Abstract
Human DNA topoisomerase 1 (Top1) is a crucial enzyme responsible for alleviating torsional stress on DNA during transcription and replication, thereby maintaining genome stability. Previous researches had found that non-working Top1 interacted extensively with chromosomal DNA in human cells. However, the reason for its retention on chromosomal DNA remained unclear. In this study, we discovered a close association between Top1 and chromosomal DNA, specifically linked to the presence of G-quadruplex (G4) structures. G4 structures, formed during transcription, trap Top1 and hinder its ability to relax neighboring DNAs. Disruption of the Top1–G4 interaction using G4 ligand relieved the inhibitory effect of G4 on Top1 activity, resulting in a further reduction of R-loop levels in cells. Additionally, the activation of Top1 through the use of a G4 ligand enhanced the toxicity of Top1 inhibitors towards cancer cells. Our study uncovers a negative regulation mechanism of human Top1 and highlights a novel pathway for activating Top1.
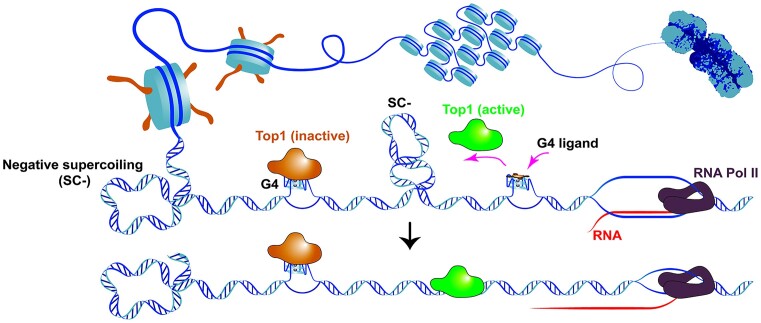
Graphical Abstract
Graphical Abstract.
Introduction
Human DNA topoisomerase 1 (Top1) is a type IB topoisomerase that catalyzes the transient breaking and rejoining of a single strand of DNA which allows the strands to pass through one another, thus altering the topology of DNA (1). Top1 readily removes both positive and negative supercoiling in DNA and resolves the torsional stress associated with DNA replication, transcription, and chromatin condensation (2).
Top1 serves as a critical regulator of gene transcription and chromosome integrity (1). Deletion of Top1 in excitatory neurons causes genomic instability and early onset neurodegeneration (3). Decreased Top1 levels induced by activation-induced cytidine deaminase (AID) result in DNA structural alterations and DNA cleavage during class switch recombination (4). Knockdown of Top1 promotes RNAPII accumulation at transcription starting sites and reduces the full expression of long genes (5,6). In cancer, abnormal expression of Top1 is closely related to poorer overall survival rate and disease-free survival rate. Elevated levels of Top1 have been detected in colon adenocarcinoma, ovarian, esophageal, stomach, breast, lung carcinomas, leukemia cells and malignant melanomas (7–9). Therefore, modulating the activity of Top1 in tumor cells to prevent cell division made it an important drug target for anticancer therapy (10).
Transcription generated positive and negative DNA supercoiling are the primary substrates for Top1. In the absence of topoisomerase, the formation of positive DNA supercoiling (SC+) in front of the DNA transcription machinery tightens the DNA duplex, hindering the movement of RNA polymerase (1). Conversely, the accumulation of negative supercoiling (SC–) behind RNA polymerase weakens Watson-Crick base pairing and facilitates the formation of non-B secondary DNA structures such as hairpins, cruciform DNA, triplex DNA (H-DNA), R-loops and G-quadruplexes (G4) (11–14). Among these non-B secondary DNA structures, G4 is unique because Top1 specifically binds to it (15–17). G4 is a four-stranded high-ordered structure formed by multiple guanines that interact via Hoogsteen hydrogen bonds. G4-forming motifs are significantly enriched at the transcription start sites (TSS) in the genomic DNA of warm-blooded animals (18,19), indicating their regulatory role in gene transcription (20,21). However, the formation of G4 requires DNA to be in a single-stranded state, and complementary C-rich DNA in double-stranded DNA does not favor G4 formation. Understanding how G4s are formed in the gene promoter region and the factors involved in this process are key questions in revealing their functions. Previous studies have shown that a moving RNA polymerase can generate dynamic negative supercoils on the double-stranded DNA behind it, triggering the formation of G4s in the upstream region of the promoter (22,23). Elimination of negative supercoils during this process using E.coli Top1 strongly inhibits G4 formation, suggesting that Top1 has the ability to regulate the biological functions of G4s by manipulating DNA topology (11). Interestingly, G4s have also been reported to inhibited Top1 activity (24,25), implying that G4s might regulate Top1 through a negative feedback mechanism.
Recent studies have demonstrated that Top1 has numerous binding sites on chromosomal DNA, particularly in gene promoters. The retention of Top1 in gene promoter is not associated with its activity, as the topoisomerase I-DNA cleavage complexes (Top1cc) are absent in these regions (5,26). It has been reported that RNA polymerase II pausing at gene promoters contributes to the accumulation and inactivation of Top1 in this region (5). However, whether this phenomenon is related to other elements in gene promoters, such as G4, remains unknown. In this work, we uncovered the global interaction between G4 and Top1 in human genomic DNA and found that G4 formation in transcribed DNA significantly inhibited the activity of Top1. Furthermore, disrupting the interaction between Top1 and G4 using G4-interacting ligands can counteract the inhibitory effect of G4 on Top1 activity. One of the ligands (ZnTTAPc) was found to enhance Top1 activity in cells, inhibit R-loop formation, and increase the cytotoxicity of Top1 inhibitors. Our work highlights a novel approach to modulate human Top1 activity.
Materials and methods
Materials
Oligonucleotides (Supplementary Tables S1–S4) and thiazolyl blue tetrazolium bromide (MTT) were purchased from Sangon Biotech (China). Camptothecin (CPT), topotecan, and anti-FLAG M2 magnetic beads were obtained from Sigma-Aldrich. Pyridostatin (PDS) and PhenDC3 were acquired from MedChemExpress. Tetrakis-(2-trimethylamino-ethylethanol) phthalocyaninato zinc tetraiodine (ZnTTAPc) was synthesized as described (27). Purified Top1 was custom ordered from Sino Biological, Inc. (China). Briefly, histidine tagged Top1 (NP_003277.1) (Met1-Phe765) were expressed in insect cells infected with recombinant baculovirus and purified using Ni columns. The purity was verified using Coomassie blue staining (Supplementary Figure S1). The pGL3-basic plasmid was purchased from Promega. G4P was purified as previously described (28). CUT&Tag kit for Illumina was purchased from Hunan Ruoyu Biotech (China). A siRNA targeting the human Top1 gene (siTop1, 5′-GGAUGAUGCUGAUUAUAAA-3′) and a nonspecific control siRNA (siNC, 5′- UUCUCCGAACGUGUCACGU-3′) were purchased from Guangzhou RiboBio (China).
Plasmids
Plasmids for in vitro transcription were constructed as previously described (11), with the double-stranded DNA (dsDNA) containing CSTB, MYC, 3*CSTB, 3*MYC G-quadruplex forming sequences and the control non-G4 sequence (Supplementary Table S1) being inserted between a T7 promoter and a SP6 promoter through EcoRI/BglII restriction sites. For Top1 overexpression, the human Top1 gene containing a 3× FLAG tag was cloned into the pIRES2-EGFP plasmid at the NheI/BamHI restriction site. Plasmid DNA was first extracted using the TIAN prep Midi Plasmid Kit (Tiangen, China), then treated with 0.2 mg/ml of proteinase K (Thermo Scientific) in a buffer containing 10 mM Tris−HCl (pH 8.0), 2 mM EDTA, 150 mM NaCl at 55°C for 30 min and recovered using the Wizard DNA Clean-Up System (Promega).
In vitro transcription
Transcription was carried out as described previously (11). When a T7 transcription was followed by an SP6 transcription, the T7 transcription was stopped by adding a final concentration of 2 μM T7 inhibitor (5′-GAAATTAATACGACTCACTATA-3′) (29). Then, 0.02 mM Fluorescein-12-UTP (Roche) and 2 U/μl SP6 RNA polymerase (Thermo Scientific) were added. The samples were incubated at 37°C for 60 min, followed by a treatment with 0.04 U/μl DNase I (Thermo Scientific) at 37°C for 15 min and an extraction with phenol/chloroform. RNA products were denatured in 80% formamide and resolved on an 8% denaturing polyacrylamide gel.
Cell lines and cell culture
HeLa, HCT116 and MDA-MB-231 cells were kindly provided by Stem Cell Bank, Chinese Academy of Sciences. HeLa cells and HCT116 cells were cultured in DMEM supplemented with 10% FBS and 1% penicillin/streptomycin. MDA-MB-231 cells were cultured in RPMI 1640 supplemented with 10% FBS and 1% penicillin/streptomycin.
Nuclear extract preparation
HeLa cells were cultured to 50–70% confluence and harvested using trypsin. The cells were centrifuged at 500 g for 5 min and washed three times with phosphate buffered saline (pH 7.4). Subsequently, 5 × 106 Cells were resuspended in 500 μl ice-cold buffer containing 10 mM Tris−HCl (pH 7.5), 150 mM NaCl, 1.5 mM MgCl2, 0.5 mM AEBSF and 0.5% NP40, and incubated on ice for 5 min. Nuclei was collected by centrifugation at 800 g for 5 min and dispensed in 100 μl of buffer containing 10 mM Tris−HCl (pH 7.5), 150 mM NaCl and 0.5 mM AEBSF. The dispersed nuclei were slowly mixed with 400 μl of CHAPS lysis buffer (10 mM Tris−HCl (pH 7.5), 1.5 mM EGTA, 0.5% CHAPS (m/V), 10% glycerol (v/v), 5 mM DTT, 0.5 mM AEBSF, 0.4 mg/ml BSA). After incubating on ice for 30 min, the nuclei lysate was centrifuged at 16 000 g for 20 min, and the supernatant was collected and stored at –80°C.
