Abstract
Knowledge of immune mechanisms responsible for the cross-protection between highly divergent viruses such as human immunodeficiency virus type 1 (HIV-1) and HIV-2 may contribute to an understanding of whether virus variability may be overcome in the design of vaccine candidates which are broadly protective across the HIV subtypes. We demonstrate that despite the significant difference in virus amino acid sequence, the majority of HIV-2-infected individuals with different HLA molecules possess a dominant cytotoxic T-cell response which is able to recognize HIV-1 Gag protein. Furthermore, HLA-B5801-positive subjects show broad cross-recognition of HIV-1 subtypes since they mounted a T-cell response that tolerated extensive amino acid substitutions within HLA-B5801-restricted HIV-1 and HIV-2 epitopes. These results suggests that HLA-B5801-positive HIV-2-infected individuals have an enhanced ability to react with HIV-1 that could play a role in cross-protection.
Human immunodeficiency virus type 1 (HIV-1) and HIV-2 are related human retroviruses that show various biological and structural differences. HIV-2 is found mainly in West Africa, whereas HIV-1 is spreading throughout the world. HIV-2 is less transmissible, and HIV-2-positive patients exhibit longer clinical latency periods than individuals infected with HIV-1 (23). A recent report has also shown that the mortality in HIV-2-infected individuals is only twice as high as in the uninfected population and, in the majority of adults, survival is not affected by HIV-2 status (31).
Although the two viruses are similar in genomic organization, various genetic and enzymatic differences have been found at many stages of the retroviral life cycle. They differ significantly in terms of amino acid sequence, the more conserved being the Pol and Gag sequences, which exhibit less than 60% homology (17).
Despite these differences, epidemiological data and animal studies have shown some evidence of cross-protection between the two viral infections. Travers et al. reported that HIV-2-infected women had a lower incidence of HIV-1 infection than did HIV-seronegative women in a cohort of commercial sexual workers in Dakar (37), and rhesus macaques immunized with a recombinant HIV-1 poxvirus vaccine are protected against HIV-2 challenge (2). These studies, though not conclusive (1, 6), suggest that differences in the virus may not necessarily preclude the development of defensive immunity to a subsequent pathogenic infection, an old-fashioned concept pioneered by Jenner, who used cowpox to vaccinate against human smallpox.
The immunological basis of cross-protection is largely unknown, and a clear understanding of the role played by the humoral or cell-mediated immune response in HIV protection is still lacking. However, mounting evidence suggests that cytotoxic T-lymphocyte (CTL) response could be the key element. Indeed, the protection afforded in animal models against simian (13) and feline (12) immunodeficiency virus infections is closely correlated with the induction of specific CTL response, and HIV-1 and HIV-2 HLA-B35-restricted cross-reactive CTLs have been postulated to confer protection against repeated HIV exposure (33).
CTLs recognize short viral peptides, 8 to 11 amino acids long, that are generated by the intracellular processing of endogenously synthesized viral antigens within the infected cells, which are expressed at the cell surface in the binding groove of HLA class I molecules. The specificity of the T-cell response is determined by the interaction of the antigen-specific T-cell receptor (TCR) with the peptide-HLA complex, and this interaction, together with non-antigen-specific signals, activates the CTLs (15).
The presence of cross-reactive CTLs able to lyse HIV-1- or HIV-2-infected cells should be dependent on the extent of conservation between the two viruses within the epitopes selected by particular HLA class I molecules. It is well known that amino acid substitutions within the epitopes can abrogate the CTL response by inhibiting either HLA binding or TCR recognition (32). However, a number of recent studies have shown that T cells can recognize apparently unrelated peptides (10, 41), and crystallographic data have shown physical limits to the TCR epitope specificity due to the limited size of contact between the TCR and the peptide (14), suggesting a flexibility in T-cell recognition of antigen (19).
Some individuals with a particular HLA profile which is responsible for presentation of the viral antigen and for selection of the T-cell repertoire may possess a CTL response not affected by mutations within the epitope, as has been demonstrated in subjects with HLA alleles B27 (28) and B35 (33). In these cases, amino acid substitutions within the HIV-1 and -2 epitopes were tolerated by the CTLs.
In this study, we have investigated the extent of cross-reacting CTLs between HIV-2 and HIV-1 in a group of HIV-2-infected subjects with different HLA class I types. We have shown that despite differences in amino acid sequence between the two viruses, the majority of HIV-2-positive subjects possess CTLs which are able to recognize HIV-1 Gag protein.
Furthermore, analysis of HLA profiles and the fine specificity of the cytotoxic response demonstrated that HLA-B5801-positive subjects show broad cross-recognition of HIV-1 isolates. These subjects mounted a CTL response that tolerated extensive amino acid substitutions within an HLA-B5801-restricted HIV-1 epitope.
MATERIALS AND METHODS
Study subjects.
Eighteen HIV-2-positive patients were recruited in The Gambia. Ethical approval was obtained from The Gambia Government/MRC Ethical Committee. All the patients were asymptomatic, with no history of antiretroviral drugs and CD4 percentages in the normal range (>30%).
Patients attending the MRC clinic were initially tested by a combined HIV-1 and HIV-2 enzyme immunoassay (Wellcozyme 1+2; Murex Diagnostics, Dartford, Kent, England). The diagnosis of HIV-2 infection was made on the basis of negative HIV-1 and positive HIV-2 competitive enzyme-linked immunosorbent assays (36) followed by type-specific peptide-based immunoenzymatic strips (PeptiLav; Diagnostic Pasteur, Marnes-la-Cougette, France). HIV-1 negativity was repeatedly reconfirmed during the period of the study. In addition, HIV-2 positivity was confirmed by PCR for HIV-2 proviral DNA (using specific long terminal repeat primer sets for HIV-2 as described in references 5 and 33).
HLA typing.
Tissue typing was performed at HLA class I and class II by amplification refractory mutation system-PCR using sequence-specific primers (8) following DNA extraction from 3 ml of whole blood by a salting-out method (Puregene DNA extraction kit; Gentra Systems Inc., Minneapolis, Minn.). The level of resolution of this method is equivalent or greater than that obtained with serological tissue typing and is especially suitable for non-Caucasian populations for whom serological typing may be difficult to perform in the field or inaccurate because the relevant antisera are not readily available. Certain specificities (HLA-A2, -A30, and -B58) were further subtyped by nested single specific primer-PCR.
rVV.
Recombinant vaccinia viruses (rVV) expressing the HIV-2ROD gag and pol genes were supplied by Transgene (Strasbourg, France). rVV expressing the HIV-2BEN nef gene was supplied by G. Voss, and a control vaccinia virus expressing an irrelevant protein (β-galactosidase) was supplied by B. Moss. rVV expressing the gag genes of HIV-1 subtypes A1, A2, and C were produced and characterized as described previously (25). Briefly, A2 and C p55 of Gag was amplified by PCR from plasmids kindly supplied by F. McCutchan (Henry M. Jackson Foundation, Bethesda, Md.), using the Pfu enzyme with the following primers: 5′-GTAACCATGGGTGCGAGAGCG-3′ and 5′-GGCCAGATCTCATGATTTGAGGGAA-3′. PCR products were then digested with the restriction enzymes BglII and NcoI and cloned into pSC11 precut with NcoI and BglII. The p55 DNA for virus A1 was originally amplified from a Ugandan patient and cloned into a modified pCDNA3 plasmid (supplied by Peter Balfe, University College Hospital, London, England). This fragment was then excised by digestion with BamHI and NcoI and subcloned into pSC11, precut with NcoI and BglII. Maxipreps of each clone were prepared by using a CsCl protocol and then sequenced by using the Sequenase version 2.0 protocol (U.S. Biochemical). TK143 cells were then infected with wild-type vaccinia virus at a multiplicity of infection of 0.5 and transfected with 15 μg of DNA resuspended in 100 μl of double-distilled H2O and 50 μl of Lipofectin reagent (Gibco, Life Technologies, Paisley, Scotland) as instructed by the manufacturer. Cells were harvested 2 days later, and rVV were isolated by three rounds of plaque purification. Stocks were then grown and titers were determined as previously described (26).
