Abstract
Apoptosis is a form of regulated cell death essential for tissue homeostasis and embryonic development. Apoptosis also plays a key role during bacterial infection, yet some intracellular bacterial pathogens (such as Shigella flexneri, whose lipopolysaccharide can block apoptosis) can manipulate cell death programs as an important survival strategy. Septins are a component of the cytoskeleton essential for mitochondrial dynamics and host defense, however, the role of septins in regulated cell death is mostly unknown. Here, we discover that septins promote mitochondrial (i.e., intrinsic) apoptosis in response to treatment with staurosporine (a pan‐kinase inhibitor) or etoposide (a DNA topoisomerase inhibitor). Consistent with a role for septins in mitochondrial dynamics, septins promote the release of mitochondrial protein cytochrome c in apoptotic cells and are required for the proteolytic activation of caspase‐3, caspase‐7, and caspase‐9 (core components of the apoptotic machinery). Apoptosis of HeLa cells induced in response to infection by S. flexneri ΔgalU (a lipopolysaccharide mutant unable to block apoptosis) is also septin‐dependent. In vivo, zebrafish larvae are significantly more susceptible to infection with S. flexneri ΔgalU (as compared to infection with wildtype S. flexneri), yet septin deficient larvae are equally susceptible to infection with S. flexneri ΔgalU and wildtype S. flexneri. These data provide a new molecular framework to understand the complexity of mitochondrial apoptosis and its ability to combat bacterial infection.
Keywords: apoptosis, cytoskeleton, mitochondria, septins, Shigella, zebrafish
1. INTRODUCTION
Apoptosis, a non‐inflammatory form of regulated cell death used to eliminate damaged cells, is essential for tissue homeostasis and embryonic development (Danial & Korsmeyer, 2004). The core apoptotic machinery involves caspases (Thornberry & Lazebnik, 1998) that reside in the cytosol as zymogens/proenzymes and consist of two major subgroups: initiator caspases (caspase‐8, caspase‐9) and effector/executioner caspases (caspase‐3, caspase‐7) (Salvesen & Ashkenazi, 2011). Apoptotic stimuli induce activation of initiator caspases through dedicated signaling pathways, initiated intrinsically (i.e., mitochondria‐mediated pathway) or extrinsically (i.e., receptor‐mediated pathway) (Ashkenazi & Dixit, 1998; Bratton & Salvesen, 2010). Following activation, initiator caspases trigger proteolytic stimulation of effector caspase‐3 and caspase‐7 zymogens that enable the execution phase of the apoptotic cell death program by cleaving functionally important proteins within a cell (Ashkenazi & Salvesen, 2014).
Septins are a poorly understood component of the cytoskeleton discovered from their role in cytokinesis (Mostowy & Cossart, 2012; Spiliotis & Nakos, 2021; Woods & Gladfelter, 2021). In humans, septins are organized into four groups based on amino acid sequence similarity: the SEPT2 group (SEPT1, SEPT2, SEPT4, SEPT5), SEPT3 group (SEPT3, SEPT9, SEPT12), SEPT6 group (SEPT6, SEPT8, SEPT10, SEPT11, SEPT14), and SEPT7 group (SEPT7). Members of each septin group assemble into hetero‐oligomeric complexes (including SEPT2‐SEPT6‐SEPT7‐SEPT9‐SEPT9‐SEPT7‐SEPT6‐SEPT2) and higher‐order structures (including filaments and rings). In the case of bacterial infection, septins are recognized for their role in host defense against the important human pathogen Shigella flexneri (Robertin & Mostowy, 2020; Van Ngo & Mostowy, 2019). Here, septins assemble into cage‐like structures that entrap dividing bacterial cells for targeting to autophagy (Krokowski et al., 2018; Lobato‐Márquez et al., 2021; Mostowy et al., 2010). Septins also interact with the cytosolic GTPase dynamin‐related protein 1 (Drp1) to enhance mitochondrial fission (Pagliuso et al., 2016; Sirianni et al., 2016). In the case of cell death, apoptosis‐related protein in the TGF‐β signaling pathway (ARTS), an unusual septin derived from alternative splicing of SEPT4, has been described to promote mitochondrial apoptosis (Larisch et al., 2000). Despite the evidence for ARTS in mitochondrial apoptosis (Edison et al., 2012; Edison et al., 2017; García‐Fernández et al., 2010; Gottfried, Rotem, Lotan, Steller, & Larisch, 2004; Larisch et al., 2000; Mamriev et al., 2020), a role for other septins in apoptosis was unknown.
Apoptosis is widely recognized as important for immune defense against bacterial invasion, yet pathogens deploy a variety of mechanisms to manipulate cell death pathways and promote infection (Naderer & Fulcher, 2018). In the case of S. flexneri, both inhibition and induction of apoptosis have been described. S. flexneri can inhibit apoptosis in epithelial cells by inhibiting the activation of caspase‐3 (Clark & Maurelli, 2007), and recent work identified that S. flexneri lipopolysaccharide (LPS) can directly bind to caspase‐3/‐7 via its O‐antigen moiety, blocking apoptosis signaling (Günther et al., 2020). On the other hand, work has shown that S. flexneri can induce apoptosis of infected cells via mitochondrial depolarization and caspase‐9 activation (Lembo‐Fazio et al., 2011). Taken together, the precise role of apoptosis in control of S. flexneri infection is complex and, in terms of host and pathogen survival, is the subject of great interest (Ashida, Suzuki, & Sasakawa, 2021).
In this study, we investigate a role for septins in mitochondrial apoptosis induced by pharmacological treatment and S. flexneri infection. We discover that septins (SEPT7 and SEPT2) are required for apoptotic cell death. Consistent with this, we show that septins promote the release of cytochrome c from mitochondria, as well as the activation of caspase‐3, caspase‐7, and caspase‐9. Finally, we highlight the role of septin‐mediated apoptosis during S. flexneri infection in vitro using human epithelial cells and in vivo using zebrafish larvae.
2. RESULTS
2.1. Septins are required for apoptotic cell death
To test a role for septins in apoptosis of human epithelial cells (HeLa), small interfering RNA (siRNA) was used to deplete SEPT7, and staurosporine (STS, a pan‐kinase inhibitor) was used to induce mitochondrial apoptosis (Figure 1a). We observed that 24 hr incubation of 1 μM STS dramatically reduced the viability of control cells (9.2 ± 1.9% cells survived), but only partially reduced the viability of SEPT7‐depleted cells (23.1 ± 2.0% cells survived) (Figure 1b). Similar results are observed for SEPT2‐depleted cells (Figure 1c, d). To visualize apoptosis in STS‐treated cells, samples were fluorescently labeled with Annexin V (to identify phosphatidylserine externalization, a hallmark of apoptotic cells) and Hoechst (to visualize nuclei) (Figure 1e). In agreement with cell viability results, the proportion of Annexin V‐positive cells is significantly reduced (4.2 ± 0.1 fold) in SEPT7‐depleted cells, as compared to control cells (Figure 1f).
FIGURE 1.
