative leader peptide is underlined with dashes, the proposed SU-TM cleavage site is marked by asterisks, the cell fusion domain is marked by number signs, the transmembrane segment is marked by plus signs, and the basic cytoplasmic tail is marked by carets. The amino acid positions of the Env proteins are to the right of the sequences.
Previously, an internal promoter in the primate foamy virus env gene that directs early transcription of the 3′ open reading frames was identified (6, 20–22, 29, 34). An analogous promoter region in BFV is apparent by sequence comparison with HFV and SFV-1 (Fig. 5). A DNA fragment containing the putative BFV internal promoter actively directs the transcription of Borf-1 in transient transfection experiments (9a).
FIG. 5.
Alignment of foamy virus internal promoters. The nucleotide position of the first base from the start of transcription is shown on the right. Conserved nucleotides are in boldface. The TATA box and the starts of HFV (⇑) and SFV-1 (↑) transcription are underlined, as is the putative transcription start in BFV.
Relative levels of BFV spliced pol mRNA, gag mRNA, and Borf-2 mRNA.
Recent experiments have indicated that low levels of spliced pol mRNA are produced during HFV infection, but the actual amount of this mRNA species was not quantified (46). Our initial experiments showed that BFV produced a spliced pol mRNA that appeared to be relatively abundant. We undertook a series of quantitative-competitive RT-PCR experiments to estimate the relative amount of BFV spliced pol mRNA relative to other viral transcripts. The experiments were designed to amplify cDNA that is unique to the spliced pol mRNA (spanning the splice junction) and to compare its level to that of viral RNAs that contain gag and Borf-2 sequences. For these experiments, total RNA was isolated from BFV-infected Cf2Th cells and quantitated. cDNA was made from the RNA by using random hexamers as primers and used as the template for competitive PCR. The competitors for each of these mRNAs were quantitated and mixed in different amounts with the cDNAs derived from viral RNA in PCRs. The relative amounts of PCR product derived from the competitor and from cDNA can be distinguished by digestion with BamHI or BglII followed by agarose gel electrophoresis. When the PCR products derived from the cDNA and competitor are equal, the amount of cDNA equals the amount of competitor.
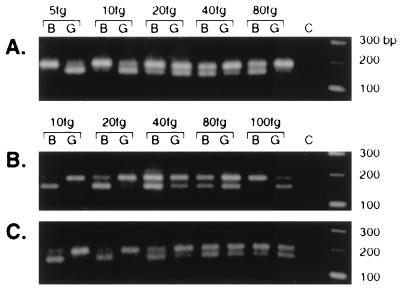
A segment near the 3′ end of the BFV genome (within Borf-2), which is presumably present in all viral mRNAs, was also amplified as an additional measurement of viral RNA levels. No amplification products were observed with RNA from uninfected cells or with RNA from infected cells when the reverse transcription step was omitted. Typical results from these experiments are shown in Fig. 6. The approximate amounts of spliced pol mRNA, gag-containing viral RNA, and viral RNA representing a region in Borf-2 are ca. 20 to 40, ca. 40 to 80, and ca. 80 to 100 fg, respectively. These results were reproducible, within a factor of 2, for RNAs from the same sample and among samples independently collected at the same time point. The amount of viral RNAs approximately doubled from day 2 to day 4 (data not shown). Since the Borf-2 sequence is present in all RNA species initiated from the LTR and the internal promoter, the expected amount of the Borf-2 mRNA should be greater than the sum of the gag and spliced pol mRNAs. The contribution of the internal promoter to the level of Borf-2 mRNA is unknown, and the confidence level of these experiments is approximately a factor of 2. The result that Borf-2 mRNA abundance was at least the sum of gag and pol mRNAs thus is consistent with our expectation and implies that the internal promoter contributed no more than 50% of the transcripts containing the 3′ end of the virus in these experiments. In summary, these results show that the splicing event that leads to BFV pol mRNA is not rare, with the ratio of BFV spliced pol mRNA to gag-containing mRNA being within the range of 1:1 to 1:4.
FIG. 6.
Competitive PCR determination of the levels of gag, spliced pol, and Borf-2 specific mRNAs from cells collected after 4 days of cocultivation. The amount of specific competitor DNA added to each PCR is shown above appropriate gel lanes. Lanes C show the no-DNA-added PCR control. Five microliters of each PCR was cut in 20 μl with either BamHI (B) or BglII (G) and run on a 3% agarose gel for analysis. (A) Spliced pol mRNA; (B) gag mRNA; (C) Borf-2 mRNA.
