Abstract
Heterozygous ARID1B variants result in Coffin–Siris syndrome. Features may include hypoplastic nails, slow growth, characteristic facial features, hypotonia, hypertrichosis, and sparse scalp hair. Most reported cases are due to ARID1B loss of function variants. We report a boy with developmental delay, feeding difficulties, aspiration, recurrent respiratory infections, slow growth, and hypotonia without a clinical diagnosis, where a previously unreported ARID1B missense variant was classified as a variant of uncertain significance. The pathogenicity of this variant was refined through combined methodologies including genome‐wide methylation signature analysis (EpiSign), Machine Learning (ML) facial phenotyping, and LIRICAL. Trio exome sequencing and EpiSign were performed. ML facial phenotyping compared facial images using FaceMatch and GestaltMatcher to syndrome‐specific libraries to prioritize the trio exome bioinformatic pipeline gene list output. Phenotype‐driven variant prioritization was performed with LIRICAL. A de novo heterozygous missense variant, ARID1B p.(Tyr1268His), was reported as a variant of uncertain significance. The ACMG classification was refined to likely pathogenic by a supportive methylation signature, ML facial phenotyping, and prioritization through LIRICAL. The ARID1B genotype–phenotype has been expanded through an extended analysis of missense variation through genome‐wide methylation signatures, ML facial phenotyping, and likelihood‐ratio gene prioritization.
Keywords: ARID1B, Coffin–Siris syndrome, episignature, FaceMatch, GestaltMatcher, LIRICAL
1. INTRODUCTION
ARID1B is a frequent cause of intellectual disability (ID; van der Sluijs et al., 2019). ARID1B‐related neurodevelopmental disorder (ARID1B‐RD) displays phenotypic heterogeneity, including Coffin–Siris syndrome 1, OMIM #135900 (ARID1B‐CSS) and ID with or without dysmorphic features (Vergano et al., 1993). ARID1B‐RD can present with variable ID, myopia, spasticity, epilepsy, feeding difficulties, and laryngomalacia, while typical features of ARID1B‐CSS also include nail hypoplasia, hypotonia, limb hypertrichosis, and characteristic facial features (van der Sluijs et al., 2019; Vergano et al., 1993). The clinical features observed in ARID1B cohorts depend on the selection criteria. Due to ascertainment bias, individuals with a priori diagnosis of ARID1B‐CSS may over‐represent more discernible features (Bogershausen & Wollnik, 2018; Santen et al., 2014; van der Sluijs et al., 2019; Vergano et al., 1993). Pathogenic variants may be inherited from a mildly affected parent (Mignot et al., 2016; van der Sluijs et al., 2019, 2021). BAF complex disruption is associated with Coffin–Siris and Nicolaides–Baraitser syndromes (SMARCA2 OMIM #600014; Wolff et al., 2012), collectively known as BAFopathies, which include developmental delay, coarse facial features, and phalangeal abnormalities (Aref‐Eshghi et al., 2018, 2019). Coffin–Siris syndrome may result from pathogenic variants in other genes, including ARID1A (CSS2 OMIM #614607), SMARCB1 (CSS3 OMIM #614608), SMARCA4 (CSS4 OMIM #614609), SMARCE1 (CSS5 OMIM #616938), ARID2 (CSS6 OMIM #617808), DPF2 (CSS7 OMIM #618027), SMARCC2 (CSS8 OMIM #618362), and SMARCD1 (CSS11 OMIM #618779).
ARID1B loss of function variants are identified commonly in ARID1B‐RD. Missense variants are reported rarely and are usually considered not to be pathogenic (Santen et al., 2014; van der Sluijs et al., 2019). Previous reports of missense ARID1B variants include a maternally inherited heterozygous variant c.6092 T > C p.(Ile2031Thr) in a proband with agenesis of the corpus callosum where the mother had mild ID (de novo in mother; Mignot et al., 2016). A likely pathogenic missense variant has been reported in a male with ID and facial dysmorphism, c.2308G > A p.(Gly770Arg; Grozeva et al., 2015). Other published ARID1B missense variants include two inherited and two de novo variants presenting with short stature (Yu et al., 2015) and two de novo variants in children with ID (Zhang et al., 2020). At the time of reporting, ClinVar documents 18 ARID1B likely pathogenic and 2 pathogenic missense variants in ARID1B (see Table S1) compared to 123 truncating variants. Additional ARID1B missense variants in ClinVar include 400 variants of uncertain significance, 125 likely benign, 49 benign, and 47 variants with conflicting interpretations (https://www.ncbi.nlm.nih.gov/clinvar/?term=ARID1B%5Bgene%5D&redir=gene).
We describe a 1‐year‐old male with moderate ID, hypotonia, feeding difficulties, nephrolithiasis, aspiration, and recurrent lower respiratory tract infections. A heterozygous ARID1B missense variant identified via trio whole exome sequencing was reported as a variant of uncertain significance (VUS). The combination of genome‐wide methylation signatures, ML Facial Phenotyping (GestaltMatcher and FaceMatch), and likelihood ratio interpretation of clinical abnormalities (LIRICAL) variant reprioritization showed utility in pathogenicity reclassification to obtain a refined diagnosis of likely pathogenic.
