Abstract
The hepatitis C virus (HCV) genome encodes two envelope glycoproteins (E1 and E2). These glycoproteins interact to form a noncovalent heterodimeric complex which is retained in the endoplasmic reticulum (ER). To identify whether E1 and/or E2 contains an ER-targeting signal potentially involved in ER retention of the E1-E2 complex, these proteins were expressed alone and their intracellular localization was studied. Due to misfolding of E1 in the absence of E2, no conclusion on the localization of its native form could be drawn from the expression of E1 alone. E2 expressed in the absence of E1 was shown to be retained in the ER similarly to E1-E2 complex. Chimeric proteins in which E2 domains were exchanged with corresponding domains of a protein normally transported to the plasma membrane (CD4) were constructed to identify the sequence responsible for its ER retention. The transmembrane domain (TMD) of E2 (C-terminal 29 amino acids) was shown to be sufficient for retention of the ectodomain of CD4 in the ER compartment. Replacement of the E2 TMD by the anchor signal of CD4 or a glycosyl phosphatidylinositol (GPI) moiety led to its expression on the cell surface. In addition, replacement of the E2 TMD by the anchor signal of CD4 or a GPI moiety abolished the formation of E1-E2 complexes. Together, these results suggest that, besides having a role as a membrane anchor, the TMD of E2 is involved in both complex formation and intracellular localization.
Hepatitis C virus (HCV) is an enveloped virus which belongs to the Flaviviridae family (15). Its genome encodes two membrane-associated envelope glycoproteins (E1 and E2). E1 and E2 glycoproteins interact to form a complex which has been proposed as a functional subunit of HCV virions (11, 17, 26, 41). Characterization of HCV glycoprotein complex formation expressed by using the vaccinia/T7 or Sindbis virus system indicates that a majority of HCV glycoproteins are misfolded (9, 11). Recently, we have produced a monoclonal antibody (MAb) which recognizes properly folded E2 and precipitates native HCV glycoprotein complexes but not misfolded aggregates (9). Properly folded E1 and E2 interact to form a heterodimer stabilized by noncovalent interactions, and the kinetics of association between E1 and E2 indicate that the formation of stable E1-E2 complexes is slow (half-time of association [t1/2] ≈ 2 h). The folding of E1 and E2 has been studied and indicates that formation of intramolecular disulfide bonds is slow for E1 (t1/2 > 1 h) whereas it is rapid for E2 (12). By using human and mouse MAbs, it has been shown that folding of a subdomain(s) of E2 correlates with acquisition of intramolecular disulfide bonds but that complete folding of E2 is slow (t1/2 ≈ 2 h) (9, 19). In addition, E1 expressed in the absence of E2 does not fold properly, suggesting that E2 plays a chaperone-like role in the folding of E1 (32).
The HCV glycoproteins are heavily modified by N-linked glycosylation and contain hydrophobic domains in their carboxy-terminal regions acting presumably as membrane anchors, giving the proteins a type I membrane topology (43). The E2 glycoprotein extends to residue 746 (position on the polyprotein), and deletions of at least 31 C-terminal amino acids lead to its secretion (47). This is in accordance with other data proposing that the hydrophobic anchor domain begins at amino acid 718 (33). However, only a shorter secreted form of E2 glycoprotein ending at amino acid 661 appears to be properly folded (32). For E1, a larger deletion (71 amino acids) seems to be necessary for its secretion, but this secreted protein is not properly folded (32).
Due to the lack of an efficient cell culture replication system, HCV particle assembly and release have not been examined directly. However, the lack of complex glycans, the endoplasmic reticulum (ER) localization of expressed HCV glycoproteins (11, 41), and the absence of these proteins on the cell surface (11, 49) suggest that initial virion morphogenesis may occur by budding into intracellular vesicles. More recently, we have confirmed that the mature E1-E2 heterodimer does not leave the ER, suggesting that E1 and/or E2 contains a signal for retention of the heterodimer in this compartment (9).
In this study, we show that E2 glycoprotein expressed alone is retained in the ER similarly to the E1-E2 heterodimer and that a signal for ER retention of E2 resides in its transmembrane domain (TMD) (C-terminal 29 amino acids). The evidence for this retention signal was derived from expression of chimeric proteins in which E2 domains were exchanged with corresponding domains of a protein normally transported to the plasma membrane (CD4).
MATERIALS AND METHODS
Cell culture.
The HepG2, HeLa, CV-1, and 143B (thymidine kinase-deficient) cell lines were obtained from the American Type Culture Collection, Rockville, Md. Cell monolayers were grown in Dulbecco’s modified essential medium (Gibco BRL) supplemented with 5% fetal bovine serum.
Plasmid constructs.
Plasmids expressing chimeric proteins were constructed by a standard method (45). Briefly, DNA sequences of protein domains were introduced into plasmid pTM1 (35) by PCR with appropriate oligonucleotides and templates. Plasmids expressing chimeric proteins were constructed in two steps by introducing successively the sequences of domains from two different proteins. A unique restriction site was introduced at the junction between the sequences derived from the two proteins for cloning facility. HCV sequences were amplified from H strain (13) clones. Plasmids pTM1/E2(371-661)-DAF and pTM1/E2(371-717)-DAF express E2 sequences with a glycosyl phosphatidylinositol (GPI) anchor. These two plasmids contain the signal sequence of E2 and the sequence of the ectodomain of E2 or part of it in fusion with the last 37 amino acids of decay-accelerating factor (DAF). Between these two sequences, there is a junction sequence encoding two additional amino acids (Thr and Arg). Plasmids pTM1/E2(371-661)-CD4(374-435) and pTM1/E2(371-717)-CD4(374-435) contain the signal sequence of E2 and the sequence of the ectodomain of E2 or part of it in fusion with the C-terminal 62 amino acids of CD4. Between these two sequences, there is a junction sequence encoding two additional amino acids (Thr and Arg). Plasmids pTM1/CD4(1-371)-E2(662-746) and pTM1/CD4(1-371)-E2(718-746) contain the signal sequence of CD4 followed by the sequence of its ectodomain in fusion with the C-terminal 85 or 29 amino acids of E2. Between these two sequences, there is a junction sequence encoding two additional amino acids (Gly and Ser). Plasmid pTM1/CD4 contains the sequence of the entire CD4 glycoprotein (amino acids 1 to 435) and its signal sequence. Plasmids containing sequences amplified by PCR were verified by sequencing.
Generation and growth of viruses.
Vaccinia virus recombinants were generated by homologous recombination essentially as described previously (24) and plaque purified twice on thymidine kinase-deficient 143B cells under bromodeoxyuridine (50 μg/ml) selection. Stocks of vTF7-3 (a vaccinia virus recombinant expressing the T7 DNA-dependent RNA polymerase) (16), the wild-type vaccinia virus strain, Copenhagen, and its thermosensitive derivative ts7 (10), and vaccinia virus recombinants expressing HCV proteins or chimeric proteins were grown and titrated on CV-1 cell monolayers.
