Abstract
As an initial approach to define the requirements for the replication of bovine viral diarrhea virus (BVDV), a member of the Flaviviridae family with a positive-strand RNA genome, full-length genomic and subgenomic RNAs were originated by in vitro transcription of diverse BVDV cDNA constructs and transfected into eucaryotic host cells. RNA replication was measured either directly by an RNase protection method or by monitoring the synthesis of viral protein. When full-length BVDV cRNA was initially applied, the synthesis of negative-strand RNA intermediates as well as progeny positive-strand RNA was detected posttransfection in the cytoplasm of the host cells. Compared to the negative-strand RNA intermediate, an excess of positive-strand RNA was synthesized. Surprisingly, a subgenomic RNA molecule, DI9c, corresponding to a previously characterized defective interfering particle, was found to support both steps of RNA replication in the absence of a helper virus as well, thus functioning as an autonomous replicon. DI9c comprises the 5′ and 3′ untranslated regions of the BVDV genome and the coding regions of the autoprotease Npro and the nonstructural proteins NS3, NS4A, NS4B, NS5A, and NS5B. Most interestingly, the NS2 polypeptide was thus determined to be nonessential for RNA replication. As expected, deletion of the genomic 3′ end as well as abolition of the catalytic function of the virus-encoded serine protease resulted in DI9c molecules that were unable to replicate. Deletion of the entire Npro gene also destroyed the ability of DI9c molecules to replicate. On the other hand, DI9c derivatives in which the 5′ third of the Npro gene was fused to a ubiquitin gene, allowing the proteolytic release of NS3 in trans, turned out to be replication competent. These results suggest that the RNA sequence located at the 5′ end of the open reading frame exerts an essential role during BVDV replication. Replication of DI9c and DI9c derivatives was found not to be limited to host cells of bovine origin, indicating that cellular factors functioning as potential parts of the viral replication machinery are well conserved between different mammalian cells. Our data provide an important step toward the ready identification and characterization of viral factors and genomic elements involved in the life cycle of pestiviruses. The implications for other Flaviviridae and, in particular, the BVDV-related human hepatitis C virus are discussed.
Flaviviruses, pestiviruses, hepatitis C viruses (HCV), and HCV-related recent isolates constitute the family Flaviviridae (30, 49, 50; reviewed in references 16 and 41). The small enveloped virions of the members of the Flaviviridae contain a positive-strand (messenger sense) linear RNA genome of about 9.5 to 12.5 kb that consists of a single open reading frame (ORF) flanked by untranslated regions at the 5′ and 3′ ends (5′-UTR and 3′-UTR, respectively). The 5′ terminus of the flavivirus genome was found to be capped by a type I cap structure (41). Conversely, initiation of translation of the pestivirus and HCV ORFs is mediated by an internal ribosomal entry site (IRES) (40, 42, 59). Translation of ORFs of the Flaviviridae yields a single polyprotein that is co- and posttranslationally processed by cellular as well as viral proteases into a number of different mature viral polypeptides.
At present, little is known about the molecular mechanisms underlying viral propagation. Preliminary information came from infection studies suggesting that the replication of the Flaviviridae follows a strategy similar to that of other positive-strand RNA viruses (reviewed in references 41 and 62). Accordingly, after entry into the host cell and decapsidation, the viral genome serves as a template for the translation of viral proteins. Supposedly in conjunction with cellular factors and functionally linked to the translation process, the viral proteins catalyze transcription of the positive-strand RNA genome into full-length complementary negative-strand RNA molecules, which then act as templates for the synthesis of progeny positive-strand viral RNA molecules.
Classical swine fever virus (CSFV), border disease virus of sheep, and bovine viral diarrhea virus (BVDV) represent the members of the genus Pestivirus which are important animal pathogens (reviewed in references 9 and 57). The gene order of pestiviruses has been determined to be 5′ Npro, C, Erns, E1, E2, p7, NS2-3, NS4A, NS4B, NS5A, NS5B 3′. Npro, which has no counterpart in the other members of the Flaviviridae, is a nonstructural autoprotease that generates its own C terminus by autoproteolytic cleavage from the viral polyprotein and thereby the N terminus of the core protein (51, 63). The structural proteins have been shown to arise from the polyprotein via proteolytic processing by host signal peptidases (reviewed in reference 57). Like the well-characterized situation for pestivirus-related HCV (2, 19, 58), the majority of cleavage events generating the nonstructural proteins are catalyzed by a serine protease; the respective domain is contained within the N-terminal part of NS3 (56, 64, 65).
Pestiviruses can be differentiated into cytopathogenic (cp) and noncytopathogenic (noncp) strains according to the effect of an infection on cells in tissue cultures. In contrast to infection with noncp strains, infection with cp pestiviruses leads to the lysis of the cellular host. cp BVDV strains apparently develop from noncp BVDV strains by rearrangement of the viral genome due to RNA recombination, i.e., deletions, duplications of certain parts of the viral genome, and insertions of parts of cellular mRNAs (reviewed in references 32 and 57). As a common feature, all mutations affect the coding region for the NS2-3 polypeptide, which is expressed in noncp isolates as an uncleaved protein. The cp phenotype was shown to correlate strictly with the appearance of the C-terminal part of NS2-3, the NS3 protein, thus considered a characteristic marker of cp BVDV. For example, molecular analyses of the cp isolate BVDV CP7 revealed an insertion of 27 nucleotides in the NS2-coding region which was demonstrated to be sufficient for mediating NS2-3 cleavage and for conferring cytopathogenicity (33, 55). Another cp isolate, CP9, was found to be composed of two viral RNA molecules, a complete noncp BVDV RNA genome and a viral RNA with a remarkable deletion encompassing the whole region coding for the structural proteins as well as the NS2-coding part of the NS2-3 gene. The presence of the truncated RNA was shown to lead to the expression of NS3 and to mediate cytopathogenicity. Moreover, it was demonstrated to significantly interfere with the replication of the helper virus RNA, thus exhibiting all the properties of a defective interfering (DI) particle (54). DI particles have been identified in most animal as well as plant virus systems (reviewed in references 24 and 45). Based on the discovery of autonomously replicating subgenomic DI particles (RNA replicons) of Sindbis virus and Semliki Forest virus, recombinants of these positive-strand RNA viruses are used as expression systems for heterologous proteins in various host cells (7, 29).
In this paper, we demonstrate that RNA molecules corresponding to the previously described DI9 of BVDV CP9, termed DI9c, are capable of functioning as fully autonomous RNA replicons, i.e., of replicating in vivo without the support of a helper virus. Consequently, that part of the BVDV encodes the five mature viral proteins, NS3 (viral protease and helicase), NS4A and NS4B, and NS5A and NS5B (putative RNA-dependent RNA polymerase) (5, 36, 61, 64), was defined as being sufficient to support complete RNA replication. These findings are considered to represent an important step toward a better understanding of the multiplication strategy of pestiviruses and to provide valuable information for the current research on related viruses.
MATERIALS AND METHODS
Cells and viruses.
MDBK (Madin-Darby bovine kidney) cells were obtained from the American Type Culture Collection (Rockville, Md.). BHK-21 cells were kindly provided by J. Cox (Federal Research Centre for Virus Diseases of Animals, Tübingen, Germany). Human Hep3b, HepG2, and HuH7 hepatocytes as well as CHO cells were gifts from G. Migliaccio (Istituto di Ricerche di Biologia Molecolare, Pomezia (Rome), Italy). Virus strains CP7 and CP9 were described previously (54, 55). Cells were grown in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal calf serum and nonessential amino acids.
Construction of recombinant plasmids.