Electrophoretic mobility shift assay (EMSA)
5′-FAM labeled core G4 DNA (Supplementary Table S2) and the two DNA strands of dsDNA containing a G4 (Supplementary Table S3) were dissolved at 20 nM in a buffer containing 20 mM Tris–HCl (pH 7.4), 75 mM KCl, 1 mM EDTA, 0.4 mg/ml BSA, denatured at 95°C for 5 min, and slowly cooled down to 25°C. DNA was then incubated with 1 μM purified Top1 at 4°C for 1 hour. Samples were resolved on 10% non-denaturing polyacrylamide gel containing 75 mM KCl at 4°C for 2 hours in 1 × TBE buffer containing 75 mM KCl. DNA was visualized by the FAM dye covalently labeled at the 5′ end of the DNA on a Chemi-Doc MP (Bio-Rad).
Biolayer interferometry (BLI)
The interactions between purified Top1 and various DNA structures (Supplementary Table S2) were assessed using streptavidin biosensors on an Octet-RH16 device (ForteBio). All DNAs were biotin-labeled at the 3′ end. Prior to the experiment, the streptavidin-coated biosensors underwent a 30-min equilibration in wash buffer (10 mM Tris−HCl, pH 7.4; 75 mM KCl; 0.5 mM EDTA; 0.1% v/v Triton X-100 and 1 mg/ml BSA). The BLI protocol was configured as follows: Baseline 1 (60 s) in column 1 (wash buffer), Loading (120 s) in column 2 (200 nM DNAs in wash buffer), Baseline 2 (60 s) in column 3 (wash buffer), Association (200 s) in column 4 (0–200 nM of Top1 in wash buffer containing 0.1 mg/ml fish sperm DNA), Dissociation (200 s) in column 5 (wash buffer containing 0.1 mg/ml fish sperm DNA). Fish sperm DNA was included in the association and dissociation phases to minimize nonspecific binding. The experiments were conducted at 30°C with shaking at 1000 rpm. The binding constants of Top1 and DNA were determined by fitting the data to the association–dissociation equation using GraphPad Prism 6 software.
DMS footprinting
DMS footprinting was performed as described previously (30).
Exonuclease digestion assay
The 5′-FAM labeled G-rich DNA and its complementary DNA (refer to Supplementary Table S3) were dissolved at 0.1 μM in a buffer containing 20 mM Tris–HCl (pH 7.4), 75 mM KCl, 1 mM EDTA and 0.4 mg/ml BSA. The mixture was denatured at 95°C for 5 minutes and slowly cooled down to 25°C. Subsequently, DNAs were incubated on ice for 1 hour in the presence or absence of 0.2 μM purified Top1. Prior to exonuclease digestion, MgCl2 and dithiothreitol (DTT) were added to the samples to reach final concentrations of 8 mM and 2 mM, respectively. The DNAs were then digested with 0.2 U/μl T4 DNA Polymerase (Thermo Scientific, USA) at 37°C for 10 min. Reactions were stopped by adding a final concentration of 50 mM EDTA and 0.1% SDS. Samples were heated at 95°C for 5 min in 80% formamide, separated on a 12% denaturing polyacrylamide gel, and analyzed using a ChemiDoc MP (Bio-Rad).
Topoisomerase I-mediated DNA relaxation assay
The DNA relaxation assay was performed according to previously reported methods (31). Reactions were carried out in a total volume of 20 μl of buffer containing 0.2 μg negatively supercoiled pGL3-basic plasmid, 1 μl nuclear extract (500 cell/μl) or 80 pM purified Top1, 35 mM Tris−HCl (pH 8.0), 72 mM KCl, 8 mM MgCl2, 5 mM DTT, 5 mM spermidine and 25 mM EDTA in the presence or absence of G4 DNAs or G4 ligands. The samples were incubated at 37°C for 15 min and then treated with 0.1% SDS and 0.1 mg/ml Proteinase K (Roche) at 60°C for 30 min. For transcription-coupled plasmid DNA relaxation, reactions were carried out in a total volume of 20 μl of buffer containing 35 mM Tris−HCl (pH 8.0), 72 mM KCl, 8 mM MgCl2, 5 mM DTT, 5 mM spermidine, 2 mM NTP, 0.002 U/μl ppase (Thermo Scientific), 1 U/μl T7 RNA polymerase (Thermo Scientific) and 1 U/μl RNase inhibitor (Thermo Scientific). After transcription at 37°C for 30 min, samples were first treated with 1 μl nuclear extract (500 cell/μl) or 80 pM purified Top1 at 37°C for 15 min, then treated with 0.5 μl RNase A (10 mg/ml, Thermo Scientific) and 1 μl EDTA (0.5 M) at 37°C for 30 min, and finally treated with 0.1% SDS and 0.1 mg/ml Proteinase K (Roche) at 60°C for 30 min. Supercoiled (SC-) and relaxed (R) plasmid DNA were separated using a 1% agarose gel. After electrophoresis, the gels were stained with 1 × nucleic acid gel stain GeneFinder (Zealway) and visualized with Gel Doc XR+ system (Bio-Rad).
Transient transfection
For transient transfection, HCT116 cells were cultured in 15 cm dishes to 70–80% confluence and then transfected with 30 μg of pNLS-G4P-IRES2-EGFP (28) using lipofectamine 3000 (Thermo Scientific) according to the manufacturer's instructions. The cells were cultured for an additional 24 hours before harvesting. For Top1 overexpression, cells were seeded on a 6-well plate (3 × 105 cells/well) and cultured for 12 hours. Subsequently, the cells were transfected with 2 μg of pIRES2-EGFP-Top1 or empty control vector using lipofectamine 3000 (Thermo Scientific). The cells were cultured for an additional 48 hours before harvesting. siRNAs transfections were performed with RNAiMAX (Thermo Scientific) following the manufacturer's instructions. The cells were cultured for an additional 72 hours before harvesting.
Slot-blot analysis of R-loop
Genomic DNAs from HeLa and HCT116 cells were isolated using a genomic DNA extraction kit (AP-MN-BL-GDNA-250G, Axygen) following the manufacturer's instructions. The DNA samples were then spotted on a nylon membrane using a Slot Blot device. The spotted DNA was crosslinked with the membrane by exposing it to ultraviolet light (UVP CL-1000 Ultraviolet Cross-linker) with a wavelength of 254 nm for 4 min. Subsequently, the membrane was blocked with blocking buffer (5% BSA in TBST) for 1 hour at room temperature. The membrane was then incubated overnight at 4°C with gentle shaking in a 1:2000 dilution of anti-DNA–RNA hybrid antibody (clone S9.6, Sigma-Aldrich). After three washes with TBST, the membrane was incubated with a 1:5000 dilution of anti-mouse IgG (H + L) antibody, HRP conjugate (KPL) for 1 hour at room temperature. Following another three washes, the signal from S9.6 was detected using the Western Bright ECL kit (Advansta) according to the manufacturer's instructions. To detect double-stranded DNA, the membrane was stripped and then incubated overnight at 4°C with gentle shaking in a 1:2000 dilution of anti-dsDNA antibody (ab27156, Abcam). The signal of double-stranded DNA was detected using the same procedure as for the R-loop signal detection. In the case of the RNase H treated control, 1 μg of genomic DNA was pre-incubated with 10 U of RNase H (NEB) for 4 hours at 37°C. The S9.6 signal was quantified and normalized using ImageJ software.
Western blot
The western blot assay was performed according to the standard protocol. The following antibodies were used: anti-Topoisomerase I (Abcam, ab109374), anti-alpha tubulin (Proteintech, 66031-1-Ig), anti-beta actin (Proteintech, 66009-1-Ig).
Chromatin immunoprecipitation (ChIP) library construction
The G4-ChIP library was constructed as described previously (28).
ChIP-seq data analysis
The clean paired-end sequencing data were analyzed as described (28). The human genome (version hg38) was used as a reference. The ChIP-seq data of Top1 and Top1-Seq in HCT116 cells were obtained from NCBI GEO datasets (GSE57628). The bigwig file of H3K4me3 ChIP-seq was downloaded from NCBI GEO datasets (GSM5502575).
Peaks of reads enrichment were identified using macs2 (32) with the following parameters: –qvalue 0.05, –keep-dup 1, and default values for the other parameters. The overlap between G4P and Top1 signals was investigated using bedtools. The number of original G4P peaks was calculated once if any overlaps were found in the Top1 peaks, for R1 and R2 respectively. Venn diagrams were plotted using the Python libraries matplotlib 3.3.4 and matplotlib-venn 0.11.6 (33). Average coverage signal of reads was generated from the bigwig files using deeptools/bigwigAverage. Profiles and heatmaps of reads were generated from the bigwig files using deeptools/computeMatrix followed by plotProfile and plotHeatmap, respectively. The bed file of NCBI RefSeq gene was downloaded from the UCSC website (http://genome.ucsc.edu/). Duplicate coordinates in the bed files were removed. A scatterplot of the genome-wide global correlation of these signals was plotted by calculating average scores for G4P parallels and Top1, using multiBigwigSummary with the parameters –removeOutliers –corMethod spearman –log1p.
Cleavage under targets and tagmentation (CUT&Tag)
The CUT&Tag method was constructed following the described protocol (34) with some modifications. A total of 105 cells were washed twice with 100 μl wash buffer (20 mM HEPES–KOH at pH 7.5, 150 mM KCl, 0.5 mM spermidine, 1 × protease inhibitors), and then resuspended with 100 μl wash buffer. Next, 10 μl of pre-activated concanavalin A coated magnetic beads were added to the cells and incubated on a rotator for 10 minutes at room temperature. The supernatant was removed, and the bead-bound cells were resuspended in 100 μl of dig-wash buffer (0.05% digitonin, 0.1% BSA, 20 mM HEPES-KOH at pH 7.5, 150 mM KCl, 0.5 mM spermidine, 2 mM EDTA, 1× protease inhibitors) containing either 9 μg/ml of recombinant anti-DNA:RNA hybrid antibody (S9.6, Abcam) or 1 μg/ml of recombinant anti-Topoisomerase I antibody (ab109374, Abcam). After one-hour rotation at room temperature, the cells were washed twice with 100 μl of dig-wash buffer and resuspended with 100 μl of dig-wash buffer containing 1 μl of goat anti-mouse IgG (AP124, Sigma) or 1 μl of goat anti-rabbit IgG (AP132, Sigma), incubated for another one hour at room temperature in the dark, and washed three times with dig-wash buffer. A 1:250 dilution of pA-Tn5 adapter complex (∼0.2 μM) was prepared in 100 μl of dig-wash buffer and added to the cells, which were then rotated for one hour at room temperature. After five washes with dig-wash buffer, the bead-bound cells were resuspended with 50 μl of tagmentation buffer (20 mM HEPES–KOH at pH 7.5, 150 mM KCl, 10 mM MgCl2, 0.5 mM spermidine, 1× protease inhibitors) and incubated at 37°C for 1 hour. To terminate the tagmentation reaction, 2.5 μl of 0.5 M EDTA, 2.5 μl of 10% SDS and 1 μl of 20 mg/ml proteinase K were added and incubated at 55°C for 30 min, followed by an additional incubation at 65°C for 15 min. The DNA was then extracted using 122 μl of Ampure XP beads (Beckman Counter) for library preparation. DNA was eluted in 40 μl of 5 mM Tris−HCl (pH 8.0).