Vaccinia virus recombinants vHCV170-809, vHCV371-809, vHCV371-661, and vHCV1-383 have been described previously (14, 32).
Antibodies.
MAbs H53 (anti-E2 [40]) and OKT4 (anti-CD4 [42]) were used in this work. Concentrated MAbs were produced in vitro by using a MiniPerm apparatus (Heraeus) as recommended by the manufacturer.
Metabolic labeling and immunoprecipitation.
Cells were infected with the appropriate vaccinia virus recombinants and metabolically labeled with Tran35S-label (ICN) as previously described (11). Labeled infected cells were then lysed with 0.5% Triton X-100 in 20 mM Tris-Cl (pH 7.5)–150 mM NaCl–2 mM EDTA. Immunoprecipitations were carried out as described previously (12).
Endoglycosidase digestions.
Immunoprecipitated proteins were eluted from protein A-Sepharose in 30 μl of dissociation buffer (0.5% sodium dodecyl sulfate [SDS] and 1% 2-mercaptoethanol) by boiling for 10 min. The protein samples were then divided into two equal portions for digestion with endo-β-N-acetylglucosaminidase H (endo H; New England Biolabs) or no digestion (control). Digestions were carried out for 1 h at 37°C in the buffer provided by the manufacturer. Digested samples were mixed with an equal volume of 2× Laemmli sample buffer and analyzed by SDS-polyacrylamide gel electrophoresis (PAGE).
PI-PLC treatment.
Phosphatidylinositol-specific phospholipase C (PI-PLC) digestions were performed on HeLa cells infected by the appropriate vaccinia virus recombinants and labeled with [35S]methionine. At 24 h postinfection, infected cells were washed twice with phosphate-buffered saline (PBS) at 4°C and resuspended by pipetting. Cells were then incubated with 0.5 U of PI-PLC (Boehringer-Mannheim) per ml in PBS for 1 h at 37°C. Cells and supernatant were harvested and analyzed by immunoprecipitation to detect glycoprotein expression.
Immunofluorescence.
Subconfluent HepG2 or CV-1 cells grown on coverslips were infected by the appropriate vaccinia virus recombinants at a multiplicity of infection of 1 PFU/cell. At 12 h postinfection, cells were fixed for 10 min with isopropanol or paraformaldehyde (4% in PBS). Cells fixed with paraformaldehyde were permeabilized or not for 30 min at room temperature with Tris-buffered saline containing 0.1% Triton X-100. Cells were stained with anti-E2 MAb H53 (dilution, 1/600) or anti-CD4 MAb OKT4 (dilution, 1/100) followed by rabbit anti-mouse (rhodamine conjugated; DAKO) or donkey anti-mouse (fluorescein isothiocyanate [FITC]-conjugated; Jackson) immunoglobulin (dilution, 1/100).
Flow cytometric analysis.
For detection of cell surface expression of chimeric proteins, HeLa cells grown in six-well plates were infected with the appropriate vaccinia virus recombinants at a multiplicity of infection of 5 PFU/cell. At 8 h postinfection, cells were washed twice with PBS at 4°C and resuspended by pipetting. Cells infected by recombinants expressing the ectodomain of E2 were stained with anti-E2 MAb H53 (dilution, 1/600) followed by FITC-conjugated rabbit anti-mouse immunoglobulin (dilution, 1/100; DAKO). Cells infected by recombinants expressing the ectodomain of CD4 were stained with FITC-conjugated anti-CD4 MAb 13B8.2 (dilution, 1/100; Immunotech). Stained cells were fixed in PBS–1% paraformaldehyde for 30 min, resuspended in 1 ml of PBS, and subjected to flow cytometric analysis with an Epics-Profile II (Coulter). For each sample, 104 cells were analyzed.
RESULTS
Intracellular localization of E2 expressed in the absence of E1.
In order to identify the signal(s) involved in ER retention of the native HCV glycoprotein complex, E1 and E2 were expressed alone. Immunofluorescence studies and analysis of its endo H sensitivity indicate that E1 expressed in the absence of E2 does not leave the ER (data not shown). However, we have shown recently that coexpression with E2 is required for the proper folding of E1 (32). Therefore, when E1 is expressed alone, we cannot differentiate an ER localization due to a specific signal from a retention caused by misfolding of the protein (20). Due to this problem and since E2 folds properly in the absence of E1 (32), only the intracellular localization of E2 was studied in this work.
A conformation-sensitive E2-reactive MAb which recognizes the properly folded form of E2 (H53 [40]) was used in this work in order to reduce the background due to the presence of misfolded proteins. In pulse-chase experiments, this MAb precipitates E2 with kinetics similar to what has been observed previously with another MAb (MAb H2 [9]). However, since its relative affinity is higher, MAb H53 instead of H2 was used in this study.
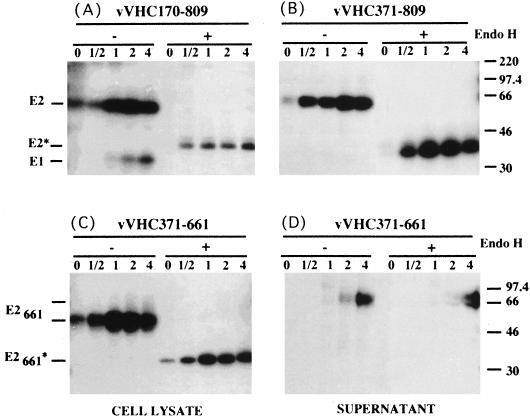
Immunofluorescence studies indicated that the intracellular localization of E2 in the presence of E1 is similar to that in the absence of E1 and that the proteins showed an ER-like distribution of fluorescence (Fig. 1). As another indicator of intracellular trafficking of E2, we also examined the acquisition of endo H resistance. Endo H removes the chitobiose core of high-mannose and some hybrid forms of N-linked sugars but not the complex forms (44). Resistance to digestion with endo H is indicative that glycoproteins have moved from the ER to at least the medial Golgi apparatus and trans-Golgi apparatus, where complex sugars are added. The sensitivity of E2 to digestion with endo H was studied in pulse-chase experiments (Fig. 2). Similar to what was observed for E2 coexpressed with E1 (Fig. 2A), no endo H-resistant form was detected for E2 expressed in the absence of E1 even after 4 h of chase (Fig. 2B). This suggests that E2 expressed in the absence of E1 does not reach the medial Golgi apparatus and trans-Golgi apparatus. To confirm that E2 glycans can be modified by enzymes present in the medial Golgi apparatus or trans-Golgi apparatus, the sensitivity to digestion with endo H was analyzed for a truncated form of E2 which is secreted (32) (Fig. 2 C and D).