The full-length BVDV CP7 cDNA clone (pA/BVDV), a variant of this clone containing an NcoI site overlapping the translational start codon of the ORF (pA/BVDV/N), and the chimeric DI9 cDNA construct (pA/BVDV/D9), all composed by use of plasmid pACYC177 (New England BioLabs), were described previously (33). Plasmids containing mutations in the cDNA encoding DI9c were constructed as follows. The mutation of Ser at position 1752 (following the nomenclature of Deng and Brock [13]) (UCG) to Ala (GCG), previously introduced into BVDV CP7 (55), was cloned via BspEI and BlpI into pA/BVDV/D9. The resulting plasmid was named pDI9cpm1 according to the terminology used throughout this work. To create pDI9cNS3, a PCR fragment of the entire coding region for NS3 from BVDV CP7, comprising an additional ATG within an NcoI restriction site upstream of the GGA coding glycine 1590 of BVDV NS3 (13), was cut with NcoI-SacI and cloned into pA/BVDV/D9 (see Fig. 5A). To create the ubiquitin-NS3 fusion gene, the mouse ubiquitin gene derived from plasmid Ub-nsp4 (28) was cut with BglII (sixth nucleotide of the ORF), made blunt with the Klenow fragment (New England BioLabs), and cut with PstI (nsp-coding region). The fragment was ligated into the ORF of plasmid pRN653 (53), prepared by cutting with EcoRI (blunted) and PstI. Due to this cloning procedure, the initial 12 nucleotides of the ubiquitin ORF are identical to the first 12 nucleotides of the BVDV CP7 Npro-coding region in the resulting plasmid (pRNubi) (see Fig. 5A). Next, a BsmBI site was engineered downstream of the last codon of the ubiquitin ORF (glycine 76) by PCR with primer SP6 as the sense primer and ubC (5′ CTG CAG CGT CTC GCA TGC CAC CGC GGA GTC GCA GCA 3′) as the antisense primer. The 5′ overhang of the generated BsmBI site was designed to be identical to an NcoI overhang and contained an ATG. Accordingly, an in-frame methionine codon was inserted downstream of the ubiquitin ORF (see Fig. 5A). An NcoI (pRNubi 5′ polylinker)-BsmBI fragment of this latter construct was ligated into pDI9cNS3 restricted with NcoI, thus creating pDI9cubi (see Fig. 5A). To create pDI9cΔNproubi, the XhoI (5′-UTR)-HincII (Npro gene) fragment of pA/BVDV (CP7) (33) was first cloned into pCITE-2A (Novagene) predigested with NcoI (blunted) and XhoI. As a consequence of HincII (blunted)-NcoI (blunted) ligation, the vector-derived NcoI site was restored. From this construct, an NcoI fragment encompassing the translation initiation codon of BVDV and 42 downstream codons of Npro followed by an NcoI site was ligated into pDI9cubi restricted with NcoI. All constructs were verified by nucleotide sequencing. For in vitro transcription of RNA with T7 RNA polymerase, all pA/BVDV derivatives were linearized with SmaI (see Results). To obtain the radioactively labeled RNA probes that were used in the RNase protection assay, the HindIII fragment at positions 10141 to 10421 of BVDV CP7 and corresponding to the 5′ terminus of the NS5B-coding region was cloned in both orientations into the HindIII site of pBluescript KS (Stratagene). For in vitro transcription with T3 RNA polymerase, the recombinant plasmids were linearized with EcoRI to generate either sense- or antisense-oriented RNA transcripts.
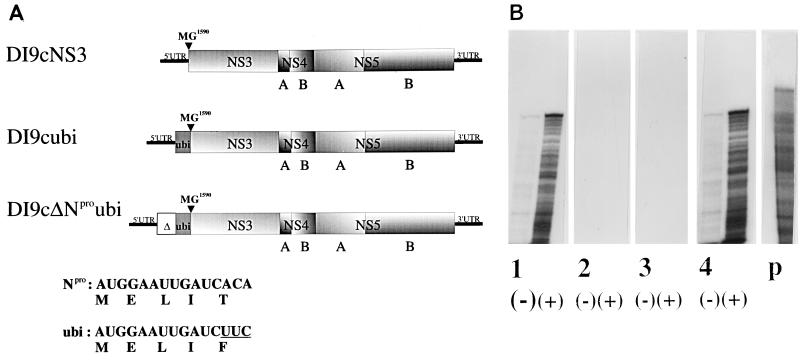
FIG. 5.
The 5′-coding region of Npro is essential for autonomous RNA replication of DI9c. (A) Top: schematic drawing of the DI9c derivatives used in this study (see the legend to Fig. 1A for details). As indicated (MG1590) and as described in detail in Materials and Methods, DI9cNS3 RNA contains an additional AUG codon upstream of GGA coding for glycine 1590 of BVDV CP7 NS3. DI9cubi RNA also contains the ubiquitin gene (ubi), and DI9cΔNproubi also contains the initial 42 codons of the Npro gene (indicated by Δ). Bottom: RNA and primary amino acid sequence comparisons for the initial five codons of BVDV CP7 cRNA and the DI9cubi ORF. (B) RNase protection assay of BHK-21 cells 24 h p.t. with different DI9c derivatives. Lanes: 1, protection of RNA derived from cells transfected with DI9c (positive control); 2, protection of RNA p.t. with DI9cNS3; 3, protection of RNA p.t. with DI9cubi; 4, protection of RNA p.t. with DI9cΔNproubi; p, sense probe (1/10 of the experimental input was loaded on the gel). (−), sense probe; (+), antisense probe.
In vitro transcription and purification of RNA.
The linearized plasmids were transcribed in vitro by a standard protocol. Transcription was stopped by the addition of 5 U of DNase I/10 μl (Boehringer GmbH, Mannheim, Germany), and the mixture was further incubated for 15 min and extracted with phenol-chloroform-isoamyl alcohol (PCA); the yielded RNA was precipitated with 0.9 M ammonium acetate–65% ethanol, avoiding coprecipitation of unincorporated nucleotides. The RNA was dried and redissolved in water, and its concentration was determined by measuring the optical density at 260 nm. Radiolabeled ([32P]GTP) transcripts were further purified by electrophoresis and elution from denaturing acrylamide gels (see below).
RNA transfection.
Transfection of MDBK cells was carried out as described previously (33). Five micrograms of viral RNA transcript was bound in 100 μl of HBSS buffer (5 g of HEPES, 8 g of NaCl, 0.37 g of KCl, 0.125 g of Na2HPO4 · 2H2O, and 1 g of dextrose per liter [pH 7]) to 100 μg of DEAE-dextran (Sigma) for 30 min on ice. About 2 × 107 cells, corresponding to the approximate number growing in a 100-mm dish, were washed once with serum-free medium and resuspended in buffer containing the RNA-DEAE complexes. After 30 min at 37°C, 20 μl of dimethyl sulfoxide was added, and the cells were washed once in HBSS and once in medium without serum and further grown for various times in fully supplemented medium. BHK-21 cells, CHO cells, and hepatocytes were washed twice in phosphate-buffered saline (PBS; 20 mM NaPO4, 130 mM NaCl) and transfected with 5 μg of RNA transcript by electroporation (2 pulses, 200 Ω, 25 μF, 900 V [CHO and hepatocytes] or 1,500 V [BHK-21]) with a model II gene pulser (Bio-Rad). The efficiency of RNA transfection was estimated by immunofluorescence (IF) (see below).
RNase protection assay.
Transfected cells grown for various times in a 100-mm plate were washed once with PBS, removed from the plate with a rubber policeman, and lysed in 375 μl of lysis buffer (see above) for 5 min on ice with occasional shaking. The nuclei were pelleted by centrifugation for 2 min at 1,000 × g. When the nuclear fraction was tested for RNA replication, the pellet was resuspended in 375 μl of high-salt buffer (20 mM Tris-Cl [pH 8], 500 mM NaCl, 1.5 mM MgCl2) and pulled five times through a 26-gauge needle, and the debris was separated by centrifugation. The supernatant was treated for 15 min at 37°C with RNase-free DNase, centrifuged again, and subjected to the same procedure as the cytoplasmic fraction. The respective supernatants were supplemented with 8 μl of 10% (wt/vol) sodium dodecyl sulfate–50 μg of proteinase K and digested for 1 h at 37°C. After two extractions with PCA, the nucleic acids were precipitated with ethanol and the washed pellet was dissolved in 180 μl of hybridization buffer [80% (vol/vol) formamide, 40 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES) (pH 6.4), 400 mM NaCl, 1 mM EDTA]. Dissolved RNA (30 μl) was denatured at 85°C for 2 min and subjected to hybridization at 45°C for at least 5 h either with or without the labeled (5 × 105 to 10 × 105 cpm, about 109 cpm/μg) BVDV sense or antisense probe. Subsequently, 350 μl of RNase digestion buffer (10 mM Tris-Cl [pH 7.5], 1 M NaCl, 5 mM EDTA) as well as RNase T1 and RNase A (at final concentrations of 25 U and 3.5 μg, respectively) was added to the mixture and digestion was performed for at least 1 h at 37°C. After proteinase K digestion, PCA extraction, and ethanol precipitation, the protected fragments were analyzed electrophoretically on 5 or 10% acrylamide–100 mM Tris–100 mM boric acid–2.5 mM EDTA gels containing 7 M urea.
