Abstract
Plasma human immunodeficiency virus type 1 (HIV-1) populations were genetically analyzed at their most variable locus, the envelope gene, during the rapid emergence of resistance to protease inhibitor monotherapy. Plasma virus populations remained genetically constant prior to drug treatment and during the 1 to 2 weeks following initiation of therapy, while viremia fell 10- to 100-fold. Concomitant with rapid plasma viremia rebounds associated with the emergence of drug-resistant virus, marked alterations were then detected at the env locus. Plasma population changes lasted only a few weeks before the reappearance of the pretreatment envelope variants. The emergence of resistance to single protease inhibitors was therefore associated with major but transient changes at a nonselected locus. Selection for resistance to single protease inhibitors thus appears to be more complex than the continued replication of a large, random, and therefore genetically representative sampling of the pretreatment plasma population. The possibility that drug-privileged anatomical sites containing distinct envelope variants and/or a small effective HIV-1 population size account for these results is discussed.
Following initially effective antiviral monotherapy with either human immunodeficiency virus type 1 (HIV-1) reverse transcriptase or protease inhibitors, plasma viremia often rapidly rebounds and the disease course resumes (25, 36, 37, 41). Such increases in plasma viremia have been correlated with the emergence of drug-resistant viruses (35). HIV-1 genomes with single-nucleotide mutations associated with drug resistance are theoretically expected (5, 6, 14) and are actually detected (19, 29) in untreated patients. Such pretreatment HIV-1 variants may seed emerging, drug-resistant, viral populations, following drug selection. This likely occurs with HIV-1 reverse transcriptase inhibitors such as 3TC and nevirapine, where single pol mutations result in high-level drug resistance. Viruses highly resistant to the protease inhibitor ritonavir (requiring multiple pro mutations) may also be derived by ongoing replication of single-nucleotide site mutants since a V82A mutation is found predominantly alone during the earliest phase of plasma viremia rebound (27).
A deterministic model of HIV-1 genetic diversity has been proposed that assumes a large effective population size and emphasizes the role of competition between constantly generated variants of slightly different replication fitness in determining the exact frequency of single-site mutants (5, 6). According to this model, any particular single-nucleotide mutation associated with drug resistance may be found on as many as 0.1 to 1% of the viral genomes in the fully differentiated quasispecies which have evolved following several years of infection. Such computations are consistent with sequence analyses of proviral variants in untreated individuals (19, 29) and with the estimated frequency of the nevirapine-conferring pol mutation (Y181C) prior to treatment (14). Assuming that 108 cells are infected per day (15, 41), a large and therefore representative variant sampling of the pretreatment population is thus expected to carry any particular single nucleotide mutation (≥0.001 × 108 = ≥105 single-site mutants replicating daily). Following the drug selection and amplification of so many drug-resistant viral lineages from the pretreatment population, no genetic changes are anticipated at a nonselected locus (i.e., env).
An alternative model of HIV-1 population genetics has proposed that an effective population number as low as 103 perpetuates HIV-1 in vivo (20, 21). This model, emphasizing the role of stochastic events in the generation of HIV-1 genetic diversity, predicts that the pretreatment frequency of single-site drug-resistant mutant genomes is unpredictable due to chance sampling effects.
To generate population genetics data pertinent to these models of drug selection in vivo, plasma virus populations were monitored at their most variable locus, the envelope gene, in patients who experienced a rapid viral-load rebound following initially effective protease inhibitor monotherapies. Using an assay capable of reproducibly detecting even minor changes in env variant frequencies, we found that plasma virus populations can undergo major, although transient, alterations at that locus during the emergence of protease inhibitor-resistant virus. Possible explanations are discussed.
MATERIALS AND METHODS
Patient selection.
Patients were selected from two protease inhibitor monotherapy drug trials by using several criteria. Patients who showed a 1- to 2-log plasma viral-load decrease within 2 weeks of initiating treatment, followed by a plasma viral-load rebound to levels at least 1 log above their plasma viremia nadir within 2 months of initiating treatment, were selected. Two patients who showed a sustained reduction in plasma viremia for more than 4 months were also selected. Patients P105, P403, P404, and P406 received 600 mg of ritonavir twice a day (25), and patients P02, P07, P08, P09, and P13 received 500 to 750 mg of nelfinavir twice a day. Regimen compliance was verified by interrogation and counting the pills left during each patient’s visit. Protease inhibitor monotherapies were continued uninterrupted during the entire period under study here, including following viral-load rebound. The ritonavir formulation used was in the form of frozen capsules which did not show the gastrointestinal side effects of the later formulation.
Plasma viral RNA isolation and reverse transcription-nPCR.
