Abstract
The 3C-like proteinase (3CLpro) of mouse hepatitis virus (MHV) is predicted to cleave at least 11 sites in the 803-kDa gene 1 polyprotein, resulting in maturation of proteinase, polymerase, and helicase proteins. However, most of these cleavage sites have not been experimentally confirmed and the proteins have not been identified in vitro or in virus-infected cells. We used specific antibodies to identify and characterize a 22-kDa protein (p1a-22) expressed from gene 1 in MHV A59-infected DBT cells. Processing of p1a-22 from the polyprotein began immediately after translation, but some processing continued for several hours. Amino-terminal sequencing of p1a-22 purified from MHV-infected cells showed that it was cleaved at a putative 3CLpro cleavage site, Gln_Ser4014 (where the underscore indicates the site of cleavage), that is located between the 3CLpro domain and the end of open reading frame (ORF) 1a. Subclones of this region of gene 1 were used to express polypeptides in vitro that contained one or more 3CLpro cleavage sites, and cleavage of these substrates by recombinant 3CLpro in vitro confirmed that amino-terminal cleavage of p1a-22 occurred at Gln_Ser4014. We demonstrated that the carboxy-terminal cleavage of the p1a-22 protein occurred at Gln_Asn4208, a sequence that had not been predicted as a site for cleavage by MHV 3CLpro. Our results demonstrate the usefulness of recombinant MHV 3CLpro in identifying and confirming cleavage sites within the gene 1 polyprotein. Based on our results, we predict that at least seven mature proteins are processed from the ORF 1a polyprotein by 3CLpro and suggest that additional noncanonical cleavage sites may be used by 3CLpro during processing of the gene 1 polyprotein.
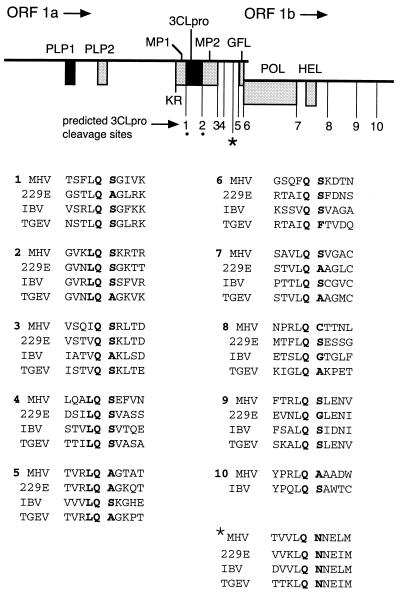
Gene 1 of mouse hepatitis virus (MHV) A59 encodes a fusion polyprotein with a predicted mass of 803 kDa (2, 10, 15). Expression of the entire polyprotein of gene 1 requires translation of two overlapping open reading frames (ORFs), 1a and 1b. Since these ORFs are in different reading frames, ORF 1b can be expressed only if a ribosomal frameshift occurs at the end of ORF 1a (4, 5, 21). The ORF 1a portion of gene 1 encodes two experimentally confirmed proteinases, papain-like proteinase 1 (PLP-1) and 3C-like proteinase (3CLpro), as well as an additional proteinase motif, PLP-2, for which no activity has yet been identified (1, 15). The MHV 3CLpro has been shown to autoproteolytically liberate itself from the nascent polyprotein in vitro and in virus-infected cells (in cyto) (18, 19). Eleven cleavage sites have been predicted to be cleaved by 3CLpro, 10 of which have a dipeptide consisting of Gln at position 1 (P1) and Ser, Asp, Gly, or Cys at P1′ (15) (Fig. 1). The putative cleavage sites are conserved among the four sequenced coronaviruses and are generally located within the polyprotein and at the putative Q_(S,A,G) dipeptide cleavage site motif (where the underscore indicates the site of cleavage). Six of the predicted MHV 3CLpro cleavage sites are located in a 1,120-amino-acid (aa) region starting at 3CLpro and ending at the carboxy terminus of the ORF 1a polyprotein (aa 3334 to 4454). This region is comprised of 3CLpro as well as a region of predominantly hydrophobic residues between aa 3636 and 3921 (MP-2), a region of unknown function between aa 3922 and 4317, and the putative growth factor-like domain extending from aa 4318 to 4454 (GFL). We were particularly interested in the 532-aa region from the carboxy terminus of the MP-2 domain to the end of GFL, since there are four predicted 3CLpro cleavage sites within this small area and no functions have been proposed for these domains.
FIG. 1.
MHV gene 1 organization and putative 3CLpro cleavage sites. The diagram shows the organization of the 22-kb gene 1 of the MHV 32-kb RNA. The locations of the PLP-1 and PLP-2 domains, the MP-1 and MP-2 hydrophobic domains, 3CLpro, the GFL domain, RNA-dependent RNA polymerase (POL), and helicase (HEL) are shown as shaded boxes. Locations of predicted MHV 3CLpro cleavage sites are numbered below the diagram. KR, Lys-Arg dipeptide also proposed as a 3CLpro cleavage site (15). The dots denote the confirmed cleavage sites flanking 3CLpro in the polyprotein. The ∗ indicates the Q_N4208 cleavage site identified and described in this paper. The sequences surrounding the confirmed or putative MHV 3CLpro cleavage sites (denoted by MHV) are aligned with the deduced amino acid sequences of HCV 229E (229E) (11), IBV (3), and TGEV (9). Alignments were performed with MacVector version 6.01.
