Abstract
African swine fever (ASF) virus is a large DNA virus that shares the striking icosahedral symmetry of iridoviruses and the genomic organization of poxviruses. Both groups of viruses have a complex envelope structure. In this study, the mechanism of formation of the inner envelope of ASF virus was investigated. Examination of thin cryosections by electron microscopy showed two internal membranes in mature intracellular virions and all structural intermediates. These membranes were in continuity with intracellular membrane compartments, suggesting that the virus gained two membranes from intracellular membrane cisternae. Immunogold electron microscopy showed the viral structural protein p17 and resident membrane proteins of the endoplasmic reticulum (ER) within virus assembly sites, virus assembly intermediates, and mature virions. Resident ER proteins were also detected by Western blotting of isolated virions. The data suggested the ASF virus was wrapped by the ER. Analysis of the published sequence of ASF virus (R. J. Yanez et al., Virology 208:249–278, 1995) revealed a reading frame, XP124L, that encoded a protein predicted to translocate into the lumen of the ER. Pulse-chase immunoprecipitation and glycosylation analysis of pXP124L, the product of the XP124L gene, showed that pXP124L was retained in the ER lumen after synthesis. When analyzed by immunogold electron microscopy, pXP124L localized to virus assembly intermediates and fully assembled virions. Western blot analysis detected pXP124L in virions isolated from Percoll gradients. The packaging of pXP124L from the lumen of the ER into the virion is consistent with ASF virus being wrapped by ER cisternae: a mechanism which explains the presence of two membranes in the viral envelope.
African swine fever (ASF) virus is a large icosahedral enveloped DNA virus that causes a lethal hemorrhagic disease in domestic pigs. The virus is endemic in areas of southern Europe and in Africa where it causes major problems for the development of pig industries. At present there are no vaccines, and the disease is controlled through the slaughter of infected animals. The economic importance of ASF virus has made the virus the focus of much research since it was first described in 1921 (32). ASF virus is unique among animal viruses, and its classification has been controversial. ASF virus shares the striking icosahedral symmetry of iridoviruses (5, 8, 13, 34), while the presence of inverted terminal repeats and covalently linked ends in the 170-kDa genome suggests similarities with poxviruses (16). The ASF virus genome encoding at least 150 proteins has been sequenced (17, 51), and the amino acid sequences of at least 11 structural proteins are known. p73 is the major structural protein (14, 28) and has sequence similarities to the capsid protein of iridoviruses (39). The ordered proteolysis of pp220 produces p150, p37/p34 and p14 (40), which together comprise 25% of the viral proteins (3). These proteins localize to the interior of the virion (3). Three proteins, J13L/p54, I1L/p17, and p22, with membrane-spanning domains localize to the viral envelope (10, 37, 41, 43). Three other structural proteins, p14.5 encoded by E120R (30), p10 encoded by K78R (35), and p5AR encoded by A104R (7), have DNA-binding properties (51) and may be involved in DNA packaging. The virus has been the subject of several detailed electron microscopy studies (2–4, 8, 9, 11, 13, 34, 47). Electron micrographs of sections taken through ASF virus assembly sites reveal fully assembled virions as 200-nm hexagons and an ordered series of assembly intermediates with one to six sides of a hexagon. Close inspection of intracellular virions identifies multiple concentric layers of differing electron densities. According to recent models, the layers represent a central electron-dense nucleocapsid core, surrounded by an inner core shell, an inner envelope, and an outer capsid layer (3). The mechanism of formation of the inner envelope of ASF virus has not been resolved.
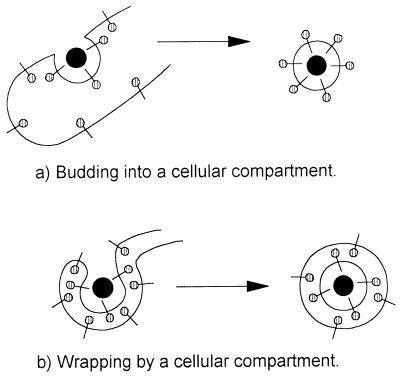
Most viruses gain a single membrane envelope by budding into intracellular membrane compartments or from the plasma membrane, as reviewed in reference 21. When viruses bud into an intracellular compartment, the domains of the membrane proteins that are initially located in the lumen of membrane compartments are exposed on the outside of the virion after release from the cell (Fig. 1a). A second mechanism of envelopment, described recently for poxviruses and herpesviruses (18, 20, 24, 38, 42, 46, 50), is more complex and involves the wrapping of virions by membrane cisternae derived from specific membrane compartments. Wrapping provides two membrane envelopes in one step and leaves the virion free in the cytoplasm. When compared with budding, wrapping reverses the orientation of membrane proteins within the virus such that the domains of membrane proteins located in the lumen of the wrapping organelle are confined to the interior of the virus after release from the cell, whereas cytoplasmic tails are exposed on the outside of the virus (Fig. 1b). Given these important consequences for understanding the mechanism of assembly of the virus and for determining the final orientation of membrane proteins in virions, we have set out to determine whether ASF virus acquires its membranes by the conventional budding mechanism or whether the virus is wrapped by intracellular membrane compartments before release from the cell.
FIG. 1.
Schematic comparison of budding and wrapping mechanisms of virus envelopment. (a) Budding. Viral nucleoprotein complexes bind to the cytoplasmic domains of virally encoded integral membrane proteins (| , membrane glycoproteins). Interactions between viral proteins lead to membrane curvature, and the virion gains a single membrane by budding into the lumen of the membrane compartment. When the virion is released from the cell, oligosaccharides ( ) are exposed on the surface of the virus, and the cytoplasmic tail of the membrane glycoprotein is buried within the virion. (b) Wrapping. Viral nucleoprotein complexes bind to the cytoplasmic domains of virally encoded integral membrane proteins. The nucleoprotein complex is then wrapped by the membrane cisternae, and the virus gains two membranes. The particle remains in the cytosol. When the virion is released from the cell by cell lysis, oligosaccharides ( ) are buried within the two membranes of the virion while the cytoplasmic tail of the membrane glycoprotein is exposed on the surface of the virus.
In this study we have taken advantage of thin cryoelectron microscopic sections to enhance the definition of viral membranes. The micrographs show two membranes within mature intracellular virions and all structural intermediates. They also show assembly intermediates in continuity with cellular membrane compartments. Consistent with our earlier study showing that p73 was enveloped by the endoplasmic reticulum (ER) (15), immunogold labelling experiments show resident proteins of the ER within membranes found at assembly sites, in virus assembly intermediates, and in mature virions. Importantly, we have identified a protein (pXP124L) encoded by ASF virus that translocates completely into the lumen of the ER and is incorporated as a structural protein of the virus. The presence of two membranes within intracellular virions and structural intermediates and the packaging of a structural protein from the lumen of the ER into the virus, strongly suggest that ASF virus is wrapped by the ER.
MATERIALS AND METHODS
Cells and viruses.
RS2 cells, a renal swine cell line, generated and maintained at the Institute for Animal Health (Pirbright Laboratory, Surrey, United Kingdom), Vero cells (ECACC 84113001), and CHO-K1 cells (ECACC 85051005) were grown at 37°C in HEPES-buffered Dulbecco’s modified Eagles medium supplemented with 10% fetal calf serum, l-glutamine (20 mM), penicillin (100 U/ml), and streptomycin (100 U/ml). Tissue culture-adapted BA71v and Uganda isolates of ASF virus and the virulent Malawi isolate have been described previously (11, 43). Vesicular stomatis virus (VSV) Indiana isolate was obtained from Nigel Ferris (Institute for Animal Health, Pirbright Laboratories).
Antibodies.
RxER was a generous gift from Daniel Louvard and Evelyne Coudrier (29; Morphogenese et signalisation cellulaire, Institut Curie, Paris, France), and P5D4, recognizing the cytoplasmic tail of VSV G protein, was from Thomas Kries (25; Department of Cell Biology, University of Geneva, Geneva, Switzerland). The monoclonal antibody 4H3 recognizes p73, the major structural protein of ASF virus, and has been described previously (15). Antibody 17KG12, recognizing the p17 product of the D117R gene in BA71v (I1L gene in Malawi) (41), was obtained from Igensa (Madrid, Spain). Rabbit antibody specific for ERP72 (31) was a gift from Peter Arvan (Albert Einstein College of Medecine, New York, N.Y.), and rabbit antibody specific for calnexin was a gift from Ineke Braakman and Ari Helenius (Department of Cell Biology, Yale University, New Haven, Conn.). Antibody recognizing ERP60 was raised in rabbits by using a peptide PIIQEEKPKKKKKAQEDL found at the C terminus of the protein (22). Antibodies recognizing pXP124L were raised in rabbits by using a peptide SNPVPHNRPHRLGRKIYEK which represents 19 amino acids found at the carboxy terminus of the protein sequence predicted from the work of Almendral et al. (1).
Electron microscopy. (i) Ultrathin resin section.
Infected cells grown in tissue culture flasks were fixed for 15 to 30 min simultaneously with 2% paraformaldehyde and 2.5% glutaraldehyde in 100 mM cacodylate (pH 7.2). The definition of membranes was enhanced by incubating fixed cells in 0.75% diaminobenzidine (Sigma-Aldrich Company Ltd, Poole, England) in Tris-HCl (50 mM, pH 7.5) containing 0.07% hydrogen peroxide for 30 min (37a). Cells were then washed, scraped from the tissue culture flask, and intensified for 1 h at room temperature by using 1% osmium tetroxide in 100 mM cacodylate (pH 7.2). Cells were progressively dehydrated in ethanol, embedded in Spurr resin (Agar Scientific Ltd, Stansted, England), and polymerized overnight at 70°C. Ultrathin sections (70 to 90 nm), cut with a Reichert, Austra, OmU3 microtome using glass knives, were contrasted directly for 20 min in 50% ethanol saturated with uranyl acetate, washed in double-distilled H2O, stained using Renyold’s lead citrate for 5 min in an NaOH-rich atmosphere, and finally washed in double-distilled H2O. The sections were examined with a JEOL 1200 EX electron microscope.
(ii) Immunogold electron microscopy.
