ABSTRACT
In the context of sustainable diet, the development of soy-based yogurt fermented with lactic acid bacteria is an attractive alternative to dairy yogurts. To decipher the metabolism of Lactobacillus delbrueckii subsp. delbrueckii during soy juice (SJ) fermentation, the whole genome of the strain CIRM-BIA865 (Ld865) was sequenced and annotated. Then Ld865 was used to ferment SJ. Samples were analyzed throughout fermentation for their cell number, carbohydrate, organic acid, free amino acid, and volatile compound contents. Despite acidification, the number of Ld865 cells did not rise, and microscopic observations revealed the elongation of cells from 3.6 µm (inoculation) to 36.9 µm (end of fermentation). This elongation was observed in SJ but not in laboratory-rich medium MRS. Using transcriptomic analysis, we showed that the biosynthesis genes of peptidoglycan and membrane lipids were stably expressed, in line with the cell elongation observed, whereas no genes implicated in cell division were upregulated. Among the main sugars available in SJ (sucrose, raffinose, and stachyose), Ld865 only used sucrose. The transcriptomic analysis showed that Ld865 implemented the two transport systems that it contains to import sucrose: a PTS system and an ABC transporter. To fulfill its nitrogen needs, Ld865 probably first consumed the free amino acids of the SJ and then implemented different oligopeptide transporters and proteolytic/peptidase enzymes. In conclusion, this study showed that Ld865 enables fast acidification of SJ, despite the absence of cell division, leads to a product rich in free amino acids, and also leads to the production of aromatic compounds of interest.
IMPORTANCE
To reduce the environmental and health concerns related to food, an alternative diet is recommended, containing 50% of plant-based proteins. Soy juice, which is protein rich, is a relevant alternative to animal milk, for the production of yogurt-like products. However, soy “beany” and “green” off-flavors limit the consumption of such products. The lactic acid bacteria (LAB) used for fermentation can help to improve the organoleptic properties of soy products. But metabolic data concerning LAB adapted to soy juice are lacking. The aim of this study was, thus, to decipher the metabolism of Lactobacillus delbrueckii subsp. delbrueckii during fermentation of a soy juice, based on a multidisciplinary approach. This result will contribute to give tracks for a relevant selection of starter. Indeed, the improvement of the organoleptic properties of these types of products could help to promote plant-based proteins in our diet.
KEYWORDS: fermentation, lactic acid bacteria, Lactobacillus delbrueckii, Lactobacillus delbrueckii subsp. delbrueckii, food, soy juice, soy milk, yogurt, genomics, transcriptomics, metabolomics
INTRODUCTION
In order to reduce the environmental and health concerns related to food, an alternative diet is recommended in Western countries: it should be more sustainable and contain 50% of plant-based proteins rather than mostly animal-based proteins (1–3). In this context, the consumption of soybean-derived products offers a valuable opportunity to reduce that of animal proteins. Indeed, soybeans have a high protein content (up to 40% of dry matter). Therefore, if soybean is grown under environmentally friendly conditions, it could be a promising ally for food transition. Soy juice, also improperly called soy milk because it resembles cow’s milk, is a water extract of soybeans. Compared to other plant-based milk substitutes, soy juice has been shown to be rich in proteins (4, 5) and quite close to that of cow’s milk in terms of protein quality according to the DIAAS (digestible indispensable amino acid score) (4, 6). Moreover, soy proteins have gelification properties that are of interest for food processing. For all these reasons, soy juice is used in the manufacture of alternatives to products that traditionally use milk, such as yogurts. Moreover, soy juice is suitable for targeted populations with specific needs, such as vegans and those who are lactose intolerant. However, soy juice-based products suffer from some drawbacks that limit their consumption, particularly in Western countries where soy-based products are not part of the food culture. First, they contain varying amounts of stachyose and raffinose, two indigestible oligosaccharides that can be source of digestive discomfort in humans such as flatulence (7–9). The second limitation is their flavor, which is described as “beany,” partly because of the oxidation of fatty acids that generates aldehydes, and is often not appreciated by consumers. Different volatile compounds are involved in this off-flavor, including hexanal, which is responsible for the hay or green bean odors in soy juice (8, 10, 11).
Lactic acid bacteria (LAB) are common in food fermentation, either as spontaneous microbiota or used as starters. Some LAB (particularly those able to degrade lactose) have a long history of use in the making of dairy-fermented foods (yogurt and other fermented milks, cheeses, etc.). However, not all LAB species and strains are able to ferment plant-based milks. For instance, Jan et al. (12) showed that L. delbrueckii subsp. bulgaricus, which can metabolize lactose but not sucrose, is a well-adapted species to grow in cow milk but not in soy juice whose sucrose is the main sugar. As a consequence, bacterial cells presented an elongated phenotype in this adverse medium. Strains which can metabolize non-digestible sugars, with peculiar aromatic properties compatible with plant notes, or with an ability to degrade off-flavor compounds, are particularly sought after. Several of these properties have been studied in some LAB species, highlighting the strain-dependency of these phenotypes in terms of their ability to degrade stachyose and raffinose (13, 14) or reduce hexanal (15, 16).
In a previous work, we performed a wide-ranging screening of 270 strains of different LAB species to investigate their ability to metabolize soy juice sugars, i.e., sucrose, raffinose, and stachyose (16). We showed that 97% of the sucrose-positive strains were also able to ferment soy juice, indicating that an ability to use sucrose is a good criterion for the selection of strains for soy juice fermentation. Among 20 strains of L. delbrueckii tested by Harlé et al. (16), Lactobacillus delbrueckii subsp. delbrueckii CIRM-BIA865 (named hereafter Ld865) was one of the most effective at acidifying soy juice. In the present work, our objective was to decipher the behavior of this sucrose-positive strain, during soy juice fermentation. For that, we sequenced the genome of Ld865 and investigated its metabolism throughout the time course of soy juice fermentation, combining different omics approaches: genomics, transcriptomics, and targeted metabolomics.
MATERIALS AND METHODS
Culture conditions and fermentation profile of Ld865
The strain Lactobacillus delbrueckii subsp.delbrueckii CIRM-BIA865 (named Ld865) was provided by the International Center for Microbial Resources-Food Associated Bacteria (CIRM-BIA, https://collection-cirmbia.fr). Ld865 was activated from frozen glycerol stock (−80°C) with two consecutive sub-cultures of 12 h inoculated at 1% (vol/vol) in de Man, Rogosa, and Sharpe [MRS (17)] broth before being used for culture in soy juice. Subcultures were incubated at 37°C without agitation under air atmosphere. The fermentation profile of Ld865 was determined using the API 50CH micro gallery (bioMérieux Diagnostics, Marcy-l'Etoile, France) according to manufacturer’s instructions.
Soy juice fermentation, kinetics of acidification, and fermented soy juice analysis
The soy juice used for the fermentations was a commercial UHT soy juice (Sojade, Triballat Noyal SAS, France) referred to as SJ below. No sugar was added. The same SJ batch was used throughout the study. SJ (1,000 mL) was inoculated with Ld865 at 1% (vol/vol) [equivalent to 5.7 × 105 colony-forming unit (CFU)/mL], immediately aliquoted in 100 mL bottles and incubated at 37°C under static conditions. The monitoring of acidification during fermentation was carried out with iCINAC System (Ysebaert, Frepillon, France) that enables automatic and precise monitoring of acidification, with one measurement every 5 min. The iCINAC was equipped with Mettler Toledo ISM probes (Ag/AgCl, E0 = 196 mV at 37°C, Paris, France). All fermentations were carried out in triplicate from independent subcultures.
For subsequent analyses of fermented soy juices (FSJs), samples were collected throughout fermentations when the targeted pH values of 6.5, 6.0, and 5.0 were reached, and after 26.5 h fermentation for the final point (pH 4.6). Samples for organic acid, carbohydrate, and amino acid (AA) analyses were cooled in ice and stored at −20°C in 40 mL straight containers with screw caps (Corning Gosselin) until analysis. For each sample, analyses were performed in duplicate.
Microbial analyses
One milliliter of FSJ was diluted in 9 mL of Tryptone salt (0.1% Tryptone, AES Laboratoire, Combourg, France; 0.85% NaCl, Labogros, Buchs, France). Serial decimal dilutions were then realized by micro-method as previously described (18). CFUs were enumerated on MRS agar plates after incubation under anaerobic conditions for 24 h at 43°C.
Carbohydrate quantification
Oligosaccharides (stachyose, raffinose), disaccharides (sucrose, lactose), and monosaccharides (glucose, fructose, and galactose) were quantified in FSJs by high-performance anion-exchange chromatography (HPAEC) and pulsed amperometric detection (PAD) on an ICS-5000 + Dionex system (Thermo Electron SAS, Courtaboeuf France), as previously described (19), with some modifications. Aliquots of FSJ (1 g) were deproteinized by adding 40 mg of salicylic acid followed by 30 min incubation at 0°C, and centrifugation (4,000 × g, 5 min, RT). Supernatants were then diluted by 150-fold in milli-Q water (Merck, Darmstadt, Germany), filtered through 0.45 µm pore diameter filters (chromafil Xtra PVDF 45/13, Macherey-Nagel, Hoerdt, France), and kept frozen at −20°C until analysis.
The system was equipped with a Dionex CarboPac PA210-Fast-4 mm column (2 × 150 mm) suited for mono-, di-, tri-, and oligosaccharide analysis, preceded by a CarboPac PA210-4 µm guard column (2 × 30 mm). Analyses were performed at 30°C and at a flow rate of 0.2 mL/min, with the following elution gradient: initial conditions, 13 mM KOH maintained for 32 min, linear rise to 42 min up to 100 mM KOH, maintained from 42 to 52 min, followed by reversion to the initial conditions with a linear decrease from 52 to 60 min. Data were acquired and processed by using the Chromeleon software. Carbohydrates were quantified using multi-standard external calibration [prepared at 0.5, 1, 2, 5, 10, 20, and 40 mg/L] (Merck, France).
Organic acid quantification
Lactic, acetic, citric, succinic, and pyruvic acids were analyzed by high-performance liquid chromatography (HPLC, Dionex, P680, Sunnyvale California) as previously described (16).
Amino acid quantification
The total and free AAs were analyzed by ion chromatography as previously described (20). Briefly, samples (0.5 mL) were treated with 25 mg of sulfosalicylic acid during 1 h at 4°C to precipitate proteins, centrifuged (1,000 × g, 10 min, RT), and the supernatant diluted 3- to 5-fold in the injection buffer (lithium citrate 0.2 M–pH 2.2) (Biochrom Ltd., Cambridge, UK).
Volatile compound analysis
Volatile compounds (aliquots of 2.5 g of fermented SJ) were analyzed by headspace (HS) gas chromatography-mass spectrometry (GC-MS) using Turbomatrix HS-40 trap, Clarus 680 gas chromatograph, and Clarus 600T quadrupole mass spectrometer (PerkinElmer, Courtaboeuf). The principle of the method was previously detailed (21), with modifications of the chromatographic conditions according to (16). Volatile compounds were identified by comparing their retention index (RI) and mass spectra, with those from the NIST 2008 Mass Spectral Library (Scientific Instrument Services, Ringoes, NJ, United States) and from literature and, when available, with those of standards (Sigma Aldrich, France) analyzed in the same conditions.
Whole-genome sequencing, assembly, and annotation
Ld865 was grown in MRS during 24 h at 37°C. Cell pellets (equivalent to 1010 CFU) were then obtained by centrifugation (5,000 × g, 10 min, RT) of cultures. DNA was extracted from pellets using DNeasy midi kit (Qiagen) according to the protocol previously described (22). Genome was sequenced by Illumina NovaSeq 2 × 150 pb and PacBio Sequel (Eurofins Genomics, Constance, Germany). Genome was assembled using Unicycler (23) on the Galaxy Genotoul bioinformatics platform [Toulouse, France (24)]. Genome sequence was integrated in the MicroScope platform hosted in the Genoscope (CEA, Evry, France) for automatic annotation (25). Manual annotation and metabolic reconstruction were performed using the MetaCyc database (26) and the Kyoto Encyclopedia of Genes and Genomes (KEGG—https://www.kegg.jp/ or https://www.genome.jp/kegg/). Sequence data have been deposited in the European Nucleotide Archive (ENA) at EMBL-EBI under accession number PRJEB50396.