To amplify the libraries, 21 μl of DNA was mixed with a uniquely barcoded i5 and a uniquely barcoded i7 primer, and amplified using Q5 high-fidelity 2 × master mix (NEB). The libraries were purified with 1.3 × volume of Ampure XP beads, and eluted in 30 μl of 10 mM Tris−HCl (pH 8.0).
Cell viability assay
The relative cell viability in response to CPT or topotecan combined with ZnTTAPc was evaluated using the standard MTT assay. HeLa, HCT116 and MDA-MB-231 cells were seeded in 96-well plates. After 24 hours of seeding, the cells were treated with CPT or topotecan at the indicated concentration (0, 0.01, 0.03, 0.1, 0.3, 1, 3 μM) combined with different concentrations of ZnTTAPc (0, 1, 3, 10 μM). After treatment for 48 hours, the mediums were replaced with fresh drugs of the same concentration and the treatment continued for 24 hours. Then, 10 μl of MTT solution (5 mg/ml in PBS) was added to each well and incubated for another 4 hours at 37°C. The medium was removed, and 150 μl of DMSO was added to each well to dissolve formamide crystals. The absorbance was measured at 490 nm using a Spark 10M (Tecan) microplate reader. Cell viability was determined using GraphPad Prism 6.0 software.
Results
Top1 exhibits extensive interaction with chromosome G4
Previous studies revealed that human Top1 could bind intramolecular and intermolecular G4s (15–17). To investigate whether the interaction between Top1 and chromosomal DNA is mediated by G4, we conducted ChIP-seq analysis using G4P protein to detect chromosome G4 in HCT116 cells. G4P is a small artificial G4-binding protein known for its specific recognition of G4s and its application in living cell G4 detection (28). The findings presented in Supplementary Figure S2 indicated that over 65% of G4P peaks in HCT116 cells contained at least one putative G4-forming sequence (PQS). Moreover, the PQS within genomic DNA exhibited noteworthy enrichment in the regions corresponding to G4P peaks, consistent with observations from previous studies (28).
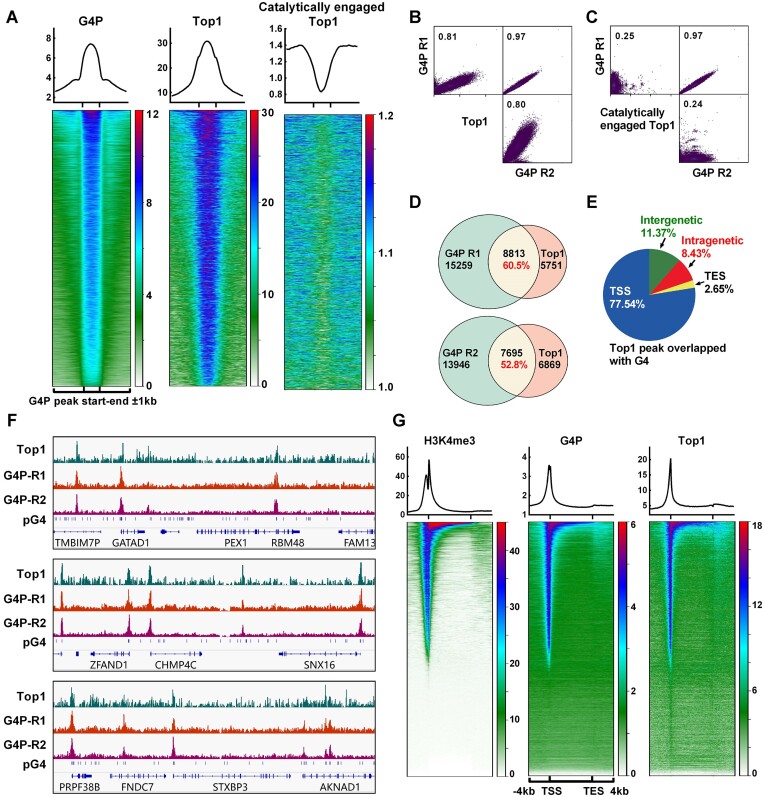
ChIP-seq data of Top1 was obtained from a published study (5). In order to gain insights into the co-localization of G4 and Top1, we sorted the G4P peak region file of HCT116 cells based on mean G4P read counts in descending order and generated heatmaps depicting the Top1 signal across this G4P-binding region file. The results revealed a high degree of overlap between the locations of Top1 and G4, with the signal intensity of Top1 positively correlated with that of G4 (Figure 1A and B). These findings suggest that Top1 may exhibit broad interactions with chromosome G4 structures. Furthermore, we compared the distribution of the catalytically engaged Top1 (5) with that of G4 and observed an inverse correlation between the location and signal intensity of it and those of G4 (Figure 1A and C). Given that catalytically engaged Top1 represents active Top1, these results indicate that G4-associated Top1 is likely inactive. These findings are consistent with previous studies demonstrating the inhibitory effect of G4 on Top1 activity (24,25).
Figure 1.
Retention of Top1 on chromosomal DNA is closely associated with G4. (A) Profile (top) and heatmap (bottom) of Top1 at the G4P peak ±1 kb regions. The heatmaps were generated using the G4P peak bed file sorted based on the mean G4P reads of HCT116 cells in descending order. The data for Top1 and the catalytically engaged Top1 were obtained from NCBI GEO Datasets (GSM2058666 and GSM1385717). (B, C) Scatter plot matrices illustrate the correlation between G4P and Top1 signals. The values in the graphs represent the Pearson correlation coefficient. (D) The overlap of peaks between G4P and Top1 is shown. (E) Distribution of Top1 peaks overlapping with G4P peaks in genomic regions. (F) Examples of Top1 and G4P binding peaks on chromatin DNA. (G) Heatmap of H3K4me3, G4P, and Top1 within the ±4 kb regions around TSS-TES. The gene regions were sorted based on the mean H3K4me3 reads of HCT116 cells in descending order. ChIP-seq data for H3K4me3 was from NCBI GEO Datasets (GSM945304).
Further analysis of the Top1 and G4P peaks revealed that over 50% of the Top1 binding sites on the chromosomal DNA overlapped with G4 (Figure 1D), and the vast majority of these loci were located in gene-associated regions (Figure 1E). Moreover, G4 and Top1 exhibited high enrichment in gene promoter regions (Figure 1E and F), suggesting a potential association with active transcription. This was further supported by the strong positive correlation between Top1 and G4 signaling with H3K4me3 signaling (Figure 1G), which was a hallmark of active promoters.
Top1 directly binds to G4 in single- or double-stranded DNA
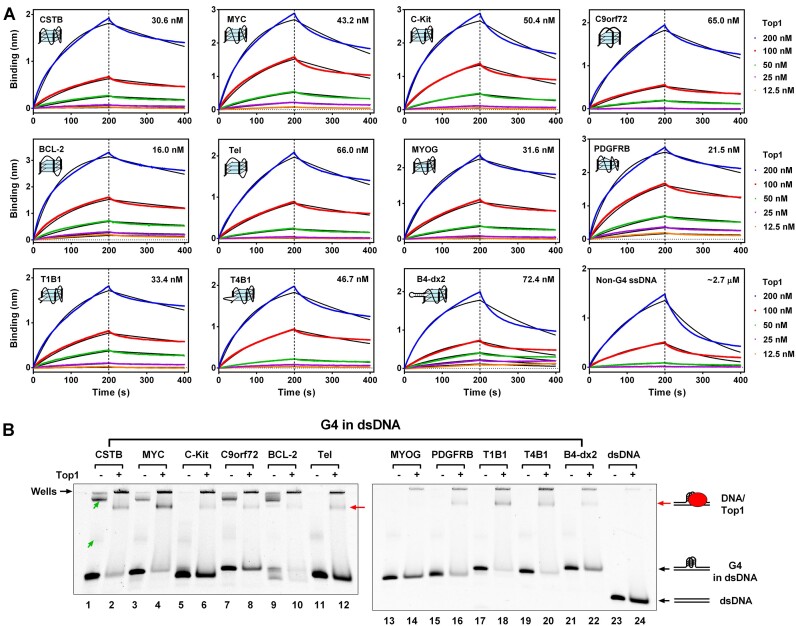
To explore whether the inactivation of Top1 in gene promoters is associated with a direct interaction between Top1 and G4, we initiated an electrophoretic mobility shift assay (EMSA) to assess the binding affinity of Top1 with G4 structures. As shown in Supplementary Figure S3, when employing G4 structures formed from single-stranded G-rich DNA (referred to as core G4) for binding with Top1, the resulting binding band between Top1 and G4 was nearly imperceptible, and the original DNA band remained unchanged after the addition of Top1. Previous studies also found weak binding affinity between Top1 and G4 structures (16), indicating that the interaction between Top1 protein and core G4s is limited in strength and cannot be clearly visualized in EMSA. Consequently, we opted for biolayer interferometry, a more sensitive technique for detecting protein and nucleic acid interactions, to evaluate the binding affinity between Top1 and the core G4s. The results in Figure 2A revealed that Top1 bound to all types of G4 with binding constants ranging from 16.0 to 72.4 nM. Its binding ability to G4 was significantly stronger than its binding ability to non-G4 single-stranded DNA (binding constant approximately 2.7 μM). In addition to G4, Top1 also bound to hairpin and i-motif DNA structures, albeit with much lower affinity than G4 (Supplementary Figure S4). Top1 exhibited strong binding to triplex DNA with a binding constant (38.3 nM) comparable to that of G4. Moreover, the binding of Top1 to triplex DNA exhibited clear Mg2+ ion-dependent characteristics (35), suggesting dependence on the formation of the triplex structure (Supplementary Figure S4).