The vaccinia virus expressing the subtype B Gag has been described elsewhere (27).
Overlapping peptides.
Peptides were purchased from Chiron Mimotopes (Clayton, Victoria, Australia). The HIV-2ROD Gag peptides were 18-mers overlapping by 12 residues. The purity of peptides was determined by reverse-phase high-pressure liquid chromatography (80 to 90% purity). Peptides were dissolved in dimethyl sulfoxide and diluted in RPMI 1640.
Production of HIV-2-specific CTL lines.
Bulk-cultured CTLs were generated as previously described (27). In brief, one-fifth of peripheral blood mononuclear cells (PBMC), separated from whole blood or thawed from previously frozen samples stored in liquid nitrogen, was added to the other four-fifths after 24 h of stimulation with phytohemagglutinin (2 μg/ml; Sigma Chemical Co., St. Louis, Mo.) to activate expression of autologous HIV. The cells were cultured in RPMI 1640 (Sigma) supplemented with antibiotics and 10% fetal calf serum (Globepharm, Esher, Surrey, England) for 5 days, after which 10% Lymphocult T (Biotest) was added to the medium. CTL lines were maintained for 2 to 3 weeks by adding medium containing 10% Lymphocult T every 3 days. Assays were performed on days 10 to 21 of culture.
Target cell preparation.
Epstein-Barr virus-transformed B-lymphoblastoid cell lines established from each subject were infected with rVV at a multiplicity of infection of 3:1 for 1 h and cultured in RPMI 1640 plus 20% fetal calf serum for 16 h. The cells were then labeled with 51Cr (100 μCi of Na251CrO4; Amersham International, Amersham, England) for 1 h and washed twice. In peptide-based assays, target cells labeled with 51Cr were pulsed either with pools of five peptides or with individual synthetic peptides for 1 h.
Cr release assay.
In 96-well U-bottom plates, target cells were aliquoted at 5,000 per well. Effector cells were added to targets at different effector-to-target (E/T) ratios. The amount of 51Cr released in the culture supernatants was quantitated after 5 to 6 h of incubation, and percent cytotoxicity was determined by using the following formula: (E − M/D − M) × 100, where E is the experimental 51Cr release, M is the 51Cr released in the presence of culture medium (which ranged between 15 and 25% of total release), and D is the total 51Cr released in the presence of 5% Triton X-100 detergent. Results were regarded as positive if recognition of the HIV rVV or peptides was greater than 10% above that of peptide or vaccinia virus controls.
Test for expression of HIV-1 rVV.
After the cytotoxic assay, the approximate efficiency of infection of the targets was checked by resuspending the remaining targets in 500 ml of 300 mM 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) in double-distilled H2O before incubation at 37°C for 1 h. A blue color was indicative of vaccinia virus expression. This was not done for targets infected with HIV-2 gag, pol, and nef rVV since they do not carry the galactosidase gene. Any assays in which the targets did not show equivalent staining by X-Gal were discarded.
RESULTS
CTL in HIV-2-positive patients predominantly recognize HIV-2 Gag protein.
HIV-2 CTL responses against the structural proteins Gag and Pol and against the nonstructural regulatory protein Nef were investigated in 18 HIV-2-infected patients. PBMC were stimulated in vitro with endogenous virus by coculture with autologous phytohemagglutinin-activated T cells and then expanded with interleukin-2. Polyclonal T-cell lines were tested against autologous target cells infected with rVV expressing the HIV-2ROD Gag, Pol, and Nef proteins. Some HIV-2-positive subjects, from whom the lines did not grow in sufficient numbers to allow a complete screen, were tested only against Gag (patient 9 [P9]) or against Gag and Pol (P6, P10, P13, and P14).
As shown in Table 1, in almost all cases, HIV-2-specific cytolytic activity which predominantly recognized the antigen derived from the gag gene product was demonstrable. Indeed, polyclonal T-cell lines derived from 15 of 18 HIV-2-positive patients were able to recognize target cells expressing the HIV-2 Gag protein, whereas recognition of the pol gene product was detectable in 5 of 16 subjects and Nef-specific cytolytic activity was present in 4 of 12 subjects.
TABLE 1.
CTL response to HIV-2 proteins in HIV-2-infected subjectsa
| Patient | HLA class I typing | % Cytotoxicity (E:T ratio = 20:1)
|
|||
|---|---|---|---|---|---|
| Vw | Vgag | Vpol | Vnef | ||
| P1 | A0205/3402 B1801/5801 C0501/1701 | 5 | 31 | 10 | 6 |
| P2 | A0201/0301 B5801/5301 C0302/4 | 3 | 58 | 8 | 21 |
| P3 | A0205/3402 B1801/5801 C0501/1701 | 7 | 35 | 9 | 8 |
| P4 | A0202/68 B0801/63 C14/1601 | 10 | 36 | 25 | 35 |
| P5 | A29/30 B5801/78 C0701 | 4 | 37 | 35 | 8 |
| P6 | A0201/3 B5801/35 C4/0302 | 1 | 27 | 6 | ND |
| P7 | A36/66 B5801 C0701 | 4 | 15 | 6 | 12 |
| P8 | A30/68 B41/65 C0802/1701 | 1 | 33 | 15 | 11 |
| P9 | ND | 8 | 55 | ND | ND |
| P10 | A0202/23 B35/52 C4/1601 | 6 | 25 | 8 | ND |
| P11 | A0201/23 B67/56 C0302 | 12 | 43 | 28 | 15 |
| P12 | A2/33 B58 C? | 2 | 30 | 2 | 2 |
| P13 | A0205/2301 B72/52 C1601/0202 | 4 | 20 | 11 | ND |
| P14 | A3301/68 B65/5301 C4/0802 | 3 | 5 | 2 | ND |
| P15 | A0205/68 B5801/72 C0202/0701 | 5 | 21 | 5 | 5 |
| P16 | A2301/26 B0801/57 C0304/0701 | 10 | 26 | 18 | 25 |
| P17 | A3/68 B72 C0202/4 | 5 | 8 | 5 | 5 |
| P18 | A0201/30 B? C4/0701 | 10 | 9 | 11 | 11 |
Cytotoxicity by CTL lines of different HIV-2 patients against autologous target cells infected with rVV expressing HIV-2 Gag (Vgag), Pol (Vpol), Nef (Vnef) proteins, and irrelevant protein (Vw). Cytotoxic values above 10% of the values obtained against target cells infected with Vw are considered positive. ND, not determined.