In this study we used a specific antiserum to identify a 22-kDa protein from MHV A59-infected cells that is processed from the region of the ORF 1a polyprotein between MP-2 and the end of ORF 1a (p1a-22). We have shown that 3CLpro is responsible for cleaving this protein at an amino-terminal Gln_Ser site that was previously predicted to be a cleavage site for the proteinase. We also have identified a new cleavage site at the carboxy terminus of the 22-kDa protein that does not conform to the canonical Gln_(Ser,Ala,Gly) motif. Together these results confirm that 3CLpro is responsible for processing at the carboxy-terminal region of the MHV ORF 1a polyprotein.
MATERIALS AND METHODS
B4 antibody production.
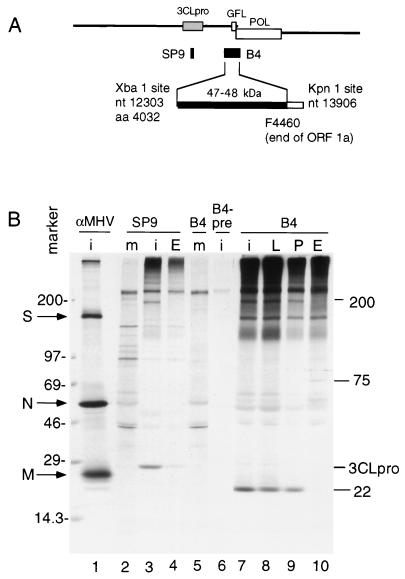
The B4 protein was expressed in Escherichia coli in the pQE-30 vector (Qiagen). The B4 subclone extended from the XbaI site at nucleotide (nt) 12303 to the KpnI site at nt 13906, based on the sequence of MHV A59 (2) (Fig. 2). This clone potentially encoded aa 4032 to 4460 of ORF 1a, the ORF 1a-ORF 1b frameshift, and 114 aa of ORF 1b, with a total calculated size of 71 kDa (including the amino acid sequence MRGSHHHHHHTDPHGTSS encoded by plasmid sequences at the 5′ end of the construct); however, the size of the His-tag-purified E. coli-expressed protein (50 kDa) corresponded exactly to the calculated mass of aa 4032 to 4460 and the additional amino-terminal amino acids, indicating that only the ORF 1a portion of the B4 construct was translated in E. coli. The His-tag-purified 50-kDa protein was concentrated and used to induce polyclonal antibodies in rabbits, resulting in the B4 antiserum. The B4 immune serum but not the preimmune serum was able to detect the B4 protein by Western blot analysis.
FIG. 2.
Identification of a 22-kDa protein in MHV-infected DBT cells. (A) The schematic shows the location of the region of gene 1 used to express the protein against which the B4 antibodies were raised. The subclone extends from the XbaI site at nt 12303 to the KpnI site at nt 13906, and the expressed protein extends from aa 4032 to the end of ORF 1a (filled bar). (B) Immunoprecipitation of proteins from MHV-infected DBT cells. Cells were infected (lanes i) or mock infected (lanes m), and proteins were labeled and harvested as described in Materials and Methods. Proteins were immunoprecipitated with antibodies against whole MHV virions (αMHV), the SP9 antisera directed against 3CLpro, or the B4 antiserum. B4-pre indicates preimmune serum from the rabbit used to induce antibodies against the B4 protein. In lanes 4, 8, 9, and 10, proteins were labeled in the presence of the proteinase inhibitors leupeptin (lane L), phenylmethylsulfonyl fluoride (lane P), or E64d (lanes E). Mass markers (in kilodaltons) and the locations of the spike (S), nucleocapsid (N), and matrix proteins (M) are indicated to the left of the gel, and the locations of 3CLpro and other newly identified proteins are shown to the right of the gel.
Radiolabeling of ORF 1a proteins and immunoprecipitation.
MHV A59 infections of DBT cells, synchronization of translation in cyto, pulse-label and pulse-chase experiments, and immunoprecipitation were all performed as previously described (7).
Cloning and expression of r3CLpro.
The cloning and expression of active MHV 3CLpro in E. coli were similar to the methods of Seybert et al. (24) and are described elsewhere (25). Briefly, the precise 3CLpro domain was expressed as a maltose binding protein-recombinant 3CLpro (MBP-r3CLpro) fusion protein in pMAL-C2 (New England Biolabs), partially purified on an amylose-affinity column, and separated from MBP with factor Xa (graciously provided by Paul Bock). The partially purified r3CLpro preparation was used for all experiments. Because the preparation was only partially purified, exact amounts of proteinase could not be determined for each experiment. However, based on total protein concentrations it was estimated that 10 to 20 μM of r3CLpro was used during cleavage reactions.
Cloning and expression of polypeptides containing putative 3CLpro cleavage sites.
Subclones of the region of ORF 1a between nt 11994 and 13118 were constructed by PCR from a cDNA of the 3′ 2.5 kb of the ORF 1a region of gene 1 (see Fig. 5). Left primers included an NcoI site, and right primers included an XhoI site, allowing for ligation into pET-23d (Novagen). This vector contains an optimal AUG in the NcoI site, allowing for full-length expression of all constructs. The recombinant plasmids were used to program expression of polypeptides in a combined in vitro transcription-translation reticulocyte lysate system (TnT; Promega) in the presence of [35S]methionine and [35S]cysteine. In this study, 0.5 μg of plasmid was in a 25-μl reaction mixture in the presence of 800 μCi of [35S]methionine per ml with incubation for 90 min at 30°C. Translation reactions were terminated by addition of sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer (13) followed by boiling for 5 min.
FIG. 5.
