Abstract
The N terminal amino acid of nonstructural protein nsP4, the viral RNA polymerase, is a tyrosine in all sequenced alphaviruses; this is a destabilizing amino acid for the N-end rule pathway and results in rapid degradation of nsP4 produced in infected cells or in reticulocyte lysates. We have constructed 11 mutants of Sindbis virus bearing Phe, Ala, Thr, Cys, Leu, Met, Asn, Gln, Glu, Arg, or Pro at the N terminus of nsP4. Translation of RNAs in reticulocyte lysates showed that cleavage at the nsP3/nsP4 site occurred efficiently for all mutants except for Glu-nsP4, which was cleaved inefficiently, and Pro-nsP4, which was not detectably cleaved, and that Tyr, Cys, Leu, Arg, and Phe destabilized nsP4 but Ala, Met, Thr, Asn, Gln, and Glu stabilized nsP4 to various extents. The viability of the mutants was examined by transfection of chicken cells at 30 or 40°C. The Phe-nsP4 mutant formed large plaques at both temperatures. The Met-nsP4 mutant was also viable but formed small plaques at 30°C and minute plaques at 40°C. The remaining mutants did not form plaques at either temperature. However, after prolonged incubation at 30°C, all the mutants except Glu-nsP4 and Pro-nsP4 produced viable viruses. In the case of Cys-, Leu-, Asn-, Gln-, or Arg-nsP4, revertants that were indistinguishable in plaque phenotype from the wild-type virus arose by same-site reversion to Tyr, Trp, Phe, or His by a single nucleotide substitution in the original mutant codon. Viable viruses also arose from the Ala-, Leu-, Cys-, Thr-, Asn-, Gln-, and Arg-nsP4 mutants that retained the original mutations at the N terminus of nsP4, but these viruses formed smaller plaques than the wild-type virus and many were temperature sensitive. Our results indicate that only nsP4s bearing N-terminal Tyr, Phe, Trp, or His have wild-type or near-wild-type activity for RNA replication and that rapid degradation of nsP4 is not a prerequisite for its function. nsP4s bearing other N-terminal residues, with the exception of Met-nsP4, have only very low or negligible activity, so that no detectable infectious virus can be produced. However, suppressor mutations can arise that enable most such nsP4s to regain significant but still suboptimal activity.
Sindbis virus is the type member of the genus Alphavirus in the family Togaviridae, with a single-stranded plus-sense RNA of 11.7 kb (22). Four nonstructural proteins required for viral RNA replication and transcription, nsP1, nsP2, nsP3, and nsP4, are translated from the 5′ two-thirds of the genome as two overlapping polyproteins, P123 and P1234, which are cleaved by a proteolytic activity present in the C-terminal half of nsP2 (10). Differential processing of these nonstructural polyproteins results in regulation of minus-strand and plus-strand RNA synthesis in infected cells, with the result that minus-strand RNA is made only early after infection (16, 20).
nsP4 is believed to be the viral RNA polymerase because it possesses certain sequence motifs characteristic of RNA polymerases (12, 13) and because a temperature-sensitive mutation in nsP4 is known that leads to cessation of RNA elongation upon a shift to a nonpermissive temperature (1, 8). Production and accumulation of nsP4 are regulated by three mechanisms in Sindbis-infected cells. First, translation of nsP4 occurs only upon readthrough of a UGA termination codon located 7 amino acids upstream from the nsP3/nsP4 boundary (21), and it occurs with an efficiency of 5 to 20% (4, 20). Wild-type virus grows more efficiently than a UGA-to-sense-codon mutant (17). Strauss and Strauss have postulated (22) that a function of the opal codon may be to produce sufficient quantities of the trans-acting protease to efficiently convert the initial replicase, which contains uncleaved P123 plus nsP4 and can synthesize only minus-strand RNA efficiently, to a replicase which contains cleaved products plus nsP4 and which can now make plus-strand RNA (16, 20). Another possibility is that the opal codon serves to regulate the ratio of capping or helicase activities present in P123 and its cleaved products to RNA polymerase activity present in nsP4. Second, nsP4 is produced only early in infection by cis cleavage of the nsP3/nsP4 bond in polyprotein P1234 (4, 19). Later in infection, accumulation of trans-acting protease leads to nascent cleavage at the nsP2/nsP3 bond such that P1234 cannot be produced and P12 and P34 are the primary translation products. P12 undergoes further cleavage, but P34 accumulates late in infection because the viral nonstructural proteinases present at this time are unable to cleave the nsP3/nsP4 bond (4). Third, nsP4 is metabolically unstable and vulnerable to a rapid degradation by the N-end rule pathway (5), in which the stability of a protein is dependent on the amino acid at its N terminus (23). The N-terminal amino acid of Sindbis virus nsP4 is Tyr, which is a destabilizing amino acid in mammalian cells, and it has been shown that while a fraction of nsP4 is stable, presumably because it is sequestered within RNA replication complexes (2), free nsP4 is rapidly degraded (9).