DISCUSSION
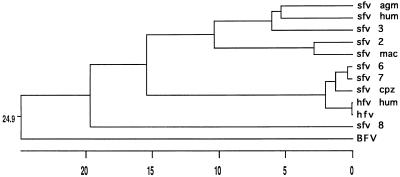
We have shown that BFV has a genomic organization very similar to that of the primate foamy viruses, which themselves are closely related to each other. This similarity extends beyond gag, pol, and env genes to the two 3′ accessory genes, Borf-1 and Borf-2. While Borf-1 and Borf-2 do not show sequence similarity with their counterparts, Borf-1 encodes a transactivator of transcription from the 5′ LTR, like its analogous gene in the primate viruses. As expected, the RT-encoding region in pol is the most highly conserved sequence in BFV and clearly places this bovine virus in the foamy virus genus (Fig. 7). Among the group of foamy viruses, HFV and SFV-1 exhibit the greatest similarity to BFV, not only in RT but in gag and env as well (Table 1).
FIG. 7.
Phylogenetic tree. Amino acid sequences representing segments of IN (44) were analyzed with software from DNAStar (41).
The replication and gene expression of foamy viruses, as documented in HFV and SFV-1 and, in some respects, in SFV-3, distinguish them from all other retroviruses. We have shown that BFV also shares several of these features. The sequence of BFV shows a putative internal promoter in env, and plasmids containing this region transactivate the LTR in transfection studies (data not shown). A spliced RNA capable of directing the synthesis of the Pol polyprotein is present in infected cells, and the splice-junction sequence is very similar to those reported for HFV and feline foamy virus (4, 46). The HFV pol mRNA was reported to be of low abundance, an observation that is consistent with a splicing mechanism that regulates the amount of Pol. We have carried out quantitative determinations of the BFV pol mRNA and found it to be present at high levels, approaching the level of viral RNA. These determinations were made early in infection, because spumavirus virions are not efficiently released; they accumulate in cells and could skew the ratio of spliced versus unspliced viral RNA in the analysis. The abundance of BFV pol mRNA suggests that if the ratio of Gag to Pol seen in other retroviruses (20:1), due to translational suppression or translational frameshifting, is present in BFV, it is not regulated by the relative amounts of their respective mRNAs. Thus, splicing does not appear to be a limiting step in the production of Pol. The BFV initiator codon for Pol translation is in a reasonable context (16) with an A residue at −3 and C residues at −4, −6, and −7. Perhaps the pol leader exerts a negative influence on translation, or perhaps the BFV Pol protein is more abundant than in other retrovirus systems.
The protein products of the primate foamy viruses and BFV have not been extensively characterized from cells or virus particles. We have identified the domains of Gag that likely encompass the mature MA, CA, and NC of BFV, based on analogy with data in reports describing HFV proteins (3, 10, 30). One clearly conserved region within the BFV gag gene encodes the GR boxes of the putative NC protein. The GR motifs have been postulated to bind genomic RNA during virion assembly and to direct transport of HFV Gag to the nucleus (42). While nuclear localization of Gag in primate foamy viruses is unique among retroviruses, its purpose is unclear. Based on immunofluorescence, BFV Gag is also likely to be targeted to the nucleus (24). The amino acid positions in the GR boxes that are conserved among all the foamy viruses may define the functional cores of these motifs. Interestingly, while the GR boxes are conserved in the foamy viruses, the GRII motif is dispensable for viral replication in vitro (47).
The extent of amino acid conservation in the Env protein of primate foamy viruses compared with that in BFV is striking. For example, in most retroviruses, Env is less conserved than is Gag, the opposite of our findings for BFV and the primate foamy viruses. Bovine leukemia virus and human T-cell leukemia virus type 1 Gag and Env proteins are ca. 35 and 20% identical, respectively. It seems rather remarkable that over 40% of the amino acid residues are identical in the BFV and HFV Env proteins (Fig. 4). One possible interpretation of this similarity is that foamy viruses bind to the same receptor and that this receptor itself is highly conserved among animal species. The use of a common receptor may have implications for the development of foamy viruses as vectors in gene therapy. It has been established that foamy viruses can infect a wide variety of cultured cells. Identification and characterization of the primate foamy virus and BFV receptors and studies of host range and tissue tropism of these viruses are important preliminaries to the development of these viruses as vectors for gene delivery.
