Abstract
Background
Extracellular vesicles (EVs) play a critical role in intercellular communication under physiological and pathological conditions, including cancer. EVs cargo reflects their cell of origin, suggesting their utility as biomarkers. EVs are detected in several biofluids, and their ability to cross the blood–brain barrier has highlighted their potential as prognostic and diagnostic biomarkers in gliomas, including glioblastoma (GBM). Studies have demonstrated the potential clinical utility of plasma‐derived EVs in glioma. However, little is known about the clinical utility of saliva‐derived EVs in GBM.
Methods
Small EVs were isolated from whole mouth saliva of GBM patients pre‐ and postoperatively. Isolation was performed using differential centrifugation and/or ultracentrifugation. EVs were characterized by concentration, size, morphology, and EVs cell‐surface protein markers. Protein cargo in EVs was profiled using mass spectrometry.
Results
There were no statistically significant differences in size and concentration of EVs derived from pre‐ and post GBM patients' saliva samples. A higher number of proteins were detected in preoperative samples compared to postoperative samples. The authors found four highly abundant proteins (aldolase A, 14‐3‐3 protein ε, enoyl CoA hydratase 1, and transmembrane protease serine 11B) in preoperative saliva samples from GBM patients with poor outcomes. Functional enrichment analysis of pre‐ and postoperative saliva samples showed significant enrichment of several pathways, including those related to the immune system, cell cycle and programmed cell death.
Conclusions
This study, for the first time, demonstrates the feasibility of isolating and characterizing small EVs from pre‐ and postoperative saliva samples from GBM patients. Preliminary findings encourage further large cohort validation studies on salivary small EVs to evaluate prognosis in GBM.
Keywords: biomarkers, extracellular vesicles, glioblastoma, mass spectrometry, proteomics, saliva
Short abstract
The current study demonstrates the feasibility of isolation and characterization of small extracellular vesicles (EVs) from saliva samples of glioblastoma (GBM) patients. It also demonstrates that the proteome content of EVs revealed GBM‐related proteins, some already described in previous studies using GBM cell‐conditioned media and plasma of GBM patients. Preliminary findings encourage further studies on salivary small EVs as a source of noninvasive prognostic biomarkers to predict outcomes in patients with GBM.
INTRODUCTION
Glioblastoma (GBM) is the most common type of brain tumor in adults. Despite aggressive treatment, patient prognosis remains poor. 1 , 2 The majority of GBM patients experience recurrences within 6 to 9 months from diagnosis. 3 , 4 , 5 During treatment and follow‐up assessments, GBM patients' response is monitored mainly using imaging techniques. 6 , 7 , 8 Nevertheless, these techniques fail to reliably confirm tumor progression. 9 , 10 Currently, there are no biomarkers to monitor response to treatment or disease progression in GBM patients. 8
Liquid biopsy, the detection and analysis of tumor derived biomarkers in body fluids, is emerging as an exciting new field in the management of brain cancer patients. 11 , 12 , 13 The use of liquid biopsy may provide tumor information and assist in detecting disease progression earlier than clinical symptoms or subsequent magnetic resonance imaging (MRI) assessment. For GBM, putative biomarkers need to cross the blood–brain barrier (BBB) to be detected in body fluids. In this context, extracellular vesicles (EVs) present an advantage over other liquid biopsy‐based biomarkers (e.g., circulating tumor cells), because EVs can cross even intact BBBs, while protecting their cargo (e.g., nucleic acids, protein, lipids). 14 , 15
EV is a generic term to define cell‐derived membrane structures 16 released under physiological or pathological conditions. EVs play a crucial role in intercellular communication. 17 EVs may be classified as exosomes that are of endosome‐origin and microvesicles (ectosomes/microparticles) that are derived from budding of plasma membranes. 18 EVs classification according to their biogenesis pathway is challenging, and no consensus has been achieved in terms of specific markers to differentiate them. 16 Therefore, the International Society for Extracellular Vesicles endorses the use of operational terms for EV subtypes. 16 In this study, instead of using the terminology “exosomes,” we opted for using “small EVs” (<200 nm). EVs can be isolated from several body fluids, including blood and saliva. 19 Although blood is a largely analyzed body fluid in a clinical setting, saliva has been gaining attention as an alternative biofluid over blood sampling. Saliva collection is easy and cost‐effective, requires no special equipment or specialized staff, and multiple samples can be collected from an individual. 20 Interestingly, the composition of saliva is altered under pathological conditions such as in cancer. 20 , 21 , 22 , 23 , 24 , 25 A limited number of studies have investigated the potential of salivary biomarkers in brain tumors. 26 , 27
Saliva harbors many proteins that can be of clinical relevance. Approximately 20%–30% of blood proteins can also be found in saliva. 21 However, when using whole mouth saliva for mass spectrometry (MS)‐based proteomics studies, sometimes low abundant proteins may be undetected due to the presence of highly abundant proteins (e.g., amylase), 28 which is also inherited to blood‐based proteomics studies. In an attempt to overcome this limitation, we conducted sequential window acquisition of all theoretical mass spectra (SWATH‐MS) to allow for higher proteomic coverage. Salivary EVs have been explored as diagnostic or prognostic biomarkers in several cancer types. 29 , 30 , 31 , 32 , 33 , 34 Currently, there are no published studies investigating the potential role of salivary small EVs in patients with GBM.
In this study, we analyzed the protein content of EVs derived from GBM patients pre‐ and posttreatment with favorable outcomes and unfavorable outcomes with an aim to investigate the prognostic potential of EV as biomarkers for GBM.
MATERIALS AND METHODS
Clinical samples collection and processing
This study was approved by the human research ethics committee (HREC/2019/QRBW/48780) of Royal Brisbane and Women's Hospital (RBWH; Brisbane Australia) and the Queensland University of Technology (QUT) (approval number: 1900000292). All documents were acknowledged by the RBWH research governance (RGO). All participant patients gave their written consent to participate in this study, and blood samples were collected from patients before and after brain surgery or needle biopsy. Pre‐ and postoperative (within 2 weeks of brain surgery) whole mouth saliva samples, hereafter referred to as “saliva,” from GBM patients (n = 18) and age‐ and sex‐matched healthy controls (n = 5) were obtained as previously described. 30 , 35 , 36 The favorable or unfavorable outcomes were calculated using the time elapsed between the preoperative saliva collection date and the date of progression, death, or the last follow‐up visit with an MRI scan image. The progression was only considered once confirmed by MRI results and physicians' notes.
Isolation of EVs
Small EVs from saliva (200 µL) were pelleted by differential centrifugation and ultracentrifugation as previously described 30 , 37 , 38 (Figure S1A). All steps were performed at 4°C. Small EVs pellets were then resuspended (50 µl) in filtered (0.22 µm) phosphate‐buffered saline (PBS) and stored at −80°C for further experiments.
Nanoparticle tracking analysis
EVs were diluted (1:200) in filtered (0.22 µm) PBS and analyzed using the NanoSight NS300 with a 405‐nm laser (NanoSight, Ltd, Malvern, UK). Three videos of 30 seconds were recorded for each sample, and a report was generated on the size distribution and concentration of particles. The camera type used was sCMOS; and the laser type was blue 405. The temperature was set to be 25.0°C during video recordings. Camera level setting ranged from 12 to 14. Analysis setting for detection threshold ranged from 2 to 3. All other settings were set to automatic.