1.1. Case report
The proband was born at 30 weeks' gestation with growth parameters adjusted for prematurity showing weight 1.63 kg (80th centile), length 40.5 cm (42nd centile), and head circumference 30.0 cm (95th centile). His parents were non‐consanguineous. He was referred at 9 weeks corrected gestational age (CGA) to assess central apneas, laryngomalacia, an atrial septal defect, and feeding difficulties with slow growth. His weight was 4.2 kg (15th centile), length 53 cm (7th centile), and head circumference of 40 cm (69th centile). He was observed to have coarse facial features, bitemporal narrowing, dolichocephaly, and apparent rhizomelia. An uncoordinated suck and hyperekplexia with normal reflexes and tone were observed. A diagnosis of nephrolithiasis was made based on echogenic material on renal ultrasound with subsequent hypercalciuria on urinary studies. Based on a maternal history of renal calculi, fluids optimization was recommended, and potassium citrate commenced. A skeletal survey, MRI brain, SNP‐microarray, and biochemical testing, including investigations for congenital disorders of glycosylation and storage disorders, were non‐diagnostic. On review at 8 months CGA, he had moderate to severe developmental delay, ongoing feeding difficulties, right‐sided conductive hearing loss, severe gastro‐esophageal reflux, and dysphagia requiring gastrostomy feeds. Supplemental oxygen was needed for central apneas. His weight was 9.5 kg (65th centile), length 65 cm (32nd centile), and head circumference 48.2 cm (>99th centile). He had nonfamilial facial features with full lips, ptosis, a short full nose, and a prominent glabella. Deep palmar and sole creases, hypodontia, and hypotonia were present. Repeat MRI brain showed mild prominence of the ventricles and extra‐axial CSF space, considered within normal limits for age and a likely small arachnoid cyst in the left posterior fossa. There was no other significant intracranial structural abnormality, and a specific clinical diagnosis was not made.
2. MATERIALS AND METHODS
DNA was extracted from peripheral blood, and exome sequencing was performed using CRE II whole exome kit (Agilent) on a NovaSeq 6000 instrument (Illumina) with variant annotation, filtering, and prioritization were performed using an in‐house genomics pipeline, Genomics Annotation and Interpretation Application (GAIA) as described previously (Sundercombe et al., 2021). Variants were prioritized using an automated PubMed search linked to the referral phenotype performed within GAIA as well as annotation from clinical genetic and variant databases, including ClinVar (http://www.ncbi.nlm.nih.gov/clinvar), HGMD (http://www.hgmd.cf.ac.uk/ac/index.php), and OMIM (http://www.omim.org).
Genome‐wide methylation testing was performed on bisulfite converted DNA using the EZ DNA Methylation Kit (Zymo Research). Following conversion, methylation analysis was performed using the Infinium MethylationEPIC beadchip array (Illumina) and processed on a NextSeq 550 (Illumina). The resulting methylated and unmethylated signal intensity data were imported into R 3.5.2 for analysis. Normalization was performed according to the Illumina normalization method with background correction using the minfi package. The methylation level for each probe was measured as a beta value calculated from the ratio of the methylated signals to the sum of unmethylated and methylated signals (Aref‐Eshghi et al., 2019). DNA methylation signature analysis was performed using the EpiSign analysis protocol as previously described (Sadikovic et al., 2021).
FaceMatch (www.facematch.org.au; Dudding‐Byth et al., 2017) and GestaltMatcher (www.gestaltmatcher.org; Hsieh et al., 2022; Hustinx et al., 2023) were utilized for ML facial phenotyping to compare facial images and gene lists generated from the genomic output from GAIA after trio exome analysis. Weighted facial matches were assessed compared to the clinical diagnosis in the proband.
LIRICAL (https://lirical.readthedocs.io/en/latest/; Robinson et al., 2020) was utilized to perform phenotype‐driven prioritization of candidate genes. The phenotypic features were standardized to the Human Phenotype Ontology (HPO) terms and used to reprioritize the list of filtered variants from the genomic pipeline.
3. RESULTS
3.1. Variant curation
Variant curation was phenotype‐driven with classification according to the American College of Medical Genetics and Genomics (ACMG) criteria. Trio exome sequencing (ES) identified a de novo heterozygous variant predicted to result in a missense change in ARID1B: Chr6(GRCh38) g.157184318 T > C, NM_001374828.1:c.3802 T > C, p.(Tyr1268His) in exon 14 of 21 where the normally encoded tyrosine is replaced by histidine. The pathogenicity categorization for the variant was initially assessed as a VUS.
