Abstract
We have identified a rat cytomegalovirus (RCMV) gene that encodes a G-protein-coupled receptor (GCR) homolog. This gene (R33) belongs to a family that includes the human cytomegalovirus UL33 gene. R33 was found to be transcribed during the late phase of RCMV infection in rat embryo fibroblasts. Unlike the mRNAs from all the other members of the UL33 family that have been studied to date, the R33 mRNA is not spliced. To study the function of the R33 gene, we constructed an RCMV strain in which the R33 open reading frame is disrupted. The mutant strain (RCMVΔR33) did not show differences in replication from wild-type RCMV upon infection of several rat cell types in vitro. However, marked differences were seen between the mutant and wild-type strain in the pathogenesis of infection in immunocompromised rats. First, the mutant strain induced a significantly lower mortality than the wild-type virus did. Second, in contrast to wild-type RCMV, the mutant strain did not efficiently replicate in the salivary gland epithelial cells of immunocompromised rats. Although viral DNA was detected in salivary glands of RCMVΔR33-infected rats up to 14 days postinfection, it could not be detected at later time points. This indicates that although the strain with R33 deleted is probably transported to the salivary glands in a similar fashion to that for wild-type virus, the mutant virus is not able to either enter or replicate in salivary gland epithelial cells. We conclude that the RCMV R33 gene plays a vital role in the pathogenesis of infection.
G-protein-coupled receptors (GCRs) are proteins that have a crucial function in signal transduction through cell membranes. Upon interaction with extracellular ligands, GCRs transduce a signal into the cell by activating a cascade of cellular processes, which is initiated by the activation of GTP-binding proteins (G proteins). Interestingly, GCRs are encoded not only by eukaryotic and prokaryotic genes but also by genes of viruses. To date, 18 putative GCR genes have been discovered within viral genomes: 16 of these genes are located within the genomes of beta- and gammaherpesviruses (17, 20, 27, 40, 41, 44, 47), and 2 are located within poxviruses (15, 35). The functions of any of these GCRs in the pathogenesis of viral disease are yet unclear.
Within the genome of human cytomegalovirus (HCMV), four genes that encode GCR homologs were identified: UL33, UL78, US27, and US28 (19, 27). The amino acid sequences derived from the last two genes were found to have the highest sequence similarity to cellular chemokine receptors (22, 25). In addition, the US28 translation product is capable of binding β-chemokines in vitro, hence triggering the mobilization of intracellular Ca2+ (32). Due to the species specificity of HCMV, it is difficult to study the function of the US27 and US28 genes in vivo. Moreover, these genes do not seem to have any counterparts within the genomes of other herpesviruses. In contrast, UL33- and UL78-like genes are conserved among all betaherpesviruses. The function of these genes can therefore be studied in vivo. Genes similar to UL33 and UL78 have been found within the genomes of murine cytomegalovirus (MCMV) (M33 and M78, respectively [44]), human herpesvirus 6 (HHV-6) (U12 and U51 respectively [27]), and human herpesvirus 7 (HHV-7) (U12 and U51, respectively [41]). Although the positions of UL78-like genes within the betaherpesvirus genomes are conserved, their sequences are highly divergent (27, 41, 44). In contrast, both the genome location and sequence of UL33-like genes are conserved among all betaherpesviruses studied to date (27, 41, 44). The HCMV UL33 gene was found to be expressed both in membranes of cultured fibroblasts and in viral envelopes (34). Both the HCMV UL33 and MCMV M33 genes are dispensable for viral replication in vitro in human and murine fibroblasts, respectively (23, 34). Recombinant MCMV strains that lack a functional M33 gene were also examined in vivo (23). In contrast to wild-type MCMV, recombinant MCMV could not be recovered from the salivary glands of infected mice. In addition, recombinant MCMV was found not to replicate after direct administration of the virus into the salivary glands (23). Although these data clearly provided evidence for an important role of UL33-like GCRs in salivary gland tropism, it remained unclear whether these receptors play a role in the dissemination of virus to various target organs. It is also unknown whether the UL33-like genes have a function in the pathogenesis of infection and mortality among hosts.
To gain more insight in the function of UL33-like genes, we set out to identify a UL33 homolog within the rat cytomegalovirus (RCMV) genome. Here we present the sequence and transcriptional analysis of this gene, which we termed R33. To investigate the role of R33 in the pathogenesis of RCMV infection, an RCMV strain that does not contain a functional R33 gene was generated. Although disruption of the R33 open reading frame (ORF) did not affect RCMV replication in different permissive cell types in vitro, it dramatically reduced the mortality among a group of RCMV-infected rats. We found that although the mutant virus was transported to the salivary glands of infected rats, the virus was not able to either enter or replicate in these glands. Taken together, these findings indicate that R33 is important not only for replication of RCMV in salivary glands but also for the pathogenesis of RCMV infection in immunocompromised rats.
MATERIALS AND METHODS
Cells and virus.
Primary rat embryo fibroblasts (REF), rat heart endothelium cell line 116 (RHEC), and monocyte/macrophage cell line R2 (MΦ) were cultured as described previously (11, 21, 54). RCMV (Maastricht strain) was propagated in REF (11). Virus titers were determined by a plaque assay by standard procedures (11). RCMV DNA was isolated from culture medium as described by Vink et al. (53).
Identification, cloning and sequence analysis of the RCMV R33 gene.
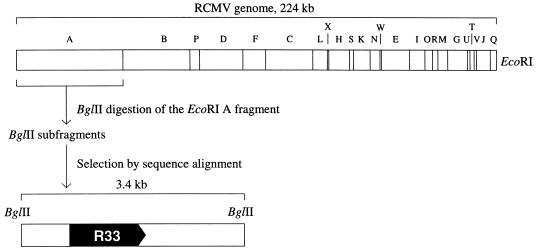
Approximately 5 μg of RCMV virion DNA was digested with EcoRI. After separation through a 0.6% low-melting-point agarose gel, the EcoRI A fragment, which is approximately 50 kb (36), was excised and purified. This fragment (Fig. 1) was subsequently digested with BglII, and the resulting fragments were cloned into the BamHI site of vector pUC119 and (partially) sequenced. The sequences were checked for the presence of HCMV UL33-homologous regions by alignment with the EMBL nucleic acid sequence database (EMBL, Heidelberg, Germany) with the FASTA software (42). Thus, a 3.4-kb RCMV BglII fragment that showed significant similarity to the HCMV UL33 gene was identified. Both strands of the 3.4-kb BglII fragment were sequenced with a T7 sequencing kit (Pharmacia Biotech, Roosendaal, The Netherlands). All sequence information was generated with overlapping plasmid constructs. Sequence analysis was performed with the Geneskipper software package (EMBL).
FIG. 1.
Restriction map of the RCMV genome (36) and the relative position of the R33 gene, which encodes a putative G protein-coupled receptor (pR33). The 3.4-kb BglII fragment that contains the R33 ORF is indicated below the genome map.
Recombination plasmid construction.
A 2.8-kb BamHI-BglII RCMV fragment containing the R33 ORF was cloned into the BamHI site of pUC119, resulting in construct p026 (Fig. 2). A recombination plasmid (p031) was constructed by replacing the 0.5-kb MluNI fragment within the R33 ORF of p026 with a 1.5-kb DNA fragment that contains the neomycin resistance gene (neo). The 1.5-kb fragment was derived from plasmid Rc/CMV (Invitrogen, Leek, The Netherlands) by digestion with BamHI and EcoRI followed by incubation with deoxynucleoside triphosphates (dNTPs) and DNA polymerase I Klenow fragment (Pharmacia Biotech) to create blunt ends. The neo ORF is flanked by a simian virus 40 (SV40) early promoter and an SV40 polyadenylation signal.
FIG. 2.
Nucleotide sequence of the R33 gene, and predicted amino acid sequence of the pR33 peptide. The open boxes indicate seven putative transmembrane domains (tm1 to tm7) and a putative N-linked glycosylation site (NXT/S). Charged amino acid residues in the N-terminal (extracellular) region and the third intracellular region (between tm5 and tm6) are enclosed in open squares. The charges of these residues are printed at the top right of each square. Black boxes indicate consensus sequences [(S/T)X(K/R)], of which the S/T residue might be phosphorylated by protein kinase C. The amino acid residues that are conserved among all pUL33-like proteins (Fig. 3) are encircled. The residues that are conserved between chemokine receptors (Fig. 3) are enclosed in black circles. The underlined nucleotide sequences indicate sequences identical or complementary to the sequences of oligonucleotides that were used in RT-PCR (Fig. 5B).
Generation of an RCMV R33 null mutant.
Approximately 107 REF were trypsinized and subsequently centrifuged for 5 min at 500 × g. The cells were resuspended in 0.25 ml of culture medium, after which 10 μg of XbaI-KpnI-digested plasmid p031 was added. The suspension was transferred to a 0.4-cm electroporation cuvette (Bio-Rad, Veenendaal, The Netherlands), and pulsed at 0.25 kV and 500 μF in a Bio-Rad Gene Pulser electroporator. The cells were subsequently seeded in 10-cm culture dishes. At 6 h after transfection, the cells were infected with RCMV at a multiplicity of infection (MOI) of 1. The culture medium was supplemented with 50 μg of G418 per ml at 16 h postinfection (p.i.). When transfected and infected REF cultures showed extensive cytopathic effect, the tissue culture medium was transferred to a fresh subconfluent REF monolayer. At 1 h after transfer, the culture medium was refreshed and supplemented with G418. The procedure of virus propagation under G418 selection and transfer of culture medium to fresh REF monolayers was repeated twice. Subsequently, recombinant virus (RCMVΔR33) was purified by two rounds of plaque purification.
