Abstract
Objective
Obesity has become a global health issue and a risk factor for hyperuricemia. However, the associations between obesity and hyperuricemia are sometimes confounding. In the present study, we performed mendelian randomization (MR) analysis to study their relationship and investigate the underlying mechanism by network pharmacology.
Method
Body mass index (BMI) and uric acid related to single nucleotide polymorphism were selected as instrumental variables for MR analysis. Three robust analytical methods are used for bidirectional MR analysis such as inverse-variance weighting, weighted median and MR-Egger regression. Then, we further performed sensitivity analysis to evaluate the horizontal pleiotropy, heterogeneities, and stability. The targets related to obesity and hyperuricemia were collected, screened and further conducted for Kyoto Encyclopedia of Genes and Genomes pathway enrichment to explore the mechanism of obesity and hyperuricemia using network pharmacology.
Results
The positive causality was indicated between BMI and hyperuricemia based on inverse variance-weighted analysis [odds ratio:1.23, 95% confidence interval: 1.11 to 1.30 for each standard deviation increase in BMI (4.6 kg/m2)]. Conversely, hyperuricemia did not influence BMI. 235 intersected targets from obesity and hyperuricemia were collected. Insulin resistance were the top 1 key target. The mechanism between obesity and hyperuricemia are associated with important pathways including adipocytokine signaling pathway, insulin resistance and cholesterol metabolism et al.
Conclusions
Our MR analysis supported the causal association between obesity and hyperuricemia based on availablegenome-wide association analysis summary statistics. Obesity leads to hyperuricemia via insulin resistance, which is a key link in the huge network pathways using network pharmacology.
Keywords: Obesity, Hyperuricemia, Association, Mendelian randomization, Network pharmacology
1. Introduction
Hyperuricemia is induced by over-production or less-excretion of uric acid as an indicator for gout, hypertension, diabetes, coronary heart disease, metabolic syndrome and other chronic diseases [1,2]. The morbidity of hyperuricemia will reach 18.5% in men and 21.0 % in women by 2025 [3]. It can be seen that hyperuricemia becomes an public issue, which is urgent to be solved. There exist the associations between obesity and hyperuricemia in many epidemiological studies [[4], [5], [6], [7], [8]]. In a population-based study of men in Japan, visceral fat accumulation was identified in 56% of men with hyperuricemia, hinting at the role of obesity in potentially causing elevated urate levels [7,8].
However, many limitations exist in conventional epidemiological designs, such as potential reverse causation, non-random selection across environmental, lifestyle, dietary, and genetic risk factors [9]. The associations seen in epidemiology are sometimes residual confounded and thus misleading [10]. The above limitations are easily surmounted in mendelian randomization (MR) analysis. MR has unique advantages for causal inference of exposure factors. It can provide an efficient approach to assess the potential causal genetic associations in epidemiological studies [[11], [12], [13], [14], [15]]. This approach is similar to randomized controlled trials in which genetic alleles are randomly classified during pregnancy [16]. The wide availability of genome-wide association analysis (GWAS) make MR a robust method for causal inference of exposure factors than conventional observational studies to test causality and preventing the adverse effects on human health of modifiable exposures. It presents a time- and cost-efficient approach to provide reliable associations especially when randomized controlled trials to examine causality are not feasible [17].
“One disease-one gene target or one-signaling pathway” mode cannot fully explain the potential mechanism of hyperuricemia and obesity. Network pharmacology is an effective approach to explore the complex mechanism among biological systems, drugs, and diseases based on network topology and bioinformatics methods. It could establish an interaction network of multi-compounds, multi-proteins and disease with holistic philosophy characteristics [[18], [19], [20]]. The network is composed of multiple targets and pathways, which systematically reveal the complex mechanism of obesity and hyperuricemia.