Quantitative PCR to evaluate population level
Ld865 was grown in soy juice or in MRS as described above. At different times of growth, 1 mL of cultures were centrifuged (8,000 × g, 10 min, 4°C) and DNA was extracted from cell pellets as previously described (27). qPCR were performed on DNA extracted using CFX 96 thermocycler (Bio-Rad, Germany) and Sybrgreen mix as previously described (27). Amplification was performed with a 15 µL final volume mixture, containing 5 µL of the DNA-containing solution. Primers targeting the single copy tuf gene of Ld865 (5′-AAGAGACTTGCTTTCAGAATACGG-3′/5′-CAACGTCCATCAGTTCTTCG-3′) were picked from Achilleos & Berthier (28) and checked on L. delbrueckii CIRM-BIA865 genome using Primer 3 software (https://primer3.ut.ee/). The efficiency of the PCR was 97%, and the melting curve confirmed the specificity of the reaction. Using a standard curve, Ld865 cell numbers were assessed in equivalent copy number/µL of culture according to MIQE guidelines (29). Results were compared to numerations obtained by serial dilution of the same cultures on plates.
RNA-seq analysis of soy juice fermented with Ld865
RNA extraction
One milliliter of FSJ sample was treated with 2 mL of RNA protect cell reagent (Qiagen, Hilden, Germany) and centrifuged (10,000 × g, 5 min, 4°C). The supernatant was discarded and the pellet frozen at −80°C with 2 mL of RNA protect until analysis. After defrosting, the cells were collected by centrifugation (10,000 × g, 5 min, 4°C). The cell pellets were then suspended in 200 µL lysis buffer (50 mM Tris–HCl, 1 mM EDTA; pH 8.0) with 20 mg/mL lysozyme (MP Biomedicals, Illkirch, France) and 50 U/mL mutanolysin (Sigma, Saint Quentin Fallavier, France) and incubated for 30 min at 24°C. Suspension was then transferred to 2 mL tubes containing 50 mg zirconium beads (diameter: 0.1 mm; BioSpec Products, Bartlesville) and 50 µL of sodium dodecyl sulfate (SDS) (20%) and shaken in a Precellys Evolution (Bertin, Montigny-le-Bretonneux, France) for two cycles of 40 s at 6,500 rpm. RNA was then extracted from the cell lysate with the RNeasy Mini kit (Qiagen). Subsequent DNase treatments (Dnase Rnase free, Ambion) were applied according to the manufacturer’s instructions. RNA concentrations were quantified by Qubit 4.0 Fluorometer and Qubit single strand assay kit (Invitrogen). Lack of DNA was also checked using Qubit 4.0 Fluorometer and Qubit double strand assay kit (Invitrogen) according to manufacturer instructions. RNA quality (RIN) was evaluated using an Agilent 2100 bioanalyzer (Agilent Technologies, Santa Clara, CA). The RNA samples were stored at −80°C until being supported by the sequencing platform.
RNA labeling, hybridization, and sequencing
All RNA samples with a RIN value greater than 7.7, indicative of a good RNA quality and integrity, and with a quantity higher than 2 µg were sequenced. RNA library preparations end sequencing were conducted at GENEWIZ, LLC (Leipzig, Germany). Whole transcriptome RNA enrichment was performed using NEBNext rRNA Depletion Kit (Bacteria) and NEBNext Ultra II RNA Library Prep Kit for Illumina by following manufacturer’s recommendations (NEB, Ipswich, MA, USA). Briefly, enriched RNAs were fragmented for 15 min at 94°C. First- and second-strand cDNA were subsequently synthesized. cDNA fragments were end repaired and adenylated at their 3′, and universal adapter was ligated to cDNA fragments, followed by index addition and library enrichment with limited cycle PCR. Sequencing libraries were validated using the Agilent TapeStation 4200 (Agilent Technologies, Palo Alto, CA, USA) and quantified by using Qubit 2.0 Fluorometer (Invitrogen, Carlsbad, CA) as well as by quantitative PCR (Applied Biosystems, Carlsbad, CA, USA). The sequencing libraries have been multiplexed and clustered on 1 lane of 1 flowcell. After clustering, the flowcell was loaded on the Illumina HiSeq 2500 instrument according to manufacturer’s instructions. The samples were sequenced using a 2 × 150 Paired End (PE) configuration. Image analysis and base calling were conducted by the HiSeq Control Software (HCS) on the HiSeq 2500 instrument. Raw sequence data (.bcl files) generated from Illumina HiSeq 2500 were converted into fastq files and de-multiplexed using Illumina CASSAVA 1.8.2 program. One mis-match was allowed for index sequence identification.
RNA-seq data analyses
Computational analysis was performed on the Galaxy Genotoul bioinformatics platform (24). Sequenced reads were trimmed and adapted using Trim Galore, and mapped against Ld865 reference genome with Bowtie 2 (30). No mismatch was allowed and only reads that mapped to Ld865 sequence were further analyzed. Reads mapped on each coding sequence (CDS) were counted using HTSeq-count [-stranded = reverse, -a 0, -t CDS, -i db_xref, -m union (31)]. The strategy of RNA-seq data analysis implemented in this study consisted to a normalization of raw counts of reads between samples, ensuring that the counts are adjusted for differences in sequencing depth between successive samples along the fermentation kinetic (32). After normalization, list of differentially expressed (DE) genes between two successive sampling times (determined by pH) was generated using SARtools (33) embedded DEseq2 (34) using a modified t-test with a P-value adjusted by Bonferoni (35) inferior to 0.05. Fold changes expressed how many times genes were modulated between two successive points. The start of fermentation was, for example, investigated by comparing gene expressions between the pHs 6.5 and 6.0.
Statistical analyses
For AA and sugar analysis, the means of replicates were compared using a Tukey post hoc test from the R package statix (P-value < 0.05). To determine whether the concentrations in volatile compounds differed according to the stage of culture, one-way analyses of variance (ANOVA) were performed using the aov R function (RStudio. Inc.). The means of replicates were compared using a Tukey post hoc test from the R package car (P-value < 0.05). A principal component analysis (PCA) was performed by using FactoMineR to illustrate the global changes in the volatile profile using log-transformed values using the FactoExtra package of R.
RESULTS
Fermentation of soy juice by Ld865
The ability of strain Ld865 to propagate in soy juice (SJ) was evaluated through changes to pH and bacterial counts during fermentation at 37°C. The initial pH of the SJ used was 7.2, and it was inoculated with Ld865 at 5.7 × 105 CFU/mL. After inoculation, the pH fell gradually over time to reach pH 5.2, i.e., the gelification pH, after about 11 h of incubation (Fig. 1). Fermentation was halted 26.5 h after inoculation, at a pH of 4.6.
Fig 1.
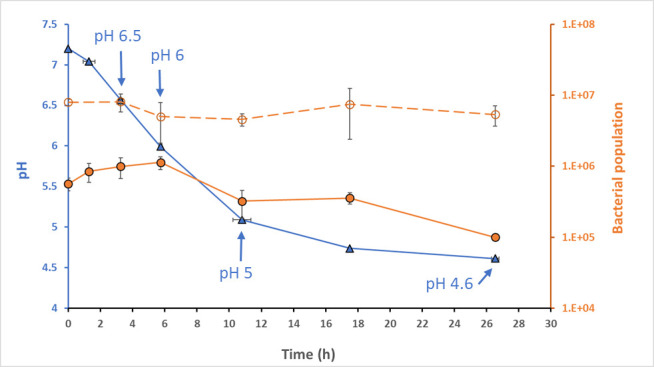
Kinetic of fermentation of soy juice with the strain Lactobacillus delbrueckii subsp. delbrueckii CIRM-BIA865. Fermentation, performed at 37°C, was followed by pH measurement (left axis, blue curve) and cell enumeration (right axis) based on two methods: (i) plate counting on MRS expressed as log CFU/mL (orange solid line) and (ii) counting Ld865 genome copy numbers by quantitative PCR (orange dotted line). The data are the mean of 3 independent experiments. The blue arrows indicate the sampling time performed throughout fermentation for transcriptomic and metabolic analysis: pH 6.5 (3.3 h of fermentation), pH 6 (5.8 h of fermentation), pH 5 (10.8 h of fermentation), and pH 4.6 (26.5 h of fermentation).
Despite the acidification of SJ, the number of Ld865 cells measured by plate counting did not increase during fermentation. The population remained close to the number of inoculated cells during the first 8 h of fermentation and then dropped to reach a final count of 1.1 × 105 CFU/mL at the end of fermentation (Fig. 1). The number of cells was also estimated through an absolute quantification based on the genome copy number. For this, the copy number of a single-copy gene (tuf gene) was quantified using real-time PCR. The numbers of Ld865 cells calculated from the DNA copy number immediately after inoculation was 7.9 × 106 cells/mL. This count higher than the one obtained by plate counting could be explained by (i) the cells organized in small chains and (ii) part of the population which is viable but no longer cultivable. Despite this difference, as observed by plate counting throughout fermentation and at the end of the experiment, the cell numbers measured by absolute quantification were stable confirming the absence of Ld865 growth (Fig. 1).
Microscopic observation of Ld865 cells during soy juice fermentation
To better understand the ability of Ld865 to acidify SJ in the absence of detectable bacterial growth, Ld865 cells were examined by microscopy. Throughout fermentation, cells displayed a rod-shaped morphology, but their length steadily increased, from about 3.6 ± 1.8 µm at the start of fermentation to reach cell length of 20.2 ± 8.7 µm, 29.25 ± 14 µm, and 36.9 ± 11.4 µm after 8, 12, and 24 h of fermentation, respectively (Fig. 2). Regardless of the fermentation time, no septum was observed in the elongated cells observed in SJ, indicating that they did not divide. To evaluate the influence of the growth medium, cell size was also evaluated during growth in MRS, in which Ld865 cells displayed a rod-shaped morphology at the start of fermentation, as in SJ (Fig. 2). But unlike SJ, their size did not vary throughout fermentation, being about 3.8 ± 1.9 µm at the start of fermentation and 4.0 ± 1.0 µm after 24 h of fermentation.
Fig 2.
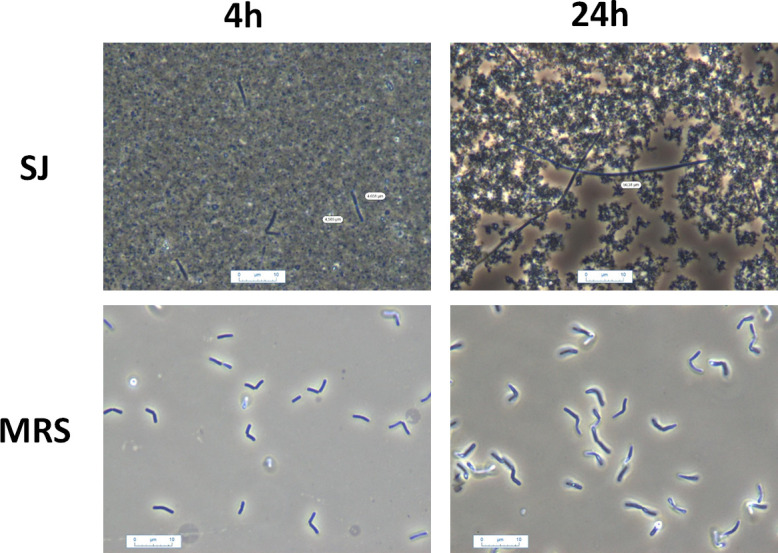
Microscopic observations of Lactobacillus delbrueckii subsp. delbrueckii CIRM-BIA865 during growth in soy juice (SJ, up pictures) or MRS (down pictures). The pictures were taken after 4 h (left) and 24 h (right) of fermentation.
Metabolic analysis of soy juice fermented with Ld865
The content in SJ of sugars, organic acids, amino acids, and volatile compounds was monitored kinetically during the fermentation process.
Sugar content
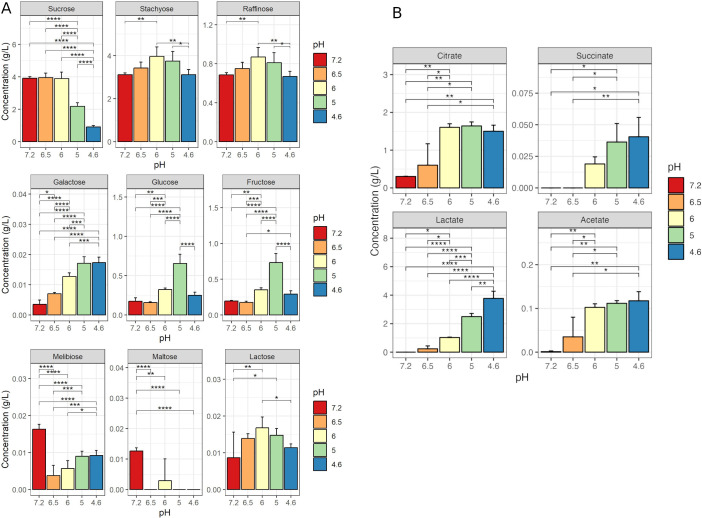
Unfermented SJ contained 3.91 g/L sucrose, 3.11 g/L stachyose, 0.69 g/L raffinose, 0.20 g/L fructose, and 0.18 g/L glucose (Fig. 3A). During fermentation, the principal sugar consumed by Ld865 was sucrose although no exhaustion was observed, and a residual content of 0.91 g/L sucrose was measured at the end of fermentation. Neither stachyose nor raffinose was consumed during fermentation. An increase in these two sugars was measured transiently, with maximal concentrations that reached 127% of their initial level at pH 6 (5.8 h of fermentation). This increase could be due to the error of the method. Likewise, melibiose, maltose, and lactose remained at low levels throughout fermentation (Fig. 3A). A constant and significant increase of galactose content is also observed during fermentation, with a final concentration remaining low (<0.018 g/L at the end of fermentation). Glucose and fructose contents were significantly higher (P < 0.05) at the end of fermentation (0.25 and 0.29 g/L, respectively) than in unfermented SJ (0.18 and 0.19 g/L, respectively) but remained low. An increase in these two sugars was measured transiently at pH 6 (5.8 h of fermentation) and pH 5 (10.8 h of fermentation), with maximal values of 0.66 g/L glucose and 0.73 g/L fructose at pH 5 and could result from the sucrose degradation.