Southern blot hybridization.
Both wild-type RCMV and RCMVΔR33 DNA were isolated, digested with BamHI, electrophoresed through a 1% agarose gel, and blotted onto a Hybond N+ nylon membrane (Amersham, ’s Hertogenbosch, The Netherlands) as described previously (8). Both a 2.4-kb BamHI fragment, containing the intact R33 ORF (R33 probe [Fig. 2]), and a 1.5-kb BamHI-EcoRI fragment containing the neo gene (neo probe [Fig. 2]) were used as probes. Hybridization and detection experiments were performed with digoxigenin DNA-labeling and chemoluminescence detection kits (Boehringer Mannheim, Almere, The Netherlands).
Isolation of poly(A)+ RNA and Northern blot analysis.
RCMV poly(A)+ RNA was isolated at immediate-early (IE), early (E), and late (L) times of infection of REF. To obtain IE mRNA, REF were treated with 100 μg of cycloheximide per ml 1 h before, during, and 16 h after infection. During the 1-h infection period, the cells were exposed to RCMV at an MOI of 1. E mRNA was isolated after infection of REFs with RCMV (MOI = 1) and treatment of the cells with 100 μg of phosphonoacetic acid per ml from 3 h p.i. until the cells were harvested at 13 h p.i. L mRNA was isolated after infection of REF with either RCMV or RCMVΔR33 (MOI = 0.01) and harvesting of cells at 72 h p.i. To obtain mRNA from mock-infected (M) cells, a similar procedure to that described for the purification of L mRNA was used, except that RCMV infection was omitted. mRNA was purified with a QuickPrep Micro mRNA purification kit (Pharmacia Biotech). Aliquots (1 μg) of mRNA were electrophoresed through agarose under denaturing conditions, as described by Brown and Mackey (9). Then the RNA was transferred to Hybond N membranes (Amersham) as described previously (9). The 402-bp XbaI-BamHI fragment, 960-bp SacI fragment, and 550-bp KpnI fragment from p026 (see Fig. 7) were used to generate probes. These fragments contain R32, R33, and R34-specific sequences, respectively (Fig. 2). Labeling with [α-32P]dATP (ICN, Zoetermeer, The Netherlands) was carried out with a random primed DNA labeling kit (Boehringer Mannheim). Hybridization and autoradiography were carried out as described previously (9).
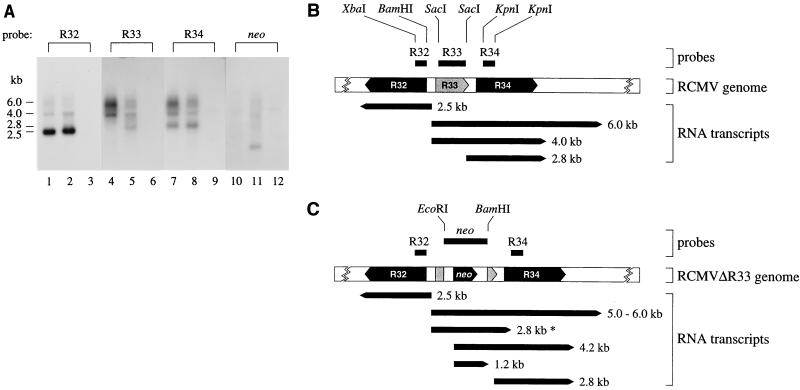
FIG. 7.
The expression of genes neighboring the R33 gene is not affected by disruption of R33. (A) To determine whether transcription of the R32 and R34 genes was affected by disruption of R33, transcription of the R33 region of the genomes of both RCMV and RCMVΔR33 was analyzed by Northern blot hybridization, using probes specific for R32, R33, R34, and neo. The figure shows autoradiographs in which lanes 1, 4, 7, and 10 represent poly(A)+ RNA from RCMV-infected REF, lanes 2, 5, 8, and 11 represent poly(A)+ RNA from RCMVΔR33-infected REF, and lanes 3, 6, 9, and 12 represent poly(A)+ RNA from mock-infected REF. The estimated lengths of the transcripts are indicated on the left in kilobases. (B) Estimated lengths and positions of transcripts from the RCMV R32, R33, and R34 genes, as derived from panel A. The lengths of the indicated R32 and R34 ORFs are estimated and based on the lengths of MCMV M32 and M34 (44); the complete DNA sequence of this region of the RCMV genome is not yet available. (C) Estimated lengths and positions of transcripts from the RCMVΔR33 R32, ’ΔR33’, R34, and neo genes, as derived from panel A. Both the 2.8-kb∗ and 4.2-kb RNA transcripts hybridize to the R33, R34 and neo probes. It is not known whether the 5′ ends of these transcripts map to the 5′ end of the R33 gene or the 5′ end of the neo gene.
RT-PCR.
L mRNA was treated with DNase I (Pharmacia Biotech) and subsequently reverse transcribed with a superscript plasmid system for cDNA synthesis and plasmid cloning (Gibco BRL, Breda, The Netherlands). The following primers (obtained from Eurogentec, Seraing, Belgium) were used for amplification of the cDNA: 5′-GATCGGATCCATGAGGGTGATTGAAGAGATTCGG-3′ (the sequence in italics is located at positions 532 to 555 in Fig. 2) and 5′-CGTAAAGCTTAGTCCCTCGCCACCGACAGG-3′ (the complement of the sequence in italics is located at positions 815 to 833 in Fig. 2). PCR mixtures contained 10 mM Tris-HCl (pH 9.0), 50 mM KCl, 1.5 mM MgCl2, 0.01% Triton X-100, 0.2 mM each dNTP, 0.5 μM each oligonucleotide primer, 0.1 U of Taq DNA polymerase (Pharmacia Biotech) per μl, and (i) H2O (negative control), (ii) first-strand cDNA synthesis reaction mixture from which reverse transcriptase (RT) was omitted (negative control), (iii) 1 μl of 10-fold-diluted first-strand DNA synthesis reaction mixture, or (iv) 1 ng of genomic RCMV DNA (positive control). The reaction tubes were placed in a GeneAmp PCR System 9600 thermal cycler (Perkin-Elmer, Nieuwerkerk aan de IJssel, The Netherlands), which was programmed to incubate samples for 5 min at 95°C followed by 35 cycles of 60 s at 95°C, 60 s at 58°C, and 60 s at 72°C. Finally, the tubes were incubated for 10 min at 72°C. The PCR products were analyzed by agarose gel electrophoresis and stained with ethidium bromide.
Replication of RCMVΔR33 in vitro.
REF, RHEC, and MΦ were grown in 96-well plates and infected with either RCMV or RCMVΔR33 at an MOI of either 0.1 or 1.0. Culture medium samples (three per virus) were taken at 1, 3, 5, and 7 days p.i. and subjected to plaque titer determination. The cells were fixed and stained with monoclonal antibodies (MAb) against RCMV E proteins (MAb RCMV 8 [12]) as described previously (54). The degree of infection was determined by counting the number of antigen-positive cells relative to the total number of cells in six different wells (four microscopic fields per well at a magnification of ×400).
Survival of RCMV-infected and RCMVΔR33-infected rats.
Male specific-pathogen-free Lewis/N RT1 rats (Central Animal Facility, Maastricht University, Maastricht, The Netherlands) were kept under standard conditions (45). Six-week-old rats (body weights ranging from 140 to 180 g) were divided into four groups of five rats. The rats were immunosuppressed by 5 Gy of total-body Röntgen irradiation 1 day before infection, as described by Stals et al. (45). Intraperitoneal infection was carried out with either 1 × 106 PFU of RCMV, 1 × 106 PFU of RCMVΔR33, 5 × 106 PFU of RCMV, or 5 × 106 PFU of RCMVΔR33. Virus stocks were derived from tissue culture medium of virus-infected REF. The number of surviving rats was recorded daily until day 28 p.i., when the surviving rats were sacrificed. Several internal organs of these rats were subjected to plaque assays as described below.
Pathogenesis of infection with RCMVΔR33.
Two groups of 15 male specific-pathogen-free Lewis/N RT1 rats (10 weeks old, with a body weight of 250 to 300 g, immunosuppressed 1 day before infection) were infected with 5 × 106 PFU of either RCMV or RCMVΔR33. On days 3, 5, 7, 10, and 14 p.i., three rats from each group were sacrificed and their internal organs were collected. These organs were subjected to both plaque assay and immunohistochemistry (10). Tissue sections (4 μm) of the (submaxillary) salivary gland, spleen, kidney, liver, lung, heart, and pancreas were stained with MAb RCMV 8 (which detects E-phase-expressed RCMV polypeptides in the nuclei of RCMV-infected cells [12]). Frozen sections (4 μm) of salivary gland, liver, and pancreas tissue from rats sacrificed on days 7 and 14 p.i. were stained with either MAb 341 (which detects CD8+ cells [49]), MAb R73 (which detects TCRαβ+ cells [29]), MAb W3/25 (which detects CD4+ cells; Serotec, Oxford, United Kingdom), MAb ED1 (which detects inflammatory macrophages [24]), MAb ED2 (which detects resident tissue macrophages [24]), MAb 323 (which detects natural killer cells [18]), or MAb OX6 (which detects class II major histocompatibility complex [MHC] proteins; Sanbio B.V., Volendam, The Netherlands).