Figure 2.
Top1 Binds to G4 In Vitro. (A) Binding affinity assessment between Top1 and core G4s using biolayer interferometry (BLI). Core G4s (Supplementary Table S2) were folded from single-stranded DNA (ssDNA) containing a G-quadruplex-forming sequence and a biotin group at the 3′ end. A non-G4 ssDNA served as a control. DNAs were loaded onto streptavidin biosensor tips via the biotin group. The indicated concentrations refer to Top1 in the association phase. The association phase was 200 s, followed by a 200-s dissociation phase. Binding is represented by wavelength shift (measured in nanometers (nm)) detected by the Octet instrument. The number on the upper right side of each figure is the binding constant. (B) Evaluation of the interaction between Top1 and G4 in double-stranded DNA (dsDNA) using electrophoretic mobility shift assay (EMSA). The dsDNA containing the G-quadruplex forming sequence was not complementary to each other within the G-rich region to enable G4 formation (Supplementary Table S3). A non-G4 dsDNA served as the control. The concentrations of Top1 and DNA were 1 and 0.1 μM, respectively. The band indicated by the red arrow represents the DNA/Top1 binding complex. Bands with a slower electrophoretic migration speed indicated by the green arrows were intermolecular G4s formed between DNA strands. The bands retained in the gel wells might be the complex of intermolecular G4s and Top1 or nonspecific binding between Top1 and DNAs.
G4s in chromosomes are mainly formed in dsDNA. DNA in complementary or flanking double-stranded regions may impact the binding of G4 by Top1. In order to test the binding ability of Top1 to G4 in this condition, we first designed a series of dsDNA constructs containing G4 structures. These dsDNAs positioned the G-rich sequence in the middle of the DNA strand without complementary DNA pairing, facilitating G4 structure formation. The DMS footprinting results in Supplementary Figure S5 demonstrated that, except for MYOG, all G-rich sequences formed G4 structures. When these G4s in dsDNA were used to bind with Top1 in EMSA, the original DNA bands showed attenuation, and the migration speed of some DNA slowed down, appearing as smeared bands. An apparent new band emerged near the gel well (Figure 2B, indicated by the red arrow), possibly representing the DNA/Top1 complex. In contrast, dsDNA containing no G4 (non-G4) and MYOG without G4 formation (Supplementary Figure S5) was not bound by Top1 (Figure 2B). Compared with the EMSA results in the presence of core G4s, which showed almost no DNA/Top1 binding phenomenon (Supplementary Figure S3), Top1 showed obvious binding to G4s in dsDNA, indicating a stronger binding ability to G4s in dsDNA. A potential explanation for this difference is that the double-stranded region flanking the G4 structure facilitates its interaction with Top1. Our exonuclease hydrolysis experiment also confirmed that when Top1 binds to G4 in dsDNA, it can protect more than 20 bases next to G4 from being hydrolyzed by exonuclease (Supplementary Figure S6).
Binding to G4 results in Top1 inactivation
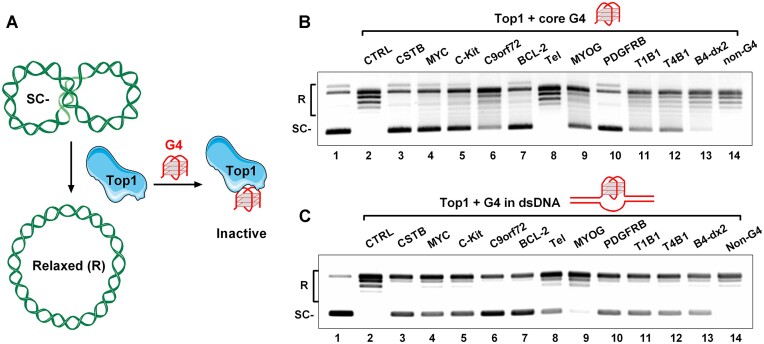
Subsequently, we extended our investigations by conducting a plasmid relaxation assay using purified human Top1 in the presence of various G4 DNAs (as shown in Figure 3A) to evaluate the inhibitory effect of G4 structures on Top1 activity. Most of the core G4s we examined in Figure 2A, but not the non-G4 single-stranded DNA, significantly inhibited the relaxation activity of Top1 (as depicted in Figure 3B). These results align with prior studies (24,25). However, there is no strict correspondence between the inhibitory ability of G4 on Top1 activity and the affinity between them. Among these G4s, parallel-structured G4s (CSTB, MYC, and C-Kit) (28,36,37), hybrid G4 (BCL-2) (38), and G-vacancy-containing G4 (PDGFRB) (22,39) exhibited potent inhibition of Top1. While antiparallel G4 (C9orf72) (40), bulged G4 (T1B1 and T4B1) (41), G-vacancy-containing G4 (MYOG) (42), and G4 containing a hairpin structure in the loop (B4-dx2) (43) displayed weaker inhibition of Top1 activity. In particular, the binding ability of the Tel hybrid G4 (42) to Top1 is similar to that of other G4s (C-Kit, C9orf72, B4-dx2) with Top1, but it has almost no ability to inhibit the activity of Top1(as seen in Figure 3B). In addition to the DNA binding site in its catalytic pocket, Top1 also possesses secondary DNA binding sites located on conserved surfaces of proteins involved in DNA topology recognition (44–46). One explanation for the differences in the ability of different G4s to affect Top1 activity is that G4s with different topological conformations may have different preferences for Top1 binding sites, and only those G4s binding to its catalytic pocket can effectively inhibit its activity.
Figure 3.
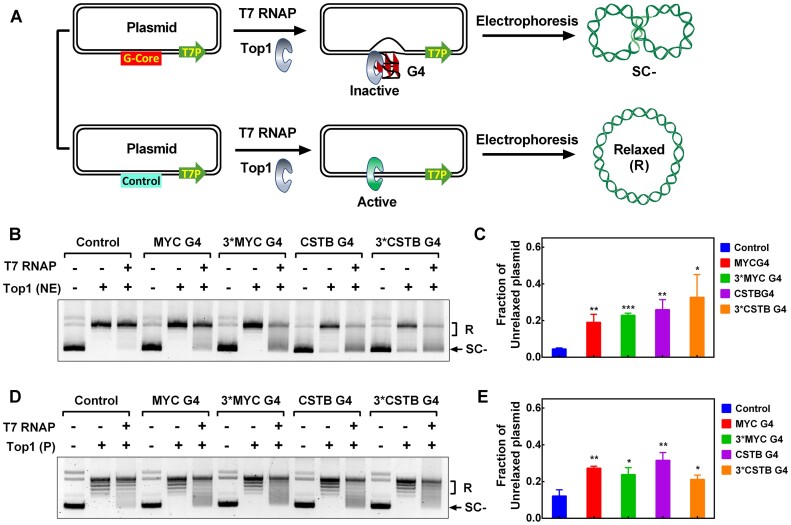
G4 exhibits a strong inhibitory effect on the activity of Top1 in relaxing negatively supercoiled DNA. (A) A schematic illustration describes the process of Top1 unwinding a supercoiled plasmid (SC-) to a relaxed plasmid (R), and the interference of this process by G4. (B) Detection of the activity of the purified human Top1 using a plasmid relaxation assay in the presence of 5 μM of 11 core G4s (CSTB, MYC, C-Kit, C9orf72, BCL-2, Tel, MYOG, PDGFRB, T1B1, T4B1, B4-dx2) and a non-G4 ssDNA (Supplementary Table S2). (C) Detection of the activity of the purified human Top1 using a plasmid relaxation assay in the presence of 1 μM of 11 G4s (CSTB, MYC, C-Kit, C9orf72, BCL-2, Tel, MYOG, PDGFRB, T1B1, T4B1, B4-dx2) in dsDNA or a non-G4 dsDNA (Supplementary Table S3). A negatively supercoiled pGL3-basic plasmid served as the substrate for Top1. The relaxed and supercoiled plasmids were labeled as R and SC–, respectively.
We also extended the plasmid relaxation assay in the presence of synthetic dsDNA containing a G4 structure. The ten G4s (CSTB, MYC, C-Kit, C9orf72, BCL-2, Tel, PDGFRB, T1B1, T4B1, B4-dx2) in dsDNA that could be bound by Top1 (Figure 2B) all showed inhibition of Top1 activity, while MYOG, which did not form G4 (Supplementary Figure S5), did not significantly inhibit Top1 activity (Figure 3C). The effect of G4 in dsDNA on the activity of Top1 was somewhat different from that of core G4. For example, the core G4 of Tel has almost no inhibition on Top1 activity, but in dsDNA, the G4 of Tel shows a significant inhibitory effect (Figure 3B and C). One reason for this result is that the interaction between G4 and Top1 in dsDNA is enhanced compared with core G4. On the other hand, it may be that the conformation of G4 in dsDNA has changed compared with core G4, especially the G4 of Tel has many different conformations and is very sensitive to its flanking sequences (47–49). In addition to G4, we also examined the effects of other DNA secondary structures, such as i-motif, hairpin, and triplex core structures, or hairpin, triplex, and bulge structures in dsDNA, on Top1 activity. We found that, similar to most G4s, the triplex structure could also inhibit the activity of Top1 (Supplementary Figure S7).
Top1 from eukaryotic cells is an ATP independent enzyme, and it does not require a divalent cation (e.g. Mg2+), for activity. These enzymatic properties allow for a clear distinction between Top1 and other cellular topoisomerases, such that the assay of Top1 activity can be performed directly using cell lysates in EDTA-containing buffer conditions (31,50). To study whether G4 can affect the activity of Top1 in complex cellular components, we performed the plasmid relaxation assay using nuclear extracts in the presence of core G4s and G4s in dsDNA. The inhibitory effect of different core G4s on Top1 activity in nuclear extracts (Supplementary Figure S8A) was very similar to that on purified Top1 (Figure 3B). G4 in dsDNA also inhibits the activity of Top1 in nuclear extracts, but its inhibitory effect is weaker than that of purified Top1. In particular, the two G4s (Tel and B4-dx2) with weak binding ability to Top1 (Figure 2A) almost lost their ability to inhibit Top1 activity (Supplementary Figure S8B). This may be due to the presence of large amounts of double-stranded DNA-binding proteins in nuclear extracts, which occupy the region next to G4 and prevent Top1 from binding to it.
