Abstract
The adenovirus fiber mediates the agglutination of erythrocytes. Based on differential hemagglutinating properties, subgenus D adenoviruses can be subdivided into clusters DI, DII, and DIII. While subgenus DI adenoviruses agglutinate rat and human erythrocytes, DII adenoviruses simply agglutinate rat erythrocytes and DIII adenoviruses display no or only weak rat erythrocyte agglutination. Amino acid sequence comparisons revealed distinct domains on the fiber knob which could be involved in hemagglutination. In order to localize and characterize the domains responsible for the interaction with rat and human erythrocytes, potential hemagglutination domains of the adenovirus type 9 (Ad9) (subgenus DI) fiber knob were introduced into Ad17 (subgenus DII) and Ad28 (subgenus DIII) fiber knobs by primer-directed mutagenesis. Furthermore, rat erythrocyte hemagglutination domains were also introduced into the Ad3 (subgenus B) fiber knob, which only agglutinated monkey erythrocytes. Altogether, 27 chimeric and mutated fiber proteins were expressed in Escherichia coli and subsequently tested for hemagglutination activity. The hemagglutination tests revealed that at least two domains can mediate the agglutination of rat erythrocytes. While one domain is located on the GH loop, the other domain extends from the C β strand to the CD loop. The domain on the GH loop was partially conserved in all adenoviruses showing an incomplete hemagglutination pattern with rat erythrocytes. The domains involved in the agglutination of human erythrocytes are located on the CD and HI loops of the subgenus DI fiber knob.
Besides being associated with a variety of diseases, including respiratory, ophthalmic, and gastrointestinal infections, adenoviruses have recently received special attention as potential viral vectors for gene therapy. Since the fiber protein is responsible for the attachment of the virion to specific receptors on the cell surface (5, 30), thus also being of significant importance for tissue tropism, a detailed understanding of the molecular structure of this protein could be helpful in developing a new, tissue-specific generation of adenovirus vectors.
The fiber protein, protruding outward from the 12 vertices of the capsid, comprises a short N-terminal tail, a shaft of variable length, and a globular C-terminal knob (12). The conserved N terminus contains the sequences responsible for association with the penton base as well as the nuclear localization signal (19, 29). The shaft consists of repeating motifs of a 15-amino-acid β structure, with the number of repeats varying among virus serotypes. A conserved amino acid sequence (TLWT) marks the boundary between the shaft and the knob domain, which is responsible for interaction with the host cell receptor. The published crystal structure of the adenovirus type 5 (Ad5) fiber knob domain allows the mapping of functional domains (40, 41). It was shown that the Ad5 knob can block virus infection (14) and that the receptor binding specificity of adenovirus fibers can be altered by exchanging the knob domains (11, 37). While subgenus C and B adenovirus serotypes recognize distinct receptors (6, 24, 38), subgenus C adenoviruses and Ad9 (subgenus D) share the same fiber receptor (33). It was recently demonstrated that a 46-kDa HeLa cell surface protein serves as a common receptor for subgenus C adenoviruses and coxsackie B viruses (3). Furthermore, it was reported that the class I major histocompatibility complex could also serve as an adenovirus receptor (21). The fiber knob also carries the type-specific γ antigen (9, 27), which determines, together with the ɛ antigen of the hexon, the serotype specificity of an adenovirus. The γ determinant is composed of at least 17 amino acids that are not restricted to a distinct region on the fiber knob (10).
Since hemagglutination (HA) by human adenoviruses was first demonstrated by Rosén in 1958 (34), it has been shown that members of the six subgenera (A to F) display different HA properties (2, 26). While, e.g., subgenus B adenoviruses only agglutinate monkey erythrocytes, subgenus D adenoviruses can be classified into three clusters: cluster DI adenoviruses agglutinate rat and human erythrocytes, cluster DII adenoviruses agglutinate only rat erythrocytes, and cluster DIII adenoviruses show no or only weak agglutination of rat erythrocytes. The agglutination of erythrocytes is fiber mediated, and specific receptors seem to be present on the erythrocyte membrane. Since intact virions carry several fibers, they can establish a bridge between erythrocytes, leading to HA. In contrast, fibers alone cannot cause HA, as they are monovalent. However, it was shown that fibers obtained from tissue cultures (28) and recombinant fibers (25) can form polymers which are able to agglutinate erythrocytes.
Amino acid sequence comparisons revealed distinct domains on the fiber knob which could be expected to mediate the agglutination of rat and human erythrocytes. To localize and characterize these domains, 27 chimeric and mutated Ad9 (subgenus DI), Ad17 (subgenus DII), Ad28 (subgenus DIII), and Ad3 (subgenus B) fiber proteins were expressed in Escherichia coli. The recombinant proteins were tested in HA tests.
MATERIALS AND METHODS
Cells and viruses.
Ad3 (prototype GB; American Type Culture Collection [ATCC]), Ad9 (prototype Hicks; ATCC), Ad17 (prototype Ch22; ATCC), and Ad28 (prototype BP-5; ATCC) were passaged several times in HeLa cells. Viral DNA was extracted from infected cells as previously described (7).
PCR amplification, construction of Ad17 and Ad28 plasmids, and DNA sequencing.
The Ad17 and Ad28 fiber genes were amplified with the previously described F1-F2 primer pair (31). After the PCR procedure (31), appropriate samples were ligated into pUC18. Several suitable clones were selected, and subsequent sequencing of both DNA strands was performed with specific internal primers by the method of Sanger and coworkers (35). As the complete Ad17 and Ad28 fiber gene nucleotide sequences have been made generally available in the EMBL database under accession no. Y14241 (Ad17) and Y14242 (Ad28), a detailed annotation of the nucleotide sequences will not be shown here. An amino acid sequence comparison with the Ad9, Ad15, and Ad19 fiber polypeptides is shown in Fig. 1.
FIG. 1.