Cells were embedded in Lowicryl resin after being dehydrated in ethanol at progressively lower temperatures (23, 48). Briefly, infected adherent cells were fixed in 3% paraformaldehyde in Sorenson’s phosphate buffer (SPB; 0.1 M NaH2PO4 · H2O–0.1 M Na2HPO4, pH 7.2) on ice. Cells were scraped off plates, pelleted, and washed in SPB before being dehydrated progressively in ethanol for 20 min per step as follows: 30% ethanol at 4°C, 50% ethanol at −20°C, 70% ethanol at −30°C, followed by 95% and 100% ethanol again at −30°C. Cells were then infiltrated progressively with Lowicryl resin (K4M or HM20; Agar Scientific Ltd). Resin was polymerized at −30°C for 20 h and 1 day at room temperature under long-wave UV light. Sections mounted on nickel grids were blocked for 1 h in 5× blocking buffer (5% bovine serum albumin, 5% normal goat serum, 50 mM glycine–phosphate-buffered saline [PBS], pH 7.4) and incubated overnight at 4°C with the first antibody diluted in 1× blocking buffer. Grids were washed in 1× blocking buffer and incubated with a secondary antibody (goat anti-rabbit immunoglobulin G [IgG] conjugated to 10- or 15-nm colloidal gold [Biocell International, Cardiff, United Kingdom]) for 1 h at room temperature. They were then washed three times in 1× blocking buffer and three times in PBS and fixed for 10 min in 2.5% glutaraldehyde in PBS before being washed four times for 2 min in double-distilled H2O. Sections were finally stained for 5 min in uranyl acetate and 1 min in lead as described above. The following quantification was done for each antibody using windows of 7.5 μm2. The number of gold beads found in the virus factory at 18 h postinfection was compared with the number of gold beads found in the nucleus (excluding the nuclear membrane) and found in the cytoplasm. At 18 h postinfection, most virus factories covered the 7.5-μm2 area. Smaller virus factories were excluded. The quantification of labelling was carried out on four grids each containing several sections. A minimum of 10 areas were included per grid. A total of 52 areas of 7.5 μm2 were counted each for the nucleus, the cytoplasm, and virus factories. As a control, and because we did not have the preimmune serum for RxER, we counted the number of beads found in the virus factories when using normal rabbit serum. This was carried out with eight different normal rabbit sera used at the same dilution as RxER. A minimum of 15 7.5-μm2 areas were counted for each serum. A one-way analysis of variance (Minitab Inc.) was used to analyze the data. Results were presented as the mean and standard deviation of the mean.
(iii) Ultrathin cryosections.
Infected cells were harvested by scraping, fixed in 0.1% glutaraldehyde in 0.1 M cacodylate buffer (pH 7.2), and embedded in 5% gelatin. Gelatin blocks were infused with 2.3 M sucrose containing 10% polyvinylpyrrolidone dissolved in 1.1 M Na2CO3. Blocks were frozen in liquid nitrogen and sectioned on a Reichert-Jung Ultracut E with an FC 4E cryoattachment at −110°C, using a 35° diatome diamond knife. Cryosections were collected on 2.3 M sucrose and directly stain mounted by the method of Tokuyasu (44, 45). Briefly, grids were washed in water, contrasted with neutral uranyl acetate for 10 min, washed again in water, contrasted with 4% aqueous uranyl acetate, and embedded in 1.1% methylcellulose (400 centipose) containing 0.2% aqueous uranyl acetate. Specimens were examined on a Hitachi H7000 microscope.
(iv) Permeabilization of cells.
Infected cells were either permeabilized by streptolysin O (SLO) or by osmotic shock with water as described by Krijnse-Locker et al. (26, 27). Briefly, to permeabilize with SLO, adherent cells were washed with ice-cold PBS and then SLO buffer (1 mM dithiothreitol, 150 mg of bovine serum albumin 200 mM HEPES). They were then incubated with SLO (200 U/ml in SLO buffer) on ice for 10 min and at 37°C for 30 min. Cells were finally washed with PBS, fixed, and embedded in Spurr resin as described above. Cells were permeabilized by incubation in water for 5 min at room temperature. The water was quickly replaced by 4% paraformaldehyde in PBS, and the cells were then embedded in Spurr resin and processed as described above.
Metabolic labelling and immunoprecipitation.
Metabolic labelling and immunoprecipitation were carried out as previously described by Cobbold et al. (15). Briefly, infected cells were starved in methionine/cysteine-free Eagles medium for 10 min and then labelled with 0.75 Mbq of 35S-Express (New England Nuclear, Boston, Mass.) per ml in the same medium. Cells were chased by replacing the labelling medium with normal culture medium. At appropriate times after incubation at 37°C, cells were washed and lysed on ice in immunoprecipitation buffer (10 mM Tris, pH 7.5, 150 mM NaCl, 10 mM iodoacetamide, 1 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, and 1 mg/ml each of leupeptin, pepstatin, chymostatin, and antipain) containing 1% Nonidet P-40. Antigens were immunoprecipitated by overnight incubation with antibodies immobilized with protein A or G coupled to Sepharose B. Proteins were resolved by sodium dodecyl sulfate (SDS)-polyacrylamide gels and detected by autoradiography.
Endoglycosidase H digestion.
The immunoprecipitate was denatured by the addition of 10 μl of 1% SDS and boiling for 3 min. Digestion was performed at 37°C overnight after the addition of 50 μl of 50 mM sodium phosphate (pH 6.0) containing 1 mU of endoglycosidase H (Boehringer Mannheim, Lewes, United Kingdom).
Triton X-114 and alkali carbonate extractions.
Triton X-114 and alkali carbonate extractions were performed as described previously (15). Infected cells were pulse labelled and removed from culture plates by trypsin. Washed cells were suspended in 0.25 M sucrose buffer in 50 mM Tris, 1 mM EDTA, pH 7.5, and homogenized by 15 passages through a 25-gauge needle. A crude membrane fraction was obtained by pelleting the postnuclear membrane fraction at 14,000 rpm for 10 min, using an Eppendorf 5402 microcentrifuge. For Triton X-114 extractions, the membranes were solubilized in 1% Triton X-114 dissolved in 150 mM NaCl containing 10 mM Tris at pH 7.5. After incubation at 4°C for 10 min, lysates were warmed to 30°C for 10 min and the detergent and aqueous phases were separated by centrifugation at 2,000 rpm for 5 min. Both phases were diluted to 1 ml with 150 mM NaCl dissolved in 10 mM Tris, pH 7.5, and analyzed for the presence of antigen by immunoprecipitation. For alkaline carbonate extractions, the membranes were resuspended in 0.1 ml of 100 mM Na2CO3, pH 11.5, and incubated on ice for 20 min. The suspension was then diluted to 1 ml with PBS, and the membranes were pelleted by centrifugation at 14,000 × g for 30 min at 4°C. The soluble and membrane fractions were analyzed for the presence of antigen by immunoprecipitation.
Virus purification.
ASF virus was purified by Percoll equilibrium centrifugation as previously described by Carrascosa et al. (11). Vero cells infected with the BA71v strain of ASF virus were cultured until an extensive cytopathic effect was observed. For analysis of metabolically labelled virus, 8.3 × 107 infected cells were labelled with 40 Mbq of 35S-Express (New England Nuclear) during infection and mixed with viruses released from 4.2 × 108 unlabelled cells. Cell debris was removed by centrifugation for 10 min at 500 × g, and the viruses were pelleted at 30,000 × g for 1 h. The virus was then suspended in 6.6 ml of PBS and mixed with isotonic Percoll (Pharmacia Biotech, St. Albans, England) to give a 45% self-forming gradient. After centrifugation in a Beckman 70 Ti rotor at 20,000 rpm for 30 min, the lower band was collected and ASF virus was recovered by dilution and recentrifugation. To verify the purity of the preparation, proteins were resolved by SDS-polyacrylamide gel electrophoresis (PAGE) and silver stained (Silver Stain Kit from Sigma Chemical Co., St. Louis, Mo.).
Western blotting.
Proteins were resolved by SDS-PAGE and transferred onto Protran BA85 cellulose nitrate membrane (Schleicher & Schuell, Dassel, Germany). The membrane was blocked overnight at 4°C in 5% Marvel PBS-Tween 20. After two washes in PBS-Tween 20, the membrane was incubated with the first antibody, washed, and incubated with goat anti-rabbit or anti-mouse antibody conjugated to horseradish peroxidase (Southern Biotechnology Associates, Inc, Birmingham, Ala.); both antibodies were diluted in 10% normal goat serum–5% Marvel PBS-Tween 20. After washes, the Western blot was revealed by using the ECL kit (Amersham Life Science, Buckinghamshire, England).
RESULTS
ASF virus has two membranes in its inner envelope.
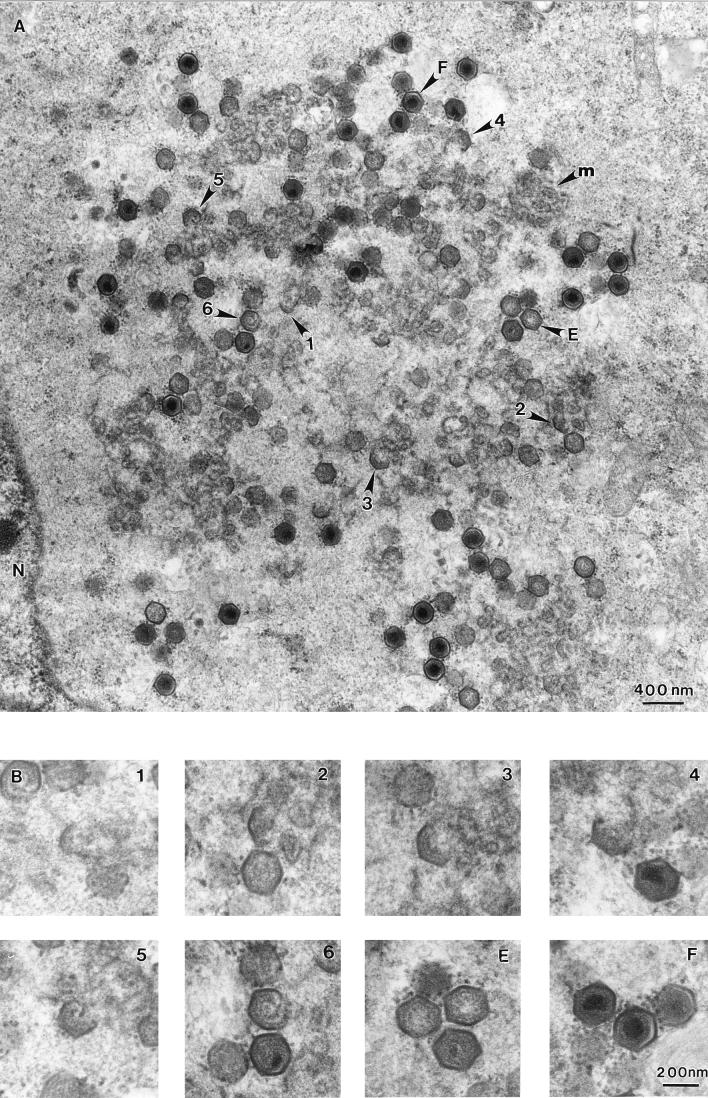
Assembly of ASF virus takes place in the cytoplasm in perinuclear locations called viroplasm or virus factories (8, 33, 34, 36, 47). Figure 2A shows a typical virus factory observed 16 h after infection of RS2 cells with the Uganda isolate of ASF virus. The cells were processed to Spurr resin, and thin sections were examined using transmission electron microscopy. The virus factory was located next to the nucleus and contained large quantities of membrane, several electron-dense structures, and virions at different stages of morphogenesis. The fully assembled intracellular forms of ASF virus were seen as hexagons in cross-section. Some virions had electron-dense centers, suggesting the presence of a nucleoprotein core, whereas others appeared empty. The virus factory also contained structural intermediates visualized as geometric forms with one to six sides of a hexagon. Panel B shows a series of structural intermediates, chosen from panel A, at higher magnification, suggesting a possible assembly progression leading to the production of icosahedral particles.
