ABSTRACT
Many female squids and cuttlefishes have a symbiotic reproductive organ called the accessory nidamental gland (ANG) that hosts a bacterial consortium involved with egg defense against pathogens and fouling organisms. While the ANG is found in multiple cephalopod families, little is known about the global microbial diversity of these ANG bacterial symbionts. We used 16S rRNA gene community analysis to characterize the ANG microbiome from different cephalopod species and assess the relationship between host and symbiont phylogenies. The ANG microbiome of 11 species of cephalopods from four families (superorder: Decapodiformes) that span seven geographic locations was characterized. Bacteria of class Alphaproteobacteria, Gammaproteobacteria, and Flavobacteriia were found in all species, yet analysis of amplicon sequence variants by multiple distance metrics revealed a significant difference between ANG microbiomes of cephalopod families (weighted/unweighted UniFrac, Bray–Curtis, P = 0.001). Despite being collected from widely disparate geographic locations, members of the family Sepiolidae (bobtail squid) shared many bacterial taxa including (~50%) Opitutae (Verrucomicrobia) and Ruegeria (Alphaproteobacteria) species. Furthermore, we tested for phylosymbiosis and found a positive correlation between host phylogenetic distance and bacterial community dissimilarity (Mantel test r = 0.7). These data suggest that closely related sepiolids select for distinct symbionts from similar bacterial taxa. Overall, the ANGs of different cephalopod species harbor distinct microbiomes and thus offer a diverse symbiont community to explore antimicrobial activity and other functional roles in host fitness.
IMPORTANCE
Many aquatic organisms recruit microbial symbionts from the environment that provide a variety of functions, including defense from pathogens. Some female cephalopods (squids, bobtail squids, and cuttlefish) have a reproductive organ called the accessory nidamental gland (ANG) that contains a bacterial consortium that protects eggs from pathogens. Despite the wide distribution of these cephalopods, whether they share similar microbiomes is unknown. Here, we studied the microbial diversity of the ANG in 11 species of cephalopods distributed over a broad geographic range and representing 15–120 million years of host divergence. The ANG microbiomes shared some bacterial taxa, but each cephalopod species had unique symbiotic members. Additionally, analysis of host–symbiont phylogenies suggests that the evolutionary histories of the partners have been important in shaping the ANG microbiome. This study advances our knowledge of cephalopod–bacteria relationships and provides a foundation to explore defensive symbionts in other systems.
KEYWORDS: symbiosis, cephalopod, accessory nidamental gland, Alphaproteobacteria, Verrucomicrobia, phylosymbiosis, microbiomes, microbial communities
INTRODUCTION
Associations with microorganisms have been crucial for the successful evolution of animals, often contributing essential roles in development and specialized functions (e.g., metabolism, digestion, protection from pathogens, and providing host camouflage) (1, 2). Many cephalopods form symbiotic relationships with bacteria, for example, the specialized light organ of bobtail squid (Sepiolidae) and photophores in some loliginid squid host bioluminescent bacteria that are members of the Vibrionaceae and are used in a camouflage behavior known as counterillumination (3, 4). Many cephalopods also have a second symbiotic organ called the accessory nidamental gland (ANG) that hosts a bacterial consortium and is part of the female reproductive system (5–10). The ANG symbiosis has been studied in the Hawaiian bobtail squid Euprymna scolopes, where the ANG bacteria are recruited from the environment each generation (11–13). During oviposition, bacteria are deposited in the jelly coats of eggs, where they provide both antibacterial and antifungal protection for developing embryos (14–16). Antibacterial and antifungal activities have also been found in bacteria isolated from the ANG and eggs of the squids Doryteuthis pealeii (17) and Uroteuthis duvaucelii (18).
The basic structure of the ANG is similar across cephalopods. The organ consists of convoluted tubules composed of epithelial cells that surround extracellular bacteria (7, 9). In E. scolopes, distinct tubules have different bacterial taxa (9). A few studies have described the development of the ANG in sepiolids (12, 13), sepiids (19), and loliginids (20, 21). In E. scolopes, the ANG begins to form around 4 weeks after hatching and reaches complete development at sexual maturity (approx. 2 months). It begins as a bi-lobed organ that is poised to recruit bacteria from the environment and eventually becomes a single uniform gland (12, 13).
Although the ANG is distributed among multiple cephalopod species, whether the microbiome of this gland is conserved, as has been described for light organ/photophore symbioses (22, 23), is unknown. Phylosymbiosis occurs when the host phylogeny reflects the host-associated microbiome community relationships at a moment in time and space and does not assume a stable evolutionary association (24, 25). Though not universal, phylosymbiosis is observed in the microbiomes of many animals and insects, except, to date, for birds (26–28) and microscopic marine invertebrates (29). In marine organisms, phylosymbiosis has been observed in corals, sponges, ascidians (30), fishes (31) and marine mammals (32), and recently in the digestive tracts of some cephalopods (33). In E. scolopes, complete ANG development depends upon the presence of environmental bacteria (12, 13), and the symbiotic bacteria are vital in egg protection (14); however, whether the ANG exhibits phylosymbiosis is unknown. Examining the ANG microbiome of multiple cephalopod species for phylosymbiotic patterns may provide a first step to determining eco-evolutionary mechanisms that drive this pattern, as has been described in other host–microbe associations (25).
The ANG of E. scolopes has a stable microbiome of Alphaproteobacteria, Gammaproteobacteria, Verrucomicrobia, and Flavobacteriia (9, 11). In other cephalopods, Alphaproteobacteria, Gammaproteobacteria, and Flavobacteriia were found in Doryteuthis pealeii of the Loliginidae family (7) and many Roseobacter spp. (Alphaproteobacteria) in Sepia officinalis of the Sepiidae family (6). Similar observations were made in 14 other cephalopod species belonging to families Loliginidae and Idiosepiidae (8). However, except for work published in E. scolopes, these previous studies relied on 16S rRNA gene clone libraries, fluorescence in situ hybridization, or characterizing a limited number of cultured strains. With high-throughput sequencing using 16S rRNA gene community analysis, we characterized the ANG bacterial diversity from four cephalopod families: Idiosepiidae (pygmy squid), Sepiidae (cuttlefish), Sepiolidae (bobtail squid), and Loliginidae (pencil squid), comprising 11 species and 7 genera to understand whether there is a core cephalopod ANG microbiome, if microbiota vary between different cephalopod groups and geographical regions, and whether phylosymbiotic signatures could be detected. This study lays the foundation for further investigations of mechanisms that drive the selection of ANG bacteria in cephalopods.
MATERIALS AND METHODS
Sample collection
ANGs from 11 species of squid and cuttlefish were collected from multiple locations (Table 1; Fig. 1). All samples were collected in accordance with appropriate animal protocols and local permit collections: E. scolopes, protocol A18-029 approved by the Institutional Animal Care and Use Committee, University of Connecticut; Euprymna tasmanica: permit number 200891 U.C. Merced; and Idiosepius pygmaeus: Queensland Department of Agriculture Fisheries and Forestry, permit number 170251. Dissection of E. tasmanica followed animal ethics guidelines at James Cook University and was approved by the James Cook University Animal Ethics Committee (JCU Animal Ethics Number A2189; Sepia officinalis were collected by certificated fishermen at the Ría de Vigo, Spain, and in the Pertuis Charentais, France. The individuals were transported in proper containers to the Experimental Culture Facilities of IIM-CSIC and UMR LIENSs, which are registered as “User and breeding center on animal experimentation” (approval number ES360570202001 and 173002, respectively). Transport, housing, and handling were carried out following the principles of animal welfare, within 2 h after fishing. Procedures for transportation, euthanasia, and dissection were carried out in accordance with the principles published in the European Animal Directive (2010/63/EU) for the protection of experimental animals and were approved by the National Competent Authority ethics committee from Spain (Research Project ES360570202001/17/EDUCFORM 07/CGM01) and France (APAFIS#20520-2019050614554709). All animals were euthanized using anesthetic such as MgCl2 (3%; wt/vol) and ethanol (1%; vol/vol) or ethanol (2%; vol/vol) dissolved in seawater, and ANGs were carefully dissected using sterilized scissors.
TABLE 1.
The taxonomy and species names of the cephalopod samples collected along with their site of collection, number of replicates for each site, and the year the samples were collected
| Family | Genus | Species | Collection site | GPS coord. | No. of replicates | Year of collection |
|---|---|---|---|---|---|---|
| Sepiolidae | Euprymna | E. scolopes | USA (Oahu, Hawaii) | 21°25′44.0″N, 157°47′32.4″W | 3 | 2015 |
| 21°16′51.42″N, 157°43′33.07″W | 3 | 2015 | ||||
| E. tasmanica | Australia (New South Wales) | 33°59′34.8″S, 151°10′31.1″E | 5 | 2016 | ||
| E. morsei | Japan (Mainland) | 34°15′08.7″N, 136°31′40.0″E | 5 | 2018 | ||
| E. berryi | 34°15′08.7″N, 136°31′40.0″E | 5 | 2018 | |||
| Eumandya | E. parva | Japan (Okinawa) | 26°30′21.6″N, 127°52′51.6″E | 8 | 2018 | |
| Loliginidae | Uroteuthis | U. duvaucelii | India (Kochi) | 9°51′28.6″N, 76°10′24.3″E | 5 | 2019 |
| Doryteuthis | D. pealeii | USA (Massachusetts) | 41°25′53.4″N, 71°12′55.1″W | 4, 4 | 2015, 2019 | |
| Sepioteuthis | S. lessoniana | Japan (Okinawa) | 26°19′52.0″N, 127°55′38.3″E | 6 | 2018 | |
| Idiosepiidae | Idiosepius | I. pygmaeus | Australia (Queensland) | 19°14′24.0″S, 146°49′12.0″E | 6 | 2019 |
| Sepiidae | Sepia | S. officinalis | France (La Rochelle) | 46°04′10.0″N, 1°13′59.5″W | 5 | 2021 |
| Spain (Vigo) | 42°09′31.0″N, 8°52′46.9″W | 6 | 2021 | |||
| S. esculenta | Japan (Hiroshima) | 34°17′38.4″N, 132°23′01.0″E | 5 | 2022 |
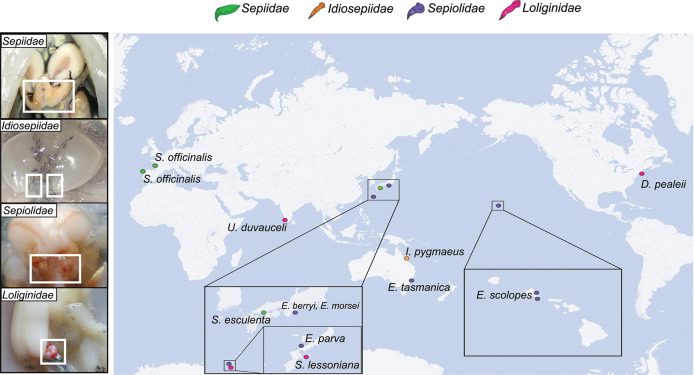
Fig 1.