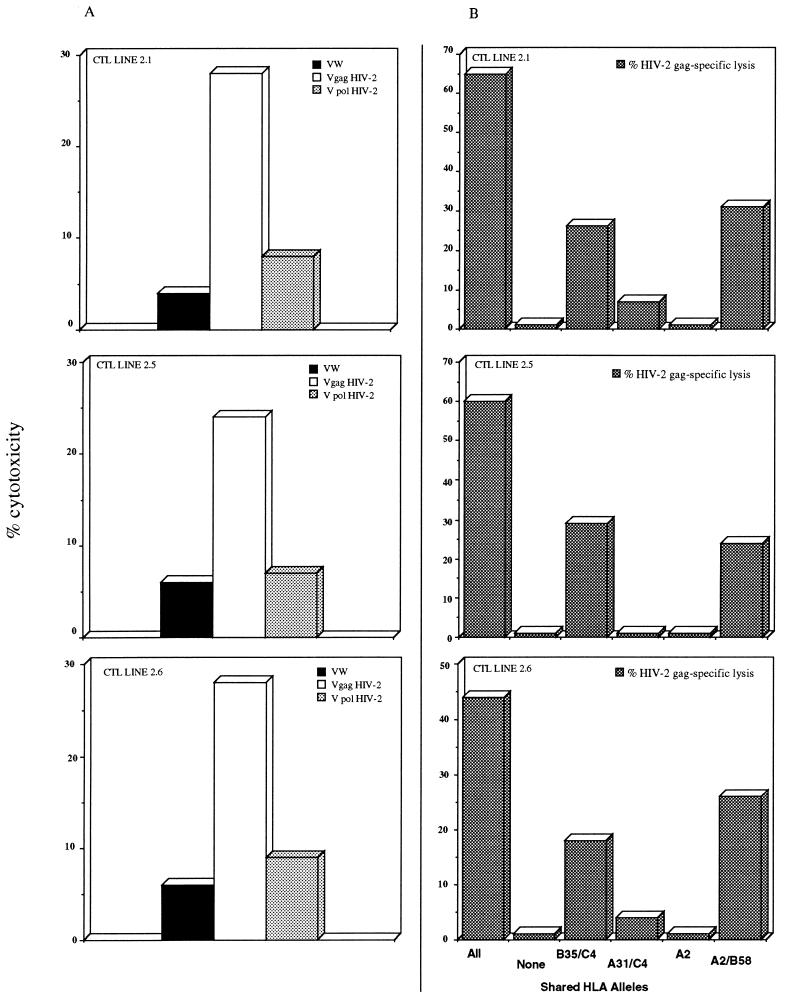
Different lines produced in the same patients (e.g., P6) gave comparable results, and the recognition of HIV-2 Gag protein by CTL was restricted by HLA class I molecules (Fig. 1).
FIG. 1.
Recognition of HIV-2 Gag protein by CTL lines of patient 6 (A) Effector cells were tested against autologous target cells at an E/T ratio of 20:1. Before 51Cr labeling, targets were infected with rVV (VW, wild-type vaccinia virus; Vgag HIV-2, rVV carrying the HIV-2 gag open reading frame; V pol HIV-2, rVV carrying the HIV-2 pol open reading frame. (B) HIV-2 Gag-specific CTL lines were tested against autologous or allogenic target cells infected with wild-type and HIV-2 Gag vaccinia virus. Specific lysis was calculated by subtracting lysis of target cells infected with wild-type vaccinia virus from lysis of those infected with HIV-2 Gag vaccinia virus. Shared HLA alleles are indicated. The complete HLA class I profile of patient 6 is A0201 A3 B5801 B35 C4 C0302. The E/T ratio was 40:1.
HIV-2 Gag-specific CTL lines recognize the Gag proteins of different HIV-1 clades.
Since Gag-specific CTLs were the predominant population in HIV-2 subjects, HIV-1 and HIV-2 cross-reactive CTL were analyzed by testing polyclonal Gag-specific CTL lines against target cells presenting Gag proteins from four different HIV-1 subtypes. rVV carrying the gag genes of four strains derived from HIV-1 subtypes isolated in Uganda (A1), Thailand (A2), Europe (B), and Uganda (C) were constructed (25).
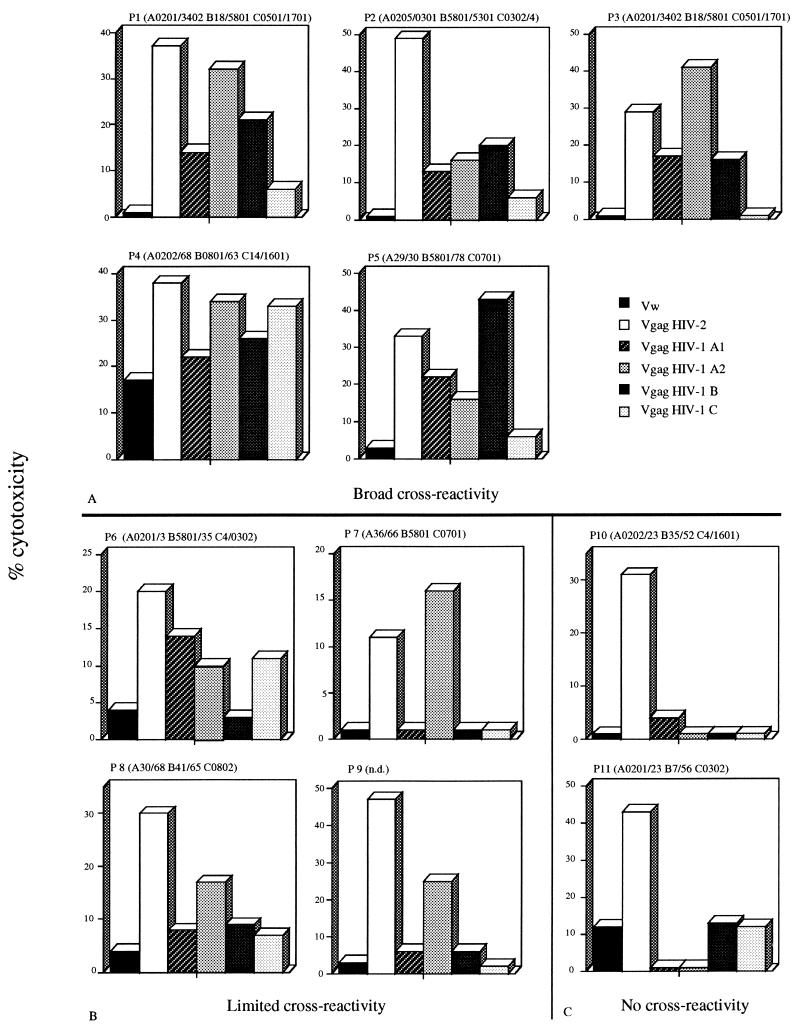
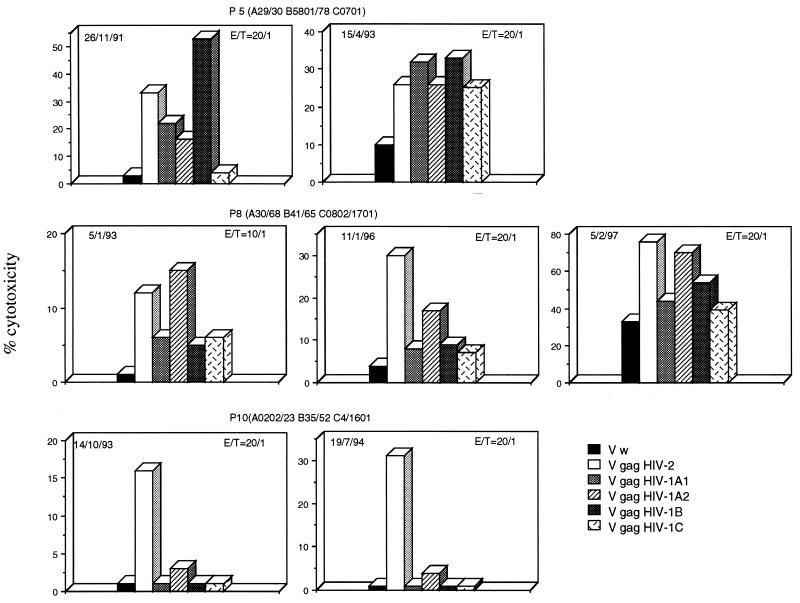
Nine of eleven of the HIV-2-infected subjects demonstrated HIV-2 Gag CTLs able to recognize antigen derived from the gag gene product of at least one HIV-1 clade (Fig. 2). Five subjects demonstrated a broad level of cross-reactivity, since CTL lines induced in these subjects were able to recognize at least three different HIV-1 clades (Fig. 2A). In four other patients (P6 to P9), we were able to demonstrate recognition of one (Fig. 2B) HIV-1 clade, whereas only two subjects (P10 and P11) had no cross-reactive CTL (Fig. 2C). CTL lines from three subjects (P3, P5, and P7) expressed a level of lysis of HIV-1 Gag comparable to that of the HIV-2 Gag protein. In the other subjects, lysis of target cells expressing HIV-1 Gag proteins was lower than lysis of the HIV-2 Gag-expressing targets. To confirm these results, we investigated whether HIV-2 Gag-specific CTL lines produced at different time points express a stable pattern of cross-reactivity. In two of three patients (P8 and P10), the results of CTL cross-reactivity at different time points showed complete consistency. In P5, there was cross-reactivity against three HIV-1 clades on one occasion and against all four on another (Fig. 3). The analysis of the HLA class I profile of the patient population studied shows that the HLA-B5801 molecule was present in four of five patients who demonstrate broad cross-reactivity.