FIG. 2.
ASF virus assembly site examined by transmission electron microscopy of ultrathin Spurr resin sections. (A) A section, taken through an RS2 cell infected for 16 h with the Uganda isolate of ASF virus, shows a viral factory next to the nucleus (N). The micrograph shows the fully assembled intracellular forms of ASF virus visualized in cross-section as hexagons with electron-dense centers (F), empty particles seen as hexagons with electron-lucent cores (E), several assembly intermediates seen as one to six sided (1 to 6), and amorphous membrane material (m). (B) Selected images, from panel A, of the one- to six-sided intermediates (1 to 6) and of empty and full particles (E and F) are shown at higher magnification.
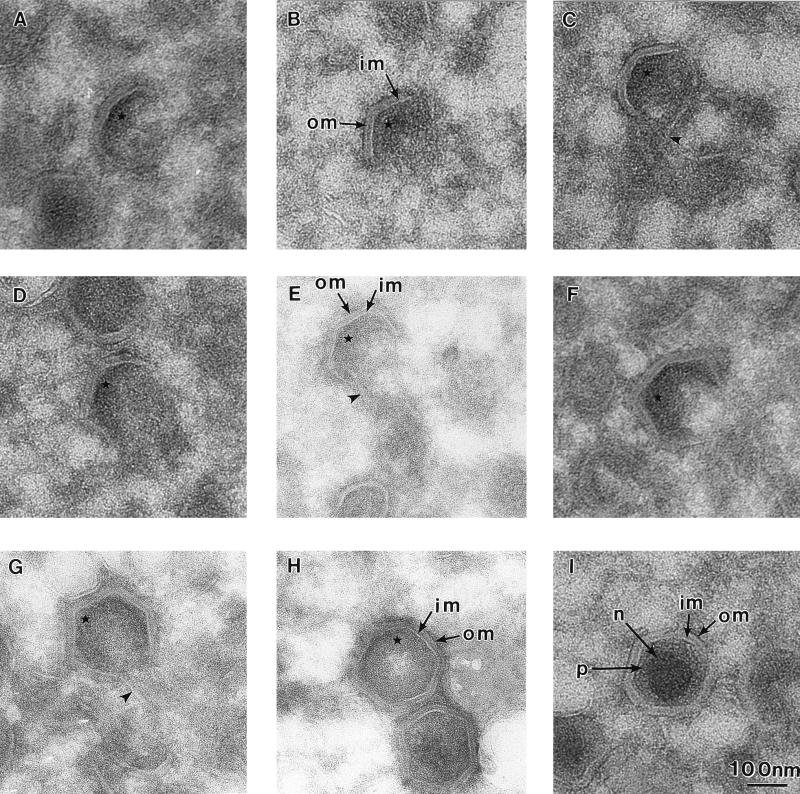
Next we examined the inner envelope in detail to see if it were composed of a single membrane layer indicative of envelopment by budding or whether it contained two membranes indicative of wrapping by membrane cisternae. The viral envelope was visualized by using thin cryosections in which membranes appear electron lucent against an electron-dense background. Figure 3 shows a series of cryosections of viral structural intermediates of increasing complexity (A to G) and fully assembled intracellular virions (H and I). Two electron-lucent membrane layers, the inner membrane and the outer membrane, were clearly visible within virions and all assembly intermediates. Also evident was a progressive increase in the level of electron-dense material on the inner face of the membranes as viral structures became more complex. This may represent the assembly of a protein complex on the inner face of the particle, as has been suggested recently by Andres et al. (3). Several concentric structures have been described for the intracellular form of ASF virus. We can distinguish, as indicated in panel I, in thin cryosections the central electron-dense nucleoid surrounded by a protein core shell and two membranes, the inner membrane and the outer membrane. The outer membrane observed here on the intracellular form of the virus is not to be confused with the additional membrane seen on the extracellular form of the virion added as the virus buds from the plasma membrane (2, 8, 34).
FIG. 3.
Intracellular ASF virus and all assembly intermediates have two membranes. RS2 cells were examined 16 h after infection with the Uganda isolate of ASF virus. The images were selected from thin cryosections taken through virus assembly sites. Note that the resolution of the membranes of the virions and assembly intermediates is improved in the cryosections compared with the resin sections presented in Fig. 2. (A to G) One- to six-sided assembly intermediates, indicating a possible assembly pathway. In several cases, continuity of the two viral membranes (im, inner membrane; om, outer membrane) with intracellular membranes was observed (arrowhead in panels C, E, and G). An increase of electron density on the concave surface of the virus (∗) paralleled the increase in complexity of the particles (A to H). (H and I) Fully assembled intracellular particles which appear empty and full, respectively. The different components of an ASF virus particle, referred to in the text, include a nucleoprotein core (n), a protein core shell (p), an inner membrane (im), and an outer membrane (om) as indicated in panel I.
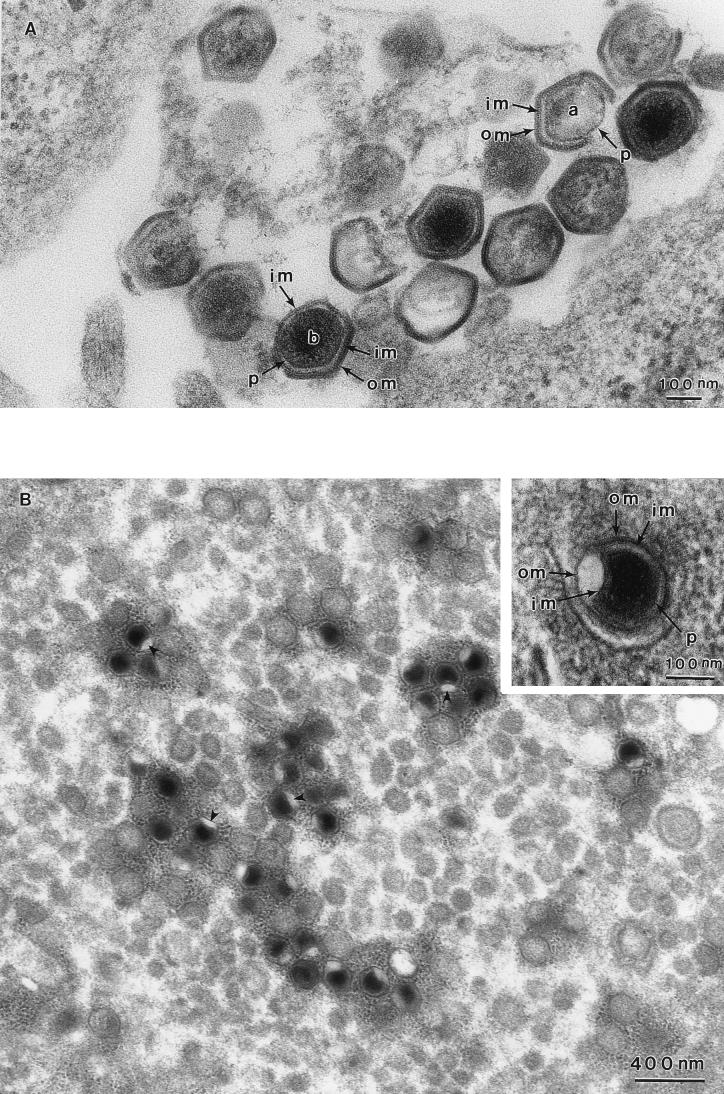
The two membranes were also seen in damaged virions occasionally found in samples embedded in Spurr resin. Figure 4A shows a section through several intracellular virions. In the virions indicated, portions of the viral shell have been broken. In one virion (a), both the outer membrane and the inner membrane were lost from two faces of the particle, exposing the protein core shell. In the other virion (b), the outer membrane has been broken away from three faces of the particle, revealing an intact inner membrane, which is still covering the protein core shell. Once again, we can appreciate the close proximity of the membranes in intact parts of the virions. Interestingly, the angular symmetry of the particle is maintained when both membranes are intact but is lost when the outer membrane has been removed, suggesting that both the outer membrane and the inner membrane, and/or proteins associated with them, are needed to maintain icosahedral symmetry.
FIG. 4.
The inner and outer membranes of ASF virus particles can be separated in damaged virions and when cells are treated with SLO. (A) Damaged intracellular virions observed in a Spurr resin section. In virion a, both the outer membrane (om) and the inner membrane (im) are broken and lost from two faces of the particle, exposing the protein core shell (p). In virion b, the outer membrane (om) is broken away from three faces of the particle, revealing an intact inner membrane (im), which is still covering the protein core shell (p). (B) These micrographs show virions present in cells treated with SLO. A dimple (arrowheads) is present in a single face of most of the fully assembled virions present in this virus factory. At a higher magnification (inset), the inner (im) and outer (om) membranes appear to be separated at the site of the dimple. The protein core shell (p) is clearly visible.
