Abstract
The mitochondrial genome resides in the mitochondria present in nearly all cell types. The porcine (Sus scrofa) mitochondrial genome is circa 16.7 kb in size and exists in the multimeric format in cells. Individual cell types have different numbers of mitochondrial DNA (mtDNA) copy number based on their requirements for ATP produced by oxidative phosphorylation. The oocyte has the largest number of mtDNA of any cell type. During oogenesis, the oocyte sets mtDNA copy number in order that sufficient copies are available to support subsequent developmental events. It also initiates a program of epigenetic patterning that regulates, for example, DNA methylation levels of the nuclear genome. Once fertilized, the nuclear and mitochondrial genomes establish synchrony to ensure that the embryo and fetus can complete each developmental milestone. However, altering the oocyte's mtDNA copy number by mitochondrial supplementation can affect the programming and gene expression profiles of the developing embryo and, in oocytes deficient of mtDNA, it appears to have a positive impact on the embryo development rates and gene expression profiles. Furthermore, mtDNA haplotypes, which define common maternal origins, appear to affect developmental outcomes and certain reproductive traits. Nevertheless, the manipulation of the mitochondrial content of an oocyte might have a developmental advantage.
Keywords: genomic balance, mitochondrial DNA, mitochondrial supplementation, nuclear transfer, oogenesis
1. INTRODUCTION
In the last few years, the role played by the mitochondrial genome in fertilization outcome and embryo development has gained increasing interest. In this context, the pig (Sus scrofa) has largely been regarded as a model of human embryology and development (Humpherson et al., 2005) and as a model for conducting human preclinical testing and trials (Bode et al., 2010; Larsen & Rolin, 2004) for which it has been deemed to be excellent in both respects (Perleberg et al., 2018). Nevertheless, the outcomes of these studies should be of significant value to pig scientists, veterinarians, and breeders who seek to maximize reproductive and other economic breeding traits to produce superior animals for the food chain and other breeding purposes. This review focuses on the role of the mitochondrial genome from the perspective of the oocyte and how it can influence fertilization outcome and development; and how its copy number is strictly regulated to ensure key developmental milestones are met. We further highlight how the nucleus and mitochondrial genome need to interact to ensure that development can progress.
2. MITOCHONDRIAL DNA (mtDNA)
The maternally inherited mitochondrial genome (Giles et al., 1980), otherwise known as mtDNA, resides in each of the large numbers of mitochondria present within many cell types. It is a double‐stranded circular genome that ranges from 16.2 kb (mice) to almost 16.7 kb (pigs) in size across mammalian species (Anderson et al., 1981; Bibb et al., 1981; Ursing & Arnason, 1998) (Figure 1a). It encodes 13 of the greater than 90 subunits of the electron transfer chain, which generates ATP through oxidative phosphorylation (OXPHOS), and is essential for cells with high energy requirements, such as neurons, and heart and skeletal muscle cells (Moyes et al., 1998). The remainder of the genes encoding the other subunits of the electron transfer chain and all other genes of the mitochondrion are encoded by the nuclear genome. mtDNA also encodes 2 ribosomal RNA (rRNA) subunits and 22 transfer RNAs (tRNAs) and has 1 noncoding region, the D‐Loop (Figure 1b), which contains regulatory regions that interact with the nuclear‐encoded mtDNA‐specific transcription and replication factors (Kucej & Butow, 2007). The D‐Loop also contains two hypervariable regions that are used by molecular geneticists to map common maternal origins and migratory routes for many species (Wallace et al., 1999). Based on their maternal origins, individuals are grouped into mtDNA haplotypes that are characterized by similar mitochondrial genomes (Wallace et al., 1999).
Figure 1.
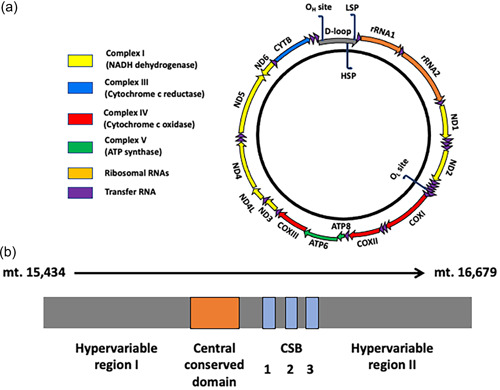
The porcine mitochondrial genome. (a) The porcine mitochondrial genome is 16.7 kb in size and encodes 13 of the subunits of the electron transfer chain, namely ND1 to 6 and ND4L, CYTB, COX I to III, and ATP6 and ATP8. It also encodes 2 rRNAs (12S and 16S rRNAs), and 22 tRNAs. It contains two noncoding regions. The smaller region is located two‐thirds of the way around the genome and houses the origin of L‐strand replication (OL). The major noncoding region is the D‐Loop, which contains the regulatory regions, the H‐strand promoter region (HSP), the L‐strand promoter region (LSP), and the origin of H‐strand replication (OH). (b) The D‐Loop (mt. 15,434 to mt. 16,679) also contains two hypervariable regions I and II, the central conserved domain, and three conserved sequence boxes (CSB1‐). rRNA, ribosomal RNA; tRNA, transfer RNA.
3. mtDNA REPLICATION
mtDNA replication is dependent on a number of nuclear‐encoded mtDNA‐specific transcription and replication factors that translocate and enter the mitochondrion, many of which possess mitochondrial‐targeting tags (Kucej & Butow, 2007). The key transcriptional factors comprise the mitochondrial RNA polymerase (POLRMT) (Tiranti et al., 1997), and mitochondrial transcription factors A (TFAM) (Fisher & Clayton, 1988), B1 (TFB1M), and B2 (TFB2M) (Falkenberg et al., 2002). mtDNA transcription precedes mtDNA replication, which is essential as the transcript that is generated undergoes cleavage to provide the primer (Hillen et al., 2017), used by the mitochondrial‐specific replicase, DNA polymerase gamma (POLG), to initiate and promote replication (Wernette & Kaguni, 1986). In mammals, the polymerase consists of two components, a catalytic subunit (encoded by POLG) and two accessory subunits (encoded by POLG2) which anchor the catalytic subunit to its template (Carrodeguas et al., 1999) (Figure 2). Replication is also dependent on the mtDNA‐specific helicase, Twinkle (TWNK), and the mitochondrial single‐stranded DNA binding protein (SSBP1) (Korhonen et al., 2003). To facilitate replication of this double‐stranded molecule, the mtDNA‐specific topoisomerase (encoded by TOP1MT) is responsible for cleaving and religating one of the strands (Zhang et al., 2007).
Figure 2.
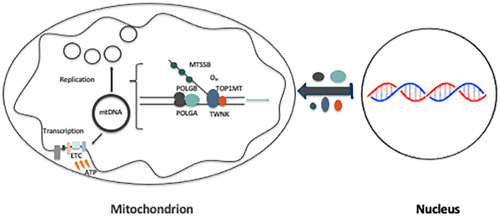
The mitochondrial replication machinery mtDNA replication is dependent on nuclear‐encoded mtDNA‐specific transcription and replication factors that translocate to the mitochondrion. Firstly, mtDNA transcription is initiated, which is a pre‐requisite for replication to proceed. The key factors involved in mtDNA replication are POLGA, POLGB, TWNK, TOP1MT, and MTSSB. Each is specific to mtDNA replication only. ETC, electron transfer chain; mtDNA, mitochondrial DNA.
4. mtDNA COPY NUMBER
Each mitochondrion possesses between 2 and 10 copies of mtDNA which are tethered to nucleoid structures comprising the nuclear‐encoded mtDNA transcription and replication machinery and anchoring proteins (Kucej & Butow, 2007). These copies of mtDNA originate from an individual's mother's primordial germ cells, the very first germ cells present just after gastrulation (Nguyen et al., 2019). This population ranges from approximately 200 copies in mouse (Cao et al., 2007; Cree et al., 2008; Wai et al., 2008) (Figure 3), possess approximately 1130 copies (Tsai et al., 2017). During oogenesis, this population increases exponentially resulting in the mature, fertilizable, metaphase II oocyte possessing between 180,000 and 500,000 copies of mtDNA (Figure 3) across a number of mammalian species including the pig (May‐Panloup, Chretien, et al., 2005; May‐Panloup, Vignon, et al., 2005; T. A. Santos et al., 2006).
Figure 3.
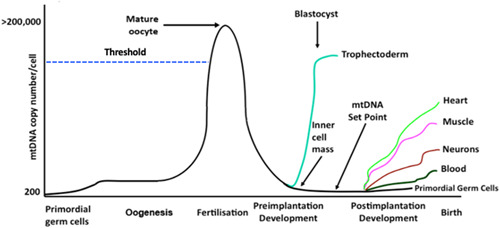
The strict regulation of mtDNA copy number during development During oogenesis, mtDNA copy number exponentially increases to reach a peak at fertilization. Failure of the maturing oocyte to increase mtDNA copy number above the threshold (dotted blue line) can result in fertilization failure or early embryo arrest. mtDNA copy number per cell progressively decreases during preimplantation development until the blastocyst stage when replication is initiated in the trophectoderm only. The inner cell mass cells continue to reduce mtDNA copy number and the “mtDNA set point” is established, which is essential for mature cells to acquire the required numbers of mtDNA copy as their precursor cells undergo differentiation into their mature, fully functional forms. mtDNA, mitochondrial DNA.
