Abstract
Using the cis-acting human cytomegalovirus (HCMV) packaging elements (pac 1 and pac 2) as DNA probes, specific DNA-protein complexes were detected by electrophoretic mobility shift assay (EMSA) in both HCMV-infected cell nuclear extracts and recombinant baculovirus-infected cell extracts containing the HCMV p130 (pUL56) protein. DNA-binding proteins, which were common in uninfected and infected cell extracts, were also detected. Mutational analysis showed that only the AT-rich core sequences in these cis-acting motifs, 5′-TAAAAA-3′ (pac 1) and 5′-TTTTAT-3′ (pac 2), were required for specific DNA-protein complex formation. The specificity of the DNA-protein complexes was confirmed by EMSA competition. Furthermore, a specific endonuclease activity was found to be associated with lysates of baculovirus-infected cells expressing recombinant p130 (rp130). This nuclease activity was time dependent, related to the amount of rp130 in the assay, and ATP independent. Nuclease activity remained associated with rp130 after partial purification by sucrose gradient centrifugation, suggesting that this activity is a property of HCMV p130. We propose a possible involvement of p130 in HCMV DNA packaging.
Human cytomegalovirus (HCMV), one of eight human herpesviruses, can cause serious illness in neonates as well as in immunocompromised adults (2). For example, transplant and AIDS patients may develop life-threatening diseases as a consequence of primary infection or reactivation of latent infection. Present therapeutic approaches are limited, and new strategies that may result from a better understanding of the molecular events involved in viral maturation are needed.
The HCMV virion consists of an envelope, an amorphous tegument, and an icosahedral nucleocapsid, which is assembled in the nuclei of infected cells. The precise molecular events of HCMV capsid assembly and subsequent DNA packaging are not well understood. It is generally accepted that viral DNA is packaged into a procapsid consisting of major capsid protein (UL86), minor capsid protein (UL85), minor capsid protein-binding protein (UL46), smallest capsid protein (UL47/48), assembly protein (UL80.5), and proteinase precursor protein (UL80a) (8). The assembly protein is removed during DNA insertion. It is unclear how the concatenated viral DNA contacts empty capsids and is cleaved and packaged into the capsid.
Recent studies with herpes simplex virus type 1 (HSV-1) mutants that were temperature sensitive suggest that cleavage of the concatenated DNA does not occur in the absence of packaging (1). One possible model would be the involvement of cleavage packaging protein(s) which could facilitate incorporation of DNA into the procapsid by attaching to a specific motif within the viral genome. With HSV-1, the UL36 gene product (ICP1) and a smaller protein (possibly encoded by UL37) are part of a complex that recognizes the HSV-specific a sequence and are required for cleavage and packaging of viral DNA from concatemers (6, 7). In addition, the HSV-1 ICP 18.5 (UL28) gene product and the pseudorabies virus (PrV) homolog (16) were also reported to play an important role in DNA packaging (1, 14). Addison et al. (1) demonstrated that empty capsids were observed under conditions nonpermissive for the expression of the HSV-1 ICP 18.5 gene product. The HSV-1 ICP 18.5 mutants failed to cleave concatenated viral DNA in noncomplementing cells, suggesting that cleavage and packaging require ICP 18.5. Similar results were reported by Mettenleiter et al. (14) for PrV mutant protein. These observations suggest that the HSV-1 UL36, UL37, and UL28 gene products are involved in cleavage and packaging of concatenated viral DNA.
In a recent study, we identified and partially characterized the gene product of HCMV UL56 (4). The HCMV UL56 gene product of 130 kDa is the homolog of the HSV-1 UL28 gene product. It is therefore postulated that UL56 possesses properties comparable to those of HSV-1 UL28, implying an involvement in cleavage and packaging of DNA. The HCMV genomic a sequence is a short sequence located at both termini of the genome and repeated in an inverted orientation at the L-S junction. The a sequence plays a key role in replication as a cis-acting signal for cleavage and packaging of progeny viral DNA and circularization of the viral genome. The HCMV a sequence contains two conserved motifs, pac 1 and pac 2, which are required for cleavage and packaging of the viral DNA (18). Both sequence motifs are located on one side of the cleavage site. The pac 1 and pac 2 motifs have an AT-rich core flanked by a GC-rich sequence. During the initial step of viral DNA packaging, a capsid-associated protein may bind to the pac sequences and may be involved in cleavage of the viral DNA concatemer.
In this study, electrophoretic mobility shift assays (EMSAs) were performed with DNA probes spanning the region of these cis-acting elements. These studies demonstrate that specific proteins from HCMV-infected nuclear extracts or baculovirus-UL56-infected cell extracts bind to the pac motifs. Using affinity-purified monospecific antibodies, we show that p130 is present in specific DNA-protein complexes containing the pac motifs of the viral genome. Furthermore, evidence is presented for a sequence-specific endonuclease activity of recombinant HCMV p130, using circular plasmid DNA bearing the a sequence as a substrate.
MATERIALS AND METHODS
Cells and viruses.
Human foreskin fibroblasts (HFF) were grown in Eagle’s minimum essential medium supplemented with 10% fetal calf serum, vitamins, nonessential amino acids, glutamine, and antibiotics (penicillin [0.5 U/ml] and gentamicin [60 μg/ml]) as described previously (4). HCMV AD169 stocks were prepared as described previously (3). Baculovirus (Autographa california nuclear polyhedrosis virus) was grown in 5B1-4 (High Five) cells or SF158 insect cells as described previously (19).