IF assay.
Transfected cells were washed twice with PBS and fixed for 20 min at 4°C to the plate with 4% (wt/vol) paraformaldehyde. After careful washing with PBS, the cells were permeabilized with 1% (wt/vol) N-octyl glucoside (Sigma) for 5 min at 4°C, washed again with PBS, and exposed to the anti-BVDV NS3 antibody (11) in PBS containing 2% (wt/vol) bovine serum albumin and 0.05% (vol/vol) Tween 20 (Sigma) for 2 h at 37°C. Unbound antibody was removed by two washes with PBS, and the cells were exposed to a secondary antibody (CY3; Jackson Laboratories) for 1 h at 37°C. The cells were washed again and examined with an Olympus BX 60 microscope.
RT-PCR.
Reverse transcription (RT)-PCR was carried out with 30 μl of preparations of total cytoplasmic RNA (corresponding to 1.5 × 106 lysed cells; see above) 24 h posttransfection by the protocol of Meyers and Thiel (34). As primers, an oligonucleotide complementary to the 5′-UTR (BVDV positions 347 to 371; sense; 5′ AAA TCT CTG CTG TAC ATG GCA CAT G 3′) and an oligonucleotide complementary to the NS3-coding region (BVDV positions 5441 to 5463; antisense; 5′ ATT CTG TCT CAC CAG TTA ACT T 3′) were used.
RESULTS
Monitoring the replication of in vitro-transcribed BVDV genomic RNA.
For many years, studies on the life cycle of the members of the Flaviviridae have been severely hampered by the lack of cDNA copies of the viral RNA genomes capable of producing infectious RNA transcripts in vitro. This handicap was recently overcome with the successful composition of stable infectious cDNA constructs of some flavivirus species (reviewed in reference 41) as well as the pestiviruses CSFV and BVDV (33, 35, 37, 43, 60). For BVDV CP7, the complete cDNA genome was cloned under the control of a bacteriophage T7 RNA polymerase promoter and with a unique restriction site for linearization at the 3′ end of the viral sequence. Transfection of full-length cRNA transcripts into bovine host cells was shown to yield infectious progeny virus and to yield a cytopathic effect (CPE) about 30 to 48 h posttransfection (p.t.) (33; data not shown).
To initiate a study about BVDV RNA replication, we first needed to set up a suitable experimental system, which should allow monitoring in vivo of the de novo synthesis of viral negative-strand RNA intermediates as well as the production of progeny positive-strand RNA molecules. Since the amount of replicating RNA in a limited number of transfected cells (see below) was expected to be rather low, we decided to monitor BVDV RNA replication by RNase protection, an assay system already applied successfully with other positive-strand RNA virus systems and proven to exhibit a high sensitivity of detection. Pilot experiments carried out with unlabeled genomic RNA transcripts and 32P-labeled complementary RNA probes provided evidence that quantities of ≥1 pg of RNA (corresponding to about 1.5 × 105 molecules of BVDV RNA) could be successfully detected with this method (data not shown).
In our first experiments, we used the full-length BVDV CP7 cDNA construct for in vitro transcription of viral cRNA. As a control, a cDNA template with a premature runoff was transcribed to generate viral genomic RNA in which the 3′ end of the NS5B (the putative RNA-dependent RNA polymerase)-coding region and the 3′-UTR were deleted (Fig. 1). The RNAs were introduced via DEAE-dextran into MDBK cells and analyzed for the presence of replication products at 24 h p.t. prior to the onset of a significant CPE (Fig. 2). The transfected cells were rapidly lysed with an isotonic buffer-containing detergent, and the intact nuclei were separated by gentle low-speed centrifugation (see Materials and Methods). Initially the cytoplasmic fraction and the nuclear fraction were both recovered for subsequent analysis. Since, as expected, viral RNA was detected exclusively in the cytoplasm of the host cells (Fig. 2A, lanes 1), only this cellular fraction was used in all further experiments. After digestion and extraction of the proteins, the pool of cytoplasmic RNAs was subjected to a stringent hybridization procedure with 32P-labeled RNA probes, which either were complementary (antisense probe) or corresponded (sense probe) to a part of the genomic NS5B-coding region and thus bound specifically to positive-strand or negative-strand BVDV RNA, respectively. Single-stranded RNA was hydrolyzed by exposing the hybridization reaction to a mixture of RNase A and RNase T1, and the double-stranded RNase-protected probe was subsequently analyzed by electrophoresis on denaturing polyacrylamide gels and then by autoradiography. In an initial control experiment, labeled sense and antisense probes were hybridized to the BVDV cDNA and treated with RNase. As shown in Fig. 2A, protection of the probe was observed as being not complete due to vector-derived sequences at its 5′ extremity which were not complementary to the viral RNA. Accordingly, the protected fragment migrated consistently faster in the analytical gel than the originally used probe. Protection analysis of a cytoplasmic RNA preparation of full-length BVDV cRNA after transfection of MDBK cells gave rise to products of the same sizes as in the positive control when both the antisense and the sense probes were used to mediate protection to RNase, the latter, however, to a significantly lower extent (Fig. 2A, lanes 2). Thus, each of the BVDV replication products, a large amount of positive-strand RNA as well as the negative-strand RNA intermediate, could be measured 24 h p.t. Although the level of detected negative-strand RNA was already considered significant in the initial experiment, the sensitivity of detection could be clearly increased by a simple modification of the experimental procedure. According to a strategy proven successful with the poliovirus system (1, 38), the cytoplasmic RNAs of BVDV CP7 cRNA-transfected cells were subjected to a cycle of hybridization and RNase treatment without an external probe. By this method, full-length negative-strand RNA molecules contained in the RNA pool were annealed to and thus protected by positive-strand counterparts. Cellular RNAs as well as unpaired surplus positive-strand RNAs were degraded. When the labeled sense probe was applied in a second protection round, a significantly lower (i.e., only equimolar) level of competing positive-strand RNA permitted the detection of negative-strand RNA with a higher efficiency (Fig. 2A, lanes 2). Neither negative- nor positive-strand RNA could be measured in cells transfected with 3′-truncated BVDV CP7 cRNA (Fig. 2A, lanes 3).
FIG. 1.
Schematic drawing of the different BVDV cRNAs used in this study. The individual viral polypeptides processed from the polyprotein are depicted with different kinds of shading. Ins, 27-base insertion characteristic of BVDV CP7 (55). The 3′-truncated RNAs are marked with the restriction cleavage sites used for linearization of the respective cDNA constructs (33; see also Materials and Methods). The Ser 1752 mutation is indicated in DI9cpm1.
FIG. 2.