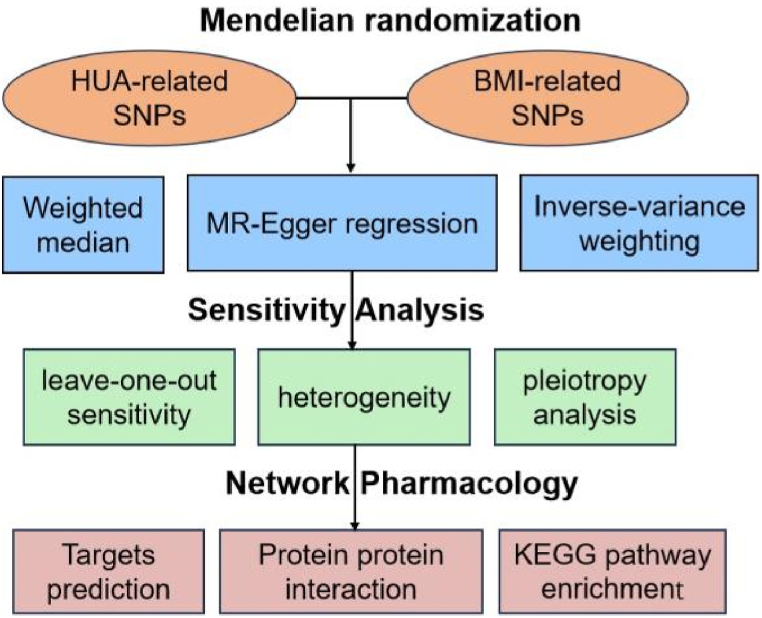
Therefore, in the study, MR analysis and network pharmacology approaches were performed to assess the potential causal relationship between obesity and hyperuricemia from the gene level and its underlying mechanism using pooled statistics from large-scale non overlapping GWAS shown in Fig. 1. The results of the present study could provide a theoretical support for clinical treatment of hyperuricemia in obese patients.
Fig. 1.
Schematic representation of the study design.
2. Materials and methods
2.1. Data collection related to MR analysis
Genetic data for obesity and hyperuricemia were obtained from a large meta-analysis of genome-wide association analysis (GWAS) (https://gwas.mrcieu.ac.uk/). The data set MRCIEU ID for obesity and hyperuricemia were bbj-a-3 and bbj-a-57, respectively. The studies included subjects recruited from Eastern Asians. The subjects were genotyped using the Illumina HumanOmniExpressExome BeadChip or a combination of Illumina HumanOmniExpress and HumanExome BeadChips. Imputation was performed by minimac (v0.1.1). A total of 173,430 subjects include 72,390 obese patients and 101,040 controls [21]. In addition, single nucleotide polymorphisms (SNP) were 6,108,953. There are 162,255 subjects including 109,029 patients with hyperuricemia and 53,226 controls [22]. Single nucleotide polymorphisms (SNP) strongly related to uric acid (UA) and BMI were identified by the original GWAS. The number of SNP is 6,108,953. In order to avoid the impact of SNP linkage disequilibrium (LD) on the analysis results, the SNP screening criteria were set to p < 5 × 10−8, LD r2 < 0.01.
2.2. MR statistical analysis
The inverse variance-weighted analysis (IVW) method was used to estimate the association between the increase in genetic prediction per 10 units of BMI index and the odds ratio of UA concentration (unit: mg/dl). Weighted median and MR-Egger regression methods were used for sensitivity analysis. Weighted median method at least 50 % of SNP loci meet the premise of effective instrumental variables. After ranking the included SNP by weight, the median of the corresponding distribution function was obtained as the analysis result. MR-egger regression can be used to evaluate the bias caused by genetic pleiotropy. If the intercept of the regression equation is close to 0, it is considered that genetic pleiotropy has little effect. Heterogeneity between each SNP was according to individual exclusion test and heterogeneity test.
2.3. Targets prediction and pathways analysis
To predict targets related to obesity and hyperuricemia, we searched the following database including Genecards (https://www.genecards.org/), Therapeutic Target Database (TTD, http://db.idrblab.net/ttd/), Online Mendelian Inheritance in Man (OMIM) database (https://www.omim.org/), Drugbank (https://go.drugbank.com) and Gene Expression Omnibus (GEO) DataSets (http://www.ncbi.nlm.nih.gov/gds/). The intersected genes were imported into String platform for network construction and Cytoscape was used to screen the potential targets and visualization. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment were performed using Cytoscape plug-in ClueGO to explore the mechanism of obesity and hyperuricemia.
2.4. Differentially expressed genes (DEG) screening criteria
We employed the normalization of our gene expression data between arrays function from the limma package in rstudio. This function is tailor-made for normalizing microarray data, effectively removing technical variations and ensuring comparability across different arrays. The criteria of differentially expressed genes (DEGs) in our study were based on p value and fold change value. We set a stringent cut-off, considering genes with an absolute logFC greater than 1 as significantly differentially expressed. This threshold was chosen to focus on genes with substantial changes in expression levels, thereby enhancing the biological relevance and potential impact of our findings. Heat and volcano map were drawn by using Heatmap and ggplot 2 package in rstudio, respectively. P < 0.05 was taken as statistically significance.