Plasmid construction and oligonucleotides.
Restriction enzymes were purchased from New England Biolabs (Beverly, Mass.) or Boehringer Mannheim (Mannheim, Germany) and used as instructed by the manufacturer. A HindIII-to-EcoRI fragment from pRC/CMV-UL56 was cloned into the same sites of the baculovirus shuttle vector pBacPAK9 (Clontech, Palo Alto, Calif.) to yield plasmid pBac-UL56. Plasmid pBacPAK8-GUS (Clontech) contains the gene coding for β-glucuronidase (GUS). Recombinant baculovirus expressing either p130 (UL56) or GUS was isolated as described by Clontech. The following single-stranded oligonucleotides and their complementary oligonucleotides were purified by polyacrylamide gel electrophoresis (PAGE) in 15% polyacrylamide gels containing 45 mM Tris-HCl (pH 8.3), 45 mM boric acid, and 1 mM EDTA: 5′-ATTTCACCCCCCCGCTAAAAACACCCCCCCGCCCCAC-3′ and 5′-CGTGCGCGCCGCACGCCGCTTTTATGCGCCGCCGCCGTCCCA-3′. Complementary oligonucleotides were combined, denatured for 2 min at 82°C, and annealed overnight at room temperature. The double-stranded oligonucleotides were designated pac 1 and pac 2, respectively. Mutant pac 1 and pac 2 DNAs were annealed as described above and elsewhere in the text. The AP1 and the SP1 consensus oligonucleotides, 5′-CGCTTGATGAGTCAGCCGGAA-3′ and 5′-ATTCGATCGGGGCGGGGCGAGC-3′, respectively, and purified AP1 and SP1 proteins were purchased from Promega (Madison, Wis.).
For generation of labeled probes, 50 ng of each double-stranded DNA was end labeled with [γ-32P]ATP and T4 polynucleotide kinase (Promega). Unincorporated radioactive ATP was removed by using Sephadex G50-150 spin columns.
Nuclear extracts.
Extracts were prepared from mock- or HCMV-infected HFF (multiplicity of infection of 5) by a modification of the method of Lee and Green (13). Cells were harvested 48 h after infection, rinsed twice with cold buffer A containing 10 mM HEPES (pH 7.9), 1.5 mM MgCl2, 10 mM KCl, and 0.5 mM dithiothreitol (DTT) with protease inhibitors (0.5 mM phenylmethylsulfonyl fluoride, 5 μg of leupeptin per ml, 2 μg of pepstatin per ml, and 1.0 mM TLCK [Nα-p-tosyl-l-lysine chloromethyl ketone]), suspended in 10 volumes buffer A, and allowed to swell at 4°C for 20 min. The cells were sedimented by centrifugation at 1,000 × g for 5 min, suspended in 1.5 volumes buffer A, and lysed with 11 strokes in a Dounce homogenizer (B pestle). The nuclei were sedimented and resuspended in buffer C containing 20 mM HEPES (pH 7.9), 25% (vol/vol) glycerol, 0.45 M NaCl, 1.5 mM MgCl2, and 0.2 mM EDTA with the same protease inhibitors as in buffer A. The salt extraction was performed for 30 min at 4°C. After centrifugation, the supernatant was dialyzed against buffer D containing 20 mM HEPES (pH 7.9), 20% (vol/vol) glycerol, 0.1 M KCl, and 0.2 mM EDTA with protease inhibitors. Precipitates were sedimented, and the supernatants were frozen in aliquots with liquid nitrogen. Whole-cell lysates were prepared from baculovirus-infected insect cells as described above but without separation of the nuclear and cytoplasmic fractions.
EMSA.
The binding reaction mixture contained nuclear extracts (1 μg) from either mock-infected or HCMV-infected HFF. Whole-cell extracts (1 μg) were used from the recombinant baculovirus-infected insect cells. In addition, each binding reaction mixture contained 2 μg of poly(dA-dT) · poly(dA-dT) in 15 μl of 1 mM DTT–20% glycerol–1 mM EDTA–10 mM Tris-HCl (pH 7.5)–15 mM NaCl. The reaction mixtures were incubated at room temperature for 20 min. Competition assays were done with unlabeled DNA at 10- or 50-fold molar excess over labeled probe DNA. For AP1 binding, poly(dI-dC) · poly(dI-dC) in 15 μl of the buffer described above was used. After addition of 32P-labeled probe (2 × 106 cpm), the reaction mixture was incubated for an additional 20 min at room temperature. Samples were subjected to electrophoresis in a 5% polyacrylamide gel (36:1 acrylamide/bisacrylamide) containing 45 mM Tris-HCl (pH 8.3), 45 mM boric acid, and 1 mM EDTA. The gels were dried and exposed to Kodak X-Omat film or Kodak Biomax film (Eastman, Kodak Company, Rochester, N.Y.).
Immunoblots and antibodies.