PCR.
Total cellular DNA was extracted from the spleen, kidney, liver, lung, heart, and pancreas with a DNA extraction kit (Gull Laboratories, Salt Lake City, Utah), and DNA concentrations were determined by spectrophotometry. The DNA samples were serially diluted from 100 to 10−8 μg. Each of the diluted DNA samples was incubated for 10 min at 95°C, immediately cooled on ice, and included in a two-round PCR. In the first PCR, primers (obtained from Eurogentec) that hybridized with the RCMV DNA polymerase gene were used (6). The sequences of the primers are 5′-AAGGGATCCGATTTCGCCAGCCTCTACC-3′ (in which the sequence in italics represents nucleotides 11726 to 11744 of GenBank accession no. U50550) and 5′-AAGGGATCCTGTCGGTGTCCCCGTACAC-3′ (in which the sequence in italics represents the sequence complementary to nucleotides 12221 to 11239 of GenBank accession no. U50550). The use of these primers results in a product of 536 bp. The nested PCR results in a product of 431 bp, with primers 5′-AAGGGATCCCCTCTGTTACTCCACCCTGC-3′ (in which the sequence in italics represents nucleotides 11767 to 11786 of GenBank accession no. U50550) and 5′-TTCGGATCCACGCCGACCTCGGAGACCAG-3′ (in which the sequence in italics represents the sequence complementary to nucleotides 12158 to 12177 of GenBank accession no. U50550). The reaction mixtures (50 μl) contained diluted target DNA, 80 μM each dNTP, 0.37 μM each oligonucleotide primer, 1 U of Taq DNA polymerase (Pharmacia Biotech), and Taq DNA polymerase reaction buffer (Pharmacia Biotech). The reaction tubes were placed in a GeneAmp PCR System 9600 thermal cycler that was programmed to incubate samples for 150 s at 95°C, 30 s at 58°C, and 60 s at 72°C followed by 35 cycles of 30 s at 95°C, 30 s at 58°C, and 60 s at 72°C. Finally, the tubes were incubated for 10 min at 72°C. A 5-μl portion of each reaction mixture from the initial PCR was transferred to 45 μl of a nested-PCR mixture. Nested PCR was run immediately after completion of the initial PCR. The PCR products were analyzed by agarose gel electrophoresis followed by staining with ethidium bromide.
GenBank accession number.
The nucleotide and amino acid sequences discussed in this paper have been deposited in the GenBank database under accession no. U91788.
RESULTS
Identification, cloning, and sequence analysis of the RCMV R33 gene.
To identify a putative RCMV homolog of the HCMV UL33 gene, we hypothesized that this homolog would be located at a position within the RCMV genome analogous to the position of UL33 within the HCMV genome. This hypothesis was based on the observation that the genomes of HCMV and RCMV are largely colinear (6, 53a). Previously, we identified the RCMV homolog of the HCMV UL54 gene (6). In the HCMV genome, the UL33 gene is approximately 35 kb distant from UL54. If the RCMV homologs of the UL33 and UL54 genes are separated by a similar distance, the UL33-homologous gene would be situated near the center of the EcoRI A fragment of the RCMV genome (Fig. 1) (6, 36). To localize the putative UL33-like RCMV gene, we purified the RCMV EcoRI A fragment and subjected it to digestion with BglII. The resulting fragments were cloned, and their sequences were determined. The presence of UL33-like sequences was investigated by alignment of the various sequences with the EMBL nucleic acid sequence database. Thus, a 3.4-kb BglII fragment that showed considerable similarity to the HCMV UL33 gene was identified. The 3.4-kb fragment was found to contain an ORF that putatively encodes a 387-amino-acid polypeptide with a predicted molecular mass of 43.2 kDa. The amino acid sequence of this polypeptide was highly similar to that of the proteins that were predicted to be encoded by the MCMV M33 gene (44) and the HCMV UL33 gene (20) (65 and 40% identity, respectively, according to a global alignment protocol by Myers and Miller [38]). This indicated that the RCMV homolog of these genes had indeed been cloned. By analogy to the nomenclature of the corresponding HCMV and MCMV genes, the RCMV UL33-like gene was termed R33.
To investigate whether the predicted amino acid sequence of the R33-derived protein (pR33) possesses features that are characteristic of GCRs, the pR33 amino acid sequence was analyzed with the computer program TMpred (ISREC Bioinformatics Group, Epalinges, Switzerland). This program can predict potential transmembrane regions in amino acid sequences by comparing the hydrophobicity profiles of these sequences with those of existing transmembrane domains. As expected, computation revealed an extracellular N terminus and seven potential transmembrane regions, each of which might be folded as an α-helix. In addition, several amino acid residues that are conserved among most GCRs were identified (reviewed by Probst et al. [43]): two cysteine residues at positions 106 and 187 (Fig. 2), which may form a disulfide bridge, and a conserved arginine-tyrosine motif (RY, positions 31 and 32) which might be involved in G-protein coupling. Within the fourth cytoplasmic domain of the R33-derived protein is located a cysteine residue at position 319. It was found for several other GCRs that cysteine residues within this cytoplasmic domain are substrates for palmitoylation (43). Another interesting feature of the pR33 amino acid sequence is the presence of two regions near the C terminus, which might be phosphorylated by protein kinase C [phosphorylation site consensus (S/T)X(K/R), at positions 321 to 323 and 326 to 328]. An unusual stretch of 11 prolines is found near the C terminus of pR33. The biological relevance of this large proline stretch is unknown.
In addition to features that are conserved among GCRs in general, the R33-derived amino acid sequence shows several features characteristic of chemokine receptors (1, 23): (i) an N-linked glycosylation site (NYT, at positions 20 to 22) and several negatively charged amino acid residues located in the extracellular N-terminal region; (ii) two cysteine residues (Cys23 and Cys277), which are likely to form a disulfide bridge, thereby joining the N-terminal region with the fourth extracellular loop; (iii) several positively charged amino acid residues within the third intracellular loop; (iv) invariant amino acids within the transmembrane regions (these amino acid residues are indicated by solid circles in Fig. 2); and (v) several serine and threonine residues in the intracellular C-terminal region (Fig. 2). UL33-like GCRs also have conserved amino acid residues in or near the transmembrane regions, which are not present in typical chemokine receptors (these residues are indicated by open circles in Fig. 2).
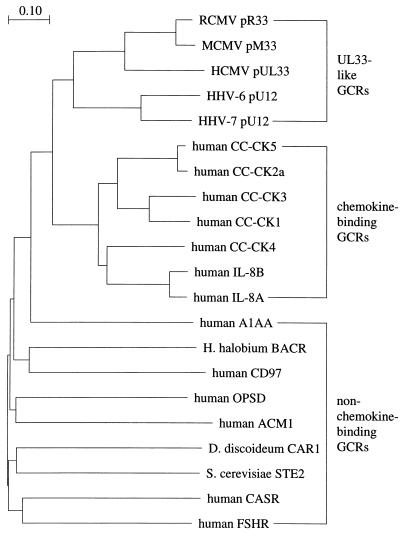
To further investigate the relationship between pR33 and chemokine receptors, we compared the amino acid sequences of the betaherpesvirus-encoded pUL33-like GCRs with sequences from a representative set of mammalian chemokine receptors as well as nine non-chemokine-binding GCRs. The entire sequence of each predicted protein was used in a CLUSTAL W multiple alignment (48). As expected, the pR33 polypeptide was found to have the highest similarity to other pUL33-like GCRs. The amino acid residues that are conserved among this group of virus-encoded GCRs are circled in Fig. 2. Many residues present in pUL33-like GCRs can also be found in chemokine receptors (enclosed in solid circles in Fig. 2). Most of the conserved residues are localized within the predicted seventh transmembrane region (tm7) of pR33. By using the regions of the aligned GCR sequences that correspond to this transmembrane region, a phylogenetic tree was calculated (Fig. 3). As shown in Fig. 3, a group of GCRs that includes the proteins encoded by the RCMV R33, MCMV M33, HCMV UL33, HHV-6 U12, and HHV-7 U12 genes can be distinguished. A second group represents the complete set of human chemokine receptors. These two groups can clearly be distinguished from the remaining (non-chemokine-binding) GCRs. The phylogenetic tree indicates that the UL33-like GCRs have a higher similarity to chemokine-binding receptors than to other, non-chemokine-binding GCRs.
FIG. 3.
pR33 is a member of the chemokine receptor-like GCR family. To compare the sequences of UL33-like GCRs, chemokine receptors, and non-chemokine-binding GCRs, a phylogenetic tree was calculated. The tree is based on a multiple alignment of amino acid sequences that are colinear with the putative seventh transmembrane region (amino acids 288 to 307) of pR33 (Fig. 2). CLUSTAL W pairwise alignment (48) was set to BLOSUM30 protein weight matrix, gap open penalty = 10, and gap extension penalty = 0.1. Multiple alignment was set to BLOSUM series, gap opening penalty = 10, gap extension penalty = 0.05, delay divergent sequences = 0.4. The virus-encoded GCRs are indicated analogous to pR33; i.e., the ORF designation is preceded by a ’p’. CC-CK, C-C chemokine receptor; IL-8, interleukin-8 receptor; A1AA, αA1-adrenergic receptor (13); H. halobium BACR, Halobacterium halobium bacteriorhodopsin (30); CD97, cluster designation 97 (28); OPSD, rhodopsin (39); ACM1, M1 muscarinic acetylcholine receptor (3); D. discoideum CAR1, Dictyostelium discoideum cyclic AMP receptor 1 (31); S. cerevisiae STE2, Saccharomyces cerevisiae pheromone α-factor receptor (14); CASR, extracellular calcium-sensing receptor (2); FSHR, follicle-stimulating hormone receptor (37).
