Abstract
Vaccinia virus produces two morphologically distinct forms of infectious virus, termed intracellular mature virus (IMV) and extracellular enveloped virus (EEV). EEV is important for virus dissemination within a host and has different surface proteins which bind to cell receptors different from those used by IMV. Six genes are known to encode EEV-specific proteins. One of these, B5R, encodes a 42-kDa glycoprotein with amino acid similarity to members of the complement control protein superfamily and contains four copies of a 50- to 70-amino-acid repeat called the short consensus repeat (SCR). Deletion of B5R causes a small-plaque phenotype, a 10-fold reduction in EEV formation, and virus attenuation in vivo. In this study, we inserted mutated versions of the B5R gene lacking different combinations of the SCRs into a virus deletion mutant lacking the B5R gene. The resultant viruses each formed small plaques only slightly larger than those of the deletion mutant; however, the virus containing only SCR 1 formed plaques slightly larger than those of viruses with SCRs 1 and 2 or SCRs 1, 2, and 3. All of these viruses produced approximately 50-fold more infectious EEV than wild-type virus and formed comet-shaped plaques under liquid overlay. Despite producing more EEV, the mutant viruses were unable to induce the polymerization of actin on intracellular virus particles. The implications of these results for our understanding of EEV formation, release, and infectivity are discussed.
Vaccinia virus (VV) is the most extensively studied poxvirus. Like other members of this family of viruses, VV has a large double-stranded DNA genome, replicates in the cytoplasm, encodes enzymes for transcription and DNA replication, and expresses many virulence factors that aid virus propagation in vivo (24).
VV morphogenesis is a complex process that produces several distinct intermediates and more than one form of mature infectious virion (1, 16). Morphogenesis is initiated by the production of crescent-shaped structures within virus factories (for a review, see reference 8). These crescents contain host lipids and virus-encoded proteins and are thought to be formed by modification of host membranes of the intermediate compartment between the endoplasmic reticulum and the Golgi stack (39). Growth of these crescents produces complete spherical particles which lack infectivity and are termed immature virions. The nucleoprotein core of immature virions condense to form electron-dense intracellular mature virus (IMV), the first infectious form of virus and the form representing the majority of infectious progeny.
IMV may either remain intracellular and be released by cell lysis, become wrapped by intracellular membranes (17, 23, 28) derived from the trans-Golgi network (TGN) (12, 34) or early endosomes (41) to form intracellular enveloped virus (IEV), or bud through the plasma membrane late during infection (43). The wrapping of IMV by TGN or early endosomal membranes requires the specific interaction of virus proteins on the IMV surface with virus proteins exposed on the cytosolic side of these cellular membranes. IEV particles can polymerize actin in a manner reminiscent of intracellular bacteria such as Shigella and Listeria to form actin tails which propel the virus particles to the cell surface (5, 6). Once at the cell surface, the outer membrane fuses with the plasma membrane to form extracellular enveloped virus (EEV), most of which remains at the cell surface and is called cell-associated enveloped virus (CEV) (3). Enveloped virus is important for virus spread between cells and for the long-range dissemination of virus in cell culture or in vivo (1–3, 26, 27, 30). EEV binds to different cellular receptors from IMV (45) and is resistant to neutralization by antibody (15, 44).
Six genes that encode EEV-specific proteins have been identified. These are A56R, encoding the hemagglutinin (gp86) (29, 35), F13L (protein p37) (13), B5R (gp42) (10, 18), A34R (gp22-24) (9), A36R (p45-50) (25), and A33R (gp23-28) (31). The functions of these proteins are being studied by using virus mutants which have specific genes either deleted, mutated, or repressed (for a review, see reference 38). None of the EEV proteins are needed for IMV formation, but they are all required for virus virulence in one model or another, and they have different effects on plaque phenotype and the formation and infectivity of EEV. The hemagglutinin does not affect plaque size but causes a syncytial phenotype (16, 36). Deletion of gp42 and p37 cause a small plaque size and a dramatic (≤10-fold) decrease in EEV formation due to a failure to wrap IMV with TGN or tubular endosomal membranes (2, 11, 47). Sequences within the transmembrane and cytoplasmic tail of B5R are important for targeting the protein to the wrapping membrane because fusion of these regions of the B5R protein to human immunodeficiency virus (HIV) gp120 caused localization of HIV gp120 onto EEV (19). Loss of p45-50 causes a more modest (three- to fivefold) reduction in plaque size and EEV formation (25). The A34R protein has multiple functions: it is required for a normal plaque size (9); it determines whether EEV remains attached to the cell surface as CEV or if it is released as EEV (4, 22); it is important for some aspect of the reinfection process, since A34R− EEV has a five- to sixfold reduction in specific infectivity (22); and it is required for the formation of intracellular actin tails (46). A mutant lacking A33R has not yet been described.
In this study, we examined the functions of the B5R protein by the construction of mutant B5R alleles and the insertion of these into a B5R deletion mutant virus. B5R is related to members of complement control protein superfamily and contains four copies of a short consensus repeat (SCR) (50 to 70 amino acids in length) that are typical of this superfamily (10, 18, 40). We have deleted either SCR 4 alone, SCRs 3 and 4, or SCRs 2, 3, and 4 and examined the properties of viruses expressing these mutant proteins. Each virus produces a small plaque which forms comets under liquid overlay, and consistent with the comet plaque phenotype, there is a dramatic increase in EEV formation. In addition, each mutant virus is unable to induce the formation of intracellular actin tails.
MATERIALS AND METHODS
Cells and viruses.
Human TK−143B cells, monkey kidney CV-1 and BS-C-1 cells, and rabbit kidney RK13 cells were grown in Dulbecco’s modified Eagle’s medium in 10% fetal bovine serum (FBS). A VV Western Reserve (WR) mutant with 75% of the B5R coding region deleted (vΔB5R) was described previously (11). Viruses were grown and titrated in BS-C-1 cells as described previously (20).
Reagents.
Rabbit polyclonal antibody to the B5R gene product (10) and mouse monoclonal antibodies (MAbs) to the VV D8L gene product (AB1.1) (25) and the A27L gene product (5B4/2F2) (7) have been described elsewhere. Phalloidin and tunicamycin were obtained from Sigma, and [3H]thymidine (70 to 86 Ci/mmol) was obtained from Amersham.
Generation of mutant B5R genes.