FIG. 1.
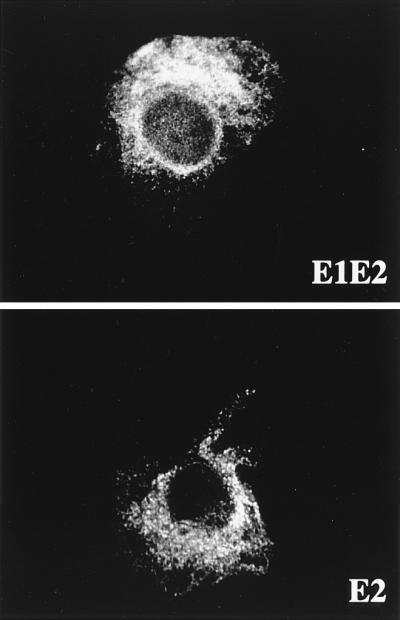
Localization by indirect immunofluorescence of E2, expressed in the presence or absence of E1. CV-1 cells were coinfected with vTF7-3 and either vHCV170-809 (E1-E2) or vHCV371-809 (E2) at a multiplicity of infection of 1 PFU/cell. Cells were fixed with isopropanol at 12 h postinfection and immunostained with anti-E2 MAb H53.
FIG. 2.
Sensitivity of E2 to endo H treatment. HepG2 cells were coinfected with vTF7-3 and vaccinia virus recombinants expressing either E1-E2 (vHCV170-809), E2 (vHCV371-809), or a truncated form of E2 (vHCV371-661) at a multiplicity of infection of 5 PFU/cell. Infected cells were pulse-labeled for 10 min and chased for the indicated times (in hours). Cell lysates were immunoprecipitated with MAb H53 and then treated or not with endo H. Samples were separated by SDS-PAGE (10% polyacrylamide). Deglycosylated proteins are indicated by asterisks. The sizes (in kilodaltons) of protein molecular mass markers are indicated on the right. The predicted sequence of E2 contains 11 N-linked potential glycosylation sites.
Together, these results, similar to what has been previously observed for native E1-E2 complexes (9), strongly suggest that HCV glycoprotein E2 expressed in the absence of E1 is retained in the ER at steady state and does not cycle farther than the cis-Golgi apparatus. This also suggests that an ER retention signal is present in E2.
The TMD of E2 is sufficient to retain, in the ER, the ectodomain of a protein which is normally expressed on the cell surface.
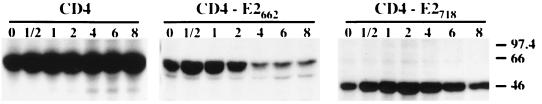
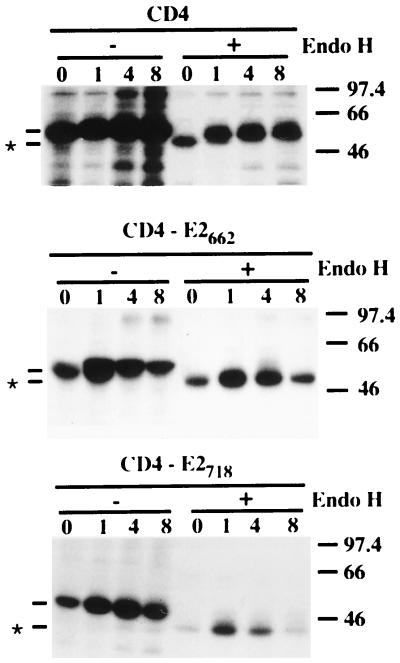
The ER localization of E2 (see above) as well as the secretion of a properly folded truncated form of E2 (32) suggests that an ER retention signal is present in the C-terminal 85 amino acids of E2. In order to test this hypothesis, a chimeric protein containing the ectodomain of CD4 (a protein normally exported to the cell surface) and the C-terminal 85 amino acids of E2 was constructed (CD4-E2662 [Fig. 3]). In addition, since secretion of E2 proteins with shorter deletions has also been observed by others (47), a second chimeric protein containing the C-terminal 29 amino acids of E2 in fusion with the ectodomain of CD4 was constructed (CD4-E2718 [Fig. 3]). Expression of these chimeric proteins was analyzed in pulse-chase experiments followed by immunoprecipitation with an anti-CD4 MAb (OKT4) and compared with the expression of CD4. As shown in Fig. 4, neither CD4 nor the chimeric proteins showed any modification in their electrophoretic mobility over time. However, the intensity of the chimeric proteins started to decrease after 2 and 4 h for CD4-E2662 and CD4-E2718, respectively, whereas it remained constant for CD4, suggesting that the chimeric proteins have a shorter half-life than CD4. This is probably due to degradation. To evaluate whether or not they leave the ER, the sensitivity of these chimeric proteins to digestion by endo H was analyzed in pulse-chase experiments (Fig. 5). The CD4 protein contains two N-linked glycans, and only one of them becomes endo H resistant (48). During the pulse, CD4 was sensitive to endo H treatment, and its resistant form was detected after 1 h of chase or more, whereas the chimeric proteins remained endo H sensitive even after 8 h of chase (Fig. 5). This suggests that CD4-E2662 and CD4-E2718 do not reach the trans-Golgi apparatus.
FIG. 3.
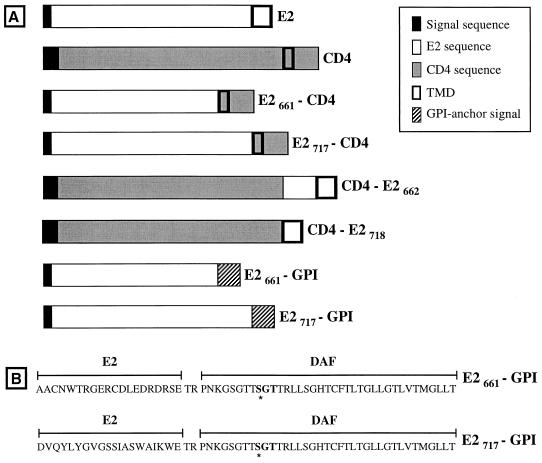
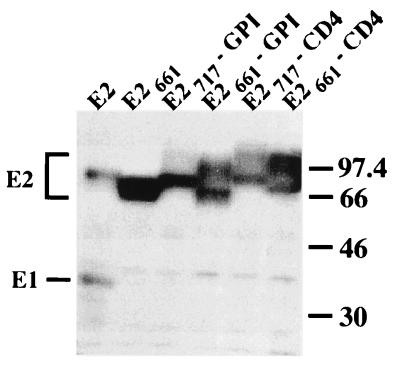
Schematic representation of the parental proteins E2 and CD4 and chimeric proteins used in this study drawn to scale. E2661-CD4, ectodomain of E2, lacking its C-terminal 56 amino acids, fused to the TMD and cytoplasmic domain of CD4; E2717-CD4, ectodomain of E2 fused to the TMD and cytoplasmic domain of CD4; CD4-E2662, ectodomain of CD4 fused to the C-terminal 85 amino acids of E2; CD4-E2718, ectodomain of CD4 fused to the TMD of E2 (C-terminal 29 amino acids); E2661-GPI, ectodomain of E2, lacking its C-terminal 56 amino acids, fused to the C-terminal 37 amino acids of DAF; E2717-GPI, ectodomain of E2 fused to the C-terminal 37 amino acids of DAF. The ectodomain of E2 is from amino acid 384 to 717 (position on the polyprotein) and the TMD of E2 from amino acid 718 to 746. The ectodomain of CD4 is from amino acid 1 to 373, its TMD is from amino acid 374 to 395, and its cytosolic domain is from amino acid 396 to 435. Details on constructions are reported in Materials and Methods. (B) C-terminal portions of E2661-GPI and E2717-GPI. The amino acid sequences identify the junction between the E2 ectodomain or its truncated form and the 37-amino-acid GPI anchor addition sequence from DAF. Between these two sequences, there is a sequence of two additional amino acids resulting from cloning. The asterisk below the Ser in the DAF sequence indicates the predicted point of attachment for the GPI anchor (34).
