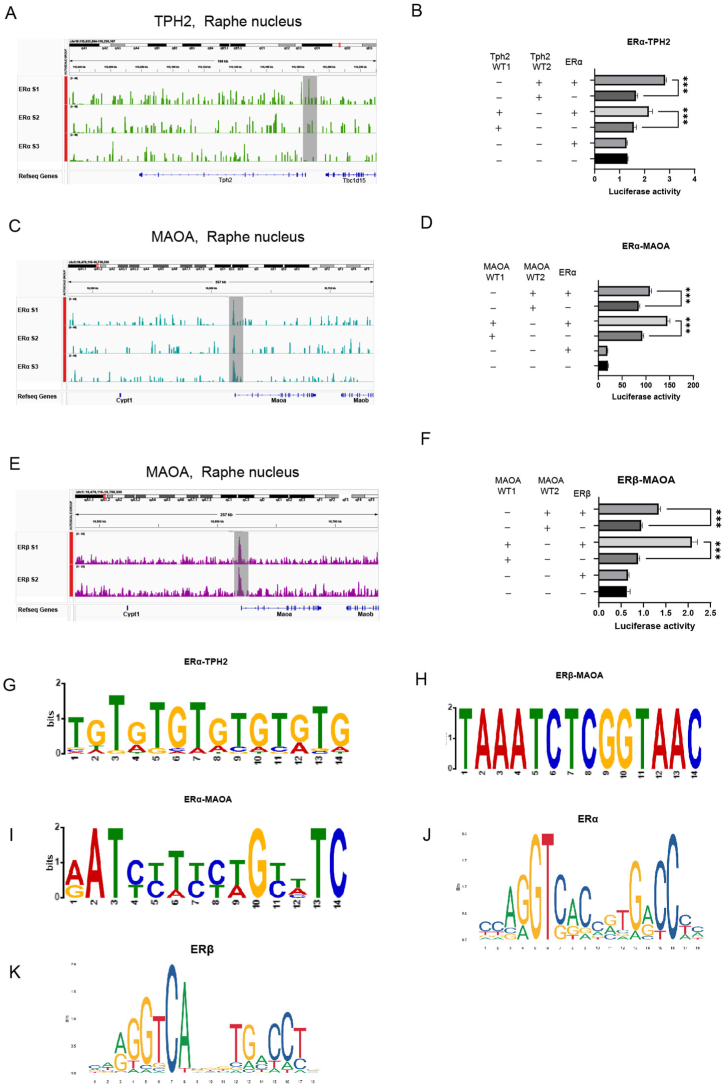
Fig. 15.
CUT&Tag profiling of oestrogen nuclear receptors as transcription factors in the RN.
(A, C, E) Integrative Genome Viewer plots showing the peak distribution of CUT&Tag signals (ERα and ERβ) at TPH2 (A) and MAOA (C, E). The promoter region of the gene is shaded in grey. The data are representative of three (A, C) or two (E) independent experiments.
(B, D, F) The transcription of ERα at TPH2 (B) and MAOA (D) and that of ERβ (F) at MAOA revealed two sites in the promoter region. CUT&Tag profiling of ERα and ERβ was performed in SY5Y cells via a dual-luciferase assay. The y-axis represents luciferase activity. The data are shown as the means ± SEMs; n = 3 biologically independent experiments per group. Student's t-test was used to analyse the differences between the two groups; ***P < 0.001.
(G, H, I) Logo representation of the motif found by CUT&Tag profiling of ERα (G and I) and ERβ (H) and its alignment with the motif of the transcription factors ERα and ERβ retrieved from the Jaspar database.
(J, K) Examples of the top known motifs of ERα (J) and ERβ (K) obtained from https://jaspar.genereg.net/.