Abstract
An in vitro assay was developed to investigate endonuclease activity of Thogoto virus, a tick-borne orthomyxovirus. Endonuclease activity relied on an interaction between the 3′ and 5′ termini of virion RNA (vRNA) and not those of cRNA. Evidence was obtained that cap structures are cleaved directly from cap donors and that cleavage does not occur after pyrimidines. A 5′ hook structure, present in the vRNA promoter but not the cRNA promoter, was introduced into cRNA promoter mutants. These mutants stimulated endonuclease activity, although at levels slightly lower than that of vRNA. The ability of the cRNA promoter to stimulate endonuclease activity when mutated to contain a 5′ hook structure indicates that this structure constitutes a switching mechanism for endonuclease activity between the vRNA and cRNA promoters.
Thogoto virus (THOV) has been classified within the new genus Thogotovirus of the family Orthomyxoviridae (25) and can infect both vertebrates and ticks. The virus is structurally and genetically similar to influenza viruses and possesses a genome consisting of six negative-sense, single-stranded RNA segments (8). Each segment possesses conserved regions of semicomplementary nucleotides at the 3′ and 5′ termini which strongly resemble those of influenza viruses (18, 20). Its gene products are related to influenza virus polymerase proteins PB2 (segment 1), PB1 (segment 2), PA (segment 3), and nucleocapsid protein (segment 5) (18, 28). THOV segment 4 encodes a surface glycoprotein which is unrelated to any influenza virus protein but instead shows remarkable sequence homology to a baculovirus surface glycoprotein (22), possibly reflecting its tick-borne mode of transmission. With respect to transcription and replication, THOV appears to have many similarities with influenza viruses (for a review, see reference 15). These processes require a virus-encoded polymerase complex containing THOV proteins analogous to the PB1, PB2, PA, and NP proteins of influenza viruses. Such complexes are present inside virions (known as viral cores) which can be purified to carry out in vitro polymerase assays (20). THOV virion RNA (vRNA) serves as a template for the synthesis of mRNA and cRNA. mRNA synthesis is primed by host-derived cap structures, and the resulting molecules are truncated and polyadenylated, whereas cRNA molecules are complete, uncapped copies of vRNA which, in turn, serve as templates in vRNA synthesis (1, 20, 27, 28). Cap snatching of THOV differs from that in influenza viruses in that only the cap structure, preferentially m7GpppAm, is stolen from host messengers (1, 19, 28).
Recently, we have characterized the vRNA and cRNA promoters of THOV (20, 21). These promoters consist of the 3′- and 5′-terminal sequences of their respective RNA molecules, which form forked panhandle structures that are required for polymerase activity. Whereas the vRNA promoter stimulates transcription with capped primers, the cRNA promoter does not, pointing to the existence of a mechanism whereby capped mRNA and uncapped vRNA can be synthesized by the same polymerase complex, regulated by the type of RNA present in the complex. Another notable difference between the vRNA and cRNA promoters is the absence of a 5′ hook structure in the unpaired region of the cRNA promoter, which is vital for in vitro polymerase activity of the vRNA promoter (20, 21). In this paper, we report an in vitro endonuclease assay and its use in investigating the role of the 5′ hook structure in endonuclease activity. We demonstrate that this hook constitutes the switching mechanism for endonuclease activity between the vRNA and cRNA promoters.
MATERIALS AND METHODS
Preparation of viral cores.
THOV viral cores were prepared as described previously (19). Briefly, monolayers of BHK-21 (baby hamster kidney) cells were infected with Thogoto/SiAr/126/72 virus at approximately 0.05 PFU/cell. Medium was harvested 30 h postinfection and clarified by low-speed centrifugation to remove cellular material. Virus was pelleted for 2 h at 15°C through a 33% (vol/vol) glycerol cushion at 28,000 rpm in a Sorvall AH629 rotor. The concentrated virus was resuspended in TMN buffer (100 mM Tris-HCl [pH 7.4], 100 mM NaCl, 10 mM MgCl2, 1 mM dithiothreitol) supplemented with 5% glycerol and 1% Nonidet P-40 and incubated for 30 min at room temperature. The disrupted virus suspension was loaded onto a discontinuous glycerol gradient (66 and 33% [vol/vol]) in TMN buffer and centrifuged for 2 h at 15°C at 30,000 rpm in a Sorvall TH641 rotor. The interface was collected, loaded onto a 33% (vol/vol) glycerol cushion in TMN buffer, and centrifuged for 1 h at 15°C at 30,000 rpm in a Sorvall TH641 rotor. Pelleted cores were resuspended in TMN with 50% glycerol and frozen until use. Protein content was estimated with the Bio-Rad protein assay against bovine serum albumin standards.
