Abstract
Mutants of feline immunodeficiency virus (FIV) resistant to (−)-β-2′,3′-dideoxy-3′-thiacytidine (3TC) were selected by culturing virus in the presence of increasing stepwise concentrations of 3TC. Two plaque-purified variants were isolated from the original mutant population, and both of these mutants were resistant to 3TC. Surprisingly, these mutants were also phenotypically resistant to 3′-azido-3′-deoxythymidine (AZT) and to the combination of 3TC and AZT. Purified reverse transcriptase (RT) from one of these plaque-purified mutants was resistant to the 5′-triphosphates of 3TC and AZT. DNA sequence analysis of the RT-encoding region of the pol gene amplified from the plaque-purified mutants revealed a Pro-to-Ser mutation at position 156 of RT. A site-directed mutant of FIV engineered to contain this Pro-156-Ser mutation was resistant to 3TC, AZT, and the combination of 3TC and AZT, confirming the role of the Pro-156-Ser mutation in the resistance of FIV to these two nucleoside analogs. This represents the first report of a lentiviral mutant resistant to the combination of AZT and 3TC due to a single, unique point mutation.
The emergence of drug-resistant variants of human immunodeficiency virus type 1 (HIV-1) is believed to be responsible for the failure of current antiviral chemotherapy to halt the clinical progression of AIDS (2, 10, 48). Drug-resistant mutants arise rapidly in HIV-1-infected individuals treated with most of the currently approved drugs, including nucleoside and nonnucleoside inhibitors of reverse transcriptase (RT) (12, 26, 27, 49, 50, 52, 53, 55) and the protease inhibitors (8, 21, 50). In addition, numerous mutants which are resistant to RT or protease inhibitors have been selected in vitro (13–15, 17, 22, 25, 39). In both laboratory and clinical isolates, resistance usually correlates with mutations in the RT- or protease-encoding regions, respectively, of the pol gene (50).
Therapeutic strategies using combinations of inhibitors have provided the greatest success in slowing the clinical decline of HIV-1-infected individuals. The most successful treatment protocol employs a triple combination approach of simultaneous treatment with (−)-β-2′,3′-dideoxy-3′-thiacytidine (3TC), 3′-azido-3′-deoxythymidine (AZT), and a protease inhibitor such as indinavir (32). A central feature of this combination is the unique interaction between 3TC and AZT. Mutants resistant to 3TC arise during therapy through the acquisition of a Met-to-Val or Met-to-Ile mutation at position 184 of RT, as first reported by Schinazi et al. (51). However, these mutations at position 184 have been demonstrated to phenotypically suppress AZT resistance mutations, thereby providing a basis for sustained drug efficacy (28, 30, 58). The combination of AZT and 3TC represents a significant improvement over conventional AZT monotherapy in suppressing virus load and in delaying the resurgence of virus titers associated with the emergence of drug resistance (28).
We have developed systems using the feline immunodeficiency virus (FIV) as a model for examining the mechanisms of viral resistance to AIDS therapy (36). The immune deficiency and neuropathogenesis resulting from infection of domestic cats with FIV are remarkably similar to AIDS in humans (1, 4, 11, 42–45, 61). FIV represents a particularly attractive model for studies of resistance to inhibitors of HIV-1 due to the similarities between HIV-1 RT and FIV RT with respect to physical properties, catalytic activities, and sensitivity to the triphosphate forms of AZT, 2′,3′-dideoxycytidine (ddC), 2′,3′-dideoxyinosine (ddI), 2′,3′-didehydro-3′-deoxythymidine (d4T), and 3TC (9, 33–35, 54). In addition, the first drug-resistant lentiviral variants selected in vitro were AZT-resistant mutants of FIV (46). We have subsequently reported FIV mutants resistant to ddI (16), ddC (31), d4T (62), (−)-β-2′,3′-dideoxy-5-fluoro-3′-thiacytidine [(−)-FTC] (54), and the combination of AZT and ddI (16).
We have recently reported mutants of FIV selected with (−)-FTC which are cross-resistant to 3TC due to a Met-to-Thr mutation at position 183 of RT (which corresponds to position 184 of HIV-1 RT) (54). These FIV mutants are similar in phenotype to the Met-184-Val and Met-184-Ile mutants of HIV-1, as all of these variants retain wild-type sensitivity to AZT. Here we report that selection with 3TC results in lentiviral variants resistant to 3TC and to the combination of 3TC and AZT. These mutants may indicate a possible mechanism by which HIV-1 can evade 3TC-AZT combination chemotherapy.
MATERIALS AND METHODS
Chemicals.
Phosphonoformic acid (PFA), dCTP, dTTP, and ddC were purchased from Sigma Chemical Co., St. Louis, Mo. AZT and the 5′-triphosphate of AZT (AZTTP) were provided by Glaxo Wellcome Co., Research Triangle Park, N.C.; d4T was provided by Bristol-Myers Squibb Co., Wallingford, Conn.; 9-(2-phosphonylmethoxyethyl)adenine (PMEA) was provided by Gilead Sciences, Inc., Foster City, Calif.; ddI was provided by the Developmental Therapeutics Branch, Division of AIDS, National Institute of Allergy and Infectious Diseases. 3TC, (−)-FTC, and the 5′-triphosphate of 3TC (3TCTP) were synthesized and fully characterized by mass spectroscopy, nuclear magnetic resonance, and high-pressure liquid chromatography as described previously (5, 7, 18). [5-3H]dCTP and [methyl-3H]dTTP were obtained from Dupont-New England Nuclear, Boston, Mass. GeneAmp PCR Core reagents were purchased from Perkin-Elmer Cetus, Norwalk, Conn. The Taq DyeDeoxy Terminator Cycle sequencing kit was purchased from Applied Biosystems, Foster City, Calif. Restriction enzymes PstI, HindIII, NsiI, and PacI were obtained from Boehringer Mannheim, Indianapolis, Ind., and T4 DNA ligase was obtained from Gibco BRL, Grand Island, N.Y. All other chemicals were reagent grade or better.
Cells and virus.