Representative photos of the ANGs from different families of cephalopods (left) and sampling locations (right). The ANG is situated inside the white inset box. Scale bars: Sepiidae 1 cm; Idiosepiidae 0.1 cm; Sepiolidae 0.5 cm; Loliginidae 0.5 cm. Right: sampling locations and species from which ANGs were collected. Colors correspond to different cephalopod families.
The ANG samples of E. scolopes, Euprymna morsei, Euprymna berryi, and D. pealeii from 2015 were flash-frozen upon collection. The ANGs from the remaining samples were dissected almost immediately or within 2 h from the time of collection and stored in DNA/RNA Shield (Zymo Research, Irvine, CA, USA) during transportation to the University of Connecticut and subsequently stored at −80°C until processing. The samples in Fig. 1 were mapped with QGIS (34). Two samples of Doryteuthis opalescens ANGs were also collected from Oregon, USA (44°40′01.2″N, 124°17′02.4″W) (CDFW Scientific Collection Permit SC-13563), but due to the insufficient replicates were not included in the analyses. We have separately described these two replicates (Fig. S1).
Validation of preservative
To confirm the efficacy of DNA/RNA Shield as a preservation buffer, we compared the ANG microbiome from E. scolopes where one half of the ANG was stored at −80°C immediately after dissection and the other half was placed in DNA/RNA Shield at room temperature for 1 week (n = 3). Samples were processed, and the microbial composition and beta diversity of the halves were compared as described below. No significant differences in the ANG microbiome based on the preservation method were observed [Bray–Curtis permutational multivariate analysis of variance (PERMANOVA): q-value =0.82].
DNA extraction, sequencing, and analysis
DNA extractions were performed as previously described for E. scolopes (11). Briefly, ANG samples and five extraction controls (containing only filter sterilized artificial seawater) were transferred to 1.5-mL tubes and homogenized with a sterile pestle in ultra-pure water to dissociate the host tissue. Samples were centrifuged at 5,000 rcf to remove host cell debris, and the supernatant with bacterial cells was lysed with bead-beating and processed for DNA extraction with the Qiagen DNEasy Blood and Tissue kit. The DNA was quantified with the Qubit dsDNA high-sensitivity assay (ThermoFisher Scientific). Negligible DNA (<0.01 ng/µm) was found in the controls, which were also processed for PCR and sequencing.
The V4 region of the bacterial 16S rRNA gene (primer 515f GTGYCAGCMGCCGCGGTAA and 806r GGACTACNVGGGTWTCTAAT) was amplified by PCR (35) and confirmed with agarose gel electrophoresis. No bands were detected in negative controls. PCR products were sequenced on a Miseq (Illumina Inc, USA) at Microbial Analysis, Resources and Services, University of Connecticut. The full amplification and sequencing protocol are available at http://www.earthmicrobiome.org/protocols-and-standards.
The raw sequences were denoised and demultiplexed using QIIME2.2020 (36). The amplicon sequence variants (ASVs) were generated using the DADA2 method (37) and aligned against the trained Greengenes reference database at a 99% or higher identity (38) (May 2013 release). Some ASVs of Opitutae and Leisingera were not assigned by Greengenes and were manually annotated with NCBI BLAST (optimized for megablast). Chloroplast, mitochondria, and sequences unassigned at the kingdom level were filtered out. The number of reads in the negative controls ranged from 0 to 15 reads/sample. Among the negative controls, one ASV assigned as Rhodospirillaceae was common between the negative controls (>50% of reads) and all replicates of Sepia officinalis (>40% of reads). However, we do not interpret the reads in S. officinalis as spurious since the number of reads far outweigh the negative controls by more than 3,000 reads. The other ASVs in negative controls were assigned as Bacilli and Rhodobacterales, but these specific ASVs were not found in any ANG samples.
The QIIME2 artifacts were converted to phyloseq objects using qiime2R (v0.99.6). The final amplicon sequence variant (ASV)–abundance matrix was analyzed by two different methods: (1) normalized by transform_sample_counts and (2) rarefied by rarefy_even_depth, both to the minimum number of reads in the sample set (1,538) in R 3.3.2 with the phyloseq (v1.26.1) package (39). Rarefaction curves were generated with alpha_rarefaction in QIIME2 (Fig. S2). No difference in alpha and beta diversity analyses was observed with the two methods; thus, the statistical analyses shown here are with transformed data. The data were analyzed for differences between families of cephalopods and between species within the same family. We calculated alpha diversity with observed, Shannon, and inverse Simpson metrics using estimate_richness and plot_richness with phyloseq. Pairwise significances between comparisons were performed with the Wilcoxon test, and group comparisons were performed with the Kruskal–Wallis test with Bonferroni-corrected P-values. Beta diversities were analyzed with Bray–Curtis and weighted and unweighted UniFrac (40). The effect of host taxonomy on beta diversity differences was calculated with PERMANOVA with beta-group-significance (QIIME2) with 999 permutations. In addition to PERMANOVA in QIIME2, pairwise.adonis (v0.4) was used to analyze pairwise differences between groups. For diversity comparisons within families, samples were subset and normalized to their minimum number of reads (sepiolids = 7,671, loliginids = 6,289, and sepiids = 1,538). Other R packages used for making figures include ggplot (41), RColorBrewer (42), dplyr (43), ggpubr, scales, and reshape2 (44).
Comparison of Euprymna scolopes ANGs from present and past studies
ANG samples from E. scolopes in this study were combined with a previous study by reference (45). Pairwise comparison of samples from this study and reference (45) found no significant difference (Bray–Curtis PERMANOVA: P-value =0.3). Thus, regardless of the version of QIIME used [QIIME2.2020 in this study and QIIME1 in reference (45)], we observed no differences in microbiomes of E. scolopes. The ANG microbiome described by reference (45) using QIIME1 observed a higher abundance of Flavobacteriia (0.05%–10%). This extended analysis of additional ANG samples was not included in phylosymbiosis analyses to ensure relatively similar sample sizes. The included E. scolopes samples were randomly selected from the larger available data set (11, 45). Of those samples selected, re-analyses of ANG microbiomes from different geographic locations on Oahu, Hawaii, were not significantly different (Bray–Curtis PERMANOVA: P-value =0.879).
Phylogenetic similarity of Opitutae and Ruegeria sequences
Multiple sequences identified as Opitutae and Ruegeria were found in the sepiolids. These sequences were submitted to NCBI with accession numbers (OQ305981–OQ305990). We used ARB SILVA’s ACT (alignment, classification, and tree service) to determine the sources (host-associated or environmental) of closely related sequences (95% similarity cut-off) to sequences from sepiolid ANGs. Pelagiococcus litoralis was used as the outgroup for the Opitutae tree and Leisingera cerula as the outgroup in the Ruegeria tree. FastTree with default parameters (GTR+CAT model) was used to generate the tree in the Newick format.
Phylosymbiosis
Phylosymbiosis was analyzed using methods as previously described (46). ASV abundances were first averaged for the replicates, and microbial cladograms were constructed using Bray–Curtis and weighted UniFrac distances to test for patterns of phylosymbiosis (26). The host tree phylogeny was obtained from Tanner et al. (47), modified to retain only the species used in this project using drop.tip with ape package (48). The host trees were modified to include sepiolid phylogenies resolved by Sanchez et al. (49). We considered ASVs present in at least 50% of the replicates and with at least 30% relative abundance in each host species as dominant and were identified with QIIME2 plugin q2-coremicrobiome. The topological congruences of the host and microbiome trees were measured with normalized matching cluster and Robinson–Foulds metrics with 10,000 random trees using a previously published script (26). Both metrics produce a score ranging from 0 (completely congruent) to 1 (completely incongruent). Mantel tests (vegan package) with 1,000 permutations were run to evaluate the Pearson and Spearman correlation of host divergence times and microbiome distance and host geographic distance and microbiome distance. The host divergence times were obtained from references (47, 49). The geographic distance matrix was generated using the Geographic distance matrix generator (v1.2.3). The distance decay plots were plotted with betapart (v1.5.4) package.