Preparation of model RNAs.
Short model RNAs were transcribed with T7 RNA polymerase from partial DNA duplexes that consisted of an upstream double-stranded T7 RNA polymerase promoter region and a 5′-terminal overhang corresponding to the transcribed sequence as described previously (19). Briefly, DNA duplexes were made by mixing two partially complementary DNA oligonucleotides in equivalent molar concentrations in the presence of 10 mM Tris-HCl (pH 7.4), 1 mM EDTA, and 100 mM NaCl, followed by annealing at 80°C for 5 min and slow cooling down to room temperature. T7 RNA transcripts were synthesized with T7 RNA polymerase (Promega) in the presence of 0.5 μM DNA duplex, 40 mM Tris-HCl (pH 7.9), 10 mM NaCl, 6 mM MgCl2, 2 mM spermidine, 10 mM dithiothreitol, 0.5 mM each nucleoside triphosphate, and 20 U of RNasin (Promega) for 2 h at 37°C. The reaction mixture was treated for 15 min at 37°C with RNase-free DNase (Sigma), extracted twice with phenol-chloroform, precipitated with ethanol in the presence of 2 M ammonium acetate, washed in 80% ethanol, and dissolved in TE buffer (10 mM Tris-Cl [pH 7.4], 1 mM EDTA). To estimate concentration the RNA was dephosphorylated with alkaline phosphatase (Boehringer Mannheim) and end labeled with T4 polynucleotide kinase (Gibco BRL) in the presence of [α-32P]ATP, followed by electrophoresis through a 22% polyacrylamide–7 M urea gel alongside a similarly labeled DNA primer of known concentration.
Preparation of synthetic cap donors.
Two methods were adopted to produce radiolabeled capped RNA transcripts. To construct cap donors in which the cap structure was labeled, model RNA transcripts (see above) were converted to 32P-radiolabeled cap-1-containing structures by concurrent capping and methylation reactions (25 μl) containing approximately 5 pmol of RNA, 2.5 U of guanylyltransferase-(guanine-7-)-methyltransferase-5′-triphosphatase enzyme complex from vaccinia virus, and 3 μl of the carboxymethyl-Sephadex fraction of vaccinia virus 2′ O-methyltransferase (2, 3, 5) in 25 mM HEPES (pH 7.5)–2.5 mM MgCl2–8 mM dithiothreitol–10 μM GTP–0.1 mM S-adenosylmethionine–5 μg of RNase-free yeast carrier tRNA–20 U of RNasin–5 μl of [α-32P]GTP (200 Ci/mmol). The reaction mixtures were incubated at 37°C for 1 h, phenol-chloroform extracted, and ethanol precipitated. To obtain cap donors radiolabeled in the chain rather than the cap structure, in vitro runoff transcripts were synthesized in the presence of unlabeled m7GpppGm (Pharmacia) and [α-32P]UTP. Capped transcripts were further purified by electrophoresis through 20% polyacrylamide gels containing 7 M urea, identified by UV shadowing, excised from the gel, and eluted overnight at 4°C in water.
In vitro endonuclease assay.