Virus produced from a molecular clone of the Petaluma strain of FIV, 34TF10 (56), was used as wild-type FIV for these studies. Wild-type and mutant strains of FIV were grown and maintained in Crandell feline kidney (CrFK) cells with L&M medium supplemented with 10% fetal bovine serum as previously described (37, 54). Following selection, FIV mutants were maintained in medium containing 3 μM 3TC, and all cultures were replenished with fresh medium and drug every 2 days.
Focal infectivity assay.
Inhibition of FIV infection by antiviral drugs was quantified by a focal infectivity assay as described previously (46). Resulting data were plotted as the percentage of control foci (no drug) versus inhibitor concentration. Concentrations of drug required to inhibit focus formation by 50% (50% effective concentrations [EC50s]) were obtained directly from the linear portion of these plots by using a computer-generated regression line (46). Within an experiment, each value represents the mean of four determinations. Results from three or more independent experiments were used to derive the EC50 ± standard error.
Selection and plaque purification of 3TC-resistant mutants.
FIV mutants reported here were obtained by selection with 3TC alone with a stepwise selection protocol as described previously (62). Briefly, virus generated from 34TF10 was initially cultured in the presence of 1 μM 3TC and then subjected to five additional rounds of infection in which the concentration of 3TC was doubled with each subsequent round, ending in a sixth round of infection at 32 μM 3TC. Each round of infection was initiated with cell-free virus from the previous round of infection. The resulting population, designated 3TR-1c, was resistant to 3TC and AZT (data not shown). This stock was then plaque purified in 3 μM 3TC as previously described (47) to obtain 3TR-3c and 3TR-7c, which were then used for further analysis.
Enzymes and enzyme assays.
RT was purified from virions of mutant FIV as previously described (34). Assays for RT activity with poly(rA)-oligo(dT) or poly(rI)-oligo(dC) as template-primer were also as reported elsewhere (33, 34). Kinetic parameters were determined by using intercept values calculated from double-reciprocal plots (9, 33). RT purified from 34TF10 virions (34) was used as the wild-type control.
Nucleic acid preparation and sequence analysis.
Total cellular DNA containing proviral DNA was extracted from infected CrFK cells and used for amplification by PCR as previously described (31, 47, 54). PCR product was directly sequenced at the Murdock Molecular Biology Facility with a Taq DyeDeoxy Terminator sequencing kit and analyzed on a model 373A automated DNA sequencer (Applied Biosystems). Sequencing was performed in the forward and reverse directions with primers at 250-bp intervals of the RT-encoding region of the pol gene.
Site-directed mutagenesis.
In order to construct a molecular clone of FIV containing the Pro-to-Ser mutation at codon 156 of RT, a 2,109-bp EcoRI-HindIII fragment, corresponding to nucleotides 1871 to 3980 of the proviral portion of pFIV-34TF10, was cloned into the pTZ18u phagemid vector (Bio-Rad Laboratories) and mutagenized with the Muta-Gene in vitro mutagenesis kit (Bio-Rad). The mutagenesis primer 5′-GATATATCAATGAACTTAA-3′ was used to introduce the Pro-156-Ser mutation (mutation underlined). Following sequence analysis to confirm the presence of the desired mutation, an 869-bp NsiI-PacI fragment corresponding to nucleotides 2674 to 3543 of pFIV-34TF10 was ligated into NsiI/PacI-digested pFIV-34TF10 and transformed into SURE-2 Ultracompetent Escherichia coli (Stratagene). Clones were sequenced in order to verify the presence of the mutation and the integrity of the pol gene, and 1 μg of the resulting plasmid DNA was purified and used to transfect CrFK cells as previously described (47) for the production of virus. All constructs were also introduced into the J5 strain of E. coli JM109 for long-term storage and plasmid propagation.
RESULTS
Selection and plaque purification of 3TC-resistant FIV mutants.
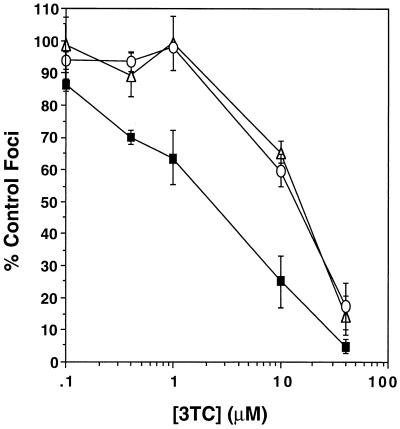
Virus produced from the 34TF10 molecular clone of FIV was passaged in the presence of increasing stepwise concentrations of 3TC, as described previously (62) and in Materials and Methods. This population, designated 3TR-1c, was plaque purified in order to minimize heterogeneity within the mutant population. Two plaque-purified mutants, designated 3TR-3c and 3TR-7c, were seven- to eightfold resistant to 3TC (Fig. 1) and were chosen for further phenotypic characterization.
FIG. 1.
Susceptibility to inhibition by 3TC of FIV 34TF10 (▪) and the plaque-purified 3TC-resistant mutants 3TR-3c (▵) and 3TR-7c (○). Results are from three or more experiments, with four determinations per experiment. Bars represent standard errors of the means.
Mutants 3TR-3c and 3TR-7c were cross-resistant to the cytidine analogs (−)-FTC and ddC, as predicted from studies of mutants of FIV selected with (−)-FTC. Surprisingly, both 3TR-3c and 3TR-7c were also cross-resistant to AZT (Table 1) and were therefore phenotypically different from all previous mutants of HIV-1 or FIV selected with 3′-thiacytidine nucleosides. Additionally, both mutants showed wild-type susceptibility to ddI, PMEA, d4T, and PFA (Table 1).
TABLE 1.