In this study, we characterized Sindbis mutants, as well as revertants of these mutants, that have different amino acids at the N terminus of nsP4, resulting in differences in the stabilities of nsP4, in an effort to elucidate the importance of the rapid degradation of nsP4 in viral replication. Our results show that efficient function of nsP4 is dependent upon the presence of an aromatic amino acid or histidine at the N terminus and that other N-terminal amino acids, whether stabilizing or destabilizing, lead to reduced activity.
MATERIALS AND METHODS
Cells and medium.
Secondary chicken embryo fibroblast monolayers were cultured in Eagle’s minimum essential medium containing 3% fetal calf serum and used for RNA transfection, virus growth, and plaque assay as described previously (20).
N-terminal mutants.
pToto1101 (18), a full-length cDNA clone of Sindbis virus from which infectious RNA transcripts can be produced in vitro, was used as a parental clone for generating mutations in the N-terminal residue of nsP4. The wild-type Tyr codon (TAC, nucleotides [nt] 5769 to 5771, where nt 1 is the first nucleotide in the genomic RNA) was changed to Ala (GCC), Met (ATG), Leu (CTC), Arg (AGG), or Phe (TTC) by cassette mutagenesis as previously described (4). Changes to Cys (TGC), Thr (ACT), Asn (AAC), Gln (CAG), Glu (GAG), or Pro (CCC) codons were carried out by in vitro mutagenesis as follows. The region in pToto1101 from nt 5758 to 6085 was amplified by PCR with a plus-sense mutagenic primer annealing to nt 5758 to 5782, 5′-GGGTAGGTGGGXXXATATTTTCGAC-3′ (where XXX is the codon for residue 1 of nsP4), and a minus-sense primer, YA16, annealing to nt 6069 to 6085, 5′-TACAATGGTTTCGGATA-3′, and the resultant 0.3-kb DNA was purified on a commercial spin column (Qiagen Inc., Chatworth, Calif.). Similarly, the region from nt 5164 to 5782 was amplified with a minus-sense mutagenic primer annealing to nt 5758 to 5782, 5′-GTCGAAAATATX′X′X′CCCACCTACCC-3′ (where X′X′X′ is complementary to the codon for residue 1 of nsP4), and a plus-sense primer, YA19, annealing to nt 5164 to 5180, 5′-ATAACACCTCGCTTGAT-3′, and the resultant 0.6-kb DNA was purified. The 0.3-kb DNA and 0.6-kb DNA were then fused by a fusion PCR approach, in which the two fragments were mixed, denatured, annealed, and subjected to a second cycle of PCR amplification with the YA16 and YA19 primers. The 0.9-kb fragment was purified and digested with SpeI (nt 5263) and EcoRI (nt 5870), and the 0.6-kb SpeI-EcoRI fragment was purified and cloned into an intermediate vector, pSIN34CE2, containing a 4.5-kb SpeI (nt 5263)-to-BssHII (nt 9805) insert from pToto1101 in a Proteus1 plasmid vector (18). The resulting insert was then removed and inserted into an SpeI- and BssHII-digested pToto1101 vector. More than three independent clones were examined for each mutant. The nsP4 N-terminal mutant constructs were called pToto1101.4X, where X indicates the mutated amino acid at the N terminus of nsP4.
The Gly→Val substitution at the P2 position of the nsP2/nsP3 cleavage site, which abolishes processing at the nsP2/nsP3 bond (19), was also combined with the nsP4 N-terminal mutations to examine the stabilities of the mutant nsP4s during in vitro translation. To make these constructs, the SpeI-BssHII fragment containing the mutated nsP4 N terminus from pToto1101.4X was cloned into an SpeI-BssHII-digested pToto1101.2V vector (20).
Analysis of in vitro translation products.
Miniprep DNA of pToto1101 constructs was prepared by a modified boiling method (11) and digested with XhoI. The linearized template DNA was transcribed with an SP6 RNA polymerase at 38°C for 1 h in the presence of a cap analog. Transcript RNA was translated in rabbit reticulocyte lysates (Promega Biotec, Madison, Wis.) in the presence of [35S]Met (1,200 Ci/mmol) at 30°C for 90 min. Labeled translation products were separated in a sodium dodecyl sulfate-polyacrylamide gel and visualized by fluorography with sodium salicylate (3).
RNA transfection.
Confluent monolayers of chicken cells in 35-mm plates were treated with DEAE-dextran and transfected with RNA transcripts diluted in phosphate-buffered saline. For direct plaque analysis, cells were overlaid with Eagle’s minimal essential medium containing 1% agarose and 3% calf serum after transfection and incubated at 30 or 40°C for 2 days before being stained with neutral red. For observation of cytopathic effects and recovery of viable viruses, the transfected cells were incubated in a liquid medium for up to 7 days at 30 or 40°C. Viruses produced during this extended incubation were examined by plaque assay on confluent monolayers of chicken cells with incubation at 30 or 40°C for 2 days before being stained.