The plasmid relaxation assay described above employed an excess addition of synthetic G4. However, it remained unclear whether G4 structures formed co-transcriptionally could act as inhibitors for Top1 and protect the local DNA from relaxation (Figure 4A). To investigate this possibility, we performed a transcription-coupled plasmid relaxation assay using a plasmid containing a T7 promoter, a SP6 promoter, and a G-core sequence (Supplementary Figure S9A). We first validated the formation of G4s in plasmids using an RNA polymerase arrest assay (11). In this experiment, G4 formation induced by downstream transcription with T7 RNA polymerase impedes transcription initiated from the upstream SP6 promoter, generating prematurely terminated (PT) transcripts (Supplementary Figure S9A). The results in Supplementary Figure S9B showed that PT bands were only produced by plasmids containing G-core sequences and displayed G4-associated K+ ion-dependent characteristics. In addition, the number of PT bands corresponded to the number of repeats of the G-core sequence in the plasmid DNA. The proportion of transcription termination bands caused by G4 is approximately 22–36.5%, indicating that at least a corresponding proportion of plasmids form G4.
Figure 4.
G4 formed during transcription hinders DNA relaxation by human Top1. (A) Schematic diagram of transcription-induced G4 formation and its effect on Top1 activity. Transcription leads to the formation of G4 structures at the upstream G-core region of a plasmid DNA through the transmission of negative supercoiling. Top1 becomes trapped by the G4 structure, thereby preventing the relaxation of the plasmid. (B) The relaxation of negatively supercoiled plasmids containing different G4 forming sequences was examined using nuclear extracts (NE). The plasmids were constructed separately by inserting a non-G4 sequence (Control), one repeat of MYC G-core sequence (MYC G4), one repeat of CSTB G-core sequence (CSTB G4), three repeats of MYC and CSTB G-core sequence (Supplementary Table S1) at the G-core region described in (A). Plasmid DNA was either transcribed or not transcribed with T7 RNA polymerase (T7 RNAP) for 30 minutes before the addition of nuclear extracts. The relaxed plasmid and unrelaxed supercoiled plasmids were labeled as R and SC-, respectively. (C) The inhibition of Top1 activity was quantified based on the fraction of unrelaxed supercoiled plasmid shown in (B). (D) The relaxation of negatively supercoiled plasmids containing different G4 forming sequences was assessed using purified Top1 (P). Plasmid DNA was either transcribed or not transcribed with T7 RNA polymerase (T7 RNAP) for 30 min before the addition of Top1. (E) The inhibition of Top1 activity was quantified based on the fraction of unrelaxed supercoiled plasmid observed in (D). The data presented represent the mean values from three independent experiments, and the error bars represent the standard deviation (SD). P values were calculated using two-tailed unpaired Student's t-test (not significant (ns), P > 0.05; * P< 0.05, ** P< 0.01, *** P< 0.001).
For the plasmid relaxation assay, the plasmid was first transcribed with T7 RNA polymerase and subsequently treated with purified Top1 or nuclear extracts. The resistance of the plasmid to Top1-mediated relaxation was assessed by calculating the fraction of unrelaxed negatively supercoiled plasmid DNA (Supplementary Figure S10). The results in Figure 4B and C indicate that approximately 20% of plasmids containing the G-core sequence cannot be efficiently unwound by Top1 from nuclear extracts after T7 transcription. In contrast, the control plasmid without the G-core sequence was almost completely relaxed under the same conditions. Similar results were obtained in the experimental group using purified Top1 (Figure 4D and E). Since the negative supercoiling produced by transcription does not significantly improve the ability of the multiple repeated G-core sequences to form G4 (Supplementary Figure S9B), there is little difference in the proportion of plasmids with different copy numbers of G-core sequences that resist Top1 unwinding after transcription (Figure 4C and E).
In summary, G4 structures can directly interact with Top1 and can even inhibit Top1 activity within complex nuclear extracts. This broad inhibitory influence of G4s on Top1 activity may elucidate the inactivity of G4-associated Top1 on chromosomal DNA.
G4 ligand disrupts G4–Top1 interaction and reactivates Top1
The inhibition of Top1 activity by G4 may be due to its occupation of the active site of Top1 or its restriction of Top1 translocation. We then speculate that disrupting G4 interaction with Top1 using G4 ligand might restore Top1 activity. To verify this hypothesis, we conducted plasmid relaxation assays using purified human Top1 in the presence of G4 DNA and G4 ligands.
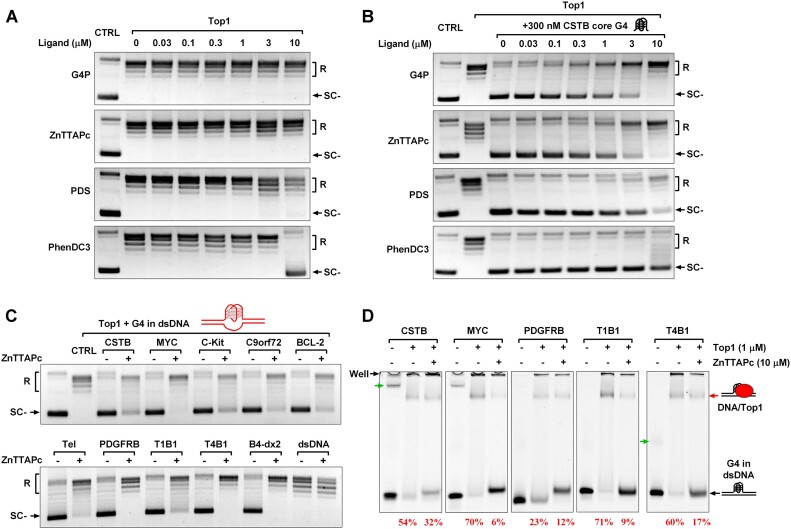
Initially, we examined whether G4 ligands alone affected Top1 activity. It was observed that G4P protein and two G4-binding compounds (ZnTTAPc and PDS) had minimal impact on Top1 activity at concentrations below 10 μM. The classical G4-binding compound PhenDC3 significantly inhibited Top1 activity at 10 μM (Figure 5A). We further studied the effect of G4 ligands on Top1 activity in the presence of CSTB core G4 and found that 300 nM CSTB core G4 could significantly inhibit the activity of Top1, but this inhibition could be reversed by G4 ligands. Among them, G4P protein and ZnTTAPc have the most obvious effects at a concentration of 10 μM, which can completely relieve the inhibition of Top1 activity by G4. Since phenDC3 itself inhibits Top1 activity, it cannot relieve the inhibition of Top1 activity by G4, even if its concentration reaches 10 μM (Figure 5B).
Figure 5.
G4 ligands disrupt the interaction between Top1 and G4 structures, alleviating the inhibitory effect of G4 on Top1 activity. (A) The impact of G4 ligands on the activity of purified human Top1. A negatively supercoiled pGL3-basic plasmid served as the substrate for Top1. The relaxed and supercoiled plasmids were labeled as R and SC-, respectively. (B) The effect of G4 ligands on the activity of purified human Top1 in the presence of 300 nM CSTB core G4. (C) The effect of ZnTTAPc on the relaxation of a negatively supercoiled pGL3-basic plasmid in the presence of 1 μM of 10 G4s (CSTB, MYC, C-Kit, C9orf72, BCL-2, Tel, PDGFRB, T1B1, T4B1, B4-dx2) in dsDNA or a non-G4 dsDNA (Supplementary Table S3). (D) Assessment of the impact of ZnTTAPc on the binding between Top1 and G4 in dsDNA through EMSA. G4 in dsDNA was prepared as described in Figure 2B. The concentrations of DNA, Top1, and ZnTTAPc were 0.1, 1 and 10 μM, respectively. The band indicated by the red arrow represents the DNA/Top1 binding complex. The percentage number corresponding to the lane below the gel image indicates the proportion of G4 DNA bound by Top1. Bands with a slower electrophoretic migration speed, indicated by the green arrows, were intermolecular G4s formed between DNA strands. The bands retained in the gel wells might be the complex of intermolecular G4 and Top1 or nonspecific binding between Top1 and DNAs.
We also repeated the plasmid relaxation assay using nuclear extracts in the presence of G4 DNA and G4 ligand. Top1 activity in nuclear extracts is more susceptible to G4 ligands than purified Top1. For example, PDS and phenDC3 strongly inhibit Top1 activity above 3 μM (Supplementary Figure S11). This may be because Top1 in nuclear extracts has many cofactors (5,51,52), and the non-specific effects of high concentrations of small molecules will affect the activity of Top1. Similar to the experimental group using purified Top1, G4P protein and ZnTTAPc did not affect the activity of Top1 in nuclear extracts within 10 μM. However, in the presence of CSTB core G4, G4P protein and ZnTTAPc only need to reach a concentration of more than 1 μM to effectively alleviate the inhibition of Top1 by G4 (Supplementary Figure S11). The possible reason for this result is that G4P protein and ZnTTAPc release G4-trapped Top1 from endogenous DNA or RNA in nuclear extracts, thus increasing the concentration of effective Top1 in the reaction.
The activation of G4-trapped Top1 using G4 ligand was also confirmed in a plasmid relaxation assay in the presence of G4s in dsDNA. The results in Figure 5C show that ZnTTAPc effectively alleviates the inhibition of Top1 activity by the ten G4s in dsDNA seen in Figure 3C.
Disruption of the G4–Top1 interaction by G4 ligands was further validated using EMSA in the presence of ZnTTAPc and dsDNAs containing CSTB, MYC, PDGFRB, T1B1, and T4B1 G4. The results in Figure 5D show that ZnTTAPc effectively reduced the proportion of Top1-bound G4 DNA. Since ZnTTAPc is a G4-specific binding ligand (53), the reduction of G4-Top1 interaction by ZnTTAPc can be attributed to ZnTTAPc competing with Top1 for G4 binding. The above results indicate that G4 ligands have the potential to relieve the inhibitory effect of G4 structure on Top1 and act as a Top1 activator.