The population present in the metaphase II oocyte is considered to be an investment in subsequent developmental events (St John, 2016). Indeed, the metaphase II oocyte possesses significantly more copies of mtDNA than any other cell type and, when compared with the nuclear genome at this stage of development, is an equal partner in terms of genomic or DNA contribution on a per nucleotide basis (St John, 2019). This investment is necessary as, apart from a minor mtDNA replication event that takes place between fertilization and the 2‐cell stage (McConnell & Petrie, 2004), there are significant reductions in copy number per cell between the 4‐ and 16‐cell stages, as evidenced in pig embryo development (Spikings et al., 2007) as each newly formed blastomere divides and, thus, possesses fewer copies of mtDNA (El Shourbagy et al., 2006) (Figure 3). These decreases are matched by low levels of expression of the nuclear‐encoded mtDNA‐replication factors which, again from studies in the pig, demonstrates the strict regulation of this process (Spikings et al., 2007). Furthermore, there appears to be an active process of extrusion of mtDNA into the embryo's neighboring environment (Hammond et al., 2017; Stigliani et al., 2014). As argued by others, the developing embryo, thus, begins to increasingly rely on aerobic glycolysis for energy production (Krisher & Prather, 2012), a combination of low levels of oxygen consumption and glycolysis, and a process that is also utilized by tumor cells and first proposed as the Warburg Hypothesis (Warburg, 1956). In the pig, this is coupled with an amino acid metabolism at the blastocyst stage that is similar to the human embryo (Humpherson et al., 2005).
At the blastocyst stage, the final stage of preimplantation development, mtDNA replication is initiated but this is restricted to the trophectoderm (Houghton, 2006; Spikings et al., 2007) (Figure 3), which gives rise to the placenta. This most likely supports the metabolic requirements of these cells to both provide the inner cell mass with its required nutrients, and to promote implantation, and once the blastocyst implants, it is able to feed on the metabolites produced by its neighboring environment to promote OXPHOS and provide nutrients throughout fetal development. Unlike nuclear DNA, the mitochondrial genome is replicated multiple times per cell cycle. This process does not take place uniformly in all mitochondria but rather in a limited number and is likely focal (Chatre & Ricchetti, 2013), which may account for the disparity in the assessment of mtDNA copy number in trophoblast biopsies and its relevance to aneuploidy and implantation rates (Fragouli et al., 2015; Treff et al., 2017).
MtDNA replication is quiescent in the inner cell mass, which contains undifferentiated, pluripotent cells (Spikings et al., 2007) that progress through gastrulation, differentiate, and give rise to the fetus (Figure 3). The persistent dilution of mtDNA copy number before gastrulation establishes the “mtDNA set point,” which is the population of mtDNA, that is, the founder population of mtDNA, that contributes to the offspring's somatic cells (Facucho‐Oliveira et al., 2007; Kelly et al., 2013). Therefore, when cells initiate specialization during organogenesis, they use this founder population of mtDNA as the template for replication. Replication then proceeds in a cell‐specific manner to provide individual cell types with sufficient copies of mtDNA to support their requirements for OXPHOS‐derived ATP. This, thus, enables mature cells to meet the energy requirements associated with their specialized functions (St John et al., 2010) (Figure 3). For example, human skeletal muscle and cardiac cells will acquire approximately 6800 and 3650 copies per cell, respectively (Miller et al., 2003). Similar studies in mini pigs show wide‐ranging differences in mtDNA copy number among tissues (G. Cagnone et al., 2016). Nevertheless, with tissue‐specific studies, it must also be remembered that the cell types within tissues would have varying numbers of mtDNA copy based on their specific requirements for OXPHOS‐derived ATP. Interestingly, the regulation of mtDNA copy number in a cell‐specific manner is best exemplified by the differences between mature human sperm (purified populations) that have fertilization potential and their mtDNA copy number limited to approximately 10 copies per cell (Amaral et al., 2007) and mature oocytes (stripped of their cumulus cells) which possess more than 200,000 copies. Ultimately, mtDNA copy number in oocytes and resulting embryos is critical for embryo development and resulting fetal development.
5. THE MITOCHONDRIAL GENETIC BOTTLENECK
At gastrulation, some pluripotent cells give rise to primordial germ cells which are populated with copies of mtDNA that are transmitted to the next generation through the metaphase II oocyte. It has been proposed that these copies pass through a filtering or purifying process, described as the mitochondrial genetic bottleneck (Marchington et al., 1998) that would normally weed out mutant copies of the mitochondrial genome to ensure that the recycling of this maternal only inherited genome passes the least deleterious effects to the next generation of offspring. However, when the mutant load is high in these cells, mtDNA disease can ensue (McFarland et al., 2007). Indeed, mtDNA disease is well‐documented in humans and is likely to have existed in other species but has been eradicated due to breeding selection programs where only the fittest or commercially most beneficial lineages survive. Nevertheless, the bottleneck appears to exist in livestock species. Early studies in cattle reported a large drift of mtDNA genotypes across generations of maternally related cows (Laipis et al., 1988; Olivo et al., 1983).
In the pig, naturally occurring mtDNA variants (somatic mutations and deletions), some of which might be deleterious, can persist at high levels in oocytes and preimplantation embryos, and be transmitted from one generation to the next through the female germline (G. Cagnone et al., 2016). To this extent, we identified four naturally occurring mtDNA variants (single base pair deletions) in a mini pig population. Each of the four variants indicated different levels in oocytes and embryos but their levels were suppressed in somatic tissues, especially those with a high requirement for OXPHOS‐derived ATP. Consequently, the levels did not surpass the threshold associated with the phenotypic onset of mtDNA disease (Russell & Turnbull, 2014; Wei & Chinnery, 2020). For example, the onset of Leber's hereditary optic neuropathy requires over 60% of the mtDNA molecules to be mutated in the affected tissue (Chinnery et al., 2001) while 85% mutant mtDNA load is necessary for the onset of Myoclonic epilepsy with ragged red fibers syndrome (Boulet et al., 1992). Interestingly, there were gender‐specific differences for heart and liver tissues for one variant and generational differences for several tissues for the same variant (G. Cagnone et al., 2016). Indeed, the consensus opinion is that mtDNA point mutations associated with mammalian aging are insufficient to cause a phenotypic response (Moore et al., 2020). For example, in hair, the exponential increase in mutation with age ranges from 0 to 1.436 ± 0.2086% of total mtDNA content (Zheng et al., 2012), which, in the context of mtDNA copy number per cell suggests little if no impact on health and well‐being. However, there is evidence from mouse models generated through ooplasmic transfer, where cytoplasm, which includes mitochondria and mtDNA, is transferred from one oocyte into a recipient, suggesting that there are differences in levels of heteroplasmy (the mixing of two genotypes) between generations (Burgstaller et al., 2018) and that the divergence of heteroplasmic molecules in germ and somatic lineages take place early in development (Johnston et al., 2015).
Nevertheless, analysis of the breeding lines of commercial pigs in Australia showed that there was no correlation between the total number of mtDNA variants harbored by each of the mtDNA haplotypes investigated and developmental competence, maturation to metaphase II, fertilization rates, blastocyst rates, and litter size (Tsai et al., 2016). However, when individual variants were assessed at a presence of greater than 25%, there was a negative correlation with oocyte developmental competence; and more specifically with the number of variants present at greater than 25% in the Cytochrome B gene. Furthermore, the level of a variant at position 16,383 in one of the regulatory regions, conserved sequence box II (Figure 1b), correlated positively with mtDNA copy number for developmentally competent oocytes. This particular variant is within the site of interaction for the Mitochondrial Transcription Elongation Factor (Jiang et al., 2019). Termination at this site results in the production of a primer that enables mtDNA replication to proceed.
In all, there is no clear‐cut evidence to suggest that the presence of naturally occurring variants in oocytes is indicative of oocyte quality or the potential of any given oocyte to give rise to offspring. However, it is worth noting that mutations to POLG can result in a host of deleterious mtDNA mutations in oocytes, as demonstrated in the Polg mutator mouse model (Ma et al., 2019). Nevertheless, these should be seen in the context of the mutations associated with the nuclear‐encoded mtDNA replication factors that can give rise to mtDNA disease (Fekete et al., 2019; Rahman & Copeland, 2019; Remtulla et al., 2019). Likewise, it is important to note that high mutant loading of pathogenic variants can lead to spontaneous abortions even at the late stages of gestation (Monnot et al., 2011). Indeed, the persistence of mtDNA in humans might be explained by the high numbers of carriers of mtDNA rearrangements (Elliott et al., 2008) and efficient medical practice to support affected individuals; and the likely elimination in livestock species since affected animals would have been selected against, based on their lack of commercial viability.
6. ALTERING mtDNA COPY NUMBER IN THE OOCYTE
Autologous mitochondrial supplementation primarily arose from the refinement of another assisted reproductive technology, namely cytoplasmic transfer (Cohen et al., 1997), due to the necessity to determine if the cytoplasmic factors introduced into the oocyte that enhanced development were specifically mitochondrial or not. A study employing a pig model used purified populations of mitochondria isolated from mature oocytes and introduced these into oocytes deficient in mtDNA (El Shourbagy et al., 2006). The initial findings showed that fertilization rates were enhanced to match those of oocytes possessing sufficient copies of mtDNA. These experiments highlight the clinical data that showed a relationship between passing a putative threshold (>150,000 copies; Figure 3) for mtDNA copy number and fertilization outcome and subsequent development (May‐Panloup, Chretien, et al., 2005; May‐Panloup, Vignon, et al., 2005; T. A. Santos et al., 2006). Further work has shown that supplementation of mtDNA deficient oocytes not only results in improved fertilization outcomes, but also enhanced blastocyst rates (G. L. M. Cagnone et al., 2016). A similar outcome has been observed when adding the growth factor neuregulin 1 to the oocyte in vitro maturation media (Mao et al., 2012), as was also the case with supplementation of the glycoside mogroside V to the maturation media (Nie et al., 2020). Both resulted in increased mtDNA copy number in mature oocytes and improved embryo developmental competence. There were also significant changes in gene expression profiles of the nuclear genome (approximately 190 genes) at the blastocyst stage of development to the extent that the supplemented populations exhibited gene expression profiles more similar to those of blastocysts from oocytes that had appropriate levels of mtDNA (G. L. M. Cagnone et al., 2016). Moreover, these changes appeared to occur throughout preimplantation development (Tsai & St John, 2018) and arose from differences in gene expression for metaphase II oocytes deficient in mtDNA compared to those with sufficient levels (Tsai et al., 2018).