The biology of foamy viruses in vivo is relatively unknown and merits investigation. With the reagents that have recently been developed, the mechanisms of foamy virus latency, persistence, and possible pathogenesis can be examined.
ACKNOWLEDGMENTS
We thank Volker Vogt for his advice and reading of the manuscript. We also thank Martin Lochelt and Rolf Flugel for providing us with the sequence of HFV prior to its being deposited in GenBank (accession no. U21247).
This work was supported in part with funds from CSREES USDA under project no. 433381 (D.L.H.) and with a grant from the Harold Wetterberg Foundation (J.W.C.).
REFERENCES
- 1.Amborski G F, Lo J L, Seger C L. Serological detection of multiple retroviral infections in cattle: bovine leukemia virus, bovine syncytial virus, and bovine visna virus. Vet Microbiol. 1989;20:247–253. doi: 10.1016/0378-1135(89)90048-5. [DOI] [PubMed] [Google Scholar]
- 2.Appleby R C. Antibodies to bovine syncytial virus in dairy cattle. Vet Rec. 1979;105:80–81. doi: 10.1136/vr.105.4.80. [DOI] [PubMed] [Google Scholar]
- 3.Bartholoma A, Muranyi W, Flugel R M. Bacterial expression of the capsid antigen domain and identification of native gag proteins in spumavirus-infected cells. Virus Res. 1992;23:27–38. doi: 10.1016/0168-1702(92)90065-h. [DOI] [PubMed] [Google Scholar]
- 4.Bodem J, Löchelt M, Winkler I, Flower R P, Delius H, Flügel R M. Characterization of the spliced pol transcript of feline foamy virus: the splice acceptor site of the pol transcript is located in gag of foamy viruses. J Virol. 1996;70:9024–9027. doi: 10.1128/jvi.70.12.9024-9027.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bothe K, Aguzzi A, Lassmann H, Rethwilm A, Horak I. Progressive encephalopathy and myopathy in transgenic mice expressing human foamy virus genes. Science. 1991;253:555–557. doi: 10.1126/science.1650034. [DOI] [PubMed] [Google Scholar]
- 6.Campbell M, Renshaw-Gegg L, Renne R, Luciw P A. Characterization of the internal promoter of simian foamy viruses. J Virol. 1994;68:4811–4820. doi: 10.1128/jvi.68.8.4811-4820.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Enders J F, Peebles T C. Propagation in tissue cultures of cytopathogenic agents from patients with measles. Proc Soc Exp Biol Med. 1954;86:277–286. doi: 10.3181/00379727-86-21073. [DOI] [PubMed] [Google Scholar]
- 8.Enssle J, Jordan I, Maurer B, Rethwilm A. Foamy virus reverse transcriptase is expressed independently from the Gag protein. Proc Natl Acad Sci USA. 1996;93:4137–4141. doi: 10.1073/pnas.93.9.4137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flugel R M, Rethwilm A, Maurer B, Darai G. Nucleotide sequence analysis of the env gene and its flanking regions of the human spumaretrovirus reveals two novel genes. EMBO J. 1987;6:2077–2084. doi: 10.1002/j.1460-2075.1987.tb02473.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9a.Garnica, J., and D. Holzschu. Unpublished data.