3. Results
3.1. Causal association of obesity and hyperuricemia by MR method
Firstly, the bidirectional causality between obesity and hyperuricemia was assessed by bidirectional MR analysis including inverse variance-weighted (IVW), MR-Egger and weighted median. As shown in Table 1, a significant causal relationship between BMI and UA (OR = 1.206,95%CI 1.114–1.305,p < 0.001) for IVW analysis. Otherwise, the causal effect of UA on BMI was not statistically significant (OR = 0.995,95%CI 0.961–1.030,p > 0.1).
Table 1.
Mendelian randomization (MR) results for inverse variance weighted (IVW), weighted median, and MR- Egger methods.
| MR method | beta | SE | p-value | N SNPs | OR | 95%CI | |
|---|---|---|---|---|---|---|---|
| BMI→ UA |
IVW | 0.187 | 0.040 | 3.45E-06 | 21 | 1.206 | 1.114–1.305 |
| Weighted median | 0.160 | 0.045 | 3.67E-04 | 21 | 1.173 | 1.075–1.281 | |
| MR-Egger | 0.081 | 0.147 | 5.86E-01 | 21 | 1.085 | 0.814–1.447 | |
| UA→ BMI |
IVW | −0.005 | 0.018 | 0.776 | 78 | 0.995 | 0.961–1.030 |
| Weighted median | −0.034 | 0.020 | 0.076 | 78 | 0.966 | 0.930–1.004 | |
| MR-Egger | −0.075 | 0.029 | 0.011 | 78 | 0.928 | 0.877–0.982 |
BMI, Body mass index (BMI); SE, standard error; SNP, single nucleotide polymorphism; N SNP, number of single nucleotide polymorphism; IVW, inverse variance weighted; CI, confidence interval. OR, odds ratio.
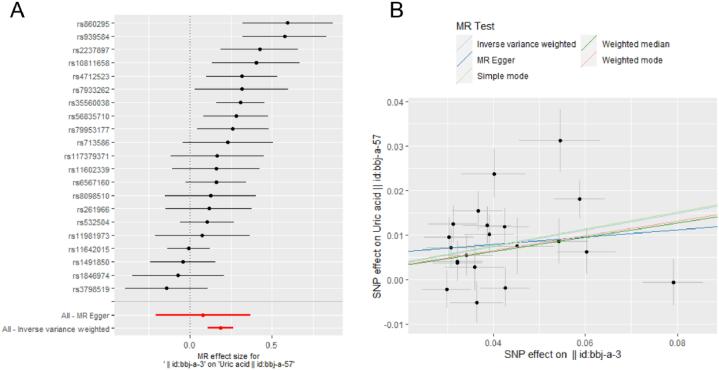
The associations for each genetic instrument with BMI and UA are presented in Fig. 2. The forest plot (Fig. 2A) and scatter plot (Fig. 2B) showed that there existed a causal relationship from BMI to UA for IVW analysis. The inverse variance weighted (IVW) result is the main MR test. The elevated BMI increases the risk of elevated UA as shown in Fig. 2A. The blue line representing the IVW method is close to horizontal (Supplementary Fig. 1), showing that there is no significant causal effect from UA to BMI.
Fig. 2.
Mendelian randomization (MR) analysis from body mass index (BMI) to uric acid (UA). Forest plot (A). Scatter plot (B). In the Forest plot, the black dots represent the logarithm odds ratio (lgOR) of UA increased by the standard deviation (SD) in BMI, which is generated using each single nucleotide polymorphism (SNP) as a separate tool variable. The red dots show the effect sizes of all SNP combinations by different MR methods. Horizontal bars represents 95% confidence interval (CI). In the scatter plot, X axis (SD unit) represents the effect of SNP on BMI, Y axis (lgOR) represents the effect of SNP on uric acid. Each black dot represents a separate SNP, The vertical and the horizontal bars represent 95% CI of the effect size of uric acid and BMI, respectively. Three slopes represent the causal estimation for three MR methods. The light blue slope is the IVW method, the dark blue slope is the MR Egger method, and the green slope is the weighted median method.