Fig 3.
Sugar (A) and organic acid (B) concentrations in soy juice (SJ) unfermented and fermented with Lactobacillus delbrueckii subsp. delbrueckii CIRM-BIA865. SJ was inoculated at 1% (vol/vol) with the strain, and fermentation was conducted at 37°C. Sampling was performed at the start of fermentation (pH 7.2) and when the pH values reached 6.5 (3.3 h of fermentation), 6.0 (5.8 h of fermentation), 5.0 (10.8 h of fermentation), and 4.6 (26.5 h of fermentation). Results are the mean values (in g/L ) between three independent fermentations and two replicates. Stars indicate statistical differences from a Tukey test: *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001; ****P ≤ 0.0001.
Organic acid content
The main organic acid detected in the unfermented SJ was citrate, at a concentration of 0.30 g/L (Fig. 3B). During SJ fermentation, citrate levels rose more than twofold; this apparent increase could result from an extraction bias due to its possible complexation with soy proteins at neutral pH and its better extraction at low. Concentrations of acetate and succinate also increased during fermentation but remained at very low levels. Lactate was the main organic acid produced during fermentation, a concentration of 3.77 g/L being detected at the end of fermentation. No pyruvate was found, regardless of the duration of fermentation.
Amino acid (AA) content
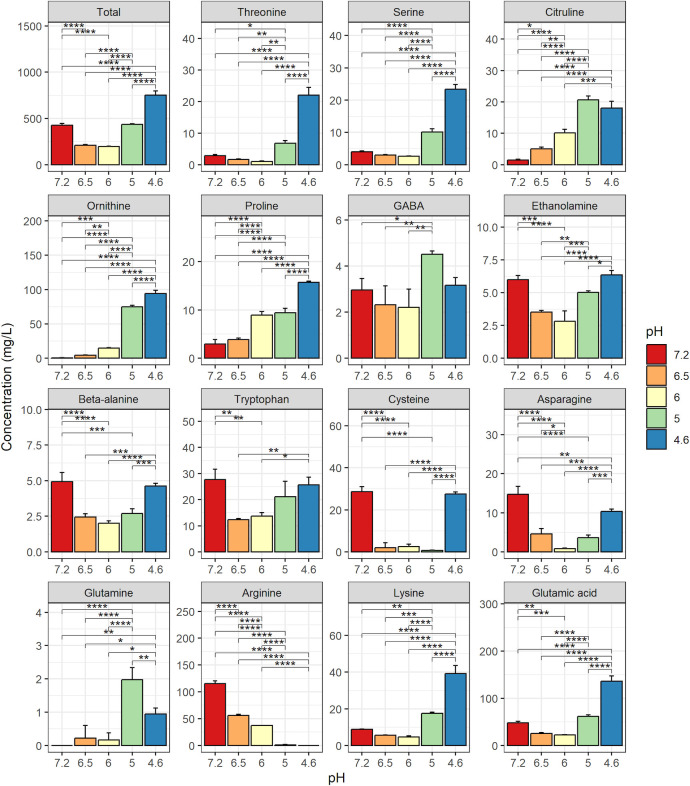
Unfermented SJ contained 35 g/L total AAs and this concentration did not vary throughout fermentation (data not shown). The unfermented SJ also contained 426 mg/L of free AAs (Fig. 4). The concentration of free AAs fell between pH 7.2 and pH 6.0 (from 426 to 198 mg/L) and then rose until the end of fermentation to reach a final concentration of 751 mg/L. Individually, at the end of fermentation, the concentrations of several AA (threonine, serine, citruline, ornithine, and proline) were significantly much higher (considering a fold change > 5) than those in the unfermented SJ. However, those of gamma-aminobutyric acid (GABA), ethanolamine, beta-alanine, tryptophan, cysteine, and asparagine were close to the initial levels. Finally, glutamine, which was not detected in the unfermented SJ, was found at a very low concentration at the end of fermentation (0.4 mg/L), and arginine was the only free AA to be exhausted by the end of fermentation, with a concentration decreasing from 116 to 0 mg/L between the start and the end of fermentation (pHs 7.2 and 4.6, respectively).
Fig 4.
Free amino-acid concentrations in soy juice (SJ) unfermented and fermented with Lactobacillus delbrueckii subsp. delbrueckii CIRM-BIA865. Sampling was performed at the start of fermentation (pH 7.2) and when the pH values reached 6.5 (3.3 h of fermentation), 6.0 (5.8 h of fermentation), 5.0 (10.8 h of fermentation), and 4.6 (26.5 h of fermentation). Results are the mean values (in mg/L) between three independent fermentations and two replicates. Stars indicate statistical differences from a Tukey test: *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001; ****P ≤ 0.0001.
Volatile compounds
Thirty-five compounds were identified in fermented or unfermented SJ: 10 acids, 8 ketones, 7 alcohols, 6 aldehydes, 1 ester, and 3 other compounds (online supplementary file 1 Table S1). The volatile profile changed during fermentation. The concentration of seven compounds, including three aldehydes, significantly decreased, whereas those of 21 compounds increased (Table 1). PCA performed on the concentrations of the 35 volatiles showed that the first axis, which accounted for more than 60% of total variance, separated the cultures as a function of fermentation stage (Fig. S1) . The first axis was positively correlated with most volatiles, the abundance of which increased overtime, whereas 2-pentylfuran and three aldehydes decreased in abundance during fermentation. Although quantitatively small, most of the differences observed were statistically significant, and four compounds displayed a fold-change (ratio between the final and initial concentrations) around or superior to 10 (i.e., an increase of 1 in log value): two fatty acids (pentanoic and hexanoic acids, associated with sweaty odors), acetoin associated with a buttery odor, and 2-nonanone with a green odor.
TABLE 1.
Volatile compounds identified in soy juice (SJ) unfermented and fermented with Lactobacillus delbrueckii subsp. delbrueckii CIRM-BIA865 throughout fermentationa
| Concentration (arbitrary units) for sample | |||||
|---|---|---|---|---|---|
| Volatile compound | Unfermented SJ | Fermented SJ | |||
| pH 7.2, 0.0 h | pH 6.5, 3.3 h | pH 6.0, 5.8 h | pH 5.0, 10.8 h | pH 4.6, 26.6 h | |
| Hexanal (a) | 8.8 ab | 9 a | 8.8 b | 8.6 c | 8.5 c |
| Hept-2-enal (a) | 7.4 b | 7.5 b | 7.5 b | 7.5 b | 7.7 a |
| Propan-1-ol (b) | 7.2 a | 7.3 a | 7.2 a | 6.9 b | 6.9 b |
| Pentanal (a) | 8.1 a | 8.1 a | 8a | 7.8 b | 7.8 b |
| 3-Methylbutanal (a) | 8.2 a | 8 b | 7.9 b | 7.8 b | 7.8 b |
| 1-Butanol (b) | 9 a | 9.1 a | 9 a | 8.9 a | 8.6 b |
| 2-Pentylfuran (g) | 9 a | 9.1 a | 9 a | 8.9 a | 8.6 b |
| 2-Ethylfuran (g) | 8.6 b | 8.9 a | 8.8 ab | 8.8 ab | 8.6 b |
| Propan-2-one (c) | 9 a | 8.9 a | 9 a | 8.8 a | 8.9 a |
| 2-Methylpropan-1-ol (b) | 6.1 a | 6.6 a | 6.4 a | 6.8 a | 6.5 a |
| 2-Butanone (c) | 8.1 a | 8.1 a | 8.1 a | 8.2 a | 8.2 a |
| Ethyl acetate (e) | 7.4 a | 7.6 a | 7.3 a | 7.4 a | 7.7 a |
| Propanoic acid (d) | 8 a | 7.9 a | 8.3 a | 7.8 a | 8.1 a |
| Benzaldehyde (a) | 7.8 a | 7.9 a | 7.8 a | 7.9 a | 7.9 a |
| Acetaldehyde (a) | 9.2 a | 9.2 a | 9.3 a | 9.2 a | 9.3 a |
| 2,5-Dimethylpyrazine (f) | 7.3 b | 7.5 ab | 7.5 ab | 7.6 a | 7.7 a |
| Formic acid (d) | 7.3 b | 7.5 ab | 7.5 ab | 7.5 ab | 7.6 a |
| Acetic acid (d) | 9.1 b | 9.5 ab | 9.5 ab | 9.4 ab | 9.7 a |
| Butanoic acid (d) | 7.3 c | 7.5 bc | 7.4 bc | 7.6 ab | 7.8 a |
| Pentanoic acid (d) | 7 c | 7.5 bc | 7.5 bc | 7.7 ab | 8.2 a |
| Hexanoic acid (d) | 7.5 c | 7.7 b | 7.7 bc | 7.8 ab | 8.2 a |
| Heptanoic acid (d) | 6.8 c | 6.9 bc | 6.9 abc | 7.1 ab | 7.3 a |
| Octanoic acid (d) | 7.1 b | 7.4 ab | 7.5 ab | 7.5 ab | 7.7 a |
| Nonanoic acid (d) | 7.4 b | 7.7 ab | 7.8 ab | 7.7 ab | 7.9 a |
| Decanoic acid (d) | 6.4 b | 6.7 ab | 6.7 ab | 6.8 ab | 6.9 a |
| Diacetyl (c) | 7.8 b | 7.8 b | 8 a | 8.1 a | 8.1 a |
| Acetoin (c) | 7.7 c | 7.9 c | 7.9 c | 8.5 b | 8.9 a |
| 3-Hydroxypropan-2-one (c) | 8.2 c | 8.3 c | 8.3 c | 8.6 b | 8.8 a |
| Oxolan-2-one (c) | 6.1 c | 6.1 bc | 6.2 abc | 6.3 abc | 6.3 a |
| Heptan-2-one (c) | 7.8 b | 8.1 ab | 8 a | 8.1 a | 8.2 a |
| Nonan-2-one (c) | 6.7 e | 7 d | 7.2 c | 7.4 b | 7.6 a |
| 1-Pentanol (b) | 7.6 b | 7.7 ab | 7.6 ab | 7.7 ab | 7.7 a |
| 1-Hexanol (b) | 7.6 c | 7.8 b | 7.9 b | 8.1 a | 8.2 a |
| Phenylmethanol (b) | 6.7 c | 6.9 b | 6.9 b | 6.9 ab | 7.1 a |
| 1-Phenylethanol (b) | 5.9 c | 6.2 b | 6.2 b | 6.3 ab | 6.5 a |
The concentrations are expressed as log 10 concentrations of the abundance of a specific m/z fragment for each compound. Letters after the concentrations indicate statistical differences from a Tukey test with an alpha error of 0.05 between the changes of concentration over time for a given compound. The letter in brackets indicates the nature of compound: aldehyde (a), alcohol (b), ketone (c), acid (d), ester (e), pyrazine (f), and furan (g).
Whole-genome analysis of Ld865
The genome of Ld865 was analyzed in order to identify metabolic pathways that were either complete or not, which could be of interest in terms of SJ fermentation. We focused on carbohydrate metabolism because it is determinant for acid lactic production and some are responsible for digestive discomfort. We also focused on nitrogen metabolism because it is essential for growth. Ld865 contained a single circular chromosome containing 2,008,473 bases pairs (bp) with a GC content of 49.7% (Table S2). The genome sequence contained 9 rRNA operons and 94 tRNAs. Among the 1,962 CDS identified, 85% (1,676 CDSs) were classified into at least one COG database category (Clusters of Orthologous Genes), including 358 CDSs in COG S/R (unknown function and general function prediction only) (Table S2). Apart from R and S, the most represented COG categories were L, i.e., replication, recombination, and repair (13.5% of CDSs), and E, i.e., amino acid transport and metabolism (7.9% of CDSs) (Fig. 5A).
Fig 5.