Reaction mixtures of 25 μl containing approximately 1 pmol of cap donor, 0.5 μg of model RNA (optional), 2 μg of core proteins, 50 mM HEPES (pH 7.5), 0.25% Triton X-100, 100 mM KCl, 10 mM MgCl2, 1 mM dithiothreitol, 5 μg of tRNA, and 20 U of RNasin were incubated for 1 h at 37°C. Subsequently, reaction mixtures were boiled for 5 min in loading buffer (50% formamide, 4% formaldehyde, 5% glycerol, 0.04% bromophenol blue, 0.04% xylene cyanol) and fractionated by 20% polyacrylamide gel electrophoresis in the presence of 7 M urea.
RESULTS
Conditions for endonuclease activity.
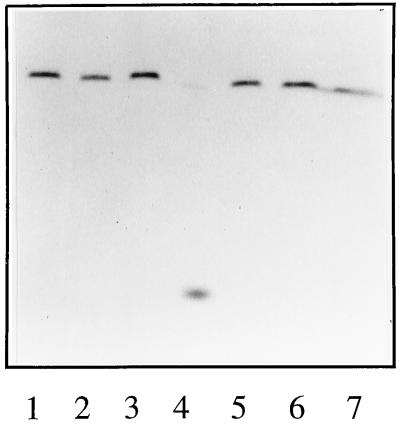
To determine the conditions for in vitro endonuclease activity of THOV, a 12-nucleotide (nt)-long RNA molecule corresponding to the first 12 nt of rabbit globin mRNA (ACACUUCCUUUU) (13) was synthesized by in vitro runoff transcription and capped in vitro with the vaccinia virus capping enzyme complex in the presence of [32P]GTP. The 13-nt-long (including the 5′-terminal m7G residue) cap donor thus obtained was tested in an in vitro endonuclease assay in the presence or absence of vRNA and cRNA promoter-like model RNAs (Fig. 1A and B). The results show that the cap donor was specifically cleaved to a single fragment in the presence of the vRNA promoter (Fig. 2, lane 4). No cleavage was observed in the absence of the 3′ or 5′ vRNA promoter arms (Fig. 2, lanes 2 and 3) or when the cRNA promoter or its 3′ or 5′ arm alone was present (Fig. 2, lanes 5 to 7). The cleaved fragment migrated slightly slower than unincorporated [32P]GTP (data not shown), suggesting that cleavage had occurred after the 5′-terminal m7GpppAm dinucleotide of the cap donor.
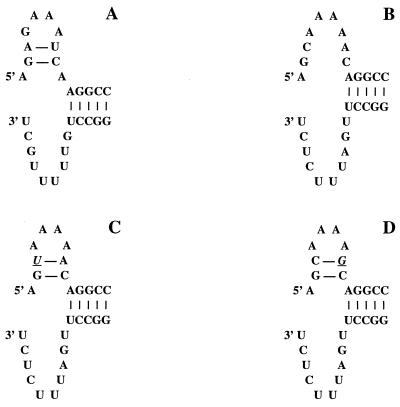
FIG. 1.
Various promoter structures of THOV, obtained by mixing short model RNAs corresponding to the 3′ and 5′ promoter arms. (A) vRNA (20); (B) cRNA (21); (C) a mutant cRNA promoter containing a hook in the 5′ promoter arm obtained by changing the residue at position 3 from cytosine to uracil; (D) a mutant cRNA promoter containing a hook in the 5′ promoter arm obtained by changing the residue at position 8 from adenine to guanine. Mutations are underlined and in italics.
FIG. 2.
Conditions for endonuclease activity with cap donors 32P labeled in the cap. Reactions were performed with cores plus cap donor supplemented with the 3′ vRNA promoter arm (lane 2), the 5′ vRNA promoter arm (lane 3), the vRNA promoter (lane 4), the 3′ cRNA promoter arm (lane 5), the 5′ cRNA promoter arm (lane 6), and the cRNA promoter (lane 7). Lane 1 contains cap donor alone.