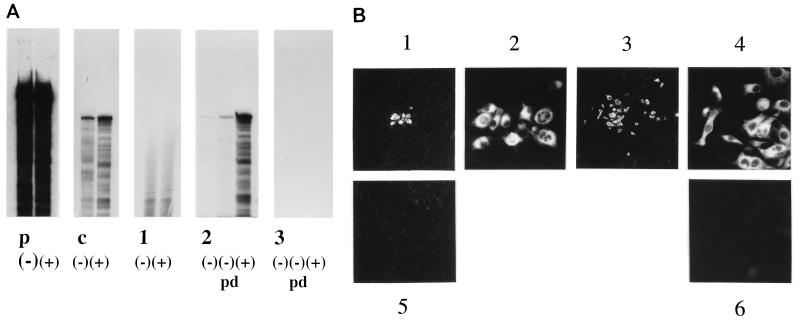
Monitoring of BVDV RNA replication. (A) RNase protection assay performed as described in Materials and Methods at 24 h p.t. with MDBK cells transfected with RNA derived from BVDV CP7 cRNA or BVDV CP7Δ3′ cRNA. Lanes: p, input sense (−) and antisense (+) probes (280 nucleotides); c, protected products obtained with sense (−) and antisense (+) probes after RNase protection with BVDV CP7 cDNA; 1, RNase protection carried out with RNA from the nuclear fraction of BVDV CP7 cRNA-transfected MDBK cells (left lane, protection with sense probe; right lane, protection with antisense probe); 2, RNase protection carried out with RNA from the cytoplasmic fraction of BVDV CP7 cRNA-transfected MDBK cells (left lane, protection with sense probe without prior cycle of prehybridization-predigestion (pd); middle lane, protection with sense probe after prior cycle of hybridization-predigestion (pd); right lane, protection with antisense probe); 3, same experiments as in lanes 2 but carried out with cytoplasmic RNA from BVDV CP7Δ3′ cRNA-transfected MDBK cells. The figure is an autoradiogram of a 10% polyacrylamide–7 M urea gel. (B) IF analysis at 24 h p.t. of MDBK cells transfected with either BVDV CP7 cRNA or BVDV CP7Δ3′ cRNA. Anti-NS3 antibody was used. 1, MDBK cells transfected with BVDV CP7 cRNA (magnification, ×100); 2, same as 1 but at a magnification of ×400; 3, different image of MDBK cells transfected with BVDV CP7 cRNA (magnification, ×100); 4, same as 3 but at a magnification of ×400; 5, MDBK cells transfected with BVDV CP7Δ3′ cRNA (magnification, ×100); 6, MDBK cells transfected with BVDV CP7Δ3′ cRNA (magnification, ×400).
These data confirmed the suitability of the RNase protection assay for our purpose—to monitor BVDV replication by direct detection of RNA products. Input RNA was not detected at either 6 or 24 h p.t. (Fig. 2A and data not shown). Thus, all positive-strand RNA determined in our experiments corresponded exclusively to de novo-synthesized progeny RNA. It was also shown that RNA replication of BVDV apparently takes place in an asymmetric way, a phenomenon that has already been reported for a number of other positive-strand RNA viruses (see below).
Replication of BVDV could be also indirectly monitored by measuring the synthesis of viral protein after transfection of cells with the in vitro-transcribed genomic cRNA. In the experiment shown in Fig. 2B, MDBK cells were transfected with either the full-length or the 3′-truncated genomic cRNA, fixed on the plate at 24 h p.t., and subjected to an IF analysis with a monoclonal antibody directed against the viral nonstructural protein NS3. When the full-length BVDV CP7 cRNA was transfected into MDBK cells, 0.5 to 5% of the cells exhibited positive IF (Fig. 2B). In accordance with the finding that at 24 h p.t. progeny BVDVs were already assembled and capable of reinfecting neighboring cells by horizontal spread, fluorescent single cells as well as formations consisting of several cells were found on the plate (Fig. 2B). Cells transfected with the 3′-truncated BVDV cRNA were negative in the IF assay (Fig. 2B). Hence, we concluded that the expression of NS3 detectable by IF requires RNA amplification rather than expression from unamplified input RNA. Data from the IF experiments are thus in agreement with the RNase protection assay data. Consequently, a positive IF pattern with an anti-NS3 antibody is an indication of viral RNA replication.
BVDV DI9c is an autonomous RNA replicon.
Simultaneously with the construction of infectious BVDV CP7 cDNA (see above), Meyers et al. (33) also generated a chimeric BVDV DI cDNA composed of sequences derived from the cp strains CP9 and CP7 (Fig. 1A). Like natural DI9, in vitro-generated transcripts of the hybrid DI cDNA (denoted as DI9c) transfected into cells previously infected with a non-cp helper virus were found to be viable, to induce a CPE in MDBK cells, and to exhibit all the characteristics of a DI particle (33; data not shown).
BVDV DI9c contains the coding regions for autoprotease Npro and the nonstructural proteins NS3 to NS5B as well as the 5′-UTR and 3′-UTR. Thus, the majority of viral factors and cis-acting RNA elements conceivably involved in viral replication should be encoded by this RNA molecule. It was therefore interesting to determine whether DI9c itself might already support either the first or even both steps of RNA replication, in other words, function as a partial or fully autonomous RNA replicon. Accordingly, MDBK cells were transfected via DEAE with in vitro-transcribed DI9c and examined at 24 h p.t. for RNA replication. Direct analysis of the RNA replication products by RNase protection (see above) revealed that in the cytoplasm of DI9c-transfected MDBK cells, de novo synthesis of negative-strand as well as positive-strand RNA indeed could be measured (Fig. 3A). From a series of parallel experiments, we estimated the amounts of RNA replication products yielded by DI9c-transfected cells to be in the same range as in cells transfected with the full-length BVDV cRNA (Fig. 2A and 3A). In keeping with these results, DI9c-transfected cells were positive in the anti-NS3 IF assay (Fig. 3B). However, in contrast to cells in which the entire BVDV genome had been introduced, mostly single cells exhibited positive IF staining. This finding was explained by the fact that DI9c does not encode viral structural proteins, so transfection of this subgenomic RNA consequently is not expected to yield infectious virions. Neighboring positive cells sometimes were detected, most likely due to passage of the replicating RNA to daughter cells during cell division (Fig. 3B). As expected, neither RNA replication nor synthesis of NS3 was observed in cells transfected with a 3′-truncated DI9c (Fig. 3B). Cross-contamination of the transfection mixtures with full-length BVDV RNA was ruled out by RT-PCR (data not shown). Taken together, these results demonstrate that BVDV DI9c encodes all factors and elements that are necessary and sufficient to catalyze its own amplification. However, it is remarkable and surprising that apparently neither the BVDV structural proteins nor NS2 is an essential part of the replication machinery.
FIG. 3.
Characterization of DI9c as an autonomous RNA replicon. (A) RNase protection analysis of MDBK and BHK-21 cells transfected with DI9c 24 h p.t. The protected fragments were analyzed on a 10% polyacrylamide–7 M urea gel. Lanes: p, input sense (−) and antisense (+) probes (1/10 of the experimental input was loaded); 1, RNase protection with cytoplasmic RNA obtained from MDBK cells transfected with DI9c using sense (−) and antisense (+) probes; 2, RNase protection with cytoplasmic RNA obtained from BHK-21 cells transfected with DI9c; 3, MDBK cells with DI9cΔ3′; 4, MDBK cells with DI9cpm1. (B) IF analysis of MDBK and BHK-21 cells transfected with DI9c, DI9cΔ3′, or DI9cpm1 24 h p.t. 1, MDBK cells with DI9c (magnification, ×100); 2 and 3, MDBK cells with DI9c (magnification, ×400); 4, BHK-21 cells with DI9c (magnification, ×100); 5, BHK-21 cells with DI9c (magnification, ×400); 6, MDBK cells with DI9cΔ3′ (magnification, ×100); 7, MDBK cells with DI9cpm1 (magnification, ×400); 8, BHK-21 cells with DI9cΔ3′ (magnification, ×100); 9, BHK-21 cells with DI9cpm1 (magnification, ×400).
Autonomous replication of BVDV DI9c is not limited to bovine host cells.
With the subgenomic DI9c RNA replicon, we found a suitable system available to initiate further investigations of the RNA replication mechanism. We wanted to address the question of whether the replication of DI9c is dependent on specific factors encoded by the bovine host cells for BVDV. Accordingly, we introduced in vitro-transcribed DI9c RNA into a range of different cell lines. Since it is known that BHK-21 (baby hamster kidney) cells can be transfected with RNA at a high efficiency (28), we first introduced DI9c via electroporation into these cells. As estimated in the IF assay, on average about 70 to 90% of BHK-21 cells could be successfully transfected by this method, and the replication products were detected by RNase protection (Fig. 3). Interestingly, the occurrence of DI9c replication also was observed in other cell lines, e.g., CHO (Chinese hamster ovary) cells and a number of human hepatocytes (data not shown). These data suggest that host proteins supposedly involved in the BVDV RNA replication pathway correspond to generally encoded rather than cell-type-specific factors. Apart from this main aspect, the significantly higher efficiency of transfection of RNA into BHK-21 cells than into MDBK cells (Fig. 3B) represented a remarkable improvement in the procedure and was thus used in all of the following experiments.
A DI9c derivative encoding a defective serine protease domain is replication deficient.