Patient plasma samples stored at −80°C (0.5 ml) were centrifuged at 24,000 × g for 1 h at 4°C. The supernatant was removed, and 220 μl of a lysis solution (157 mM HEPES [pH 7.5], 12.5 mM EDTA, 1.6% lithium lauryl sulfate, 19 μg of sheared salmon sperm DNA per ml, 8 μg of MS2 RNA per ml, 630 mM LiCl, 1 mg of proteinase K per ml) was added. Tubes were vortexed for 10 s and heated to 53°C for 20 min. A 10-μl volume of yeast tRNA (1 mg/ml) and 22 μl of 3 M sodium acetate were added. The lysate was then extracted with 200 μl of phenol saturated with 0.1 M Tris [pH 7.5] plus 40 μl of 49:1 chloroform-isoamyl alcohol, and the mixture was gently mixed on a rotating platform at room temperature for 15 min. Eppendorf tubes were spun for 5 min at 12,000 rpm (Eppendorf 5415C). A 250-μl volume of isopropanol was added to the aqueous fraction, which was kept at −20°C overnight. RNA was precipitated at 12,000 rpm for 10 min, and the RNA pellet was rinsed with 70% ethanol. The air-dried RNA pellets were resuspended in 25 μl of reverse transcription buffer and reverse transcribed with 12.5 pmol of antisense primer 5′-GCGCCC-3′ (3), 3 U of RNasin (Promega, Madison, Wis.) and 100 U of superscript II (RNase H-negative reverse transcriptase; Gibco BRL, Gaithersburg, Md.) for 30 min at 42°C. The nested PCR (nPCR) for env V3-to-V5 region used 2 μl of cDNA as the template and outer primer pair KK1-ED12 and inner primer pair ES7-ES8. All of the primers used have been described previously (11), except KK1 (GCACAGTACAATGTACACATGGAA; positions 6529 to 6552 on the IIIB isolate BH10 sequence) (34). The combined efficiency of viral RNA extraction and cDNA synthesis was determined by end point titration (using nPCR) of the cDNA derived from plasma samples containing known numbers of viral RNA molecules (determined by the Chiron branched DNA assay [bDNA]).
HTA, data recording, and quantitation.
DNA heteroduplexes formed between closely related HIV-1 env sequence variants (i.e., from the same person) display moderate electrophoretic migration retardations which are generally more sensitive to minor sequence changes than are the highly retarded mobilities of DNA heteroduplexes formed between widely divergent HIV-1 env variants (i.e., from different individuals) (7, 9–11). The ability of DNA heteroduplex tracking analysis (HTA) to detect genetic changes in plasma HIV-1 populations was therefore maximized by using patient-specific HTA probes closely related to the quasispecies under study. A unique HTA probe was generated from each patient by end point dilution of cDNA from their last time point plasma sample, followed by nPCR. The clonality (homogeneity) of the dilution end point nPCR product was verified by heteroduplex mobility analysis (7, 9). The first-round PCR product from the end point dilution was then used to initiate a second-round PCR using 5′ 33P-labeled ES8 and 5′ biotin-labeled ES7 primers. The single-stranded radiolabeled HTA probe was purified by binding the biotin moiety of the second-round PCR product to streptavidin-coated magnetic beads and releasing the complementary strand (the HTA probe) by using a 0.1 N NaOH solution as previously described (7, 9). The clonal, radiolabeled, single-stranded HTA probe was then reannealed to a 100-fold excess of target DNA derived by RT-nPCR from the longitudinally collected, reverse-transcribed plasma HIV-1 RNA. To increase the number of plasma virus RNA genomes simultaneously analyzed by HTA, two independent nPCRs were generated from each plasma sample and both PCR products were pooled prior to hybridization with the single-stranded HTA probe. Images from the PhosphorImager ImageQuant program (see Fig. 1 and 2) were converted to eight-bit Tiff files, opened using Photoshop 3.0 (Adobe Systems, Mountain View, Calif.), and transferred to the graphics program Canvas 3.53 (Deneba Systems, Miami, Fla.).
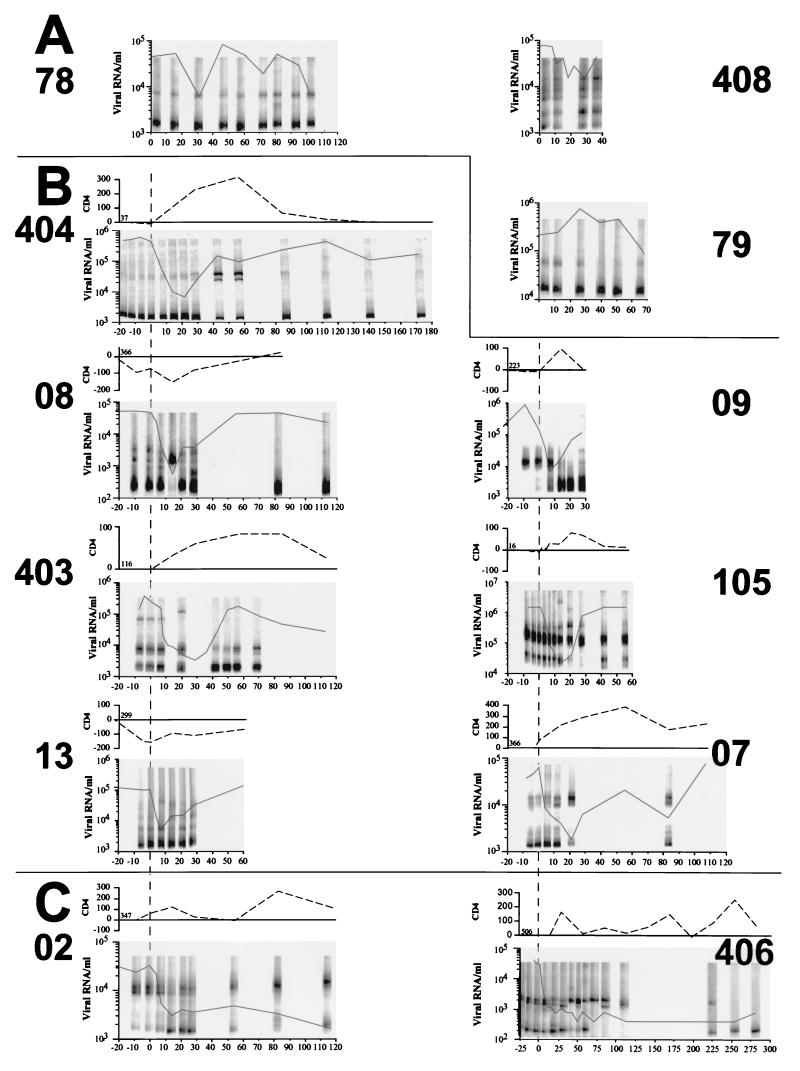
FIG. 1.