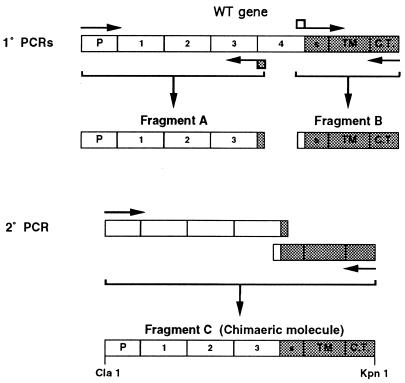
Mutant B5R genes which lacked SCR 4, SCRs 3 and 4, or SCRs 2, 3, and 4 were constructed by splicing by overlap extension (SOE) (14) and gene cloning. The DNA template for PCR was plasmid pSTH4, which contains the entire B5R gene and flanking sequences cloned into pUC13 (10). Each mutant gene was constructed by separately amplifying the gene in two pieces and then joining these pieces together by SOE to a produce the complete mutant gene with a 5′ ClaI site and a 3′ KpnI site (Fig. 1). Each gene was then cloned into pEM6 so that the B5R promoter and coding region were inserted into the VV thymidine kinase (TK) gene. pEM6 is a plasmid containing a version of the B5R gene cloned into the TK gene and flanked by ClaI and KpnI sites (full details to be published elsewhere). The 5′ part of each mutant gene started 115 nucleotides upstream of the initiation codon of the open reading frame, so as to include the entire B5R promoter region (10), and extended downstream to either the end of the first, second, or third SCR domain at amino acid 75, 128, or 185 of the VV strain WR wild-type (WT) protein, respectively. The 3′ part of the gene started from the translation termination codon and continued upstream to include the cytoplasmic tail, the transmembrane anchor, and 39-amino-acid spacer region before the SCRs, starting at amino acid 242 of the WT protein. The internal oligonucleotides for each fragment contained overlaps enabling these individual PCR fragments to be assembled into a single gene by SOE using the outermost oligonucleotides as primers. After assembly, the PCR fragment was digested with ClaI and KpnI, cloned into pEM6, which had been digested with the same enzymes, and sequenced to confirm the fidelity of the DNA. The plasmids containing SCR 1, SCRs 1 and 2, or SCRs 1, 2, and 3 were called pEM20, pEM19, and pEM18, respectively. The WT B5R gene was also assembled and inserted into the TK locus as a control for ectopic expression of the mutant B5R genes. The WT gene was generated by ligating two fragments which contained either the 5′ or 3′ region of the B5R gene. The 5′ region including the promoter was excised from plasmid pEM9 with ClaI and HpaI, the latter enzyme cutting the open reading frame at codon 285. pEM9 is a plasmid containing the 5′ region of the B5R gene cloned within the TK locus. This fragment was cloned into plasmid pEM11 which had been digested with the same enzymes. pEM11 is a plasmid containing the 3′ region of the B5R gene flanked by the TK locus (details to be published elsewhere). Thus, the entire WT gene was assembled within the TK locus. This plasmid was called pEM21.
FIG. 1.
Schematic showing the formation of a mutant B5R gene lacking SCR 4 by SOE. Similar strategies were used to produce mutant B5R genes lacking SCRs 3 and 4 or SCRs 2, 3, and 4. P, promoter; S, spacer region between SCR 4 and transmembrane domain; TM, transmembrane domain; C.T., cytoplasmic tail.
Construction of recombinant VV.
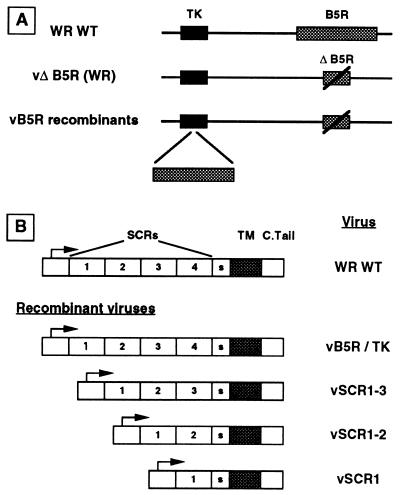
The WT or mutant B5R genes were inserted into the TK locus of a mutant virus lacking the B5R gene (vΔB5R) (11) by infecting CV-1 cells with vΔB5R at 0.5 PFU/cell and then transfecting these cells 2 h postinfection (hpi) separately with plasmid pEM18, pEM19, pEM20, or pEM21. Progeny virus was harvested 48 hpi, and TK− viruses were isolated by plaque assay on human TK−143B cells in the presence of bromodeoxyuridine as described previously (20). Recombinant plaques were distinguished from spontaneous TK− isolates by screening with oligonucleotides spanning the insertion site within the TK gene (37). Viruses were plaque purified three times under selection before stocks were grown and titrated on BS-C-1 cells. Recombinant viruses were called vEM11, vEM12, vEM13, and vEM14. These contained SCR 1 (vEM11), SCRs 1 and 2 (vEM12), SCRs 1, 2, and 3 (vEM13) linked to the 39-amino-acid spacer region and transmembrane and cytoplasmic tail, or the entire WT gene (vEM14). In this report, these viruses are referred to as vSCR1, vSCR1-2, vSCR1-3, and vB5R/WT, respectively.
Immunoblotting.
BS-C-1 cells were infected at 10 PFU/cell, and extracts were prepared from cells at 16 hpi as described previously (25). After resolution by electrophoresis on sodium dodecyl sulfate–10% polyacrylamide gels proteins were electrophoretically transferred to nitrocellulose membranes (42) and detected by incubation with specific antibodies. To detect B5R, the filter was incubated with rabbit anti-B5R serum (10) diluted 1:2,500. To confirm that cells had been equally infected with the different viruses, filters were stripped and reprobed with mouse MAb AB1.1 directed against the D8L gene product (25).
Analysis of comet formation.
Monolayers of RK13 cells were infected with 50 PFU and incubated under liquid overlay, minimal essential medium (MEM) containing 2.5% FBS, for between 2 and 5 days. Medium was removed, and the cells were stained with 0.1% crystal violet in 15% ethanol.
Infectivity assays.
To measure the infectious virus present into the culture supernatant, RK13 cells were infected at 1 PFU/cell and harvested at 24 hpi. Culture medium was removed and clarified by centrifugation at 2,000 rpm (Beckman GPR benchtop centrifuge) and titrated directly in either the absence or the presence of MAb 5B4/2F2 (7) to neutralize IMV as described previously (45). Viruses were diluted prior to addition of antibody to maintain similar virus/antibody ratios. Virus infectivity present in cells was assayed by scraping the infected cells into phosphate-buffered saline (PBS), combining this with the pellets derived from clarification of culture supernatant (see above), and collecting cells by centrifugation (as described above). After three cycles of freeze-thawing and sonication, the infectivity present was determined by plaque assay on BS-C-1 cells (20).
CsCl density gradient centrifugation.
Flasks of RK13 cells (175 cm2) were infected at 10 PFU/cell and labeled with 100 μCi of [3H]thymidine from 1.75 hpi. At 24 hpi, the culture medium was collected and clarified by centrifugation at 2,000 rpm for 10 min in a Beckman GPR benchtop centrifuge, and the virus was pelleted from the supernatant at 14,000 rpm for 80 min at 4°C in a Beckman SW41 ultracentrifuge rotor. The virus pellet was gently resuspended in 1 ml of 10 mM Tris-HCl (pH 9) and layered onto a CsCl density gradient. The gradient was formed and then centrifuged as described previously (25). Fractions (0.5 ml) were collected, and their density and radioactivity were determined by refractometry and scintillation counting. The virus infectivity in the fresh, clarified supernatant and in the infected cells (which were detached from the flask and collected by centrifugation) was determined by plaque assay on BS-C-1 cells.
Immunocytochemistry.
BS-C-1 cells were seeded onto 20-mm-diameter glass microscope slides (Chance Proper Ltd., West Midlands, England) to attain 20 to 30% confluence. Binding of viruses (10 PFU/cell) was performed on ice for 1 h to synchronize infections. Cells were then washed three times in MEM (37°C) and incubated at 37°C for various periods. Cells were fixed in PBS containing 4% paraformaldehyde at room temperature for 20 min and permeabilized with 0.1% Triton X-100 in PBS before staining. Immunofluorescence was analyzed and recorded by using a Bio-Rad MRC1024 confocal laser scanning microscope, and the resulting images were processed with the Adobe Photoshop program.