3.2. MR sensitive analysis
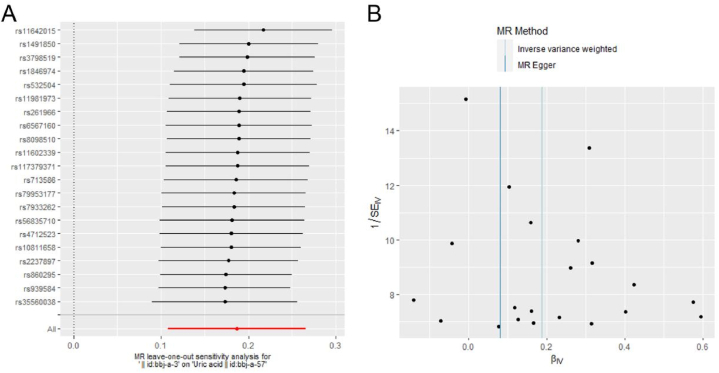
Secondly, we analyzed a series of sensitivity tests including leave-one-out sensitivity, heterogeneity and pleiotropy analysis. In a leave-one-out sensitivity analysis, we found that no single SNP was strongly driving the overall effect of BMI on UA. As shown in Fig. 3, all horizontal bars in the forest plot (Fig. 3 A) are on the right of zero and funnel plot (Fig. 3B) presents approximate symmetry indicating that the MR results are stable and robust. However, all horizontal bars in the supplementary forest plot (Supplementary Fig. 2) step over zero, which means that no matter which SNP is removed, there is no significant causal effect from UA to BMI.
Fig. 3.
Leave-one- out sensitivity analysis from body mass index (BMI) to uric acid (UA). A. forest plot. B. Funnel plot. The black dots represent the logarithm odds ratio (lgOR) of UA increased by the standard deviation (SD) in BMI, which is generated using each SNP as a separate tool variable. The red dots show the effect sizes of all SNP combinations by different MR methods. Horizontal bars represents 95% CI.
In pleiotropy test (Table 2), there is no statistical difference according to the Egger intercept of MR-Egger (p > 0.1), which showed that there is no level pleiotropy. In the heterogeneity test, Q is far less than 0.05, and there is strong heterogeneity between instrumental variable. Then, we also use the random effect model to estimate the effect of MR. In the random effect model, there is a causal relationship of BMI on UA (p < 0.05).
Table 2.
Sensitive analysis by heterogeneity test, random effects models and pleiotropy test.
| Sensitive analysis | MR method | Se | p-value | Q-value |
|---|---|---|---|---|
| Heterogeneity test | MR-Egger | NA | NA | 2.84E-05 |
| Heterogeneity test | Inverse variance weighted | NA | NA | 2.87E-05 |
| Random effect | Inverse variance weighted | 0.040269 | 3.45E-06 | NA |
| Pleiotropy test | MR-Egger intercept | 0.006217 | 0.463662 | NA |
SE, standard error; N SNP, number of single nucleotide polymorphism, NA, no applicable.
3.3. Predicting targets of obesity and hyperuricemia
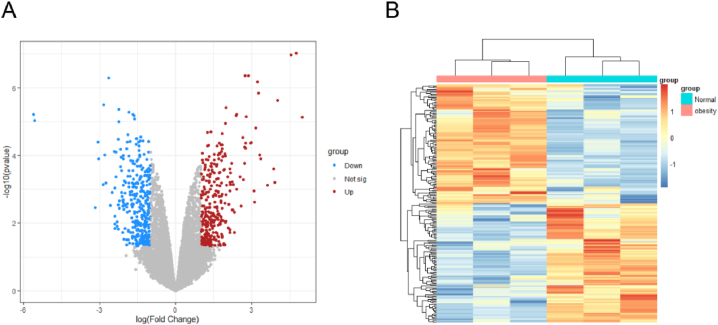
Firstly, the clinical obesity gene profiles were collected from public available micro-array data in the GEO database gene chip (GSE128021). We identified 1348 differential expressed genes (DEGs) in obesity as compared to individuals with normal BMI. (Fig. 4A and B). Secondly, we integrated the obesity targets from DrugBank, OMIM and TTD databases with DEGs to avoid data redundancy. A total of 2380 obesity targets and 800 high hyperuricemia targets were identified. Finally, 235 common targets were overlapped between hyperuricemia targets and obesity targets (Fig. 4).