Transmission electron microscopy
EV morphology was assessed using transmission electron microscopy (TEM). Samples were mixed by vigorous vortexing. Five (µl) drops of sample were placed onto a parafilm, and the mounting grid was placed over the droplet. The mount was then incubated with 2% uranyl acetate (negative staining). EVs were imaged on a JEOL JEM‐1400 TEM at 100 kV mounted with a 2K TVIPS CCD camera at the Central Analytical Research Facility (CARF)‐QUT.
Western blot
Total protein concentration was quantified using Pierce BCA Protein Assay Kit (Thermo Fisher Scientific). Western blot was performed as previously described. 39 Equal amounts of EVs protein (5 µg) were loaded onto gels, and a GBM cell line (U251MG) was used as a positive control. U251MG cell line was gifted by Prof. Bryan W. Day (QIMR, Brisbane, Australia). The following primary and secondary antibodies were used: CD63 (Santa Cruz, #15363), CD9 (Cell Signaling, #13174), GM‐130 (Cell Signaling, #12480), CD81 (Santa Cruz, #166029) Calnexin (Abclonal, #A4846), Aldolase A (C‐10) (Santa Cruz, #390733), ECH1 (B‐3) (Santa Cruz, #515270), 14‐3‐3 ε (8C3) (Santa Cruz, #23957), and anti‐rabbit or anti‐mouse IgG‐HRP secondary antibody (Cell Signaling, #7074 or #7076). All primary antibodies were diluted 1:1000 and secondary 1:2000.
SWATH mass spectrometry analysis
Sample processing and SWATH‐MS analyses were performed as previously described by our group. 35 In brief, samples were processed using filter‐aided sample processing, 40 digested using trypsin, desalted using StageTips containing strong cation exchange membrane and analyzed using liquid chromatography–tandem mass spectrometry LC–MS/MS. Ten microliters of indexed retention time (iRT) peptides were spiked into the peptide samples to allow for the recalibration of retention times in subsequent LC‐MS/MS analyses. 41 The MS data generated was searched against the Human SwissProt/UniProt database (March 2021) using ProteinPilot (SCIEX) software and the Paragon Algorithm, as previously described. 42 The spectral library consisted of 507 proteins and 11,582 peptides at 95% confidence. SWATH‐MS data analysis was performed using the SWATH Microapp (v2.0) plug‐in for PeakView (v2.2, SCIEX) software. The criteria for protein quantification were: six peptides detected per protein, six transitions detected per peptide, 95% confidence threshold, 1% false discovery rate (FDR), 6‐minute peak detection window, and 50 ppm XIC extraction window. Retention time calibration using iRT peptides was performed before ion extraction. 43 Additional information and more details are described in the Supporting Material.
Bioinformatic analysis
Gene Ontology (GO) enrichment analysis of differentially abundant proteins was performed using the Database for Annotation, Visualization and Integrated Discovery Bioinformatics Resources 6.8 (https://david.ncifcrf.gov/). GO annotation was classified into two categories, namely biological process and molecular functions. An adjusted p value <.05 was considered significant. The GO Protein class analysis was generated using Panther classification 44 (http://www.pantherdb.org/). Pathway analyses were performed using the Kyoto Encyclopedia of Genes and Genomes (KEGG) (http://www.genome.jp/kegg/) and Reactome (https://reactome.or) to identify enriched pathways and generate a report, respectively. A protein–protein interaction (PPI) network was built using Cytoscape (http://www.cytoscape.org) based on findings from the STRING database (https://stringdb.org).
Statistical analysis
Statistical analyses were performed using GraphPad Prism Software and package MSStats version 2.0 45 in R. 35 Protein significance analysis was performed by applying a linear mixed‐effects model using MSStats, as described previously. 35 The protein abundance levels between patients with unfavorable and favorable outcomes were compared using the Mann‐Whitney test (GraphPad Prism). Differences between more than two groups were evaluated by one‐way ANOVA (GraphPad Prism). A p value <.05 was defined as statistically significant. The partial least squares‐discriminant analysis (PLS‐DA) was performed using the package mix0mics in R. Volcano plots and receiver operator characteristic (ROC) curve analysis were generated in GraphPad Prism.
RESULTS
Demographic and clinical information from patients
Demographic and clinical information from GBM patients and healthy controls were collected (Table S1). The average age of GBM patients was 60 years (ranging from 37 to 82 years) and for the control group was 63.5 years (ranging from 58 to 71 years). The healthy control group consisted of three women and two men, whereas the GBM cohort included nine women and nine men. Nearly all patients were classified as IDH‐wild type GBM (n = 17) and one patient was classified with IDH1R132H mutation. At the time of diagnosis, which was before the new 2021 World Health Organization classification of central nervous system tumors, patients presenting IDH1R132H mutation had their tumors still classified as GBM. Within the cohort of GBM patients, 11 were confirmed with disease recurrence or were deceased within 9 months from diagnosis (unfavorable outcome), whereas four had no recurrence or confirmed recurrence after 9 months from diagnosis (favorable outcome). For three patients, follow‐up information was obtained less than 9 months from diagnosis, so although no progression was evident, these GBM patients were excluded from further analysis on the prognostic significance of small EVs (Table S1). Because of the known prognostic differences in patients with IDH mutation and the lack of information on the prognosis according to our 9‐month cutoff, the only patient with IDH mutation was excluded from the favorable/unfavorable analysis.
Isolation and characterization of salivary small EVs in GBM patients
Following isolation of salivary small EVs (Figure S1A), size distribution and concentrations were assessed by nanoparticle tracking analysis (NTA) (Figure 1A). The average size of salivary EVs from healthy controls, pre‐, and postoperative samples was 149.6 nm, 163.3, and 143.3 nm, respectively. The average concentration (particles/ml of saliva) of EVs was 8.8 × 109 in preoperative samples and 5.6 × 109 in postoperative samples whereas in healthy controls it was 3.95 × 109. Interestingly, although not statistically significant, there is a decrease in EV concentration in GBM patients after surgery (p = .1867). Additionally, TEM images corroborated the presence of small EVs, showing their typical cup‐shaped structure (Figure 1B). Immunoblotting of small EV markers was performed in a subset of samples (n = 7) (Figure 1C), showing positive bands for CD9, CD81, and CD63 (Figure 1C). Negative markers GM130 and Calnexin were absent in all isolated salivary EV samples, suggesting no contamination of larger EVs or cells 16 (Figures 1C and S1B). Taken together, these results confirm the successful isolation of small EVs from saliva of GBM patients.
FIGURE 1.
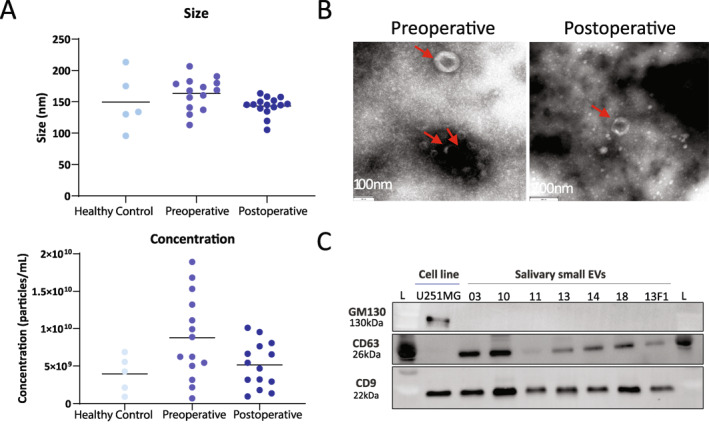
(A) Size and concentration of salivary small extracellular vesicles in healthy controls, and pre‐ and postoperative glioblastoma (GBM) patients. (B) Morphology of salivary small extracellular vesicles imaged by transmission electron microscopy. Representative images of the cup‐shaped morphology of extracellular vesicles (red arrow) isolated from pre‐ (left) and postoperative (right) samples. (C) Immunoblotting for positive (CD9 and CD63) and negative (GM130) markers of small extracellular vesicles isolated from saliva of GBM patients. F1 indicates postoperative sample; L, molecular weight ladder; U251MG, commercial GBM cell line.