Electron microscopy.
Cells were infected at 5 PFU/cell with the indicated viruses, harvested at 16 hpi, and processed for electron microscopy as described previously (32).
RESULTS
Viruses with a mutant B5R gene.
The B5R protein is required for the wrapping of IMV by intracellular membranes, EEV formation, normal plaque size, and virus virulence. B5R is a typical type I membrane glycoprotein with an amino-terminal signal sequence followed by an extracellular hydrophilic domain containing four SCRs, a transmembrane anchor sequence, and a short cytoplasmic domain. To dissect the roles of these different domains in the virus replication cycle, we have constructed mutant B5R genes which lack different combinations of SCRs. These mutants contain either SCR 1, SCRs 1 and 2, or SCRs 1, 2, and 3 linked to the other regions of the B5R gene (Fig. 2). Each of these genes was assembled by SOE (Fig. 1), cloned into a transfer vector enabling insertion into the VV TK locus, sequenced, and used to construct recombinant viruses, using the B5R deletion mutant vΔB5R as the parental virus (11). As a control, the WT B5R gene was also inserted into the TK locus of vΔB5R. These viruses are referred to here as vSCR1, vSCR1-2, vSCR1-3, and vB5R/TK. The genomes of these recombinant viruses were carefully analyzed by agarose gel electrophoresis of HindIII- and SalI/EcoRI-digested DNA, Southern blotting, and PCR using oligonucleotides specific for the B5R gene and regions flanking the TK gene. These analyses confirmed that the natural B5R locus contained the deleted B5R allele of the parental virus vΔB5R, the ectopic WT or mutant B5R genes had been inserted into the TK gene, there were no additional copies of B5R elsewhere in the virus genome, and all restriction fragments which did not include the TK or B5R loci were indistinguishable for all viruses (data not shown).
FIG. 2.
Structures of recombinant VV genomes (A) and mutant B5R genes (B). TM, transmembrane domain; S, spacer; C.Tail, cytoplasmic tail.
Protein expression by mutant viruses.
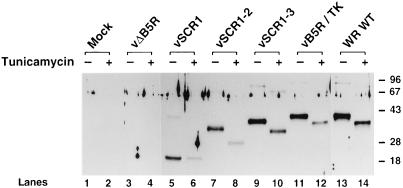
To examine the size and location of the B5R protein made by the mutant viruses, we infected BS-C-1 cells with each virus and analyzed infected cells extracts by immunoblotting with a rabbit antibody against the B5R protein (10). Figure 3 shows that WT WR- and vB5R/TK-infected cells produced a 42-kDa B5R protein which was slightly less abundant in vB5R/TK-infected cells and absent in vΔB5R- or mock-infected cells. Viruses vSCR1-3, vSCR1-2, and vSCR1 produced B5R proteins of decreasing size as expected, and in each case trace amounts of higher-molecular-weight complexes were present, as noted previously for the WT B5R protein (10, 18). Incubation of infected cells in the presence of tunicamycin, to inhibit addition of carbohydrate linked to asparagine residues, reduced the size of all B5R proteins except that made by vSCR1. Presumably this B5R protein was not modified by N-linked carbohydrate because, unlike vSCR1-2 and vSCR1-3, it did not contain the NX(S/T) motifs in SCR 2.
FIG. 3.
Immunoblot showing the B5R proteins made in cells infected with the different viruses. BS-C-1 cells were infected with the indicated viruses at 10 PFU/cell in the presence or absence of tunicamycin (10 μg/ml), and extracts were prepared at 16 hpi. After electrophoresis through a 10% polyacrylamide gel, the proteins were transferred to nitrocellulose and B5R protein was detected by incubation with rabbit antibody against B5R (1:1,000 dilution) (10) followed by a horseradish peroxidase-conjugated species-specific secondary antibody and ECL reagents (Amersham). Molecular weight markers are shown in kilodaltons.
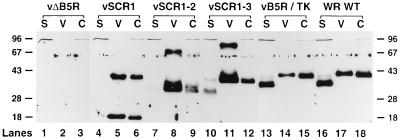
B5R is a membrane glycoprotein that becomes part of the outer envelope of EEV and is easily detected in virions released from the infected cell. A fraction of the mature B5R protein is also proteolytically digested to produce a soluble protein of 35 kDa (21). The presence of virus-associated and soluble B5R protein was therefore examined in the supernatant of cells infected separately with each of the virus mutants (Fig. 4). WT WR and vB5R/TK viruses each synthesized a 42-kDa protein that was present in infected cells and released virus particles (lanes 14, 15, 17, and 18). These viruses also produced a soluble 35-kDa protein as expected (lanes 13 and 16). No B5R proteins were made in vΔB5R-infected cells (lanes 1 to 3) or mock-infected cells (data not shown). vSCR1, vSCR1-2, and vSCR1-3 each released B5R protein into the cell culture supernatant that was the same size as that present in infected cells. This protein was shown to be virion associated rather than soluble since it was pelleted by centrifugation (lanes 5, 6, 8, 9, 11, and 12). This finding suggested, unexpectedly, that EEV was being made by these mutant viruses, and the intensity of the B5R proteins detected by immunoblotting indicated that the levels of EEV released were as high as for WT virus, if not higher. A corresponding increase in another EEV-specific protein (p37) was also observed in extracellular virus released from cells infected with these mutants (data not shown).
FIG. 4.
Immunoblot showing that mutant B5R proteins are released from infected cells. RK13 cells were infected with the indicated viruses at 10 PFU/cell and incubated in MEM with 2.5% FBS for 24 h. The supernatants of infected cells were clarified by low-speed centrifugation (2,000 rpm in a Beckman GPR benchtop centrifuge for 10 min) to pellet detached cells and cell debris and then high-speed centrifugation (14,000 rpm; SW28 rotor; Beckman ultracentrifuge; 80 min) to pellet virus particles (lanes V) from soluble protein (lanes S). An infected cell extract (C) was prepared as for Fig. 3. B5R proteins were resolved by electrophoresis and detected as described for Fig. 3. Molecular weight markers are shown in kilodaltons.
The mutants differed from WT in two other respects. First, a much greater proportion of virion-associated B5R protein was found in complexes than in monomers. Evidently the formation of these complexes does not require SCR 2, 3, or 4. Second, each mutant virus released proportionally less soluble protein than did WT virus. For vSCR1 and vSCR1-2, only trace amounts of soluble protein were detected (lanes 4 and 7), and these proteins were the same size as those in cells and virions and might represent residual virus particles. For vSCR1-3, slightly more soluble protein was present (lane 10), but this was still considerably less than for WT virus (lane 16), and the soluble protein was mostly smaller than B5R protein present in cells and virions. These data suggest that SCRs 3 and 4 influence the efficiency of proteolytic cleavage. The size of the 35-kDa proteolytic fragment made by WT virus suggests that the cleavage is likely to occur between SCR 4 and the transmembrane domain. If this is the case, the reduced cleavage in the absence of SCR 4, and the virtual abrogation of cleavage when SCRs 3 and 4 are deleted, suggests that these SCRs affecting the folding of the protein that influences access by the protease.
Plaque phenotypes of mutant viruses.