RT-PCR and nucleotide sequencing.
RNA was extracted from 0.4 ml of the primary transfection medium with phenol-chloroform and precipitated with ethanol. cDNA was synthesized by reverse transcription with minus-sense primer YA16, and the region from nt 5164 to 6085 was amplified with primers YA16 and YA19. The 0.9-kb DNA was digested with SpeI and EcoRI, and the resultant 0.6-kb fragment was cloned into an SpeI- and EcoRI-digested pSIN34CE2. The nucleotide sequence of the nsP4 N-terminal region was determined from more than three independent clones by sequencing with Sequenase, using a minus-sense primer annealing to nt 5857 to 5876.
Testing of nsP4 mutations in a wild-type background.
The Trp-nsP4 and His-nsP4 mutations were introduced into pToto1101 by replacing the BamHI fragment (nt 4634 to 7335) of pToto1101 with that of double-stranded cDNA made to the revertant virus RNA by the method of Gubler and Hoffman (7). The Trp-nsP4 and His-nsP4 mutations were also combined with the 2V mutation by replacing the BamHI fragment of pToto1101.2V with that from the mutant cDNA.
RESULTS
Stabilities of mutant nsP4s in vitro.
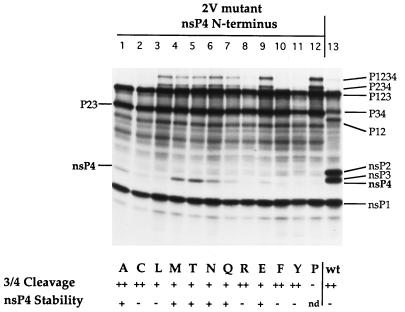
To examine the stabilities of the nsP4s bearing different amino acids at the N terminus, RNA transcripts from pToto1101 derivatives containing the nsP4 N-terminal mutations were translated in rabbit reticulocyte lysates (Fig. 1). The transcripts also contained the 2V mutation, which abolishes cleavage at the nsP2/nsP3 site, so that a P23 polyprotein was produced instead of nsP2 and nsP3, the latter of which interferes with visualization of nsP4 because of their similar migration rates in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (Fig. 1, lane 13). We have previously shown that production of nsP4 is not influenced by the 2V mutation (19).
FIG. 1.
Analysis of Sindbis virus nsP4 N-terminal mutants by translation in rabbit reticulocyte lysates. The N terminus of nsP4, which is Tyr in the wild type (lane 11), was changed to Ala (lane 1), Cys (lane 2), Leu (lane 3), Met (lane 4), Thr (lane 5), Asn (lane 6), Gln (lane 7), Arg (lane 8), Glu (lane 9), Phe (lane 10), or Pro (lane 12) in pToto1101.2V, which has a Gly→Val mutation at the P2 position of the nsP2/nsP3 cleavage site. Lane 13 shows translation products from the wild-type pToto1101 transcripts. In vitro transcripts of the different RNAs were translated in rabbit reticulocyte lysates at 30°C for 90 min in the presence of [35S]methionine. The cleavage efficiency at the nsP3/nsP4 site and the stability of the mutant nsP4s, as determined from the upper panel, are shown below. Cleavage efficiency: ++, efficient; +, less efficient; −, not detectable. nsP4 stability: +, stable; −, unstable; nd, not determined.
Inspection of Fig. 1 shows that the extent of cleavage at the nsP3/nsP4 bond and the amount of nsP4 present differ depending upon the N terminus of nsP4. The extent of cleavage, and thus the amount of nsP4 initially produced, can be estimated from the amounts of residual P1234 and P234 containing uncleaved nsP4 (note that the P123 and P23 bands serve as internal controls for the extent of translation in each reaction). Cleavage at the nsP3/nsP4 bond in the Ala-, Cys-, and Phe-nsP4 translation products (Fig. 1, lanes 1, 2, and 10) was as efficient as that in the Tyr-nsP4 translation product (lane 11), with little or no detectable uncleaved P1234 or P234 remaining; cleavage of Arg-nsP4 was only slightly less efficient, with trace amounts of uncleaved precursors (lane 8); cleavage of Leu-, Met-, Thr-, Asn-, and Gln-nsP4 mutant translation products (lanes 3 through 7) was detectably less efficient, with significant amounts of uncleaved precursors P1234 and P234 still present; cleavage of Glu-nsP4 was still less efficient (lane 9); and cleavage of Pro-nsP4 was not detectable (lane 12).