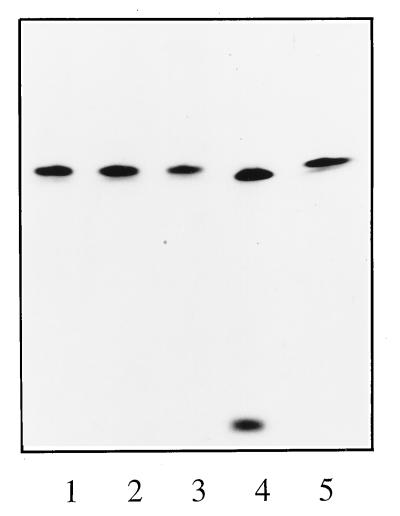
To rule out the possibility that the cap donor was cleaved further along the molecule followed by exonuclease digestion to generate a cap-A-sized product, a cap donor was synthesized with an unlabeled cap and a 32P-labeled chain. An RNA molecule corresponding to the first 12 nt of rabbit globin mRNA with an extra 5′-terminal G residue (GACACUUCCUUUU) was synthesized by in vitro runoff transcription in the presence of m7GpppGm and [32P]UTP. The resulting 13-nt-long (including the 5′-terminal m7G residue) cap donor was tested in an in vitro endonuclease assay in the presence or absence of vRNA and cRNA promoter-like model RNAs. The cap donor was specifically cleaved to a single product in the presence of the vRNA promoter (Fig. 3, lane 4). No other low-molecular-weight products were evident, indicating that the cap structure was removed with no labeled nucleotides. Residual, uncut RNA is shown as a band comigrating with the untreated cap donor (Fig. 3, lanes 1 and 4). Similar to the results shown in Fig. 2, no cleavage was observed in the absence of the 3′ or 5′ vRNA promoter arm (Fig. 3, lanes 2 and 3) or when the cRNA promoter was present (Fig. 3, lane 5). The internally labeled cap donor was reduced in size by seemingly only 1 nt, implying that only the 5′-terminal m7G residue was removed. This is unlikely, as such a residue cannot serve as a primer for capped mRNA, and would imply that the THOV polymerase complex has pyrophosphatase activity. Therefore, the more likely explanation is that the cleavage product constituted a 12-nt-long molecule generated by cleavage of the 13-nt-long cap donor directly after the 5′-terminal m7GpppGm dinucleotide. The lack of a 5′ phosphate on this product would explain its slower migration. Overall, the data show that as for in vitro polymerase activity, in vitro endonuclease activity requires the presence of both termini of vRNA. This result is consistent with the fact that the vRNA promoter, but not the cRNA promoter, can stimulate transcription with capped primers (19–21). The data agree with reports for influenza A virus endonuclease activity, which also requires both termini of vRNA (10).
FIG. 3.
Conditions for endonuclease activity with cap donors 32P labeled in the chain. Reactions were performed with viral cores plus cap donor supplemented with the 3′ vRNA promoter arm (lane 2), the 5′ vRNA promoter arm (lane 3), the vRNA promoter (lane 4), and the cRNA promoter (lane 5). Lane 1 contains the cap donor alone; lane M contains end-labeled 13- and 14-nt RNA markers.
Requirements of the cap donor for endonuclease activity.
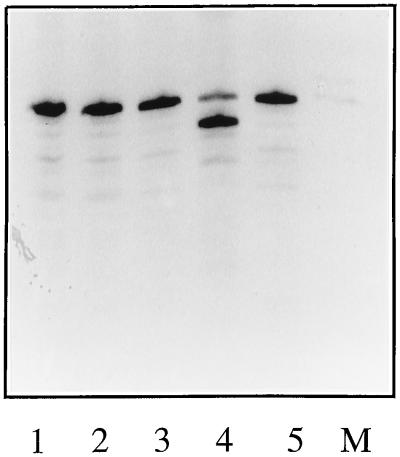
The results shown above support the unique cap-snatching mechanism of THOV and agree with previous data which demonstrated that cap-A and cap-G structures (both purine residues) can be cleaved from cap donors to act as primers in mRNA synthesis (19). To test whether THOV could cleave after a pyrimidine residue, or further than 1 nt away from the cap structure, we synthesized a series of 13-nt-long (including the 5′-terminal m7G residue) cap donors with the first purine at a variable distance from the cap (Table 1). In this experiment, cap donors were 32P labeled both in the cap and the chain. These cap donors were tested in an in vitro endonuclease assay in the presence of the vRNA promoter. The results show that only the 12A cap donor was cleaved to give two products (Fig. 4, lane 4). Cleavage was not observed with any of the other cap donors (Fig. 4, lanes 1 to 3). These results indicate that the THOV polymerase complex will cleave cap donors only after a purine residue. Position 1 purine-containing donors appear to be stringently required, as no cleavage occurred after purines positioned more than 1 nt downstream of the cap (Fig. 4, lane 3).