The two viral membranes could be separated when infected cells were permeabilized with SLO before fixation (panel B). Surprisingly, the treatment caused a dimple to form in one face of the virion. At a higher magnification (inset), separation of the two membranes at the site of permeabilization is clearly visible. The precise mechanism of selective destabilization of one and only one vertex of the virion (panel B, arrowheads) is unknown, but the asymmetry in the structure may reflect the mechanism of release of the virion from the ER membrane as will be discussed later.
The two membranes of the intracellular ASF virus are in continuity with cellular membranes.
The two membranes seen within intracellular ASF virus particles could arise if ASF virus were to gain the first membrane by budding into the lumen of a membrane compartment and then gain a second membrane by budding out of the compartment into the cytosol. After close examination of many virus assembly sites by electron microscopy, we and others (3, 4, 9, 13) have been unable to detect virus assembly intermediates, or icosahedral virus particles, within the lumen of intracellular membrane compartments. In addition, we have been unable to detect assembly intermediates with single membrane envelopes in thin cryosections. Both observations argue indirectly against acquisition of two membranes by two budding steps during transit through a membrane compartment. As suggested above, ASF virus could gain two membranes simultaneously by being wrapped by an intracellular membrane compartment. A wrapping pathway has been described for vaccinia virus and herpesvirus. Strong evidence for wrapping in these cases has been the observation of continuity between viral envelopes and intracellular membrane compartments (42, 46).
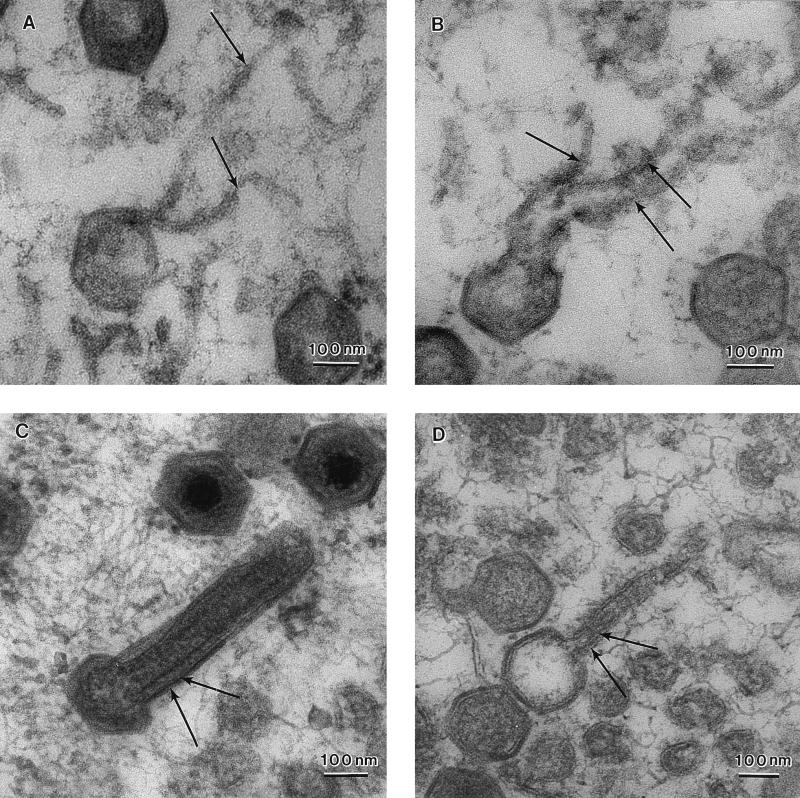
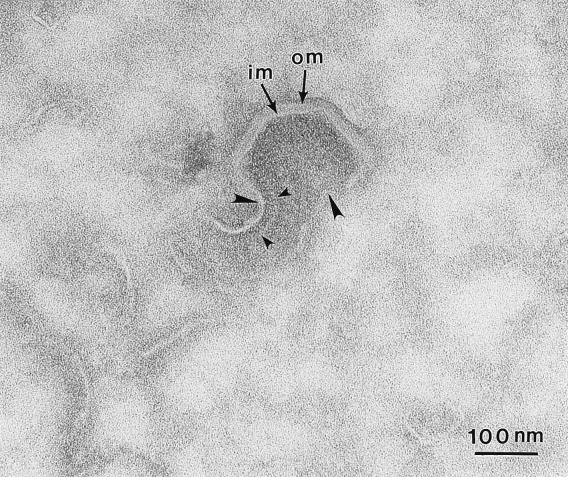
In the next experiments, cells were permeabilized by osmotic shock to release cytosolic proteins and increase the resolution of membrane structures within viral assembly sites (Fig. 5). Panels A and B show four- and five-sided assembly intermediates of ASF virus. In each case the free edges of the particle are attached to tubular/membranous material (arrows), suggesting continuity of the assembly intermediate with cellular membrane compartments. Two membranes could occasionally be seen joined to the same free edge of the assembling particle (top of panels A and B). Having observed these structures in permeabilized cells, a search for similar assembly intermediates in Spurr resin sections of nontreated cells was carried out. In panels C and D, nearly complete five-sided particles are shown, both the closing edges of the virion are again attached to membranes (arrows) extending into the cytosol. Figure 6 shows a thin cryosection which has captured a four-sided assembly intermediate. The viral membranes are seen extending into the cell (large arrowheads). Interestingly, a spiculelike protein coat (small arrowheads) is clearly visible on the concave surface of the virus particle and on the membrane extending into the cytosol. The image again suggests that assembly of a protein coat on the concave face may drive the formation of icosahedral particles.
FIG. 5.
The membranes of ASF virus assembly intermediates are in continuity with host membrane compartments. RS2 cells were examined 16 h after infection with the Uganda isolate of ASF virus. (A and B) Thin sections of infected cells permeabilized by osmotic shock to increase the visualization of membranes embedded in Spurr resin. Note that the free edges of four- (A) and five- (B) sided assembly intermediates are seen attached to membranous material extending into the cytoplasm (arrows). Two membranes are clearly seen attached to the same free edge (upper edge on the micrographs) of the assembly intermediate. (C and D) Similar structures were observed in nontreated cells. The membranes (arrows) are however harder to visualize due to the compact nature of the assembly intermediate.
FIG. 6.
A spiculelike protein coat forms on the concave side of assembly intermediates. This micrograph shows a cryosection of a four-sided intermediate. The two membranes (im and om) are seen in the assembling virion and are extending from both edges into the cytosol (large arrowheads). Note the spiculelike protein coat on the concave face of the particle (small arrowheads).
Resident membrane proteins of the ER are found in virus factories and in virions secreted from cells.
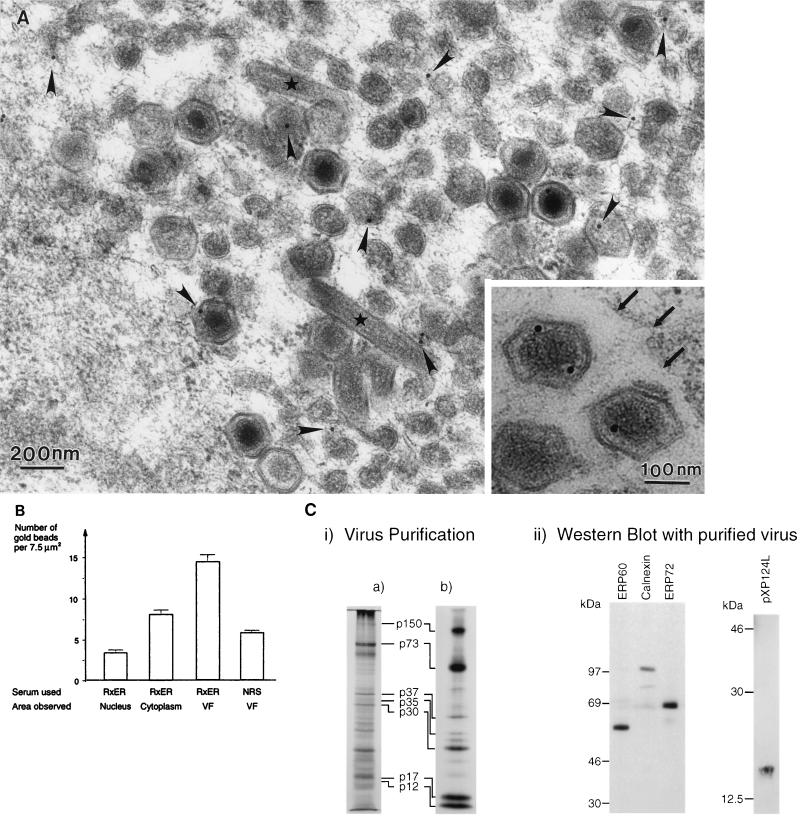
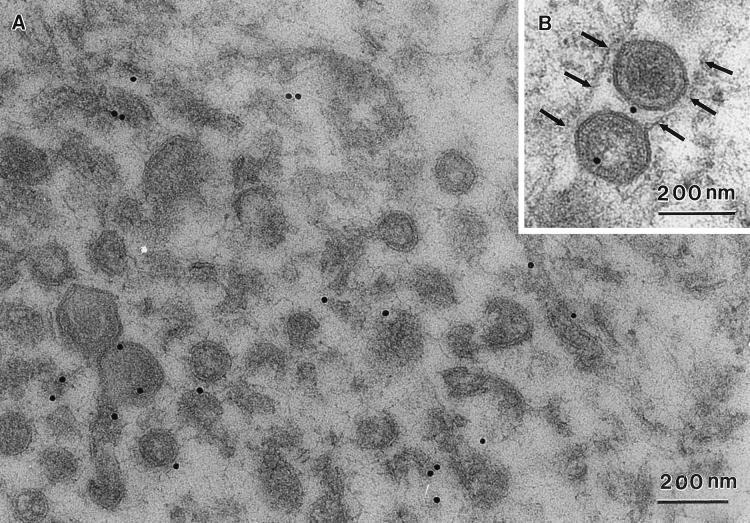
We have shown previously that p73, the major structural protein of ASF virus, is enveloped by the ER (15). The relationship between the ER and virus assembly sites was therefore analyzed in more detail, using immunogold electron microscopy. The rabbit polyclonal antibody RxER recognizes four membrane proteins of the ER (29). Figure 7A and B shows that these ER proteins were found within virus assembly sites. Immunogold labelling was observed on the membranes within the virus factory, on the membranes of virus assembly intermediates, and on fully assembled virions. The inset shows the presence of label on two virions secreted from a cell. The labelling of these particles by the RxER antibody shows that the ER membrane proteins are retained within virions after release from cells. This finding is consistent with earlier work by Carrascosa et al. (11), showing the presence of host proteins in highly purified preparations of ASF virus. A statistical analysis of the immunogold labelling is shown (Fig. 7B). The background labelling produced by the RxER antibody was assessed by analysis of 7.5-μm2 areas taken over the nucleus, an organelle expected to be devoid of ER markers. The graph shows that the level of labelling of viral factories with the ER-specific antibody was fivefold greater than the signal seen over the cell nucleus, indicating the presence of ER proteins at virus assembly sites. As expected, the cytoplasm was labelled with RxER. The signal was threefold higher than seen for the nucleus, indicating the presence of ER membranes in the cytosol. Less than one gold bead was observed for each 7.5-μm2 area when the first antibody was omitted and the gold conjugate was used alone. In a second control, the background labelling produced by normal rabbit serum was calculated by counting the number of gold beads present in 7.5-μm2 areas taken over virus factories. The labelling produced by normal rabbit serum was one-third of the signal given by the antibody specific for ER proteins, again suggesting specific labelling of virus assembly sites by the ER-specific antibody.
