Abstract
The main limitation of both the rabbit and mouse models of rotavirus infection is that human rotavirus (HRV) strains do not replicate efficiently in either animal. The identification of individual genes necessary for conferring replication competence in a heterologous host is important to an understanding of the host range restriction of rotavirus infections. We recently reported the identification of the P type of the spike protein VP4 of four lapine rotavirus strains as being P[14]. To determine whether VP4 is involved in host range restriction in rabbits, we evaluated infection in rotavirus antibody-free rabbits inoculated orally with two P[14] HRVs, PA169 (G6) and HAL1166 (G8), and with several other HRV strains and animal rotavirus strains of different P and G types. We also evaluated whether the parental rhesus rotavirus (RRV) (P5B[3], G3) and the derived RRV-HRV reassortant candidate vaccine strains RRV × D (G1), RRV × DS-1 (G2), and RRV × ST3 (G4) would productively infect rabbits. Based on virus shedding, limited replication was observed with the P[14] HRV strains and with the SA11 Cl3 (P[2], G3) and SA11 4F (P6[1], G3) animal rotavirus strains, compared to the homologous ALA strain (P[14], G3). However, even limited infection provided complete protection from rotavirus infection when rabbits were challenged orally 28 days postinoculation (DPI) with 103 50% infective doses of ALA rabbit rotavirus. Other HRVs did not productively infect rabbits and provided no significant protection from challenge, in spite of occasional seroconversion. Simian RRV replicated as efficiently as lapine ALA rotavirus in rabbits and provided complete protection from ALA challenge. Live attenuated RRV reassortant vaccine strains resulted in no, limited, or productive infection of rabbits, but all rabbits were completely protected from heterotypic ALA challenge. The altered replication efficiency of the reassortants in rabbits suggests a role for VP7 in host range restriction. Also, our results suggest that VP4 may be involved in, but is not exclusively responsible for, host range restriction in the rabbit model. The replication efficiency of rotavirus in rabbits also is not controlled by the product of gene 5 (NSP1) alone, since a reassortant rotavirus with ALA gene 5 and all other genes from SA11 was more severely replication restricted than either parental rotavirus strain.
Rotaviruses are the leading cause of acute viral gastroenteritis in humans and animals throughout the world. Rotaviruses belong to the Reoviridae family and are characterized by a genome consisting of 11 segments of double-stranded RNA (dsRNA), enclosed in a triple-layered protein capsid (28). Serotype designations are based on independent neutralization determinants on the two outer capsid proteins VP4 (P serotypes, for protease-sensitive protein) and VP7 (G serotypes, for glycoprotein) (28). Serotype specificity determined by cross-neutralization assays using hyperimmune sera against the whole virus is mainly defined by VP7, and 14 G serotypes have been identified (28). Recently, antisera or monoclonal antibodies raised to VP4 and sequence analysis of VP4 identified 12 P serotypes and 20 P genotypes, respectively (28, 39). Rotavirus VP4 protein is responsible for a number of important biological functions, such as the enhancement of infectivity by proteolytic cleavage of VP4 into VP8* and VP5*, hemagglutination, restricted growth in cell culture, virulence, initial virus attachment to cells, and protease sensitivity associated with plaque formation (1, 4, 25, 34, 40, 51).
The use of animal models, including the rabbit and mouse models, has been essential to the understanding of rotavirus infection, pathology, disease, immunity, and testing of prospective vaccines in children (21). The limitations of the rabbit and adult mouse models of rotavirus infection for vaccine testing are as follows: (i) human rotavirus (HRV) strains do not efficiently replicate in either animal, (ii) clinical disease is not observed, and (iii) only homologous virus strains (isolated from the same species) replicate efficiently and spread horizontally to uninoculated control animals, whereas heterologous virus strains (isolated from a different species) do not (6, 15, 16, 29, 31, 35, 37, 44, 50, 55). We and others developed a rabbit model of rotavirus infection that is useful for defining basic parameters of active immunity, immunogenicity, and protective efficacy of vaccines (12, 15–21, 36, 61). Rabbits are productively infected with homologous lapine rotavirus strains up to at least the age of 5 years, which allows examination of active and long-term immunity for vaccine studies (13, 15–17, 36, 61). Group A lapine rotavirus strains have been isolated in Canada, Japan, Italy, and the United States, and those that have been characterized are serotype G3 (8, 11, 15, 53, 56, 61). Recently, the P type of four different strains was identified as genotype P[14] (11). Previously, limited infection of rabbits with a heterologous strain had been obtained only with SA11 Cl3 (P[2], G3) (15).
Attempts to identify host range and virulence determinants for rotavirus have implicated different constellations of genes, including genes 2, 3, 4, 5, 8, 9, 10, and 11 (5, 23, 30, 33, 37, 38, 41, 43, 44, 60, 62, 65). Although host range restriction and virulence may be multigenic, two genes, 4 and 5, are of interest because they cluster according to species of origin, suggesting a role in host range restriction. The finding that genome segment 5 (NSP1) sequences cluster according to species of origin (24, 39, 65) and that, in the mouse model, gene 5 segregates with transmission of virus among littermates (5), led to the hypothesis that NSP1 is involved in host range restriction. VP4 sequence analyses of rotavirus strains isolated from different species revealed that specific VP4 types also generally correlate with the species of origin of each rotavirus strain (43, 60). Therefore, once we identified the P type of four lapine rotaviruses as P[14], we tested two P[14] HRV strains, PA169 (G6) and HAL1166 (G8) (32) to determine if VP4 is involved in host range restriction. We also tested several other HRV strains, live attenuated reassortant candidate vaccine strains [rhesus rotavirus (RRV) × D (G1), RRV × DS-1 (G2), and RRV × ST3 (G4)], and animal rotavirus strains of different P and G types to determine if they could productively infect rabbits. In addition, to evaluate whether the single rotavirus gene 5 is responsible for replication efficiency in rabbits, rabbits were inoculated with a reassortant rotavirus with the lapine ALA gene 5 and all the other genes from the simian rotavirus SA11 Cl3 strain.
MATERIALS AND METHODS
Animals.
Two- to 36-month-old rotavirus antibody-free New Zealand White rabbits used for inoculations were reared at the Veterans Affairs Medical Center in Houston as previously reported (15–17).
Viruses.
All virus strains were cultivated in fetal rhesus monkey kidney cells (MA104) in the presence of trypsin as previously described (15, 26). The lapine ALA (P[14], G3) strain was passaged 10 times in MA104 cells prior to inoculation of rabbits as described previously (11, 15). All other strains were plaque purified three times. Virus titers were determined by plaque-forming assay or focus fluorescent assay (FFA) and expressed as PFU per milliliter or focus-forming units (FFU) per milliliter, respectively (10, 15). The G and P serotypes, species of origin, titers of the inocula, and sources of rotavirus strains used in this study are summarized in Table 1.
TABLE 1.
Origins, serotypes, titers, and sources of rotavirus strains used in this study
| Virus | Origin | Serotype
|
Dose of inoculum (PFU)b | Source | |
|---|---|---|---|---|---|
| Pa | G | ||||
| ALA | Lapine | [14] | 3 | 3.5 × 105 | M. Thouless, University of Washington, Seattle, Wash. |
| SA11 Cl3c | Simian | [2] | 3 | 1 × 108 | H. Malherbe, Gull Laboratories, Salt Lake City, Utah |
| SA11 4Fd | Simian | 6[1] | 3 | 8.8 × 108 | H. Pereira, Instituto Oswaldo Cruz, Rio de Janeiro, Brazil |
| RRV | Simian | 5B[3] | 3 | 2.4 × 108 | H. B. Greenberg, Stanford University, Palo Alto, Calif. |
| OSUe | Porcine | 9[7] | 5 | 1 × 108f | L. J. Saif, Ohio State University, Wooster, Ohio |
| NCDV-Lincoln | Bovine | P6[1] | 6 | 5 × 107 | N. R. Blacklow, University of Massachusetts, Worcester, Mass. |
| H-2 | Equine | P11[12] | 3 | 4.3 × 108 | Y. Hoshino, National Institutes of Health, Bethesda, Md. |
| FI-23 | Equine | P11[12] | 14 | 2.7 × 106 | M. E. Conner, Baylor College of Medicine, Houston, Tex. |
| Wa | Human | 1A[8] | 1 | 9.8 × 106 | H. F. Clark, Wistar Institute, Philadelphia, Pa. |
| K8 | Human | 3A[9] | 1 | 3.2 × 104 | S. Urasawa, Sapporo Medical University, Sapporo, Japan |
| DS-1 | Human | 1B[4] | 2 | 9 × 104 | Y. Hoshino, National Institutes of Health, Bethesda, Md. |
| Ito | Human | 1A[8] | 3 | 1.2 × 107 | Y. Inaba, National Institute of Animal Health, Ibaraki, Japan |
| PA169 | Human | 3B[14] | 6 | 6 × 107 | Y. Hoshino, National Institutes of Health, Bethesda, Md. |
| HAL1166 | Human | 3B[14] | 8 | 6 × 105 | Y. Hoshino, National Institutes of Health, Bethesda, Md. |
| 69M | Human | 4[10] | 8 | 3.7 × 105 | S. Matsuno, National Institute of Health, Tokyo, Japan |
| WI61 | Human | 1A[8] | 9 | 1.2 × 107 | H. F. Clark, Wistar Institute, Philadelphia, Pa. |
| PA169 × DS-1 | Human × humang | 3B[14] | 2 | 1.1 × 107 | Y. Hoshino, National Institutes of Health, Bethesda, Md. |
| RRV × D | Simian × humanh | 5B[3] | 1 | 1.1 × 109 | H. B. Greenberg, Stanford University, Palo Alto, Calif. |
| RRV × DS-1 | Simian × humani | 5B[3] | 2 | 3 × 108 | H. B. Greenberg, Stanford University, Palo Alto, Calif. |
| RRV × ST3 | Simian × humanj | 5B[3] | 4 | 1.4 × 107 | H. B. Greenberg, Stanford University, Palo Alto, Calif. |
| SA11 Cl3 × ALA | Simian × lapinek | [2] | 3 | 1.9 × 107 | This report |
The P genotype is surrounded by brackets and preceded by the P serotype (when it is known) (28).