Distinct bands of Ala-, Met-, Thr-, Asn-, Gln-, and Glu-nsP4 were present in the fluorograph (Fig. 1, lanes 1, 4 to 7, and 9), but no detectable bands of Cys-, Leu-, Arg-, Phe-, or wild-type Tyr-nsP4 were present (lanes 2, 3, 8, 10, and 11), indicating that these five polypeptides were completely degraded in the experiment (no Pro-nsP4 was present because cleavage to produce it was not detectable). Taking cleavage efficiencies into consideration, we concluded that Met-, Thr-, Asn-, Gln-, and Glu-nsP4 were most stable, Ala-nsP4 was moderately stable, and Cys-, Leu-, Arg-, and Phe-nsP4, as well as the wild-type Tyr-nsP4, were unstable in rabbit reticulocyte lysates.
Plaque phenotype of nsP4 N-terminal mutants.
We examined the ability of the mutants carrying different N-terminal residues on nsP4 to form a plaque, and the results are shown in Fig. 2. In this experiment, cells were transfected with RNA, overlaid with medium containing agarose, incubated at 30 or 40°C, and stained with neutral red. The Phe-nsP4 mutant and the wild-type Tyr-nsP4 formed large plaques at both 30 and 40°C. The Met-nsP4 mutant formed small plaques at 30°C and minute plaques at 40°C but produced similar numbers of plaques at the two temperatures, so that it is not temperature sensitive for plaque formation. The other nine mutants did not form visible plaques at either temperature, and the mutations are thus lethal by this criterion.
FIG. 2.
Growth of mutants with different N-terminal residues in nsP4. The N terminus of nsP4 (Tyr = Y in the wild-type virus) was changed to Phe (F), Met (M), Ala (A), Cys (C), Leu (L), Thr (T), Asn (N), Gln (Q), Arg (R), Glu (E), or Pro (P). RNA transcripts were transfected into chicken cells, and the transfected cells were overlaid with medium containing agarose, incubated at 30 or 40°C, and stained with neutral red to visualize plaques. The sizes of the plaques formed are shown in millimeters; a minus sign indicates that no plaques were visible. A second set of cells were transfected with the different mutants, overlaid with liquid medium, and incubated at 30 or 40°C for 7 days or until cytopathology was obvious. +, cytopathology developed and viable virus could be isolated from the culture; −, no cytopathology was visible after 7 days, and no virus could be demonstrated in the culture.
Recovery of viable viruses from transfected cells.
Although no distinct plaques were formed after RNA transfection and agarose overlay, cells transfected with Ala-, Cys-, Leu-, Thr-, Asn-, Gln-, or Arg-nsP4 mutant transcripts showed cytopathology after incubation in liquid medium at 30°C for 2 to 7 days, depending on the experiment, and viable viruses could be recovered from these culture fluids (Fig. 2). To study the appearance of viable viruses, several plates of cells were independently transfected with RNA from each of the mutants so as to generate multiple independent stocks of viruses from each mutant, and these independent stocks were characterized in various ways (Table 1).
TABLE 1.
Phenotypes of revertant virusesa
| Mutant N terminus | Revertantb stock no. | Assay at 30°C
|
Assay at 40°C
|
Revertantc N terminus | ||
|---|---|---|---|---|---|---|
| Plaque size | Titer | Plaque size | Titer | |||
| Ala | 1 | Small | 109 | Minute | 108 | Ala |
| 2 | Small | 109 | Small | 107 | Ala | |
| 3 | Small | 109 | Small | 107 | Ala | |
| 4 | Small | 109 | Minute | 108 | Ala | |
| Cys | 1 | Large | 109 | None | Trp | |
| 2 | Large | 109 | Large | 109 | Tyr | |
| 3 | Large | 109 | Large | 109 | Tyr | |
| Leu | 1 | Large | 109 | Large | 109 | His |
| 2 | Large | 109 | Large | 109 | Phe | |
| 3 | Small | 109 | Small | 108 | Leu | |
| 4 | Small | 109 | Small | 108 | Leu | |
| 5 | Small | 109 | None | Leu | ||
| 6 | Large | 109 | Large | 109 | Phe | |
| 7 | Small | 109 | Minute | 109 | Leu | |
| Thr | 1 | Small | 109 | Small | 108 | Thr |
| 2 | Small | 109 | Minute | 105 | Thr | |
| 3 | Small | 109 | Minute | 105 | Thr | |
| 4 | Small | 109 | Minute | 105 | Thr | |
| Asn | 1 | Large | 109 | Large | 109 | Tyr |
| 2 | Large | 109 | Large | 109 | Tyr | |
| 3 | Small | 108 | None | Asn | ||
| 4 | Large | 109 | Large | 109 | Tyr | |
| Gln | 1 | Small | 108 | Small | 105 | Gln |
| 2 | Small | 108 | Small | 105 | Gln | |
| 3 | Small | 108 | Small | 105 | Gln | |
| 4 | Large | 109 | Large | 109 | His | |
| Arg | 1 | Large | 109 | Large | 109 | Trp |
| 2 | Large | 109 | Large | 109 | Trp | |
| 3 | Large | 109 | Large | 109 | Trp | |
| 4 | Small | 109 | Small | 106 | Arg | |
| 5 | Large | 109 | Large | 109 | Trp | |
Chicken cells were transfected with RNA from the various mutants and incubated at 30°C in liquid medium until cytopathology developed. The resulting revertant stock was serially diluted and subjected to titer determination by plaque assay on chicken cells at either 30 or 40°C. The plaque titers obtained at the two temperatures and the sizes of plaques formed are shown. None, no plaques detected.