R33 transcription.
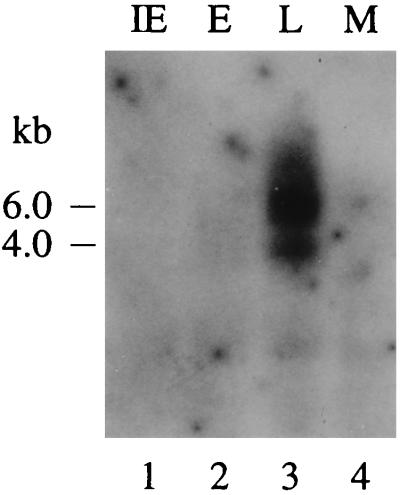
To examine the expression of R33 at IE, E, and L times of infection in REF, we set out to identify R33 transcripts by Northern blot analysis. As shown in Fig. 4, R33-specific transcripts could be detected only in the L phase of infection. Similar expression patterns have previously been reported for the UL33 and M33 genes, which were both transcribed exclusively during the L phase of HCMV and MCMV infection, respectively (23, 55). Two major R33 transcripts, of approximately 4.0 and 6.0 kb, can be distinguished in Fig. 4. Since the length of the R33 ORF is only 1,161 bp and a consensus polyadenylation signal is lacking at the 3′ end of the gene, it is likely that the R33 transcripts contain not only R33 sequences but also sequences from one or more neighboring genes. Northern blot hybridization data (shown below) support the hypothesis of cotranscription.
FIG. 4.
The RCMV R33 gene is transcribed at late times of infection in REF. The figure shows an autoradiograph of a Northern blot that was hybridized with an R33-specific probe. Lanes 1, 2, and 3 represent the IE, E, and L phases of infection, respectively. In lane 4, mRNA from mock-infected (M) cells was separated. The estimated lengths of the transcripts are indicated on the left.
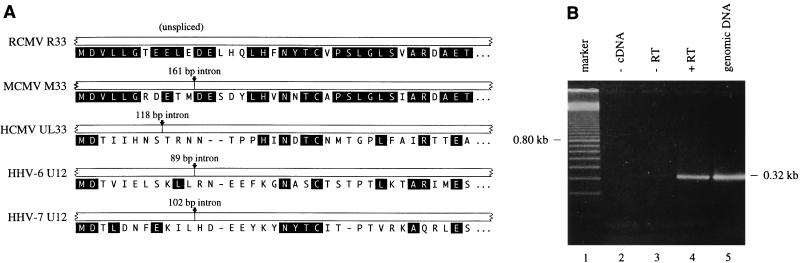
Transcripts from both UL33 and M33 are spliced near the 5′ ends, within regions that encode the N-terminal parts of pUL33 and pM33, respectively (23). It was proposed that transcripts of UL33-like genes from the betaherpesviruses HHV-6 and HHV-7 are spliced in a similar fashion (23). This proposal was based on an alignment of N-terminal amino acid sequences of UL33-like GCRs: a higher degree of similarity was seen between these N-terminal sequences if the presence of an intron was suggested in the corresponding genes of HHV-6 and HHV-7 (23). Interestingly, RCMV R33 transcripts are probably not spliced, since the N-terminal amino acid sequence that is predicted from the unspliced R33 sequence is highly similar to the amino acid sequences derived from the spliced UL33 and M33 transcripts (Fig. 5A). To investigate the presence of a potential intron near the predicted start codon of the R33 gene, the R33 mRNA was analyzed by RT-PCR (Fig. 5B). As anticipated, the PCR products that were generated with either R33 cDNA that was derived from L-phase mRNA or genomic RCMV DNA of a similar length (approximately 320 bp; Fig. 5B, lanes 4 and 5). A control reaction, in which the L-phase mRNA was not treated with RT before PCR, did not generate any PCR products (lane 3), indicating that the amplified products were derived from mRNA rather than contaminating genomic DNA. These results indicate that splicing does not occur within a region spanning from 199 bp upstream of the R33 start codon to 103 bp downstream of the start codon (nucleotides 532 to 833 in Fig. 2).
FIG. 5.
R33 transcripts are not spliced near the 5′ end. (A) Alignment of the N termini of UL33-like GCRs and the position of introns within the 5′ region of the corresponding genes. Amino acid residues that are conserved between pR33 and at least one of the other pUL33-like proteins are indicated as white letters in black boxes. (B) To identify a potential intron near the 5′ end of the R33 gene, an RT-PCR was performed on poly(A)+ RNA of RCMV-infected cells (lane 4, + RT). As negative controls, either target DNA (lane 2, − cDNA) or RT transcriptase (lane 3, − RT) was omitted. Genomic RCMV DNA was included as a positive control for the PCR (lane 5, genomic DNA). Lane 1 contains a molecular mass reference (100-bp marker). A photograph of an ethidium bromide-stained agarose gel is shown.
Generation of an RCMV R33 null mutant.
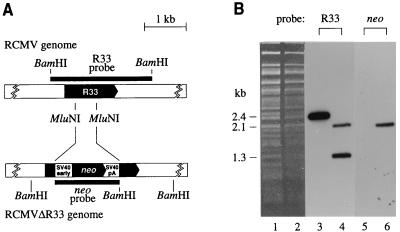
To investigate the role of R33 in the pathogenesis of RCMV disease, a mutant RCMV strain (RCMVΔR33), in which the R33 gene was disrupted by replacing the 0.5-kb MluNI fragment from the R33 ORF with a 1.5-kb neomycin expression cassette, was constructed (Fig. 6A). The deletion/insertion mutation was first introduced into a plasmid containing the R33 gene. The R33 gene within the RCMV genome was subsequently replaced by the mutated R33 gene via homologous recombination, after transfection of fibroblasts with the recombination plasmid followed by infection with RCMV. Selection for recombinant virus was established by supplementing the growth medium with G418. After plaque purification, the purity of the recombinant virus was checked by both Southern blot analysis and PCR. Virion DNA from both RCMV and RCMVΔR33 was purified and digested with BamHI. After agarose gel electrophoresis and transfer of the DNA to a filter, hybridization was done with either an R33-specific probe or a neo-specific probe. Cleavage of RCMV DNA with BamHI should generate a single 2.4-kb fragment containing R33 sequences, whereas cleavage of RCMVΔR33 DNA should result in two fragments (of 1.3 and 2.1 kb) containing R33 sequences (Fig. 6A). In addition, the 2.1-kb BamHI fragment of RCMVΔR33 should contain neo sequences. As shown in Fig. 6B, BamHI digestion of RCMVΔR33 DNA indeed resulted in two R33-hybridizing fragments, of 1.3 and 2.1 kb (lane 4). As predicted, the 2.1-kb RCMVΔR33 BamHI fragment also hybridized to the neo probe (lane 6). Since a 2.4-kb R33-hybridizing BamHI fragment was not detected in lane 4, we conclude that the recombinant virus is pure. The integrity of the RCMVΔR33 genome was also confirmed both by comparing seven different restriction endonuclease patterns of wild-type and mutant RCMV genomes and by PCR with various primer combinations (data not shown).
FIG. 6.
Generation of an RCMV R33 null mutant. (A) To determine the role of R33 in RCMV infection, a mutant RCMV strain was generated by replacing part of the R33 ORF by a neomycin resistance gene (neo). ORFs are indicated by black arrows. Black rectangles above and below each genome indicate the position of the DNA probes that were used for Southern blot hybridization. SV40 early, simian virus 40 early promoter; SV40 pA, polyadenylation signal. Note that the indicated MluNI sites are lost in the RCMVΔR33 genome. (B) Southern blot hybridization of RCMV and RCMVΔR33 virion DNA. A photograph of an ethidium bromide-stained gel containing BamHI-digested genomic DNA from RCMV (lane 1) and RCMVΔR33 (lane 2) is shown. After transfer to a nylon filter, the DNA from lanes 1 and 2 was hybridized to either the R33 probe (lanes 3 and 4, respectively) or the neo probe (lanes 5 and 6, respectively).
RCMVΔR33 transcripts.