Rabbits were orally inoculated with 1 to 3 ml of virus.
SA11 was first isolated by Malherbe and Strickland-Cholmley (42), and SA11 Cl3 was plaque purified three times before characterization (7, 27).
SA11 4F was first isolated by H. Pereira (52) and was subcloned in our laboratory for characterization (7).
OSU porcine rotavirus is piglet gut passage P-64 (2).
Titer of OSU piglet passage was determined by FFA.
Reassortant whose gene 8 is derived from the human rotavirus strain DS-1, while all its other genes are derived from the human strain PA169 (32).
Reassortant whose segment 9 is derived from the human strain D, while all its other genes are derived from the simian strain RRV (47).
Reassortant whose segments 8 and 5 are derived from the human strain DS-1, while all its other genes are derived from the simian strain RRV (46, 47).
Reassortant whose segment 9 is derived from the human strain ST3, while all its other genes are derived from the simian strain RRV (48).
Reassortant (R-54) whose segment 5 is derived from the lapine strain ALA, while all its other genes are derived from the simian strain SA11 Cl3.
Production of SA11 × ALA reassortant strains.
Reassortants of rotavirus strains SA11 Cl3 and ALA were made to test the hypothesis that gene 5 from the replication-efficient lapine ALA rotavirus would confer that phenotype on the simian SA11 Cl3 virus, which normally has limited replication (15). The SA11 Cl3 strain was plaque purified three times as described elsewhere (27) (Table 1). The uncloned ALA virus stock was passaged 10 times in MA104 cells, and in order to determine if it was electrophoretically homogeneous, the uncloned ALA virus stock was plated onto MA104 cells and 100 plaques were picked. Each plaque was passaged once in MA104 cells to make P1 stocks. Then, each P1 stock was 32P labeled by infecting MA104 cells and adding 32Pi. We concluded that our ALA stock was homogeneous and could be used as a parental virus without further purification because 61 of 100 amplified individual plaques showed labeled dsRNA electropherotypes identical to those of ALA and 39 of 100 plaques apparently lacked virus, as no labeled dsRNA was detected (data not shown). Since it was determined that the ALA virus stock was electrophoretically homogeneous, the uncloned ALA virus stock was used to generate the single-gene reassortant virus between SA11 Cl3 and ALA. Coinfections were carried out at several multiplicities of infection as previously described (54). Progeny virus was plated without selection, and individual plaques were picked. Screening of reassortant strains was performed as described by Gombold and Ramig (33). A single reassortant, R-54, contained the single ALA gene 5, and the remainder of its genes were from SA11 Cl3. The reassortant R-54 was isolated from a 5 × 10−5 dilution of the viral progeny obtained from the coinfection of each parental virus at a multiplicity of infection of 2 (data not shown). The R-54 reassortant was plaque purified three times before it was inoculated into rabbits.
Animal inoculations and procedures.
To prevent complications due to Clostridium infections (3, 15), several weeks prior to the initiation of rotavirus studies, rabbits were vaccinated twice intramuscularly with a Clostridium spiroforme toxoid developed and kindly supplied by R. Carman (TechLabs, Inc., Blacksburg, Va.). Rabbits were orally inoculated with individual HRV or animal rotavirus strains of different P and G serotypes essentially as described previously (15, 16). For heterologous (nonlapine) rotavirus strains, we inoculated rabbits with the highest-titered virus stock available (Table 1), because high virus titer has been reported to be important for the induction of protective immunity in heterologous hosts (29, 31). Negative-control rabbits were mock inoculated with 1.5 ml of phosphate-buffered saline (PBS). Rabbits were maintained in open cages for primary and challenge inoculations, except for rabbits inoculated with the high-titered simian rotavirus strain RRV and the live attenuated reassortant vaccine rotavirus strains, which were maintained in individual negative-pressure isolation units until challenge inoculation.
All rabbits were challenged orally with approximately 103 50% infectious doses (ID50) (3.5 × 105 PFU/ml) of lapine ALA rotavirus 28 days postinoculation (DPI) as described previously (15–17, 22). Total protection from challenge was defined as no fecal shedding of virus antigen as detected by enzyme-linked immunosorbent assay (ELISA). To obtain an estimate of the level of protection that would reflect both the duration and the magnitude of virus antigen shedding, we calculated the percent protection from challenge with homologous lapine ALA rotavirus for individual animals by comparing the area under the curve of virus shedding (optical density [OD] plotted against the number of days postchallenge [DPC]) for each individual animal to the mean of the areas under the curves for the PBS control animals. The group means of percent protection were then calculated.
Collection of samples.
For detection of rotavirus-specific antibody responses, serum, intestinal lavage, and fecal samples were collected at 0 and 28 DPI and 28 DPC as described previously (15–17). Processing of fecal samples for antibody detection was performed essentially as described for virus antigen, except that fecal samples were diluted to a 5% solution of cold (4°C) PBS containing soybean trypsin inhibitor (0.1 mg/ml), Merthiolate (0.1 mg/ml), and 0.1% Tween 20. To detect virus antigen shedding, fecal samples were collected 0 to 10 DPI and 0 to 14 DPC and were processed as described elsewhere (15).
ELISA to measure rotavirus excretion and total antibody responses.
The antigen ELISA was performed as described previously (15–17, 22). An ELISA to measure total (immunoglobulin A [IgA], IgM, and IgG) antibody to rotavirus was performed as described previously (15–17, 22). A positive reaction was defined as an OD reading of ≥0.1 for the antigen ELISA or a value of ≥0.1 after subtraction of the OD values of the antigen-negative well (mock) from those of the antigen-positive well for the antibody ELISA. Statistical analyses comparing antibody titers between groups were performed by using the Mann-Whitney U test in the SPSS version 7.0 for Windows (SPSS, Inc., Chicago, Ill.).
Analysis of rotavirus infection in rabbits.
Virus antigen shedding detected after inoculation of heterologous viruses was defined as limited replication if the amount and days of shedding were approximately fourfold lower than those obtained with homologous strains (productive infection) and antibody conversion occurred. Minimal replication of a virus in rabbits was defined as antibody conversion after inoculation of the heterologous strain, but no detectable virus antigen shedding.
FFA to determine infectious virus titers and FFNA to measure neutralizing antibodies.
FFAs were performed as described previously (10) on fecal samples collected following heterologous or homologous rotavirus inoculation of rabbits to compare infectious virus shedding relative to ELISA virus antigen shedding. Focus fluorescent neutralization assays (FFNAs) were performed essentially as described previously (10), with the end point determined as the serum dilution producing a ≥66% reduction in the number of fluorescent foci.
Virus isolation and analysis of RNA by polyacrylamide gel electrophoresis.
To confirm that rabbits shed only the virus with which they were inoculated, selected fecal suspensions from virus-inoculated rabbits which were positive for rotavirus by ELISA were tested directly or after amplification in MA104 by polyacrylamide gel electrophoresis. Fecal samples which had low levels of virus were amplified in MA104 cells. The samples were filtered and activated with 20 μg of trypsin (Worthington, Freehold, N.J.)/ml and then were inoculated into roller tubes of MA104 cells in the presence of 1 μg of trypsin/ml as described elsewhere (10). Nucleic acids of representative input and recovered virus from fecal material or amplification in cell culture were extracted and subjected to electrophoresis in a 7% polyacrylamide gel, and genome segments were visualized by silver staining (10).
Sequential passage of P[14] HRV strains in rabbits.
To determine whether in vivo passage of the P[14] HRV strains PA169 and HAL1166 would improve infectivity, rabbits were inoculated orally with Genetron (10%) extracts of fecal material obtained from the rabbits inoculated with these strains (first passage); a total of two passages for PA169 and three passages for HAL1166 were performed. The titer of virus in each inoculum was determined by FFA as described previously (10). Genetron treatment did not affect the infectious virus titer of the fecal material prior to each passage level. The infectious virus doses of the second and third passage of HAL1166 were 2.5 × 104 and 9.5 × 102 PFU, respectively, whereas the infectious virus does of the second passage of PA169 was 2 × 102 PFU.
RESULTS
Infection of rabbits inoculated with homologous, heterologous, and reassortant candidate vaccine strains.
As is seen in homologous rotavirus infection, no rabbit inoculated with heterologous rotaviruses developed diarrhea (12, 13, 15–17, 36). Productive infection with lapine ALA rotavirus can be established in rabbits up to at least the age of 5 years (13). Since no significant differences are seen in either the immune response or the amount of virus shed following infection in rabbits aged 2 to 54 weeks (13), a broad age range of rabbits was used for this study. The ages of rabbits used for this study varied from 2 to 36 months. All rabbits were 2 to 5 months old, except for one of three rabbits inoculated with 69M and those inoculated with Ito, PA169 × DS-1, RRV, or the reassortant vaccine candidates, which ranged from 12 to 36 months.