- 10.Giron M-L, Colas S, Wybier J, Rozain F, Emanoil-Ravier R. Expression and maturation of human foamy virus Gag precursor polypeptides. J Virol. 1997;71:1635–1639. doi: 10.1128/jvi.71.2.1635-1639.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goepfert P A, Wang G, Mulligan M J. Identification of an ER retrieval signal in a retroviral glycoprotein. Cell. 1995;82:543–544. doi: 10.1016/0092-8674(95)90026-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Herchenroder O, Renne R, Loncar D, Cobb E K, Murthy K K, Schneider J, Mergia A, Luciw P A. Isolation, cloning, and sequencing of simian foamy viruses from chimpanzees (SFVcpz): high homology to human foamy virus (HFV) Virology. 1994;201:187–199. doi: 10.1006/viro.1994.1285. [DOI] [PubMed] [Google Scholar]
- 13.Jackson M R, Nilsson T, Peterson P A. Identification of a consensus motif for retention of transmembrane proteins in the endoplasmic reticulum. EMBO J. 1990;9:3153–3161. doi: 10.1002/j.1460-2075.1990.tb07513.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jorden I, Enssle J, Guettler E, Mauer B, Rethwilm A. Expression of human foamy virus reverse transcriptase involves a spliced pol mRNA. Virology. 1996;224:314–319. doi: 10.1006/viro.1996.0534. [DOI] [PubMed] [Google Scholar]
- 15.Khan E, Mack J P G, Katz R A, Kulkosky J, Skalka A M. Retroviral integrase domains: DNA binding and the recognition of LTR sequences. Nucleic Acids Res. 1990;19:851–860. doi: 10.1093/nar/19.4.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kozak M. An analysis of 5′-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 1987;15:8125–8148. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kupiec J J, Kay A, Hayat M, Ravier R, Peries J, Galibert F. Sequence analysis of the simian foamy virus type 1 genome. Gene. 1991;101:185–194. doi: 10.1016/0378-1119(91)90410-d. [DOI] [PubMed] [Google Scholar]
- 18.Linial M L, Miller A D. Retroviral RNA packaging: sequence requirements and implications. Curr Top Microbiol Immunol. 1990;157:125–152. doi: 10.1007/978-3-642-75218-6_5. [DOI] [PubMed] [Google Scholar]
- 19.Lochelt M, Fluegel R M. The molecular biology of human and primate spuma retroviruses. In: Levy J A, editor. The Retroviridae. Vol. 4. New York, N.Y: Plenum Press; 1995. pp. 239–292. [Google Scholar]
- 20.Löchelt M, Flügel R M, Aboud M. The human foamy virus internal promoter directs the expression of the functional Bel 1 transactivator and Bet protein early after infection. J Virol. 1994;68:638–645. doi: 10.1128/jvi.68.2.638-645.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lochelt M, Muranyi W, Flugel R M. Human foamy virus genome possesses an internal, Bel-1-dependent and functional promoter. Proc Natl Acad Sci USA. 1993;90:7317–7321. doi: 10.1073/pnas.90.15.7317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lochelt M, Yu S F, Linial M L, Flugel R M. The human foamy virus internal promoter is required for efficient gene expression and infectivity. Virology. 1995;206:601–610. doi: 10.1016/s0042-6822(95)80077-8. [DOI] [PubMed] [Google Scholar]
- 23.Loh P C. Spumaviruses. In: Levy J A, editor. The Retroviridae. Vol. 2. New York, N.Y: Plenum Press; 1993. pp. 361–397. [Google Scholar]
- 24.Malmquist W A, Van Der Maaten M J, Boothe A D. Isolation, immunodiffusion, immunofluorescence, and electron microscopy of a syncytial virus of lymphosarcomatous and apparently normal cattle. Cancer Res. 1969;29:188–200. [PubMed] [Google Scholar]
- 25.Marino S, Kretschmer C, Bradner S, Cavard C, Zider A, Briand P, Flsenmann S, Wagner E F, Aguzzi A. Activation of HIV transcription by human foamy virus in transgenic mice. Lab Invest. 1995;73:103–110. [PubMed] [Google Scholar]
- 26.Maurer B, Bannert H, Darai G, Flügel R M. Analysis of the primary structure of the long terminal repeat and the gag and pol genes of the human spumaretrovirus. J Virol. 1988;62:1590–1597. doi: 10.1128/jvi.62.5.1590-1597.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McClure M O, Bieniasz P D, Schulz T F, Chrystie I L, Simpson G, Aguzzi A, Hoad J G, Cunningham A, Kirkwood J, Weiss R A. Isolation of a new foamy retrovirus from orangutans. J Virol. 1994;68:7124–7130. doi: 10.1128/jvi.68.11.7124-7130.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McCulloch R K, Choong C S, Hurley D M. An evaluation of competitor type and size for use in the determination of mRNA by competitive PCR. PCR Methods Appl. 1995;4:219–226. doi: 10.1101/gr.4.4.219. [DOI] [PubMed] [Google Scholar]