FIG. 2.
Cross-recognition of HIV-2 and HIV-1 Gag protein by HIV-2 Gag-specific CTL lines. Effector cells were tested against autologous target cells infected with rVV expressing the Gag proteins of HIV-2 and HIV-1 clades A1, A2, B, and C (see the legend to Fig. 1). (A) Patients with broad cross-reactivity; (B and C) patients with limited (B) and no (C) cross-reactivity. HLA class I profiles of the different subjects are shown at the top of each panel. The E/T ratio varied according to the number of cells that grew from each culture but was usually between 20:1 and 50:1. Recognition of different HIV-1 clades was regarded as positive if lysis of different HIV-1 Gag rVV was more than 10% above the level of control vaccinia virus.
FIG. 3.
CTL lines show a stable pattern of HIV-1 Gag cross-recognition. CTL lines were produced from PBMC isolated on the date indicated. Effector CTL were tested against autologous target cells infected with the indicated rVV. The E/T ratio is indicated and varied according to the number of cells that grew from each culture.
Fine specificity of the HIV-2 Gag-specific CTL response.
To determine whether HIV-1 and HIV-2 cross-reactivity was due to recognition of viral sequences conserved between the two viruses which were presented by selected HLA alleles, we screened the epitope specificity of the Gag-specific CTL responses of three subjects.
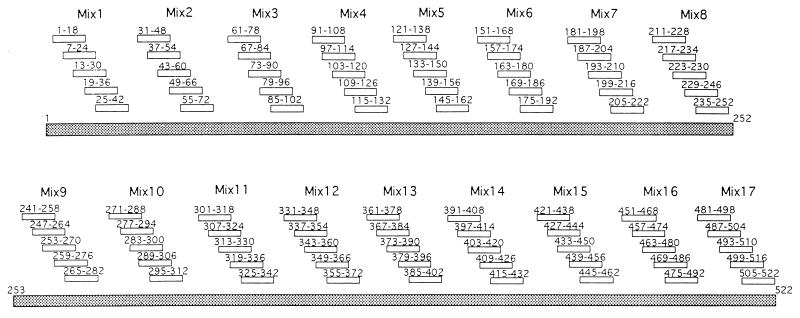
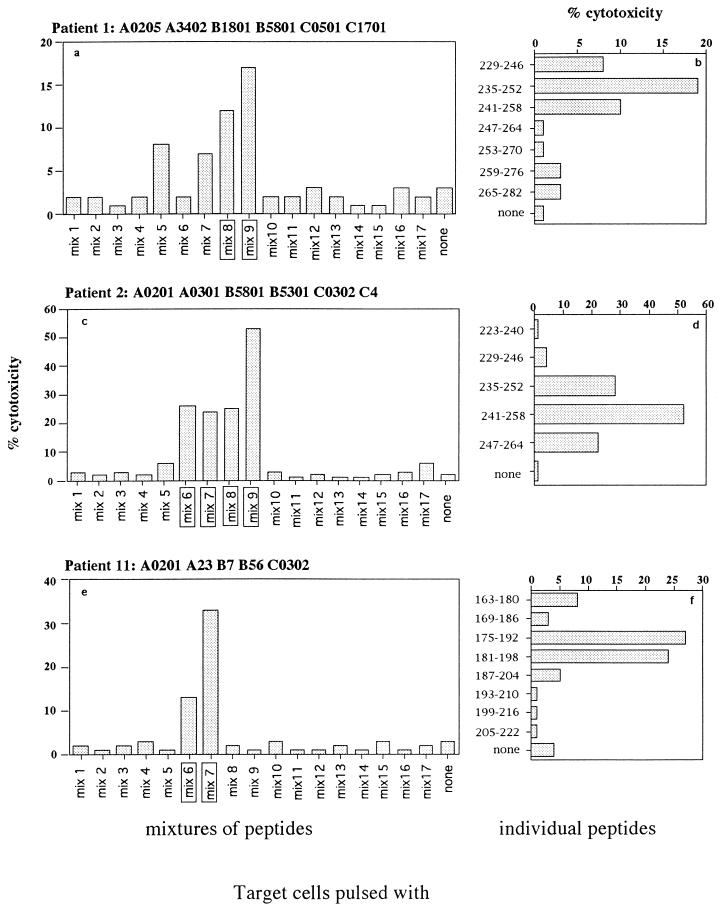
Two of the patients (P1 [A0205 A3402 B1801 B5801 C0501 C1701] and P2 [A0201 A0301 B5801 B5301 C0302 C4]) were able to recognize A1, A2, and B HIV-1 clades; the third patient (P11 [A0201 A23 B67 B56 C0302]) did not recognize any HIV-1 Gag proteins. To define the sequences recognized by the Gag-specific CTL lines, 85 overlapping synthetic peptides spanning the p16 and p28 sequences of HIV-2 Gag were pooled in 17 mixtures of five peptides each (Fig. 4). In the first step, the mixtures were used to sensitize autologous target cells; then peptides present in the mixtures that had sensitized target cells for CTL lysis were used individually.
FIG. 4.
Sequences of the 18-mer synthetic peptides covering the whole HIV-2ROD Gag protein. The mixtures of peptides used for sensitization of target cells in the fine specificity assays are indicated.