In contrast to productive homologous lapine P[14] ALA rotavirus infection (Fig. 1A), limited or no replication (based on the amount of virus shedding) was observed with the heterologous P[14] HRV strains PA169 and HAL1166 (Fig. 1B). One of three rabbits inoculated with HAL1166 and one of three inoculated with PA169 had detectable fecal virus antigen which lasted 1 to 2 days starting at 3 to 4 DPI, whereas for each of these strains, the other two rabbits inoculated did not have detectable virus antigen shedding. A single-gene-reassortant strain, PA169 × DS-1, whose cognate gene 9 (gene 8) is derived from the human strain DS-1 and all of whose other genes are from the human strain PA169, failed to replicate in rabbits (n = 2) (data not shown). The titers of ALA at the peak of virus shedding typically ranged from 1.5 × 105 to 9.8 × 105 FFU/ml. However, the titer of infectious heterologous HAL1166 rotavirus shed by one rabbit at 3 DPI (Fig. 1B) was 2.5 × 104 FFU/ml, while that of PA169 at 4 DPI (Fig. 1B) was 20 FFU/ml.
FIG. 1.
Fecal virus antigen-shedding curves of individual rabbits inoculated with ALA (A), HRV strain HAL1166 (▪, •, ▴) or PA169 (⧫, □, ○) (B), simian rotavirus strain SA11 Cl3 (▪, •, ▴) or SA11 4F (⧫, □) (C), and simian strain RRV (▪, •) or the modified Jennerian reassortant candidate vaccine strain RRV × D (▴, ⧫), RRV × DS-1 (□, ○), or RRV × ST3 (▵) (D). Fecal rotavirus antigen shedding was assessed by ELISA from 0 to 10 DPI and expressed as net OD450 readings. Readings of ≥0.1 are considered positive.
Rabbits inoculated with simian rotavirus SA11 Cl3 (n = 3) or SA11 4F (n = 2) shed virus antigen for 2 to 3 days (mean duration of fecal shedding, 2.6 or 2.0 days, respectively) from 4 to 8 DPI, with a peak occurring at the 1st or 2nd day of shedding (Fig. 1C). Although virus antigen excretion was observed in all SA11 Cl3- and SA11 4F-inoculated rabbits, the level and the duration of virus shedding were limited in comparison to those for lapine ALA-infected rabbits (Fig. 1A). Titers of infectious simian SA11 Cl3 and SA11 4F rotavirus strains shed on peak days of antigen shedding (Fig. 1C) ranged from 1.2 × 103 to 5.3 × 104 FFU/ml. There was a 1- to 2-day delay in onset and peak of heterologous-virus antigen shedding compared to those for ALA-infected rabbits (Fig. 1A).
Unlike the simian strains SA11 Cl3 or SA11 4F, rabbits (n = 2) were productively infected with simian RRV (P5B[3], G3). Fecal virus antigen shedding comparable to that seen with productive homologous lapine ALA rotavirus infection (Fig. 1A) was detected in both RRV-inoculated rabbits from 3 to 8 DPI, with a peak of shedding occurring at 5 to 6 DPI (Fig. 1D). Replacement of the RRV gene 9 by the cognate gene 9 of human origin altered the ability of the reassortants to replicate in rabbits. Oral inoculation of rabbits with live attenuated reassortant candidate vaccine strains resulted in no (RRV × ST3), limited (RRV × D), or productive (RRV × DS-1; although one of the two rabbits failed to shed virus) infection of rabbits (Fig. 1D). Titers of infectious simian RRV and reassortant candidate vaccine rotavirus strains on peak days of virus shedding ranged from 1.55 × 103 to 2 × 105 and from 1.9 × 102 to 7 × 104 FFU/ml, respectively.
Based on the lack of detectable virus antigen shedding, rabbits were not productively infected by the HRV strains Wa, K8, DS-1, Ito, 69M, and WI61, by the equine rotavirus strains H-2 and FI-23, by the bovine rotavirus strain NCDV-Lincoln, or by the porcine rotavirus strain OSUwt (data not shown).
Immune response to homologous and heterologous viruses and to reassortant vaccine strains.
Infection of rabbits with the heterologous strains was also monitored by the ability of the infecting virus to induce a primary serologic and intestinal antibody response. All preinoculation serum and intestinal samples were rotavirus antibody negative at a dilution of 1:50 (data not shown). All rabbits inoculated with heterologous rotavirus strains that exhibited either productive (RRV and RRV × DS-1) or limited (HAL1166, PA169, SA11, Cl3, SA11 4F, and RRV × D) replication based on fecal virus antigen shedding developed serum and intestinal antibody responses similar in magnitude to those obtained with a homologous virus infection (Table 2). Rabbits inoculated with rotavirus strain 69M, Ito, H-2, FI-23, or OSUwt, as well as those inoculated with the reassortant virus strain PA169 × DS-1 or RRV × ST3 developed low-titered serologic or intestinal antibodies to rotavirus, suggesting that low-level (abortive) replication occurred in the absence of detectable virus shedding (Table 2). For each of the two P[14] HRV strains (HAL1166 and PA169), although virus shedding was not observed in two of three rabbits inoculated with each HRV strain, one of the rabbits that did not shed virus, as well as the rabbit that did, developed high antibody titers in both the serum and the intestine, indicating that two of the three rabbits were infected. No rotavirus-specific antibodies were detected in rabbits inoculated with PBS, Wa, K8, DS-1, WI61, or NCDV-Lincoln.
TABLE 2.
Rotavirus antibody responses of rabbits orally inoculated with selected heterologous (nonlapine) rotavirus strains
| Virus | Origina | Serotype | No. of respon- dersb/no. of inoculated rabbits | Total (IgM, IgG, IgA) antibody GMTc (range)
|
Serum neutralizing antibody GMT (range) against indicated virusd
|
||
|---|---|---|---|---|---|---|---|
| Serum | Intestinal | Immunizing strain | Lapine ALA strain | ||||
| PBS | 0/6 | 25 | 2.5 | 50 | |||
| ALA | Lapine | P[14], G3 | 5/5 | 409,600 (409,600–409,600) | 1,280 (640–5,120) | 6,400 (3,200–12,800) | 6,400 (3,200–12,800) |
| SA11 Cl3 | Simian | P[2], G3 | 3/3 | 25,600 (1,600–204,800) | 1,613 (1,280–2,560) | 635 (400–1,600) | 400 (200–800) |
| SA11 4Fe | Simian | P6[1], G3 | 2/2 | 72,408 (51,200–102,400) | 320 (160–640) | 3,200 (3,200) | 566 (400–800) |
| RRV | Simian | P5B[3], G3 | 2/2 | 204,800 (102,400–409,600) | 640 (640) | 3,200 (1,600–6,400) | 1,131 (800–1,600) |
| OSUwt | Porcine | P9[7], G5 | 1/1 | 12,800 | 20 | 800 | 100 |
| NCDV-Lincoln | Bovine | P6[1], G6 | 0/2 | 25 | 2.5 | ND | ND |
| H-2 | Equine | P11[12], G3 | 2/2 | 36,204 (25,600–51,200) | 80 (40–60) | 1,600 (1,600–1,600) | 567 (400–800) |
| FI-23 | Equine | P11[12], G14 | 1/2 | 400 (<50–6,400) | 2.5 | 100 (<100–200) | 50 |
| Wa | Human | P1A[8], G1 | 0/2 | 25 | 2.5 | ND | ND |
| K8 | Human | P3A[9], G1 | 0/2 | 25 | 2.5 | ND | ND |
| DS-1 | Human | P1B[4], G2 | 0/2 | 25 | 2.5 | ND | ND |
| Ito | Human | P1A[8], G3 | 1/2 | 50 (<50–100) | 2.5 | ND | ND |
| PA169 | Human | P3B[14], G6 | 2/3 | 4,032 (<50–51,200) | 254 (<5–5,120) | 79 (<100–100) | 50 |
| HAL1166 | Human | P3B[14], G8 | 2/3 | 2,540 (<50–204,800) | 101 (<5–5,120) | 159 (<100–800) | 100 (<100–400) |
| 69M | Human | P4[10], G8 | 2/3 | 200 (<50–1,600) | 8 (<5–80) | 50 | 50 |
| WI61 | Human | P1A[8], G9 | 0/1 | 25 | 2.5 | ND | ND |
| PA169 × DS-1 | Human × human | P3B[14], G2 | 2/2 | 1,131 (100–12,800) | 5 (5) | 141 (<100–400) | 50 |
| RRV × D | Simian × human | P5B[3], G1 | 2/2 | 51,200 (25,600–102,400) | 905 (320–2,560) | 2,263 (800–6,400)f | 50 |
| RRV × DS-1 | Simian × human | P5B[3], G2 | 2/2 | 144,816 (102,400–204,800) | 640 (320–1,280) | 1,132 (800–1,600)g | 50 |
| RRV × ST3 | Simian × human | P5B[3], G4 | 1/1 | 51,200 | 40 | 400h | 50 |
Origins of reassortants PA169 × DS-1, RRV × D, RRV × DS-1, and RRV × ST3 are explained in Table 1, footnotes g through j, respectively.