Fig. 4.
Obesity -related targets using clinical micro-array data. (A). Volcano map of Gene expression levels. Green, red and gray represent down-regulated genes, up-regulated genes and no significant genes, respectively. (B). Heat map of differential expressed genes between normal control and obesity group.
3.4. Key targets and pathway enrichment analysis
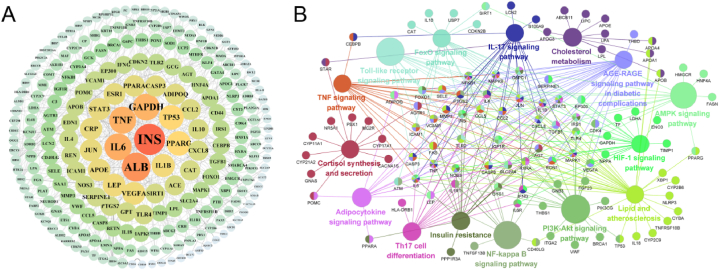
To further investigate the complex mechanism, we imported 230 intersecting targets into the STRING platform and constructed a protein-protein interaction (PPI) network. The PPI network contained 235 protein target nodes connected by 3692 edges, with an average node degree of 31.41 (shown in Fig. 5A). The top 5 key proteins of insulin resistance (INS), albumin (ALB), interlukin-6 (IL-6), tumor necrosis (TNF) and Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were screened to be key targets. INS is the top 1 key target.
Fig. 5.
The underlying mechanism of obesity and hyperuricemia. (A) Protein-protein interaction (PPI) network obtained from STRING database and constructed by Cytoscape. Each node size and color depth are proportional to their node degree. Edge width is proportional to the edge betweenness. (B) Grouping of KEGG enrichment analysis of the 235 intersected targets of obesity leading to hyperuricemia. Functionally related groups partially overlap. KEGG pathway is represented as a node. The nodes of a group are labeled in the same color. Two groups share the nodes with two colors.
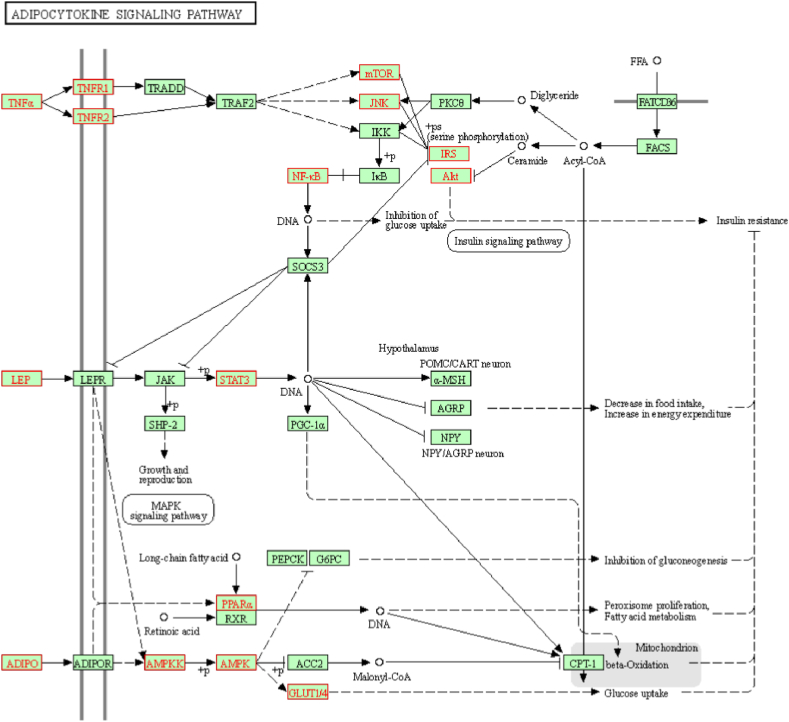
In order to explain the complex mechanism of obesity leading to hyperuricemia, Grouping of KEGG enrichment analysis were carried out for the 235 targets using cluego plug-in of Cytoscape 3.8.2. KEGG pathways with p value < 0.05 were screened (Fig. 5B) including adipocytokine signaling pathway, insulin resistance, PI3K-AKT signaling pathway, TNF signaling pathway, AMPK signaling pathway, FOXO signaling pathway, AGE-RAGE signaling pathway in diabetic complications, and so on. The targets related to the pathway are also presented on Fig. 5B. KEGG pathway is represented as a node. The nodes of a group are labeled in the same color. Two groups share the nodes with two colors. In the adipocytokine signaling pathways as shown in Fig. 6, TNF-α, leptin (LEP) and adiponectin are important targets, which could activate series targets of AMPK, NF-kB, mTOR, AKT, and STAT3, ultimately leading to insulin resistance. The pathways of adipocytokine signaling pathways, PI3K-AKT signaling pathway, TNF signaling pathway, AMPK signaling pathway, FOXO signaling pathway, AGE-RAGE signaling pathway in diabetic complications is also related to insulin resistance. It is speculated that insulin resistance is a key link among the pathway network of obesity leading to hyperuricemia.