Proteomic profiling of small EVs in pre‐ and postoperative saliva samples from GBM patients
Following the characterization step confirming the enrichment of small EVs in our samples, a total of 12.5 µg of protein was used for data‐dependent acquisition (DDA)‐MS and SWATH‐MS analysis. The workflow is shown in Figure 2A. A total of 507 proteins were identified by the DDA analysis in the small EVs fraction isolated from saliva samples of GBM patients. Of these, 238 (47.0%) were found only in preoperative samples, 215 (42.4%) were detected in both conditions, and only 54 (10.6%) were found exclusively postoperatively (Figure 2B).
FIGURE 2.
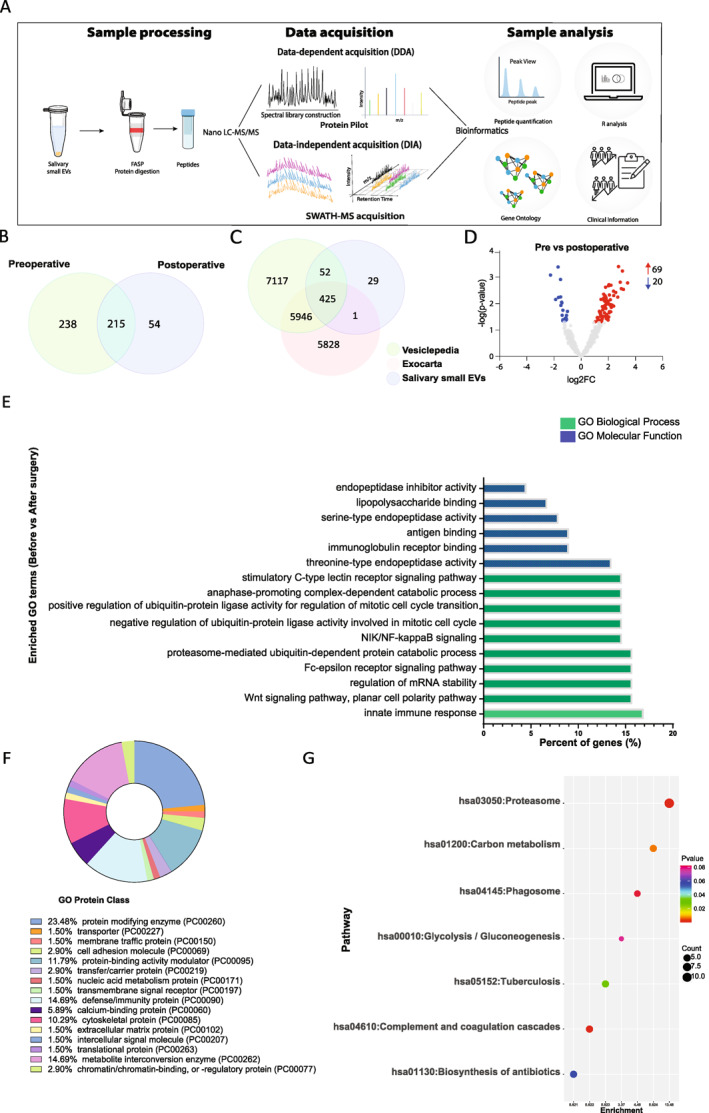
(A) The study workflow. Following isolation of small extracellular vesicles (EVs), proteins were aliquoted and protein digestion was performed. The peptides were concentrated, and mass spectrometry was performed for data‐dependent and data‐independent acquisition (SWATH‐MS). Protein Pilot software was used for peptide identification. Bioinformatic analyses were performed and correlated with clinical information from patients. (B) Venn diagram of identified proteins in salivary EVs pre‐ and postoperative (C) Venn diagram of all proteins identified in salivary small EV samples of glioblastoma (GBM) patients compared to proteins annotated in two EV databases, Exocarta and Vesiclepedia. (D) Volcano plot identifying proteins from salivary small EVs by their log2‐fold changes (log2FC) against their corresponding adjusted p value in patients before and after surgery. Red and blue dots represent proteins with significantly higher and lower abundance, respectively. (E) Gene Ontology analyses of top 10 biological processes (green) and molecular functions (blue) of differentially abundant proteins from pre‐ and postoperative GBM patients generated using Database for Annotation, Visualization and Integrated Discovery. (F) Donut Chart of Gene Ontology Protein class analysis. Graph generated using Panther classification 44 (http://www.pantherdb.org/). (G) Bubble chart showing Kyoto Encyclopedia of Genes and Genomes pathway analysis of differentially abundant proteins in pre‐ and postoperative salivary EV samples.
The identified proteins were compared to the ones reported in known EVs databases. We cross‐referenced our findings with ExoCarta (v5) 46 , 47 , 48 and Vesiclepedia (v4.1) 49 , 50 (Figure 2C). We observed that 478 of 507 (94%) proteins identified in our study have been previously reported in at least one of the databases. There were 29 exclusive identifications in our spectral library listed in Table S2. In addition, quantitative analysis was performed on the identified proteins using SWATH‐MS. A total of 89 significant differentially abundant proteins (DAPs) were identified between pre‐ and postoperative saliva samples from GBM patients (Table S3). Among them, 69 were more abundant in patients before surgery, whereas 20 were less abundant (Figure 2D).
