Abstract
Pediatric high-grade gliomas (pHGG) are devastating and incurable brain tumors with recurrent mutations in histone H3.3. These mutations promote oncogenesis by dysregulating gene expression through alterations of histone modifications. We identify aberrant DNA repair as an independent mechanism, which fosters genome instability in H3.3 mutant pHGG, and opens new therapeutic options. The two most frequent H3.3 mutations in pHGG, K27M and G34R, drive aberrant repair of replication-associated damage by non-homologous end joining (NHEJ). Aberrant NHEJ is mediated by the DNA repair enzyme polynucleotide kinase 3′-phosphatase (PNKP), which shows increased association with mutant H3.3 at damaged replication forks. PNKP sustains the proliferation of cells bearing H3.3 mutations, thus conferring a molecular vulnerability, specific to mutant cells, with potential for therapeutic targeting.
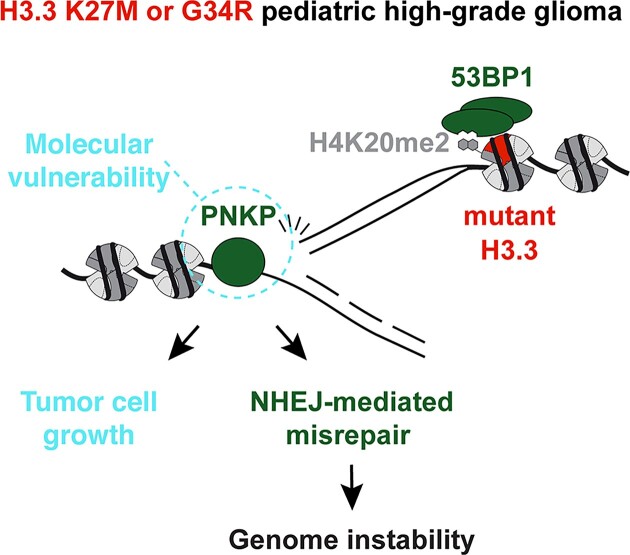
Graphical Abstract
Graphical Abstract.
Introduction
Pediatric high-grade gliomas (pHGGs) are deadly brain tumors with less than ten percent survival within two years of diagnosis. Despite aggressive radio/chemotherapy regimens, pHGGs remain incurable and are the leading cause of cancer-related death in children (1). The toxicity of existing therapies and the emergence of resistance hinder the efficacy of current therapeutic protocols and call for the development of alternative, targeted therapeutic strategies that exploit specific molecular features of cancer cells (1,2). One such feature is exemplified by heterozygous, dominant point mutations in the H3F3A gene, which encodes the H3.3 histone variant. These recurrent mutations are active players in the oncogenic process in pHGG (3), with three mutually exclusive mutations resulting in single amino-acid substitutions at lysine 27 to methionine (H3.3 K27M), giving rise to midline tumors of the central nervous system, or at glycine 34 to arginine or valine (H3.3 G34R/V) in cerebrocortical tumors (4–6). These mutations perturb histone post-translational modifications (PTMs) in cis (7,8) or in trans (9,10), thus interfering with genome-wide gene expression programs, stalling the differentiation of interneuron progenitor cells at different stages (11–14) and fueling cell transformation (reviewed in (3,15,16)).
Besides their effect on gene expression, pHGG H3.3 mutations also promote genome instability (4,6,7,17–20), which is one of the major drivers of cellular transformation (21). In particular, H3.3 mutant pHGGs display increased copy number alterations and chromosomal rearrangements compared to wild-type (WT) H3.3 tumors (4,17,22). Mechanistically, H3 G34 mutations in fission yeast and mammals and the non-pHGG H3.3 K36M mutation in human cells hijack the response to DNA damage (7,10,19,20,23) by dominantly interfering with the DNA repair-promoting function of WT H3.3 (24,25) or by downregulating DNA damage response genes (18). Moreover, K27M and G34 mutations alter H3.3 association with several DNA repair factors in human cells (7,26,27). These observations expand the role of mutated H3.3 proteins beyond their ability to dysregulate gene expression and pave the way for the identification of additional oncogenic functions. However, mechanistic understanding of the impact of pHGG H3.3 mutations on DNA repair and genome integrity in human cells is still lacking, and whether and how this function can be exploited for therapy remains underexplored.
Materials and methods
Human cell lines
U2OS (human osteosarcoma, female) and HeLa cells (human cervical carcinoma, female) were cultured in Dulbecco's modified Eagle's medium DMEM Gluta-Max (Life Technologies) supplemented with 10% foetal bovine serum (Eurobio) and antibiotics (100 U/ml penicillin, 100 μg/ml streptomycin, Life Technologies) and maintained at 37°C under 5% CO2 in a humified incubator. U2OS cells stably expressing SNAP-tagged H3.1 or H3.3 wild-type or mutant, and U2OS LacO H3.3-SNAP cells with integrated 256 tandem LacO repeats and stably expressing SNAP-tagged wild-type H3.3 (28) were cultured in the same medium supplemented with 100 μg/ml G418 (Life Technologies). Flp-In T-REx HEK293 cells were cultured in DMEM (Gibco), supplemented with 10% foetal bovine serum (Wisent) and antibiotics (100 U/ml penicillin, 100 μg/ml streptomycin, Life Technologies), at 37°C in 5% CO2. SF9402 and SF9427 (wild-type H3.3) cell lines were from (29). SU-DIPG-XVII (H3.3 K27M) were from (30). HSJD-002-GBM cells (H3.3 G34R) were from A. Carcaboso (Institut de Recerca Sant Joan de Deu, Barcelona, Spain). SF9402, SF9427, SU-DIPG-XVII and HSJD-002-GBM were cultured in Tumor Stem Medium (31), which contains DMEM/F12 1:1 (Invitrogen), Neurobasal-A (Invitrogen), 10 mM HEPES (Invitrogen), 1 × MEM sodium pyruvate (Invitrogen), 1× MEM non-essential amino acids (Invitrogen), 1% GlutaMax (Invitrogen), 20 ng/ml human basic fibroblast growth factor (CliniSciences), 20 ng/ml human epidermal growth factor (CliniSciences), 20 ng/ml human platelet-derived growth factor (PDGF)-A and PDGF-B (CliniSciences), 10 ng/ml heparin (StemCell Technologies) and 1× B27 without Vitamin A (Invitrogen). pHGG lines were generally grown in suspension flasks as tumorspheres, except when they underwent transfection and proliferation assay, for which they were dissociated and plated on plates coated with laminin (10 μg/ml, Sigma-Aldrich). MGBM1 cells (H3.3 G34R) were cultured in DMEM Gluta-Max supplemented with 10% foetal bovine serum and antibiotics (100 U/ml penicillin, 100 μg/ml streptomycin, Life Technologies) (32). All glioma cells were maintained at 37°C under 5% CO2 in a humified incubator and verified for expression of the expected H3.3 proteins by Western blot analysis with antibodies raised against H3.3 K27M or G34R (Supplementary Table S1). No further cell line authentication was performed. Non-cancerous astrocytes CRL-8621 (ATCC) were grown in Eagle's minimum essential medium (EMEM, ATCC 30-2003) supplemented with 10% foetal bovine serum (Eurobio) and antibiotics (100 U/ml penicillin, 100 μg/ml streptomycin, Life Technologies).
Generation of U2OS stable cell lines
U2OS cells stably expressing C-terminal, SNAP-tagged H3.3, either wild-type, K27M, G34R, G34V, G34W or K36M mutated were generated by transfection of plasmid encoding wild-type or mutated H3.3 and selection of clones in limiting dilution in medium supplemented with G418 (Life Technologies) starting 48 hours after transfection. To verify the presence of mutations in the clones, genomic DNA was extracted and subjected to PCR amplification with dedicated primers (Supplementary Table S1). The amplification product was verified by Sanger sequencing (GATC Biotech). Single clones harboring each H3.3 mutation were expanded and evaluated for levels of expression of the exogenous SNAP-tagged H3.3 proteins and for the presence of histone PTM alterations described in tumor samples.
Yeast strains
Schizosaccharomyces pombe strains (Supplementary Table S1) containing point mutations in histone H3, K27M in hht2+, G34R and G34V in hht3+, were generated by a PCR-based module method. pnk1Δ and xrc4Δ strains were derived from the fission yeast deletion library and the gene deletions were verified by PCR. All other strains were constructed through genetic crosses. For serial dilution plating assays (spot assays), ten-fold dilutions of a mid-log phase culture were plated on the indicated medium and grown for 3 days at 30°C. For proliferation assays, overnight liquid S. pombe cultures were grown to saturation in YES media (yeast extract with supplement). Saturated cultures were equilibrated to an OD600 of 1.0, arrayed in a 96-well microtiter plate, and pinned in quadruplicate to achieve a 384-colony density (i.e. 4 technical replicates for each position of the microtiter plate) using a Singer RoToR robot (Singer Instruments, Inc., Somerset, UK). Strains were grown on YES solid agar media with the indicated camptothecin concentrations. Plates with pinned colonies were incubated at 30°C and scanned every 96 min for growth curves. Colony density was measured using screenmill (33) and the values of the four technical replicates were averaged. Care was taken to minimize systematic bias in experiments (e.g. by distributing strains evenly throughout the 96-well plate to minimize position and neighboring strain effects).
Drug treatments and inhibitors
Camptothecin (CPT) was used at 0.1 μM for 3 h, or at 1 μM for 1 or 3 h for iPOND in human cells, and at 5 or 10 μM in yeast cells; hydroxyurea (HU) at 2 mM for 3 h; mitomycin C (MMC) at 200 ng/ml for 24 h and 25 ng/ml for 48 h for repair foci analyses and metaphase spreads, respectively; bleomycin (Bleo) at 20 μg/ml for 3 h. The PARP inhibitor Olaparib (AZD2281) was used at the indicated concentrations. An overnight treatment with 2 mM thymidine followed by 3 h release in fresh medium was used to enrich cells in S phase for iPOND experiments (70–75% of cells were in S phase as evaluated by FACS). The EZH2 inhibitor GSK126 (EZH2i) was used at 1 μM for 72 h. The SUV4-20H1/2 inhibitor A196 was used at 2.5 μM for 48 h.
Site-directed mutagenesis
The H3F3A and H3F3B human cDNA sequences (GenScript) were cloned by using ClaI and EcoRI restriction enzymes into the pSNAPm plasmid (New England Biolabs), with the SNAP tag in the C-terminus of the insert. These plasmids were subjected to directed mutagenesis to introduce the cancer-associated mutations K27M, G34R/V/W and K36M (4,5,34). GFP-PNKP kinase (K378M) and phosphatase (D171N) mutants were generated by GenScript from GFP-PNKP wild-type plasmid (Supplementary Table S1). Generation of the mutated plasmids was verified by Sanger sequencing (GATC Biotech).
Immunofluorescence, image acquisition and analysis
Cells grown on glass coverslips (Thorlabs) were either fixed directly with 2% paraformaldehyde (PFA) and permeabilized with 0.2% Triton X-100 in PBS or pre-extracted before fixation with 0.5% Triton X-100 in CSK buffer (Cytoskeletal buffer: 10 mM PIPES pH 7.0, 100 mM NaCl, 300 mM sucrose, 3 mM MgCl2) to remove soluble proteins (not bound to chromatin) and then fixed with 2% PFA. Samples were blocked in 5% bovine serum albumin (BSA, Sigma-Aldrich) in PBS supplemented with 0.1% Tween 20 (Euromedex) before incubation with primary antibodies and secondary antibodies conjugated to Alexa Fluor 488 or 568 (Invitrogen). Coverslips were mounted in Vectashield medium with DAPI (Vector Laboratories) and observed with a Leica DMI6000 epifluorescence microscope using a Plan-Apochromat 40×/1.3 or 63×/1.4 oil objective. Images were captured using a CCD camera (Photometrics) and Metamorph software. Images were mounted with Adobe Photoshop applying the same treatment of fluorescence levels to all images from the same experiment. Fiji software was used for image analyses using custom macros. Nuclei were delineated based on DAPI staining, and CPT-damaged replication forks based on γH2A.X staining. S phase, replicating cells were discriminated based on EdU staining. The position of the LacO array was determined based on mCherry-LacR signal. DNA repair and PLA foci were identified and counted by using the find maxima function (Fiji software), on the best focal plane for DNA repair foci and on maximum intensity z-projections in the case of PLA foci. At least 70 cells/sample were scored in each experiment. Results of automatic foci counting were graphed as number of foci per cell or as number of cells with more than 5 or 10 DNA repair foci that was set as a threshold.
Ethynyl-deoxyUridine (EdU) labeling of S phase cells
For discrimination of S phase cells, 10 μM 5-ethynyl-2′-deoxyUridine (EdU, Sigma-Aldrich) was incorporated into cells for 15 min prior to DNA damage treatment and fixation. EdU was revealed using Click-It EdU Imaging kit (Invitrogen) according to manufacturer's instructions.