TABLE 1.
Cap donors with first purine at different positions from the cap
| Name | Sequence
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cap | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | |
| 12A | m7Gppp | Am | A | A | A | A | A | A | A | A | A | A | A |
| 1U11A | m7Gppp | Um | A | A | A | A | A | A | A | A | A | A | A |
| 2U10A | m7Gppp | Um | U | A | A | A | A | A | A | A | A | A | A |
| 6U6A | m7Gppp | Um | U | U | U | U | U | A | A | A | A | A | A |
FIG. 4.
Requirements of the cap donor for endonuclease activity. Cap donors were 32P labeled in the cap and in the chain. Reactions were performed with viral cores plus the vRNA promoter supplemented with cap donor 6U6A (lane 1), 2U10A (lane 2), 1U11A (lane 3), and 12A (lane 4). Lane 5 contains cap donor 12A alone. See Table 1 for details of cap donors.
A 5′ hook structure in the cRNA promoter rescues endonuclease activity.
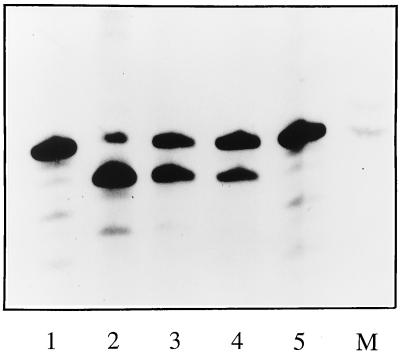
Having established conditions for in vitro endonuclease activity, we tested the effects of mutations in the cRNA promoter on endonuclease activity. In particular, we examined cRNA promoters in which a potential 5′ hook structure similar to that found in the vRNA promoter was created. This was achieved by introducing a second intrastrand base pair between residues 3 and 8 of the 5′ promoter arm. In one mutant, the residue at position 3 was changed from cytosine to uracil, allowing a base pair to form with the adenine at position 8 (Fig. 1C). In a second mutant, residue 8 was changed from adenine to guanine to allow base pairing with the position 3 cytosine (Fig. 1D). These vRNA-like cRNA promoters were tested for in vitro endonuclease activity, using 13-nt-long cap donors labeled in the chain. Again, endonuclease activity was observed in the presence of the vRNA promoter (Fig. 5, lane 2) but not the wild-type cRNA promoter (Fig. 5, lane 5), which agrees with the results presented above. Both 5′ hook-containing cRNA mutants stimulated endonuclease activity, although at slightly reduced levels than vRNA (Fig. 5, lanes 3 and 4), demonstrating that in vitro endonuclease activity relies on the presence of a 5′ hook in the promoter.
FIG. 5.
Endonuclease activity stimulated by cRNA promoter mutants. Cap donors were 32P labeled in the chain. Reactions were performed with viral cores plus cap donor supplemented with the vRNA promoter (lane 2), the mutant cRNA promoters A (lane 3) and B (lane 4), and the wild-type cRNA promoter (lane 5). Lane 1 contains cap donor alone; lane M contains end-labeled 13- and 14-nt RNA markers. See Fig. 1 for details of the promoters and mutants thereof.