In comparison with the full-length cRNA, an autonomous RNA replicon offers several advantages for the investigation of the diverse functional elements involved in the replication process by reverse genetics. As an initial approach, a point mutation was introduced into the DI9c cDNA, changing Ser 1752, the catalytic residue of the NS3 serine protease, into an alanine (Fig. 1A). This mutation, pm1, was previously shown to completely abolish the enzymatic activity of transiently expressed pestivirus NS3 protease in vivo (55, 64). Analysis of DI9cpm1 RNA-transfected cells at 24 h p.t. revealed that this DI particle was replication deficient; i.e., neither RNA synthesis nor replication-associated viral protein synthesis could be detected (Fig. 3). Proteolytic processing of the DI RNA-encoded polyprotein NS3-NS4A-NS4B-NS5A-NS5B was therefore concluded as being an essential prerequisite for the initiation of RNA replication.
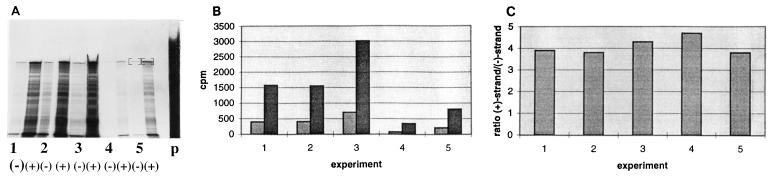
Determining the ratio of positive- and negative-strand RNAs in DI9c-transfected cells.
The above-described experiments demonstrated that the replication of DI9c proceeds independently from positive-strand RNA packaging and virion assembly. Accordingly, we assumed that in cells containing the replicating RNA, at a late stage p.t. the de novo synthesis of viral RNA and viral proteins should come to an equilibrium and the ratio of positive-strand RNA to negative-strand RNA should be constant. To substantiate this hypothesis, the ratio of positive-strand RNA to negative-strand RNA was determined in a number of independent transfection experiments by RNase protection 24 and 48 h p.t. (the latter not shown). Although the efficiency of RNA transfection varied significantly between different experiments, the ratio was found reproducibly to be in the range of 4:1 (Fig. 4). As expected, the same value was obtained independently whether RNAs were quantified 24 or 48 h p.t. (the latter not shown).
FIG. 4.
Determining the ratio of positive-strand RNA to negative-strand RNA in DI9c-transfected cells. BHK-21 cells were transfected in five independent experiments with DI9c, and the cytoplasmic RNA was extracted and subjected to an RNase protection assay 24 h p.t. To facilitate quantitation, positive and negative strands were both assayed with the same antisense probe, the latter by detection of positive strands which were protected by equimolar amounts of negative strands in a prehybridization-predigestion cycle (see Materials and Methods and Results). (A) The protected fragments were separated on a 10% polyacrylamide–7 M urea gel. p, amount of input probe that (with respect to the viral RNA) was added in excess into each protection assay mixture. (−), detected negative-strand RNA; (+), detected positive-strand RNA. (B) The major bands on the gel were extracted (indicated in panel A, lanes 5) and quantified by Cerenkov counting. Light grey columns represent the amounts of protected negative-strand RNA; dark grey columns represent the amounts of protected positive-strand RNA. (C) The ratio of positive-strand RNA to negative-strand RNA was calculated for each of the five experiments.
The 5′ extremity of the Npro-coding region is essential for DI9c replication.
BVDV DI particles characterized so far contain either the whole Npro gene or a portion of it linked in frame to the 5′ terminus of the NS3-coding region (27, 54). Conversely, the DI particles of the related CSFV completely lack the Npro gene (34). We decided to examine the role of the Npro protein and/or the Npro-coding region in replication. In a first attempt, the whole Npro gene of DI9c cDNA was removed and a translation initiation codon was fused to the NS3-coding region. Thus, a BVDV RNA was generated with a genomic organization colinear to that of naturally occurring CSFV DI particles (Fig. 5A). Surprisingly, this DI9c derivative (DI9cNS3) was found to be replication deficient, as judged by an IF assay (data not shown) and RNase protection (Fig. 5B, lanes 2). Next, the Npro-coding region was entirely substituted with a monomeric ubiquitin gene cassette (ubi), and the carboxy-terminal glycine 76 of ubi and a methionine were combined in frame with the N terminus of NS3 (DI9cubi) (Fig. 5A; see also Materials and Methods). Previous studies showed that cleavage of the ubi-NS3 fusion protein and generation of authentic NS3 are mediated by cellular ubiquitin carboxy-terminal hydrolases in trans (53). The release of the ubi monomer was verified by transient expression experiments with the T7 vaccinia virus system (17; data not shown). The introduction of DI9cubi into MDBK or BHK-21 cells, however, showed that complete replacement of the Npro-coding region also yielded a BVDV minigenome unable to replicate (Fig. 5B, lanes 3). Two interpretations were conceivable for this result: the viral Npro protein may have an additional function essential for RNA replication; alternatively, the genomic region coding for Npro or a part thereof contains an important RNA sequence element. Such an element should, however, be larger than the initial 12 nucleotides which, due to the cloning procedure, are identical in the BVDV CP7 Npro gene and the applied ubiquitin ORF (Fig. 5A; see also Materials and Methods). Accordingly, we engineered a mutant of DI9c (DI9cΔNproubi) in which the 5′ end of the ORF comprising the initial 126 nucleotides of the Npro-coding region of CP7 was fused in frame to the terminus of the ubi gene of clone DI9cubi (Fig. 5A). Proteolytic removal of the Npro-ubi fusion protein and the release of NS3 were again experimentally verified by T7 vaccinia virus-driven expression (data not shown). As shown in Fig. 5B, lanes 4, transfection of the DI9cΔNproubi RNA into target cells evidently gave rise to both steps of RNA replication. The possibility of contamination of the DI9cΔNproubi RNA by other replicating RNAs (such as DI9c) was ruled out by RT-PCR, which was carried out on total cytoplasmic RNA 24 h p.t. (data not shown). These data suggest that the 5′-terminal Npro-coding region is crucial for DI9c RNA replication.
DISCUSSION
cDNA copies of positive-strand viral RNA genomes that can be used to generate infectious RNA transcripts are considered powerful tools for detailed studies on the molecular features of virus infection, replication, and assembly via so-called reverse genetics. Encouraged by the recent successful engineering of a BVDV cDNA from cloned sequences, our original aim was to establish an efficient experimental system that allows investigation of the RNA replication process of a pestivirus. Initial experiments with full-length genomic RNA transcripts of BVDV CP7 revealed that viral replication could be monitored in host cells with two different methods of detection: RNase protection for direct determination of the in vivo-synthesized replication products and IF to measure replication-associated synthesis of the viral NS3 antigen. In the course of the reported experiments, both assay systems were found to detect neither input (transfected) BVDV cRNA nor protein expression of input RNA only. Accordingly, viral genomes lacking the RNA-dependent RNA polymerase-encoding region or containing certain mutations lethal for replication were shown to be negative in the respective assays.
De novo synthesis of viral RNA was determined to occur only in the cytoplasm of BVDV-transfected target cells and to proceed asymmetrically, with an excess of newly synthesized positive-strand RNA with respect to the negative-strand RNA intermediate. These data are in good accordance with the replication model of positive-strand RNA viruses, further confirming on the molecular level that BVDV is a typical example of such viruses. Our transfection experiments with cDNA-derived viral RNA are also congruent with the data of Chu and Westaway for Kunjin virus (8) as well as recent infection experiments of Gong et al., who monitored the synthesis of RNA intermediates in a one-step growth curve for BVDV (18).
The most important result of this study is the surprising finding that a subgenomic BVDV RNA, DI9c, comprising only 65% of the complete BVDV genome was demonstrated to support both steps of the replication pathway in vivo, therefore encoding all the proteins, motifs, and RNA sequences essentially involved in these diverse catalytic processes. With regard to its genetic organization, the DI9c RNA is a cDNA-derived homolog of the DI particle DI9, which was originally characterized as part of a cp BVDV CP9 isolate from cattle and which was found to grow in cell cultures together with an intact helper virus (54).