Random plasmid integration assay
Cells grown in 6-well plates were transfected with siRNAs and, later the same day, the cells were transfected with 2 μg/well gel-purified FspI-BspDI-linearized pEGFP-C1-IRES-puro plasmid. The cells were transfected once more with siRNAs the following day. Cells were collected 48 h later, counted and seeded in 10 cm diameter dishes either lacking or containing 0.375 μg/ml puromycin. The transfection efficiency was determined on the same day by FACS analysis of EGFP-positive cells. The cell dishes were incubated at 37°C to allow colony formation and medium was refreshed on days 4 and 8. On days 10–12, the cells were stained with 0.5% Crystal Violet (Sigma-Aldrich)/20% ethanol solution to score colonies with >50 cells. Random plasmid integration events on the puromycin-containing plates were normalized to the plating efficiency (plate without puromycin) and to the transfection efficiency.
Colony forming assays
Cells seeded at low density were exposed to genotoxic treatment at the indicated concentrations (acute treatment of 1 h for CPT and chronic treatment for PARPi). Colonies were stained 12 days later with 0.5% crystal violet/20% ethanol and counted. Results were normalized to plating efficiencies (untreated cells).
Metaphase spreads
To prepare metaphase spreads, Colcemid (Gibco) was added to the culture medium at 0.1 μg/ml for 3 h before collecting the cells. Cells were washed in PBS, trypsinized, centrifuged at 1200 rpm for 5 min and resuspended in 75 mM KCl for 15 min at 37°C. Cells were then fixed with fresh ethanol/acetic acid (v:v = 3:1) at –20°C overnight. The next day, cells were centrifuged at 1200 rpm for 5 min and resuspended in fresh fixative before dropping onto slides and then air dried. Slides were mounted with Vectashield Antifade Mounting Medium containing DAPI (Vector laboratories) and metaphase spreads were examined using an Axio Observer Z1 epifluorescence microscope equipped with an ORCA-ER camera (Hamamatsu). Mitomycin (MMC) was added at a concentration of 25 ng/ml for 48 h before harvesting cells for metaphase spread preparation. At least 30 metaphase spreads were scored per sample in each experiment for the presence of radial chromosomes.
Mutational signature analysis on primary pHGG samples
pHGG samples for single nucleotide variant (SNV) mutational signature analysis were acquired from previously published data available under EGAS00001000575, EGAS00001001139, EGAS00001000572 and EGAS00001000192. Novel data was generated from samples obtained from the DIPG-BATs clinical trial (NCT01182350), the Dana-Farber Tissue Bank or collaborating institutions, under protocols approved by the institutional review board of the Dana-Farber/Harvard Cancer Center with informed consent (DFCI protocols 10417, 10201 and DFCI 19293). DNA was extracted from single Diffuse Midline Glioma cores, pHGG biopsies and autopsy samples using Qiagen AllPrep DNA/RNA extraction kits. For whole-genome sequencing, genomic DNA was fragmented and prepared for sequencing to 60× depth on an Illumina HiSeq 2000 instrument. Reads from both novel and published data were aligned to the reference genome hg19/ GRCh37 with BWA83, duplicate-marked, and indexed using SAMtools and Picard. Base quality score was bias adjusted for flowcell, lane, dinucleotide context, and machine cycle and recalibrated, and local realignment around insertions or deletions (indels) was achieved using the Genome Analysis Toolkit. SNV signature analysis was performed using Palimpsest (https://github.com/FunGeST/Palimpsest) on a VCF containing somatic mutations identified by Mutect2.
Extraction of cellular proteins and western blot analysis
Total extracts were obtained by scraping cells in Laemmli buffer (50 mM Tris–HCl pH 6.8, 1.6% sodium dodecyl sulfate (SDS), 8% glycerol, 4% β-mercaptoethanol, 0.0025% bromophenol blue) followed by 5–10 min denaturation at 95°C. For western blot analysis, extracts along with molecular weight markers (Precision plus protein Kaleidoscope standards, Bio-Rad) were run on 4–20% Mini-PROTEAN TGX gels (Bio-Rad) in running buffer (200 mM glycine, 25 mM Tris, 0.1% SDS) and transferred onto nitrocellulose membranes (Protran) with a Trans-Blot SD semi-dry or wet transfer cell (Bio-Rad). Proteins of interest were probed using the appropriate primary and horse radish peroxidase (HRP)-conjugated secondary antibodies (Jackson Immunoresearch), detected using SuperSignal West Pico or Femto chemiluminescence substrates (Pierce). The resulting signal was visualized on hyperfilms MP (Amersham) with a film processor (SRX105, Konica) or using the Odyssey Fc-imager (LI-COR Biosciences). ImageJ and Image Studio Lite softwares were used for densitometric quantification of protein bands.
Quantification of mutant histones by mass spectrometry
Total extracts from U2OS cells expressing SNAP-tagged H3.3, wild-type or mutant, were loaded on 4–20% Mini-PROTEAN TGX gels (Bio-Rad). Histone and SNAP-tagged histone pieces were excised from the gel after Coomassie staining. Gel pieces containing histones were washed with 100 mM ammonium bicarbonate, dehydrated with acetonitrile, chemically propionylated with propionic anhydride and digested overnight with trypsin. Tryptic peptides were extracted sequentially with 70% acetonitrile/0.25% TFA and acetonitrile, filtered using C8-StageTips, vacuum concentrated and reconstituted in 15 μl of 0.1% FA.
For LC–MS/MS purposes, desalted peptides were injected in an Ultimate 3000 RSLCnano system (Thermo) and separated in a 25-cm analytical column (75 μm ID, 1.6 μm C18, IonOpticks) with a 50-min gradient from 2 to 37% acetonitrile in 0.1% formic acid. The effluent from the HPLC was directly electrosprayed into a Qexactive HF (Thermo) operated in data dependent mode to automatically switch between full scan MS and MS/MS acquisition with the following parameters: survey full scan MS spectra (from m/z 375–1600) were acquired with resolution R = 60 000 at m/z 400 (AGC target of 3 × 106). The 10 most intense peptide ions with charge states between 2 and 5 were sequentially isolated to a target value of 1 × 105, and fragmented at 27% normalized collision energy. Typical mass spectrometric conditions were: spray voltage, 1.5 kV; no sheath and auxiliary gas flow; heated capillary temperature, 250ºC; ion selection threshold, 33 000 counts.
Data analysis was performed with the Skyline (version 21.2) by using doubly and triply charged peptide masses for extracted ion chromatograms. Automatic selection of peaks was manually curated based on the relative retention times and fragmentation spectra with results from Proteome Discoverer 1.4. Integrated peak values were exported for further calculations. The relative abundance of an observed modified peptide was calculated as percentage of the overall peptide. H3.1 and H3.3 27–40 peptides were used for the quantification of mutant H3.3 over total H3.
GFP pull-down
U2OS cells transiently expressing GFP or GFP-PNKP were collected in PBS 2 days after plasmid transfection (two 10 cm diameter plates per condition). Cell pellets were resuspended in 1 ml ice-cold lysis buffer (50 mM Tris pH 7.5, 150 mM NaCl, 0.5% NP-40, 2.5 mM MgCl2, Roche EDTA-free Protease inhibitor and PhosStop phosphatase inhibitor) supplemented with benzonase (0.5 U/μl final, Merck Millipore) and samples were incubated for 1 h at 4°C under constant mixing. The cell lysates were cleared by centrifugation at full speed for 10 min at 4°C and incubated overnight at 4°C with 15 μl of pre-washed GFP-Trap agarose beads (ChromoTek) in lysis buffer. Input samples were collected from cell lysates before the addition of beads. After five washes in lysis buffer adjusted to 250 mM NaCl, beads were boiled for 5 min at 95°C in Laemmli buffer. Pull-down and input samples were analyzed by western blot.
siRNA and plasmid transfections
siRNAs purchased from Eurofins MWG Operon or Sigma-Aldrich (Supplementary Table S1) were transfected into cells using Lipofectamine RNAiMAX (Invitrogen) following manufacturer's instructions. Cells were analyzed and/or harvested 48–72 h post-transfection except for proliferation assays, where cells were analyzed over a 7-day period after transfection. Cells were transfected with recombinant plasmid DNA (Supplementary Table S1) using Lipofectamine 2000 (Invitrogen) according to manufacturer's instructions. For complementation experiments, cells were concomitantly transfected with siRNA targeting PNKP 3′UTR (50 nM final) and plasmid DNA (0.5 μg/ml final) using Lipofectamine 2000 (Invitrogen) according to manufacturer's instructions.
SNAP labeling of newly synthesized histones
For labeling newly synthesized SNAP-tagged histones (35), parental histones were quenched with 10 μM SNAP-cell Block (NEB) for 30 min in culture medium followed by 30-min wash in fresh medium and a 2-h chase. To mark S phase cells/replication forks, EdU was incorporated for 30 min at the end of the chase period. The new SNAP-tagged histones synthesized during the chase were fluorescently labelled with 4 μM of the green-fluorescent reagent SNAP-cell Oregon green (New England Biolabs) during a 15-min pulse step followed by 30-min wash in fresh medium. Alternatively, when combined with Proximity Ligation Assay (PLA), cells were pre-extracted using 0.5% Triton X-100 in cytoskeleton (CSK) buffer and fixed in 2% paraformaldehyde after the quench-chase steps. New SNAP-tagged histones were then pulse-labeled for 30 min with 5 μM final SNAP-biotin (New England Biolabs) diluted 1:200 in 10% Duolink blocking buffer (Sigma-Aldrich) in PBS. After washings, soluble proteins were removed by permeabilization with 0.5% Triton X-100 in cytoskeleton (CSK) buffer, and cells were fixed and processed for immunostaining or PLA.
Proximity ligation assay (PLA)
PLA (36) was performed to detect colocalization foci between PNKP and γH2A.X or between newly synthesized H3.3-SNAP and γH2A.X at camptothecin-damaged replication forks. The Duolink® In Situ PLA® detection kit (Sigma) was used following manufacturer's recommendations. Briefly, cells on glass coverslips (Thorlabs) were incubated 1 h at 37°C in Duolink blocking buffer (Sigma-Aldrich) and then with primary antibodies directed against the target proteins (anti-PNKP, anti-biotin to detect new H3.3-SNAP-biotin and anti-γH2A.X to detect sites of DNA damage) diluted in antibody dilution reagent (Sigma-Aldrich). Anti-biotin and anti-γH2A.X were incubated for 1 h at room temperature and anti-PNKP overnight at 4°C. Coverslips were then incubated for 1 h at 37°C with secondary antibodies each harboring a PLA probe (Duolink In Situ PLA MINUS/PLUS probes, Sigma-Aldrich). The PLA probes that bind to the constant regions of the primary antibodies contain a unique DNA strand. If the proteins of interest interact with each other, the DNA probes hybridize to make circular DNA during the 30 min ligation step at 37°C. The resulting circle DNA can be amplified (1 h 40 min amplification at 37°C, Duolink In Situ Detection Reagents Green, Sigma-Aldrich) and visualized by fluorescently labeled complementary oligonucleotide probes incorporation. Coverslips were mounted in Duolink In Situ Mounting Medium with DAPI (Sigma-Aldrich). To study PLA foci in S phase cells, EdU labeling by click chemistry was performed before the blocking step.
Isolation of proteins on nascent DNA (iPOND)
iPOND was performed largely as described previously (37), with the following modifications. A total of 3 × 107 logarithmically growing cells per sample were labeled with 10 μM EdU for 15 min. Following EdU incorporation, cells were treated or not with camptothecin, fixed with 1% formaldehyde for 15 min at room temperature, followed by 5-min incubation with 0.125 M glycine to quench the formaldehyde. Cells were harvested by scraping, washed three times with PBS, flash frozen in liquid nitrogen and kept at −80°C. Within 2 weeks, samples were processed for EdU-based pulldown and purification of replication fork-associated proteins. Briefly, click chemistry reactions were performed on pre-permeabilized samples to conjugate biotin to the EdU-labeled DNA by using Biotin Picolyl azide (Sigma Aldrich). Sonication was performed with a Bioruptor Pico sonicator (Diagenode) and DNA shearing was evaluated on an agarose gel. Shearing for optimal detection of the proteins of interest was set to an average DNA fragment size of 800 bp. Total input samples were taken after sonication and clearing of samples and kept at –20°C until loading on SDS-PAGE gels. Streptavidin beads (Dynabeads MyOne Streptavidin-C1, Life technologies) were used to capture the biotin-conjugated DNA–protein complexes. Captured complexes were washed extensively using SDS and high-salt wash buffers. Purified replication fork proteins were eluted under reducing conditions by boiling in Laemmli sample buffer for 5 min. Total input and capture samples corresponding to equal amounts of cells were resolved on SDS-PAGE gels and analyzed by western blot.
Flow cytometry and cell cycle analysis
Cells were fixed in ice-cold 70% ethanol before DNA staining with 50 μg/ml propidium iodide (Sigma-Aldrich) in PBS containing 0.05% Tween 20 and 0.5 mg/ml RNase A (USB/Affymetrix). DNA content was analyzed by flow cytometry using a FACSCalibur Flow Cytometer (BD Biosciences) and FlowJo Software (TreeStar).