Susceptibilities of FIV 34TF10 and plaque-purified 3TC-resistant mutants of FIV to antiviral compounds as determined by focal infectivity assay
| Compound | Mean EC50 (μM) ± SEMa for:
|
Fold increaseb for:
|
|||
|---|---|---|---|---|---|
| 34TF10 | 3TR-3c | 3TR-7c | 3TR-3c | 3TR-7c | |
| 3TC | 1.5 ± 0.3 | 12 ± 0.8 | 11 ± 0.6 | 8.0 | 7.3 |
| (−)-FTC | 1.0 ± 0.1 | 4.4 ± 0.3 | 6.7 ± 0.2 | 4.4 | 6.7 |
| AZT | 1.3 ± 0.2 | 6.2 ± 1.4 | 5.8 ± 0.6 | 4.8 | 4.5 |
| ddC | 2.0 ± 0.1 | 7.0 ± 0.8 | 7.5 ± 1.9 | 3.5 | 3.8 |
| ddI | 3.2 ± 0.5 | 2.4 ± 0.7 | 3.0 ± 0.3 | 0.8 | 0.9 |
| d4T | 13 ± 1.8 | 16 ± 2.3 | 15 ± 0.6 | 1.2 | 1.2 |
| PMEA | 1.1 ± 0.1 | 1.0 ± 0.0 | 0.8 ± 0.0 | 0.9 | 0.7 |
| PFA | 140 ± 4.9 | 120 ± 9.5 | 100 ± 1.1 | 0.9 | 0.7 |
Values are from three or more experiments, with four determinations per experiment.
Increase over value for 34TF10.
RT.
Purified RT from 3TR-3c was compared to wild-type FIV RT for susceptibility to inhibition by the 5′-triphosphates of 3TC (3TCTP) and AZT (AZTTP). Inhibition by both 3TCTP and AZTTP was competitive with respect to dCTP and dTTP, respectively. Km and Ki values are summarized in Table 2. 3TR-3c RT was 8.7-fold resistant to inhibition by 3TCTP, based on comparison of Ki/Km ratios for the mutant and wild-type RTs. 3TR-3c RT was also resistant to AZTTP, with a twofold increase in Ki/Km ratio over the wild-type value.
TABLE 2.
Kinetic constants for RTs from wild-type FIV and 3TR-3c
| Inhibitor |
Ki (nM)a for RT of:
|
Ki/Km for RT of:
|
Fold increase (Ki/Km)b | ||
|---|---|---|---|---|---|
| 34TF10 | 3TR-3c | 34TF10 | 3TR-3c | ||
| 3TCTP | 154 ± 21 | 2,660 ± 380 | 0.013 | 0.113 | 8.7 |
| AZTTP | 11.8 ± 0.3 | 31.0 ± 0.6 | 0.0010 | 0.0024 | 2.4 |
Values are reported as the means ± standard errors of the means of at least two experiments with three determinations per experiment. The mode of inhibition by 3TCTP or AZTTP was competitive with respect to substrate. Template primers used were poly(rA)-oligo(dT) for AZTTP and poly(rI)-oligo(dC) for 3TCTP. The Km for dCTP was 12.1 ± 0.4 μM for 34TF10 RT and 23.5 ± 1.5 μM for 3TR-3c RT. The Km for dTTP was 11.4 ± 1.8 μM for 34TF10 RT and 13.2 ± 2.0 for 3TR-3c RT.
Increase of Ki/Km for 3TR-3c over Ki/Km for 34TF10 RT.
Nucleotide sequence analysis.
DNA sequence analyses of the RT-encoding regions of the pol genes from 3TR-3c and 3TR-7c were performed in both the forward and the reverse directions. The resulting sequences were compared to that of the 34TF10 molecular clone of FIV. Both plaque-purified mutants contained a C-to-T transition at position 2801 which results in a Pro-to-Ser mutation at codon 156 of FIV RT (Fig. 2). Both isolates shared additional mutations at positions 348 (Ile to Thr, T to C at position 3378) and 469 (Asp to Glu, T to A at position 3742) of RT. 3TR-7c was also shown to contain a unique mutation at amino acid 227 (Thr to Ala, A to G at position 3014) which was not present in 3TR-3c.
FIG. 2.
Nucleotide and deduced amino acid sequences of the region of the FIV pol gene surrounding position 2801. The corresponding amino acid sequence from HIV-1 is also shown for comparison. Note that HIV-1 and FIV RTs exhibit extensive homology in this region and that the FIV sequence is displaced by one residue relative to the HIV-1 sequence. Nucleotide sequence data for 3TR-3c and 3TR-7c were compared to the sequence data for FIV 34TF10. Mutations are shown in boldface.
In order to determine the phenotypic stability of 3TR-3c and 3TR-7c, both mutants were passaged for three rounds of infection in the absence of 3TC by protocols described previously (47). Both plaque-purified mutants remained significantly resistant to 3TC even in the absence of drug. However, resistance to 3TC decreased approximately twofold by the third round of infection, with 3TR-3c and 3TR-7c displaying EC50s of 4.5 ± 0.7 and 5.5 ± 1.2 μM, respectively.
Site-directed mutagenesis.
Based on the three-dimensional crystal structure of HIV-1 RT and the high degree of homology between HIV-1 and FIV RTs in the areas surrounding positions 156 and 183 (corresponding to amino acids 157 and 184 of HIV-1 RT), we would predict that amino acid 156 lies near amino acid 183 in FIV RT. This prediction places Pro-156 in close physical proximity to a position in the YMDD motif which is known to confer resistance to 3TC when mutated in HIV-1 (Met-184) (6, 13, 14, 51, 52, 58), FIV (Met-183) (54), and hepatitis B virus (Met-550) (29). Therefore, we chose to substitute the Pro-156-Ser change into the FIV 34TF10 molecular clone by site-directed mutagenesis to determine the role of this mutation in resistance to AZT and 3TC. The resulting mutant, designated FIVPro156Ser, was eight- to ninefold resistant to 3TC, with an EC50 of 10.8 ± 1.1 μM. The site-directed mutant was also four- to fivefold resistant to AZT, with an EC50 of 7.5 ± 0.4 μM.
Resistance to the combination of 3TC and AZT.