High Five cells were infected at a multiplicity of infection of 2 with either wild-type virus or recombinant baculovirus-UL56. The cells were harvested at 48 h after infection, sonicated, and centrifuged at 2,000 × g for 5 min. The supernatant and the pellet fractions were analyzed by sodium dodecyl sulfate (SDS)-PAGE, and the fractionated polypeptides were transferred to nitrocellulose filters as described previously (4). Antisera were raised against p130 by injection of 10 μg of DNA of the eucaryotic expression vector pRC/CMV-UL56, which contains the gene coding for p130. The immunization protocol has been previously described (12). Affinity purification of anti-p130 antibody was done as follows. A β-galactosidase fusion protein, fusproll, containing a hydrophilic amino acid stretch from amino acids 324 to 524 of p130 (4, 5) was coupled to activated immunoaffinity supports Affi-Gel 10 and Affi-Gel 15 (Bio-Rad Laboratories, Richmond, Calif.; 1:2 Affi-Gel 10/Affi-Gel 15) as described by Bio-Rad. The anti-p130 antiserum was incubated overnight with the prepared affinity matrix. After the matrix was washed twice with ice-cold start buffer containing 0.02% NaN3, the bound antibody was eluted in 200 mM glycine-HCl (pH 2.5). The purified anti-p130 antibody was neutralized with 100 mM Tris-HCl (pH 8.0).
Nuclease activity.
Supercoiled plasmid DNA containing the a sequence (pON205) or control plasmid (pUC9) was amplified in Escherichia coli DH5α and purified by Qiagen midipreparation (Qiagen, Hilden, Germany) as specified by the supplier. Extracts from recombinant or wild-type baculovirus-infected cells or uninfected cells were incubated with 2 μg of plasmid DNA in a final volume of 50 μl in 10 mM Tris-HCl (pH 7.5)–10 mM MgCl2–1 mM DTT–50 mM NaCl for 1 h at 37°C. DNA was fractionated by electrophoresis in a 1% agarose gel.
Sucrose gradient sedimentation.
Extracts from recombinant baculovirus-infected cells were prepared as described above. Extracts were layered on a 5-ml 5 to 20% sucrose gradient with a 500 μl of 40% sucrose cushion at the bottom. The gradient was sedimented at 35,000 rpm in a Beckman SW55 rotor for 14 h. Nine 500-μl fractions were collected and analyzed by immunoblotting.
RESULTS
Binding of an HCMV-infected cell protein to DNA bearing pac 1 and pac 2 motifs.
To determine whether an infected cell-specific protein binds to the pac motifs, nuclear extracts and 32P-labeled probes bearing the cis-acting packaging signals were prepared for EMSA. Preliminary experiments using large DNA probes obtained from plasmid pON205 (18) by restriction endonuclease digestion suggested that HCMV-infected cell nuclear extracts had a protein that bound to a-sequence-containing DNA. Therefore, the smaller pac 1 and pac 2 oligonucleotides were prepared as described in the Materials and Methods and as shown in Fig. 1A.
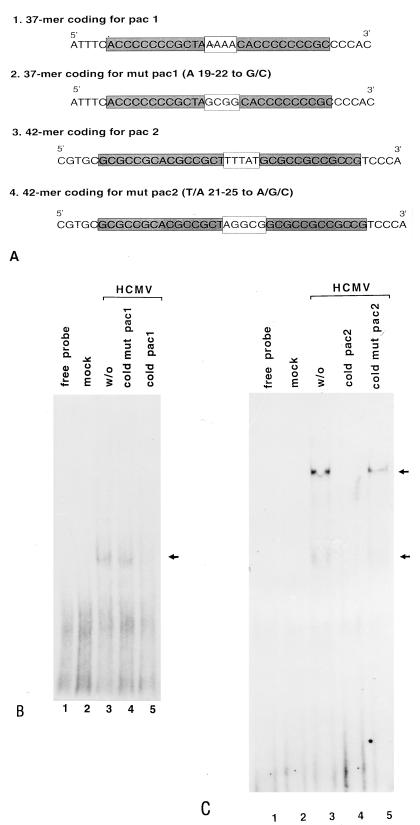
FIG. 1.
Effects of mutations in the packaging motifs on competition as well as on DNA-protein complex formation. EMSA was performed as described in Materials and Methods. (A) DNA sequences. pac 1, wild-type DNA containing the pac 1 motif; mut pac 1, DNA containing the mutagenized pac 1 motif; pac 2, wild-type DNA fragment containing the pac 2 motif; mut pac 2, DNA containing the mutated pac 2 motif. (B) Autoradiogram of EMSA competition with mut pac 1. Lanes: 1, DNA probe alone; 2, DNA probe plus mock-infected cell extracts; 3, DNA probe plus HCMV-infected cell nuclear extracts; 4, same as lane 3 plus 50-fold molar excess of unlabeled mut pac 1; 5, same as lane 3 plus 50-fold molar excess of unlabeled pac 1. (C) Autoradiogram of EMSA competition with mut pac 2. Lanes: 1, DNA probe alone; 2, DNA probe plus mock-infected cell nuclear extracts; 3, DNA probe plus HCMV-infected cell nuclear extracts; 4, same as lane 3 plus 50-fold molar excess of unlabeled wild-type pac 2; 5, same as lane 3 plus 50-fold molar excess of unlabeled mut pac 2. Arrows on the right indicate specific complexes.