G4 ligand activates Top1 and reduces R-loop levels in human cells
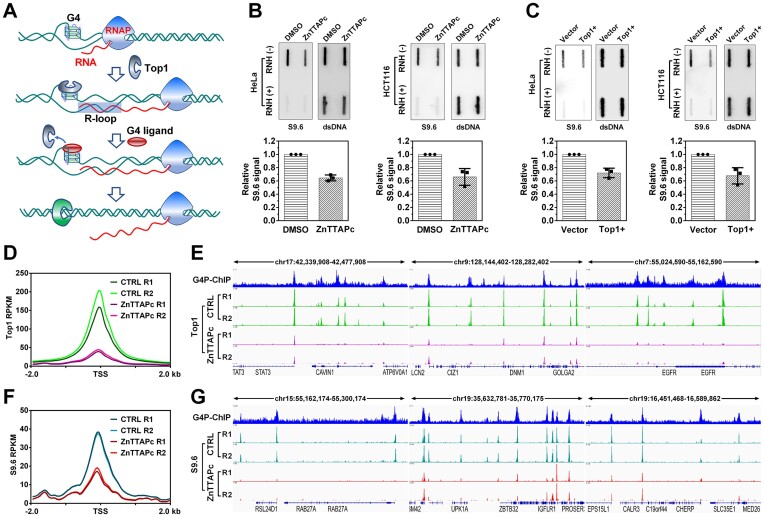
Previous studies have indicated that Top1 plays a role in relaxing DNA supercoiling associated with transcription and preventing R-loop formation in human cells (54,55). If Top1 is captured and inactivated by G4 in gene promoters, it can result in the accumulation of negative supercoils and R-loop structures on chromosomal DNA. Therefore, displacing free Top1 from G4 using a G4 ligand could theoretically reduce R-loop formation (Figure 6A).
Figure 6.
ZnTTAPc releases Top1 trapped by chromosome G4 and reduces intracellular R-loop levels. (A) Illustration of the interaction between G4, G4 ligand, Top1 and R-loop. Transcription-induced negative supercoiling on DNA strands promotes the formation of G4 structures and R-loops. Top1 becomes trapped by G4 and loses its ability to relax the negatively supercoiled DNA. Displacing Top1 on G4 using a G4 ligand allows it to relax the DNA, thereby inhibiting R-loop formation. (B) Detection of R-loop in the genomic DNA from HeLa and HCT116 cells treated or untreated with 10 μM ZnTTAPc using slot blot. R-loop levels were quantified and normalized by calculating the ratio of signals obtained from the R-loop-specific antibody (S9.6) and the double-stranded DNA (dsDNA) antibody. (C) Detection of R-loop on genomic DNA from HeLa and HCT116 cells transfected with a plasmid encoding the Top1 gene (Top1+) or an empty control vector, using slot blot analysis. (D) Top1 distribution around the transcription start sites (TSS) of genes. Top1 binding sites were identified in the chromosomes of HCT116 cells, treated or untreated with 10 μM ZnTTAPc, using the CUT&Tag assay. (E) Examples of specific G4 formation sites (G4P-ChIP) and Top1 binding sites in the chromosomal DNA of HCT116 cells. (F) R-loop distribution around TSS in genes. R-loops were detected in the chromosomes of HCT116 cells, treated or untreated with 10 μM ZnTTAPc, using the CUT&Tag assay. (G) Examples of G4 (G4P-ChIP) and R-loop formation sites in the chromosomal DNA of HCT116 cells.
As depicted in Figure 6B, treatment of HeLa and HCT116 cells with 10 μM ZnTTAPc resulted in a significant decrease in R-loop signals detected by slot blot using the S9.6 antibody. The effect of ZnTTAPc on R-loop levels was similar to that of Top1 overexpression (Figure 6C) and opposite to that of Top1 knockdown (Supplementary Figure S12). Since ZnTTAPc did not alter the protein level of Top1 (Supplementary Figure S13), and the reduction of R-loops on genomic DNA is not associated with the changes in gene expression after ZnTTAPc treatment (Supplementary Figure S14), its impact on R-loops is likely achieved by increasing Top1 activity.
To further confirm that the reduction of R-loop under ZnTTAPc treatment is related to Top1, we preformed CUT&Tag assays to detect the signals of Top1 and R-loop in the chromosomal DNA of HCT116 cells. Compared with the untreated samples, the Top1 signal around the gene promoter in the ZnTTAPc-treated samples were significantly reduced (Figure 6D). Moreover, the examples in Figure 6E show that the Top1 binding sites with reduced peak signal are closely related to the G4 formation sites. This suggests that ZnTTAPc reduces the retention of Top1 on chromosome G4, allowing it to move freely. Correspondingly, the signals of R-loop in gene promoters in ZnTTAPc-treated samples were also greatly decreased compared to untreated samples (Figure 6F and G). These results indicate that ZnTTAPc releases Top1 on chromosome G4, allowing it to eliminate DNA negative supercoiling and thereby inhibit R-loop formation in cells.
G4 ligand enhances the cytotoxicity of Top1 inhibitor
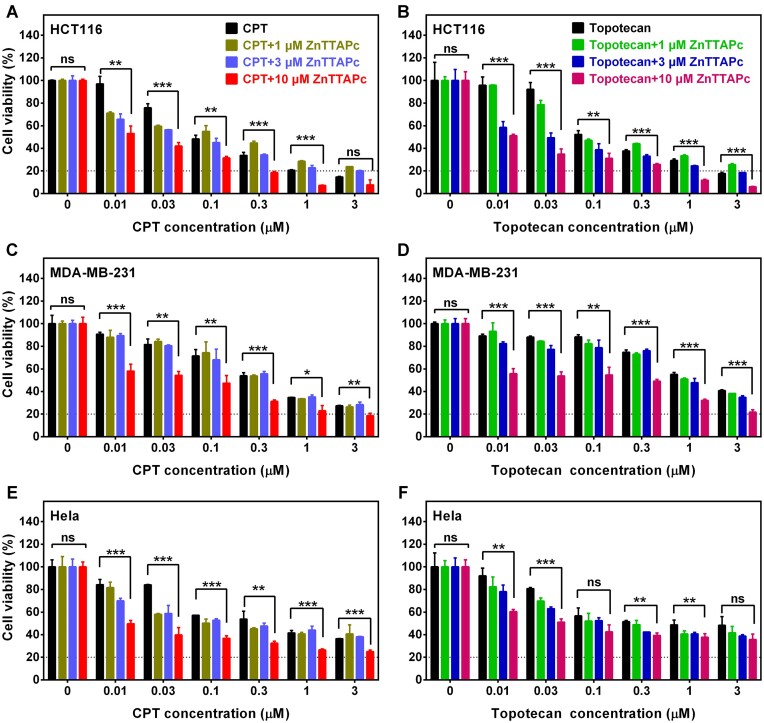
Active Top1 is a well-known target for chemotherapeutic drugs such as camptothecin (CPT) and its derivatives (topotecan, etc.). CPT interacts with the intermediate topoisomerase I-DNA cleavage complex, preventing DNA re-ligation, and inducing lethal DNA strand breaks (56). As previously shown, being trapped by G4 would reduce the concentration of free Top1. Therefore, activating Top1 using G4 ligands could theoretically enhance the cytotoxicity of Top1 inhibitors.
To verify this hypothesis, we assessed the cellular viability of HCT116, MDA-MB-231 and HeLa cells in the presence of ZnTTAPc and Top1 inhibitors using MTT assays. As the concentration of CPT and topotecan increased, the viability of HCT116 cells exhibited a drug concentration-dependent decrease. ZnTTAPc alone had minimal effect on the viability of HCT116 cells, even at a concentration of 10 μM. However, when cells were co-treated with ZnTTAPc and CPT or topotecan, ZnTTAPc significantly enhanced the cytotoxicity of CPT and topotecan (Figure 7A and B), especially at a ZnTTAPc concentration of 10 μM. Similarly, 10 μM ZnTTAPc significantly enhanced the cytotoxicity of Top1 inhibitors in MDA-MB-231 and HeLa cells (Figure 7C–F). The impact of ZnTTAPc on different Top1 inhibitors varied, reflecting the diverse toxicities of these inhibitors across different cells. For instance, 10 μM ZnTTAPc combined with a Top1 inhibitor at or above 0.3 μM reduced HCT116 cell viability to less than 20%. However, achieving this effect in other cells (Figure 7E and F) might necessitate a 10-fold increase in the Top1 inhibitor (Figure 7C and D). This discrepancy can be attributed to various factors. Firstly, distinct cells exhibit varying tolerances to Top1 inhibitors. Secondly, the quantity of G4 formed in different cells may differ, leading to distinct amounts of Top1 captured by G4 that can be released by ZnTTAPc. In summary, these findings indicate the potential use of G4 ligands as drug sensitizers for Top1 inhibitors in tumor therapy.
Figure 7.
ZnTTAPc synergistically enhances the cytotoxicity of Top1 inhibitors in cancer cells. (A, B) Cell viability assay of HCT116 cells treated with CPT (0–3 μM) or topotecan (0–3 μM), in combination with ZnTTAPc (0, 1, 3, 10 μM) for a duration of 72 hours. (C, D) Cell viability assay of MDA-MB-231 cells treated with CPT (0–3 μM) or topotecan (0–3 μM), in combination with ZnTTAPc (0, 1, 3, 10 μM) for a duration of 72 hours. (E, F) Cell viability assay of HeLa cells treated with CPT (0–3 μM) or topotecan (0–3 μM), in combination with ZnTTAPc (0, 1, 3, 10 μM) for a duration of 72 hours. Error bars represent the mean ± SD of three independent experiments. P values were calculated using two-tailed unpaired Student's t-test (not significant (ns), P > 0.05; * P< 0.05, ** P< 0.01, *** P< 0.001).