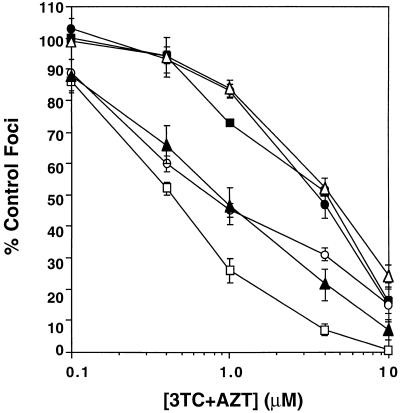
We have also determined the susceptibility of FIVPro156Ser to the combination of 3TC and AZT when present simultaneously and in equimolar concentrations in the focal infectivity assay. Inhibition data for the site-directed mutant were determined in parallel with the plaque-purified mutants (3TR-3c and 3TR-7c), wild-type 34TF10, and the site-directed 3TC-resistant mutants, FIVMet183Val and FIVMet183Thr. The results of these assays are shown in Fig. 3 and summarized in Table 3. FIVMet183Val and FIVMet183Thr displayed a twofold decrease in susceptibility to the combination of 3TC and AZT. These data are consistent with previous results which illustrate that these mutants are resistant to 3TC but remain sensitive to AZT. In contrast, FIVPro156Ser and the plaque-purified mutants 3TR-3c and 3TR-7c were seven- to eightfold resistant to the combination of 3TC and AZT. These results illustrate that the Pro-156-Ser mutation in FIV confers resistance to the combination of AZT and 3TC.
FIG. 3.
Susceptibility to inhibition by the combination of 3TC and AZT of 34TF10 (□), 3TR-3c (▪), 3TR-7c (•), FIVMet183Val (○), FIVMet183Thr (▴), and FIVPro156Ser (▵). Values in the plot represent equimolar concentrations of 3TC plus AZT. For example, a 1 μM value for the drug combination represents 1 μM 3TC plus 1 μM AZT. Results are from three experiments with four determinations per experiment. Bars represent standard errors of the means and are omitted when the error is too small to be shown.
TABLE 3.
Susceptibilities of FIV 34TF10, plaque-purified 3TC-resistant mutants, and site-directed mutants to the combination of 3TC and AZTa
| Virus | Mean EC50 (μM) ± SEMb | Fold increasec |
|---|---|---|
| 34TF10 | 0.48 ± 0.04 | |
| 3TR-3c | 3.21 ± 0.39 | 6.7 |
| 3TR-7c | 3.20 ± 0.26 | 6.6 |
| FIVMet183Val | 1.10 ± 0.29 | 2.3 |
| FIVMet183Thr | 0.92 ± 0.23 | 1.9 |
| FIVPro156Ser | 3.87 ± 0.85 | 8.0 |
3TC and AZT present in equimolar concentrations as described in the legend to Fig. 3.
Values are from three or more experiments, with four determinations per experiment.
Increase over value for 34TF10.
DISCUSSION
The FIV mutants that we have selected with 3TC are unique in that they are not only resistant to 3TC but also resistant to AZT and to the combination of 3TC and AZT. These mutants carry a unique Pro-to-Ser mutation at position 156 of RT which is responsible for resistance to this drug combination. Previously reported 3TC-resistant mutants of HIV-1 and FIV contain mutations in the methionine codon of the YMDD motif of RT (Met to Val/Ile/Thr in HIV-1 and Met to Val/Thr in FIV) (6, 13, 14, 23, 51, 54, 58). These mutations confer resistance to 3TC but not to AZT. Similarly, mutations in HIV-1 previously reported to confer resistance to AZT do not confer resistance to 3TC. The results presented here represent the first report of a lentiviral mutant containing a novel point mutation in RT which confers resistance to 3TC and AZT individually and in combination.
We have recently reported mutants of FIV which were selected with (−)-FTC and contained a Met-to-Thr mutation at position 183 of RT (54). These mutants were resistant to (−)-FTC and 3TC but remained susceptible to AZT. It is intriguing that two closely related nucleoside analogs would yield different patterns of drug resistance. These genotypic and phenotypic differences may result from a single chemical change from a 5-proton to a 5-fluoro on the pyrimidine base of the oxathiolane nucleoside used for selection. Alternatively, these differing outcomes may be the result of the differing selection protocols used to obtain these mutants [high concentration of (−)-FTC versus stepwise selection with 3TC]. Further work with 3TC with a high drug concentration for the selection is planned.
Previous studies of HIV-1 mutants resistant to the combination of AZT and ddI have illustrated that mutants resistant to combinations of antiviral drugs can arise as the result of point mutations which are not observed for mutants selected with either drug alone (19). The Pro-156-Ser mutation, shown here to confer resistance to the combination of 3TC and AZT in FIV, was also not predicted from the common mutations in HIV-1 or FIV yielding resistance to AZT or 3TC alone. Thus, point mutations which result in multidrug resistance can be quite different from those which confer resistance to monotherapy.
It has been noted elsewhere that the levels of viral resistance to 3TC observed for the FIV mutants resistant to 3TC alone (54), or the combination of 3TC and AZT, differ by an order of magnitude from the levels of resistance seen for HIV-1 mutants obtained both from clinical isolates and in vitro selections. These differences probably result from differences in the phenotypic assays used to determine the susceptibility profiles of FIV and HIV-1 mutants. The focal infectivity assay used to determine the drug susceptibilities of FIV isolates generates data resulting from the inhibition of a single round of viral replication and is based upon the direct quantitation of infectious virions. In contrast, the p24-based assays commonly used to determine drug susceptibilities of HIV-1 isolates are performed over multiple rounds of replication, and the relative level of resistance (fold resistance) is magnified over several cycles of replication.
We have previously described AZT-resistant mutants of FIV which revert very rapidly (within a single round of infection) in the absence of AZT (47). For both the Pro-156-Ser mutants described here, and for Met-183-Thr variants of FIV resistant to 3TC alone (54), the viral isolates remained significantly resistant to 3TC following three rounds of infection in the absence of drug. However, with each of these 3TC-resistant mutants of FIV, a decrease in EC50 was observed by the third round of infection in the absence of drug, suggesting that wild-type virus may have begun to emerge in the populations. This is most likely due to a selective disadvantage of the variants when replicating in the absence of 3TC. Studies of HIV-1 isolates resistant to 3TC have shown that Met-184-Val, Met-184-Thr, and Met-184-Ile mutants may have impaired replication rates relative to that of wild-type HIV-1 in primary peripheral blood mononuclear cell cultures (3, 23). We have noted that 3TC-resistant mutants of FIV also replicate slower and yield consistently lower titers than wild-type FIV when cultured on CrFK cells (data not shown). Detailed comparisons of the replication kinetics of these viruses in CrFK cells and in primary human lymphocyte cultures are in progress.