Using the pac 1 probe, we detected specific DNA-protein complexes with HCMV-infected HFF nuclear extracts (Fig. 1B, lane 3). The binding of protein(s) to the pac 1 probe was specific because nonradioactive pac 1 DNA competed for the specific complexes (Fig. 1B, lane 5). Likewise, proteins in infected cell nuclear extracts bound specifically to the pac 2 probe (Fig. 1C, lane 3). A 50-fold molar excess of nonradioactive pac 2 competed with the pac 2 probe for specific proteins (Fig. 1C, lane 4). To determine whether a nonspecific GC-rich DNA sequence was sufficient for protein binding in infected cell nuclear extracts, substitutions were introduced for the A and T residues of pac 1 and pac 2, respectively (Fig. 1A). EMSA with mutant DNA probes as well as competition EMSA was performed. EMSA showed that wild-type pac 1 or pac 2 DNA competed for protein binding using the pac 2 probe, but mutant DNA did not (Fig. 1B, lane 4; Fig. 1C, lane 5). We were also unable to detect complex formation by EMSA with mutated pac probes and infected cell nuclear extracts. DNA containing a nonspecific motif for SP1 or AP1 failed to compete (data not shown). We conclude that the DNA binding is not due to the GC-rich sequence in the pac probes and the AT-rich sequence is required for binding of proteins from HCMV-infected cell nuclear extracts.
EMSA with recombinant baculovirus-expressed rp130.
To demonstrate that the binding of p130 to pac motifs is independent of other HCMV proteins, a recombinant baculovirus expressing p130 (rp130) was isolated. The expression of rp130 from recombinant baculovirus-UL56 was assayed by immunoblotting. Wild-type baculovirus and recombinant baculovirus-UL56-infected cells were harvested at 48 h postinfection and subjected to denaturing gel electrophoresis followed by immunoblotting as described in Materials and Methods. The insoluble sediment as well as the supernatant from recombinant baculovirus-UL56-infected cells showed a specific reaction with the antibody against p130 (Fig. 2, lane 1 and 2), while that of wild-type baculovirus-infected cells did not (Fig. 2). We conclude that at least some of the HCMV rp130 is soluble and consequently suitable for EMSA.
FIG. 2.
Immunoblot of HCMV rp130 antigen for detection of the HCMV rp130 synthesized in insect cells infected with recombinant baculovirus-UL56. Infected cells were fractionated, and aliquots of each fraction were separated by SDS-PAGE followed by immunoblotting with a specific antiserum against p130. Lanes: 1, supernatant fraction from recombinant baculovirus-UL56-infected H5 cells; 2, sediment from recombinant baculovirus-infected H5 cells; 3, supernatant from wild-type baculovirus-infected H5 cells; 4, sediment from wild-type baculovirus-UL56-infected H5 cells. Molecular mass standards (M) are indicated on the left; the position of rp130 proteins is indicated by an arrow.
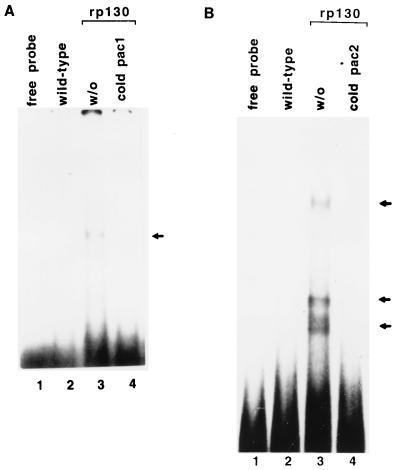
To determine if rp130 from baculovirus-UL56-infected cell extracts binds specifically to DNA bearing the packaging signal (pac 1 and pac 2), we performed EMSA as described in Materials and Methods. Using the pac 1 probe, we detected one major specific DNA-protein complex with rp130 (Fig. 3A, lane 3). Likewise, one minor and two major specific DNA-protein complexes were detected with the pac 2 probe (Fig. 3B, lane 2). Control baculovirus-infected cell extracts did not retard the mobility of the pac DNA probe (Fig. 3A, lane 2). The specific DNA-protein complex formation was reduced in the presence of unlabeled pac competitor DNA (Fig. 3A, lane 4; Fig. 3B, lane 4). We propose that HCMV rp130 binds the pac sequences.
FIG. 3.
EMSA with baculovirus rp130-containing cell extract and pac DNA probe. (A) Lane 1, free pac 1 probe; lane 2, EMSA with extract from wild-type baculovirus-infected cells; lane 3, EMSA with extract containing rp130; lane 4, same as lane 3 plus 50-fold molar excess of unlabeled pac 1. (B) Lanes: 1, free pac 2 probe; 2, EMSA with extracts from wild-type baculovirus-infected cells; 3, EMSA with extract containing rp130; 4, same as lane 2 plus 50-fold molar excess of unlabeled pac 2. All the results represent an experiment on the same gel. Specific DNA-protein complexes are indicated by arrows.
Nuclease activity of rp130.