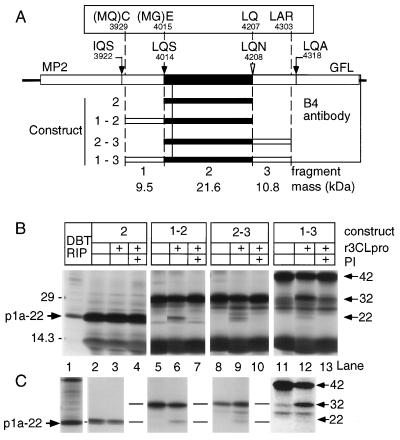
Cloning, expression, and trans cleavage of p1a-22 in vitro. (A) Schematic of the carboxy-terminal region of the ORF 1a polyprotein. The putative p1a-22 domain is shown as filled rectangles. The locations of the sequenced LQ_S cleavage site and the putative IQ_S and LQ_A cleavage sites are shown by filled arrows, and the location of the LQ_N motif is shown by the open arrowhead. The boxed amino acid residues show the amino- and carboxy-terminal residues expressed from the cloned constructs. Residues in parentheses (MQ or MG) were contributed by the primers used to clone the fragments by PCR. The fragment number (1 through 3) and the calculated masses of the individual fragments are shown at the bottom of the diagram, and the constructs are numbered according to the fragments comprising them. (B) The results of the expression of constructs shown in panel A, along with products of cleavage by r3CLpro, are shown. Lane 1 shows p1a-22 immunoprecipitated from MHV-infected DBT cells (DBT RIP), and the location of p1a-22 along with molecular mass standards (in kilodaltons) is indicated to the left. Proteins expressed from construct 2, 1-2, 2-3, or 1-3 were incubated in the presence (+) or absence of r3CLpro and proteinase inhibitor E64 (PI). The masses of expressed proteins and cleavage products are shown to the right of the gel. (C) The products of expression and r3CLpro cleavage were immunoprecipitated with B4 antiserum. All lanes correspond to the proteins seen directly above them in panel B. The levels of contrast of lanes 11 and 12 were equally altered (Photoshop version 4.0) to show the minor 22-kDa cleavage product in lane 12 that was easily seen on the original fluorogram.
trans-cleavage assays.
Radiolabeled polypeptides were incubated with eluate from the amylose-affinity column containing r3CLpro, MBP, and Factor Xa at 30°C for 4 h. Typically, 1 to 2 μl of in vitro translation reaction mixture in reticulocyte lysate was incubated with 1 to 5 μl of r3CLpro-containing solution. Reactions were terminated by addition of SDS-PAGE sample buffer and boiling for 5 min, prior to electrophoresis on SDS–5 to 18% gradient polyacrylamide gels.
Amino-terminal radiosequencing.
The 22-kDa protein was radiolabeled with [35S]Met, [3H]Val, or [3H]Leu, immunoprecipitated from a total of 2.7 × 107 MHV-infected DBT cells with the B4 antiserum, and electrophoresed on an SDS–5 to 18% gradient polyacrylamide gel. The 22-kDa protein was visualized by autoradiography and was transferred to a polyvinylidene difluoride membrane in buffer containing 100 mM CAPS (3-[cyclohexylamino]-1-propanesulfonic acid) and 10% methanol. The protein was subjected to Edman degradation (20 cycles) on an Applied Biosystems, Inc., model 470 sequencer, and individual fractions were assessed for radioactivity by scintillation counting.
RESULTS
Detection of gene 1 proteins in MHV-infected DBT cells.
The B4 antiserum was used to immunoprecipitate gene 1 proteins from MHV A59-infected DBT cells (in cyto). The B4 antiserum was raised in rabbits against a fusion protein expressed in E. coli that incorporated aa 4032 to 4460 of the ORF 1a polyprotein. In initial studies, the MHV proteins were radiolabeled for 2 h beginning at 8 h postinfection and proteins were immunoprecipitated from lysates of whole cells with B4 antiserum (Fig. 2). The previously described SP9 antiserum was used as a control, along with preimmune serum from the same rabbit used to raise the B4 antibodies. Polyclonal antibodies raised against sucrose-gradient-purified MHV virions (αMHV) were used to identify structural proteins that might coprecipitate with B4.
αMHV detected prominent spike (S), nucleocapsid (N), and matrix (M) proteins, while SP9 precipitated 3CLpro (Fig. 2B, lanes 1 and 3). The new B4 antiserum detected several proteins that were not precipitated either by B4 in mock-infected cells or by B4 preimmune serum in infected cells. The most prominent protein had an apparent mass of 22 kDa (p1a-22) (Fig. 2B, lane 7). B4 also detected a protein with an apparent mass of 200 kDa, with mobility identical to that of a protein precipitated by the anti-3CLpro antibody, SP9 (lane 3). The presence of the proteinase inhibitors leupeptin and phenylmethylsulfonyl fluoride (lanes 8 and 9) caused some reduction in the amount of p1a-22 detected, and E64d almost completely eliminated detectable p1a-22 and also abolished the 200-kDa protein (lanes 4 and 10). These results demonstrated that p1a-22 was processed from the region between MP-2 and the end of the ORF 1a polyprotein and that this processing was sensitive to known inhibitors of 3CLpro. The precipitation of the 200-kDa protein by both SP9 and B4 and its sensitivity to E64d strongly suggested that it was a proteolytic precursor that incorporated both 3CLpro and the newly identified p1a-22.
The B4 antiserum coprecipitated the structural S glycoprotein, as has been seen with other gene 1 antibodies (7). We presume that this is due to colocalization of structural and nonstructural proteins in replication complexes that are subsequently coprecipitated under the conditions used for immunoprecipitation. Finally, a 75-kDa protein was detected by B4 only in the presence of E64d and thus might be a precursor to p1a-22; however, as the 75-kDa protein was not detected during pulse-label or pulse-chase experiments, it may be a precursor that is not seen during normal translation and processing of the gene 1 polyprotein.
