Abstract
Co-transcriptional processing of nascent pre-mRNAs by the spliceosome is vital to regulating gene expression and maintaining genome integrity. Here, we show that the deficiency of functional U5 small nuclear ribonucleoprotein particles (snRNPs) in Drosophila imaginal cells causes extensive transcriptome remodeling and accumulation of highly mutagenic R-loops, triggering a robust stress response and cell cycle arrest. Despite compromised proliferative capacity, the U5 snRNP-deficient cells increased protein translation and cell size, causing intra-organ growth disbalance before being gradually eliminated via apoptosis. We identify the Xrp1-Irbp18 heterodimer as the primary driver of transcriptional and cellular stress program downstream of U5 snRNP malfunction. Knockdown of Xrp1 or Irbp18 in U5 snRNP-deficient cells attenuated JNK and p53 activity, restored normal cell cycle progression and growth, and inhibited cell death. Reducing Xrp1-Irbp18, however, did not rescue the splicing defects, highlighting the requirement of accurate splicing for cellular and tissue homeostasis. Our work provides novel insights into the crosstalk between splicing and the DNA damage response and defines the Xrp1-Irbp18 heterodimer as a critical sensor of spliceosome malfunction and mediator of the stress-induced cellular senescence program.
Graphical Abstract
Graphical Abstract.
Introduction
Pre-mRNA splicing catalyzed by the spliceosome is central to the regulation of gene expression in eukaryotes. Its physical and functional coupling with transcription ensures the production of cell- and condition-specific transcript variants required for development and the function of different cell types in multicellular organisms (1–3). Defects in the splicing machinery caused by mutations in spliceosome components or their dysregulation have been linked to tissue- or even cell-type-specific pathologies in humans and correlated with cellular senescence and aging (4–8). The diversity of processes that depend on functional spliceosomes, and the phenotypes inflicted by spliceosome dysfunction, underscores the complexity of the molecular machine and highlights the need for an in-depth understanding of its integral role in gene regulation in an in vivo context.
The spliceosome is a large ribonucleoprotein complex composed of five small nuclear ribonucleoprotein particles (snRNPs), each of which comprises specific uridine-rich small nuclear RNA (U snRNA) and a set of core and auxiliary proteins (9). In multicellular eukaryotes, most introns are removed by major (U2) spliceosomes, which contain U1, U2, U4/U6 and U5 snRNPs. The minor (U12) spliceosome, comprising U11, U12, Y4atac, U5 and U6atac (10,11), processes <0.5% of introns (12). The U5 snRNP is the only snRNP common to both spliceosome types, playing a vital role in spatial organization of the catalytic center, positioning of critical U snRNA and substrate exon residues, and timely activation of the spliceosome (9,13–14). The U5 snRNP consists of U5 snRNA, Sm-protein ring, several accessory proteins and core factors, including the pre-mRNA processing factor 8 (PRPF8/Prp8) (15). Despite its universal requirement for pre-mRNA splicing, mutations in Prp8 cause retina-specific disease, an autosomal dominant form of the Retinitis Pigmentosa type 13 (RP13) (6,16). The U5 snRNP joins the spliceosome as part of the U4/U6•U5 tri-snRNP, following a highly coordinated, multistep assembly process that takes part in both the cytoplasm and the nucleus. The cytoplasmic phase requires Ecdysoneless (Ecd), an evolutionarily conserved protein that serves as an adapter of the PAQosome (particle for arrangement of quaternary structure) complex (17–20). PAQosome facilitates the assembly of various crucial multiprotein complexes, including U4 and U5 snRNPs, small nucleolar and telomerase ribonucleoproteins, RNA polymerases and phosphatidylinositol 3-kinase-related kinases (PIKKs) complexes such as ataxia-telangiectasia mutated (ATM), ATM- and Rad3-Related (ATR), mammalian target of rapamycin (mTOR) and tuberous sclerosis complex (TSC) (21). Ecd promotes U5 snRNP biogenesis through interactions with Prp8, the Sm ring protein SmD3 and U5 snRNA (17). In the absence of Ecd, Prp8 is destabilized, and U5 snRNP biogenesis is stalled, leading to apoptosis and ultimately lethality in the Drosophila model (17,22). Similar to dysfunction of other U5 snRNP core components (16,23–25), Ecd deficiency causes genome-wide changes in gene expression and splicing, and has been linked to a variety of phenotypes, including reduced proliferation (26), aberrant nuclear mRNA export (27) and activation of both unfolded protein- and oxidative stress-responses (17,28). However, knowledge on the cascade of events and mechanisms that trigger or contribute to loss of cellular homeostasis, and ultimately death, remains incomplete.
Here, we exploit the genetic accessibility of the Drosophila model to elucidate the course of events arising from scarcity of the U5 snRNP inflicted by knockdown of the biogenesis factor Ecd or an RP13-associated mutation in Prp8. We identify Xrp1 and its dimerizing partner Irbp18 as key drivers of transcriptional changes and cellular stress responses to U5 snRNP malfunction. While Xrp1/Irbp18 knockdown alleviated the harmful consequences of prolonged stress signaling, it did not restore spliceosome fidelity, highlighting the primary role and the requirement of U5 snRNP in maintaining cellular homeostasis by controlling pre-mRNA splicing.
Materials and methods
Fly stocks and husbandry
Drosophila stocks and crosses were maintained at 25°C (unless specified otherwise) on a diet consisting of 0.8% agar, 8% cornmeal, 1% soymeal, 1.8% dry yeast, 8% malt extract and 2.2% sugar-beet syrup, supplemented with 0.625% propionic acid and 0.15% Nipagin. The following fly stocks were used: w1118 (BDSC; RRID:BDSC_3605), ecd1 (29), prp8del14 (23), UAS-Prp8[S2178F](23), nub-Gal4, UAS-myr-mRFP (nub>mRFP) (17), nub-Gal4 (17), UAS-ecdRNAi (22), UAS-xrp1RNAi #1 (BDSC; RRID:BDSC_34521), UAS-xrp1RNAi #2 (VDRC, ID 107860), UAS-Irbp18RNAi (VDRC, ID 101871), UAS-p35 (BDSC; RRID:BDSC_5072), tub-Gal80TS (BDSC; RRID:BDSC_7017), UAS-LT3-Dam (30), UAS-LT3-Dam-RNase H1WT (this study, see below), ubi-FUCCI (BDSC; RRID:BDSC_55124), TRE-DsRED-2R (31), xrp1-lacZ (BDSC; RRID:BDSC_11569), gstD1-GFP (32), p53::GFP[FlyFos] (VDRC, ID 318453), egr::GFP[FlyFos] (VDRC, ID 318615), nos-phiC31, attP2 (BDSC; RRID:BDSC_25710), Irbp18::GFP[FlyFos] (VDRC, ID 318564), Irbp::GFP[FlyFos] (VDRC, ID 318033). See Supplementary Table S1 for all stock and recombinant line genotypes used in this study.
Tissue dissection and immunostaining
Wing imaginal discs (WDs) were dissected on day 7 after egg laying (AEL) (unless specified otherwise) in phosphate buffered saline (PBS) and fixed using 4% paraformaldehyde (PFA) in PBS containing 0.1% TritonX-100 (PBS-T) for 25 min at room temperature. Blocking was performed with 0.3% bovine serum albumin (BSA) in PBS-T for 30 min at room temperature. Incubation with primary antibodies diluted in the blocking solution was performed overnight at 4°C while nutating. The following primary antibodies were used in this study: anti-RFP (rabbit, 1:1000; MBL #PM005; RRID:AB_591279), anti-GFP (rabbit, 1:500; Thermo Fisher Scientific #G10362; RRID:AB_2536526), anti-GFP (goat, 1:500; Abcam #ab6673; RRID:AB_305643), anti-p53 antibody mix (mouse, 1:200; DSHB #p53 25F4, 7A4; RRID:AB_579787, RRID:AB_579786), anti-phospho-Histone H2AvD (rabbit, 1:300; Rockland #600–401-914S; RRID:AB_11183655), anti-Ecd C-terminal (rat, 1:700) (22), anti-Ecd N-terminal (rat, 1:700) (22), anti-Fibrillarin (rabbit, 1:200; Abcam #ab5821; RRID:AB_2105785), anti-Lamin (mouse, 1:1000; DSHB #adl67.10; RRID:AB_528336), anti-Prp8 (rabbit, 1:1000) (23), anti-phospho-histone H3 (rabbit, 1:500; Cell Signaling Technology #9701; RRID:AB_331535), anti-Dcp-1 (rabbit, 1:500; Cell Signaling Technology #9578; RRID:AB_2721060), anti-MMP1 antibody mix (mouse, 1:100; DSHB #14A3D2, #3A6B4, #3B8D12; RRID:AB_579782, RRID:AB_579780, RRID:AB_579781), anti-puromycin (mouse, 1:1000; Millipore #MABE343; RRID:AB_2566826), anti-beta-Galactosidase (mouse, 1:100; DSHB #jie7; RRID:AB_528101). After five washes in PBS-T, staining with the appropriate secondary antibodies, diluted 1:1000 in blocking solution, was performed overnight at 4°C while nutating: donkey anti-rabbit Alexa Fluor 488 (Jackson ImmunoResearch #111–545-144; RRID:AB_2338052), donkey anti-goat Cy2 (Jackson ImmunoResearch #705–225-147; RRID:AB_2307341), donkey anti-rabbit Cy5 (Jackson ImmunoResearch #711–175-152; RRID:AB_2340607), donkey anti-mouse Cy5 (Jackson ImmunoResearch #715–175-151; RRID:AB_2340820), donkey anti-rat Cy5 (Jackson ImmunoResearch #712–175-153; RRID:AB_2340672), donkey anti-mouse Cy3 (Jackson ImmunoResearch #715–165-151; RRID:AB_2315777), donkey anti-rat FITC (Jackson ImmunoResearch #712–095-153; RRID:AB_2340652).
Image acquisition and processing
Images of Drosophila WDs were acquired with the Olympus FV-1000 confocal microscope using 20× UPlan S-Apo (NA 0.85), 40× UPlanApo (NA 1.30) and 60× UPlanApo (NA 1.35) objectives. All micrographs shown are maximum Z-projections generated with FluoView FV-10ASW software using 1.2 or 1.4 μm step size unless specified otherwise (Olympus, RRID:SCR_014215). Panel assembly and brightness/contrast adjustments were done using Adobe Photoshop CC (Adobe Systems Inc., RRID:SCR_014199). Images used for quantification of fluorescence intensity were acquired using the same settings. WD size was measured by manual selection of the entire WD, or the nubbin domain marked by mRFP and expressed as μm2. Quantification of pH3 foci was performed on maximum projections of confocal Z-stacks using the ‘Analyze Particles’ command within ImageJ FIJI with a minimum size cutoff of 3 μm2 after image thresholding using the maximum entropy method (33). Apoptosis was measured as the total area of Dcp-1 signal within the nubbin domain in maximum projections of confocal Z-stacks, after thresholding (default method). Levels of endogenous p53 or GFP-tagged p53 are represented as ratios of mean gray values of the fluorescence within and outside of the nubbin domain. Nucleolar sizes were determined based on immunostaining with a Fibrillarin antibody. Images were first de-noised (‘Remove speckles’ function), Gaussian blur was applied (radius value of 2), and background was subtracted (rolling ball radius of 20 pixels). Images were thresholded and the area of nucleoli was measured. Cell sizes were measured by manual selection of the membrane mRFP signal driven by the nub-Gal4 driver. For each biological replicate, all cells spanning the diagonal corners of a micrograph were measured. All quantification related processing and measurements were performed with ImageJ FIJI (RRID:SCR_002285). Statistical significance was determined and data plotted using GraphPad Prism (RRID:SCR_002798).
Pupation and eclosion rate
Female flies (nub>mRFP) crossed to males from different RNAi stocks were allowed to lay eggs for 24 h. Early third instar larvae were transferred to fresh vials and kept at 25°C. The number of pupae and eclosed adults was calculated as a percentage of the transferred larvae and formed pupae, respectively.
Puromycin incorporation assay and quantification
Third instar wandering larvae (day 7 AEL) were inverted in PBS, and the intestine and fat body were removed. The tissues were transferred to a solution of 5 μg/ml of puromycin (Thermo Fisher Scientific #A1113803) in Shield and Sang M3 insect media (Sigma-Aldrich #S8398) containing 10% FBS (Gibco, Life Technologies) for 20 min. Samples were then fixed in 4% PFA in PBS-T for 40 min and processed for immunostaining as described above. Regions of interests were outlined based on the nub>mRFP signal and the mean gray values of puromycin antibody signal measured. The mean gray values of the hinge and notum regions of the WDs were used to calculate the fluorescence signal intensity ratio.
EdU incorporation assay
The EdU incorporation assay was performed using the Click-iT Plus EdU cell proliferation kit for imaging (ThermoFisher Scientific #C10638). Third instar wandering larvae (day 7 AEL) were inverted in PBS and incubated in 15 μM EdU in PBS for 20 min at room temperature with gentle shaking. Upon washing twice with 3% BSA in PBS, samples were fixed in 3.7% formaldehyde for 15 min at room temperature. Samples were then washed and permeabilized using PBS-T. After washing twice with 3% BSA in PBS, the samples were incubated in Click-It reaction cocktail for 30 minutes at room temperature protected from light, according to manufacturer’s recommendations. The samples were washed once, mounted and imaged as described above.
RNA isolation and RNA sequencing
Approximately 150 WDs were dissected from third instar larvae (day 7 AEL) in PBS and snap-frozen in liquid nitrogen. Total RNA was isolated according to standard TRI Reagent protocol (Sigma Aldrich #T9424), followed by DNase I treatment (Invitrogen #AM2238) and repurification as described in (34). For RNA sequencing, 2 μg of total RNA was used for library preparation (Illumina TruSeq Stranded total RNA Ribo-Zero) with four biological replicates per experimental group. Pair-end sequencing (100 bp) was performed using the Illumina HiSeq 2000 platform. Image analysis and base calling were carried out with the Illumina RTA software at run time.