FIG. 4.
Expression of CD4, CD4-E2662, and CD4-E2718 analyzed in pulse-chase experiments. HepG2 cells were coinfected with vTF7-3 and the appropriate vaccinia virus recombinant at a multiplicity of infection of 5 PFU/cell. Infected cells were pulse-labeled for 10 min and chased for the indicated times (in hours). Cell lysates were immunoprecipitated with MAb OKT4 (anti-CD4). Samples were separated by SDS-PAGE (10% polyacrylamide). The sizes (in kilodaltons) of protein molecular mass markers are indicated on the right.
FIG. 5.
Sensitivities of CD4-E2662 and CD4-E2718 to endo H treatment. HepG2 cells were coinfected with vTF7-3 and the appropriate vaccinia virus recombinant at a multiplicity of infection of 5 PFU/cell. Infected cells were pulse-labeled for 10 min and chased for the indicated times (in hours). Cell lysates were immunoprecipitated with MAb OKT4 and then treated or not with endo H. Samples were separated by SDS-PAGE (10% polyacrylamide). Deglycosylated proteins are indicated by asterisks. The sizes (in kilodaltons) of protein molecular mass markers are indicated on the right.
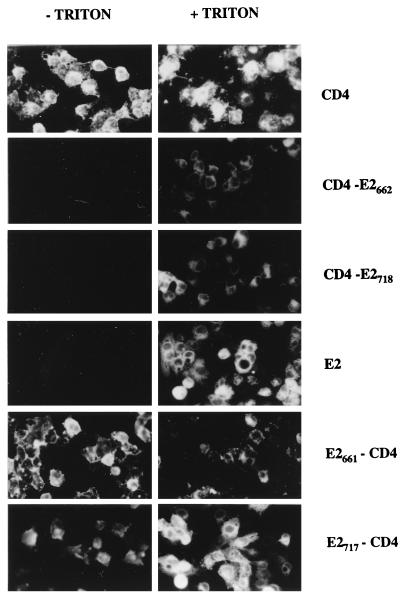
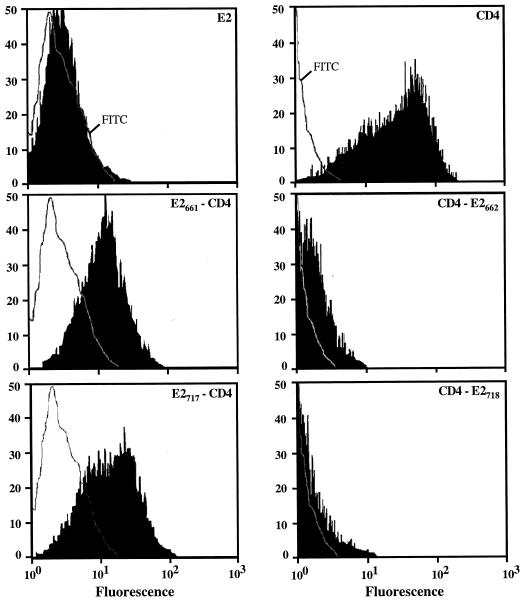
Cell surface expression of these proteins was analyzed by immunofluorescence and flow cytometry. Cells expressing CD4, CD4-E2662, or CD4-E2718 and fixed with paraformaldehyde were all positive as determined by immunofluorescence after permeabilization with Triton X-100, whereas only CD4-expressing cells were detected in the absence of detergent (Fig. 6). The absence of detectable CD4-E2662 and CD4-E2718 on the surface of cells infected by vaccinia virus recombinants expressing these proteins was confirmed by flow cytometry (Fig. 7).
FIG. 6.
Cell surface expression of chimeric proteins analyzed by indirect immunofluorescence. HepG2 cells were coinfected with vTF7-3 and the appropriate vaccinia virus recombinant at a multiplicity of infection of 1 PFU/cell. Cells were fixed with paraformaldehyde at 12 h postinfection, permeabilized or not with Triton X-100, and immunostained with anti-E2 (E2, E2661-CD4, and E2717-CD4) or anti-CD4 (CD4, CD4-E2662, and CD4-E2718) antibodies.
FIG. 7.
Cell surface expression of chimeric proteins analyzed by flow cytometry. HeLa cells were coinfected with vTF7-3 and the appropriate vaccinia virus recombinant at a multiplicity of infection of 5 PFU/cell. At 8 h postinfection, cells were immunostained with anti-E2 (E2, E2661-CD4, and E2717-CD4) MAb H53 followed by FITC-conjugated rabbit anti-mouse immunoglobulin or with FITC-conjugated anti-CD4 MAb 13B8.2 (CD4, CD4-E2662, and CD4-E2718). Stained cells were fixed in PBS–1% paraformaldehyde before flow cytometric analysis. The level of cell surface expression is indicated by the shift of the solid histogram (MAb H53 plus FITC conjugate or MAb 13B8.2) to the right from the open control histogram (FITC conjugate alone for E2, E2661-CD4, and E2717-CD4; MAb 13B8.2 on uninfected cells for CD4, CD4-E2662, and CD4-E2718).
Together, these data indicate that the TMD of E2 plays a major role in its retention in the ER.
Replacement of the TMD of E2 by the anchor and cytoplasmic domains of CD4 leads to E2 expression on the cell surface.