The Pro-156 position of FIV RT corresponds to the Pro-157 of HIV-1 RT (Fig. 2). Structural models of HIV-1 RT based on crystallographic data have shown that Pro-157 is located within the N-terminal portion of the αE helix (positions 155 to 174 of the p66 subunit) and is proximal to Met-184 (20, 24, 41). Based on the 57% identity and 87% homology of amino acid sequence within this region (residues 148 to 162 of HIV-1 RT [Fig. 2]) and the overall high degree of homology between HIV-1 and FIV RTs (38, 56), we predict that the Pro-156 residue of FIV RT is proximal to Met-183 in the active site of the FIV enzyme. Therefore, we speculate that the resistance to AZT and 3TC conferred by the Pro-156-Ser mutation may result from an alteration in deoxynucleoside triphosphate binding and/or template-primer positioning during reverse transcription (41, 57, 60).
Studies of RTs from 3TC-resistant HIV-1 variants may provide clues to the biochemical consequences resulting from mutations near the active site of the enzyme. The Met-184-Val HIV-1 mutant displays an increased fidelity of nucleotide insertion in gel-based, kinetic assays (40, 59). Additionally, RTs from both the Met-184-Val and Met-184-Ile mutants of HIV-1 produce shorter cDNA products in vitro than does the wild-type enzyme, suggesting that these mutant RTs may be less processive enzymes (3). Due to its location, the Pro-156-Ser mutation in FIV reported here may also have effects on the fidelity and/or processivity of the mutant enzyme.
Combination chemotherapy involving the use of AZT and 3TC, often in conjunction with a protease inhibitor, has provided a substantial therapeutic advantage over conventional monotherapy. The data presented here illustrate that lentiviral mutants resistant to the combination of 3TC and AZT can be selected in vitro and may represent a potential mechanism for the development of multiple drug resistance during combination therapy for HIV-1. In addition, these data suggest that it may not be necessary for HIV-1 to accumulate a large number of point mutations in order to become resistant to the triple combination of 3TC, AZT, and a protease inhibitor. However, the mutants reported here appear to be replication impaired in cell culture and therefore might be expected to result in lower viral loads in vivo. The role of such mutants in pathogenesis will be addressed experimentally with the FIV cat model.
ACKNOWLEDGMENTS
This work was supported by Public Health Service grants AI28189 (T.W.N.) and AI38755 (B.D.P. and T.W.N.) from the National Institute of Allergy and Infectious Diseases and by the Department of Veterans Affairs and the Georgia Research Center on AIDS and HIV Infection (R.F.S.).
We thank Robert M. Lloyd, Jr., for providing technical information and Joan Strange and the Murdock Molecular Biology Facility for DNA sequence analysis and oligonucleotide synthesis.
REFERENCES
- 1.Ackley C D, Yamamoto J K, Levy N, Pedersen N C, Cooper M D. Immunologic abnormalities in pathogen-free cats experimentally infected with feline immunodeficiency virus. J Virol. 1990;64:5652–5655. doi: 10.1128/jvi.64.11.5652-5655.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arts E J, Wainberg M A. Mechanisms of nucleoside analog antiviral activity and resistance during human immunodeficiency virus reverse transcription. Antimicrob Agents Chemother. 1996;40:527–540. doi: 10.1128/aac.40.3.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Back N K T, Nijhuis M, Keulen W, Boucher C A B, Essink B B O, van Kuilenburg A B P, van Gennip A H, Berkhout B. Reduced replication of 3TC-resistant HIV-1 variants in primary cells due to a processivity defect of the reverse transcriptase enzyme. EMBO J. 1996;15:4040–4049. [PMC free article] [PubMed] [Google Scholar]
- 4.Barlough J E, Ackley C D, George J W, Levy N, Acevedo R, Moore P F, Rideout B A, Cooper M D, Pedersen N C. Acquired immune dysfunction in cats with experimentally induced feline immunodeficiency virus infection: comparison of short-term and long-term infections. J Acquired Immune Defic Syndr. 1991;4:219–227. [PubMed] [Google Scholar]
- 5.Beach J W, Jeong L S, Alves A J, Pohl D, Kim H O, Chang C-N, Doong S-L, Schinazi R F, Cheng Y-C, Chu C K. Synthesis of enantiomerically pure (2′R,5′S)-(−)-1-[2-(hydroxymethyl)-oxathiolan-5-yl]cytosine as a potent antiviral agent against hepatitis B virus (HBV) and human immunodeficiency virus (HIV) J Org Chem. 1992;57:2217–2219. [Google Scholar]
- 6.Boucher C A B, Cammack N, Schipper P, Schuurman R, Rouse P, Wainberg M A, Cameron J M. High-level resistance to (−) enantiomeric 2′-deoxy-3′-thiacytidine in vitro is due to one amino acid substitution in the catalytic site of human immunodeficiency virus type 1 reverse transcriptase. Antimicrob Agents Chemother. 1993;37:2231–2234. doi: 10.1128/aac.37.10.2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Choi C K, Beach J W, Wilson L J, Yeola S, Liotta D C, Schinazi R F. In situ complexation directs the stereochemistry of N-glycosylation in the synthesis of oxathiolanyl and dioxythiolanyl nucleoside analogs. J Am Chem Soc. 1991;113:9337–9379. [Google Scholar]
- 8.Condra J H, Schieif W A, Blahy O M, Gabryelski L J, Graham D J, Quintero J C, Rhodes A, Robbins H L, Roth E, Shivaprakash M, Titus D, Yang T, Teppler H, Squires K E, Deutsch P J, Emini E A. In vivo emergence of HIV-1 variants resistant to multiple protease inhibitors. Nature (London) 1995;374:569–571. doi: 10.1038/374569a0. [DOI] [PubMed] [Google Scholar]