The outcomes of these studies open the debate as to whether the supplementation process is a cellular, metabolic or genomic event. This has arisen in the context of the use of mitochondria isolated from oogonial stem cells or egg precursor cells (White et al., 2012). The proponents of the approach argue that the additional mitochondria are added to fuel development (Woods et al., 2018), that is, they are adding extra units of energy. However, the process only introduces approximately 780 copies of mtDNA (G. L. M. Cagnone et al., 2016). Normally mtDNA deficient oocytes possess approximately 50,000 copies of mtDNA. If each mitochondrion possesses two copies of mtDNA per mitochondrion (Wai et al., 2008), increasing the mitochondrial number by 390 mitochondria is likely to have very little impact. The more likely outcome is that the addition of extra copies of mtDNA from sister oocytes transformed the minor replication event between fertilization and the 2‐cell stage into a major replication event (G. L. M. Cagnone et al., 2016). Indeed, this replication event increases mtDNA copy number by 4.4‐fold in mtDNA‐deficient oocytes and ensures sufficient mtDNA is present at the blastocyst stage to support subsequent developmental events. A similar outcome was also observed at the blastocyst stage in the aforementioned studies using neuregulin 1 and mogroside V (Mao et al., 2012; Nie et al., 2020); and through a protease inhibitor MG132 and resveratrol on in vitro matured oocytes (Sato et al., 2014). Furthermore, the DNA methylation profiles in the CpG island of POLG are altered with increases in expression for this gene during this short window of mtDNA replication (Tsai et al., 2018). These events take place before the major round of embryonic genome activation in the pig at the 4‐cell stage (K. Lee et al., 2014). In addition, knockdown of POLG2 leads to reduced maturation and blastocyst formation in pig oocytes along with reduced mtDNA copy number (S. K. Lee et al., 2015).
Taken to together, these events, along with changes in nuclear gene expression, suggest that these are genomic rather than metabolic events, although certain gene networks associated within metabolism are affected (G. L. M. Cagnone et al., 2016). Indeed, the use of a model that employs nonselected pig metaphase II oocytes (i.e., does not discriminate between mtDNA deficient and mtDNA‐normal oocytes) shows that mitochondrial supplementation alone induces significant changes in DNA methylation and nuclear gene expression by the blastocyst stage (Okada et al., 2022). Consequently, it appears that mitochondrial supplementation triggers a resetting of the genomic balance that was established between the two genomes during oocyte maturation (St John, 2019). Interestingly, though, it does not appear to affect genes associated with imprinting (Okada et al., 2022).
7. mtDNA REPLICATION AND DNA DEMETHYLATION PROFILES DURING OOGENESIS AND IN THE EARLY EMBRYO
In the context of oogenesis, it appears that the regulation of the synergy between the two genomes is a key to the developmental outcome. During the proliferation and migration of primordial germ cells, DNA methylation is globally erased (Smallwood & Kelsey, 2012). Large‐scale de novo DNA methylation takes place during oogenesis coupled with the significant increases in mtDNA copy number. Following fertilization, there is differential genome‐wide DNA demethylation that takes place in the newly formed embryo based on paternal and maternal contributions (Smallwood & Kelsey, 2012; Stewart et al., 2016). Indeed, early development is marked by significant changes to DNA methylation, other epigenetic regulators and gene expression (von Meyenn et al., 2016). For example, during preimplantation development, the TET family of enzymes ensures the erasure of parental DNA methylation patterns up to the blastocyst stage and the de novo methyltransferases, that is, DNMT3a and DNMT3b, mediate de novo DNA methylation during preimplantation and postimplantation development. At the same time, DNMT1 maintains the predestined profiles (Guo et al., 2014; F. Santos et al., 2002).
The synchronous changes between the nuclear and mitochondrial genomes are key to the concept of cells achieving “genomic balance” (Figure 4) in order that cells function effectively at different stages of development and when mature (St John, 2019). Perturbing genomic balance could result in disease, as shown in models of tumorigenesis with cells being unable to mediate differentiation which requires replication of mtDNA copy to support advanced cellular function (Dickinson et al., 2013). However, genomic balance can be reset through DNA methylation agents such as 5‐Azacytidine and Vitamin C, which can prevent tumorigenesis and allow cells to undergo differentiation with expected increases in mtDNA copy number (Dickinson et al., 2013; Sun & John, 2018; Sun et al., 2018). Likewise, failure of oocytes to acquire sufficient copies of mtDNA leads to fertilization failure or developmental arrest during preimplantation development (May‐Panloup, Chretien, et al., 2005; May‐Panloup, Vignon, et al., 2005; T. A. Santos et al., 2006) though this can be countered through mitochondrial supplementation (G. L. M. Cagnone et al., 2016). Interestingly, the addition of homocysteine, an intermediate in the one‐carbon metabolism that donates methyl groups for methylation processes involved in epigenetic gene regulation, results in reduced mtDNA copy number and affected the quality of porcine oocytes which then recovered through the addition of 5‐Azacytidine (Jia et al., 2019).
Figure 4.
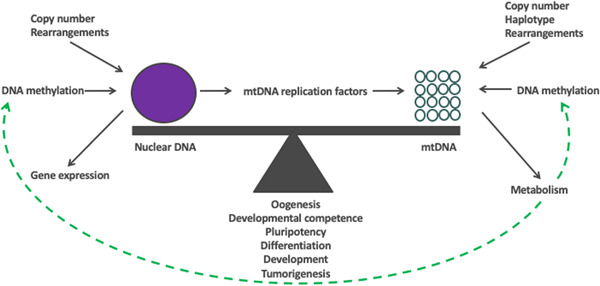
Genomic balance. At key stages in development, a cell strikes a balance between its two genomes in order that it can progress to the next stage of development. This process is mediated by the constant exchange of regulatory information between the nucleus and the mitochondrial genome. This ensures that the cell at any given stage acquires sufficient copies of mtDNA in order that the cell can undertake its specialized function using as much or as little ATP derived through OXPHOS. At the same time, the nuclear genome contributes to genomic balance through epigenetic changes by altering, for example, the levels of DNA methylation, that control gene expression. Other factors include DNA rearrangements, such as mutations and deletions, and copy number variants that will have an impact on phenotype. The mitochondrial genome will contribute through the levels of mtDNA copy number available that will influence the means of cellular metabolism available to the cell, which, is also influenced by the cell's mtDNA haplotype. This results in metabolic factors being released that regulate DNA methylation and other epigenetic modifiers, which act on both the nuclear and mitochondrial genomes. mtDNA, mitochondrial DNA; OXPHOS, oxidative phosphorylation.
8. mtDNA HAPLOTYPES
Over millions of years, different maternal lineages have evolved and, based on their mtDNA sequences, they cluster into groupings known as mtDNA haplotypes (Ruiz‐Pesini et al., 2004). Consequently, a single mtDNA haplotype is defined by groups of mtDNA sequences from individuals that cluster together and define a group's common maternal origins. mtDNA haplotypes can confer advantages and disadvantages to the individual (Gerber et al., 2001; Innocenti et al., 2011). They are thought to predispose individuals to disease (Blanco et al., 2018; Fuku et al., 2007; Koo et al., 2019; Liou et al., 2016; Marom et al., 2017; Shen et al., 2022); influence tolerance to warm and cold climates (Ruiz‐Pesini et al., 2004; Wallace et al., 2003) and high altitude (Ji et al., 2012); sperm motility (Ruiz‐Pesini et al., 2000); the size of the ovarian reserve (May‐Panloup et al., 2014); and overall reproductive capacity and fertility (Sutarno et al., 2002; Tsai et al., 2016). They also influence growth and physical performance (Nagao et al., 1998), milk quality (Brown et al., 1989), and key economic breeding values (EBVs) associated with animal production (St John & Tsai, 2018).
In the context of reproductive capacity, Australian commercial pigs belong to one of five mtDNA haplotypes (A–E) indicative of their origins arising from one of five female founders (St John & Tsai, 2018; Tsai et al., 2016). Three of these founders originated from Asia while two were of European origin. Between the five mtDNA haplotypes, there are approximately 200 single nucleotide variants, which affect both protein coding genes and noncoding regions. These genomic variations result in differences in litter size based on mtDNA haplotypes. Females from haplotypes A and B were predisposed to producing fewer piglets per litter than C, D, and E, while females from haplotypes C (p < 0.05), D (p < 0.01), and E (p < 0.05) were more likely to produce at least three litters of ≥15 piglets per parity than A (Tsai et al., 2016). Furthermore, there were differences in developmental efficiencies among the mtDNA haplotypes. By determining the efficiency of each developmental stage relative to the number of piglets born, haplotype C was the most efficient at converting mature and fertilized oocytes into offspring, and haplotype D was the most efficient at blastocyst conversion. However, haplotypes A and B were the least efficient at converting mature and fertilized oocytes and blastocysts into live offspring. In addition, given the relationship between mtDNA copy number and the ability of an oocyte to mature and develop once fertilized (Spikings et al., 2007), developmentally competent oocytes from each mtDNA haplotype exhibited differences in mtDNA copy number (Tsai et al., 2016). Haplotype D had the highest copy number and haplotype B had the lowest. Similarly, others have seen differences between female pigs having either high or low levels of mtDNA (K. Lee et al., 2014), which suggests a haplotype effect. In all, these outcomes indicate that each haplotype has a different reproductive strategy that impacts on litter size across generations, and is, thus, a heritable trait. In addition, other EBVs such as fat density, muscle depth, fat to leanness ratios, lifetime daily gain, and teat quality were influenced by mtDNA haplotype. There were also gender‐specific effects on teat quality (St John & Tsai, 2018).