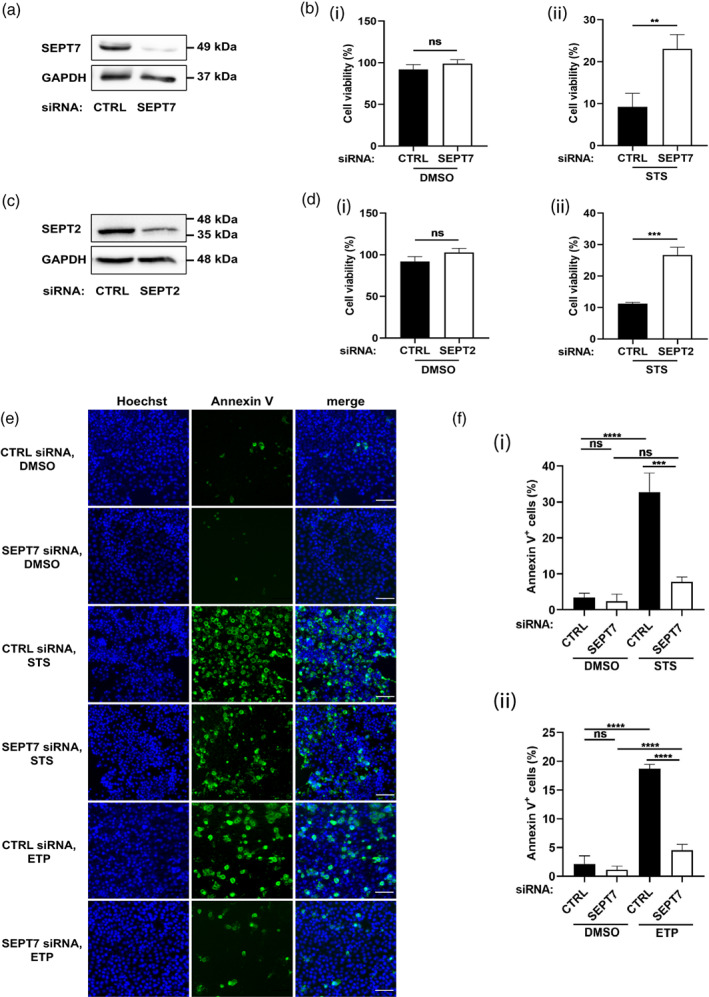
Septin‐depleted cells undergo significantly less apoptosis. (a) HeLa cells were transfected for 72 hr with control (CTRL) siRNA or a siRNA targeting SEPT7. Effect of siRNA on SEPT7 expression. A representative Western blot of HeLa cell lysates is shown. GAPDH was used as a loading control. (b) Impact of siRNA targeting SEPT7 on viability of HeLa cells in (i) untreated or (ii) STS‐treated conditions. Cell viability is measured using MTT assays as described in Materials and Methods. Data are mean ± SEM from three independent experiments. **p = .0069 by two‐tailed Student's t test. (c) HeLa cells were transfected for 72 hr with control (CTRL) siRNA or a siRNA targeting SEPT2. Effect of siRNA on SEPT2 expression. A representative Western blot of HeLa cell lysates is shown. (d) Impact of siRNA targeting SEPT2 on viability of HeLa cells in (i) untreated or (ii) STS‐treated conditions. Cell viability is measured using MTT assays as described in Materials and Methods. Data are mean ± SEM from three independent experiments. ***p = .0004 by two‐tailed Student's t test. (e) Control and SEPT7‐depleted cells were treated with DMSO or 1 μM STS or 500 μM ETP for 10 hr and labeled with Alexa488‐Annexin V (Annexin V) and Hoechst. Representative widefield microscopy images showing Annexin V in green and Hoechst in blue. Scale bar = 100 μm. (f) The % of Annexin V positive cells is calculated in Fiji by dividing the number of cells that displayed Annexin V labeling by the total number of cells identified by nuclear Hoechst labeling, then multiplying by 100. Each bar represents the mean % ± SEM from three independent experiments (a minimum of 1,000 cells were counted per condition separated in three independent experiments). ****p < .0001by two‐way ANOVA.
We next used etoposide, a drug that inhibits topoisomerase II causing errors in DNA replication, to induce mitochondrial apoptosis. Consistent with results obtained for STS‐treated cells, depletion of SEPT7 significantly reduced (4.2 ± 0.8 fold) the proportion of Annexin V‐positive cells, as compared to control cells (Figure 1e, f). Together, these results show that septins promote mitochondrial apoptosis of HeLa cells.
2.2. Septins are required for cytochrome c release
Cytochrome c is located in the mitochondrial intermembrane space under normal conditions, but under apoptotic conditions is released to the cytosol for the activation of initiator caspases (Bock & Tait, 2020; Dorstyn, Akey, & Kumar, 2018). Considering that septins are well known to play a key role in mitochondrial dynamics (Pagliuso et al., 2016; Sirianni et al., 2016), we hypothesized that septins can also promote the release of cytochrome c when mitochondrial apoptosis is stimulated. We treated control and SEPT7‐depleted cells with STS and examined cytochrome c release using confocal microscopy and immunostaining of cytochrome c (as well as Tom20, a mitochondrial marker) (Figure 2a). In this case, the number of SEPT7‐depleted cells releasing cytochrome c is significantly reduced (3.2 ± 0.4 fold), as compared to control cells (Figure 2b).
FIGURE 2.
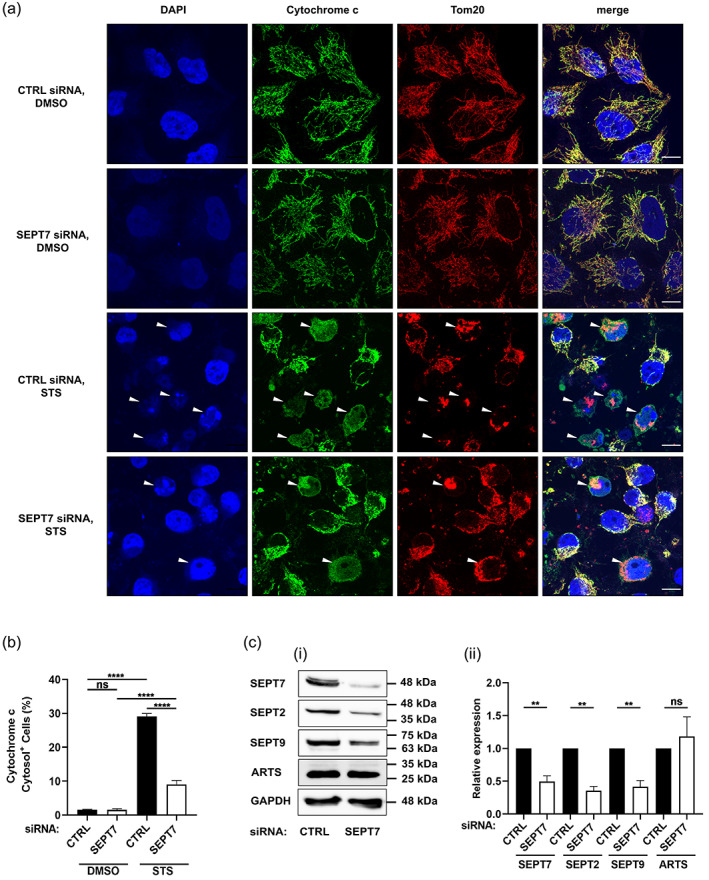
Cytochrome c release is significantly reduced in SEPT7‐depleted cells. HeLa cells were transfected with control or SEPT7 siRNA, treated with DMSO or 1 μM STS for 10 hr before being fixed and labeled with an anti‐cytochrome c and anti‐Tom20 antibodies and DAPI. (a) Representative confocal microscopy images showing cytochrome c in green, Tom20 in red and DAPI in blue. Arrowheads indicate cells releasing cytochrome c. Scale bar = 10 μm. (b) The percentage of cells releasing cytochrome c is calculated in ImageJ by counting cells that exhibited cytosolic (rather than mitochondrial) cytochrome c labeling, and dividing by the total number of cells, then multiplying by 100. Each bar represents the mean % ± SEM from three independent experiments (a minimum of 1,000 cells were counted per condition separated in three independent experiments). ****p < .0001 by two‐way ANOVA. (c) SEPT7 and ARTS act in different pathways. (i) HeLa cells were treated with control or SEPT7 siRNA for 72 hr. Whole‐cell lysates were immunoblotted for SEPT7, SEPT2, SEPT9, and ARTS. GAPDH was used as a loading control. Data are representative Western blots of three independent experiments. (ii) Densitometry analysis of the bands was performed using ImageJ. Data are mean ± SEM from three independent experiments. **p < .01 by two‐tailed Student's t test
Previous work has shown that ARTS, a variant of SEPT4, promotes mitochondrial apoptosis (Larisch et al., 2000). To investigate if SEPT7 depletion compromises the activation of apoptosis via ARTS, we quantified protein levels of ARTS and other septins in SEPT7‐depleted conditions. Consistent with previous work, we showed that the depletion of SEPT7 significantly reduced SEPT2, SEPT7, and SEPT9 protein levels (Estey et al., 2010; Lobato‐Marquez et al., 2019), but ARTS protein levels are not affected by SEPT7 depletion (Figure 2c). These results indicate that the requirement of SEPT7 in mitochondrial apoptosis is not due to the regulation of ARTS.