FIG. 7.
Marker proteins of the ER localize to ASF virus assembly sites and secreted virions. RS2 cells were examined 16 h after infection with the Uganda isolate of ASF virus. Cells were fixed and embedded in Lowicryl resin. (A) Immunogold labelling. Lowicryl sections were incubated with RxER, an antibody recognizing four ER membrane proteins, and visualized using goat anti-rabbit IgG coupled to 15-nm colloidal gold. Label (arrowheads) was observed within the virus factory on membranous material, virus assembly intermediates, and fully assembled virions. Note also the cigar-shaped structures (∗) occasionally seen in virus assembly sites. The inset shows labelling by the RxER antibody of secreted virions. The outer surface of the cell is indicated at the top right corner of the micrograph (large arrows). (B) Statistical analysis of immunolabelling. The specificity of the RxER labelling of virus assembly sites (VF) was assessed by comparison of immunolabelling of nuclei and cytoplasm in the same sections. The number of gold beads present per 7.5 μm2 are indicated. Preimmune sera were not available for the RxER antibody, a selection of 12 normal rabbit sera (NRS) were therefore used at the same dilution for control studies. The number of gold beads present per 7.5-μm2 area covering the virus factory are shown. The data represent the mean of the number of beads per 7.5-μm2 area. The error bars represent the standard deviation of the mean. (C) Biochemical analysis of virions isolated on Percoll gradients. (i) Virus isolation. Vero cells were infected with the BA71v strain of ASF virus. Virions were isolated using self-forming Percoll density gradients, and proteins present were analyzed by SDS-PAGE under reducing conditions. The left-hand panel shows a 12.5% gel after silver staining. The right-hand panel shows metabolically labelled proteins resolved by using a 10% gel and visualized by autoradiography. The major structural proteins of the virus described previously (4, 11) are indicated. (ii) Western blot analysis. Proteins present in Percoll-purified ASF virus were resolved by SDS-PAGE and analyzed by Western blotting. SDS-PAGE (7.5% polyacrylamide) was used to separate proteins for detection of the resident ER proteins ERP60, calnexin, and ERP72; 12.5% gels were used to detect pXP124L. The migration of molecular mass markers is shown.
The inset to Fig. 7A shows immunogold labelling of virions by the ER-specific antibody. To further test for the presence of ER proteins in virions, ASF virus was isolated by Percoll gradient centrifugation and probed by Western blot (Fig. 7C). Percoll-purified virions are reported to contain up to 34 different proteins (11). The silver-stained gel reveals a protein distribution similar to that described previously (4, 11, 43), characterized by a major protein at 70 kDa (p73), a cluster of proteins between 30 and 40 kDa, bands of variable intensity between 30 and 20 kDa, and strong signals for p17 and p12 at the bottom of the gel. The same pattern, but with enhanced labelling of p150, was observed for metabolically labelled virions. Importantly, pp220, the protein precursor of p150, was absent from the preparations, showing that the virus sample was not contaminated with immature intracellular forms. The right-hand panel shows a Western blot of the same preparation probed with antibodies against calnexin, the major protein detected by the RxER antibody (49), and two lumenal ER proteins, ERP60 and ERP72. Consistent with the immunogold labelling experiment, the blot shows that all three antigens were present in the virus preparation.
An integral membrane protein of ASF virus, p17, localizes to membrane structures within virus factories and cosediments with ER marker proteins on density gradients.
The experiments described above demonstrated the presence of ER membranes in virus assembly sites and located resident proteins of these organelles in virions and assembly intermediates. If the ER is involved in the envelopment of ASF virus, then viral structural proteins, particularly viral membrane proteins, would be expected to localize to the ER prior to and during assembly of the virus. Our previous experiments (15) support this, since we have shown that newly synthesized p17, a nonglycosylated integral membrane protein of ASF virus encoded by the I1L gene, colocalizes with ER marker proteins after subcellular membrane fractionation. In the next experiment, the localization of p17 in infected cells was analyzed by immunogold electron microscopy. Figure 8 shows strong labelling of the virus factory by the antibody specific for p17. Localization of p17 was not restricted to fully assembled six-sided structures, but p17 was also found in membranes within the virus factory and on the one- to six-sided assembly intermediates and in virions secreted from cells (inset). The level of labelling, calculated from analysis of 7.5-μm2 areas taken over virus factories, was threefold higher (63 beads) than that observed for the RxER antibody. Very little labelling (three beads/area) was observed over nuclei, and on average less than one gold bead was observed in 7.5-μm2 areas taken over virus factories when the first antibody was omitted and the gold conjugate was added alone. The sedimentation of p17 in an ER membrane fraction and the immunolocalization of p17 in the membranes concentrating in virus assembly sites is again consistent with the involvement of the ER in the envelopment of ASF virus.
FIG. 8.
Localization of ASF virus integral membrane protein, p17, within virus assembly sites and virions. RS2 cells were examined 16 h after infection with the Uganda isolate of ASF virus. The cells were fixed, embedded in Lowicryl, and immunolabelled with the 17KG12 antibody specific for p17, followed by goat anti-mouse IgG conjugated to 15-nm gold. As seen for the resident proteins of the ER (Fig. 7) p17 was localized to membranes, assembly intermediates, and fully assembled virions. The inset shows labelling of a secreted virion.
An ASF virus protein encoded by the XP124L gene localizes to the lumen of the ER and to the membranes of viral factories and assembled virions.
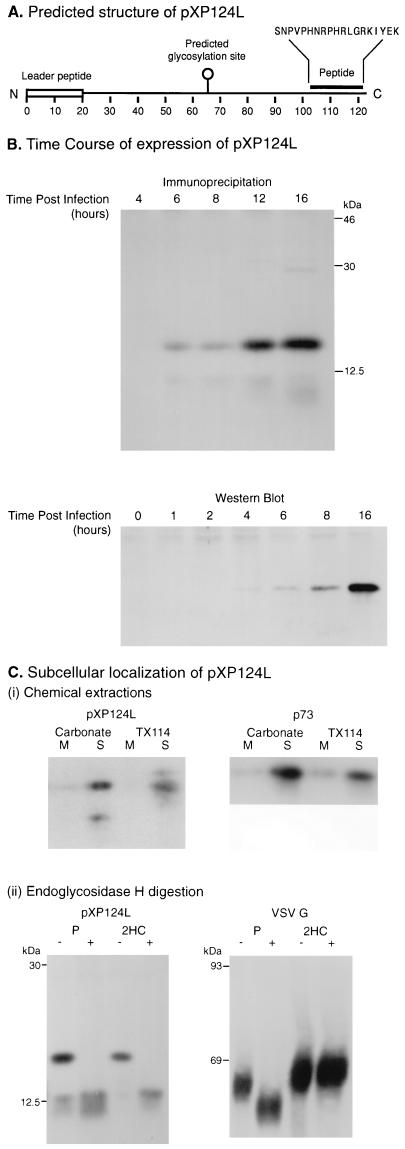
Figure 7 showed that membrane proteins of the ER were present in membranes within the virus factories and at low, but significant, levels within virions. These data suggest that the envelope of ASF virus originates from the ER, but they do not distinguish between the budding or wrapping mechanisms of envelopment outlined in Fig. 1. Convincing evidence for wrapping of ASF virions by the ER would be the detection of soluble lumenal ER proteins between the two membranes in virions and in membranes found at virus assembly sites. Western blot analysis of virions secreted from cells detected the lumenal ER proteins ERP72 and ERP60; however, preliminary immunogold experiments using antibodies specific for these proteins showed low levels in the viral factories (data not shown). Low levels of labelling may arise because, relative to the ER, the membranes in the factories are closely apposed to each other, thereby reducing the lumenal space available for host proteins. An alternative approach to demonstrate membrane wrapping would be to study viral structural proteins that have the potential to be packaged from the ER lumen into the virion. The DNA sequence of ASF virus revealed several proteins with single N-terminal hydrophobic stretches of 20 amino acids, characteristic of signal peptides able to translocate the proteins into the lumen of the ER. One such reading frame, XP124L, encoded a protein of 124 amino acids with an N-terminal signal peptide (Fig. 9A). Importantly, the reading frame lacked a second hydrophobic stop transfer sequence that could act as a membrane-spanning domain, suggesting that the protein would translocate completely into the lumen of the ER. The gene also encoded a single N-linked glycosylation site at position 64, allowing the location of the protein within the ER-Golgi membrane system to be analyzed by analysis of N-linked oligosaccharides. Almendral et al. (1) have detected mRNA encoding the protein in cells infected with ASF virus, suggesting that the protein is synthesized during viral infection.
FIG. 9.