Figure 5a shows that CTL line of P1 lysed target cells pulsed with the mixtures covering regions 211–252 (mix 8) and 241-282 (mix 9); the assay performed with target cells pulsed with individual peptides showed a clear recognition of peptide 235-252 (Fig. 5b). It is noteworthy that an HLA-B57 and -B58 HIV-1 clade B epitope (underlined) has been demonstrated in p24 region 240-249 (TSTLQEQIGW) (16); alignment of Gag sequences of HIV-2 and HIV-1 showed that this HIV-1 epitope is represented by HIV-2ROD sequence 241-250 (TSTVEEQIQW). P2 presented a multispecific CTL response focused on different epitopes within the Gag protein. The Gag-specific CTL line lysed target cells pulsed with four different mixtures (mixes 6, 7, 8, and 9), with a dominant CTL response directed against mix 9 (Fig. 5c). Individual peptides overlapping mixes 6 and 7 (not shown), 7 and 8 (not shown), and 8 and 9 (Fig. 5d) were used to sensitize target cells and demonstrate lysis in targets sensitized with peptides 175-192 and 181–198 (an epitope likely in region 181-192) (not shown) and with peptides 235-252 and 241-258 (an epitope likely in region 241-252).
FIG. 5.
Fine specificity of the Gag-specific CTL response. CTL lines of P1 (a and b), P2 (c and d), and P11 (e and f) were tested against autologous target cells pulsed with peptide mixtures (a, c, and e) or individual peptides selected from the responding mixtures (b, d, and f). The concentration of the peptides used for sensitized target cells was always 1 μM; the E/T ratio was 10:1.
The CTL line from P11, who did not demonstrate cross-recognition between HIV-1 and -2, lysed target cells pulsed with the mixtures covering regions 151-192 (mix 6) and 181-222 (mix 7) of p28 (Fig. 5e). Since the two peptide mixtures overlapped, it was likely that epitope recognition of the Gag-specific CTL line was in region 181-192. The cytotoxic assay performed with individual peptides to sensitize target cells confirmed that lysis was present only in cells pulsed with peptides 175-192 and 181-198 (Fig. 5f), suggesting that the cytotoxic epitope is located within sequence 181-192.
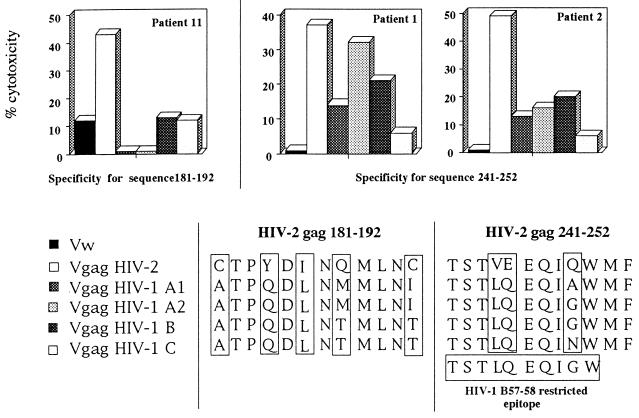
Taken together, the results of these fine specificity studies show that the two subjects able to recognize region 241-252 demonstrated CTL lysis of target cells expressing HIV-1 Gag proteins of clades A1, A2, and B (Fig. 6), whereas P11, with a Gag-specific CTL response focused in region 181-192, was not able to recognize the Gag proteins of different HIV-1 clades (Fig. 6).
FIG. 6.
Different patterns of cross-reactivity expressed by CTL lines recognizing HIV-2 Gag sequences 181–192 and 241–252 and comparison of amino acid sequences of HIV-2ROD and HIV-1 clade A1, A2, B, and C Gag expressed by different rVV in these two regions. Amino acids which differ from the HIV-2 index sequence are indicated within the boxes. The amino acid sequence of the HIV-1 B57-B58-restricted Gag epitope 240–249 is also shown.
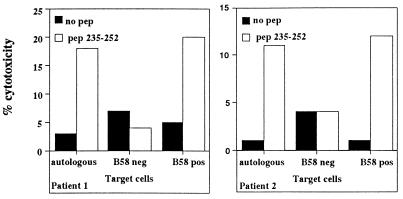
Furthermore, HLA restriction experiments carried out with CTL lines able to recognize HIV-2 peptide 235-252 demonstrated that presentation of the peptide to Gag-specific CTL lines was mediated by the HLA-B5801 molecule (Fig. 7). Analysis of the amino acid sequence of the HIV-2 and HIV-1 Gag proteins expressed by the different rVV show that in region 181-192, the two viruses differed in five amino acids, but also in HIV-2 region 241-252, three distinct amino acid substitutions, at positions 244, 245, and 249, were present and interestingly grouped in region 241-250, corresponding to the HIV-1 Gag epitope 240-249.
FIG. 7.
HLA-restricted recognition of HIV-2 Gag peptide 235–252. HIV-2 Gag peptide 235–252-specific CTL lines of P1 and P2 were tested against autologous target cells, target cells matched only at HLA-B5801, or unmatched target cells pulsed or unpulsed with the peptide at a concentration of 1 μM. The E/T ratio was 10:1. Lysis of the different targets is indicated.
Nevertheless, the amino acid residues involved in the peptide binding to the HLA-B5801 molecules were conserved (serine at position 2 and tryptophan at position 9) (11), and the recognition of target cells expressing HIV-1 Gag clades A1, A2, and B suggests that the amino acid substitutions present in the HIV-1 sequences could be tolerated by the TCRs of HIV-2-specific CTL.
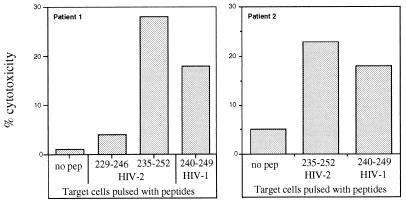
This hypothesis was analyzed by testing the recognition of the HIV-1 (clades B and A2) Gag peptide 240-249 by HIV-2 Gag peptide 235-252-specific CTL lines. CTL lines specific for the HIV-2 sequence recognized the target cells pulsed with the HIV-1 peptide 240-249 (Fig. 8).
FIG. 8.
Recognition of B57-B58-restricted HIV-1 Gag epitope 240–249 by CTL lines specific for HIV-2 Gag peptide 235–252. Lysis of target cells incubated with the HIV-1 and HIV-2 peptides (1 μM) is indicated. The E/T ratio was 10:1.
Induction of the CTL response by HIV-1 peptide in an HIV-2-infected subject.
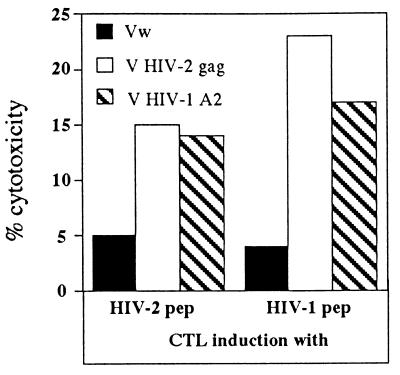
The requirements of lysis or proliferation of CTL are different, and cytotoxicity can be elicited by triggering a small number of TCRs (38). The capacity to induce a CTL response specific for HIV-2 and -1 by HIV-2 Gag peptide 241-258 (containing sequence 241–250 [TSTVEEQIQWMFRPQNPV]) and by HIV-1 Gag peptide 240-249 (TSTLQEQIGW) was measured in the PBMC of subject P2. Figure 9 shows that the HIV-1 peptide was able to induce a CTL response that recognizes both HIV-1 and HIV-2 Gag proteins. Although we cannot completely rule out that this subject had been primed by previous HIV-1 challenge, the fact that this subject is not a commercial sexual worker makes such a possibility less likely.
FIG. 9.
HIV-1-specific CTL induction by HIV-1 peptide 240–249. PBMC of patient 2 (HLA-B5801 positive) were stimulated either with HIV-1 Gag peptide 240–249 or HIV-2 Gag peptide 235–252 for 7 days and then tested against HLA-B5801-positive target cells infected with rVV expressing unrelated, HIV-2, and HIV-1 A2 Gag proteins as described in Materials and Methods.