Discussion
Human Top1 is an essential enzyme responsible for relieving supercoiling and torsional tension in DNA arising during DNA replication and transcription. Previous studies have shown that certain synthetic G4 structures can bind to and inactivate human Top1 in vitro (15–17,24,25). In this study, we demonstrated that the interaction between Top1 and G4 is a widespread phenomenon on genomic DNA. G4 structures present in gene promoters effectively trap free Top1, rendering it inactive and impeding its free movement, thus preventing the relaxation of promoter DNA. This finding sheds light on one of the key reasons why single-stranded DNA (57) and DNA secondary structures, such as R-loops (12), form in gene promoter regions, as their formation is highly dependent on negative DNA supercoiling.
Human Top1 was found to be activated by BRD2 (58) and Ser2 phosphorylated RNAPII (5). However, the factors responsible for repressing free Top1 remain unknown. Human Top1 is not strictly dependent on divalent ions, and in the absence of limiting factors, it would relax all the DNA in cells. The inhibition of free Top1 by chromosome-associated G4 structures unveils a negative regulatory mechanism for Top1. This mechanism helps maintain the promoter regions of specific genes in a negatively supercoiled state, ensuring continuous and active gene expression.
The elucidation of how chromosome G4 structures repress Top1 activity provides insights into Top1 activation. Our study demonstrates both in vitro and in cell experiments that a G4 ligand, ZnTTAPc, can activate Top1 by preventing its interaction with G4 structures. As a Top1 activator, ZnTTAPc effectively reduces R-loop levels in cells and enhances the cytotoxicity of Top1 inhibitors such as CPT. This method of Top1 activation is more efficient and practical compared to the overexpression of Top1.
The activation of Top1 by G4 ligands represents an unexplored aspect that holds great potential for the development of dual-target inhibitors targeting both G4 and Top1. An ideal G4/Top1 dual-target inhibitor should possess two fundamental capabilities: firstly, it should selectively recognize G4 structures and disrupt the interaction between Top1 and G4; secondly, it should not directly interact with Top1 but instead target the Top1cc complex. A class of compounds known as indenoisoquinoline topoisomerase inhibitors fulfills these criteria, making them promising candidates for G4/Top1 dual-target inhibitors. These inhibitors exhibit high affinity for G4 structures (59) and effectively inhibit the re-ligation of Top1cc (60). Their mode of action is akin to the combined effect of ZnTTAPc and CPT in cells, which may explain their exceptional ability to induce cancer cell death.
The underlying molecular mechanism through which G4 inhibits Top1 activity warrants deeper exploration. Our findings suggest that the inhibitory effect of G4 on Top1 is not solely contingent on their affinity. Notably, in addition to G4, triplex DNA also showed interaction with Top1 and inhibited its activity (Supplementary Figures S4 and S7). This implies the existence of an unknown binding site or interaction mode between DNA secondary structures and Top1, which significantly affects Top1 activity. This property is expected to be revealed through crystal structure analysis in the future.
Supplementary Material
Acknowledgements
The authors thank Prof. Danzhou Yang for her valuable suggestions.
Contributor Information
Hui-ting Liang, School of Biomedical Sciences, Hunan University, Changsha 410082, China; School of Pharmaceutical Sciences, Shenzhen Campus of Sun Yat-Sen University, Shenzhen 518107, China.
Jiang-yu Yan, School of Biomedical Sciences, Hunan University, Changsha 410082, China.
Hao-jun Yao, School of Biomedical Sciences, Hunan University, Changsha 410082, China.
Xue-nan Zhang, School of Biomedical Sciences, Hunan University, Changsha 410082, China.
Zhi-ming Xing, School of Biomedical Sciences, Hunan University, Changsha 410082, China.
Lin Liu, School of Public Health (Shenzhen), Shenzhen Campus of Sun Yat-sen University, Shenzhen 518107, China.
Yao-qing Chen, School of Public Health (Shenzhen), Shenzhen Campus of Sun Yat-sen University, Shenzhen 518107, China.
Guo-rui Li, School of Biomedical Sciences, Hunan University, Changsha 410082, China.
Jing Huang, School of Biomedical Sciences, Hunan University, Changsha 410082, China.
Yi-de He, School of Pharmaceutical Sciences, Shenzhen Campus of Sun Yat-Sen University, Shenzhen 518107, China.
Ke-wei Zheng, School of Biomedical Sciences, Hunan University, Changsha 410082, China.
Data availability
Original sequencing (fastq) and processed (bigwig) G4P-ChIP datasets, and CUT&Tag datasets for R-loop have been deposited in and can be downloaded from the NCBI Gene Expression Omnibus (GEO) under accession code GSE239694.
Supplementary data
Supplementary Data are available at NAR Online.
Funding
National Natural Science Foundation of China [22277149, 21708042, 22207131]. Funding for open access charge: National Natural Science Foundation of China.
Conflict of interest statement. None declared.
References
- 1. Pommier Y., Nussenzweig A., Takeda S., Austin C. Human topoisomerases and their roles in genome stability and organization. Nat. Rev. Mol. Cell Biol. 2022; 23:407–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Leppard J.B., Champoux J.J. Human DNA topoisomerase I: relaxation, roles, and damage control. Chromosoma. 2005; 114:75–85. [DOI] [PubMed] [Google Scholar]
- 3. Fragola G., Mabb A.M., Taylor-Blake B., Niehaus J.K., Chronister W.D., Mao H., Simon J.M., Yuan H., Li Z., McConnell M.J. et al. Deletion of topoisomerase 1 in excitatory neurons causes genomic instability and early onset neurodegeneration. Nat. Commun. 2020; 11:1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kobayashi M., Aida M., Nagaoka H., Begum N.A., Kitawaki Y., Nakata M., Stanlie A., Doi T., Kato L., Okazaki I.M. et al. AID-induced decrease in topoisomerase 1 induces DNA structural alteration and DNA cleavage for class switch recombination. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:22375–22380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Baranello L., Wojtowicz D., Cui K., Devaiah B.N., Chung H.J., Chan-Salis K.Y., Guha R., Wilson K., Zhang X., Zhang H. et al. RNA polymerase II regulates topoisomerase 1 activity to favor efficient transcription. Cell. 2016; 165:357–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. King I.F., Yandava C.N., Mabb A.M., Hsiao J.S., Huang H.S., Pearson B.L., Calabrese J.M., Starmer J., Parker J.S., Magnuson T. et al. Topoisomerases facilitate transcription of long genes linked to autism. Nature. 2013; 501:58–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Grunnet M., Calatayud D., Schultz N.A., Hasselby J.P., Mau-Sorensen M., Brunner N., Stenvang J. TOP1 gene copy numbers are increased in cancers of the bile duct and pancreas. Scand. J. Gastroenterol. 2015; 50:485–494. [DOI] [PubMed] [Google Scholar]
- 8. Dancey J., Eisenhauer E.A. Current perspectives on camptothecins in cancer treatment. Br. J. Cancer. 1996; 74:327–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Giovanella B.C., Stehlin J.S., Wall M.E., Wani M.C., Nicholas A.W., Liu L.F., Silber R., Potmesil M. DNA topoisomerase I–targeted chemotherapy of human colon cancer in xenografts. Science. 1989; 246:1046–1048. [DOI] [PubMed] [Google Scholar]
- 10. Bjornsti M.A., Kaufmann S.H. Topoisomerases and cancer chemotherapy: recent advances and unanswered questions. F1000Res. 2019; 8:F1000 Faculty Rev-1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zheng K.W., He Y.D., Liu H.H., Li X.M., Hao Y.H., Tan Z. Superhelicity constrains a localized and R-loop-dependent formation of G-quadruplexes at the upstream region of transcription. ACS Chem. Biol. 2017; 12:2609–2618. [DOI] [PubMed] [Google Scholar]
- 12. Kouzine F., Wojtowicz D., Baranello L., Yamane A., Nelson S., Resch W., Kieffer-Kwon K.R., Benham C.J., Casellas R., Przytycka T.M. et al. Permanganate/S1 nuclease footprinting reveals non-B DNA structures with regulatory potential across a mammalian genome. Cell Syst. 2017; 4:344–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. El Hage A., French S.L., Beyer A.L., Tollervey D Loss of topoisomerase I leads to R-loop-mediated transcriptional blocks during ribosomal RNA synthesis. Genes Dev. 2010; 24:1546–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mirkin S.M. Discovery of alternative DNA structures: a heroic decade (1979–1989). Front. Biosci. 2008; 13:1064–1071. [DOI] [PubMed] [Google Scholar]
- 15. Marchand C., Pourquier P., Laco G.S., Jing N., Pommier Y. Interaction of human nuclear topoisomerase I with guanosine quartet-forming and guanosine-rich single-stranded DNA and RNA oligonucleotides. J. Biol. Chem. 2002; 277:8906–8911. [DOI] [PubMed] [Google Scholar]
- 16. Arimondo P.B., Riou J.F., Mergny J.L., Tazi J., Sun J.S., Garestier T., Helene C. Interaction of human DNA topoisomerase I with G-quartet structures. Nucleic Acids Res. 2000; 28:4832–4838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Keller J.G., Hymoller K.M., Thorsager M.E., Hansen N.Y., Erlandsen J.U., Tesauro C., Simonsen A.K.W., Andersen A.B., Vandso Petersen K., Holm L.L. et al. Topoisomerase 1 inhibits MYC promoter activity by inducing G-quadruplex formation. Nucleic Acids Res. 2022; 50:6332–6342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Xiao S., Zhang J.Y., Zheng K.W., Hao Y.H., Tan Z. Bioinformatic analysis reveals an evolutional selection for DNA:RNA hybrid G-quadruplex structures as putative transcription regulatory elements in warm-blooded animals. Nucleic Acids Res. 2013; 41:10379–10390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Eddy J., Vallur A.C., Varma S., Liu H., Reinhold W.C., Pommier Y., Maizels N. G4 motifs correlate with promoter-proximal transcriptional pausing in human genes. Nucleic Acids Res. 2011; 39:4975–4983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Varshney D., Spiegel J., Zyner K., Tannahill D., Balasubramanian S. The regulation and functions of DNA and RNA G-quadruplexes. Nat. Rev. Mol. Cell Biol. 2020; 21:459–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bochman M.L., Paeschke K., Zakian V.A. DNA secondary structures: stability and function of G-quadruplex structures. Nat. Rev. Genet. 2012; 13:770–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chen J.N., He Y.D., Liang H.T., Cai T.T., Chen Q., Zheng K.W. Regulation of PDGFR-beta gene expression by targeting the G-vacancy bearing G-quadruplex in promoter. Nucleic Acids Res. 2021; 49:12634–12643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Xia Y., Zheng K.W., He Y.D., Liu H.H., Wen C.J., Hao Y.H., Tan Z. Transmission of dynamic supercoiling in linear and multi-way branched DNAs and its regulation revealed by a fluorescent G-quadruplex torsion sensor. Nucleic Acids Res. 2018; 46:7418–7424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Li D., Chen X., Yan R., Jiang Z., Zhou B., Lv B. G-quadruplex-containing oligodeoxynucleotides as DNA topoisomerase I inhibitors. Int. J. Biol. Macromol. 2022; 223:281–289. [DOI] [PubMed] [Google Scholar]