Proximity biotinylation
Flp-In T-REx HEK293 cells were induced for 24 h with 1 μg/ml doxycycline and 50 μM biotin and snap frozen before lysis in 10 ml lysis buffer (50 mM Tris–HCl pH 7.5, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% Triton X-100, 0.1% SDS, 1:500 protease inhibitor cocktail (Sigma-Aldrich), 1:1000 benzonase nuclease (Novagen)). Samples were rotated at 4°C for 1 h, briefly sonicated and centrifuged at 45 000 × g for 30 min at 4°C. Cleared supernatants were added to 30 μl of packed, pre-equilibrated Streptavidin sepharose beads (GE Healthcare) and incubated for 3 h at 4°C with end-over-end rotation. Beads were centrifuged at 2000 rpm for 2 min, and in a fresh tube washed twice with 1 ml of lysis buffer and twice with 1 ml of 50 mM ammonium bicarbonate pH 8.3. Beads were transferred in ammonium bicarbonate to a fresh centrifuge tube and washed two more times. On-bead tryptic digestion was performed with 1 μg MS-grade TPCK trypsin (Promega, Madison, WI) dissolved in 200 μl of 50 mM ammonium bicarbonate pH 8.3 overnight at 37°C. The following morning, 0.5 μg MS-grade TPCK trypsin was added, and beads were incubated two additional hours at 37°C. Beads were pelleted by centrifugation at 2000 × g for 2 min, and the supernatant was transferred to a fresh Eppendorf tube. Beads were washed twice with 150 μl of 50 mM ammonium bicarbonate, and the washes were pooled with the first eluate. The sample was lyophilized and resuspended in buffer A (0.1% formic acid). 1/5th of the sample was analyzed per mass spectrometry run.
Mass spectrometry was performed and analyzed as previously described (38). Briefly, high performance liquid chromatography was conducted using a 2 cm pre-column (Acclaim PepMap 50 mm × 100 μm inner diameter (ID)), and 50 cm analytical column (Acclaim PepMap, 500 mm × 75 μm diameter; C18; 2 μm; 100 Å, Thermo Fisher Scientific, Waltham, MA), running a 120 min reversed-phase buffer gradient at 225 nl/min on a Proxeon EASY-nLC 1000 pump in-line with a Thermo Q-Exactive HF quadrupole-Orbitrap mass spectrometer. A parent ion scan was performed using a resolving power of 60 000, then up to the 20 most intense peaks were selected for MS/MS (minimum ion count of 1000 for activation) using higher energy collision induced dissociation (HCD) fragmentation. Dynamic exclusion was activated such that MS/MS of the same m/z (within a range of 10 ppm; exclusion list size = 500) detected twice within 5 s were excluded from analysis for 15 s. For protein identification, Thermo .RAW files were converted to the .mzXML format using Proteowizard (39), then searched using X!Tandem (40) and COMET (41) against the human Human RefSeq Version 45 database (containing 36113 entries). Data were analyzed using the trans-proteomic pipeline (TPP) (42,43) via the ProHits software suite (v3.3) (44). Search parameters specified a parent ion mass tolerance of 10 ppm, and an MS/MS fragment ion tolerance of 0.4 Da, with up to two missed cleavages allowed for trypsin. Variable modifications of +16@M and W, +32@M and W, +42@N-terminus and + 1@N and Q were allowed. Proteins identified with an iProphet cut-off of 0.9 (corresponding to ≤1% FDR) and at least two unique peptides were analyzed with SAINT Express v.3.3.1. Ten control runs (from cells expressing the FlagBirA* epitope tag) were collapsed to the two highest spectral counts for each prey and compared to the two biological and two technical replicates of histone BioID. High confidence interactors were defined as those with Bayesian false discovery rate (BFDR) ≤0.01.
Human cell proliferation assays
The effect of PNKP knockdown on cell proliferation in human cells was measured as follows: 24 h after siRNA transfection, cells were seeded in 60-mm diameter tissue culture plates (20 000 cells/plate for U2OS, 40 000 to 80 000 for pHGG cells). pHGG cell lines were transfected with siRNAs twice, both 48 and 24 h before seeding. Cell viability was assessed after 3, 5 and 7 days in culture by staining with trypan blue (Invitrogen) or counting with an automated cell counter for viable cells (Fluidlab R-300 cell counter, Anvajo).
Quantification and statistical analysis
Statistical analyses were carried out using Graphpad Prism software. P values for mean comparisons between two groups were calculated with a Student's t-test with Welch's correction when necessary. Multiple comparisons were performed by one- or two-way ANOVA with Bonferroni, Tukey's, Holm–Sidak or Dunnett's post-tests or using the non-parametric Kruskal–Wallis's test in case of non-Gaussian distributions. Comparisons of proliferation curves were based on non-linear regression with a polynomial quadratic model or exponential decay model (human cells) or with a sigmoidal dose response model with variable slope (yeast cells). ns: non-significant, ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001, ****P< 0.0001. Statistical parameters including sample size (n) and dispersion of the data (SD or SEM) are indicated in the figure legends, as also analysis details. Figures were prepared with prism.
Results
H3.3 mutants drive aberrant DNA repair in S phase
To study the impact of various H3.3 mutations on DNA repair in human cells in an isogenic context, we generated U2OS cell lines stably expressing SNAP-tagged WT or individual mutant H3.3 proteins (bearing K27M, G34R/V pHGG mutations, and G34W, K36M non-pHGG mutations) in an H3.3 WT background (Supplementary Figure S1A). The engineered cell lines have comparable expression of the different H3.3-SNAP proteins (Supplementary Figure S1B, C). They also recapitulate some of the histone PTM changes (H3K27me3 and H3K36me3) and the mutant H3.3 to WT H3 ratio, around 2–4%, that characterize H3.3 mutant pHGGs (9,10) (Supplementary Figure S1B-D). In this system, we analyzed the focal accumulation of DNA repair factors upon treatment with different genotoxic agents. Similar to the positive control H3.3 K36M (10), upon replication stress, two pHGG mutants, H3.3 K27M and G34R, showed impaired foci formation of the RAD51 recombinase (RAD51) (Figure 1A), and also, to a lesser extent, of Fanconi Anemia Complementation Group D2 (FANCD2)(Supplementary Figure S2A), both of which are involved in pathways that preferentially repair replication-associated DNA damage (45). In addition to impaired FANCD2 focus formation in the mutants, we observed a modest reduction of the ratio between ubiquitylated and non-ubiquitylated FANCD2 in cells expressing H3.3 G34R (Supplementary Figure S2B), which likely results in impaired retention of FANCD2 on chromatin and suggests an upstream defect in the Fanconi Anemia repair pathway. We evaluated a possible compensatory activation of the non-homologous end joining (NHEJ) repair pathway in the same cell lines and observed increased foci formation of TP53-binding protein 1 (53BP1), a positive regulator of NHEJ (46), in cells expressing H3.3 K27M, G34R or the non-pHGG G34W oncomutant (Figure 1B). The altered recruitment of repair factors was significant upon interference with replication fork (RF) progression (47) by camptothecin (CPT), hydroxyurea (HU) or mitomycin C (MMC), but not upon treatment with the radiomimetic agent bleomycin, which triggers DNA damage throughout the cell cycle (Figure 1A,B, Supplementary Figure S2A, C, D), indicating that H3.3 K27M and G34R mutants skew the repair of RF-associated DNA lesions. Importantly, the observed defect is not due to a different cell cycle distribution of cells expressing mutant H3.3 (Supplementary Figure S2E), to differential signaling of DNA damage, as shown by comparable levels of γH2A.X (Supplementary Figure S2F), nor to differential abundances of repair proteins (Supplementary Figure S2G, H). To functionally analyze NHEJ activity, we exploited the random plasmid integration assay, which confirmed increased NHEJ activity in cells expressing the pHGG mutants H3.3 K27M and G34R, but not G34V (Figure 1C, Supplementary Figure S2I). Functional differences between the G34R and G34V histone H3 mutants in DNA damage repair were also observed in fission yeast (20,23), a highly amenable model system exploited in DNA repair studies, albeit showing a different repair pathway balance as compared to higher eukaryotes. We used this model system to assess the possible conservation of the increased, aberrant NHEJ activity observed in cells expressing pHGG mutant histones. Fission yeast contains three identical histone H3 genes and we introduced the K27M, G34R and G34V mutations into one of the H3 genes, while leaving the other two intact. Proliferation analyses showed that deletion of the core NHEJ factor Xrc4 (xrc4Δ) sensitized H3 WT and G34V strains to CPT damage, consistent with a protective role of NHEJ in these strains, which was not observed in K27M and G34R strains (Figure 1D). xrc4Δ even rescued the CPT sensitivity of G34R yeast cells, as observed both in proliferation and serial dilution plating assays, indicating that aberrant NHEJ drives the sensitivity of this strain to CPT. While yeast strains expressing pHGG mutant histones, particularly the G34R mutant strain, are hypersensitive to CPT treatment compared to the WT strain (Figure 1D), this feature is not shared by human cells expressing H3.3 pHGG mutants (Supplementary Figure S2J). Building on the observed RAD51 focus formation defect in H3.3 K27M or G34R human U2OS cells, we investigated their sensitivity to PARP inhibition (48). In line with the RAD51 focus formation defect, H3.3 K27M or G34R but not G34V expressing cells exhibited mild hypersensitivity to PARP inhibition, albeit not to the same extent as H3.3 K36M mutant cells (Supplementary Figure S2K). Together, these findings illustrate that aberrant DNA repair in H3.3 K27M and G34R cells is associated with specific patterns of sensitivity to genotoxic stress.
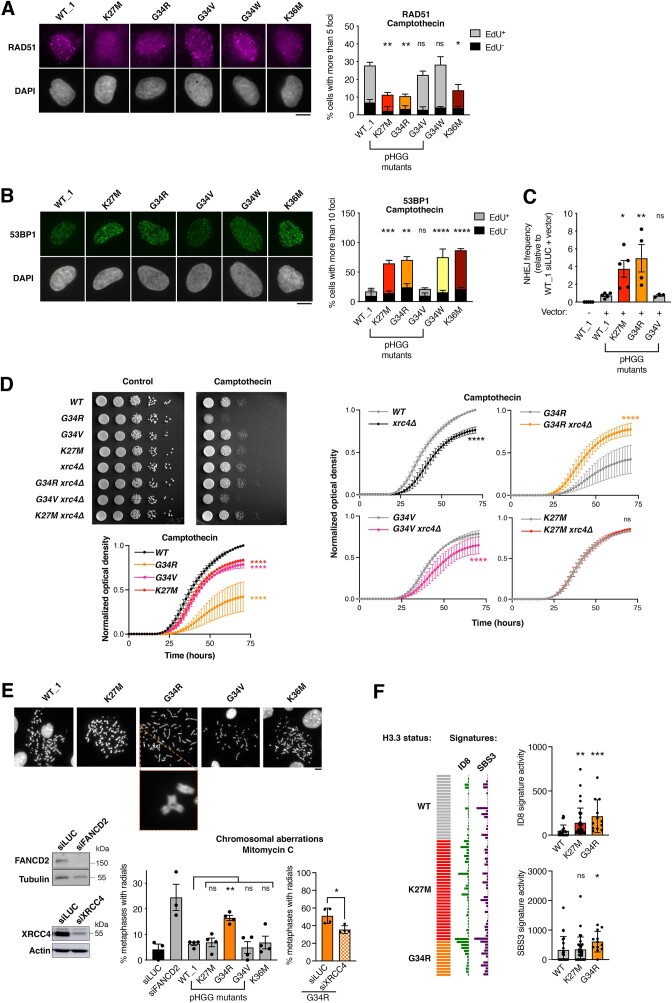
Figure 1.
H3.3 K27M and G34R pHGG mutations hijack DNA repair in S phase and harbor genomic instability features of aberrant NHEJ. (A, B) Analysis of RAD51 (A) and 53BP1 (B) repair foci by immunofluorescence in U2OS cells stably expressing wild-type H3.3 (WT_1) or the indicated mutants and treated with camptothecin (3 h, 0.1 μM). Representative images of repair foci in EdU+ cells are shown. Bar graphs depict the percentage of EdU+ and EdU− cells harboring more than 5 (RAD51) or 10 (53BP1) foci. Mean ± SEM from two to three independent experiments, with n > 117 per sample for each experiment. (C) Analysis of NHEJ activity by random plasmid integration assay in U2OS cells stably expressing wild-type (WT_1) or mutant H3.3. Vector –, negative untransfected control. (D) Serial dilution analyses and proliferation curves of S. pombe strains expressing wild-type or mutant H3, depleted for the core NHEJ factor Xrc4 (xrc4Δ) and grown in standard growth medium (control) or in the presence of camptothecin (10 μM). Mean ± SEM from three independent experiments. (E) Scoring of radials in metaphase spreads of U2OS cells stably expressing wildtype (WT_1) or mutant H3.3 and treated with Mitomycin C (48 h, 25 ng/ml). Cells transfected with siRNA against FANCD2 (siFANCD2) and the core NHEJ factor XRCC4 (siXRCC4) are used as positive and negative controls (siLUC, siLuciferase, control). A representative example of a radial chromosome is shown in the inset. The western blots show siRNA efficiency (Tubulin, Actin, loading controls). Mean ± SEM from three to four independent experiments, with n > 24 per sample for each experiment. (F) Oncoprint representation of DNA repair-driven mutational signatures (ID8, indels 8; SBS3, single-base substitutions 3) in whole-genome sequences of pre-treatment, TP53-mutated, primary pHGG samples harboring wild-type H3.3 (WT), H3.3 K27M or G34R. Statistical significance is calculated by two-way ANOVA (A, B), one-way ANOVA (C, E), non-linear regression analysis with a sigmoidal dose-response model (D) or the non-parametric Kruskal–Wallis test (F). *P< 0.05; **P< 0.01; ***P< 0.001; ns: P> 0.05. Scale bars, 10 μm.