Differential gene expression and splicing analyses
Initial quality control of the raw data was performed using FastQC (RRID:SCR_014583). Illumina sequencing adapters were removed from the sequencing reads with the cutadapt tool version 3.5 (35) (RRID:SCR_011841). For differential gene expression analysis, kallisto v0.46.1 (36) (RRID:SCR_016582) was used for pseudoalignment of obtained reads to the complete Drosophila melanogaster transcriptome (BDGP6.28, Ensembl release 102) and transcript-level abundance quantification using 100 bootstrap samples. Summarization of the obtained estimated counts to gene-level was performed using tximport v1.22.0 (37) (RRID:SCR_016752). Differential gene expression was determined using DESeq2 v1.34.0 (38) (RRID:SCR_015687). Heatmaps comparing expression levels of selected genes between samples were constructed from DESeq2 normalized counts and log2FC values using Morpheus (https://software.broadinstitute.org/morpheus/). Isoform-specific expression was assessed by sleuth v0.30.0 (39). For differential splicing analysis, the reads were aligned to the Drosophila melanogaster genome (BDGP6.28, Ensembl release 102) using STAR v2.7.9a (40) (RRID:SCR_004463) in basic two-pass mode. The resulting alignments were loaded into NxtIRFcore v1.2.1 for intron retention analysis on individual biological replicates in R. The resulting output tables were then used with the IRFinder v2.0.0 (41) DESeq2 wrapper for differential intron retention analyses using default settings. To analyze events other than intron retention, the data were analyzed using SpliceWiz v.1.1.5. (42) and a reduced stringency utilizing nominal instead of Adj. P-values. A splice junction was considered if it was present in at least one of the biological replicates, and at least 10 reads covering the junction.
Gene Ontology (GO) term enrichment analysis was performed using ShinyGO v0.76 (43) using a custom background gene list containing all genes detected by the RNA-seq analysis. Fisher’s exact test with false discovery rate correction was used. Metascape v3.5 (44) was used to generate protein–protein interaction and MCODE networks (45) using the default settings.
Quantitative and semi-quantitative reverse transcription PCR (RT-PCR) intron retention assay
cDNA was synthesized from 2 μg of total RNA using GoScript Reverse Transcriptase (Promega #A5003) with random hexamers according to the manufacturer’s protocol. Quantitative polymerase chain reaction (PCR) was performed in three technical replicates and at least three biological replicates. Expression values were normalized to the levels of tbp transcripts (FlyBase ID: FBtr0071679). For calculation of the fold changes, ΔΔCT method was used (46). Statistical significance of gene expression changes was determined using two-way ANOVA with Sidak’s correction for multiple comparisons.
For semi-quantitative PCR, a distinct set of biological samples independent from those used for RNA sequencing was generated. The cDNA synthesis followed the same protocol. Fragments of interest were amplified using the EmeraldAmp GT PCR Master Mix (Takara #RR310B) with either intron- or exon-specific forward primers combined with a common exon–exon junction-specific reverse primer. This primer pairing allowed detection of unspliced or spliced products, respectively. In the case of prp19 due to constraints related to exon length, either an exon–exon or intron–exon junction forward primer was combined with a universal reverse primer binding exon 2 to amplify spliced or unspliced products, respectively. The PCR products were resolved on a 3% agarose gels and stained with ethidium bromide. Intensity profiles of the resulting bands were plotted using Image FIJI. The area under the profile curves was then measured to calculate the intron–exon ratios. Statistical significance was determined and data plotted using GraphPad Prism. See Supplementary Table S2 for primer sequences.
In vivo Targeted DamID
The coding sequence of RNase H1 (isoform RA) was amplified from cDNA and cloned into the pUASt-attB-LT3-NDam vector (30) using the NotI and XhoI restriction enzymes (NEB #R0146S and #R0189S, respectively). Transgenic flies were obtained by insertion of the pUASt-attB-LT3-NDam-RNase H1 construct into the attP2 site on the third chromosome (68A4) using PhiC31 integrase-mediated transgenesis (BDSC; RRID:BDSC_25710). The expression of the UAS-based Dam-fusion constructs in the wing pouch using the nub-Gal4 driver was induced by shifting the developing larvae from room temperature to 29°C for 24 h. This resulted in the inactivation of the temperature sensitive Gal80TS while simultaneously rendering the mutant Ecd protein produced from the homozygous ecd1 loci non-functional (17). Third instar larvae were dissected on day 7 AEL. Samples were collected in four biological replicates, each containing 120 WDs. Sample processing was performed according to Vogel et al. (47) with modifications described in Mundorf et al. (48). Preparation for sequencing was performed as previously described (48). Sequencing libraries were generated using the Illumina TruSeq DNA kit library preparation protocol and sequenced on an Illumina HiSeq 2000 machine as pair-end 50 bp reads. The resulting sequencing reads files were trimmed using the cutadapt tool version 3.5 (35) (RRID:SCR_011841) and processed using the damidseq_pipeline v1.4.6 (49) to obtain GFF files containing the normalized log2 ratio (Dam-fusion/Dam) for each GATC fragment in the Drosophila genome. Dam-RNase H1WT binding was assessed using a peak finding algorithm based on the one described by Wolfram et al. (50). False discovery rate (FDR) was calculated for peaks (formed of two or more consecutive GATC fragments) for the individual replicates. Peaks with FDR < 0.01 were classified as significant. Additionally, a mean log2 ratio threshold of 0.5 for each peak was implemented for peak-to-gene association. Significant peaks present in all four replicates were used to form a final peak file. Any gene within 5 kb of a peak (with no other genes in between) was identified as a peak-associated gene. Read alignment files were converted into bedGraph format for coverage visualization using the bamCoverage tool with a bin size of 10 bp. Paired reads were extended, and data were normalized by 1× depth (reads per genome coverage, RPGC) with effective genome size set to 142 573 017 bp. Averages of Dam-Fusion/Dam ratios for each GATC fragment from four biological replicates were used to generate the corresponding genome browser tracks. To identify differential binding events in each condition, the difference between binding intensities for each genomic GATC fragment (represented as log2 Dam-RNase H1/Dam-only ratios) was calculated. Significant peaks with higher intensities in each condition compared to the other were then associated with the nearest gene (maximum distance of 5 kb). The genomic feature distribution was obtained using the ChipSeeker R package (51).
Results
Spliceosome malfunction induces stress signaling and progressive apoptosis
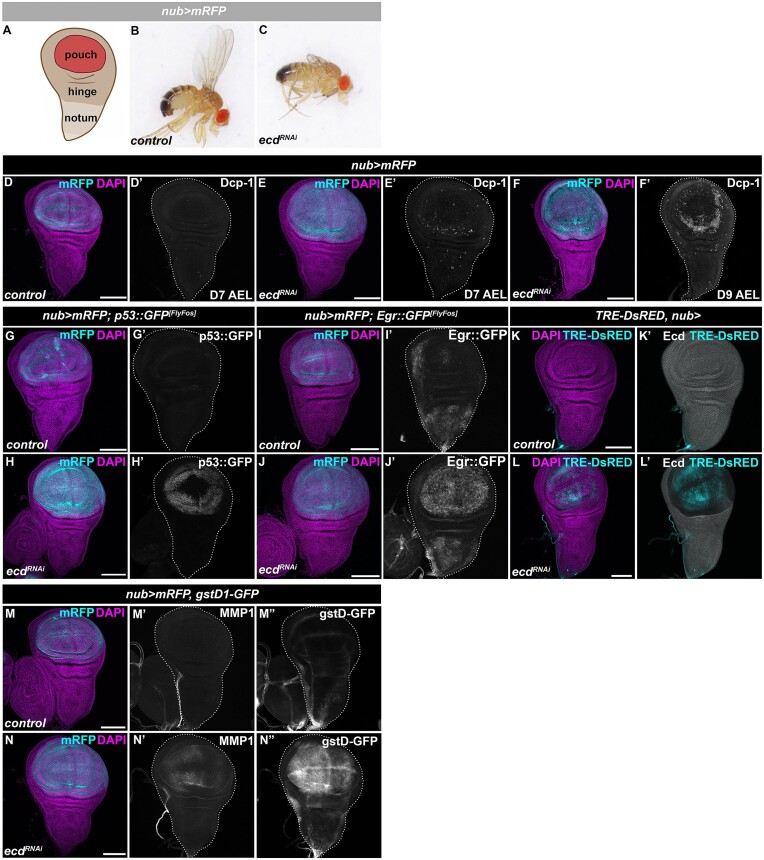
Ecd has emerged as a crucial mediator of U5 snRNP biogenesis (17–19,22). To characterize the sequence of events triggered by spliceosome dysfunction on the cellular and tissue level, we reduced Ecd function in the pouch region of the developing WD by expressing transgenic ecd RNAi (UAS-ecdRNAi), under the control of the nubbin-Gal4 driver (Figure 1A) (hereafter referred to as nub>). Importantly, suppressing ecd expression in actively cycling imaginal cells of the WD pouch domain allowed normal development until the late third instar larval stage. However, in contrast to control larvae (nub>/+), the larval-to-pupal transition of nub>ecdRNAi animals was delayed, with the majority dying during metamorphosis. The few adults that managed to eclose lacked wing blades (Figure 1B and C).
Figure 1.
Ecd deficiency results in increased cell death and stress response signaling involving JNK and p53. (A) Schematic representation of the Drosophila WD comprising pouch, hinge and notum generated using BioRender.com (Biorender, RRID:SCR_018361). The nub-Gal4 driver is active the pouch region highlighted by mRFP expression. (B,C) Representative image of control (nub>mRFP) (B) and a rare nub>mRFP ecdRNAi adult escaper male fly lacking the wing blade (C). (D–F) Immunostaining shows increased Dcp-1 signal in pouch region of nub>mRFP ecdRNAi WD on day 7 (E’) with further enhancement on day 9 AEL (F’), compared to D7 AEL control (nub>mRFP) WD (D’). Note, the Dcp-1 signal is not ubiquitous across the nubbin domain but enriched around the perimeter of the nub>mRFP ecdRNAi wing pouch (E’, F’). (G–J) Immunostaining shows increased levels of GFP-tagged p53 and Egr FlyFos transgenes in nub>mRFP ecdRNAi WDs at 7 days AEL (H,J) compared to control WDs (G,I). (K, L) JNK activity is increased while Ecd levels are reduced in pouch domain of nub>ecdRNAi (L) WD compared to control (nub>) WDs (K) as determined by immunostaining for TRE-DsRED transgenic reporter and Ecd protein. (M,N) Immunostaining shows increased levels of MMP1 (N’) and expression of a gstD1-GFP transgenic reporter (N’’) in pouch region of nub>mRFP ecdRNAi WDs compared to control WDs (M’, M’’). The confocal micrographs of WDs show maximum intensity projections of multiple confocal sections. DAPI and mRFP label nuclei and the nubbin domain, respectively. WDs are outlined by white dotted lines based on DAPI staining; scale bars: 100 μm. See also Supplementary Figure S1.
In agreement with previous studies (17,22,52) and the adult wingless phenotype (Figure 1C), Ecd deficiency caused progressive apoptosis within the pouch domain, as demonstrated by immunostaining of activated Dcp-1, the main effector caspase of Drosophila (Figure 1D–F and Supplementary Figure 1A–D). Compared to a moderate enrichment of apoptotic cells in nub>ecdRNAi WDs on day 7 AEL relative to control, large patches of Dcp-1 positive cells were observed in delayed 9-day-old larvae (Figure 1D–F and Supplementary Figure 1A,B). In contrast to the limited cell death on day 7 AEL, wing pouch cells with reduced Ecd levels showed a strong activation of stress-induced signaling pathways and their downstream effectors. Using immunostaining of endogenous or a transgenic FlyFos-based tagged protein, we detected upregulation of p53 (Figure 1G, H and Supplementary Figure 1E–G), an evolutionarily conserved regulator of genome stability, cell survival, senescence and apoptosis (53–56). The enrichment of p53 protein in the nub>ecdRNAi wing pouch cells coincided with the activation of the Jun-N-terminal Kinase (JNK) signaling pathway, as demonstrated by the increased expression of a tumor necrosis factor (TNF) ligand Eiger (Egr) (Figure 1I, J). Egr serves as a primary upstream activator of JNK signaling (57), as evidenced by the TRE-DsRED transgenic transcriptional reporter (31) (Figure 1K, L). Additionally, the accumulation of a JNK-regulated target gene, Matrix metalloprotease 1 (MMP1) (58), was observed (Figure 1M’, N’). Moreover, Ecd-deficient cells upregulated gstD1-GFP (Figure 1M", N"), a transcriptional reporter sensitive to changes in cellular redox homeostasis and proteotoxic stress (32,59,60). These results highlight the activation of p53- and JNK-mediated signaling among the early responses to reduced Ecd levels. Furthermore, gstD1-GFP and p53 upregulation was also observed in imaginal disc cells overexpressing the pathogenic RP13-linked mutant variant of Prp8 protein (Drosophila Prp8[S2178F]/human PRPF8[S2118F]) (Supplementary Figure 1H–M), suggesting a shared response to U5 snRNP malfunction, inflicted either by scarcity of the biogenesis factor or mutation in the core splicing factor. Importantly, blocking apoptosis by overexpression of p35, a pan-caspase inhibitor, prevented physical cell elimination (Supplementary Figure 1A–D) but did not restore the cellular homeostasis (Supplementary Figure 1E–G, J–M) required for normal adult appendage development. Together, this suggests that rather than acting as robust apoptotic inducers that ensure rapid cell elimination, p53 and JNK may work in concert to mount cytoprotective responses promoting cellular senescence-like state to delay cell death.