RESULTS
ANG bacterial communities differ between cephalopod families
The total number of amplicon reads from all 70 samples was 2,153,636, ranging from 1,538 to 123,544 reads per sample, with an average read depth of 22,879. Rarefaction curves, plotting observed ASVs against the sequencing depth from 1,538 reads, displayed a plateau for most species, indicating that the sequencing depth was adequate to capture the bacterial diversity (Fig. S2). The normalized data set had 1,221 unique ASVs. Significant differences were observed with Wilcoxon tests in alpha diversity measures between families of cephalopods (Fig. 2A). There was no significant difference in richness and evenness between species of loliginids and sepiids, but within sepiolids, E. scolopes had a significantly higher diversity (Kruskal–Wallis rank sum test, Bonferroni-corrected P > 0.05, Fig. S3). ANG communities from the loliginids had the highest richness while those from the sepiolids had the lowest (Fig. 2A).
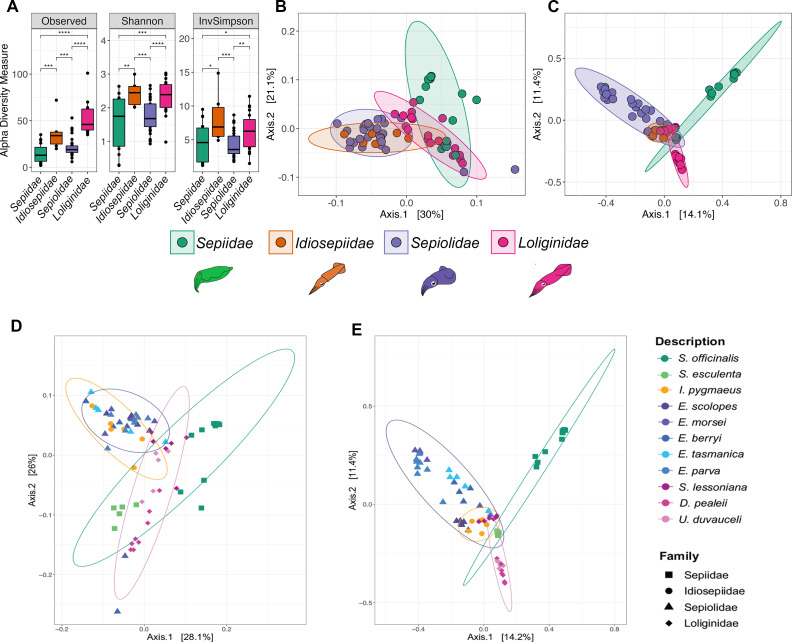
Fig 2.
Alpha and beta diversity of ASVs between cephalopod families. (A) Alpha diversity indices of ASVs by cephalopod family. Asterisks indicate significant differences between groups (Wilcoxon test, Bonferroni-corrected ****P < 0.0001, ***P = 0.001, **P = 0.01, and *P = 0.05). Principal coordinate analysis of (B) weighted UniFrac distances and (C) Bray–Curtis distances across cephalopod families and (D) weighted UniFrac and (E) Bray–Curtis distances across cephalopod families and species, where ellipses represent 95% confidence intervals between families.
PERMANOVA of beta diversity metrics (weighted UniFrac, unweighted UniFrac, and Bray–Curtis) revealed that the overall microbiomes of ANGs between families of cephalopods were significantly different (Fig. 2B and C; Table 2). All comparisons of species/families with unweighted UniFrac were significantly different (q < 0.01). Ordination plots of principal component analyses with weighted UniFrac and Bray–Curtis distances also showed close clustering of samples within the same family (Fig. 2B through E). There was an overlap between the ANG microbiomes of I. pygmaeus (sole representative of Idiosepiidae) and some sepiolids. Pairwise PERMANOVA with weighted UniFrac of I. pygmaeus with each sepiolid revealed similarity to the ANG microbiomes of E. tasmanica and E. morsei, but not other sepiolids (Table 2). Significant differences were observed with weighted UniFrac and Bray–Curtis distances within loliginids and sepiids (Table 2). A smaller significant difference (q-value =0.03) within sepiolids was observed with weighted UniFrac and a larger difference with Bray–Curtis (q-value =0.001, Table 2). Pairwise comparison within sepiolid species revealed a significant difference between all species using Bray–Curtis (q-value =0.001), in weighted UniFrac (q-value =0.032), and between combinations of Eumandya parva, Euprymna berryi, and E. scolopes (Table S1). Other pairwise comparisons of sepiolid species were not significant using weighted UniFrac (q-value >0.11, Table S1). These analyses indicate that while most cephalopod species harbored distinct ANG communities, some of the more closely related sepiolids harbored similar ANG microbiomes.
TABLE 2.
Comparison of ANG microbiomes with permutational analysis of variance of weighted UniFrac and Bray–Curtis distancesa
| Comparisons | Weighted UniFrac | Bray–Curtis | ||||
|---|---|---|---|---|---|---|
| q-value | Effect size (%) | Pseudo-F | q-value | Effect size (%) | Pseudo-F | |
| Between families | 0.001 | 32 | 14 | 0.001 | 24 | 7.1 |
| Within loliginids | 0.004 | 57 | 12 | 0.003 | 48 | 7.5 |
| Within sepiids | 0.01 | 78 | 25 | 0.01 | 70 | 32 |
| Within sepiolids | 0.032 | 25 | 1.9 | 0.001 | 35 | 6.8 |
| I. pygmaeus × E. morsei | 0.04 | 19 | 1.8 | 0.005 | 35 | 4.8 |
| I. pygmaeus × E. tasmanica | 0.03 | 23 | 2.5 | 0.004 | 35 | 4.8 |
| D. pealeii 2015 × 2019 | 0.5 | 15 | 1.2 | 0.2 | 17 | 1.5 |
| S. officinalis France × Spain | 0.1 | 20 | 2.1 | 0.05 | 22 | 2.7 |
The q-values in shaded boxes represent a non-significant difference.
The year of collection and location of the same species did not affect the microbiome composition. For example, no difference in the ANG microbiome was observed between D. pealeii collected in 2015 and 2019 (Table 2). Also, the most abundant ASVs (Rhodobacteraceae, Alphaproteobacteria) remained consistently present in all D. pealeii samples regardless of the time of collection (Fig. 4). There was also no significant difference with weighted UniFrac in the ANG microbiomes of S. officinalis collected from coasts of France and Spain, separated by 748 km (Table 2). However, S. officinalis samples did vary by collection site when analyzed via unweighted UniFrac (q-value =0.002, R2 = 37%) and Bray–Curtis (q-value =0.05, Table 2), and alpha diversity was also significantly higher in S. officinalis collected from France than those collected from Spain (P = 0.01, Fig. S3).
Bacterial composition of the ANGs from different cephalopods
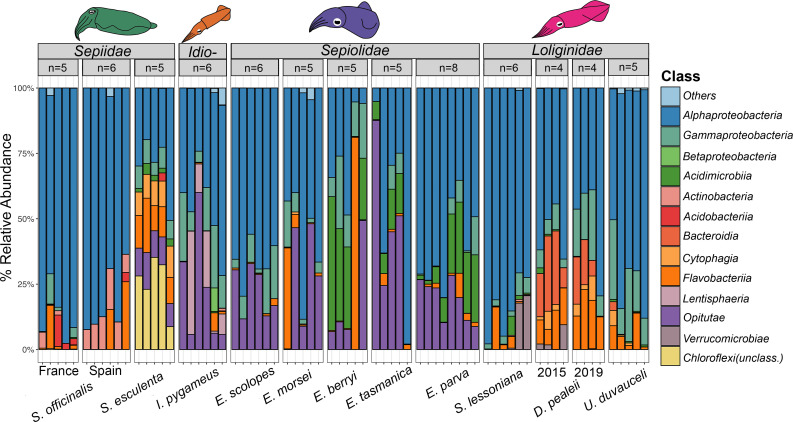
One thousand one hundred eighty-six ASVs were identified from 15 bacterial phyla across all host species. Alphaproteobacteria dominated the microbiomes of most ANGs, with a smaller abundance of Gammaproteobacteria (Fig. 3). Class Alphaproteobacteria was found in all species at varying abundances (Fig. 4). Rhodobacteraceae occurred at the highest abundance in the sepiolids (as members of Ruegeria) (30% ± 23%), followed by loliginids (28% ± 5%), I. pygmaeus (13% ± 12%), and S. esculenta (7% ± 13%). Rhodospirillaceae were dominant in the ANGs of Sepia officinalis from both France (60% ± 25%) and Spain (72 ± 26%), and Hyphomicrobiaceae were dominant in S. lessoniana (59% ± 20%) (Fig. 4). From the phylum Verrucomicrobia, members of the Opitutae class were present in 90% of the replicates of the sepiolids at varying abundances, highest in E. tasmanica (40% ± 28%) and E. scolopes (32.3% ± 10%), followed by E. morsei (28% ± 23%), E. parva (20% ± 8%), and E. berryi (15% ± 19%), and all replicates of I. pygmaeus (24% ± 20%) (Fig. 3 and 4). Four of six replicates of I. pygmaeus also possessed Lentisphaeria (19% ± 10%) (Fig. 3), which is a sister clade to Verrucomicrobia. Acidimicrobiia (phylum Actinobacteria) were found in more than 80% of replicates of E. berryi (28% ± 18%), E. parva (15% ± 10%), and E. tasmanica (12% ± 5%) (Fig. 3). Actinobacteria were also found in S. officinalis from both France (1% ± 2%) and Spain (10% ± 4%) and S. esculenta (2% ± 0.9%) (Fig. 3). Members of the Bacteroidia class were dominant in the loliginid Doryteuthis pealeii (33% ± 10%) but relatively absent in the other loliginids (Fig. 4), although other members of the Bacteroidetes phylum were more widespread (Fig. 3). The ANG of S. esculenta was the only species that possessed an unclassified Chloroflexi (23% ± 5%) (Fig. 3). The relatively low abundance ASVs (<0.1%) were mostly unclassified Proteobacteria and Bacteroidetes (Fig. S4). We also identified two unclassified bacterial ASVs in S. officinalis and E. morsei that were previously found in fish food made of D. opalescens (accession number: JC192016) (Fig. S4).