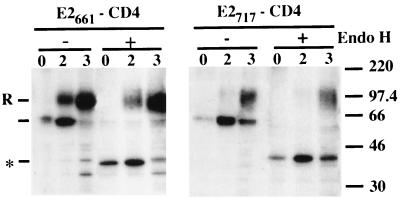
To evaluate the absence of an additional retention signal in the ectodomain of E2, other chimeric proteins containing the ectodomain of E2 ending at amino acid 661 (E2661-CD4) or 717 (E2717-CD4) in fusion with the C-terminal 62 amino acids of CD4 were constructed (Fig. 3). These chimeric proteins were analyzed as described above with a conformation-sensitive MAb (H53) which recognizes an epitope present in the ectodomain of E2. In pulse-chase experiments, two bands were detected after immunoprecipitation of the chimeric proteins (Fig. 8). The glycoprotein E2 and the fast-migrating forms E2661-CD4 and E2717-CD4 showed similar molecular masses. The intensity of this band increased during the first 2 and 4 h of chase for E2661-CD4 and E2717-CD4, respectively, and then decreased. The slow-migrating band, not detected for E2, appeared after 1 and 2 h for E2661-CD4 and E2717-CD4, respectively, and peaked after 4 h of chase before starting to decrease in intensity. The slow migration of this band is probably due to modification of the glycans present in the chimeric proteins. Indeed, it was endo H resistant (Fig. 9), suggesting that E2661-CD4 and E2717-CD4 reach at least the medial Golgi apparatus or trans-Golgi apparatus. It has to be noted that the slow-migrating band had a lower intensity for E2717-CD4 (see Discussion).
FIG. 8.
Expression of E2, E2661-CD4, and E2717-CD4 analyzed in pulse-chase experiments. HepG2 cells were coinfected with vTF7-3 and the appropriate vaccinia virus recombinant at a multiplicity of infection of 5 PFU/cell. Infected cells were pulse-labeled for 10 min and chased for the indicated times (in hours). Cell lysates were immunoprecipitated with MAb H53 (anti-E2). Samples were separated by SDS-PAGE (10% polyacrylamide). The sizes (in kilodaltons) of protein molecular mass markers are indicated on the right.
FIG. 9.
Sensitivities of E2661-CD4 and E2717-CD4 to endo H treatment. HepG2 cells were coinfected with vTF7-3 and the appropriate vaccinia virus recombinant at a multiplicity of infection of 5 PFU/cell. Infected cells were pulse-labeled for 10 min and chased for the indicated times (in hours). Cell lysates were immunoprecipitated with MAb H53 and then treated or not with endo H. Samples were separated by SDS-PAGE (10% polyacrylamide). Deglycosylated proteins are indicated by an asterisk, and endo H-resistant forms are indicated by an “R.” The sizes (in kilodaltons) of protein molecular mass markers are indicated on the right.
Cell surface expression of these chimeric proteins was analyzed by immunofluorescence and flow cytometry. Cells expressing E2, E2661-CD4, or E2717-CD4 and fixed with paraformaldehyde were all positive as determined by immunofluorescence after permeabilization with Triton X-100, whereas only E2661-CD4- or E2717-CD4-expressing cells were detected in the absence of detergent (Fig. 6). Cell surface expression of these chimeric proteins was confirmed by flow cytometry (Fig. 7).
Together, these results indicate that replacement of the TMD of E2 by the anchor and cytoplasmic domains of CD4 leads to E2 expression on the cell surface.
Replacement of the E2 TMD by a GPI moiety leads to expression of E2 on the cell surface.
Many cellular proteins are embedded in membranes via a GPI anchor (8). To evaluate the influence of a GPI anchor on the cellular localization of the ectodomain of E2, chimeric proteins with a signal for a GPI anchor were constructed. Replacement of the normal TMD and cytoplasmic domain of several other type I integral membrane glycoproteins with the C-terminal 37 amino acids of DAF confers GPI anchor addition (7, 23, 25, 27). E2661-GPI and E2717-GPI were constructed by appending the 37-amino-acid signal of DAF to the ectodomain of E2 (E2717-GPI) or a shorter form thereof (E2661-GPI) (Fig. 3).
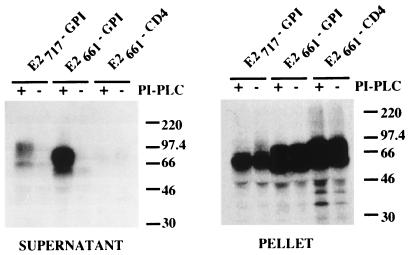
In order to show that a GPI molecule had indeed been added and was responsible for anchoring the ectodomain of E2 in the cell membrane, the sensitivities of these chimeric proteins to PI-PLC digestion were evaluated (Fig. 10). In the absence of PI-PLC treatment E2661-GPI and E2717-GPI were not detected in the supernatant, whereas after PI-PLC treatment released proteins could be precipitated by our E2-reactive MAb but not in the control expressing E2661-CD4. Intracellular expression of the recombinant proteins has been confirmed by immunoprecipitation with the cell lysate as antigen (Fig. 10). These data indicate that E2661-GPI and E2717-GPI are anchored to the membrane via a GPI moiety and that their ectodomains can be released by digestion with PI-PLC. These two proteins were studied in pulse-chase experiments to test their sensitivities to digestion by endo H and examined by immunofluorescence to analyze their expression on the cell surface. Results were very similar to what was observed for E2661-CD4 and E2717-CD4 (data not shown), indicating that the addition of a GPI moiety to the ectodomain of E2 leads to the expression of E2 on the cell surface.
FIG. 10.
Sensitivities of E2661-GPI and E2717-GPI to PI-PLC treatment. HeLa cells coinfected with vTF7-3 and the appropriate vaccinia virus recombinant were labeled from 4 to 20 h postinfection. Cells were then washed and incubated in PBS in the presence (+) or absence (−) of PI-PLC for 1 h at 37°C. Supernatants and cell pellets were harvested separately and prepared for immunoprecipitation with MAb H53. Samples were separated by SDS-PAGE (10% polyacrylamide). The sizes (in kilodaltons) of protein molecular mass markers are indicated on the right.
Replacement of the TMD of E2 abolishes the formation of E1-E2 complexes.
Results obtained in this work strongly suggest that an ER retention signal is present in the TMD of E2. However, we cannot exclude the presence of another signal in E1 which could participate in ER retention of the native E1-E2 complex. The presence of a potential retention signal has not been studied in this work, since coexpression with E2 is required for the proper folding of E1 (32). As an alternative way to study whether a potential retention signal in E1 could also be involved in retaining the native E1-E2 complex in the ER, E1 was coexpressed with the chimeric forms of E2 produced in this work. However, E1 did not interact with any of these chimeric proteins as shown by immunoprecipitation (Fig. 11), suggesting that the TMD of E2 is involved in HCV glycoprotein interactions, as previously proposed (32, 47).
FIG. 11.