Ecd scarcity induces signs of DNA damage and increases RNase H1 binding to DNA
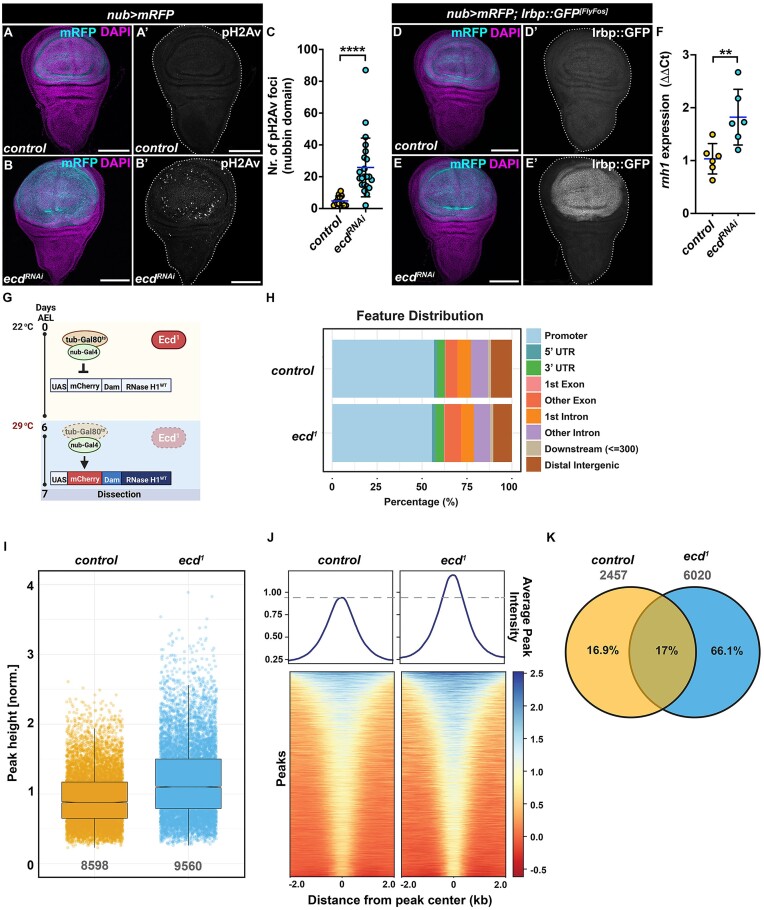
One of the emerging hallmarks of spliceosome dysfunction is DNA damage and loss of genome integrity (61–64). This damage manifests as DNA double-strand breaks (DSBs) and the accumulation of RNA:DNA hybrid structures, known as R loops, in which the nascent transcript anneals to the DNA template strand (65). The fact that loss of Ecd interferes with U5 snRNP biogenesis and induces the major DNA damage sensor p53 indicated that Ecd-deficient cells could suffer from DNA damage and ectopic R-loop formation. Indeed, immunostaining revealed increased number of pH2Av foci, marking DSBs, in Ecd depleted cells of the wing pouch compared to the rest of the tissue and control WDs (Figure 2A–C). Furthermore, Ecd-deficient cells showed upregulation of the Inverted repeat-binding protein (Irbp) (Figure 2D, E), the Drosophila homolog of the human XRCC6 (Ku70 protein), which forms heterodimers with Ku80 to participate in DSB repair via nonhomologous end joining (NHEJ) (66,67). Importantly, strong enrichment of Irbp::GFP was also observed in wing pouch cells overexpressing the pathogenic Prp8[S2178F] protein (Supplementary Figure 1L, M). In addition, we detected increased expression of RNase H1 (rnh1) mRNA, an enzyme that is recruited to, and degrades the RNA moiety of R-loops (68), in WDs bearing Ecd-deficient cells (Figure 2F). To map genome-wide distribution of RNase H1, as a proxy for R-loop formation, we performed Targeted DamID (TaDa) (30) on control and ecd1 homozygous mutant WDs expressing Dam-RNase H1 fusion protein in the nubbin domain (Figure 2G). The Dam only expressing control and ecd1 mutant WD samples served as normalization controls for the experiment. In both control and ecd1 mutant imaginal tissues, the majority of Dam-RNase H1 peaks were associated within promoter-proximal regions (Figure 2H). However, ecd1 mutants showed an increase in both the mean intensity and total number of peaks (Figure 2I, J). Furthermore, the analysis revealed a 2.45-fold higher number of genes associated with increased Dam-RNase H1 binding in ecd1 mutants than in controls (Figure 2K and Supplementary Dataset S1). Collectively, these data show that spliceosome malfunction inflicted by Ecd loss results in DNA damage. The increased expression and binding of RNase H1 in Ecd-deficient cells further suggest aberrant R-loop formation, which may contribute to the dysregulation of gene expression and genome instability (65,69).
Figure 2.
DNA damage induced by Ecd loss correlates with increased RNase H1 binding to DNA. (A–C) Representative confocal micrographs (A, B) and quantification (C) demonstrate an increase in phosphorylation of H2Av in wing pouch of nub>mRFP ecdRNAi (B) (n = 22) compared to control WDs (A) (n = 13) at day 7 AEL as visualized with an antibody against phosphorylated histone variant H2Av (pH2Av); scale bars: 100 μm. For quantification (C), the number of pH2Av foci was quantified within the nubbin domain. Unpaired nonparametric two-tailed Mann–Whitney test was used to calculate P-values. (D–E) Wing pouch-specific knockdown of ecd (nub>mRFP ecdRNAi) increases expression of Irbp::GFP FlyFos reporter (E’) compared to control WDs (D’) at day 7 AEL; scale bars: 100 μm. (F) Fold change in expression of endogenous rnh1 transcript in nub>mRFP ecdRNAi WDs relative to control as determined by RT-qPCR analysis (ΔΔCt), n = 6. Levels of TATA-binding protein (tbp) transcripts were used for normalization. Unpaired Student’s t-test assuming equal variances was used to determine statistical significance. (G) Schematic representation of the TaDa experimental design taking advantage of the temperature-sensitive ecd1 mutant allele and an α-tubulin promoter-driven transcriptional repressor Gal80ts (tub-Gal80ts). Gal80ts blocks expression of UAS-mCherry-Dam-RNase H1WT or UAS-mCherry-Dam fusion construct by the nub-Gal4 driver in ecd1 homozygous mutant or control larvae when kept at the permissive temperature (25°C). On day 6 AEL, the larvae were transferred to a non-permissive temperature (29°C) to simultaneously abolish the function of Ecd and induce expression of the UAS-Dam transgenes. WDs samples were dissected on day 7 AEL. The scheme was generated using BioRender.com. (H) Distribution of significant Dam-RNase H1 peaks in control and ecd1 conditions, in relation to different genomic features expressed as percentage of all features. (I) Scatter plot of intensities of all significant Dam-RNase H1 peaks in control and ecd1 WDs. The summary of the data is shown as a boxplot and interquartile range (IQR). Whiskers show 1.5*IQR. (J) Dam-RNase H1 peak intensity profiles across a 4 kb region centered on all significant peaks detected in both control and ecd1 WDs. The heatmap color and profile plots show the higher average Dam-RNase H1 peak intensity across all significant peaks in ecd1 WDs. (K) Venn diagram shows the overlap between genes in proximity of Dam-RNase H1 binding events that are higher in either control or ecd1 WDs. The majority of GATC fragments throughout the genome are characterized by higher Dam-RNase H1 peak intensities in ecd1 compared to the control. The overlap between two groups represents gene-proximal local redistribution of Dam-RNase H1 binding events. The confocal micrographs of WDs show maximum intensity projections of multiple confocal sections unless stated otherwise. DAPI and mRFP label nuclei and the nubbin domain, respectively. WDs are outlined by white dotted lines based on DAPI staining. Data in charts represent means ± SD, **P < 0.01, ****P < 0.0001. See also Supplementary Figure S1 and Supplementary Dataset S1.
Ecd deficiency results in G2-phase cell cycle arrest and aberrant growth
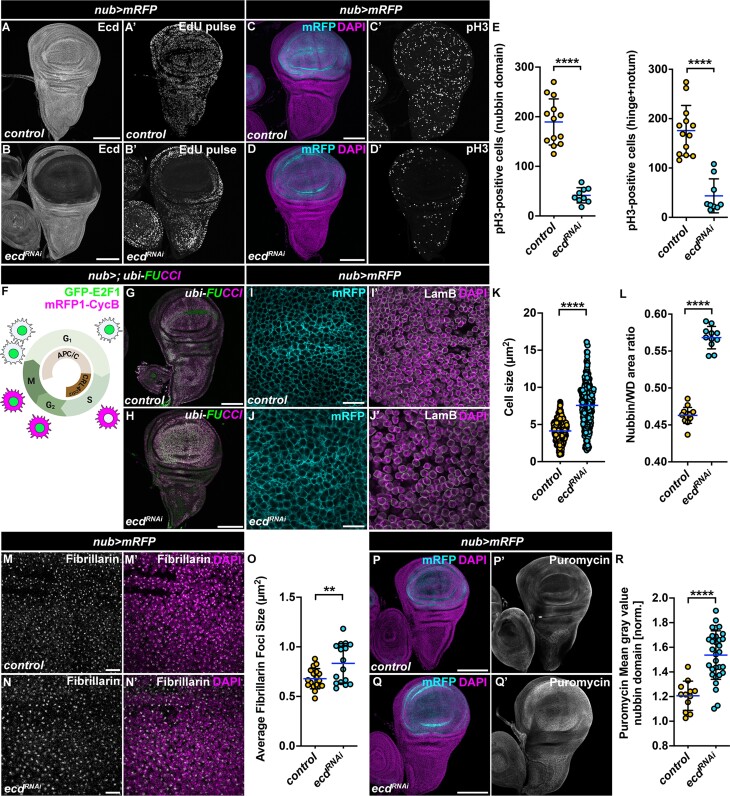
In the physiological context, accumulation of DNA damage triggers DNA-damage checkpoints that cause transient or permanent cell cycle arrest depending on the extent of damage and the capacity of cell to repair it. To determine the replicative and proliferative capacity of WD cells with compromised Ecd function, we performed EdU labeling along with immunostaining for the mitotic marker, phosphorylated Histone H3 (pH3). We observed significant reduction in the number of EdU- and pH3-positive cells in the nubbin compartment of the nub>ecdRNAi WDs compared to controls (Figure 3A–E), indicative of cell cycle stalling or arrest. However, the decrease in mitosis also occurred in hinge and notum region of nub>ecdRNAi WDs (Figure 3E), suggesting possible intra-organ and systemic regulation of proliferation (70,71). To define the precise cell cycle stage in which the nub>ecdRNAi WD cells arrest, we utilized the transgenic fly ubi-FUCCI system (fluorescent ubiquitination-based cell cycle indicator), based on fluorochrome-tagged degrons from the Cyclin B (mRFP1-CycB1-266) and E2F1 (GFP-E2F11-230) proteins, expressed from the ubiquitin promoter (72). Under normal conditions CycB is targeted for destruction by the Anaphase-promoting Complex (APC/C) during mitosis and early G1 phase, while E2F1 is degraded by CRL4Cdt2- during S phase. Accumulation of both mRFP1- CycB1-266 and GFP-E2F11-230 marks cells in G2, a phase associated with cell growth and elevated protein synthesis (Figure 3F). Given the delayed apoptosis and high JNK activity that has been shown to promote G2-phase stalling (73), we hypothesized that Ecd-deficient cells would be positive for both fluorescent degrons. Indeed, in contrast to control WDs, which comprised cells in all phases of the cell cycle in a developmentally controlled cell-cycle pattern (Figure 3G), the majority of nub>ecdRNAi WD cells were arrested in G2 phase (Figure 3H). The accumulation of G2 stalled/arrested cells was also evident in the wing pouches overexpressing the RP13-linked mutant Prp8[S2178F] protein (Supplementary Figure S1N–O). Despite reduced mitotic activity (Figure 3C–E), Ecd-deficient cells retained their growth potential and grew significantly larger compared control nub>WD cells (Figure 3I–K). This manifested as an increased size of the nubbin domain of nub>ecdRNAi WDs relative to the combined size of the hinge and notum (Figure 3L). Interestingly, ecd knockdown cells also contained larger nucleoli (Figure 3M–O), as determined by immunostaining of Fibrillarin, a component of C/D box small nucleolar ribonucleoprotein (snoRNP) particles (74). As changes in nucleoli and cell size have been correlated with the level of ribosomal biogenesis/translation (75,76), we asked if ecd silencing altered the rate of protein synthesis using puromycin-based labeling (77). Intriguingly, immunostaining of WDs following a pulse treatment of puromycin showed significantly higher signal in Ecd-depleted wing pouch cells compared to the rest of the tissue and to control WDs (Figure 3P–R). Together, these results demonstrate that the activation of stress signaling in Ecd-deficient cells correlates with their arrest in the G2 phase of the cell cycle and attenuation of mitotic activity within the wing pouch, extending to neighboring parts of the tissue. Despite genomic instability, the G2 arrested Ecd-deficient cells increase in size, likely due to elevated translational activity, which contributes to intra-organ growth disbalance affecting organ proportions.
Figure 3.
Cell cycle arrest caused by reduced Ecd is accompanied by increased cell growth and translation. (A, B) In vivo EdU incorporation is reduced in nub>mRFP ecdRNAi WDs (B’) compared to control (A’). Immunostaining with an Ecd-specific antibody shows efficient reduction of Ecd protein in the pouch region of WDs following overexpression of an UAS-ecdRNAi transgene relative to the rest of the tissue (B) and control WD (A); scale bars: 100 μm. (C–E) Immunostaining of WDs 7-days AEL with an anti-pH3 antibody revealed overall reduced mitotic activity in nub>mRFP ecdRNAi WD (D) compared to controls (C); scale bars: 100 μm. Quantification of pH3-positive cells in the nubbin domain and hinge + notum regions in control (n = 13) and nub>mRFP ecdRNAi (n = 9) WDs confirms negative impact, cell autonomous and non-autonomous, of ecd knockdown on cell proliferation (E). An unpaired Student’s t-test assuming equal variances was used to determine statistical significance. (F–H) Schematic of the fly ubi-FUCCI (ubi-GFP-E2F11-230 and ubi-mRFP1-NLS-CycB1-266) cell cycle reporter shows G1 (GFP), S (RFP) and G2/M (GFP and RFP) phases of the cell cycle (F). Compared to control (nub>; ubi-FUCCI) (G), many Ecd-deficient cells in the pouch region of nub>ecdRNAi; ubi-FUCCI WD arrest in G2 phase of the cell cycle (H). (I–K) High magnification images of the nubbin domain of control (I) and nub>mRFP ecdRNAi WDs (J) at day 7 AEL. DAPI and Lamin B mark the nucleus and nuclear lamina, respectively. The images are projections of 2–3 consecutive confocal sections; scale bars: 10 μm. mRFP labeling of cell membranes was used for quantification of cell sizes (K), see Materials and Methods, (n = 7 WDs per genotype). (L) Wing pouch-specific knockdown of ecd (nub>mRFP ecdRNAi) (n = 14) leads to wing pouch overgrowth in relation to the hinge and notum of the WDs compared to control (nub>mRFP) (n = 13). The data represent a ratio of the nubbin domain and the entire WD at 7 days AEL. An unpaired two-tailed Student’s t-test assuming equal variances was used to determine statistical significance. (M–O) Representative images (M, N) and quantification (O) show enlarged nucleoli in wing pouch cells of nub>mRFP ecdRNAi WDs (n = 16) (N) relative to control (M) (n = 18) at day 7 AEL. Nucleoli were visualized by immunostaining with an anti-Fibrillarin antibody. DAPI labels nuclei; scale bars: 20 μm. Quantification shows the average nucleolar size per replicate in both conditions. A two-tailed Student’s t-test assuming equal variances was used to determine statistical significance. (P–R) Representative confocal micrographs (P, Q) and quantification (R) show increased puromycin incorporation (20-min pulse) in wing pouch of nub>mRFP ecdRNAi (n = 31) (Q) compared to control WDs (P) (n = 12) at day 7 AEL as visualized with an anti-puromycin antibody; scale bars: 100 μm. An unpaired Student’s t-test assuming equal variances was used to determine statistical significance. The images are projections of multiple confocal sections of WDs at day 7 AEL unless stated otherwise. DAPI and mRFP label nuclei and the nubbin domain, respectively. Data in charts represent means ± SD, **P < 0.01, ****P < 0.0001. See also Supplementary Figure S3.