Fig 3.
Percent relative abundance of taxa with ASVs greater than 0.1% of normalized data set. Representative figures of cephalopods are above the names of the cephalopod families (where Idio- indicates Idiosepiidae). “n” represents the number of biological replicates from each species.
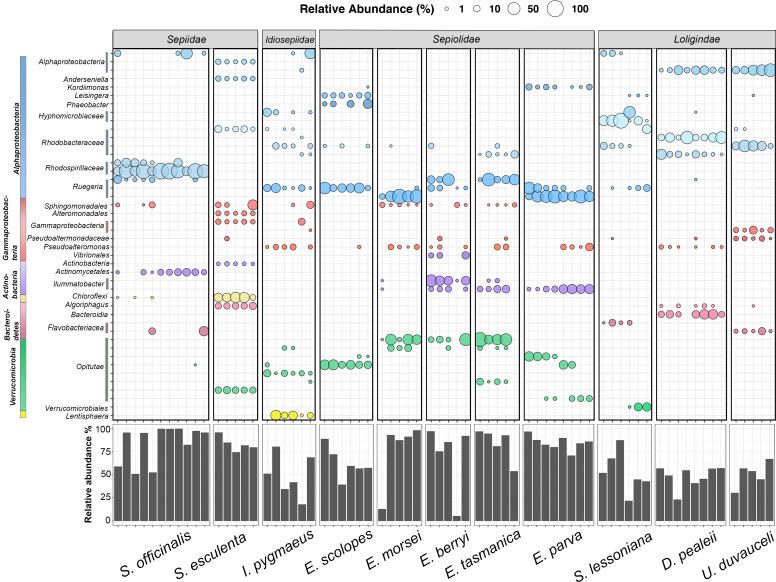
Fig 4.
Bubble plot of the top 44 most abundant ASVs that made up approximately 40%–50% of each cephalopod species. The ASVs were identified to the closest taxonomic level assigned. Similar colors of the bubbles represent ASVs from the same class/phylum. The size of the bubble reflects the relative abundance of the ASV in each sample. The bar plot at the bottom corresponds to the percent relative abundance of the ASVs in the bubble plot for each sample.
Two samples of Doryteuthis opalescens ANG (analyzed separately due to the low sample size) had a high abundance of Alphaproteobacteria (22% ± 3%) and Gammaproteobacteria (60% ± 15%) but lacked other taxa like Bacteroidia and Flavobacteriia that were found in D. pealeii ANGs (Fig. S1). One ASV identified as an Alphaproteobacteria (accession number: OQ435377) was common to all the Doryteuthis samples (Fig. S1). This shared ASV was further identified with the NCBI database as a Hyphomicrobiales of genus Anderseniella.
For each cephalopod species, 5–10 ASVs made up ~50% of the relative abundance of their microbiomes (Fig. 4). Although some ASVs were shared between species, no single ASV was found common across all samples. For example, an Alphaproteobacteria ASV was shared between the loliginids D. pealeii and U. duvaucelii. Also, a Ruegeria ASV was shared between E. morsei and E. parva, Illumatobacter sp. was found in majority of E. berryi, E. tasmanica, and E. parva, and an Opitutae ASV was found in most E. morsei, E. berryi, and E. tasmanica samples. Overall, closely related bacteria were found between sepiolids. However, similar observations were not made with different Sepia species (S. esculenta and S. officinalis), which had significantly different microbiomes (Fig. 4; Table 2).
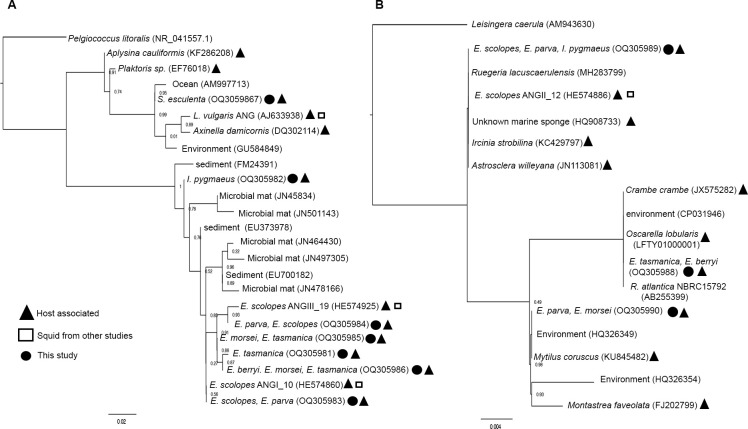
The alignment of Opitutae and Ruegeria ASVs found in sepiolids, I. pygmaeus, and S. esculenta with other closely related bacteria from the ARB databases (SILVA) (50) revealed that these ASVs were associated with both hosts and environmental samples (Fig. 5). The Opitutae ASVs from sepiolids were more closely related to each other than other sequences in the ARB databases (Fig. 5). Sepiolid Opitutae ASVs were also related to uncultured Verrucomicrobia from the ANG of the European squid Loligo vulgaris (Fig. 5). The ASVs from this study had a >99% sequence identity with ANG bacteria from E. scolopes from previous studies (Fig. 5) (9). Phylogenetic trees of the sepiolid Ruegeria ASVs classified them as closely related to different species such as R. lacuscaerulensis and R. atlantica (Fig. 5). These varying results may suggest that the Opitutae in different sepiolids are more closely related to each other or, more likely, that there are insufficient cultured representatives available for Opitutae sequences compared to those for Ruegeria spp.
Fig 5.
Phylogenetic tree of abundant Opitutae spp. and Ruegeria spp. in ANG microbiomes. Maximum likelihood phylogenetic tree from FastTree (Newick format) of 240 bp of 16S rRNA gene sequences identified as (A) bacterial class Opitutae spp. of phylum Verrucomicrobia in sepiolids, Sepia esculenta, and Idiosepius pygmaeus and (B) genus Ruegeria spp. in sepiolids and I. pygmaeus. The symbols represent the sources of the sequences.
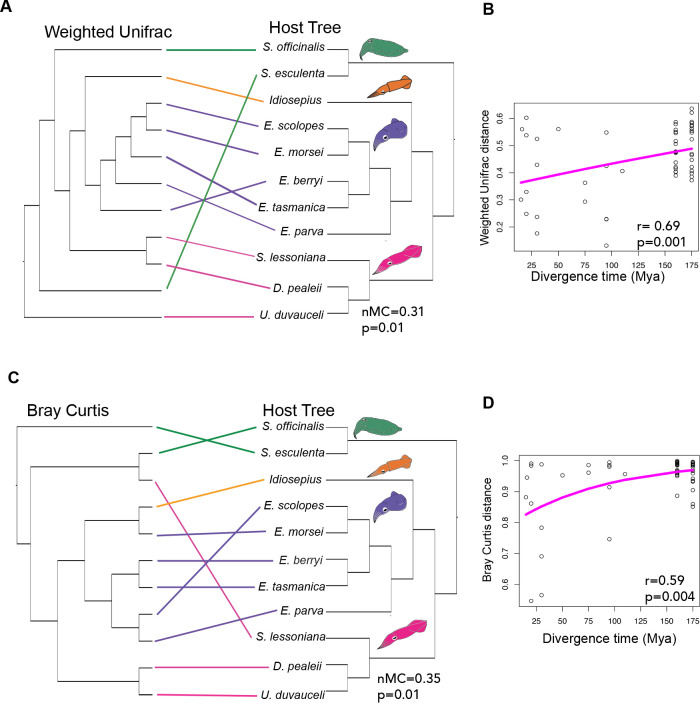
Phylosymbiosis: positive correlation between ANG microbiomes and host phylogeny
The hierarchical clustering of ANG microbiomes with weighted UniFrac and Bray–Curtis revealed significant congruence with host phylogeny using normalized matching cluster [nMC, ranges from 0 (complete congruence) to 1 (complete incongruence)] methods (Bray–Curtis nMC = 0.35, P = 0.01; weighted UniFrac nMC = 0.31, P = 0.01) (Fig. 6A and C). However, there was no statistically significant congruence with the Robinson–Foulds method (Bray–Curtis and weighted UniFrac nRF >0.6, P = 0.005). The nMC method is more refined to account for subtree congruence than the Robinson–Foulds method (51). We further filtered the microbiome for abundant bacterial ASVs and identified a stronger congruence with both Bray–Curtis (nMC = 0.28, P = 0.0004) and weighted UniFrac (nMC = 0.29, P-value =0.0007) (Fig. S5) and observed similar results with Robinson–Foulds (nRF >0.6, P = 0.005). These results suggest that the more abundant ANG bacteria may be driving phylosymbiosis.
Fig 6.
Comparison of the dendrograms of hosts and associated ANG microbiomes. The microbiome trees were generated with (A) weighted UniFrac distances and (C) Bray–Curtis distances. The topological congruence of the microbiome and host trees were calculated with nMC. (B, D) The rate of microbiome divergence across phylogenetic distance (B: weighted UniFrac, D: Bray–Curtis) with divergence times in Mya. The pink line represents distance decay. The P-values and Mantel r statistics are shown in the bottom right corners of the plot.