Genetic changes at the env locus of plasma HIV-1 populations. Longitudinal plasma viral load (number of HIV RNA copies/ml), CD4 counts per microliter, and plasma virus population env composition as determined by HTA are shown. Viral loads were determined by bDNA assay. CD4 levels are relative to baseline pretreatment levels, which are indicated numerically. A clonal, single-stranded, radiolabeled HIV-1 env DNA strand was derived from each patient and reannealed with a 100-fold excess of the product of nPCRs initiated with cDNA derived from longitudinally collected plasma HIV-1 RNA (see Materials and Methods). The radiolabeled DNA heteroduplexes were resolved on a 5% polyacrylamide gel. HTA patterns are shown only below the single-stranded DNA mobility position where all DNA heteroduplexes were found. The same scale is used on the time (x) axis for all patients except P406. (A) Untreated patients. (B) Treated patients with a rapid viral-load rebound. Day 0 indicates initiation of monotherapy. (C) Treated patients with a sustained viral-load reduction. Day 0 indicates initiation of monotherapy.
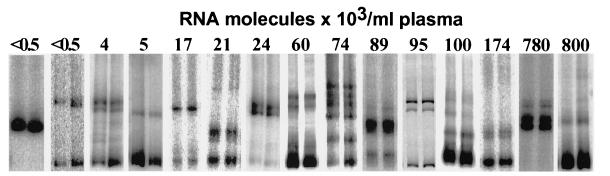
FIG. 2.
Correct sampling of plasma virus population diversity using HTA. Two aliquots of cDNA from plasma samples with different viral loads were used to initiate independent, duplicate nPCRs. The frequency of distinct env variants in each separate nPCR was then analyzed by HTA (see Materials and Methods).
Mathematical analysis.
The ImageQuant program (Molecular Dynamics, Sunnyvale, Calif.) was used to determine the relative signal intensities of the pre- and postdrug treatment env variants in P404 and P09 by quantifying the HTA signal with different electrophoretic mobilities. The HTA band signal intensity reflects the variant frequency (7), allowing the relative amounts of env variants to be calculated and relative fitness to be determined. As previously shown (15, 41), the exponential slope (S) of the pretreatment variant decay during days 0 to 14 of therapy represents a minimal estimate of the death rate constant (d) of infected cells producing virus. A maximal estimate of the half-life (t1/2) of actively infected cells is thus t1/2 = ln(2)/S. Similarly, by measuring the exponential slope (R) of a viral rebound, one can calculate the doubling time (t2) of the emerging variant population as follows: t2 = ln(2)/R.
The relative fitness (w) of two viral variants is defined as the ratio between the growth of the less fit virus (V1) and the growth of the more fit virus (V2) during the period of one viral generation (g) as follows: w = [V1(t + g)/V1(t)]/[V2(t + g)/V2(t)]. We assume here that the less and more fit viral variants grow (or decay) exponentially with rates e1 and e2 (where e2 > e1), respectively. Therefore, by measuring the exponential slopes of the two populations, relative fitness can be calculated according to the formula w = exp[g(e1 − e2)]. For the viral generation time (g), we used the previously calculated maximal estimate of 2.6 ± 0.8 days (32). Therefore, the values reported here are minimal estimates of relative fitness.
RESULTS
Plasma viremia following antiprotease monotherapy.
Nine individuals who had undergone HIV-1 protease inhibitor monotherapy (ritonavir or nelfinavir) were selected for this study. All of the patients experienced a rapid drop in plasma viremia to approximately 1 to 10% of pretreatment levels within 2 weeks of initiating treatment, reflecting effective blocking of new infection by these drugs and rapid turnover of the major pool of virus-producing cells (Fig. 1B and C) (15, 32, 41). Plasma viral loads then rebounded to levels at least 10-fold above the nadir within 14 to 28 days for two of the three patients with the highest pretreatment viremia (P105 and P09) and 28 to 60 days for five other patients with generally lower starting plasma viral loads (P404, P403, P07, P08, and P13). Also selected were two individuals (P02 and P406) who showed a sustained viral-load reduction, including one whose viremia remained below the bDNA assay detection level of 500 HIV RNA copies/ml for >200 days. The pretreatment viral loads of these two long-term responders were the lowest of the group selected.
Reproducible sampling of plasma HIV-1 populations.