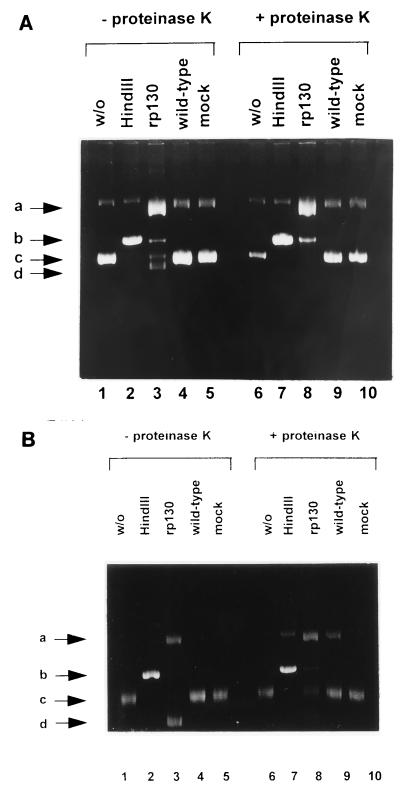
To determine whether HCMV p130 also has the capacity to cleave DNA containing the packaging motifs, experiments were undertaken to examine nuclease activity of rp130, using as a substrate circular plasmid DNA pON205, which contains a 1.8-kbp fragment that spans the L-S junction and includes a single a sequence (18). Separation by agarose gel electrophoresis showed that plasmid DNA molecules incubated in the absence of rp130 were present as fast-migrating supercoiled form (band c) and, in a lesser amount, as open circular form (band a) (Fig. 4A, lane 1). After treatment with restriction enzyme HindIII, which recognizes a single restriction endonuclease site on pON205, the band b fast-migrating forms were converted to linear molecules (b and b) (Fig. 4A, lane 2). In the presence of rp130, plasmid DNA was converted to open circular molecules with retarded migration (band a) and to linear forms (band b) (Fig. 4A, lane 3). In addition, DNA forms migrating faster than the supercoiled form were produced (band d) (Fig. 4A, lane 3). Mock-infected extracts or extracts from wild-type-infected cells had no effect (Fig. 4A, lanes 4 and 5). To verify that the two additional DNA forms were a result of DNA-protein complexes, samples were incubated with proteinase K (final concentration, 1 μg/μl) for another hour at 37°C (Fig. 4A, lanes 6 to 10). After treatment, only open circular and linear molecules were detected in the rp130 reaction mixture, thus indicating that the fast-migrating forms resulted from interaction of the protein with the DNA (Fig. 4A, lane 8).
FIG. 4.
Nicking activity of HCMV rp130. (A) Lane 1, plasmid pON205 in the absence of protein; lane 2, pON205 treated with restriction enzyme HindIII; lane 3, pON205 incubated with rp130; lane 4, pON205 incubated with wild-type-infected extracts; lane 5, pON205 incubated in the presence of mock-infected extracts; lane 6, same as lane 1 but treated with proteinase K; lane 7, same as lane 2 after treatment with proteinase K; lane 8, same as lane 3 plus treatment with proteinase K; lane 9, same as lane 4 treated with proteinase K; lane 10, same as lane 5 but incubated with proteinase K. (B) Lanes: 1, control plasmid pUC9 alone; 2, pUC9 treated with restriction enzyme HindIII; 3, pUC9 incubated with rp130; 4, pUC9 incubated with wild-type-infected extracts; 5, pUC9 plus mock-infected extracts; 6, same as lane 1 plus treatment with proteinase K; 7, same as lane 2 after proteinase K treatment; 8, same as lane 3 treated with proteinase K; 9, same as lane 4 incubated with proteinase K; 10, same as lane 5 after treatment with proteinase K. The arrows indicated four different plasmid DNA forms: open circular molecules (a), linear forms (b), supercoiled DNA (c), and additional fast-migrating molecules (d).
The specificity of the reaction was confirmed by using a plasmid without the a sequence (pUC9). Again, untreated plasmid DNA preparations consisted of a fast-migrating supercoiled form (band c) and of slowly migrating open circular molecules (band a) (Fig. 4B, lane 1). In the presence of HindIII, all supercoiled as well as nicked molecules were converted to linear forms (band b) (Fig. 4B, lane 2). The parental plasmid pUC9 was, however, not cleaved by rp130. Again, a faster-migrating DNA form comparable to the one of the reaction with pON205 was observed (band d) (Fig. 4B, lane 3). In the case of pUC9, these fast-migrating molecules were converted to open circle and supercoiled forms by proteinase K treatment (Fig. 4B, lane 8). Taken together, these observations suggested that HCMV p130 is able to specifically cleave DNA containing the a sequence.
Influence of incubation time and protein concentration on enzyme activity.
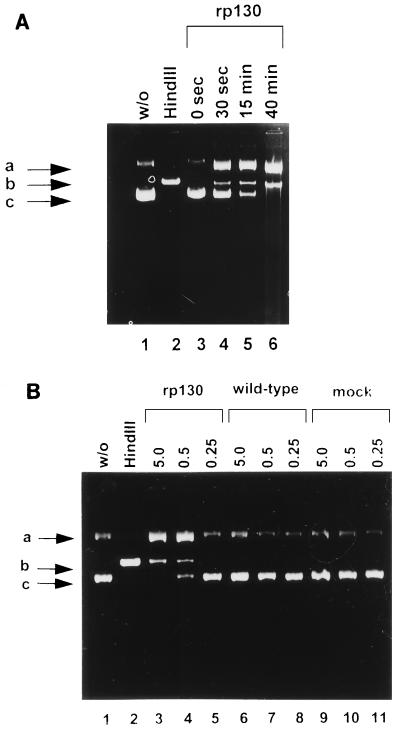
To examine the time course of nuclease activity, the reaction mixtures were incubated at 37°C and reactions were stopped by addition of proteinase K after various time intervals. The yield of nicked (band a) and linear (band b) molecules increased during the course of the incubation, while the supercoiled form disappeared (Fig. 5A, lanes 3 to 6).
FIG. 5.