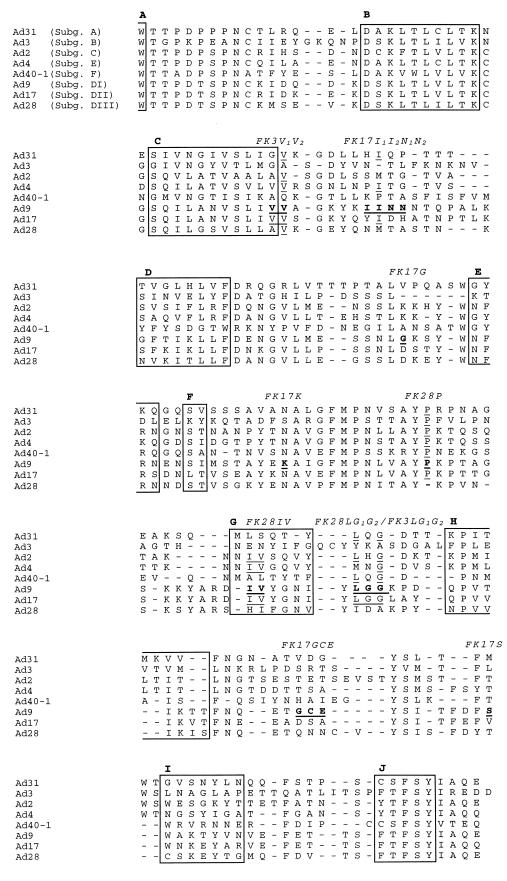
Comparison of the predicted amino acid sequences of the Ad9, Ad19, Ad15, Ad17, and Ad28 fiber polypeptides. The numbering on the left takes into account the deletions which resulted from the alignment. The structural domains tail, shaft, and knob and the eight repeating motifs in the shaft region are marked by arrows. Amino acids identical to the amino acids presented in the preceding sequence are marked by asterisks; plus signs indicate homology to Ad9; number signs indicate homology to Ad19; and deletions are represented by dashes. Subg., subgenus. The amino acid residues which were exchanged between Ad9, Ad3, Ad17, and Ad28 are underlined and written in bold (the Ad3 knob sequence is shown in Fig. 2).
Construction of recombinant plasmids.
The Ad3 (36), Ad9 (31), Ad17, and Ad28 fiber DNA sequences were used to evaluate primer pairs for amplifying the fiber knob region. As the last shaft motif is necessary for trimerization (14, 20), the amplified regions consisted of the knob domain and the eighth (last) shaft motif. Since the PCR products were subsequently cloned into a pQE31 or pQE32 expression vector (Qiagen), which provides an in-frame start codon, the forward primers (Kf-x) did not have an ATG initiation codon; the reverse primers (Kr) ended with the termination codon TGA or TAA: Kf-3, 5′-GGTCTTACATTTGACTC-3′; Kr3, 5′-TCAGTCATCTTCTCTAA-3′; Kf-9, 5′-GGAGACTTGGTAGCATT-3′; Kr9/28, 5′-TCATTCTTGGGCGATAT-3′; Kf-17, 5′-GGATACTTGGTAGCATG-3′; Kr17, 5′-TTATTGTTGGGCAATAT-3′; and Kf-28, 5′-GGAGATTTGGTGGCCTG-3′.
Amplified products of the expected sizes were ligated into pUC18 for sequencing. In each case, a clone having a perfect match with the published or previously determined sequences was selected for cloning into a pQE expression vector (pQE31 or pQE32) followed by expression in E. coli. The expressed proteins possessed an N-terminal affinity tag consisting of the amino acid residues RGS and six consecutive histidine residues [RGS(H)6]. The fiber knob proteins are referred to as FK3, FK9, FK17, and FK28, respectively (F for fiber; K for knob region).
Construction of chimeric fiber genes.
In order to confirm that the fiber tail and shaft do not contribute to the domains responsible for HA, the four chimeric fiber proteins FTS9/K17, FTS17/K9, FTS9/K28, and FTS28/K9 were generated (F for fiber; TS for tail and shaft region; K for knob region). The shaft-knob junction was located at the conserved TLWT amino acid residues (Fig. 1). Ad9, Ad17, and Ad28 DNAs were amplified by PCR with different primer pairs (forward primers: Ff-9/17, Ff-28, Kf9/28, and Kf17; reverse primers: TSr9, TSr17, TSr28, Kr9/28, and Kr17). The primers used for the amplification of the tail and shaft regions were as follows: Ff-9/17, 5′-TCAAAGAGGCTCCGGGT-3′; Ff-28, 5′-ACAAAGAGGCTCCGGGT-3′; TSr9, 5′-TAGGGTGCGCTTATCTT-3′; TSr17, 5′-AAGTGTGCGCGTGTCAT-3′; and TSr28, 5′-TAGAGTGCGCCTGTCAT-3′. The primers used for amplification of the knob regions were as follows: Kf9/28, 5′-TGGACAACTCCAGACAC-3′; Kf17, 5′-TGGACAACACCAGACAC-3′; and Kr9/28 and Kr17 (see above).
The chimeric fiber genes were created by ligating the Ff-x/TSr PCR products with the corresponding Kf/Kr PCR products (i.e., Ff-9/17/TSr9 with Kf17/Kr17, generating the chimera FTS9/K17). After ligation, a second PCR was carried out with the Ff-x/Kr primer pairs. As described above, the amplified full-length chimeric fiber genes were ligated into pUC18. The nucleotide sequence of the cloned insert was determined, and in each case, a clone having the correct sequence was selected for cloning into a pQE expression vector and subsequent expression in E. coli.
Construction of mutant fiber genes by primer-directed mutagenesis.
In order to find the domains involved in the agglutination of rat and human erythrocytes, mutated Ad3, Ad9, Ad17, and Ad28 fiber knobs were constructed. Distinct amino acid exchanges in the knob domains were made by primer-directed mutagenesis (18). Two PCR products (i.e., Kf-17/K17Src and K17S/Kr17) overlapping in sequence and containing the same primer-introduced mutation were generated and purified by gel electrophoresis. After denaturation and annealing of the two PCR products, subsequent reamplification with the Kf-x/Kr (outside) primer pairs, resulting in the enrichment of the complete mutated fiber knob region, was performed. Twelve mutagenic primer pairs were evaluated for the Ad3 and Ad28 fiber knobs (introducing potential domains for the agglutination of rat erythrocytes), and 12 mutagenic primer pairs were evaluated for the Ad17 fiber knob (introducing potential domains for the agglutination of human erythrocytes). Four primer pairs were synthesized to exchange HA domains on the Ad9 fiber knob with the corresponding non-HA regions of the Ad3, Ad17, and Ad28 fiber knobs.