GO and signaling pathway enrichment analyses in salivary small EVs pre‐ and postoperative
To investigate the functional significance of DAPs between pre‐ and postoperative saliva samples, functional enrichment analyses were performed, including GO enrichment and KEGG pathway analysis. Several biological processes (BP) were found to be significantly enriched (Figure 2E; Table S4). The three most notable BP included innate immune response (FDR p = 9.40 × 10−07), Wnt signaling pathway (FDR p = 2.20 × 10−13), and proteasome‐mediated ubiquitin‐dependent protein catabolic process (FDR p = 1.10 × 10−09). For the annotated molecular function (MF), proteins were significantly enriched in threonine‐type endopeptidase activity (FDR p = 1.50 × 10−18), immunoglobulin receptor binding (FDR p = 3.10 × 10−09), lipopolysaccharide binding (FDR p = 3.80 × 10−06), antigen binding (FDR p = 3.40 × 10−05), endopeptidase inhibitor activity (FDR p = 3.40 × 10−02), and serine‐type endopeptidase activity (FDR p = 4.90 × 10−02) (Figure 2E, Table S5). The interaction network generated of BP and MF is shown in Figure S2. Some cluster interactions can be observed within proteins related to the immune system (e.g., humoral immune system, defense response to bacterium, and antimicrobial humoral response). Other proteins presented disconnected members, unrelated to the other proteins (e.g., extracellular matrix disassembly, protein activation cascade, and regulation of protein processing). In addition, we have classified protein class, showing that 23.48% of proteins were enriched in protein modifying enzyme (i.e., a protein that directly covalently modifies another protein), whereas 14.69% in defense/immunity protein (Figure 2F). KEGG pathways involved were also classified (Figure 2G), and the most representative functional pathway was related to the proteasome (FDR p = 2.46 × 10−13), which was also observed in the clustering of proteasome‐related proteins in the PPI network (Figure S3). In addition, significant enrichment of pathways of complement and coagulation cascades (p = 1.20 × 10−3), carbon metabolism (p = 7.15 × 10−3), and tuberculosis (p = 3.20 × 10−2) was detected (Figure 2G). Reactome pathway enrichment revealed 147 overrepresented pathways (FDR p < 0.05) (Figure S3). Among them, the top 10 (FDR p = 7.88 × 10−15) were neutrophil degranulation, regulation of activated PAK‐2p34 by proteasome‐mediated degradation, auto degradation of Cdh1 by Cdh1:APC/C, CF‐β‐TrCP mediated degradation of Emi1, CDT1 association with the CDC6:ORC:origin complex, APC/C:Cdc20‐mediated degradation of Securin, Vif‐mediated degradation of APOBEC3G, degradation of AXIN, Orc1 removal from chromatin, CDK‐mediated phosphorylation, and removal of Cdc6. Overall, through the functional enrichment analyses, we have observed that the majority of proteins were associated with the immune system, including the innate immune system (e.g., neutrophil degranulation and FCERI‐mediated nuclear factor–κB activation) and also the adaptive immune system with antigen processing: ubiquitination and proteasome degradation, and downstream TCR signaling. In addition, pathways related to the cell cycle and programmed cell death were also enriched in the isolated small EVs (Figure S4).
Prognostic potential of salivary small EVs in GBM
To investigate the relationship between the protein content of small EVs and patients' clinical outcomes, we separated the GBM cohort into patients with favorable outcomes (progression‐free survival [PFS] ≥9 months) and unfavorable outcomes (PFS <9 months). This conservative cutoff was established considering that the average time for a GBM patient to present disease recurrence is usually within 6 to 9 months from diagnosis. 3 , 4 , 5
The vesicles size was not altered according to the patients' prognosis (Figure 3A). However, there is a significant difference (p = .0242) when comparing EV concentration between favorable and unfavorable outcomes preoperatively (Figure 3B). We further performed a PLS‐DA of the total salivary‐EV proteome signatures in pre‐ (Figure 3C) and postoperative (Figure 3D) samples of patients with favorable (blue) or unfavorable (red) outcomes. The resulting score plots of the analysis showed a clear separation between patients with favorable and unfavorable prognoses in the preoperative condition. However, postoperatively, we observed a partial overlap between patients. Next, we identified the DAPs in each group pre‐ (Figure 3E) and postoperatively (Figure 3F). Before surgery, a total of 66 DAPs were detected, among them, 64 were more abundant in patients with unfavorable outcomes, whereas two were less abundant. After surgery, 15 DAPs were identified, five more abundant and 10 less abundant in patients with unfavorable outcomes. A list of all DAPs is available in Table S10. The GO analysis of these DAPs pre‐ and postoperative is shown in Tables S6 and S8 (BP) and Tables S7 and S9 (MF).
FIGURE 3.
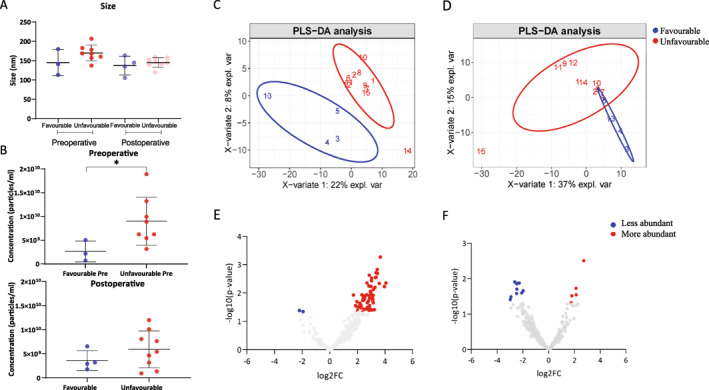
(A) Size of salivary small extracellular vesicles of glioblastoma (GBM) patients with favorable and unfavorable outcomes in pre‐ and postoperative samples. (B) Concentration of salivary small extracellular vesicles of GBM patients with favorable and unfavorable outcomes in pre‐ and postoperative samples. The Mann‐Whitney test (GraphPad Prism) was used to determine significance (*p < .05). Partial least squares‐discriminant analysis score plots of proteome signatures in the total salivary small extracellular vesicles (EVs) from GBM patients with favorable prognosis (blue) and unfavorable prognosis (red) (C) pre‐ and (D) postoperative. Volcano plots of all proteins from salivary small EVs of patients with unfavorable and favorable prognoses (E) pre‐ and (F) postoperative. Plots correspond to proteins' log2‐fold changes against their corresponding adjusted p value. Red dots represent more abundant proteins. Blue dots correspond to less abundant proteins.
Considering the more representative separation from the PLS‐DA of preoperative samples (Figure 3C), we selected four proteins for further investigation, namely aldolase A (ALDOA), 14‐3‐3 protein ε (1433E), transmembrane protease serine 11B (TM11B), and enoyl CoA hydratase 1 (ECH1) (Figure 4A). Our criteria for selection included (1) a fold change of at least 1.5, (2) p value <.05, and (3) biological relevance. All protein candidates presented increased abundance in patients with unfavorable outcomes compared to patients with favorable outcomes. ALDOA, 1433E, and ECH1 abundance was also verified by Western blotting considering their biological relevance. For ALDOA (Figure 4B), we confirmed that patients with unfavorable outcomes presented with visually stronger bands compared to patients with a good outcome. 1433E and ECH1 were not detected in the Western blot (Figure S5). This result might be due to the low abundance of both proteins (target protein abundance relative to total protein content = 1 × 10−5). Additionally, we performed a ROC curve analysis of ALDOA, which showed a sensitivity of 90.91% and specificity of 100.0% (Figure 4C).
FIGURE 4.
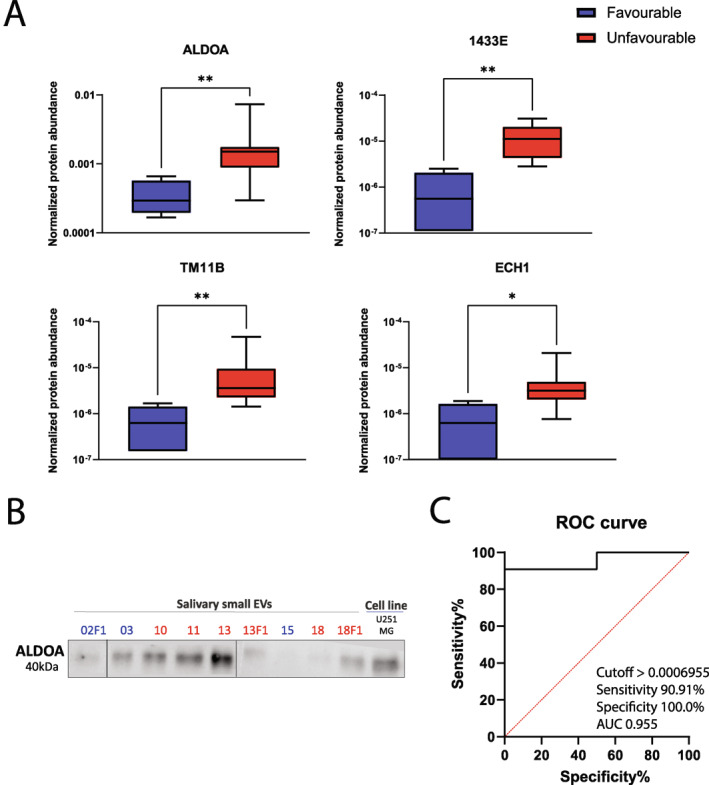
(A) Box‐and‐whisker plots of normalized protein abundance of four protein biomarker candidates preoperatively (aldolase A [ALDOA], 14‐3‐3 protein ε, enoyl CoA hydratase 1, and transmembrane protease serine 11B). The Mann‐Whitney test (GraphPad Prism) was used to determine significance (*p < .05; **p < .01). (B) Verification of ALDOA using an independent method, Western blotting, in patients with favorable (numbers colored in blue) and unfavorable (numbers colored in red) prognoses. (C) Receiver operator characteristic curve analysis of ALDOA in preoperative salivary extracellular vesicles from glioblastoma patients with favorable and unfavorable outcomes.