Mantel tests of host divergence times (47) and microbiome distances revealed significant positive Spearman and Pearson correlations with weighted UniFrac (P = 0.001, r = 0.7–0.8) and Bray–Curtis (P = 0.004, r = 0.6–0.63). This correlation can also be visualized with a distance decay plot (Fig. 6B and D). Similar results were observed for the correlation of host and microbiome distance of dominant ASVs with the Mantel test (weighted UniFrac P: 0.001, r = 0.76–0.83; Bray–Curtis P = 0.001, r = 0.062–0.065). A much lower significant negative correlation was observed between geographic distance and weighted UniFrac microbiome distance (Mantel test: P = 0.049, r = −0.04) and was not significant with the Bray–Curtis distance (Mantel test P = 0.38, r = 0.05) (Fig. S6). The r-value in the Mantel test ranges from −1 to +1, where −1 implies a negative correlation, +1 is a positive correlation, and 0 signifies no correlation. Thus, we observed a positive correlation between host phylogeny and ANG microbiomes (suggesting phylosymbiosis) but no correlation with geographic distance.
DISCUSSION
In this study, we surveyed the microbiomes of a female-specific symbiotic organ, the ANG, in multiple host species from four cephalopod families. Overall, the cephalopod ANG microbiomes have a lower bacterial diversity compared to the mammalian gut and oral microbiomes (52–54), marine sponges (55), and corals (30). High abundances of Alphaproteobacteria were found in all species of cephalopods similar to the observations made in previous studies of single host species (7, 8, 11) while lower abundances of Gammaproteobacteria were also found in all host species. However, at lower bacterial taxonomic levels, the ANG bacteria varied between host families and between host species. We also provide further evidence that ANGs from mature adults were consistent over time and space. The ANG microbiomes of D. pealeii were similar from samples collected in 2015 and 2019, and S. officinalis from waters of France and Spain were similar (Table 2). These findings agree with observations made for E. scolopes (11, 45) where ANG communities were consistent between collection years and from different sampling locations on Oahu, HI, USA. Overall, these combined studies suggest a consistent species-specific ANG microbiome. A consistent microbiome across time and space may imply a strong selection pressure to acquire specific ANG bacteria during colonization.
The alpha diversity of ANG symbionts between cephalopod families varied significantly. The loliginids have a richer and more even bacterial community compared to the sepiids and sepiolids (Fig. 2). This difference may be due to many reasons, such as the range of host migration, the necessity for increased protection from diverse pathogens, or other factors. The loliginids have a large migratory range, so hosts may have increased exposure to different microbial niches in different environments (56–58). Beta diversity analyses showed significant differences in microbiomes between families (Table 2). Bray–Curtis and weighted and unweighted UniFrac analyses suggest that microbiomes between sepiids, loliginids, sepiolids, and I. pygmaeus were different. Further, microbiomes within sepiids and loliginids were also different, which may suggest unique microbiomes between species in these families. Though the microbiomes of the ANGs from closely related sepiolids were more similar and clustered together with weighted UniFrac, they were different with the Bray–Curtis analysis (Table S1; Fig. 2). Since weighted UniFrac considers taxonomic relationships of bacterial communities and Bray–Curtis does not (59), similar microbiomes within sepiolids as analyzed with weighted UniFrac suggest that related but not identical bacterial communities are present in the ANGs of this group.
Most of the sepiolids possessed a relatively high abundance of Ruegeria (Alphaproteobacteria in the Roseobacter clade) and Opitutae (Verrucomicrobia). Species of Ruegeria and Opitutae made up ~50% of the relative abundance of most sepiolid ANG microbiomes (Fig. 3 and 4). The presence of these taxa in the ANG of E. scolopes was also confirmed using fluorescence in situ hybridization (9). A previous study found that the relative abundance of Opitutae had a negative correlation with squid size in E. scolopes, while the relative abundance of Alphaproteobacteria had a positive correlation to squid size, suggesting that members of the Opitutae may initially colonize the ANG in this species (12). Follow-up work also suggests a role for these bacteria in initiating the development of the ANG in E. scolopes (13). The high Opitutae prevalence in all the sepiolids and I. pygmaeus suggests a conserved function for this bacterial family in these cephalopod groups (Fig. 3 and 5). To date, no member of this group has been successfully cultured, but the data from this study suggest that the Opitutae may play an important role in the ANG symbiosis of some cephalopods.
Cephalopods evolved ~540 Mya in the Cambrian, and species used in this study diverged ~120–15 Mya (47). With this diverse data set, we found that recently diverged cephalopod species had similar microbiomes compared to those that diverged more than 30 Mya (Fig. 6; Table S1). The sepiids diverged approximately 120 Mya, followed by idiosepiids (~95 Mya), sepiolids, and loliginids (~80 Mya). Within the cephalopod families, the sepiolids analyzed for this study diverged from each other ~15–30 Mya (Table S1), the sepiids S. officinalis and S. esculenta ~50 Mya, and the loliginids ~80–100 Mya (47, 49). The Mantel test and the dendrogram-based approaches are thought to have high sensitivity in detecting the host phylogenetic correlation, with the Mantel test being more sensitive (59). The Mantel test here indicated an increase in host divergence times correlated with an increase in microbiome divergence. The differences observed in the microbiomes of species within the loliginid and sepiid families are likely due to the increased divergence times (Fig. 6A and C). A similar pattern was found in Nasonia sp. wasps, where host species that diverged 0.4 Mya had a more similar microbiome than those that diverged 1.0 Mya (60). Differences between the sepiid ANG microbiomes may also be explained by variable egg-laying behaviors. Sepia officinalis deposits ink into their egg cases during laying, whereas S. esculenta does not (61). As cephalopod ink is known to have antimicrobial properties (62, 63), functional differences in egg protection may also explain the selection of different microbiomes found in these two cuttlefish species. Phylosymbiosis was also observed in the gut microbiomes of other cephalopod species, but these putative symbionts and their function are largely unknown (33).
Phylosymbiosis of multiple organs within the same host has only been examined for a few animals, e.g., the gut and/or skin of mammals (64, 65), bats (52), and fishes (31). However, the strength of phylosymbiosis varies depending on the organ type and host exposure to the environment (59). Some cephalopods also provide two different internal symbiotic organs (the light organ and the ANG) with specialized roles. The light organ or photophore in many cephalopods houses bioluminescent bacteria belonging to the Vibrionaceae (Gammaproteobacteria) in sepiolids and loliginids (4) that provide camouflage via counterillumination against nocturnal predators (66, 67). Like the ANG, bacteria in light organs also have phylogenetic congruence with their squid hosts (67). Colonization experiments of different strains of Vibrio fischeri revealed that native or closely related strains are more effective at colonizing the light organ (67, 68), and the requirements for light organ colonization are well studied in E. scolopes (23). Thus, bacterial adaptation may also have a role in phylosymbiosis. Further experiments such as the transplantation of ANG bacteria between species of cephalopods may reveal the functional benefits of phylosymbiosis. Recent studies that describe the development of the ANG in E. scolopes suggest that such gnotobiotic experiments under controlled conditions may be possible to test these hypotheses (13). The potential to examine phylosymbiosis between these light organs and ANGs, which in contrast to the gut microbiome are posited to have only a restricted open period for colonization, could provide a unique insight into varying selective pressures moving forward.
Host–microbe relationships that are conserved over evolutionary time may indicate a significant functional role (25). There are many reasons that phylosymbiosis can arise such as coevolution, cospeciation, ecological drift, or shifts in the host geographic range (25). Though phylosymbiosis is observed in many species, its functional significance for associations is still unclear (25). Transfer experiments of microbiota between Nasonia wasp species demonstrated an impact on larval growth and adult survival, which suggests that selective pressure underpins phylosymbiosis in the case of Nasonia spp. (24, 46). While both the ANG and light organ symbioses are defensive, the selective pressures associated with the loss of function of these symbioses are difficult to test in the laboratory. Because the light organ and ANG are not nutritionally obligate symbioses for the host, the lack of an organ does not affect host survivability. However, in animals raised under aposymbiotic and symbiotic conditions, bacterial colonization is necessary for morphogenesis and/or full development of the organs (3, 13, 23). The ANG and its bacteria may have a similar relationship like that of the light organ and vibrio species in multiple cephalopods as both these relationships are important for host defense.
Defensive symbioses are common in both terrestrial and aquatic environments (69). Bacterial symbionts, metabolites, and/or biosynthetic gene clusters responsible for antimicrobial activity have been identified from the ANG and/or eggs of E. scolopes (14–16), D. pealeii (17), and U. duvaucelii (18). Compounds and strains of Alpha- and Gammaproteobacteria and Flavobacteriia isolated from E. scolopes demonstrated various degrees of inhibition against three strains of the fungus Fusarium keratoplasticum, the yeast Candida albicans, and other marine bacteria (14–16). The fungal-inhibiting species included Ruegeria sp., which here was found in all sepiolids, I. pygmaeus, and S. officinalis. Leisingera sp. isolated from E. scolopes eggs produces the antibacterial compound indigoidine (16), but species from this genus were not found in any other sepiolids. Pseudoalteromonas spp. from E. scolopes also strongly inhibited fungi and other bacteria (14, 15) and here were also detected in I. pygmaeus, E. morsei, E. parva, and D. pealeii. Members of Actinobacteria are known for their potent secondary metabolites (70) and were found here in multiple cephalopod ANGs including E. berryi, E. tasmanica, E. parva, and the sepiids. Future studies will explore the antimicrobial potential of ANG symbionts from other cephalopods and potentially culture new and uncharacterized strains.