The mutated fiber knobs were named after the introduced mutation; e.g., the original Ad17 amino acid residues YIDH (Fig. 1 and 2) were changed to the Ad9 residues IINN (the second Ad9 I was conserved in both sequences). The resulting protein was named FK17I1I2N1N2 (subscript numerals were used to distinguish between identical amino acid residues). The FK9IDA&GA and FK9YIDH&V proteins have two mutated domains. The amino acids exchanged are marked in Fig. 1 and 2.
FIG. 2.
Fiber knob amino acid alignment of human adenoviruses representing subgenera (Subg.) A to F. Deletions are represented by dashes. Potential HA domains in the Ad9 fiber knob are underlined and written in bold. Amino acid residues identical to Ad9 amino acid residues are underlined. FK3V1V2 to FK17S represent mutated fiber knob proteins. The boxes indicate the β strands (A to J) corresponding to the published structure of the Ad5 knob domain. The regions (loops; not indicated) between the β strands were labeled by Xia et al. (40, 41) after the β strands that they connect, e.g., AB loop and CD loop; the DG loop also includes the short β strands E and F.
The sequences of the primers used to create the primer-directed mutations were as follows (the mutated nucleotides are underlined; the reverse complementary primers are not shown): K28P, 5′-CATAACAGCTTATCCAAAACCCGTCAATTC-3′; K28IV, 5′-GCTATGCCAGAAGTATAGTTTTTGGAAATGTATATATTG-3′; K28LG1G2, 5′-GGAAATGTATATCTTGGTGGAAAGCCATATAATCC-3′; K3LG1G2, 5′-GGTCAATGCTACCTTGGTGGAAGCGATGGTGCCC-3′; K3L, 5′-GGTCAATGCTACCTTAAAGCAAGCGATGGTGCCC-3′; K3G1, 5′-GGTCAATGCTACTACGGTGCAAGCGATGGTGCCC-3′; K3G2, 5′-GGTCAATGCTACTACAAAGGAAGCGATGGTGCCC-3′; K3V1V2, 5′-GGATATGTAACGCTAATGGTCGTATCAGACTACGTTAACACC-3′; K3V1, 5′-GGATATGTAACGCTAATGGTCGCCTCAGACTACGTTAACACC-3′; K3V2, 5′-GGATATGTAACGCTAATGGGAGTATCAGACTACGTTAACACC-3′; K9IDA, 5′-GGAAACATCTACATTGATGCTAAGCCAGATCAACCA-3′; K9GA, 5′-GGCTAATGTGTCATTAATTGGAGCCGCTGGTAAGTACAAAATTATC-3′; K17I1I2N1N2, 5′-GTCAGGAAAATATCAAATTATAAATAACGCTACAAATCCAAC-3′; K17I1I2N1, 5′-GTCAGGAAAATATCAAATTATAAATCACGCTACAAATCCAAC-3′; K17I2N1N2, 5′-GTCAGGAAAATATCAATACATAAATAACGCTACAAATCCAAC-3′; K17I2N1, 5′-GTCAGGAAAATATCAATACATAAATCACGCTACAAATCCAAC-3′; K17I2N2, 5′-GTCAGGAAAATATCAATACATAGACAACGCTACAAATCCAAC-3′; K17I1I2, 5′-GTCAGGAAAATATCAAATTATAGACCACGCTACAAATCCAAC-3′; K17G, 5′-CCAAGTTCAAACCTTGGTTCCACATATTGGAACTTTAG-3′; K17K, 5′-GTATCTGAGGCATATAAAAAAGCAGTTGAATTTATG-3′; K17GCE, 5′-CTTTTAATGAAGAAGCAGGATGTGAATACTCTATAACATTTG-3′; K17S, 5′-CTATAACATTTGAATTTAGTTGGAATAAAGAATATGCC-3′; K9YIDH, 5′-GATGGTAAGTACAAATACATAGACCACAATACTCAACCAGCTC-3′; and K9V, 5′-CTATCACATTTGATTTTGTATGGGCCAAGACTTATG-3′. Altogether, a total of 23 fiber knob mutants were created by applying primer-directed mutagenesis. Ligation into pUC18 and subsequent sequencing were followed by cloning into a pQE vector and expression in E. coli.
Expression of the fiber constructs.
For expression of the recombinant fiber constructs in E. coli (strain M15), bacteria were grown in 2× YT (yeast extract-tryptone) medium and induced with 2 mM isopropyl-β-d-thiogalactopyranoside for 4 h at 37°C. Cells from a 500-ml culture were harvested by centrifugation at 3,000 × g for 15 min, and the pellets were resuspended in sonication buffer (50 mM NaH2PO4 [pH 8.0], 300 mM NaCl) at 2 volumes per g (wet weight). After being frozen and thawed, the cells were sonicated thoroughly. The lysate was centrifuged at 13,000 × g for 10 min to pellet cell debris. The fiber protein-containing supernatant was harvested. All recombinant fiber proteins were soluble. Since expression of the chimeric fiber proteins yielded less recombinant protein than expression of the knob proteins, the former proteins were concentrated sixfold with microconcentrators (Centricon-30; Amicon).
The expression of each fiber protein was controlled by combined denaturing-nondenaturing sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) with 8, 12, or 15% polyacrylamide gels. For denatured sample preparation, the lysates were suspended in sample buffer containing 0.1 M dithiothreitol (final concentration) and boiled prior to being loaded onto SDS-PAGE gels. For native sample preparation, the buffer contained no dithiothreitol and the samples were not boiled. The Western blot procedure was performed by standard techniques (23). The gels were blotted onto nitrocellulose membranes (Gibco BRL). An antibody (RGS-His antibody; Qiagen) directed against the RGS(H)6 epitope served as the primary antibody, and an anti-mouse antibody conjugated to alkaline phosphatase (Boehringer GmbH, Mannheim, Germany) served as the secondary antibody. The color reaction was developed with 5-bromo-4-chloro-3-indolylphosphate toluidinium (Boehringer).