Fig. 6.
The adipocytokine signaling pathway.
4. Discussion
Hyperuricemia has become a major public health problem, especially for obese people who are subject to hyperuricemia clinically. A 55 percents increase in the relative risk of gout per 5 units increase in BMI [23]. BMI is a widely accepted and practical measure to assess obesity in many epidemiological studies. It provides a quantifiable and accessible method to estimate adiposity, correlating significantly with more direct measures of body fat. It is often used as a exposure variable in the MR analysis. Thus, in our MR analysis, we employed BMI as the exposure variable for obesity based on its prevalence in large-scale studies and its effectiveness in capturing the essence of obesity as a risk factor for hyperuricemia. The MR results showed that BMI could cause hyperuricemia.
Hyperuricemia is difficult to cure and easy to rebound. The most common drugs used for the management of hyperuricemia are uricostatic agents (eg, allopurinol, oxypurinol, febuxostat) and uricosuric agents (eg, probenecid, benzbromarone) [24]. However, allopurinol [25] has some side effects such as liver and kidney dysfunction or damage [26], which ultimately affect the excretion and synthesis of uric acid. It takes long-term use of allopurinol especially for obese patients. Although the drugs can improve the temporary symptoms of elevated urate, they do not solve the real cause of hyperuricemia. Once the obese patients stop taking medication, levels of UA will still rise. To prevent obese patients from developing hyperuricemia, it is essential to study the relationship between the obesity and hyperuricemia. Thus, we further used network pharmacology to explain its underlying mechanism. Firstly, we did PPI analysis with the intersected genes of obesity and hyperuricemia. The top 1 target is insulin resistance. Insulin resistance is a key process in the progression of obesity leading to hyperuricemia. If we can prevent the occurrence of insulin resistance, thereby blocking the activation of downstream pathways and targets, then we may be able to truly prevent hyperuricemia in obese patients from the source. GAPDH is an important enzyme in uric acid synthesis, which is regulated by insulin. Insulin resistance causes the diminished activity of GAPDH resulting in an increase in UA production [27].
The “one disease-one target-one drug” is not accorded with the complex huge network of insulin resistance in the most profound manner [28]. Thus, in the present study, we further performed KEGG analysis to target multi-pathways and network proteins such as adipocytokine signaling pathway, insulin resistance and so on, which were closely related to the causal association between obesity and hyperuricemia. A bidirectional MR result assessed the causal relationships among insulin resistance, hyperuricemia and gout [29]. Advanced glycation end products (AGEs) contribute to insulin resistance via oxidative stress generation and chronic inflammation [30]. Network pharmacology-based treatments substantially lower the dose of each drug as compared with mono-therapy and still achieve the same or even a more significant therapeutic effect [[31], [32], [33]]. Obese hyperuricemia patients may be truly cured through regulating insulin resistance to prevent the progression of hyperuricemia.
5. Conclusion
Our mendelian randomization analysis supported the causal association between obesity and hyperuricemia using available GWAS summary statistics. Obesity leads to hyperuricemia through the key link of insulin resistance based on the results of network pharmacology.
Data availability
The data is available from the corresponding author.
CRediT authorship contribution statement
Kailai Panlu: Writing – original draft, Investigation, Formal analysis, Data curation. Zizun Zhou: Visualization, Investigation. Lin Huang: Methodology, Formal analysis, Data curation. Lei Ge: Data curation, Methodology, Visualization. Chengping Wen: Supervision, Project administration, Funding acquisition. Huiqing Lv: Writing – review & editing, Writing – original draft, Supervision.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement and funding
The research was funded by National Key R&D Program of China (No. 2022YFC3501204) and the Key Supported Projects of the Joint Fund of the National Natural Science Foundation of China (No. U21A20402) and Zhejiang Science and Technology Plan (No. 2023C03040).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e27074.