We also evaluated the hypothesis that geography may contribute to differences observed in ANG microbiomes between host families. While we do not have many multiple representatives of the same species collected from different geographical locations, S. officinalis collected from France and Spain were separated by 730 km, yet their ANG microbiomes were very similar, even at the ASV level. Many cuttlefish, including S. officinalis, exhibit a genetic pattern of isolation-by-distance, where the populations have increased genetic differences with increased geographies (<300 km), i.e., they have low mobility (71, 72). This could mean that populations of S. officinalis from France were different from those in Spain. Previous research also found that two genetically isolated populations of E. scolopes had very similar ANG bacterial compositions (45). These findings also support the hypothesis that geography may not significantly influence the selection of bacteria at the broader taxonomic level. In sepiolids, species or strains of bioluminescent vibrios in the light organ correlated to the geographical distribution of the hosts (73). Similarly, the ANG microbiome data suggest that related but distinct microbiomes are selected by closely related cephalopods. The species of Ruegeria and Opitutae were found in the ANGs of sepiolids, and the ASV differences may be attributed to differences in geography or strain availability during ANG colonization. The similar microbiome in sepiolids from different geographic regions separated by 1,000–8,000 km also suggests that the influence of geography may not be significant at the broader taxonomic levels but may impact microbiome composition at the species/strain level. E. morsei and E. berryi were collected from the same location, but these two species had distinct (Bray–Curtis analyses) but similar (weighted UniFrac analyses) microbiomes (Table S1). If ANG bacteria in E. morsei and E. berryi are recruited during a discreet colonization window as has been described for E. scolopes (12, 13), then geographic differences during host development may account for varying bacterial diversity. Future studies raising different sepiolids in the same substrate and water conditions may help answer this question. Given the role of ANG bacteria in egg protection, bacteria with similar antimicrobial function may be selected by the host. Functional redundancy in the infant human gut for Bifidobacterium longum involved in milk metabolism has been observed (74). Furthermore, like the similarities observed for the microbiome species in the mammalian gut (53, 74), host factors shared by different cephalopod species may help select similar symbionts. Future studies that compare the full-length 16S gene along with metagenomes may reveal strain diversity and functional redundancy in ANG microbiomes between cephalopod species.
Cephalopods obtain their ANG bacteria via horizontal transmission each generation; thus, environmental bacterial communities are important to consider (11–13, 21). Although we did not collect environmental seawater samples in this study, previous work showed that the majority of operational taxonomic units (OTUs) found in the ANG of E. scolopes were also detected in the seawater and sediment of the host’s natural environment (11). Similar microbiomes within S. officinalis collected from different locations and within sepiolids may suggest a strong selective pressure for certain bacteria. Life histories may also influence how different cephalopods obtain their ANG bacteria. Cephalopod families also have different life history patterns. Some sepiolids and sepiids bury themselves in sand, whereas loliginids live a pelagic lifestyle (61, 75, 76). However, sepiolids and sepiids do not have similar microbiomes, so lifestyle is unlikely to be the sole contributor to bacterial selection.
The 16S rRNA microbiome analyses of the ANG microbiome from 11 species of cephalopods revealed shared bacterial communities at higher taxonomic levels. Finer analyses demonstrated distinct microbiomes associated with cephalopod families and species. The high bacterial diversity associated with the ANG defensive symbiosis may provide a unique opportunity to screen for potentially novel antimicrobial compounds and other secondary metabolites. We observed differences in the microbiomes as the host evolutionary distance increased. Sepiolids collected from geographically distant regions separated by thousands of kilometers had similar microbiomes, suggesting a conserved functional role of the ANG bacterial community and an evolutionary relationship between hosts and symbionts. Moreover, a growing number of cephalopod genomics and symbiont metagenomic/transcriptomic resources (77–79) will shed light on the evolution of the ANG and its functional role in hosts.
ACKNOWLEDGMENTS
We thank D. Rokhsar at the Okinawa Institute of Science and Technology Graduate University and University of California Berkeley and members of the Ikeda laboratory at the University of the Ryukyus for their assistance with collections and the use of laboratory space. We thank Kendra Maas of UConn’s Microbial Analysis, Resource, and Services for the assistance with sequencing. We also thank members of the Nyholm lab for the helpful feedback on the manuscript.
This work was funded by the Gordon and Betty Moore Foundation, the National Science Foundation, and the University of Connecticut Office of Vice President for Research to S.V.N. and a Lerner–Gray Grant for Marine Research to S.J.M.
Contributor Information
Allison H. Kerwin, Email: akerwin@mcdaniel.edu.
Spencer V. Nyholm, Email: spencer.nyholm@uconn.edu.
Knut Rudi, Norwegian University of Life Sciences, Ås, Norway.
DATA AVAILABILITY
The 16S rRNA gene sequences generated with Illumina Miseq were deposited in the NCBI sequence read archive under BioProject accession number PRJNA880911. The sequences of the E. scolopes samples collected from 21°25′44.0″N, 157°47′32.4″W and 21°16′51.42″N, 157°43′33.07″W were previously deposited in the European Nucleotide Archive accession numbers PRJEB23264 (ERR2191588, ERR2200327, and ERR2191589) and PRJEB14655 (ERR1777318, ERR1777324, and ERR1777391), respectively. The accession numbers for Opitutae spp. are OQ305981-OQ305987 and Ruegeria spp. are OQ305988-OQ305990. The accession number for the sequence of Alphaproteobacteria shared between all the Doryteuthis spp. is OQ435377.
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/aem.00990-23.
Relative abundance of ASVs for Doryteuthis opalescens and D. pealeii.
Rarefaction curves of amplicon sequence variants.
Alpha diversity measures for three cephalopod families.
Relative abundance of low abundance taxa.
Dendograms of hosts and associated abundant ANG microbiomes.
Distance decay plot of microbiome distance and host geographic distance.
Paired beta diversity measures between sepiolid species.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Malard F, Dore J, Gaugler B, Mohty M. 2021. Introduction to host microbiome symbiosis in health and disease. Mucosal Immunol 14:547–554. doi: 10.1038/s41385-020-00365-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. McFall-Ngai MJ. 2014. The importance of microbes in animal development: Lessons from the squid-Vibrio Symbiosis. Annu Rev Microbiol 68:177–194. doi: 10.1146/annurev-micro-091313-103654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Nyholm SV, McFall-Ngai MJ. 2021. A lasting symbiosis: how the hawaiian bobtail squid finds and keeps its bioluminescent bacterial partner. Nat Rev Microbiol 19:666–679. doi: 10.1038/s41579-021-00567-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Pankey MS, Minin VN, Imholte GC, Suchard MA, Oakley TH. 2014. PNAS plus: from the cover: predictable transcriptome evolution in the convergent and complex bioluminescent organs of squid. Proc Natl Acad Sci U S A 111:E4736–42. doi: 10.1073/pnas.1416574111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lum‐Kong A, Hastings TS. 1992. The accessory nidamental glands of Loligo forbesi (cephalopoda: loliginidae): characterization of symbiotic bacteria and preliminary experiments to investigate factors controlling sexual maturation. J Zoology 228:395–403. doi: 10.1111/j.1469-7998.1992.tb04443.x [DOI] [Google Scholar]
- 6. Grigioni S, Boucher-Rodoni R, Demarta A, Tonolla M, Peduzzi R. 2000. Phylogenetic characterisation of bacterial symbionts in the accessory nidamental glands of the sepioid Sepia officinalis (cephalopoda: decapoda). Mar Biology 136:217–222. doi: 10.1007/s002270050679 [DOI] [Google Scholar]
- 7. Barbieri E, Paster BJ, Hughes D, Zurek L, Moser DP, Teske A, Sogin ML. 2001. Phylogenetic characterization of epibiotic bacteria in the accessory nidamental gland and egg capsules of the squid Loligo pealei (cephalopoda: loliginidae). Environ Microbiol 3:151–167. doi: 10.1046/j.1462-2920.2001.00172.x [DOI] [PubMed] [Google Scholar]
- 8. Pichon D, Gaia V, Norman MD, Boucher-Rodoni R. 2005. Phylogenetic diversity of epibiotic bacteria in the accessory nidamental glands of squids (cephalopoda: loliginidae and idiosepiidae). Marine Biology 147:1323–1332. doi: 10.1007/s00227-005-0014-5 [DOI] [Google Scholar]
- 9. Collins AJ, LaBarre BA, Won BSW, Shah MV, Heng S, Choudhury MH, Haydar SA, Santiago J, Nyholm SV. 2012. Diversity and partitioning of bacterial populations within the accessory nidamental gland of the squid Euprymna scolopes. Appl Environ Microbiol 78:4200–4208. doi: 10.1128/AEM.07437-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lindgren AR, Pankey MS, Hochberg FG, Oakley TH. 2012. A multi-gene phylogeny of cephalopoda supports Convergent morphological evolution in association with multiple habitat shifts in the marine environment. BMC Evol Biol 12:129. doi: 10.1186/1471-2148-12-129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kerwin AH, Nyholm SV. 2017. Symbiotic bacteria associated with a bobtail squid reproductive system are detectable in the environment, and stable in the host and developing eggs. Environ Microbiol 19:1463–1475. doi: 10.1111/1462-2920.13665 [DOI] [PubMed] [Google Scholar]
- 12. Kerwin AH, McAnulty SJ, Nyholm SV. 2021. Development of the accessory nidamental gland and associated bacterial community in the hawaiian bobtail squid, Euprymna scolopes. Biol Bull 240:205–218. doi: 10.1086/713965 [DOI] [PubMed] [Google Scholar]