DISCUSSION
We have demonstrated for the first time the feasibility of isolating and characterizing small EVs from saliva samples (pre‐ and post‐treatment) of GBM patients. Pre‐ and post‐treatment saliva samples demonstrated no significant differences in relation to particle size and concentration of small EVs. However, patients with unfavorable outcomes presented with a higher concentration of EV particles in preoperative samples compared to patients with favorable outcomes. We also found four differentially abundant proteins (ALDOA, 1433E, TM11B, and ECH1) in saliva samples from GBM patients with unfavorable outcomes compared to patients with favorable outcomes. One of the highly abundant proteins, ALDOA, is an enzyme that plays a major role in glycolysis and the maintenance of glucose. 51 A study has shown that the knockdown of genes regulating glycolysis, including ALDOA, in GBM cells reduced cell viability, migration, and invasion. 52 , 53 In contrast, overexpression of ALDOA has been correlated with higher cellular proliferation leading to poor prognosis in pancreatic cancer, 54 lung cancer, 55 , 56 colorectal cancer, 57 liver cancer, 58 gastric cancer, 59 and kidney cancers. 60 1433E (YWHAE) regulates the cell cycle and signaling pathways and genetic abnormalities in astrocytoma formation. Furthermore, there is evidence linking the downregulation of 14‐3‐3 ζ protein, a distinct isoform of the YWHA protein family, with human GBM cells becoming more responsive to apoptosis induction. 61 For TM11B, the knockdown of another transmembrane protease serine family member, TMPRSS3, inhibited cell proliferation, migration, invasion, and induced apoptosis of glioma cells. 62
We adhered to the International Society for Extracellular Vesicles (ISEV) recommendations (to use at least two complementary techniques to characterize small EVs: TEM, Western blotting, and NTA) when isolating and characterizing small EVs from saliva samples. 16 Using TEM, we observed a double‐layer membrane structure with a cup‐shaped morphology, corresponding to the size range of small EVs (<200 nm) in saliva samples. Our findings corroborate previously published data. 63 , 64 , 65 In addition, we observed that the concentration of small EVs was higher in the preoperative samples compared to postoperative saliva samples and healthy controls (Figure 1A). A reduction in small EVs' concentration post‐surgery may be attributed to a reduction of EVs being released by the tumor following surgery. Similarly, Osti et al. 66 have identified a higher concentration of EV in GBM patients compared to healthy controls, brain metastasis, and extra‐axial brain tumors patients. The authors have also observed a decrease in plasmatic EVs in GBM patients post‐surgery compared to pre‐surgery, which corroborates our data.
Recent studies have reported that the protein content of small EVs reflects the phenotypic signature of GBM cells. 17 , 67 This would mean that identifying the protein cargo associated with small EVs may help to address the lack of prognostic biomarkers for GBM. 68 Although EVs have been investigated in other body fluids derived from GBM patients, 69 , 70 , 71 , 72 thus far, there are no studies in literature on small EVs isolated from saliva of GBM patients. We identified 507 proteins from pre‐ and post‐treatment saliva samples. The majority of the proteins detected in our study have been identified in two publicly available EV databases (Exocarta and Vesiclepedia). In addition, we have also found unique proteins in salivary EVs of GBM patients and these were predominantly immunoglobulins and proteins involved in the TGF‐β signaling pathway. We reported a total of 89 DAPs in salivary small EVs between pre‐ and postoperative GBM patients that is comparable to the 102 DAPs reported from plasma‐derived EVs of GBM patients. 63 , 64 , 65 The main GO annotations enriched in our proteome were associated with the immune system.
Proteins identified in salivary EVs from GBM patients have been found in previous findings of GBM‐derived blood EVs and conditioned media harvested from GBM cells. Seven of the DAPs identified in our study were included in a list of “potential glioblastoma markers” using EVs from GBM cell lines 73 and 11 were included in a list of “GBM EV protein signature” 66 using plasmatic EVs. 66 , 73 , 74 Mainly, these proteins are involved in the complement and coagulation cascade and regulation of iron metabolism. 66 They have been reported to be differentially abundant in EVs 66 derived from plasma and serum of GBM patients. 75 A common marker across all studies is C3. C3 is a member of the complement system and plays a critical role in the innate immunity. 76 PPIA, which regulates protein folding and trafficking, has been reported to be upregulated and to play a key role in the progression of a number of cancer types. 77 These results suggest that we have successfully enriched in small EVs associated with GBM.
Although we have adhered to current ISEV guidelines for isolation and characterization of small EVs, it is important to highlight that there is no consensus on a standard technique to isolate and characterize EVs. Another limitation of our study is the small patient cohort due to the rarity of GBM. Although our results show an enrichment of salivary small EV proteins associated with a “GBM signature” reported from other studies using plasmatic EVs or GBM‐cell conditioned media, it is essential to interpret our data with caution. Because not only tumor cells secrete small EVs, but nearly all cells in our body are constantly releasing small EVs, our results from EVs' cargo may also be reflecting the changes due to systemic inflammation and not only from tumor‐derived EVs. Most of the patients were receiving medication, including corticosteroids, which may influence both the vesicles release and their protein pattern, as previously reported in the literature. 78 Further research should be undertaken to investigate their prognostic potential in GBM. A noninvasive assessment of GBM using saliva, independent of tumor tissue obtained at brain surgery, sheds light on novel possibilities for GBM subtyping detection, monitoring of disease progression, and serial sample collections to analyze the tumor behavior post‐treatment.
Our preliminary data demonstrate the feasibility of isolating and characterizing small EVs from pre‐ and postoperative saliva samples from GBM patients. There were no statistically significant differences in size and concentration of small EVs between both time points, however, a distinct protein profile was observed. Our preliminary findings encourage further large cohort validation studies on salivary small EVs to evaluate prognosis in GBM.
AUTHOR CONTRIBUTIONS
Juliana Müller Bark: Conceptualization, formal analysis, investigation, methodology, project administration, writing–original draft, and writing–review and editing. Lucas Trevisan França de Lima: Formal analysis, investigation, methodology, writing–original draft, and writing–review and editing. Xi Zhang: Formal analysis, supervision, and writing–review and editing. Daniel Broszczak: Data curation, formal analysis, and writing–review and editing. Paul J. Leo: Conceptualization, supervision, and writing–review and editing. Rosalind L. Jeffree: Conceptualization, resources, supervision, and writing–review and editing. Benjamin Chua: Conceptualization, supervision, and writing–review and editing. Bryan W. Day: Conceptualization, supervision, and writing–review and editing. Chamindie Punyadeera: Conceptualization, formal analysis, funding acquisition, methodology, project administration, resources, supervision, and writing–review and editing.