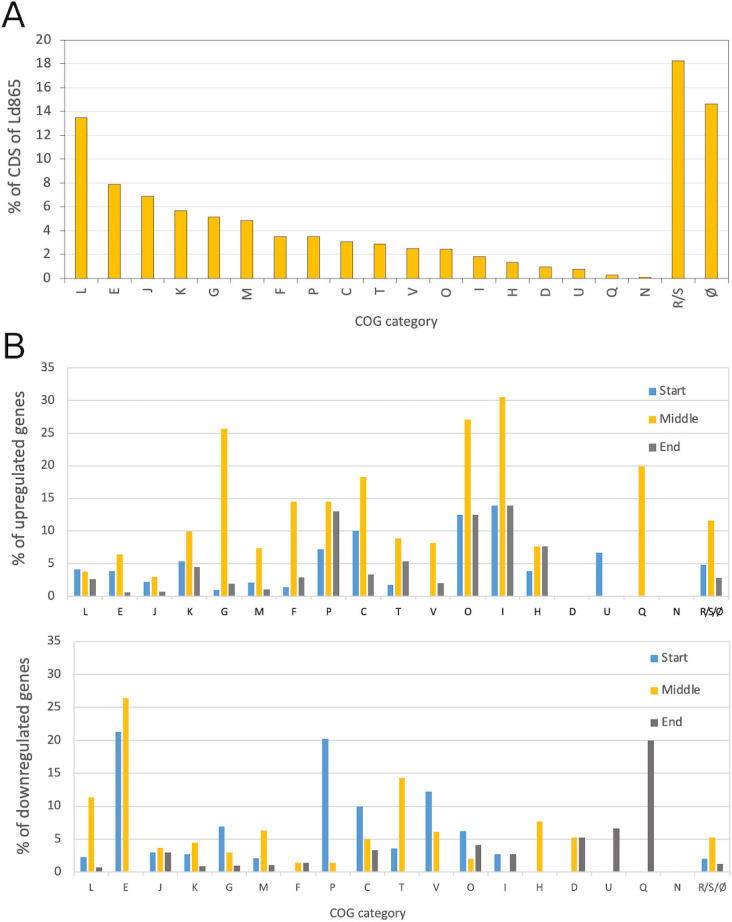
COG classification of Lactobacillus delbrueckii subsp. delbrueckii CIRM-BIA865 coding sequences (A) and transcription level during soy juice fermentation (B). (A) COG database categories of coding sequences (CDS) in Lactobacillus delbrueckii subsp. delbrueckii CIRM-BIA865 (Ld865) whole genome. Data are expressed as percent of total CDSs of the genome. The genome of Ld865 contains 1962 CDS, and 84.5% of the CDSs can be classified in a COG category. (B) For each COG category, percentage of genes differentially expressed (DE), between three successive steps of fermentation of soy juice by Ld865. Up: genes upregulated; down: genes downregulated. Blue bars: between pHs 6.5 and 6.0 (start of fermentation); Orange bars: between pHs 6.0 and 5.0 (middle of fermentation); Blue bars: between pHs 5.0 and 4.6 (end of fermentation). 100% represent all the genes classified in the COG. For example, between pHs 6.5 and 6.0 (start of fermentation), 3.9% and 21.3% of all the CDSs belonging to COG E are up and downregulated regulated, respectively. (A and B) The letter codes for COG (Clusters of Orthologous Genes) database categories are C, energy production and conversion; D, cell division and chromosome partitioning; E, amino acid transport and metabolism; F, nucleotide transport and metabolism; G, carbohydrate transport and metabolism; H, coenzyme metabolism; I, lipid metabolism; J, translation, ribosomal structure, and biogenesis; K, transcription; L, DNA replication, recombination, and repair; M, cell envelope biogenesis, outer membrane; N, cell motility and secretion; O, post-translational modification, protein turnover, and chaperones; P, inorganic ion transport and metabolism; Q, secondary metabolite biosynthesis, transport, and catabolism; T, signal transduction mechanisms; U, intracellular trafficking, secretion, and vesicular transport; V, defense mechanisms; S, function unknown; R, general function prediction only; and Ø, no category attributed.
Carbohydrate metabolism
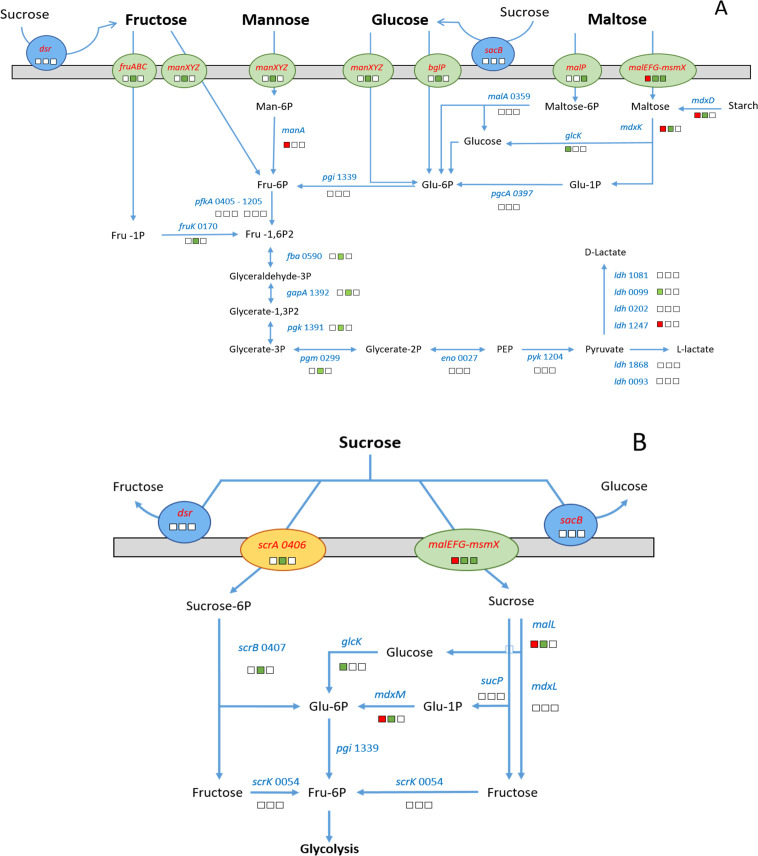
The Ld865 genome sequence contained genes coding for glycolysis, and transport and catabolism of several carbohydrates (101 genes), thus suggesting that Ld865 is able to consume a wide range of carbohydrates as carbon and energy sources. These genes included components required for the use of extracellular glucose, fructose, mannose, maltose, and trehalose (Fig. 6A). Concerning sucrose, the main sugar of soy juice, we identified four potential degradation pathways (Fig. 6B). The first pathway implicated the PTS-family protein ScrA (LDD865_V1_0406) involved in the transport and phosphorylation of sucrose into sucrose-6-phosphate, the sucrose-6-phosphate hydrolase ScrB (LDD865_V1_0407) involved in the conversion of sucrose-6-phosphate into β-d-fructose and glucose-6-phosphate, and the fructokinase ScrK (LDD865_V1_0054) involved in the conversion of β-d-fructose into fructose-6P, which can also fuel glycolysis. The second pathway involved the ABC transporter MsmX-MalEFG (from LDD865_V1_1778 to LDD865_V1_1781), the sucrose phosphorylase SucP (LDD865_V1_0063) which catalyzes the phosphorolysis of sucrose into Glucose-1P and d-fructose, the two α-glucosisades MalL (LDD865_V1_1785) and MdxL (LDD865_V1_1787), and the β-phosphoglucomutase MdxM (LDD865_V1_1782). Note that most of the genes coding for components in this potential sucrose assimilation pathway clustered together on the Ld865 chromosome. Finally, for the third and fourth pathways, the hydrolysis of sucrose could take place extracellularly and implicate enzymes predicted as extracellular: a glucansucrase (Dsr, LDD865_V1_0588) and a levansucrase (SacB, LDD865_V1_0265). These enzymes can cleave external sucrose into fructose and glucose; one moiety may be implemented in EPS biosynthesis, whereas the other may be further assimilated by dedicated metabolic pathways (Fig. 6A).
Fig 6.
Carbohydrate transport and metabolism pathway of Lactobacillus delbrueckii subsp. delbrueckii CIRM-BIA865, and gene expression profile throughout fermentation of soy juice. (A) global carbohydrate metabolism and (B) sucrose metabolism. The pathways were reconstructed based on the manual annotation of Ld865 genome. The expression of Ld865 genes was investigated by comparing genes differentially expressed (DE) throughout fermentation. The gene expression is represented by squares, red and green for DE genes down- and up-regulated, respectively. White squares are used for non-DE genes. The first square represents the gene expression between pH values of 6.5 (3.3 h of fermentation) and 6 (5.8 h of fermentation) (start of fermentation), the second, between 6 (5.8 h of fermentation) and 5 (10.8 h of fermentation) (middle of fermentation), and the third between 5 (10.8 h of fermentation) and 4.6 (26.5 h of fermentation) (end of fermentation). Orange, green, and blue ellipses forms represent PTS system transport, ABC transporter, and extracellular enzyme, respectively.
We also deciphered the inability of Ld865 to utilize as carbon sources stachyose, raffinose, and melibiose, the other main sugars in soy juice, as well as galactose and lactose. Accordingly, the Ld865 genome lacked (i) an α-galactosidase encoding gene required for stachyose, raffinose, and melibiose utilization; (ii) a β-galactosidase encoding gene, involved in lactose utilization; and (iii) galactokinase and UDP-glucose-hexose-1-phosphate uridylyltransferase encoding genes (enzymes EC 2.7.1.6 and EC 2.7.7.12) required for galactose assimilation via the Leloir pathway.
Nitrogen metabolism
The genome of Ld865 contained 156 genes involved in the biosynthesis and transport of various AAs as well as in the utilization of extracellular proteins and peptides as AA sources. The genes coding for the biosynthesis pathways of all AAs were present in the genome of Ld865, with the exception of those for the biosynthesis of serine, glycine, tryptophan, histidine, phenylalanine, tyrosine, and branched-chain amino acids, which were absent or uncomplete, suggesting Ld865 auxotrophy for these AAs. Ld865 was also probably able to acquire AAs from the extracellular environment. Indeed, we identified genes coding for ABC transporters specific to glutamine (glnH1PMH2Q and glnP2Q2), cystine (tcyBCA), methionine (metQ, metPNQ2), arginine (artQR-), and branched-chain AAs (livJHMGF). Also, permeases with different specificities, including a glutamate:GABA antiporter (gadC), an arginine/ornithine antiporter (arcD), a dl-alanine permease (serP2), a serine permease (serP1), two branched chain AA symporters (brnQ and brnQ2) were detected in the Ld865 genome (Table 2). Finally, Ld865 possessed a proteolytic system composed of proteases, peptidases, and peptide transport systems, potentially capable of supplying the cells in AAs from proteins present in the growth medium. First, 10 putative protease encoding genes, including the cell wall protease prtB gene (LDD865_V1_0941), were identified (Table 2). The serine proteinase PrtB is implicated in the degradation of milk proteins into peptides in dairy fermentations and is found in other strains and LAB species (36–38). The peptides released by PrtB can then be taken up by two oligopeptide transport systems belonging to the ABC transporter family, oppAA2BCDD1F (named opp1, from LDD865_v1_0763 to LDD865_v1_0770) and oppAA2BCDF (named opp2, from LDD865_v1_1722 to LDD865_v1_1731), as previously described for other LABs. Finally, once transported into the cytoplasm, peptides can be degraded into AAs by intracellular peptidases. We identified 38 genes coding for putative peptidases in the Ld865 genome, including specific and general peptidases as well as endopeptidases (Table 2). Lastly, it is interesting to note the presence of the arginine deiminase pathway (ADI pathway) that could be used to generate ATP via the breakdown of arginine. This pathway is encoded by the three genes arcABC, clustered together with arcD, the previously mentioned arginine/ornithine antiporter on the Ld865 chromosome.
TABLE 2.