DISCUSSION
Use of an in vitro endonuclease assay to investigate further the cap-snatching mechanism of THOV confirmed previously reported data (19) that mRNA synthesis is initiated preferentially with m7GpppAm cap structures but can also initiate with m7GpppGm. This finding agrees with data obtained by 5′-terminal sequence analysis of THOV mRNA, which showed that the majority of full-length mRNA molecules start with cap-A (1, 28). Interestingly, similar observations were made for 5′-terminal sequences of influenza virus mRNA (16, 27). Here, we provide additional evidence that these cap structures are cleaved directly off cap donors and that purine, but not pyrimidine, residues appear to be substrates. Thus, THOV cap binding and endonuclease activities appear to be directed toward the same residue. This is clearly not the case for influenza virus, which cleaves its host messenger-derived primers some 10 to 15 nt downstream of the cap (4, 16, 23, 24). Despite this difference in the cap-snatching mechanisms of THOV and influenza virus, clear analogies exist. In influenza virus, cleavage of mRNA occurs almost exclusively after purines with a preference for A residues (3, 6, 9, 12, 26). Moreover, it was shown that the endonuclease requires only a single nucleotide located 3′ of the cleavage site in order to act as a substrate (7). In THOV, priming with cap structures involves base pairing with the template vRNA. Priming with cap-A structures resulted in in vitro transcripts 1 nt longer than those generated in the presence of cap-G, resulting from base pairing with the 3′ ultimate U and 3′ penultimate C residues of the template, respectively (19). Recent studies with influenza virus also indicated that base pairing between primer and template RNA affects the position of priming (7, 10, 11). A conflict still remains with the observation that initiation of influenza virus transcription could occur in vitro with capped poly(U) molecules (17), indicating that base pairing was not required. However, U residues can form non-Watson-Crick base pairs with other nucleotides (14), which could be responsible for priming. Notable in this respect is that capped poly(C) molecules did not result in significant priming (17). The ability of cap analogs to initiate THOV transcription was shown to be positively correlated to the methyl contents of the cap structure: m7GpppGm structures were better primers than m7GpppG, while unmethylated GpppG had no priming activity (19). Similar observations have been reported for influenza virus (5). Thus, the only notable difference between THOV and influenza virus cap snatching appears to be the cleavage site choice of the enzyme.
THOV cores show cap-snatching activity in the presence of the vRNA promoter but not the cRNA promoter, indicating that the signals for cap snatching are found in the conserved vRNA terminal regions. The cRNA and vRNA promoters are very similar, indicating that subtle differences between these structures hold the key to the endonuclease switching mechanism. The unpaired 3′ arm of the cRNA promoter is 1 nt longer than its 5′ arm, whereas in the vRNA promoter the 5′ arm is longer (Fig. 1). There are several sequence differences between the two promoters, and the vRNA promoter adopts a 5′ hook structure which is absent in the cRNA promoter (Fig. 1) (19–21). These earlier studies showed that base pairing between and within promoter arms was more important to promoter activity than the identity of the nucleotides. The 5′ hook structure, which is absent in the cRNA promoter, was therefore a prime candidate for involvement in endonuclease activity. Here, we have provided experimental evidence that this is indeed the case: a 5′ hook structure introduced to the cRNA promoter efficiently rescued endonuclease activity in vitro (Fig. 5). Mutants of the vRNA promoter where the 5′ hook was destroyed were inactive in endonucleolytic cleavage (data not shown), indicating that the 5′ hook normally present in the vRNA promoter is responsible for stimulating cap-snatching activity. These results suggest that a novel endonuclease switching mechanism, involving the 5′ hook structure, determines whether capped mRNA is synthesized from vRNA or uncapped vRNA is synthesized from cRNA.
The results reported here have important implications for influenza virus research. Despite differences between THOV and influenza viruses, the transcription and replication mechanisms appear to be very similar. Moreover, striking conformational similarities exist between the vRNA promoters, as is demonstrated by the fact that THOV cores are able to transcribe influenza A virus vRNA-like promoters in vitro (20). Hence, it is conceivable that a similar endonuclease switching mechanism exists in influenza A virus, which may have potential in new antiviral strategies.
ACKNOWLEDGMENTS
We thank G. G. Brownlee, O. Haller, F. Weber, A. Portela, and J. Ortin for helpful discussions.
This work was supported by a grant from the EU Human Capital and Mobility Network program (ERBCHRXCT940453). M.B.L. was supported by a Biological and Biotechnological Sciences Research Council studentship.