CONFLICT OF INTEREST STATEMENT
Chamindie Punyadeera reports holding a patent for biomarkers for glioblastoma prognosis. The other authors declare no conflicts of interest.
Supporting information
Supporting Information S1
ACKNOWLEDGMENTS
The authors thank Trang Le, Jenny Edmunds, Charmaine Micklewright, Jacqui Keller (clinical trials coordinators at RBWH‐Brisbane, Australia), the Central Analytical Research Facility (CARF), QUT, Rebecca Fieth (CARF‐QUT), and Raj Gupta (CARF‐QUT). Figures S1 and 2A were created using Servier Medical Art templates, licensed under a Creative Commons Attribution 3.0 Unported License (https://smart.servier.com). Juliana Müller Bark is funded by ATM LATAM QUT Postgraduate Research Scholarship. This work was funded by the Royal Brisbane and Women's Hospital (RBWH) Foundation projects grant. Chamindie Punyadeera is currently receiving funding from Cancer Australia (APP1145657 and 2012560), NIH R21, NHMRC Ideas Grant (APP 2002576 and APP), and RBWH Foundation Project Grants. Bryan W. Day received funding from The Sid Faithfull Group and Cure Brain Cancer Foundation to conduct this study.
Open access publishing facilitated by Griffith University, as part of the Wiley ‐ Griffith University agreement via the Council of Australian University Librarians.
Müller Bark J, Trevisan França de Lima L, Zhang X, et al. Proteome profiling of salivary small extracellular vesicles in glioblastoma patients. Cancer. 2023;129(18):2836‐2847. doi: 10.1002/cncr.34888
REFERENCES
- 1. Stupp R, Mason WP, van den Bent MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352(10):987‐996. doi: 10.1056/nejmoa043330 [DOI] [PubMed] [Google Scholar]
- 2. Jeffree RL. Current management of cerebral gliomas. Aust J Gen Pract. 2020;49(4):194‐199. doi: 10.31128/ajgp-09-19-5063 [DOI] [PubMed] [Google Scholar]
- 3. Twelves C, Sabel M, Checketts D, et al. A phase 1b randomised, placebo‐controlled trial of nabiximols cannabinoid oromucosal spray with temozolomide in patients with recurrent glioblastoma. Br J Cancer. 2021;124(8):1379‐1387. doi: 10.1038/s41416-021-01259-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Weller M, Cloughesy T, Perry JR, Wick W. Standards of care for treatment of recurrent glioblastoma—are we there yet? Neuro Oncol. 2013;15(1):4‐27. doi: 10.1093/neuonc/nos273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Shergalis A, Bankhead A, 3rd , Luesakul U, Muangsin N, Neamati N. Current challenges and opportunities in treating glioblastoma. Pharmacol Rev. 2018;70(3):412‐445. doi: 10.1124/pr.117.014944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Arevalo OD, Soto C, Rabiei P, et al. Assessment of glioblastoma response in the era of bevacizumab: longstanding and emergent challenges in the imaging evaluation of pseudoresponse. Front Neurol. 2019;10:460. doi: 10.3389/fneur.2019.00460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Stupp R, Brada M, van den Bent MJ, Tonn JC, Pentheroudakis G, Group EGW. High‐grade glioma: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow‐up. Ann Oncol. 2014;25(Suppl 3):iii93‐101. doi: 10.1093/annonc/mdu050 [DOI] [PubMed] [Google Scholar]
- 8. Szopa W, Burley TA, Kramer‐Marek G, Kaspera W. Diagnostic and therapeutic biomarkers in glioblastoma: current status and future perspectives. BioMed Res Int. 2017;2017:8013575. doi: 10.1155/2017/8013575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Delgado‐Lopez PD, Rinones‐Mena E, Corrales‐Garcia EM. Treatment‐related changes in glioblastoma: a review on the controversies in response assessment criteria and the concepts of true progression, pseudoprogression, pseudoresponse and radionecrosis. Clin Transl Oncol. 2018;20(8):939‐953. doi: 10.1007/s12094-017-1816-x [DOI] [PubMed] [Google Scholar]
- 10. Ellingson BM, Chung C, Pope WB, Boxerman JL, Kaufmann TJ. Pseudoprogression, radionecrosis, inflammation or true tumor progression? challenges associated with glioblastoma response assessment in an evolving therapeutic landscape. J Neuro Oncol. 2017;134(3):495‐504. doi: 10.1007/s11060-017-2375-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Muller Bark J, Kulasinghe A, Chua B, Day BW, Punyadeera C. Circulating biomarkers in patients with glioblastoma. Br J Cancer. 2019;122(3):295‐305. doi: 10.1038/s41416-019-0603-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Shankar GM, Balaj L, Stott SL, Nahed B, Carter BS. Liquid biopsy for brain tumors. Expert Rev Mol Diagn. 2017;17(10):943‐947. doi: 10.1080/14737159.2017.1374854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Best MG, Sol N, Zijl S, Reijneveld JC, Wesseling P, Wurdinger T. Liquid biopsies in patients with diffuse glioma. Acta Neuropathol. 2015;129(6):849‐865. doi: 10.1007/s00401-015-1399-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ramirez SH, Andrews AM, Paul D, Pachter JS. Extracellular vesicles: mediators and biomarkers of pathology along CNS barriers. Fluids Barriers CNS. 2018;15(1):19. doi: 10.1186/s12987-018-0104-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. García‐Romero N, Carrión‐Navarro J, Esteban‐Rubio S, et al. DNA sequences within glioma‐derived extracellular vesicles can cross the intact blood‐brain barrier and be detected in peripheral blood of patients. Oncotarget. 2017;8(1):1416‐1428. doi: 10.18632/oncotarget.13635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Théry C, Witwer KW, Aikawa E, et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J Extracell Vesicles. 2018;7(1):1535750. doi: 10.1080/20013078.2018.1535750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kalluri R. The biology and function of exosomes in cancer. J Clin Invest. 2016;126(4):1208‐1215. doi: 10.1172/jci81135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Colombo M, Raposo G, Thery C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu Rev Cell Dev Biol. 2014;30(1):255‐289. doi: 10.1146/annurev-cellbio-101512-122326 [DOI] [PubMed] [Google Scholar]
- 19. Raposo G, Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol. 2013;200(4):373‐383. doi: 10.1083/jcb.201211138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pfaffe T, Cooper‐White J, Beyerlein P, Kostner K, Punyadeera C. Diagnostic potential of saliva: current state and future applications. Clin Chem. 2011;57(5):675‐687. doi: 10.1373/clinchem.2010.153767 [DOI] [PubMed] [Google Scholar]
- 21. Schulz BL, Cooper‐White J, Punyadeera CK. Saliva proteome research: current status and future outlook. Crit Rev Biotechnol. 2013;33(3):246‐259. doi: 10.3109/07388551.2012.687361 [DOI] [PubMed] [Google Scholar]