ASF virus-encoded protein pXP124L is a soluble protein retained within the lumen of the ER. (A) Predicted structure of pXP124L. The diagram indicates the structure of pXP124L predicted from the published DNA sequence of the XP124L gene (1, 51), showing the hydrophobic leader peptide, the predicted N-linked glycosylation site, and the synthetic peptide used to raise antiserum. (B) Time course of expression of pXP124L. Vero cells infected with the BA71v isolate of ASF virus were analyzed at increasing time after infection for expression of the XP124L gene product by quantitative Western blotting (bottom) or by metabolic labelling followed by immunoprecipitation (top). Proteins were resolved by SDS-PAGE (12.5% polyacrylamide). (C) (i) Chemical extraction. pXP124L is a soluble protein. Vero cells infected with the BA71v isolate of ASF virus for 16 h were pulse-labelled for 30 min. A crude membrane fraction prepared from the cells was extracted by using alkaline sodium carbonate or Triton X-114. Soluble (S) and membrane fractions (M) were immunoprecipitated with the antipeptide antibody recognizing pXP124L or 4H3 recognizing p73 and, analyzed using SDS-PAGE (12.5% polyacrylamide) followed by autoradiography. (ii) Endoglycosidase H digestion. pXP124L is retained in the ER. Vero cells infected with the BA71v isolate of ASF virus for 16 h or CHO-K1 cells infected with VSV for 5 h were pulse-labelled for 30 min (P) and then chased for 2 h (2HC). Cell lysates were immunoprecipitated by using antibodies specific for pXP124L or VSV G protein, and half of each precipitate was digested with endoglycosidase H (+ lanes). Proteins were resolved by using SDS-PAGE (12.5% polyacrylamide) followed by autoradiography.
An antibody was raised against a peptide corresponding to amino acids 103 to 120 at the C terminus of the protein (Fig. 9A). To test for expression of the protein, cells infected with ASF virus were pulse-labelled at increasing times after infection and immunoprecipitated using the anti-peptide antibody (Fig. 9B, top) or lysed and analyzed by quantitative Western blotting (Fig. 9B, bottom). In each experiment, the protein (pXP124L) was first detected between 4 and 6 h postinfection, with levels reaching a maximum at 16 h. For most proteins, translocation into the ER is followed by removal of the signal peptide by signal peptidase. For some proteins, however, the signal peptide is not cleaved and functions as a membrane anchor. To ensure that pXP124L was soluble within the lumen of the ER, a crude membrane fraction prepared from metabolically labelled cells infected with ASF virus was extracted with Triton X-114 or alkaline carbonate. The distribution of pXP124L between the fractions corresponding to integral membrane or cytosolic fractions was determined by immunoprecipitation. The right panel of Fig. 9C(i) shows that p73, a major structural protein of ASF virus that lacks membrane-targeting sequences, separates to the soluble fraction in each case. The left panel shows that pXP124L was also recovered from the soluble fraction of alkaline carbonate or Triton X-114 extractions, in each case indicating that the signal sequence of pXP124L does not act as a membrane anchor and that pXP124L was soluble within the lumen of the ER.
Interestingly, the labelled protein migrated at 14.5 kDa on SDS-PAGE, approximately 2 kDa larger than the 12.5 kDa of the protein backbone predicted from the XP124L gene. The 2-kDa increase in size was consistent with the presence of a single N-linked high-mannose oligosaccharide. To test for posttranslational glycosylation, cells infected with ASF virus for 16 h were pulse-labelled for 30 min and chased for increasing times prior to immunoprecipitation. Half of each sample was digested with endoglycosidase H. Comparison of the first two lanes of the gel presented in the left panel of Fig. 9 C(ii) shows that the migration of the pulse-labelled pXP124L increased after digestion with endoglycosidase H, indicating the removal of N-linked oligosaccharides. The deglycosylated form migrated at 12.5 kDa, the size predicted for the protein backbone. A minor band at 12.5 kDa was detected in the pulse lane, suggesting that a small quantity of pXP124L was not glycosylated after translocation into the ER. This band was not, however, seen on the Western blot (see panel B, bottom), indicating that steady-state levels of the nonglycosylated form were low. In fact, the smaller form of pXP124L appeared unstable since it was not observed in the chase lanes. Endoglycosidase H removes N-linked oligosaccharides added to protein in the ER but does not cleave oligosaccharides that have been processed by Golgi enzymes. These properties of the enzyme are shown in the analysis of CHO-K1 cells infected with VSV (VSV G panel in Fig. 9C). The pulse-labelled VSV G protein present in the ER was sensitive to endoglycosidase H, but became resistant to digestion after a 2-h chase, indicating transport to the Golgi apparatus. When the same experiment was repeated for pXP124L, the protein remained sensitive to endoglycosidase H during the same 2-h time course, indicating retention in the ER.
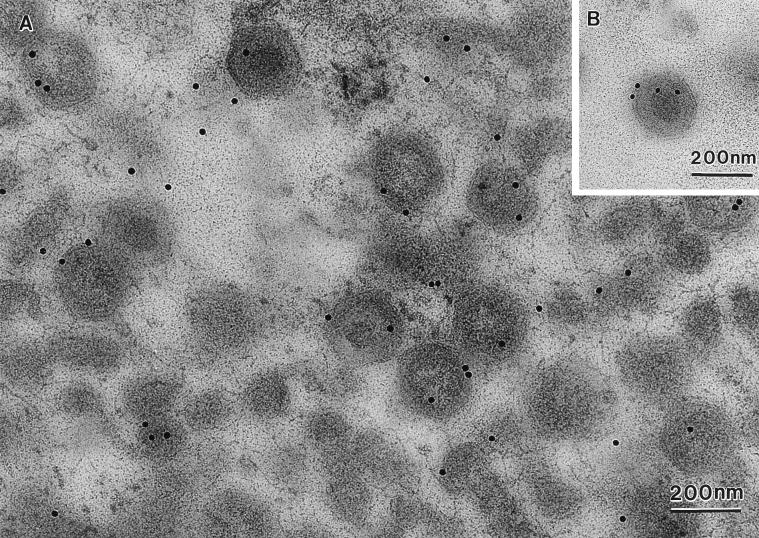
The above results showed that pXP124L was soluble within the lumen of the ER. We have argued that packaging of a soluble lumenal ER protein into ASF virus requires that the two membranes of the virus are in continuity with the ER during virus envelopment and that this would satisfy the criteria necessary to establish the wrapping of ASF virions by ER cisternae. The distribution of pXP124L at virus assembly sites was therefore studied using immunogold electron microscopy. Figure 10 shows a viral factory section probed with the antibody. Significantly, pXP124L was localized to membranes within the factory, ASF virus assembly intermediates, and fully assembled virions. The inset to Fig. 10 shows that pXP124L could also be detected in virions secreted from cells. A statistical analysis showed that on average 14.2 beads per 7.5-μm2 area were located over the virus factory, a level fivefold higher than that seen over the nucleus (3.0 beads) or over the virus factory when sections were incubated with preimmune serum (3.0 beads). To provide further evidence that pXP124L is present in virions, virus isolated from infected cells was probed by Western blot. Fig. 7C shows the presence of pXP124L in the virus preparation when the blot was probed with the antibody specific for pXP124L. The detection of pXP124L at virus assembly sites and within the assembled virus strongly suggests that the two membranes of the intracellular ASF virus originate from extensions of the ER.
FIG. 10.
pXP124L localizes to membranes of the viral factory and to assembled virions. RS2 cells were examined 16 h after infection with the Uganda isolate of ASF virus. The cells were fixed, embedded in Lowicryl, and immunolabelled with rabbit antipeptide antibody specific for pXP124L followed by goat anti-rabbit IgG conjugated to 15-nm gold. The section through the virus assembly site shows pXP124L localized to membranous structures, assembly intermediates, and fully assembled virions. The inset shows immunolabelling of secreted virions, with the plasma membrane being indicated with large arrows.
DISCUSSION
It has been known for several years that ASF virus is composed of concentric layers formed by protein coats and membranes. In both the intracellular and extracellular forms of the virus, a multilayer envelope surrounds the nucleoprotein core, whereas a loose outer membrane is acquired as the virus buds from the plasma membrane. The role of the outer membrane in the virus life cycle remains unclear. It is often lost during purification and is not necessary for infection (5, 34).
Our present study concerns the mechanism of formation of the inner envelope. In particular, we have investigated whether the membranes of the inner envelope are acquired through the wrapping of virions by membrane cisternae or formed while ASF virus buds into intracellular membrane compartments. When virions are wrapped by membrane cisternae, they acquire two membranes in one step and remain in the cytosol. During budding, virions gain a single membrane and are delivered into the lumen of intracellular membrane compartments (Fig. 1). We and others (3, 4, 9, 13) have been unable to detect ASF virions in the lumen of intracellular membrane compartments. These data argue indirectly against a budding pathway.
The precise number of membranes in the inner envelope of the intracellular form of ASF virus has been the subject of much debate. Several studies show a single inner membrane above the core shell. The failure to resolve two lipid bilayers within the inner envelope may be because the lipid bilayers of ASF virus are tightly juxtaposed and appear as a single membrane, as has been shown for the two inner membranes of vaccinia virus (38, 42) and human cytomegalovirus (46). Work by Arzuza et al. (4), on the other hand, has demonstrated two inner membrane envelopes in ASF virus, but it has been suggested that these images may represent recently endocytosed virions and that the extra envelope is the limiting membrane of a cytoplasmic vesicle (3). In this present study we have tried to resolve this issue and have used thin cryosections to better identify membranes in the virus. Importantly, we see two lipid membrane layers in the intracellular form of ASF virus and in all assembly intermediates. The membranes of assembly intermediates were in continuity with cellular membrane compartments, suggesting that ASF virus received two membranes from intracellular membrane cisternae, data consistent with envelopment by membrane wrapping. Immunogold electron microscopy revealed that the membranes that accumulate at assembly sites and the membranes of ASF virus assembly intermediates were labelled with antibodies specific for the ER. Moreover, resident proteins of the ER were detected in purified virions by Western blotting. In addition p17, a structural membrane protein of ASF virus that we had previously localized to ER membranes by subcellular membrane fractionation (15), was concentrated in membranes at assembly sites, assembly intermediates, and fully assembled virions. Taken together with results of our earlier study showing envelopment of p73, the major structural protein of ASF virus, by the ER, the data suggest that ASF virus is wrapped by the ER.