HA tests.
The domains responsible for the agglutination of rat and human erythrocytes were determined by HA tests. For the HA tests, virions or recombinant proteins were diluted in serial twofold steps in 96-well plates containing 25 μl of McIlvaine NaCl buffer (0.1 M citric acid, 0.2 M Na2HPO4 [pH 7.2]; diluted 1:50 with 0.87% NaCl). To each dilution, 25 μl of a 2% suspension of rat or human erythrocytes was added. The sedimentation pattern was determined after incubation for 1 h at room temperature. HA tests with rat erythrocytes were also used, in addition to SDS-PAGE, to quantify the amounts of recombinant proteins tested for the agglutination of human erythrocytes.
RESULTS
Amino acid sequence comparisons.
As the Ad9 and Ad17 fiber knobs showed an amino acid identity of 73%, while Ad9 and the previously published Ad15 (subgenus DII) fiber knob sequences showed a homology of only 54% (31), Ad17 was selected for the construction of mutated knob proteins. Ad9 and Ad17 showed amino acid identities with Ad28 in the knob region of 58 and 57%, respectively. The Ad17 and Ad28 fibers exhibited the typical subgenus D fiber features, with eight repeating shaft motifs and the predicted subgenus D-specific YARNQNI amino acid residues in the tail region (amino acid residues 19 to 25; Fig. 1). Amino acid sequence comparisons between all so-far-analyzed subgenus D fibers (1, 8, 31) and fibers of subgenera A, B, C, E, and F (4, 13, 15, 16, 22, 32, 36) revealed several potential erythrocyte binding domains on the fiber knob. Representative sequences are shown in Fig. 1 and 2.
Cloning and expression of the recombinant fiber proteins.
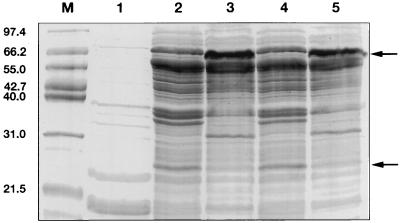
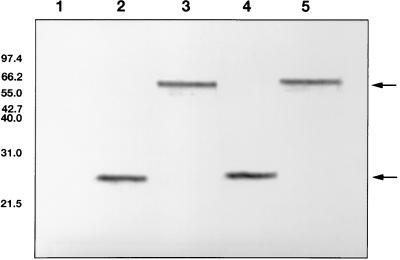
PCR amplification of the Ad3, Ad9, Ad17, and Ad28 fiber knob regions (also containing the last shaft motif) generated amplimers of 609 bp (Ad3), 585 bp (Ad9 and Ad17), and 576 bp (Ad28). The mutant fiber knob PCR amplimers also were of the expected lengths. Amplification of the Ad9, Ad17, and Ad28 tail and shaft fragments ligated with the Ad9, Ad17, and Ad28 knob fragments, respectively, yielded products, each of approximately 1,100 bp, which corresponded to the full-length fiber genes. The expressed knob proteins were visualized as approximately 23-kDa proteins in denaturing SDS-PAGE and as approximately 66-kDa proteins in nondenaturing SDS-PAGE and Western blotting (Fig. 3 and 4). The 23-kDa bands represent the monomeric form of the fiber knob, and the 66-kDa bands represent the trimeric form. The chimeric fiber proteins (FTS9/K17, FTS17/K9, FTS9/K28, and FTS28/K9) showed molecular masses of approximately 43 kDa (monomer) and 130 kDa (trimer) in SDS-PAGE and Western blotting (data not shown). These results agreed with the molecular masses predicted by the amino acid sequences of the fiber proteins [including the RGS(H)6 epitope]. In lysates of uninduced cultures, no fiber proteins were observed.
FIG. 3.
Coomassie blue-stained SDS-PAGE (15% polyacrylamide) gel of expressed recombinant fiber knob proteins. Lanes: M, molecular mass markers (the sizes of the markers are indicated on the left in kilodaltons); 1, uninduced E. coli M15 cells (transformed with pQE); 2, fiber knob protein FK17GCE (sample boiled prior to loading); 3, fiber knob protein FK17GCE (sample not boiled); 4, fiber knob protein FK17S (sample boiled prior to loading); 5, fiber knob protein FK17S (sample not boiled). The arrows indicate the positions of the monomeric and trimeric fiber knob proteins.
FIG. 4.
Western blot (of 15% polyacrylamide gel). Lanes: 1, uninduced E. coli M15 cells (transformed with pQE); 2, fiber knob protein FK17GCE (sample boiled prior to loading); 3, fiber knob protein FK17GCE (sample not boiled); 4, fiber knob protein FK17S (sample boiled prior to loading); 5, fiber knob protein FK17S (sample not boiled). The sizes of the markers are indicated on the left in kilodaltons. The arrows indicate the positions of the monomeric and trimeric fiber knob proteins.
Rat erythrocyte binding domains.
All three subgenus D adenovirus fiber knob proteins (FK9, FK17, and FK28) reacted with rat erythrocytes (Table 1). However, while FK9 and FK17 yielded high HA titers, the HA titer of FK28 was severalfold lower. As expected, the Ad3 fiber knob protein (FK3) showed no rat erythrocyte HA. The chimeric, complete fiber proteins FTS9/K17, FTS17/K9, and FTS28/K9 reacted according to the fiber knobs. This result demonstrated that their tail and shaft regions did not participate in the agglutination of rat erythrocytes. While the FK28 HA titer was already significantly lower than the HA titers of the other fiber knob proteins, the chimeric protein FTS9/K28 showed no agglutination at all. This finding might be explained by the lower rates of expression of the chimeric fibers and/or less effective polymerization of the FTS9/K28 fiber protein. However, even HA results obtained with complete Ad28 virions do not seem to be consistent: while Wigand (39) could not detect rat erythrocyte HA, Hierholzer and Dowdle (17) demonstrated rat erythrocyte HA.