Transcriptome profiling of Ecd-deficient cells highlights upregulation of translation, stress and DNA damage response
To define the molecular network orchestrating the responses to Ecd scarcity, we profiled the transcriptome of control (nub>) and nub>ecdRNAi WDs using mRNA-seq (Supplementary Figure S2A, B). In total, we detected transcripts associated with 17621 genes (Supplementary Dataset S2). Most differentially expressed genes (DEGs) in nub>ecdRNAi were protein coding genes (Figure 4A, B) of which 2912 were downregulated and 3166 upregulated compared to controls (uniquely identified or overlapping genes, pAdj< 0.05) (Figure 4A and Supplementary Dataset S2). The second most numerous and regulated transcript biotype corresponded to transposable elements (TEs) (Figure 4B). In total, we detected transcripts from 399 TEs, 273 of which were significantly up- and 126 down- regulated in ecdRNAi compared to controls (Supplementary Figure S2C and Supplementary Dataset S2).
Figure 4.
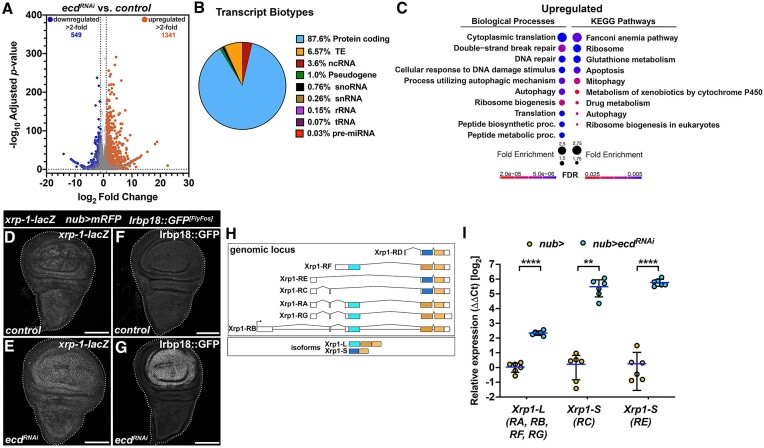
The transcriptome is remodeled in response to reduced Ecd levels, including upregulation of Xrp1 and Irbp18 C/EBP class of bZIP transcription factors. (A) The volcano plot depicts the significantly regulated genes in nub>mRFP ecdRNAi WDs compared to controls (nub>mRFP) detected by RNA sequencing. Genes up- and down-regulated > 2-fold are highlighted in color. (B) Pie chart shows the proportion of each transcript biotype detected as significantly regulated (pAdj< 0.05) in response to reduced Ecd levels in nub>mRFP ecdRNAi WDs compared to controls (nub>mRFP). (C) GO enrichment plot for significantly upregulated genes (pAdj< 0.05) in nub>mRFP ecdRNAi WDs compared to controls (nub>mRFP). Plots show fold enrichment of the top 10 significant functional GO terms. (D–G) Immunostaining shows elevated expression of xrp1-lacZ and Irbp18::GFP FlyFos reporter constructs nub>mRFP ecdRNAi (E, G) compared to control WDs (D, F) at day 7 AEL. The images are projections of multiple confocal sections. WDs are outlined by white dotted lines based on DAPI staining; scale bars: 100 μm. (H, I) Schematic of the xrp1 genomic locus encoding 7 different transcripts giving rise to long (Xrp1-L) and short (Xrp1-S) protein isoforms (H). RT-qPCR analysis revealed strong upregulation of xrp1 transcripts in nub>mRFP ecdRNAi WDs relative to control samples (nub>mRFP), particularly RC and RE coding for Xrp1-S protein isoform (I). Levels of tbp transcript were used for normalization. Unpaired Student’s t-test assuming equal variances was used to determine statistical significance. Data represent means ± SD, n = 6, **P < 0.01, ****P < 0.0001. Statistical significance of differences between isoform expression was determined using two-way ANOVA with Sidak’s correction for multiple comparisons. See also Supplementary Figure S2 and Supplementary Dataset S2.
The GO enrichment analysis of DEGs significantly upregulated in nub>ecdRNAi WDs showed a clear enrichment for functional categories associated with translation, ribosome and peptide biosynthesis, DNA damage and repair, autophagy and glutathione metabolism (Figure 4C). In contrast, significantly downregulated genes were overrepresented for a diverse array of biological processes and pathways, including proteasome and spliceosome, and signaling pathways vital to imaginal disc and epithelium development, tissue morphogenesis and neurogenesis (Supplementary Figure S2D).
To identify the potential drivers of the transcriptional changes in Ecd-deficient cells, we analyzed the expression of 757 known or predicted transcription factors (78) (https://www.mrc-lmb.cam.ac.uk/genomes/FlyTF/). Given the substantial changes to the transcriptome, unsurprisingly, over half of the examined TFs (385) were differentially expressed in response to ecd silencing using pAdj< 0.05 as a cut off (Supplementary Dataset S2). Of the 181 TFs whose expression increased, structure-specific recognition protein (Ssrp), inverted repeat binding protein 18 (Irbp18) and Xrp1 were the three most significantly regulated. Intriguingly, all three proteins have been implicated in DNA repair and/or maintenance of genome stability (79–81). Ssrp is a high mobility group (HMG) box transcription factor that controls chromatin organization as part of the Facilitates Chromatin Transcription (FACT) complex (82,83). Xrp1 and Irbp18 are basic region leucine zipper (bZIP) proteins of the CCAAT/enhancer-binding protein (C/EBP) family whose heterodimers have been shown to be critical for repairing DSBs following P-element transposition (79,81). Recently, Xrp1-Irbp18 have been described as key regulators of proteotoxic stress responses controlling PERK-dependent expression of antioxidant genes and elimination of looser cells during cell competition (59,60,84).
To confirm the increased expression of Xrp1 and Irbp18 in Ecd-deficient cells detected in our transcriptomic analysis, we utilized a xrp1-lacZ transcriptional reporter and an Irbp18::GFP FlyFos transgenic line, respectively. Immunostaining showed a clear induction of Xrp1 and Irbp18 expression in wing pouch cells of the nub>ecdRNAi WDs relative to the rest of the tissue and control WDs (Figure 4D–G). Moreover, the RT-qPCR analysis revealed that of the multiple functional Xrp1 isoforms (Figure 4H) transcripts encoding RC and RE short isoforms were upregulated to a greater extent than longer isoforms in response to ecd knockdown relative to control (Figure 4I).
In summary, our genome-wide RNA profiling revealed a high degree of transcriptional remodeling in wing imaginal tissue in response to ecd loss. The molecular signature determined by our bioinformatic analysis is in strong agreement with the experimental findings described above, both highlighting a targeted upregulation of the translational machinery, stress response and DNA damage/repair responses among the key processes affected by Ecd deficiency. The upregulation of Xrp1 and Irbp18 further suggested that these two transcription factors may act as the key mediators of the responses and phenotypes in Ecd-depleted cells.
Xrp1-Irbp18 drives the stress and growth responses in spliceosome compromised cells
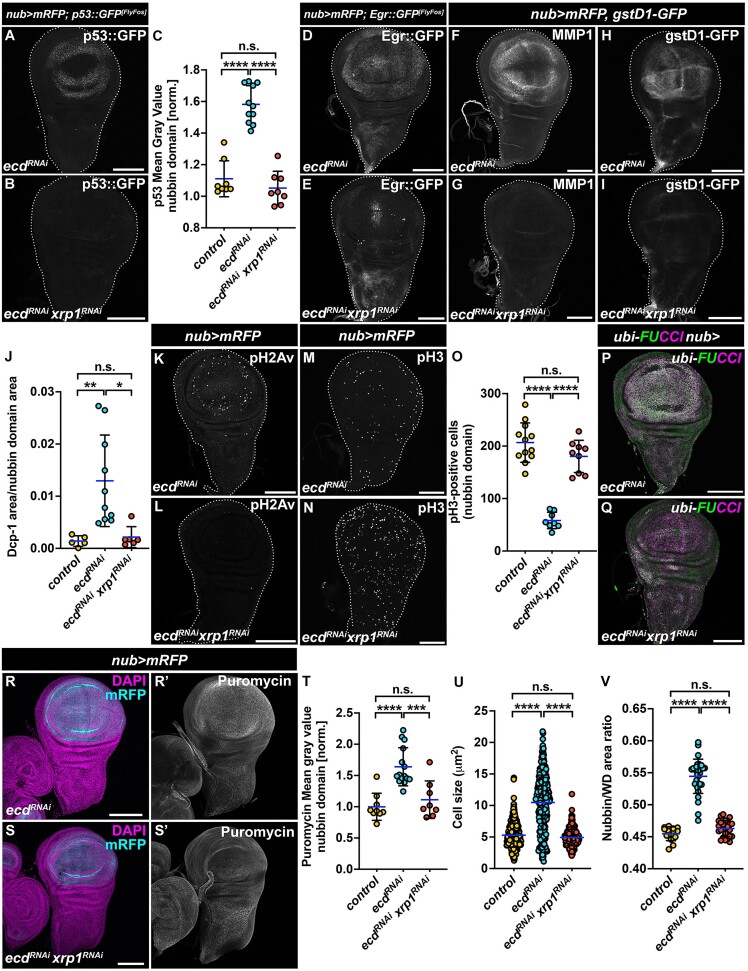
To determine the requirement of Xrp1 and Irbp18 for the Ecd deficient phenotypes, we knocked down the two TFs individually in wing imaginal cells lacking Ecd. Simultaneous knockdown of Xrp1 not only abolished the increase in p53, Egr, MMP1 and GstD1 expression (Figure 5A–I) but also attenuated the apoptosis and DNA damage observed in nub>ecdRNAi WDs, as shown by immunostaining for Dcp-1 and pH2Av, respectively (Figure 5J–L). In addition, it restored the number of pH3 positive cells to control levels (Figure 5M–O), alleviated the G2 cell cycle arrest (Figure 5P, Q), reduced the rate of protein synthesis (Figure 5R–T), normalized cell size to control levels (Figure 5U) and prevented wing pouch overgrowth (Figure 5V). Importantly, inhibition of Irbp18, whose upregulation in Ecd-deficient cells is largely dependent on Xrp1 (Supplementary Figure S3A–D), mimicked the effect of Xrp1 loss (Supplementary Figure S3E–P). This suggests that Xrp1 likely acts in concert with Irbp18 to drive the hallmarks of cellular senescence. These results highlight a central role for Xrp1 in activating the canonical JNK and p53 signaling and in regulating downstream cellular responses to Ecd deficiency, including cell proliferation, cellular translation and growth, DNA damage and apoptosis. Notably, by overexpressing a pathogenic Prp8[S2178F]mutant protein, we were able to recapitulate both the major responses to Ecd deprivation and their dependence on Xrp1 (Supplementary Figure S4A–P). The similarity in cellular and molecular phenotypes resulting from either Ecd loss or expression of the pathogenic form of Prp8 suggests that the Xrp1-mediated stress response is not exclusive to Ecd disruption but represents a more general consequence of U5 snRNP and spliceosome dysfunction.
Figure 5.
Xrp1 controls stress responses in Ecd-deficient cells but is not responsible for lethality. (A–J) Immunostaining shows that the increased p53::GFP (A), Egr::GFP (D), MMP1 (F) and gstD1-GFP (H) expression in nub>mRFP ecdRNAi WDs is abolished by simultaneous knockdown of xrp1 (B, E, G, I), while Dcp-1 levels are reverted to those in control WDs. (J) Quantification of p53::GFP signal in the nubbin domain of control (nub>mRFP), nub>ecdRNAi and nub>ecdRNAi xrp1RNAi WDs (C). One-way ANOVA with Tukey’s multiple comparisons test was used to calculate P-values. (K, L) Phosphorylation of H2Av is Xrp1-dependent, as knockdown of xrp1 leads to the loss of fluorescent signal corresponding to phosphorylated H2Av (pH2Av) in Ecd-deficient WDs (nub>mRFP ecdRNAi xrp1RNAi). (N, O) Representative confocal micrographs (M, N) and quantification (O) show reduced pH3 signal in nub>ecdRNAi WDs (n = 9) that is normalized to control counts (n = 12) when xrp1RNAi is co-expressed with ecdRNAi (n = 9). One-way ANOVA with Tukey’s multiple comparisons test was used to calculate P-values for quantification of pH3-positive cells in the nubbin domain. (P, Q) G2 phase cell cycle arrest of wing pouch cells induced by Ecd deficiency (P) is reverted in nub>ecdRNAi xrp1RNAi WDs (Q) as determined by immunostaining of ubi-FUCCI cell cycle reporter. (R, T) Representative confocal micrographs of WDs 7-days AEL stained with the anti-puromycin antibody (R, S) and fluorescent signal quantification (T) show that xrp1 silencing (nub>mRFP ecdRNAi xrp1RNAi) (n = 8) reverts the increased translation rates caused by ecd knockdown (nub>mRFP ecdRNAi (n = 15) WDs (T) to control levels (n = 9). One-way ANOVA with Tukey’s multiple comparisons test was used to calculate P-values. (U, V) Wing pouch-specific knockdown of xrp1 using nub>mRFP driver completely abolishes increase in cell size (U) and overgrowth of the nubbin domain (V) caused by Ecd scarcity. Membrane-targeted mRFP signal was used to measure cell sizes localized on diagonal in the nubbin domain of control (nub>mRFP, n = 7), nub>mRFP ecdRNAi (n = 7) and nub>mRFP ecdRNAi xrp1RNAi (n = 7) WDs (U). A ratio of the nubbin domain and the entire WD were determined based on the mRFP and DAPI signals (V). Control (nub>mRFP, n = 14), nub>mRFP ecdRNAi (n = 31) and nub>mRFP ecdRNAi xrp1RNAi (n = 26). One-way ANOVA with Tukey’s multiple comparisons test was used to calculate P-values. The images are projections of multiple confocal sections of WDs 7-days AEL. DAPI and mRFP label nuclei and the nubbin domain, respectively (R, S). WDs are outlined by white dotted lines based on DAPI staining; scale bars: 100 μm. Data in charts represent means ± SD, **P < 0.01, ***P < 0.001, ****P < 0.0001, n.s. = non-significant. See also Supplementary Figures S3 and S4.