Identification of genes coding for proteins composing the proteolytic system of Lactobacillus delbrueckii subsp. delbrueckii CIRM-BIA865
| Locus tag | Gene name | Product | EC number localizationa | |
|---|---|---|---|---|
| Peptidases | ||||
| LDD865_v1_0120 | _ | Putative dipeptidase | 3.4.-.- | C |
| LDD865_v1_0121 | _ | Putative dipeptidase | 3.4.-.- | C |
| LDD865_v1_0126 | pepN | Aminopeptidase N | 3.4.11.2 | C |
| LDD865_v1_0149 | pepD | Dipeptidase A | 3.4.-.- | C |
| LDD865_v1_0181 | pepF | Oligoendopeptidase F | 3.4.24.- | C |
| LDD865_v1_0300 | _ | Metalloendopeptidase | 3.4.24.- | M |
| LDD865_v1_0311 | pip | Proline iminopeptidase | 3.4.11.5 | C |
| LDD865_v1_0324 | pip2 | Proline iminopeptidase | 3.4.11.5 | C |
| LDD865_v1_0381 | _ | Putative endopeptidase | _ | C |
| LDD865_v1_0419 | pepV | Dipeptidase (beta-Ala-Xaa dipeptidase) | 3.4.13.- | C |
| LDD865_v1_0446 | pepC3 | Aminopeptidase C | 3.4.22.40 | C |
| LDD865_v1_0474 | _ | Putative peptidase | _ | C |
| LDD865_v1_0564 | pepQ | Xaa-Pro dipeptidase | 3.4.13.9 | C |
| LDD865_v1_0582 | dacA | d-alanyl-d-alanine carboxypeptidase | 3.4.16.4 | C |
| LDD865_v1_0592 | pbpF | Penicillin-sensitive transpeptidase | 2.4.1.129, 3.4.16.4 | M |
| LDD865_v1_0697 | pepX | Xaa-Pro dipeptidyl-peptidase | 3.4.14.11 | C |
| LDD865_v1_0732 | pepQ | Xaa-Pro aminopeptidase | 3.4.13.9 | C |
| LDD865_v1_0900 | _ | Acylaminoacyl-peptidase | 3.4.19.1 | C |
| LDD865_v1_0917 | pepT | Peptidase T | 3.4.11.4 | C |
| LDD865_v1_0919 | sipT | Signal peptidase I | 3.4.21.89 | M |
| LDD865_v1_0953 | pcp | Pyrrolidone-carboxylate peptidase | 3.4.19.3 | C |
| LDD865_v1_0958 | _ | ld-carboxypeptidase, muramoyltetrapeptide carboxypeptidase | 3.4.17.13 | C |
| LDD865_v1_1009 | pip | Proline iminopeptidase | 3.4.11.5 | C |
| LDD865_v1_1071 | lspA | Signal peptidase II | 3.4.23.36 | M |
| LDD865_v1_1079 | ponA | Penicillin-insensitive transglycosylase/penicillin-sensitive transpeptidase | 2.4.1.129, 3.4.16.4 | M |
| LDD865_v1_1080 | _ | Acylaminoacyl-peptidase | 3.4.19.1 | C |
| LDD865_v1_1265 | _ | serine-type endopeptidase | 3.4.21.53 | M |
| LDD865_v1_1401 | dmpA | D-aminopeptidase | 3.4.11.9 | C |
| LDD865_v1_1402 | dppA | D-aminopeptidase | 3.4.11.- | C |
| LDD865_v1_1423 | _ | Zn-dependent peptidase | 3.4.24.- | C |
| LDD865_v1_1424 | _ | Putative Zn-dependent peptidase | _ | C |
| LDD865_v1_1468 | mapA | Methionine aminopeptidase | 3.4.11.18 | C |
| LDD865_v1_1506 | pepA | Glutamyl aminopeptidase | 3.4.11.7 | C |
| LDD865_v1_1507 | pepDA | Dipeptidase A | 3.4.13.19 | C |
| LDD865_v1_1631 | pepT2 | Peptidase T | 3.4.11.4 | C |
| LDD865_v1_1719 | pepC1 | Aminopeptidase C | 3.4.22.40 | C |
| LDD865_v1_1720 | pepC2 | Aminopeptidase C | 3.4.22.40 | C |
| LDD865_v1_1755 | pepO | Neutral endopeptidase | 3.4.24.- | C |
| Proteases | ||||
| LDD865_v1_0805 | eep | zinc metalloprotease | 3.4.24.- | M |
| LDD865_v1_0867 | hsIV | two-component ATP-dependent protease. ATP-dependent protease HslV | 3.4.25.2 | C |
| LDD865_v1_0941 | prtP | Peptidase lactocepin | 3.4.21.96 | M |
| LDD865_v1_0944 | htpX | Zn-dependent protease | 3.4.24.- | M |
| LDD865_v1_0952 | PrsW | Intramembrane metalloprotease | _ | M |
| LDD865_v1_1397 | clpP | ATP-dependent Clp protease, proteolytic subunit | 3.4.21.92 | C |
| LDD865_v1_1634 | ftsH | ATP-dependent zinc metalloprotease FtsH (cell division protease FtsH) | 3.4.24.- | M |
| LDD865_v1_1821 | htpX | Protease HtpX Zn-dependent with chaperone function | 3.4.24.- | M |
| LDD865_v1_1835 | htrA | Serine protease Do-like HtrA | 3.4.21.107 | M |
| LDD865_v1_1847 | lexA | SOS-response repressor and protease | 3.4.21.88 | M |
| Transport systems | ||||
| LDD865_v1_1497 to 1501, 1958 to 1959 | glnH1PMH2Q, glnP2Q2 | Glutamine ABC transporter | ||
| LDD865_v1_1686 to 1688 | tcyBCA | Cystine ABC transporter | ||
| LDD865_v1_0102, 1241 to 1243 | metQ, metPNQ2 | Methionine ABC transporter | ||
| LDD865_v1_0302 to 0304 | artQR | Arginine ABC transporter | ||
| LDD865_v1_0722 to 0726 | livJHMGF | Branched-chain AA ABC transporter | ||
| LDD865_v1_0685 | gadC | Glutamate:GABA antiporter | ||
| LDD865_v1_0350 | arcD | Arginine/ornithine antiporter | ||
| LDD865_v1_0459 | serP2 | dl-alanine permease | ||
| LDD865_v1_0418 | serP1 | Serine permease | ||
| LDD865_v1_1529, 1876 | brnQQ2 | Branched chain AA symporters | ||
C: protein predicted as cytoplasmic, M: protein predicted as membrane
Transcriptomic analysis of Ld865 during soy juice fermentation
The expression of Ld865 genes was investigated by comparing genes differentially expressed (DE) throughout fermentation. Four sampling points were analyzed: at pH 6.5 (3.3 h of fermentation), pH 6 (5.8 h of fermentation), pH 5 (10.8 h of fermentation), and pH 4.6 (26.5 h of fermentation). The differential expression of each gene was calculated between two consecutive points, i.e., between pH values of 6 and 6.5 (start of fermentation), between 5 and 6 (middle of fermentation), and finally between 4.6 and 5 (end of fermentation) (Table S3). Throughout fermentation, 186, 354, and 91 genes were found to be DE at the start, middle, and end of fermentation, respectively (considering an adjusted P-value < 0.05, and |Log2(FC)| >1). DE genes were found in almost all COG categories but not with the same distribution (Fig. 5B). Between pH 6.5 and 6 (start of fermentation), the COG with the highest percentage of DE genes were COG P, E, and C, with 27.5%, 25.2%, and 20% of DE genes, respectively. Between pH 6 and 5, the most modulated COGs were E and I (32.9% and 30.5% of genes from these categories were DE, respectively), followed by O and G (about 29%). It is during this phase of fermentation that the most DE genes were counted. Finally, between pH 5 and 4.6, the end of fermentation, none category presented a percentage of DE genes superior to 20%. Taking account of these data and of the physiological and biochemical data previously described, we further focused the transcriptomic analysis on COG categories C (energy production and conversion), G (carbohydrate transport and metabolism), and E (AA transport and metabolism) to investigate metabolism during growth in soy juice. We also focused on COG categories M (cell envelope biogenesis), I (lipid transport and metabolism), D (cell division and chromosome partitioning), and F (nucleotide transport and metabolism), with the aim to decipher also the elongation of the cells during growth in SJ.
Carbohydrate metabolism, energy production and conversion
During the middle stage of fermentation, numerous genes implicated in carbohydrate metabolism were DE and mainly upregulated (Fig. 6 and Table S3). This notably included genes involved in sucrose transport (scrA of the PTS system, and msmX-malEFG of the ABC transporter) and catabolism (scrB, malL, and mdxDKM). The dsr gene, coding for a glucansucrase, was also upregulated but at a low level of transcription (FC of 1.6). In addition, the expression of genes coding for the transport and catabolism of other sugars, i.e., fructose, glucose/fructose/mannose, and maltose, was upregulated. Finally, several genes implicated in glycolysis pathway were also upregulated (fba, tpiA, gapA, pgk, pgm). Note that several transcriptional regulators related to sugar assimilation were also DE. The genes encoding the transcriptional activators TreR (LDD865_v1_0253) and MdxR (LDD865_v1_1786) that might positively regulate expression of the bglG and the malL-mdxDKM-msmX-malEFG cluster, respectively, were upregulated.
Membrane and cell wall biogenesis
During the start of fermentation, 4.2% of CDS belonging to COG M were DE (four genes), including a gene encoding for a putative peptidoglycan-binding protein (LDD865_V1_1845) which is up-regulated . Then, during the middle of fermentation, this rate reached 13.7%, with 6 and 7 among genes down- and upregulated, respectively. Among these, we identified gene implicated in the PG biosynthesis pathway (dacA), as well as glmS, a gene implicated in UDP-N-acetyl-glucosamine, a precursor of PG (Table S3). Fatty acids are important components in the phospholipids that make up the lipid bilayer of cell membranes. Belonging to the fatty acid biosynthesis pathway, numerous genes implicated in the initiation and elongation steps were upregulated, mainly between pH 6.0 and 5.0 (fabG LDD865_V1_1422 and LDD865_V1_1147; fabI LDD865_V1_1140 for elongation step, and accABCD and fabHA in initiation). Furthermore, several genes implicated in the transport of cofactors necessary for the biosynthesis of fatty acids are also upregulated throughout fermentation: the biotin/riboflavin transport genes ribU (LDD865_V1_1194) and bioY (LDD865_V1_0088).
Machinery of DNA replication, cell division
Among the genes belonging to COG D, two genes were found to be downregulated: crcB (LDD865_V1_1367) between pH 6 and 5, and crcB2 (LDD865_V1_1368) between pH 5 and 4.6. The functions of these genes are not well characterized, and no other gene implicated in cell division was found to be DE.
Nitrogen metabolism
Overall, the genes coding for components of the proteolytic system and proteins involved in the biosynthesis and transport of AAs were downregulated or constitutively expressed during the early and middle stages of fermentation. At the start of fermentation, 25.1% of CDS belonging to COG E were DE (i.e., 39 CDS). They are divided into 33 and 6 down- and upregulated genes, respectively. During the middle of fermentation, the rate of DE genes for this category reached 32.9%, with 41 and 10 down- and upregulated genes, respectively. Downregulated genes notably included those encoding the PrtB proteinase (LDD865_V1_0941), components of the oligopeptide transport systems Opp1 and Opp2 and components of amino acid transporters (e.g., TcyABC for cystine, ArtR for arginine, LivJHMGF for branched chain amino acids). Among amino acid biosynthesis genes, the lysine biosynthesis pathway from l-aspartate is mainly downregulated from the start of fermentation (for the genes asd, lysA, dapX, dapH, dapH, dapA), and during the middle of fermentation (for the asd, lysA, and dapX genes). However, we note that few Ld865 nitrogen metabolic genes were upregulated. These exceptions concerned genes involved in the synthesis of proline (proA) and those involved in glutamine and arginine synthesis glnA, argG, and argH), as well as peptidase genes (pepF, pip2, dmpA, pepD). The arcABC genes coding for the arginine deiminase (ADI) pathway are also upregulated at the start of fermentation, as well as the associated arginine/ornithine antiporter arcD. At the end of fermentation, in the COG E category, only the gene LDD865_v1_1580 encoding for an oligopeptitide ABC transporter was DE and upregulated.
Nucleotide transport and metabolism
Several genes implicated in the biosynthesis of pyrimidine were found to be upregulated. Notably, during the middle stage of fermentation, the expression of four out of the five genes involved in the production of uridine-5′-phosphate from the pyrimidine precursor carbamoyl phosphate (pyrBCEF: LDD865_v1_1055, LDD865_v1_1054, LDD865_v1_0600, LDD865_v1_601) was upregulated, with FC ranging from 4.94 to 12.78. Moreover, different pathways leading to the production of the precursor carbamoyl phosphate were also upregulated. They included previously mentioned genes coding for the import and degradation of arginine into carbamoyl phosphate (arcABCD) during the start of fermentation and the conversion of glutamine into carbamoyl phosphate (pyrAA, pyrAB) during the middle of fermentation.
Stress-related metabolism
During fermentation, especially in the middle and at the end, numerous genes implicated in stress response were up-regulated (Table S3). This includes genes coding for chaperone systems DnaJ/DnaK/GrpE and GroES/GroEL. This also includes genes coding for ATP-dependent Clp chaperone–proteases stress response proteins ClpA, ClpP, and ClpE.