- 22. Malamud D. Saliva as a diagnostic fluid. Dent Clin North Am. 2011;55(1):159‐178. doi: 10.1016/j.cden.2010.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhang C‐Z, Cheng X‐Q, Li J‐Y, et al. Saliva in the diagnosis of diseases. Int J Oral Sci. 2016;8(3):133‐137. doi: 10.1038/ijos.2016.38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nonaka T, Wong DTW. Saliva‐exosomics in cancer: molecular characterization of cancer‐derived exosomes in saliva. Enzymes. 2017;42:125‐151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Trevisan França de Lima L, Müller Bark J, Rasheduzzaman M, Ekanayake Weeramange C, Punyadeera C. Chapter 10 ‐ Saliva as a matrix for measurement of cancer biomarkers. In: Ramanathan LV, Fleisher M, Duffy MJ, eds. Cancer Biomarkers. Elsevier; 2022:297‐351. [Google Scholar]
- 26. Suma H, Prabhu K, Shenoy R, Annaswamy R, Rao S, Rao A. Estimation of salivary protein thiols and total antioxidant power of saliva in brain tumor patients. J Cancer Res Therapeut. 2010;6(3):278‐281. doi: 10.4103/0973-1482.73357 [DOI] [PubMed] [Google Scholar]
- 27. García‐Villaescusa A, Morales‐Tatay JM, Monleón‐Salvadó D, et al. Using NMR in saliva to identify possible biomarkers of glioblastoma and chronic periodontitis. PLoS One. 2018;13(2):e0188710. doi: 10.1371/journal.pone.0188710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Han Y, Jia L, Zheng Y, Li W. Salivary exosomes: emerging roles in systemic disease. Int J Biol Sci. 2018;14(6):633‐643. doi: 10.7150/ijbs.25018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Langevin S, Kuhnell D, Parry T, et al. Comprehensive microRNA‐sequencing of exosomes derived from head and neck carcinoma cells in vitro reveals common secretion profiles and potential utility as salivary biomarkers. Oncotarget. 2017;8(47):82459‐82474. doi: 10.18632/oncotarget.19614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tang KD, Wan Y, Zhang X, et al. Proteomic alterations in salivary exosomes derived from human papillomavirus‐driven oropharyngeal cancer. Mol Diagn Ther. 2021;25(4):505‐515. doi: 10.1007/s40291-021-00538-2 [DOI] [PubMed] [Google Scholar]
- 31. Machida T, Tomofuji T, Maruyama T, et al. miR‐1246 and miR‐4644 in salivary exosome as potential biomarkers for pancreatobiliary tract cancer. Oncol Rep. 2016;36(4):2375‐2381. doi: 10.3892/or.2016.5021 [DOI] [PubMed] [Google Scholar]
- 32. Lau C, Kim Y, Chia D, et al. Role of pancreatic cancer‐derived exosomes in salivary biomarker development. J Biol Chem. 2013;288(37):26888‐26897. doi: 10.1074/jbc.m113.452458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sun Y, Huo C, Qiao Z, et al. Comparative proteomic analysis of exosomes and microvesicles in human saliva for lung cancer. J Proteome Res. 2018;17(3):1101‐1107. doi: 10.1021/acs.jproteome.7b00770 [DOI] [PubMed] [Google Scholar]
- 34. Sun Y, Xia Z, Shang Z, et al. Facile preparation of salivary extracellular vesicles for cancer proteomics. Sci Rep. 2016;6(1):24669. doi: 10.1038/srep24669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhang X, Walsh T, Atherton JJ, Kostner K, Schulz B, Punyadeera C. Identification and validation of a salivary protein panel to detect heart failure early. Theranostics. 2017;7(18):4350‐4358. doi: 10.7150/thno.21727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sun CX, Bennett N, Tran P, et al. A pilot study into the association between oral health status and human papillomavirus‐16 infection. Diagnostics. 2017;7(1):11. doi: 10.3390/diagnostics7010011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Raposo G, Nijman HW, Stoorvogel W, et al. B lymphocytes secrete antigen‐presenting vesicles. J Exp Med. 1996;183(3):1161‐1172. doi: 10.1084/jem.183.3.1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Théry C, Amigorena S, Raposo G, Clayton A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr Protoc Cell Biol. 2006;30(1). Chapter 3:Unit 3.22. doi: 10.1002/0471143030.cb0322s30 [DOI] [PubMed] [Google Scholar]
- 39. Kapeleris J, Müller Bark J, Ranjit S, et al. Modeling reoxygenation effects in non‐small cell lung cancer cell lines and showing epithelial‐mesenchymal transition. J Cancer Res Clin Oncol. 2022;148(12):3501‐3510. doi: 10.1007/s00432-022-04242-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhang X, Sadowski P, Punyadeera C. Evaluation of sample preparation methods for label‐free quantitative profiling of salivary proteome. J Proteonomics. 2020;210:103532. doi: 10.1016/j.jprot.2019.103532 [DOI] [PubMed] [Google Scholar]
- 41. Escher C, Reiter L, MacLean B, et al. Using iRT, a normalized retention time for more targeted measurement of peptides. Proteomics. 2012;12(8):1111‐1121. doi: 10.1002/pmic.201100463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zang T, Broszczak DA, Cuttle L, Broadbent JA, Tanzer C, Parker TJ. The blister fluid proteome of pediatric burns. J Proteonomics. 2016;146:122‐132. doi: 10.1016/j.jprot.2016.06.026 [DOI] [PubMed] [Google Scholar]
- 43. Escher C, Reiter L, MacLean B, et al. Using iRT, a normalized retention time for more targeted measurement of peptides. Proteomics. 2012;12(8):1111‐1121. doi: 10.1002/pmic.201100463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mi H, Ebert D, Muruganujan A, et al. PANTHER version 16: a revised family classification, tree‐based classification tool, enhancer regions and extensive API. Nucleic Acids Res. 2021;49(D1):D394‐D403. doi: 10.1093/nar/gkaa1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Choi M, Chang CY, Clough T, et al. MSstats: an R package for statistical analysis of quantitative mass spectrometry‐based proteomic experiments. Bioinformatics. 2014;30(17):2524‐2526. doi: 10.1093/bioinformatics/btu305 [DOI] [PubMed] [Google Scholar]
- 46. Keerthikumar S, Chisanga D, Ariyaratne D, et al. ExoCarta: a web‐based compendium of exosomal cargo. J Mol Biol. 2016;428(4):688‐692. doi: 10.1016/j.jmb.2015.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Mathivanan S, Fahner CJ, Reid GE, Simpson RJ. ExoCarta 2012: database of exosomal proteins, RNA and lipids. Nucleic Acids Res. 2012;40(D1):D1241‐D4. doi: 10.1093/nar/gkr828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Mathivanan S, Simpson RJ. ExoCarta: a compendium of exosomal proteins and RNA. Proteomics. 2009;9(21):4997‐5000. doi: 10.1002/pmic.200900351 [DOI] [PubMed] [Google Scholar]
- 49. Pathan M, Fonseka P, Chitti SV, et al. Vesiclepedia 2019: a compendium of RNA, proteins, lipids and metabolites in extracellular vesicles. Nucleic Acids Res. 2019;47(D1):D516‐d9. doi: 10.1093/nar/gky1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kalra H, Simpson RJ, Ji H, et al. Vesiclepedia: a compendium for extracellular vesicles with continuous community annotation. PLoS Biol. 2012;10(12):e1001450. doi: 10.1371/journal.pbio.1001450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Rose IA, O'Connell EL. Studies on the interaction of aldolase with substrate analogues. J Biol Chem. 1969;244(1):126‐134. doi: 10.1016/s0021-9258(19)78201-5 [DOI] [PubMed] [Google Scholar]