During the wrapping of viruses, the lumenal content of the wrapping cisternae are contiguous with the space between the two membranes and are eventually incorporated into the virus particle (Fig. 1). This allows proteins in the lumen of the wrapping organelle to be packaged into the virion. The sequence of the XP124L gene of ASF virus revealed a single N-terminal leader peptide, suggesting that the protein would translocate completely into the lumen of the ER. The protein, pXP124L, would therefore have the potential to be packaged into the virion from the ER lumen. The solubility of pXP124L in the ER was confirmed biochemically by showing the release of pXP124L from membranes by alkaline carbonate and the extraction of the protein into the aqueous phase of Triton X-114. Moreover, N-linked oligosaccharides attached to the newly synthesized protein remained sensitive to endoglycosidase H after a 2-h chase, showing that pXP124L was retained in the ER after synthesis. We have shown previously that the envelopment of ASF virus takes approximately 2 h (15); pXP124L was therefore retained in the ER for the time taken to form the viral envelope. When analyzed by immunogold electron microscopy, the pXP124L protein was found in fully assembled virions and in assembly intermediates; pXP124L was also detected by immunoblotting of virions isolated from cells. Taken together, the data strongly suggested that pXP124L was packaged into the virion from the lumen of the ER and that the two membranes of the inner envelope of ASF virus were formed from extensions of ER. These data are consistent with ASF virus being wrapped by the ER.
Another observation made in this study (Fig. 3 and 6), and in the recent work of Andres et al. (3), is the presence of a protein coat on the concave face of assembly intermediates, formed either of spicules (Fig. 6) or of a regular array of globular subunits (3). The different resolution of spicules or globules could be due to the different electron microscopy techniques used, which could contrast similar elements in slightly different ways. Andres et al. (3) have shown that antibodies specific for the ASF virus polyprotein pp220 and the processed forms of the polyprotein, p150, p34, and p37/14, label the concave surface of assembling particles and fully assembled virions. It is possible that these proteins may be involved in generating icosahedral symmetry. Indeed the ordered processing of the polyprotein may provide p150, p37/14, and p34 in the correct stoichiometry for ordered assembly into the virion, as has been suggested by Simon-Mateo et al. (40). It has been suggested that this inner protein shell functions as a matrix during ASF virus assembly.
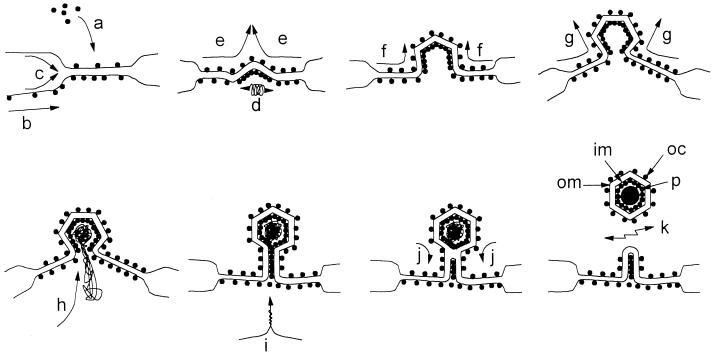
Figure 11 shows a model for ASF virus assembly that incorporates our present data and the data provided by several groups (3, 4, 9, 12, 13) describing the distribution of proteins and membranes in virus assembly sites. The ER is a continuous membrane network that extends throughout the cytoplasm of cells. The model does not, therefore, require virions to be wrapped by the tip or leading edge of a membrane cisternae. Instead, we think that viral structural proteins assemble on the ER membrane. Proteins present in the lumen of the ER, such as pXP124L, now have access to the space between the two inner envelopes and can be packaged into virions. Cooperative interactions between viral proteins on both sides of the membrane and across the ER, via interaction with lumenal proteins, may constrict the ER cisternae. This may exclude some host lumenal ER proteins and explain the low level of ER proteins detected in virus assembly sites. As assembly proceeds, further interactions between viral proteins cause progressive bending of the membrane cisternae, producing the one- to six-sided assembly intermediates seen in cross-sections of virus assembly sites. Progressive assembly in this way allows the genome to be packaged at a late stage during formation of the icosahedron. In the final steps, two membrane fusion events involving the inner and outer lipid bilayers allow the release of the virion from the ER. A lack of fusion, possibly due to limiting quantities of an essential protein, may explain the production of extended cigar-shaped virions often seen at virus assembly sites especially late in infection (Fig. 5C and D). Interestingly, when infected cells were permeabilized by SLO, we consistently noticed a distortion of one face of the virion (Fig. 4B). The results point to an assymetry in ASF virus particles. The area of weakness may represent the site of particle closure formed when the virus leaves the ER.
FIG. 11.
A model for the assembly and envelopment of the intracellular form of ASF virus. Viral structural proteins synthesized in the cytosol, for example p73 and the products of the pp220 and pp60 polyproteins, assemble on the ER membrane (a). Proteins present in the lumen of the ER and integral membrane proteins can also be packaged into assembly sites (b and c). Cooperative interactions between proteins on both sides of the ER cisternae constrict the ER lumen (d and e) and cause progressive bending of the ER cisternae into the one- to six-sided assembly intermediates seen in cross-sections of virus assembly sites (f and g). The genome may be packaged at a late stage during formation of the icosahedron (h). In the final steps, two membrane fusion events involving the inner and outer lipid bilayers allow release of the virion from the ER (i, j, and k). The outer (om) and inner membrane (im) layers of the intracellular form are indicated. Note that this mechanism allows the outer capsid (oc) and the inner core shell (p) to be assembled from the same pool of cytoplasmic structural proteins.
Earlier work by Carrascosa et al. (12), demonstrating a capsid layer outside the membrane envelope, is consistent with a model of membrane wrapping. It is generally believed that the capsid of ASF virus contains p73. Protein p73 is a major structural protein accounting for 30% of the viral protein (3, 11, 14, 28), and antibodies specific for p73 neutralize the infectivity of ASF virus, suggesting that the antigen is exposed on the surface of the virion (6, 19). Importantly, p73 lacks membrane-targeting sequences that would translocate the protein across a membrane. Figure 11 shows that wrapping provides the only means of incorporating soluble cytosolic proteins, such as p73 or the products of the pp220 or pp60 polyproteins, into a capsid outside the membrane envelope. This is because during membrane wrapping soluble proteins have access to both faces of the membrane cisternae, forming the envelope of ASF virus. A budding pathway, on the other hand, transfers the enveloped particle into the lumen of the organelle providing the membrane. The outer envelope of the virus is then separated from the cytosol, and cytosolic proteins such as p73 would be unable to assemble an outer capsid above the membrane.
ACKNOWLEDGMENTS
We thank Daniel Louvard and Evelyne Coudrier (Morphogenese et signalisation cellulaire, Institut Curie, Paris, France) for RxER antibody and Thomas Kries (Department of Cell Biology, University of Geneva, Geneva, Switzerland) for P5D4. We also thank Terry Wise (AAHL) for assistance and M. Parkhouse (IAH) for support of the project.
The British Society for Immunology provided financial support from the Alan Williams Travel Fund for Sharon M. Brookes to travel to Australia.
REFERENCES
- 1.Almendral J M, Almazan F, Blasco R, Vinuela E. Multigene families in African swine fever virus: family 110. J Virol. 1990;64:2064–2072. doi: 10.1128/jvi.64.5.2064-2072.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alves De Matos A P, Marcal M R, Moura Nunes F J, Castro Portugal F L, Vigario J D. Ultrastructural and cytochemical aspects of the maturation of the ASFV particle. Rep Trab Inst Nac Vet. 1980;12:79–84. [Google Scholar]
- 3.Andres G, Simon-Mateo C, Vinuela E. Assembly of African swine fever virus: role of polyprotein pp220. J Virol. 1997;71:2331–2341. doi: 10.1128/jvi.71.3.2331-2341.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arzuza O, Urzainqui A, Diaz-Ruiz J R, Tabares E. Morphogenesis of African swine fever virus in monkey kidney cells after reversible inhibition of replication by cycloheximide. Arch Virol. 1992;124:343–354. doi: 10.1007/BF01309814. [DOI] [PubMed] [Google Scholar]
- 5.Black D N, Brown F. Purification and physicochemical characteristics of ASFV. J Gen Virol. 1976;32:509–518. doi: 10.1099/0022-1317-32-3-509. [DOI] [PubMed] [Google Scholar]
- 6.Borca M V, Irusta P, Carrillo C, Afonso C L, Burrage T, Rock D L. African swine fever virus structural protein p72 contains a conformational neutralizing epitope. Virology. 1994;201:413–418. doi: 10.1006/viro.1994.1311. [DOI] [PubMed] [Google Scholar]
- 7.Borca M V, Irusta P M, Kutish G F, Carrillo C, Afonso C L, Burrage T, Neilan J G, Rock D L. A structural DNA binding protein of African swine fever virus with similarity to bacterial histone-like proteins. Arch Virol. 1996;141:301–313. doi: 10.1007/BF01718401. [DOI] [PubMed] [Google Scholar]
- 8.Breese S S, Deboer C J. Electron microscope observations of African swine fever virus in tissue culture cells. Virology. 1966;28:420–428. doi: 10.1016/0042-6822(66)90054-7. [DOI] [PubMed] [Google Scholar]
- 9.Brookes S M, Dixon L K, Parkhouse R M E. Assembly of African swine fever virus: quantitative ultrastructural analysis in vitro and in vivo. Virology. 1996;224:84–92. doi: 10.1006/viro.1996.0509. [DOI] [PubMed] [Google Scholar]
- 10.Camacho A, Vinuela E. Protein p22 of African swine fever virus: an early structural protein that is incorporated into the membrane of infected-cells. Virology. 1991;181:251–257. doi: 10.1016/0042-6822(91)90490-3. [DOI] [PubMed] [Google Scholar]