REFERENCES
- 1.Albo C, Martín J, Portela A. The 5′ ends of Thogoto virus (Orthomyxoviridae) mRNAs are homogeneous in both length and sequence. J Virol. 1996;70:9013–9017. doi: 10.1128/jvi.70.12.9013-9017.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barbosa E, Moss B. mRNA (nucleoside-2′-)-methyl-transferase from vaccinia virus. Purification and substrate specificity. J Biol Chem. 1987;253:7698–7702. [PubMed] [Google Scholar]
- 3.Beaton A R, Krug R M. Selected host cell capped RNA fragments prime influenza viral RNA transcription in vivo. Nucleic Acids Res. 1981;9:4423–4436. doi: 10.1093/nar/9.17.4423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bouloy M, Plotch S J, Krug R M. Globin mRNAs are primers for the transcription of influenza viral RNA in vitro. Proc Natl Acad Sci USA. 1978;75:4886–4890. doi: 10.1073/pnas.75.10.4886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bouloy M, Plotch S J, Krug R M. Both the 7-methyl and the 2′ O-methyl groups in the cap of mRNA strongly influence its ability to act as primer for influenza virus RNA transcription. Proc Natl Acad Sci USA. 1980;77:3952–3956. doi: 10.1073/pnas.77.7.3952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caton A J, Robertson J S. Structure of the host-derived sequences present at the 5′ ends of influenza virus mRNA. Nucleic Acids Res. 1980;8:2591–2603. doi: 10.1093/nar/8.12.2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chung T D Y, Cianci C, Hagen M, Terry B, Matthews J T, Krystal M, Colonno R. Biochemical studies on capped RNA primers identify a class of oligonucleotide inhibitors of the influenza virus RNA polymerase. Proc Natl Acad Sci USA. 1994;91:2372–2376. doi: 10.1073/pnas.91.6.2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clerx J P M, Fuller F, Bishop D H L. Tick-borne viruses structurally similar to Orthomyxoviruses. Virology. 1983;127:205–219. doi: 10.1016/0042-6822(83)90384-7. [DOI] [PubMed] [Google Scholar]
- 9.Dhar R, Channock R M, Lai C-J. Nonviral oligonucleotides at the 5′ terminus of cytoplasmic influenza viral mRNA deduced from cloned complete genomic sequences. Cell. 1980;21:495–500. doi: 10.1016/0092-8674(80)90486-9. [DOI] [PubMed] [Google Scholar]
- 10.Hagen M, Chung T D Y, Butcher J A, Krystal M. Recombinant influenza virus polymerase: requirement of both 5′ and 3′ viral ends for endonuclease activity. J Virol. 1994;68:1509–1515. doi: 10.1128/jvi.68.3.1509-1515.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hagen M, Tiley L, Chung T D Y, Krystal M. The role of template-primer interactions in cleavage and initiation by the influenza virus polymerase. J Gen Virol. 1995;76:603–611. doi: 10.1099/0022-1317-76-3-603. [DOI] [PubMed] [Google Scholar]
- 12.Hay A J, Skehel J J, McCauley J. Characterization of influenza virus RNA complete transcripts. J Virol. 1982;116:517–522. doi: 10.1016/0042-6822(82)90144-1. [DOI] [PubMed] [Google Scholar]
- 13.Heindell H C, Liu A, Paddock G V, Studnicka G M, Salser W A. The primary sequence of rabbit alpha-globin mRNA. Cell. 1978;15:43–54. doi: 10.1016/0092-8674(78)90081-8. [DOI] [PubMed] [Google Scholar]
- 14.Holbrook S R, Cheong C, Tinoco I, Kim S H. Crystal structure of an RNA double helix incorporating a track of non-Watson-Crick base pairs. Nature (London) 1991;353:579–581. doi: 10.1038/353579a0. [DOI] [PubMed] [Google Scholar]
- 15.Krug R M, Alonso-Caplen F V, Julkunen I, Katze M G. Expression and replication of the influenza virus genome. In: Krug R M, editor. The influenza viruses. New York, N.Y: Plenum Press; 1989. pp. 89–152. [Google Scholar]