To convincingly detect variant frequency changes in such genetically complex populations as HIV-1 quasispecies, adequate variant sampling needs to be demonstrated. To circumvent the technical and theoretical problems associated with the sequencing of PCR-derived subclones (10, 23, 26, 39) and the practical limitation of sequencing the large numbers of plasmid subclones required to adequately sample a large number of genetically complex populations, a nonsequencing method of sequence variant population analysis, HTA, was used. HIV-1 RNA was purified from plasma samples, cDNA was synthesized, and the 600-bp env V3-to-V5 region was amplified by nPCR (see Materials and Methods). The combined efficiency of viral RNA extraction and cDNA synthesis was determined to be 10 to 50% (see Materials and Methods). The nPCR DNA products were then reannealed with tracer amounts of clonal, patient-specific HTA probes (see Materials and Methods), and the mobility of the radiolabeled DNA heteroduplexes was analyzed by electrophoresis through polyacrylamide gels. With this method, every radiolabeled DNA heteroduplex band exhibiting a distinct electrophoretic mobility through polyacrylamide gel reflects at least one different env sequence variant (7, 9). The intensity of each heteroduplex band also reflects the frequency of the corresponding variant in the population (7). Changes in HTA banding patterns can therefore be used to document changes in the env sequence variants making up the HIV-1 populations.
The reproducibility of the variant population sampling (i.e., the frequency distribution of the distinct env variants coamplified) was determined by comparing the HTA patterns obtained with the products from two independent nPCRs initiated with separate aliquots of cDNA from the same plasma samples. Identical HTA patterns were obtained with the products of duplicate, independent nPCR amplifications (Fig. 2). The same frequencies of distinct env variants could therefore be reproducibly amplified, demonstrating proper population sampling and validating the use of this technique for the genetic analysis of plasma HIV-1 RNA populations.
Genetically stable plasma virus populations in the absence of antiretroviral treatment.
By use of the same procedure, the V3-to-V5 region of the envelope gene was amplified from viral RNA from longitudinally collected plasma samples from three individuals who had not initiated antiviral therapies during the course of sample collection or the preceding 4 months (P408, P78, and P79). In these untreated individuals, viral loads remained relatively stable, fluctuating less than 10-fold, and the HTA patterns remained constant, reflecting ongoing replication of the same combinations of env variants (Fig. 1A). Plasma samples longitudinally collected prior to therapy in subsequently treated patients also showed perpetuation of the same env populations (Fig. 1B and C). Therefore, in the absence of antiviral therapy, during the short periods analyzed, plasma HIV-1 populations did not undergo large changes in the frequency distributions of their env variants.
Rapid and transient changes in plasma virus populations during emergence of drug resistance.
During the first week following initiation of monotherapy, while the plasma viral load dropped 1 to 2 logs, the frequency distribution of env variants also remained constant (Fig. 1B and C). Concomitant with a viral-load rebound, major changes in the genetic composition of plasma virus populations were then detected in P404, P09, and P08 and, to a lesser extent, P07 (Fig. 1B). A reduction in overall genetic diversity levels (number of HTA bands) was not consistently observed. Instead, changes in the relative frequency of different env variants were seen. In patient P404, diversity actually increased from a single variant to three major variants (Fig. 1B). By 15 days of monotherapy, P09 showed almost complete replacement of his initial env quasispecies with a new form. Emerging env variants were present at very low levels at day 0 of treatment in P404 and P09 plasma (Fig. 1B and 3). In P08, plasma virus population diversity was only reduced at the nadir of virus production, when a previously minor variant rapidly rose to prominence; a week later, the multiple variants seen prior to treatment had re-emerged (Fig. 1B). In P07, a pre-existing variant also rose to temporary predominance. More subtle population changes were seen in P105 and P403, with minor variants no longer detected at several adjacent time points during viral-load rebound. Only minor changes were observed in P13, whose plasma virus env composition remained largely unchanged during a 20-fold viral-load drop and a rapid rebound. At later times, for all patients from whom samples were available, the plasma virus populations reverted to their pretreatment envelope variant compositions, as reflected by reappearance of the pretreatment HTA patterns by days 42 (P105) and 112 (P404). The unavailability of P09 samples beyond 28 days of monotherapy prevented evaluation of the long-term duration of env genetic changes in this subject.
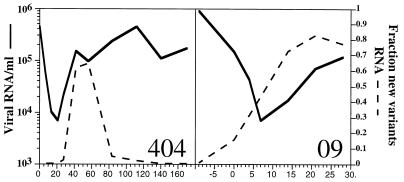
FIG. 3.
Frequency changes of different env variants following initiation of monotherapy and plasma viral loads. Time in days with respect to initiation of therapy is shown on the x axis.
Quasispecies changes in two patients with a sustained reduction in viremia.
Although P02 showed only a 10-fold decrease in plasma viremia, this reduction was sustained for at least 120 days. Ongoing plasma viremia of 2 × 103 to 5 × 103 HIV RNA copies/ml reflected partial viral suppression of HIV-1. P02 also showed a transient change in his plasma virus population starting 2 weeks after initiation of therapy, and the virus then reverted to its pretreatment env composition.
P406 manifested a long-term response to ritonavir, with the viral load falling from 3 × 104 to <500 RNA copies/ml by day 57, followed by low (500 to 1,000 HIV RNA copies/ml) but intermittently detectable viremia until day 85. Plasma viremia then fell below the bDNA assay detection limit until the last sample available at day 281, when HIV-1 RNA was again detected at a low level. Other antiviral drugs were then added to this subject’s regimen, his viremia fell and has remained below the detection limit for >1.5 years (data not shown). In this patient, HIV-1 cDNA could be amplified by nPCR from all of the plasma samples tested except those collected during the long interval when viremia was <500 RNA copies/ml. Between days 57 and 71, the plasma virus population lost a major variant. Between days 113 and 253, the population changed again, with the rapid electrophoretic mobility variant reappearing while the other variants were no longer detected. Thus, a reduction in plasma viral population diversity was observed in a long-term responder to ritonavir monotherapy.