2.3. Septins are required for activation of caspase‐3/ ‐7/, and ‐9
Activation of effector caspases (caspase‐3 and caspase‐7) occurs in two steps, beginning with initial cleavage by active caspase‐9 to form the p20 subunit. After this cleavage, caspase‐3 or caspase‐7 removes its own prodomain to generate the active p17 subunit. To understand how septins promote mitochondrial apoptosis, we measured the effect of septin depletion on STS‐ or etoposide‐induced activation of caspase‐9 by Western blotting (Figure 3a, b). In both cases, SEPT7 depletion caused a significant decrease in levels of active caspase‐9, as compared to control cells. Consistent with a role for septins in mitochondrial apoptosis, SEPT7 depletion caused a significant decrease in levels of active caspase‐3 and caspase‐7, as compared to control cells (Figure 3a, b). Similar results are observed for SEPT2‐depleted cells (Figure 3c). Importantly, SEPT7 depletion had no significant effect on levels of caspase‐4, an inflammatory caspase with no role in STS‐mediated mitochondrial apoptosis (Figure 3d).
FIGURE 3.
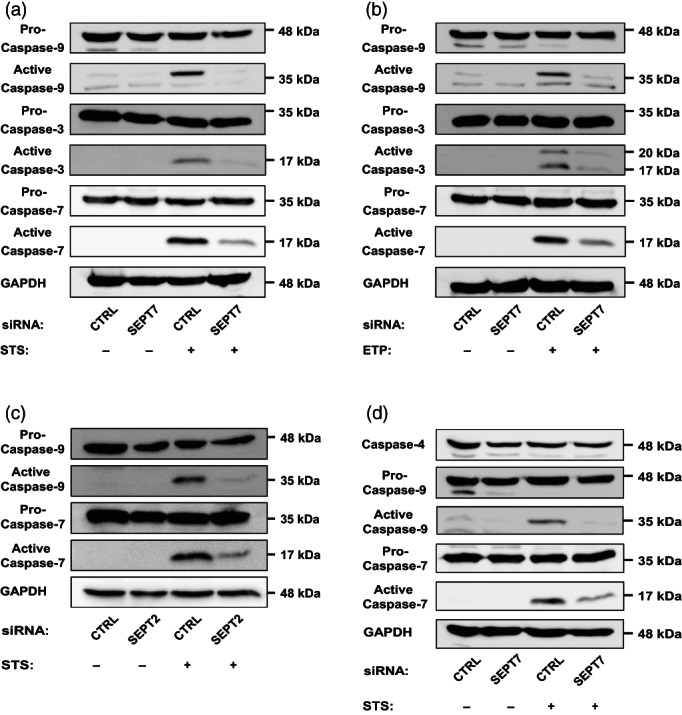
Initiator and executioner caspase cleavage is significantly reduced when septins are depleted. (a), (b) HeLa cells were transfected with control or SEPT7 siRNA for 72 hr. Cells were then treated with DMSO or (a) 1 μM STS or (b) 500 μM ETP for 10 hr. Whole‐cell lysates were immunoblotted against caspase‐3, cleaved caspase‐3, caspase‐7, caspase‐9, and GAPDH (used as loading control). Data are representative of three independent experiments. (c) SEPT2 is required for caspase activation. HeLa cells were transfected with control or SEPT2 siRNA for 72 hr. Cells were then treated with DMSO or 1 μM STS for 10 hr. Whole‐cell lysates were immunoblotted with antibodies to caspase‐7, caspase‐9, and GAPDH. Data are representative of three independent experiments. (d) HeLa cells were transfected with control or SEPT7 siRNA for 72 hr. Cells were then treated with DMSO or 1 μM STS for 10 hr. Whole‐cell lysates were immunoblotted with antibodies to caspase‐4, caspase‐7, caspase‐9, and GAPDH. Data are representative of three independent experiments
Next, we imaged HeLa cells labeled for active caspase‐3 using confocal microscopy (Figure 4a). Consistent with results obtained by Western blot, SEPT7‐depleted cells showed a significant decrease (3.0 ± 0.6 fold) in the number of apoptotic cells labeled for active caspase‐3, as compared to control cells (Figure 4b). Similar results are obtained when we labeled for active caspase‐3 and performed flow cytometry analysis on siRNA‐treated cells (Figure 4c). Together, these results highlight an important role for septins in the activation of caspase‐3/ ‐7/, and ‐9 during mitochondrial apoptosis.
FIGURE 4.
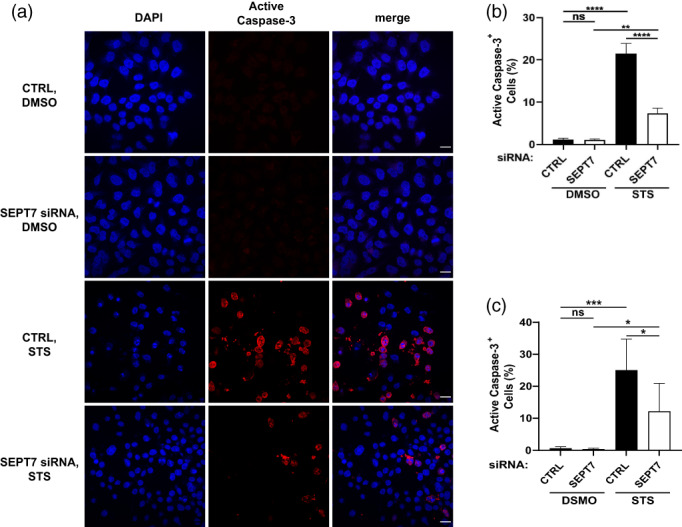
SEPT7 is required for the activation of caspase‐3. (a) Control and SEPT7‐depleted cells were treated with DMSO or STS, and stained with cleaved anti‐caspase‐3 antibody and DAPI. Representative confocal microscopy images showing cleaved caspase‐3 in red and DAPI in blue. Scale bar = 100 μm. (b) The percentage of cleaved caspase‐3‐stained cells was calculated in Fiji by dividing the number of cells that displayed cleaved caspase‐3 staining by the total number of cells, then multiplying by 100. Each bar represents the mean % ± SEM from three independent experiments (a minimum of 1,000 cells were counted per condition separated in three independent experiments). **p < .01, ****p < .0001 by two‐way ANOVA. (c) HeLa cells (CTRL or SEPT7 siRNA transfected) were treated with 1 μM staurosporine for 5 hr, followed by staining of active caspase‐3 and analyzed by flow cytometry (a minimum of cells were counted per condition separated in three independent experiments). *p < 0.05, ***p < 0.001 by two‐way ANOVA
2.4. Testing the role of septin‐mediated apoptosis during S. flexneri infection
New work has shown that S. flexneri LPS can bind to caspase‐3/ ‐7 via its O‐antigen moiety, blocking apoptosis signaling (Günther et al., 2020). Considering this, we used a S. flexneri mutant lacking the O‐antigen and outer core components of LPS (ΔgalU) (Lobato‐Márquez et al., 2021) to test for caspase activation during infection of HeLa cells. Consistent with a role for bacterial LPS in blocking apoptosis, S. flexneri ΔgalU fails to inhibit the activation of caspase‐3/ ‐7/ and ‐9 as efficiently as wildtype S. flexneri (Figure 5a). Strikingly, the activation of caspase‐3/ ‐7/ ‐9 in response to S. flexneri ΔgalU infection is dependent on SEPT7 (Figure 5a).