9. NUCLEAR TRANSFER AND mtDNA TRANSMISSION
Nuclear transfer primarily involves the transfer of a nucleus, often within a cell, that is required to be propagated into an enucleated oocyte and the reconstructed oocyte is activated (Campbell, 1999). The reconstructed oocyte will then progress to an embryo and divide in much the same manner as a fertilized oocyte. This concept originated from the work of Gurdon and colleagues who were the first to use nuclear transfer to produce frogs with a somatic cell (Gurdon et al., 1958), and, thus, gave rise to the term somatic cell nuclear transfer (SCNT) (Figure 5). This was followed by the transfer of cultured embryonic‐derived (Campbell et al., 1996) and somatic cells to produce the first mammalian offspring, namely Dolly the Sheep (Wilmut et al., 1997); and blastomeres to produce rhesus macaque monkeys (Meng et al., 1997), cattle (Bondioli et al., 1990; Prather et al., 1987), and pigs (Prather et al., 1989).
Figure 5.
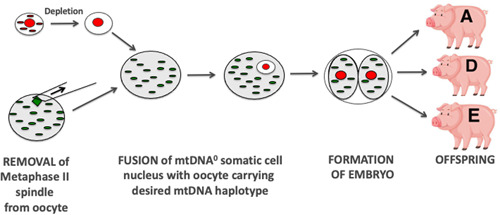
Utilization of SCNT to promote pig breeding lines. The somatic cell is depleted of its mtDNA and introduced into an oocyte containing mtDNA from a specific mtDNA haplotype (e.g., A, D, or E). Once activated, it can develop into an embryo and give rise to offspring. This allows for embryos and offspring to be studied to determine if mtDNA haplotypes influence phenotypic traits with the aim of generating enhanced lines of breeding pigs. mtDNA, mitochondrial DNA; SCNT, somatic cell nuclear transfer.
A key problem is that the mtDNA present in the somatic or embryonic cell accompanies the nucleus (sometimes referred to as “mtDNA carryover”) and, once fused to the enucleated oocyte, it is incorporated into the reconstructed oocyte and can be potentially transmitted to the offspring (Steinborn et al., 1998). The levels of transmission of accompanying mtDNA are random. In large animal models of SCNT, namely sheep, pigs, and cattle, the offspring can inherit from 0% to 59% of their total mtDNA content from the somatic cell (Burgstaller et al., 2007; Takeda et al., 2003, 2006). Where there is a mixing of two diverse mtDNA genotypes (heteroplasmy), there is the potential for the offspring to inherit two mtDNA genomes, namely from the donor cell and recipient oocyte and, thus, from two individuals. If they are of the same or very similar mtDNA haplotypes, the issue of compatibility is not necessarily significant. However, if the genotypes are more distant then compatibility between the haplotypes is an issue (St John et al., 2005; St John et al., 2010). This arises, as within a species, there is variability in the sequences from different mtDNA haplotypes for the encoding genes. This would result in the amino acids contributing to each of the 13 mtDNA‐encoded subunits of the electron transfer chain having different configurations, as shown in a study of pig serial nuclear transfer (St John et al., 2005). Consequently, when two mitochondrial genomes are present, electron transfer chains can be assembled that have different efficiencies for generating ATP through OXPHOS as demonstrated in a pig model of SCNT where discordant interaction between the nucleus and mtDNA affected OXPHOS gene expression (Park et al., 2015); and mouse cybrid (cells derived from the fusion of an mtDNA depleted cell with an enucleated cell) (McKenzie et al., 2003) and pig cybrid (Yu et al., 2015) models.
The question that needs to be addressed is whether mtDNA carryover can be completely avoided. In SCNT, it is possible to deplete the somatic cell's mtDNA using an mtDNA depletion agent to produce embryos (Lloyd et al., 2006; Srirattana & John, 2017) and live offspring that inherit their mtDNA from the oocyte only (J. H. Lee et al., 2010), as in the case with natural conception (Figure 5). Consequently, it is possible to generate animals with a mixture of traits present in the nuclear genome and match these with desired traits associated with the mitochondrial genome that would give enhanced breeding lines. In the pig, this might constitute an animal with desired meat quality that would be transmitted through the nuclear genome and matching with an mtDNA haplotype associated with enhanced oocyte quality and litter size (Figure 5). In countries where the import of new pig genetics through breeding lines is not allowed, for example, Australia, this offers an opportunity to increase and diversify breeding lines and expand pig genetics by alerting genomic combinations that might require large numbers of breeding rounds to achieve.
10. CONCLUSION
In the context of understanding the role of the mitochondrial genome in the oocyte and embryo, the pig has proven to be an invaluable model. It has shown how mtDNA copy number is strictly regulated in the oocyte and developing embryo and the importance of oocyte mtDNA copy number and variants to the developmental outcome. Furthermore, it highlights how mtDNA haplotypes influence the developmental outcome and how the association of mtDNA haplotypes and traits could be exploited to produce animals with enhanced phenotypes. Nonetheless, the knowledge gained from the pig oocyte and embryo with regard to mtDNA variants and genetics is not only relevant in terms of generating human preclinical models. It has specific relevance to pig reproductive capacity. In many respects, the outcomes are waiting for the pig industry to use them to enhance pig production, capacity, and quality.
AUTHOR CONTRIBUTIONS
Justin C. St John: Conceptualization; funding acquisition; writing—original draft; formal analysis; writing—review & editing. Takashi Okada: Writing—review & editing; writing—original draft; investigation; formal analysis. Eryk Andreas: Writing—original draft; writing—review & editing; investigation; formal analysis. Alexander Penn: Investigation; writing—original draft; writing—review & editing; formal analysis.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
ACKNOWLEDGMENTS
We are grateful to the National Health and Medical Research Council for funding grants GNT1136065, GNT1160106, and GNT2000723 which supported this study. This study was supported by the National Health and Medical Research Council grants GNT 1160106 and GNT 1136065. Open access publishing facilitated by The University of Adelaide, as part of the Wiley ‐ The University of Adelaide agreement via the Council of Australian University Librarians.Open access publishing facilitated by The University of Adelaide, as part of the Wiley ‐ The University of Adelaide agreement via the Council of Australian University Librarians.
St John, J. C. , Okada, T. , Andreas, E. , & Penn, A. (2023). The role of mtDNA in oocyte quality and embryo development. Molecular Reproduction and Development, 90, 621–633. 10.1002/mrd.23640
Open access publishing facilitated by The University of Queensland, as part of the Wiley ‐ The University of Queensland agreement via the Council of Australian University Librarians.
REFERENCES
- Amaral, A. , Ramalho‐Santos, J. , & St John, J. C. (2007). The expression of polymerase gamma and mitochondrial transcription factor A and the regulation of mitochondrial DNA content in mature human sperm. Human Reproduction, 22(6), 1585–1596. 10.1093/humrep/dem030 [DOI] [PubMed] [Google Scholar]
- Anderson, S. , Bankier, A. T. , Barrell, B. G. , de Bruijn, M. H. , Coulson, A. R. , Drouin, J. , Eperon, I. C. , Nierlich, D. P. , Roe, B. A. , Sanger, F. , Schreier, P. H. , Smith, A. J. , Staden, R. , & Young, I. G. (1981). Sequence and organization of the human mitochondrial genome. Nature, 290(5806), 457–465. http://www.ncbi.nlm.nih.gov/pubmed/7219534 [DOI] [PubMed] [Google Scholar]
- Bibb, M. J. , Etten, V. , Wright, R. A. , Walberg, C. T., M. W. , & Clayton, D. A. (1981). Sequence and gene organization of mouse mitochondrial DNA. Cell, 26(2, Pt 2), 167–180. 10.1016/0092-8674(81)90300-7 [DOI] [PubMed] [Google Scholar]
- Blanco, F. J. , Valdes, A. M. , & Rego‐Perez, I. (2018). Mitochondrial DNA variation and the pathogenesis of osteoarthritis phenotypes. Nature Reviews Rheumatology, 14(6), 327–340. 10.1038/s41584-018-0001-0 [DOI] [PubMed] [Google Scholar]
- Bode, G. , Clausing, P. , Gervais, F. , Loegsted, J. , Luft, J. , Nogues, V. , & Sims, J. (2010). The utility of the minipig as an animal model in regulatory toxicology. Journal of Pharmacological Toxicol Methods, 62(3), 196–220. 10.1016/j.vascn.2010.05.009 [DOI] [PubMed] [Google Scholar]
- Bondioli, K. R. , Westhusin, M. E. , & Looney, C. R. (1990). Production of identical bovine offspring by nuclear transfer. Theriogenology, 33(1), 165–174. 10.1016/0093-691X(90)90607-U [DOI] [Google Scholar]
- Boulet, L. , Karpati, G. , & Shoubridge, E. A. (1992). Distribution and threshold expression of the tRNA(Lys) mutation in skeletal muscle of patients with myoclonic epilepsy and ragged‐red fibers (MERRF). American Journal of Human Genetics, 51(6), 1187–1200. http://www.ncbi.nlm.nih.gov/pubmed/1334369 [PMC free article] [PubMed] [Google Scholar]