Effects of the incubation time and protein concentration on the enzyme activity. (A) Nuclease reaction mixtures were incubated at 37°C for the indicated time, and reactions were stopped by addition of proteinase K. Lanes: 1, pON205 alone; 2, pON205 treated with HindIII; 3, pON205 plus incubation of rp130 for 0 s; 4, incubation with rp130 for 30 s; 5, incubation for 15 min; 6, incubation for 40 min. (B) Nuclease reactions were performed for 40 min at 37°C prior to proteinase K treatment. Lanes: 1, pON205 alone; 2, pON205 treated with HindIII; 3, pON205 plus incubation with 5 μg of rp130; 4, incubation with 0.5 μg of rp130; 5, incubation with 0.25 μg of rp130; 6, pON205 plus incubation with 5 μg of extract of wild-type baculovirus; 7, incubation with 0.5 μg of wild-type extract; 8, incubation with 0.25 μg of wild-type extract; 9, pON205 plus 5 μg of mock-infected extract; 10, incubation with 0.5 μg of mock-infected extract; 11, incubation with 0.25 μg of mock-infected extract. Positions of open circular DNA molecules (a), linear molecules (b), and supercoiled molecules (c) are indicated.
To further examine the specificity of the nuclease activity, the reaction was performed with different amounts of recombinant protein. As shown in Fig. 5B, nuclease activity was clearly concentration dependent: in the presence of 0.25 μg of protein no reaction was detectable (Fig. 5B, lane 5), while some, although incomplete, activity was observed by increasing the protein concentration to 0.5 μg (Fig. 5B, lane 4). The protein concentration had no effect on the control assays (Fig. 5B, lanes 6 to 11). Taken together, the results indicate that the specific endonuclease activity was closely related to the amount of rp130.
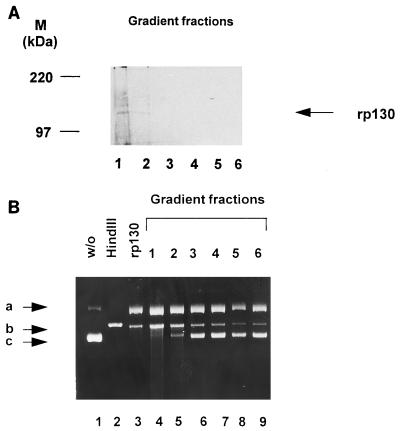
To determine if the nuclease activity is specifically linked to rp130, a single-step purification was used. Supernatant from baculovirus-UL56-infected cells was sedimented through a 5 to 20% sucrose gradient, and aliquots of gradient fractions were separated by SDS-PAGE, transferred to a polyvinylidene difluoride membrane, and probed with pAbUL56. Interestingly, rp130 sedimented rapidly and was detected only in the first two fractions (Fig. 6A, lane 1 and 2). The same fractions were used for a nuclease activity assay. Plasmid DNA was incubated together with an aliquot of each fraction prior to treatment with proteinase K. The protein of the first two fractions was able to convert supercoiled molecules (band c) into open circular (band a) and linear (band b) forms (Fig. 6B, lanes 4 and 5). These experiments showed that the nuclease activity is a property of HCMV p130.
FIG. 6.
Single-step purification of rp130 and direct association with enzyme activity. (A) Recombinant-infected cells were lysed and layered on a 5 to 20% sucrose gradient. Fractions were collected, separated by SDS-PAGE (8% gel) and transferred to a polyvinylidene difluoride membrane. Detection was performed with an anti-p130 antibody. Lane 1, fraction 1; lane 2, fraction 2; lane 3, fraction 3; lane 4, fraction 4; lane 5, fraction 5; lane 6, fraction 6. Molecular mass markers (M) are shown on the left; the position of rp130 is indicated by the arrow on the right. (B) Nuclease activity was assayed with an aliquot of each fraction. Lanes: 1, pON205 alone; 2, pON205 treated with HindIII; 3, pON205 plus extract containing rp130; 4, plasmid plus protein of fraction 1; 5, incubation with an aliquot of fraction 2; 6, incubation with an aliquot of fraction 3; 7, incubation with an aliquot of fraction 3; 8, incubation with an aliquot of fraction 4; 8, incubation with an aliquot of fraction 5; 9, incubation with an aliquot of fraction 6. The arrows indicate three different plasmid forms: open circular (a), linear (b), and supercoiled (c).
DISCUSSION
We have shown by EMSA that proteins in HCMV-infected cell nuclear extracts bind specifically to the cis-acting packaging motifs, pac 1 and pac 2, of the viral DNA a sequence. Little is known about proteins that bind to these packaging motifs. A report by Kemble and Mocarski (11) demonstrated that a host cell protein bound specifically to the pac 2 element. This DNA-protein complex may represent the complex that is in our experiments common to both mock- and HCMV-infected extracts. The DNA-protein binding conditions used in this study also detected infected-cell-specific protein binding to the pac motifs.
We also provide evidence by EMSA that baculovirus-UL56-infected whole-cell extracts containing HCMV rp130 also form specific DNA-protein complexes with the DNA pac motifs, but wild-type baculovirus extracts do not. Using a specific anti-p130 antibody, we obtained evidence for the presence of HCMV p130 in some DNA-protein complexes containing the pac motifs (unpublished observation).