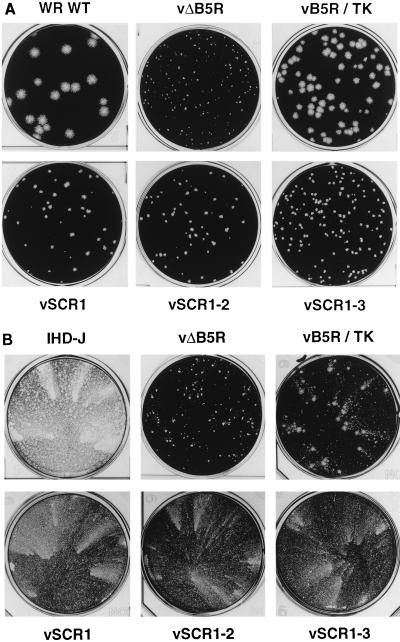
The plaque phenotypes produced by the different viruses on BS-C-1 cells are shown in Fig. 5A. As noted previously (11), the deletion mutant vΔB5R produced a very small plaque that was clearly visible only after 4 to 5 days of incubation. vB5R/TK, containing the ectopic WT B5R gene, produced a plaque that was marginally smaller than WT plaques. In comparison, the plaques formed by viruses with mutated B5R SCRs were considerably smaller, albeit larger than plaques formed by vΔB5R. Notably, vSCR1 plaques were appreciably larger than those of vSCR1-2 and vSCR1-3. Although small, this difference was reproducibly observed. Collectively, these data show that the SCRs influence plaque size in a complex manner: loss of only SCR 4 causes a drastic reduction in plaque size compared to WT, yet as long as the spacer, transmembrane, and cytoplasmic domains are retained, the plaque size increases as more SCRs are deleted.
FIG. 5.
Plaque morphologies of virus mutants. (A) Monolayers of BS-C-1 cells were infected with the indicated viruses, incubated in 1.5% carboxymethyl cellulose in MEM with 2.5% FBS for 96 h, and then stained with crystal violet. (B) Monolayers of BS-C-1 cells were infected with the indicated viruses, incubated in MEM with 2.5% FBS for 96 h, and then stained with crystal violet.
VV strains which produce larger amounts of EEV, such as IHD-J, produce comet-shaped plaques if incubated under liquid overlay (1). These represent the unidirectional spread of virus from the primary plaque (comet head), and the mechanism by which they are formed is not understood. Figure 5B shows that VV strain IHD-J produces large comets, whereas vB5R/TK produces only small comets by 96 hpi (similar in morphology to WT WR [data not shown]). Virus vΔB5R also fails to produce comets, as previously reported (11), consistent with the low levels of EEV produced. Surprisingly, vSCR1, vSCR1-2, and vSCR1-3 produced clear comets, suggesting enhanced EEV release, despite their small plaque phenotype (Fig. 5A). This indicated that SCRs 2, 3, and 4 are not required to produce extracellular virus and that when they are present they prevent virus release.
Deletion of SCRs greatly increases EEV.
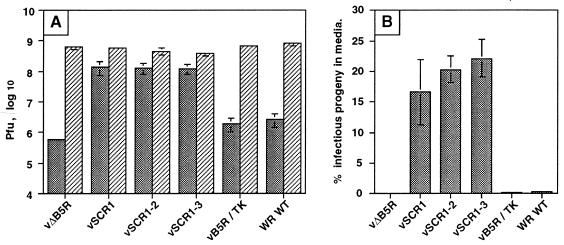
To measure EEV release more accurately, the infectivity present in the cell supernatant was determined for each virus and compared with that in infected cells. All viruses produced similar levels of infectivity in cells. As expected for WT WR and vB5R/TK, there was very little infectious virus released into the supernatant compared to that retained in infected cells, and the level of extracellular virus was reduced further with vΔB5R (Fig. 6A). However, for viruses lacking one or more SCR, there was an approximately 50-fold increase in infectious virus released. This is demonstrated more clearly by plotting the infectivity released into the cell culture supernatant as a percentage (mean ± standard error of the mean [SEM]) of total infectivity (Fig. 6B). Extracellular virus represents <1% of total infectivity for WT WR and vB5R/TK and approximately 0.1% for vΔB5R, but between 17 and 23% of infectivity was extracellular for the SCR mutants. This level approaches that found with VV strain IHD-J. This result was consistent with the increased amounts of virion-associated B5R protein in the supernatant of cells infected with these viruses (Fig. 4) and the comet-shaped plaques that they formed (Fig. 5B).
FIG. 6.
B5R mutant viruses release enhanced levels of virus into the culture supernatant. RK13 cells were infected with the indicated viruses at 1 PFU/cell. At 24 hpi, the culture supernatant was collected and clarified by low-speed centrifugation (2,000 rpm in a Beckman GPR benchtop centrifuge for 10 min), and then the infectious virus present in this fraction was determined by duplicate plaque assay on monolayers of BS-C-1 cells. The infected cells were scraped from the monolayer into PBS, combined with the pellets derived from clarification of the culture supernatant, pelleted as described above, and lysed by freeze-thawing and sonication. Infectious virus present in the sonicate was determined by plaque assay on BS-C-1 cells. Plaque assays were stained between 48 and 120 hpi. (A) Data are presented as the titer of virus present in cells (hatched bars) or the culture supernatant (stippled bars) ± SEM (n = 3); (B) data are expressed as the percentage of total infectious virus that was released into the culture supernatant ± SEM (n = 3).
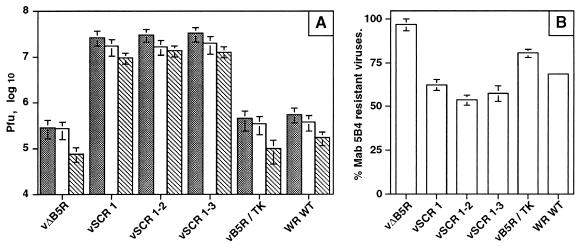
To demonstrate that the enhanced infectivity in the culture supernatant reflected increased release of enveloped virus, rather than IMV released by cell lysis, we determined the proportion of infectivity in the supernatant that was neutralized by MAb 5B4/2F2, directed against the IMV 14-kDa surface protein (7). Figure 7A shows the total infectivity in the supernatant, the infectivity resistant to MAb 5B4, and the infectivity neutralized by the antibody; Fig. 7B shows the percentage of infectivity in the supernatant that is resistant to neutralization by 5B4/2F2 and represents EEV. These data show that the proportions of infectivity represented by IMV are 35 to 45% for the SCR mutants and 30 or 20% for WT WR or vB5R/TK, respectively. Although only a small amount of extracellular virus was produced from vΔB5R-infected cells, it was notable that 97% of this amount was resistant to neutralization by MAb 5B4/2F2. The concentration of MAb 5B4/2F2 used in these experiments was able to neutralize 92% of purified IMV (data not shown).
FIG. 7.
The virus released into the culture supernatant is resistant to neutralization by MAb 5B4/2F2. The experiment described in the legend to Fig. 6 was repeated, and this time the titer of infectious virus present in the culture supernatant was determined in the presence or absence of MAb 5B4/2F2 as described previously (45). (A) Stippled bars, total virus; open bars, virus resistant to neutralization by MAb 5B4/2F2; hatched bars, virus neutralized by MAb 5B4/2F2. Titers are shown ± SEM (n = 2). (B) Data expressed as the percentage of infectious virus present in the culture supernatant that is resistant to neutralization by MAb 5B4/2F2 ± SEM (n = 2).