- 9.Cronn R C, Remington K M, Preston B D, North T W. Inhibition of reverse transcriptase from feline immunodeficiency virus by analogs of 2′-deoxyadenosine-5′-triphosphate. Biochem Pharmacol. 1992;44:1375–1381. doi: 10.1016/0006-2952(92)90539-u. [DOI] [PubMed] [Google Scholar]
- 10.D’Aquila R T, Johnson V A, Welles S L, Japour A J, Kuritzkes D R, DeGruttola V, Reichelderfer P S, Coombs R W, Crumpacker C S, Kahn J O, Richman D D. Zidovudine resistance and HIV-1 disease progression during antiretroviral therapy. Ann Intern Med. 1995;122:401–408. doi: 10.7326/0003-4819-122-6-199503150-00001. [DOI] [PubMed] [Google Scholar]
- 11.English R V, Nelson P, Johnson C M, Nasisse M, Tompkins W A, Tompkins M B. Development of clinical disease in cats experimentally infected with feline immunodeficiency virus. J Infect Dis. 1994;170:543–552. doi: 10.1093/infdis/170.3.543. [DOI] [PubMed] [Google Scholar]
- 12.Fitzgibbon J E, Howell R M, Haberzettl C A, Sperber S J, Gocke D J, Dubin D T. Human immunodeficiency virus type 1 pol gene mutations which cause decreased susceptibility to 2′,3′-dideoxycytidine. Antimicrob Agents Chemother. 1992;36:153–157. doi: 10.1128/aac.36.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gao Q, Gu Z, Hiscott J, Dionne G, Wainberg M A. Generation of drug-resistant variants of human immunodeficiency virus type 1 by in vitro passage in increasing concentrations of 2′,3′-dideoxycytidine and 2′,3′-dideoxy-3′-thiacytidine. Antimicrob Agents Chemother. 1993;37:130–133. doi: 10.1128/aac.37.1.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gao Q, Gu Z, Parniak M A, Cameron I, Cammack N, Boucher C, Wainberg M A. The same mutation that encodes low-level human immunodeficiency virus type 1 resistance to 2′,3′-dideoxyinosine and 2′,3′-dideoxycytidine confers high-level resistance to the (−) enantiomer of 2′,3′-dideoxy-3′-thiacytidine. Antimicrob Agents Chemother. 1993;37:1390–1392. doi: 10.1128/aac.37.6.1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gao Q, Gu Z, Parniak M A, Li X, Wainberg M A. In vitro selection of variants of human immunodeficiency virus type 1 resistant to 3′-azido-3′-deoxythymidine and 2′,3′-dideoxyinosine. J Virol. 1992;66:12–19. doi: 10.1128/jvi.66.1.12-19.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gobert J M, Remington K M, Zhu Y-Q, North T W. Multiple-drug-resistant mutants of feline immunodeficiency virus selected with 2′,3′-dideoxyinosine alone and in combination with 3′-azido-3′-deoxythymidine. Antimicrob Agents Chemother. 1994;38:861–864. doi: 10.1128/aac.38.4.861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gu Z, Gao Q, Fang H, Salomon H, Parniak M A, Goldberg E, Cameron J, Wainberg M A. Identification of a mutation at codon 65 in the IKKK motif of reverse transcriptase that encodes human immunodeficiency virus resistance to 2′,3′-dideoxycytidine and 2′,3′-dideoxy-3′-thiacytidine. Antimicrob Agents Chemother. 1994;38:275–281. doi: 10.1128/aac.38.2.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoong L K, Strange L E, Liotta D C, Koszalka G W, Burns C L, Schinazi R F. Enzyme-mediated enantioselective preparation of pure enantiomers of the antiviral agent 2′,3′-dideoxy-5-fluoro-3′-thiacytidine [(−)-FTC] and related compounds. J Org Chem. 1992;57:5563–5585. [Google Scholar]
- 19.Iversen A K N, Shafer R W, Wehrly K, Winters M A, Mullins J I, Chesebro B, Merigan T C. Multidrug-resistant human immunodeficiency virus type 1 strains resulting from combination antiretroviral chemotherapy. J Virol. 1996;70:1086–1090. doi: 10.1128/jvi.70.2.1086-1090.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jacobo-Molina A, Ding J, Nanni R G, Clark A D, Jr, Lu X, Tantillo C, Williams R L, Kamer G, Ferris A L, Clark P, Hizi A, Hughes S H, Arnold E. Crystal structure of human immunodeficiency virus reverse transcriptase complexed with double-stranded DNA at 3.0 Å resolution shows bent DNA. Proc Natl Acad Sci USA. 1993;90:6320–6324. doi: 10.1073/pnas.90.13.6320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jacobsen H, Yasargil K, Winslow D L, Craig J C, Krohn A, Duncan I B, Mous J. Characterization of human immunodeficiency virus type 1 mutants with decreased sensitivity to proteinase inhibitor Ro 31-8959. Virology. 1995;206:527–534. doi: 10.1016/s0042-6822(95)80069-7. [DOI] [PubMed] [Google Scholar]
- 22.Kaplan A H, Michael S F, Wehbie R S, Knigge M F, Paul D A, Everit L, Kempf D J, Norbeck D W, Erickson J W, Swanstrom R. Selection of multiple human immunodeficiency virus type 1 variants that encode viral proteases with decreased sensitivity to an inhibitor of the viral protease. Proc Natl Acad Sci USA. 1994;91:5597–5601. doi: 10.1073/pnas.91.12.5597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Keulen W, Back N K T, van Wijk A, Boucher C, Berkhout B. Initial appearance of 184Ile variant in lamivudine-treated patients can be explained by the mutational bias of human immunodeficiency virus reverse transcriptase. J Virol. 1997;71:3346–3350. doi: 10.1128/jvi.71.4.3346-3350.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kohlstaedt L A, Wang J, Friedman J M, Rice P A, Steitz T A. Crystal structure at 3.5 Å of HIV-1 reverse transcriptase complexed with an inhibitor. Science. 1992;256:1783–1790. doi: 10.1126/science.1377403. [DOI] [PubMed] [Google Scholar]