- Brown, D. R. , Koehler, C. M. , Lindberg, G. L. , Freeman, A. E. , Mayfield, J. E. , Myers, A. M. , Schutz, M. M. , & Beitz, D. C. (1989). Molecular analysis of cytoplasmic genetic variation in Holstein cows. Journal of Animal Science, 67(8), 1926–1932. http://www.ncbi.nlm.nih.gov/pubmed/2571604 [DOI] [PubMed] [Google Scholar]
- Bui, H. T. , Van Thuan, N. , Kwon, D. N. , Choi, Y. J. , Kang, M. H. , Han, J. W. , Kim, T. , & Kim, J. H. (2014). Identification and characterization of putative stem cells in the adult pig ovary. Development, 141(11), 2235–2244. 10.1242/dev.104554 [DOI] [PubMed] [Google Scholar]
- Burgstaller, J. P. , Kolbe, T. , Havlicek, V. , Hembach, S. , Poulton, J. , Piálek, J. , Steinborn, R. , Rülicke, T. , Brem, G. , Jones, N. S. , & Johnston, I. G. (2018). Large‐scale genetic analysis reveals mammalian mtDNA heteroplasmy dynamics and variance increase through lifetimes and generations. Nature Communications, 9(1), 2488. 10.1038/s41467-018-04797-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgstaller, J. P. , Schinogl, P. , Dinnyes, A. , Muller, M. , & Steinborn, R. (2007). Mitochondrial DNA heteroplasmy in ovine fetuses and sheep cloned by somatic cell nuclear transfer. BMC Developmental Biology, 7, 141. 10.1186/1471-213X-7-141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cagnone, G. , Tsai, T. S. , Srirattana, K. , Rossello, F. , Powell, D. R. , Rohrer, G. , & St John, J. C. (2016). Segregation of naturally occurring mitochondrial DNA variants in a mini‐pig model. Genetics, 202(3), 931–944. 10.1534/genetics.115.181321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cagnone, G. L. M. , Tsai, T. S. , Makanji, Y. , Matthews, P. , Gould, J. , Bonkowski, M. S. , Elgass, K. D. , Wong, A. S. A. , Wu, L. E. , McKenzie, M. , Sinclair, D. A. , & John, J. C. S. (2016). Restoration of normal embryogenesis by mitochondrial supplementation in pig oocytes exhibiting mitochondrial DNA deficiency. Scientific Reports, 6, 23229. 10.1038/srep23229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell, K. H. (1999). Nuclear transfer in farm animal species. Seminars in Cell and Developmental Biology, 10(3), 245–252. 10.1006/scdb.1999.0310 [DOI] [PubMed] [Google Scholar]
- Campbell, K. H. , McWhir, J. , Ritchie, W. A. , & Wilmut, I. (1996). Sheep cloned by nuclear transfer from a cultured cell line. Nature, 380(6569), 64–66. 10.1038/380064a0 [DOI] [PubMed] [Google Scholar]
- Cao, L. , Shitara, H. , Horii, T. , Nagao, Y. , Imai, H. , Abe, K. , Hara, T. , Hayashi, J. I. , & Yonekawa, H. (2007). The mitochondrial bottleneck occurs without reduction of mtDNA content in female mouse germ cells. Nature Genetics, 39(3), 386–390. 10.1038/ng1970 [DOI] [PubMed] [Google Scholar]
- Carrodeguas, J. A. , Kobayashi, R. , Lim, S. E. , Copeland, W. C. , & Bogenhagen, D. F. (1999). The accessory subunit of Xenopus laevis mitochondrial DNA polymerase gamma increases processivity of the catalytic subunit of human DNA polymerase gamma and is related to class II aminoacyl‐tRNA synthetases. Molecular and Cellular Biology, 19(6), 4039–4046. http://www.ncbi.nlm.nih.gov/pubmed/10330144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatre, L. , & Ricchetti, M. (2013). Prevalent coordination of mitochondrial DNA transcription and initiation of replication with the cell cycle. Nucleic Acids Research, 41(5), 3068–3078. 10.1093/nar/gkt015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnery, P. F. , Andrews, R. M. , Turnbull, D. M. , & Howell, N. N. (2001). Leber hereditary optic neuropathy: Does heteroplasmy influence the inheritance and expression of the G11778A mitochondrial DNA mutation? American Journal of Medical Genetics, 98(3), 235–243. [DOI] [PubMed] [Google Scholar]
- Cohen, J. , Scott, R. , Schimmel, T. , Levron, J. , & Willadsen, S. (1997). Birth of infant after transfer of anucleate donor oocyte cytoplasm into recipient eggs. The Lancet, 350(9072), 186–187. [DOI] [PubMed] [Google Scholar]
- Cree, L. M. , Samuels, D. C. , de Sousa Lopes, S. C. , Rajasimha, H. K. , Wonnapinij, P. , Mann, J. R. , Dahl, H. H. , & Chinnery, P. F. (2008). A reduction of mitochondrial DNA molecules during embryogenesis explains the rapid segregation of genotypes. Nature Genetics, 40(2), 249–254. 10.1038/ng.2007.63 [DOI] [PubMed] [Google Scholar]
- Dickinson, A. , Yeung, K. Y. , Donoghue, J. , Baker, M. J. , Kelly, R. D. , McKenzie, M. , Johns, T. G. , & St John, J. C. (2013). The regulation of mitochondrial DNA copy number in glioblastoma cells. Cell Death and Differentiation, 20(12), 1644–1653. 10.1038/cdd.2013.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott, H. R. , Samuels, D. C. , Eden, J. A. , Relton, C. L. , & Chinnery, P. F. (2008). Pathogenic mitochondrial DNA mutations are common in the general population. American Journal of Human Genetics, 83(2), 254–260. 10.1016/j.ajhg.2008.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Shourbagy, S. H. , Spikings, E. C. , Freitas, M. , & St John, J. C. (2006). Mitochondria directly influence fertilisation outcome in the pig. Reproduction, 131(2), 233–245. 10.1530/rep.1.00551 [DOI] [PubMed] [Google Scholar]
- Facucho‐Oliveira, J. M. , Alderson, J. , Spikings, E. C. , Egginton, S. , & St John, J. C. (2007). Mitochondrial DNA replication during differentiation of murine embryonic stem cells. Journal of Cell Science, 120(22), 4025–4034. 10.1242/jcs.016972 [DOI] [PubMed] [Google Scholar]
- Falkenberg, M. , Gaspari, M. , Rantanen, A. , Trifunovic, A. , Larsson, N. G. , & Gustafsson, C. M. (2002). Mitochondrial transcription factors B1 and B2 activate transcription of human mtDNA. Nature Genetics, 31(3), 289–294. 10.1038/ng909 [DOI] [PubMed] [Google Scholar]
- Fekete, B. , Pentelényi, K. , Rudas, G. , Gál, A. , Grosz, Z. , Illés, A. , Idris, J. , Csukly, G. , Domonkos, A. , & Molnar, M. J. (2019). Broadening the phenotype of the TWNK gene associated Perrault syndrome. BMC Medical Genetics, 20(1), 198. 10.1186/s12881-019-0934-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher, R. P. , & Clayton, D. A. (1988). Purification and characterization of human mitochondrial transcription factor 1. Molecular and Cellular Biology, 8(8), 3496–3509. http://www.ncbi.nlm.nih.gov/pubmed/3211148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floros, V. I. , Pyle, A. , Dietmann, S. , Wei, W. , Tang, W. , Irie, N. , Payne, B. , Capalbo, A. , Noli, L. , Coxhead, J. , Hudson, G. , Crosier, M. , Strahl, H. , Khalaf, Y. , Saitou, M. , Ilic, D. , Surani, M. A. , & Chinnery, P. F. (2018). Segregation of mitochondrial DNA heteroplasmy through a developmental genetic bottleneck in human embryos. Nature Cell Biology, 20(2), 144–151. 10.1038/s41556-017-0017-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fragouli, E. , Spath, K. , Alfarawati, S. , Kaper, F. , Craig, A. , Michel, C. E. , Kokocinski, F. , Cohen, J. , Munne, S. , & Wells, D. (2015). Altered levels of mitochondrial DNA are associated with female age, aneuploidy, and provide an independent measure of embryonic implantation potential. PLoS Genetics, 11(6), e1005241. 10.1371/journal.pgen.1005241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuku, N. , Park, K. S. , Yamada, Y. , Nishigaki, Y. , Cho, Y. M. , Matsuo, H. , Segawa, T. , Watanabe, S. , Kato, K. , Yokoi, K. , Nozawa, Y. , Lee, H. K. , & Tanaka, M. (2007). Mitochondrial haplogroup N9a confers resistance against type 2 diabetes in Asians. American Journal of Human Genetics, 80, 407–415. 10.1086/512202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber, A. S. , Loggins, R. , Kumar, S. , & Dowling, T. E. (2001). Does nonneutral evolution shape observed patterns of DNA variation in animal mitochondrial genomes? Annual Review of Genetics, 35, 539–566. 10.1146/annurev.genet.35.102401.09110635/1/539, [pii] [DOI] [PubMed] [Google Scholar]
- Giles, R. E. , Blanc, H. , Cann, H. M. , & Wallace, D. C. (1980). Maternal inheritance of human mitochondrial DNA. Proceedings of the National Academy of Sciences of the United States of America, 77(11), 6715–6719. http://www.ncbi.nlm.nih.gov/pubmed/6256757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, F. , Li, X. , Liang, D. , Li, T. , Zhu, P. , Guo, H. , Wu, X. , Wen, L. , Gu, T. P. , Hu, B. , Walsh, C. P. , Li, J. , Tang, F. , & Xu, G. L. (2014). Active and passive demethylation of male and female pronuclear DNA in the mammalian zygote. Cell Stem Cell, 15(4), 447–459. 10.1016/j.stem.2014.08.003 [DOI] [PubMed] [Google Scholar]
- Gurdon, J. B. , Elsdale, T. R. , & Fischberg, M. (1958). Sexually mature individuals of Xenopus laevis from the transplantation of single somatic nuclei. Nature, 182(4627), 64–65. http://www.ncbi.nlm.nih.gov/pubmed/13566187 [DOI] [PubMed] [Google Scholar]