FIGURE 5.
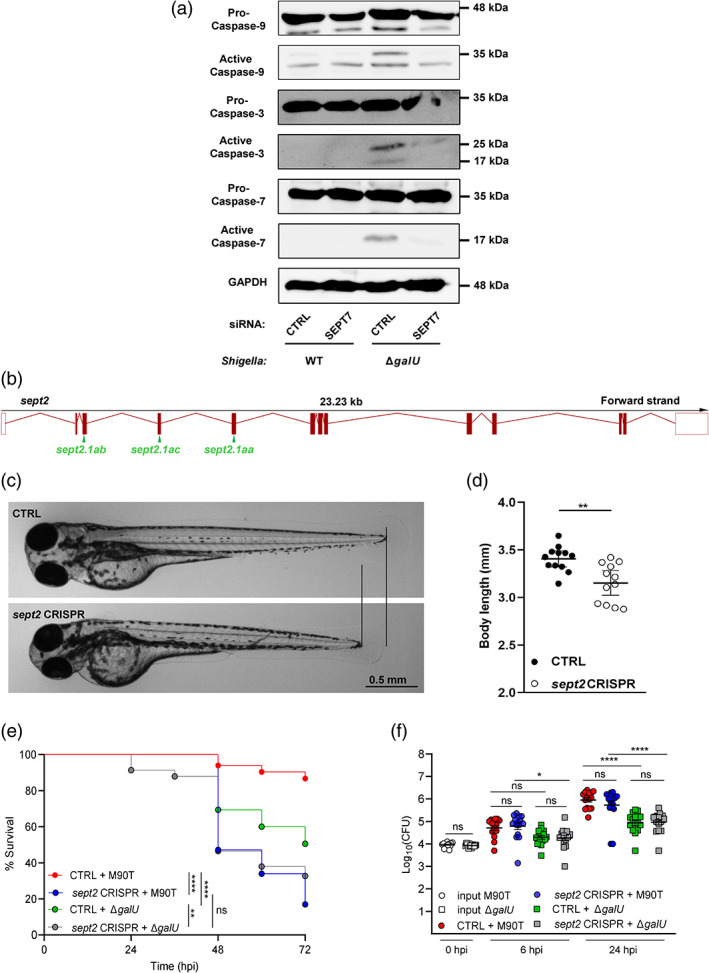
Septins are required for apoptosis induced by Shigella infection. (a) HeLa cells were transfected with control or SEPT7 siRNA for 72 hr. Cells were then infected with S. flexneri M90T or the S. flexneri ΔgalU mutant for 4 hr. Whole‐cell lysates were immunoblotted against caspase‐3, caspase‐7, and caspase‐9. GAPDH was used as a loading control. Data are representative of three independent experiments. (b) Schematic representation of CRISPR/Cas9 targeting of sept2. Three crisprRNA (crRNA, green arrows) were designed, targeting three consecutive exons (red boxes) of the zebrafish sept2 gene. (c) Representative images of control (CTRL) and sept2 crispants. Lines indicate a difference in overall body length between CTRL and sept2 crispants. Images were acquired using Leica M205 FA stereomicroscope. (d) Quantification of difference in body length between CTRL and sept2 crispants. Bars represent the mean ± SEM from three independent experiments (n = 12 larvae per bar). **p < .01 by Student's t test. (e) Survival assays of CTRL and sept2 crispants injected with approximately 9,000 CFU of either S. flexneri WT or S. flexneri ΔgalU. **p < .01, ****p < .0001 by log–rank (Mantel–Cox) test. Curves are cumulative of 3 independent experiments (n = 53–85 larvae per curve). Statistical tests for survival curves were performed using the log–rank (Mantel–Cox) test (which intrinsically take into consideration the overall trend of survival over the different times of observation), and are indicated for overall survival. (f) CFU enumeration of CTRL and sept2 crispants injected with approximately 9,000 CFU of either S. flexneri WT or S. flexneri ΔgalU. CFU were collected at 0, 6, and 24 hr post‐infection, and are indicated on the X‐axis. Bars represent the mean ± SEM from three independent experiments (n = 15 larvae per bar). *p < .05, ****p < .0001 by one‐way ANOVA. Statistical tests for CFU counts were performed using one‐way ANOVA, and are indicated for the individual timepoints
To investigate the role of septin‐mediated apoptosis during infection in vivo, we used the well‐established S. flexneri‐zebrafish infection model (Duggan & Mostowy, 2018; Gomes & Mostowy, 2020; Torraca & Mostowy, 2018). Zebrafish septins are highly homologous to human septins and all septin groups (SEPT2, SEPT3, SEPT6, SEPT7) are represented in zebrafish, though some septins are duplicated in zebrafish (Figure S1A). For example, three homologs of human SEPT7 are present in zebrafish (sept7a, sept7b, sept15). In contrast, only one zebrafish sept2 gene has been identified, and it has high homology to human SEPT2 (Figure S1A). Moreover, RNAseq data from our lab (Torraca et al., 2019) suggested that sept2 is the most expressed septin in zebrafish at the whole animal level (Figure S1B, Table S1). Considering this, to investigate the role of septin‐mediated apoptosis in vivo, we targeted zebrafish Sept2 using a highly effective CRISPR‐Cas9 method (Kroll et al., 2021) resulting in F0 knockout (Figure 5b, Figure S1C).
At 3 days post‐fertilization (dpf), sept2 crispants displayed some growth impairment, as shown by a reduced body length (Figure 5c‐d); only larvae capable of normal hatching and free from major developmental abnormalities were selected for infection studies. sept2 crispants are significantly more susceptible to wildtype S. flexneri infection as compared to control larvae (Figure 5e–f), highlighting an in vivo role for septins in host defense. Control larvae are significantly more susceptible to S. flexneri ΔgalU as compared to control larvae infected with wildtype S. flexneri, but sept2 crispants (i.e., where mitochondrial apoptosis is compromised) are not significantly more susceptible to S. flexneri ΔgalU as compared to sept2 crispants infected with wildtype S. flexneri (Figure 5e‐f).
3. CONCLUDING REMARKS
Investigations using cellular and animal models have shown that septins are crucial for sensing bacteria and for promoting effector mechanisms to eliminate them (Mostowy & Shenoy, 2015; Robertin & Mostowy, 2020; Van Ngo & Mostowy, 2019). Here, we discover that septins are required for mitochondrial apoptosis because they promote cytochrome c release and activation of caspase‐3, caspase‐7, and caspase‐9. We highlight the role of septin‐mediated apoptosis during S. flexneri infection in vitro using human epithelial cells and in vivo using zebrafish larvae. Further investigations into septin biology will help to decipher the molecular mechanisms underlying human diseases associated with mitochondrial apoptosis, including susceptibility to infection.