- 52. Sanzey M, Abdul Rahim SA, Oudin A, et al. Comprehensive analysis of glycolytic enzymes as therapeutic targets in the treatment of glioblastoma. PLoS One. 2015;10(5):e0123544. doi: 10.1371/journal.pone.0123544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Sun J, He D, Fu Y, et al. A novel lncRNA ARST represses glioma progression by inhibiting ALDOA‐mediated actin cytoskeleton integrity. J Exp Clin Cancer Res. 2021;40(1):187. doi: 10.1186/s13046-021-01977-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ji S, Zhang B, Liu J, et al. ALDOA functions as an oncogene in the highly metastatic pancreatic cancer. Cancer Lett. 2016;374(1):127‐135. doi: 10.1016/j.canlet.2016.01.054 [DOI] [PubMed] [Google Scholar]
- 55. Chang Y‐C, Chan Y‐C, Chang W‐M, et al. Feedback regulation of ALDOA activates the HIF‐1α/MMP9 axis to promote lung cancer progression. Cancer Lett. 2017;403:28‐36. doi: 10.1016/j.canlet.2017.06.001 [DOI] [PubMed] [Google Scholar]
- 56. Chang YC, Chiou J, Yang YF, et al. Therapeutic targeting of aldolase a interactions inhibits lung cancer metastasis and prolongs survival. Cancer Res. 2019;79(18):4754‐4766. doi: 10.1158/0008-5472.can-18-4080 [DOI] [PubMed] [Google Scholar]
- 57. Yamamoto T, Kudo M, Peng W‐X, et al. Identification of aldolase A as a potential diagnostic biomarker for colorectal cancer based on proteomic analysis using formalin‐fixed paraffin‐embedded tissue. Tumour Biol. 2016;37(10):13595‐13606. doi: 10.1007/s13277-016-5275-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Li X, Jiang F, Ge Z, et al. Fructose‐bisphosphate Aldolase A regulates hypoxic adaptation in hepatocellular carcinoma and involved with tumor malignancy. Dig Dis Sci. 2019;64(11):3215‐3227. doi: 10.1007/s10620-019-05642-2 [DOI] [PubMed] [Google Scholar]
- 59. Jiang Z, Wang X, Li J, Yang H, Lin X. Aldolase A as a prognostic factor and mediator of progression via inducing epithelial‐mesenchymal transition in gastric cancer. J Cell Mol Med. 2018;22(9):4377‐4386. doi: 10.1111/jcmm.13732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Na N, Li H, Xu C, et al. High expression of Aldolase A predicts poor survival in patients with clear‐cell renal cell carcinoma. Therapeut Clin Risk Manag. 2017;13:279‐285. doi: 10.2147/tcrm.s123199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Zhu J, Wang H, Huang YQ, et al. Comprehensive analysis of a long non‐coding RNA‐associated competing endogenous RNA network in glioma. Oncol Lett. 2020;20(4):63. doi: 10.3892/ol.2020.11924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Huo J‐F, Chen X‐B. Knockdown of TMPRSS3 inhibits cell proliferation, migration/invasion and induces apoptosis of glioma cells. J Cell Biochem. 2019;120(5):7794‐7801. doi: 10.1002/jcb.28054 [DOI] [PubMed] [Google Scholar]
- 63. Kibria G, Ramos EK, Lee KE, et al. A rapid, automated surface protein profiling of single circulating exosomes in human blood. Sci Rep. 2016;6(1):36502. doi: 10.1038/srep36502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Palanisamy V, Sharma S, Deshpande A, Zhou H, Gimzewski J, Wong DT. Nanostructural and transcriptomic analyses of human saliva derived exosomes. PLoS One. 2010;5(1):e8577. doi: 10.1371/journal.pone.0008577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Jung MK, Mun JY. Sample preparation and imaging of exosomes by transmission electron microscopy. J Vis Exp. 2018(131). doi: 10.3791/56482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Osti D, Del Bene M, Rappa G, et al. Clinical significance of extracellular vesicles in plasma from glioblastoma patients. Clin Cancer Res. 2019;25(1):266‐276. doi: 10.1158/1078-0432.ccr-18-1941 [DOI] [PubMed] [Google Scholar]
- 67. Lane R, Simon T, Vintu M, et al. Cell‐derived extracellular vesicles can be used as a biomarker reservoir for glioblastoma tumor subtyping. Commun Biol. 2019;2(1):315. doi: 10.1038/s42003-019-0560-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Redzic JS, Ung TH, Graner MW. Glioblastoma extracellular vesicles: reservoirs of potential biomarkers. Pharmgenomics Pers Med. 2014;7:65‐77. doi: 10.2147/pgpm.s39768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Akers JC, Ramakrishnan V, Kim R, et al. miR‐21 in the extracellular vesicles (EVs) of cerebrospinal fluid (CSF): a platform for glioblastoma biomarker development. PLoS One. 2013;8(10):e78115. doi: 10.1371/journal.pone.0078115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Cumba Garcia LM, Peterson TE, Cepeda MA, Johnson AJ, Parney IF. Isolation and analysis of plasma‐derived exosomes in patients with glioma. Front Oncol. 2019;9(651). doi: 10.3389/fonc.2019.00651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Anastasi F, Greco F, Dilillo M, et al. Proteomics analysis of serum small extracellular vesicles for the longitudinal study of a glioblastoma multiforme mouse model. Sci Rep. 2020;10(1):20498. doi: 10.1038/s41598-020-77535-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Hallal S, Ebrahim Khani S, Wei H, et al. Deep Sequencing of small RNAs from neurosurgical extracellular vesicles substantiates miR‐486‐3p as a circulating biomarker that distinguishes glioblastoma from lower‐grade astrocytoma patients. Int J Mol Sci. 2020;21(14):4954. doi: 10.3390/ijms21144954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Naryzhny S, Volnitskiy A, Kopylov A, et al. Proteome of glioblastoma‐derived exosomes as a source of biomarkers. Biomedicines. 2020;8(7):216. doi: 10.3390/biomedicines8070216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Hallal S, Azimi A, Wei H, et al. A comprehensive proteomic SWATH‐MS workflow for profiling blood extracellular vesicles: a new avenue for glioma tumour surveillance. Int J Mol Sci. 2020;21(13):4754. doi: 10.3390/ijms21134754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Bouwens TA, Trouw LA, Veerhuis R, Dirven CM, Lamfers ML, Al‐Khawaja H. Complement activation in Glioblastoma multiforme pathophysiology: evidence from serum levels and presence of complement activation products in tumor tissue. J Neuroimmunol. 2015;278:271‐276. doi: 10.1016/j.jneuroim.2014.11.016 [DOI] [PubMed] [Google Scholar]
- 76. Dunkelberger JR, Song W‐C. Complement and its role in innate and adaptive immune responses. Cell Res. 2010;20(1):34‐50. doi: 10.1038/cr.2009.139 [DOI] [PubMed] [Google Scholar]
- 77. Nigro P, Pompilio G, Capogrossi MC. Cyclophilin A: a key player for human disease. Cell Death Dis. 2013;4(10):e888‐e. doi: 10.1038/cddis.2013.410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Kusuda Y, Kondo Y, Miyagi Y, et al. Long‐term dexamethasone treatment diminishes store‐operated Ca2+ entry in salivary acinar cells. Int J Oral Sci. 2019;11(1):1. doi: 10.1038/s41368-018-0031-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information S1