Kinetics of expression of p1a-22.
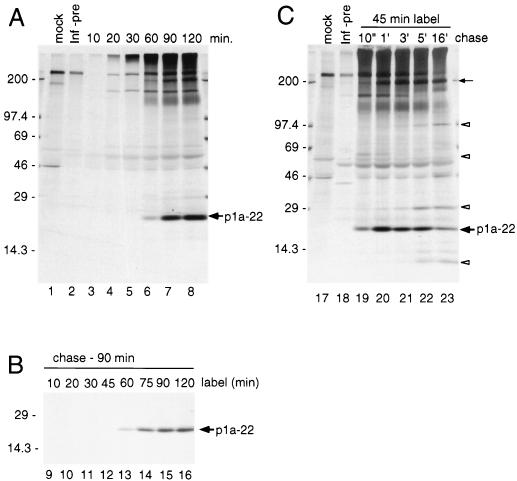
We next determined the kinetics of expression and processing of p1a-22 in cyto during pulse-label and pulse-chase translation (Fig. 3). Viral proteins were radiolabeled following high-salt translational synchronization to ensure that detected proteins were de novo translation products rather than proteins that were partially translated prior to addition of radiolabel (6, 23). The proteins were immunoprecipitated with B4 antiserum. In infected cells, p1a-22 and the 200-kDa protein were first observed at 60 min of labeling and accumulated without detectable intermediates (Fig. 3A). We next performed an experiment using different labeling times and a constant chase of 90 min (Fig. 3B) to define the time of incorporation of label into p1a-22 during translation. Incorporation was first detected between 45 and 60 min, similar to the time of first appearance during pulse-labeling, indicating that at least some cleavage of p1a-22 occurred as soon as translation was completed (Fig. 3B, lanes 12 to 13).
FIG. 3.
Kinetics of translation and processing of p1a-22. (A) Pulse-label translation. Proteins were radiolabeled with [35S]methionine at 7 h postinfection, and samples were withdrawn from 10 to 120 min and used for immunoprecipitation by B4 antiserum, followed by electrophoresis and fluorography on SDS–5 to 18% gradient polyacrylamide gels. Lysates of mock-infected cells were immunoprecipitated by B4 antiserum (lane mock), and lysates of infected cells were immunoprecipitated with B4 preimmune serum (lane Inf-pre). Molecular mass standards (in kilodaltons) are to the left of the gel, and the location of p1a-22 is shown to the right of the gel. (B) Translation with different durations of labeling and with constant chase. Proteins were labeled in MHV-infected DBT cells for periods from 10 to 120 min and then chased with media containing excess unlabeled methionine and cycloheximide for an additional 90 min. The locations of markers and p1a-22 are shown. (C) Pulse-chase translation. Proteins were radiolabeled for 45 min and then chased in media lacking [35S]Met but containing a 10-fold excess of unlabeled methionine for 10 min (10") to 16 h (16′). All samples were immunoprecipitated and analyzed as described above for panel A. All labeling is as described for panel A. The location of the 200-kDa protein is indicated by the small arrow, and open arrowheads indicate the locations of proteins seen only after prolonged chase.
Pulse-chase labeling of MHV gene 1 proteins in cells was performed to define the pattern of processing of p1a-22 and to determine if there was a clear precursor-product relationship between the 200-kDa protein and p1a-22. The pulse-chase experiment involved a 45-min radiolabel after high-salt synchronization, followed by cold chase from 10 min to 16 h (Fig. 3C). Because the experiments were not initiated until 6 h postinfection, the 5- and 16-h chase points represent 11 and 22 h postinfection, respectively. By the 5-h chase, the monolayer was almost 100% involved in virus-induced syncytia, and by 16 h of chase, portions of the monolayer were detached and some cell destruction had occurred. p1a-22 was easily detected at the end of the 10-min chase (Fig. 3C, lane 19), consistent with the pulse-labeling time of 60 min. p1a-22 was most prominent between the 1- and 5-h chases but was still easily detected after 16 h of chase. The continued accumulation of p1a-22 for up to 3 h of chase clearly demonstrated that although some processing of p1a-22 was immediately cotranslational, a significant amount was delayed. Finally, the detection of p1a-22 at very late chase indicated that new molecules of p1a-22 continued to be processed even at very late times postinfection or that previously processed molecules were resistant to degradation.
The extended chase demonstrated that the 200-kDa protein had a pattern of detection by B4 similar to that of p1a-22 and also provided evidence for processing of additional proteins not detected during pulse-label experiments. A 69-kDa protein was detected at 10 min and 1 h of chase (Fig. 3C, lanes 19 to 20) but not at subsequent times. Proteins with apparent masses of 100, 30, and ∼10 kDa were first seen at the 5-h chase and could still be detected at the 16-h chase (Fig. 3C, lanes 22 and 23). It is possible to propose many potential relationships among these proteins (see Discussion), but testing with additional antibodies in this region will be necessary to completely define the relationship of p1a-22 to the 200-kDa protein and the other proteins seen during prolonged chase. However, the experiment did indicate that processing of this region of the ORF 1a polyprotein continued throughout infection and that the pattern of precursors and products such as p1a-22 may be quite complex.