Multiple independent RNA transfections were carried out to generate independent revertant stocks.
The N-terminal amino acid in the revertant viruses was determined by sequencing cDNA clones obtained by RT-PCR. Changed amino acids are in bold type.
The plaque phenotypes of the recovered viruses varied considerably among the virus stocks, but the viruses could be broadly divided into wild-type and non-wild-type categories (Table 1). Some of the revertants were wild type or pseudo-wild type in phenotype; stocks of these viruses had a titer of >109 when assayed at either 30 or 40°C, and large plaques were formed at both temperatures. At least one of the viruses recovered from the Cys-, Leu-, Asn-, Gln-, and Arg-nsP4 mutants had this wild-type phenotype. Other revertants were non-wild type, forming small plaques at both 30 and 40°C and usually exhibiting temperature sensitivity; stocks of these viruses had a titer of ≥108 when assayed at 30°C but a titer up to 4 orders of magnitude lower when assayed at 40°C. At least one of the viruses recovered from the Cys-, Leu-, Asn-, Gln-, and Arg-nsP4 mutants had such a phenotype, and only such non-wild-type viruses were recovered from the Ala- and Thr-nsP4 mutants in four independent transfection experiments with each mutant.
Cells transfected with the Pro- and Glu-nsP4 transcripts did not develop significant cytopathic effects. Viable viruses were not detected in the culture fluid either by plaque assay or by secondary passage, indicating that the Pro- and Glu-nsP4 mutations were absolutely lethal. Since cleavage at the nsP3/nsP4 bond in the Pro-nsP4 mutant translation products was not detectable, the lethality of this mutation is probably due to the lack of production of nsP4, rather than to inactivation of nsP4 by the N-terminal Pro residue, since we have shown previously that cleavage of the nsP3/nsP4 site is essential for viral RNA replication (20). In the Glu-nsP4 mutant, cleavage at the nsP3/nsP4 bond does occur in vitro, albeit inefficiently, and it seems probable that the mutation is lethal because Glu-nsP4 is not active, although it is possible that the mutation is lethal because insufficient nsP4 is produced.
nsP4 N-terminal amino acid in rescued viruses.
Since the original Ala-, Cys-, Leu-, Thr-, Asn-, Gln-, and Arg-nsP4 mutants did not form plaques at either 30 or 40°C, we assumed that the viable viruses recovered at 30°C resulted from reversion of the original mutation or from suppression by a second-site mutation. To determine whether the original mutations were still present in the recovered viruses, RNA was extracted from virus preparations and a region encoding the nsP3/nsP4 cleavage site was amplified by RT-PCR, cloned, and sequenced. More than three independent RT-PCR clones were sequenced from each revertant stock to ensure that a consensus sequence was obtained. The results are shown in Table 1.
In all the recovered viruses that had a wild-type plaque phenotype (that is, forming equal numbers of large plaques at both 30 and 40°C), the original mutations had been replaced with Tyr, Phe, Trp, or His, which in each case resulted from a single nucleotide substitution (Fig. 3). Thus, Cys (UGC) reverted to Tyr (UAC) in two independent revertants, Leu (CUC) changed to His (CAC) in one stock or to Phe (UUC) in two stocks, Asn (AAC) changed to Tyr (UAC) in three stocks, Gln (CAG) changed to His (CAU) in one stock, and Arg (AGG) changed to Trp (UGG) in four revertant stocks. Revertant stock 1 from the Cys-nsP4 mutant also had Trp (UGG) at the nsP4-N-terminus. This virus formed large plaques at 30°C but no viable plaques at 40°C, and we assume that this virus had a new mutation that rendered it temperature sensitive as discussed below.
FIG. 3.
Possible amino acid substitutions at the N terminus of the nsP4 mutants. For eight of the nsP4 mutants that were examined, the amino acid substitutions that are possible with a single nucleotide change in the mutant codon are shown. Boxed substitutions (in boldface type) were observed in one or more revertants, whereas none of the other substitutions were found. The shaded substitutions are possible changes to the six amino acids that were not directly tested at the N terminus of nsP4; the fact that these substitutions were never observed in revertant viruses indicates that these six amino acids are not acceptable N-terminal residues. Y, pyrimidine; P, purine (used in third codon positions where degeneracy exists).