Effect of E2 TMD replacement on the formation of E1-E2 complexes. HepG2 cells were coinfected with vTF7-3, vHCV1-383 (expressing E1), and the appropriate vaccinia virus recombinant at a multiplicity of infection of 5 PFU/cell. Infected cells were labeled from 4 to 20 h postinfection. Cell lysates were immunoprecipitated with MAb H53. Samples were separated by SDS-PAGE (10% polyacrylamide). The sizes (in kilodaltons) of protein molecular mass markers are indicated on the right.
DISCUSSION
Enveloped viruses acquire their envelopes by budding through one of several host cellular membranes. A large number of enveloped viruses bud through the plasma membrane. Several viruses, however, bud at internal membranes, such as those of the ER (e.g., rotaviruses), the ER-Golgi apparatus intermediate compartment (coronaviruses), or the Golgi complex (Bunyaviridae) (18, 38). In these cases, virus particles are released from the infected cells either after cell lysis (e.g., rotaviruses) or after transport of virus-containing vesicles to the cell surface (e.g., coronaviruses and bunyaviruses), in which case virus particles are released after fusion of these vesicles with the plasma membrane. For viruses which bud intracellularly, there needs to be an accumulation of the viral membrane glycoproteins, which form the spikes, in the appropriate compartment (38). A strategy that most of these viruses have developed is to endow the spike proteins with signals for compartment-specific targeting and retention, similar to normal compartment-specific cellular proteins (2–4, 21, 29, 30, 50). Based on this observation, ER retention of HCV glycoprotein complexes suggests that budding of HCV particles could occur in the ER. This hypothesis is reinforced by the observation that flaviviruses acquire their envelopes by budding at internal membranes (probably modified ER) (reviewed in reference 38). The identification of retention signals is therefore important for understanding the mechanisms of virus budding and protein compartmentalization. In this study, we showed that E2 glycoprotein expressed in the absence of E1 is retained in the ER and we have identified a signal for ER retention of E2 in its TMD (C-terminal 29 amino acids).
HCV glycoprotein E2 contains an ER retention signal. The lack of complex glycans, the intracellular distribution of E2 analyzed by immunofluorescence, and the absence of its expression on the cell surface indicate that E2 is present in the ER at steady state and does not cycle farther than the cis-Golgi apparatus. This is similar to what has been observed for the native E1-E2 complex (9). Because of the inefficient folding of HCV glycoproteins (9, 12), retention of E2 in the ER could be due to misfolding. As a general rule, newly synthesized proteins that have acquired a properly folded structure are transported from the ER to their final destinations, whereas incompletely folded or misfolded proteins are retained and eventually degraded (20). This conformation-based sorting phenomenon has been called “quality control” (22). However, a conformation-sensitive MAb which recognizes the native form of E2 was used in this work. In addition, recent studies indicate that E2 expressed in the absence of E1 folds properly (32). This suggests that retention of E2 in the ER is not due to the pressure of the quality control but rather to a targeting signal present in E2.
The ER retention signal of E2 maps to its TMD. Replacement of the TMD of E2 with the anchor sequence of CD4 or a GPI moiety was sufficient for its export to the cell surface. In addition, a chimeric protein containing the ectodomain of CD4 fused to the TMD of E2 was retained in the ER. This indicates that the C-terminal 29 amino acids of E2 contain the information necessary for ER retention. However, the ratio of chimeric E2 proteins leaving the ER was improved when the C-terminal 56 amino acids of the ectodomain of E2 were removed. This suggests that this sequence can reinforce E2 retention in the ER. Alternatively, as recent data obtained by workers in our laboratory suggested (32), the presence of these 56 residues could reduce the efficiency of E2 folding, which, in turn, could lead to less efficient export out of the ER.
HCV glycoprotein E2 is retained in the ER by a targeting signal which does not have the features of a classical ER retention motif. Because the default pathway for the vectorial flow of proteins is supposed to lead from the ER to the plasma membrane (39), mechanisms are necessary to retain resident proteins within intracellular organelles. Several specific signals for the retention and retrieval of ER proteins have been identified. Retrieval from the Golgi complex of soluble ER resident proteins is mediated by recognition of the amino acid sequence KDEL by a specific receptor (37). For transmembrane proteins, intracytoplasmic sequences play a role in ER retention and retrieval. A dilysine motif at the C terminus of type I proteins allows for retrieval from the Golgi complex to the ER in a coat promoter-dependent manner (reviewed in reference 36); a diarginine N-terminal motif may play an analogous role for type II proteins (46), and a tyrosine-based motif has also been shown to function as a cytoplasmic ER retention signal (31). Here we report that the TMD of a type I protein can also be implicated in ER retention, as has been shown for some other proteins (1, 5, 51). Recently, it has been proposed that in the absence of dominant luminal or cytosolic associations, proteins are distributed based on interactions between their TMDs and the surrounding lipid environment (6, 51). Since cholesterol is known to increase membrane thickness and to decrease deformability, and because its concentration in lipid bilayers increases along the secretory pathway, it has been implicated as being the consequence of lipid-based sorting along the secretory pathway (6, 51). Our data on ER retention of E2 suggest that E2 could fit in this model.
The ER retention signal present in E2 could be sufficient to retain E1-E2 complexes in the ER, but we cannot exclude the presence of another signal in E1. Since coexpression with E2 is required for the proper folding of E1 (32), we cannot discriminate an ER localization of E1 determined by a specific signal from a retention due to misfolding of the protein (20). An alternative approach to solve this problem is to coexpress E1 with E2 in which the retention signal has been deleted. However, the interaction between E1 and E2 is abolished when E1 is coexpressed with E2 in which the TMD has been deleted (32, 47) or when it is coexpressed with chimeric proteins in which the TMD of E2 has been replaced by the anchor signal of CD4 or a GPI moiety (this work). Other approaches will therefore be necessary to analyze the presence of a potential retention signal in E1.
The TMD of HCV glycoprotein E2 is multifunctional. Besides anchoring E2 in the cell membrane and potentially in the HCV envelope (33), the TMD of E2 has other functions. Its C-terminal half is the signal sequence for the p7 polypeptide (28). It plays an important role in the interaction between HCV glycoproteins to form native E1-E2 complexes (references 32 and 47 and this work). Finally, as we show in this work, it is also responsible for the retention of E2 in the ER. This finding reinforces our hypothesis that HCV acquires its envelope by budding through ER membranes.
ACKNOWLEDGMENTS
We thank Françoise Jacob-Dubuisson for critical reading of the manuscript and Sophana Ung for excellent technical assistance. We are grateful to D. R. Littman and C. M. Rice for the gifts of plasmids containing the sequences for DAF and CD4, respectively.
This work was supported by the following grants: an ATIPE grant from the CNRS, a grant from the ARC (grant 1039), and a grant from the INSERM/AFS (INSERM grant 5FS10).