The splicing defects caused by Ecd deficiency are Xrp1 independent
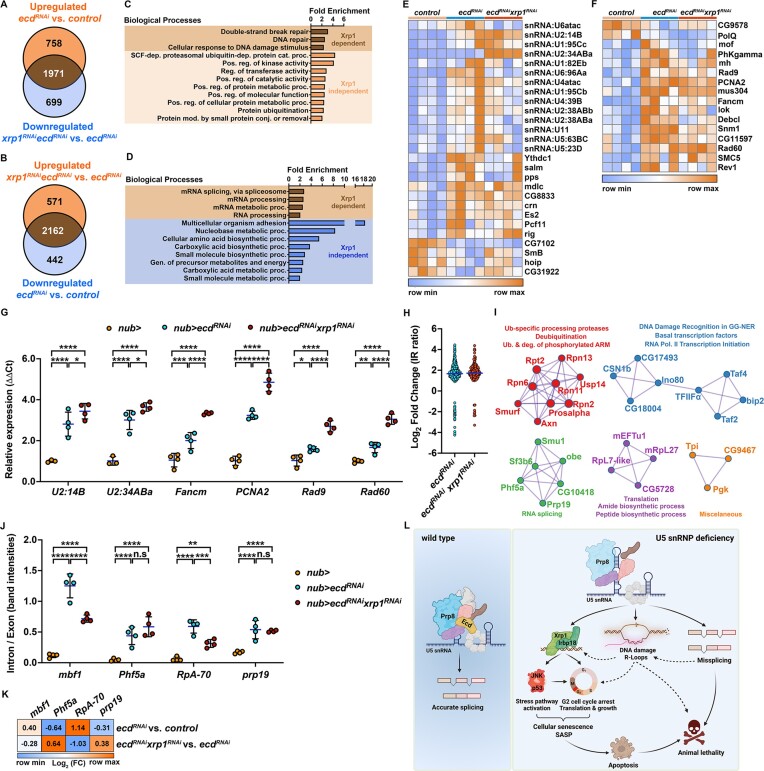
The comprehensive rescue of cellular behaviors provided by Xrp1 or Irbp18 silencing was associated with a significant improvement in pupation rates, reaching control levels compared to a reduction caused by Ecd deficiency alone (Supplementary Figure S3Q). However, the absence of neither of the two TFs mitigated the pupal lethality of animals with targeted ecd knockdown in the nubbin domain (Supplementary Figure S3R). This suggests that Ecd plays an essential role distinct from governing the Xrp1-Irbp18 function and that Xrp1-Irbp18 upregulation is triggered as part of the adaptive response to damage inflicted by Ecd scarcity. Considering the involvement of Ecd in U5 snRNP and spliceosome assembly, we hypothesized that Ecd deficiency provokes changes to gene expression and splicing, independent of Xrp1-Irbp18 that are incompatible with successful development. To this end, we profiled and compared transcriptomes of control (nub>), nub>ecdRNAi and nub>ecdRNAi xrp1RNAi WDs using bulk mRNA-seq generated independently of that above (Supplementary Figure S5 and Supplementary Dataset S3). A highly significant overlap of DEGs in response to reduced Ecd between the two transcriptomic experiments (72%, P ≤ 0.0001, hypergeometric test) underscored the robustness of the molecular phenotype. Of the 2729 genes upregulated in response to Ecd deficiency compared to control (uniquely identified or overlapping genes, pAdj< 0.05), expression of 72% was significantly reduced upon simultaneous knockdown of xrp1 in comparison to ecdRNAi alone (Figure 6A and Supplementary Dataset S3). The Xrp1 molecular footprint was even more pronounced among 2604 genes downregulated by ecd knockdown with 83% being re-expressed in nub>ecdRNAi xrp1RNAi cells (Figure 6B). In agreement with the phenotypic rescue, and the role of Xrp1 in stress responses and in the maintenance of genome stability (79,81,85–86), Xrp1-dependent DEGs showed enrichment for functions related to responses to DNA damage and repair but also pre-mRNA splicing and RNA processing (Figure 6C, D and Supplementary Dataset S3). In contrast, the Xrp1-independent group of upregulated and downregulated genes revealed overrepresentation of genes implicated in the regulation of protein and enzyme activities, including proteasomal degradation (Figure 6C and Supplementary Dataset S3) and nucleobase metabolic and biosynthetic processes, respectively (Figure 6D and Supplementary Dataset S3).
Figure 6.
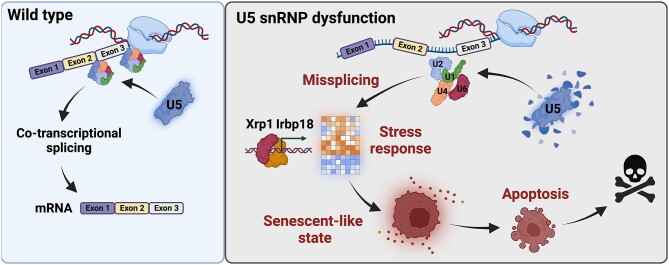
Splicing defects inflicted by Ecd deficiency are Xrp1-independent. (A, B) Venn diagrams show an overlap between genes upregulated (A) and downregulated (B) in Ecd-deficient WDs (nub>mRFP ecdRNAi) compared to controls (nub>mRFP), and those downregulated (A) and upregulated (B) in double deficient (nub>mRFP ecdRNAi xrp1RNAi) WDs compared to ecd knockdown alone. Genes at the intersection of the two groups are considered Xrp1-dependent. (C, D) GO enrichment plots for significantly upregulated (C) and downregulated (D) genes (pAdj< 0.05) in nub>mRFP ecdRNAi WDs compared to controls (nub>mRFP) whose expression is Xrp1-dependent and independent. Plots show fold enrichment of significant functional GO terms as identified by ShinyGO v0.76. Only GO categories with ≥ 5 genes and Fold Enrichment ≥ 2 are shown. Redundant GO terms were eliminated. (E, F) Expression of Xrp1-independent genes implicated in the cellular response to DNA damage stimulus (F) and pre-mRNA splicing (E) in the WDs samples of indicated genotypes. Heatmaps represents the Z-scores of normalized counts across rows. (G) Quantitative RT-PCR of candidate transcripts involved in splicing and the DNA damage response, identified by the RNA-seq analysis as potentially Xrp1-independent. Knockdown of Xrp1 (nub>mRFP ecdRNAi xrp1RNAi) did not restore the levels of the transcripts to control (nub>mRFP) levels. Data are fold changes in expression relative to control (ΔΔCt method). Levels of TATA-binding protein (tbp) transcripts were used for normalization. Two-way ANOVA with Tukey’s multiple comparisons test was used to calculate P-values. Lines represent means ± SD. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. (H) Fold changes (log2) of intron retention ratios significantly different (pAdj < 0.05) between control (nub>mRFP) and Ecd-deficient (nub>mRFP ecdRNAi) or double-deficient WDs (nub>mRFP ecdRNAi xrp1RNAi) for each detected intron retention event. The line represents the mean. (I) Protein–protein interaction network enrichment analysis for genes misspliced in Ecd-deficient (Xrp1 independent) cells. Neighborhoods of densely connected proteins as determined by the MCODE algorithm applied to the protein interaction networks generated by Metascape (44). Functional enrichment analysis was applied to the MCODE networks (45), and the three most significant GO terms were retained. (J) Semi-quantitative PCR on candidate genes shows increased intron retention in Ecd-deficient WDs (nub>mRFP ecdRNAi) compared to the controls (nub>mRFP). The splicing defects persist upon xrp1 knockdown (nub>mRFP ecdRNAi xrp1RNAi). The intron/exon ratio was calculated based on intron- and exon-specific band intensities (see Supplementary Figure S5C). Two-way ANOVA with Tukey’s multiple comparisons test was used to calculate P-values. Data represent means ± SD. **P < 0.01, ***P < 0.001, ****P < 0.0001, n.s. = non-significant. (K) Fold changes (log2) of candidate genes with identified intron retention events by the IRFinder algorithm as compared to the control (nub>mRFP) WDs, or between the two experimental groups (nub>mRFP ecdRNAi and nub>mRFP ecdRNAi xrp1RNAi). Knockdown of xrp1 restored the expression of candidate genes deregulated in Ecd-deficient WDs close to control levels. (L) Graphical summary. Intact U5 snRNP ensures constitutive and alternative splicing to generate transcript variants necessary for cell function and normal animal development. U5 snRNP dysfunction, caused either by maturation defects due to Ecd loss or Prp8 mutations, leads to aberrant splicing, gene dysregulation, DNA damage and R-loop accumulation, which trigger transcriptional upregulation of Xrp1 and Irbp18. The Xrp1-Irbp18 heterodimer acts as a key regulator of cellular stress signaling and the downstream responses, promoting senescence-like phenotype, including cell cycle arrest, increased translation rates, cell growth and SASP. Chronic activation of Xrp1-Irbp18 ultimately leads to apoptosis and organism death. In the absence of Xrp1-Irbp18-mediated response, loss of functional U5 snRNP remains lethal, likely due to dysregulation and aberrant splicing of genes essential for cellular homeostasis, accompanied by accumulation of R-loops and DNA damage. The model was generated using BioRender.com. See also Supplementary Datasets S3, S4, S5 and Supplementary Figures S5, S6, S7.
Although pre-mRNA splicing, RNA metabolism and DNA damage response (DDR) emerged among the main processes controlled by Xrp1, a handful of splicing factors, spliceosome components and key regulators of genome stability remained dysregulated in nub>ecdRNAi xrp1RNAi WDs (Figure 6E, F). RT-qPCR, using an independent set of biological samples, validated this Xrp1-independent upregulation, including the expression of the two U2:14B and U2:34ABa snRNA isoforms, Fanconi anemia group M helicase (Fancm), Proliferating cell nuclear antigen 2 (PCNA2), Rad60 and Rad9 (Figure 6G and Supplementary Dataset S3). In fact, the expression of all six genes was further increased upon simultaneous knockdown of xrp1 compared to nub>ecdRNAi WDs (Figure 6G). Using SpliceWiz (42), we analyzed Ecd-deficient samples for different types of splicing events and identified intron retention (IR) as one of the most common changes (Supplementary Figure S6 and Supplementary Dataset S4). Focusing on retention events, we performed comparative splicing analysis using NxtIRFcore and IRFinder2 (41), which revealed notable changes in pre-mRNA processing in both nub>ecdRNAi and nub>ecdRNAi xrp1RNAi compared to control WDs (Supplementary Dataset S5). We found 413 differential splicing events (pAdj < 0.05) in ecdRNAiand 224 in ecdRNAi xrp1RNAisamples, with many missplicing events being shared between the two genotypes. For example, 120 out of 212 introns and known exons retained in nub>ecdRNAi compared to control were also identified in nub>ecdRNAi xrp1RNAi (Figure 6H and Supplementary Dataset S5). Strikingly, the GO enrichment analysis of the protein–protein interaction network generated from Xrp1-independent misspliced genes identified five MCODE networks consisting of components related to RNA splicing, ubiquitin-mediated proteasomal degradation, DNA damage recognition, transcription and translation (Figure 6I and Supplementary Dataset S5). The retention events were validated by the semi-quantitative RT-PCR on selected candidate genes involved in replication (RpA-70), regulation of stress-induced gene expression (mbf1), RNA splicing (prp19, Phf5a), and genome maintenance and transcription (prp19). Consistent with the genome-wide splicing analysis, all four candidates were misspliced in response to reduced Ecd (Figure 6J and Supplementary Figure S7A, B) and this aberrant pattern persisted upon simultaneous knockdown of ecd and xrp1 (nub>ecdRNAi xrp1RNAi), although not to the same extent for mbf1 and RpA-70 (Figure 6J). Importantly, the expression of the tested candidates was restored to near control levels by xrp1 knockdown (Figure 6K and Supplementary Dataset S3), thus minimizing the potential confounding effect of differential gene expression on splicing. Taken together, these data provide evidence for a vital role of Ecd in controlling both the expression and pre-mRNA splicing of key sensors and regulators of genome, transcriptome and proteome integrity, independent of Xrp1-Irbp18.
Discussion
The U5 snRNP forms the heart of the active spliceosome and is indispensable for splicing all introns in multicellular eukaryotes. A complete absence of individual U5 snRNP components causes early embryonic lethality (26,87–89) and loss of function clones cannot be recovered or are swiftly eliminated from mitotically active tissue (17,22,87,90). However, mutations in, and deregulation of, U5 snRNP components have been linked to tissue-specific pathologies as diverse as retinopathies, craniofacial disorders and cancer (6,91–96). In this study, we used the developing wing imaginal epithelium of Drosophila to determine the cascade of events triggered by U5 snRNP scarcity caused either by targeted knockdown of the PAQosome adaptor and U5 snRNP biogenesis factor Ecd, or overexpression of an RP13-associated pathogenic variant of Prp8. Our results show that the absence of functional U5 snRNP, in the mitotically active cells of the nubbin domain, triggers a stress response program which engages the protective function of p53- and JNK-mediated signaling until their sustained activity becomes toxic and induces apoptosis. We establish Xrp1 and its dimerizing partner Irbp18 as the primary upstream inducers of p53 and JNK signaling, mediating cellular responses to the damage caused by U5 snRNP malfunction. In mammals, C/EBP transcription factors have been recognized as important regulators of genes associated with senescence and senescence-associated secretory phenotype (SASP) (97). Here, we provide genetic evidence that increased expression of Xrp1-Irbp18 in Ecd-deficient cells drives cell behaviors characteristic of cellular senescence and SASP, including G2-phase cell cycle arrest, DNA damage, increased translation rate, cell growth and paracrine secretion (98). Importantly, besides governing canonical stress signaling and cellular properties of Ecd-deficient cells, Xrp1-Irbp18 activation reduces proliferation also in neighboring imaginal territories, distorting organ proportions. We propose that this non-autonomous signaling could be mediated through a secretion of a systemic Drosophila insulin-like peptide 8 (Dilp8) (70) whose expression dramatically increased in Ecd-deficient cells (Supplementary Dataset S2). Silencing Xrp1-Irbp18 abolishes the stress-induced senescent phenotypes of Ecd-deficient cells, revealing pervasive missplicing of critical regulators of transcription, splicing, DNA damage and proteostasis, which may, in turn, trigger upregulation of Xrp1-Irbp18, leading to animal lethality (Figure 6L).
Underscoring a functional and physical coupling of transcription and splicing (1,99,100), our molecular profiling of WDs comprising Ecd-deficient cells revealed a vast dysregulation of both transcription and splicing. Our analysis showed that changes in the nub>ecdRNAi transcriptome were dominated by aberrant expression of protein-coding genes. However, we also detected widespread de-repression of transposons, mostly residing in heterochromatic regions, indicating a possible disruption of chromatin organization and/or the mechanisms required for TE silencing. Interestingly, excessive TE activation has been observed in senescent human cells and in response to aging in both Drosophila and mammals (101–104). Moreover, a recent analysis of RNA-seq datasets of senescent or aged cells in both humans and mice correlated increased transposon expression with transcriptional readthrough and intron retention (105).