In all the other viruses recovered, which formed small plaques at 30°C and small to minute plaques or no plaques at 40°C, the N-terminal amino acid of nsP4 was the same as in the original mutant. This includes all four viruses obtained from the Ala-nsP4 mutant and all four viruses from the Thr-nsP4 mutant, as well as one or more stocks of the Leu-, Asn-, Gln-, and Arg-nsP4. These viruses must have second-site suppressor mutations elsewhere in the genome that allow nsP4 to function with an otherwise lethal N-terminal amino acid. The function of nsP4 remains impaired, however, because the plaque morphology is different and the viruses do not form plaques well at 40°C.
The Trp-nsP4 and His-nsP4 mutations were introduced into pToto1101 to test the effect of these mutations in a wild-type background. Transfection of cells with RNA transcripts from three independent clones of the two mutants resulted in the formation of large plaques at both 30 and 40°C, demonstrating that these amino acids are acceptable residues at the N terminus of nsP4 for wild-type or near-wild-type function and ruling out the possibility that other unmapped mutations in the genome of the recovered viruses are responsible for the wild-type phenotype. The Trp- and His-nsP4 mutations were also combined with the 2V mutation, and RNA transcripts were translated in rabbit reticulocyte lysates. Cleavage at the nsP3/nsP4 bond occurred efficiently in both mutants, and the resulting Trp- and His-nsP4 were unstable (data not shown).
DISCUSSION
Requirement for Tyr, Trp, Phe, or His at the nsP4 N terminus.
These studies show that only nsP4s bearing Tyr, Trp, Phe, or His at the N terminus possess wild-type or near-wild-type function. These amino acids have in common an unsaturated ring, which is fully conjugated in the case of the three aromatic amino acids. Virus with N-terminal Met, which shares with these amino acids the property of being bulky and hydrophobic, is also viable but forms small plaques and is thus attenuated. All the other amino acids tested at the N terminus are lethal, but for at least Ala, Leu, Thr, Asn, Gln, and Arg (and probably Cys), the lethality can be suppressed by mutations elsewhere in the genome. The resulting virus does not replicate as efficiently as the wild type, however, as shown by the small-plaque phenotype and the temperature sensitivity of most of these viruses. Pro is probably lethal because cleavage to produce nsP4 is very limited or nonexistent and RNA replication is too limited to allow revertants to arise. Glu is also lethal and seems to impair function so completely that suppressors cannot arise. The remaining six amino acids not tested in this study, Ser, Ile, Lys, Val, Asp, and Gly, appear to be unacceptable for nsP4 function, because they could all have arisen by a single nucleotide substitution in one or more of the mutants tested (Fig. 3), and the fact that no revertants with these amino acids were found suggests that they are also lethal. Although these six amino acids must be lethal, it is unknown whether they could be suppressed by mutations elsewhere in the genome.
The finding that there is a requirement for one of a set of specific amino acids at the N terminus of nsP4 for activity is consistent with previous findings that cleavage to release nsP4 from the precursor polyprotein is absolutely required for virus replication in transfected cells (20) or for RNA synthesis in a reconstituted system in which transfected cDNA constructs were used to express Sindbis virus polyproteins (14–16). Furthermore, the effects of N-terminal Tyr, Met, Arg, Ala, and Leu on the activity of nsP4 in the reconstituted RNA synthesis system are consistent with our present results: Tyr-nsP4 had wild-type function, Met-nsP4 had very limited function, and Arg-, Ala-, and Leu-nsP4 had no detectable RNA synthesis activity (16). At present it is unknown whether Tyr (or acceptable alternatives) at the N terminus of nsP4 is required for the initiation of minus-strand RNA, for the initiation of plus-strand RNA, for the procession of initiated chains, or for all nsP4 activities. It is noteworthy that in the reconstituted system of Lemm and Rice (15), Met-nsP4 (produced either by substituting the N-terminal Tyr with Met or by adding a Met N-terminal to the Tyr) was able to synthesize minus-strand RNA, albeit inefficiently, but was unable to synthesize plus-strand RNA.
Although Tyr, Trp, Phe, and His allow near-wild-type function of nsP4 as judged by plaque phenotype and vigorous growth at both 30 and 40°C, they do not appear to be interchangeable. Tyr is conserved among all alphaviruses examined to date, and it is clear that this N-terminal residue has a selective advantage in nature. It is noteworthy that in the Cys and Asn mutants, where reversion to Tyr is possible by a single nucleotide substitution, five of seven revertants examined had reverted to Tyr (Fig. 3; Table 1). Similarly, for the Arg mutant, four of five revertants had changed to Trp. In contrast, for the Leu and Gln mutants, which can revert to Phe or His by a single nucleotide change but not to Tyr or Trp, only 4 of 11 viable revertants had changed to Phe or His while 7 had second-site suppressors. Although the numbers are small and other explanations are possible, this may suggest that viruses encoding nsP4s bearing Tyr or Trp have a greater selective advantage over viruses containing suppressor mutations than do viruses encoding nsP4s bearing Phe or His.