- Hammond, E. R. , McGillivray, B. C. , Wicker, S. M. , Peek, J. C. , Shelling, A. N. , Stone, P. , Chamley, L. W. , & Cree, L. M. (2017). Characterizing nuclear and mitochondrial DNA in spent embryo culture media: Genetic contamination identified. Fertility and Sterility, 107(1), 220–228 e225. 10.1016/j.fertnstert.2016.10.015 [DOI] [PubMed] [Google Scholar]
- Hillen, H. S. , Parshin, A. V. , Agaronyan, K. , Morozov, Y. I. , Graber, J. J. , Chernev, A. , Schwinghammer, K. , Urlaub, H. , Anikin, M. , Cramer, P. , & Temiakov, D. (2017). Mechanism of transcription anti‐termination in human mitochondria. Cell, 171(5), 1082–1093 e1013. 10.1016/j.cell.2017.09.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houghton, F. D. (2006). Energy metabolism of the inner cell mass and trophectoderm of the mouse blastocyst. Differentiation, 74(1), 11–18. [DOI] [PubMed] [Google Scholar]
- Humpherson, P. G. , Leese, H. J. , & Sturmey, R. G. (2005). Amino acid metabolism of the porcine blastocyst. Theriogenology, 64(8), 1852–1866. 10.1016/j.theriogenology.2005.04.019 [DOI] [PubMed] [Google Scholar]
- Innocenti, P. , Morrow, E. H. , & Dowling, D. K. (2011). Experimental evidence supports a sex‐specific selective sieve in mitochondrial genome evolution. Science, 332(6031), 845–848. 10.1126/science.1201157 [DOI] [PubMed] [Google Scholar]
- Ji, F. , Sharpley, M. S. , Derbeneva, O. , Alves, L. S. , Qian, P. , Wang, Y. , Chalkia, D. , Lvova, M. , Xu, J. , Yao, W. , Simon, M. , Platt, J. , Xu, S. , Angelin, A. , Davila, A. , Huang, T. , Wang, P. H. , Chuang, L. M. , Moore, L. G. , … Wallace, D. C. (2012). Mitochondrial DNA variant associated with Leber hereditary optic neuropathy and high‐altitude Tibetans. Proceedings of the National Academy of Sciences of the United States of America, 109(19), 7391–7396. 10.1073/pnas.1202484109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia, L. , Zeng, Y. , Hu, Y. , Liu, J. , Yin, C. , Niu, Y. , Wang, C. , Li, J. , Jia, Y. , Hong, J. , & Zhao, R. (2019). Homocysteine impairs porcine oocyte quality via deregulation of one‐carbon metabolism and hypermethylation of mitochondrial DNA. Biology of Reproduction, 100(4), 907–916. 10.1093/biolre/ioy238 [DOI] [PubMed] [Google Scholar]
- Jiang, S. , Koolmeister, C. , Misic, J. , Siira, S. , Kühl, I. , Silva Ramos, E. , Miranda, M. , Jiang, M. , Posse, V. , Lytovchenko, O. , Atanassov, I. , Schober, F. A. , Wibom, R. , Hultenby, K. , Milenkovic, D. , Gustafsson, C. M. , Filipovska, A. , & Larsson, N. G. (2019). TEFM regulates both transcription elongation and RNA processing in mitochondria. EMBO Reports, 20(6):e48101. 10.15252/embr.201948101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston, I. G. , Burgstaller, J. P. , Havlicek, V. , Kolbe, T. , Rülicke, T. , Brem, G. , Poulton, J. , & Jones, N. S. (2015). Stochastic modelling, Bayesian inference, and new in vivo measurements elucidate the debated mtDNA bottleneck mechanism. eLife, 4, e07464. 10.7554/eLife.07464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly, R. D. , Sumer, H. , McKenzie, M. , Facucho‐Oliveira, J. , Trounce, I. A. , Verma, P. J. , & St John, J. C. (2013). The effects of nuclear reprogramming on mitochondrial DNA replication. Stem Cell Reviews, 9(1), 1–15. 10.1007/s12015-011-9318-7 [DOI] [PubMed] [Google Scholar]
- Koo, B. S. , Song, Y. , Lee, S. , Sung, Y. K. , Shin, K. J. , Cho, N. H. , & Jun, J. B. (2019). Association of Asian mitochondrial DNA haplogroup B with new development of knee osteoarthritis in Koreans. International Journal of Rheumatic Diseases, 22(3), 411–416. 10.1111/1756-185X.13453 [DOI] [PubMed] [Google Scholar]
- Korhonen, J. A. , Gaspari, M. , & Falkenberg, M. (2003). TWINKLE has 5' → 3' DNA helicase activity and is specifically stimulated by mitochondrial single‐stranded DNA‐binding protein. Journal of Biological Chemistry, 278(49), 48627–48632. 10.1074/jbc.M306981200 [DOI] [PubMed] [Google Scholar]
- Krisher, R. L. , & Prather, R. S. (2012). A role for the Warburg effect in preimplantation embryo development: metabolic modification to support rapid cell proliferation. Molecular Reproduction and Development, 79(5), 311–320. 10.1002/mrd.22037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucej, M. , & Butow, R. A. (2007). Evolutionary tinkering with mitochondrial nucleoids. Trends in Cell Biology, 17(12), 586–592. 10.1016/j.tcb.2007.08.007 [DOI] [PubMed] [Google Scholar]
- Laipis, P. J. , Van de Walle, M. J. , & Hauswirth, W. W. (1988). Unequal partitioning of bovine mitochondrial genotypes among siblings. Proceedings of the National Academy of Sciences of the United States of America, 85(21), 8107–8110. 10.1073/pnas.85.21.8107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen, M. O. , & Rolin, B. (2004). Use of the Gottingen minipig as a model of diabetes, with special focus on type 1 diabetes research. Institute for Laboratory Animal Research Journal, 45(3), 303–313. http://www.ncbi.nlm.nih.gov/pubmed/15229377 [DOI] [PubMed] [Google Scholar]
- Lee, J. H. , Peters, A. , Fisher, P. , Bowles, E. J. , St John, J. C. , & Campbell, K. H. (2010). Generation of mtDNA homoplasmic cloned lambs. Cellular Reprogramming, 12(3), 347–355. 10.1089/cell.2009.0096 [DOI] [PubMed] [Google Scholar]
- Lee, K. , Hamm, J. , Whitworth, K. , Spate, L. , Park, K. W. , Murphy, C. N. , & Prather, R. S. (2014). Dynamics of TET family expression in porcine preimplantation embryos is related to zygotic genome activation and required for the maintenance of NANOG. Developmental Biology, 386(1), 86–95. 10.1016/j.ydbio.2013.11.024 [DOI] [PubMed] [Google Scholar]
- Lee, S. K. , Zhao, M. H. , Kwon, J. W. , Li, Y. H. , Lin, Z. L. , Jin, Y. X. , & Cui, X. S. (2014). The association of mitochondrial potential and copy number with pig oocyte maturation and developmental potential. Journal of Reproduction and Development, 60(2), 128–135. 10.1262/jrd.2013-098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, S. K. , Zhao, M. H. , Zheng, Z. , Kwon, J. W. , Liang, S. , Kim, S. H. , Kim, N. H. , & Cui, X. S. (2015). Polymerase subunit gamma 2 affects porcine oocyte maturation and subsequent embryonic development. Theriogenology, 83(1), 121–130. 10.1016/j.theriogenology.2014.08.019 [DOI] [PubMed] [Google Scholar]
- Liou, C. W. , Chuang, J. H. , Chen, J. B. , Tiao, M. M. , Wang, P. W. , Huang, S. T. , Huang, T. L. , Lee, W. C. , Weng, S. W. , Huang, P. H. , Chen, S. D. , Chen, R. S. , Lu, C. S. , & Lin, T. K. (2016). Mitochondrial DNA variants as genetic risk factors for parkinson disease. European Journal of Neurology, 23(8), 1289–1300. 10.1111/ene.13020. [DOI] [PubMed] [Google Scholar]
- Lloyd, R. E. , Lee, J. H. , Alberio, R. , Bowles, E. J. , Ramalho‐Santos, J. , Campbell, K. H. , & St John, J. C. (2006). Aberrant nucleo‐cytoplasmic cross‐talk results in donor cell mtDNA persistence in cloned embryos. Genetics, 172(4), 2515–2527. 10.1534/genetics.105.055145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, H. , Hayama, T. , Van Dyken, C. , Darby, H. , Koski, A. , Lee, Y. , Gutierrez, N. M. , Yamada, S. , Li, Y. , Andrews, M. , Ahmed, R. , Liang, D. , Gonmanee, T. , Kang, E. , Nasser, M. , Kempton, B. , Brigande, J. , McGill, T. J. , Terzic, A. , … Mitalipov, S. (2019). Biology of Reproduction, 102(3), 607–619. 10.1093/biolre/ioz202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao, J. , Whitworth, K. M. , Spate, L. D. , Walters, E. M. , Zhao, J. , & Prather, R. S. (2012). Regulation of oocyte mitochondrial DNA copy number by follicular fluid, EGF, and neuregulin 1 during in vitro maturation affects embryo development in pigs. Theriogenology, 78(4), 887–897. 10.1016/j.theriogenology.2012.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchington, D. R. , Macaulay, V. , Hartshorne, G. M. , Barlow, D. , & Poulton, J. (1998). Evidence from human oocytes for a genetic bottleneck in an mtDNA disease. American Journal of Human Genetics, 63(3), 769–775. 10.1086/302009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marom, S. , Friger, M. , & Mishmar, D. (2017). MtDNA meta‐analysis reveals both phenotype specificity and allele heterogeneity: a model for differential association. Scientific Reports, 7, 43449. 10.1038/srep43449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- May‐Panloup, P. , Chretien, M. F. , Jacques, C. , Vasseur, C. , Malthiery, Y. , & Reynier, P. (2005). Low oocyte mitochondrial DNA content in ovarian insufficiency. Human Reproduction, 20(3), 593–597. 10.1093/humrep/deh667 [DOI] [PubMed] [Google Scholar]
- May‐Panloup, P. , Desquiret, V. , Morinière, C. , Ferré‐L'hôtellier, V. , Lemerle, S. , Boucret, L. , Lehais, S. , Chao de la Barca, J. M. , Descamps, P. , Procaccio, V. , & Reynier, P. (2014). Mitochondrial macro‐haplogroup JT may play a protective role in ovarian ageing. Mitochondrion, 18, 1–6. 10.1016/j.mito.2014.08.002 [DOI] [PubMed] [Google Scholar]