Importantly, the molecular signature of Ecd-deficient WDs correlated with our observed phenotypes. The accumulation of DNA damage markers (e.g. pH2Av) was accompanied by the upregulation of several genes directly involved in DNA damage sensing and repair pathways. These pathways included homologous recombination (HR, mre11, Rad50, nbs, RPA3), non-homologous end joining (NHEJ, Irbp, Ku80, Xrp1, Irbp18, DNAlig3, DNAlig4, RPA1), global-genome nucleotide excision repair (GG-NER, mrn, mus201 Xpc, Ercc1) and Fanconi anemia pathway required for resolving of highly toxic interstrand crosslinks (ICLs, FANCI, FANCM, DNApol-iota, DNApol-zeta). The increased translation rate was supported by the upregulation of genes encoding small and large ribosomal subunits, as well as regulators of ribosome biosynthesis and cytoplasmic translation. Conversely, genes associated with the spliceosome, proteasome function, protein processing, and signaling pathways controlling epithelial morphogenesis and differentiation were downregulated. The upregulation of autophagy-related genes could indicate a compensation for proteasome impairment (106) and correlates with accelerated lysosome-dependent degradation of Prp8 in the absence of Ecd (17). Importantly, autophagy is one of the effector mechanisms of senescence required for acquisition of the senescence phenotype (107). Interestingly, p53 can promote autophagy (108,109) and switches from a pro-apoptotic inducer to a pro-survival factor in G2-arrested cells (110). Together, this suggests that Xrp1-dependent p53 upregulation could exert a cytoprotective role in splicing-deficient cells by inducing autophagy and delaying proteotoxicity caused by increased translation and proteasome impairment, thereby promoting survival and maintenance of irreversibly damaged cells.
Strikingly, >70% of genes differentially expressed in response to reduced Ecd levels were dependent on Xrp1, underscoring Xrp1 as the major driver of stress-induced senescent cellular behaviors. However, given the extensive deregulation of transcription factors in nub>ecdRNAi wing discs, we propose that the normalization of gene expression upon xrp1 knockdown results from both loss of regulation by Xrp1, and a rewiring of the TF network, including p53 and JNK-controlled transcriptional targets. Consistent with its role in protecting genome integrity (53,79,81), the upregulation of DDR and repair genes in response to U5 snRNP scarcity required Xrp1. In contrast, pre-mRNA splicing via spliceosome and RNA metabolism were highlighted among processes downregulated by Xrp1. These results indicate that Xrp1 induction in Ecd-deficient cells may be two sided, promoting DDR to alleviate the primary insult to genome integrity while aggravating spliceosome dysfunction. It is plausible that the chronic shortage of properly processed transcripts and/or translation of aberrantly spliced mRNAs could enhance the stress load in Ecd-deficient cells. Intriguingly, some of the DDR and repair genes (e.g. Irbp18, Ku80, RpA-70andHis2Av) and splicing factors (e.g. prp19, x16) have been identified as interacting partners of the large and small Xrp1 protein isoforms (85). This is important to consider when interpreting our results. While the downregulation of pH2Av foci upon xrp1 silencing in Ecd-deficient cells could signal alleviation of DNA damage, it is also plausible that the DNA damage persists and cannot be properly recognized because of the reduced expression and/or failed recruitment of DNA damage factors in the absence of Xrp1.
There is now a large body of experimental evidence linking deregulated splicing to DNA damage and genomic instability with pre-mRNA processing and genomic stability interacting through various mechanisms, including aberrant splicing of DDR factors or the accumulation of RNA/DNA duplexes known as R-loops (111). While R-loops prove beneficial in fine-tuning gene expression, their ectopic formation and persistence due to spliceosome dysfunction are a significant source of DNA damage and genomic instability from budding yeast to mammals (61,63,64,112,113). The increase in the number and strength of RNase H1 chromatin binding events identified by our TaDa profiling, together with the Xrp1-independent upregulation of several genome caretakers required for R-loop removal, such as Fanconi anemia group M helicase (Fancm), Rad60, Proliferating cell nuclear antigen 2 (PCNA2) and Smc5 (114–117), supports the notion that R-loops may accumulate in response to Ecd loss. Whether R-loop formation is a direct consequence of missplicing and how they may contribute to the phenotypes of Ecd-deficient cells is a matter for future investigation. Importantly, splicing analyses revealed Xrp1-independent missplicing of genes pivotal to transcription, splicing, DNA damage and proteostasis. In this context, missplicing of prp19 and components of the Ino80 chromatin remodeling complex (Ino80C, Ino80 and CG18004-Ino80 complex subunit E) are particularly intriguing. Prp19 encodes an evolutionarily conserved splicing factor with E3 ubiquitin ligase activity. Prp19 was originally described as a part of the NineTeen Complex (NTC), which is required for the catalytic activation of the spliceosome (118). However, Prp19 also plays an essential role in transcriptional elongation and DNA repair by promoting the recruitment and stabilization of the THO/TREX (TRanscription-EXport) complex and DNA repair factors (119). Ino80C is indispensable for nucleosome remodeling during transcription, replication and DNA damage repair (120,121). Proper function of THO/TREX and Ino80C has been shown to prevent the formation of deleterious DNA-RNA hybrids (122–124). Given the pivotal role of Prp19 and Ino80C at the interface of DNA replication, transcription and RNA metabolism, their impaired function in Ecd-deficient cells may contribute to molecular defects and organismal death prior to and independent of the activation of Xrp1-driven stress responses. Interestingly, the mammalian Prp19 ortholog, PRPF19, was found to be among the most downregulated splicing factors during replicative senescence, and its knockdown was sufficient to induce premature p53-dependent senescence of human fibroblasts through altered splicing of the p53 inhibitor MDM4 (125).
Our study contributes novel insights into the emerging role of Xrp1 as a stress-inducible transcription factor that governs molecular and cellular phenotypes in response to various stress insults, ranging from irradiation and P-element transposition (53,79,81), ER and proteotoxic stress (59,60,86,126–128) to spliceosome malfunction (our study). Consistent with the published studies, our results show that Xrp1 levels must be tightly regulated as its overexpression or chronic activation is sufficient to induce cell death (60,70,84,127–129). However, we also demonstrate that the cellular context, in which Xrp1 operates is essential for the phenotypic outcome. Xrp1 has been implicated in the neurotoxicity and motor defects in a Drosophila model of amyotrophic lateral sclerosis (ALS) based on loss of an RNA binding protein Cabeza (Caz) or overexpression of its mutant human ortholog FUS (85). More recently, Xrp1 was defined as a master regulator of Minute cell competition caused by ribosomal protein haploinsufficiency (‘Minute’ - Rp/+ cells) (60,70,86,126,129). In all cases, Xrp1 upregulation drives significant transcriptome remodeling and the Xrp1-controlled gene sets show marked overlap among Ecd-deficient and Rp/+ imaginal discs as well as caz mutant larval brains (85,86 and our data). The phenotypic consequences of Xrp1 induction are, however, different, and sometimes opposing. In Rp/+ cells, Xrp1 upregulation reduced global translation rate and cell growth by engaging in proteotoxic stress-induced PERK-eIF2α signaling resulting in apoptotic elimination of cells with lower fitness. Reducing Xrp1 levels restored both translation and growth of Rp/+ cells allowing their survival surrounded by wild type neighbors. In contrast, in U5 snRNP-deficient cells, Xrp1 triggers senescence program promoting translation and growth, while apoptosis is delayed until irreparable damage and breakdown of proteostasis override the pro-survival mechanisms. Supporting our findings, recent single cell profiling studies define Xrp1 as a shared molecular hallmark of senescence cell clusters in wounded and tumor bearing imaginal tissue (130). We propose that the differential impact of Xrp1 upregulation on protein synthesis and growth may lie in different stress signals to which Xrp1 initially responds and the cell cycle stage or cellular milieu in which it operates. Unlike Ecd-deficient cells, Rp/+ cells might be prone to proteotoxicity due to aggregation of orphan ribosomal proteins (127,128) that will poise Xrp1 activity towards amplification of PERK-eIF2α signaling turning off translation. When surrounded by wild type cells, Rp/+ cells will be eliminated. Importantly, stimulating autophagy in RP-deficient cells proved beneficial reducing proteotoxic stress and tissue damage (128). Although experimental evidence pinpointed RpS12 protein to activate Xrp1 in Rp/+ cells (70,86,131,132), the molecular mechanism remains unknown. The trigger for Xrp1 upregulation in cells experiencing U5 snRNP shortage remains to be determined. Given that Prp19 has been detected within Xrp1 interactome (85), it is tempting to speculate that Prp19 missplicing could affect Xrp1 levels and/or activity. Based on our results, we further suggest that the simultaneous activation of p53 and JNK signaling downstream of Xrp1 is crucial to the senescent phenotype of Ecd-deficient cells. Activation of p53 and JNK enforce G2 cell cycle arrest that is favorable to increased protein synthesis, growth, DNA damage repair and proteostasis network rewiring while reduces sensitivity to apoptosis (73,110). We envision that aggravation of proteotoxic stress due to chronic Xrp1 activity and/or lifting the cell cycle break would drive cell death.
In conclusion, our study highlights the critical role of the functional spliceosome in maintaining cellular, tissue, and organismal homeostasis. We have identified Xrp1 as a key driver of stress responses triggered by U5 snRNP scarcity and implicated cellular senescence, governed by the C/EBP family bZIP transcription factors, in pathologies caused by U5 snRNP dysfunction. Given the emerging evidence linking pre-mRNA splicing to the aging process (133–137), dysregulation of splicing factors observed in aging might fuel the accumulation of senescent cells, promoting the decline of cellular and tissue functions, and the development of age-related disease. The striking similarity between the phenotypes induced by Ecd deficiency and the pathogenic Prp8 mutant protein, together with their dependence on Xrp1, underscores the involvement of Ecd in U5 snRNP biogenesis. Whether Ecd loss affects PAQosome-mediate assembly of client protein complexes other than U5 snRNP and how their dysfunction contributes to stress-induced senescence represents an interesting avenue for further investigation.
Supplementary Material
Acknowledgements
The authors thank Alexandra Trifunovic Laboratory, the Bloomington Drosophila Stock Center supported by NIH [NIH P40OD018537] (BDSC, Bloomington, IN, USA), the Drosophila Genomics Resource Center supported by NIH [2P40OD010949] (DGRC, Bloomington, IN, USA), the Zurich ORFeome Project (FlyORF, Zurich, Switzerland), the Vienna Drosophila Resource Center (VDRC, Vienna, Austria), and the Developmental Studies Hybridoma Bank (DSHB, Iowa City, IA, USA) for fly stocks, plasmids, and antibodies. The authors are grateful to Tina Bresser, Agnieszka Sokol and Nils Teuscher for generation of transgenic lines, fly stock maintenance and technical assistance. We also thank the RNA team of the Cologne Center for Genomics (CCG) for the preparation and sequencing of the RNA libraries.
Authors contributions: D.S. and M.U. conceived the study. D.S., L.T. and M.U. carried out the investigation. D.S., L.T. and M.U. performed formal analysis. D.S. wrote the initial draft. D.S., L.T. and M.U. wrote the manuscript and generated the visualizations therein. M.U. provided supervision, project administration, and funding.
Contributor Information
Dimitrije Stanković, Institute for Genetics and Cologne Excellence Cluster on Cellular Stress Responses in Aging-Associated Diseases (CECAD), University of Cologne, Cologne 50931, Germany.
Luke S Tain, Institute for Genetics and Cologne Excellence Cluster on Cellular Stress Responses in Aging-Associated Diseases (CECAD), University of Cologne, Cologne 50931, Germany.
Mirka Uhlirova, Institute for Genetics and Cologne Excellence Cluster on Cellular Stress Responses in Aging-Associated Diseases (CECAD), University of Cologne, Cologne 50931, Germany.
Data availability
The raw and processed next generation sequencing data have been deposited in NCBI’s Gene Expression Omnibus (RRID:SCR_005012) and are accessible through GEO Series accession numbers GSE213325 for the Targeted DamID data (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE213325), GSE203127 and GSE213298 for the RNA-Seq data (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE213298), (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE203127).
Supplementary data
Supplementary Data are available at NAR Online.
Funding
Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) [CECAD, EXC 2030, UH 243/6-1 to M.U.]. Funding for open access charge: University of Cologne; Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) [UH 243/6-1 to M.U.].
Conflict of interest statement. The authors declare no conflicts of interests.