Buoyant density and specific infectivity of released EEV.
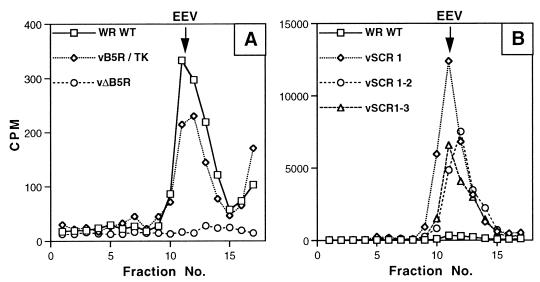
To determine the buoyant density of the EEV released from cells infected by the mutant viruses, virions were metabolically labeled with [3H]thymidine and purified from the culture medium by CsCl density gradient centrifugation. Figure 8B demonstrates that these mutants released EEV of normal density (1.23 g/ml) and confirmed the greatly enhanced levels of EEV compared to WT WR or vB5R/TK (Fig. 8A). The slightly greater amount of EEV released by vSCR1 was not found in a repeat experiment. The presence of very little IMV in the CsCl gradients (at 1.27 g/ml) is not inconsistent with the demonstration that approximately one-third of extracellular virus is neutralized by IMV-specific MAb (Fig. 7), because EEV with a damaged outer envelope can be neutralized by IMV antibody while still retaining EEV density in CsCl gradients (15, 45). The ratio of the infectivity present in clarified tissue culture supernatant and the counts per minute present in EEV fractions of CsCl density gradients showed that the specific infectivities of EEV from vSCR1, vSCR1-2, and vSCR1-3 were 1.4-, 1.7-, and 2.0-fold higher than that of EEV of vB5R/TK. This finding showed that SCRs 2, 3, and 4 are not required for EEV infectivity and that a slight increase in specific infectivity was observed in their absence.
FIG. 8.
CsCl density gradient contrifugation of EEV. RK13 cells were infected with the indicated virus and labeled with [3H]thymidine, and EEV was purified by CsCl density gradients as described in Materials and Methods. (A) EEV from cells infected with WT WR, vB5R/TK, and vΔB5R; (B) EEV from WT WR, vSCR1, vSCR1-2 and vSCR1-3. Note the different scales. The radioactivity from the peak fractions representing EEV was compared to the virus infectivity in the fresh culture supernatant to give a specific infectivity value for each virus. This value was compared to that of vB5R/TK or WT WR viruses, and ratio of the specific infectivity (mutant/WT) is given in the text.
Wrapping of IMV by intracellular membranes.
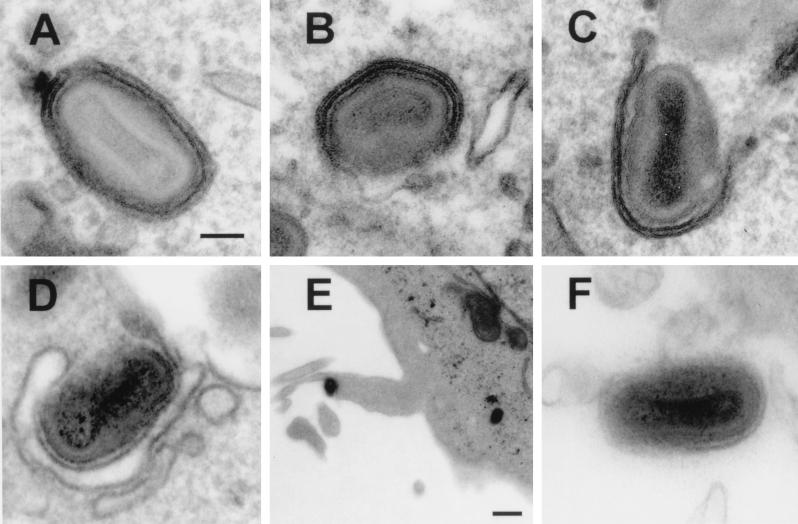
Increased titers of infectious virus in the supernatant might be due to enhanced wrapping of IMV by intracellular membranes, more efficient transport of IEV to the cell periphery, or increased release of EEV from the cell. To address these possibilities, we examined first whether IMV particles were wrapped by intracellular membranes to form IEV. Figure 9 shows that cells infected with WR WT and each virus lacking one or more SCR contained virus particles that were wrapped by intracellular membranes. This was also seen with virus vB5R/TK (data not shown), and cells infected with this virus contained virus particles projecting from the cell on macrovilli that presumably are due to actin tails (Fig. 9E). The proportion of IMV particles that were wrapped, or in the process of being wrapped, out of several hundred intracellular particles was determined for each virus. For WT WR, 45 of 280 (16%) of particles were wrapped. For vSCR1, vSCR1-2, and vSCR1-3, 66 of 227 (29%), 63 of 335 (19%), and 85 of 323 (26%), respectively, were wrapped. The membranes of some of the IEVs shown in Fig. 9 are incomplete, but these mutants were not defective in completion of the wrapping process. This was established by examining 100 virions from each virus that were associated with a wrapping membrane and scoring the proportion of these which were partially or completely wrapped. For WR, 71% were fully wrapped, while for vSCR1, vSCR1-2, and vSCR1-3 67, 66, and 60%, respectively, were completely wrapped. For vΔB5R, there was a major defect in wrapping as previously reported (11, 47) and only 2 of 157 virions were adjacent to intracellular membranes; in these cases, it was uncertain if these virions were in direct contact. Despite this finding, some EEV was detected in the supernatant of cells infected with vΔB5R (Fig. 6 and 7), and this virus was enveloped (Fig. 9) and was resistant to neutralization by an IMV neutralizing antibody (Fig. 7). The outer envelope seemed more tightly wrapped around the IMV particle than for WT virus; possibly this was related to the greater resistance to neutralization.
FIG. 9.
Electron microscopy of cells infected with the indicated viruses. Note the formation of intracellular virus particles that have been, or are being, wrapped by intracellular membranes in cells infected with WR WT, vSCR1, vSCR1-2, and vSCR1-3 (A to D). (E) Enveloped particle leaving a B5R/TK-infected cell on an actin tail. (F) EEV particle produced from a vΔB5R-infected cell. Bars: (A to D and F) 100 nm; (E) 500 nm.
B5R SCRs are required for formation of intracellular actin tails.