It is not possible to compare the relative efficacies of CTL induction by the two virus sequences since we used for our assay peptides of different lengths (HIV-2 18-mer peptide and HIV-1 9-mer peptide) that may substantially differ in HLA binding. Nevertheless, the data show that the HIV-1 peptides containing three amino acid substitutions with respect to the prototype HIV-2 sequence could still activate a CTL response, suggesting that an HIV-1 challenge in an HIV-2-infected subject could activate an HIV-1-specific CTL response.
DISCUSSION
In this study, we used polyclonal CTL lines selected from PBMC of HIV-2-infected subjects by stimulation with the whole autologous HIV-2 to define whether a dominant HIV-2-specific CTL response has the ability to recognize HIV-1. The lines were tested initially against the product of HIV-2 gag, nef, and pol genes. The gag and pol genes of HIV-1 and -2 display about 60% nucleotide sequence homology. It is thus more likely that homologies between the linear amino acid sequence of the two viruses presented to CTL are present in these two proteins. The CTL response against Env protein, which has only 40% amino acid homology and a high degree of intraclade variability, was not measured.
A clear immunodominance of the CTL response against HIV-2 Gag protein was present in our patient population, confirming previous work involving HIV-2-infected patients (5). Whether the ability to mount a Gag-specific CTL response in HIV-2 infection correlates with lower rate of disease progression as has been demonstrated in long-term asymptomatic HIV-1 infection needs more careful analysis of a large HIV-2 patient population (20).
The analysis of the ability of HIV-2 Gag-specific CTL lines to recognize different HIV-1 subtypes derived from different geographical areas demonstrates that a majority (9 of 11) of HIV-2 subjects have cross-reactive CTL able to lyse target cells presenting Gag proteins of different HIV-1 clades.
The molecular basis of the cross-recognition could be explained by the HLA profile of the individuals, since different HLA molecules may select for sequence with high homology between the heterologous HIV isolates. On the other hand, CTL cross-recognition of peptides with little homology has been shown not only in HIV-1 and HIV-2 infections (28, 33, 39) but also in other viral infections (4, 22, 35). In all of these cases, amino acid substitutions within the epitopes were well tolerated.
The analysis of HLA profiles and the fine specificity of the HIV-2 Gag-specific CTL response in our HIV-2-infected patients shows that four of the five patients with broad cross-reactivity toward HIV-1 subtypes are HLA-B5801 positive and that this HLA molecule is able to select an HIV-2-specific CTL response which recognizes an HIV-1 sequence. This peptide, corresponding to HIV-1 Gag region 240-249, is immunodominant and associated with slow progression of the disease in HLA-B57 and -B5801 HIV-1-positive patients (16).
The HIV-1 240–249 and HIV-2 241-250 Gag regions show the presence of three amino acid substitutions (Fig. 6) which do not affect the residues at positions 2 and 10 involved in the peptide binding to HLA molecules (11, 16) and are not able to abolish recognition of the HIV-1 sequence by the HIV-2-specific CTLs. These results demonstrate that cross-reactive CTLs do not need to be specific for complete sequence homologies; some amino acid substitutions within the cytotoxic epitopes can be tolerated.
Even though we favor the idea of broad cross-reactivity due to the flexibility of TCR recognition, we must point out that our experiments were carried out with polyclonal CTL lines, as we did not want to narrow the T-cell repertoire present in our patients. For this reason, we cannot completely rule out the possibility that distinct populations of CTL with separate specificities for HIV-1 and HIV-2 are responsible for the cross-recognition. This interpretation implies that in the majority of the HIV-2-positive patients we were able to expand a preexisting HIV-1-specific memory CTL population, as has been demonstrated in a mouse model of virus infections (34) in which extensive cross-reactivity between distantly related viruses was due to reactivation of memory CTLs as a result of previous exposure.
However, even in this model, a degree of cross-reactivity at the clonal level was present, and it seems very unlikely that 9 of 11 of our HIV-2-infected subjects have been challenged by HIV-1, have completely controlled the infection, and have developed an HIV-1 memory CTL response. Although a definitive demonstration of wide cross-reactivity needs a clonal T-cell analysis, our data show that a large proportion of HIV-2-infected subjects have an ongoing cytotoxic response that has the ability to recognize HIV-1 and that tolerated some amino acid substitutions within the epitopes.
It is of note that in our experiments, target cells expressing different HIV-1 Gag proteins with identical amino acid sequences in the epitopes presented in association with HLA-B58 were not equally recognized by CTLs. For example CTLs from P1 recognize HIV-1 clades A2 and B differently, even though their sequences in Gag region 241-252 are identical (Fig. 6). Variations in the sequences flanking defined cytotoxic epitopes, probably affecting peptide transport or processing, have been demonstrated to affect CTL recognition (9). It is thus possible that a different level of loading of peptide 241-252 in HLA-B5801 molecules is achieved by different HIV-1 clades, influencing the level of CTL recognition. Furthermore, P1 and P2, who recognize the same epitope in the context of the HLA-B5801 molecule, show different levels of cross-reactivity toward HIV-1 Gag proteins. However, the CTL assays were performed with polyclonal CTL lines, and while P1 shows an HIV-2 Gag-specific CTL response focused on the single cross-reactive sequence Gag 241–252, P2 show a multispecific CTL response. CTL specific for the non-cross-reactive (HIV-2 Gag 181-192) and cross-reactive (HIV-2 Gag 241-252) sequences are present in this line (Fig. 5c). Only a fraction of HIV-2 Gag-specific CTLs are likely to be responsible for the cross-reactivity demonstrated by P2, which would account for the lower level of HIV-1 Gag protein recognition. An alternative interpretation of these differing patterns of cross-reactivity is that different CTLs are likely to be selected in different individuals in response to the same HLA-peptide complex (24). These CTLs would be differentially influenced by the amino acid substitutions present in the corresponding HIV-1 Gag sequence.
A recent study of HIV-1-infected subjects failed to demonstrate the presence of CTLs which are cross-reactive between HIV-1 and HIV-2 (25). The difference between HIV-1- and HIV-2-infected subjects may be in the different T-cell repertoires present in the two viral infections, which have low heterogeneity in HIV-1 infection (18, 29) and may limit recognition of epitope variants with mutations in regions interacting with the TCR (30). If HIV-2-infected patients demonstrate a broader T-cell repertoire, this finding might explain not only the different degree of cross-recognition but also the different clinical outcome of the two infections.
In this study, we also found that stimulation of PBMC of HIV-2-infected subjects by an HIV-1 peptide led to the detection of a CTL population able to recognize HIV-1, suggesting that the HIV-2-infected subjects after HIV-1 challenge may expand a CTL population with a potential protective effect against HIV-1 infection.
In conclusion, this work suggests that immunity against HIV-2 may enhance the immune system’s ability to react with HIV-1. However, the demonstration of cross-reactive CTL against HIV-1 in HIV-2-positive subjects does not necessarily mean that these cells play a role in protection. Indeed, we cannot rule out the possibility that recognition of the altered HIV-1 peptide has an antagonistic effect on the CTL response (7, 21) or can stimulate different cytokines (40) with a negative effect on the antiviral immune response.
However, it is important to point out that the HLA-B5801 and -B35 molecules that appear to preferentially select cross-reactive CTLs are present in about 50 to 60% of the Gambian and Senegalese populations (3). Work by Travers et al. has suggested that approximately more than half of the women with HIV-2 infection are protected against HIV-1 (37).