DISCUSSION
The aim of the present work was to decipher the behavior of L. delbrueckii subsp. delbrueckii CIRM-BIA865 during SJ fermentation based on a multi-omic approach including genomic, transcriptomic, and targeted metabolomic. In the current context of food transition toward a diet containing less animal protein, the development of soy juice-yogurts has a potential role to play. However, because these plant-based “yogurt like” products are much more recent than dairy ones, notably in Western countries, there is a lack of genomic, biochemical, physiological, and metabolic information on newly developed starters that are able to ferment these substrates. In the previous work, out of the 20 strains of L. delbrueckii studied, 12 acidified SJ (16), and 4 of these were qualified as “fast acidifying,” including Ld865, as they acidified SJ below pH 6 within 10 h.
The Ld865 strain was first isolated from plants. Analysis of its 16S rRNA gene, combined with determination of its carbohydrate fermentation profile (Table S4) indicated that Ld865 belongs to the subspecies delbrueckii. Ld865 is now the fourth strain in this subspecies whose entire genome has been sequenced (along with the type strain DSM20074, TUA4408L, and NBRC 3202). The genome content is in agreement with those of other complete genomes of the subspecies in terms of size, CDS number, and GC content (39). As previously observed in L. delbrueckii species, the GC3 value is high (64.1%) which is in line with its thermophilic growth since Hurst & Merchant reported a GC3 comprised between 48% and 67% for thermophilic species (40).
Among other features, the delbrueckii subspecies is distinguished from the other subspecies (i.e., bulgaricus, lactis, indicus, jakobsenii, and sunkii) by its inability to metabolize lactose (39, 41–43), which explains why this subspecies is not used in dairy fermentation. It is also known to be able to metabolize sucrose but not raffinose and stachyose (44). Here, we confirmed and extended this knowledge by showing that Ld865 is also able to catabolize fructose and glucose but not galactose (Table S4). It should be noted that Baek et al. (39) found the inability to degrade trehalose as a characteristic specific of the subspecies, whereas we found Ld865 as trehalose-positive.
Concerning the gene content in line with the metabolism of these carbohydrates, we did not identify any beta-galactosidase encoding gene, necessary for lactose utilization. Nor did we find any genes coding for a lactose-permease. This could explain the inability of Ld865 to use lactose. Likewise, its inability to use stachyose and raffinose could be due to the absence of an alpha-galactosidase gene. Concerning the ability of Ld865 to degrade sucrose, we identified the potential genetic determinants of different sucrose degradation pathways. A first one implemented the ScrA sucrose-PTS system and the ScrB and ScrK enzymes, while a second implemented a potential sucrose ABC transporter, i.e., the MsmX-MalEFG system, a cytoplasmic sucrose phosphorylase, two potential oligo-1,6-glucosidases (alpha-glucosidase), and a β-phosphoglucomutase. These two systems are commonly found in LAB (45–47). Finally, we found a predicted glucansucrase (Dsr) and a predicted levansucrase (SacB), two extracellular enzymes, that can hydrolysis sucrose into fructose and glucose. These extracellular sucrose degradation mechanisms are commonly found in L. delbrueckii; particularly in the lactis and delbrueckii subspecies and more generally in LAB, acting in both a strain- and species-dependent manner (45, 46).
This study showed that during fermentation, among the three main sugars present in SJ (sucrose, stachyose, and raffinose), Ld865 only fermented sucrose, which is consistent with the genetic properties described above. Based on gene expression profiles, sucrose is probably imported and metabolized via the PTS-pathway, as described for Streptococcus thermophilus (47). However, interestingly, Ld865 also implemented the MalEFG-MsmX ABC transporter to import external sucrose, which probably makes the strain more efficient in the use the external sucrose. Indeed, this ABC transporter has been described as an efficient transport system for both maltose and sucrose, in the species Thermus thermophilus and Streptococcus mutans (48, 49). The glucansucrase Dsr could hydrolyze external sucrose and have contributed to the increase in the external fructose content detected in SJ during the middle stage of fermentation. Finally, the extracellular levansucrase SacB is probably not implicated in sucrose degradation because sacB was not DE and was expressed at a very low level during SJ fermentation.
The proteolytic system of LAB comprises proteinases, peptidases, and specific AA and peptide transport systems. The genomic sequence of Ld865 revealed the presence of at least 50 genes encoding for potential proteases or peptidases, and numerous transport systems dedicated to AA or small peptide transport. For comparison, Zheng et al. (50) identified 45 genes encoding for proteases/peptidases in L. delbrueckii subsp. bulgaricus 2038, and Gao et al. (51) identified 33 genes encoding for peptidases and AA uptake system in L. plantarum Y44. Moreover, we identified in the Ld865 genome the genes involved in the biosynthesis of several AAs. The AA biosynthesis and supply capability, including the proteolytic system of Ld865, is, thus, similar to that found in dairy LAB. Transcriptomic analysis, combined with metabolomic data, enabled us to decipher the nitrogen metabolism of Ld865 during SJ fermentation. Apart from glutamine, SJ contains all free AAs, most of them at high levels. In the early stage of fermentation, we observed a decrease of all AAs, except proline, while their concentration increased at later stages, except for glutamine and arginine. This fall in AA levels arose from their consumption by Ld865, while their increase could result from the proteolytic activity of Ld865 and their export from the cells after peptide internalization and hydrolysis. The downregulation of numerous genes involved in AA biosynthesis and supply, detected during early fermentation, was in line with a preferential use of exogenous AAs, a source more energetically advantageous than their synthesis. In the same way, although downregulated, the high level of expression of many ABC transporters, permeases, and antiporter systems between pH 6.5 and 6 also supports the hypothesis of a preferential use of external AAs. The cell wall protease prtB gene, which presented a high level of transcription, could also contribute to fuel the nitrogen needs of Ld865 during SJ fermentation. A crucial role for cell wall protease in SJ fermentation had already been shown with S. thermophilus PrtS, a homolog of PrtB (47).
Then, during further fermentation, the majority of the transport systems were downregulated, as well as prtB. This could indicate that Ld865 imported fewer AAs/peptides into its cytoplasm and needed less AAs from the external medium. Concomitantly, different peptidase-encoding genes were up- and downregulated at specific times during fermentation, suggesting an adaptation of the peptidase system to supply AAs for cell metabolism. Finally, the origin of the increase in free AAs observed in the SJ from pH 6 is uncertain. Presumably, this increase was due to cell lysis at the end of fermentation, which could the release into the medium intracellular proteolytic enzymes that hydrolyze proteins into AAs. This phenomenon has been widely described in dairy fermentations, and particularly in cheese (52, 53). This rise in AAs during fermentation might also be of interest because some AAs can be precursors of aroma compounds (54, 55).
We, thus, showed that the entire proteolytic system of Ld865 was active during SJ fermentation (AA transporters, proteases/peptidases, and AA biosynthesis) and possibly contributed to the nitrogen requirement of the strain. Lastly, it is interesting to note the particular case of arginine metabolism, which is the only AA depleted at the end of fermentation. In parallel, the concentrations of ornithine and citrulline increased. The transcriptomic data revealed an upregulation of the genes of the ADI pathway during the first stage of fermentation. Altogether these results suggest a conversion of arginine into citrulline and ornithine, CO2, and ammonium via the ADI pathway, which protects the cells against acidity, and provide energy, as observed in many bacteria including LAB (56, 57). This pathway also participates in ATP generation, as described by Pols et al. (58).
Regarding flavor compounds, some of the volatiles identified, i.e., acetaldehyde, butane-2,3-dione (diacetyl), 3-hydroxybutan-2-one (acetoin), and 2-butanone, are considered major flavor compounds in dairy yogurt (59). All result from the activity of LAB. Acetaldehyde, a compound essential to the typical aroma of yogurt and associated with a fresh and green note, is produced by LAB in a strain-dependent manner (60). In the present study, acetaldehyde is detected and, thus, could contribute to the flavor of the product, but its content did not significantly vary during fermentation (Table 1), suggesting that Ld865 does not produce this compound. Diacetyl and its reduced derivative acetoin are associated with a sweet and buttery aromatic note but also yogurt. Interestingly, acetoin and diacetyl concentrations showed a 4- and 1.4-fold increase during fermentation (Table 1). The concentration of other potential flavor compounds of yogurt, propan-2-one (acetone), and 2-butanone, associated with a sweet and fruity note, did not vary during fermentation. In addition, some fatty acids increased in concentration, the main being butanoic, pentanoic, and hexanoic acids. These compounds could be associated with cheesy notes, as observed in Lben, a traditional Moroccan fermented milk (61). Finally, fermentation with Ld865 reduced the concentration of hexanal and 2-pentylfuran, compounds that are responsible of “off-flavors.” In summary, the strain Ld865 exhibited a low potential to produce yogurt flavor compounds. However, it produced some flavor compounds of interest, even if in low concentrations and also reduced hexanal and 2-pentylfuran.
During fermentation, we observed that despite the metabolic activity of Ld865 (i.e., sucrose fermentation, metabolite production, and proteolytic activity), the cell number of Ld865 did not increase. However, concomitantly with acidification, cell length increased (about 10 times). This phenomenon was specifically observed during SJ fermentation, and not in the nutrient-rich MRS broth medium. Transcriptomic analysis showed that no known genes implicated in DNA replication and cell division were DE during fermentation, whereas several genes implicated in PG and membrane lipid synthesis were upregulated. Ld865 cell elongation may, thus, result from the synthesis of new sidewalls but without initiation of the division process (DNA replication and division). These results argue for a control of Ld865 cell division at the translational and/or post-translational levels. Several studies have also reported that septal and peripheral PG synthesis can be uncoupled by post-transcriptional controls in Lactococcus lactis, an ovococcus that can undergo coccus-to-rod transition and further filamentation during growth in a synthetic medium (62, 63). Under favorable environmental conditions, growing bacteria usually exhibit a relatively constant cell size and the bacterial growth is accompanied by cell division; growth is, thus, linked to an increase in the bacterial population. However, cell length, which is a fine-tuned process [for a review of cell size control, see reference (64)], has been shown to be sensitive to growth conditions, and notably environmental changes and stresses. Cell elongation was first documented in Salmonella (65), where cell size differed according to the availability of nutrients in the growth medium (glucose-rich medium). Several studies subsequently identified different components of the central metabolism as key metabolites for coordination between growth and division. In Bacillus subtilis, cell division was shown to depend on pyruvate utilization (66). Weart et al. (67) showed that B. subtilis alpha-phosphoglucomutase, involved in the interconversion of glucose-6-phosphate and glucose-1-phosphate, affects cell size by controlling the availability of the substrate for the UgtP enzyme, which prevents Z-ring formation in a nutrient-dependent manner. An impact on the cell shape of phosphoglucomutase has also been documented in Escherichia coli and L. lactis (68, 69). Moreover, an aberrant morphology of the L. lactis beta-phosphoglucomutase-mutant was observed in maltose but not when the cells were grown on glucose or lactose (69). Ludszuweit et al. (70) also observed in L. delbrueckii subsp. delbrueckii that medium composition influenced cell size, and bacteria grown in filter-sterilized MRS broth were smaller than those cultivated in heat-sterilized MRS. Unfavorable environmental conditions such as pH, temperature, antibiotic, osmotic stress, or DNA damage have also been described as being implicated in cell elongation observed in L. delbrueckii, L. lactis, Lactobacillus, and E. coli (63). We were not able to identify the type(s) of the component(s) and/or the conditions that impaired the division process during our study, but the many stress genes that are upregulated during fermentation indicated that bacterial cells have to face stressful conditions. It has been hypothesized that the cell elongation which leads to an increase in the total bacterial cell surface area might be a response to maximizing nutritional uptake in cells exposed to nutrient-limiting environments (71). Finally, it is interesting to note that the morphological changes observed during growth in SJ were not specific to the delbrueckii subspecies, since a L. delbrueckii subsp. bulgaricus strain, grown in SJ permeate, also displayed a cell elongation phenotype (12). Contrary to Ld865, the strain used in this work did not acidify, which was expected as the species is not able to use the sugars of soy juice. The authors conclude that bacteria were under stress, and the stress cause(s) remain to be identified. The identification of SJ factors involved in Ld865 cell elongation deserves now further investigation.