Rapid emergence and high fitness of new variants.
The relative amounts of pretreatment and emerging env variants were determined in the two patients with the simplest patterns of plasma virus population changes, P404 and P09 (Fig. 3). Together with plasma viral loads and CD4 cell counts, these values were used to derive the following viral replication parameters. The concentration of the pretreatment variant in plasma decayed exponentially, giving maximal estimates of the t1/2 values of the cells producing these variants of 2.3 and 1.4 days for P404 and P09, respectively, in agreement with previous estimates (15, 32, 41). The emerging env variants grew exponentially, with t2 values of 1.9 and 3.3 days for P404 (days 28 to 42) and P09 (days 7 to 21), respectively.
The relative fitness (w) under drug selection of the pretreatment and emerging genomes carrying different env variants was determined. Minimal estimates of the relative fitness of pre- and postdrug selection genomes were w = 0.61 ± 0.1 and 0.51 ± 0.1 for patients P09 and P404, respectively. At 56 to 84 days after initiation of treatment, the emerging genomes in P404 were themselves largely replaced by genomes bearing the env variant that had dominated before treatment. During that interval, the relative fitness of the post- and predrug selection env variants carrying genomes was 0.72 ± 0.05.
DISCUSSION
According to a deterministic model of HIV-1 population genetics, the frequency of any particular mutation in the differentiated viral quasispecies found a few years following primary infection is primarily a function of its production rate and the relative replication fitness it confers on the genome (5, 6). Mutation rates are highly dependent on sequence context (2, 17, 24), with an average rate of 3 × 10−5 bp/replication cycle (24). The fitness cost associated with some drug resistance-associated mutations in the absence of drug selection has been estimated, from their frequency in untreated patients (19, 29) and competition between variants in vivo (13), to be approximately 1% (6). Using such assumptions and a steady-state value of 108 cells infected per day, 3 × 105 of the proviruses generated daily are expected to carry any particular single-site drug resistance-associated mutation (5, 6). Following the application of drug selection, the continued replication of such a large drug-resistant mutant population would be expected to maintain the genetic composition of the quasispecies at other (nonselected) loci. The plasma virus population changes seen at the env locus during the rapid emergence of protease inhibitor-resistant viruses therefore reflect a more complex process. Two explanations are proposed to account for these results.
One possibility involves variable levels of drug efficacy at different anatomical sites of virus production. HIV-1 populations with genetically distinct envelopes have been reported in different anatomical sites from the same person. Simultaneously collected peripheral blood mononuclear cells and brain tissue (12, 18, 30, 33, 38, 42), cerebrospinal fluid (40), semen (8, 44), spleen tissue (12), lung tissue (16), and lymph nodes (1, 38) have been shown to harbor distinct env variants. Compartmentalization of distinct pol variants have also been observed in brain versus spleen samples, and such local populations have been termed virodemes (42). Hence, complex viral populations in plasma may be made up of variants released from production sites containing distinct env variants, where protease inhibitor efficacy may differ for reasons such as inefficient transport and more rapid metabolism. A minimally ritonavir-resistant single-site protease mutant such as V82A, usually detected early in plasma virus during viral-load rebounds (27), should replicate better in compartments with reduced drug activity and be released into the plasma more readily than similar mutants at more drug-accessible sites. Since protease inhibitors block proteolytic processing in virus-producing cells, virions produced in a drug-privileged site would still be infectious at sites with better protease inhibitor activity. Further replication and mutation of single-site mutants under strong drug selection pressure may then lead to the generation of higher-level resistance mutants preferentially linked with tissue-specific env variants. The env changes observed in virus populations in plasma may therefore reflect the selective amplification of virus originating from tissues in which these protease inhibitors’ activity is reduced. It is also expected that reduced drug selection within drug-privileged sites would result in the delayed appearance of high-level drug resistance-associated mutations in proviruses found at these sites relative to that seen in more drug-accessible sites. The delayed appearance in autopsied brain tissue relative to spleen or cardiac blood DNA of proviral zidovudine (AZT) resistance-associated mutations (38, 42) may similarly reflect reduced AZT selective pressure on the viral population in the brain.
A second possibility involves the selection and amplification of so few drug-resistant lineages that the frequency distribution of env variants is altered (i.e., a genetic bottleneck). The loss of minority env variants detected in P105 and P403 might reflect the passage of their quasispecies through an effective population number small enough to result in the loss of low-frequency variants. In P404, P09, P08, and, to a lesser degree, P07, previously low-frequency variants were preferentially amplified. The amplification of previously rare env variants is unexpected according to the deterministic model, which predicts no population change. These results are more compatible with a random sampling of drug-resistant variants with the consequent amplification of previously rare env variants. Such founder effect following selection with a single antiviral drug and the rapid viral-load rebound seen in the patients studied here is more likely if one assumes an effective population numbering in the thousands as postulated by a recent stochastic model of HIV-1 evolution (20, 21).