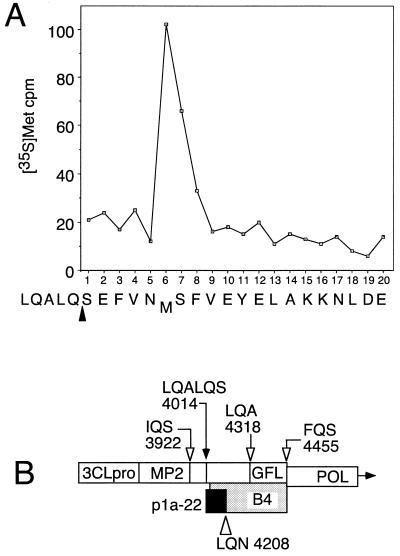
Identification of the amino terminus of p1a-22.
Since p1a-22 was the most abundant and stable protein detected by B4, we chose first to determine the location of p1a-22 in the gene 1 polyprotein. We hypothesized that one of the predicted 3CLpro cleavage sites in this region of the polyprotein was the amino-terminal cleavage site of p1a-22. We therefore labeled MHV proteins with [35S]Met, [3H]Val, or [3H]Leu, immunoprecipitated p1a-22 from lysates of virus-infected cells using the B4 antibodies, separated the proteins by SDS-PAGE, and transferred p1a-22 to PVDF membranes for sequencing by Edman degradation and scintillation counting of individual residues. Only [35S]Met gave adequate label intensity for subsequent evaluation (Fig. 4A). Analysis of the first 20 residues of p1a-22 demonstrated that a methionine residue was present only at the sixth position. The deduced amino acid sequence between nt 11700 and 14100 of MHV A59 gene 1 contained 11 places where this pattern could be obtained; only one of these, beginning at Ser4014, conformed to a putative 3CLpro cleavage site, specifically LQ_S4014. This site was 18 residues amino terminal to the first amino acid in the protein used to induce the B4 antiserum. If p1a-22 began at LQ_S4014, it would extend well into the B4 antibody region (Fig. 4B). The LQ_S motif was unusual in that it was preceded in the MHV gene 1 protein by LQA, also a possible 3CLpro cleavage site. Although the LQ_S motif is completely conserved among MHV, infectious bronchitis virus (IBV), human coronavirus 229E (HCV 229E), and transmissible gastroenteritis virus (TGEV), the LQA sequence is present only in the MHV gene 1 protein sequence (3, 9, 11). An alignment of the sites of these viruses is shown in Fig. 1 at cleavage site 4.
FIG. 4.
Amino-terminal radiosequencing of p1a-22 from virus-infected cells. (A) p1a-22 was sequenced as described in Materials and Methods. The count of [35S]Met per minute per fraction is shown on the y axis, and the fraction number is shown on the x axis. The presumptive sequence and cleavage site (arrowhead) are shown below the fraction numbers. (B) Schematic of the carboxy-terminal region of the ORF 1a polyprotein shows the confirmed amino terminus of p1a-22 (LQALQ_S4014; filled arrow) and other predicted 3CLpro cleavage sites (open arrows). The extent of the protein used to induce B4 antibodies is indicated by the shaded bar, and the probable extent of the p1a-22 domain is indicated by the black box. The location of the putative LQ_N carboxy-terminal cleavage site of p1a-22 is shown by the open triangle. POL, the RNA-dependent RNA polymerase.
Determination of the carboxy terminus of p1a-22 and identification of a new 3CLpro cleavage site.
Based on the apparent mass of p1a-22 and the identified amino terminus, we hypothesized that the carboxy-terminal cleavage of p1a-22 might occur in the sequence VVLQ_N4208NEL. The LQ_NNE sequence is completely conserved in MHV, IBV, HCV 229E, and TGEV, and the analogous LQ_N site has recently been shown to be a cleavage site for IBV 3CLpro (16). The carboxy terminus of p1a-22 could not be directly determined, because there was no other abundant protein detected by B4 in infected cells that clearly represented the carboxy-terminal cleavage product. We therefore pursued in vitro approaches to define the carboxy terminus of p1a-22. We constructed a series of subclones from this region of MHV gene 1 that were predicted to express proteins containing one or two 3CLpro cleavage sites and tested if cleavage by r3CLpro could occur in vitro at the LQ_N4208 sequence (Fig. 5).
The constructs carried DNA that encoded either the amino acids between S4014 and Q4207, (construct 2) or this domain plus either or both regions flanking it in the polyprotein (constructs 1-2, 2-3, and 1-3) (Fig. 5A). The constructs did not incorporate the predicted cleavage sites, IQ_S3922 and LQ_A4318. In vitro translation of construct 2 resulted in a protein that exactly comigrated with p1a-22 immunoprecipitated by B4 from MHV-infected cells (Fig. 5B, lanes 1 and 2). Incubation of the in vitro-translated 22-kDa polypeptide with r3CLpro did not result in any detectable cleavage events (Fig. 5B, lane 3). The 22-kDa protein expressed in vitro was precipitated by B4 (Fig. 5C, lanes 2 and 3). These results indicated that our hypothesis about the general coding domain of p1a-22 was correct.