Previous studies of a mutant HSV-1 with temperature-sensitive expression or deletion of ICP 18.5 (UL28) and a respective PrV mutant showed an accumulation of empty capsids and a failure to cleave concatenated viral DNA (1, 14, 15). Using electron microscopy to examine the HSV-1 mutants, Addison et al. (1) showed that at the nonpermissive temperature, concatenated viral DNA together with empty capsids was found exclusively in the nucleus. Further analysis of two UL28 deletion mutants by Tengelsen et al. (20) demonstrated that the UL28 gene product is required for HSV-1 DNA cleavage and encapsidation. Lastly, Mettenleiter et al. (14) confirmed that the PrV ICP 18.5 homolog is essential for replication because the mutant was unable to productively replicate in noncomplementing cells. After infection of noncomplementing cells with the mutant, an accumulation of empty capsids in the nucleus was found. Taken together, the data led the authors to conclude that mutation of the HSV-1 ICP 18.5 protein and the PrV ICP 18.5 homolog exhibit a viral DNA packaging defect.
Recombinant ICP 18.5 protein from baculovirus-infected cell extracts and possibly from infected cell extracts appears to be insoluble; therefore, these proteins are not suitable for DNA-protein complex formation (9a). In contrast, we find some of the HCMV p130 protein produced in insect cells to be soluble, while the majority of the viral gene product is insoluble and found in three different electrophoretic forms in the pellet of the insect cellular extract. Only the slow electrophoretic form of the rp130 is soluble, suggesting that this fraction participates in complex formation. This may explain why EMSAs with HSV-infected cell extracts or baculovirus-UL28 infected cell extracts have been negative, while HCMV-infected or recombinant baculovirus-UL56 extracts were able to form specific complexes with the pac DNA probes.
Since the regulatory cis sites for transcription of the gB gene are within the coding sequence of the HSV-1 ICP 18.5 (UL28) gene, it has been suggested that the ICP 18.5 gene product affects the transport of viral glycoproteins to the cell surface (17). Since these data were based on analysis of a virus which contained more than one temperature-sensitive mutation, the evidence of this function has not been substantiated. Furthermore, another mutant which has a mutation in ICP 18.5 showed normal exposure of viral glycoproteins at the cell surface (9). Although the exact role of HSV-1 ICP 18.5 is not known, our data for HCMV p130 suggest that the HSV gene product could be a DNA-binding protein.
The viral DNA packaging process may require a transient DNA-protein binding to an empty capsid. Thomsen et al. (21) showed by using a set of recombinant HSV-1 capsid proteins that in the absence of ICP 18.5 (UL28), mature capsids were not formed. A similar observation was made in a cell-free system where the identical proteins were used (15).
We further demonstrated that the recombinant HCMV p130 has an enzymatic activity that converts supercoiled plasmid DNA containing the a sequence into open circular as well as linear molecules. We showed that the reaction has specific kinetics and that it is independent of ATP (data not shown). A nicking activity was described by Im and Muzyczka (10) for adeno-associated virus (AAV) origin-binding protein Rep68. These authors provide evidence that Rep68 has an ATP-dependent site-specific endonuclease activity, the ability to bind to a specific site within the AAV DNA, and a DNA helicase activity. These Rep68 protein activities are apparently required for AAV DNA replication. Furthermore, the Rep homolog of human herpesvirus 6 is postulated to have nicking activity (22). Our observation also implies that p130 may be involved in cleaving viral DNA. This enzyme activity could be required for virus assembly, e.g., the last step in DNA packaging, where two a sequences need to be cut. However, it is clear that this process is more complex, and therefore p130, possibly together with other viral proteins, is responsible for this event.
In this report, we show that HCMV p130 can interact with specific HCMV DNA packaging motifs and is able to cleave DNA bearing these motifs. These findings suggest a possible function of p130 in HCMV DNA packaging. This issue remains to be established by HCMV p130 (UL56) mutations and by baculovirus assembly and packaging assays.
ACKNOWLEDGMENTS
We thank C. Grosskurth for expert technical assistance and members of the laboratories for critical reading of the manuscript. We thank E. Mocarski for plasmid pON205.
This study was supported by Public Health Service grant AI-13562 to Mark F. Stinski and by the Deutsche Forschungsgemeinschaft (SFB 286 Teilprojekt A3). Elke Bogner was the recipient of an Alexander von Humboldt Foundation postdoctoral fellowship.