The similarity of results obtained with SEPT7 and SEPT2 strongly suggests that septin hetero‐oligomers (and not septin monomers or dimers) are required for their role in apoptosis. This is in contrast to the apoptotic role of ARTS which is viewed to act independently of other septins. ARTS is localized to the mitochondrial outer membrane (Edison et al., 2012). In response to apoptotic stimuli, ARTS translocates to the cytosol and directly binds and antagonizes anti‐apoptotic XIAP, leading to the release of non‐lethal active caspases (e.g., caspase 9) from XIAP. In turn, these caspases cleave substrates (e.g., Bid), resulting in mitochondrial outer membrane permeabilization (MOMP) and apoptosis. Work has shown that ARTS acts as a scaffold to bring XIAP into close proximity with anti‐apoptotic Bcl‐2 and promote its degradation (Edison et al., 2017). Considering work from our lab and others (Krokowski et al., 2018; Lobato‐Márquez et al., 2021; Mageswaran et al., 2021; Mostowy et al., 2010; Pagliuso et al., 2016; Sirianni et al., 2016) it is tempting to speculate that septins (including SEPT7 and SEPT2, but not ARTS) are recruited to mitochondrial constriction sites that present micron‐scale curvature. Here, septins may function as a scaffold to recruit Drp1 to mitochondrial fission sites (in healthy cells) or apoptotic foci (in apoptotic cells), and promote interactions of Drp1 with other proteins involved in MOMP (e.g., Bax). Consistent with this, new work has shown that Drp1 interacts with Bax at apoptotic foci resulting in MOMP and apoptosis (Jenner et al., 2022).
To test the role of septins in apoptosis in vivo, we took advantage of the Shigella‐zebrafish infection model (Duggan & Mostowy, 2018; Gomes & Mostowy, 2020; Torraca & Mostowy, 2018). We observe that, as compared to control larvae, sept2 crispants are not more susceptible to S. flexneri ΔgalU (i.e., bacteria that do induce apoptosis) as compared to wildtype S. flexneri (i.e., bacteria that do not induce apoptosis). In contrast, control larvae are more susceptible to S. flexneri ΔgalU as compared to wildtype S. flexneri. Considering that survival of sept2 crispants is not affected in this case, these data are in agreement with our hypothesis that septins are required for S. flexneri ΔgalU to induce host cell death via apoptosis. However, given the pleiotropic role of septins in vivo, these results involving zebrafish infection are likely not solely because of Sept2's role in apoptosis. As compared to control larvae, sept2 crispants are significantly more susceptible to wildtype S. flexneri. In agreement with previous work from our lab studying septins in zebrafish (Mazon‐Moya et al., 2017; Mostowy et al., 2013), these data suggest that the role of Sept2 in restricting bacterial infection may also be acting through septin cages, bacterial autophagy, and inflammation control. Relatively little is known about the role of apoptosis during S. flexneri infection in vivo. It will thus be of great interest to further study the link between septins and apoptosis using a zebrafish model of Shigella infection.
4. MATERIALS AND METHODS
4.1. Mammalian cell lines, bacterial strains, and media
The human epithelial cell line HeLa (ATCC CCL‐2) was grown at 37 °C and 5% CO2 in Dulbecco's modified Eagle medium (DMEM, #5796‐500ML, Sigma) supplemented with 10% fetal bovine serum (FBS, #10500‐064‐500ML, Gibco).
S. flexneri M90T (Mostowy et al., 2010) or S. flexneri ΔgalU (lacking O‐antigen and outer core components of LPS) (Lobato‐Márquez et al., 2021) were grown in trypticase soy (TCS) agar containing 0.01% (w/v) congo red to select for red colonies, indicative of a functional type III secretion system (T3SS). TCS liquid cultures were inoculated with individual red colonies of S. flexneri M90T or S. flexneri ΔgalU and were grown overnight at 37 °C with shaking. The following day, bacterial cultures were diluted in fresh prewarmed TCS (1:50 v/v) and cultured until an optical density (OD600) of 0.6.
4.2. siRNA transfections and chemical treatments
HeLa cells (7 × 104) were plated in 6‐well plates (Thermo Scientific) for 24 hr and then transfected with selected siRNAs as previously described (Lobato‐Márquez et al., 2019; Mostowy et al., 2010; Sirianni et al., 2016). siRNA transfection was performed in DMEM with oligofectamine (Invitrogen) according to the manufacturer’s instructions. Cells were tested 72 hr after siRNA transfection. Control siRNA (ID#14614) and predesigned siRNA for SEPT2 (ID#14709 and ID#14614), SEPT4 (ID#142770), SEPT7 (ID#10323) or SEPT9 (ID#18228) are all from Ambion.
Cells were treated with 1 μM staurosporine (STS, #ALX‐380‐014‐C250, Enzo) or 500 μM etoposide (ETP, #E1383‐25MG, Sigma) for 10 hr. Equivalent volumes of Dimethyl sulfoxide (DMSO) were used as controls.
4.3. Measurement of cell viability
Viability of HeLa cells (Figures 1b and S1B) was measured by MTT (3‐[4,5‐deimethylthiazol‐2‐yl]‐2,5‐diphenyltetrazolium bromide) assays (Kumar, Nagarajan, & Uchil, 2018). After DMSO or STS treatments, cells were washed with pre‐warmed PBS and placed in serum‐free DMEM. MTT was added to each well to a final concentration of 1 mg/mL and plates were incubated for 45 min at 37 °C in 5% CO2. DMSO was added to each well (final volume 50%) and plates were agitated gently for 10 min at room temperature to dissolve the precipitate of the blue dyes. Samples were then diluted 1:1 with DMSO and measured the absorbance at the wavelength of 600 nm.
4.4. Apoptosis assays
For apoptosis assays (Figure 1c), approximately 2.5 × 104 cells were seeded into Lab‐Tek 8‐well glass‐bottom chamber slides (#177445, Thermo Scientific) and transfected with siRNA 72 hr prior to DMSO, STS or ETP treatments. Alexa488‐Annexin V (#A13201, Invitrogen), and Hoechst (#62249, Thermo Scientific) were added directly to the media and incubated for 15 min at 37 °C in 5% CO2. All live imaging was performed at 37 °C in 5% CO2 and captured using a 10x lens on a Zeiss Axio Observer Z1 widefield epifluorescence microscope driven by ZEN Blue 2.3 software (Carl Zeiss).
4.5. Antibodies and Western blotting
Rabbit polyclonal antibodies used were anti‐caspase‐3 (ID#9662S, Cell Signaling), anti‐caspase‐9 (ID#9502S, Cell Signaling), anti‐cleaved caspase‐3 (ID#9661S, Cell Signaling), anti‐SEPT7 (ID#18991, IBL), anti‐SEPT9 (ID#A302‐353A) and anti‐Tom20 (ID#42406S, Cell Signaling). Mouse antibodies used were anti‐ARTS (ID#A4471‐1ML, Sigma), anti‐caspase‐4 (ID#M029‐3, MBL), anti‐caspase‐7 (ID#9494S, Cell Signaling), anti‐cytochrome c (ID#12963A, Cell Signaling), anti‐GAPDH (ID#ab8245, Abcam), and anti‐SEPT2 (ID#60075‐1‐Ig, Proteintech Europe). Secondary antibodies used were goat anti‐mouse (ID#P0260, Dako) or anti‐rabbit (ID#P0448, Dako) antibodies, both horseradish peroxidase‐conjugated. GAPDH was used as a loading control. All antibodies were diluted in blocking solution (Tris Buffered Saline containing 0.1% Tween 20) supplemented with 5% fatty acid‐free milk.
For immunoblotting, HeLa cells were solubilized in radioimmunoprecipitation assay (RIPA) buffer (ID#R0278‐50ML, Sigma) supplemented with protease inhibitors (cOmplete ULTRA tablets, #05892970001 Roche) and phosphatase inhibitors (PhosSTOP, #04906845001, Roche). Protein concentration of lysates was determined using a bicinchoninic acid (BCA) assay kit (#23225, Pierce), and equal protein amounts of each sample were migrated on 10 or 12% SDS/polyacrylamide gels. All Western blotting experiments involved the transfer of samples to polyvinylidene difluoride membranes (PVDF, #IPVH00010, MerckMillipore), incubation with primary antibodies or secondary antibodies coupled to horseradish peroxidase, and detection using enhanced chemiluminescence (ECL) Plus reagent (#32132, Pierce). Chemiluminescence was imaged using a ChemiDoc™ MP imaging system (Bio‐Rad).