Contributor Information
Chengping Wen, Email: chengpw2010@126.com.
Huiqing Lv, Email: lvhuiqing@zcmu.edu.cn.
Appendix A. Supplementary data
The following is/are the supplementary data to this article.
References
- 1.Merriman T.R., Dalbeth N. The genetic basis of hyperuricaemia and gout. Joint Bone Spine. 2011;78(1):35–40. doi: 10.1016/j.jbspin.2010.02.027. [DOI] [PubMed] [Google Scholar]
- 2.Yanai H., et al. Molecular biological and clinical understanding of the pathophysiology and treatments of hyperuricemia and its association with metabolic syndrome, cardiovascular diseases and chronic kidney disease. Int. J. Mol. Sci. 2021;22(17) doi: 10.3390/ijms22179221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu J., et al. Hyperuricemia and clustering of cardiovascular risk factors in the Chinese adult population. Sci. Rep. 2017;7(1):5456. doi: 10.1038/s41598-017-05751-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee J., et al. Visceral fat obesity is highly associated with primary gout in a metabolically obese but normal weighted population: a case control study. Arthritis Res. Ther. 2015;17(1):79. doi: 10.1186/s13075-015-0593-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lyu L.C., et al. A case-control study of the association of diet and obesity with gout in Taiwan. Am. J. Clin. Nutr. 2003;78(4):690–701. doi: 10.1093/ajcn/78.4.690. [DOI] [PubMed] [Google Scholar]
- 6.Thottam G.E., Krasnokutsky S., Pillinger M.H. Gout and metabolic syndrome: a tangled web. Curr. Rheumatol. Rep. 2017;19(10):60. doi: 10.1007/s11926-017-0688-y. [DOI] [PubMed] [Google Scholar]
- 7.Ali N., et al. Prevalence of hyperuricemia and the relationship between serum uric acid and obesity: a study on Bangladeshi adults. PLoS One. 2018;13(11) doi: 10.1371/journal.pone.0206850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tamba S., et al. Relationship between the serum uric acid level, visceral fat accumulation and serum adiponectin concentration in Japanese men. Intern. Med. 2008;47(13):1175–1180. doi: 10.2169/internalmedicine.47.0603. [DOI] [PubMed] [Google Scholar]
- 9.Huskins W.C., Fowler V.G., Jr., Evans S. Adaptive designs for clinical trials: application to healthcare epidemiology research. Clin. Infect. Dis. 2018;66(7):1140–1146. doi: 10.1093/cid/cix907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smith G.D., Ebrahim S. 'Mendelian randomization': can genetic epidemiology contribute to understanding environmental determinants of disease? Int. J. Epidemiol. 2003;32(1):1–22. doi: 10.1093/ije/dyg070. [DOI] [PubMed] [Google Scholar]
- 11.Geng T., et al. Childhood BMI and adult type 2 diabetes, coronary artery diseases, chronic kidney disease, and cardiometabolic traits: a mendelian randomization analysis. Diabetes Care. 2018;41(5):1089–1096. doi: 10.2337/dc17-2141. [DOI] [PubMed] [Google Scholar]
- 12.Ding M., et al. Dairy consumption, systolic blood pressure, and risk of hypertension: mendelian randomization study. BMJ. 2017;356:j1000. doi: 10.1136/bmj.j1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lawlor D.A., et al. Mendelian randomization: using genes as instruments for making causal inferences in epidemiology. Stat. Med. 2008;27(8):1133–1163. doi: 10.1002/sim.3034. [DOI] [PubMed] [Google Scholar]
- 14.Huang T., et al. Association of homocysteine with type 2 diabetes: a meta-analysis implementing Mendelian randomization approach. BMC Genom. 2013;14:867. doi: 10.1186/1471-2164-14-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Benn M., Nordestgaard B.G. From genome-wide association studies to Mendelian randomization: novel opportunities for understanding cardiovascular disease causality, pathogenesis, prevention, and treatment. Cardiovasc. Res. 2018;114(9):1192–1208. doi: 10.1093/cvr/cvy045. [DOI] [PubMed] [Google Scholar]
- 16.Didelez V., Sheehan N. Mendelian randomization as an instrumental variable approach to causal inference. Stat. Methods Med. Res. 2007;16(4):309–330. doi: 10.1177/0962280206077743. [DOI] [PubMed] [Google Scholar]