- 25. Shuai L., Deng M., Zhang D., Zhou Y., Zhou X. Quadruplex-duplex motifs as new topoisomerase I inhibitors. Nucleosides Nucleotides Nucleic Acids. 2010; 29:841–853. [DOI] [PubMed] [Google Scholar]
- 26. Das S.K., Kuzin V., Cameron D.P., Sanford S., Jha R.K., Nie Z., Rosello M.T., Holewinski R., Andresson T., Wisniewski J. et al. MYC assembles and stimulates topoisomerases 1 and 2 in a “topoisome”. Mol. Cell. 2022; 82:140–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhang L., Huang J., Ren L., Bai M., Wu L., Zhai B., Zhou X. Synthesis and evaluation of cationic phthalocyanine derivatives as potential inhibitors of telomerase. Bioorg. Med. Chem. 2008; 16:303–312. [DOI] [PubMed] [Google Scholar]
- 28. Zheng K.W., Zhang J.Y., He Y.D., Gong J.Y., Wen C.J., Chen J.N., Hao Y.H., Zhao Y., Tan Z. Detection of genomic G-quadruplexes in living cells using a small artificial protein. Nucleic Acids Res. 2020; 48:11706–11720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zheng K.W., Wu R.Y., He Y.D., Xiao S., Zhang J.Y., Liu J.Q., Hao Y.H., Tan Z. A competitive formation of DNA:RNA hybrid G-quadruplex is responsible to the mitochondrial transcription termination at the DNA replication priming site. Nucleic Acids Res. 2014; 42:10832–10844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zheng K.W., Chen Z., Hao Y.H., Tan Z. Molecular crowding creates an essential environment for the formation of stable G-quadruplexes in long double-stranded DNA. Nucleic Acids Res. 2010; 38:327–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nitiss J.L., Soans E., Rogojina A., Seth A., Mishina M. Topoisomerase assays. Curr. Protoc. Pharmacol. 2012; Chapter 3:Unit 3 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W. et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008; 9:R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hunter J.D. Matplotlib: a 2D graphics environment. Comput. Sci. Eng. 2007; 9:90–95. [Google Scholar]
- 34. Kaya-Okur H.S., Wu S.J., Codomo C.A., Pledger E.S., Bryson T.D., Henikoff J.G., Ahmad K., Henikoff S. CUT&Tag for efficient epigenomic profiling of small samples and single cells. Nat. Commun. 2019; 10:1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Del Mundo I.M.A., Zewail-Foote M., Kerwin S.M., Vasquez K.M. Alternative DNA structure formation in the mutagenic human c-MYC promoter. Nucleic Acids Res. 2017; 45:4929–4943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Fernando H., Reszka A.P., Huppert J., Ladame S., Rankin S., Venkitaraman A.R., Neidle S., Balasubramanian S. A conserved quadruplex motif located in a transcription activation site of the human c-kit oncogene. Biochemistry-Us. 2006; 45:7854–7860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Stump S., Mou T.C., Sprang S.R., Natale N.R., Beall H.D. Crystal structure of the major quadruplex formed in the promoter region of the human c-MYC oncogene. PLoS One. 2018; 13:e0205584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Dai J.X., Dexheimer T.S., Chen D., Carver M., Ambrus A., Jones R.A., Yang D.Z. An intramolecular G-quadruplex structure with mixed parallel/antiparallel G-strands formed in the human BCL-2 promoter region in solution. J. Am. Chem. Soc. 2006; 128:1096–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wang K.B., Dickerhoff J., Wu G.H., Yang D.Z. PDGFR-β promoter forms a vacancy G-quadruplex that can Be filled in by dGMP: solution structure and molecular recognition of guanine metabolites and drugs. J. Am. Chem. Soc. 2020; 142:5204–5211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhou B., Liu C.D., Geng Y.Y., Zhu G. Topology of a G-quadruplex DNA formed by hexanucleotide repeats associated with ALS and FTD. Sci. Rep. 2015; 5:16673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mukundan V.T., Phan A.T. Bulges in G-quadruplexes: broadening the definition of G-quadruplex-forming sequences. J. Am. Chem. Soc. 2013; 135:5017–5028. [DOI] [PubMed] [Google Scholar]
- 42. Li X.M., Zheng K.W., Zhang J.Y., Liu H.H., He Y.D., Yuan B.F., Hao Y.H., Tan Z. Guanine-vacancy-bearing G-quadruplexes responsive to guanine derivatives. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:14581–14586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Nguyen T.Q.N., Lim K.W., Phan A.T. Duplex formation in a G-quadruplex bulge. Nucleic Acids Res. 2020; 48:10567–10575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Subramani R., Juul S., Rotaru A., Andersen F.F., Gothelf K.V., Mamdouh W., Besenbacher F., Dong M., Knudsen B.R. A novel secondary DNA binding site in human topoisomerase I unravelled by using a 2D DNA origami platform. ACS Nano. 2010; 4:5969–5977. [DOI] [PubMed] [Google Scholar]
- 45. D’Annessa I., Coletta A., Sutthibutpong T., Mitchell J., Chillemi G., Harris S., Desideri A. Simulations of DNA topoisomerase 1B bound to supercoiled DNA reveal changes in the flexibility pattern of the enzyme and a secondary protein-DNA binding site. Nucleic Acids Res. 2014; 42:9304–9312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lau M.S., Hu Z., Zhao X., Tan Y.S., Liu J., Huang H., Yeo C.J., Leong H.F., Grinchuk O.V., Chan J.K. et al. Transcriptional repression by a secondary DNA binding surface of DNA topoisomerase I safeguards against hypertranscription. Nat. Commun. 2023; 14:6464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ambrus A., Chen D., Dai J., Bialis T., Jones R.A., Yang D Human telomeric sequence forms a hybrid-type intramolecular G-quadruplex structure with mixed parallel/antiparallel strands in potassium solution. Nucleic Acids Res. 2006; 34:2723–2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Dai J., Punchihewa C., Ambrus A., Chen D., Jones R.A., Yang D Structure of the intramolecular human telomeric G-quadruplex in potassium solution: a novel adenine triple formation. Nucleic Acids Res. 2007; 35:2440–2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Bao H.L., Liu H.S., Xu Y. Hybrid-type and two-tetrad antiparallel telomere DNA G-quadruplex structures in living human cells. Nucleic Acids Res. 2019; 47:4940–4947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Nitiss J.L., Kiianitsa K., Sun Y., Nitiss K.C., Maizels N. Topoisomerase assays. Curr. Protoc. 2021; 1:e250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Gobert C., Bracco L., Rossi F., Olivier M., Tazi J., Lavelle F., Larsen A.K., Riou J.F. Modulation of DNA topoisomerase I activity by p53. Biochemistry. 1996; 35:5778–5786. [DOI] [PubMed] [Google Scholar]
- 52. Goswami A., Qiu S., Dexheimer T.S., Ranganathan P., Burikhanov R., Pommier Y., Rangnekar V.M. Par-4 binds to topoisomerase 1 and attenuates its DNA relaxation activity. Cancer Res. 2008; 68:6190–6198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zheng K.W., Zhang D., Zhang L.X., Hao Y.H., Zhou X.A., Tan Z. Dissecting the strand folding orientation and formation of G-quadruplexes in single- and double-stranded nucleic acids by ligand-induced photocleavage footprinting. J. Am. Chem. Soc. 2011; 133:1475–1483. [DOI] [PubMed] [Google Scholar]
- 54. Promonet A., Padioleau I., Liu Y., Sanz L., Biernacka A., Schmitz A.L., Skrzypczak M., Sarrazin A., Mettling C., Rowicka M. et al. Topoisomerase 1 prevents replication stress at R-loop-enriched transcription termination sites. Nat. Commun. 2020; 11:3940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Manzo S.G., Hartono S.R., Sanz L.A., Marinello J., De Biasi S., Cossarizza A., Capranico G., Chedin F. DNA topoisomerase I differentially modulates R-loops across the human genome. Genome Biol. 2018; 19:100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Pommier Y. DNA topoisomerase I inhibitors: chemistry, biology, and interfacial inhibition. Chem. Rev. 2009; 109:2894–2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Wu T., Lyu R., You Q., He C. Kethoxal-assisted single-stranded DNA sequencing captures global transcription dynamics and enhancer activity in situ. Nat. Methods. 2020; 17:515–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kim J.J., Lee S.Y., Gong F., Battenhouse A.M., Boutz D.R., Bashyal A., Refvik S.T., Chiang C.M., Xhemalce B., Paull T.T. et al. Systematic bromodomain protein screens identify homologous recombination and R-loop suppression pathways involved in genome integrity. Genes Dev. 2019; 33:1751–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wang K.B., Elsayed M.S.A., Wu G., Deng N., Cushman M., Yang D. Indenoisoquinoline topoisomerase inhibitors strongly bind and stabilize the MYC promoter G-quadruplex and downregulate MYC. J. Am. Chem. Soc. 2019; 141:11059–11070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Cushman M. Design and synthesis of indenoisoquinolines targeting topoisomerase I and other biological macromolecules for cancer chemotherapy. J. Med. Chem. 2021; 64:17572–17600. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Original sequencing (fastq) and processed (bigwig) G4P-ChIP datasets, and CUT&Tag datasets for R-loop have been deposited in and can be downloaded from the NCBI Gene Expression Omnibus (GEO) under accession code GSE239694.