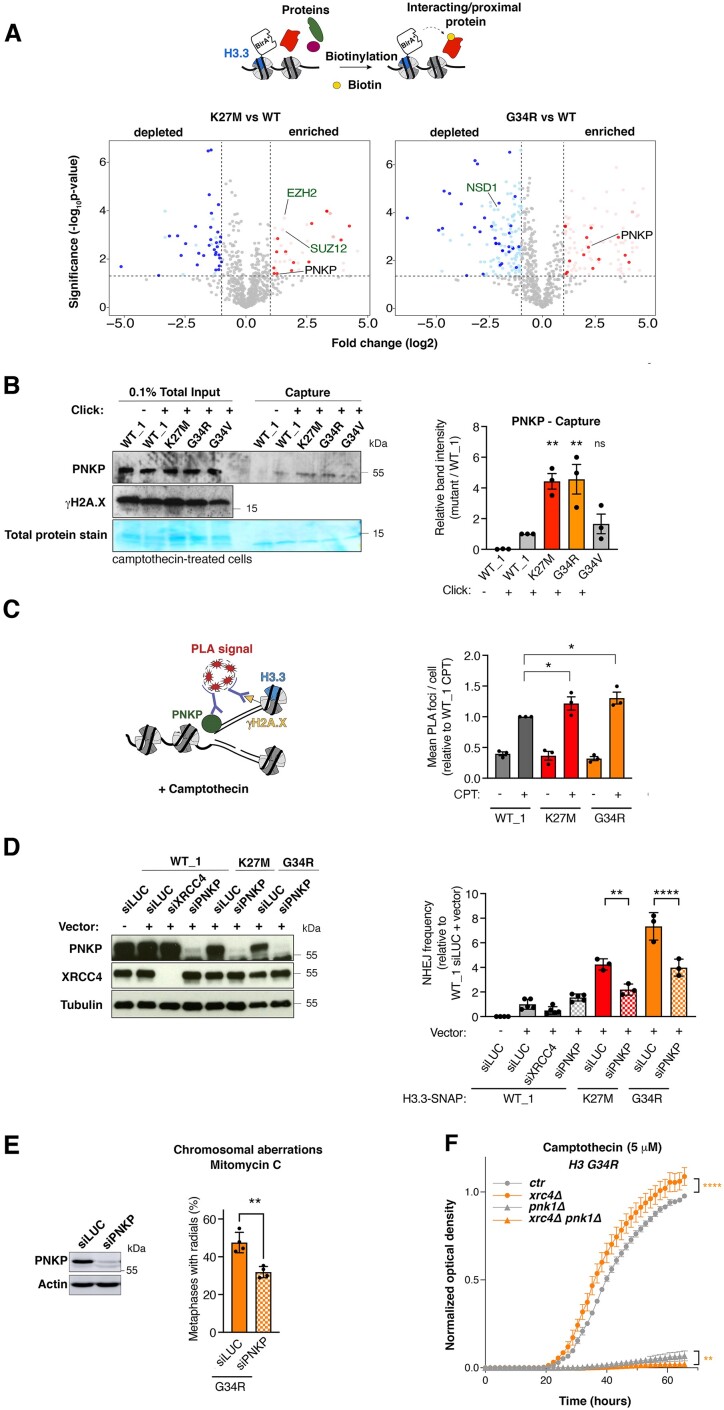
To test whether aberrant DNA repair in H3.3 K27M and G34R cells was associated with genome instability in human cells, we examined the occurrence of radials, i.e. chromosomal aberrations that derive from mis-joining of broken chromatids through aberrant NHEJ (46). We observed a marked accumulation of radials in H3.3 G34R U2OS cells upon MMC treatment, which were reduced upon knock-down of the core NHEJ factor XRCC4 (Figure 1E). To conclusively link the aberrant DNA repair in H3.3 K27M and G34R cells to genome instability onset in a glioma context, we analyzed the presence of mutational signatures generated by defects in DNA double-strand break repair pathways (49) in a panel of whole genome-sequenced, pre-treatment, TP53-mutant primary pHGGs. Both H3.3 K27M and G34R pHGGs presented higher levels of mutational signatures deriving from aberrant NHEJ (ID8) compared to WT H3.3 pHGGs, and H3.3 G34R also displayed a stronger signature activity of defective homologous recombination (SBS3) (Figure 1F). Collectively, our data support a model where the pHGG mutations H3.3 K27M and G34R skew the repair of S phase DNA damage towards aberrant NHEJ, thus sustaining a specific pattern of genome instability.
Aberrant NHEJ is independent of H3K27/K36me3 loss
To test whether pHGG H3.3 mutants skew the repair of S phase damage through gain- or loss-of-function mechanisms, we first evaluated the impact of siRNA-mediated depletion of H3.3 on RAD51 and 53BP1 focus formation in CPT-damaged U2OS cells. Depletion of H3.3 did not affect the proportion of cells in S phase (Supplementary Figure S3A) nor γH2A.X induction in response to CPT (Supplementary Figure S3B). Contrary to H3.3 mutations, H3.3 loss did not alter RAD51 and 53BP1 focus formation in response to CPT (Figure 2A), despite an increase in 53BP1 nuclear levels (Supplementary Figure S3C). Thus, H3.3 K27M and G34R mutations do not phenocopy H3.3 loss but rather confer a new function to histone H3.3 upon CPT-induced damage, corroborating the gain-of-function hypothesis.
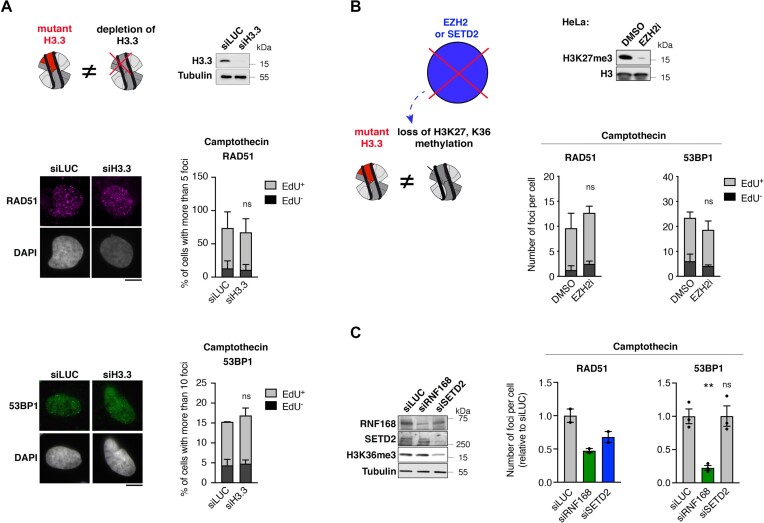
Figure 2.
pHGG H3.3 mutants hinder DNA repair through a gain-of-function mechanism independently of hypomethylation at lysines 27 and 36 of histone H3. (A) Analysis of RAD51 and 53BP1 foci by immunofluorescence in U2OS cells transfected with siRNAs against Luciferase (siLUC, control) or H3.3 (siH3.3) and treated with camptothecin (3 h, 0.1 μM). The western blot shows siRNA efficiency (Tubulin, loading control). Representative images of RAD51 and 53BP1 foci in EdU+ cells are shown. Bar graphs depict the percentage of EdU+ and EdU− cells harboring more than 5 (RAD51) or 10 (53BP1) foci. Mean ± SEM from two independent experiments, with n > 113 per sample for each experiment. (B) Analysis of RAD51 and 53BP1 foci by immunofluorescence in HeLa cells treated with DMSO or the EZH2 inhibitor GSK126 (EZH2i, 72 h, 1 μM) and damaged with camptothecin (3 h, 0.1 μM). The western blot shows the efficiency of EZH2 inhibition by analyzing H3K27me3 levels. Bar graphs depict the number of RAD51 or 53BP1 foci per cell in EdU+ and EdU− cell populations. Mean ± SEM from three independent experiments, with n > 125 per sample for each experiment. (C) Analysis of RAD51 and 53BP1 foci by immunofluorescence in U2OS cells transfected with the indicated siRNAs (siLUC, negative control; siRNF168, positive control known to inhibit both RAD51 and 53BP1 foci formation) and treated with camptothecin (3 h, 0.1 μM). siRNA efficiencies and H3K36me3 levels are analyzed by western blot (Tubulin, loading control). Bar graphs depict the mean number of RAD51 or 53BP1 foci per cell relative to siLUC. Mean ± SEM from two and three independent experiments, with n > 131 for each experiment. Statistical significance is calculated by two-way ANOVA (A, B) or one-way ANOVA (C). *P< 0.05; **P< 0.01; ***P< 0.001; ns: P> 0.05. Scale bars, 10 μm.
Dysregulation of gene expression programs by H3.3 K27M and G34R mutations is mediated by reduced trimethylation at lysines 27 (9,50) and 36 (7,13) of histone H3 (H3K27me3 and H3K36me3), respectively. To study whether the observed DNA repair defect is mediated by analogous perturbations of histone PTMs, we reduced H3K27me3 and H3K36me3 by inhibiting or depleting the corresponding lysine methyltransferases. The H3K27 methyltransferase Enhancer of Zeste 2 (EZH2) is endogenously inhibited in U2OS cells (51,52), thus preventing further reduction of H3K27me3 upon expression of H3.3 K27M (Supplementary Figure S1B and Supplementary Figure S3D). Yet, aberrant DNA repair is still observed in H3.3 K27M U2OS cells (Figure 1A, B), arguing against a contribution of H3K27me3 reduction to this repair defect. We confirmed this hypothesis in HeLa cells by chemical inhibition of EZH2 (EZH2i) (Figure 2B), which did not recapitulate the DNA repair defect observed in H3.3 K27M-expressing U2OS cells (Figure 1A, B). Similarly, reducing H3K36me3 by depleting SET Domain Containing 2 (SETD2) did not result in increased 53BP1 foci formation but solely in the expected reduction of RAD51 foci formation (19,53,54) in CPT-treated U2OS cells (Figure 2C). Together, these experiments demonstrate that H3.3 K27M and G34R mutants skew DNA repair towards NHEJ in S phase by conferring a gain-of-function to histone H3.3, independently of hypomethylation of H3K27 and K36.
Mutant H3.3 histones are deposited at damaged forks
The impact of H3.3 mutations on the repair of S phase DNA damage and the reported de novo deposition of H3.3 at sites of DNA damage (24,25,55) prompted us to investigate whether WT and pHGG H3.3 mutant proteins were de novo deposited at damaged RFs. First, we verified that, contrary to the replicative histone variant H3.1, WT and mutant H3.3 histones were not deposited at undamaged RFs, as shown by SNAP-tag-based imaging of newly synthesized histones (Supplementary Figure S3E). We next exploited fluorescent labeling of SNAP-tagged, newly synthesized histone H3.3 in a model of RF blockage, where stably integrated Lac operon (LacO) arrays generate an obstacle to DNA polymerase progression when bound by the Lac repressor (LacR) (Figure 3A) (56). Upon RF blockage, monitored by γH2A.X accumulation (Supplementary Figure S3F), we found a local enrichment of newly synthesized H3.3 on the LacO array specifically in S phase cells (Figure 3A), revealing a previously uncharacterized de novo deposition of H3.3 at sites of a replication block. To further study the deposition of newly synthesized H3.3 proteins in both WT and mutant cells, we setup a novel imaging-based method, SNAP-PLA (Proximity Ligation Assay), that measures by PLA (36) the colocalization between biotin-labeled, newly synthesized SNAP-tagged histones and γH2A.X at CPT-damaged RFs (Figure 3B). Thus, we could detect de novo deposition of WT H3.3 specifically in CPT-damaged, S phase cells (Figure 3B and Supplementary Figure S3G), recapitulating data obtained in the LacO system (Figure 3A) and validating the SNAP-PLA approach. Moreover, we detected de novo deposition of H3.3 K27M and G34R mutant proteins at damaged RFs, which was comparable to that of WT H3.3, while the de novo deposition H3.3 G34V was significantly reduced (Figure 3B). We validated these findings through the proteomic-based isolation of proteins on nascent DNA (iPOND) upon RF damage with CPT. The enrichment of SNAP-tagged H3.3 at CPT-damaged RFs was enhanced in S phase synchronized cells, supporting an S phase-specific deposition of H3.3 at damaged RFs (Supplementary Figure S3H). Using this method, we confirmed the deposition of H3.3 K27M and G34R mutant proteins at CPT-damaged RFs (Figure 3C). The deposition of H3.3 G34V was detectable in this assay but with reduced levels compared to WT H3.3 (Figure 3C). These findings unravel a local function of H3.3 in RF protection and repair and suggest that the deposition of pHGG H3.3 mutant proteins may locally affect the chromatin landscape and/or the recruitment of repair factors at damaged RFs, ultimately skewing fork repair.