- May‐Panloup, P. , Vignon, X. , Chretien, M. F. , Heyman, Y. , Tamassia, M. , Malthiery, Y. , & Reynier, P. (2005). Increase of mitochondrial DNA content and transcripts in early bovine embryogenesis associated with upregulation of mtTFA and NRF1 transcription factors. Reproductive Biology and Endocrinology, 3, 65. 10.1186/1477-7827-3-65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell, J. M. , & Petrie, L. (2004). Mitochondrial DNA turnover occurs during preimplantation development and can be modulated by environmental factors. Reproductive BioMedicine Online, 9(4), 418–424. http://www.ncbi.nlm.nih.gov/pubmed/15511342 [DOI] [PubMed] [Google Scholar]
- McFarland, R. , Taylor, R. W. , & Turnbull, D. M. (2007). Mitochondrial disease‐‐its impact, etiology, and pathology. Current Topics in Developmental Biology, 77, 113–155. 10.1016/S0070-2153(06)77005-3 [DOI] [PubMed] [Google Scholar]
- McKenzie, M. , Chiotis, M. , Pinkert, C. A. , & Trounce, I. A. (2003). Functional respiratory chain analyses in murid xenomitochondrial cybrids expose coevolutionary constraints of cytochrome b and nuclear subunits of complex III. Molecular Biology and Evolution, 20(7), 1117–1124. 10.1093/molbev/msg132 [DOI] [PubMed] [Google Scholar]
- Meng, L. , Ely, J. J. , Stouffer, R. L. , & Wolf, D. P. (1997). Rhesus monkeys produced by nuclear transfer. Biology of Reproduction, 57(2), 454–459. 10.1095/biolreprod57.2.454 [DOI] [PubMed] [Google Scholar]
- Miller, F. J. , Rosenfeldt, F. L. , Zhang, C. , Linnane, A. W. , & Nagley, P. (2003). Precise determination of mitochondrial DNA copy number in human skeletal and cardiac muscle by a PCR‐based assay: Lack of change of copy number with age. Nucleic Acids Research, 31(11), e61. http://www.ncbi.nlm.nih.gov/pubmed/12771225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monnot, S. , Gigarel, N. , Samuels, D. C. , Burlet, P. , Hesters, L. , Frydman, N. , Frydman, R. , Kerbrat, V. , Funalot, B. , Martinovic, J. , Benachi, A. , Feingold, J. , Munnich, A. , Bonnefont, J. P. , & Steffann, J. (2011). Segregation of mtDNA throughout human embryofetal development: m.3243A>G as a model system. Human Mutation, 32(1), 116–125. 10.1002/humu.21417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore, T. M. , Zhou, Z. , Strumwasser, A. R. , Cohn, W. , Lin, A. J. , Cory, K. , Whitney, K. , Ho, T. , Ho, T. , Lee, J. L. , Rucker, D. H. , Hoang, A. N. , Widjaja, K. , Abrishami, A. D. , Charugundla, S. , Stiles, L. , Whitelegge, J. P. , Turcotte, L. P. , Wanagat, J. , & Hevener, A. L. (2020). Age‐induced mitochondrial DNA point mutations are inadequate to alter metabolic homeostasis in response to nutrient challenge. Aging cell, 19, e13166. 10.1111/acel.13166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyes, C. D. , Battersby, B. J. , & Leary, S. C. (1998). Regulation of muscle mitochondrial design. Journal of Experimental Biology, 201(3), 299–307. http://www.ncbi.nlm.nih.gov/pubmed/9503641 [PubMed] [Google Scholar]
- Nagao, Y. , Totsuka, Y. , Atomi, Y. , Kaneda, H. , Lindahl, K. F. , Imai, H. , & Yonekawa, H. (1998). Decreased physical performance of congenic mice with mismatch between the nuclear and the mitochondrial genome. Genes and Genetic Systems, 73(1), 21–27. http://www.ncbi.nlm.nih.gov/pubmed/9546205 [DOI] [PubMed] [Google Scholar]
- Nguyen, D. H. , Jaszczak, R. G. , & Laird, D. J. (2019). Heterogeneity of primordial germ cells. Current Topics in Developmental Biology, 135, 155–201. 10.1016/bs.ctdb.2019.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie, J. , Yan, K. , Sui, L. , Zhang, H. , Zhang, H. , Yang, X. , Lu, S. , Lu, K. , & Liang, X. (2020). Mogroside V improves porcine oocyte in vitro maturation and subsequent embryonic development. Theriogenology, 141, 35–40. 10.1016/j.theriogenology.2019.09.010 [DOI] [PubMed] [Google Scholar]
- Okada, T. , McIlfatrick, S. , Hin, N. , Aryamanesh, N. , Breen, J. , & St John, J. C. (2022). Mitochondrial supplementation of Sus scrofa metaphase II oocytes alters DNA methylation and gene expression profiles of blastocysts. Epigenetics & Chromatin, 15(1), 12. 10.1186/s13072-022-00442-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivo, P. D. , de Walle, V. , Laipis, M. J., P. J. , & Hauswirth, W. W. (1983). Nucleotide sequence evidence for rapid genotypic shifts in the bovine mitochondrial DNA D‐loop. Nature, 306(5941), 400–402. 10.1038/306400a0 [DOI] [PubMed] [Google Scholar]
- Park, J. , Lai, L. , Samuel, M. S. , Wax, D. , Prather, R. S. , & Tian, X. (2015). Disruption of mitochondrion‐to‐nucleus interaction in deceased cloned piglets. PLoS One, 10(6), e0129378. 10.1371/journal.pone.0129378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perleberg, C. , Kind, A. , & Schnieke, A. (2018). Genetically engineered pigs as models for human disease. Disease Models & Mechanisms, 11(1), dmm030783. 10.1242/dmm.030783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prather, R. S. , Barnes, F. L. , Sims, M. M. , Robl, J. M. , Eyestone, W. H. , & First, N. L. (1987). Nuclear transplantation in the bovine embryo: assessment of donor nuclei and recipient oocyte. Biology of Reproduction, 37(4), 859–866. 10.1095/biolreprod37.4.859 [DOI] [PubMed] [Google Scholar]
- Prather, R. S. , Sims, M. M. , & First, N. L. (1989). Nuclear transplantation in early pig embryos. Biology of Reproduction, 41(3), 414–418. 10.1095/biolreprod41.3.414 [DOI] [PubMed] [Google Scholar]
- Rahman, S. , & Copeland, W. C. (2019). POLG‐related disorders and their neurological manifestations. Nature Reviews Neurology, 15(1), 40–52. 10.1038/s41582-018-0101-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remtulla, S. , Emilie Nguyen, C. T. , Prasad, C. , & Campbell, C. (2019). Twinkle‐associated mitochondrial DNA depletion. Pediatric Neurology, 90, 61–65. 10.1016/j.pediatrneurol.2018.08.007 [DOI] [PubMed] [Google Scholar]
- Ruiz‐Pesini, E. , Lapena, A. C. , Diez‐Sanchez, C. , Perez‐Martos, A. , Montoya, J. , Alvarez, E. , & Enriquez, J. A. (2000). Human mtDNA haplogroups associated with high or reduced spermatozoa motility. American Journal of Human Genetics, 67(3), 682–696. 10.1086/303040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz‐Pesini, E. , Mishmar, D. , Brandon, M. , Procaccio, V. , & Wallace, D. C. (2004). Effects of purifying and adaptive selection on regional variation in human mtDNA. Science, 303(5655), 223–226. 10.1126/science.1088434 [DOI] [PubMed] [Google Scholar]
- Russell, O. , & Turnbull, D. (2014). Mitochondrial DNA disease‐molecular insights and potential routes to a cure. Experimental Cell Research, 325(1), 38–43. 10.1016/j.yexcr.2014.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos, F. , Hendrich, B. , Reik, W. , & Dean, W. (2002). Dynamic reprogramming of DNA methylation in the early mouse embryo. Developmental Biology, 241(1), 172–182. 10.1006/dbio.2001.0501 [DOI] [PubMed] [Google Scholar]
- Santos, T. A. , El Shourbagy, S. , & St John, J. C. (2006). Mitochondrial content reflects oocyte variability and fertilization outcome. Fertility and Sterility, 85(3), 584–591. 10.1016/j.fertnstert.2005.09.017 [DOI] [PubMed] [Google Scholar]
- Sato, D. , Itami, N. , Tasaki, H. , Takeo, S. , Kuwayama, T. , & Iwata, H. (2014). Relationship between mitochondrial DNA copy number and SIRT1 expression in porcine oocytes. PLoS One, 9(4), e94488. 10.1371/journal.pone.0094488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, F. C. , Weng, S. W. , Tsai, M. H. , Su, Y. J. , Li, S. C. , Chang, S. J. , Chen, J. F. , Chang, Y. H. , Liou, C. W. , Lin, T. K. , Chuang, J. H. , Lin, C. Y. , & Wang, P. W. (2022). Mitochondrial haplogroups have a better correlation to insulin requirement than nuclear genetic variants for type 2 diabetes mellitus in Taiwanese individuals. Journal of Diabetes Investigation, 13(1), 201–208. 10.1111/jdi.13629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smallwood, S. A. , & Kelsey, G. (2012). De novo DNA methylation: a germ cell perspective. Trends in Genetics, 28(1), 33–42. 10.1016/j.tig.2011.09.004 [DOI] [PubMed] [Google Scholar]
- Spikings, E. C. , Alderson, J. , & St John, J. C. (2007). Regulated mitochondrial DNA replication during oocyte maturation is essential for successful porcine embryonic development. Biology of Reproduction, 76(2), 327–335. 10.1095/biolreprod.106.054536 [DOI] [PubMed] [Google Scholar]
- Srirattana, K. , & John, J. C. (2017). Manipulating the mitochondrial genome to enhance cattle embryo development. G3: Genes|Genomes|Genetics, 7(7), 2065–2080. 10.1534/g3.117.042655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinborn, R. , Zakhartchenko, V. , Wolf, E. , Muller, M. , & Brem, G. (1998). Non‐balanced mix of mitochondrial DNA in cloned cattle produced by cytoplast‐blastomere fusion. FEBS Letters, 426(3), 357–361. 10.1016/s0014-5793(98)00351-2 [DOI] [PubMed] [Google Scholar]