- 11.Carrascosa A L, Delval M, Santaren J F, Vinuela E. Purification and properties of African swine fever virus. J Virol. 1985;54:337–344. doi: 10.1128/jvi.54.2.337-344.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carrascosa J L, Carazo J M, Carrascosa A L, Garcia N, Santisteban A, Vinuela E. General morphology and capsid fine structure of African swine fever virus particles. Virology. 1984;132:160–172. doi: 10.1016/0042-6822(84)90100-4. [DOI] [PubMed] [Google Scholar]
- 13.Carrascosa J L, Gonzalez P, Carrascosa A L, Garcia-Barreno B, Enjuanes L, Vinuela E. Localization of structural proteins in African swine fever virus particles by immunoelectron microscopy. J Virol. 1986;58:377–384. doi: 10.1128/jvi.58.2.377-384.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cistue C, Tabares E. Expression in vivo and in vitro of the major structural protein (VP73) of African swine fever virus. Arch Virol. 1992;123:111–124. doi: 10.1007/BF01317142. [DOI] [PubMed] [Google Scholar]
- 15.Cobbold C, Whittle J T, Wileman T. Involvement of the endoplasmic reticulum in the assembly and envelopment of African swine fever virus. J Virol. 1996;70:8382–8390. doi: 10.1128/jvi.70.12.8382-8390.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Delavega I, Gonzalez A, Blasco R, Calvo V, Vinuela E. Nucleotide sequence and variability of the inverted terminal repetitions of African swine fever virus DNA. Virology. 1994;201:152–156. doi: 10.1006/viro.1994.1277. [DOI] [PubMed] [Google Scholar]
- 17.Dixon L K, Twigg S R F, Baylis S A, Vydelingum S, Bristow C, Hammond J M, Smith G L. Nucleotide sequence of a 55 kbp region from the right end of the genome of a pathogenic African swine fever virus isolate (Malawi LIL20/1) J Gen Virol. 1994;75:1655–1684. doi: 10.1099/0022-1317-75-7-1655. [DOI] [PubMed] [Google Scholar]
- 18.Gershon A A, Sherman D L, Zhu Z L, Gabel C A, Ambron R T, Gershon M D. Intracellular transport of newly synthesized varicella-zoster virus: final envelopment in the trans-Golgi network. J Virol. 1994;68:6372–6390. doi: 10.1128/jvi.68.10.6372-6390.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gomez-Puertas P, Rodriguez F, Oviedo J M, Ramiro-Ibanez F, Ruiz-Gonzalvo F, Alonso C, Escribano J M. Neutralizing antibodies to different proteins of African swine fever virus inhibit both virus attachment and internalization. J Virol. 1996;70:5689–5694. doi: 10.1128/jvi.70.8.5689-5694.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Granzow H, Weiland F, Jons A, Klupp B G, Karger A, Mettenleiter T C. Ultrastructural analysis of the replication cycle of pseudorabies virus in cell culture: a reassessment. J Virol. 1997;71:2072–2082. doi: 10.1128/jvi.71.3.2072-2082.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Griffiths G A, Rottier P. Cell biology of viruses that assemble along the biosynthetic pathway. Semin Cell Biol. 1992;3:367–381. doi: 10.1016/1043-4682(92)90022-N. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haugejorden S M, Srinivasan M, Green M. Analysis of the retention signals of 2 resident luminal endoplasmic reticulum proteins by in vitromutagenesis. J Biol Chem. 1991;266:6015–6018. [PubMed] [Google Scholar]
- 23.Hyatt A D. Immunogold labelling techniques. In: Harris R, editor. Electron microscopy in biology: a practical approach. Oxford, United Kingdom: IRL Press; 1991. pp. 56–81. [Google Scholar]
- 24.Jones F, Grose C. Role of cytoplasmic vacuoles in varicella-zoster virus glycoprotein trafficking and virion envelopment. J Virol. 1988;62:2701–2711. doi: 10.1128/jvi.62.8.2701-2711.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kreis T E. Microinjected antibodies against the cytoplasmic domain of vesicular stomatitis virus glycoprotein block its transport to the cell surface. EMBO J. 1986;5:931–941. doi: 10.1002/j.1460-2075.1986.tb04306.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krijnse-Locker J, Parton R G, Fuller S D, Griffiths G, Dotti C G. The organization of the endoplasmic reticulum and the intermediate compartment in cultured rat hippocampal neurons. Mol Biol Cell. 1995;6:1315–1332. doi: 10.1091/mbc.6.10.1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krijnse-Locker J, Schleich S, Rodriguez D, Goud B, Snijder E J, Griffiths G. The role of a 21-kDa viral membrane protein in the assembly of vaccinia virus from the intermediate compartment. J Biol Chem. 1996;271:14950–14958. doi: 10.1074/jbc.271.25.14950. [DOI] [PubMed] [Google Scholar]
- 28.Lopez-Otin C, Freije J M P, Parra F, Mendez E, Vinuela E. Mapping and sequence of the gene coding for protein-p72, the major capsid protein of African swine fever virus. Virology. 1990;175:477–484. doi: 10.1016/0042-6822(90)90432-q. [DOI] [PubMed] [Google Scholar]
- 29.Louvard D, Reggio H, Warren G. Antibodies to the Golgi complex and the rough endoplasmic reticulum. J Cell Biol. 1982;92:92–107. doi: 10.1083/jcb.92.1.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martinez-Pomares L, Simon-Mateo C, Lopez-Otin C, Vinuela E. Characterization of the African swine fever virus structural protein p14.5: a DNA binding protein. Virology. 1997;229:201–211. doi: 10.1006/viro.1996.8434. [DOI] [PubMed] [Google Scholar]
- 31.Mazzarella R A, Srinivasan M, Haugejorden S M, Green M. Erp72, an abundant luminal endoplasmic reticulum protein, contains 3 copies of the active site sequences of protein disulfide isomerase. J Biol Chem. 1990;265:1094–1101. [PubMed] [Google Scholar]
- 32.Montgomery R E. One form of swine fever occuring in British East Africa (Kenya colony) J Comp Pathol. 1921;34:159–191. [Google Scholar]
- 33.Moulton J, Coggins L. Synthesis and cytopathogenesis of ASFV in porcine cell cultures. Am J Vet Res. 1968;29:219–232. [PubMed] [Google Scholar]
- 34.Moura-Nunes J F, Vigario J D, Terrinha A M. Ultrastucture study of African swine fever virus replication in cultures of swine bone marrow cells. Arch Virol. 1975;49:59–66. doi: 10.1007/BF02175596. [DOI] [PubMed] [Google Scholar]
- 35.Munoz M, Freije J M P, Salas M L, Vinuela E, Lopez-Otin C. Structure and expression in Escherichia coli of the gene coding for protein p10 of African swine fever virus. Arch Virol. 1993;130:93–107. doi: 10.1007/BF01318999. [DOI] [PubMed] [Google Scholar]
- 36.Pan I C, Shimizu M, Hess W R. Replication of African swine fever virus in cell culture. Am J Vet Res. 1980;41:1357–1367. [PubMed] [Google Scholar]
- 37.Rodriguez F, Alcaraz C, Eiras A, Yanez R J, Rodriguez J M, Alonso C, Rodriguez J F, Escribano J M. Characterization and molecular basis of heterogeneity of the African swine fever virus envelope protein p54. J Virol. 1994;68:7244–7252. doi: 10.1128/jvi.68.11.7244-7252.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schmelz M, Sodeik B, Ericsson M, Wolffe E J, Shida H, Hiller G, Griffiths G. Assembly of vaccinia virus: the second wrapping cisterna is derived from the trans Golgi network. J Virol. 1994;68:130–147. doi: 10.1128/jvi.68.1.130-147.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schnitzler P, Darai G. Identification of the gene encoding the major capsid protein of fish lymphocystis disease virus. J Gen Virol. 1993;74:2143–2150. doi: 10.1099/0022-1317-74-10-2143. [DOI] [PubMed] [Google Scholar]
- 40.Simon-Mateo C, Andres G, Vinuela E. Polyprotein processing in African swine fever virus: a novel strategy of gene expression for a DNA virus. EMBO J. 1993;12:2977–2987. doi: 10.1002/j.1460-2075.1993.tb05960.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Simon-Mateo C, Freije J M P, Andres G, Lopez-Otin C, Vinuela E. Mapping and sequence of the gene encoding protein p17, a major African swine fever virus structural protein. Virology. 1995;206:1140–1144. doi: 10.1006/viro.1995.1039. [DOI] [PubMed] [Google Scholar]
- 42.Sodeik B, Doms R W, Ericsson M, Hiller G, Machamer C E, Vanthof W, Vanmeer G, Moss B, Griffiths G. Assembly of vaccinia virus: role of the intermediate compartment between the endoplasmic reticulum and the Golgi stacks. J Cell Biol. 1993;121:521–541. doi: 10.1083/jcb.121.3.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sun H L, Jacobs S C, Smith G L, Dixon L K, Parkhouse R M E. African swine fever virus gene j131 encodes a 25–27 kda virion protein with variable numbers of amino-acid repeats. J Gen Virol. 1995;76:1117–1127. doi: 10.1099/0022-1317-76-5-1117. [DOI] [PubMed] [Google Scholar]
- 44.Tokuyasu K T. A technique for ultracryotomy of cell suspensions and tissues. J Cell Biol. 1973;57:551–565. doi: 10.1083/jcb.57.2.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tokuyasu K T. Application of cryoultramicrotomy to immunocytochemistry. J Microsc. 1986;143:139–149. doi: 10.1111/j.1365-2818.1986.tb02772.x. [DOI] [PubMed] [Google Scholar]
- 46.Tooze J, Hollinshead M, Reis B, Radsak K, Kern H. Progeny vaccinia and human cytomegalovirus particles utilize early endosomal cisternae for their envelopes. Eur J Cell Biol. 1993;60:163–178. [PubMed] [Google Scholar]
- 47.Vigario J D, Relvas M E, Ferraz F P, Ribeiro J M, Pereira C G. Identification and localization of genetic material of ASFV by autoradiography. Virology. 1967;33:173–175. doi: 10.1016/0042-6822(67)90108-0. [DOI] [PubMed] [Google Scholar]
- 48.Villiger W. Lowicryl resins. In: Hayat M A, editor. Colloidal gold: principles, methods and applications. Vol. 3. New York, N.Y: Academic Press, Inc.; 1991. pp. 59–71. [Google Scholar]
- 49.Wada I, Rindress D, Cameron P H, Ou W J, Doherty J J, Louvard D, Bell A W, Dignard D, Thomas D Y, Bergeron J J M. Ssr-alpha and associated calnexin are major calcium binding proteins of the endoplasmic reticulum membrane. J Biol Chem. 1991;266:19599–19610. [PubMed] [Google Scholar]
- 50.Whealy M E, Card J P, Meade R P, Robbins A K, Enquist L W. Effect of brefeldin A on alphaherpesvirus membrane protein glycosylation and virus egress. J Virol. 1991;65:1066–1081. doi: 10.1128/jvi.65.3.1066-1081.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yanez R J, Rodriguez J M, Nogal M L, Yuste L, Enriquez C, Rodriguez J F, Vinuela E. Analysis of the complete nucleotide sequence of African swine fever virus. Virology. 1995;208:249–278. doi: 10.1006/viro.1995.1149. [DOI] [PubMed] [Google Scholar]