TABLE 1.
HA tests with rat erythrocytes
| Proteina | HA titer |
|---|---|
| Virion Ad9 | 1:128 |
| FK9 | 1:128 |
| FK17 | 1:128 |
| FK28 | 1:8 |
| FK3 | −b |
| FTS28/K9 | 1:128 |
| FTS9/K17 | 1:128 |
| FTS9/K28 | − |
| FK28P | 1:8 |
| FK28IV | 1:8 |
| FK28L1G1G2 | 1:32 |
| FK3L1G1G2 | 1:16 |
| FK3L1 | − |
| FK3G1 | 1:8 |
| FK3G2 | 1:16 |
| FK3V1V2 | 1:8 |
| FK3V1 | 1:4 |
| FK3V2 | − |
| FK9IDA | 1:8 |
| FK9IDA&GA | 1:4 |
FK, fiber knob proteins; FTS, chimeric fiber proteins.
−, no HA.
Three Ad28 regions which completely differed from the corresponding regions of subgenus DI and subgenus DII adenoviruses were selected for primer-directed mutagenesis (Fig. 1). In FK28P, a proline residue (P) was added to the Ad28 knob region; in FK28IV, the histidine-isoleucine (HI) residues were exchanged with isoleucine-valine (IV) residues; and in FK28LG1G2, the isoleucine-aspartic acid-alanine (IDA) residues were exchanged with leucine-glycine-glycine (LGG) residues. While the FK28P and FK28IV knob proteins showed no differences from the unmutated FK28 knob protein in HA titers (Table 1), FK28LG1G2 showed an increase in the HA titer, which indicated the introduction of an additional HA domain. When planning the experiments, we presumed that FK28, like FTS9/K28, would not show agglutination of rat erythrocytes. As we wanted clear-cut results (HA or no HA), we decided to confirm the previous results with the corresponding mutated Ad3 knob protein and to use only the Ad3 knob for further mutations. By exchanging the tyrosine-lysine-alanine (YKA) residues in the Ad3 knob with LGG residues (FK3LG1G2), we demonstrated that this domain is involved in the agglutination of rat erythrocytes (Table 1). While each of the two G residues could independently mediate rat erythrocyte HA, as demonstrated by HA with Ad3 fiber knobs having single amino acid mutations (FK3G1 and FK3G2), FK3L did not show HA activity.
Since FK28 showed rat erythrocyte HA even though it did not possess the LGG property, at least one additional HA domain could be postulated. To confirm our prediction, the LGG residues in the Ad9 knob were exchanged with Ad28 IDA residues. As expected, the FK9IDA protein could still agglutinate rat erythrocytes. Subsequently, we selected a domain for primer-directed mutagenesis which was partially conserved in Ad28 but not conserved in Ad3 (Fig. 2). As shown in Table 1, the Ad3 knob in which the glycine-alanine (GA) residues were altered to valine-valine (VV) was able to agglutinate rat erythrocytes. Further mutations with single amino acid exchanges revealed that only FK3V1 showed rat erythrocyte HA. Since the second valine (V2) of the VV domain, which was the only conserved amino acid in the corresponding Ad28 region, was not sufficient for HA (FK3V2; Table 1) and since FK9IDA&GA still exhibited HA activity, a further HA domain for the agglutination of rat erythrocytes could be expected.
The crystal structure of the Ad5 fiber knob revealed that the knob contains two β sheets, a V sheet, and an R sheet (40, 41). The surface of the V sheet (consisting of the J, C, B, and A β strands), which is highly conserved among different adenovirus serotypes, points toward the virion side in the trimer structure of the knob, while the R sheet (consisting of the D, I, H, and G β strands) is probably involved in the binding of cellular receptors. Six prominent loops connect the β strands. A sequence alignment of the subgenus D adenovirus knobs with the Ad5 knob allowed the localization of the domains involved in HA (Fig. 5). The VV residues are located on the C β strand (V1) and the CD loop (V2), and the LGG residues are located on the GH loop (Fig. 2 and 5).
FIG. 5.
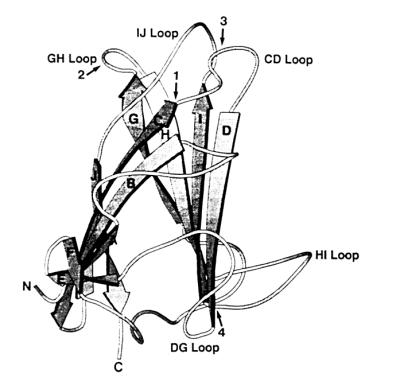
Localization of the HA domains within the polypeptide chain of the knob monomer. The positions of the HA domains were determined by comparing the subgenus D fiber knob sequences to the Ad5 knob structure published by Xia et al. (41). Loops, β strands (A to J), and chain termini are labeled. Arrows 1 and 2 indicate the domains involved in the agglutination of rat erythrocytes, and arrows 3 and 4 indicate the domains involved in the agglutination of human erythrocytes.
Human erythrocyte binding domains.
As predicted, only fiber knob protein FK9 and the chimeric fiber proteins FTS17/K9 and FTS28/K9, containing the Ad9 knob, reacted with human erythrocytes, while FK17, FK28, FK3, and the corresponding chimeric fiber proteins FTS9/K17 and FTS9/K28 showed no agglutination of human erythrocytes. The data obtained with the chimeric fiber proteins showed, as expected from the above-described results, that the fiber knob is also sufficient for the agglutination of human erythrocytes and that the tail and the shaft are not involved in HA (Table 2).
TABLE 2.
HA tests with human erythrocytes
| Proteina | HA titer |
|---|---|
| Virion Ad9 | 1:128 |
| FK9 | 1:128 |
| FK17 | −b |
| FTS17/K9 | 1:128 |
| FTS9/K17 | − |
| FK17I1I2N1N2 | 1:32 |
| FK17I1I2N1 | 1:32 |
| FK17I2N1N2 | − |
| FK17I2N1 | − |
| FK17I2N2 | − |
| FK17I1I2 | 1:16 |
| FK17G | − |
| FK17K | − |
| FK17GCE | − |
| FK17S | 1:64 |
| FK9YIDH&V | 1:8 |
FK, fiber knob proteins; FTS, chimeric fiber proteins.