The ARID1B:p.(Tyr1268His) variant has not been reported in the literature and is absent in population databases (gnomAD) [PM2]. While ARID1B is not uniformly under missense constraint (Z = 2.59, o/e = 0.79[0.75–0.83]), there is a localized missense constraint of the region containing the variant in the ARID/BRIGHT DNA binding domain (PF01388) with a MetaDome tolerance score of dn/ds = 0.1 (highly intolerant to missense change; see Figure S1a; Wiel et al., 2019). There is a cluster of likely pathogenic or pathogenic variants in exon 14 with an absence of benign missense variation [PM1] (Figure S1b). Protein prediction modeling using the Protein Hope platform showed that the mutant histidine variant was smaller and less hydrophobic than the native tyrosine. In silico prediction tools supported a deleterious effect [PP3_moderate] (Mutscore 0.968, VARITY_R 0.89, ClinPred 0.9993, REVEL 0.891, Varcards 21:23). Missense variants are uncommonly reported as pathogenic or likely pathogenic in ARID1B‐RD [BP1].
Given the phenotypic correlation with ARID1B‐CSS, the PS2_supporting criterion was included in the pathogenicity categorization, downgraded due to the significant genetic heterogeneity of ID. Using the ACMG classification criteria for pathogenicity categorization, the de novo missense ARID1B:p.(Tyr1268His) variant reported as a VUS with a posterior probability of pathogenicity of 67.5% based on one moderate [PM1], three supporting [PM2_supporting, PS2_supporting, and PP3] and one benign supporting criteria [BP1] (Tavtigian et al., 2018).
Additional independent phenotyping was performed using EpiSign, ML facial technology, and LIRICAL to determine whether the Bayesian components of phenotype specificity or the number of genes to be considered could be altered in the ACMG criteria.
3.2. Episignature
Mutations in ARID1B, and several other genes that are part of the BAF complex, result in a distinct DNA methylation profile in patients with BAF‐complex disorders (Aref‐Eshghi et al., 2018). This episignature shows a strong association with high sensitivity and specificity for BAF‐complex disorders, allowing its use as a diagnostic biomarker. The biological causation has not been precisely determined. However, one plausible explanation is disruption in a transcriptional regulatory complex leads to disruptions in gene expression and chromatin states during early development that remain detectable in differentiated tissues in affected patients. The methylation profile was consistent with a BAFopathy episignature (Aref‐Eshghi et al., 2018), with the variant clustering with other individuals with BAFopathies (see Figure 1). The BAF episignature cohort is genetically heterogeneous including both truncating and pathogenic missense variants(Aref‐Eshghi et al., 2018). There is insufficient evidence currently of sub clustering based on the variant type within each gene. This result reduced the genetic heterogeneity for the de novo classification PS2, allowing upgrading the strength of PS2_supporting to PS2_moderate, as there are currently approximately 90 genes with an episignature, and no additional variants in BAFopathy genes were identified.
FIGURE 1.
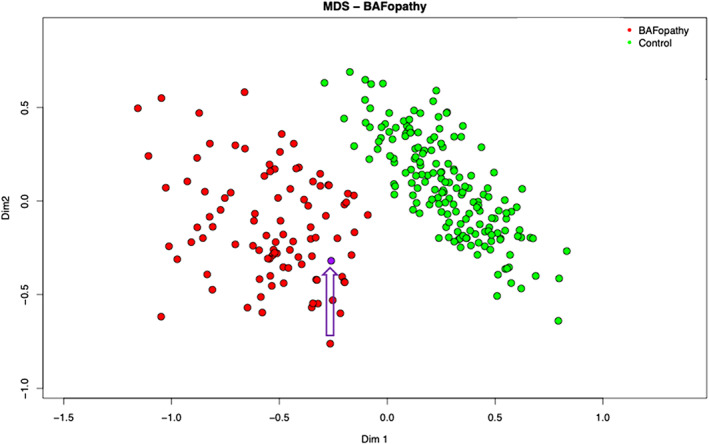
Episignature image, proband in purple with arrow, BAFopathy positive episignatures in red.
3.3. ML‐facial phenotyping
FaceMatch produces a similarity score to a series of photographs converted to a distance via a linear transformation and ranked. Distances above 0.85 are non‐informative, 0.8 to 0.85 indicate a mild similarity, and below 0.8 is a possible match. The face alone did not generate a FaceMatch correlation; however, when filtered against only the filtered genes from the trio ES, ARID1B was calculated to be the top‐ranked gene with a distance of 0.8193 (see Figure 2a).
FIGURE 2.