Conclusion
In order to get a soy-based yogurt, several conditions have to be fulfilled: (i) the fermentation has to fit industrial criteria of production and (ii) the final product has to be good for the consumer, in terms of organoleptic and nutritional properties. The strain Ld865 was chosen because a precedent study showed it fills the first criterion. Concerning the second criterion, we showed that fermentation with Ld865 reduced the concentration of hexanal and 2-pentylfuran, two compounds responsible of “off-flavors,” albeit to a limited extent. However, compounds of interest in yogurt-like products were also produced, such as acetoin and diacetyl. Despite the acidification properties of Ld865, cells lengthened but did not divide during fermentation, indicating that SJ constitutes a stressful environment for the cells. Identification of the relevant stress factors might contribute to counteracting them and, thus, further improve the acidification rate. Among the available SJ sugars, Ld865 only used sucrose as carbon source. The inability of Ld865 to use stachyose and raffinose, two oligosaccharides potentially responsible for intestinal discomfort among consumers (72, 73), may hinder its use as single strain starter in soybean yogurt-like products. To overcome this, it would be interesting to associate Ld865 with a strain endowed with the alpha-galactosidase activity necessary to ferment raffinose and stachyose. Generally speaking, a specific difficulty in the development of a starter culture adapted to the production of soy juice yogurts is that, like other vegetable juices, but unlike cow’s milk, the composition of soy juice (a water extract of soybean) can vary considerably, depending on the manufacturing process. Regardless of the process implemented, soy juice always offers an important source of protein (about 35 g/L) and lipids (about 18 g/L) (USDA, 2019. https://fdc.nal.usda.gov/), but it may contain very variable amounts of sugar, ranging from 1 to 35 g/L (73, 74) including 50% sucrose and 50% of raffinose and stachyose. For these reasons, the fermentation results obtained with a specific SJ cannot be generalized to others, and the starter bacteria should be adapted accordingly. Finally, in the present study, it is interesting to note that fermentation leads to an increase in free amino acids, from 426 before fermentation to 751 mg/L in fermented SJ. The fermentation of non-dairy “milk,” thus, represents a major opportunity to increase the nutritional quality of plant-based beverages. It offers a challenge for research to identify ad hoc starters in order to ferment these new plant-based milk substitutes based on a better knowledge of their metabolism in such substrates.
ACKNOWLEDGMENTS
LABGeM (CEA/Genoscope & CNRS UMR8030), the France Génomique, and French Bioinformatics Institute national infrastructures (funded as part of the Investissement d'Avenir program managed by Agence Nationale pour la Recherche, contracts ANR-10-INBS-09 and ANR-11-INBS-0013) are acknowledged for their support within the MicroScope annotation platform. The authors are grateful to the Toulouse Occitanie bioinformatics platform (Bioinfo Genotoul, https://doi.org/10.15454/1.5572369328961167E12) for providing help and/or computing and/or storage resources.
Contributor Information
Stéphanie-Marie Deutsch, Email: stephanie-marie.deutsch@inrae.fr.
Danilo Ercolini, Universita degli Studi di Napoli Federico II, Portici, Italy.
DATA AVAILABILITY
The sequence of the Ld865 genome sequenced in the study was deposited in the European Nucleotide Archive (ENA) at EMBL-EBI under accession number PRJEB50396. The transcriptome data generated in the study were deposited in the ArrayExpress platform under accession numbers E-MTAB-12095. The data supporting the biochemical results are available at https://entrepot.recherche.data.gouv.fr, under DOI 10.57745/AOBH6L.
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/aem.01936-23.
PCA of volatile compounds.
Volatil compounds data (Table S1) and fermentation profile (Table S4).
Lactobacillus delbrueckii subsp. delbrueckii CIRM-BIA865 Genome annotation.
Transcriptomic data.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. de Boer J, Aiking H. 2018. Prospects for pro-environmental protein consumption in Europe: cultural, culinary, economic and psychological factors. Appetite 121:29–40. doi: 10.1016/j.appet.2017.10.042 [DOI] [PubMed] [Google Scholar]
- 2. Willett W, Rockström J, Loken B, Springmann M, Lang T, Vermeulen S, Garnett T, Tilman D, DeClerck F, Wood A, et al. 2019. Food in the anthropocene: the EAT–Lancet commission on healthy diets from sustainable food systems. The Lancet 393:447–492. doi: 10.1016/S0140-6736(18)31788-4 [DOI] [PubMed] [Google Scholar]
- 3. Springmann M, Clark M, Mason-D’Croz D, Wiebe K, Bodirsky BL, Lassaletta L, de Vries W, Vermeulen SJ, Herrero M, Carlson KM, Jonell M, Troell M, DeClerck F, Gordon LJ, Zurayk R, Scarborough P, Rayner M, Loken B, Fanzo J, Godfray HCJ, Tilman D, Rockström J, Willett W. 2018. Options for keeping the food system within environmental limits. Nature 562:519–525. doi: 10.1038/s41586-018-0594-0 [DOI] [PubMed] [Google Scholar]
- 4. Jeske S, Zannini E, Arendt EK. 2017. Evaluation of physicochemical and glycaemic properties of commercial plant-based milk substitutes. Plant Foods Hum Nutr 72:26–33. doi: 10.1007/s11130-016-0583-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Fructuoso I, Romão B, Han H, Raposo A, Ariza-Montes A, Araya-Castillo L, Zandonadi RP. 2021. An overview on nutritional aspects of plant-based beverages used as substitutes for cow's milk. Nutrients 13:2650. doi: 10.3390/nu13082650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chalupa-Krebzdak S, Long CJ, Bohrer BM. 2018. Nutrient density and nutritional value of milk and plant-based milk alternatives. Int Dairy J 87:84–92. doi: 10.1016/j.idairyj.2018.07.018 [DOI] [Google Scholar]
- 7. Guillon F, Champ M-J. 2002. Carbohydrate fractions of legumes: uses in human nutrition and potential for health. Br J Nutr 88 Suppl 3:S293–S306. doi: 10.1079/BJN2002720 [DOI] [PubMed] [Google Scholar]
- 8. Kaneko S, Kumazawa K, Nishimura O. 2011. Studies on the key aroma compounds in soy milk made from three different soybean cultivars. J Agric Food Chem 59:12204–12209. doi: 10.1021/jf202942h [DOI] [PubMed] [Google Scholar]
- 9. Curiel JA, Coda R, Centomani I, Summo C, Gobbetti M, Rizzello CG. 2015. Exploitation of the nutritional and functional characteristics of traditional Italian legumes: the potential of sourdough fermentation. Int J Food Microbiol 196:51–61. doi: 10.1016/j.ijfoodmicro.2014.11.032 [DOI] [PubMed] [Google Scholar]
- 10. Lv Y-C, Song H-L, Li X, Wu L, Guo S-T. 2011. Influence of blanching and grinding process with hot water on beany and non-beany flavor in soymilk. J Food Sci 76:S20–S25. doi: 10.1111/j.1750-3841.2010.01947.x [DOI] [PubMed] [Google Scholar]
- 11. Shi X, Li J, Wang S, Zhang L, Qiu L, Han T, Wang Q, Chang S-C, Guo S. 2015. Flavor characteristic analysis of soymilk prepared by different soybean cultivars and establishment of evaluation method of soybean cultivars suitable for soymilk processing. Food Chem 185:422–429. doi: 10.1016/j.foodchem.2015.04.011 [DOI] [PubMed] [Google Scholar]
- 12. Jan G, Tarnaud F, Rosa do Carmo FL, Illikoud N, Canon F, Jardin J, Briard-Bion V, Guyomarc’h F, Gagnaire V. 2022. The stressing life of Lactobacillus delbrueckii subsp. bulgaricus in soy milk. Food Microbiol 106:104042. doi: 10.1016/j.fm.2022.104042 [DOI] [PubMed] [Google Scholar]
- 13. Mital BK, Steinkraus KH. 1979. Fermentation of soy milk by lactic acid bacteria. J Food Prot 42:895–899. doi: 10.4315/0362-028X-42.11.895 [DOI] [PubMed] [Google Scholar]
- 14. Wang Y-C, Yu R-C, Yang H-Y, Chou C-C. 2003. Sugar and acid contents in soymilk fermented with lactic acid bacteria alone or simultaneously with bifidobacteria. Food Microbiol 20:333–338. doi: 10.1016/S0740-0020(02)00125-9 [DOI] [Google Scholar]
- 15. Lorn D, Nguyen T-K-C, Ho P-H, Tan R, Licandro H, Waché Y. 2021. Screening of lactic acid bacteria for their potential use as aromatic starters in fermented vegetables. Int J Food Microbiol 350:109242. doi: 10.1016/j.ijfoodmicro.2021.109242 [DOI] [PubMed] [Google Scholar]
- 16. Harlé O, Falentin H, Niay J, Valence F, Courselaud C, Chuat V, Maillard M-B, Guédon É, Deutsch S-M, Thierry A. 2020. Diversity of the metabolic profiles of a broad range of lactic acid bacteria in soy juice fermentation. Food Microbiol 89:103410. doi: 10.1016/j.fm.2019.103410 [DOI] [PubMed] [Google Scholar]
- 17. De Man JC, Rogosa M, Sharpe ME. 1960. A medium for the cultivation of lactobacilli. J Appl Bacteriol 23:130–135. doi: 10.1111/j.1365-2672.1960.tb00188.x [DOI] [Google Scholar]
- 18. Baron F, Cochet M-F, Ablain W, Grosset N, Madec MN, Gonnet F, Jan S, Gautier M. 2006. Rapid and cost-effective method for microorganism enumeration based on miniaturization of the conventional plate-counting technique. Lait 86:251–257. doi: 10.1051/lait:2006005 [DOI] [Google Scholar]
- 19. Canon F, Maillard M-B, Henry G, Thierry A, Gagnaire V, Björkroth J. 2021. Positive interactions between lactic acid bacteria promoted by nitrogen-based nutritional dependencies. Appl Environ Microbiol 87:e0105521. doi: 10.1128/AEM.01055-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lorieau L, Halabi A, Ligneul A, Hazart E, Dupont D, Floury J. 2018. Impact of the dairy product structure and protein nature on the proteolysis and amino acid bioaccessiblity during in vitro digestion. Food Hydrocoll 82:399–411. doi: 10.1016/j.foodhyd.2018.04.019 [DOI] [Google Scholar]
- 21. Pogačić T, Maillard M-B, Leclerc A, Hervé C, Chuat V, Yee AL, Valence F, Thierry A. 2015. A methodological approach to screen diverse cheese-related bacteria for their ability to produce aroma compounds. Food Microbiol 46:145–153. doi: 10.1016/j.fm.2014.07.018 [DOI] [PubMed] [Google Scholar]
- 22. Falentin H, Deutsch S-M, Loux V, Hammani A, Buratti J, Parayre S, Chuat V, Barbe V, Aury J-M, Jan G, Le Loir Y. 2016. Permanent draft genome sequence of the probiotic strain Propionibacterium freudenreichii CIRM-BIA 129 (ITG P20). Stand Genomic Sci 11:6. doi: 10.1186/s40793-015-0120-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wick RR, Judd LM, Gorrie CL, Holt KE. 2017. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol 13:e1005595. doi: 10.1371/journal.pcbi.1005595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. GenoToul Bioinfo . 2018. GenoToul bioinformatics facility. Available from: 10.15454/1.5572369328961167E12 [DOI]
- 25. Vallenet D, Calteau A, Dubois M, Amours P, Bazin A, Beuvin M, Burlot L, Bussell X, Fouteau S, Gautreau G, Lajus A, Langlois J, Planel R, Roche D, Rollin J, Rouy Z, Sabatet V, Médigue C. 2020. MicroScope: an integrated platform for the annotation and exploration of microbial gene functions through genomic, pangenomic and metabolic comparative analysis. Nucleic Acids Res 48:D579–D589. doi: 10.1093/nar/gkz926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Caspi R, Billington R, Fulcher CA, Keseler IM, Kothari A, Krummenacker M, Latendresse M, Midford PE, Ong Q, Ong WK, Paley S, Subhraveti P, Karp PD. 2018. The MetaCyc database of metabolic pathways and enzymes. Nucleic Acids Res 46:D633–D639. doi: 10.1093/nar/gkx935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Falentin H, Postollec F, Parayre S, Henaff N, Le Bivic P, Richoux R, Thierry A, Sohier D. 2010. Specific metabolic activity of ripening bacteria quantified by real-time reverse transcription PCR throughout Emmental cheese manufacture. Int J Food Microbiol 144:10–19. doi: 10.1016/j.ijfoodmicro.2010.06.003 [DOI] [PubMed] [Google Scholar]
- 28. Achilleos C, Berthier F. 2017. Evaluation of qPCR and plate counting for quantifying thermophilic starters in cheese. Food Microbiol. 65:149–159. doi: 10.1016/j.fm.2017.01.024 [DOI] [PubMed] [Google Scholar]
- 29. Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, Vandesompele J, Wittwer CT. 2009. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem 55:611–622. doi: 10.1373/clinchem.2008.112797 [DOI] [PubMed] [Google Scholar]