−, no HA.
Five regions of the Ad17 knob were selected for primer-directed mutagenesis. These regions were conserved within subgenus DI adenoviruses but differed from the corresponding regions of all other subgenera, including subgenera DII and DIII (Fig. 1 and 2). All subgenus DI and DII fiber proteins were able to agglutinate rat erythrocytes. The virions and unmutated proteins showed an HA titer of 1:128 for human erythrocytes (Table 2). Two of the five mutated fiber knob proteins, FK17I1I2N1N2 and FK17S, were able to agglutinate human erythrocytes. In the fiber knob protein FK17I1I2N1N2, the Ad17 tyrosine-isoleucine-aspartic acid-histidine (YIDH) domain was mutated to isoleucine-isoleucine-asparagine-asparagine (IINN); in FK17S, a valine (V) residue was exchanged with a serine (S) residue. In order to investigate if all of the amino acids in the IINN domain were necessary for HA or if fewer amino acids were sufficient to mediate HA, five mutated fiber knob proteins were created. HA tests with fiber knob proteins possessing different combinations of two amino acid exchanges (FK17I1I2, FK17I2N1, and FK17I2N2) revealed that only the protein with two isoleucine residues (FK17I1I2) was able to mediate the agglutination of human erythrocytes. Since FK17I1I2 showed HA, it was not surprising that FK17I1I2N1 also showed HA. FK17I2N1N2 did not agglutinate human erythrocytes. To confirm our results, the protein FK9YIDH&V (in which the Ad9 IINN and S residues were exchanged with the corresponding Ad17 residues) was created. In comparison with that of the unmutated Ad9 fiber knob protein, the HA activity of this protein was considerably lower, indicating that HA domains were altered.
The IINN residues are located on the CD loop and the S residue is located on the HI loop of the fiber knob (Fig. 2 and 5). In Table 3, the biochemical nature of the amino acids in the HA domains is shown. Exchanging Ad3, Ad17, and Ad28 amino acid residues with Ad9 amino acid residues mostly changed the charge and/or polarity of the domains, which could alter the HA properties. Especially for nonpolar hydrophobic residues (e.g., valine and isoleucine), it can be proposed that when they are found on the outside of a protein, they must be there for a specific purpose.
TABLE 3.
Amino acid exchanges in the HA domains
| Serotype | Loop regions
|
||
|---|---|---|---|
| CD | GH | HI | |
| Ad9 | V1 V2 I1 N1 N2 | L G1 G2 | S |
| Ad17 | Ya Db Hb | Va | |
| Ad28 | Ic Da Ac | ||
| Ad3 | Gd Ad | Ya Ka Ac | |
Nonpolar ↔ polar (amino acid exchanges between biochemically distinct groups).
Uncharged polar ↔ charged polar (amino acid exchanges between biochemically distinct groups).
Nonpolar ↔ nonpolar and nonpolar hydrophobic ↔ nonpolar hydrophobic (amino acid exchanges between biochemically identical groups).
Nonpolar hydrophobic ↔ nonpolar (amino acid exchanges between biochemically distinct groups).
DISCUSSION
The focus of this study was to localize and characterize the domains on the adenovirus knob that mediate the agglutination of rat and human erythrocytes. Potential HA domains of the Ad9 fiber knob were introduced into the Ad3, Ad17, and Ad28 fiber knobs by primer-directed mutagenesis. The mutated fiber knob proteins were expressed in E. coli and tested for HA activity. Three of the domains involved in HA are located on the CD, GH, and HI loops. The other domain extends from the C β strand to the CD loop. While most of the conserved residues occur in the β-sandwich motif, the surface loops are variable, allowing changes in HA properties without disturbing the overall structure. On the other hand, amino acid exchanges located in the β strands could alter the conformation of the entire knob domain and thus also affect the folding of the knob loops. It must still be elucidated whether the four analyzed amino acid domains directly interact with erythrocytes or whether HA activity is a result of conformational changes in the loops or the entire knob. It was surprising that the IJ loop, which is located between the CD and GH loops in the folded fiber knob monomer (40, 41), did not seem to exhibit HA activity. Since the Ad17 amino acid sequence in this region is identical to the corresponding Ad9 sequence, it was evident that the loop does not carry an HA domain for human erythrocytes. Based on a sequence comparison with all of the so-far-analyzed subgenus D fiber knobs, it can also be predicted that it does not carry an HA domain for rat erythrocytes. A closer look at the crystal structure of the trimerized Ad5 knob polypeptides (40, 41) revealed that the IJ loop is not as exposed on the surface as the CD, GH, and HI loops, which could explain why it is relatively conserved and does not exhibit HA activity. So far, the function of the conserved subgenus DI GCE sequence (Fig. 1, amino acid residues 343 to 345) on the exposed HI loop (Fig. 2 and 5) remains unclear.
The number of amino acid residues sufficient for mediating HA varied between one and two residues. Since the IINN amino acid residues on loop CD were also partially conserved (data not shown) in the subgenus D immunological variant strains Ad9/Hx and 15/Hx (8), which are unable to agglutinate human erythrocytes, it could be assumed that mutations without the first isoleucine (I1) would not change the HA properties. This assumption was confirmed by our results. Comparison of the HA titers of the Ad28 proteins showed that the LGG domain seems more effective in mediating HA than the VV domain; this result might indicate that the domain located on the GH loop plays a more important role in rat erythrocyte HA than the other domain, which is located partially on the C β strand and partially on the CD loop. This finding correlates with the decrease in HA activity of FK9IDA in comparison with the unmutated virions and proteins, while the additional exchange of the VV domain with the corresponding Ad3 region (FK9IDA&GA) did not lead to a significant decrease in HA. It is possible that the combined domains are necessary for a complete HA pattern, especially since analysis of the corresponding domains of subgenus A, C, F, and E serotypes revealed that the G2 amino acid residue was conserved in all adenoviruses showing incomplete agglutination of rat erythrocytes.