REFERENCES
- 1.Addison C, Rixon F J, Preston V G. Herpes simplex virus type 1 UL28 gene product is important for the formation of mature capsids. J Virol. 1990;71:2377–2384. doi: 10.1099/0022-1317-71-10-2377. [DOI] [PubMed] [Google Scholar]
- 2.Alford C A, Britt W J. Cytomegalovirus. In: Roizman B, Whiteley R J, Lopez C, et al., editors. The human herpesviruses. New York, N.Y: Raven Press, Ltd.; 1993. pp. 227–255. [Google Scholar]
- 3.Bogner E, Reschke M, Reis B, Reis E, Britt W, Radsak K. Recognition of compartmentalized intracellular analogs of glycoprotein H of human cytomegalovirus. Arch Virol. 1992;126:67–80. doi: 10.1007/BF01309685. [DOI] [PubMed] [Google Scholar]
- 4.Bogner E, Reschke M, Reis B, Mockenhaupt T, Radsak K. Identification of the gene product encoded by ORF UL56 of human cytomegalovirus genome. Virology. 1993;196:290–293. doi: 10.1006/viro.1993.1477. [DOI] [PubMed] [Google Scholar]
- 5.Chee M, Bankier A, Beck S, Bohni R, Brown C, Cerny R, Horsnell T, Huchison C, Kouzarides T, Martignetti E, Preddie E, Satchnell S, Tomlinson P, Weston K, Barell B. Analysis of the protein coding content of the sequence of human cytomegalovirus AD169. Curr Top Microbiol Immunol. 1990;154:125–169. doi: 10.1007/978-3-642-74980-3_6. [DOI] [PubMed] [Google Scholar]
- 6.Chou J, Roizman B. Characterization of DNA sequence-common and sequence-specific proteins binding to cis-acting sites for cleavage of the terminal a sequence of the herpes simplex virus 1 genome. J Virol. 1989;63:1059–1068. doi: 10.1128/jvi.63.3.1059-1068.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deiss L P, Chou J, Frenkel N. Functional domains within the a sequence involved in the cleavage-packaging of herpes simplex virus DNA. J Virol. 1986;59:605–618. doi: 10.1128/jvi.59.3.605-618.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gibson W. Structure and assembly of the virion. Intervirology. 1996;39(5–6):389–400. doi: 10.1159/000150509. [DOI] [PubMed] [Google Scholar]
- 9.Holland L E, Sandri-Goldin R M, Goldin A L, Glorioso J C, Levine M. Transcriptional and genetic analysis of the herpes simplex virus type 1 genome: coordinates 0.29 to 0.45. J Virol. 1984;49:947–959. doi: 10.1128/jvi.49.3.947-959.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9a.Homa, F. Personal communication.
- 10.Im D S, Muzyczka N. The AAV origin binding protein rep68 is an ATP-dependent site-specific endonuclease with DNA helicase activity. Cell. 1990;61:447–457. doi: 10.1016/0092-8674(90)90526-k. [DOI] [PubMed] [Google Scholar]
- 11.Kemble G W, Mocarski E S. A host cell protein binds to a highly conserved sequence element (pac-2) within the cytomegalovirus a sequence. J Virol. 1989;63:4715–4728. doi: 10.1128/jvi.63.11.4715-4728.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klenk A, Geyer E, Zahner H, Trojan H J. Isolation of antigen from Litomosoides carinii macrofilariae detecting serum antibodies due to Onchocerca volvolus. Z Parasitenkd. 1983;69:377–386. doi: 10.1007/BF00927879. [DOI] [PubMed] [Google Scholar]
- 13.Lee K A W, Green M R. Small-scale preparation of extracts from radiolabeled cells efficient in pre-mRNA splicing. Methods Enzymol. 1990;181:20–23. doi: 10.1016/0076-6879(90)81108-7. [DOI] [PubMed] [Google Scholar]
- 14.Mettenleiter T C, Saalmüller A, Weiland F. Pseudorabies virus protein homologous to herpes simplex virus type 1 ICP 18.5 is necessary for capsid maturation. J Virol. 1993;67:1236–1245. doi: 10.1128/jvi.67.3.1236-1245.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Newcomb W W, Homa F L, Thomsen D R, Ye Z, Brown J C. Cell-free assembly of the herpes simplex virus capsid. J Virol. 1994;68:6059–6063. doi: 10.1128/jvi.68.9.6059-6063.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pederson N E, Enquist L. Overexpression in bacteria and identification in infected cells of the pseudorabies virus protein homologous to herpes simplex virus type 1 ICP 18.5. J Virol. 1991;65:3746–3758. doi: 10.1128/jvi.65.7.3746-3758.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pellet P E, Jenkins F J, Ackermann M, Sarmeinto M, Roizman B. Transcription initiation sites and nucleotide sequence of a herpes simplex 1 gene conserved in the Epstein-Barr virus genome and reported to affect the transport of viral glycoproteins. J Virol. 1986;60:1134–1140. doi: 10.1128/jvi.60.3.1134-1140.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Spaete R R, Mocarski E S. The a sequence of the cytomegalovirus genome functions as a cleavage/packaging signal for herpes simplex virus defective genomes. J Virol. 1985;54:817–824. doi: 10.1128/jvi.54.3.817-824.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Summers M D, Smith G E. A manual of methods for baculovirus vectors and insect cell culture procedures. Texas Agricultural Experiment Station Bulletin no. 1555. College Station, Tex: Texas Agricultural Experiment Station; 1987. [Google Scholar]
- 20.Tengelsen L A, Pederson N E, Shaver P R, Wathen M W, Homa F L. Herpes simplex virus type 1 DNA cleavage and encapsidation require the product of the UL28 gene: isolation and characterization of two UL28 deletion mutants. J Virol. 1993;67:3470–3480. doi: 10.1128/jvi.67.6.3470-3480.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thomsen D R, Roof L L, Homa F L. Assembly of herpes simplex virus (HSV) intermediate capsids in insect cells infected with recombinant baculoviruses expressing HSV capsid proteins. J Virol. 1994;68:2442–2457. doi: 10.1128/jvi.68.4.2442-2457.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thomson B J, Weindler F W, Gray D, Schwaab V, Heilbronn R. Human herpesvirus 6 (HHV-6) is a helper virus for adeno-associated virus type 2 (AAV-2) and the AAV-2 rep gene homologue in HHV-6 can mediate AAV-2 DNA replication and regulate gene expression. Virology. 1994;204:304–311. doi: 10.1006/viro.1994.1535. [DOI] [PubMed] [Google Scholar]