To investigate the effect of disruption of the R33 gene on transcription of both the R33 gene and genes neighboring the disrupted gene in the RCMVΔR33 genome, a Northern blot hybridization experiment was performed. The genes upstream and downstream of the R33 gene were found to have considerable sequence similarity to the MCMV M32 and M34 genes, respectively (data not shown). These RCMV genes are therefore referred to as R32 and R34, respectively. Probes were generated from R32-, R33-, R34-, and neo-specific DNA fragments (Fig. 7B) and were hybridized with poly(A)+ RNA extracted from either RCMV- or RCMVΔR33-infected fibroblasts (Fig. 7A). RCMV generates one major transcript from the R32 gene, with a length of approximately 2.5 kb (Fig. 7A, lane 1). Minor R32 transcripts of approximately 4.0 and 6.0 kb can be seen. Disruption of the R33 gene did not result in dramatic changes in R32 transcription (compare lanes 1 and 2). Transcription of R34 of RCMV resulted in three major transcripts of approximately 2.8, 4.0, and 6.0 kb (lane 7). Since the 4.0- and 6.0-kb R34 transcripts comigrate with the R33 transcripts (compare lanes 4 and 7), these RNAs are likely to represent cotranscripts of both the R33 and the R34 genes. Although there is no detectable difference between RCMV and RCMVΔR33 in expression of the 2.8-kb R34 transcript, modest differences can be seen for the larger transcripts (lanes 7 and 8). Most notably, the 4.0-kb transcript seems to be replaced by a 4.2-kb species. A similar observation can be made for the R33 4.0-kb transcript (lane 4 and 5), which further supports the notion that this species represents an R33-R34 cotranscript. In the genome of RCMVΔR33, part of the R33 gene is replaced by the neo gene. The RCMVΔR33 transcripts that hybridized to R33 as well as R34 sequences also hybridized to the neo probe (lane 11). In addition, RCMVΔR33 expressed a unique 1.2-kb neo transcript (lane 11). Another transcript unique to RCMVΔR33 is a 2.8-kb species that contained R33 as well as neo sequences (lanes 5 and 11). The lengths and predicted positions of each of the transcripts that are generated in the R32 to R34 region of the genomes of RCMV and RCMVΔR33 are summarized in Fig. 7B. Although there are clear differences between RCMV and RCMVΔR33 in the transcription of R33 or “ΔR33,” the major transcripts of the R32 and R34 genes are unaffected. Therefore, we conclude that the genes which are neighboring the disrupted R33 ORF are functional.
The R33 gene is not essential for virus replication in various cell types in vitro.
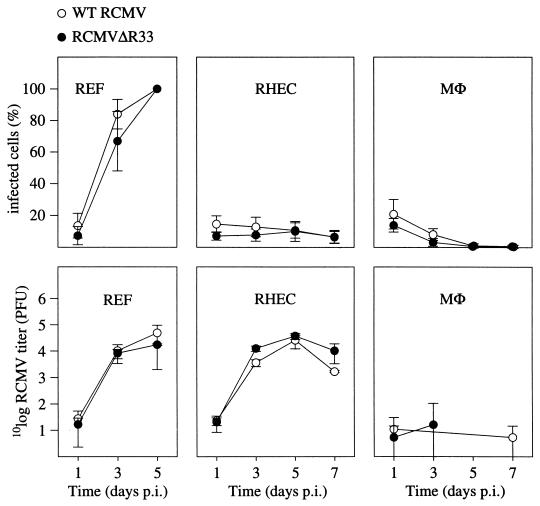
To study whether RCMVΔR33 and RCMV have different replication characteristics in vitro, three different cell types which are thought to play a role in CMV infection in vivo were infected with both viruses and the ratio of infected cells over uninfected cells was determined at various time points. In addition, the amount of infectious virus that was produced by each cell type was investigated. The cell types used in these experiments were REF, RHEC, and MΦ. As shown in Fig. 8, the ratios of infected to uninfected cells did not differ significantly between RCMV- and RCMVΔR33-infected cells, regardless of the cell type. Also, no significant differences were seen between the recombinant virus and RCMV in the virus titers produced by each cell type. These data indicate that R33 is not essential for viral replication in REF, RHEC, and MΦ in vitro.
FIG. 8.
The R33 gene is not essential for virus replication in vitro. REF, MHEC, and MΦ were infected with either RCMV or RCMVΔR33, and the replicative potential of these viruses was assessed by immunofluorescence and plaque assay. The upper graphs show the infected-cell/total-cell ratios at various time points p.i. The lower graphs show virus titers that were determined in culture medium up to 7 days p.i. Standard deviations are indicated by vertical bars. REF were monitored up to 5 days p.i., when 100% of the cells showed cytopathic effect. On days 5 and 7 p.i., virus could not be detected in medium samples that were taken from cultures of infected MΦ. Data from these time points are therefore not included in the graph.
R33 has a critical function in the pathogenesis of RCMV infection in vivo.
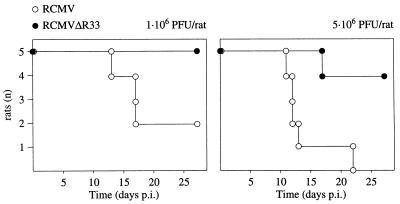
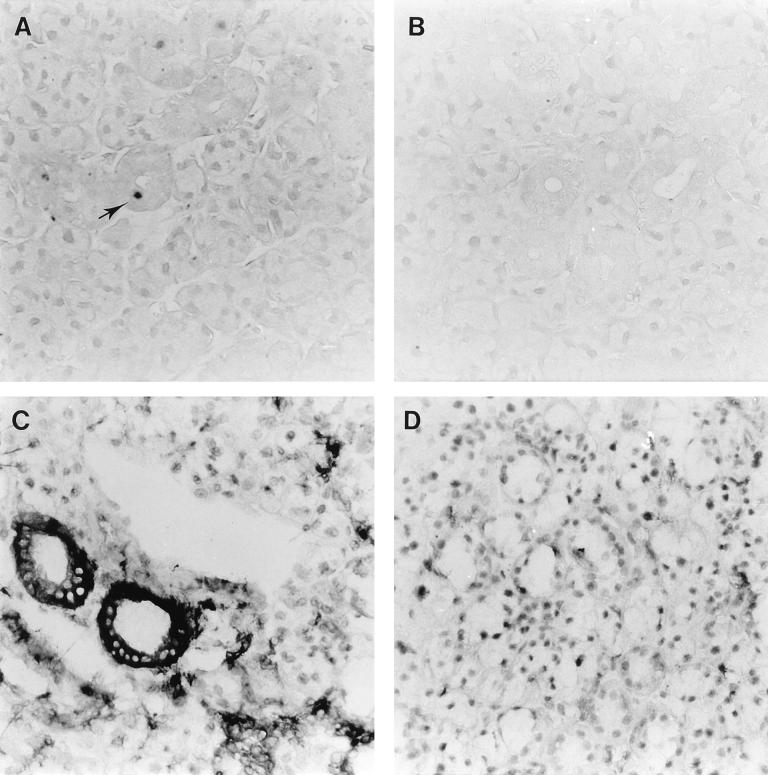
The role of R33 in the pathogenesis of RCMV disease was investigated by infection of groups of immunosuppressed rats with either RCMV or RCMVΔR33. In an initial experiment, 6-week-old immunosuppressed Lewis/N RT1 rats were inoculated with either 1 × 106 or 5 × 106 PFU of RCMV or RCMVΔR33. The number of surviving rats in each group was monitored until 28 days p.i. Surprisingly, a dramatically higher survival was observed in the group of RCMVΔR33-infected rats than in the group of RCMV-infected rats (Fig. 9). This suggests that R33 plays an important role in the pathogenesis of RCMV disease. On day 28 p.i., the surviving rats were sacrificed and several organs were subjected to plaque assay. Although in most organs no significant differences in virus titers were observed between RCMV- and RCMVΔR33-infected rats, virus could not be detected in salivary glands from RCMVΔR33-infected rats whereas virus could easily be detected in salivary glands from RCMV-infected rats (data not shown). To study this observation in more detail, a follow-up experiment was performed in which two groups of 15 10-week-old rats were infected with 5 × 106 PFU of either RCMV or RCMVΔR33. On days 3, 5, 7, 10, and 14 p.i., three rats from each group were sacrificed and the presence of virus in internal organs was analyzed by both immunohistochemistry and plaque assay. For 14 days p.i., virus titers did not differ significantly between organs from RCMV- and RCMVΔR33-infected rats. Virus titers ranged from <1 to 3.3 × 102 PFU/ml in organs from RCMV-infected rats and from <1 to 1.0 × 103 PFU/ml in organs from RCMVΔR33-infected rats. In contrast, although high titers of virus (>104 PFU/ml) were found within salivary glands of RCMV-infected rats on day 10 p.i. and at later time points, virus could not be recovered from the salivary glands of RCMVΔR33-infected rats (Table 1). In addition, RCMV antigens were not detected in salivary gland sections of RCMVΔR33-infected rats (Table 1; Fig. 10B), whereas these antigens could easily be detected in the salivary gland epithelial cells of RCMV-infected rats (Table 1; Fig. 10A). These data indicate that R33 is important for entry and/or replication of RCMV in salivary glands of the rat.
FIG. 9.
Survival of two groups of immunocompromised rats after intraperitoneal inoculation with either wild-type RCMV or RCMVΔR33. Two groups of rats (five in each group) were infected with either 1 × 106 or 5 × 106 PFU of virus. Survival was recorded up to 28 days p.i.
TABLE 1.
Virus titersa and immunohistochemical detection of RCMV and RCMVΔR33 in salivary glands
| Virus | 10-week-old rats, 5 × 106 PFU of RCMV
|
6 week-old rats, 1 × 106 PFU of RCMV
|
|||||
|---|---|---|---|---|---|---|---|
| 3, 5, and 7 days p.i.
|
10 days p.i.
|
14 days p.i.
|
Titer at 28 days p.i. | ||||
| Titera | IPOXb | Titer | IPOX | Titer | IPOX | ||
| RCMV | <1 | 0/3 | 3.6 ± 0.3 | 2/3 | >4 | 3/3 | >4 |
| RCMVΔR33 | <1 | 0/3 | <1 | 0/3 | <1 | 0/3 | <1 |
Log10 virus titers are shown as mean ± standard deviation (PFU per milliliter).