Emergence of viable viruses.
The emergence of viable viruses from many of the nonviable nsP4 N-terminal mutants must mean that limited RNA replication does occur after transfection with these mutant RNAs and that although the rate of replication is too low to permit the formation of a plaque, it is sufficient to allow mutations during replication that result in the appearance of revertants that overgrow the culture. Such RNAs have been called quasi-infectious by Gmyl et al. (6).
Revertants appeared only upon growth at 30°C; growth at 40°C is evidently more stringent, and presumably insufficient RNA replication occurs to permit revertants to arise. All of the same-site reversion events were the result of a single nucleotide change in the codon encoding the mutant amino acid (Fig. 3). The reason why no N-terminal revertant viruses were established from the Ala- or Thr-nsP4 mutants is apparently that these codons require two nucleotide substitutions to revert to an aromatic amino acid or His. The Glu mutant is of interest in this regard. It is not possible to revert to one of the four acceptable amino acids by a single nucleotide change (Fig. 3), and thus a viable virus would have to arise by suppressing the effect of the N-terminal Glu. Such a suppressor might be difficult to obtain because of the poor cleavage of Glu-nsP4 as well as the possibly reduced activity of the Glu-nsP4 enzyme.
The appearance of revertant 1 from Cys-nsP4 is also worthy of comment. This revertant had N-terminal Trp, which by itself results in a wild-type phenotype, but this revertant was temperature sensitive. We suggest that this revertant originally arose as the result of a second-site suppressor that also rendered the virus temperature sensitive, as is the case for most of the revertants that carry suppressor mutations. During growth of this revertant, a second reversion to Trp could have allowed it to grow even better at 30°C so that it forms large plaques at this temperature, in contrast to the small plaques formed by other suppressor revertants, but remains temperature sensitive.
It is interesting that there are so many different ways in which the nonviable mutants can revert to a viable phenotype. We have isolated 31 independent revertants starting from different N-terminal mutants, and these possess a variety of phenotypes that result from a variety of mutations that resulted in viable viruses. Although the wild-type sequence has a selective advantage, these results illustrate the inherent flexibility of the alphavirus genome, and of RNA viruses in general, and the ability to accommodate change.
Rapid turnover and nsP4 function.
Our results show that only Tyr, Trp, Phe, or His as nsP4 N-terminal residues lead to full activity, and these residues are all destabilizing for the N-end rule pathway. The other 16 residues, many of which were tested directly but some only indirectly by a failure to appear in revertants, lead to nsP4 that has reduced activity. The fact that Cys or Arg at the N terminus destabilizes nsP4 but renders it inactive whereas Met stabilizes nsP4 but results in partial activity indicates that there is no correlation between instability of nsP4 and its activity and that rapid turnover of nsP4 is a result of the requirement for an aromatic amino acid or histidine for full activity. We suggest that the primary role of the N-terminal tyrosine is to interact with other residues in nsP4, so that it folds properly, or to be involved in the interaction of nsP4 with other viral nonstructural proteins or cellular factors and that these interactions may be required for the recognition of viral promoters during RNA synthesis.
ACKNOWLEDGMENTS
We are grateful to Matthew Metts and Brian Kim for help in characterization of revertant viruses and to Ellen Strauss for help in preparing the manuscript.
This work was supported by grant AI 10793 from the NIH.