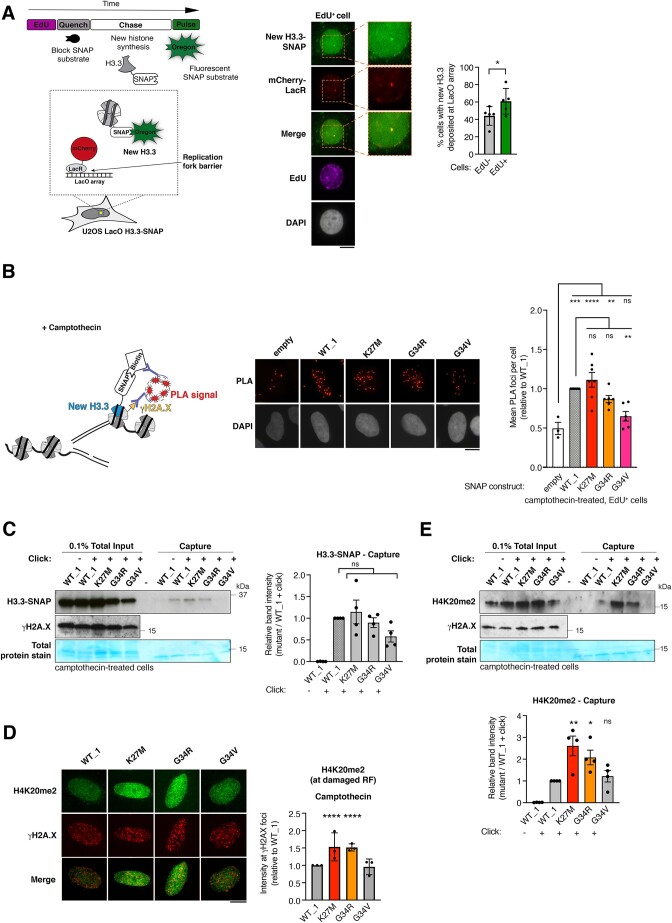
Figure 3.
H3.3 K27M and G34R pHGG mutants are de novo deposited and associated with increased H4K20me2 at damaged replication forks. (A) Scheme of the assay to monitor de novo deposition of wild-type H3.3-SNAP at the LacR-occupied LacO array fork barrier in U2OS LacO cells stably expressing SNAP-tagged H3.3 and transfected with mCherry-LacR. Images of a representative cell and 2.5× zoom on the LacO array. Quantification of new H3.3-SNAP accumulation (% cells presenting an enrichment) at LacR-occupied LacO array in EdU+ and EdU− cells. Mean ± SEM from five independent experiments, with n > 20 per sample for each experiment. The H3.3 enrichment detected in EdU-negative cells likely reflects cells that were in S phase when the mCherry-LacR was already expressed and bound to the LacO array but before EdU addition since cells were transfected 24h before harvesting. (B) Schematic representation of the SNAP-PLA assay to visualize the colocalization of γH2A.X with newly synthesized SNAP-tagged H3.3 (labeled with biotin) at RFs damaged with camptothecin (3 h, 0.1 μM). Representative images and quantification of SNAP-PLA colocalization foci between new H3.3 and γH2A.X in EdU+ U2OS cells stably expressing wild-type (WT_1) or mutant H3.3-SNAP, or SNAP tag only as a control (empty). Mean ± SEM from three to seven independent experiments, with n > 130 per sample for each experiment. (C) Western blot analysis of input and capture samples from iPOND experiments performed in U2OS cells expressing wild-type (WT_1) or mutant H3.3-SNAP, synchronized in S phase and damaged with camptothecin (1 h, 1 μM). Click -, negative control (no biotin). Total protein stain shows the position of the streptavidin monomer, detectable at similar levels in all capture samples. Bar graphs depict H3.3-SNAP band intensity in capture samples relative to WT_1. Mean ± SEM from four independent experiments. (D) Immunofluorescence analysis of H4K20me2 levels at γH2A.X foci in EdU-positive U2OS cells stably expressing wild-type (WT_1) or mutant H3.3 and treated with camptothecin (3 h, 0.1 μM). Quantification of H4K20me2 intensity relative to WT_1. Mean ± SD from three independent experiments, with n > 17 per sample for each experiment. (E) Western blot analysis of input and capture samples from iPOND experiments performed in U2OS cells expressing SNAP-tagged wild-type (WT_1) or mutant H3.3, synchronized in S phase and damaged with camptothecin (1 h, 1 μM). Click -, negative control (no biotin). Bar graphs depict H4K20me2 band intensity in capture samples relative to WT_1. Mean ± SEM from three independent experiments. Statistical significance is calculated by paired t-test (A), one-way ANOVA (B–E). *P< 0.05; **P< 0.01; ***P< 0.001; ns: p> 0.05. Scale bars, 10 μm.
To evaluate the impact of pHGG H3.3 mutant proteins on the chromatin landscape at damaged RFs, we analyzed by immunofluorescence the levels of H4K20me2, a histone mark known to drive 53BP1 foci formation on damaged chromatin (57). H4K20me2 nuclear levels were consistently higher in H3.3 K27M and G34R mutant cells (Supplementary Figure S3I), which was not due to different cell cycle distributions between WT and mutant cells (Supplementary Figure S2E) and we observed higher H4K20me2 at CPT-damaged RFs marked by γH2A.X in S phase cells in the same mutants (Figure 3D). These findings are in line with the increased capacity of those cells to form 53BP1 foci (Figure 1B). Importantly, we also detected increased H4K20me2 at CPT-damaged RFs by iPOND in cells expressing H3.3 K27M and G34R compared to cells expressing WT H3.3 or the G34V mutant (Figure 3E). Since the methyltransferases SETD8 and SUV4-20H1/2 methylate H4K20 in human cells (58), we employed loss-of-function approaches against these enzymes to test their contribution to the increased H4K20me2 levels observed in H3.3 pHGG mutant cells. We focused our analysis on S-phase cells to circumvent any effect of those treatments on the cell cycle, H4K20me2 being a cell cycle-regulated mark. The H4K20me2 increase in cells expressing H3.3 G34R and K27M mutants compared to WT H3.3 was abrogated by SETD8 depletion while it remained detectable upon SUV4-20H1/2 inhibition (Supplementary Figure S3J, K). These results indicate that aberrant H4K20 monomethylation by SETD8 is the major step driving increased H4K20me2 in mutant cells. They also suggest that the H3.3 G34R and K27M mutants may affect H4K20 methylation by stimulating the recruitment and/or the activity of SETD8 methyltransferase. Together, these data establish that the deposition of H3.3 K27M and G34R mutant proteins at damaged RFs is associated with dysregulated methylation of histone H4 that may, in turn, affect fork repair by impacting the recruitment of repair factors.
PNKP associates with mutant H3.3
To identify DNA repair factors that preferentially associate with the H3.3 K27M and G34R mutants, we employed proximity-dependent biotinylation (BioID) (59,60) in HEK293 human cells ectopically expressing WT, K27M or G34R H3.3 fused to the mutant BirA* biotin ligase, followed by mass spectrometry analysis (Figure 4A, Supplementary Table S2 and (27)). Validating this approach, we detected the expected preferential association of EZH2 with the H3.3 K27M mutant (9), while Nuclear Receptor Binding SET Domain Protein 1 (NSD1), responsible for H3K36 mono and dimethylation, showed reduced association to H3.3 G34R, in accordance with the reduced methylation of H3K36 by NSD1 in the presence of G34R (7) (Figure 4A). Out of the 21 proteins that preferentially associated with both K27M and G34R compared to WT H3.3, 6 were connected to the DNA damage response (HOXA10, GSK3B, PNKP, UBE2T, C3orf37/HMCES, CGGBP1). Among those, we focused our attention on the DNA end processing enzyme Polynucleotide Kinase 3′-Phosphatase (PNKP, Figure 4A), which contributes to NHEJ among other DNA repair pathways by transferring a phosphate group between broken DNA ends before ligation (61). In addition, PNKP was identified as an H3.3 G34R interactor in a previous study (26) and plays a central role in neurodevelopment (62). PNKP total levels were not increased in cells expressing H3.3 K27M or G34R (Supplementary Figure S4A). Nevertheless, we observed a stronger association of exogenously expressed GFP-tagged PNKP with both H3.3 mutants compared to WT H3.3 in pulldown experiments, which corroborates the BioID data (Supplementary Figure S4B). Similarly, we observed increased binding of PNKP to CPT-damaged RFs in H3.3 K27M and G34R cells by iPOND (Figure 4B) and by PLA between PNKP and γH2A.X (Figure 4C and Supplementary Figure S4C), further substantiating the preferential association of this DNA repair enzyme with chromatin harboring those H3.3 mutant proteins.
Figure 4.
PNKP mediates aberrant NHEJ in H3.3 K27M and G34R mutant cells. (A) Identification of proteins associated with wild-type (WT) and mutant H3.3 (K27M, G34R) by proximity-dependent biotinylation (BioID) in HEK293 cells expressing BirA*-tagged H3.3 proteins. Volcano plots show interactors enriched (red) or depleted (blue) in H3.3 K27M (left) or G34R (right) samples compared to WT H3.3 sample, with each dot representing an interactor. Significant interactors whose log2 fold change is >1 and whose P-value is <0.05 (–log10P-value > 1.30) are highlighted in colors and common interactors between H3.3 K27M and G34R are shown in dark colors. Positive controls are depicted in green. (B) Western blot analysis of input and capture samples from iPOND experiments performed in U2OS cells expressing wild-type (WT_1) or mutant H3.3-SNAP, synchronized in S phase and damaged with camptothecin (1 h, 1 μM). Click –, negative control (no biotin). Total protein stain shows the position of the streptavidin monomer, detectable at similar levels in all capture samples. Bar graphs depict PNKP band intensity in capture samples relative to WT_1. Mean ± SEM from three independent experiments. The representative experiment shown is the same as in Figure 3C. (C) PNKP association with CPT-damaged replication forks analyzed by PLA between PNKP and γH2A.X in EdU-positive cells. A scheme of the experiment is shown on the left. Mean ± SEM from three independent experiments. (D) Analysis of NHEJ activity by random plasmid integration assay in U2OS cells stably expressing wild-type (WT_1) or mutant H3.3 and transfected with siRNAs against Luciferase (siLUC, control) or PNKP (siPNKP). Samples siLUC–, siLUC+ and siXRCC4+ are the same shown in Figure S2I graph. Western blot analysis shows siRNA efficiency (Tubulin, loading control). Vector –, negative untransfected control. (E) Scoring of radials in metaphase spreads of U2OS H3.3 G34R cells transfected with siRNA against Luciferase (siLUC, control) or PNKP (siPNKP) and treated with Mitomycin C (48 h, 25 ng/ml). The western blot shows siRNA efficiency (Actin, loading control). Mean ± SEM from four independent experiments, with n > 24 per sample for each experiment. (F) Proliferation curves of S. pombe strains expressing H3G34R, depleted for the core NHEJ factor Xrc4 (xrc4Δ) and for Pnk1 (pnk1Δ) and grown in the presence of camptothecin (5 μM). Statistical significance is calculated by one-way ANOVA (B–D), by non-linear regression analysis with a sigmoidal dose-response model (D). *P< 0.05; **P< 0.01; ***P< 0.001; ns: P> 0.05.
PNKP promotes aberrant repair in mutant cells
We hypothesized that the preferential association of PNKP with H3.3 K27M and G34R may drive the aberrant NHEJ observed in cells expressing these mutants. Thus, we measured NHEJ activity by random plasmid integration assay upon knockdown of PNKP in U2OS cells expressing WT or mutant H3.3. In contrast to the depletion of the core NHEJ factor XRCC4, PNKP knockdown did not affect NHEJ activity in WT H3.3 cells. However, knock-down of PNKP significantly reduced NHEJ in H3.3 K27M and G34R cells (Figure 4D), and radial chromosome formation in H3.3 G34R cells (Figure 4E), showing that PNKP promotes aberrant NHEJ repair and contributes to genome instability in these cells. Similarly, in fission yeast, co-deletion of the PNKP ortholog pnk1 did not rescue CPT-sensitivity in the G34R xrc4Δ strain, arguing that the aberrant xrc4-mediated NHEJ in a G34R background is dependent on pnk1 (Figure 4F). Together, these data demonstrate that PNKP contributes to aberrant NHEJ in H3.3 mutant cells and that this process is evolutionarily conserved. Notably, PNKP depletion did not significantly reduce H4K20me2 nuclear levels and 53BP1 foci formation in S-phase upon CPT damage in H3.3 K27M and G34R mutant cells (Supplementary Figure S4D-E). In contrast, 53BP1 depletion reduced PNKP accumulation at CPT-damaged replication forks in H3.3 mutant cells as shown by PLA between PNKP and γH2A.X (Supplementary Figure S4F). These findings support a model where PNKP accumulation promoting aberrant NHEJ at damaged forks in H3.3 mutant cells is downstream of H4K20me2 and 53BP1.