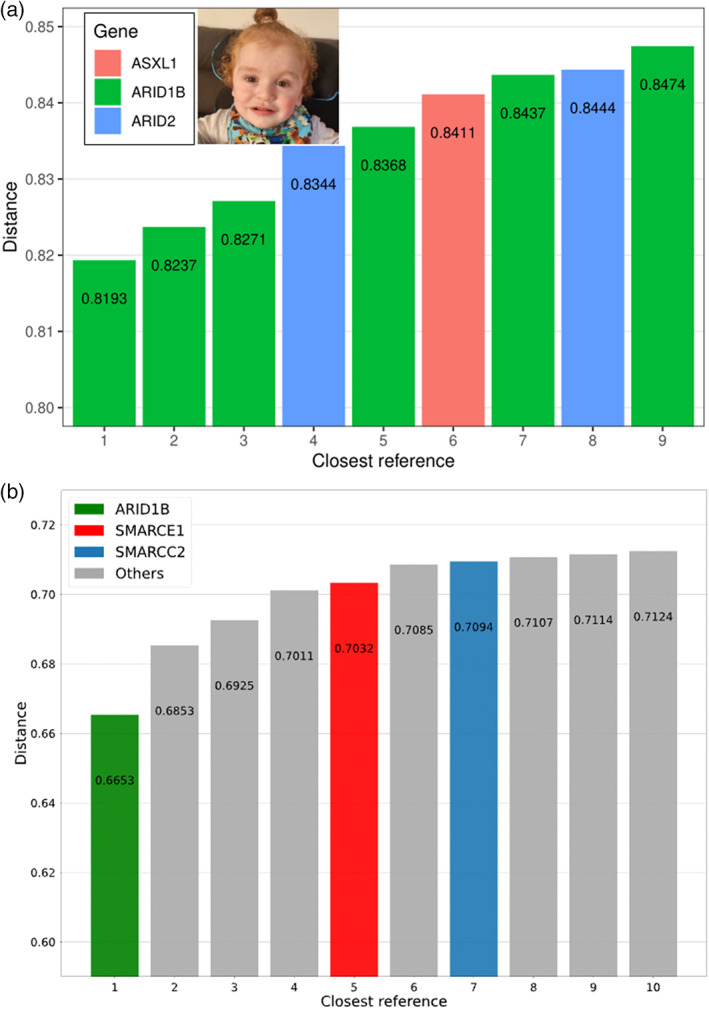
(a) Results from FaceMatch, rank results indicate the order in increasing distance, and each bar match to a reference image within the 67 reference individuals in the FaceMatch database with OMIM syndrome and gene in the list of interest. The number of reference matches with distance below the threshold of 0.85. (b) Results from GestaltMatcher: rank results to the top‐10 similar patients among 7459 patients' images with 449 different disorders in GestaltMatcher Database. The ranks of patients' genes are ARID1B, ZBTB20, MTOR, KCNK9, SMARCE1, RAC1, SMARCC2, MAP3K7, ACTG1, and CREBBP.
GestaltMatcher uses deep‐learning algorithms to quantify facial image similarities to 7459 patients' images with 449 different disorders in GestaltMatcher Database (GMDB; Lesmann et al., 2023). In GMDB, there were 130 images with CSS. ARID1B was GestaltMatcher's top prioritized gene for the described individual (see Figure 2b). Therefore, ARID1B was the only matched possibility in the genomic pipeline‐filtered genes. Based on these results, the pathogenic supporting term for a consistent phenotype PP4 was added.
3.4. LIRICAL
LIRICAL uses a likelihood ratio (LR) to estimate the posttest diagnostic probability, the LR for each observed HPO phenotype, and the predicted pathogenicity of observed genotypes (Robinson et al., 2020). The referral terms chosen were hypotonia (HP:0001252), atrial septal defect (HP:0001631), postnatal growth retardation (HP:0008897), aspiration (HP:0002835), recurrent infections due to aspiration (HP:0004891), feeding difficulties in infancy (HP:0008872), and moderate global developmental delay (HP:0011343). LIRICAL identified ARID1B as the top‐ranked gene in the exome sequencing output with a posttest probability of 96%. This prioritization increased the Bayesian contribution of phenotype and genotype, further supporting pathogenicity.
4. RECLASSIFICATION
The revised phenotypic correlation included the pathogenic supporting term for a consistent phenotype [PP4] and upgrading of PS2 [PS2_moderate]. After the initial reporting of the case, based on weighting recommendations for PP3, due to a REVEL score of 0.891, PP3 was upgraded to [PP3_moderate] (Pejaver et al., 2022). The de novo missense ARID1B variant was reclassified as likely pathogenic with a posterior probability of pathogenicity of 97.5% based on three moderate [PM1, PP3_moderate, and PS2_moderate] and three supporting criteria [PM2_supporting, PP2, and PP4]. BP1 was used given the predominant LOF ARID1B mutational mechanism (Pejaver et al., 2022).
5. DISCUSSION
This report highlights the challenges in assigning pathogenicity to de novo missense variants in a gene typically characterized by LOF. ARID1B haploinsufficiency could result from missense variants, although variants within exon 1 are unlikely to impact protein function due to frequent benign missense variation in that region (Bogershausen & Wollnik, 2018). Published likely pathogenic missense variants typically cluster near the C‐terminus (Bogershausen & Wollnik, 2018). Many ARID1B missense variants cluster near the highly conserved missense variant identified in this study (Grozeva et al., 2015; Mignot et al., 2016; Quinodoz et al., 2022). Classifying a novel ARID1B missense variant as likely pathogenic demonstrates the importance of considering missense variation in diseases typically associated with LOF or haploinsufficiency.