References
- 1. Tellier M., Maudlin I., Murphy S. Transcription and splicing: a two-way street. Wiley Interdiscip Rev. RNA. 2020; 11:e1593. [DOI] [PubMed] [Google Scholar]
- 2. Boumpas P., Merabet S., Carnesecchi J. Integrating transcription and splicing into cell fate: transcription factors on the block. Wiley Interdiscip Rev. RNA. 2023; 14:e1752. [DOI] [PubMed] [Google Scholar]
- 3. Herzel L., Ottoz D.S.M., Alpert T., Neugebauer K.M. Splicing and transcription touch base: co-transcriptional spliceosome assembly and function. Nat. Rev. Mol. Cell Biol. 2017; 18:637–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Scotti M.M., Swanson M.S. RNA mis-splicing in disease. Nat. Rev. Genet. 2016; 17:19–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bhadra M., Howell P., Dutta S., Heintz C., Mair W.B. Alternative splicing in aging and longevity. Hum. Genet. 2020; 139:357–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wood K.A., Eadsforth M.A., Newman W.G., O’Keefe R.T The role of the U5 snRNP in genetic disorders and cancer. Front. Genet. 2021; 12:636620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Deschênes M., Chabot B. The emerging role of alternative splicing in senescence and aging. Aging Cell. 2017; 16:918–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Harries L.W. Dysregulated RNA processing and metabolism: a new hallmark of ageing and provocation for cellular senescence. FEBS J. 2023; 290:1221–1234. [DOI] [PubMed] [Google Scholar]
- 9. Will C.L., Luhrmann R. Spliceosome structure and function. Cold Spring Harb. Perspect. Biol. 2011; 3:a003707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tarn W.Y., Steitz J.A. A novel spliceosome containing U11, U12, and U5 snRNPs excises a minor class (AT-AC) intron in vitro. Cell. 1996; 84:801–811. [DOI] [PubMed] [Google Scholar]
- 11. Tarn W.-Y., Steitz J.A. Highly diverged U4 and U6 small nuclear RNAs required for splicing rare AT-AC introns. Science. 1996; 273:1824–1832. [DOI] [PubMed] [Google Scholar]
- 12. Moyer D.C., Larue G.E., Hershberger C.E., Roy S.W., Padgett R.A. Comprehensive database and evolutionary dynamics of U12-type introns. Nucleic Acids Res. 2020; 48:7066–7078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Matera A.G., Wang Z. A day in the life of the spliceosome. Nat. Rev. Mol. Cell Biol. 2014; 15:108–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wahl M.C., Will C.L., Luhrmann R. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009; 136:701–718. [DOI] [PubMed] [Google Scholar]
- 15. Grainger R.J., Beggs J.D. Prp8 protein: at the heart of the spliceosome. RNA. 2005; 11:533–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ruzickova S., Stanek D Mutations in spliceosomal proteins and retina degeneration. RNA Biol. 2017; 14:544–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Erkelenz S., Stankovic D., Mundorf J., Bresser T., Claudius A.K., Boehm V., Gehring N.H., Uhlirova M. Ecd promotes U5 snRNP maturation and Prp8 stability. Nucleic Acids Res. 2021; 49:1688–1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Malinova A., Cvackova Z., Mateju D., Horejsi Z., Abeza C., Vandermoere F., Bertrand E., Stanek D., Verheggen C. Assembly of the U5 snRNP component PRPF8 is controlled by the HSP90/R2TP chaperones. J. Cell Biol. 2017; 216:1579–1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cloutier P., Poitras C., Durand M., Hekmat O., Fiola-Masson E., Bouchard A., Faubert D., Chabot B., Coulombe B. R2TP/Prefoldin-like component RUVBL1/RUVBL2 directly interacts with ZNHIT2 to regulate assembly of U5 small nuclear ribonucleoprotein. Nat. Commun. 2017; 8:15615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Klimešová K., Vojáčková J., Radivojević N., Vandermoere F., Bertrand E., Verheggen C., Staněk D TSSC4 is a component of U5 snRNP that promotes tri-snRNP formation. Nat. Commun. 2021; 12:3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lynham J., Houry W.A. The role of Hsp90-R2TP in macromolecular complex assembly and stabilization. Biomolecules. 2022; 12:1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Claudius A.K., Romani P., Lamkemeyer T., Jindra M., Uhlirova M. Unexpected role of the steroid-deficiency protein ecdysoneless in pre-mRNA splicing. PLoS Genet. 2014; 10:e1004287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Stankovic D., Claudius A.K., Schertel T., Bresser T., Uhlirova M. A Drosophila model to study retinitis pigmentosa pathology associated with mutations in the core splicing factor Prp8. Dis. Model Mech. 2020; 13:dmm043174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yang C., Georgiou M., Atkinson R., Collin J., Al-Aama J., Nagaraja-Grellscheid S., Johnson C., Ali R., Armstrong L., Mozaffari-Jovin S. et al. Pre-mRNA processing factors and retinitis pigmentosa: RNA splicing and beyond. Front. Cell Dev. Biol. 2021; 9:700276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rubio-Peña K., Fontrodona L., Aristizábal-Corrales D., Torres S., Cornes E., García-Rodríguez F.J., Serrat X., González-Knowles D., Foissac S., Porta-De-La-Riva M. et al. Modeling of autosomal-dominant retinitis pigmentosa in Caenorhabditis elegans uncovers a nexus between global impaired functioning of certain splicing factors and cell type-specific apoptosis. RNA. 2015; 21:2119–2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kim J.H., Gurumurthy C.B., Naramura M., Zhang Y., Dudley A.T., Doglio L., Band H., Band V. Role of mammalian Ecdysoneless in cell cycle regulation. J. Biol. Chem. 2009; 284:26402–26410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Saleem I., Mirza S., Sarkar A., Raza M., Mohapatra B., Mushtaq I., Kim J.H., Mishra N.K., Alsaleem M.A., Rakha E.A. et al. The mammalian ecdysoneless protein interacts with RNA helicase DDX39A To regulate nuclear mRNA export. Mol. Cell. Biol. 2021; 41:e0010321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Olou A.A., Sarkar A., Bele A., Gurumurthy C.B., Mir R.A., Ammons S.A., Mirza S., Saleem I., Urano F., Band H. et al. Mammalian ECD protein is a novel negative regulator of the PERK arm of the unfolded protein response. Mol. Cell. Biol. 2017; 37:e00030-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Garen A., Kauvar L., Lepesant J.A. Roles of ecdysone in Drosophila development. Proc. Natl. Acad. Sci. USA. 1977; 74:5099–5103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Southall T.D., Gold K.S., Egger B., Davidson C.M., Caygill E.E., Marshall O.J., Brand A.H. Cell-type-specific profiling of gene expression and chromatin binding without cell isolation: assaying RNA Pol II occupancy in neural stem cells. Dev. Cell. 2013; 26:101–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chatterjee N., Bohmann D A versatile ΦC31 based reporter system for measuring AP-1 and Nrf2 signaling in Drosophila and in tissue culture. PLoS One. 2012; 7:e34063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sykiotis G.P., Bohmann D Keap1/Nrf2 signaling regulates oxidative stress tolerance and lifespan in Drosophila. Dev. Cell. 2008; 14:76–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kapur J.N., Sahoo P.K., Wong A.K.C. A new method for gray-level picture thresholding using the entropy of the histogram. Computer Vision Graph. Image Process. 1985; 29:273–285. [Google Scholar]
- 34. Mundorf J., Uhlirova M. The Drosophila imaginal disc tumor model: visualization and quantification of gene expression and tumor invasiveness using genetic mosaics. J. Vis. Exp. 2016; 116:e54585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal. 2011; 17:3. [Google Scholar]
- 36. Bray N.L., Pimentel H., Melsted P., Pachter L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 2016; 34:525–527. [DOI] [PubMed] [Google Scholar]
- 37. Soneson C., Love M.I., Robinson M.D. Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Res. 2015; 4:1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014; 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pimentel H., Bray N.L., Puente S., Melsted P., Pachter L. Differential analysis of RNA-seq incorporating quantification uncertainty. Nat. Methods. 2017; 14:687–690. [DOI] [PubMed] [Google Scholar]
- 40. Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013; 29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Middleton R., Gao D., Thomas A., Singh B., Au A., Wong J.J.L., Bomane A., Cosson B., Eyras E., Rasko J.E.J. et al. IRFinder: assessing the impact of intron retention on mammalian gene expression. Genome Biol. 2017; 18:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wong A.C., Wong J.J.-L., Rasko J.E., Schmitz U. SpliceWiz: easy, optimized, and accurate alternative splicing analysis in R. 2022; bioRxiv doi:06 July 2022, preprint: not peer reviewed 10.1101/2022.07.05.498887. [DOI]
- 43. Ge S.X., Jung D., Yao R. ShinyGO: a graphical gene-set enrichment tool for animals and plants. Bioinformatics. 2020; 36:2628–2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zhou Y., Zhou B., Pache L., Chang M., Khodabakhshi A.H., Tanaseichuk O., Benner C., Chanda S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019; 10:1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Bader G.D., Hogue C.W.V. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinf. 2003; 4:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001; 25:402–408. [DOI] [PubMed] [Google Scholar]
- 47. Vogel M.J., Peric-Hupkes D., van Steensel B. Detection of in vivo protein-DNA interactions using DamID in mammalian cells. Nat. Protoc. 2007; 2:1467–1478. [DOI] [PubMed] [Google Scholar]
- 48. Mundorf J., Donohoe C.D., McClure C.D., Southall T.D., Uhlirova M. Ets21c governs tissue renewal, stress tolerance, and aging in the drosophila intestine. Cell Rep. 2019; 27:3019–3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Marshall O.J., Brand A.H. damidseq_pipeline: an automated pipeline for processing DamID sequencing datasets. Bioinformatics. 2015; 31:3371–3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wolfram V., Southall T.D., Brand A.H., Baines R.A. The LIM-homeodomain protein islet dictates motor neuron electrical properties by regulating K(+) channel expression. Neuron. 2012; 75:663–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Yu G., Wang L.G., He Q.Y. ChIPseeker: an R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics. 2015; 31:2382–2383. [DOI] [PubMed] [Google Scholar]
- 52. Redfern C.P.F., Bownes M. Pleiotropic effects of the ‘ecdysoneless-1’ mutation of Drosophila melanogaster. Mol. Gen. Genet. MGG. 1983; 189:432–440. [Google Scholar]
- 53. Brodsky M.H., Weinert B.T., Tsang G., Rong Y.S., McGinnis N.M., Golic K.G., Rio D.C., Rubin G.M. Drosophila melanogaster MNK/Chk2 and p53 regulate multiple DNA repair and apoptotic pathways following DNA damage. Mol. Cell. Biol. 2004; 24:1219–1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Jin S., Martinek S., Joo W.S., Wortman J.R., Mirkovic N., Sali A., Yandell M.D., Pavletich N.P., Young M.W., Levine A.J. Identification and characterization of a p53 homologue in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA. 2000; 97:7301–7306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Vousden K.H., Prives C. Blinded by the light: the growing complexity of p53. Cell. 2009; 137:413–431. [DOI] [PubMed] [Google Scholar]
- 56. Ollmann M., Young L.M., Di Como C.J., Karim F., Belvin M., Robertson S., Whittaker K., Demsky M., Fisher W.W., Buchman A. et al. Drosophila p53 is a structural and functional homolog of the tumor suppressor p53. Cell. 2000; 101:91–101. [DOI] [PubMed] [Google Scholar]
- 57. Igaki T., Kanda H., Yamamoto-Goto Y., Kanuka H., Kuranaga E., Aigaki T., Miura M. Eiger, a TNF superfamily ligand that triggers the Drosophila JNK pathway. EMBO J. 2002; 21:3009–3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Uhlirova M., Bohmann D JNK- and Fos-regulated Mmp1 expression cooperates with Ras to induce invasive tumors in Drosophila. EMBO J. 2006; 25:5294–5304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Brown B., Mitra S., Roach F.D., Vasudevan D., Ryoo H.D. The transcription factor Xrp1 is required for PERK-mediated antioxidant gene induction in Drosophila. eLife. 2021; 10:e74047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Langton P.F., Baumgartner M.E., Logeay R., Piddini E. Xrp1 and Irbp18 trigger a feed-forward loop of proteotoxic stress to induce the loser status. PLoS Genet. 2021; 17:e1009946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Tresini M., Warmerdam D.O., Kolovos P., Snijder L., Vrouwe M.G., Demmers J.A., van I.W.F., Grosveld F.G., Medema R.H., Hoeijmakers J.H. et al. The core spliceosome as target and effector of non-canonical ATM signalling. Nature. 2015; 523:53–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Naro C., Bielli P., Pagliarini V., Sette C. The interplay between DNA damage response and RNA processing: the unexpected role of splicing factors as gatekeepers of genome stability. Front. Genet. 2015; 6:142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Paulsen R.D., Soni D.V., Wollman R., Hahn A.T., Yee M.-C., Guan A., Hesley J.A., Miller S.C., Cromwell E.F., Solow-Cordero D.E. et al. A genome-wide siRNA screen reveals diverse cellular processes and pathways that mediate genome stability. Mol. Cell. 2009; 35:228–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Li X., Manley J.L. Inactivation of the SR protein splicing factor ASF/SF2 results in genomic instability. Cell. 2005; 122:365–378. [DOI] [PubMed] [Google Scholar]
- 65. Aguilera A., García-Muse T. R loops: from transcription byproducts to threats to genome stability. Mol. Cell. 2012; 46:115–124. [DOI] [PubMed] [Google Scholar]
- 66. Beall E.L., Admon A., Rio D.C. A Drosophila protein homologous to the human p70 Ku autoimmune antigen interacts with the P transposable element inverted repeats. Proc. Natl. Acad. Sci. USA. 1994; 91:12681–12685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Jacoby D.B., Wensink P.C. Yolk protein factor 1 is a Drosophila homolog of Ku, the DNA-binding subunit of a DNA-dependent protein kinase from humans. J. Biol. Chem. 1994; 269:11484–11491. [PubMed] [Google Scholar]
- 68. Crossley M.P., Bocek M., Cimprich K.A. R-loops as cellular regulators and genomic threats. Mol. Cell. 2019; 73:398–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Niehrs C., Luke B. Regulatory R-loops as facilitators of gene expression and genome stability. Nat. Rev. Mol. Cell Biol. 2020; 21:167–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Boulan L., Andersen D., Colombani J., Boone E., Leopold P. Inter-organ growth coordination is mediated by the Xrp1-Dilp8 axis in Drosophila. Dev. Cell. 2019; 49:811–818. [DOI] [PubMed] [Google Scholar]
- 71. Sanchez J.A., Mesquita D., Ingaramo M.C., Ariel F., Milan M., Dekanty A. Eiger/TNFalpha-mediated Dilp8 and ROS production coordinate intra-organ growth in Drosophila. PLoS Genet. 2019; 15:e1008133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Zielke N., Korzelius J., van Straaten M., Bender K., Schuhknecht G.F.P., Dutta D., Xiang J., Edgar B.A. Fly-FUCCI: a versatile tool for studying cell proliferation in complex tissues. Cell Rep. 2014; 7:588–598. [DOI] [PubMed] [Google Scholar]
- 73. Cosolo A., Jaiswal J., Csordas G., Grass I., Uhlirova M., Classen A.K. JNK-dependent cell cycle stalling in G2 promotes survival and senescence-like phenotypes in tissue stress. eLife. 2019; 8:e41036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Rodriguez-Corona U., Sobol M., Rodriguez-Zapata L.C., Hozak P., Castano E. Fibrillarin from archaea to human. Biol. Cell. 2015; 107:159–174. [DOI] [PubMed] [Google Scholar]
- 75. Buchwalter A., Hetzer M.W. Nucleolar expansion and elevated protein translation in premature aging. Nat. Commun. 2017; 8:328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Wu Y., Pegoraro A.F., Weitz D.A., Janmey P., Sun S.X. The correlation between cell and nucleus size is explained by an eukaryotic cell growth model. PLoS Comput. Biol. 2022; 18:e1009400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Aviner R. The science of puromycin: from studies of ribosome function to applications in biotechnology. Comput. Struct. Biotechnol. J. 2020; 18:1074–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Adryan B., Teichmann S.A. FlyTF: a systematic review of site-specific transcription factors in the fruit fly Drosophila melanogaster. Bioinformatics. 2006; 22:1532–1533. [DOI] [PubMed] [Google Scholar]