- Stewart, K. R. , Veselovska, L. , & Kelsey, G. (2016). Establishment and functions of DNA methylation in the germline. Epigenomics, 8(10), 1399–1413. 10.2217/epi-2016-0056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- St John, J. C. (2016). Mitochondrial DNA copy number and replication in reprogramming and differentiation. Seminars in Cell and Developmental Biology, 52, 93–101. 10.1016/j.semcdb.2016.01.028 [DOI] [PubMed] [Google Scholar]
- St John, J. C. (2019). Genomic balance: two genomes establishing synchrony to modulate cellular fate and function. Cells, 8(11), 1306. 10.3390/cells8111306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- St John, J. C. , Facucho‐Oliveira, J. , Jiang, Y. , Kelly, R. , & Salah, R. (2010). Mitochondrial DNA transmission, replication and inheritance: a journey from the gamete through the embryo and into offspring and embryonic stem cells. Human Reproduction Update, 16(5), 488–509. 10.1093/humupd/dmq002 [DOI] [PubMed] [Google Scholar]
- St John, J. C. , Moffatt, O. , & D'Souza, N. (2005). Aberrant heteroplasmic transmission of mtDNA in cloned pigs arising from double nuclear transfer. Molecular Reproduction and Development, 72(4), 450–460. 10.1002/mrd.20370 [DOI] [PubMed] [Google Scholar]
- St John, J. C. , & Tsai, T. S. (2018). The association of mitochondrial DNA haplotypes and phenotypic traits in pigs. BMC Genetics, 19(1), 41. 10.1186/s12863-018-0629-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stigliani, S. , Persico, L. , Lagazio, C. , Anserini, P. , Venturini, P. L. , & Scaruffi, P. (2014). Mitochondrial DNA in Day 3 embryo culture medium is a novel, non‐invasive biomarker of blastocyst potential and implantation outcome. Molecular Human Reproduction, 20(12), 1238–1246. 10.1093/molehr/gau086 [DOI] [PubMed] [Google Scholar]
- Sun, X. , & John, J. C. (2018). Modulation of mitochondrial DNA copy number in a model of glioblastoma induces changes to DNA methylation and gene expression of the nuclear genome in tumours. Epigenetics & Chromatin, 11(1), 53. 10.1186/s13072-018-0223-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, X. , Johnson, J. , & John, J. C. (2018). Global DNA methylation synergistically regulates the nuclear and mitochondrial genomes in glioblastoma cells. Nucleic Acids Research, 46(12), 5977–5995. 10.1093/nar/gky339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutarno, C. , Greeff, J., J. M. , & Lymbery, A. J. (2002). Mitochondrial DNA polymorphisms and fertility in beef cattle. Theriogenology, 57(6), 1603–1610. http://www.ncbi.nlm.nih.gov/pubmed/12035972 [DOI] [PubMed] [Google Scholar]
- Takeda, K. , Akagi, S. , Kaneyama, K. , Kojima, T. , Takahashi, S. , Imai, H. , Yamanaka, M. , Onishi, A. , & Hanada, H. (2003). Proliferation of donor mitochondrial DNA in nuclear transfer calves (Bos Taurus) derived from cumulus cells. Molecular Reproduction and Development, 64(4), 429–437. 10.1002/mrd.10279 [DOI] [PubMed] [Google Scholar]
- Takeda, K. , Tasai, M. , Iwamoto, M. , Akita, T. , Tagami, T. , Nirasawa, K. , Hanada, H. , & Onishi, A. (2006). Transmission of mitochondrial DNA in pigs and progeny derived from nuclear transfer of Meishan pig fibroblast cells. Molecular Reproduction and Development, 73(3), 306–312. 10.1002/mrd.20403 [DOI] [PubMed] [Google Scholar]
- Tiranti, V. , Savoia, A. , Forti, F. , D'Apolito, M. F. , Centra, M. , Rocchi, M. , & Zeviani, M. (1997). Identification of the gene encoding the human mitochondrial RNA polymerase (h‐mtRPOL) by cyberscreening of the expressed sequence tags database. Human Molecular Genetics, 6(4), 615–625. https://www.ncbi.nlm.nih.gov/pubmed/9097968 [DOI] [PubMed] [Google Scholar]
- Treff, N. R. , Zhan, Y. , Tao, X. , Olcha, M. , Han, M. , Rajchel, J. , Morrison, L. , Morin, S. J. , & Scott RT, Jr. (2017). Levels of trophectoderm mitochondrial DNA do not predict the reproductive potential of sibling embryos. Human Reproduction, 32(4), 954–962. 10.1093/humrep/dex034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai, T. S. , Johnson, J. , White, Y. , & John, J. C. (2017). The molecular characterization of porcine egg precursor cells. Oncotarget, 8(38), 63484–63505. 10.18632/oncotarget.18833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai, T. S. , Rajasekar, S. , & St John, J. C. (2016). The relationship between mitochondrial DNA haplotype and the reproductive capacity of domestic pigs (Sus scrofa domesticus). BMC Genetics, 17(1), 67. 10.1186/s12863-016-0375-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai, T. S. , & St John, J. C. (2018). The effects of mitochondrial DNA supplementation at the time of fertilization on the gene expression profiles of porcine preimplantation embryos. Molecular Reproduction and Development, 85(6), 490–504. 10.1002/mrd.22985 [DOI] [PubMed] [Google Scholar]
- Tsai, T. S. , Tyagi, S. , & St John, J. C. (2018). The molecular characterisation of mitochondrial DNA deficient oocytes using a pig model. Human Reproduction, 33(5), 942–953. 10.1093/humrep/dey052 [DOI] [PubMed] [Google Scholar]
- Ursing, B. M. , & Arnason, U. (1998). The complete mitochondrial DNA sequence of the pig (Sus scrofa). Journal of Molecular Evolution, 47(3), 302–306. http://www.ncbi.nlm.nih.gov/pubmed/9732457 [DOI] [PubMed] [Google Scholar]
- von Meyenn, F. , Berrens, R. V. , Andrews, S. , Santos, F. , Collier, A. J. , Krueger, F. , Osorno, R. , Dean, W. , Rugg‐Gunn, P. J. , & Reik, W. Comparative principles of DNA methylation reprogramming during human and mouse in vitro primordial germ cell specification. Developmental Cell, 39(1), 104–115. 10.1016/j.devcel.2016.09.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wai, T. , Teoli, D. , & Shoubridge, E. A. (2008). The mitochondrial DNA genetic bottleneck results from replication of a subpopulation of genomes. Nature Genetics, 40(12), 1484–1488. 10.1038/ng.258 [DOI] [PubMed] [Google Scholar]
- Wallace, D. C. , Brown, M. D. , & Lott, M. T. (1999). Mitochondrial DNA variation in human evolution and disease. Gene, 238(1), 211–230. 10.1016/s0378-1119(99)00295-4 [DOI] [PubMed] [Google Scholar]
- Wallace, D. C. , Ruiz‐Pesini, E. , & Mishmar, D. (2003). mtDNA variation, climatic adaptation, degenerative diseases, and longevity. Cold Spring Harbor Symposia on Quantitative Biology, 68, 479–486. http://www.ncbi.nlm.nih.gov/pubmed/15338651 [DOI] [PubMed] [Google Scholar]
- Warburg, O. (1956). On respiratory impairment in cancer cells. Science, 124(3215), 269–270. http://www.ncbi.nlm.nih.gov/pubmed/13351639 [PubMed] [Google Scholar]
- Wei, W. , & Chinnery, P. F. (2020). Inheritance of mitochondrial DNA in humans: implications for rare and common diseases. Journal of Internal Medicine, 287(6), 634–644. 10.1111/joim.13047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wernette, C. M. , & Kaguni, L. S. (1986). A mitochondrial DNA polymerase from embryos of Drosophila melanogaster. Purification, subunit structure, and partial characterization. Journal of Biological Chemistry, 251, 14764–14770. [PubMed] [Google Scholar]
- White, Y. A. , Woods, D. C. , Takai, Y. , Ishihara, O. , Seki, H. , & Tilly, J. L. (2012). Oocyte formation by mitotically active germ cells purified from ovaries of reproductive‐age women. Nature Medicine, 18(3), 413–421. 10.1038/nm.2669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilmut, I. , Schnieke, A. E. , McWhir, J. , Kind, A. J. , & Campbell, K. H. (1997). Viable offspring derived from fetal and adult mammalian cells. Nature, 385(6619), 810–813. 10.1038/385810a0 [DOI] [PubMed] [Google Scholar]
- Woods, D. C. , Khrapko, K. , & Tilly, J. L. (2018). Influence of maternal aging on mitochondrial heterogeneity, inheritance, and function in oocytes and preimplantation embryos. Genes (Basel), 9(5), 265. 10.3390/genes9050265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, G. , Xiang, H. , Tian, J. , Yin, J. , Pinkert, C. A. , Li, Q. , & Zhao, X. (2015). Mitochondrial haplotypes influence metabolic traits in porcine transmitochondrial cybrids. Scientific Reports, 5, 13118. 10.1038/srep13118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, H. , Meng, L. H. , & Pommier, Y. (2007). Mitochondrial topoisomerases and alternative splicing of the human TOP1mt gene. Biochimie, 89(4), 474–481. 10.1016/j.biochi.2006.11.002 [DOI] [PubMed] [Google Scholar]
- Zheng, Y. , Luo, X. , Zhu, J. , Zhang, X. , Zhu, Y. , Cheng, H. , Xia, Z. , Su, N. , Zhang, N. , & Zhou, J. (2012). Mitochondrial DNA 4977 bp deletion is a common phenomenon in hair and increases with age. Bosnian Journal of Basic Medical Sciences/Udruženje basičnih mediciniskih znanosti, 12(3), 187–192. 10.17305/bjbms.2012.2480 [DOI] [PMC free article] [PubMed] [Google Scholar]