Mei and Wadell (25) showed for subgenus B:2 adenoviruses that domains involved in the agglutination of monkey erythrocytes are located on the GH and HI loops. This finding supports our results, since two of the domains involved in HA (one for the agglutination of rat and one for the agglutination of human erythrocytes) were also localized on these loops. At first we were surprised that single amino acids could change HA activity, but for B:2 adenoviruses (25) and also for influenza viruses (42) it has been demonstrated that single amino acid changes can lead to an altered HA specificity. We previously reported that most of the amino acids contributing to the Ad9 and Ad19 type-specific γ determinants, which can be distinguished in HA inhibition tests, were mainly distributed on the fiber knob loops, with a concentration on the CD loop (10). Since the analyzed HA domains are located adjacent to the amino acid residues of the γ determinants on the fiber loops, and especially on the CD loop, our previous suggestion that antibodies binding to the γ determinants could conceal the HA domains (thus preventing HA) is further supported.
In summary, the presented data demonstrated that it is possible to change the HA specificity of adenoviruses by exchanging domains from hemagglutinating and nonhemagglutinating adenoviruses. They also showed that functional knob domains can be altered without disturbing the overall conformation. Since HA activity is not just a property of adenoviruses, our studies could also be of interest for virologists dealing with other hemagglutinating viruses.
ACKNOWLEDGMENT
We thank Johann Deisenhofer, University of Texas Southwestern Medical Center, for kindly permitting us to relate our results to the structure of the Ad5 fiber knob monomer.
REFERENCES
- 1.Arnberg N, Mei Y-F, Wadell G. Fiber genes of adenoviruses with tropism for the eye and the genital tract. Virology. 1997;227:239–244. doi: 10.1006/viro.1996.8269. [DOI] [PubMed] [Google Scholar]
- 2.Bauer H, Wigand R. Eigenschaften der Adenovirus-Hämagglutinine. Z Hyg Infektionskr. 1963;149:96. [PubMed] [Google Scholar]
- 3.Bergelson J M, Cunningham J A, Droguett G, Kurt-Jones E A, Krithivas A, Hong J S, Horwitz M S, Crowell R L, Finberg R W. Isolation of a common receptor for coxsackie B viruses and adenoviruses 2 and 5. Science. 1997;275:1320–1323. doi: 10.1126/science.275.5304.1320. [DOI] [PubMed] [Google Scholar]
- 4.Chroboczek J, Ruigrok R W H, Cusack S. Adenovirus fiber. In: Doerfler W, Boehm P, editors. The molecular repertoire of adenoviruses. I. Berlin, Germany: Springer-Verlag; 1995. pp. 163–200. [Google Scholar]
- 5.Defer C, Belin M-T, Caillet-Boudin M-L, Boulanger P. Human adenovirus-host cell interactions: comparative study with members of subgroups B and C. J Virol. 1990;64:3661–3673. doi: 10.1128/jvi.64.8.3661-3673.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Di Guilmi A M, Barge A, Kitts P, Gout E, Chroboczek J. Human adenovirus serotype 3 (Ad3) and the Ad3 fiber protein bind to a 130-kDa membrane protein on HeLa cells. Virus Res. 1995;38:71–81. doi: 10.1016/0168-1702(95)00043-p. [DOI] [PubMed] [Google Scholar]
- 7.Doerfler W. Nonproductive infection of baby hamster kidney cells (BHK21) with adenovirus type 12. Virology. 1969;38:587–606. doi: 10.1016/0042-6822(69)90179-2. [DOI] [PubMed] [Google Scholar]
- 8.Eiz B, Adrian T, Pring-Åkerblom P. Immunological adenovirus variant strains of subgenus D: comparison of the hexon and fiber sequences. Virology. 1995;213:313–320. doi: 10.1006/viro.1995.0004. [DOI] [PubMed] [Google Scholar]
- 9.Eiz B, Adrian T, Pring-Åkerblom P. Recombinant fiber proteins of human adenoviruses Ad9, Ad15, and Ad19: localization of the haemagglutination and the type-specific determinant. Res Virol. 1997;148:5–10. doi: 10.1016/s0923-2516(97)81905-x. [DOI] [PubMed] [Google Scholar]
- 10.Eiz B, Pring-Åkerblom P. Molecular characterization of the type-specific γ-determinant located on the adenovirus fiber. J Virol. 1997;71:6576–6581. doi: 10.1128/jvi.71.9.6576-6581.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gall J, Kass-Eisler A, Leinwand L, Falck-Pedersen E. Adenovirus type 5 and 7 capsid-chimera: fiber replacement alters receptor tropism without affecting primary immune neutralization epitopes. J Virol. 1996;70:2116–2123. doi: 10.1128/jvi.70.4.2116-2123.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Green M, Wrigley N G, Russell W C, Martin S R, McLachlan A D. Evidence for a cross-β-sheet structure in the adenovirus fibre. EMBO J. 1983;2:1357–1365. doi: 10.1002/j.1460-2075.1983.tb01592.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gruber W C, Russell D J, Tibbetts C. Fiber gene and genomic origin of human adenovirus type 4. Virology. 1993;196:603–611. doi: 10.1006/viro.1993.1516. [DOI] [PubMed] [Google Scholar]
- 14.Henry L J, Xia D, Wilke M E, Deisenhofer J, Gerard R D. Characterization of the knob domain of the adenovirus type 5 fiber protein expressed in Escherichia coli. J Virol. 1994;68:5239–5246. doi: 10.1128/jvi.68.8.5239-5246.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hérissé J, Galibert F. Nucleotide sequence of the EcoRI E fragment of adenovirus 2 genome. Nucleic Acids Res. 1981;9:1229–1240. doi: 10.1093/nar/9.5.1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hérissé J, Rigolet M, Dupont de Dienchin S, Galibert F. Nucleotide sequence of adenovirus 2 DNA fragment encoding for the carboxylic region of the fibre protein and the entire E4 region. Nucleic Acids Res. 1981;9:4023–4042. doi: 10.1093/nar/9.16.4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hierholzer J C, Dowdle W R. Hemagglutination properties of adenovirus types 20, 25 and 28 (34817) Proc Soc Exp Biol Med. 1970;134:482–488. doi: 10.3181/00379727-134-34817. [DOI] [PubMed] [Google Scholar]