REFERENCES
- 1.Ahn K, Szczesna-Skorupa E, Kemper B. The amino-terminal 29 amino acids of cytochrome P450 2C1 are sufficient for retention in the endoplasmic reticulum. J Biol Chem. 1993;268:18726–18733. [PubMed] [Google Scholar]
- 2.Alconada A, Bauer U, Hoflack B. A tyrosine-based motif and a casein kinase II phosphorylation site regulate the intracellular trafficking of the varicella-zoster virus glycoprotein I, a protein localized in the trans-Golgi network. EMBO J. 1996;15:6096–6110. [PMC free article] [PubMed] [Google Scholar]
- 3.Andersson A M, Melin L, Bean A, Pettersson R F. A retention signal necessary and sufficient for Golgi localization maps to the cytoplasmic tail of a Bunyaviridae (Uukuniemi virus) membrane glycoprotein. J Virol. 1997;71:4717–4727. doi: 10.1128/jvi.71.6.4717-4727.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Armstrong J, Patel S. The Golgi sorting domain of coronavirus E1 protein. J Cell Sci. 1991;98:567–575. doi: 10.1242/jcs.98.4.567. [DOI] [PubMed] [Google Scholar]
- 5.Bonifacino J S, Cosson P, Shah N, Klausner R D. Role of potentially charged transmembrane residues in targeting proteins for retention and degradation within the endoplasmic reticulum. EMBO J. 1991;10:2783–2793. doi: 10.1002/j.1460-2075.1991.tb07827.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bretcher M S, Munro S. Cholesterol and Golgi apparatus. Science. 1993;261:1280–1281. doi: 10.1126/science.8362242. [DOI] [PubMed] [Google Scholar]
- 7.Caras I, Weddell G, Davitz M, Nussenzweig V, Martin D W. Signal for attachment of a phospholipid membrane anchor in decay accelerating factor. Science. 1987;238:1280–1283. doi: 10.1126/science.2446389. [DOI] [PubMed] [Google Scholar]
- 8.Cross G A M. Glycolipid anchoring of plasma membrane proteins. Annu Rev Cell Biol. 1990;6:1–39. doi: 10.1146/annurev.cb.06.110190.000245. [DOI] [PubMed] [Google Scholar]
- 9.Deleersnyder V, Pillez A, Wychowski C, Blight K, Xu J, Hahn Y S, Rice C M, Dubuisson J. Formation of native hepatitis C virus glycoprotein complexes. J Virol. 1997;71:697–704. doi: 10.1128/jvi.71.1.697-704.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Drillien R, Spehner D, Kirn A. Complementation and genetic linkage between vaccinia virus temperature-sensitive mutants. Virology. 1982;119:372–381. doi: 10.1016/0042-6822(82)90096-4. [DOI] [PubMed] [Google Scholar]
- 11.Dubuisson J, Hsu H H, Cheung R C, Greenberg H B, Russell D G, Rice C M. Formation and intracellular localization of hepatitis C virus envelope glycoprotein complexes expressed by recombinant vaccinia and Sindbis viruses. J Virol. 1994;68:6147–6160. doi: 10.1128/jvi.68.10.6147-6160.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dubuisson J, Rice C M. Hepatitis C virus glycoprotein folding: disulfide bond formation and association with calnexin. J Virol. 1996;70:778–786. doi: 10.1128/jvi.70.2.778-786.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feinstone S, Alter H J, Dienes H P, Shimizu Y, Popper H, Blackmore D, Sly D, London W T, Purcell R H. Non-A, non-B hepatitis in chimpanzees and marmosets. J Infect Dis. 1981;144:588–598. doi: 10.1093/infdis/144.6.588. [DOI] [PubMed] [Google Scholar]
- 14.Fournillier-Jacob A, Cahour A, Escriou N, Girard M, Wychowski C. Processing of the E1 glycoprotein of hepatitis C virus expressed in mammalian cells. J Gen Virol. 1996;77:1055–1064. doi: 10.1099/0022-1317-77-5-1055. [DOI] [PubMed] [Google Scholar]
- 15.Francki R I B, Fauquet C M, Knudson D L, Brown F, editors. Classification and nomenclature of viruses. Fifth Report of the International Committee on Taxonomy of Viruses. Vienna, Austria: Springer-Verlag; 1991. p. 223. [Google Scholar]
- 16.Fuerst T R, Niles E G, Studier F W, Moss B. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc Natl Acad Sci USA. 1986;83:8122–8126. doi: 10.1073/pnas.83.21.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grakoui A, Wychowski C, Lin C, Feinstone S M, Rice C M. Expression and identification of hepatitis C virus polyprotein cleavage products. J Virol. 1993;67:1385–1395. doi: 10.1128/jvi.67.3.1385-1395.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Griffiths G, Rottier P. Cell biology of viruses that assemble along the biosynthetic pathway. Semin Cell Biol. 1992;3:367–381. doi: 10.1016/1043-4682(92)90022-N. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Habersetzer, F., A. Fournillier, J. Dubuisson, D. Rosa, S. Abrignani, C. Wychowski, I. Nakano, C. Trépo, C. Desgranges, and G. Inchauspé. Unpublished data. [DOI] [PubMed]
- 20.Hammond C, Helenius A. Quality control in the secretory pathway. Curr Opin Cell Biol. 1995;7:523–529. doi: 10.1016/0955-0674(95)80009-3. [DOI] [PubMed] [Google Scholar]
- 21.Hobman T C, Woodward L, Farquahr M G. Targeting of a heterodimeric membrane complex to the Golgi complex: rubella virus E2 glycoprotein contains a transmembrane Golgi retention signal. Mol Biol Cell. 1995;6:7–20. doi: 10.1091/mbc.6.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hurtley S M, Helenius A. Protein oligomerization in the endoplasmic reticulum. Annu Rev Cell Biol. 1989;5:277–307. doi: 10.1146/annurev.cb.05.110189.001425. [DOI] [PubMed] [Google Scholar]
- 23.Kemble G W, Henis Y I, White J M. GPI- and transmembrane-anchored influenza hemagglutinin differ in structure and receptor binding activity. J Cell Biol. 1993;122:1253–1265. doi: 10.1083/jcb.122.6.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kieny M-P, Lathe R, Drillien R, Spehner D, Skory S, Schmitt D, Wiktor T, Koprowski H, Lecocq J-P. Expression of rabies virus glycoprotein from a recombinant vaccinia virus. Nature. 1984;312:163–166. doi: 10.1038/312163a0. [DOI] [PubMed] [Google Scholar]