- 30. Langmead B, Schatz MC, Lin J, Pop M, Salzberg SL. 2009. Searching for SNPs with cloud computing. Genome Biol 10:R134. doi: 10.1186/gb-2009-10-11-r134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Anders S, Pyl PT, Huber W. 2015. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics 31:166–169. doi: 10.1093/bioinformatics/btu638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Anders S, Huber W. 2010. Differential expression analysis for sequence count data. Genome Biol 11:R106. doi: 10.1186/gb-2010-11-10-r106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Varet H, Brillet-Guéguen L, Coppée J-Y, Dillies M-A. 2016. SARTools: a DESeq2- and EdgeR-based R pipeline for comprehensive differential analysis of RNA-Seq data. PLoS One 11:e0157022. doi: 10.1371/journal.pone.0157022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. doi: 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Dunn OJ. 1961. Multiple comparisons among means. J Am Stat Assoc 56:52–64. doi: 10.1080/01621459.1961.10482090 [DOI] [Google Scholar]
- 36. Kieliszek M, Pobiega K, Piwowarek K, Kot AM. 2021. Characteristics of the proteolytic enzymes produced by lactic acid bacteria. Molecules 26:1858. doi: 10.3390/molecules26071858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hou J, Liu F, Ren D, Han W, Du Y. 2015. Effect of culturing conditions on the expression of key enzymes in the proteolytic system of Lactobacillus bulgaricus. J Zhejiang Univ Sci B 16:317–326. doi: 10.1631/jzus.B1400230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kunji ERS, Mierau I, Hagting A, Poolman B, Konings WN. 1996. The proteotytic systems of lactic acid bacteria. Antonie Van Leeuwenhoek 70:187–221. doi: 10.1007/BF00395933 [DOI] [PubMed] [Google Scholar]
- 39. Baek M, Kim KW, Yi H. 2023. Subspecies-level genome comparison of Lactobacillus delbrueckii. Sci Rep 13:3171. doi: 10.1038/s41598-023-29404-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hurst LD, Merchant AR. 2001. High guanine–cytosine content is not an adaptation to high temperature: a comparative analysis amongst prokaryotes. Proc R Soc Lond B 268:493–497. doi: 10.1098/rspb.2000.1397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Weiss N, Schillinger U, Kandler O. 1983. Lactobacillus lactis, Lactobacillus leichmannii and Lactobacillus bulgaricus, subjective synonyms of Lactobacillus delbrueckii, and description of Lactobacillus delbrueckii subsp. lactis comb. nov. and Lactobacillus delbrueckii subsp. bulgaricus comb. nov. Syst Appl Microbiol 4:552–557. doi: 10.1016/S0723-2020(83)80012-5 [DOI] [PubMed] [Google Scholar]
- 42. Delley M, Germond J-E. 2002. Differentiation of Lactobacillus helveticus, Lactobacillus delbrueckii subsp bulgaricus, subsp lactis and subsp delbrueckii using physiological and genetic tools and reclassification of some strains from the ATCC collection. Syst Appl Microbiol 25:228–231. doi: 10.1078/0723-2020-00106 [DOI] [PubMed] [Google Scholar]
- 43. Dellaglio F, Felis GE, Castioni A, Torriani S, Germond J-E. 2005. Lactobacillus delbrueckii subsp. indicus subsp. nov., isolated from Indian dairy products. Int J Syst Evol Microbiol 55:401–404. doi: 10.1099/ijs.0.63067-0 [DOI] [PubMed] [Google Scholar]
- 44. De Vos P, Garrity GM, Jones D, Krieg NR, Ludwig W, Rainey FA, Schleifer K-H, Whitman WB. 2009. The FirmicutesBergey’s manual of systematic bacteriology. Springer. http://hdl.handle.net/1854/LU-799073. [Google Scholar]
- 45. El Kafsi H, Binesse J, Loux V, Buratti J, Boudebbouze S, Dervyn R, Kennedy S, Galleron N, Quinquis B, Batto J-M, Moumen B, Maguin E, van de Guchte M. 2014. Lactobacillus delbrueckii ssp. lactis and ssp. bulgaricus: a chronicle of evolution in action. BMC Genomics 15:407. doi: 10.1186/1471-2164-15-407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Gänzle MG, Follador R. 2012. Metabolism of oligosaccharides and starch in lactobacilli: a review. Front Microbiol 3:340. doi: 10.3389/fmicb.2012.00340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Boulay M, Al Haddad M, Rul F. 2020. Streptococcus thermophilus growth in soya milk: sucrose consumption, nitrogen metabolism, soya protein hydrolysis and role of the cell-wall protease PrtS. Int J Food Microbiol 335:108903. doi: 10.1016/j.ijfoodmicro.2020.108903 [DOI] [PubMed] [Google Scholar]
- 48. Silva Z, Sampaio M-M, Henne A, Böhm A, Gutzat R, Boos W, da Costa MS, Santos H. 2005. The high-affinity maltose/trehalose ABC transporter in the extremely thermophilic bacterium Thermus thermophilus HB27 also recognizes sucrose and palatinose. J Bacteriol 187:1210–1218. doi: 10.1128/JB.187.4.1210-1218.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kilic AO, Honeyman AL, Tao L. 2007. Overlapping substrate specificity for sucrose and maltose of two binding protein-dependent sugar uptake systems in Streptococcus mutans. FEMS Microbiol Lett 266:218–223. doi: 10.1111/j.1574-6968.2006.00522.x [DOI] [PubMed] [Google Scholar]
- 50. Zheng H, Liu E, Hao P, Konno T, Oda M, Ji Z-S. 2012. In silico analysis of amino acid biosynthesis and proteolysis in Lactobacillus delbrueckii subsp. bulgaricus 2038 and the implications for bovine milk fermentation. Biotechnol Lett 34:1545–1551. doi: 10.1007/s10529-012-1006-4 [DOI] [PubMed] [Google Scholar]
- 51. Gao Y, Liu Y, Sun M, Zhang H, Mu G, Tuo Y. 2020. Physiological function analysis of Lactobacillus plantarum Y44 based on genotypic and phenotypic characteristics. J Dairy Sci 103:5916–5930. doi: 10.3168/jds.2019-18047 [DOI] [PubMed] [Google Scholar]
- 52. Valence F, Deutsch S-M, Richoux R, Gagnaire V, Lortal S. 2000. Autolysis and related proteolysis in Swiss cheese for two Lactobacillus helveticus strains. J Dairy Res 67:261–271. doi: 10.1017/s0022029900004118 [DOI] [PubMed] [Google Scholar]
- 53. Husson-Kao C, Mengaud J, Gripon JC, Benbadis L, Chapot-Chartier MP. 2000. Characterization of Streptococcus thermophilus strains that undergo lysis under unfavourable environmental conditions. Int J Food Microbiol 55:209–213. doi: 10.1016/s0168-1605(00)00166-5 [DOI] [PubMed] [Google Scholar]
- 54. Yvon M, Rijnen L. 2001. Cheese flavour formation by amino acid catabolism. Int Dairy J 11:185–201. doi: 10.1016/S0958-6946(01)00049-8 [DOI] [Google Scholar]
- 55. Ardö Y. 2006. Flavour formation by amino acid catabolism. Biotechnol Adv 24:238–242. doi: 10.1016/j.biotechadv.2005.11.005 [DOI] [PubMed] [Google Scholar]
- 56. Vrancken G, Rimaux T, Weckx S, De Vuyst L, Leroy F. 2009. Environmental pH determines citrulline and ornithine release through the arginine deiminase pathway in Lactobacillus fermentum IMDO 130101. Int J Food Microbiol 135:216–222. doi: 10.1016/j.ijfoodmicro.2009.07.035 [DOI] [PubMed] [Google Scholar]
- 57. Cunin R, Glansdorff N, Piérard A, Stalon V. 1986. Biosynthesis and metabolism of arginine in bacteria. Microbiol Rev 50:314–352. doi: 10.1128/mr.50.3.314-352.1986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Pols T, Singh S, Deelman-Driessen C, Gaastra BF, Poolman B. 2021. Enzymology of the pathway for ATP production by arginine breakdown. FEBS J. 288:293–309. doi: 10.1111/febs.15337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Cheng H. 2010. Volatile flavor compounds in yogurt: a review. Crit Rev Food Sci Nutr 50:938–950. doi: 10.1080/10408390903044081 [DOI] [PubMed] [Google Scholar]
- 60. Liu W, Yu J, Sun Z, Song Y, Wang X, Wang H, Wuren T, Zha M, Menghe B, Heping Z. 2016. Relationships between functional genes in Lactobacillus delbrueckii ssp. bulgaricus isolates and phenotypic characteristics associated with fermentation time and flavor production in yogurt elucidated using multilocus sequence typing. J Dairy Sci 99:89–103. doi: 10.3168/jds.2015-10209 [DOI] [PubMed] [Google Scholar]
- 61. Sarhir ST, Amanpour A, Bouseta A, Selli S. 2019. Key odorants of a Moroccan fermented milk product “Lben” using aroma extract dilution analysis. J Food Sci Technol 56:3836–3845. doi: 10.1007/s13197-019-03854-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Deghorain M, Fontaine L, David B, Mainardi J-L, Courtin P, Daniel R, Errington J, Sorokin A, Bolotin A, Chapot-Chartier M-P, Hallet B, Hols P. 2010. Functional and morphological adaptation to peptidoglycan precursor alteration in Lactococcus lactis. J Biol Chem 285:24003–24013. doi: 10.1074/jbc.M110.143636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Pérez-Núñez D, Briandet R, David B, Gautier C, Renault P, Hallet B, Hols P, Carballido-López R, Guédon E. 2011. A new morphogenesis pathway in bacteria: unbalanced activity of cell wall synthesis machineries leads to coccus-to-rod transition and filamentation in ovococci: filamentation of Lactococcus lactis. Mol Microbiol 79:759–771. doi: 10.1111/j.1365-2958.2010.07483.x [DOI] [PubMed] [Google Scholar]
- 64. Chien A-C, Hill NS, Levin PA. 2012. Cell size control in bacteria. Curr Biol 22:R340–R349. doi: 10.1016/j.cub.2012.02.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Schaechter M, Maaloe O, Kjeldgaard NO. 1958. Dependency on medium and temperature of cell size and chemical composition during balanced grown of Salmonella typhimurium. J Gen Microbiol 19:592–606. doi: 10.1099/00221287-19-3-592 [DOI] [PubMed] [Google Scholar]
- 66. Monahan LG, Hajduk IV, Blaber SP, Charles IG, Harry EJ, Gottesman S. 2014. Coordinating bacterial cell division with nutrient availability: a role for glycolysis. mBio 5:e00935-14. doi: 10.1128/mBio.00935-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Weart RB, Lee AH, Chien A-C, Haeusser DP, Hill NS, Levin PA. 2007. A metabolic sensor governing cell size in bacteria. Cell 130:335–347. doi: 10.1016/j.cell.2007.05.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Lu M, Kleckner N. 1994. Molecular cloning and characterization of the pgm gene encoding phosphoglucomutase of Escherichia coli. J Bacteriol 176:5847–5851. doi: 10.1128/jb.176.18.5847-5851.1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Levander F, Andersson U, Rådström P. 2001. Physiological role of β-phosphoglucomutase in Lactococcus lactis. Appl Environ Microbiol 67:4546–4553. doi: 10.1128/AEM.67.10.4546-4553.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Ludszuweit M, Schmacht M, Keil C, Haase H, Senz M. 2020. Impact of media heat treatment on cell morphology and stability of L. acidophilus, L. johnsonii and L. delbrueckii subsp. delbrueckii during fermentation and processing. Fermentation 6:94. doi: 10.3390/fermentation6040094 [DOI] [Google Scholar]
- 71. Young KD. 2006. The selective value of bacterial shape. Microbiol Mol Biol Rev 70:660–703. doi: 10.1128/MMBR.00001-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Donkor ON, Henriksson A, Vasiljevic T, Shah NP. 2007. α-galactosidase and proteolytic activities of selected probiotic and dairy cultures in fermented soymilk. Food Chem 104:10–20. doi: 10.1016/j.foodchem.2006.10.065 [DOI] [Google Scholar]
- 73. Singh BP, Vij S. 2018. α-galactosidase activity and oligosaccharides reduction pattern of indigenous lactobacilli during fermentation of soy milk. Food Biosci 22:32–37. doi: 10.1016/j.fbio.2018.01.002 [DOI] [Google Scholar]
- 74. Garro MS, de Valdez GF, Oliver G, de Giori1 GS. 1998. Growth characteristics and fermentation products of Streptococcus salivarius subsp. thermophilus, Lactobacillus casei and L . fermentum in soymilk. Z Lebensm Unters Forsch 206:72–75. doi: 10.1007/s002170050217 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PCA of volatile compounds.
Volatil compounds data (Table S1) and fermentation profile (Table S4).
Lactobacillus delbrueckii subsp. delbrueckii CIRM-BIA865 Genome annotation.
Transcriptomic data.
Data Availability Statement
The sequence of the Ld865 genome sequenced in the study was deposited in the European Nucleotide Archive (ENA) at EMBL-EBI under accession number PRJEB50396. The transcriptome data generated in the study were deposited in the ArrayExpress platform under accession numbers E-MTAB-12095. The data supporting the biochemical results are available at https://entrepot.recherche.data.gouv.fr, under DOI 10.57745/AOBH6L.