In comparison with a full-length virus, a subgenomic viral replicon provides some important advantages for genetic and biochemical studies. First, a major problem for the investigation of RNA viruses, namely, the genetic instability due to RNA recombination and the absence of a polymerase proofreading activity, is mostly circumvented in the short-term experimental approaches that make use of the replicon. Moreover, experiments with subgenomic RNA offer the opportunity to examine RNA replication as an isolated process. Separation of RNA replication from virion assembly and maturation is reflected by an equilibrium of positive- and negative-strand RNAs, with a relatively small excess of positive-strand RNA 24 h p.t. and later in DI9c-transfected cells (Fig. 4). However, the 4:1 ratio determined here has to be considered a DI particle-specific value which cannot be applied to an infection with the complete BVDV genome. DI9c provides a valuable system for the further examination of the replication process as well as for the possible future development of a BVDV replicon-based system for foreign gene expression.
DI9c was found to replicate in numerous different eucaryotic cell types. Data obtained predominantly with bacteriophage Qβ (reviewed in reference 6) and some well-characterized positive-strand RNA plant virus systems (reviewed in reference 12) suggest that these viruses may, as a rule, recruit cellular factors fulfilling specific functions in viral genome replication. Given that, as in poliovirus, translation of the viral polyprotein and viral replication are also closely linked in BVDV and given that host factors are definitely involved in the replication pathway, it is tempting to speculate that these factors may correspond to universal components of the translation apparatus, the largest RNA-protein complex present in the cytoplasm (1, 3, 4).
From our initial genetic experiments, in which the NS3 protease of DI9c was inactivated, we concluded that processing of the DI particle-encoded NS polyprotein is a necessary requirement to permit the first step of RNA replication. Of course, this conclusion does not imply that only fully mature viral nonstructural proteins NS3 to NS5B fulfill diverse enzymatic functions. Hence, additional experiments similar to those that have been done with alphaviruses (28, 48) are needed to determine the in vivo functions of mature polypeptides as well as of putative functional processing intermediates.
The BVDV DI9c replicon lacks the genes encoding the polypeptides that presumably form the virion, i.e., the core protein, the envelope proteins Erns (exhibiting an interesting RNase activity) (46), E1, and E2, and the p7 protein (15). The fact that all of the structural proteins turned out to be dispensable for the replication process is in keeping with similar results obtained for poliovirus (10, 21, 25, 39) and for alphaviruses, for which DI particle-like replicons were shown to encompass only the genomic part encoding the viral nonstructural proteins (reviewed in reference 52). In view of these data, it is intriguing that DI9c apparently replicates in the absence of the N-terminal part of nonstructural protein NS2-3, namely, the NS2 protein. The functional role of NS2 in the life cycle of pestiviruses and the related HCV is still elusive. Proteolytic processing of BVDV NS2-3 was shown to be a direct consequence of genomic rearrangements and to be related to numerous severe consequences for the pathogenesis of a viral infection (summarized in reference 57). Conversely, in HCV, NS2/3 is processed by a viral autoprotease spanning the carboxy terminus of the NS2 protein and the serine protease domain located in the amino-terminal region of NS3 (20, 22). For some HCV isolates, uncleaved NS2 was shown to influence the proper biogenesis of the downstream polypeptides (14). Santolini et al. demonstrated that the HCV NS2 polypeptide is integrated into the endoplasmic reticulum membrane and appears to be associated with the envelope proteins of the virus (44). Similar observations made by Selby et al. (47) therefore prompted the speculation that NS2 might function as a structural rather than a nonstructural viral component. On the other hand, NS2 of HCV was also reported to coprecipitate with NS5A and NS5B (23) and thus, along with these viral proteins, ought to be considered part of the replication complex. In sum, further experiments are needed to shed some light on the functional role of pestivirus as well as HCV NS2 proteins.
Deletion of the entire Npro-coding region resulted in RNAs that were incapable of replicating. However, DI RNAs that contained the 5′ extremity of the ORF (126 nucleotides), thus encoding only 42 amino acids of Npro and a complete ubiquitin monomer that could substitute for the proteolytic function of Npro, were surprisingly found to be replication competent (Fig. 5). Even though a putative function of the residual Npro peptide cannot be ruled out, these data more likely suggest that an essential genomic sequence element is harbored within the Npro-coding region, like cis-acting RNA elements at the respective genomic locations in the flavivirus Kunjin (26) and HCV (31). For the flavivirus, the function of the ORF 5′ end was stated to be unrelated to translation initiation, indicating that an important replication signal is encoded by the RNA sequence. For HCV, 366 nucleotides of the core-encoding sequence were determined most likely to be part of the IRES. Since in pestiviruses initiation of translation of the polyprotein is also directed by an IRES element (40, 42), our experimental data may indeed indicate that the RNA sequence of the BVDV ORF represents an essential structural and functional part of the IRES. Kupfermann et al. recently reported the molecular characterization of DI13, a DI RNA that encodes only 13 amino acids of Npro fused to 10 amino acids of E1 and NS3, the latter missing 5 amino acids at its amino terminus (27). Independent DI isolates of the pestivirus CSFV were reproducibly characterized to contain a translation initiation codon linked to glycine 1590, the supposed N terminus of NS3. These observations suggest that the sequence requirements at the genomic 5′ end of BVDV might be less than 126 nucleotides. A CSFV-analogous BVDV DI particle (DI9cNS3) as well as a replicon containing ubiquitin instead of Npro (DI9cubi) were incapable of autonomous replication, even though in the latter 12 of the initial nucleotides of the Npro-coding region and the applied artificial ubiquitin gene cassette are identical. In view of this scenario, it can be stated that for BVDV CP7, a hypothetical cis-acting RNA element localized at the 5′ end of the ORF should be between 12 and 126 nucleotides. Additional studies are needed to further confirm and elucidate the functional role of such an RNA element during the replication of BVDV CP7. Since it cannot be ruled out that the described CSFV DI particles may require the presence of a helper virus for replication, comparative experiments are currently in progress to understand the discrepancies in the genomic organization of different pestivirus DI particles.
The information that only 5 of 12 intact polypeptide-encoding regions of BVDV CP7 were defined as being sufficient to catalyze the complete RNA replication pathway of the virus will facilitate future efforts to elucidate the basic mechanism of the viral multiplication strategy. Another fascinating aspect concerns the genomic organization of the region encoding NS3 to NS5B, which in BVDV and human HCV is virtually colinear, fueling speculation that the replication strategies of both viruses follow similar rules. Since a suitable infectious system is still lacking for HCV, our data may also contribute to a better understanding of the life cycle of this human pathogen.
ACKNOWLEDGMENTS
We thank Astrid Kaiser for some technical assistance.
This study was supported by the SFB 535 ‘Invasionsmechanismen und Replikationsstrategien von Krankheitserregern’ from the Deutsche Forschungsgemeinschaft. S.-E.B. is supported by the Infektionsforschung-Stipendienprogramm (2131) of the BMBF (Bundesministerium Bildung und Forschung) administrated by the Deutsches Krebsforschungszentrum (DKFZ).