Clinical phenotyping shows ascertainment bias, with extreme phenotypes facilitating both syndrome recognition and gene identification. Many syndromes have facial dysmorphism as key diagnostic features; however, reliance on clinical diagnosis biases means that traditional phenotyping provides a weak ACMG framework Bayesian likelihood modification. ML‐facial phenotyping can increase the specificity of facial phenotyping (Dudding‐Byth et al., 2017). GestaltMatcher and FaceMatch can match individuals with known or unknown disorders with functionality to explore facial similarity (Dudding‐Byth et al., 2017; Hsieh et al., 2022). Syndrome distinctiveness in GestaltMatcher correlated with expert opinion (Hsieh et al., 2022). Similarly, the diagnostic rate was >80% for all syndromes in the FaceMatch pilot study and diagnostically more accurate than three senior clinical geneticists (p < 0.00001; Dudding‐Byth et al., 2017).
Human‐independent phenotyping methodologies provide non‐biased syndrome correlation to facilitate the interpretation of DNA sequencing data (Dudding‐Byth et al., 2017; Hsieh et al., 2022), with a role in classifying ARID1B variants (van der Sluijs et al., 2021). Both GestaltMatcher and FaceMatch had ARID1B as the top‐ranked gene in the variants from the genomic output. These results are consistent with facial phenotyping providing the highest diagnostic accuracy when combined with a filtered gene list.
Genome‐wide DNA methylation patterns may provide additional evidence for variant classification in genes affected by chromatin remodeling or epigenetic reprogramming (Aref‐Eshghi et al., 2019; Sadikovic et al., 2021; Rooney & Sadikovic, 2022). EpiSign analysis was particularly important for reclassification as it provided an independent functional link between an atypical ARID1B missense variant and a cluster of ARID1B episignatures.
The correct diagnosis was within the LIRICAL first three ranks in 92.9% of 262 Mendelian diseases, with a mean posttest probability of 67.3% (Robinson et al., 2020). The HPO terms used are identified commonly in many syndromes, but when applied to a filtered gene list, they became highly specific. Global developmental delay, significant gastro‐esophageal reflux, and feeding difficulties are frequent in ID and CSS (van der Sluijs et al., 2019; Vergano et al., 1993); however, none of the other gene variants considered from the exome analysis scored on these phenotypes. While the nephrolithiasis and hypercalciuria were considered familial in nature, nephrolithiasis are also reported in ARID1B patients (van der Sluijs et al., 2019). Although hypertrichosis, sparse scalp hair, and fifth‐digit hypoplasia were not observed in the proband, full lips were consistent with ARID1B‐CSS, adding Bayesian support for the pathogenicity of the variant.
6. CONCLUSION
The ARID1B variant has been re‐classified through an extended analysis, providing a diagnosis and accurate reproductive counseling. EpiSign, ML‐facial phenotyping, and LIRICAL demonstrated the utility of methylation signatures, phenotyping independent of clinical assessments, and the reprioritization of variants from a genomic pipeline for variant reclassification. Identifying an ARID1B VUS should prompt discussion with the referring clinician about the possibility of an ARID1B‐RD and consider further investigation using phenotyping and episignature analyses.
AUTHOR CONTRIBUTIONS
Caitlin Forwood: conceptualization contributed clinical background, original draft, and revisions. Jason Pinner: lead clinician contributing to case report, conceptualization, manuscript revision. Katie Ashton: genomic analysis and reporting, manuscript revision. Ying Zhu and Futao Zhang: bioinformatic pipeline development, data analysis and integration with LIRICAL. Kerith‐Rae Dias, Krystle Standen, Carey‐Anne Evans, and Louise Carey: data analysis and application of EpiSign. Michael Cardamone, Carolyn Shalhoub, and Hala Katf: clinicians who contributed clinical information. Carlos Rivera and Tracy Dudding‐Byth: development of FaceMatch and yielded results with the genomic output, manuscript revision. Tzung‐Chien Hsieh and Peter Krawitz: development of GestaltMatcher and yielded results with the genomic output, manuscript revision. Peter Robinson: designed LIRICAL and yielded the results with the genomic output. Bekim Sadikovic: development of EpiSign, results interpretation/application and manuscript revision. Michael Buckley and Tony Roscioli: conceptualization, supervision of data and manuscript, genomic analysis, and data analysis/integration. All co‐authors reviewed and revised the manuscript.
CONFLICT OF INTEREST STATEMENT
The authors declare no conflicts of interest.