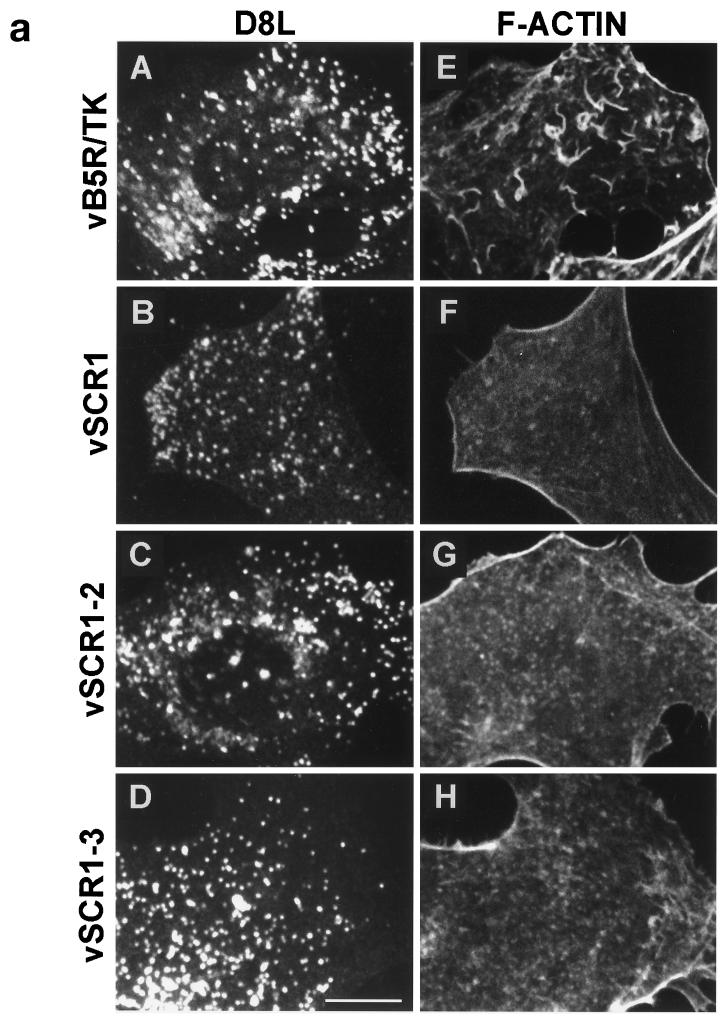
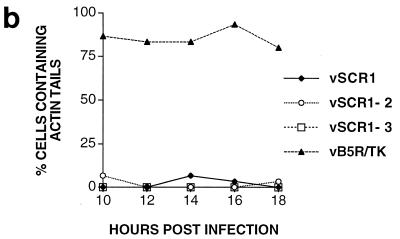
Some IEV particles induce the polymerization of actin on one side of the virion to produce actin tails that propel the virus to the cell surface (5). Failure to envelop IMV with membranes prevents this process (5). As the B5R SCR mutants produce IEV, an increased amount of EEV, but small plaques, we investigated if these mutants could induce the formation of intracellular actin tails. Figure 10a shows that the SCR mutant viruses and vB5R/TK make many virus particles that move from virus factories to the cell periphery (D8L). However, whereas vB5R/TK formed many intracellular actin tails (F-actin), none of the B5R SCR mutants did so. To eliminate the possibility that the SCR deletion viruses form actin tails at other times during infection, cells were examined from 10 to 18 hpi (Fig. 10b). After staining infected cells with MAb AB1.1 and phalloidin, the number of infected cells containing virions with actin tails was determined. At these times, between 76 and 82% of cells infected with vB5R/TK had virions with actin tails, but very few cells infected with vSCR mutants showed any actin tails. This defect was not due to a failure to make IEV (Fig. 9), indicating that these IEVs can move out to the cell surface in a manner independent of actin tail formation. This result was reminiscent of the phenotype of A34R deletion mutants which produce enhanced amount of EEV (22) and fail to form intracellular actin tails (46).
FIG. 10.
(a) Fluorescence microscopy of infected cells. BS-C-1 cells were infected with the indicated viruses at 10 PFU/cell. At 14 hpi, cells were permeabilized and stained with mouse MAb AB1.1 directed against the D8L gene product of IMV (25) and rhodamine isothiocyanate-phalloidin to detect F-actin. Note the actin tails in IEV particles in panel E. Bar = 10 μm. (b) Quantitation of actin tails made by the cells infected with the indicated viruses. Cells were infected and stained as for panel A at the indicated times. Between 30 and 40 cells containing approximately 300 D8L-positive particles were counted for each virus at each time point.
DISCUSSION
This report presents a mutational analysis of the extracellular domain in the B5R protein that forms part of the outer envelope of EEV. The SCRs are demonstrated to affect virus plaque size and the formation of intracellular actin tails. In addition, greatly enhanced levels of infectious EEV are released when these SCRs are deleted.
We constructed recombinant VVs that expressed the WT or mutant forms of the B5R protein from the TK locus of a VV mutant from which the majority of the endogenous B5R gene had been deleted. These viruses expressed B5R proteins of the predicted sizes that were present in infected cells and became associated with EEV particles released into the culture supernatant. Use of tunicamycin, an inhibitor of N-linked glycosylation, demonstrated that all B5R proteins containing SCR 2 were glycosylated, whereas expression of only SCR 1 produced a protein lacking N-linked carbohydrate. However, neither glycosylation nor SCRs 2, 3, and 4 are required for incorporation of B5R into EEV particles, indicating that other regions of the protein are needed for targeting to the membranes used to wrap IMV. This result is consistent with the recent demonstration that a chimeric molecule containing the B5R transmembrane and cytoplasmic domain fused to HIV gp120 can be incorporated into EEV (19). B5R proteins lacking SCRs 2, 3, and 4, SCRs 3 and 4, or only SCR 4 had an increased propensity to form higher-molecular-weight aggregates, rather than being monomeric, in EEV particles. This finding indicated that the complex formation, which involves the formation of disulfide bonds (10, 18), does not require these domains and therefore must occur through SCR 1 or either or both of the two cysteine residues near the junction between the cytoplasmic tail and the transmembrane domain. Little of the mutant B5R proteins was cleaved to produce a soluble B5R protein found with the WT protein (21). Possibly the correct spatial organization of SCRs 3 and 4 or presentation of a particular motif is required to permit cleavage.
Analysis of the plaques formed by the mutant viruses showed that loss of SCR 4 was sufficient to produce a very small plaque phenotype, although this was marginally larger than that of the B5R deletion mutant. Deletion of SCRs 3 and 4 made little difference compared to deletion of SCR 4 alone, but deletion of SCRs 2, 3, and 4 caused a further slight increase in plaque size. This was unexpected and is presently without explanation. Although plaques formed by the mutant viruses were all small compared to WT plaques, they all produced comets, indicating enhanced EEV release. This was confirmed with infectivity measurements of virus released from infected cells. EEV levels were approximately 50-fold higher than that of WT virus and nearly 300- to 400-fold greater than that of vΔB5R. Therefore, SCR 4 of the B5R protein specifically, and possibly SCRs 2 and 3 also, is required for retaining EEV on the cell surface. Deletion of the A34R EEV glycoprotein caused a similar result; despite having a very small plaque phenotype, 25-fold-higher levels of EEV were released by the A34R deletion mutant (22) and increased levels of EEV were noted with a mutant A34R protein containing a K151E substitution (4). The infectivity of the A34R− EEV was diminished five- to sixfold, but there was no such infectivity defect with EEV of the B5R SCR mutants; with these B5R mutants, the ratio of infectivity in the fresh supernatant and counts per minute present in EEV purified in CsCl density gradients showed a slight increase in specific infectivity.
The B5R protein is required for the envelopment of IMV by cytoplasmic membranes (11, 47) to form IEV. IEV is needed for the formation of virus-associated actin tails (5) which propel IEV through the cell prior to EEV release. Yet viruses containing mutant B5R proteins with only one, two, or three SCRs released 50-fold more infectious virus than WT, and this virus was EEV not IMV, as shown by its buoyant density and resistance to neutralization by an IMV-specific MAb. Despite this enhanced EEV release, the B5R SCR mutants failed to make actin tails. This result is again reminiscent of the situation with an A34R deletion mutant that does not make actin tails (46) but releases enhanced levels of EEV (22). Taken together, these data demonstrate that actin tail formation is not required for the formation of EEV. Electron microscopy showed that IEV particles are formed by the A34R− virus (46) and by the B5R mutants described here, and so IEV particles can move to the surface in an actin tail-independent manner. It is also possible that some enveloped virus are formed by direct budding of IMV through the plasma membrane as reported late in infection with VV strain IHD-W (43). This may also explain the formation by vΔB5R of EEV that are resistant to neutralization by MAb 5B4/2F2 despite the failure to wrap IMV by intracellular membranes.