Rabbits which seroconverted following primary rotavirus inoculation.
Measured by ELISA at 28 DPI. For titers of <50 and <5, 25 and 2.5, respectively, were used to calculate the geometric mean titer (GMT). A GMT of 25 or 2.5, for serum or intestinal antibodies, respectively, was considered negative.
Serum neutralizing antibody was measured (by FFNA at 28 DPI) only for rabbits that developed an antibody response as measured by ELISA. For titers of <100, 50 was used to calculate the GMT. A GMT of 50 was considered negative. ND, not done.
Rabbits were sequentially inoculated with SA11 4F and OSU (28 days apart). Antibody titers increased fourfold 28 days following OSUwt inoculation (data not shown).
The HRV strain Wa (P1A[8], G1) was used to measure VP7-homotypic neutralizing serum antibodies.
The HRV strain DS-1 (P1B[4], G4) was used to measure VP7-homotypic neutralizing serum antibodies.
The HRV strain VA70 (P1A[8], G4) was used to measure VP7-homotypic neutralizing serum antibodies.
Sera from rabbits inoculated with heterologous strains or live attenuated reassortant candidate vaccine strains that developed an antibody response measured by ELISA were assayed by FFNA to determine whether neutralizing antibodies were produced. Prior to inoculation, all sera lacked neutralizing antibodies at a dilution of 1:50 (data not shown). The majority of rabbits had serum neutralizing antibodies (dilutions, 1:100 to 1:6,400) to the immunizing rotavirus strain (Table 2) and to other strains with the same P or G type as the immunizing strain (data not shown).
Transmission of heterologous viruses to controls.
We previously reported lack of horizontal transmission of heterologous SA11 Cl3, while homologous lapine ALA rotavirus is readily transmitted to uninoculated control rabbits housed in open neighboring cages in the same room (15). We monitored whether heterologous viruses (HAL1166, PA169, SA11 Cl3, and SA11 4F) that showed limited replication in rabbits would be transmitted to mock-inoculated controls. In each experiment, virus- and PBS-inoculated rabbits were housed in open neighboring cages in order to determine if horizontal transmission of heterologous viruses occurred. Neither virus shedding nor antibody conversion was detected in any of the control animals inoculated with PBS, suggesting that the efficiency of horizontal spread of heterologous viruses among rabbits is poor, possibly due to the small amount of virus shed. Transmission of RRV and the reassortant candidate vaccine strains could not be assessed, since all inoculated rabbits were housed in individual isolators under negative pressure.
Protection against viral shedding from challenge induced by heterologous and live attenuated reassortant candidate vaccine rotavirus strains.
Following challenge with lapine ALA rotavirus, all the rabbits that received PBS as the first inoculum (n = 6) shed virus antigen with a mean duration of 5.25 days (range, 3 to 9 days) (Fig. 2A). Rabbits that seroconverted following inoculation with heterologous viruses exhibited various degrees of protection from ALA challenge (Table 3). In the groups of rabbits that were inoculated with the P[14] HRV strains HAL1166 and PA169, the rabbits that seroconverted (two of three) following primary inoculation with either virus were protected from infection (82 to 100%), while the one seronegative rabbit in each group was not protected from infection (0 to 12%) (Fig. 2B and Table 3). Rabbits inoculated with the reassortant strain PA169 × DS-1 were not protected from infection (13%) (Table 3). Among the non-P[14] HRV strain-inoculated rabbits, only one rabbit that seroconverted following primary inoculation with 69M (P4[10], G8) was partially protected from infection (59%) (Fig. 2C and Table 3) following challenge; the amount of virus shed and the duration of shedding were reduced in comparison to those of the control ALA-infected rabbits (Fig. 2A). Rabbits inoculated with the HRV strain Wa were not challenged. All rabbits inoculated with the simian RRV strain or the reassortant candidate vaccine strain RRV × D, RRV × DS-1, or RRV × ST3 were completely protected from virus challenge infection (Fig. 2D). Rabbits inoculated with simian SA11 Cl3 and SA11 4F, equine H-2, and porcine OSUwt strains were completely protected from infection following ALA challenge, whereas those inoculated with equine FI-23 and bovine NCDV-Lincoln were not protected (data not shown). Table 3 summarizes the percent protection from challenge and the serum and intestinal antibody responses at 28 DPC.
FIG. 2.
Fecal virus antigen-shedding curves after lapine ALA challenge of individual rabbits previously inoculated with PBS (▪, •, ▴, ⧫, □, ○) (A), HRV strain HAL1166 (▪, •, ▴) or PA169 (⧫, □, ○) (B), HRV strain DS-1 (▪, •), 69M (▴, ⧫, □), WI61 (○), K8 (▵, ◊) or Ito (☒, □•) (C), or the simian strain RRV (▪, •) or the modified Jennerian reassortant vaccine strain RRV × D (▴, ⧫), RRV × DS-1 (□, ○), or RRV × ST3 (▵). Fecal rotavirus antigen shedding was assessed by ELISA from 0 to 14 DPC and expressed as net OD450 readings. Readings of ≥0.1 are considered positive. No virus was shed by any rabbit at 11 to 14 DPC (data not shown).
TABLE 3.
Rotavirus antibody and percent protection of rabbits challenged with 103 ID50 of homologous lapine ALA rotavirus strain
| Virus | Origina | Serotype | No. of protectedb/ challenged rabbits | % Protection from challenge (range) | Total (IgM, IgG, IgA) antibody GMT (range)c
|
|
|---|---|---|---|---|---|---|
| Serum | Intestinal | |||||
| PBS | 0/6 | 0 (0) | 275,640 (102,400–409,600) | 951 (320–5,120) | ||
| ALA | Lapine | P[14], G3 | 5/5 | 100 (100–100) | 470,507 (409,600–819,200) | NDd |
| SA11 Cl3 | Simian | P[2], G3 | 3/3 | 100 (100–100) | 204,800 (51,200–819,200) | 2,560 (1,280–5,120) |
| SA11 4Fe | Simian | P6[1], G3 | 2/2 | 100 (100–100) | 289,631 (204,800–409,600) | 1,280 (1,280) |
| RRV | Simian | P5B[3], G3 | 2/2 | 100 (100–100) | 102,400 (51,200–204,800) | 160 (80–320) |
| OSUwt | Porcine | P9[7], G5 | 1/1 | 100 | 102,400 | 1,280 |
| NCDV-Lincoln | Bovine | P6[1], G6 | 0/2 | 0 (0) | 204,800 (204,800) | 905 (640–1,280) |
| H-2 | Equine | P11[12], G3 | 2/2 | 96 (92–100) | 18,102 (12,800–25,600) | 40 (40) |
| FI-23 | Equine | P11[12], G14 | 0/2 | 1 (0–2) | 289,631 (102,400–819,200) | 905 (640–1,280) |
| Wa | Human | P1A[8], G1 | 0/0f | |||
| K8 | Human | P3A[9], G1 | 0/2 | 11 (8–13) | 204,800 (204,800) | 453 (320–640) |
| DS-1 | Human | P1B[4], G2 | 0/2 | 0 (0) | 204,800 (204,800) | 1,280 (640–2,560) |
| Ito | Human | P1A[8], G3 | 0/2 | 16 (0–32) | 409,600 (204,800–819,200) | 113 (80–160) |
| PA169 | Human | P3B[14], G6 | 2/3 | 65 (12–100) | 129,016 (51,200–204,800) | 506 (320–1,280) |
| HAL1166 | Human | P3B[14], G8 | 2/3 | 63 (0–100) | 102,400 (51,200–204,800) | 404 (320–640) |
| 69Mg | Human | P4[10], G8 | 1/3 | 20 (0–59) | 409,600 (204,800–819,200) | 640 (160–2,560) |
| WI61 | Human | P1A[8], G9 | 0/1 | 0 | 409,600 | 320 |
| PA169 × DS-1 | Human × human | P3B[14], G2 | 0/2 | 13 (0–26) | 579,261 (409,600–819,200) | 905 (640–1,280) |
| RRV × D | Simian × human | P5B[3], G1 | 2/2 | 100 (100–100) | 51,200 (51,200) | 113 (80–160) |
| RRV × DS-1 | Simian × human | P5B[3], G2 | 2/2 | 100 (100–100) | 72,408 (51,200–102,400) | 113 (80–160) |
| RRV × ST3 | Simian × human | P5B[3], G4 | 1/1 | 100 | 204,800 | 320 |
| SA11 Cl3 × ALA | Simian × lapine | P[2], G3 | 0/0f | |||
Origins of reassortants PA169 × DS-1, RRV × D, RRV × DS-1, RRV × ST3, and SA11 Cl3 × ALA are explained in Table 1, footnotes g through k, respectively.
Rabbits which exhibited at least 50% reduction in virus shedding following challenge by comparing the area under the curve for each individual animal to the mean of the areas under the curves for control rabbits.
Measured by ELISA at 28 DPC.
ND, not done. Titers of intestinal antibody to rotavirus 28 DPC do not differ significantly from the titers prior to challenge (15–17).
Rabbits were sequentially inoculated with SA11 4F and OSU (28 days apart) and were challenged 56 DPI of SA11 4F. Antibody titers increased after OSU inoculation but not after challenge.
Rabbits were not challenged.
Only one rabbit was partially protected.