- 79. Francis M.J., Roche S., Cho M.J., Beall E., Min B., Panganiban R.P., Rio D.C. Drosophila IRBP bZIP heterodimer binds P-element DNA and affects hybrid dysgenesis. Proc. Natl. Acad. Sci. USA. 2016; 113:13003–13008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Hsu T., King D.L., LaBonne C., Kafatos F.C. A Drosophila single-strand DNA/RNA-binding factor contains a high-mobility-group box and is enriched in the nucleolus. Proc. Natl. Acad. Sci. 1993; 90:6488–6492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Akdemir F., Christich A., Sogame N., Chapo J., Abrams J.M. p53 directs focused genomic responses in Drosophila. Oncogene. 2007; 26:5184–5193. [DOI] [PubMed] [Google Scholar]
- 82. Belotserkovskaya R., Oh S., Bondarenko V.A., Orphanides G., Studitsky V.M., Reinberg D FACT facilitates transcription-dependent nucleosome alteration. Science. 2003; 301:1090–1093. [DOI] [PubMed] [Google Scholar]
- 83. Shimojima T., Okada M., Nakayama T., Ueda H., Okawa K., Iwamatsu A., Handa H., Hirose S. Drosophila FACT contributes to Hox gene expression through physical and functional interactions with GAGA factor. Genes Dev. 2003; 17:1605–1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Blanco J., Cooper J.C., Baker N.E. Roles of C/EBP class bZip proteins in the growth and cell competition of Rp (‘Minute’) mutants in Drosophila. eLife. 2020; 9:e50535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Mallik M., Catinozzi M., Hug C.B., Zhang L., Wagner M., Bussmann J., Bittern J., Mersmann S., Klämbt C., Drexler H.C.A. et al. Xrp1 genetically interacts with the ALS-associated FUS orthologue caz and mediates its toxicity. J. Cell Biol. 2018; 217:3947–3964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Lee C.-H., Kiparaki M., Blanco J., Folgado V., Ji Z., Kumar A., Rimesso G., Baker N.E. A regulatory response to ribosomal protein mutations controls translation, growth, and cell competition. Dev. Cell. 2018; 46:456–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Gaziova I., Bonnette P.C., Henrich V.C., Jindra M. Cell-autonomous roles of the ecdysoneless gene in Drosophila development and oogenesis. Development. 2004; 131:2715–2725. [DOI] [PubMed] [Google Scholar]
- 88. Bujakowska K., Maubaret C., Chakarova C.F., Tanimoto N., Beck S.C., Fahl E., Humphries M.M., Kenna P.F., Makarov E., Makarova O. et al. Study of gene-targeted mouse models of splicing factor gene Prpf31 implicated in human autosomal dominant retinitis pigmentosa (RP). Invest Ophth Vis Sci. 2009; 50:5927–5933. [DOI] [PubMed] [Google Scholar]
- 89. Keightley M.C., Crowhurst M.O., Layton J.E., Beilharz T., Markmiller S., Varma S., Hogan B.M., de Jong-Curtain T.A., Heath J.K., Lieschke G.J. In vivo mutation of pre-mRNA processing factor 8 (Prpf8) affects transcript splicing, cell survival and myeloid differentiation. FEBS Lett. 2013; 587:2150–2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Andersen D.S., Tapon N. Drosophila MFAP1 is required for pre-mRNA processing and G2/M progression. J. Biol. Chem. 2008; 283:31256–31267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Mirza S., Kalluchi A., Raza M., Saleem I., Mohapatra B., Pal D., Ouellette M.M., Qiu F., Yu L., Lobanov A. et al. Ecdysoneless protein regulates viral and cellular mRNA splicing to promote cervical oncogenesis. Mol. Cancer Res. 2022; 20:305–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Zhao X., Mirza S., Alshareeda A., Zhang Y., Gurumurthy C.B., Bele A., Kim J.H., Mohibi S., Goswami M., Lele S.M. et al. Overexpression of a novel cell cycle regulator ecdysoneless in breast cancer: a marker of poor prognosis in HER2/neu-overexpressing breast cancer patients. Breast Cancer Res. Treat. 2012; 134:171–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Krausová M., Staněk D snRNP proteins in health and disease. Semin. Cell Dev. Biol. 2018; 79:92–102. [DOI] [PubMed] [Google Scholar]
- 94. Xu S.H., Zhu S., Wang Y., Huang J.Z., Chen M., Wu Q.X., He Y.T., Chen, Yan G.R ECD promotes gastric cancer metastasis by blocking E3 ligase ZFP91-mediated hnRNP F ubiquitination and degradation. Cell Death. Dis. 2018; 9:479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Kurtovic-Kozaric A., Przychodzen B., Singh J., Konarska M.M., Clemente M.J., Otrock Z.K., Nakashima M., Hsi E.D., Yoshida K., Shiraishi Y. et al. PRPF8 defects cause missplicing in myeloid malignancies. Leukemia. 2015; 29:126–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Adler A.S., McCleland M.L., Yee S., Yaylaoglu M., Hussain S., Cosino E., Quinones G., Modrusan Z., Seshagiri S., Torres E. et al. An integrative analysis of colon cancer identifies an essential function for PRPF6 in tumor growth. Genes Dev. 2014; 28:1068–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Kumari R., Jat P. Mechanisms of cellular senescence: cell cycle arrest and senescence associated secretory phenotype. Front. Cell Dev. Biol. 2021; 9:645593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Gorgoulis V., Adams P.D., Alimonti A., Bennett D.C., Bischof O., Bishop C., Campisi J., Collado M., Evangelou K., Ferbeyre G. et al. Cellular senescence: defining a path forward. Cell. 2019; 179:813–827. [DOI] [PubMed] [Google Scholar]
- 99. Nojima T., Rebelo K., Gomes T., Grosso A.R., Proudfoot N.J., Carmo-Fonseca M. RNA polymerase II phosphorylated on CTD serine 5 interacts with the spliceosome during co-transcriptional splicing. Mol. Cell. 2018; 72:369–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Saldi T., Cortazar M.A., Sheridan R.M., Bentley D.L. Coupling of RNA polymerase II transcription elongation with pre-mRNA Splicing. J. Mol. Biol. 2016; 428:2623–2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Colombo A.R., Elias H.K., Ramsingh G. Senescence induction universally activates transposable element expression. Cell Cycle. 2018; 17:1846–1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. De Cecco M., Criscione S.W., Peckham E.J., Hillenmeyer S., Hamm E.A., Manivannan J., Peterson A.L., Kreiling J.A., Neretti N., Sedivy J.M. Genomes of replicatively senescent cells undergo global epigenetic changes leading to gene silencing and activation of transposable elements. Aging Cell. 2013; 12:247–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Rigal J., Martin Anduaga A., Bitman E., Rivellese E., Kadener S., Marr M.T. Artificially stimulating retrotransposon activity increases mortality and accelerates a subset of aging phenotypes in Drosophila. eLife. 2022; 11:e80169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Li W., Prazak L., Chatterjee N., Grüninger S., Krug L., Theodorou D., Dubnau J. Activation of transposable elements during aging and neuronal decline in Drosophila. Nat. Neurosci. 2013; 16:529–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Pabis K., Barardo D., Selvarajoo K., Gruber J., Kennedy B.K. A concerted increase in readthrough and intron retention drives transposon expression during aging and senescence. eLife. 2023; 12:RP87811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Dikic I. Proteasomal and autophagic degradation systems. Annu. Rev. Biochem. 2017; 86:193–224. [DOI] [PubMed] [Google Scholar]
- 107. Young A.R., Narita M., Ferreira M., Kirschner K., Sadaie M., Darot J.F., Tavare S., Arakawa S., Shimizu S., Watt F.M. et al. Autophagy mediates the mitotic senescence transition. Genes Dev. 2009; 23:798–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Kenzelmann Broz D., Spano Mello S., Bieging K.T., Jiang D., Dusek R.L., Brady C.A., Sidow A., Attardi L.D. Global genomic profiling reveals an extensive p53-regulated autophagy program contributing to key p53 responses. Genes Dev. 2013; 27:1016–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Robin M., Issa A.R., Santos C.C., Napoletano F., Petitgas C., Chatelain G., Ruby M., Walter L., Birman S., Domingos P.M. et al. Drosophila p53 integrates the antagonism between autophagy and apoptosis in response to stress. Autophagy. 2019; 15:771–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Ruiz-Losada M., González R., Peropadre A., Gil-Gálvez A., Tena J.J., Baonza A., Estella C. Coordination between cell proliferation and apoptosis after DNA damage in Drosophila. Cell Death Differ. 2022; 29:832–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Wickramasinghe V.O., Venkitaraman A.R. RNA processing and genome stability: cause and consequence. Mol. Cell. 2016; 61:496–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Bonnet A., Grosso A.R., Elkaoutari A., Coleno E., Presle A., Sridhara S.C., Janbon G., Géli V., de Almeida S.F., Palancade B. Introns protect eukaryotic genomes from transcription-associated genetic instability. Mol. Cell. 2017; 67:608–621. [DOI] [PubMed] [Google Scholar]
- 113. Chakraborty P., Huang J.T.J., Hiom K. DHX9 helicase promotes R-loop formation in cells with impaired RNA splicing. Nat. Commun. 2018; 9:4346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Kim S., Kang N., Park S.H., Wells J., Hwang T., Ryu E., Kim B.-g., Hwang S., Kim S.-j., Kang S. et al. ATAD5 restricts R-loop formation through PCNA unloading and RNA helicase maintenance at the replication fork. Nucleic Acids Res. 2020; 48:7218–7238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Chang E.Y., Tsai S., Aristizabal M.J., Wells J.P., Coulombe Y., Busatto F.F., Chan Y.A., Kumar A., Dan Zhu Y., Wang A.Y. et al. MRE11-RAD50-NBS1 promotes Fanconi Anemia R-loop suppression at transcription-replication conflicts. Nat. Commun. 2019; 10:4265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Lafuente-Barquero J., Luke-Glaser S., Graf M., Silva S., Gómez-González B., Lockhart A., Lisby M., Aguilera A., Luke B. The Smc5/6 complex regulates the yeast Mph1 helicase at RNA-DNA hybrid-mediated DNA damage. PLoS Genet. 2017; 13:e1007136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Hodson C., van Twest S., Dylewska M., O’Rourke J.J., Tan W., Murphy V.J., Walia M., Abbouche L., Nieminuszczy J., Dunn E. et al. Branchpoint translocation by fork remodelers as a general mechanism of R-loop removal. Cell Rep. 2022; 41:111749. [DOI] [PubMed] [Google Scholar]
- 118. David C.J., Boyne A.R., Millhouse S.R., Manley J.L. The RNA polymerase II C-terminal domain promotes splicing activation through recruitment of a U2AF65-Prp19 complex. Genes Dev. 2011; 25:972–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Chanarat S., Sträßer K. Splicing and beyond: the many faces of the Prp19 complex. Biochim. Biophys. Acta. Mol. Cell Res. 2013; 1833:2126–2134. [DOI] [PubMed] [Google Scholar]
- 120. Gurova K., Chang H.W., Valieva M.E., Sandlesh P., Studitsky V.M. Structure and function of the histone chaperone FACT - Resolving FACTual issues. Biochim. Biophys. Acta Gene Regul. Mech. 2018; 1861:892–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Poli J., Gasser S.M., Papamichos-Chronakis M. The INO80 remodeller in transcription, replication and repair. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2017; 372:20160290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Prendergast L., McClurg U.L., Hristova R., Berlinguer-Palmini R., Greener S., Veitch K., Hernandez I., Pasero P., Rico D., Higgins J.M.G. et al. Resolution of R-loops by INO80 promotes DNA replication and maintains cancer cell proliferation and viability. Nat. Commun. 2020; 11:4534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Huertas P., Aguilera A. Cotranscriptionally formed DNA:RNA hybrids mediate transcription elongation impairment and transcription-associated recombination. Mol. Cell. 2003; 12:711–721. [DOI] [PubMed] [Google Scholar]
- 124. Domínguez-Sánchez M.S., Barroso S., Gómez-González B., Luna R., Aguilera A. Genome instability and transcription elongation impairment in human cells depleted of THO/TREX. PLos Genet. 2011; 7:e1002386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Yano K., Takahashi R.U., Shiotani B., Abe J., Shidooka T., Sudo Y., Yamamoto Y., Kan S., Sakagami H., Tahara H. PRPF19 regulates p53-dependent cellular senescence by modulating alternative splicing of MDM4 mRNA. J. Biol. Chem. 2021; 297:100882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Ochi N., Nakamura M., Nagata R., Wakasa N., Nakano R., Igaki T. Cell competition is driven by Xrp1-mediated phosphorylation of eukaryotic initiation factor 2α. PLoS Genet. 2021; 17:e1009958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Baumgartner M.E., Dinan M.P., Langton P.F., Kucinski I., Piddini E. Proteotoxic stress is a driver of the loser status and cell competition. Nat. Cell Biol. 2021; 23:136–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Recasens-Alvarez C., Alexandre C., Kirkpatrick J., Nojima H., Huels D.J., Snijders A.P., Vincent J.P. Ribosomopathy-associated mutations cause proteotoxic stress that is alleviated by TOR inhibition. Nat. Cell Biol. 2021; 23:127–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Baillon L., Germani F., Rockel C., Hilchenbach J., Basler K. Xrp1 is a transcription factor required for cell competition-driven elimination of loser cells. Sci. Rep. 2018; 8:17712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Floc’hlay S., Balaji R., Stankovic D., Christiaens V.M., Bravo Gonzalez-Blas C., De Winter S., Hulselmans G.J., De Waegeneer M., Quan X., Koldere D. et al. Shared enhancer gene regulatory networks between wound and oncogenic programs. eLife. 2023; 12:e81173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Ji Z., Kiparaki M., Folgado V., Kumar A., Blanco J., Rimesso G., Chuen J., Liu Y., Zheng D., Baker N.E. Drosophila RpS12 controls translation, growth, and cell competition through Xrp1. PLoS Genet. 2019; 15:e1008513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Kale A., Ji Z., Kiparaki M., Blanco J., Rimesso G., Flibotte S., Baker N.E. Ribosomal protein S12e has a distinct function in cell competition. Dev. Cell. 2018; 44:42–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Tollervey J.R., Wang Z., Hortobágyi T., Witten J.T., Zarnack K., Kayikci M., Clark T.A., Schweitzer A.C., Rot G., Curk T. et al. Analysis of alternative splicing associated with aging and neurodegeneration in the human brain. Genome Res. 2011; 21:1572–1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Mazin P., Xiong J., Liu X., Yan Z., Zhang X., Li M., He L., Somel M., Yuan Y., Phoebe Chen Y.-P. et al. Widespread splicing changes in human brain development and aging. Mol. Syst. Biol. 2013; 9:633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Lee B.P., Pilling L.C., Emond F., Flurkey K., Harrison D.E., Yuan R., Peters L.L., Kuchel G.A., Ferrucci L., Melzer D. et al. Changes in the expression of splicing factor transcripts and variations in alternative splicing are associated with lifespan in mice and humans. Aging Cell. 2016; 15:903–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Holly A.C., Melzer D., Pilling L.C., Fellows A.C., Tanaka T., Ferrucci L., Harries L.W. Changes in splicing factor expression are associated with advancing age in man. Mech. Ageing Dev. 2013; 134:356–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Harries L.W., Hernandez D., Henley W., Wood A.R., Holly A.C., Bradley-Smith R.M., Yaghootkar H., Dutta A., Murray A., Frayling T.M. et al. Human aging is characterized by focused changes in gene expression and deregulation of alternative splicing. Aging Cell. 2011; 10:868–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw and processed next generation sequencing data have been deposited in NCBI’s Gene Expression Omnibus (RRID:SCR_005012) and are accessible through GEO Series accession numbers GSE213325 for the Targeted DamID data (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE213325), GSE203127 and GSE213298 for the RNA-Seq data (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE213298), (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE203127).