- 25.Larder B A, Coates K E, Kemp S D. Zidovudine-resistant human immunodeficiency virus selected by passage in cell culture. J Virol. 1991;65:5232–5236. doi: 10.1128/jvi.65.10.5232-5236.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Larder B A, Darby G, Richman D D. HIV with reduced sensitivity to zidovudine (AZT) isolated during prolonged therapy. Science. 1989;243:1731–1734. doi: 10.1126/science.2467383. [DOI] [PubMed] [Google Scholar]
- 27.Larder B A, Kemp S D. Multiple mutations in HIV-1 reverse transcriptase confer high-level resistance to zidovudine (AZT) Science. 1989;246:1155–1158. doi: 10.1126/science.2479983. [DOI] [PubMed] [Google Scholar]
- 28.Larder B A, Kemp S D, Harington P R. Potential mechanism for sustained antiretroviral efficacy of AZT-3TC combination therapy. Science. 1995;269:696–699. doi: 10.1126/science.7542804. [DOI] [PubMed] [Google Scholar]
- 29.Ling R, Mortimer D, Ahmed M, Boxall E H, Elias E, Dusheiko G M, Harrison T J. Selection of mutations in the hepatitis B virus polymerase during therapy of transplant recipients with lamivudine. Hepatology. 1996;24:711–713. doi: 10.1002/hep.510240339. [DOI] [PubMed] [Google Scholar]
- 30.Mathez D, Schinazi R F, Liotta D, Leibowitch J. Infectious amplification of wild-type human immunodeficiency virus from patients’ lymphocytes and modulation by reverse transcriptase inhibitors in vitro. Antimicrob Agents Chemother. 1993;37:875–881. doi: 10.1128/aac.37.10.2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Medlin H K, Zhu Y-Q, Remington K M, Phillips T R, North T W. Selection and characterization of a mutant of feline immunodeficiency virus resistant to 2′,3′-dideoxycytidine. Antimicrob Agents Chemother. 1996;40:953–957. doi: 10.1128/aac.40.4.953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mellors J W. Closing in on human immunodeficiency virus-1. Nat Med. 1996;2:274–275. doi: 10.1038/nm0396-274. [DOI] [PubMed] [Google Scholar]
- 33.North T W, Cronn R C, Remington K M, Tandberg R T. Direct comparisons of inhibitor sensitivities of reverse transcriptases from feline and human immunodeficiency viruses. Antimicrob Agents Chemother. 1990;34:1505–1507. doi: 10.1128/aac.34.8.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.North T W, Cronn R C, Remington K M, Tandberg R T, Judd R C. Characterization of reverse transcriptase from feline immunodeficiency virus. J Biol Chem. 1990;265:5121–5128. [PubMed] [Google Scholar]
- 35.North T W, Hansen G L, Zhu Y-Q, Griffin J A, Shih C-K. Expression of reverse transcriptase from feline immunodeficiency virus in Escherichia coli. Antimicrob Agents Chemother. 1994;38:388–391. doi: 10.1128/aac.38.2.388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.North T W, LaCasse R A. Testing HIV-1 drugs in the FIV model. Nat Med. 1995;1:410–411. doi: 10.1038/nm0595-410. [DOI] [PubMed] [Google Scholar]
- 37.North T W, North G L T, Pedersen N C. Feline immunodeficiency virus: a model for reverse transcriptase-targeted chemotherapy for acquired immune deficiency syndrome. Antimicrob Agents Chemother. 1989;33:915–919. doi: 10.1128/aac.33.6.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Olmsted R A, Hirsch V M, Purcell R H, Johnson P R. Nucleotide sequence analysis of feline immunodeficiency virus: genome organization and relationship to other lentiviruses. Proc Natl Acad Sci USA. 1989;86:8088–8092. doi: 10.1073/pnas.86.20.8088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Otto M J, Garber S, Winslow D L, Reid C D, Aldrich P, Jadhav P K, Pattersen C E, Hodge C N, Cheng Y-S E. In vitro isolation and identification of human immunodeficiency virus (HIV) variants with reduced sensitivity to C-2 symmetrical inhibitors of HIV type 1 protease. Proc Natl Acad Sci USA. 1993;90:7543–7547. doi: 10.1073/pnas.90.16.7543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pandey V N, Kaushik N, Rege N, Sarafianos S G, Yadav P N S, Modak M J. Role of methionine 184 of human immunodeficiency virus type-1 reverse transcriptase in the polymerase function and fidelity of DNA synthesis. Biochemistry. 1996;35:2168–2179. doi: 10.1021/bi9516642. [DOI] [PubMed] [Google Scholar]
- 41.Patel P H, Jacobo-Molina A, Ding J, Tantillo C, Clark A D, Jr, Raag R, Nanni R G, Hughes S H, Arnold E. Insights into DNA polymerization mechanisms from structure and function analysis of HIV-1 reverse transcriptase. Biochemistry. 1995;34:5351–5363. doi: 10.1021/bi00016a006. [DOI] [PubMed] [Google Scholar]
- 42.Pedersen N C. The feline immunodeficiency virus. In: Levy J A, editor. The Retroviridae. Vol. 2. New York, N.Y: Plenum Press; 1993. pp. 181–219. [Google Scholar]
- 43.Pedersen N C, Ho E W, Brown M L, Yamamoto J K. Isolation of a T-lymphotrophic virus from domestic cats with an immunodeficiency-like syndrome. Science. 1987;235:790–793. doi: 10.1126/science.3643650. [DOI] [PubMed] [Google Scholar]
- 44.Phillips T R, Prospero-Garcia O, Puaoi D L, Lerner D L, Fox H S, Olmsted R A, Bloom F E, Henriksen S J, Elder J H. Neurological abnormalities associated with feline immunodeficiency virus infection. J Gen Virol. 1994;75:979–987. doi: 10.1099/0022-1317-75-5-979. [DOI] [PubMed] [Google Scholar]