The regions of the B5R protein deleted in the mutants described here are located within the lumen of intracellular organelles and are not in a position to interact directly with the cytosolic surface where actin tail polymerization takes place. Despite this, actin tail formation is inhibited. Similarly, deletion of the A34R protein (46) or the A36R protein (33) prevents actin tail formation, although these proteins present only very short peptides on the cytosolic face. These data might be explained by a model that requires lumenal interaction of the A34R, A36R, and B5R proteins (possibly with other proteins too) that creates a complex for actin nucleation on the cytosolic face of the membrane.
Enveloped virus is required for the spread of virus from cell to cell (3), but plaque size does not simply depend on whether EEV is retained as CEV, since the WR and IHD-J strains, which have either high EEV and low CEV (IHD-J) or low EEV and high CEV (WR), have similar-sized plaques. Why then are the plaques formed by the B5R SCR domain mutants or vΔA34R so small given that these viruses make so much EEV? For the vΔA34R deletion mutant, this might be influenced by the diminished infectivity of EEV, but this is not true for the B5R mutants which produce EEV comparable in infectivity to EEV produced by WT virus. A possible explanation is that actin tail formation is needed for direct spread to neighboring cells and without this plaque size is diminished. This idea fits the data for the B5R mutants and vΔA34R but fits less well with data for the vΔA36R mutant that also fails to make actin tails (33) but has a plaque significantly larger than those of vΔA34R and vΔB5R (although still smaller than WT plaques) (25). Perhaps the actin tails enable the enveloped virus (CEV or EEV) to exit from cells in a manner suitable for their efficient spread to adjacent cells, although other factors are likely to influence plaque size also.
In summary, we describe viruses with mutations in the extracellular domain of the B5R protein which prevent actin tail formation yet induce the formation of comet shaped plaques and enhanced levels of infectious EEV. These data contribute to our understanding of the role of B5R protein in plaque formation and in EEV formation and infectivity.
ACKNOWLEDGMENTS
This work was supported by the U.K. Medical Research Council and The Wellcome Trust. E.M. was the recipient of an MRC Research Studentship.
We thank C. Czerny for MAb 5B4/2F2.
REFERENCES
- 1.Appleyard G, Hapel A J, Boulter E A. An antigenic difference between intracellular and extracellular rabbitpox virus. J Gen Virol. 1971;13:9–17. doi: 10.1099/0022-1317-13-1-9. [DOI] [PubMed] [Google Scholar]
- 2.Blasco R, Moss B. Extracellular vaccinia virus formation and cell-to-cell virus transmission are prevented by deletion of the gene encoding the 37,000-dalton outer envelope protein. J Virol. 1991;65:5910–5920. doi: 10.1128/jvi.65.11.5910-5920.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blasco R, Moss B. Role of cell-associated enveloped vaccinia virus in cell-to-cell spread. J Virol. 1992;66:4170–4179. doi: 10.1128/jvi.66.7.4170-4179.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blasco R, Sisler J R, Moss B. Dissociation of progeny vaccinia virus from the cell membrane is regulated by a viral envelope glycoprotein: effect of a point mutation in the lectin homology domain of the A34R gene. J Virol. 1993;67:3319–3325. doi: 10.1128/jvi.67.6.3319-3325.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cudmore S, Cossart P, Griffiths G, Way M. Actin-based motility of vaccinia virus. Nature. 1995;378:636–638. doi: 10.1038/378636a0. [DOI] [PubMed] [Google Scholar]
- 6.Cudmore S, Reckmann I, Griffiths G, Way M. Vaccinia virus: a model system for actin-membrane interactions. J Cell Sci. 1996;109:1739–1747. doi: 10.1242/jcs.109.7.1739. [DOI] [PubMed] [Google Scholar]
- 7.Czerny C P, Mahnel H. Structural and functional analysis of orthopoxvirus epitopes with neutralizing monoclonal antibodies. J Gen Virol. 1990;71:2341–2352. doi: 10.1099/0022-1317-71-10-2341. [DOI] [PubMed] [Google Scholar]
- 8.Dales S, Pogo B G T. Biology of poxviruses. Virol Monogr. 1981;18:1–101. doi: 10.1007/978-3-7091-8625-1. [DOI] [PubMed] [Google Scholar]
- 9.Duncan S A, Smith G L. Identification and characterization of an extracellular envelope glycoprotein affecting vaccinia virus egress. J Virol. 1992;66:1610–1621. doi: 10.1128/jvi.66.3.1610-1621.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Engelstad M, Howard S T, Smith G L. A constitutively expressed vaccinia virus gene encodes a 42 kDa glycoprotein related to complement control factors that forms part of the extracellular envelope. Virology. 1992;188:801–810. doi: 10.1016/0042-6822(92)90535-w. [DOI] [PubMed] [Google Scholar]
- 11.Engelstad M, Smith G L. The vaccinia virus 42 kDa envelope protein is required for envelopment and egress of extracellular virus and for virulence. Virology. 1993;194:627–637. doi: 10.1006/viro.1993.1302. [DOI] [PubMed] [Google Scholar]
- 12.Hiller G, Weber K. Golgi-derived membranes that contain an acylated viral polypeptide are used for vaccinia virus envelopment. J Virol. 1985;55:651–659. doi: 10.1128/jvi.55.3.651-659.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hirt P, Hiller G, Wittek R. Localization and fine structure of a vaccinia virus gene encoding an envelope antigen. J Virol. 1986;58:757–764. doi: 10.1128/jvi.58.3.757-764.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Horton R M, Hunt H D, Ho S N, Pullen J K, Pease L R. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene. 1989;77:61–68. doi: 10.1016/0378-1119(89)90359-4. [DOI] [PubMed] [Google Scholar]
- 15.Ichihashi Y. Extracellular enveloped vaccinia virus escapes neutralization. Virology. 1996;217:478–485. doi: 10.1006/viro.1996.0142. [DOI] [PubMed] [Google Scholar]
- 16.Ichihashi Y, Dales S. Biogenesis of poxviruses: interrelationship between hemagglutinin production and polykaryocytosis. Virology. 1971;46:533–543. doi: 10.1016/0042-6822(71)90057-2. [DOI] [PubMed] [Google Scholar]
- 17.Ichihashi Y, Matsumoto S, Dales S. Biogenesis of poxviruses: role of A-type inclusions and host cell membranes in virus dissemination. Virology. 1971;46:507–532. doi: 10.1016/0042-6822(71)90056-0. [DOI] [PubMed] [Google Scholar]