4.6. Infection of human cells
HeLa cells (7 × 104) were seeded in 6‐well plates (Thermo Scientific) and treated with siRNAs as described above. Cell cultures were infected with S. flexneri strains at a multiplicity of infection (MOI, bacteria: cell) of 100:1 (S. flexneri wildtype and S. flexneri ΔgalU). In the case of S. flexneri wildtype, bacteria and cells were immediately centrifuged at 110 × g for 10 min at room temperature. Then, plates were placed at 37 °C and 5% CO2 for 30 min. Infected cultures were washed 2× with phosphate‐buffered saline (PBS) pH 7.4 and incubated with fresh DMEM containing 10% FBS and 50 mg/mL gentamicin at 37 °C and 5% CO2 for 4 hr.
4.7. Confocal microscopy
Hela cells (7 × 104) growing on 22 × 22‐mm glass coverslips were transfected with siRNA for 72 hr (Figures 2c and 3a). After DMSO or STS treatments, cells were fixed in 3% paraformaldehyde (PFA) in PBS for 20 min. Cells were subsequently washed 3X in PBS pH 7.4 and permeabilized 5 min with 0.1% Triton X‐100 in PBS. Cells were then washed 3X in PBS and incubated in blocking buffer (1% bovine serum albumin [BSA] in PBS) for a minimum of 30 min. Cells were probed with primary antibodies diluted in blocking buffer overnight at 4°C. Cells were washed and treated with Alexa Fluor conjugated secondary antibodies for 1 hr, followed by washes and mounting in Prolong Gold anti‐fade reagent (#P36935, Invitrogen). Microscopy images were acquired using a 63x/1.4 C‐Plan Apo oil immersion lens on a Zeiss LSM 880 confocal microscope driven by ZEN Black software (v2.3). Confocal images were processed using Airyscan processing (Weiner filter) using “Auto Filter” and “3D Processing” options.
4.8. Zebrafish infections and CRISPR‐Cas9 editing
Animal experiments were performed according to the Animals (Scientific Procedures) Act 1986 and approved by the Home Office (Project license: PPL P4E664E3C). Wildtype zebrafish (AB strain) were used for all experiments. Larvae were reared at 28.5 °C in Petri dishes containing embryo medium, consisting of 0.5x E2 water supplemented with 0.5 ppm (ppm) of methylene blue (Sigma‐Aldrich, St. Louis, Missouri). For injections and in vivo imaging, anesthesia was obtained with buffered 200 μg/mL tricaine (Sigma‐Aldrich) in embryo medium. For CRISPR‐Cas9 editing, three Alt‐R® crisprRNA (crRNA, Integrated DNA technologies) were designed, targeting three consecutive exons of the zebrafish sept2 gene. For each target, a crRNA/tracrRNA complex was obtained by combining crRNA and Alt‐R® trans‐activating crRNA (tracrRNA) at 60 μM concentration each, in IDTE buffer (10 mM Tris HCl, 0.1 mM EDTA, pH 7.5), following the manufacturer's guidelines. Complexes were aliquoted and stored at −80 °C. 1 μL of Alt‐R® Streptococcus pyogenes Cas9 nuclease V3 protein (10 μg/μL, Integrated DNA technologies) was combined with 5 μL of Cas9 buffer (20 mM HEPES; 150 mM KCl, pH 7.5), 0.1 μL of phenol red solution (10%) and 1.5 μL of each of the three crRNA/tracRNA complexes. The solution was incubated at 37 °C for 10 min, returned on ice and microinjected (1 nL/egg) in the yolk sac of zebrafish eggs at a single‐cell stage. As a control, mock injections were performed with an equivalent solution, lacking Cas9 and crRNA/tracrRNA complexes. Efficient targeting was validated by PCR and gel electrophoresis. Sequences of crRNA and PCR primers are reported in Table S2. S. flexneri M90T or S. flexneri ΔgalU were prepared for injections as in (Torraca et al., 2019). Briefly, bacteria were grown to log phase, spun down, washed in PBS, and resuspended to a density equivalent to an OD600 = 18 in an injection buffer containing 2% polyvinylpyrrolidone (Sigma‐Aldrich) and 0.5% phenol red (Sigma‐Aldrich) in PBS. 1 nL (corresponding to ~9,000 CFU) of bacterial suspension was microinjected in the hindbrain ventricle (HBV) of 3 days post‐fertilization (dpf) zebrafish larvae. Bacterial enumeration was performed a posteriori by mechanical disruption of infected larvae in PBS (Sigma‐Aldrich) and plating of serial dilutions onto Congo red‐TSA plates.
4.9. Flow cytometry
Following siRNA transfection, HeLa cells were treated with 1 μM staurosporine for 5 hr. Cells were harvested by trypsinization (Sigma, T3924) and collected after stopping the trypsin reaction with 10% (vol/vol) FBS containing medium, followed by washing three times with DPBS (Fisher, 14,190–094), centrifugation (1,500 × g for 5 min), and fixation with 4% PFA for 15 min at RT. After permeabilization with methanol (100%) (Fisher, M/3950/17), overnight at −20 °C, cells were stained (at room temperature for 2 hr) with anti‐cleaved caspase‐3 antibody (diluted 1:500 in dilution buffer (DPBS with 1% (vol/wt) BSA (Sigma, A9647) and 0.3% Triton X‐100. After 2X washing with DPBS, samples were incubated with Alexa Fluor conjugated secondary antibody for 1 hr at room temperature. Cells were analyzed by flow cytometry on a LSRII (BD Biosciences). Data were analyzed with the software FlowJo version 10.7.1.
4.10. Quantification and statistics
Image processing and quantifications were performed in Fiji (https://imagej.net/software/fiji/). The Fiji Cell Counter plugin was used (for live apoptosis assays and for fixed cytochrome c or cleaved caspase‐3 assays) by counting the total number of cells in the Hoechst or DAPI channel and the number of cells positive for Annexin V fluorescence, cytosolic cytochrome c staining or cleaved caspase‐3 staining.
Statistical analysis was performed in GraphPad Prism (v8.4, La Jolla, USA). Data are presented as mean ± SE of the mean (SEM) from at least three independent experiments per treatment. ANOVA or Student's t test were used to compare values, with p < .05 considered as significant. Statistical differences for zebrafish survival curves were analyzed using a Log‐rank (Mantel–Cox) test. All statistical details including statistical tests, significance, number of experimental replicates, and cells quantified can be found in the figure legends.
All figures were designed using Adobe Illustrator CC 2018.
AUTHOR CONTRIBUTIONS
Serge Mostowy conceived and supervised this study.
Hoan Van Ngo, Stevens Robertin, Dominik Brokatzky, Magdalena K. Bielecka, Damián.
Lobato‐Márquez, Vincenzo Torraca, and Serge Mostowy designed the experiments.
Hoan Van Ngo, Stevens Robertin, and Dominik Brokatzky performed experiments using tissue culture cells.
Magdalena K. Bielecka and Vincenzo Torraca performed experiments using zebrafish.
All authors performed data analysis, and took part in the interpretation of results and preparation of materials for the manuscript.
Serge Mostowy wrote the manuscript with comments from all authors.
CONFLICT OF INTEREST
The authors declare no conflicts of interest.