Changes in envelope genes during the emergence of drug resistance were rapid but only transient. Once a highly drug-resistant population is established, as reflected by high-level viremia, the selective forces directed against the protease locus are expected to decline, while those directed against other loci are expected to resume predominant roles. Selective pressures directed against the env locus of emerging drug-resistant plasma virus populations may therefore favor the return of the pretreatment env variants and thus account for the transient nature of the genetic changes observed. Genetic recombination between highly drug-resistant pro and env sequences may greatly accelerate such a multilocus adaptation (28), allowing drug-resistant viruses to recolonize major drug-susceptible sites of virus production using optimally adapted env sequences. Latently infected cells (4) or long-lived, virus-producing cells such as macrophages (31) may contribute such env variants. The transient nature of the env genetic changes observed in plasma RNA populations likely explains why previous studies analyzing proviral sequences (22) or plasma samples collected 6 months after initiation of AZT monotherapy (43) failed to detect a significant impact of antiviral treatment on the evolution of the env gene.
Continued genetic changes were seen in the plasma virus population of a long-term responder to ritonavir (P406). Such fluctuations may reflect differences in the t1/2 values of various long-lived (31), virus-releasing cell populations containing distinct env variants and reactivation of latently infected cells (4). Analysis of more long-term responders to antiviral therapies is needed to determine if reduction in the genetic diversity of the residual population is a common consequence of long-term virus suppression.
In conclusion, protease inhibitor monotherapies with either ritonavir or nelfinavir and selection for drug resistance had rapid, pronounced, but transient effects on the plasma virus composition at the env locus. In some patients (P403 and P105), minor env variants were lost, an indication of random sampling of a number of variants smaller than that expected for single-site mutants in a current deterministic model (6). In other patients (P404, P09, P08, and P07), previously rare env variants were amplified, an observation most compatible with the existence of a site(s) of viral replication at least partially refractory to protease inhibitors, harboring, as has been repeatedly reported for different tissues, env variants distinct from those in blood. The lack of long-term impact on the genetic diversity at the env locus following rapid viral-load rebound also indicates that no benefit, in terms of reduced evolutionary potential at a linked locus, was gained by using such monotherapies.
ACKNOWLEDGMENTS
We thank the participants of these protease inhibitor trials and Andrew J. Leigh Brown and Laurence Doukhan for helpful discussions.
This work was supported by the Aaron Diamond Foundation, a CFAR Development Award (AI27742), and NIH R29 grant (AI40408) to E.L.D.
REFERENCES
- 1.Ball J K, Holmes E C, Whitwell H, Desselberger U. Genomic variation of human immunodeficiency virus type 1 (HIV-1): molecular analyses of HIV-1 in sequential blood samples and various organs obtained at autopsy. J Gen Virol. 1994;75:67–79. doi: 10.1099/0022-1317-75-4-867. [DOI] [PubMed] [Google Scholar]
- 2.Bebenek K, Abbotts J, Wilson S H, Kunkel T A. Error-prone polymerization by HIV-1 reverse transcriptase. Contribution of template-primer misalignment, miscoding, and termination probability to mutational hot spots. J Biol Chem. 1993;268:10324–10334. [PubMed] [Google Scholar]
- 3.Boom R, Sol C J A, Salimans M M M, Jansen C L, Wertheim-van Dillen P M E, van der Noordaa J. Rapid and simple method for purification of nucleic acids. J Clin Microbiol. 1990;28:495–503. doi: 10.1128/jcm.28.3.495-503.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chun T W, Carruth L, Finzi D, Shen X, DiGiuseppe J A, Taylor H, Hermankova M, Chadwick K, Margolick J, Quinn T C, Kuo Y H, Brookmeyer R, Zeiger M A, Barditch-Crovo P, Siliciano R F. Quantification of latent tissue reservoirs and total body viral load in HIV-1 infection. Nature. 1997;387:183–188. doi: 10.1038/387183a0. [DOI] [PubMed] [Google Scholar]
- 5.Coffin J M. HIV population dynamics in vivo: implications for genetic variation, pathogenesis, and therapy. Science. 1995;267:483–489. doi: 10.1126/science.7824947. [DOI] [PubMed] [Google Scholar]
- 6.Coffin J M. Population dynamics of HIV drug resistance. In: Richman D D, editor. Antiviral drug resistance. New York, N.Y: John Wiley & Sons, Inc.; 1996. pp. 279–303. [Google Scholar]
- 7.Delwart E L, Gordon C J. Tracking changes in HIV-1 envelope quasispecies using DNA heteroduplex analysis. Methods Companion Methods Enzymol. 1997;12:348–354. doi: 10.1006/meth.1997.0489. [DOI] [PubMed] [Google Scholar]