To confirm that the 22-kDa protein could be generated by 3CLpro cleavage in vitro, we translated construct 1-2, which contained DNA encoding the 22-kDa protein, the amino-terminal LQ_S4014 site of p1a-22, and an additional amino-terminal fragment with a calculated mass of 9.5 kDa. Transcription and translation of construct 1-2 resulted in a prominent protein of 32 kDa, identical to the calculated mass for this polypeptide. Incubation of the 32-kDa protein with r3CLpro resulted in the appearance of a 22-kDa cleavage product, and this cleavage was inhibited by the addition of the proteinase inhibitor E64 (Fig. 5B, lanes 5 to 7). Both of the 32- and 22-kDa proteins were precipitated by B4 (Fig. 5C, lanes 5 and 6). The 2-3 construct contained DNA encoding the 22-kDa protein, the putative carboxy-terminal LQ_N4208 cleavage site, and a calculated 10.8-kDa carboxy-terminal fragment. In vitro translation of construct 2-3 also expressed an approximately 32-kDa protein; the 22-kDa cleavage product detected after incubation with r3CLpro was inhibited by E64 (Fig. 5B, lanes 8 to 10). Again, the 22-kDa protein was precipitated by B4 (Fig. 5C, lanes 8 and 9). The 1-3 construct containing DNA encoding both cleavage sites and both the amino- and the carboxy-terminal fragment was translated to yield a 42-kDa precursor that was cleaved into 32- and 22-kDa products by r3CLpro (Fig. 5B, lanes 11 to 13). The cleavage of p1a-22 from the 1-3 construct was the least efficient of any of the constructs tested, and cleavage was inhibited by E64. We did not detect the amino-terminal 9.5-kDa cleavage fragment, either in the total lysate or in immunoprecipitated samples. The region of the gel containing these proteins was obscured in the total lysate samples, and as expected, the amino-terminal 9.5-kDa fragment was not detected by B4 since it was outside of the region against which the antibody was directed. A protein migrating at <14.3 kDa that is consistent with the carboxy-terminal 10.8-kDa cleavage product was seen following cleavage of proteins from constructs 2-3 and 1-3, but it was very faint and could not be precisely sized on this gel. The lack of significant detection of the carboxy-terminal fragment may be due to poor detection by B4 because of a lack of epitopes in this region or due to the fact that the fragment contains only two methionine residues, in contrast to the five methionine residues in p1a-22.
None of the protein precursors translated from the subclones was cleaved to completion by r3CLpro. It is possible that the proteins expressed from the subclones used in this study did not present the cleavage sites in the way they do during translation in cyto. It is also possible that the less efficient cleavage at the LQ_S4014 and LQ_N4208 sites was a true reflection of regulated or inefficient cleavage of p1a-22 in cyto. This latter possibility is supported by our in cyto data demonstrating that p1a-22 continued to accumulate for 1 to 3 h following removal of radiolabel (Fig. 3).
DISCUSSION
We have identified a 22-kDa protein (p1a-22) cleaved from the ORF 1a portion of the gene 1 polyprotein by 3CLpro in vitro and in virus-infected cells. p1a-22 is cleaved at its amino terminus at the previously predicted LQ_S4014 site and at its carboxy terminus at LQ_N4208, a site not previously predicted for MHV. We have previously shown that 3CLpro is autoproteolytically cleaved from the MHV gene 1 polyprotein (18–20), and similar results have been obtained for 3CLpro of HCV 229E and IBV (17, 26); however, this study is the first demonstration of cleavage by 3CLpro at additional sites in the MHV polyprotein. Cleavage by r3CLpro at the sites flanking p1a-22 was less efficient than at the sites flanking 3CLpro in the polyprotein, suggesting that cleavage of p1a-22 may be regulated in part by the ability of 3CLpro to recognize and cleave these sites. This result also suggests that liberation of 3CLpro from the polyprotein may be the initial step in processing of the majority of the polyprotein and is in agreement with our previous results demonstrating that 3CLpro acts principally in trans at sites in the polyprotein. 3C proteinases or 3CLpro’s of picornaviruses, comoviruses, and potyviruses all demonstrate differential cleavage at sites in their polyproteins, and these patterns of processing appear to be important for regulating the activities of the proteins during viral RNA synthesis (8, 14). We propose that MHV also regulates gene 1 proteins by differential cleavage.
Cleavage at the sites flanking p1a-22 was more efficient when only the amino- or carboxy-terminal fragment was expressed with p1a-22 rather than when both were present, suggesting that p1a-22 processing was influenced by both the specificity of 3CLpro for the cleavage site and the context of the protein within the larger polyprotein. Both of the p1a-22 cleavage sites have features that differ from other confirmed or predicted coronavirus 3CLpro cleavage sites. The LQ_S4014 motif is immediately preceded by the tripeptide LQA, in itself a potential 3CLpro cleavage site that also comprises P5 to P3 of the LQ_S4014 cleavage site. This places a Gln at P4 in LQ_S4014, quite different from the small residues (Ser, Thr, Val, and Ala) occupying P4 in almost all other cleavage sites (Fig. 1). It will be interesting to see if the LQA site can also be cleaved by 3CLpro.
The LQ_N4208 site diverges from the previously predicted Q_(S,A,G) site for P1_P1′ of 3CLpro cleavage sites but is also the most conserved of the confirmed or predicted coronavirus 3CLpro cleavage sites, with the entire LQ_NNE (P2-to-P3′) sequence present in all four sequenced coronaviruses (Fig. 1). This site has been shown to be cleaved by 3CLpro of IBV during gene 1 protein processing (16). This degree of conservation suggests that regulation of cleavage of the proteins flanking the LQ_N site may be a critical feature of coronavirus gene 1 expression. The ability of r3CLpro to cleave LQ_N4208 also demonstrates that the cleavage site specificity of MHV 3CLpro is broader than previously predicted. Finally, it suggests that other divergent cleavage sites for 3CLpro in addition to the remaining predicted sites may be present in the gene 1 polyprotein. Interestingly, the MHV gene 1 polyprotein is also unique among the coronaviruses in possessing a putative Q_C cleavage site dipeptide at the carboxy terminus of the helicase domain in ORF 1b, although this site has not been experimentally confirmed (15).