REFERENCES
- 1.Andino R, Rieckhof G E, Achacoso P L, Baltimore D. Poliovirus RNA synthesis utilizes an RNP complex formed around the 5′-end of viral RNA. EMBO J. 1993;12:3587–3598. doi: 10.1002/j.1460-2075.1993.tb06032.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartenschlager R, Ahlborn-Laake L, Mous J, Jacobsen H. Nonstructural protein 3 of the hepatitis C virus encodes a serine-type proteinase required for cleavage at the NS3/4 and NS4/5 junctions. J Virol. 1993;67:3835–3844. doi: 10.1128/jvi.67.7.3835-3844.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barton D J, Flanegan J B. Coupled translation and replication of poliovirus RNA in vitro: synthesis of functional 3D polymerase and infectious virus. J Virol. 1993;67:822–831. doi: 10.1128/jvi.67.2.822-831.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barton D J, Black P, Flanegan J B. Complete replication of poliovirus in vitro: preinitiation RNA replication complexes require soluble cellular factors for the synthesis of VPg-linked RNA. J Virol. 1995;69:5516–5527. doi: 10.1128/jvi.69.9.5516-5527.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Behrens S-E, Tomei L, De Francesco R. Identification and properties of the RNA-dependent RNA polymerase of hepatitis C virus. EMBO J. 1995;15:12–22. [PMC free article] [PubMed] [Google Scholar]
- 6.Blumenthal T, Carmichael G G. RNA replication: function and structure of Qβ-replicase. Annu Rev Biochem. 1979;48:525–548. doi: 10.1146/annurev.bi.48.070179.002521. [DOI] [PubMed] [Google Scholar]
- 7.Bredenbeek P, Rice C M. Animal RNA virus expression systems. Semin Virol. 1992;3:297–310. [Google Scholar]
- 8.Chu P W G, Westaway E G. Replication strategy of Kunjin virus: evidence for recycling role of replicative form RNA as template in semiconservative and asymmetric replication. Virology. 1985;140:68–79. doi: 10.1016/0042-6822(85)90446-5. [DOI] [PubMed] [Google Scholar]
- 9.Collett M S. Molecular genetics of pestiviruses. Comp Immunol Microbiol Infect Dis. 1992;15:145–154. doi: 10.1016/0147-9571(92)90087-8. [DOI] [PubMed] [Google Scholar]
- 10.Collis P S, O’Donell B J, Barton D, Rogers J A, Flanegan J B. Replication of poliovirus RNA and subgenomic RNA transcripts in transfected cells. J Virol. 1992;66:6480–6488. doi: 10.1128/jvi.66.11.6480-6488.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Corapi W V, Donis R O, Dubovi E J. Characterization of a panel of monoclonal antibodies and their use in the study of the antigenic diversity of bovine viral diarrhea virus. Am J Vet Res. 1990;51:1388–1394. [PubMed] [Google Scholar]
- 12.David C, Gargouri-Bouzid R, Haenni A-L. RNA replication of plant viruses containing an RNA genome. Prog Nucleic Acid Res Mol Biol. 1992;42:157–227. doi: 10.1016/s0079-6603(08)60576-0. [DOI] [PubMed] [Google Scholar]
- 13.Deng R, Brock K V. Molecular cloning and nucleotide sequence of a pestivirus genome, noncytopathogenic bovine viral diarrhea virus strain SD-1. Virology. 1992;191:867–879. doi: 10.1016/0042-6822(92)90262-n. [DOI] [PubMed] [Google Scholar]
- 14.D’Souza E D A, O’Sullivan E, Amphlett E M, Rowlands D J, Sangar D V, Clarke B E. Analysis of NS3-mediated processing of the hepatitis C virus nonstructural region in vitro. J Gen Virol. 1994;75:3469–3476. doi: 10.1099/0022-1317-75-12-3469. [DOI] [PubMed] [Google Scholar]
- 15.Elbers K, Tautz N, Becher P, Rümenapf T, Thiel H-J. Processing in the pestivirus E2-NS2 region: identification of the nonstructural proteins p7 and E2-p7. J Virol. 1996;70:4131–4135. doi: 10.1128/jvi.70.6.4131-4135.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Francki R I, Fauquet D L, Knudson D L, Brown F. Classification and nomenclature of viruses. Arch Virol Suppl. 1991;2:223–233. [Google Scholar]
- 17.Fuerst T R, Niles E G, Studier F W, Moss B. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc Natl Acad Sci USA. 1986;83:8122–8126. doi: 10.1073/pnas.83.21.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gong Y, Trowbridge R, Macnaughton T B, Westaway E G, Shannon A D, Gowans E J. Characterization of RNA synthesis during a one-step growth curve and of the replication mechanism of bovine viral diarrhoea virus. J Gen Virol. 1996;77:2729–2736. doi: 10.1099/0022-1317-77-11-2729. [DOI] [PubMed] [Google Scholar]
- 19.Grakoui A, McCourt D W, Wychowski C, Feinstone S M, Rice C M. Characterization of the hepatitis C virus-encoded serine proteinase: determination of proteinase-dependent polyprotein cleavage sites. J Virol. 1993;67:2832–2843. doi: 10.1128/jvi.67.5.2832-2843.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grakoui A, McCourt D W, Wychowski C, Feinstone S M, Rice C M. A second hepatitis C virus-encoded proteinase. Proc Natl Acad Sci USA. 1993;90:10583–10587. doi: 10.1073/pnas.90.22.10583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hagino-Yamagashi K, Nomoto A. In vitro construction of poliovirus defective interfering particles. J Virol. 1989;63:5386–5392. doi: 10.1128/jvi.63.12.5386-5392.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hijikata M, Mizushima H, Akagi T, Mori S, Kakiuchi N, Kato N, Tanaka T, Kimura K, Shimotohno K. Two distinct proteinase activities required for the processing of a putative nonstructural precursor protein of hepatitis C virus. J Virol. 1993;67:4665–4675. doi: 10.1128/jvi.67.8.4665-4675.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hijikata M, Mizushima H, Tanji Y, Komoda Y, Hirowatari Y, Akagi T, Kato N, Kimura K, Shimotohno K. Proteolytic processing and membrane association of putative nonstructural proteins of hepatitis C virus. Proc Natl Acad Sci USA. 1993;90:10773–10777. doi: 10.1073/pnas.90.22.10773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Holland J J. Defective viral genomes. In: Fields B N, Knipe D M, editors. Fundamental virology. 2nd ed. New York, N.Y: Raven Press, Ltd.; 1991. pp. 151–165. [Google Scholar]
- 25.Kaplan G, Racaniello V R. Construction and characterization of poliovirus subgenomic replicons. J Virol. 1988;62:1687–1696. doi: 10.1128/jvi.62.5.1687-1696.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Khromykh A A, Westaway E G. Subgenomic replicons of the flavivirus Kunjin: construction and applications. J Virol. 1997;71:1497–1505. doi: 10.1128/jvi.71.2.1497-1505.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kupfermann H, Thiel H-J, Dubovi E J, Meyers G. Bovine viral diarrhea virus: characterization of a cytopathogenic defective interfering particle with two internal deletions. J Virol. 1996;70:8175–8181. doi: 10.1128/jvi.70.11.8175-8181.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lemm J A, Rümenapf T, Strauss E G, Strauss J H, Rice C M. Polypeptide requirements for assembly of functional Sindbis virus replication complexes: a model for the temporal regulation of minus- and plus-strand RNA synthesis. EMBO J. 1994;13:2925–2934. doi: 10.1002/j.1460-2075.1994.tb06587.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liljestrom R, Garoff H. A new generation of animal cell expression vectors based on the Semliki Forest virus replicon. Bio/Technology. 1991;9:1356–1361. doi: 10.1038/nbt1291-1356. [DOI] [PubMed] [Google Scholar]
- 30.Linnen J, Wages J, Zhang-Keck Z-Y, Fry K E, Krawczinsky K Z, Alter H, Koonin E, Gallagher M, Alter M, Hadziyannis S, Karayiannis P, Fung K, Nakatsuji Y, Shih J W-K, Young L, Piatak M, Hoover C, Fernandez J, Chen S, Zou J-C, Morris T, Hyams K C, Ismay S, Lifson J D, Hess G, Foung S K H, Thomas H, Bradley D, Margolis H, Kim J P. Molecular cloning and disease association of hepatitis G virus: a transfusion-transmissible agent. Science. 1996;271:505–508. doi: 10.1126/science.271.5248.505. [DOI] [PubMed] [Google Scholar]
- 31.Lu H H, Wimmer E. Poliovirus chimeras replicating under the translational control of genetic elements of hepatitis C virus reveal unusual properties of the internal ribosomal entry site of hepatitis C virus. Proc Natl Acad Sci USA. 1996;93:1412–1417. doi: 10.1073/pnas.93.4.1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meyers G, Thiel H-J. Molecular characterization of pestiviruses. Adv Virus Res. 1995;47:53–118. doi: 10.1016/s0065-3527(08)60734-4. [DOI] [PubMed] [Google Scholar]