Following challenge, animals that were not protected or were partially protected (0 to 59%) from challenge developed serum and intestinal rotavirus-specific antibody titers comparable to those of ALA-infected control animals (Table 3). Anamnestic antibody responses (≥4-fold increases in titer) were not observed in rabbits that were 100% protected from challenge, except in the rabbits initially inoculated with OSUwt or RRV × ST3. Development of an anamnestic response suggests that lapine ALA rotavirus replication occurred in these animals.
Rabbits that were partially protected or were not protected from challenge in the absence of neutralizing antibodies to ALA rotavirus developed neutralizing antibodies to ALA rotavirus at levels similar to those of control animals (data not shown). However, the presence of neutralizing antibodies to ALA rotavirus prior to challenge was not required in order to obtain total protection from challenge; only one rabbit inoculated with a non-serotype G3 strain (HAL1166) developed low levels (1:800) of neutralizing antibodies to ALA rotavirus (probably directed to the homologous P[14] VP4) (Table 2). Rabbits that were 100% protected from challenge did not develop an anamnestic neutralizing antibody response, except for rabbits initially inoculated with OSUwt or RRV × ST3. Rabbits that developed an anamnestic anti-G3 antibody response after challenge also developed >4-fold increases in neutralizing antibodies to heterologous G types (data not shown).
Electropherotype of excreted virus.
To confirm that the excreted virus was the same virus as the primary inoculum, we examined the RNA electropherotype of the virus recovered directly or amplified in cell culture from fecal samples from rabbits that shed virus in their stools. In all cases, the virus recovered from the stools or from the tissue culture adaptation was identical to the virus inoculum (data not shown).
Passage of P[14] HRV strains.
Inoculation of rabbits with HRV P[14] strains resulted in limited infection, but the human origin and the adaptation to growth in cell culture of these strains may have led to attenuation for rabbits. Therefore, we performed experiments to test whether serial passage in rabbits would increase the replication efficiency of the HRV P[14] strains. HAL1166 (P3B[14], G8) was serially passaged three times and PA169 (P3B[14], G6) was serially passaged two times in rabbits. The amount of virus shed in feces decreased to undetectable levels by the third or second passage of HAL1166 or PA169, respectively. As virus shedding decreased, seroconversion did not occur and rabbits were no longer protected from ALA challenge. In the second passage of the fecal material from one rabbit that shed HAL1166 after the first passage, one of two rabbits shed low levels (OD at 450 nm [OD450], ≤0.3) of virus antigen from days 4 to 8 after inoculation. At 28 DPI, antibody titers were 204,800 and 5,120 in serum and intestine, respectively, and subsequently, the animal was protected from challenge (data not shown). However, when the fecal material from this rabbit was passaged a third time, the virus did not replicate, as evidenced by no reactivity in either antigen or antibody ELISAs, and these animals were not protected from subsequent challenge (data not shown).
Evaluation of rotavirus gene 5 in replication efficiency in vivo.
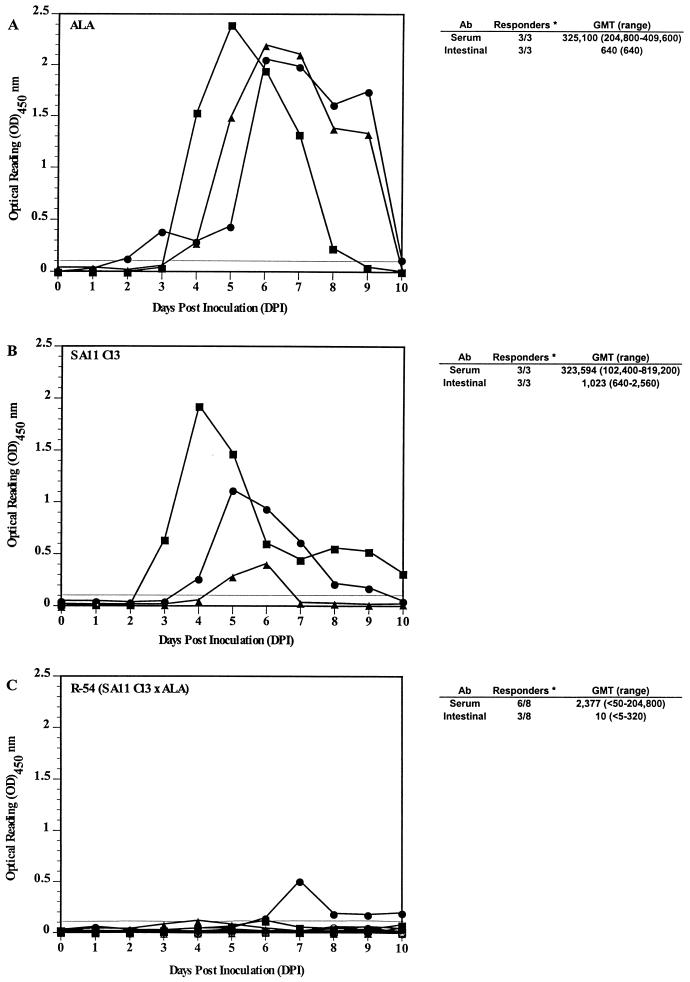
To directly compare the replication efficiency of the SA11 × ALA reassortant (R-54) containing the single ALA gene 5, groups of rabbits were each inoculated with ALA, SA11 Cl3, or R-54. As expected, rabbits (three of three) inoculated with ALA shed virus antigen for a mean duration of 6 days (Fig. 3A). The SA11 Cl3 rotavirus-infected rabbits (three of three) showed limited replication based on fecal virus antigen shedding. The mean number of days of shedding (5.3 days) was higher than that in the previous experiment (2.6 days [Fig. 1B]), but the difference was not significant (P = 0.369 by the Mann-Whitney U test) (Fig. 3B). The titers of infectious SA11 Cl3 virus shed on the peak days of antigen shedding ranged from <5 × 102 to 2.4 × 104 PFU/ml, while the titers of ALA at the peak of virus shedding typically ranged from 105 to 106 PFU/ml. Only three of eight rabbits inoculated with the SA11 × ALA reassortant (R-54) shed very low levels of virus antigen, for a mean duration of 1.4 days (Fig. 3C). The identity of the virus shed in feces was confirmed by RNA electropherotype (data not shown). Protection from ALA challenge was not assessed.
FIG. 3.
Fecal virus antigen-shedding curves of rabbits inoculated with ALA rotavirus (▪, •, ▴) (A), simian rotavirus strain SA11 Cl3 (▪, •, ▴) (B), and single-gene-5 SA11 Cl3 × ALA reassortant R-54 (▪, •, ▴, ⧫, □, ○, ▵, ◊) (C). Fecal rotavirus antigen shedding was assessed by ELISA from 0 to 10 DPI and expressed as net OD450 readings. Readings of ≥0.1 are considered positive. Rotavirus-specific total (IgM, IgG, and IgA) antibody responses at 28 DPI were measured by ELISA (15). GMT, geometric mean titer. Asterisk indicates rabbits which either seroconverted or shed virus following primary inoculation.
The levels of the serologic and intestinal immune responses induced by reassortant R-54 were significantly lower than those induced by the parental SA11 Cl3 strain (Fig. 3) (P = 0.02 and P = 0.015, respectively, by the Mann-Whitney U test). These results indicated that introduction of ALA gene 5 into an SA11 genetic background failed to convert the SA11 to an ALA phenotype for replication. The reassortant R-54 was more replication restricted in rabbits than the parental SA11 Cl3.
DISCUSSION
Previously, in the rabbit model, productive infection was observed with the homologous P[14], G3 lapine rotavirus strains (11, 15, 21, 36, 61). Limited rotavirus infection of rabbits was obtained with the heterologous P[2], G3 simian strain SA11 Cl3, but a non-G3 serotype of rotavirus capable of infecting rabbits had not been identified (15, 21). The genetic basis of host range restriction for rotavirus is not clearly understood, and different genes, including genes 4 and 5, have been implicated in several studies (5, 24, 30, 33, 37, 38, 43, 44, 60, 62, 65). In this study, evaluation of host range restriction in the rabbit model with a number of heterologous rotavirus strains of different P and G types demonstrates, for the first time, that HRV strains and heterologous animal rotavirus strains, including reassortant candidate vaccine strains (47, 48), replicate in rabbits. The ability of the heterologous viruses to replicate was dependent on the virus strains, and no clear association of replication efficiency with a particular genotype could be made. Differences in the replication efficacy of these nonlapine strains tested may be attributed to (i) the viral dose delivered to rabbits, (ii) adaptation to tissue culture, (iii) VP4 type, (iv) VP7 type, (v) a combination of specific VP4 and VP7 types, (vi) other rotavirus genes responsible for host range restriction, or (vii) a multigenic effect.
Since slight decreases in the dose of the heterologous virus in the mouse model resulted in a significant reduction of replication efficacy and immunogenicity (29), the differences we observed in the replication efficacy of the heterologous rotavirus strains tested in rabbits may be due to the inoculum dose delivered to rabbits. Indeed, most of the nonlapine rotaviruses (SA11 Cl3, SA11 4F, OSUwt, H-2, RRV, RRV × D, and RRV × DS-1) administered at a minimal dose of 108 PFU or FFU induced 100% protection against shedding after ALA challenge irrespective of their G or P types. However, the P[14] HRV strains, HAL1166 and PA169, and the reassortant candidate vaccine strain RRV × ST3, which were administered at doses of 105 to 107 PFU, were also capable of inducing complete protection from ALA challenge, suggesting that dose may not be the only factor that plays an important role in inducing protective immunity. It is possible that some virus strains, like the HRV strains with low titers (K8 and DS-1), failed to replicate in rabbits because a low number of infectious particles led to abortive replication. Alternatively, the ability of the heterologous viruses to replicate may require a specific constellation of genes; a given combination of genes may influence the replication outcome in a heterologous host.