PNKP as a therapeutic target in pHGG
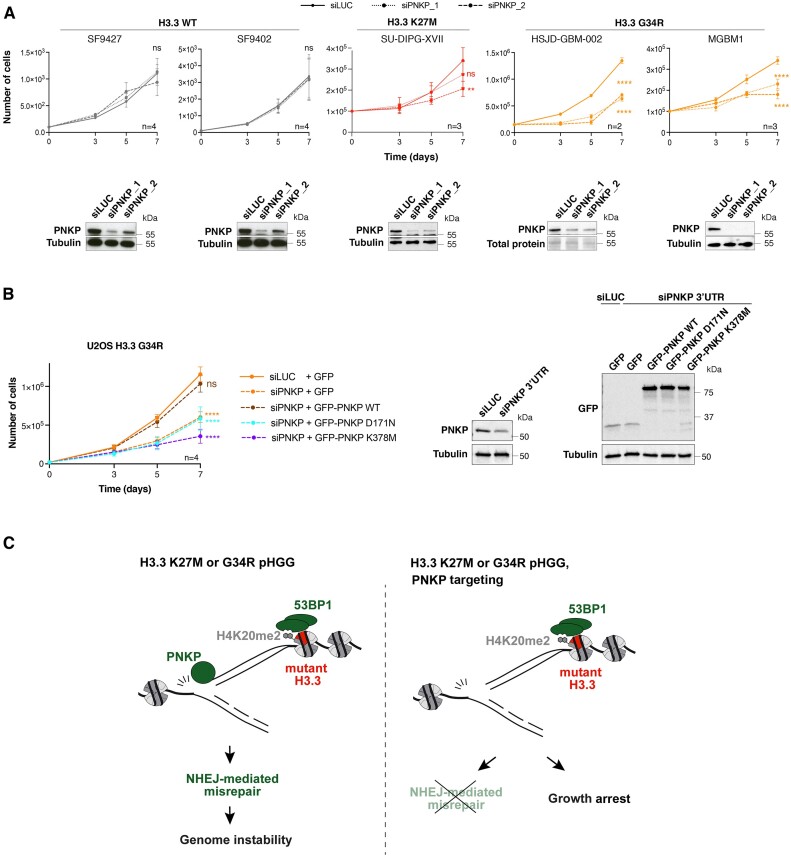
We next assessed the importance of aberrant PNKP function for H3.3 mutant cell growth. PNKP knockdown specifically impaired the growth of H3.3 K27M and H3.3 G34R U2OS cells, but not of WT H3.3 or H3.3 G34V mutant cells (Supplementary Figure S5A). To expand our findings to state-of-the-art pHGG cellular models, we exploited a panel of patient-derived glioma cells lines (Supplementary Figure S5B) and two different siRNAs against PNKP. We corroborated the specific effect of PNKP knockdown on the proliferation of glioma cells harboring endogenous H3.3 K27M or G34R mutations (Figure 5A). PNKP knockdown significantly reduced the growth of all H3.3 mutant cell lines examined in contrast to H3.3 WT glioma cell lines and non-cancerous astrocytes, which were unaffected (Figure 5A and Supplementary Figure S5C). Collectively, these data put forward PNKP as a potential therapeutic target in pHGG cells expressing specific H3.3 mutations.
Figure 5.
PNKP represents a therapeutic target in pediatric pHGG. (A) Proliferation assays in patient-derived pHGG cell lines harboring wild-type or mutant H3.3 and transfected with siRNAs against Luciferase (siLUC, control) or PNKP (siPNKP_1 and siPNKP_2). Mean ± SEM from n independent experiments. The western blots show siRNA efficiencies (Tubulin or total protein stain, loading control). (B) Proliferation assays in U2OS H3.3 G34R transfected with an siRNA against PNKP 3′UTR (siLUC, control) and expressing the indicated GFP-tagged constructs. Mean ± SEM from n independent experiments. The western blots show PNKP knock-down efficiency and the expression levels of each construct (Tubulin, loading control). (C) Current model depicting PNKP-mediated misrepair of S phase DNA damage in H3.3 K27M and G34R mutant cells (left) and the consequences of PNKP targeting in these cells (right). Statistical significance is calculated by a polynomial quadratic model (A, B). *P< 0.05; **P< 0.01; ***P< 0.001; ns: P> 0.05.
We next sought to determine if the vulnerability to PNKP loss related to its function in NHEJ or to other genome maintenance activities, such as base damage repair and/or a recently proposed function in Okazaki fragment maturation (63,64). To determine if NHEJ was responsible for sustaining H3.3 mutant cell growth, we examined whether the loss of the core NHEJ factor XRCC4 would recapitulate the growth defect observed upon PNKP loss. However, XRCC4 depletion had only a very marginal impact on cell growth if at all, and thus did not phenocopy the effect of PNKP loss, as observed both in yeast and human cells (Supplementary Figure S5D, E). While PNKP drives aberrant NHEJ in H3.3 mutant cells, the vulnerability to PNKP loss is therefore NHEJ-independent.
To explore a possible involvement of base damage repair in sustaining H3.3 mutant cell growth, we examined the impact of depleting the base excision repair factor and PNKP interactor XRCC1. XRCC1 depletion did not phenocopy the effect of PNKP loss on H3.3 mutant cell growth (Supplementary Figure S5F). Expression of the H3.3 mutants also did not result in more single-stranded DNA, which is prone to base damage, as shown by the number of foci of the single-strand DNA binding complex Replication Protein A (RPA) that is comparable to that of cells expressing wild-type H3.3 (Supplementary Figure S5G). Thus, the vulnerability to PNKP loss is independent of base damage repair and presumably relates to PNKP recently proposed function in Okazaki fragment maturation (64).
To conclusively link the anti-proliferative effect to PNKP targeting, we performed complementation experiments in U2OS H3.3 G34R cells treated with an siRNA against PNKP 3′UTR and observed that exogenous expression of GFP-tagged PNKP rescued cell growth in PNKP knocked-down cells (Figure 5B). In contrast, expressing PNKP mutated in the kinase (K378M) or phosphatase (D171N) catalytic domains (63) did not restore cell growth (Figure 5B). These results demonstrate that both catalytic activities of PNKP are required to sustain cell growth in H3.3 mutant cells.
Discussion
By dissecting how H3.3 mutants skew DNA repair, we show that mutations in core chromatin components induce genome instability by dysregulating the recruitment of repair factors at damaged RF (Figure 5C). Aberrant repair of damaged RF, leading to mitotic aberrations (47) as recently reported in cells expressing the H3.3 K27M mutant (17), is the likely cause of genome instability. However, the contribution of oncohistone-dependent genome instability to pHGG oncogenesis should be evaluated in future studies, as well as the influence of co-occurring genetic alterations (22) on aberrant repair.
By identifying a new, local function of H3.3 in the repair of damaged RFs, the present study expands our knowledge about histone variant deposition at these sites (65,66). De novo H3.3 deposition may operate through a DNA synthesis-independent mechanism on the regressed arms of damaged RFs, which have been shown to be chromatinized (67).
H3.3 K27M and G34R affect RF repair through a mechanism that is distinct from their interference with gene expression programs (3,15,18). The aberrant use of NHEJ in S phase indeed does not rely on H3K27me3 and H3K36me3 alterations, but may involve other PTM changes in mutant nucleosomes, possibly through the activity of histone modifying enzymes, which could in turn affect the binding of repair factors. For instance, the observed increase in H4K20me2 at RFs likely contributes to the enhanced 53BP1 foci formation and the channeling of repair towards NHEJ. We still miss mechanistic details on how H3.3 mutants promote aberrant PNKP binding to chromatin, contributing to aberrant NHEJ. Future work will determine if H4K20 methyltransferases stimulate PNKP association with RFs in H3.3 mutant cells. Since PNKP does not harbor known histone binding motifs, we favor an indirect mechanism of recruitment to chromatin bearing mutant H3.3 histones. The increased engagement of PNKP may also explain the lack of CPT sensitivity in human cells expressing H3.3 K27M and G34R because PNKP is known to be involved in the repair of CPT-induced damage (68).
The similarity of DNA repair phenotypes between H3.3 K27M and G34R mutant cells may indicate a common gain or loss of histone modifying activity on nucleosomes. The consistently stronger phenotype observed with the G34R mutant reflects the more efficient response to PNKP loss-of-function in H3.3 G34R mutant cells. In contrast, H3.3 G34R and G34V mutants display strikingly opposite DNA repair phenotypes, conserved from yeast to human, the molecular basis of which is still elusive. Perhaps the bulkier and positively charged arginine side chain of the G34R mutant protein causes a more drastic disruption of the H3.3 interactome. In addition, the G34V mutant is much rarer than G34R so maybe less oncogenic.
The K27M mutation is also found in the H3.1 histone variant (H3.1 K27M) in some pHGG and, as observed with H3.3 mutants, PNKP also shows a preferential association with H3.1 K27M compared to H3.1 WT (Supplementary Figure S5H), suggesting that common mechanisms may be at play. However H3.1 K27M inhibits NHEJ in human fibroblasts (69), which is opposite to the phenotype of H3.3 K27M in U2OS cells. Although a true comparison requires that the H3.1 K27M and H3.3 K27M phenotypes be studied in the same cellular background, differences in their DNA repair function can be anticipated since they show distinct distribution patterns in chromatin (70), present different co-occurring mutations (22,71) and clinical features in pHGG (72,73).
Targeting DNA repair defects is the most developed approach so far to induce tumor cell death through synthetic lethality, even if weakly exploited beyond BRCA-mutated tumors (74). By providing molecular understanding of the aberrant DNA repair capacities of H3.3 mutant pHGGs, we propose here that a synthetic interaction approach for the treatment of these tumors is warranted by targeting the PNKP enzyme. Since PNKP inhibition is already exploited to kill other cancers (75), we suggest that it be used to sensitize pHGG cells to current radio/chemotherapeutic regimens (1), for which there is very limited response. Our findings indicate that both the kinase and phosphatase activities of PNKP can be targeted to affect the growth of H3.3 mutant cells. Moreover, the specificity towards H3.3 K27M and G34R mutant cells may limit toxicity-related side effects in future treatments and opens up the possibility of employing a similar strategy in other cancers bearing the same H3.3 mutations (3).
Supplementary Material
Acknowledgements
We thank members of our laboratory, M. Touat and F. Bielle for stimulating discussions, S. Ait-Si-Ali and P-A. Defossez for critical reading of the manuscript, A. Carcaboso, N. Jabado, C.D. James, P. Knoepfler and H. van Attikum for sharing pHGG cell lines and reagents. We acknowledge the imaging platform of the Epigenetics and Cell Fate Centre.
Author contributions: B.R., S.P., G.G. and O.C. performed most of the experiments. J.D. and S.-K.B. generated U2OS cell lines expressing wild-type and mutant H3.3. B.K., R.S., B.R. and E.C. performed BioID experiments under the supervision of C.H. C-M.S., R.J.D.R. and T.T. performed fission yeast experiments with supervision from S.J. and R.R. T.W., V.B. and S.H. supervised by V.N. performed metaphase spread analyses. C.B. helped B.R. with complementation experiments. A.C. and F.D. supervised by P.B. and R.B. performed mutational signature analyses. I.F. and A.I. performed mass spectrometry-based quantification of H3.3 mutants. B.R. and S.E.P. conceived the study, supervised the work and wrote the manuscript. All authors edited and approved the final version of the manuscript.
Notes
Present address: Beatrice Rondinelli, CNRS UMR9019 Genome Integrity and Cancers, Université Paris-Saclay, Gustave Roussy Institute, Villejuif, France.
Present address: Chun-Min Shan, State Key Laboratory of Plant Genomics, Institute of Microbiology, Chinese Academy of Sciences, Beijing 100101, China.
Present address: Viviana Barra, Department of Biological, Chemical and Pharmaceutical Sciences and Technologies, University of Palermo, 90128 Palermo, Italy.
Present address: Sabah Hamadat, Inovarion, 75005 Paris, France.
Contributor Information
Giulia Giacomini, Epigenetics & Cell Fate Centre, CNRS/Université Paris Cité, Paris, France.
Sandra Piquet, Epigenetics & Cell Fate Centre, CNRS/Université Paris Cité, Paris, France.
Odile Chevallier, Epigenetics & Cell Fate Centre, CNRS/Université Paris Cité, Paris, France.
Juliette Dabin, Epigenetics & Cell Fate Centre, CNRS/Université Paris Cité, Paris, France.
Siau-Kun Bai, Epigenetics & Cell Fate Centre, CNRS/Université Paris Cité, Paris, France.
Byungjin Kim, Arthur and Sonia Labatt Brain Tumour Research Centre, The Hospital for Sick Children, Toronto, Canada.
Robert Siddaway, Arthur and Sonia Labatt Brain Tumour Research Centre, The Hospital for Sick Children, Toronto, Canada.
Brian Raught, Princess Margaret Cancer Centre, University Health Network, 101 College Street, Toronto, ON M5G, 1L7, Canada; Department of Medical Biophysics, University of Toronto, Toronto, Canada.
Etienne Coyaud, Princess Margaret Cancer Centre, University Health Network, 101 College Street, Toronto, ON M5G, 1L7, Canada; Department of Medical Biophysics, University of Toronto, Toronto, Canada; Université de Lille, Inserm, CHU Lille, U1192 - Protéomique Réponse Inflammatoire Spectrométrie de Masse - PRISM, F-59000 Lille, France.