- 33.Meyers G, Tautz N, Becher P, Thiel H-J, Kümmerer B M. Recovery of cytopathogenic and noncytopathogenic bovine viral diarrhea viruses from cDNA constructs. J Virol. 1996;70:8606–8613. doi: 10.1128/jvi.70.12.8606-8613.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meyers G, Thiel H-J. Cytopathogenicity of classical swine fever virus caused by defective interfering particles. J Virol. 1995;69:3683–3689. doi: 10.1128/jvi.69.6.3683-3689.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meyers G, Thiel H-J, Rümenapf T. Classical swine fever virus: recovery of infectious viruses from cDNA constructs and generation of recombinant cytopathogenic defective interfering particles. J Virol. 1996;70:1588–1595. doi: 10.1128/jvi.70.3.1588-1595.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miller R H, Purcell R H. Hepatitis C virus shares amino acid sequence similarity with pestiviruses and flaviviruses as well as members of two plant virus supergroups. Proc Natl Acad Sci USA. 1990;87:2057–2061. doi: 10.1073/pnas.87.6.2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moormann R J M, van Gennip H G P, Miedema G K L, Hulst M M, van Rijn P A. Infectious RNA transcribed from an engineered full-length cDNA template of the genome of a pestivirus. J Virol. 1996;70:763–770. doi: 10.1128/jvi.70.2.763-770.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Novak J E, Kirkegaard K. Improved method for detecting poliovirus negative strands used to demonstrate specificity of positive-strand encapsidation and the ratio of positive to negative strands in infected cells. J Virol. 1991;65:3384–3387. doi: 10.1128/jvi.65.6.3384-3387.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Percy N, Barclay W S, Sullivan M, Almond J W. A poliovirus replicon containing the chloramphenicol acetyltransferase gene can be used to study the replication and encapsidation of poliovirus RNA. J Virol. 1992;66:5040–5046. doi: 10.1128/jvi.66.8.5040-5046.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Poole T L, Wang C, Popp R A, Potgieter L N D, Siddiqui A, Collett M S. Pestivirus translation initiation occurs by internal ribosome entry. Virology. 1995;206:750–754. doi: 10.1016/s0042-6822(95)80003-4. [DOI] [PubMed] [Google Scholar]
- 41.Rice C M. Flaviviridae: the viruses and their replication. In: Fields B N, editor. Virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 931–961. [Google Scholar]
- 42.Rijnbrand R, van der Straaten T, van Rijn P A, Spaan W J M, Bredenbeek P J. Internal entry of ribosomes is directed by the 5′ noncoding region of classical swine fever virus and is dependent on the presence of an RNA pseudoknot upstream of the initiation codon. J Virol. 1997;71:451–457. doi: 10.1128/jvi.71.1.451-457.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ruggli N, Tratschin J D, Mittelholzer C, Hofmann M A. Nucleotide sequence of classical swine fever strain Alfort/187 and transcription of infectious RNA from stably cloned full-length cDNA. J Virol. 1996;70:3478–3487. doi: 10.1128/jvi.70.6.3478-3487.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Santolini E, Pacini L, Fipaldini C, Migliaccio G, La Monica N. The NS2 protein of hepatitis C virus is a transmembrane polypeptide. J Virol. 1995;69:7461–7471. doi: 10.1128/jvi.69.12.7461-7471.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schlesinger S. The generation and amplification of defective interfering RNAs. In: Domingo E, Holland J J, Ahlquist P, editors. RNA genetics. II. Boca Raton, Fla: CRC Press, Inc.; 1988. pp. 167–185. [Google Scholar]
- 46.Schneider R, Unger G, Stark R, Schneider-Scherzer E, Thiel H-J. Identification of a structural glycoprotein of an RNA virus as a ribonuclease. Science. 1993;261:1169–1171. doi: 10.1126/science.8356450. [DOI] [PubMed] [Google Scholar]
- 47.Selby M J, Glazer E, Masiarz F, Houghton M. Complex processing and protein:protein interactions in the E2:NS2 region of HCV. Virology. 1994;204:114–122. doi: 10.1006/viro.1994.1515. [DOI] [PubMed] [Google Scholar]
- 48.Shirako Y, Strauss J H. Regulation of Sindbis virus RNA replication: uncleaved P123 and nsP4 function in minus-strand RNA synthesis, whereas cleaved products from P123 are required for efficient plus-strand RNA synthesis. J Virol. 1994;68:1874–1885. doi: 10.1128/jvi.68.3.1874-1885.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Simons J N, Pilot-Matias T J, Leary T P, Dawson G H, Desai S M, Schlauder G G, Muerhoff A S, Erker J C, Buijk S L, Chalmers M L, VanSant C L, Mushahwar I K. Identification of two flavivirus-like genomes in the GB hepatitis agent. Proc Natl Acad Sci USA. 1995;92:3401–3405. doi: 10.1073/pnas.92.8.3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Simons J N, Leary T P, Dawson G J, Pilot-Matias T J, Muerhoff A S, Schlauder G G, Desai S M, Mushahwar I K. Isolation of novel virus-like sequences associated with human hepatitis. Nat Med. 1995;1:564–569. doi: 10.1038/nm0695-564. [DOI] [PubMed] [Google Scholar]
- 51.Stark R, Meyers G, Rümenapf T, Thiel H-J. Processing of pestivirus polyprotein: cleavage site between autoprotease and nucleocapsid protein of classical swine fever virus. J Virol. 1993;67:7088–7095. doi: 10.1128/jvi.67.12.7088-7095.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Strauss J H, Strauss E G. The alphaviruses: gene expression, replication, and evolution. Microbiol Rev. 1994;58:491–562. doi: 10.1128/mr.58.3.491-562.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tautz N, Meyers G, Thiel H-J. Processing of poly-ubiquitin in the polyprotein of an RNA virus. Virology. 1993;197:74–85. doi: 10.1006/viro.1993.1568. [DOI] [PubMed] [Google Scholar]
- 54.Tautz N, Thiel H-J, Dubovi E J, Meyers G. Pathogenesis of mucosal disease: a cytopathogenic pestivirus generated by an internal deletion. J Virol. 1994;68:3289–3297. doi: 10.1128/jvi.68.5.3289-3297.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tautz N, Meyers G, Stark R, Dubovi E J, Thiel H-J. Cytopathogenicity of a pestivirus correlates with a 27-nucleotide insertion. J Virol. 1996;70:7851–7858. doi: 10.1128/jvi.70.11.7851-7858.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tautz N, Elbers K, Stoll D, Meyers G, Thiel H-J. Serine protease of pestiviruses: determination of cleavage sites. J Virol. 1997;71:5415–5422. doi: 10.1128/jvi.71.7.5415-5422.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thiel H-J, Plagemann P G W, Moennig V. Pestiviruses. In: Fields B N, editor. Virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 1059–1074. [Google Scholar]
- 58.Tomei L, Failla C, Santolini E, De Francesco R, La Monica N. NS3 is a serine protease required for processing of hepatitis C virus polyprotein. J Virol. 1993;67:4017–4026. doi: 10.1128/jvi.67.7.4017-4026.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tsukiyama-Kohara K, Iizuka N, Kohara M, Nomoto A. Internal ribosome entry site within hepatitis C virus RNA. J Virol. 1992;66:1476–1483. doi: 10.1128/jvi.66.3.1476-1483.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vassilev V B, Collett M S, Donis R O. Authentic and chimeric full-length genomic cDNA clones of bovine viral diarrhea virus that yield infectious transcripts. J Virol. 1997;71:451–457. doi: 10.1128/jvi.71.1.471-478.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Warrener P, Collett M S. Pestivirus NS3 (p80) protein possesses RNA helicase activity. J Virol. 1995;69:1720–1726. doi: 10.1128/jvi.69.3.1720-1726.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Westaway E G. Flavivirus replication strategy. Adv Virus Res. 1987;33:45–90. doi: 10.1016/s0065-3527(08)60316-4. [DOI] [PubMed] [Google Scholar]
- 63.Wiskerchen M, Belzer S K, Collett M S. Pestivirus gene expression: the first protein product of the bovine viral diarrhea virus large open reading frame, p20, possesses proteolytic activity. J Virol. 1991;65:4508–4514. doi: 10.1128/jvi.65.8.4508-4514.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wiskerchen M, Collett M S. Pestivirus gene expression: protein p80 of bovine viral diarrhea virus is a proteinase involved in polyprotein processing. Virology. 1991;184:341–350. doi: 10.1016/0042-6822(91)90850-b. [DOI] [PubMed] [Google Scholar]
- 65.Xu J, Mendez E, Caron P R, Lin C, Murcko M A, Collett M S, Rice C M. Bovine viral diarrhea virus NS3 serine protease: polyprotein cleavage sites, cofactor requirements, and molecular model of an enzyme essential for pestivirus replication. J Virol. 1997;71:5312–5322. doi: 10.1128/jvi.71.7.5312-5322.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]