- 13. McAnulty SJ, Kerwin AH, Koch E, Nuttall B, Suria AM, Collins AJ, Schleicher TR, Rader BA, Nyholm SV. 2023. Failure to launch”: development of a reproductive organ linked to symbiotic bacteria. mBio 14:e0213122. doi: 10.1128/mbio.02131-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kerwin AH, Gromek SM, Suria AM, Samples RM, Deoss DJ, O’Donnell K, Frasca S, Sutton DA, Wiederhold NP, Balunas MJ, Nyholm SV. 2019. Shielding the next generation: symbiotic bacteria from a reproductive organ protect bobtail squid eggs from fungal fouling. mBio 10:e02376-19. doi: 10.1128/mBio.02376-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Suria AM, Tan KC, Kerwin AH, Gitzel L, Abini-Agbomson L, Bertenshaw JM, Sewell J, Nyholm SV, Balunas MJ. 2020. Hawaiian bobtail squid symbionts inhibit marine bacteria via production of specialized metabolites, including new bromoalterochromides BAC-D/D′. mSphere 5:e00166-20. doi: 10.1128/mSphere.00166-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gromek SM, Suria AM, Fullmer MS, Garcia JL, Gogarten JP, Nyholm SV, Balunas MJ. 2016. Leisingera sp. JC1, a bacterial isolate from Hawaiian Bobtail squid eggs, produces indigoidine and differentially inhibits vibrios. Front Microbiol 7:1342. doi: 10.3389/fmicb.2016.01342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Barbieri E, Barry K, Child A, Wainwright N. 1997. Antimicrobial activity in the microbial community of the accessory nidamental gland and egg cases of Loligo pealei (cephalopoda: loliginidae). Biol Bull 193:275–276. doi: 10.1086/BBLv193n2p275 [DOI] [PubMed] [Google Scholar]
- 18. Gomathi P, Nair R. 2008. Antibacterial activity in the accessory nidamental gland extracts of the Indian squid, Loligo duvauceli. IJMS 39:100–104. [Google Scholar]
- 19. Döring W. 1908. Über Bau und entwicklung des weiblichen geschlechts- apparates BEI myopsiden cephalopodan. Z Wiss Zool 91:112–189. [Google Scholar]
- 20. Tseng P-W, Li H-W, Chen C, Tseng Y-C, Chang C-F, Wu G-C. 2023. Transcriptomic profile of Symbiotic accessory nidamental gland during female maturation in bigfin reef squid. Front Mar Sci 9. doi: 10.3389/fmars.2022.1026742 [DOI] [Google Scholar]
- 21. Kaufman MR, Ikeda Y, Patton C, van Dykhuizen G, Epel D. 1998. Bacterial symbionts colonize the accessory nidamental gland of the squid Loligo opalescens via horizontal transmission. Biol Bull 194:36–43. doi: 10.2307/1542511 [DOI] [PubMed] [Google Scholar]
- 22. Visick KL, Stabb EV, Ruby EG. 2021. A lasting symbiosis: how Vibrio fischeri finds a squid partner and persists within its natural host. Nat Rev Microbiol 19:654–665. doi: 10.1038/s41579-021-00557-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nyholm SV, McFall-Ngai MJ. 2004. The winnowing: establishing the squid–Vibrio symbiosis. Nat Rev Microbiol 2:632–642. doi: 10.1038/nrmicro957 [DOI] [PubMed] [Google Scholar]
- 24. van Opstal EJ, Bordenstein SR. 2019. Phylosymbiosis impacts adaptive traits in Nasonia wasps. mBio:e00887–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lim SJ, Bordenstein SR. 2020. An introduction to phylosymbiosis. Proc Biol Sci 287:20192900. doi: 10.1098/rspb.2019.2900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Trevelline BK, Sosa J, Hartup BK, Kohl KD. 2020. A bird’s-eye view of phylosymbiosis: weak signatures of phylosymbiosis among all 15 species of cranes. Proc Biol Sci 287:20192988. doi: 10.1098/rspb.2019.2988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bodawatta KH, Koane B, Maiah G, Sam K, Poulsen M, Jønsson KA. 2021. Species-specific but not phylosymbiotic gut microbiomes of new guinean passerine birds are shaped by diet and flight-associated gut modifications. Proc Biol Sci 288:20210446. doi: 10.1098/rspb.2021.0446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Song SJ, Sanders JG, Delsuc F, Metcalf J, Amato K, Taylor MW, Mazel F, Lutz HL, Winker K, Graves GR, et al. 2020. Comparative analyses of vertebrate gut microbiomes reveal convergence between birds and bats. mBio 11:e02901-19. doi: 10.1128/mBio.02901-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Boscaro V, Holt CC, Van Steenkiste NWL, Herranz M, Irwin NAT, Àlvarez-Campos P, Grzelak K, Holovachov O, Kerbl A, Mathur V, Okamoto N, Piercey RS, Worsaae K, Leander BS, Keeling PJ. 2022. Microbiomes of microscopic marine invertebrates do not reveal signatures of phylosymbiosis. Nat Microbiol 7:810–819. doi: 10.1038/s41564-022-01125-9 [DOI] [PubMed] [Google Scholar]
- 30. O’Brien PA, Tan S, Yang C, Frade PR, Andreakis N, Smith HA, Miller DJ, Webster NS, Zhang G, Bourne DG. 2020. Diverse coral reef invertebrates exhibit patterns of phylosymbiosis. ISME J 14:2211–2222. doi: 10.1038/s41396-020-0671-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Doane MP, Morris MM, Papudeshi B, Allen L, Pande D, Haggerty JM, Johri S, Turnlund AC, Peterson M, Kacev D, et al. 2020. The skin microbiome of elasmobranchs follows phylosymbiosis, but in teleost fishes, the microbiomes converge. Microbiome 8:93. doi: 10.1186/s40168-020-00840-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Apprill A, Miller CA, Van Cise AM, U’Ren JM, Leslie MS, Weber L, Baird RW, Robbins J, Landry S, Bogomolni A, Waring G. 2020. Marine mammal skin microbiotas are influenced by host phylogeny. R Soc Open Sci 7:192046. doi: 10.1098/rsos.192046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kang W, Kim PS, Tak EJ, Sung H, Shin N-R, Hyun D-W, Whon TW, Kim HS, Lee J-Y, Yun J-H, Jung M-J, Bae J-W. 2022. Host phylogeny, habitat, and diet are main drivers of the cephalopod and mollusk gut microbiome. Anim Microbiome 4:30. doi: 10.1186/s42523-022-00184-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. QGIS.org . 2022. QGIS geographic information system. QGIS Association. Available from: http://www.qgis.org [Google Scholar]
- 35. Walters W, Hyde ER, Berg-Lyons D, Ackermann G, Humphrey G, Parada A, Gilbert JA, Jansson JK, Caporaso JG, Fuhrman JA, Apprill A, Knight R. 2016. Improved bacterial 16S rRNA gene (V4 and V4-5) and fungal internal transcribed spacer marker gene primers for microbial community surveys. mSystems 1:e00009-15. doi: 10.1128/mSystems.00009-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith GA, Alexander H, Alm EJ, Arumugam M, Asnicar F, et al. 2019. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol 37:852–857. doi: 10.1038/s41587-019-0252-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP. 2016. Dada2: high resolution sample inference from illumina amplicon data. Nat Methods 13:581–583. doi: 10.1038/nmeth.3869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL. 2006. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 72:5069–5072. doi: 10.1128/AEM.03006-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. McMurdie PJ, Holmes S. 2013. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8:e61217. doi: 10.1371/journal.pone.0061217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lozupone C, Knight R. 2005. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol 71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wickham H. 2016. ggplot2. ggplot2: elegant graphics for data analysis. Springer-Verlag New York, Cham. Available from: http://link.springer.com/10.1007/978-3-319-24277-4 [Google Scholar]
- 42. Neuwirth E Maintainer. 2022. “Package “RColorBrewer” title ColorBrewer palettes”
- 43. Wickham H, François R, Henry L, Müller K. 2022. Package dplyr: a grammar of data manipulation
- 44. Wickham H. n.d. Reshaping data with the reshape package. J Stat Soft 21. doi: 10.18637/jss.v021.i12 [DOI] [Google Scholar]
- 45. Kerwin AH, Nyholm SV. 2018. Reproductive system symbiotic bacteria are conserved between two distinct populations of Euprymna scolopes from Oahu. mSphere 3:e00531-17. doi: 10.1128/mSphere.00531-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Brooks AW, Kohl KD, Brucker RM, van Opstal EJ, Bordenstein SR, Relman D. 2016. Phylosymbiosis: relationships and functional effects of microbial communities across host evolutionary history. PLoS Biol 14:e2000225. doi: 10.1371/journal.pbio.2000225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tanner AR, Fuchs D, Winkelmann IE, Gilbert MTP, Pankey MS, Ribeiro ÂM, Kocot KM, Halanych KM, Oakley TH, da Fonseca RR, Pisani D, Vinther J. 2017. Molecular clocks indicate turnover and diversification of modern coleoid cephalopods during the mesozoic marine revolution. Proc Biol Sci 284:20162818. doi: 10.1098/rspb.2016.2818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Paradis E, Schliep K. 2019. ape 5.0: an environment for modern phylogenetics and evolutionary analyses in {R}. Bioinformatics 35:526–528. doi: 10.1093/bioinformatics/bty633 [DOI] [PubMed] [Google Scholar]
- 49. Sanchez G, Fernández-Álvarez FÁ, Taite M, Sugimoto C, Jolly J, Simakov O, Marlétaz F, Allcock L, Rokhsar DS. 2021. Phylogenomics illuminates the evolution of bobtail and bottletail squid (order sepiolida). Commun Biol 4:819. doi: 10.1038/s42003-021-02348-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO. 2013. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41:D590–6. doi: 10.1093/nar/gks1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Bogdanowicz D, Giaro K. 2013. On a matching distance between rooted phylogenetic trees. Inter J Applied Mathematics and Comp Sci 23:669–684. doi: 10.2478/amcs-2013-0050 [DOI] [Google Scholar]
- 52. Lutz HL, Jackson EW, Webala PW, Babyesiza WS, Kerbis Peterhans JC, Demos TC, Patterson BD, Gilbert JA. 2019. Ecology and host identity outweigh evolutionary history in shaping the bat microbiome. mSystems 4:e00511-19. doi: 10.1128/mSystems.00511-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Campbell TP, Sun X, Patel VH, Sanz C, Morgan D, Dantas G. 2020. The microbiome and resistome of chimpanzees, gorillas, and humans across host lifestyle and geography. ISME J 14:1584–1599. doi: 10.1038/s41396-020-0634-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Nearing JT, DeClercq V, Van Limbergen J, Langille MGI. 2020. Assessing the variation within the oral Microbiome of healthy adults. mSphere 5:e00451-20. doi: 10.1128/mSphere.00451-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Freeman CJ, Easson CG, Matterson KO, Thacker RW, Baker DM, Paul VJ. 2020. Microbial symbionts and ecological divergence of caribbean sponges: a new perspective on an ancient association. ISME J 14:1571–1583. doi: 10.1038/s41396-020-0625-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Burford BP, Wild LA, Schwarz R, Chenoweth EM, Sreenivasan A, Elahi R, Carey N, Hoving HJT, Straley JM, Denny MW. 2022. Rapid range expansion of a marine ectotherm reveals the demographic and ecological consequences of short-term variability in seawater temperature and dissolved oxygen. Am Nat 199:523–550. doi: 10.1086/718575 [DOI] [PubMed] [Google Scholar]
- 57. Payne J, O’Dor R. 2006. Comparing squid optimal cost of transport speed to actual field migrations: New data from 40-G Loligo Opalescens. GLOBEC Report. 24:16–18.