It is of note that oral inoculation of rabbits with RRV (P5B[3], G3) resulted in productive infection. The RRV inoculation dose (2.4 × 108 PFU) was approximately 1,000-fold higher than the ALA dose (3.5 × 105 PFU). It is not known whether a lower dose of RRV would result in productive infection in rabbits. Further experiments will be required to determine if RRV and ALA are shed equally under identical dosing levels.
Attempts to correlate the immune response with protection from ALA lapine challenge revealed that only those heterologous viruses that induced a significant level of local immune response conferred protection from subsequent challenge. Similar to results in the mouse model, heterologous (nonlapine) viruses may lack sufficient replication competence to complete even one round of replication and therefore fail to produce enough viral antigen to elicit an immune response in rabbits (6, 29, 44, 45). Low or undetectable levels of fecal antibodies resulted in no protection from challenge. Although only total (IgM, IgG, and IgA) intestinal antibodies were determined, intestinal anti-rotavirus IgG and IgA are both induced following oral rotavirus infection with lapine ALA rotavirus (41a). Previous studies with mice have indicated that the presence of intestinal but not serum antibodies correlates with protection (29), whereas other studies indicate that the presence of either serum IgA or intestinal antibodies following rotavirus infection in mice correlates with protection (44). In our study with rabbits, the presence of intestinal antibody was absolutely required for protection, but the presence of high total (IgM, IgG, and IgA) antibody titers in the serum in conjunction with intestinal antibodies (H-2, OSUwt, and RRV × ST3) conferred greater protection from challenge than low antibody titers in serum (69M). We do not routinely detect rotavirus-specific IgA in rabbit sera, possibly due to extremely low IgA antibody concentrations in rabbit sera (49). Heterologous animal and reassortant candidate vaccine strains with productive, limited, or minimal replication induced a range of systemic and local immune responses resulting in complete protection from challenge that correlated with the presence of serum and intestinal antibodies, but not with the presence of neutralizing antibodies against the challenge ALA strain.
Among the HRV strains tested, genotype P[14] HRV strains (HAL1166 and PA169) were capable of replicating in rabbits, albeit at a much lower level than homologous lapine P[14], G3 strains. However, only four of six rabbits inoculated with the P[14] HRV strains were infected, and a wide range of immune responses and protective efficacy against ALA challenge was produced. It was not surprising that the magnitude of the immune response to the lapine ALA rotavirus was greater than that to the P[14] HRV strains HAL1166 and PA169. Neither of the heterologous viruses PA169 and HAL1166 replicated to high titers in rabbits, and it has been reported for mice that the magnitude of the immune response to heterologous viruses probably reflects the amount of virus replication in the small intestine (29). Therefore, the magnitude of the immune response to the heterologous virus infection may differ among individual rabbits, just as in the mouse model (29). The attempt to increase the ability of the tissue culture-adapted P[14] HRV strains to infect rabbits by rabbit-to-rabbit passage was not successful. The failure of HAL1166 and PA169 to be serially passaged in rabbits may be related to the decreasing doses of virus administered at each passage level. The HRV Wa strain was adapted to grow after serial passages in a heterologous host (gnotobiotic pigs), but the Wa virus passaged in piglets was obtained from the original human fecal sample (63, 64). Therefore, it is possible that serial passage of “wild-type” virus from the original human fecal material of the P[14] HRV strains might be capable of productive infection in rabbits. Of the other non-P[14] HRV strains tested, only 69M exhibited limited replication resulting in partial protection (59%) from ALA challenge in one of three rabbits inoculated.
Although it is not clear whether the replication of heterologous virus strains in rabbits was due to the P type of the viruses, our results suggest that virus strains whose VP4 types are closely related (43) are capable of at least minimal replication (P genotypes 1, 2, 3, 7, 10, and 12). However, based on amino acid identities, genotype P[9] is the P type closest to genotype P[14], yet the HRV K8 strain (P3A[9], G1) failed to replicate in rabbits. Failure of HRV K8 to replicate in rabbits was possibly due to the low titer of the inoculum. Representative viruses of the next closest P types to P[14], 1, 2, 3, 7, 10, and 12 (11), showed evidence of infection in rabbits. Studies with mice indicate that if VP4 is involved in host range restriction, it is probably not the only gene product to be involved, since viruses with VP4 types very distinct from murine VP4 types also exhibit some degree of limited replication in mice (29, 44, 50, 55). Nonetheless, among the heterologous strains that replicate most efficiently in mice, RRV (P5B[3]) and SA11 Cl3 (P[2]) (both serotype G3, like the murine strains) appear to be the most replication-efficient heterologous strains (29, 55); coincidentally, these two P types are the ones that have the highest level of VP4 homology with the murine P[16] or P[20] types (23, 57, 59, 62). Alternatively, different combinations of VP7 and VP4 types may determine expression of some epitopes in VP4 (9) necessary for virus attachment to cells (1).
Recently, the cognate VP7 gene has also been implicated in host range restriction (37). Although the contribution of VP7 type to replication capabilities in a heterologous host is far more difficult to discern, in the case of the simian strain RRV, a single replacement of the simian VP7 gene with a human gene (47, 48) altered the replication capabilities of two reassortants, RRV × D and RRV × ST3, suggesting that VP7 may play a role in host range restriction in rabbits. However, RRV × DS-1 replicated in one of two rabbits as efficiently as lapine ALA rotavirus. Although permissive replication might be related to the high titers of the RRV and RRV × DS-1 viruses, the fact that a higher dose of the RRV × D reassortant strain failed to result in permissive replication suggests that the nature of the VP7 gene influences the replication outcome, or it may be the result of heterologous VP4 and VP7 interactions. Therefore, VP7 may be involved in, but is not exclusively responsible for, host range restriction in rabbits, since a high dose (1.2 × 107 PFU) of Ito, a G3 HRV strain related to the G3 ALA strain, failed to replicate in two of two rabbits. It is of note that both HRV strains 69M and HAL1166, which underwent minimal and limited replication, respectively, are G8 (a serotype closely related to G3), but whether the G8 type provided some replication capabilities in the heterologous host remains to be determined.
Since amino acid sequences of the genome segment 5 product, NSP1, are grouped according to species of origin, this gene has also been implicated in host range restriction (24, 41, 65). However, in the rabbit model, a reassortant containing genome segment 5 from the homologous lapine ALA rotavirus strain and the other 10 genes from the heterologous SA11 Cl3 rotavirus strain replicated less than either parental strain and was not able to spread to uninoculated controls. The results from our experiments indicate that in rabbits, gene 5 is not solely responsible for species specificity or replication efficiency in vivo and that in order to obtain reassortants that confer replication efficiency in rabbits, reassortants with other or multiple, as yet unidentified genes will require further testing. In the mouse model, genome segment 5 (NSP1) was found to segregate with permissive replication and horizontal transmission to a significant level, although the association was not absolute (i.e., other genes may be involved) (5, 30, 44). However, this association has not been tested directly in mice with a single gene 5 reassortant. Analysis of the phylogenetic NSP1 tree reveals that murine strains (EHP and EW) are more closely related (44 to 51%) to the simian strains (RRV and SA11 Cl3) than to any other strains (37 to 41%) (24, 41). These two simian strains share similarities to murine strains in their VP4s, VP7s, and NSP1s which may explain the replication exhibited by the simian strains in mice. The sequence of NSP1 of lapine strains remains unknown, but it will be interesting to determine which strains may share NSP1 sequence similarity. It is of note that genes 4 and 5 of equine strains are also very closely related to those of the murine-simian group (41, 43, 57). Snodgrass and Iša (58) have reported that equine rotavirus strains cause clinical disease in neonatal mice independently of the equine P type, so it is possible that replication was afforded by the conserved gene 5 product, which closely resembles that of murine origin.
Our results suggest that VP4 may be involved in, but is not exclusively responsible for, host range restriction in the rabbit model. Additionally, the facts that RRV was the heterologous virus that exhibited the best replication efficiency in rabbits and that the replication of RRV was less in the reassortants that contained certain human VP7s suggest that VP7, or VP4 and VP7 interactions, may play a role. Understanding of the function of VP4 in host range restriction may lead to more effective vaccine strains. In fact, multivalent reassortant vaccines with both human G and P types may be more effective (14). Since the different virus strains tested have distinct infectious doses and replication efficiencies, results from studies with animal models are relevant to the current human vaccine trials. Protection of individual rabbits vaccinated with the monovalent RRV (modified Jennerian) vaccine strains did not correlate with serotype-specific serum neutralizing antibodies but rather with local intestinal antibodies directed primarily to VP6 (16) in our assay. Similar results were obtained in the mice orally immunized with the reassortant RRV candidate vaccines (31). If the differences in protection are due to limited replication efficiency or host restriction of the heterologous vaccine virus strains, a better strategy for live orally administered vaccine will be to use live attenuated (nonvirulent) human virus strains that can replicate efficiently in the homologous host. Alternatively, an initial oral immunization with the reassortant vaccine strains may be used followed by a parenteral immunization with a virus-like particle subunit vaccine to ensure a broader and a more efficient protective immune response.