Chun-Min Shan, Department of Biological Sciences, Columbia University, New York, NY 10027, USA.
Robert J D Reid, Department of Genetics & Development, Columbia University Irving Medical Center, New York, NY 10032, USA.
Takenori Toda, Department of Biological Sciences, Columbia University, New York, NY 10027, USA.
Rodney Rothstein, Department of Genetics & Development, Columbia University Irving Medical Center, New York, NY 10032, USA; Department of Systems Biology, Columbia University Irving Medical Center, New York, NY 10032, USA.
Viviana Barra, CNRS UMR9019 Genome Integrity and Cancers, Université Paris-Saclay, Gustave Roussy Institute, Villejuif, France.
Therese Wilhelm, CNRS UMR9019 Genome Integrity and Cancers, Université Paris-Saclay, Gustave Roussy Institute, Villejuif, France.
Sabah Hamadat, CNRS UMR9019 Genome Integrity and Cancers, Université Paris-Saclay, Gustave Roussy Institute, Villejuif, France.
Chloé Bertin, CNRS UMR9019 Genome Integrity and Cancers, Université Paris-Saclay, Gustave Roussy Institute, Villejuif, France.
Alexander Crane, Department of Medical Oncology, Dana-Farber Cancer Institute, Boston, USA.
Frank Dubois, Department of Medical Oncology, Dana-Farber Cancer Institute, Boston, USA.
Ignasi Forne, Protein Analysis Unit, BioMedical Center, Faculty of Medicine, Ludwig-Maximilians-University, Martinsried, Germany.
Axel Imhof, Protein Analysis Unit, BioMedical Center, Faculty of Medicine, Ludwig-Maximilians-University, Martinsried, Germany.
Pratiti Bandopadhayay, Department of Pediatric Oncology, Dana-Farber Boston Children's Cancer and Blood Disorders Center, Boston, USA.
Rameen Beroukhim, Department of Medical Oncology, Dana-Farber Cancer Institute, Boston, USA.
Valeria Naim, CNRS UMR9019 Genome Integrity and Cancers, Université Paris-Saclay, Gustave Roussy Institute, Villejuif, France.
Songtao Jia, Department of Biological Sciences, Columbia University, New York, NY 10027, USA.
Cynthia Hawkins, Arthur and Sonia Labatt Brain Tumour Research Centre, The Hospital for Sick Children, Toronto, Canada.
Beatrice Rondinelli, Epigenetics & Cell Fate Centre, CNRS/Université Paris Cité, Paris, France.
Sophie E Polo, Epigenetics & Cell Fate Centre, CNRS/Université Paris Cité, Paris, France.
Data availability
Raw mass spectrometry data is available for download from https://massive.ucsd.edu/, accession numbers MSV000087736 and MSV000089173 (27) and from ProteomeXchange with identifier PXD041132. Whole genome sequencing data is available under EGAS00001000575, EGAS00001001139, EGAS00001000572, EGAS00001000192, and under dbGaP accession number phs002380.v1.p1 (76). Other raw data files are deposited on figshare: https://doi.org/10.6084/m9.figshare.23259920.v1.
Supplementary data
Supplementary Data are available at NAR Online.
Funding
Research in R.R. lab was supported by the National Institutes of Health [R35GM118180]; research in S.J. lab was supported by the National Institutes of Health [R35GM126910]; work in A.I. lab was funded by grants from the Deutsche Forschungsgemeinschaft (DFG) [213249687 (CRC1064) and 325871075 (CRC1309)]; work in P.B. lab was funded by Alex's Lemonade Stand Foundation and Prayers from Maria; work in R.B. lab was funded by the Gray Matters Brain Cancer Foundation and the Brown Fund for Innovation in Cancer Informatics; work in V.N. lab was supported by the European Research Council [ERC-2014-StG-638898]; work in C.H. lab was supported by the Canadian Cancer Society Research Institute [702296 and 706160]; Canadian Institutes of Health Research [PJT-156407]; research in S.E.P. lab was supported by the European Research Council [ERC-2013-StG-336427 and ERC-2018-CoG-818625]; French National Research Agency [ANR-12-JSV6-0002-01 and ANR-18-CE12-0017-01]; Labex ‘Who am I?’ [ANR-11-LABX-0071, ANR-18-IDEX-0001]; B.R. was a Marie Curie fellow [H2020-MSCA-IF-2017-799101]; G.G. is supported by a PhD grant from the Fondation pour la Recherche Medicale [ECO202106013684]; S.E.P. is an EMBO Young Investigator. Funding for open access charge: European Research Council (ERC-2018-CoG-818625).
Conflict of interest statement. P.B. receives grant funding from Novartis Institute of Biomedical Research for unrelated research, has received grant funding from Deerfield Therapeutics for unrelated research, and has served on a SAB advisory panel for QED Therapeutics. All other authors declare no competing interests.
References
- 1. Pinto L., Baidarjad H., Entz-Werlé N., Van Dyck E. Impact of chromatin dynamics and DNA repair on genomic stability and treatment resistance in pediatric high-grade gliomas. Cancers (Basel). 2021; 13:5678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lin K., Gueble S.E., Sundaram R.K., Huseman E.D., Bindra R.S., Herzon S.B. Mechanism-based design of agents that selectively target drug-resistant glioma. Science. 2022; 377:502–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Deshmukh S., Ptack A., Krug B., Jabado N. Oncohistones: a roadmap to stalled development. FEBS J. 2021; 289:1315–1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Schwartzentruber J., Korshunov A., Liu X.-Y., Jones D.T.W., Pfaff E., Jacob K., Sturm D., Fontebasso A.M., Quang D.-A.K., Tönjes M. et al. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature. 2012; 482:226–231. [DOI] [PubMed] [Google Scholar]
- 5. Wu G., Broniscer A., McEachron T.A., Lu C., Paugh B.S., Becksfort J., Qu C., Ding L., Huether R., Parker M. et al. Somatic histone H3 alterations in pediatric diffuse intrinsic pontine gliomas and non-brainstem glioblastomas. Nat. Genet. 2012; 44:251–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sturm D., Witt H., Hovestadt V., Khuong-Quang D.A., Jones D.T.W., Konermann C., Pfaff E., Tönjes M., Sill M., Bender S. et al. Hotspot mutations in H3F3A and IDH1 define distinct epigenetic and biological subgroups of glioblastoma. Cancer Cell. 2012; 22:425–437. [DOI] [PubMed] [Google Scholar]
- 7. Fang J., Huang Y., Mao G., Yang S., Rennert G., Gu L., Li H., Li G.-M. Cancer-driving H3G34V/R/D mutations block H3K36 methylation and H3K36me3–MutSα interaction. Proc. Natl. Acad. Sci. U.S.A. 2018; 115:9598–9603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Shi L., Shi J., Shi X., Li W., Wen H. Histone H3.3 G34 mutations alter Histone H3K36 and H3K27 methylation In cis. J. Mol. Biol. 2018; 430:1562–1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lewis P.W., Muller M.M., Koletsky M.S., Cordero F., Lin S., Banaszynski L.A., Garcia B.A., Muir T.W., Becher O.J., Allis C.D. Inhibition of PRC2 activity by a gain-of-function H3 mutation found in pediatric glioblastoma. Science. 2013; 340:857–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fang D., Gan H., Lee J.-H., Han J., Wang Z., Riester S.M., Jin L., Chen J., Zhou H., Wang J. et al. The histone H3.3K36M mutation reprograms the epigenome of chondroblastomas. Science. 2016; 352:1344–1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jessa S., Blanchet-Cohen A., Krug B., Vladoiu M., Coutelier M., Faury D., Poreau B., De Jay N., Hébert S., Monlong J. et al. Stalled developmental programs at the root of pediatric brain tumors. Nat. Genet. 2019; 51:1702–1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Filbin M.G., Tirosh I., Hovestadt V., Shaw M.L., Escalante L.E., Mathewson N.D., Neftel C., Frank N., Pelton K., Hebert C.M. et al. Developmental and oncogenic programs in H3K27M gliomas dissected by single-cell RNA-seq. Science. 2018; 360:331–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bjerke L., Mackay A., Nandhabalan M., Burford A., Jury A., Popov S., Bax D.A., Carvalho D., Taylor K.R., Vinci M. et al. Histone H3.3 mutations drive pediatric glioblastoma through upregulation of MYCN. Cancer Discov. 2013; 3:512–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chen C.C.L., Deshmukh S., Jessa S., Hadjadj D., Lisi V., Andrade A.F., Faury D., Jawhar W., Dali R., Suzuki H. et al. Histone H3.3G34-mutant interneuron progenitors Co-opt PDGFRA for gliomagenesis. Cell. 2020; 183:1617–1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Phillips R.E., Soshnev A.A., Allis C.D. Epigenomic reprogramming as a driver of malignant glioma. Cancer Cell. 2020; 38:647–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sahu V., Lu C. Oncohistones: hijacking the histone code. Annu. Rev. Cancer Biol. 2022; 6:293–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bočkaj I., Martini T.E.I., de Camargo Magalhães E.S., Bakker P.L., Meeuwsen-de Boer T.G.J., Armandari I., Meuleman S.L., Mondria M.T., Stok C., Kok Y.P. et al. The H3.3K27M oncohistone affects replication stress outcome and provokes genomic instability in pediatric glioma. PLoS Genet. 2021; 17:e1009868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Haase S., Banerjee K., Mujeeb A.A., Hartlage C.S., Nunez F.M., Nuñez F.J., Alghamri M.S., Kadiyala P., Carney S., Barissi M. et al. H3.3-G34 mutations impair DNA repair and promote cGAS/STING-mediated immune responses in pediatric high-grade glioma models. J. Clin. Invest. 2022; 132:e154229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pfister S.X., Ahrabi S., Zalmas L.-P., Sarkar S., Aymard F., Bachrati C.Z., Helleday T., Legube G., La Thangue N.B., Porter A.C.G. et al. SETD2-Dependent histone H3K36 trimethylation is required for homologous recombination repair and genome stability. Cell Rep. 2014; 7:2006–2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yadav R.K., Jablonowski C.M., Fernandez A.G., Lowe B.R., Henry R.A., Finkelstein D., Barnum K.J., Pidoux A.L., Kuo Y.-M., Huang J. et al. Histone H3G34R mutation causes replication stress, homologous recombination defects and genomic instability in S. pombe. eLife. 2017; 6:e27406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hanahan D. Hallmarks of cancer: new dimensions. Cancer Discov. 2022; 12:31–46. [DOI] [PubMed] [Google Scholar]
- 22. Mackay A., Burford A., Carvalho D., Izquierdo E., Fazal-Salom J., Taylor K.R., Bjerke L., Clarke M., Vinci M., Nandhabalan M. et al. Integrated molecular meta-analysis of 1,000 pediatric high-grade and diffuse intrinsic pontine glioma. Cancer Cell. 2017; 32:520–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lowe B.R., Yadav R.K., Henry R.A., Schreiner P., Matsuda A., Fernandez A.G., Finkelstein D., Campbell M., Kallappagoudar S., Jablonowski C.M. et al. Surprising phenotypic diversity of cancer-associated mutations of gly 34 in the histone H3 tail. eLife. 2021; 10:e65369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Luijsterburg M.S., de Krijger I., Wiegant W.W., Shah R.G., Smeenk G., de Groot A.J.L., Pines A., Vertegaal A.C.O., Jacobs J.J.L., Shah G.M. et al. PARP1 Links CHD2-mediated chromatin expansion and H3.3 deposition to DNA repair by non-homologous end-joining. Mol. Cell. 2016; 61:547–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Juhász S., Elbakry A., Mathes A., Löbrich M. ATRX promotes DNA repair synthesis and sister chromatid exchange during homologous recombination. Mol. Cell. 2018; 71:11–24. [DOI] [PubMed] [Google Scholar]
- 26. Lim J., Park J.H., Baude A., Yoo Y., Lee Y.K., Schmidt C.R., Park J.B., Fellenberg J., Zustin J., Haller F. et al. The histone variant H3.3 G34W substitution in giant cell tumor of the bone link chromatin and RNA processing. Sci. Rep. 2017; 7:13459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Siddaway R., Canty L., Pajovic S., Milos S., Coyaud E., Sbergio S.-G., Vadivel Anguraj A.K., Lubanszky E., Yun H.Y., Portante A. et al. Oncohistone interactome profiling uncovers contrasting oncogenic mechanisms and identifies potential therapeutic targets in high grade glioma. Acta Neuropathol. 2022; 144:1027–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Adam S., Dabin J., Chevallier O., Leroy O., Baldeyron C., Corpet A., Lomonte P., Renaud O., Almouzni G., Polo S.E. Real-time tracking of parental histones reveals their contribution to chromatin integrity following DNA damage. Mol. Cell. 2016; 64:65–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hashizume R., Andor N., Ihara Y., Lerner R., Gan H., Chen X., Fang D., Huang X., Tom M.W., Ngo V. et al. Pharmacologic inhibition of histone demethylation as a therapy for pediatric brainstem glioma. Nat. Med. 2014; 20:1394–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chen K.-Y., Bush K., Klein R.H., Cervantes V., Lewis N., Naqvi A., Carcaboso A.M., Lechpammer M., Knoepfler P.S. Reciprocal H3.3 gene editing identifies K27M and G34R mechanisms in pediatric glioma including NOTCH signaling. Commun. Biol. 2020; 3:363–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nagaraja S., Vitanza N.A., Woo P.J., Taylor K.R., Liu F., Zhang L., Li M., Meng W., Ponnuswami A., Sun W. et al. Transcriptional dependencies in diffuse intrinsic pontine glioma. Cancer Cell. 2017; 31:635–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bender S., Tang Y., Lindroth A.M., Hovestadt V., Jones D.T.W., Kool M., Zapatka M., Northcott P.A., Sturm D., Wang W. et al. Reduced H3K27me3 and DNA hypomethylation are major drivers of gene expression in K27M mutant pediatric high-grade gliomas. Cancer Cell. 2013; 24:660–672. [DOI] [PubMed] [Google Scholar]