Supporting information
FIGURE S1: (a) Tyrosine 1185 residue MetaDome tolerance score of dn/ds = 0.1 (highly intolerant to missense change) (b) Screenshot from Mutscore with pathogenic/likely pathogenic variants from ClinVar shown in yellow and a whole of exon 13 with an absence of benign missense variation (note different transcript used NM_001346813.1) (c) Hidden Markov Model logogram displaying conservation as a graphical representation based on the percentage height that an amino acid occupies. Red vertical lines are places where the sequences poorly align or gaps in the sequence. The large Y represents Tyrosine and supports the evolutionary conservation of the amino acid.
TABLE S1: ClinVar LP/P missense variants.
ACKNOWLEDGEMENTS
We would like the family for their involvement in this study. Open access publishing facilitated by University of New South Wales, as part of the Wiley ‐ University of New South Wales agreement via the Council of Australian University Librarians.
Forwood, C. , Ashton, K. , Zhu, Y. , Zhang, F. , Dias, K.‐R. , Standen, K. , Evans, C.‐A. , Carey, L. , Cardamone, M. , Shalhoub, C. , Katf, H. , Riveros, C. , Hsieh, T.‐C. , Krawitz, P. , Robinson, P. N. , Dudding‐Byth, T. , Sadikovic, B. , Pinner, J. , Buckley, M. F. , & Roscioli, T. (2023). Integration of EpiSign, facial phenotyping, and likelihood ratio interpretation of clinical abnormalities in the re‐classification of an ARID1B missense variant. American Journal of Medical Genetics Part C: Seminars in Medical Genetics, 193C:e32056. 10.1002/ajmg.c.32056
All variants are confirmed to be HGVS compliant.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available on request from the corresponding author. The data are not publicly available due to privacy or ethical restrictions.
REFERENCES
- Aref‐Eshghi, E. , Bend, E. G. , Colaiacovo, S. , Caudle, M. , Chakrabarti, R. , Napier, M. , Brick, L. , Brady, L. , Carere, D. A. , Levy, M. A. , Kerkhof, J. , Stuart, A. , Saleh, M. , Beaudet, A. L. , Li, C. , Kozenko, M. , Karp, N. , Prasad, C. , Siu, V. M. , … Sadikovic, B. (2019). Diagnostic utility of genome‐wide DNA methylation testing in genetically unsolved individuals with suspected hereditary conditions. American Journal of Human Genetics, 104(4), 685–700. 10.1016/j.ajhg.2019.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aref‐Eshghi, E. , Bend, E. G. , Hood, R. L. , Schenkel, L. C. , Carere, D. A. , Chakrabarti, R. , Nagamani, S. C. S. , Cheung, S. W. , Campeau, P. M. , Prasad, C. , Siu, V. M. , Brady, L. , Tarnopolsky, M. A. , Callen, D. J. , Innes, A. M. , White, S. M. , Meschino, W. S. , Shuen, A. Y. , Pare, G. , … Sadikovic, B. (2018). BAFopathies' DNA methylation epi‐signatures demonstrate diagnostic utility and functional continuum of Coffin–Siris and Nicolaides–Baraitser syndromes. Nature Communications, 9(1), 4885. 10.1038/s41467-018-07193-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogershausen, N. , & Wollnik, B. (2018). Mutational landscapes and phenotypic spectrum of SWI/SNF‐related intellectual disability disorders. Frontiers in Molecular Neuroscience, 11, 252. 10.3389/fnmol.2018.00252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudding‐Byth, T. , Baxter, A. , Holliday, E. G. , Hackett, A. , O'Donnell, S. , White, S. M. , Attia, J. , Brunner, H. , de Vries, B. , Koolen, D. , Kleefstra, T. , Ratwatte, S. , Riveros, C. , Brain, S. , & Lovell, B. C. (2017). Computer face‐matching technology using two‐dimensional photographs accurately matches the facial gestalt of unrelated individuals with the same syndromic form of intellectual disability. BMC Biotechnology, 17(1), 90. 10.1186/s12896-017-0410-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grozeva, D. , Carss, K. , Spasic‐Boskovic, O. , Tejada, M. I. , Gecz, J. , Shaw, M. , Corbett, M. , Haan, E. , Thompson, E. , Friend, K. , Hussain, Z. , Hackett, A. , Field, M. , Renieri, A. , Stevenson, R. , Schwartz, C. , Floyd, J. A. , Bentham, J. , Cosgrove, C. , … Raymond, F. L. (2015). Targeted next‐generation sequencing analysis of 1,000 individuals with intellectual disability. Human Mutation, 36(12), 1197–1204. 