- 29.Mergia A. Simian foamy virus type 1 contains a second promoter located at the 3′ end of the env gene. Virology. 1994;199:219–222. doi: 10.1006/viro.1994.1114. [DOI] [PubMed] [Google Scholar]
- 30.Netzer K, Rethwilm A, Maurer B, ter Meulen V. Identification of the major immunogenic structural proteins of human foamy virus. J Gen Virol. 1990;71:1237–1241. doi: 10.1099/0022-1317-71-5-1237. [DOI] [PubMed] [Google Scholar]
- 31.Neumann-Haefelin D, Fleps U, Renne R, Schweizer M. Foamy viruses. Intervirology. 1993;35:196–207. doi: 10.1159/000150310. [DOI] [PubMed] [Google Scholar]
- 32.Pahl A, Flugel R M. Characterization of the human spuma retrovirus integrase by site-directed mutagenesis, by complementation analysis, and by swapping the zinc finger domain of HIV-1. J Biol Chem. 1995;270:2957–2966. doi: 10.1074/jbc.270.7.2957. [DOI] [PubMed] [Google Scholar]
- 33.Rao J K M, Erickson J W, Wlodawer A. Structural and evolutionary relationships between retroviral and eucaryotic aspartic proteinases. Biochemistry. 1991;30:4663–4671. doi: 10.1021/bi00233a005. [DOI] [PubMed] [Google Scholar]
- 34.Renne R, Fleps U, Luciw P A, Neumann-Haefelin D. Transactivation of the two promoters of SFV-3 by different mechanisms. Virology. 1996;221:362–367. doi: 10.1006/viro.1996.0387. [DOI] [PubMed] [Google Scholar]
- 35.Renne R, Friedl E, Schweizer M, Fleps U, Turek R, Neumann-Haefelin D. Genomic organization and expression of simian foamy virus type 3 (SFV-3) Virology. 1992;186:597–608. doi: 10.1016/0042-6822(92)90026-l. [DOI] [PubMed] [Google Scholar]
- 36.Renshaw R W, Casey J W. Analysis of the 5′ long terminal repeat of bovine syncytial virus. Gene. 1994;141:221–224. doi: 10.1016/0378-1119(94)90575-4. [DOI] [PubMed] [Google Scholar]
- 37.Renshaw R W, Casey J W. Transcriptional mapping of the 3′ end of the bovine syncytial virus genome. J Virol. 1994;68:1021–1028. doi: 10.1128/jvi.68.2.1021-1028.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Renshaw R W, Gonda M A, Casey J W. Structure and transcriptional status of bovine syncytial virus in cytopathic infections. Gene. 1991;105:179–184. doi: 10.1016/0378-1119(91)90149-6. [DOI] [PubMed] [Google Scholar]
- 39.Rethwilm, A. 1996. Unexpected replication pathways of foamy viruses. J. Acquired Immune Defic. Syndr. Hum. Retrovirol. 13(Suppl. 1):S248–S253. [DOI] [PubMed]
- 40.Russell D W, Miller A D. Foamy virus vectors. J Virol. 1996;70:217–222. doi: 10.1128/jvi.70.1.217-222.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Saitou N, Nei M. The neighbor joining method: a new method of reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–525. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 42.Schliephake A W, Rethwilm A. Nuclear localization of foamy virus Gag precursor protein. J Virol. 1994;68:4946–4954. doi: 10.1128/jvi.68.8.4946-4954.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schmidt M, Rethwilm A. Replicating foamy virus-based vectors directing high level expression of foreign genes. Virology. 1995;210:167–178. doi: 10.1006/viro.1995.1328. [DOI] [PubMed] [Google Scholar]
- 44.Schweizer M, Neumann-Haefelin D. Phylogenetic analysis of primate foamy viruses by comparison of pol sequences. Virology. 1995;207:577–582. doi: 10.1006/viro.1995.1120. [DOI] [PubMed] [Google Scholar]
- 45.Tschopp R R, Brandner S, Marino S, Bothe K, Horak I, Rethwilm A, Aguzzi A. Analysis of the determinants of neurotropism and neurotoxicity of HFV in transgenic mice. Virology. 1996;216:338–346. doi: 10.1006/viro.1996.0069. [DOI] [PubMed] [Google Scholar]
- 46.Yu S F, Baldwin D N, Gwynn S R, Yendapalli S, Linial M L. Human foamy virus replication: a pathway distinct from that of retroviruses and hepadnaviruses. Science. 1996;271:1579–1582. doi: 10.1126/science.271.5255.1579. [DOI] [PubMed] [Google Scholar]
- 47.Yu S F, Edelmann K, Strong R K, Moebes A, Rethwilm A, Linial M L. The carboxy terminus of the human foamy virus Gag protein contains separable nucleic acid binding and nuclear transport domains. J Virol. 1996;70:8255–8262. doi: 10.1128/jvi.70.12.8255-8262.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]