- 33. Bryant E.E., Šunjevarić I., Berchowitz L., Rothstein R., Reid R.J.D. Rad5 dysregulation drives hyperactive recombination at replication forks resulting in cisplatin sensitivity and genome instability. Nucleic Acids Res. 2019; 47:9144–9159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Behjati S., Tarpey P.S., Presneau N., Scheipl S., Pillay N., Van Loo P., Wedge D.C., Cooke S.L., Gundem G., Davies H. et al. Distinct H3F3A and H3F3B driver mutations define chondroblastoma and giant cell tumor of bone. Nat. Genet. 2013; 45:1479–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bodor D.L., Rodríguez M.G., Moreno N., Jansen L.E.T. Analysis of protein turnover by quantitative SNAP-based pulse-chase imaging. Current Protocols in Cell Biology. 2012; Hoboken, NJ, USA: John Wiley & Sons, Inc; Chapter 8. [DOI] [PubMed] [Google Scholar]
- 36. Söderberg O., Gullberg M., Jarvius M., Ridderstråle K., Leuchowius K.-J., Jarvius J., Wester K., Hydbring P., Bahram F., Larsson L.-G. et al. Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat. Methods. 2006; 3:995–1000. [DOI] [PubMed] [Google Scholar]
- 37. Sirbu B.M., Couch F.B., Cortez D. Monitoring the spatiotemporal dynamics of proteins at replication forks and in assembled chromatin using isolation of proteins on nascent DNA. Nat. Protoc. 2012; 7:594–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gupta G.D., Coyaud É., Gonçalves J., Mojarad B.A., Liu Y., Wu Q., Gheiratmand L., Comartin D., Tkach J.M., Cheung S.W.T. et al. A dynamic protein interaction landscape of the Human centrosome-cilium interface. Cell. 2015; 163:1484–1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kessner D., Chambers M., Burke R., Agus D., Mallick P. ProteoWizard: open source software for rapid proteomics tools development. Bioinformatics. 2008; 24:2534–2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Craig R., Beavis R.C. TANDEM: matching proteins with tandem mass spectra. Bioinformatics. 2004; 20:1466–1467. [DOI] [PubMed] [Google Scholar]
- 41. Eng J.K., Jahan T.A., Hoopmann M.R. Comet: an open-source MS/MS sequence database search tool. Proteomics. 2013; 13:22–24. [DOI] [PubMed] [Google Scholar]
- 42. Pedrioli P.G.A. Trans-proteomic pipeline: a pipeline for proteomic analysis. Methods Mol. Biol. 2010; 604:213–238. [DOI] [PubMed] [Google Scholar]
- 43. Deutsch E.W., Mendoza L., Shteynberg D., Farrah T., Lam H., Tasman N., Sun Z., Nilsson E., Pratt B., Prazen B. et al. A guided tour of the Trans-Proteomic Pipeline. Proteomics. 2010; 10:1150–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Liu G., Zhang J., Larsen B., Stark C., Breitkreutz A., Lin Z.-Y., Breitkreutz B.-J., Ding Y., Colwill K., Pasculescu A. et al. ProHits: integrated software for mass spectrometry-based interaction proteomics. Nat. Biotechnol. 2010; 28:1015–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Scully R., Panday A., Elango R., Willis N.A. DNA double-strand break repair-pathway choice in somatic mammalian cells. Nat. Rev. Mol. Cell Biol. 2019; 20:698–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zhao B., Rothenberg E., Ramsden D.A., Lieber M.R. The molecular basis and disease relevance of non-homologous DNA end joining. Nat. Rev. Mol. Cell Biol. 2020; 21:765–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Berti M., Cortez D., Lopes M. The plasticity of DNA replication forks in response to clinically relevant genotoxic stress. Nat. Rev. Mol. Cell Biol. 2020; 21:633–651. [DOI] [PubMed] [Google Scholar]
- 48. Groelly F.J., Fawkes M., Dagg R.A., Blackford A.N., Tarsounas M. Targeting DNA damage response pathways in cancer. Nat. Rev. Cancer. 2023; 23:78–94. [DOI] [PubMed] [Google Scholar]
- 49. Alexandrov L.B., Kim J., Haradhvala N.J., Huang M.N., Tian Ng A.W., Wu Y., Boot A., Covington K.R., Gordenin D.A., Bergstrom E.N. et al. The repertoire of mutational signatures in human cancer. Nature. 2020; 578:94–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Venneti S., Garimella M.T., Sullivan L.M., Martinez D., Huse J.T., Heguy A., Santi M., Thompson C.B., Judkins A.R. Evaluation of histone 3 lysine 27 trimethylation (H3K27me3) and enhancer of zest 2 (EZH2) in pediatric glial and glioneuronal tumors shows decreased H3K27me3 in H3F3A K27M mutant glioblastomas. Brain Pathol. 2013; 23:558–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ragazzini R., Pérez-Palacios R., Baymaz I.H., Diop S., Ancelin K., Zielinski D., Michaud A., Givelet M., Borsos M., Aflaki S. et al. EZHIP constrains Polycomb Repressive Complex 2 activity in germ cells. Nat. Commun. 2019; 10:3858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Jain S.U., Do T.J., Lund P.J., Rashoff A.Q., Diehl K.L., Cieslik M., Bajic A., Juretic N., Deshmukh S., Venneti S. et al. PFA ependymoma-associated protein EZHIP inhibits PRC2 activity through a H3 K27M-like mechanism. Nat. Commun. 2019; 10:2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Carvalho S., Vítor A.C., Sridhara S.C., Martins F.B., Raposo A.C., Desterro J.M.P., Ferreira J., de Almeida S.F. SETD2 is required for DNA double-strand break repair and activation of the p53-mediated checkpoint. eLife. 2014; 3:e02482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Aymard F., Bugler B., Schmidt C.K., Guillou E., Caron P., Briois S., Iacovoni J.S., Daburon V., Miller K.M., Jackson S.P. et al. Transcriptionally active chromatin recruits homologous recombination at DNA double-strand breaks. Nat. Struct. Mol. Biol. 2014; 21:366–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Adam S., Polo S., Almouzni G. Transcription recovery after DNA damage requires chromatin priming by the H3. 3 histone chaperone HIRA. Cell. 2013; 155:94–106. [DOI] [PubMed] [Google Scholar]
- 56. Beuzer P., Quivy J.P., Almouzni G. Establishment of a replication fork barrier following induction of DNA binding in mammalian cells. Cell Cycle. 2014; 13:1607–1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Botuyan M.V., Lee J., Ward I.M., Kim J.-E., Thompson J.R., Chen J., Mer G. Structural basis for the methylation state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in DNA repair. Cell. 2006; 127:1361–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Tuzon C.T., Spektor T., Kong X., Congdon L.M., Wu S., Schotta G., Yokomori K., Rice J.C. Concerted activities of distinct H4K20 methyltransferases at DNA double-strand breaks regulate 53BP1 nucleation and NHEJ-directed repair. Cell Rep. 2014; 8:430–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Roux K.J., Kim D.I., Raida M., Burke B. A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J. Cell Biol. 2012; 196:801–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Scott W.A., Campos E.I. Interactions with histone H3 & tools to study them. Front. Cell Dev. Biol. 2020; 8:701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Dumitrache L.C., McKinnon P.J. Polynucleotide kinase-phosphatase (PNKP) mutations and neurologic disease. Mech. Ageing Dev. 2017; 161:121–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Shen J., Gilmore E.C., Marshall C.A., Haddadin M., Reynolds J.J., Eyaid W., Bodell A., Barry B., Gleason D., Allen K. et al. Mutations in PNKP cause microcephaly, seizures and defects in DNA repair. Nat. Genet. 2010; 42:245–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Kalasova I., Hailstone R., Bublitz J., Bogantes J., Hofmann W., Leal A., Hanzlikova H., Caldecott K.W. Pathological mutations in PNKP trigger defects in DNA single-strand break repair but not DNA double-strand break repair. Nucleic Acids Res. 2020; 48:6672–6684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Tsukada K., Imamura R., Saikawa K., Saito M., Kase N., Miyake T., Ishiai M., Matsumoto Y., Shimada M. Phosphorylation of PNKP mediated by CDKs promotes end-processing of Okazaki fragments during DNA replication. 2021;
- 65. Kim J., Sturgill D., Sebastian R., Khurana S., Tran A.D., Edwards G.B., Kruswick A., Burkett S., Hosogane E.K., Hannon W.W. et al. Replication stress shapes a protective chromatin environment across fragile genomic regions. Mol. Cell. 2018; 69:36–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Xu X., Ni K., He Y., Ren J., Sun C., Liu Y., Aladjem M.I., Burkett S., Finney R., Ding X. et al. The epigenetic regulator LSH maintains fork protection and genomic stability via MacroH2A deposition and RAD51 filament formation. Nat. Commun. 2021; 12:3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Schmid J.A., Berti M., Walser F., Raso M.C., Schmid F., Krietsch J., Stoy H., Zwicky K., Ursich S., Freire R. et al. Histone ubiquitination by the DNA damage response is required for efficient DNA replication in unperturbed S phase. Mol. Cell. 2018; 71:897–910. [DOI] [PubMed] [Google Scholar]
- 68. Pommier Y. Topoisomerase I inhibitors: camptothecins and beyond. Nat. Rev. Cancer. 2006; 6:789–802. [DOI] [PubMed] [Google Scholar]
- 69. Zhang Y., Chang J.-F., Sun J., Chen L., Yang X.-M., Tang H.-Y., Jing Y.-Y., Kang X., He Z.-M., Wu J.-Y. et al. Histone H3K27 methylation modulates the dynamics of FANCD2 on chromatin to facilitate NHEJ and genome stability. J. Cell Sci. 2018; 131:jcs215525. [DOI] [PubMed] [Google Scholar]
- 70. Sarthy J.F., Meers M.P., Janssens D.H., Henikoff J.G., Feldman H., Paddison P.J., Lockwood C.M., Vitanza N.A., Olson J.M., Ahmad K. et al. Histone deposition pathways determine the chromatin landscapes of H3.1 and H3.3 K27M oncohistones. eLife. 2020; 9:e61090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Hauser P. Classification and treatment of pediatric gliomas in the molecular era. Children (Basel). 2021; 8:739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Castel D., Philippe C., Calmon R., Le Dret L., Truffaux N., Boddaert N., Pagès M., Taylor K.R., Saulnier P., Lacroix L. et al. Histone H3F3A and HIST1H3B K27M mutations define two subgroups of diffuse intrinsic pontine gliomas with different prognosis and phenotypes. Acta Neuropathol. 2015; 130:815–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Werbrouck C., Evangelista C.C.S., Lobón-Iglesias M.-J., Barret E., Le Teuff G., Merlevede J., Brusini R., Kergrohen T., Mondini M., Bolle S. et al. TP53 Pathway alterations drive radioresistance in diffuse intrinsic pontine gliomas (DIPG). Clin. Cancer Res. 2019; 25:6788–6800. [DOI] [PubMed] [Google Scholar]
- 74. Setton J., Zinda M., Riaz N., Durocher D., Zimmermann M., Koehler M., Reis-Filho J.S., Powell S.N. Synthetic lethality in cancer therapeutics: the next Generation. Cancer Discov. 2021; 11:1626–1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Mereniuk T.R., El Gendy M.A.M., Mendes-Pereira A.M., Lord C.J., Ghosh S., Foley E., Ashworth A., Weinfeld M. Synthetic lethal targeting of PTEN-deficient cancer cells using selective disruption of polynucleotide kinase/phosphatase. Mol. Cancer Ther. 2013; 12:2135–2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Dubois F.P.B., Shapira O., Greenwald N.F., Zack T., Wala J., Tsai J.W., Crane A., Baguette A., Hadjadj D., Harutyunyan A.S. et al. Structural variants shape driver combinations and outcomes in pediatric high-grade glioma. Nat Cancer. 2022; 3:994–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw mass spectrometry data is available for download from https://massive.ucsd.edu/, accession numbers MSV000087736 and MSV000089173 (27) and from ProteomeXchange with identifier PXD041132. Whole genome sequencing data is available under EGAS00001000575, EGAS00001001139, EGAS00001000572, EGAS00001000192, and under dbGaP accession number phs002380.v1.p1 (76). Other raw data files are deposited on figshare: https://doi.org/10.6084/m9.figshare.23259920.v1.