In contrast to the limited cleavage of p1a-22 detected in vitro, p1a-22 was readily detected by the B4 antibody in virus-infected cells. Based on amounts of protein detected, p1a-22 was more abundant than 3CLpro and almost as prominent as the amino-terminal ORF 1a protein, p28 (data not shown). It was surprising that new molecules of p1a-22 continued to be processed from existing precursors until very late in infection, even after cells completely involved in virus-induced syncytium formation were lost from the monolayer. Concurrently, much larger specific precursors, such as the 200-kDa protein, were also detected throughout prolonged chase. This result suggests that accumulated gene 1 polyprotein precursors may play an important role as reservoirs for mature proteins such as p1a-22 that are then slowly cleaved by 3CLpro. This suggestion is consistent with our observation that the sites flanking p1a-22 were less efficiently cleaved by 3CLpro and also with our previous observation that inhibition of gene 1 protein processing at any time of infection abrogates viral RNA synthesis (12).
It is not yet possible to assign a function to p1a-22. The domain is conserved among the sequenced coronaviruses, as are the cleavage sites flanking it (16). The fact that the B4 antibody coprecipitated both N and S glycoproteins suggests that p1a-22 may colocalize with these proteins, possibly in membrane-bound replication complexes. Analysis of the deduced p1a-22 amino acid sequence did not reveal putative functional motifs, possible membrane-spanning domains, or homology with confirmed or predicted proteins in protein databases. Thus, if p1a-22 associates with other proteins in membrane complexes, it would likely be by protein-protein interactions or by direct binding of the protein to RNA. The 200-kDa protein that was detected by both B4 antiserum and SP9 (anti 3CLpro) may be involved in the process of protein localization, since it may contain at least the second hydrophobic domain (MP-2) and likely the first as well (MP-1). These domains have been shown to confer a requirement for membranes on the activity of 3CLpro in vitro (22). It is possible that the 200-kDa protein may be inserted in membranes of the replication complex and that processing may occur in that context, resulting in colocalization of mature proteins such as 3CLpro and p1a-22. We are currently conducting experiments to test this possibility.
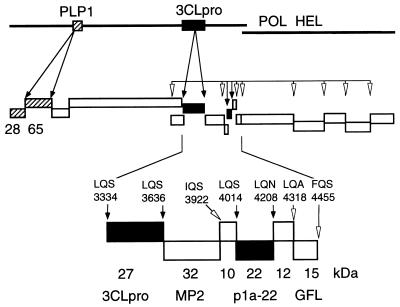
We propose that 12 proteins are proteolytically processed by 3CLpro during MHV gene 1 translation and that the region of the ORF 1a polyprotein between the MP-2 domain and the end of ORF 1a is likely composed of four small proteins with masses of 10, 12, 15, and 22 kDa (Fig. 6). When the experiments performed in cyto with B4 antiserum are reviewed in light of this model, it is possible that the 100-, 69-, 32-, and 10-kDa proteins observed during prolonged chase (Fig. 3B) may be various intermediate cleavage products from this region of the gene 1 polyprotein and the adjacent MP-2 and 3CLpro domains. We will use antibodies against the individual proposed protein domains carboxy terminal to 3CLpro in ORF 1a to probe for these proteins in cells and will use r3CLpro to precisely define the cleavage sites in ORF 1a and ORF 1b. We anticipate that we will thereby gain a complete understanding of the complex pattern of translation of MHV gene 1 and the processing of its polyprotein and will be able to determine the interactions and functions of the gene 1 proteins during virus replication.
FIG. 6.
MHV gene 1 polyprotein processing. This model is based on known and predicted cleavage sites and proteins identified in cells and in vitro. PLP-1 and proteins known to be cleaved in trans by PLP-1 (p28 and p65) are shown as hatched boxes. 3CLpro and proteins known to be cleaved by 3CLpro (3CLpro and p1a-22) are shown as filled boxes. Other predicted proteins are shown as open boxes. Experimentally confirmed cleavage sites are noted by filled arrows, while putative cleavage sites are indicated by open arrowheads. The enlarged region at the bottom of the figure shows the apparent masses of the confirmed 3CLpro and p1a-22 proteins and the location of their cleavage sites (filled boxes and arrows). The calculated masses of other predicted proteins at the end of ORF 1b are shown as open boxes and arrows (18–20). POL, RNA-dependent RNA polymerase; HEL, helicase.
ACKNOWLEDGMENTS
This work was supported by Public Health Service grant R01-AI-26603 from the National Institute of Allergy and Infectious Diseases.
Protein sequencing was performed in the shared resource of the Vanderbilt University Cancer Center (IP30CA68485).