- 18.Higuchi R. Recombinant PCR. In: Innis M A, Gelfand D H, Sninsky J J, White T J, editors. PCR protocols. A guide to methods and applications. San Diego, Calif: Academic Press, Inc.; 1991. pp. 177–183. [Google Scholar]
- 19.Hong J S, Engler J A. The amino terminus of the adenovirus fiber protein encodes the nuclear localization signal. Virology. 1991;185:758–767. doi: 10.1016/0042-6822(91)90547-o. [DOI] [PubMed] [Google Scholar]
- 20.Hong J S, Engler J A. Domains required for assembly of adenovirus type 2 fiber trimers. J Virol. 1996;70:7071–7078. doi: 10.1128/jvi.70.10.7071-7078.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hong S S, Karayan L, Tournier J, Curiel D T, Boulanger P A. Adenovirus type 5 fiber knob binds to MHC class I α2 domain at the surface of human epithelial and B lymphoblastoid cells. EMBO J. 1997;16:2294–2306. doi: 10.1093/emboj/16.9.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kidd A H, Erasmus M J. Sequence characterization of the adenovirus 40 fiber gene. Virology. 1989;172:134–144. doi: 10.1016/0042-6822(89)90115-3. [DOI] [PubMed] [Google Scholar]
- 23.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 24.Louis N, Fender P, Barge A, Kitts P, Chroboczek J. Cell-binding domain of adenovirus serotype 2 fiber. J Virol. 1994;68:4104–4106. doi: 10.1128/jvi.68.6.4104-4106.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mei Y-F, Wadell G. Epitopes and hemagglutination binding domain on subgenus B:2 adenovirus fibers. J Virol. 1996;70:3688–3697. doi: 10.1128/jvi.70.6.3688-3697.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Norrby E. Biological significance of structural adenovirus components. Curr Top Microbiol Immunol. 1968;43:1. doi: 10.1007/978-3-642-46118-7_1. [DOI] [PubMed] [Google Scholar]
- 27.Norrby E. The structural and functional diversity of adenovirus capsid components. J Gen Virol. 1969;5:221–236. doi: 10.1099/0022-1317-5-2-221. [DOI] [PubMed] [Google Scholar]
- 28.Norrby E, Wadell G, Marusyk H. Fiber-associated incomplete and complete hemagglutinins of adenovirus type 6. Arch Gesamte Virusforsch. 1969;28:239–244. doi: 10.1007/BF01249389. [DOI] [PubMed] [Google Scholar]
- 29.Novelli A, Boulanger P. Deletion analysis of functional domains in baculovirus-expressed adenovirus type 2 fiber. Virology. 1991;185:365–376. doi: 10.1016/0042-6822(91)90784-9. [DOI] [PubMed] [Google Scholar]
- 30.Philipson L, Lonberg-Holm K, Pettersson U. Virus-receptor interaction in an adenovirus system. J Virol. 1968;2:1064–1075. doi: 10.1128/jvi.2.10.1064-1075.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pring-Åkerblom P, Adrian T. Characterization of adenovirus subgenus D fiber genes. Virology. 1995;206:564–571. doi: 10.1016/s0042-6822(95)80073-5. [DOI] [PubMed] [Google Scholar]
- 32.Pring-Åkerblom P, Adrian T. Sequence characterization of the adenovirus 31 fibre and comparison with serotypes of subgenera A to F. Res Virol. 1995;146:343–354. doi: 10.1016/0923-2516(96)80597-8. [DOI] [PubMed] [Google Scholar]
- 33.Roelvink P W, Kovesdi I, Wickham T J. Comparative analysis of adenovirus fiber-cell interaction: adenovirus type 2 (Ad2) and Ad9 utilize the same cellular fiber receptor but use different binding strategies for attachment. J Virol. 1996;70:7614–7621. doi: 10.1128/jvi.70.11.7614-7621.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rosén L. Hemagglutination of adenoviruses. Virology. 1958;5:574. doi: 10.1016/0042-6822(58)90050-3. [DOI] [PubMed] [Google Scholar]
- 35.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Signäs C, Akusjärvi G, Pettersson U. Adenovirus 3 fiber polypeptide gene: implications for the structure of the fiber protein. J Virol. 1985;53:672–678. doi: 10.1128/jvi.53.2.672-678.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stevenson S C, Rollence M, Marshall-Neff J, McClelland A. Selective targeting of human cells by a chimeric adenovirus vector containing a modified fiber protein. J Virol. 1997;71:4782–4790. doi: 10.1128/jvi.71.6.4782-4790.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stevenson S C, Rollence M, White B, Weaver L, McClelland A. Human adenovirus serotypes 3 and 5 bind to two different cellular receptors via the fiber head domain. J Virol. 1995;69:2850–2857. doi: 10.1128/jvi.69.5.2850-2857.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wigand R. Adenoviruses types 20, 25, and 28: atypical members of group II. J Gen Virol. 1970;6:325–328. doi: 10.1099/0022-1317-6-2-325. [DOI] [PubMed] [Google Scholar]
- 40.Xia D, Henry L J, Gerard R D, Deisenhofer J. Crystal structure of the receptor-binding domain of adenovirus type 5 fiber protein at 1.7 Å resolution. Structure (London) 1994;2:309–319. doi: 10.1016/s0969-2126(94)00126-x. [DOI] [PubMed] [Google Scholar]
- 41.Xia D, Henry L, Gerard R D, Deisenhofer J. Structure of the receptor binding domain of adenovirus type 5 fiber protein. In: Doerfler W, Boehm P, editors. The molecular repertoire of adenoviruses. I. Berlin, Germany: Springer-Verlag; 1995. pp. 39–46. [Google Scholar]
- 42.Yang P, Bansal A, Chongguang L, Air G M. Hemagglutinin specificity and neuraminidase coding capacity of neuraminidase-deficient influenza viruses. Virology. 1997;229:155–165. doi: 10.1006/viro.1996.8421. [DOI] [PubMed] [Google Scholar]