REFERENCES
- 1.Barton D J, Sawicki S G, Sawicki D L. Demonstration in vivo of temperature-sensitive elongation of RNA in Sindbis virus mutant ts6. J Virol. 1988;62:3597–3602. doi: 10.1128/jvi.62.10.3597-3602.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barton D J, Sawicki S G, Sawicki D L. Solubilization and immunoprecipitation of alphavirus replication complexes. J Virol. 1991;65:1496–1506. doi: 10.1128/jvi.65.3.1496-1506.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chamberlain J P. Fluorographic detection of radioactivity in polyacrylamide gels with the water-soluble fluor, sodium salicylate. Anal Biochem. 1979;98:132–135. doi: 10.1016/0003-2697(79)90716-4. [DOI] [PubMed] [Google Scholar]
- 4.de Groot R J, Hardy W R, Shirako Y, Strauss J H. Cleavage-site preferences of Sindbis virus polyproteins containing the nonstructural proteinase: evidence for temporal regulation of polyprotein processing in vivo. EMBO J. 1990;9:2631–2638. doi: 10.1002/j.1460-2075.1990.tb07445.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Groot R J, Rümenapf T, Kuhn R J, Strauss E G, Strauss J H. Sindbis virus RNA polymerase is degraded by the N-end rule pathway. Proc Natl Acad Sci USA. 1991;88:8967–8971. doi: 10.1073/pnas.88.20.8967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gmyl A P, Pilipenko E V, Maslova S V, Belov G A, Agol V I. Functional and genetic plasticities of the poliovirus genome: quasi-infectious RNAs modified in the 5′-untranslated region yield a variety of pseudorevertants. J Virol. 1993;67:6309–6316. doi: 10.1128/jvi.67.10.6309-6316.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gubler U, Hoffman B J. A simple and very efficient method for generating cDNA libraries. Gene. 1983;25:263–269. doi: 10.1016/0378-1119(83)90230-5. [DOI] [PubMed] [Google Scholar]
- 8.Hahn Y S, Grakoui A, Rice C M, Strauss E G, Strauss J H. Mapping of RNA− temperature-sensitive mutants of Sindbis virus: complementation group F mutants have lesions in nsP4. J Virol. 1989;63:1194–1202. doi: 10.1128/jvi.63.3.1194-1202.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hardy W R, Strauss J H. Processing of the nonstructural polyproteins of Sindbis virus: study of the kinetics in vivo using monospecific antibodies. J Virol. 1988;62:998–1007. doi: 10.1128/jvi.62.3.998-1007.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hardy W R, Strauss J H. Processing the nonstructural polyproteins of Sindbis virus: nonstructural proteinase is in the C-terminal half of nsP2 and functions both in cis and in trans. J Virol. 1989;63:4653–4664. doi: 10.1128/jvi.63.11.4653-4664.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Holmes D S, Quigley M. A rapid boiling method for the preparation of bacterial plasmids. Anal Biochem. 1981;114:193–197. doi: 10.1016/0003-2697(81)90473-5. [DOI] [PubMed] [Google Scholar]
- 12.Kamer G, Argos P. Primary structural comparison of RNA-dependent polymerases from plant, animal and bacterial viruses. Nucleic Acids Res. 1984;12:7269–7282. doi: 10.1093/nar/12.18.7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koonin E V. The phylogeny of RNA-dependent RNA polymerases of positive-strand RNA viruses. J Gen Virol. 1991;72:2197–2206. doi: 10.1099/0022-1317-72-9-2197. [DOI] [PubMed] [Google Scholar]
- 14.Lemm J A, Rice C M. Assembly of functional Sindbis virus RNA replication complexes: requirement for coexpression of P123 and P34. J Virol. 1993;67:1905–1915. doi: 10.1128/jvi.67.4.1905-1915.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lemm J A, Rice C M. Roles of nonstructural polyproteins and cleavage products in regulating Sindbis virus RNA replication and transcription. J Virol. 1993;67:1916–1926. doi: 10.1128/jvi.67.4.1916-1926.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lemm J A, Rümenapf T, Strauss E G, Strauss J H, Rice C M. Polypeptide requirements for assembly of functional Sindbis virus replication complexes: a model for the temporal regulation of minus-strand and plus-strand RNA-synthesis. EMBO J. 1994;13:2925–2934. doi: 10.1002/j.1460-2075.1994.tb06587.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li G, Rice C M. Mutagenesis of the in-frame opal termination codon preceding nsP4 of Sindbis virus: studies of translational readthrough and its effect on virus replication. J Virol. 1989;63:1326–1337. doi: 10.1128/jvi.63.3.1326-1337.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rice C M, Levis R, Strauss J H, Huang H V. Production of infectious RNA transcripts from Sindbis virus cDNA clones: mapping of lethal mutations, rescue of a temperature-sensitive marker, and in vitro mutagenesis to generate defined mutants. J Virol. 1987;61:3809–3819. doi: 10.1128/jvi.61.12.3809-3819.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shirako Y, Strauss J H. Cleavage between nsP1 and nsP2 initiates the processing pathway of Sindbis virus nonstructural polyprotein P123. Virology. 1990;177:54–64. doi: 10.1016/0042-6822(90)90459-5. [DOI] [PubMed] [Google Scholar]
- 20.Shirako Y, Strauss J H. Regulation of Sindbis virus RNA replication: uncleaved P123 and nsP4 function in minus-strand RNA synthesis whereas cleaved products from P123 are required for efficient plus-strand RNA synthesis. J Virol. 1994;68:1874–1885. doi: 10.1128/jvi.68.3.1874-1885.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Strauss E G, Rice C M, Strauss J H. Sequence coding for the alphavirus nonstructural proteins is interrupted by an opal termination codon. Proc Natl Acad Sci USA. 1983;80:5271–5275. doi: 10.1073/pnas.80.17.5271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Strauss J H, Strauss E G. The alphaviruses: gene expression, replication, and evolution. Microbiol Rev. 1994;58:491–562. doi: 10.1128/mr.58.3.491-562.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Varshavsky A. The N-end rule. Cell. 1992;69:725–735. doi: 10.1016/0092-8674(92)90285-k. [DOI] [PubMed] [Google Scholar]