Immune peroxidase assay (IPOX) data are represented as the number of rats of which the salivary glands were found RCMV positive/total number of rats tested.
FIG. 10.
Expression of RCMV (early) proteins and rat class II MHC proteins in salivary glands of RCMV- and RCMVΔR33-infected rats. The figure shows micrographs of 4-μm sections of rat salivary glands infected with either RCMV (A and C) or RCMVΔR33 (B and D). Tissue sections were stained with either MAb against viral (E) antigens (MAb RCMV8 [A and B]) or MAb against class II MHC proteins (OX-6 [C and D]). The tissue sections were photographed at a magnification of ×400.
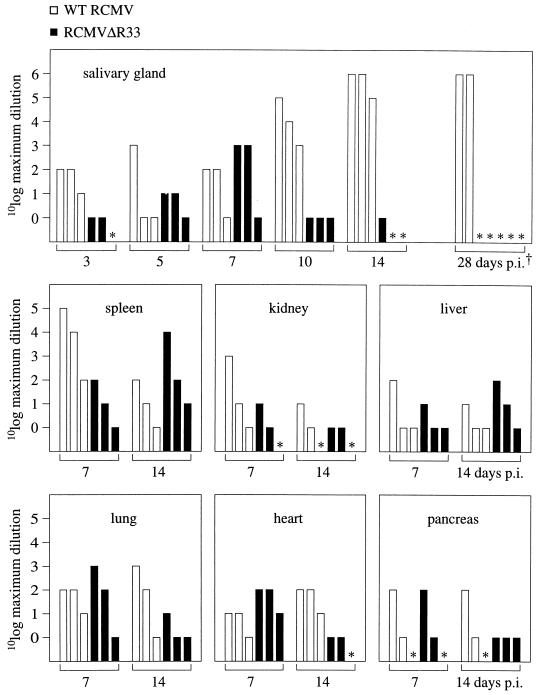
To investigate whether RCMVΔR33 is transported to the salivary glands, we set out to study dissemination of the virus in infected rats by PCR, which has a higher sensitivity than both plaque assay and immunohistochemistry. For this purpose, several internal organs of infected rats were subjected to PCR to detect viral DNA. DNA was purified from salivary gland, spleen, kidney, liver, lung, heart, and pancreas tissue derived from rats that were sacrificed on days 7, 10, 14, and 28 p.i. After purification, the DNA was serially diluted. By subjecting the DNA dilutions to PCR, a rough estimate of the quantity of viral DNA in each organ could be made. The sensitivity of a PCR assay was approximately 300 RCMV genome copies per μg of tissue DNA (approximately 1 RCMV genome copy per 800 cells [data not shown]). The virus DNA levels did not differ between most organs from either RCMV- or RCMVΔR33-infected animals (Fig. 11). In contrast, marked differences were observed between viral DNA levels in the salivary glands of RCMV- and RCMVΔR33-infected rats on days 10, 14, and 28 p.i. (Fig. 11). While RCMV DNA could easily be detected at all indicated time points, RCMVΔR33 DNA could be detected in salivary glands only up to 14 days p.i. and not at later time points. These data suggest that although RCMVΔR33 is presumably transported to the salivary glands, it does not enter and/or replicate in salivary gland epithelial cells.
FIG. 11.
PCR detection of RCMV DNA in several organs of RCMV- and RCMVΔR33-infected rats. To detect viral DNA in organs from either RCMV- or RCMVΔR33-infected rats, PCR was performed on serial dilutions of DNA that was purified from these organs. At each indicated time point, the results are shown for three RCMV-infected rats (open bars) and three RCMVDR33-infected rats (black bars). The height of each bar indicates the maximum dilution of a given DNA isolate with which a positive PCR result could still be obtained. ∗, below detection level (the number of virus genome copies in these samples is smaller than 300). We were able to detect a minimum of approximately 300 genome copies in 1 μg of tissue DNA (data not shown). †, The five samples indicated by ∗ on day 28 p.i. were infected with RCMVΔR33. The samples taken on day 28 p.i. were obtained from the experiment in Fig. 9, in which the rats were infected with 1 × 106 PFU of virus. All the other samples were taken from rats that were infected with 5 × 106 PFU of virus.
Van Dam et al. (52) recently showed that infection of rats with RCMV resulted in an increase of both influx of inflammatory cells into the salivary glands and class II MHC expression by salivary gland cells. To study the effect of RCMVΔR33 infection on the presence of infiltrating cells in the salivary glands, immunohistochemical stainings were performed on sections from rats that were sacrificed at day 7 and 14 p.i. The sections were scored for the presence of CD8+ T cells, CD4+ cells (T cells and monocytes), inflammatory and resident tissue macrophages, and NK cells. Additionally, the expression of class II MHC proteins on the surface of salivary gland epithelial cells was studied. Although significant differences in infiltrating-cell populations were not observed between salivary glands of RCMV- and RCMVΔR33-infected rats (data not shown), a clear difference was seen in the class II MHC expression, which was higher in salivary glands of RCMV-infected rats than in salivary glands of RCMVΔR33-infected rats (Fig. 10C and D). Although virtually all of the salivary gland epithelial cells of wild-type RCMV-infected rats were positive upon staining with the anti-class II MHC MAb, only a minority of these cells were positive upon staining with MAb RCMV 8. The difference between these staining patterns might be caused by either a lower sensitivity of the RCMV 8 MAb relative to the anti-class II MHC MAb or the release of interferons by the RCMV-infected cells, which might induce the expression of class II MHC proteins in most of the neighboring, noninfected epithelial cells. In conclusion, our results support the notion that, in contrast to infection with RCMV, infection of rats with RCMVΔR33 does not result in active virus replication in the salivary glands.
DISCUSSION
The RCMV R33 gene is part of a family of betaherpesvirus genes that are likely to encode GCRs. The other members of this gene family are HCMV UL33 and MCMV M33 and the U12 genes of HHV-6 and HHV-7 (20, 27, 41, 44). Both the sequence and the genome location are conserved among the UL33-like genes. As expected, the amino acid sequence that was predicted to be encoded by the RCMV R33 ORF is more similar to the amino acid sequences derived from HCMV UL33 and MCMV M33 (40 and 65% identity, respectively) than to those derived from the U12 genes (24 and 26% identity with the proteins of HHV-6 and HHV-7, respectively). Phylogenetic analysis based on multiple alignment of predicted amino acid sequences showed that the pUL33-like GCRs are more closely related to chemokine receptors than to non-chemokine-binding GCRs. In agreement with this, the R33-derived amino acid sequence shows several features characteristic of chemokine receptors (1, 23).
Sixteen beta- and gammaherpesvirus-encoded GCRs have been recognized to date (17, 20, 27, 40, 41, 44, 47). In addition, the BILF1 gene from Epstein-Barr virus (5) may encode a GCR, since its predicted amino acid sequences has 25% identity to the equine herpesvirus 2 ORF E6-encoded GCR (47). Three of the herpesvirus-encoded GCRs have been studied in detail and were found to be capable of binding chemokines and stimulating cellular downstream effectors. The herpesvirus saimiri ECRF3 gene product was found to be activated by α chemokines (1), resulting in mobilization of intracellular Ca2+. The HCMV US28 gene product could be activated by β chemokines (26), whereas the Kaposi’s sarcoma-related herpesvirus ORF 74 gene product could be activated by both α and β chemokines (4). Ligands have not yet been identified for members of the UL33 family. Whether or not UL33-like GCRs can be activated by binding to either chemokines or ligands from another class is not yet known. Similar to what was found for the Kaposi’s sarcoma-associated herpes virus ORF 74 gene product, it is possible that UL33-like GCRs can be constitutively active, irrespective of ligand binding.
Similarly to the results reported for the UL33 and M33 genes (23, 55), R33 was expressed at the late phase of infection as a cotranscript with at least one of the genes downstream of R33 (R34). However, in contrast to the UL33 and M33 transcripts, the RCMV R33 mRNA is not spliced near the 5′ terminus. On the basis of amino acid sequence alignment, it was inferred that the U12 genes of both HHV-6 and HHV-7 contain a 5′ intron similar to that of the UL33 and M33 genes (23). These data indicate that among the members of the UL33-like family that have been identified to date, the R33 gene is the only gene that does not contain an intron near the start codon.
RCMV and MCMV replicate efficiently in salivary glands of infected rats (11) and mice (23), respectively. These viruses can easily be detected in the salivary glands by conventional techniques such as immunohistochemistry or plaque assay. In contrast, recombinant MCMV strains lacking a functional M33 gene could not be detected in mouse salivary glands by a plaque assay, which indicated that M33 is important for either dissemination or replication in the salivary glands (23). Similarly, the R33 gene of RCMV was found to be important for either entry or replication in salivary glands. Although RCMVΔR33 could not be detected in salivary glands of infected rats by either plaque assay or immunohistochemistry, viral DNA could be detected up to 14 days p.i. by PCR. At later time points, RCMVΔR33 DNA could no longer be detected in the salivary glands. These data show that although recombinant virus (DNA) is able to reach the salivary glands, it is unable to persist in the epithelial cells of this tissue. It is possible that the ability to detect recombinant RCMV DNA in salivary glands early after infection is not the actual result of infection of salivary gland cells but instead the result of the presence of recombinant RCMV (DNA)-containing circulating cells (e.g., detached endothelial cells or monocytes/macrophages [46]) that pass through blood vessels of the salivary glands. The finding that R33 is essential for virus replication in salivary glands is also supported by the inability to detect class II MHC expression in the salivary gland epithelium of RCMVΔR33-infected rats. Class II MHC expression is evoked by RCMV infection of rat heart endothelium (50), liver and kidney cells (51), and salivary gland epithelium (Fig. 10).