Analysis of the frequencies of the HLA-B5801 and -B35 alleles in the HIV-2-infected and dually infected populations with the same relative risk for HIV-1 infection could be important for understanding whether the cross-reactive CTLs play a decisive role in the protection and should help in the design of a rational candidate vaccine that may broadly protect against HIV-1 infection.
ACKNOWLEDGMENTS
We thank the donors in this study for their participation and K. Joof for his hard work in the collection of blood samples. We are grateful for the technical assistance of P. T. N’Gom, A. Bayang, and M. N’Jie. We thank K. McAdam, Director of the MRC Laboratories, The Gambia, for his support.
This work was supported by an MRC Project Grant and partly by Human Science Shinkou Zaidan, Japan.
REFERENCES
- 1.Aaby P, Poulsen A-G, Larsen O, Christiansen C B, Jensen H, Melbye M, Naucler A, Dias F. Does HIV-2 infection provide cross-protection against HIV-1 infection? AIDS. 1997;7:939–940. . (Letter.) [PubMed] [Google Scholar]
- 2.Abimiku A G, Franchini G, Tartaglia J, Aldrich K, Myagkikh M, Markham P D, Chong P, Klein M-P, Kieny M, Paoletti E, Gallo R C, Robert-Guroff M. HIV-1 recombinant poxvirus vaccine induces cross-protection against HIV-2 challenge in rhesus macaques. Nat Med. 1995;1:321–329. doi: 10.1038/nm0495-321. [DOI] [PubMed] [Google Scholar]
- 3.Allsopp C E M, Harding R M, Taylor C, Bunce M, Kwiatkowski D, Anstey N, Brewster D, McMichael A J, Greenwood B M, Hill A V S. Interethnic genetic differentiation in Africa: HLA class I antigens in The Gambia. Am J Hum Genet. 1992;50:411–421. [PMC free article] [PubMed] [Google Scholar]
- 4.Anderson R W, Bennick J R, Yewdell J W, Maloy W L, Coligan J E. Influenza basic polymerase 2 peptides are recognized by influenza nucleoprotein-specific cytotoxic T lymphocytes. Mol Immunol. 1992;29:1089–1095. doi: 10.1016/0161-5890(92)90041-u. [DOI] [PubMed] [Google Scholar]
- 5.Ariyoshi K, Cham F, Berry N, Jaffar S, Sabally S, Corrah T, Whittle H. HIV-2-specific cytotoxic T-lymphocyte activity is inversely related to proviral load. AIDS. 1995;9:555–559. doi: 10.1097/00002030-199506000-00004. [DOI] [PubMed] [Google Scholar]
- 6.Ariyoshi K, Schim van der Loeff M, Sabally S, Cham F, Corrah T, Whittle H. Does HIV-2 infection provide cross-protection against HIV-1 infection? AIDS. 1997;11:1053–1054. . (Letter.) [PubMed] [Google Scholar]
- 7.Bertoletti A, Sette A, Chisari F V, Penna A, Levrero M, De Carli M, Fiaccadori F, Ferrari C. Natural variants of cytotoxic epitopes are T-cell receptor antagonist for antiviral cytotoxic T cells. Nature. 1994;369:407–410. doi: 10.1038/369407a0. [DOI] [PubMed] [Google Scholar]
- 8.Bunce M, O’Neil C M, Barnardo M C, Krausa P, Browning M J, Morris P J, Welsh K I. Phototyping: comprehensive DNA typing for HLA-A, B, C, DRB1, DRB3, DRB4, DRB5 & DQB1 by PCR with 144 primer mixes utilizing sequence-specific primers (PCR-SSP) Tissue Antigens. 1995;46:355–367. doi: 10.1111/j.1399-0039.1995.tb03127.x. [DOI] [PubMed] [Google Scholar]
- 9.Eisenlohr L C, Yewdell J W, Bennink J R. Flanking sequences influence the presentation of an endogenously synthesized peptide to cytotoxic T lymphocytes. J Exp Med. 1992;175:481–487. doi: 10.1084/jem.175.2.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Evavold B D, Sloan-Lancaster J, Wilson K J, Rothbard J B, Allen P M. Specific T cell recognition of minimally homologous peptides: evidence for multiple endogenous ligands. Immunity. 1995;2:655–663. doi: 10.1016/1074-7613(95)90010-1. [DOI] [PubMed] [Google Scholar]
- 11.Falk K, Rotzschke O, Takiguchi M, Ganu V, Stevanovic S, Jung G, Rammensee H G. Peptide motifs of HLA-B58, B60, B61, and B62 molecules. Immunogenetics. 1995;41:165–168. doi: 10.1007/BF00182333. [DOI] [PubMed] [Google Scholar]
- 12.Flynn J N, Keating P, Hosie M J, Mackett M, Stephens E B, Beatty J A, Neil J C, Jarrett O. Env-specific CTL predominate in cats protected from feline immunodeficiency virus infection by vaccination. J Immunol. 1995;157:3658–3665. [PubMed] [Google Scholar]
- 13.Gallimore A, Cranage M, Cook N, Almond N, Bootman J, Rud E, Silvera P, Dennis M, Corcoran T, Stott J, Gotch F M. Early suppression of SIV replication by CD8+ nef-specific cytotoxic T cells in vaccinated macaques. Nat Med. 1995;1:1167–1173. doi: 10.1038/nm1195-1167. [DOI] [PubMed] [Google Scholar]
- 14.Garboczi D N, Ghosh P, Utz U, Fan Q R, Biddison W E, Wiley D C. Structure of the complex between human T-cell receptor, viral peptide and HLA-A2. Nature. 1996;384:134–141. doi: 10.1038/384134a0. [DOI] [PubMed] [Google Scholar]
- 15.Germain R N. MHC-dependent antigen processing and peptide presentation: providing ligands for T lymphocyte activation. Cell. 1994;76:287–298. doi: 10.1016/0092-8674(94)90336-0. [DOI] [PubMed] [Google Scholar]
- 16.Goulder P J R, Bunce M, Krausa P, McIntyre K, Crowley S, Morgan B, Edwards A, Giangrande P, Philips R E, McMichael A J. Novel, cross-restricted, conserved and immunodominant CTL epitopes in slow progressors in HIV-1 infection. AIDS Res Hum Retroviruses. 1996;12:1891–1896. doi: 10.1089/aid.1996.12.1691. [DOI] [PubMed] [Google Scholar]
- 17.Guyader M, Emerman M, Sonigo P, Clavel F, Montangier L, Alizon M. Genome organization and transactivation of the human immunodeficiency virus type 2. Nature. 1987;326:662–669. doi: 10.1038/326662a0. [DOI] [PubMed] [Google Scholar]
- 18.Kalams S A, Johnson R P, Trocha A K, Dynan M J, Ngo H S, D’Aquila R T, Kurnick J T, Walker B D. Longitudinal analysis of T cell receptor (TCR) gene usage by human immunodeficiency virus 1 envelope-specific cytotoxic T lymphocyte clones reveals a limited TCR repertoire. J Exp Med. 1994;179:1261–1271. doi: 10.1084/jem.179.4.1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kersh G J, Allen P M. Essential flexibility in the T-cell recognition of antigen. Nature. 1996;380:495–498. doi: 10.1038/380495a0. [DOI] [PubMed] [Google Scholar]