- 8.Delwart E L, Mullins J I, Gupta P, Learn G H J, Jr, Holodniy M, Katzenstein D, Walker B D, Singh M K. Human immunodeficiency virus type 1 populations in blood and semen. J Virol. 1998;72:617–623. doi: 10.1128/jvi.72.1.617-623.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Delwart E L, Pan H, Sheppard H W, Wolpert D, Neumann A U, Korber B, Mullins J I. Slower evolution of HIV-1 quasispecies during progression to AIDS. J Virol. 1997;71:7498–7508. doi: 10.1128/jvi.71.10.7498-7508.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Delwart E L, Sheppard H W, Walker B D, Goudsmit J, Mullins J I. Human immunodeficiency virus type 1 evolution in vivo tracked by DNA heteroduplex mobility assays. J Virol. 1994;68:6672–6683. doi: 10.1128/jvi.68.10.6672-6683.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Delwart E L, Shpaer E G, McCutchan F E, Louwagie J, Grez M, Rübsamen-Waigmann H, Mullins J I. Genetic relationships determined by a heteroduplex mobility assay: analysis of HIV env genes. Science. 1993;262:1257–1261. doi: 10.1126/science.8235655. [DOI] [PubMed] [Google Scholar]
- 12.Epstein L G, Kuiken C, Blumberg B M, Hartman S, Sharer L R, Clement M, Goudsmit J. HIV-1 V3 domain variation in brain and spleen of children with AIDS: tissue-specific evolution within host-determined quasispecies. Virology. 1991;180:583–590. doi: 10.1016/0042-6822(91)90072-j. [DOI] [PubMed] [Google Scholar]
- 13.Goudsmit J, De Ronde A, Ho D D, Perelson A S. Human immunodeficiency virus fitness in vivo: calculations based on a single zidovudine resistance mutation at codon 215 of reverse transcriptase. J Virol. 1996;70:5662–5664. doi: 10.1128/jvi.70.8.5662-5664.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Havlir D V, Eastman S, Gamst A, Richman D D. Nevirapine-resistant human immunodeficiency virus: kinetics of replication and estimated prevalence in untreated patients. J Virol. 1996;70:7894–7899. doi: 10.1128/jvi.70.11.7894-7899.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ho D D, Neumann A U, Perelson A S, Chen W, Leonard J M, Markowitz M. Rapid turnover of plasma virions and CD4 lymphocytes in HIV-1 infection. Nature. 1995;373:123–126. doi: 10.1038/373123a0. [DOI] [PubMed] [Google Scholar]
- 16.Itescu S, Simonelli P F, Winchester R J, Ginsberg H S. Human immunodeficiency virus type 1 strains in the lungs of infected individuals evolve independently from those in peripheral blood and are highly conserved in the C-terminal region of the envelope V3 loop. Proc Natl Acad Sci USA. 1994;91:11378–11382. doi: 10.1073/pnas.91.24.11378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ji J, Loeb L A. Fidelity of HIV-1 reverse transcriptase copying a hypervariable region of the HIV-1 env gene. Virology. 1994;199:323–330. doi: 10.1006/viro.1994.1130. [DOI] [PubMed] [Google Scholar]
- 18.Korber B T M, Kunstman K J, Patterson B K, Furtado M, McEvilly M M, Levy R, Wolinsky S M. Genetic differences between blood- and brain-derived viral sequences from human immunodeficiency virus type 1-infected patients: evidence of conserved elements in the V3 region of the envelope protein of brain-derived sequences. J Virol. 1994;68:7467–7481. doi: 10.1128/jvi.68.11.7467-7481.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lech W J, Wang G, Yang Y L, Chee Y, Dorman K, McCrae D, Lazzeroni L C, Erickson J W, Sinsheimer J S, Kaplan A H. In vivo sequence diversity of the protease of human immunodeficiency virus type 1: presence of protease inhibitor-resistant variants in untreated subjects. J Virol. 1996;70:2038–2043. doi: 10.1128/jvi.70.3.2038-2043.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leigh-Brown A. Analysis of HIV-1 env gene sequences reveals evidence for a low effective number in the viral population. Proc Natl Acad Sci USA. 1997;94:1862–1865. doi: 10.1073/pnas.94.5.1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leigh-Brown A, Richman D D. HIV-1: gambling on the evolution of drug resistance. Nat Med. 1997;3:268–271. doi: 10.1038/nm0397-268. [DOI] [PubMed] [Google Scholar]
- 22.Leigh-Brown A L, Cleland A. Independent evolution of the env and pol genes of HIV-1 during zidovudine therapy. AIDS. 1996;10:1067–1073. [PubMed] [Google Scholar]
- 23.Lui S L, Rodrigo A G, Shankarappa R, Learn G H, Hsu L, Davidov O, Zhao L P, Mullins J I. HIV quasispecies and resampling. Science. 1996;273:415–416. doi: 10.1126/science.273.5274.415. [DOI] [PubMed] [Google Scholar]
- 24.Mansky L M, Temin H M. Lower in vivo mutation rate of human immunodeficiency virus type 1 than that predicted from the fidelity of purified reverse transcriptase. J Virol. 1995;69:5087–5094. doi: 10.1128/jvi.69.8.5087-5094.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Markowitz M, Saag M, Powderly W G, Hurley A M, Hsu A, Valdes J M, Henry D, Sattler F, La Marca A, Leonard J M, Ho D D. A preliminary study of ritonavir, an inhibitor of HIV-1 protease, to treat HIV-1 infection. N Engl J Med. 1995;333:1534–1539. doi: 10.1056/NEJM199512073332204. [DOI] [PubMed] [Google Scholar]
- 26.Meyerhans A, Vartanian J-P, Wain-Hobson S. DNA recombination during PCR. Nucleic Acids Res. 1990;18:1687–1691. doi: 10.1093/nar/18.7.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Molla A, Korneyeva M, Gao Q, Vasavanonda S, Schipper P J, Mo H M, Markowitz M, Chernyavskiy T, Niu P, Lyons N, Hsu A, Granneman G R, Ho D D, Boucher C A, Leonard J M, Norbeck D W, Kempf D J. Ordered accumulation of mutations in HIV protease confers resistance to ritonavir. Nat Med. 1996;2:760–766. doi: 10.1038/nm0796-760. [DOI] [PubMed] [Google Scholar]