Salivary gland tropism has also been shown to be affected by mutations in other CMV genes. The MCMV Vancouver strain could not be detected in the salivary glands of infected mice by a plaque assay (7). Since this virus strain was found to have both a 9.4-kb deletion and a 0.9-kb insertion relative to the parental MCMV Smith strain (7), salivary gland tropism might be affected through the combined deletion and/or disruption of several genes. Another set of genes of the MCMV K181 strain (ORF m137 to m143 [16]) was shown to be essential for replication in salivary glands. Combined disruptions of at least two of these genes were found to (partially) affect salivary gland tropism (16). Manning et al. (33) showed the importance of yet another gene in salivary gland tropism of MCMV, the salivary gland growth gene 1 (sgg1). Deletion of this gene reduced the replicative potential of MCMV, although recombinant virus could still be detected in the salivary glands by a plaque assay. The biological significance of the complete loss of salivary gland tropism through disruption of R33 and M33 from RCMV and MCMV, respectively, will have to be assessed in future studies.
A remarkable finding from our study is the increased survival that was seen among groups of RCMVΔR33-infected rats compared to groups of RCMV-infected rats. Since this increased survival is probably not solely the result of lack of RCMVΔR33 replication in the salivary glands, it is likely that disruption of the R33 gene also affects RCMV replication in other organs or tissues, such as lymph nodes and bone marrow. It is possible that the observed differences in behavior between RCMV and RCMVΔR33 in vivo might be the result of expression of Neo by the recombinant virus. However, since the expression of Neo did not have an influence on virus growth in rat fibroblasts in vitro, it was inferred that the putative expression of Neo by the recombinant virus in vivo does not dramatically influence the behavior of the virus.
In a first comparison of organs from RCMV- and RCMVΔR33-infected rats other than the salivary glands, no obvious differences were seen in virus load and expression of RCMV (E) proteins. In a future study, the pathogenesis of RCMV and RCMVΔR33 infection will be studied in greater detail, e.g., by investigating the expression of viral IE, E and L proteins and by performing a detailed analysis of the host immune response to viral challenge.
ACKNOWLEDGMENTS
We thank Erik Beuken for generating BglII subclones of the EcoRI A fragment, Jeroen Kloover and Harry van der Heijden for immunological staining of tissue sections, and Rien Blok for critical reading of the manuscript.
REFERENCES
- 1.Ahuja S K, Murphy P M. Molecular piracy of mammalian interleukin-8 receptor type B by herpesvirus saimiri. J Biol Chem. 1993;268:20691–20694. [PubMed] [Google Scholar]
- 2.Aida K, Koishi S, Tawata M, Onaya T. Molecular cloning of a putative Ca(2+)-sensing receptor cDNA from human kidney. Biochem Biophys Res Commun. 1995;214:524–529. doi: 10.1006/bbrc.1995.2318. [DOI] [PubMed] [Google Scholar]
- 3.Allard W J, Sigal I S, Dixon R A. Sequence of the gene encoding the human M1 muscarinic acetylcholine receptor. Nucleic Acids Res. 1987;15:10604. doi: 10.1093/nar/15.24.10604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arvatakis L, Geras-Raaka E, Varma A, Gershengorn M C, Cesarman E. Human herpesvirus KSHV encodes a constitutively active G-protein-coupled receptor linked to cell proliferation. Nature. 1997;385:347–350. doi: 10.1038/385347a0. [DOI] [PubMed] [Google Scholar]
- 5.Baer R J, Bankier A T, Biggin M D, Deininger P L, Farrell P J, Gibson T J, Hatful G F, Hudson G S, Satchwell S C, Sequin C, Tuffnell P S, Barrell B G. DNA sequence and expression of the B95-8 Epstein-Barr virus genome. Nature. 1984;310:207–211. doi: 10.1038/310207a0. [DOI] [PubMed] [Google Scholar]
- 6.Beuken E, Slobbe R, Bruggeman C A, Vink C. Cloning and sequence analysis of the genes encoding DNA polymerase, glycoprotein B, ICP 18.5 and major DNA-binding protein of rat cytomegalovirus. J Gen Virol. 1996;77:1559–1562. doi: 10.1099/0022-1317-77-7-1559. [DOI] [PubMed] [Google Scholar]
- 7.Boname J M, Chantler J K. Characterization of a strain of murine cytomegalovirus which fails to grow in the salivary glands of mice. J Gen Virol. 1992;73:2021–2029. doi: 10.1099/0022-1317-73-8-2021. [DOI] [PubMed] [Google Scholar]
- 8.Brown T. Analysis of DNA sequences by blotting and hybridization. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1993. pp. 4.2.1–4.2.15. [Google Scholar]
- 9.Brown T, Mackey K. Analysis of RNA by Northern and slot blot hybridization. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1997. pp. 4.9.1–4.9.16. [Google Scholar]
- 10.Bruggeman C A, Meijer H, Dormans P H J, Debie W H M, Grauls G E L M, van Boven C P A. Isolation of a cytomegalovirus-like agent from wild rats. Arch Virol. 1982;73:231–241. doi: 10.1007/BF01318077. [DOI] [PubMed] [Google Scholar]
- 11.Bruggeman C A, Meijer H, Bosman F, van Boven C P A. Biology of rat cytomegalovirus infection. Intervirology. 1985;24:1–9. doi: 10.1159/000149612. [DOI] [PubMed] [Google Scholar]
- 12.Bruning J, Debie W H M, Dormans P H J, Meijer H, Bruggeman C A. The development and characterization of monoclonal antibodies against rat cytomegalovirus induced agents. Arch Virol. 1987;94:55–70. doi: 10.1007/BF01313725. [DOI] [PubMed] [Google Scholar]
- 13.Bruno J F, Whittaker J, Song J F, Berelowitz M. Molecular cloning and sequencing of a cDNA encoding a human alpha 1A adrenergic receptor. Biochem Biophys Res Commun. 1991;179:1485–1490. doi: 10.1016/0006-291x(91)91740-4. [DOI] [PubMed] [Google Scholar]
- 14.Burkholder A C, Hartwell L H. The yeast alpha-factor receptor: structural properties deduced from the sequence of the STE2 gene. Nucleic Acids Res. 1985;13:8463–8475. doi: 10.1093/nar/13.23.8463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cao J X, Gershon P D, Black D N. Sequence analysis of HindIII Q2 fragment of capripoxvirus reveals a putative gene encoding a G-protein-coupled chemokine receptor homologue. Virology. 1995;209:207–212. doi: 10.1006/viro.1995.1244. [DOI] [PubMed] [Google Scholar]
- 16.Cavanaugh V J, Stenberg R M, Staley T L, Virgin IV H W, MacDonald M R, Paetzold S, Farrell H E, Rawlinson W D, Campbell A E. Murine cytomegalovirus with a deletion of genes spanning HindIII-J and -I displays altered cell and tissue tropism. J Virol. 1996;70:1365–1374. doi: 10.1128/jvi.70.3.1365-1374.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cesarman E, Nador R G, Bai F, Bohenzky R A, Russo J J, Moore P S, Chang Y, Knowles D M. Kaposi’s sarcoma-associated herpesvirus contains G protein-coupled receptor and cyclin D homologs which are expressed in Kaposi’s sarcoma and malignant lymphoma. J Virol. 1996;70:8218–8223. doi: 10.1128/jvi.70.11.8218-8223.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chambers W H, Vujanovic N L, DeLeo A B, Olsowy M W, Herberman R B, Hiserodt J C. Monoclonal antibody to a triggering structure expressed on rat natural killer cells and adherent lymphokine-activated killer cells. J Exp Med. 1989;169:1373–1789. doi: 10.1084/jem.169.4.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chee M S, Bankier A T, Beck S, Bohni R, Brown C M, Cerny R, Horsnell T, Hutchison III C A, Kouzarides T, Martignetti J A, Preddie E, Satchwell S C, Tomlinson P, Weston K M, Barrell B G. Analysis of the protein-coding content of the sequence of human cytomegalovirus strain AD169. Curr Top Microbiol Immunol. 1990;154:125–169. doi: 10.1007/978-3-642-74980-3_6. [DOI] [PubMed] [Google Scholar]
- 20.Chee M S, Satchwell S C, Preddie E, Weston K M, Barrell B G. Human cytomegalovirus encodes three G protein-coupled receptor homologues. Nature. 1990;344:774–777. doi: 10.1038/344774a0. [DOI] [PubMed] [Google Scholar]
- 21.Damoiseaux J G M C, Döpp E A, Calame W, Chao D, MacPherson G G, Dijkstra C D. Rat macrophage lysosomal membrane antigen recognized by monoclonal antibody ED1. Immunology. 1994;83:140–147. [PMC free article] [PubMed] [Google Scholar]
- 22.Davis-Poynter N J, Farrell H E. Masters of deception: a review of herpesvirus immune evation strategies. Immunol Cell Biol. 1996;74:513–522. doi: 10.1038/icb.1996.84. [DOI] [PubMed] [Google Scholar]