REFERENCES
- 1.Baker S C, Yokomori K, Dong S, Carlisle R, Gorbalenya A E, Koonin E V, Lai M M. Identification of the catalytic sites of a papain-like cysteine proteinase of murine coronavirus. J Virol. 1993;67:6056–6063. doi: 10.1128/jvi.67.10.6056-6063.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bonilla P J, Gorbalenya A E, Weiss S R. Mouse hepatitis virus strain A59 RNA polymerase gene ORF 1a: heterogeneity among MHV strains. Virology. 1994;198:736–740. doi: 10.1006/viro.1994.1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boursnell M F G, Brown T D K, Foulds I J, Green P F, Tomley F M, Binns M M. Completion of the sequence of the genome of the coronavirus avian infectious bronchitis virus. J Gen Virol. 1987;68:57–77. doi: 10.1099/0022-1317-68-1-57. [DOI] [PubMed] [Google Scholar]
- 4.Breedenbeek P J, Pachuk C J, Noten A F H, Charite J, Luytjes W, Weiss S R, Spaan W J M. The primary structure and expression of the second open reading frame of the polymerase gene of the coronavirus MHV-A59; a highly conserved polymerase is expressed by an efficient ribosomal frameshifting mechanism. Nucleic Acids Res. 1990;18:1825–1832. doi: 10.1093/nar/18.7.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brierley I, Boursnell M E G, Binns M M, Billimoria B, Blok V C, Brown T D K, Inglis S C. An efficient ribosomal frame-shifting signal in the polymerase-encoding region of the coronavirus IBV. EMBO J. 1987;6:3779–3785. doi: 10.1002/j.1460-2075.1987.tb02713.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Denison M R, Hughes S A, Weiss S R. Identification and characterization of a 65-kDa protein processed from the gene 1 polyprotein of the murine coronavirus MHV-A59. Virology. 1995;207:316–320. doi: 10.1006/viro.1995.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Denison M R, Zoltick P W, Hughes S A, Giangreco B, Olson A L, Perlman S, Leibowitz J L, Weiss S R. Intracellular processing of the N-terminal ORF 1a proteins of the coronavirus MHV-A59 requires multiple proteolytic events. Virology. 1992;189:274–284. doi: 10.1016/0042-6822(92)90703-R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dougherty W G, Semler B L. Expression of virus-encoded proteinase: functional and structural similarities with cellular enzymes. Microbiol Rev. 1993;57:781–822. doi: 10.1128/mr.57.4.781-822.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eleouet J F, Rasschaert D, Lambert P, Levy L, Vende P, Laude H. Complete sequence (20 kilobases) of the polyprotein-encoding gene 1 of transmissible gastroenteritis virus. Virology. 1995;206:817–822. doi: 10.1006/viro.1995.1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gorbalenya A E, Koonin E V, Donchenko A P, Blinov V M. Coronavirus genome: prediction of putative functional domains in the nonstructural polyprotein by comparative amino acid sequence analysis. Nucleic Acids Res. 1989;17:4847–4861. doi: 10.1093/nar/17.12.4847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Herold J, Raabe T, Schelle P B, Siddell S G. Nucleotide sequence of the human coronavirus 229E RNA polymerase locus. Virology. 1993;195:680–691. doi: 10.1006/viro.1993.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim J C, Spence R A, Currier P F, Lu X T, Denison M R. Coronavirus protein processing and RNA synthesis is inhibited by the cysteine proteinase inhibitor e64d. Virology. 1995;208:1–8. doi: 10.1006/viro.1995.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 14.Lawson M A, Semler B L. Picornavirus protein processing— enzymes, substrates, and genetic regulation. Curr Top Microbiol Immunol. 1990;161:49–80. [PubMed] [Google Scholar]
- 15.Lee H-J, Shieh C-K, Gorbalenya A E, Koonin E V, LaMonica N, Tuler J, Bagdzhadhzyan A, Lai M M C. The complete sequence (22 kilobases) of murine coronavirus gene 1 encoding the putative proteases and RNA polymerase. Virology. 1991;180:567–582. doi: 10.1016/0042-6822(91)90071-I. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu D, Xu H, Brown T. Proteolytic processing of the coronavirus infectious bronchitis virus 1a polyprotein: identification of a 10-kilodalton polypeptide and determination of its cleavage sites. J Virol. 1997;71:1814–1820. doi: 10.1128/jvi.71.3.1814-1820.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu D X, Brown T D K. Characterisation and mutational analysis of an ORF 1a-encoding proteinase domain responsible for proteolytic processing of the infectious bronchitis virus 1a/1b polyprotein. Virology. 1995;209:420–427. doi: 10.1006/viro.1995.1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu X T, Lu Y Q, Denison M R. Intracellular and in vitro translated 27-kDa proteins contain the 3C-like proteinase activity of the coronavirus MHV-A59. Virology. 1996;222:375–382. doi: 10.1006/viro.1996.0434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu Y, Lu X, Denison M R. Identification and characterization of a serine-like proteinase of the murine coronavirus MHV-A59. J Virol. 1995;69:3554–3559. doi: 10.1128/jvi.69.6.3554-3559.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lu Y Q, Denison M R. Determinants of mouse hepatitis virus 3C-like proteinase activity. Virology. 1997;230:335–342. doi: 10.1006/viro.1997.8479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pachuk C J, Breedenbeek P J, Zoltick P W, Spaan W J M, Weiss S R. Molecular cloning of the gene encoding the putative polymerase of mouse hepatitis coronavirus, strain A59. Virology. 1989;171:141–148. doi: 10.1016/0042-6822(89)90520-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pinon J, Mayreddy R, Turner J, Khan F, Bonilla P, Weiss S. Efficient autoproteolytic processing of the MHV-A59 3C-like proteinase from the flanking hydrophobic domains requires membranes. Virology. 1997;230:309–322. doi: 10.1006/viro.1997.8503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saborio J L, Pong S-S, Koch G. Selective and reversible inhibition of initiation of protein synthesis in mammalian cells. J Mol Biol. 1974;85:195–211. doi: 10.1016/0022-2836(74)90360-x. [DOI] [PubMed] [Google Scholar]
- 24.Seybert A, Ziebuhr J, Siddell S G. Expression and characterization of a recombinant murine coronavirus 3C-like proteinase. J Gen Virol. 1997;78:71–75. doi: 10.1099/0022-1317-78-1-71. [DOI] [PubMed] [Google Scholar]
- 25.Sims, A. C., X. T. Lu, and M. R. Denison. Unpublished data.
- 26.Ziebuhr J, Herold J, Siddell S G. Characterization of a human coronavirus (strain 229E) 3C-like proteinase activity. J Virol. 1995;69:4331–4338. doi: 10.1128/jvi.69.7.4331-4338.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]