ACKNOWLEDGMENTS
We express our gratitude to S. Crawford for helpful discussions during the preparation of the manuscript. We are extremely grateful to R. Semiens and J. Alvarado for excellent work in the maintenance of rabbits. The technical assistance of J. Esparza is greatly acknowledged. We thank M. Thouless, Y. Inaba, H. B. Greenberg, Y. Hoshino, G. Gerna, L. J. Saif, S. Urasawa, S. Matsuno, and H. F. Clark for providing rotavirus strains used in this study.
This work was supported by Public Health Service grant AI 24998 from the National Institute of Allergy and Infectious Diseases and by World Health Organization grant MIMV 2718130.
REFERENCES
- 1.Arias C F, Romero P, Alvarez V, López S. Trypsin activation pathway of rotavirus infectivity. J Virol. 1996;70:5832–5839. doi: 10.1128/jvi.70.9.5832-5839.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bohl E H, Theil K W, Saif L J. Isolation and serotyping of porcine rotaviruses and antigenic comparison with other rotaviruses. J Clin Microbiol. 1984;19:105–111. doi: 10.1128/jcm.19.2.105-111.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Borriello S P, Carman R J. Association of iota-like toxin and Clostridium spiroforme with both spontaneous and antibiotic-associated diarrhea and colitis in rabbits. J Clin Microbiol. 1983;17:414–418. doi: 10.1128/jcm.17.3.414-418.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bridger J, Burke B, Beards G M, Desselberger U. The pathogenicity of two porcine rotaviruses differing in their in vitro growth characteristics and genes 4. J Gen Virol. 1992;73:3011–3015. doi: 10.1099/0022-1317-73-11-3011. [DOI] [PubMed] [Google Scholar]
- 5.Broome R L, Vo P T, Ward R L, Clark H F, Greenberg H B. Murine rotavirus genes encoding outer capsid proteins VP4 and VP7 are not major determinants of host range restriction and virulence. J Virol. 1993;67:2448–2455. doi: 10.1128/jvi.67.5.2448-2455.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burns J W, Krishnaney A A, Vo P T, Rouse R V, Anderson L J, Greenberg H B. Analyses of homologous rotavirus infection in the mouse model. Virology. 1995;207:143–153. doi: 10.1006/viro.1995.1060. [DOI] [PubMed] [Google Scholar]
- 7.Burns J W, Chen D, Estes M K, Ramig R F. Biological and immunological characterization of a simian rotavirus SA11 variant with an altered genome segment 4. Virology. 1989;169:427–435. doi: 10.1016/0042-6822(89)90168-2. [DOI] [PubMed] [Google Scholar]
- 8.Castrucci G, Ferrari M, Frigeri F, Cilli V, Perecca L, Donelli G. Isolation and characterization of cytopathic strains of rotavirus from rabbits. Arch Virol. 1985;83:99–104. doi: 10.1007/BF01310967. [DOI] [PubMed] [Google Scholar]
- 9.Chen D, Estes M K, Ramig R F. Specific interactions between rotavirus outer capsid proteins VP4 and VP7 determine expression of a cross-reactive, neutralizing VP4-specific epitope. J Virol. 1992;66:432–439. doi: 10.1128/jvi.66.1.432-439.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ciarlet M, Ludert J E, Liprandi F. Comparative amino acid sequence analysis of the major outer capsid protein (VP7) of porcine rotaviruses with G3 and G5 serotype specificities isolated in Venezuela and Argentina. Arch Virol. 1995;140:437–451. doi: 10.1007/BF01718422. [DOI] [PubMed] [Google Scholar]
- 11.Ciarlet M, Estes M K, Conner M E. Comparative amino acid sequence analysis of the outer capsid protein VP4 from four lapine rotavirus strains reveals identity with genotype P[14] human rotavirus. Arch Virol. 1997;142:1059–1069. doi: 10.1007/s007050050142. [DOI] [PubMed] [Google Scholar]
- 12.Ciarlet, M., S. E. Crawford, C. Barone, A. Bertolotti-Ciarlet, M. K. Estes, and M. E. Conner. Unpublished data.
- 13.Ciarlet, M., M. A. Gilger, C. Barone, M. McArthur, M. K. Estes, and M. E. Conner. Unpublished data. [DOI] [PubMed]
- 14.Clark, H. F., P. A. Offit, R. W. Ellis, D. Krah, A. R. Shaw, J. J. Eiden, M. Pichichero, and J. J. Treanor. 1996. WC3 reassortant vaccines in children. Arch. Virol. 12(Suppl.):187–198. [DOI] [PubMed]
- 15.Conner M E, Estes M K, Graham D Y. Rabbit model of rotavirus infection. J Virol. 1988;62:1625–1633. doi: 10.1128/jvi.62.5.1625-1633.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Conner M E, Gilger M A, Estes M K, Graham D Y. Serologic and mucosal immune response to rotavirus infection in the rabbit model. J Virol. 1991;65:2562–2571. doi: 10.1128/jvi.65.5.2562-2571.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Conner M E, Crawford S E, Barone C, Estes M K. Rotavirus vaccine administered parenterally induces protective immunity. J Virol. 1993;67:6633–6641. doi: 10.1128/jvi.67.11.6633-6641.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Conner M E, Matson D O, Estes M K. Rotavirus vaccines and vaccination potential. Curr Top Microbiol Immunol. 1994;185:286–337. doi: 10.1007/978-3-642-78256-5_10. [DOI] [PubMed] [Google Scholar]
- 19.Conner M E, Crawford S E, Barone C, Estes M K. Rotavirus or virus-like particles administered parenterally induce active immunity. In: Brown F, Chanock R, Ginsberg H, Norrby E, editors. Vaccines 94. Proceedings of Modern Approaches to New Vaccines Including Prevention of AIDS. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1994. pp. 351–355. [Google Scholar]
- 20.Conner, M. E., C. D. Zarley, B. Hu, S. Parsons, D. Drabinski, S. Greiner, R. Smith, B. Jiang, B. Corsaro, V. Barniak, H. P. Madore, S. E. Crawford, and M. K. Estes. 1996. Virus-like particles as a rotavirus subunit vaccine. J. Infect. Dis. 174(Suppl. 1):S88–S92. [DOI] [PubMed]
- 21.Conner M E, Ramig R F. Enteric diseases. In: Nathanson N, Ahmed R, González-Scarano F, Griffin D E, Homes K V, Murphy F A, Robinson H L, editors. Viral pathogenesis. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 713–743. [Google Scholar]
- 22.Conner M E, Graham D Y, Estes M K. Determination of the duration of a primary immune response and the ID50 of ALA rabbit rotavirus in rabbits. Arch Virol. 1997;142:2281–2294. doi: 10.1007/s007050050242. [DOI] [PubMed] [Google Scholar]
- 23.Dunn S J, Burns J W, Cross T L, Vo P T, Ward R L, Bremont M, Greenberg H B. Comparison of VP4 and VP7 of five murine rotavirus strains. Virology. 1994;203:250–259. doi: 10.1006/viro.1994.1482. [DOI] [PubMed] [Google Scholar]
- 24.Dunn S J, Cross T L, Greenberg H B. Comparison of the rotavirus nonstructural protein NSP1 (NS53) from different species by sequence analysis and Northern blot hybridization. Virology. 1994;203:178–183. doi: 10.1006/viro.1994.1471. [DOI] [PubMed] [Google Scholar]
- 25.Espejo R T, López S, Arias C F. Structural polypeptides of simian rotavirus SA11 and the effect of trypsin. J Virol. 1981;37:156–160. doi: 10.1128/jvi.37.1.156-160.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Estes M K, Graham D Y, Smith E M, Gerba C P. Rotavirus stability and inactivation. J Gen Virol. 1979;43:403–409. doi: 10.1099/0022-1317-43-2-403. [DOI] [PubMed] [Google Scholar]
- 27.Estes M K, Graham D Y, Ramig R F, Ericson B L. Heterogeneity in the structural glycoprotein (VP7) of simian rotavirus SA11. Virology. 1982;122:8–14. doi: 10.1016/0042-6822(82)90372-5. [DOI] [PubMed] [Google Scholar]
- 28.Estes M K. Rotaviruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields Virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 1625–1655. [Google Scholar]
- 29.Feng N, Burns J W, Bracey L, Greenberg H B. Comparisons of the mucosal and systemic humoral immune responses and subsequent protection in mice orally inoculated with homologous or heterologous rotaviruses. J Virol. 1994;68:7766–7773. doi: 10.1128/jvi.68.12.7766-7773.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Feng N, Yu J, Vo P T, Chung D, Vo V, Greenberg H B. Abstracts of the 15th Annual Meeting of the American Society for Virology. 1996. Genetics of attenuated murine rotavirus mucosal immunogenicity in mouse, abstr. W29-9; p. 134. [Google Scholar]
- 31.Franco M A, Feng N, Greenberg H B. Molecular determinants of immunity and pathogenicity of rotavirus infection in the mouse model. J Infect Dis. 1996;174:S47–S50. doi: 10.1093/infdis/174.supplement_1.s47. [DOI] [PubMed] [Google Scholar]
- 32.Gerna G, Sears J, Hoshino Y, Steele A D, Nakagomi O, Sarasini A, Flores J. Identification of a new VP4 serotype of human rotaviruses. Virology. 1994;200:66–71. doi: 10.1006/viro.1994.1163. [DOI] [PubMed] [Google Scholar]
- 33.Gombold J L, Ramig R F. Analysis of reassortment of genome segments in mice mixedly infected with rotavirus SA11 and RRV. J Virol. 1986;57:110–116. doi: 10.1128/jvi.57.1.110-116.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Greenberg H B, Flores J, Kalica A R, Wyatt R G, Jones R. Gene coding assignments for growth restriction neutralization and subgroup specificities of the Wa and DS-1 strains of human rotavirus. J Gen Virol. 1983;64:313–320. doi: 10.1099/0022-1317-64-2-313. [DOI] [PubMed] [Google Scholar]
- 35.Greenberg H B, Vo P T, Jones R. Cultivation and characterization of three strains of murine rotavirus. J Virol. 1986;57:585–590. doi: 10.1128/jvi.57.2.585-590.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hambraeus B A M, Hambraeus L E J, Wadell G. Animal model of rotavirus infection in rabbits—protection obtained without shedding of viral antigen. Arch Virol. 1989;107:237–251. doi: 10.1007/BF01317920. [DOI] [PubMed] [Google Scholar]
- 37.Hoshino Y, Saif L J, Kang S-Y, Sereno M M, Chen W-K, Kapikian A Z. Identification of group A rotavirus genes associated with virulence of a porcine rotavirus and host range restriction of a human rotavirus in the gnotobiotic piglet model. Virology. 1995;209:274–280. doi: 10.1006/viro.1995.1255. [DOI] [PubMed] [Google Scholar]
- 38.Hua J, Mansell E A, Patton J T. Comparative analysis of the NS53 gene: conservation of the basic and cysteine-rich regions in the protein and possible stem-loop structures in the RNA. Virology. 1993;196:372–378. doi: 10.1006/viro.1993.1492. [DOI] [PubMed] [Google Scholar]
- 39.Iša P, Snodgrass D R. Serological and genomic characterization of equine rotavirus VP4 proteins identifies three different P serotypes. Virology. 1994;201:364–372. doi: 10.1006/viro.1994.1302. [DOI] [PubMed] [Google Scholar]
- 40.Kalica A R, Flores J, Greenberg H B. Identification of the rotaviral gene that codes for hemagglutination and protease-enhanced plaque formation. Virology. 1983;125:194–205. doi: 10.1016/0042-6822(83)90073-9. [DOI] [PubMed] [Google Scholar]
- 41.Kojima K, Taniguchi K, Kobayashi N. Species-specific and interspecies relatedness of NSP1 sequences in human, porcine, bovine, feline, and equine rotavirus strains. Arch Virol. 1996;141:1–12. doi: 10.1007/BF01718584. [DOI] [PubMed] [Google Scholar]
- 41a.Lee, K. K., M. Ciarlet, and M. E. Conner. Unpublished data.