- 17.Sekula P., et al. Mendelian randomization as an approach to assess causality using observational data. J. Am. Soc. Nephrol. 2016;27(11):3253–3265. doi: 10.1681/ASN.2016010098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang R., et al. Network pharmacology databases for traditional Chinese medicine: review and assessment. Front. Pharmacol. 2019;10:123. doi: 10.3389/fphar.2019.00123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Y., et al. Emerging applications of metabolomics to assess the efficacy of traditional Chinese medicines for treating type 2 diabetes mellitus. Front. Pharmacol. 2021;12 doi: 10.3389/fphar.2021.735410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li S., Zhang B. Traditional Chinese medicine network pharmacology: theory, methodology and application. Chin. J. Nat. Med. 2013;11(2):110–120. doi: 10.1016/S1875-5364(13)60037-0. [DOI] [PubMed] [Google Scholar]
- 21.Akiyama M., et al. Genome-wide association study identifies 112 new loci for body mass index in the Japanese population. Nat. Genet. 2017;49(10):1458–1467. doi: 10.1038/ng.3951. [DOI] [PubMed] [Google Scholar]
- 22.Kanai M., et al. Genetic analysis of quantitative traits in the Japanese population links cell types to complex human diseases. Nat. Genet. 2018;50(3):390–400. doi: 10.1038/s41588-018-0047-6. [DOI] [PubMed] [Google Scholar]
- 23.Aune D., Norat T., Vatten L.J. Body mass index and the risk of gout: a systematic review and dose-response meta-analysis of prospective studies. Eur. J. Nutr. 2014;53(8):1591–1601. doi: 10.1007/s00394-014-0766-0. [DOI] [PubMed] [Google Scholar]
- 24.Telbisz A., et al. Effects of the lipid environment, cholesterol and bile acids on the function of the purified and reconstituted human ABCG2 protein. Biochem. J. 2013;450(2):387–395. doi: 10.1042/BJ20121485. [DOI] [PubMed] [Google Scholar]
- 25.Benn C.L., et al. Physiology of hyperuricemia and urate-lowering treatments. Front. Med. 2018;5:160. doi: 10.3389/fmed.2018.00160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stamp L.K., Chapman P.T., Palmer S.C. Allopurinol and kidney function: an update. Joint Bone Spine. 2016;83(1):19–24. doi: 10.1016/j.jbspin.2015.03.013. [DOI] [PubMed] [Google Scholar]
- 27.Choi S., et al. Piperine reverses high fat diet-induced hepatic steatosis and insulin resistance in mice. Food Chem. 2013;141(4):3627–3635. doi: 10.1016/j.foodchem.2013.06.028. [DOI] [PubMed] [Google Scholar]
- 28.Davies K., Bukhari M.A.S. Recent pharmacological advances in the management of gout. Rheumatology. 2018;57(6):951–958. doi: 10.1093/rheumatology/kex343. [DOI] [PubMed] [Google Scholar]
- 29.McCormick N., et al. Assessing the causal relationships between insulin resistance and hyperuricemia and gout using bidirectional mendelian randomization. Arthritis Rheumatol. 2021;73(11):2096–2104. doi: 10.1002/art.41779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tomino Y., Hagiwara S., Gohda T. AGE-RAGE interaction and oxidative stress in obesity-related renal dysfunction. Kidney Int. 2011;80(2):133–135. doi: 10.1038/ki.2011.86. [DOI] [PubMed] [Google Scholar]
- 31.Nogales C., et al. Network pharmacology: curing causal mechanisms instead of treating symptoms. Trends Pharmacol. Sci. 2022;43(2):136–150. doi: 10.1016/j.tips.2021.11.004. [DOI] [PubMed] [Google Scholar]
- 32.Casas A.I., et al. From single drug targets to synergistic network pharmacology in ischemic stroke. Proc. Natl. Acad. Sci. U.S.A. 2019;116(14):7129–7136. doi: 10.1073/pnas.1820799116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou W., et al. Network pharmacology to unveil the mechanism of Moluodan in the treatment of chronic atrophic gastritis. Phytomedicine. 2022;95 doi: 10.1016/j.phymed.2021.153837. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data is available from the corresponding author.