- 25.Kost T A, Kessler J A, Patel I R, Gray J G, Overton L K, Carter S G. Human immunodeficiency virus infection and syncytium formation in HeLa cells expressing glycophospholipid-anchored CD4. J Virol. 1991;65:3276–3283. doi: 10.1128/jvi.65.6.3276-3283.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lanford R E, Notvall L, Chavez D, White R, Frenzel G, Simonsen C, Kim J. Analysis of hepatitis C virus capsid, E1, and E2/NS1 proteins expressed in insect cells. Virology. 1993;197:225–235. doi: 10.1006/viro.1993.1583. [DOI] [PubMed] [Google Scholar]
- 27.Lin A, Devaux B, Green A, Sagerstrom C, Elliott J, Davis M. Expression of T cell antigen receptor heterodimers in a lipid-linked form. Science. 1990;249:677–679. doi: 10.1126/science.1696397. [DOI] [PubMed] [Google Scholar]
- 28.Lin C, Lindenbach B D, Prágai B M, McCourt D W, Rice C M. Processing in the hepatitis C virus E2-NS2 region: identification of p7 and two distinct E2-specific products with different C termini. J Virol. 1994;68:5063–5073. doi: 10.1128/jvi.68.8.5063-5073.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Locker J K, Klumperman J, Oorschot V, Horzinek M C, Geuze H J, Rottier P J M. The cytoplasmic tail of mouse hepatitis virus M protein is essential but not sufficient for its retention in the Golgi complex. J Biol Chem. 1994;269:28263–28269. [PubMed] [Google Scholar]
- 30.Machamer C E, Rose J K. A specific transmembrane domain of a coronavirus E1 glycoprotein is required for its retention in the Golgi region. J Cell Biol. 1987;105:1205–1214. doi: 10.1083/jcb.105.3.1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mallabiabarrena A, Jimenez M A, Rico M, Alarcon B. A tyrosine-containing motif mediates ER retention of CD3-ɛ and adopts a helix-turn structure. EMBO J. 1995;14:2257–2268. doi: 10.1002/j.1460-2075.1995.tb07220.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Michalak J-P, Wychowski C, Choukhi A, Meunier J-C, Ung S, Rice C M, Dubuisson J. Characterization of truncated forms of hepatitis C virus glycoproteins. J Gen Virol. 1997;78:2299–2306. doi: 10.1099/0022-1317-78-9-2299. [DOI] [PubMed] [Google Scholar]
- 33.Mizushima H, Hijikata M, Asabe S-I, Hirota M, Kimura K, Shimotohno K. Two hepatitis C virus glycoprotein E2 products with different C termini. J Virol. 1994;68:6215–6222. doi: 10.1128/jvi.68.10.6215-6222.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moran P, Raab H, Kohr W J, Caras I W. Glycophospholipid membrane anchor attachment. J Biol Chem. 1991;266:1250–1257. [PubMed] [Google Scholar]
- 35.Moss B, Elroy-Stein O, Mizukami T, Alexander W A, Fuerst T R. New mammalian expression vectors. Nature. 1990;348:91–92. doi: 10.1038/348091a0. [DOI] [PubMed] [Google Scholar]
- 36.Nilsson T, Warren G. Retention and retrieval in the endoplasmic reticulum and the Golgi apparatus. Curr Opin Cell Biol. 1994;6:517–521. doi: 10.1016/0955-0674(94)90070-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pelham H R B. Sorting and retrieval between the endoplasmic reticulum and Golgi apparatus. Curr Opin Cell Biol. 1995;7:530–535. doi: 10.1016/0955-0674(95)80010-7. [DOI] [PubMed] [Google Scholar]
- 38.Pettersson R F. Protein localization and virus assembly at intracellular membranes. Curr Top Microbiol Immunol. 1991;170:67–104. doi: 10.1007/978-3-642-76389-2_3. [DOI] [PubMed] [Google Scholar]
- 39.Pfeffer S R, Rothman J E. Biosynthetic protein transport and sorting by the endoplasmic reticulum and Golgi. Annu Rev Biochem. 1987;56:829–852. doi: 10.1146/annurev.bi.56.070187.004145. [DOI] [PubMed] [Google Scholar]
- 40.Pillez, A., and J. Dubuisson. Unpublished results.
- 41.Ralston R, Thudium K, Berger K, Kuo C, Gervase B, Hall J, Selby M, Kuo G, Houghton M, Choo Q-L. Characterization of hepatitis C virus envelope glycoprotein complexes expressed by recombinant vaccinia viruses. J Virol. 1993;67:6753–6761. doi: 10.1128/jvi.67.11.6753-6761.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reinherz E L, Kung P C, Goldstein G, Schlossman S F. Separation of functional subsets of human T cells by a monoclonal antibody. Proc Natl Acad Sci USA. 1979;76:4061–4065. doi: 10.1073/pnas.76.8.4061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rice C M. Flaviviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 931–959. [Google Scholar]
- 44.Robbins P W, Timble R B, Wirth D F, Hering C, Maley F, Maley G F, Das R, Gibson B W, Royal N, Biemann K. Primary structure of the Streptomyces enzyme endo-β-N-acetylglucosaminidase H. J Biol Chem. 1984;259:7577–7583. [PubMed] [Google Scholar]
- 45.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 46.Schutze M-P, Peterson P A, Jackson M R. An N-terminal double-arginine motif maintains type II membrane proteins in the endoplasmic reticulum. EMBO J. 1994;13:1696–1705. doi: 10.1002/j.1460-2075.1994.tb06434.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Selby M J, Glazer E, Masiarz F, Houghton M. Complex processing and protein:protein interactions in the E2:NS2 region of HCV. Virology. 1994;204:114–122. doi: 10.1006/viro.1994.1515. [DOI] [PubMed] [Google Scholar]
- 48.Shin J, Dunbrack R L, Lee S, Strominger J L. Signals for retention of transmembrane proteins in the endoplasmic reticulum studied with CD4 truncation mutants. Proc Natl Acad Sci USA. 1991;88:1918–1922. doi: 10.1073/pnas.88.5.1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Spaete R R, Alexander D, Rugroden M E, Choo Q-L, Berger K, Crawford K, Kuo C, Leng S, Lee C, Ralston R, Thudium K, Tung J W, Kuo G, Houghton M. Characterization of the hepatitis E2/NS1 gene product expressed in mammalian cells. Virology. 1992;188:819–830. doi: 10.1016/0042-6822(92)90537-y. [DOI] [PubMed] [Google Scholar]
- 50.Weisz O A, Swift A M, Machamer C E. Oligomerization of a membrane protein correlates with its retention in the Golgi complex. J Cell Biol. 1993;122:1185–1196. doi: 10.1083/jcb.122.6.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang M, Ellenberg J, Bonifacino J S, Weissman A M. The transmembrane domain of a carboxy-terminal anchored protein determines localization to the endoplasmic reticulum. J Biol Chem. 1997;272:1970–1975. doi: 10.1074/jbc.272.3.1970. [DOI] [PubMed] [Google Scholar]