- 20.Klein M R, van Baalen C A, Holwerda A M, Kerkhof Garde S R, Bende R J, Keet I P M, Eeftinck-Schattenkerk J-K M, Osterhaus A D M E, Schuitemaker H, Miedema F. Kinetics of gag-specific cytotoxic T lymphocyte responses during the clinical course of HIV-1 infection: a longitudinal analysis of rapid progressor and long-term asymptomatics. J Exp Med. 1995;181:1365–1372. doi: 10.1084/jem.181.4.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klenerman P, Rowland-Jones S, McAdam S, Edwards J, Daenke S, Lalloo D, Koppe B, Edwards A, Philips R E, McMichael A J. Naturally occuring HIV-1 gag variants antagonise cytotoxic T-cell activity. Nature. 1994;369:403–407. doi: 10.1038/369403a0. [DOI] [PubMed] [Google Scholar]
- 22.Kulkarni A B, Morse III H C, Bennick J R, Yewdell J W, Murphy B R. Immunization of mice with vaccinia virus-M2 recombinant induces epitope-specific and cross-reactive Kd-restricted CD8+ cytotoxic T cells. J Virol. 1993;67:4086–4094. doi: 10.1128/jvi.67.7.4086-4092.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marlink R. Lessons from the second AIDS virus, HIV-2. AIDS. 1996;10:689–699. doi: 10.1097/00002030-199606001-00002. [DOI] [PubMed] [Google Scholar]
- 24.Maryanski J L, Jongeneel C V, Bucher P, Casanova J L, Walker P R. Single-cell PCR analysis of TCR repertoires selected by antigen in vivo: a high magnitude CD8 response is comprised of very few clones. Immunity. 1996;4:47–55. doi: 10.1016/s1074-7613(00)80297-6. [DOI] [PubMed] [Google Scholar]
- 25.McAdam, S., P. Kaleebu, P. Krausa, P. Goulder, N. French, B. Collin, T. Blanchard, J. Whitworth, A. McMichael, and F. Gotch. Cross-clade recognition of P55 by cytotoxic T lymphocytes in HIV-1. Submitted for publication. [DOI] [PubMed]
- 26.Moss B, Flexener C. Vaccinia virus expression vectors. Annu Rev Immunol. 1987;5:305–324. doi: 10.1146/annurev.iy.05.040187.001513. [DOI] [PubMed] [Google Scholar]
- 27.Nixon D F, Townsend A R M, Elvin J G, Rizza C R, Gallwey J, McMichael A J. HIV-1 gag-specific CTL defined with recombinant vaccinia viruses and synthetic peptides. Nature. 1988;336:484–487. doi: 10.1038/336484a0. [DOI] [PubMed] [Google Scholar]
- 28.Nixon D F, Heut S, Rothbard J, Kieny M-P, Delchambre M, Thiriart C, Rizza C R, Gotch F M, McMichael A J. An HIV-1 and HIV-2 cross-reactive cytotoxic T-cell epitope. AIDS. 1990;4:841–845. doi: 10.1097/00002030-199009000-00002. [DOI] [PubMed] [Google Scholar]
- 29.Pantaleo G, Demarest J F, Soudeyns H, Graziosi C, Denis F, Adelsberger J W, Borrow P, Saag M S, Shaw G M, Sekaly R P, Fauci A S. Major expansion of CD8+ T cells with a predominant Vb usage during the primary immune response to HIV. Nature. 1994;370:463–467. doi: 10.1038/370463a0. [DOI] [PubMed] [Google Scholar]
- 30.Pircher H, Moskphidis A, Rohrer U, Burki K, Hengartner H, Zinkernagel R M. Viral escape by selection of cytotoxic T cell-resistant virus variants in vivo. Nature. 1990;346:629–633. doi: 10.1038/346629a0. [DOI] [PubMed] [Google Scholar]
- 31.Poulsen A-G, Aaby P, Larsen O, Jensen H, Naucler A, Lisse I M, Christiansen C B, Dias F, Melbye M. 9-year HIV-2-associated mortality in an urban community in Bissau, West Africa. Lancet. 1997;349:911–914. doi: 10.1016/S0140-6736(96)04402-9. [DOI] [PubMed] [Google Scholar]
- 32.Rothbard J, Pemberton R, Bomdmer H, Askonas B A, Taylor W R. Identification of residues necessary for clonal specific recognition of a cytotoxic T cell determinant. EMBO J. 1989;8:2321–2327. doi: 10.1002/j.1460-2075.1989.tb08359.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rowland-Jones S, Sutton J, Ariyoshi K, Dong T, Gotch F, McAdam S, Whitby D, Sabally S, Gallimore A, Corrah T, Takiguchi M, Schultz T, McMichael A, Whittle H. HIV-specific cytotoxic T cells in HIV-exposed but uninfected Gambian women. Nat Med. 1995;1:59–64. doi: 10.1038/nm0195-59. [DOI] [PubMed] [Google Scholar]
- 34.Selin L K, Nahill S R, Welsh R M. Cross-reactivities in memory cytotoxic T lymphocyte recognition of heterologous viruses. J Exp Med. 1994;179:1933–1943. doi: 10.1084/jem.179.6.1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shimojo N, Maloy W L, Anderson R W, Biddison W E, Coligan J E. Specificity of peptide binding by the HLA-A2.1 molecule. J Immunol. 1989;143:2939–2945. [PubMed] [Google Scholar]
- 36.Tedder R S, O’Conner T, Hughes A, N’Jie H, Corrah T, Whittle H. Envelope cross-reactivity in Western blot for HIV-1 and HIV-2 may not indicate dual infection. Lancet. 1988;ii:927–930. doi: 10.1016/s0140-6736(88)92598-6. [DOI] [PubMed] [Google Scholar]
- 37.Travers K, Mboup S, Marlink R, Gueye-Ndiaye A, Siby T, Thior I, Traore I, Dieng-Sarr A, Sankale J-L, Mullins C, Ndoye I, Hsieh C-C, Essex M, Kanki P. Natural protection against HIV-1 infection provided by HIV-2. Science. 1995;268:1612–1615. doi: 10.1126/science.7539936. [DOI] [PubMed] [Google Scholar]
- 38.Valitutti S, Muller S, Dessing M, Lanzavecchia A. Different responses are elicited in cytotoxic T lymphocytes by different levels of T cell receptor occupancy. J Exp Med. 1996;183:1917–1921. doi: 10.1084/jem.183.4.1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wilson C C, Kalams S A, Wilkes B M, Ruhl D J, Gao F, Hahn B H, Celine Hanson I, Luzuriaga K, Wolinsky S, Koup R, Buchbinder S P, Johnson R P, Walker B D. Overlapping epitopes in human immunodeficiency virus type 1 gp120 presented by HLA A, B, and C molecules: effect of viral variation on cytotoxic T-lymphocyte recognition. J Virol. 1997;71:1256–1264. doi: 10.1128/jvi.71.2.1256-1264.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Windhagen A, Scholz C, Hollsberg P, Fakaura H, Sette A, Hafler D A. Modulation of cytokine patterns of human autoreactive T cell clones by a single amino acid substitution of their peptide ligand. Immunity. 1995;2:373–380. doi: 10.1016/1074-7613(95)90145-0. [DOI] [PubMed] [Google Scholar]
- 41.Wucherpfennig K W, Strominger J L. Molecular mimicry in T cell-mediated autoimmunity: viral peptides activate human T cell clones specific for myelin basic protein. Cell. 1995;80:695–705. doi: 10.1016/0092-8674(95)90348-8. [DOI] [PMC free article] [PubMed] [Google Scholar]