- 28.Moutouh L, Corbeil J, Richman D D. Recombination leads to the rapid emergence of HIV-1 dually resistant mutants under selective drug pressure. Proc Natl Acad Sci USA. 1996;93:6106–6111. doi: 10.1073/pnas.93.12.6106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Najera I, Holguin A, Quinones-Mateu M E, Munoz-Fernandez M A, Najera R, Lopez-Galindez C, Domingo E. pol gene quasispecies of human immunodeficiency virus: mutations associated with drug resistance in virus from patients undergoing no drug therapy. J Virol. 1995;69:23–31. doi: 10.1128/jvi.69.1.23-31.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pang S, Vinters H V, Akashi T, O’Brien W A, Chen I S. HIV-1 env sequence variation in brain tissue of patients with AIDS-related neurologic disease. J Acquired Immune Defic Syndrome. 1991;4:1082–1092. [PubMed] [Google Scholar]
- 31.Perelson A S, Essunger P, Cao Y, Vesanen M, Hurley A, Saksela K, Markowitz M, Ho D D. Decay characteristics of HIV-1-infected compartments during combination therapy. Nature. 1997;387:188–91. doi: 10.1038/387188a0. [DOI] [PubMed] [Google Scholar]
- 32.Perelson A S, Neumann A U, Markowitz M, Leonard J M, Ho D D. HIV-1 dynamics in vivo: virion clearance rate, infected cell life-span, and viral generation time. Science. 1996;271:1582–1586. doi: 10.1126/science.271.5255.1582. [DOI] [PubMed] [Google Scholar]
- 33.Power C, McArthur J C, Johnson R T, Griffin D E, Glass J D, Perryman S, Chesebro B. Demented and nondemented patients with AIDS differ in brain-derived human immunodeficiency virus type 1 envelope sequences. J Virol. 1994;68:4643–4649. doi: 10.1128/jvi.68.7.4643-4649.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ratner L, Haseltine W, Patarca R, Livak K J, Starcich B, Josephs S F, Doran E R, Rafalski J A, Whitehorn E A, Baumeister K, et al. Complete nucleotide sequence of the AIDS virus, HTLV-III. Nature. 1985;313:277–284. doi: 10.1038/313277a0. [DOI] [PubMed] [Google Scholar]
- 35.Richman D D. Drug resistance in relation to pathogenesis. AIDS. 1995;9:S49–S53. [PubMed] [Google Scholar]
- 36.Richman D D, Havlir D, Corbeil J, Looney D, Ignacio C, Spector S A, Sullivan J, Cheeseman S, Barringer K, Pauletti D, Shih C-K, Myers M, Griffin J. Nevirapine resistance mutations of human immunodeficiency virus type 1 selected during therapy. J Virol. 1994;68:1660–1666. doi: 10.1128/jvi.68.3.1660-1666.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schuurman R, Nijhuis M, van Leeuwen R, Schipper P, de Jong D, Collis P, Danner S A, Mulder J, Loveday C, Christopherson C, Kwok S, Sninsky J, Boucher C. Rapid changes in human immunodeficiency virus type 1 RNA load and appearance of drug-resistant virus populations in persons treated with lamivudine (3TC) J Infect Dis. 1995;171:1411–1419. doi: 10.1093/infdis/171.6.1411. [DOI] [PubMed] [Google Scholar]
- 38.Sheehy N, Desselberger U, Whitwell H, Ball J K. Concurrent evolution of regions of the envelope and polymerase genes of human immunodeficiency virus type 1 during zidovudine (AZT) therapy. J Gen Virol. 1996;77:1071–1081. doi: 10.1099/0022-1317-77-5-1071. [DOI] [PubMed] [Google Scholar]
- 39.Simmonds P, Balfe P, Peutherer J F, Ludlam C A, Bishop J O, Leigh Brown A J. Human immunodeficiency virus-infected individuals contain provirus in small numbers of peripheral mononuclear cells and at low copy numbers. J Virol. 1990;64:864–872. doi: 10.1128/jvi.64.2.864-872.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Steuler H, Storch-Hagenlocher B, Wildemann B. Distinct populations of human immunodeficiency virus type 1 in blood and cerebrospinal fluid. AIDS Res Hum Retroviruses. 1992;8:53–59. doi: 10.1089/aid.1992.8.53. [DOI] [PubMed] [Google Scholar]
- 41.Wei X, Ghosh S K, Taylor M E, Johnson V A, Emini E A, Deutsch P, Lifson J D, Bonhoeffer S, Novak M A, Hahn B H, Saag M S, Shaw G M. Viral dynamics in human immunodeficiency virus type 1 infection. Nature. 1995;373:117–126. doi: 10.1038/373117a0. [DOI] [PubMed] [Google Scholar]
- 42.Wong J K, Ignacio C C, Torriani F, Havlir D, Fitch N J S, Richman D D. In vivo compartementalization of human immunodeficiency virus: evidence from the examination of pol sequences from autopsy tissues. J Virol. 1997;71:2059–2071. doi: 10.1128/jvi.71.3.2059-2071.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang Y-M, Dawson S C, Landsman D, Lane H C, Salzman N P. Persistence of four related human immunodeficiency virus subtypes during the course of zidovudine therapy: relationship between virion RNA and proviral DNA. J Virol. 1994;68:425–432. doi: 10.1128/jvi.68.1.425-432.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhu T, Wang N, Carr A, Nam D S, Moor-Jankowski R, Cooper D A, Ho D D. Genetic characterization of human immunodeficiency virus type 1 in blood and genital secretions: evidence for viral compartmentalization and selection during sexual transmission. J Virol. 1996;70:3098–3107. doi: 10.1128/jvi.70.5.3098-3107.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]