- 23.Davis-Poynter N J, Lynch D M, Vally H, Shellam G R, Rawlinson W D, Barrell B G, Farrell H E. Identification and characterization of a G protein-coupled receptor homolog encoded by murine cytomegalovirus. J Virol. 1997;71:1521–1529. doi: 10.1128/jvi.71.2.1521-1529.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dijkstra C D, Dopp A E, Joling P, Kraal G. The heterogeneity of mononuclear phagocytes in lymphoid organs: distinct macrophage subpopulations in the rat recognized by monoclonal antibodies ED1, ED2 and ED3. Immunology. 1985;54:589–599. [PMC free article] [PubMed] [Google Scholar]
- 25.Gao J L, Kuhns D B, Tiffany H L, McDermott D, Li X, Francke U A, Murphy P M. Structure and functional expression of the human macrophage inflammatory protein 1α/RANTES receptor. J Exp Med. 1993;177:1421–1427. doi: 10.1084/jem.177.5.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gao J L, Murphy P M. Human cytomegalovirus open reading frame US28 encodes a functional beta chemokine receptor. J Biol Chem. 1994;269:28539–28542. [PubMed] [Google Scholar]
- 27.Gompels U A, Nicholas J, Lawrence G, Jones M, Thomson B J, Martin M E, Efstathiou S, Craxton M, Macaulay H A. The DNA sequence of human herpesvirus-6: structure, coding content and genome evolution. Virology. 1995;209:29–51. doi: 10.1006/viro.1995.1228. [DOI] [PubMed] [Google Scholar]
- 28.Hamann J, Eichler W, Hamann D, Kerstens H M, Poddighe P J, Hoovers J M, Hartmann E, Strauss M, van Lier R A. Expression, cloning and chromosomal mapping of the leukocyte activation antigen CD97, a new seven-span transmembrane molecule of the secretion receptor superfamily with an unusual extracellular domain. J Immunol. 1995;155:1942–1950. [PubMed] [Google Scholar]
- 29.Hünig T, Wallny H J, Hartley J K, Lawetzky A, Tiefenthaler G. A monoclonal antibody to a constant determinant of the rat T cell antigen receptor that induces T cell activation. J Exp Med. 1989;169:73–86. doi: 10.1084/jem.169.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Khorana H G, Gerber G E, Herlihy W C, Gray C P, Anderegg R J, Nihei K, Biemann K. Amino acid sequence of bacteriorhodopsin. Proc Natl Acad Sci USA. 1979;76:5046–5050. doi: 10.1073/pnas.76.10.5046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Klein P S, Sun T J, Saxe III C L, Kimmel A R, Johnson R L, Devreotes P N. A chemoattractant receptor controls development in Dictyostelium discoideum. Science. 1988;241:1467–1472. doi: 10.1126/science.3047871. [DOI] [PubMed] [Google Scholar]
- 32.Kuhn D E, Beall C J, Kolattukudy P E. The cytomegalovirus US28 protein binds multiple CC chemokines with high affinity. Biochem Biophys Res Commun. 1995;211:325–330. doi: 10.1006/bbrc.1995.1814. [DOI] [PubMed] [Google Scholar]
- 33.Manning W C, Stoddart C A, Lagenaur L A, Abenes G B, Mocarski E S. Cytomegalovirus determinant of replication in salivary gland. J Virol. 1992;66:3794–3802. doi: 10.1128/jvi.66.6.3794-3802.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Margulies B J, Browne H, Gibson W. Identification of the human cytomegalovirus G protein-coupled receptor homologue encoded by UL33 in infected cells and enveloped virus particles. Virology. 1996;225:111–125. doi: 10.1006/viro.1996.0579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Massung R F, Jayarama V, Moyer R W. DNA sequence analysis of conserved and unique regions of swinepox virus: identification of genetic elements supporting phenotypic observations including a novel G protein-coupled receptor homologue. Virology. 1993;197:511–528. doi: 10.1006/viro.1993.1625. [DOI] [PubMed] [Google Scholar]
- 36.Meijer H, Dreesen J C, van Boven C P. Molecular cloning and restriction endonuclease mapping of the rat cytomegalovirus genome. J Gen Virol. 1986;67:1327–1342. doi: 10.1099/0022-1317-67-7-1327. [DOI] [PubMed] [Google Scholar]
- 37.Minegishi T, Nakamura K, Takakura Y, Ibuki Y, Igarashi M. Cloning and sequencing of human FSH receptor cDNA. Biochem Biophys Res Commun. 1991;175:1125–1130. doi: 10.1016/0006-291x(91)91682-3. [DOI] [PubMed] [Google Scholar]
- 38.Myers E W, Miller W. Optimal alignments in linear space. Comput Appl Biosci. 1988;4:11–17. doi: 10.1093/bioinformatics/4.1.11. [DOI] [PubMed] [Google Scholar]
- 39.Nathans J, Hogness D S. Isolation and nucleotide sequence of the gene encoding human rhodopsin. Proc Natl Acad Sci USA. 1984;81:4851–4855. doi: 10.1073/pnas.81.15.4851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nicholas J, Cameron K R, Honess R W. Herpesvirus saimiri encodes homologues of G protein-coupled receptors and cyclins. Nature. 1992;355:362–365. doi: 10.1038/355362a0. [DOI] [PubMed] [Google Scholar]
- 41.Nicholas J. Determination and analysis of the complete nucleotide sequence of human herpesvirus 7. J Virol. 1996;70:5975–5989. doi: 10.1128/jvi.70.9.5975-5989.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pearson W R. Rapid and sensitive sequence comparison with FASTP and FASTA. Methods Enzymol. 1990;183:63–98. doi: 10.1016/0076-6879(90)83007-v. [DOI] [PubMed] [Google Scholar]
- 43.Probst W C, Snyder L A, Schuster D I, Brosius J, Sealfon S C. Sequence alignment of the G-protein coupled receptor superfamily. DNA Cell Biol. 1992;11:1–20. doi: 10.1089/dna.1992.11.1. [DOI] [PubMed] [Google Scholar]
- 44.Rawlinson W D, Farrell H E, Barrell B G. Analysis of the complete DNA sequence of murine cytomegalovirus. J Virol. 1996;70:8833–8849. doi: 10.1128/jvi.70.12.8833-8849.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stals F S, Bosman F, van Boven C P, Bruggeman C A. An animal model for therapeutic intervention studies of CMV infection in the immunocompromised host. Arch Virol. 1990;114:91–107. doi: 10.1007/BF01311014. [DOI] [PubMed] [Google Scholar]
- 46.Stoddart C A, Cardin R D, Boname J M, Manning W C, Abenes G B, Mocarski E S. Peripheral blood mononuclear phagocytes mediate dissemination of murine cytomegalovirus. J Virol. 1994;68:6243–6253. doi: 10.1128/jvi.68.10.6243-6253.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Telford E A, Watson M S, Aird H C, Perry J, Davison A J. The DNA sequence of equine herpesvirus 2. J Mol Biol. 1995;249:520–528. doi: 10.1006/jmbi.1995.0314. [DOI] [PubMed] [Google Scholar]
- 48.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Torres-Nagel N, Kraus E, Brown M, Tiefenthaler G, Mitnacht R, Williams A F, Hünig T. Differential thymus dependence of rat CD8 isoform expression. Eur J Immunol. 1992;22:2841–2848. doi: 10.1002/eji.1830221113. [DOI] [PubMed] [Google Scholar]
- 50.Ustinov J A, Loginov R J, Bruggeman C A, van der Meide P H, Häyry P J, Lautenschlager I T. Cytomegalovirus induces class II expression in rat heart endothelial cells. J Heart Lung Transplant. 1993;12:644–651. [PubMed] [Google Scholar]
- 51.Ustinov J A, Loginov R J, Bruggeman C A, Suni J, Häyry P J, Lautenschlager I T. CMV-induced class II antigen expression in various rat organs. Transplant Int. 1994;7:302–308. doi: 10.1007/BF00327161. [DOI] [PubMed] [Google Scholar]
- 52.van Dam J G, Damoiseaux J G M C, van der Heijden H A M D, Grauls G, van Breda Vriesman P J C, Bruggeman C A. Infection with rat cytomegalovirus in the immunocompromised host is associated with the appearance of a T-cell population with reduced CD8 and T cell receptor expression. J Clin Exp Med. 1997;110:349–357. doi: 10.1046/j.1365-2249.1997.4321449.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vink C, Beuken E, Bruggeman C A. Structure of the rat cytomegalovirus genome termini. J Virol. 1996;70:5221–5229. doi: 10.1128/jvi.70.8.5221-5229.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53a.Vink, C., E. Beuken, and C. A. Bruggeman. Unpublished results.
- 54.Vossen R C R M, Derhaag J G, Slobbe-van Drunen M E P, Duijvestijn A M, van Dam-Mieras M C E, Bruggeman C A. A dual role for endothelial cells in cytomegalovirus infection? A study of cytomegalovirus infection in a series of rat endothelial cell lines. Virus Res. 1996;46:65–74. doi: 10.1016/s0168-1702(96)01375-5. [DOI] [PubMed] [Google Scholar]
- 55.Welch A R, McGregor L M, Gibson W. Cytomegalovirus homologs of cellular G protein-coupled receptor genes are transcribed. J Virol. 1991;65:3915–3918. doi: 10.1128/jvi.65.7.3915-3918.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]