- 45.Podel M N, Oglesbee M, Mathes L, Krakowka S, Olmstead R, Lafredo L. AIDS-associated encephalopathy with experimental feline immunodeficiency virus infection. J Acquired Immune Defic Syndr. 1993;6:758–771. [PubMed] [Google Scholar]
- 46.Remington K M, Chesebro B, Wehrly K, Pedersen N C, North T W. Mutants of feline immunodeficiency virus resistant to 3′-azido-3′-deoxythymidine. J Virol. 1991;65:308–312. doi: 10.1128/jvi.65.1.308-312.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Remington K M, Zhu Y-Q, Phillips T R, North T W. Rapid phenotypic reversion of zidovudine-resistant feline immunodeficiency virus without loss of drug-resistant reverse transcriptase. J Virol. 1994;68:632–637. doi: 10.1128/jvi.68.2.632-637.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Richman D D. Resistance of clinical isolates of human immunodeficiency virus to antiretroviral agents. Antimicrob Agents Chemother. 1993;37:1207–1213. doi: 10.1128/aac.37.6.1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Richman D D, Havlir D, Corbeil J, Looney D, Ignacio C, Spector S A, Sullivan J, Cheeseman S, Barringer K, Pauletti D, Shih C K, Myers M, Griffin J. Nevirapine resistance mutations of human immunodeficiency virus type 1 selected during therapy. J Virol. 1992;68:1660–1666. doi: 10.1128/jvi.68.3.1660-1666.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schinazi R F, Larder B A, Mellors J W. Mutations in retroviral genes associated with drug resistance. Int Antiviral News. 1996;4:95–107. [Google Scholar]
- 51.Schinazi R F, Lloyd R M, Jr, Nguen M-H, Cannon D L, McMillan A, Ilksoy N, Chu C K, Liotta D C, Bazmi H Z, Mellors J W. Characterization of human immunodeficiency viruses resistant to oxathiolane-cytosine nucleosides. Antimicrob Agents Chemother. 1993;37:875–881. doi: 10.1128/aac.37.4.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schuurman R, Nijhuis M, van Leeuen R, Schipper P, de Jong D, Collis P, Danner S, Mulder J, Loveday C, Christopherson C, Kwok S, Sninsky J, Boucher C A B. Rapid changes in human immunodeficiency virus type 1 RNA load and appearance of drug-resistant populations in persons treated with lamivudine (3TC) J Infect Dis. 1995;171:1411–1419. doi: 10.1093/infdis/171.6.1411. [DOI] [PubMed] [Google Scholar]
- 53.Shafer R W, Kozal M J, Winters M A, Iversen A K N, Katzenstein D A, Ragni M V, Meyer W A, Gupta P, Rasheed S, Coombs R, Katzman M, Ficus S, Merigan T C. Combination therapy with zidovudine and didanosine selects for drug-resistant human immunodeficiency virus type 1 strains with unique patterns of pol gene mutations. J Infect Dis. 1994;169:722–729. doi: 10.1093/infdis/169.4.722. [DOI] [PubMed] [Google Scholar]
- 54.Smith R A, Remington K M, Lloyd R M, Jr, Schinazi R F, North T W. A novel Met-to-Thr mutation in the YMDD motif of reverse transcriptase from feline immunodeficiency virus confers resistance to oxathiolane nucleosides. J Virol. 1997;71:2357–2363. doi: 10.1128/jvi.71.3.2357-2362.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.St. Clair M H, Martin J L, Tudor-Williams G, Bach M C, Vavro C L, King D M, Kellam P, Kemp S D, Larder B A. Resistance to ddI and sensitivity to AZT induced by a mutation in HIV-1 reverse transcriptase. Science. 1991;253:1557–1559. doi: 10.1126/science.1716788. [DOI] [PubMed] [Google Scholar]
- 56.Talbott R L, Sparger E E, Lovelace K M, Fitch W M, Pedersen N C, Luciw P A, Elder J H. Nucleotide sequence and genomic organization of feline immunodeficiency virus. Proc Natl Acad Sci USA. 1989;86:5743–5747. doi: 10.1073/pnas.86.15.5743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tantillo C, Ding J, Jacobo-Molina A, Nanni R G, Boyer P L, Hughes S H, Pauwels R, Andries K, Janssen P A J, Arnold E. Locations of anti-AIDS drug binding sites and resistance mutations in the three-dimensional structure of HIV-1 reverse transcriptase. J Mol Biol. 1994;243:369–387. doi: 10.1006/jmbi.1994.1665. [DOI] [PubMed] [Google Scholar]
- 58.Tisdale M, Kemp S D, Parry N M, Larder B A. Rapid in vitro selection of human immunodeficiency virus type 1 resistant to 3′-thiacytidine inhibitors due to a mutation in the YMDD region of reverse transcriptase. Proc Natl Acad Sci USA. 1993;90:5653–5656. doi: 10.1073/pnas.90.12.5653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wainberg M A, Drosopoulos W C, Salomon H, Hsu M, Borkow G, Parniak M A, Gu Z, Song Q, Manne J, Islam S, Castriota G, Prasad V R. Enhanced fidelity of 3TC-selected mutant HIV-1 reverse transcriptase. Science. 1996;271:1282–1285. doi: 10.1126/science.271.5253.1282. [DOI] [PubMed] [Google Scholar]
- 60.Wilson J E, Aulabaugh A, Caligan B, McPherson S, Wakefield J K, Jablonski S, Morrow C D, Reardon J E, Furman P A. Human immunodeficiency virus type-1 reverse transcriptase; contribution of Met-184 to binding of nucleoside 5′-triphosphate. J Biol Chem. 1996;271:13656–13662. doi: 10.1074/jbc.271.23.13656. [DOI] [PubMed] [Google Scholar]
- 61.Yamamoto J K, Sparger E, Ho E W, Andersen P R, O’Connor T P, Mandell C P, Lowenstine L, Munn R, Pedersen N C. Pathogenesis of experimentally induced feline immunodeficiency virus infection in cats. Am J Vet Res. 1988;49:1246–1258. [PubMed] [Google Scholar]
- 62.Zhu Y-Q, Remington K M, North T W. Mutants of feline immunodeficiency virus resistant to 2′,3′-dideoxy-2′,3′-didehydrothymidine. Antimicrob Agents Chemother. 1996;40:1983–1987. doi: 10.1128/aac.40.9.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]