- 42.Malherbe H H, Strickland-Cholmley M. Simian virus SA11 and the related O agent. Arch Gesamte Virusforsch. 1967;22:235–245. doi: 10.1007/BF01240518. [DOI] [PubMed] [Google Scholar]
- 43.Mattion N M, Cohen J, Estes M K. Rotavirus proteins. In: Kapikian A Z, editor. Viral infections of the gastrointestinal tract. 2nd ed. New York, N.Y: Marcel Dekker Inc.; 1994. pp. 169–249. [Google Scholar]
- 44.McNeal M M, Broome R L, Ward R L. Active immunity against rotavirus infection in mice is correlated with viral replication and titers of serum rotavirus IgA following vaccination. Virology. 1994;204:642–650. doi: 10.1006/viro.1994.1579. [DOI] [PubMed] [Google Scholar]
- 45.McNeal M M, Ward R L. Long-term production of rotavirus antibody and protection against reinfection following a single infection of neonatal mice with murine rotavirus. Virology. 1995;211:474–480. doi: 10.1006/viro.1995.1429. [DOI] [PubMed] [Google Scholar]
- 46.Méndez E, Arias C F, López S. Binding to sialic acids is not an essential step for the entry of animal rotaviruses to epithelial cells in culture. J Virol. 1993;67:5253–5259. doi: 10.1128/jvi.67.9.5253-5259.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Midthun K, Greenberg H B, Hoshino Y, Kapikian A Z, Wyatt R G, Chanock R M. Reassortant rotaviruses as potential live rotavirus vaccine candidates. J Virol. 1985;53:949–954. doi: 10.1128/jvi.53.3.949-954.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Midthun K, Hoshino Y, Kapikian A Z, Chanock R M. Single gene substitution rotavirus reassortants containing the major neutralization protein (VP7) of human rotavirus serotype 4. J Clin Microbiol. 1986;24:822–826. doi: 10.1128/jcm.24.5.822-826.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nisonoff A, Hopper J E, Spring S B. The rabbit immunoglobulins. In: Dixon F J, Kunkel H G, editors. The antibody molecule. New York, N.Y: Academic Press; 1975. pp. 321–331. [Google Scholar]
- 50.Offit P A, Clark H F, Kornstein M J, Plotkin S A. A murine model for oral infection with a primate rotavirus (simian SA11) J Virol. 1984;51:233–236. doi: 10.1128/jvi.51.1.233-236.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Offit P A, Blavat G, Greenberg H B, Clark H F. Molecular basis of rotavirus virulence: role of gene segment 4. J Virol. 1986;57:46–49. doi: 10.1128/jvi.57.1.46-49.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pereira H G, Azeredo R S, Fialho A M, Vidal M N P. Genomic heterogeneity of simian rotavirus SA11. J Gen Virol. 1984;65:815–818. doi: 10.1099/0022-1317-65-4-815. [DOI] [PubMed] [Google Scholar]
- 53.Petric M, Middletown P J, Grant C, Tam J S, Hewitt C M. Lapine rotavirus: preliminary studies on epizoology and transmission. Can J Comp Med. 1978;42:143–147. [PMC free article] [PubMed] [Google Scholar]
- 54.Ramig R F. Isolation and genetic characterization of temperature-sensitive mutants of simian rotavirus SA11. Virology. 1982;120:93–105. doi: 10.1016/0042-6822(82)90009-5. [DOI] [PubMed] [Google Scholar]
- 55.Ramig R F. The effects of host age, virus dose, and virus strain on heterologous rotavirus infection of suckling mice. Microb Pathog. 1988;4:189–202. doi: 10.1016/0882-4010(88)90069-1. [DOI] [PubMed] [Google Scholar]
- 56.Sato K, Inaba Y, Miura Y, Tokuhisa S, Matumoto M. Antigenic relationships between rotaviruses from different species as studied by neutralization and immunofluorescence. Arch Virol. 1982;73:45–50. doi: 10.1007/BF01341726. [DOI] [PubMed] [Google Scholar]
- 57.Sereno M M, Gorziglia M I. The outer capsid protein VP4 of murine rotavirus strain Eb represents a tentative new P type. 1994. Virology. 1994;199:500–504. doi: 10.1006/viro.1994.1153. [DOI] [PubMed] [Google Scholar]
- 58.Snodgrass D R, Iša P. Serological and genomic characterization of the VP4 spike protein of equine rotavirus. Equine Infect Dis. 1994;7:71–77. doi: 10.1006/viro.1994.1302. [DOI] [PubMed] [Google Scholar]
- 59.Tajima T, Suzuki E, Ushijima H, Araki K, Kim B, Shinozaki T, Fujii R. Isolation of murine rotavirus in cell cultures: brief report. Arch Virol. 1984;82:119–123. doi: 10.1007/BF01309375. [DOI] [PubMed] [Google Scholar]
- 60.Taniguchi K, Urasawa T, Urasawa S. Species specificity and interspecies relatedness in VP4 genotypes demonstrated by VP4 sequence analysis of equine, feline, and canine rotavirus strains. Virology. 1994;200:390–400. doi: 10.1006/viro.1994.1203. [DOI] [PubMed] [Google Scholar]
- 61.Thouless M E, DiGiacomo R F, Deeb B J, Howard H. Pathogenicity of rotavirus in rabbits. J Clin Microbiol. 1988;26:943–947. doi: 10.1128/jcm.26.5.943-947.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ushijima H, Morikawa S, Hasegawa A, Mukoyama A, Suzuki E, Yamamoto T, Nishio O. Abstracts of the Fifth International Symposium of dsRNA Viruses. 1995. Characterization of VP4 and VP7 of murine rotavirus (YR-1) isolated in Japan, abstr. P08; p. 37. [Google Scholar]
- 63.Ward L, Rosen B I, Yuan L, Saif L J. Pathogenesis of an attenuated and a virulent strain of group A human rotavirus in neonatal gnotobiotic pigs. J Gen Virol. 1996;77:1431–1441. doi: 10.1099/0022-1317-77-7-1431. [DOI] [PubMed] [Google Scholar]
- 64.Wyatt R G, James W D, Bohl E H, Theil K W, Saif L J, Kalica A R, Greenberg H B, Kapikian A Z, Chanock R M. Human rotavirus type 2: cultivation in vitro. Science. 1980;207:189–191. doi: 10.1126/science.6243190. [DOI] [PubMed] [Google Scholar]
- 65.Xu L, Tian Y, Tarlow O, Harbour D, McCrae M A. Molecular biology of rotaviruses. IX. Conservation and divergence in genome segment 5. J Gen Virol. 1994;75:3413–3421. doi: 10.1099/0022-1317-75-12-3413. [DOI] [PubMed] [Google Scholar]