10.1002/humu.22901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh, T. C. , Bar‐Haim, A. , Moosa, S. , Ehmke, N. , Gripp, K. W. , Pantel, J. T. , Danyel, M. , Mensah, M. A. , Horn, D. , Rosnev, S. , Fleischer, N. , Bonini, G. , Hustinx, A. , Schmid, A. , Knaus, A. , Javanmardi, B. , Klinkhammer, H. , Lesmann, H. , Sivalingam, S. , … Krawitz, P. M. (2022). GestaltMatcher facilitates rare disease matching using facial phenotype descriptors. Nature Genetics, 54(3), 349–357. 10.1038/s41588-021-01010-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hustinx, A. , Hellmann, F. , Sumer, O. , Javanmardi, B. , Andre, E. , Krawitz, P. , & Hsieh, T.‐C. (2023). Improving deep facial phenotyping for ultra‐rare disorder verification using model ensembles 2023 IEEE/CVF winter conference on applications of computer vision (WACV). https://doi.ieeecomputersociety.org/10.1109/WACV56688.2023.00499
- Lesmann, H. , Lyon, G. J. , Caro, P. , Abdelrazek, I. M. , Moosa, S. , Pantel, J. T. , Hagen, M. t. , Rosnev, S. , Kamphans, T. , Meiswinkel, W. , Li, J.‐M. , Klinkhammer, H. , Hustinx, A. , Javanmardi, B. , Knaus, A. , Uwineza, A. , Knopp, C. , Marchi, E. , Elbracht, M. , … Hsieh, T.‐C. (2023). GestaltMatcher database—A FAIR database for medical imaging data of rare disorders. medRxiv. 10.1101/2023.06.06.23290887 [DOI] [Google Scholar]
- Mignot, C. , Moutard, M. L. , Rastetter, A. , Boutaud, L. , Heide, S. , Billette, T. , Doummar, D. , Garel, C. , Afenjar, A. , Jacquette, A. , Lacombe, D. , Verloes, A. , Bole‐Feysot, C. , Nitschke, P. , Masson, C. , Faudet, A. , Lesne, F. , Bienvenu, T. , Alby, C. , … Heron, D. (2016). ARID1B mutations are the major genetic cause of corpus callosum anomalies in patients with intellectual disability. Brain, 139(11), e64. 10.1093/brain/aww181 [DOI] [PubMed] [Google Scholar]
- Pejaver, V. , Byrne, A. B. , Feng, B. J. , Pagel, K. A. , Mooney, S. D. , Karchin, R. , O'Donnell‐Luria, A. , Harrison, S. M. , Tavtigian, S. V. , Greenblatt, M. S. , Biesecker, L. G. , Radivojac, P. , Brenner, S. E. , & ClinGen Sequence Variant Interpretation Working Group . (2022). Calibration of computational tools for missense variant pathogenicity classification and ClinGen recommendations for PP3/BP4 criteria. American Journal of Human Genetics, 109(12), 2163–2177. 10.1016/j.ajhg.2022.10.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinodoz, M. , Peter, V. G. , Cisarova, K. , Royer‐Bertrand, B. , Stenson, P. D. , Cooper, D. N. , Unger, S. , Superti‐Furga, A. , & Rivolta, C. (2022). Analysis of missense variants in the human genome reveals widespread gene‐specific clustering and improves prediction of pathogenicity. American Journal of Human Genetics, 109(3), 457–470. 10.1016/j.ajhg.2022.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson, P. N. , Ravanmehr, V. , Jacobsen, J. O. B. , Danis, D. , Zhang, X. A. , Carmody, L. C. , Gargano, M. A. , Thaxton, C. L. , Core, U. N. C. B. , Karlebach, G. , Reese, J. , Holtgrewe, M. , Kohler, S. , McMurry, J. A. , Haendel, M. A. , & Smedley, D. (2020). Interpretable clinical genomics with a likelihood ratio paradigm. American Journal of Human Genetics, 107(3), 403–417. 10.1016/j.ajhg.2020.06.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rooney, K. , & Sadikovic, B. (2022). DNA methylation episignatures in neurodevelopmental disorders associated with large structural copy number variants: Clinical implications. International Journal of Molecular Sciences, 23(14), 7862 . 10.3390/ijms23147862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadikovic, B. , Levy, M. A. , Kerkhof, J. , Aref‐Eshghi, E. , Schenkel, L. , Stuart, A. , McConkey, H. , Henneman, P. , Venema, A. , Schwartz, C. E. , Stevenson, R. E. , Skinner, S. A. , DuPont, B. R. , Fletcher, R. S. , Balci, T. B. , Siu, V. M. , Granadillo, J. L. , Masters, J. , Kadour, M. , … Alders, M. (2021). Clinical epigenomics: Genome‐wide DNA methylation analysis for the diagnosis of Mendelian disorders. Genetics in Medicine, 23(6), 1065–1074. 10.1038/s41436-020-01096-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santen, G. W. , Clayton‐Smith, J. , & ARID1B‐CSS Consortium . (2014). The ARID1B phenotype: What we have learned so far. American Journal of Medical Genetics. Part C, Seminars in Medical Genetics, 166C(3), 276–289. 10.1002/ajmg.c.31414 [DOI] [PubMed] [Google Scholar]