- 16.Krug R M, Broni B B, Bouloy M. Are the 5′ ends of influenza viral mRNA synthesized in vivo donated by host mRNAs? Cell. 1979;18:239–334. doi: 10.1016/0092-8674(79)90052-7. [DOI] [PubMed] [Google Scholar]
- 17.Krug R M, Broni B, LaFiandra A J, Morgan M A, Shatkin A J. Priming and inhibitory activities of RNAs for the influenza viral transcriptase do not require base pairing with the virion template RNA. Proc Natl Acad Sci USA. 1980;77:5874–5878. doi: 10.1073/pnas.77.10.5874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leahy M B, Dessens J T, Weber F, Kochs G, Nuttall P A. The fourth genus in the Orthomyxoviridae: sequence analysis of two Thogoto virus polymerase proteins and comparison to influenza viruses. Virus Res. 1997;50:215–224. doi: 10.1016/s0168-1702(97)00072-5. [DOI] [PubMed] [Google Scholar]
- 19.Leahy M B, Dessens J T, Nuttall P A. In vitro polymerase activity of Thogoto virus: evidence for a unique cap snatching mechanism in a tick-borne orthomyxovirus. J Virol. 1997;71:8347–8351. doi: 10.1128/jvi.71.11.8347-8351.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leahy M B, Dessens J T, Nuttall P A. Striking conformational similarities between the transcription promoters of Thogoto and influenza A viruses: evidence for intrastrand base pairing in the 5′ promoter arm. J Virol. 1997;71:8352–8356. doi: 10.1128/jvi.71.11.8352-8356.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leahy, M. B., J. T. Dessens, D. C. Pritlove, and P. A. Nuttall. The Thogoto orthomyxovirus cRNA promoter functions as a panhandle but does not stimulate cap snatching in vitro. J. Gen. Virol., in press. [DOI] [PubMed]
- 22.Morse M A, Marriott A C, Nuttall P A. The glycoprotein of Thogoto virus (a tick-borne orthomyxo-like virus) is related to the baculovirus glycoprotein GP64. Virology. 1992;186:640–646. doi: 10.1016/0042-6822(92)90030-s. [DOI] [PubMed] [Google Scholar]
- 23.Plotch S J, Bouloy M, Krug R M. Transfer of 5′-terminal cap of globin mRNA to influenza viral complementary RNA during transcription in vitro. Proc Natl Acad Sci USA. 1979;76:1618–1622. doi: 10.1073/pnas.76.4.1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Plotch S J, Bouloy M, Ulmanen I, Krug R M. A unique cap (m7GpppXm)-dependent influenza virion endonuclease cleaves capped RNAs to generate the primers that initiate viral RNA transcription. Cell. 1981;23:847–858. doi: 10.1016/0092-8674(81)90449-9. [DOI] [PubMed] [Google Scholar]
- 25.Pringle C R. Virus taxonomy 1996—a bulletin from the Xth International Conference of Virology in Jerusalem. Arch Virol. 1996;141:2251–2256. doi: 10.1007/BF01718231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shaw M W, Lamb R A. A specific sub-set of host cell mRNAs prime influenza virus mRNA synthesis. Virus Res. 1984;1:455–467. doi: 10.1016/0168-1702(84)90003-0. [DOI] [PubMed] [Google Scholar]
- 27.Staunton D, Nuttall P A, Bishop D H L. Sequence analyses of Thogoto viral RNA segment 3: evidence for a distant relationship between an arbovirus and members of the Orthomyxoviridae. J Gen Virol. 1989;70:2811–2817. doi: 10.1099/0022-1317-70-10-2811. [DOI] [PubMed] [Google Scholar]
- 28.Weber F, Haller O, Kochs G. Nucleoprotein viral RNA and mRNA of Thogoto virus: a novel “cap-stealing” mechanism in tick-borne orthomyxoviruses? J Virol. 1996;70:8361–8367. doi: 10.1128/jvi.70.12.8361-8367.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]