- 58. Zeidberg LD. 2004. Allometry measurements from in situ video recordings can determine the size and swimming speeds of juvenile and adult squid Loligo opalescens (cephalopoda: myopsida). J Exp Biol 207:4195–4203. doi: 10.1242/jeb.01276 [DOI] [PubMed] [Google Scholar]
- 59. Mazel F, Davis KM, Loudon A, Kwong WK, Groussin M, Parfrey LW. 2018. Is host filtering the main driver of phylosymbiosis across the tree of life? mSystems 3:e00097-18. doi: 10.1128/mSystems.00097-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Brucker RM, Bordenstein SR. 2013. The hologenomic basis of speciation: gut bacteria cause hybrid lethality in the genus Nasonia. Science 341:667–669. doi: 10.1126/science.1240659 [DOI] [PubMed] [Google Scholar]
- 61. O’Brien CE, Mezrai N, Darmaillacq A-S, Dickel L. 2017. Behavioral development in embryonic and early juvenile cuttlefish (Sepia officinalis). Dev Psychobiol 59:145–160. doi: 10.1002/dev.21476 [DOI] [PubMed] [Google Scholar]
- 62. Derby CD. 2014. Cephalopod ink: production, chemistry, functions and applications. Mar Drugs 12:2700–2730. doi: 10.3390/md12052700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Fahmy SR, Ali EM, Ahmed NS. 2014. Therapeutic effect of sepia ink extract against invasive pulmonary aspergillosis in mice. J Basic & Applied Zoology 67:196–204. doi: 10.1016/j.jobaz.2014.08.001 [DOI] [Google Scholar]
- 64. Ross AA, Müller KM, Weese JS, Neufeld JD. 2018. Comprehensive skin microbiome analysis reveals the uniqueness of human skin and evidence for phylosymbiosis within the class mammalia. Proc Natl Acad Sci U S A 115:E5786–E5795. doi: 10.1073/pnas.1801302115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Groussin M, Mazel F, Sanders JG, Smillie CS, Lavergne S, Thuiller W, Alm EJ. 2017. Unraveling the processes shaping mammalian gut Microbiomes over evolutionary time. Nature communications 8:14319. doi: 10.1038/ncomms14319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Jones BW, Nishiguchi MK. 2004. Counterillumination in the hawaiian bobtail squid, Euprymna scolopes berry (mollusca: cephalopoda). Marine Biology 144:1151–1155. doi: 10.1007/s00227-003-1285-3 [DOI] [Google Scholar]
- 67. Nishiguchi MK, Ruby EG, McFall-Ngai MJ. 1998. Competitive dominance among strains of luminous bacteria provides an unusual form of evidence for parallel evolution in sepiolid squid-vibrio symbioses. Appl Environ Microbiol 64:3209–3213. doi: 10.1128/AEM.64.9.3209-3213.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Bongrand C, Moriano-Gutierrez S, Arevalo P, McFall-Ngai M, Visick KL, Polz M, Ruby EG. 2020. Using colonization assays and comparative genomics to discover symbiosis behaviors and factors in Vibrio fischeri. mBio 11:e03407-19. doi: 10.1128/mBio.03407-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Flórez LV, Biedermann PHW, Engl T, Kaltenpoth M. 2015. Defensive symbioses of animals with prokaryotic and eukaryotic microorganisms. Nat Prod Rep 32:904–936. doi: 10.1039/c5np00010f [DOI] [PubMed] [Google Scholar]
- 70. Lee LH, Goh BH, Chan KG. 2020. Editorial: actinobacteria: prolific producers of bioactive metabolites. Front Microbiol 11:1612. doi: 10.3389/fmicb.2020.01612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Pérez-Losada M, Nolte MJ, Crandall KA, Shaw PW. 2007. Testing hypotheses of population structuring in the northeast Atlantic ocean and mediterranean sea using the common cuttlefish Sepia officinalis . Mol Ecol 16:2667–2679. doi: 10.1111/j.1365-294X.2007.03333.x [DOI] [PubMed] [Google Scholar]
- 72. Semmens JM, Pecl GT, Gillanders BM, Waluda CM, Shea EK, Jouffre D, Ichii T, Zumholz K, Katugin ON, Leporati SC, Shaw PW. 2007. Approaches to resolving cephalopod movement and migration patterns. Rev Fish Biol Fisheries 17:401–423. doi: 10.1007/s11160-007-9048-8 [DOI] [Google Scholar]
- 73. Zamborsky DJ, Nishiguchi MK. 2011. Phylogeographical patterns among mediterranean sepiolid squids and their vibrio symbionts: environment drives specificity among sympatric species. Appl Environ Microbiol 77:642–649. doi: 10.1128/AEM.02105-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, Magris M, Hidalgo G, Baldassano RN, Anokhin AP, Heath AC, Warner B, Reeder J, Kuczynski J, Caporaso JG, Lozupone CA, Lauber C, Clemente JC, Knights D, Knight R, Gordon JI. 2012. Human gut Microbiome viewed across age and geography. Nature 486:222–227. doi: 10.1038/nature11053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Guerra A. 2006. Ecology of Sepia officinalis. Vie Milieu - Life Environ 56:97–107. [Google Scholar]
- 76. Grigoriou P, Richardson CA. 2008. The effect of ration size, temperature, and body weight on specific dynamic action of the common cuttlefish sepia officinalis. Mar Biol 154:1085–1095. doi: 10.1007/s00227-008-1002-3 [DOI] [Google Scholar]
- 77. Wellcome Sanger Institute . Aquatic symbiosis genomics project. Available from: https://www.sanger.ac.uk/collaboration/aquatic-symbiosis-genomics-project/
- 78. Albertin CB, Medina-Ruiz S, Mitros T, Schmidbaur H, Sanchez G, Wang ZY, Grimwood J, Rosenthal JJC, Ragsdale CW, Simakov O, Rokhsar DS. 2022. Genome and transcriptome mechanisms driving cephalopod evolution. Nat Commun 13:2427. doi: 10.1038/s41467-022-29748-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Belcaid M, Casaburi G, McAnulty SJ, Schmidbaur H, Suria AM, Moriano-Gutierrez S, Pankey MS, Oakley TH, Kremer N, Koch EJ, Collins AJ, Nguyen H, Lek S, Goncharenko-Foster I, Minx P, Sodergren E, Weinstock G, Rokhsar DS, McFall-Ngai M, Simakov O, Foster JS, Nyholm SV. 2019. Symbiotic organs shaped by distinct modes of genome evolution in cephalopods. Proc Natl Acad Sci U S A 116:3030–3035. doi: 10.1073/pnas.1817322116 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Relative abundance of ASVs for Doryteuthis opalescens and D. pealeii.
Rarefaction curves of amplicon sequence variants.
Alpha diversity measures for three cephalopod families.
Relative abundance of low abundance taxa.
Dendograms of hosts and associated abundant ANG microbiomes.
Distance decay plot of microbiome distance and host geographic distance.
Paired beta diversity measures between sepiolid species.
Data Availability Statement
The 16S rRNA gene sequences generated with Illumina Miseq were deposited in the NCBI sequence read archive under BioProject accession number PRJNA880911. The sequences of the E. scolopes samples collected from 21°25′44.0″N, 157°47′32.4″W and 21°16′51.42″N, 157°43′33.07″W were previously deposited in the European Nucleotide Archive accession numbers PRJEB23264 (ERR2191588, ERR2200327, and ERR2191589) and PRJEB14655 (ERR1777318, ERR1777324, and ERR1777391), respectively. The accession numbers for Opitutae spp. are OQ305981-OQ305987 and Ruegeria spp. are OQ305988-OQ305990. The accession number for the sequence of Alphaproteobacteria shared between all the Doryteuthis spp. is OQ435377.