- Sundercombe, S. L. , Berbic, M. , Evans, C. A. , Cliffe, C. , Elakis, G. , Temple, S. E. L. , Selvanathan, A. , Ewans, L. , Quayum, N. , Nixon, C. Y. , Dias, K. R. , Lang, S. , Richards, A. , Goh, S. , Wilson, M. , Mowat, D. , Sachdev, R. , Sandaradura, S. , Walsh, M. , … Roscioli, T. (2021). Clinically responsive genomic analysis pipelines: Elements to improve detection rate and efficiency. The Journal of Molecular Diagnostics, 23(7), 894–905. 10.1016/j.jmoldx.2021.04.007 [DOI] [PubMed] [Google Scholar]
- Tavtigian, S. V. , Greenblatt, M. S. , Harrison, S. M. , Nussbaum, R. L. , Prabhu, S. A. , Boucher, K. M. , Biesecker, L. G. , & ClinGen Sequence Variant Interpretation Working, G . (2018). Modeling the ACMG/AMP variant classification guidelines as a Bayesian classification framework. Genetics in Medicine, 20(9), 1054–1060. 10.1038/gim.2017.210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Sluijs, P. J. , Alders, M. , Dingemans, A. J. M. , Parbhoo, K. , van Bon, B. W. , Dempsey, J. C. , Doherty, D. , den Dunnen, J. T. , Gerkes, E. H. , Milller, I. M. , Moortgat, S. , Regier, D. S. , Ruivenkamp, C. A. L. , Schmalz, B. , Smol, T. , Stuurman, K. E. , Vincent‐Delorme, C. , de Vries, B. B. A. , Sadikovic, B. , … Santen, G. W. E. (2021). A case series of familial ARID1B variants illustrating variable expression and suggestions to update the ACMG criteria. Genes (Basel), 12(8), 1275. 10.3390/genes12081275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Sluijs, P. J. , Jansen, S. , Vergano, S. A. , Adachi‐Fukuda, M. , Alanay, Y. , AlKindy, A. , Baban, A. , Bayat, A. , Beck‐Wodl, S. , Berry, K. , Bijlsma, E. K. , Bok, L. A. , Brouwer, A. F. J. , van der Burgt, I. , Campeau, P. M. , Canham, N. , Chrzanowska, K. , Chu, Y. W. Y. , Chung, B. H. Y. , … Santen, G. W. E. (2019). The ARID1B spectrum in 143 patients: From nonsyndromic intellectual disability to Coffin–Siris syndrome. Genetics in Medicine, 21(6), 1295–1307. 10.1038/s41436-018-0330-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergano, S. A. , van der Sluijs, P. J. , & Santen, G. (1993). ARID1B‐related disorder. In Adam M. P., Mirzaa G. M., Pagon R. A., Wallace S. E., Bean L. J. H., Gripp K. W., & Amemiya A. (Eds.), GeneReviews((R)). University of Washington, Seattle. https://www.ncbi.nlm.nih.gov/pubmed/31132234 [PubMed] [Google Scholar]
- Wiel, L. , Baakman, C. , Gilissen, D. , Veltman, J. A. , Vriend, G. , & Gilissen, C. (2019). MetaDome: Pathogenicity analysis of genetic variants through aggregation of homologous human protein domains. Human Mutation, 40(8), 1030–1038. 10.1002/humu.23798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff, D. , Endele, S. , Azzarello‐Burri, S. , Hoyer, J. , Zweier, M. , Schanze, I. , Schmitt, B. , Rauch, A. , Reis, A. , & Zweier, C. (2012). In‐frame deletion and missense mutations of the C‐terminal helicase domain of SMARCA2 in three patients with Nicolaides–Baraitser syndrome. Molecular Syndromology, 2(6), 237–244. 10.1159/000337323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, Y. , Yao, R. , Wang, L. , Fan, Y. , Huang, X. , Hirschhorn, J. , Dauber, A. , & Shen, Y. (2015). De novo mutations in ARID1B associated with both syndromic and non‐syndromic short stature. BMC Genomics, 16(1), 701. 10.1186/s12864-015-1898-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, T. , Wu, Q. , Yang, J. , Wu, D. , Shen, Y. , Yang, R. , & Huang, X. (2020). Identification of a de novo missense variant of ARID1B gene in a child with mental retardation. Zhonghua Yi Xue Yi Chuan Xue Za Zhi, 37(10), 1154–1157. 10.3760/cma.j.cn511374-20191006-00508 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
FIGURE S1: (a) Tyrosine 1185 residue MetaDome tolerance score of dn/ds = 0.1 (highly intolerant to missense change) (b) Screenshot from Mutscore with pathogenic/likely pathogenic variants from ClinVar shown in yellow and a whole of exon 13 with an absence of benign missense variation (note different transcript used NM_001346813.1) (c) Hidden Markov Model logogram displaying conservation as a graphical representation based on the percentage height that an amino acid occupies. Red vertical lines are places where the sequences poorly align or gaps in the sequence. The large Y represents Tyrosine and supports the evolutionary conservation of the amino acid.
TABLE S1: ClinVar LP/P missense variants.
Data Availability Statement
The data that support the findings of this study are available on request from the corresponding author. The data are not publicly available due to privacy or ethical restrictions.