- 18.Isaacs S N, Wolffe E J, Payne L G, Moss B. Characterization of a vaccinia virus-encoded 42-kilodalton class I membrane glycoprotein component of the extracellular virus envelope. J Virol. 1992;66:7217–7224. doi: 10.1128/jvi.66.12.7217-7224.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Katz E, Wolffe E J, Moss B. The cytoplasmic domains of the vaccinia virus B5R protein target a chimeric human immunodeficiency virus type 1 glycoprotein to the outer envelope of nascent vaccinia virions. J Virol. 1997;71:3178–3187. doi: 10.1128/jvi.71.4.3178-3187.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mackett M, Smith G L, Moss B. The construction and characterization of vaccinia virus recombinants expressing foreign genes. In: Glover D M, editor. DNA cloning: a practical approach. Vol. 2. Oxford, England: IRL Press; 1985. pp. 191–211. [Google Scholar]
- 21.Martinez-Pomares L, Stern R J, Moyer R W. The ps/hr gene (B5R open reading frame homolog) of rabbitpox virus controls pock color, is a component of extracellular enveloped virus, and is secreted into the medium. J Virol. 1993;67:5450–5462. doi: 10.1128/jvi.67.9.5450-5462.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McIntosh A A G, Smith G L. Vaccinia virus glycoprotein A34R is required for infectivity of extracellular enveloped virus. J Virol. 1996;70:272–281. doi: 10.1128/jvi.70.1.272-281.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morgan C. Vaccinia virus reexamined: development and release. Virology. 1976;73:43–58. doi: 10.1016/0042-6822(76)90059-3. [DOI] [PubMed] [Google Scholar]
- 24.Moss B. Poxviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. Vol. 2. New York, N.Y: Lippincott Raven Press; 1996. pp. 2637–2671. [Google Scholar]
- 25.Parkinson J E, Smith G L. Vaccinia virus gene A36R encodes a Mr 43-50 K protein on the surface of extracellular enveloped virus. Virology. 1994;204:376–390. doi: 10.1006/viro.1994.1542. [DOI] [PubMed] [Google Scholar]
- 26.Payne L G. Significance of extracellular enveloped virus in the in vitro and in vivo dissemination of vaccinia virus. J Gen Virol. 1980;50:89–100. doi: 10.1099/0022-1317-50-1-89. [DOI] [PubMed] [Google Scholar]
- 27.Payne L G, Kristensson K. Extracellular release of enveloped vaccinia virus from mouse nasal epithelial cells in vivo. J Gen Virol. 1985;66:643–646. doi: 10.1099/0022-1317-66-3-643. [DOI] [PubMed] [Google Scholar]
- 28.Payne L G, Kristensson K. Mechanism of vaccinia virus release and its specific inhibition by N1-isonicotinoyl-N2-3-methyl-4-chlorobenzoylhydrazine. J Virol. 1979;32:614–622. doi: 10.1128/jvi.32.2.614-622.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Payne L G, Norrby E. Presence of haemagglutinin in the envelope of extracellular vaccinia virus particles. J Gen Virol. 1976;32:63–72. doi: 10.1099/0022-1317-32-1-63. [DOI] [PubMed] [Google Scholar]
- 30.Rodriguez J F, Smith G L. IPTG-dependent vaccinia virus: identification of a virus protein enabling virion envelopment by Golgi membrane and egress. Nucleic Acids Res. 1990;18:5347–5351. doi: 10.1093/nar/18.18.5347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roper R L, Payne L G, Moss B. Extracellular vaccinia virus envelope glycoprotein encoded by the A33R gene. J Virol. 1996;70:3753–3762. doi: 10.1128/jvi.70.6.3753-3762.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sanderson C M, Parkinson J E, Hollinshead M, Smith G L. Overexpression of the vaccinia virus A38L integral membrane protein promotes Ca2+ influx into infected cells. J Virol. 1996;70:905–914. doi: 10.1128/jvi.70.2.905-914.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sanderson, C. M., M. Way, F. Frischknecht, and G. L. Smith. Unpublished data.
- 34.Schmelz M, Sodeik B, Ericsson M, Wolffe E J, Shida H, Hiller G, Griffiths G. Assembly of vaccinia virus: the second wrapping cisterna is derived from the trans Golgi network. J Virol. 1994;68:130–147. doi: 10.1128/jvi.68.1.130-147.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shida H. Nucleotide sequence of the vaccinia virus hemagglutinin gene. Virology. 1986;150:451–462. doi: 10.1016/0042-6822(86)90309-0. [DOI] [PubMed] [Google Scholar]
- 36.Shida H, Dales S. Biogenesis of vaccinia: molecular basis for the hemagglutinin-negative phenotype of the IHD-W strain. Virology. 1982;117:219–237. doi: 10.1016/0042-6822(82)90521-9. [DOI] [PubMed] [Google Scholar]
- 37.Smith G L. Gene expression from vaccinia virus vectors. In: Davison A J, Elliot R, editors. Molecular virology: a practical approach. Oxford, England: Oxford University Press; 1993. pp. 257–283. [Google Scholar]
- 38.Smith, G. L., and A. Vanderplascchen. Extracellular enveloped vaccinia virus: entry, egress and evasion. In L. Enjuanes, W. Spaan, and S. Siddell (ed.), Advances in experimental biology, in press. Plenum Press, New York, N.Y. [PubMed]
- 39.Sodeik B, Doms R W, Ericsson M, Hiller G, Machamer C E, van’t Hof W, van Meer G, Moss B, Griffiths G. Assembly of vaccinia virus: role of the intermediate compartment between the endoplasmic reticulum and the Golgi stacks. J Cell Biol. 1993;121:521–541. doi: 10.1083/jcb.121.3.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Takahashi-Nishimaki F, Funahashi S, Miki K, Hashizume S, Sugimoto M. Regulation of plaque size and host range by a vaccinia virus gene related to complement system proteins. Virology. 1991;181:158–164. doi: 10.1016/0042-6822(91)90480-y. [DOI] [PubMed] [Google Scholar]
- 41.Tooze J, Hollinshead M, Reis B, Radsak K, Kern H. Progeny vaccinia viruses and human cytomegalovirus particles utilize early endosomal cisternae for their envelopes. Eur J Cell Biol. 1993;60:163–178. [PubMed] [Google Scholar]
- 42.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tsutsui K. Release of vaccinia virus from FL cells infected with IHD-W strain. J Electron Microsc. 1983;32:125–140. [PubMed] [Google Scholar]
- 44.Vanderplasschen A, Hollinshead M, Smith G L. Antibodies against vaccinia virus do not neutralise extracellular enveloped virus but prevent virus release from infected cells and comet formation. J Gen Virol. 1997;78:2041–2048. doi: 10.1099/0022-1317-78-8-2041. [DOI] [PubMed] [Google Scholar]
- 45.Vanderplasschen A, Smith G L. A novel virus binding assay using confocal microscopy: demonstration that the intracellular and extracellular vaccinia virions bind to different cellular receptors. J Virol. 1997;71:4032–4041. doi: 10.1128/jvi.71.5.4032-4041.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wolffe E, Katz E, Weisberg A, Moss B. The A34R glycoprotein gene is required for induction of specialized actin-containing microvilli and efficient cell-to-cell transmission of vaccinia virus. J Virol. 1997;71:3905–3915. doi: 10.1128/jvi.71.5.3904-3915.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wolffe E J, Isaacs S N, Moss B. Deletion of the vaccinia virus B5R gene encoding a 42-kilodalton membrane glycoprotein inhibits extracellular virus envelope formation and dissemination. J Virol. 1993;67:4732–4741. doi: 10.1128/jvi.67.8.4732-4741.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]