Supporting information
Figure S1 Phylogeny and expression levels of zebrafish septins and Sept2 CRISPR targeting validation. (A) Phylogeny tree of septin proteins from humans (Hsa, Homo sapiens), zebrafish (Dre, Danio rerio) and yeast (Sce, Saccharomyces cerevisiae). Sequences were obtained from the Ensembl database (https://www.ensembl.org/). When multiple splicing isoforms were reported, the Ensembl canonical isoform or the isoform predicted to be the principal isoform was used in the alignment. (B) Expression level of septin genes in zebrafish in reads per kilobase million (RPKM). Data on basal septin expression extracted from RNAseq datasets (Torraca et al., 2019). (C) Gel electrophoresis of PCR products, demonstrating targeting of CRISPR/Cas9 for the three selected targets. S2: sept2 crispant; C: Control larva.
Table S1 Zebrafish septins and their corresponding expression value. Expression levels are represented by the mean and standard deviation of the reads per kilobase million (RPKM) from three biological replicates. Data on basal septin expression extracted from RNAseq datasets (Torraca et al., 2019).
Table S2. crisprRNA target sequence and PCR primers used in this study.
ACKNOWLEDGEMENTS
We thank Paras Anand, Daniel Fisch and Eva Frickel for helpful discussions. H.V.N. was supported by a Newton International Fellowship (NIF\R1\180035). D.B. is supported by the DFG Walter Benjamin Programme (BR 6637/1‐1). D.L.‐M. was funded by the European Union's Horizon 2020 research and innovation program under the Marie Skłodowska ‐ Curie grant agreement no. H2020‐MSCA‐IF‐2016‐752022. V.T. was supported by an LSHTM/Wellcome Institutional Strategic Support Fund (ISSF) Fellowship (204928/Z/16/Z). Research in the Mostowy laboratory is supported by a European Research Council Consolidator Grant (772853 ‐ ENTRAPMENT), Wellcome Trust Senior Research Fellowship (206444/Z/17/Z) and the Lister Institute of Preventive Medicine.
Van Ngo, H. , Robertin, S. , Brokatzky, D. , Bielecka, M. K. , Lobato‐Márquez, D. , Torraca, V. , & Mostowy, S. (2023). Septins promote caspase activity and coordinate mitochondrial apoptosis. Cytoskeleton, 80(7‐8), 254–265. 10.1002/cm.21696
Hoan Van Ngo and Stevens Robertin contributed equally to this study.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon request.
REFERENCES
- Ashida, H. , Suzuki, T. , & Sasakawa, C. (2021). Shigella infection and host cell death: A double‐edged sword for the host and pathogen survival. Current Opinion in Microbiology, 59, 1–7. 10.1016/j.mib.2020.07.007 [DOI] [PubMed] [Google Scholar]
- Ashkenazi, A. , & Dixit, V. M. (1998). Death receptors: Signaling and modulation. Science (New York, N.Y.), 281(5381), 1305–1308. 10.1126/science.281.5381.1305 [DOI] [PubMed] [Google Scholar]
- Ashkenazi, A. , & Salvesen, G. (2014). Regulated cell death: Signaling and mechanisms. Annual Review of Cell and Developmental Biology, 30, 337–356. 10.1146/annurev-cellbio-100913-013226 [DOI] [PubMed] [Google Scholar]
- Bock, F. J. , & Tait, S. (2020). Mitochondria as multifaceted regulators of cell death. Nature Reviews Molecular Cell Biology, 21(2), 85–100. 10.1038/s41580-019-0173-8 [DOI] [PubMed] [Google Scholar]
- Bratton, S. B. , & Salvesen, G. S. (2010). Regulation of the Apaf‐1‐caspase‐9 apoptosome. Journal of Cell Science, 123(Pt 19), 3209–3214. 10.1242/jcs.073643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark, C. S. , & Maurelli, A. T. (2007). Shigella flexneri inhibits staurosporine‐induced apoptosis in epithelial cells. Infection and Immunity, 75(5), 2531–2539. 10.1128/IAI.01866-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danial, N. N. , & Korsmeyer, S. J. (2004). Cell death: Critical control points. Cell, 116(2), 205–219. 10.1016/s0092-8674(04)00046-7 [DOI] [PubMed] [Google Scholar]
- Dorstyn, L. , Akey, C. W. , & Kumar, S. (2018). New insights into apoptosome structure and function. Cell Death and Differentiation, 25(7), 1194–1208. 10.1038/s41418-017-0025-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duggan, G. M. , & Mostowy, S. (2018). Use of zebrafish to study Shigella infection. Disease Models & Mechanisms, 11(2), dmm032151. 10.1242/dmm.032151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edison, N. , Curtz, Y. , Paland, N. , Mamriev, D. , Chorubczyk, N. , Haviv‐Reingewertz, T. , … Larisch, S. (2017). Degradation of Bcl‐2 by XIAP and ARTS promotes apoptosis. Cell Reports, 21(2), 442–454. 10.1016/j.celrep.2017.09.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edison, N. , Zuri, D. , Maniv, I. , Bornstein, B. , Lev, T. , Gottfried, Y. , … Larisch, S. (2012). The IAP‐antagonist ARTS initiates caspase activation upstream of cytochrome C and SMAC/Diablo. Cell Death and Differentiation, 19(2), 356–368. 10.1038/cdd.2011.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estey, M. P. , Di Ciano‐Oliveira, C. , Froese, C. D. , Bejide, M. T. , & Trimble, W. S. (2010). Distinct roles of septins in cytokinesis: SEPT9 mediates midbody abscission. The Journal of Cell Biology, 191(4), 741–749. 10.1083/jcb.201006031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García‐Fernández, M. , Kissel, H. , Brown, S. , Gorenc, T. , Schile, A. J. , Rafii, S. , … Steller, H. (2010). Sept4/ARTS is required for stem cell apoptosis and tumor suppression. Genes & Development, 24(20), 2282–2293. 10.1101/gad.1970110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes, M. C. , & Mostowy, S. (2020). The case for modeling human infection in zebrafish. Trends in Microbiology, 28(1), 10–18. 10.1016/j.tim.2019.08.005 [DOI] [PubMed] [Google Scholar]
- Gottfried, Y. , Rotem, A. , Lotan, R. , Steller, H. , & Larisch, S. (2004). The mitochondrial ARTS protein promotes apoptosis through targeting XIAP. The EMBO Journal, 23(7), 1627–1635. 10.1038/sj.emboj.7600155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Günther, S. D. , Fritsch, M. , Seeger, J. M. , Schiffmann, L. M. , Snipas, S. J. , Coutelle, M. , … Kashkar, H. (2020). Cytosolic gram‐negative bacteria prevent apoptosis by inhibition of effector caspases through lipopolysaccharide. Nature Microbiology, 5(2), 354–367. 10.1038/s41564-019-0620-5 [DOI] [PubMed] [Google Scholar]
- Jenner, A. , Peña‐Blanco, A. , Salvador‐Gallego, R. , Ugarte‐Uribe, B. , Zollo, C. , Ganief, T. , … Garcia‐Saez, A. J. (2022). DRP1 interacts directly with BAX to induce its activation and apoptosis. EMBO Journal, 13, e108587. 10.15252/embj.2021108587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, P. , Nagarajan, A. , & Uchil, P. D. (2018). Analysis of cell viability by the MTT assay. Cold Spring Harbor Protocols, 2018(6). 10.1101/pdb.prot095505 [DOI] [PubMed] [Google Scholar]
- Krokowski, S. , Lobato‐Márquez, D. , Chastanet, A. , Pereira, P. M. , Angelis, D. , Galea, D. , … Mostowy, S. (2018). Septins recognize and entrap dividing bacterial cells for delivery to lysosomes. Cell Host & Microbe, 24(6), 866–874.e4. 10.1016/j.chom.2018.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroll, F. , Powell, G. T. , Ghosh, M. , Gestri, G. , Antinucci, P. , Hearn, T. J. , … Rihel, J. (2021). A simple and effective F0 knockout method for rapid screening of behaviour and other complex phenotypes. eLife, 10, e59683. 10.7554/eLife.59683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larisch, S. , Yi, Y. , Lotan, R. , Kerner, H. , Eimerl, S. , Tony Parks, W. , … Roberts, A. B. (2000). A novel mitochondrial septin‐like protein, ARTS, mediates apoptosis dependent on its P‐loop motif. Nature Cell Biology, 2(12), 915–921. 10.1038/35046566 [DOI] [PubMed] [Google Scholar]
- Lembo‐Fazio, L. , Nigro, G. , Noël, G. , Rossi, G. , Chiara, F. , Tsilingiri, K. , … Bernardini, M. L. (2011). Gadd45α activity is the principal effector of Shigella mitochondria‐dependent epithelial cell death in vitro and ex vivo. Cell Death & Disease, 2(2), e122. 10.1038/cddis.2011.4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobato‐Márquez, D. , Krokowski, S. , Sirianni, A. , Larrouy‐Maumus, G. , & Mostowy, S. (2019). A requirement for septins and the autophagy receptor p62 in the proliferation of intracellular Shigella. Cytoskeleton (Hoboken, N.J.), 76(1), 163–172. 10.1002/cm.21453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobato‐Márquez, D. , Xu, J. , Güler, G. Ö. , Ojiakor, A. , Pilhofer, M. , & Mostowy, S. (2021). Mechanistic insight into bacterial entrapment by septin cage reconstitution. Nature Communications, 12(1), 4511. 10.1038/s41467-021-24721-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mageswaran, S. K. , Grotjahn, D. A. , Zeng, X. , Barad, B. A. , Medina, M. , Hoang, M. H. , … Jensen, G. J. (2021). Nanoscale details of mitochondrial fission revealed by cryo‐electron tomography. bioRxiv 12.13.472487. 10.1101/2021.12.13.472487 [DOI] [Google Scholar]
- Mamriev, D. , Abbas, R. , Klingler, F. M. , Kagan, J. , Kfir, N. , Donald, A. , … Larisch, S. (2020). A small‐molecule ARTS mimetic promotes apoptosis through degradation of both XIAP and Bcl‐2. Cell Death & Disease, 11(6), 483. 10.1038/s41419-020-2670-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazon‐Moya, M. J. , Willis, A. R. , Torraca, V. , Boucontet, L. , Shenoy, A. R. , Colucci‐Guyon, E. , & Mostowy, S. (2017). Septins restrict inflammation and protect zebrafish larvae from Shigella infection. PLoS Pathogens, 13(6), e1006467. 10.1371/journal.ppat.1006467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostowy, S. , & Cossart, P. (2012). Septins: The fourth component of the cytoskeleton. Nature Reviews Molecular Cell Biology, 13(3), 183–194. 10.1038/nrm3284 [DOI] [PubMed] [Google Scholar]
- Mostowy, S. , & Shenoy, A. R. (2015). The cytoskeleton in cell‐autonomous immunity: Structural determinants of host defence. Nature Reviews Immunology, 15(9), 559–573. 10.1038/nri3877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostowy, S. , Bonazzi, M. , Hamon, M. A. , Tham, T. N. , Mallet, A. , Lelek, M. , … Cossart, P. (2010). Entrapment of intracytosolic bacteria by septin cage‐like structures. Cell Host & Microbe, 8(5), 433–444. 10.1016/j.chom.2010.10.009 [DOI] [PubMed] [Google Scholar]
- Mostowy, S. , Boucontet, L. , Mazon Moya, M. J. , Sirianni, A. , Boudinot, P. , Hollinshead, M. , … Colucci‐Guyon, E. (2013). The zebrafish as a new model for the in vivo study of Shigella flexneri interaction with phagocytes and bacterial autophagy. PLoS Pathogens, 9(9), e1003588. 10.1371/journal.ppat.1003588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naderer, T. , & Fulcher, M. C. (2018). Targeting apoptosis pathways in infections. Journal of Leukocyte Biology, 103(2), 275–285. 10.1189/JLB.4MR0717-286R [DOI] [PubMed] [Google Scholar]
- Pagliuso, A. , Tham, T. N. , Stevens, J. K. , Lagache, T. , Persson, R. , Salles, A. , … Stavru, F. (2016). A role for septin 2 in Drp1‐mediated mitochondrial fission. EMBO Reports, 17(6), 858–873. 10.15252/embr.201541612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertin, S. , & Mostowy, S. (2020). The history of septin biology and bacterial infection. Cellular Microbiology, 22(4), e13173. 10.1111/cmi.13173 [DOI] [PubMed] [Google Scholar]
- Salvesen, G. S. , & Ashkenazi, A. (2011). Snapshot: Caspases. Cell, 147(2), 476–476.e1. 10.1016/j.cell.2011.09.030 [DOI] [PubMed] [Google Scholar]
- Sirianni, A. , Krokowski, S. , Lobato‐Márquez, D. , Buranyi, S. , Pfanzelter, J. , Galea, D. , … Mostowy, S. (2016). Mitochondria mediate septin cage assembly to promote autophagy of Shigella . EMBO Reports, 17(7), 1029–1043. 10.15252/embr.201541832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiliotis, E. T. , & Nakos, K. (2021). Cellular functions of actin‐ and microtubule‐associated septins. Current Biology: CB, 31(10), R651–R666. 10.1016/j.cub.2021.03.064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornberry, N. A. , & Lazebnik, Y. (1998). Caspases: Enemies within. Science (New York, N.Y.), 281(5381), 1312–1316. 10.1126/science.281.5381.1312 [DOI] [PubMed] [Google Scholar]
- Torraca, V. , & Mostowy, S. (2018). Zebrafish infection: From pathogenesis to cell biology. Trends in Cell Biology, 28(2), 143–156. 10.1016/j.tcb.2017.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torraca, V. , Kaforou, M. , Watson, J. , Duggan, G. M. , Guerrero‐Gutierrez, H. , Krokowski, S. , … Mostowy, S. (2019). Shigella sonnei infection of zebrafish reveals that O‐antigen mediates neutrophil tolerance and dysentery incidence. PLoS Pathogens, 15(12), e1008006. 10.1371/journal.ppat.1008006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Ngo, H. , & Mostowy, S. (2019). Role of septins in microbial infection. Journal of Cell Science, 132(9), jcs226266. 10.1242/jcs.226266 [DOI] [PubMed] [Google Scholar]
- Woods, B. L. , & Gladfelter, A. S. (2021). The state of the septin cytoskeleton from assembly to function. Current Opinion in Cell Biology, 68, 105–112. 10.1016/j.ceb.2020.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Phylogeny and expression levels of zebrafish septins and Sept2 CRISPR targeting validation. (A) Phylogeny tree of septin proteins from humans (Hsa, Homo sapiens), zebrafish (Dre, Danio rerio) and yeast (Sce, Saccharomyces cerevisiae). Sequences were obtained from the Ensembl database (https://www.ensembl.org/). When multiple splicing isoforms were reported, the Ensembl canonical isoform or the isoform predicted to be the principal isoform was used in the alignment. (B) Expression level of septin genes in zebrafish in reads per kilobase million (RPKM). Data on basal septin expression extracted from RNAseq datasets (Torraca et al., 2019). (C) Gel electrophoresis of PCR products, demonstrating targeting of CRISPR/Cas9 for the three selected targets. S2: sept2 crispant; C: Control larva.
Table S1 Zebrafish septins and their corresponding expression value. Expression levels are represented by the mean and standard deviation of the reads per kilobase million (RPKM) from three biological replicates. Data on basal septin expression extracted from RNAseq datasets (Torraca et al., 2019).
Table S2. crisprRNA target sequence and PCR primers used in this study.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon request.


